Abstract
Introduction
The hippocampus, a medial temporal lobe structure central to learning and memory, is particularly vulnerable in preterm-born neonates. To date, segmentation of the hippocampus for preterm-born neonates has not yet been performed early-in-life (shortly after birth when clinically stable). The present study focuses on the development and validation of an automatic segmentation protocol that is based on the MAGeT-Brain (Multiple Automatically Generated Templates) algorithm to delineate the hippocampi of preterm neonates on their brain MRIs acquired at not only term-equivalent age but also early-in-life.
Methods
First, we present a three-step manual segmentation protocol to delineate the hippocampus for preterm neonates and apply this protocol on 22 early-in-life and 22 term images. These manual segmentations are considered the gold standard in assessing the automatic segmentations. MAGeT-Brain, automatic hippocampal segmentation pipeline, requires only a small number of input atlases and reduces the registration and resampling errors by employing an intermediate template library. We assess the segmentation accuracy of MAGeT-Brain in three validation studies, evaluate the hippocampal growth from early-in-life to term-equivalent age, and study the effect of preterm birth on the hippocampal volume. The first experiment thoroughly validates MAGeT-Brain segmentation in three sets of 10-fold Monte Carlo cross-validation (MCCV) analyses with 187 different groups of input atlases and templates. The second experiment segments the neonatal hippocampi on 168 early-in-life and 154 term images and evaluates the hippocampal growth rate of 125 infants from early-in-life to term-equivalent age. The third experiment analyzes the effect of gestational age (GA) at birth on the average hippocampal volume at early-in-life and term-equivalent age using linear regression.
Results
The final segmentations demonstrate that MAGeT-Brain consistently provides accurate segmentations in comparison to manually derived gold standards (mean Dice's Kappa > 0.79 and Euclidean distance <1.3 mm between centroids). Using this method, we demonstrate that the average volume of the hippocampus is significantly different (p < 0.0001) in early-in-life (621.8 mm3) and term-equivalent age (958.8 mm3). Using these differences, we generalize the hippocampal growth rate to 38.3 ± 11.7 mm3/week and 40.5 ± 12.9 mm3/week for the left and right hippocampi respectively. Not surprisingly, younger gestational age at birth is associated with smaller volumes of the hippocampi (p = 0.001).
Conclusions
MAGeT-Brain is capable of segmenting hippocampi accurately in preterm neonates, even at early-in-life. Hippocampal asymmetry with a larger right side is demonstrated on early-in-life images, suggesting that this phenomenon has its onset in the 3rd trimester of gestation. Hippocampal volume assessed at the time of early-in-life and term-equivalent age is linearly associated with GA at birth, whereby smaller volumes are associated with earlier birth.
Keywords: Segmentation, Hippocampus, Preterm neonates, Early-in-life, MAGeT-Brain, MRI
Highlights
-
•
We develop a MAGeT-Brain based automatic protocol to segment hippocampus in preterm neonates.
-
•
MAGeT-Brain can accurately segment hippocampus in preterm neonates, even at early-in-life.
-
•
Hippocampal asymmetry with a larger right side is demonstrated on early-in-life images.
-
•
Smaller hippocampal volumes are associated with earlier birth in preterm neonates.
1. Introduction
Preterm birth is increasingly prevalent, with recent world-wide estimates at nearly 15 million babies born at less than 37 weeks gestation with an estimated 20% being born very preterm at less than 32 weeks (Chang et al., 2013). Very preterm-born children delivered at 24–32 weeks of gestation are at significantly high risk for developmental delays with 4–19% of them developing cerebral palsy (Moster et al., 2008; Saigal and Doyle, 2008), and 30%–60% exhibiting cognitive impairments, poor executive function and working memory, learning disabilities and motor dysfunction (Aarnoudse-Moens et al., 2009; Holsti et al., 2002; Mulder et al., 2010; Taylor et al., 2004; Vohr et al., 2003). This evolving constellation of motor and cognitive impairments recognized in preterm-born children is consistent with impaired cerebral growth impacting gray and white matter (Back and Miller, 2014; Chau et al., 2013). Given the increasing recognition of widespread brain dysmaturation in preterm neonates, there is an urgent need to quantify the growth of specific brain structures from early-in-life (Back and Miller, 2014).
Slower than expected brain growth, as detected by magnetic resonance imaging (MRI), is observed in very preterm-born children into the adolescent period (Mathur et al., 2009). In particular, the hippocampus, which is central to the developmental of learning and memory, has consistently been shown to be smaller in preterm-born children relative to term born controls (Gimenez et al., 2008; Nosarti et al., 2002; Thompson et al., 2008; Cheong et al., 2013; de Kieviet et al., 2012). This relative decrease in the rate of hippocampal development in children born very preterm has been associated with impaired cognition and working memory at 2 years of age (Thompson et al., 2008; Beauchamp et al., 2008) and school-age (Aarnoudse-Moens et al., 2009, 2012; Ford et al., 2011; Rogers et al., 2012). As the growth spurt of the hippocampus starts in the perinatal period (Insausti et al., 2010), it is of great importance to understand the hippocampal growth and development in very preterm neonates especially from early-in-life to term-equivalent age.
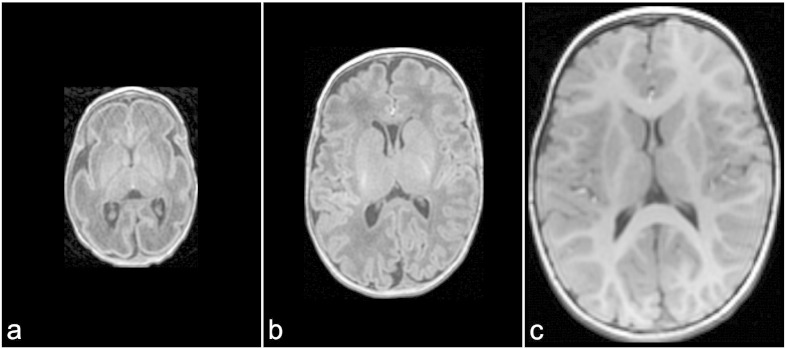
Accurate segmentation of brain structures is the key to successful volumetric analysis of hippocampal development, which may be of particular clinical significance in predicting development outcomes (Peterson et al., 2000; Choe et al., 2013). Automatic segmentation of neonatal brain images faces several challenges. Tissue contrast on T1- or T2-weighted images, an image property that many algorithms rely upon for automated segmentation, is the inverse of what is typically observed in infants and adults (Fig. 1). In addition, the brains of preterm neonates are at a much earlier stage in the gyrification process than are full-term neonates (Fig. 1). A surge in synapse formation and activity occurs at the third trimester, during which the volumes of cerebral cortical gray and myelinated white matter increase about 4- to 5-fold (Limperopoulos et al., 2010). Due to many of these factors, quantitative volumetric and/or morphometric assessment of the hippocampus for preterm infants have only been performed at term-equivalent age or later, primarily using manual segmentation (Beauchamp et al., 2008; Gousias et al., 2012; Lodygensky et al., 2005, 2008; Peterson et al., 2000; Rogers et al., 2012; Thompson et al., 2008, 2009, 2013, 2014).
Fig. 1.
Axial images from T1-weighted MR of the same preterm-born child at different ages demonstrating rapid brain growth and maturation which occurred from 29 weeks GA to 2 years of age. a. 29 weeks gestational age (GA); b. 40 weeks GA; c. 2 years. The white and gray matter contrast on images acquired at 29 and 40 weeks of GA is the inverse of that on the 2 year image.
Courtesy of Dr. Margot Taylor
To date, there are several automatic brain tissue classification techniques developed for neonates (Anbeek et al., 2013; Cardoso et al., 2013; Gui et al., 2012; Prastawa et al., 2005; Shi et al., 2011; Wang et al., 2014; Weisenfeld and Warfield, 2009; Xue et al., 2007; Yu et al., 2010). However only a limited number of algorithms have been developed to achieve detailed delineation of specific brain structures, such as the corpus callosum, basal ganglia, thalamus, hippocampus and amygdala. Nishida et al. developed a semi-automated segmentation method, which was capable of segmenting a brain into 30 brain regions (Nishida et al., 2006). They validated their results in 12 neonates aged 31.1–42.6 weeks using visual inspection rather than by comparison to gold-standard manual segmentations. Gousias and colleagues compared the use of multi-atlas and model-based segmentation (based on a maximum-probability representation) on the MR images of 5 term and 15 preterm infants scanned at term (Gousias et al., 2013) using label-based encephalic ROI templates (ALBERTs) of 50 manually segmented anatomical brain regions as inputs (Gousias et al., 2012). Makropoulos and colleagues extended this work and that by Xue et al. (2007) by implementing a multi-structure Expectation-maximization (EM) based segmentation technique for neonatal brain parcellation (Makropoulos et al., 2014) using both the subject-specific tissue classification information with the nonlinearly transformed labels of the ALBERTs.
Although manual labeling of brain structures by trained experts is generally considered the gold standard (Konrad et al., 2009), this process is labor-intensive, time-consuming, and highly subjective in ambiguous regions. In contrast, automatic segmentation methods are more objective, consistent and efficient especially for studying larger sample sizes (Morey et al., 2009). Most semi-automatic and automatic algorithms that have been published were in an effort to segment the adult hippocampus (Akhondi-Asl et al., 2011; Chupin et al., 2009a; Chupin et al., 2009b; Collins and Pruessner, 2010; Coupe et al., 2011; Csernansky et al., 1998; Fischl et al., 2002; Gao et al., 2012; Haller et al., 1997; Hao et al., 2014; Heckemann et al., 2006; Heckemann et al., 2010; Hu et al., 2011; Jorge Cardoso et al., 2013; Khan, 2011; Lötjönen et al., 2010; Pipitone et al., 2014; Shen and Davatzikos, 2002; Shen et al., 2002; Tong et al., 2013; Van Leemput et al., 2009; Wu et al., 2014; Yushkevich et al., 2010). Algorithms based on deformable models usually require prior knowledge of the geometry and statistical shape information of the interested structure, manually identified landmarks, and/or probabilistic atlases to initialize and guide the segmentation (Akhondi-Asl et al., 2011; Chupin et al., 2009a; Chupin et al., 2009b; Gao et al., 2012; Hu et al., 2011; Shen et al., 2002; Shen and Davatzikos, 2002). Classical model-based segmentation approaches employ manually labeled single brain atlas to delineate the hippocampus by propagating the atlas labels to target brain images using nonlinear registration (Haller et al., 1997; Csernansky et al., 1998). To overcome the inherent limitations of the model-based segmentation and improve segmentation accuracy, methods based on multiple atlases and label fusion techniques have been developed and widely adopted in hippocampal volumetric studies (Chakravarty et al., 2013; Collins and Pruessner, 2010; Coupe et al., 2011; Fischl et al., 2002; Hao et al., 2014; Heckemann et al., 2006; Heckemann et al., 2010; Jorge Cardoso et al., 2013; Khan et al., 2011; Lötjönen et al., 2010; Pipitone et al., 2014; Tong et al., 2013; Wu et al., 2014; Yushkevich et al., 2010). Furthermore, various atlas selection (Aljabar et al., 2009; Collins and Pruessner, 2010; Heckemann et al., 2006; Tong et al., 2013; Wolz et al., 2010) and label fusion (Coupe et al., 2011; Hao et al., 2014; Jorge Cardoso et al., 2013; Khan et al., 2011; Warfield et al., 2004; Wu et al., 2014) strategies have been explored to optimize the multi-atlas based segmentations.
The present study focuses on the development and validation of an automatic segmentation protocol to delineate the hippocampi of preterm-born neonates on their brain MRIs acquired at not only term-equivalent age but also early-in-life, in the weeks after birth when clinically stable. This automatic segmentation pipeline is based on the MAGeT-Brain algorithm (Chakravarty et al., 2013; Pipitone et al., 2014), which requires only a small number of input atlases. This pipeline is capable of minimizing the registration and resampling errors by propagating the labels to a template library that includes a subset or all of the input subject images. Neither manual nor automatic hippocampal segmentations have been achieved on brain images of preterm neonates as early as 27 weeks of post-menstrual age. Here, we present novel work presenting the accurate segmentation of the neonatal hippocampus for very preterm infants.
We first describe the protocol for manually segmenting the hippocampi of preterm neonates and present the manual hippocampal segmentation results. Then we introduce the MAGeT-Brain algorithm (Chakravarty et al., 2013; Pipitone et al., 2014), and perform a thorough multi-fold Monte Carlo cross-validation (MCCV) to evaluate the hippocampal segmentation accuracy of our proposed technique with 187 different parameter settings (a total of 164,560 segmentation data points were evaluated). We then apply this automatic segmentation pipeline using the optimal parameter setting to delineate the hippocampi on 168 early-in-life and 154 term-equivalent images of 197 preterm neonates to obtain the hippocampal volume data at these two time points. Given that hippocampal vulnerability is recognized in preterm neonates scanned at term-age or later, we examine whether earlier gestational age at birth predicted a greater impairment in hippocampal growth.
2. Materials
2.1. Study population
The University of British Columbia Clinical Research Ethics Board reviewed and approved the study. Parental informed consent was obtained for the data acquisition of each participating patient.
Enrolled as part of a longitudinal study on brain development in preterm neonates between April 2006 and September 2010, a total number of 197 very preterm neonates (97 boys) born between 24 to 32 weeks gestation age, without antenatal infections, congenital malformation or syndrome, or ultrasound evidence of a large parenchymal hemorrhagic infarction greater than 2 cm at BC Women's Hospital, Canada were included in this study (Chau et al., 2013). The mean gestational age of all the preterm neonates was 28.01 weeks with a standard deviation of 2.2.
2.2. Magnetic resonance imaging
The anatomical images of neonates were acquired with a specialized single-channel neonatal head coil (Advanced Imaging Research, Cleveland, OH) on a Siemens 1.5 T Avanto scanner (Erlangen, Germany) at two time points, once shortly after birth when clinically stable and again at term-equivalent age. An MR-compatible incubator (Lammers Medical Technology, Luebeck, Germany) was employed to accommodate the infants who were not sedated for the scan. Sequence parameters for the 3D volumetric T1-weighted images were: repetition time (TR) 36 ms; echo time (TE) 9.2 ms; flip angle 30° and voxel size 1 mm × 1.04 mm × 1.04 mm for both sagittal and coronal acquisitions. The 7 sagittal T1-weighted images were pre-processed to be in the same orientation of the 315 coronal acquisitions using MINC image processing tools (http://www.bic.mni.mcgill.ca/ServicesSoftware/HomePage).
All images were reviewed and rated by an experienced neuroradiologist (K.J.P.), blinded to the neonate's medical history, for the identification of brain injury severity. All images were scored for white matter injury (WMI) based on a 3-point scale (none = 0, minimal = 1, and moderate–severe = 2–3 combined), which was demonstrated to be predictive of adverse neurodevelopmental outcome (Chau et al., 2013). Areas exhibiting foci of abnormal T1 signal hyperintensity with no marked T2 signal hypointensity, or low-intensity T1 foci were considered for WMI scoring. K.J.P also rated intraventricular hemorrhage (IVH) on these images using Papile's system (none = 0, mild = 1–2, moderate–severe = 3–4) (Papile et al., 1978).
3. Manual segmentation of the hippocampus
Blinded with respect to gestational age at birth and functional outcome measures, an expert (J.L.W.) in manual segmentation of the hippocampus delineated the left and right hippocampi on both the early-in-life and term-equivalent images of 22 randomly selected neonates (44 images in total). The 22 neonates (7 boys), were born at a mean gestational age of 27.7 weeks (SD 1.9), and scanned early-in-life at 32.1 weeks (SD 1.9) and again at term-equivalent age at 40.4 weeks (SD 2.1). Manual segmentations were considered the gold standard for volumetric assessment and were employed as the atlases in conducting automatic hippocampal segmentation with the MAGeT-Brain method (see Section 5: Experiments).
Data were visualized and segmented using the Display software package (part of the MINC image processing toolbox). Display provides simultaneous coronal, sagittal, and axial views of the brain and can create a 3D surface of a structure. The tricubic interpolant option was used for image visualization purposes as we found that a more accurate and consistent segregation of the hippocampus from the surrounding structures within the medial temporal lobe was achievable under these conditions. The images were inspected one final time with the interpolant removed to ensure that voxels suffering from partial volume effects were not erroneously considered within the hippocampus segmentation. It should be noted that the images were not re-oriented and the segmentations were performed in the native space and on the data in its native format.
3.1. Manual segmentation protocols
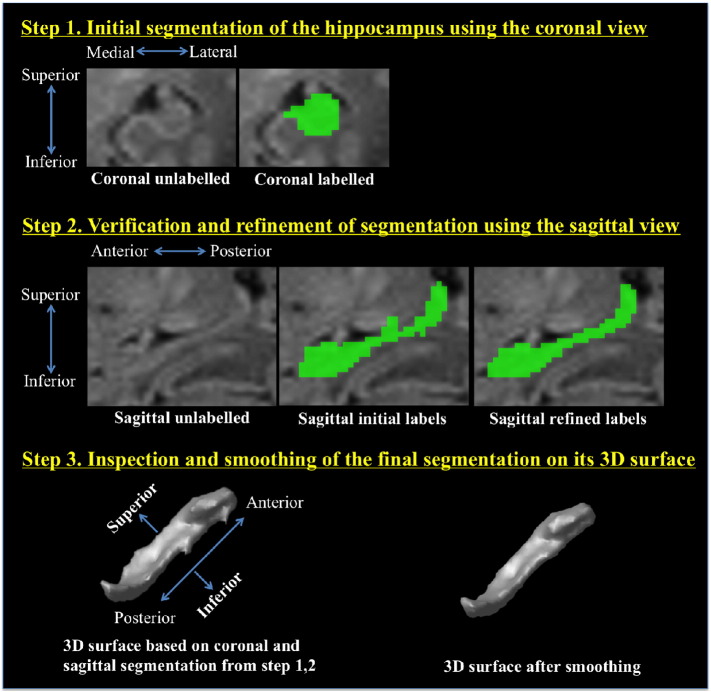
A 3-step segmentation protocol (Fig. 2) was applied to each of the early-in-life and term images. This protocol adapts the histological definitions of Duvernoy et al. (2005), as well as existing whole hippocampal segmentation protocols for MR images (Boccardi et al., 2015; Pruessner et al., 2000; Winterburn et al., 2013) to the preterm infant brain. The manual segmentation protocol below is the first that has been specifically described to accommodate the unique and potentially variable neuroanatomy in preterm infants.
Fig. 2.
3-Step manual segmentation protocol. The estimation of the hippocampal boundaries was initially performed using the coronal view and then refined using the sagittal view. 3D surface model of the segmented hippocampus was constructed for the final inspection of segmentation.
3.1.1. Step 1: initial estimation of the hippocampal boundaries using the coronal view (Fig. 2)
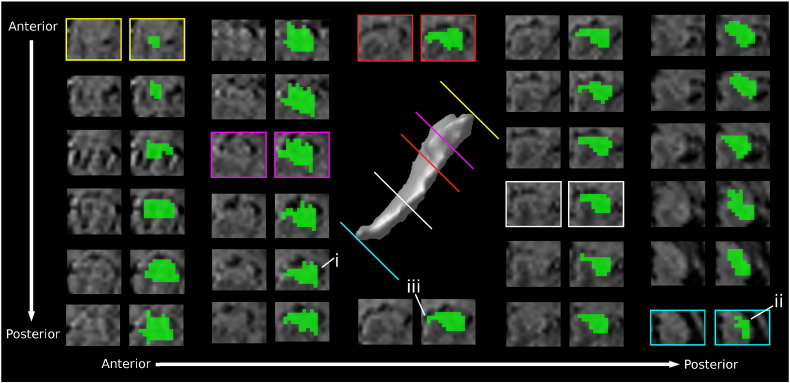
The hippocampus was present in approximately 25 coronal slices, extending from the amygdala in the anterior extremity to the fimbria in the posterior extremity. Segmentations were based on the intensity differences between the white matter of the temporal lobe (low intensity) and the gray matter of the hippocampus (high intensity). Given the paucity of myelin in the neonatal brain, this is opposite to what is usually seen on regular T1-weighted MRI of children and adults. Along with the free water content decreases of brain tissues and the myelination of white matter during the first several postnatal months, the intensities of white matter and gray matter on T1 weighted neonatal MR images gradually alter and invert to become similar to those on children and adult images. Representative segmentations for each coronal slice of an example subject are shown in Fig. 3 and are meant to act as a guide for other groups interested in replicating our protocol. The lateral border throughout can be approximated from the inferior horn of the lateral ventricle (Fig. 3, i) or the lateral ventricle (Fig. 3, ii). The medial border is defined by the location where the subiculum of the hippocampus meets the ambient cistern (Fig. 3, iii). Voxels at the outer edges of the hippocampal structure may suffer from partial volume effects. This is particularly true for the alveus and fimbria, white matter structures that lie on the superior edge of the hippocampus. Voxels that appeared to be composed of mixed structures were assigned a hippocampus label only if they were perceived to contain more hippocampus than other tissue classes (according to the qualitative judgment of the rater). The hippocampal head was often difficult to distinguish from the surrounding amygdala in the coronal view. In such cases, the head was left unsegmented and was defined later in the sagittal view, where its boundaries were much less ambiguous.
Fig. 3.
Representative segmentations of the hippocampus for each coronal slice of an example subject. i. The lateral border defined by the inferior horn of the lateral ventricle; ii. the lateral ventricle and iii. the location where the subiculum of the hippocampus meets the ambient cistern defines the medial border.
3.1.2. Step 2: refinement of segmentation using the sagittal view (Fig. 2)
The initial segmentations completed in the coronal view were verified in the sagittal view, and any incorrectly labeled voxels were revised. Any previously unlabeled voxels in and around the hippocampal region were reconsidered and given appropriate labels in the sagittal view. The sagittal view was also used to segment the hippocampal head if its segmentation was impossible to complete in the coronal view.
3.1.3. Step 3: inspection of 3D surface (Fig. 2)
The segmented hippocampus was represented as a 3D surface in Display using the marching cubes algorithm (Lorensen and Cline, 1987). The surface was adopted to identify abnormal hippocampal shape. It is expected that the surface of the hippocampus is relatively smooth throughout, so any areas that protrude excessively were trimmed. Any regions that were considered morphologically incontiguous were inspected at the homologous location in the tri-planar view of the segmentation. These voxels were further verified in the segmentation based on the actual intensity and contrast characteristics of the image.
3.2. Protocol reliability testing
Four early-in-life and four term image were randomly selected and re-segmented to assess the reliability of our 3-step manual segmentation protocol. Re-segmentations of both the left and right hippocampi in these images were performed at least 6 months after the original segmentations to ensure that rater memory was not a confounding factor. In essence, this type of test–retest metric can be used as an upper bound for the accuracy of the hippocampus segmentations. Protocol reliability was measured using the Dice's Kappa metric, which evaluates the spatial and volumetric overlap of the original and re-segmented label volumes. The Dice's Kappa can be represented by the following equation:
where k is the Dice's Kappa, A and B are the number of voxels belonging to a specific segmentation, and a perfect overlap of labels A and B has k = 1.
3.3. Results: manual segmentation of the hippocampus
The volumetric results for both the early-in-life and term images of these 22 subjects are shown in Table 1. Both the left and the right hippocampi on the term images are significantly larger than those on the early-in-life images (p < 0.001).
Table 1.
The manually segmented hippocampal volumes of the 22 subjects.
| 22 subjects | Left | Right | Average |
|---|---|---|---|
| Early-in-life (mm3) | Mean: 719.1 ± 158.0 Range: 518.7–1220.7 |
Mean: 722.4 ± 147.4 Range: 455.9–1023.2 |
Mean: 720.7 ± 146.1 494.7–1122.0 |
| Term (mm3) | Mean: 1077.5 ± 160.2 Range: 769.3–1449.7 |
Mean: 1085.3 ± 137.8 Range: 827.9–1354.0 |
Mean: 1081.4 ± 141.9 820.3–1407.3 |
Table 2 lists the results of the intra-rater reliability of the 3-step manual segmentation protocol measured with the Dice's Kappa. The results (>0.81 based on the range and >0.85 based on the mean) indicate that high stability and intra-rater reliability can be achieved using this manual segmentation protocol. Higher intra-rater reliability was obtained in segmenting the hippocampus on the early-in-life images and the manual rater's hippocampal tracing accuracy was consistent overtime.
Table 2.
The Dice's Kappa for the left and right hippocampi of 4 early-in-life and 4 term images that were manually segmented by J.L.W. in two sessions.
| Dice's Kappa: Mean (range) | Left | Right |
|---|---|---|
| Early-in-life | 0.91 (0.90–0.91) | 0.89 (0.86–0.91) |
| Term | 0.86 (0.83–0.87) | 0.86 (0.82–0.89) |
4. MAGeT-Brain segmentation
Multiple Automatically Generated Templates (MAGeT) Brain (Chakravarty et al., 2013) is the automatic image segmentation pipeline employed in this work. Unlike the conventional multi-atlas segmentation technique, MAGeT-Brain requires only a few manually segmented images as the atlases. This approach creates an intermediate dataset, which is called the template library to serve the same role as multiple atlases in segmentation, as would be used in a more traditional multi-atlas segmentation routine.
MAGeT-Brain first selects a subset or the entire set of images from the targeted subject images and uses them as the templates images. It propagates each of the labeled atlases to all the template images to generate the template library. The template image selection criteria can be modified to best represent subject populations with cases and controls or satisfy the measurement of certain neuroanatomical features according to specific study objectives. The purpose of this intermediate stage is to use the variability of the population under study to improve the final segmentation output. Then, each subject image is non-linearly registered to all the template images with the automatic normalization tools (ANTs) (Avants et al., 2008; Avants et al., 2011). Through this process, we can automatically grow the number of candidate segmentations to as many as the number of the atlases times that of the templates for each subject. To limit errors due to resampling, nonlinear registration, or irreconcilable differences in the neuroanatomy, the multiple labels acquired from the previous step for each subject are fused using a voxel voting procedure to achieve the final labeling of each subject. Majority voting was adopted in label fusion, where the most popularly voted label at each voxel determines the final labeling or segmentation (Collins and Pruessner, 2010), because it carries the advantage of computational efficiency.
More sophisticated atlas selection (Aljabar et al., 2009; Collins and Pruessner, 2010; Heckemann et al., 2006; Heckemann et al., 2010; Tong et al., 2013; Wolz et al., 2010) and label fusion methods (Coupe et al., 2011; Hao et al., 2014; Jorge Cardoso et al., 2013; Khan et al., 2011; Wang et al., 2011; Warfield et al., 2004; Wu et al., 2014; Yushkevich et al., 2010) that can be used in automatic segmentations are available. However, we did not see significant benefit of using cross-correlation weighted and normalized mutual information weighted voting over the simple majority voting fusion method in improving the segmentation accuracy of MAGeT-Brain from our previously published studies (Pipitone et al., 2014; Park et al., 2014). Previous studies also demonstrated that when given the same number of atlases, MAGeT-Brain is more accurate than multi-atlas segmentation and is more reliable than the probabilistic atlas approach (Diedrichsen et al., 2009).
Recent studies have demonstrated that MAGeT Brain can provide accurate segmentations of subcortical structures (striatum, thalamus, and pallidum) (Chakravarty et al., 2013; Raznahan et al., 2014; Shaw et al., 2014), the adult hippocampus and its subfields (Pipitone et al., 2014), the cerebellum and its lobules (Park et al., 2014), and the adolescent pituitary gland (Wong et al., 2014).
5. Experiments
Brain structures experience significant growth and myelination in the perinatal period. Rapid development and maturation of fiber tracts and individual brain regions that occur during this time result in striking morphological differences that are clearly demonstrated on neonatal brain MR images acquired at preterm (Fig. 1a) and term-equivalent age (Fig. 1b). To account for the morphological variability of brains at different ages, we assessed the reliability and accuracy of MAGeT-Brain in hippocampal segmentation from volumetric T1-weighted scans of preterm-born neonates using the following experiments.
Experiment 1 thoroughly evaluates the hippocampal segmentation accuracy of MAGeT-Brain on preterm-born neonates at early-in-life and term age over different parameter settings.
Experiment 2 studies the hippocampal volume and growth from early-in-life to term-equivalent age of preterm neonates using MAGeT-Brain with the optimal parameter setting obtained from Experiment 1.
Experiment 3 investigates if there is any statistically significant association of the premature birth with the average hippocampal volumes from early-in-life to term-equivalent age.
5.1. Experiment 1: Monte Carlo cross-validation of MAGeT-brain hippocampal segmentation
The optimal parameter setting of MAGeT-Brain for the hippocampal segmentations of preterm-born infants was validated with three trials of comprehensive 10-fold Monte Carlo cross-validation (MCCV) (Shao, 1993) on the early-in-life group, the term group, and the mixed group (with both early-in-life and term images). In these three MCCV studies, the manually segmented hippocampi on the 22 early-in-life and 22 term images were considered the gold standard.
5.1.1. Experiment 1: methods
In the first MCCV study on the 22 early-in-life images, MAGeT-Brain computed the hippocampal segmentations of these images automatically with randomly selected 5–15 (11 different numbers of) input atlases and 5–21 (17 different numbers of) templates from the atlas and template libraries. The input atlas library contained the manual segmentations of the 21 early-in-life images and the template library included their brain images. The subject under study was excluded from both the atlas and template libraries to ensure completely unbiased segmentation results. Each of the 22 images was segmented with 11 × 17 = 187 different parameter settings 10 times using MAGeT-Brain, and a total of 10 × 22 × 11 × 17 = 41,140 validation data were generated. The automatic segmentations were compared to the gold standard.
The second MCCV for the term group was conducted using a protocol similar to that described above to assess the performance of MAGeT-Brain in segmenting the hippocampi on term images with atlas library size varying from 5 to 15 images, and template library size varying from 5 to 21 images excluding the subject under study. The evaluation was based on a total of 41,140 term segmentation data that were produced with 187 parameters settings on the 22 term images over 10 folds.
The third MCCV was carried out with expanded atlas and template libraries, which were composed of all 44 hippocampal atlases and 44 images (22 early-in-life and 22 term). MAGeT-Brain pipeline was applied to segment the hippocampi of these 44 images with parameter settings of 5–15 input atlases and 5–21 templates excluding the subject under study. We obtained a total of 10 × 44 × 17 × 11 = 82,280 segmentations in this study, and analyzed the validation results using the same method as in the first and second MCCV studies.
For each of these three studies, Dice's Kappa of each segmentation with every parameter setting in each fold was calculated. The Euclidean distance between the centroid of each MAGeT-Brain segmented hippocampus and that of its manual counterpart was also derived. Ideally the difference between the automatic segmentation and the gold standard manual segmentation should be minimal, i.e. the Kappa value should be close to 1 and the Euclidean distance close to 0. The segmentation accuracy for each of the 187 parameter settings was evaluated using the average of the 22 (studies 1 and 2) or 44 (study 3) segmentations of the 10 folds. Based on these evaluation results, the optimal parameter setting of the atlas and the template libraries was determined.
The volume-related fixed and proportional biases of MAGeT-Brain segmentation with the optimal parameter setting across the range of hippocampal sizes were evaluated using the Bland–Altman plots (Bland and Altman, 1986) for all three studies. Each manually segmented hippocampal volume was compared with the mean volume segmented using MAGeT-Brain with all 187 settings over 10 folds. Moreover, the level of agreement of MAGeT-Brain segmented hippocampi with the manual segmentations in terms of centroid location was also analyzed using the Bland-Altman plots.
5.1.2. Experiment 1: results
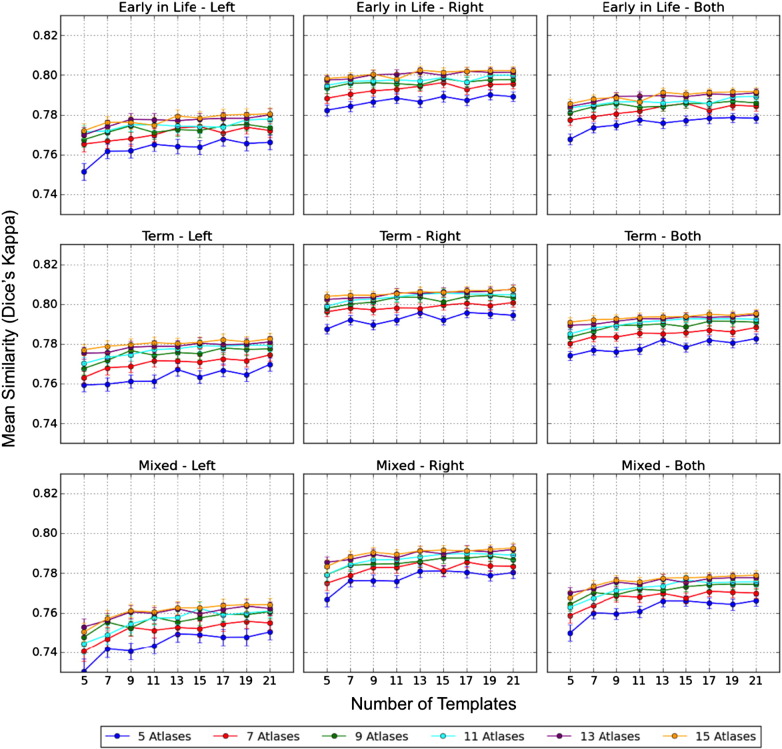
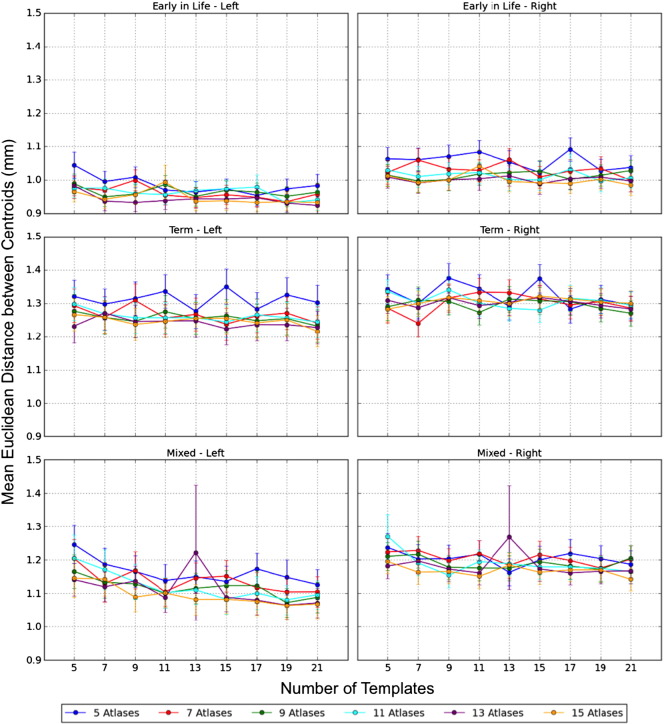
A high degree of overlap is demonstrated between the MAGeT-Brain segmented hippocampal volumes and the gold standard manual segmentations. The maximum Kappa is obtained when using 15 atlases and 21 templates to segment the hippocampi on all three groups. As illustrated in Fig. 4, the segmentation accuracy of MAGeT-Brain improves along with the increased number of atlases and templates, however the improvement diminishes when there are more than 11 images in the atlas library and 13 images in the template library. Although we included both even and odd numbers of images in the atlases and templates libraries in the three MCCV studies, we have shown the results obtained with the odd number of inputs in Figs 4 and 5 for clearer display.
Fig. 4.
Mean Dice's Kappa between the gold standard manual segmentations and MAGeT-Brain hippocampal segmentations with 187 different parameter settings (5–15 atlases and 5–21 templates) over 10 folds for 22 early-in-life images (top), 22 term images (middle), and 42 mixed images (bottom). Error bars indicate standard error.
Fig. 5.
Mean Euclidean distance between centroids of manually segmented hippocampi and those of MAGeT-Brain segmentations with 187 parameter settings over 10 folds for 22 early-in-life images (top), 22 term images (middle), and 42 mixed images (bottom). Error bars indicate standard error.
We find that the selection of input atlases and templates affects MAGeT-Brain segmentation accuracy. When images that are acquired at similar age are selected as atlases and templates, MAGeT-Brain segmentations have better concordance with the gold-standard manual segmentations (Fig. 4), while using mixed early-in-life and term atlases and templates results in less accurate segmentations, due to the existence of vast morphological variability on brain images scanned at these two time points (Fig. 1a, b).
The Euclidean distances between centroids of manually segmented hippocampi and those of MAGeT-Brain segmentations reflect a high degree of agreement in terms of locations (Fig. 5). Mean Euclidean distance decreases with the increased number of atlases, however the size of the template library does not have a significant effect on the Euclidean distance between the centroids of manual and MAGeT-Brain segmentations.
When we performed MAGeT-Brain segmentations using atlases and templates selected from the 44 mixed images (22 early-in-life and 22 term), two of the 44 images had some failed hippocampal segmentations (Kappa < 0.6) due to the severe image distortion around the hippocampal areas on these two images and the significant morphological difference between these two images and their randomly selected input atlases and templates. We noticed that 37.5% of a total of 3740 segmentation data for these two images had failed in the mixed group study when the MR-post-menstrual age (MR-PMA) of the input atlases and templates was significantly different from that of these two images. The number of failed segmentations accounts for 1.7% of all the 82,280 segmentation data in this group. Therefore, the segmentation results of these two images were excluded from the mixed group study. This indicates that MR-PMA appropriate atlases and templates should be used to avoid segmentation failure and achieve accurate MAGeT-Brain hippocampal segmentations.
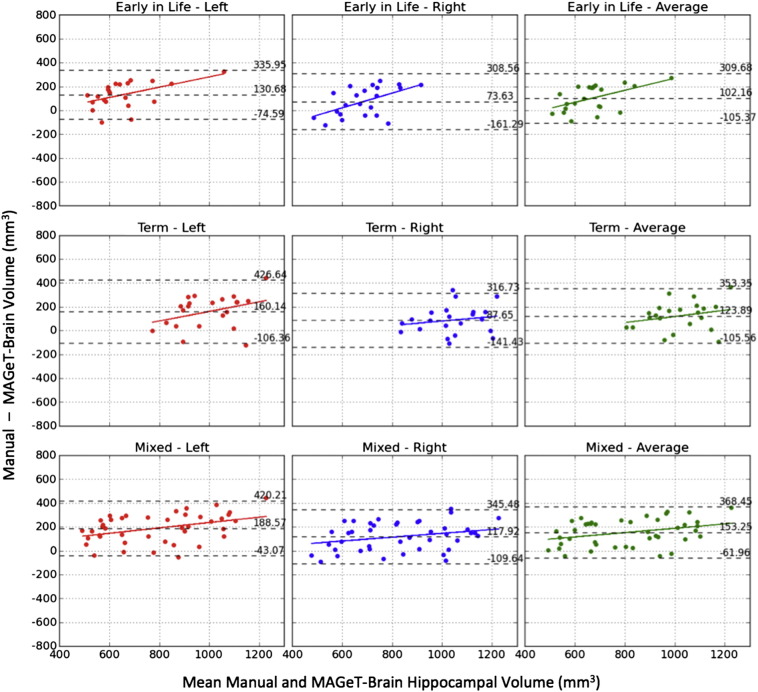
The Bland–Altman plots illustrate the levels of fixed and proportional bias observed in MAGeT-Brain segmentations in comparison with manual segmentation hippocampal volumes (Fig. 6). MAGeT-Brain appears to slightly under-estimate larger hippocampi and over-estimate smaller hippocampi, and shows a conservative fixed bias when compared to the manual segmentations. In addition, a fixed underestimation of ~10% of the structure volume is also observed.
Fig. 6.
Bland–Altman plots of MAGeT-Brain vs. manually segmented hippocampal volumes for 22 early-in-life images (top), 22 term images (middle), and 42 mixed images (bottom). The overall mean difference in volume and limits of agreement (±1.96SD) are shown by dashed horizontal lines. Linear fit lines are shown for each group.
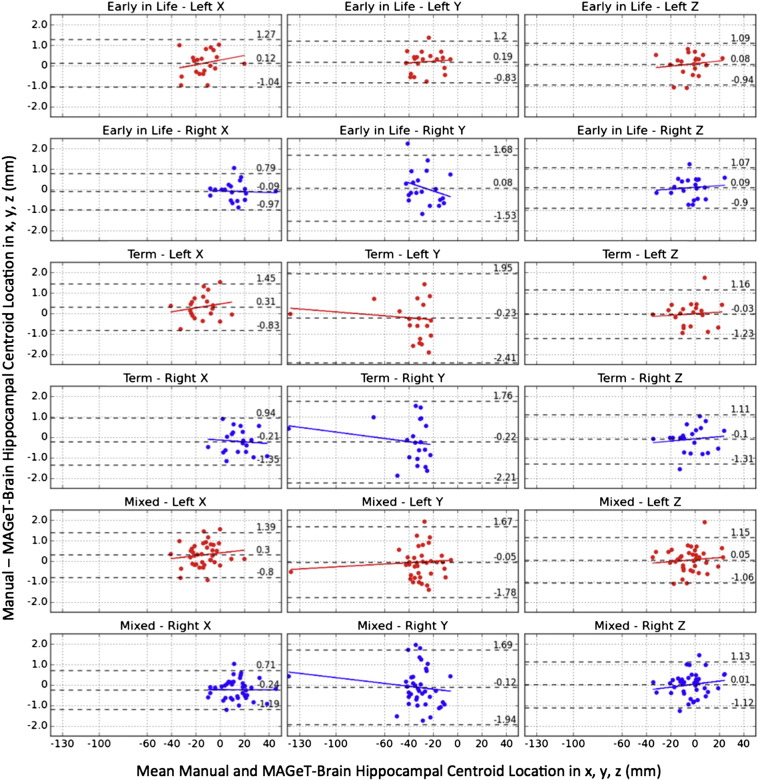
Fig. 7 shows the hippocampal centroid location related biases and thoroughly reveals the error distribution of the centroids of MAGeT-Brain segmented hippocampi in x, y, and z directions. The mean differences in x direction between the centroids of the manually segmented hippocampi and those of the MAGeT-Brain segmentations are all positive on the left side and negative on the right side for the early-in-life, term, and mixed groups, indicating that in general the location of MAGeT-Brain segmented hippocampi in x direction tends to be slightly more lateral than the manual segmentations on both left and right sides. MAGeT-Brain segmentations are more medial for more laterally located hippocampi, and more lateral for those more medially located ones. In the y direction, MAGeT-Brain segmented hippocampi are located more posteriorly than the manual segmentations for the early-in-life group and more anteriorly for the term and mixed groups. In the z direction, for the more superiorly located hippocampi, MAGeT-Brain is more likely to segment them more inferiorly on both sides for all three groups. The mean difference in the z direction is also the smallest in all three directions. Since the majority of the images were acquired coronally, the sagittal acquisitions in all three groups (2 early-in-life, 2 term, 4 mixed) were excluded in the centroid location study to maintain consistent coordinates.
Fig. 7.
Bland–Altman plots of the centroid location of MAGeT-Brain vs. manually segmented hippocampi (left & right, x, y, z) in the early-in-life (20 images), term (20 images), and mixed (38 images) groups. The overall mean difference in each coordinate direction and limits of agreement (±1.96SD) are shown by dashed horizontal lines. Linear fit lines are shown for each group.
5.2. Experiment 2: hippocampal segmentation on early-in-life and term images
In this experiment, we applied the MAGeT-Brain pipeline to automatically delineate the hippocampi on 168 early-in-life images (mean post-menstrual age [PMA] 32 weeks) and 154 term-equivalent images (mean PMA 40 weeks) acquired at BC Women's Hospital.
5.2.1. Experiment 2: methods
First, employing 15 of the 22 manually segmented hippocampi obtained on early-in-life images as input atlases and 21 randomly selected early-in-life images as templates, we calculated volumes of the left and right hippocampi on each individual image and obtained the mean of the entire early-in-life group consisting of 168 images with MAGeT-Brain. We then applied a similar automatic segmentation protocol to label hippocampi on the 154 term-equivalent images. The input atlases in the term study were replaced by 15 manual hippocampal segmentations of term images, and the templates were 21 randomly selected term images. Calculation and analysis of the MAGeT-Brain segmented hippocampal volumes were conducted for each image and the whole group without the interference of their own manual segmentations.
A total of 125 subjects in our study cohort had both early-in-life and term images. We calculated the hippocampal growth rates within this time window for these 125 subjects by dividing the volume difference by the time interval between the two scans.
where Sterm and Searly are the automatic hippocampal segmentations at term and early-in-life for each subject.
5.2.2. Hippocampal volumes and growth rates
Table 3 summarizes the mean volumes of the left and right hippocampi that were automatically delineated using the MAGeT-Brain pipeline from the 168 early-in-life and 154 term-equivalent images, as well as the hippocampal growth rates of 125 subjects between two scans. There was one subject who had negative hippocampal growth on the right side from early-in-life to term. We noticed that this infant had extensive brain injury, including white matter injury, intraventricular hemorrhage, and ventriculomegaly, which may have significantly impaired hippocampal development.
Table 3.
The automatically segmented hippocampal volumes on 168 early-in-life images, 154 term images, and the hippocampal growth rates of 125 subjects who had both early-in-life and term images.
| MAGeT-Brain | Left | Right | Average |
|---|---|---|---|
| Volume: 168 subjects at early-in-life (mm3) | Mean: 595.6 ± 121.2 Range: 358.1–1042.8 |
Mean: 648 ± 130 Range: 371.1–1217.4 |
Mean: 621.8 ± 122.4 Range: 364.6–1130.1 |
| Volume: 154 subjects at term age (mm3) | Mean: 925.2 ± 151.4 Range: 551.2–1297.7 |
Mean: 992.4 ± 156.3 Range: 595.7–1282.6 |
Mean: 958.8 ± 150.6 Range: 582.7–1273.3 |
| Growth rate: 125 subjects (mm3/week) | Mean: 38.3 ± 11.7 Range: 1.1–84.9 |
Mean: 40.5 ± 12.9 Range: −2.7–75.8 |
Mean: 39.4 ± 11.5 Range: 0.6–80.3 |
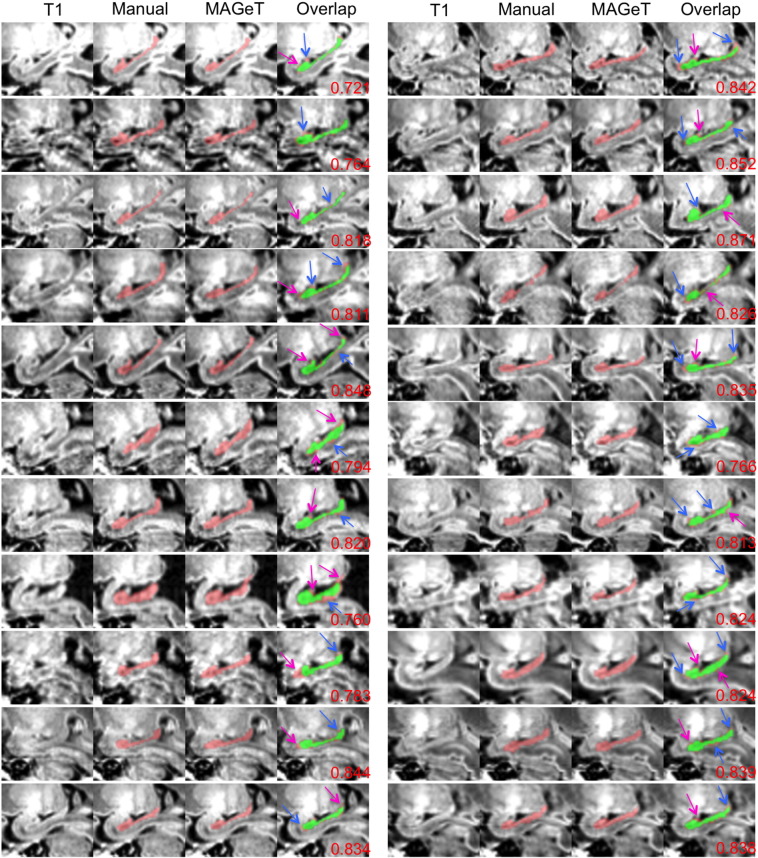
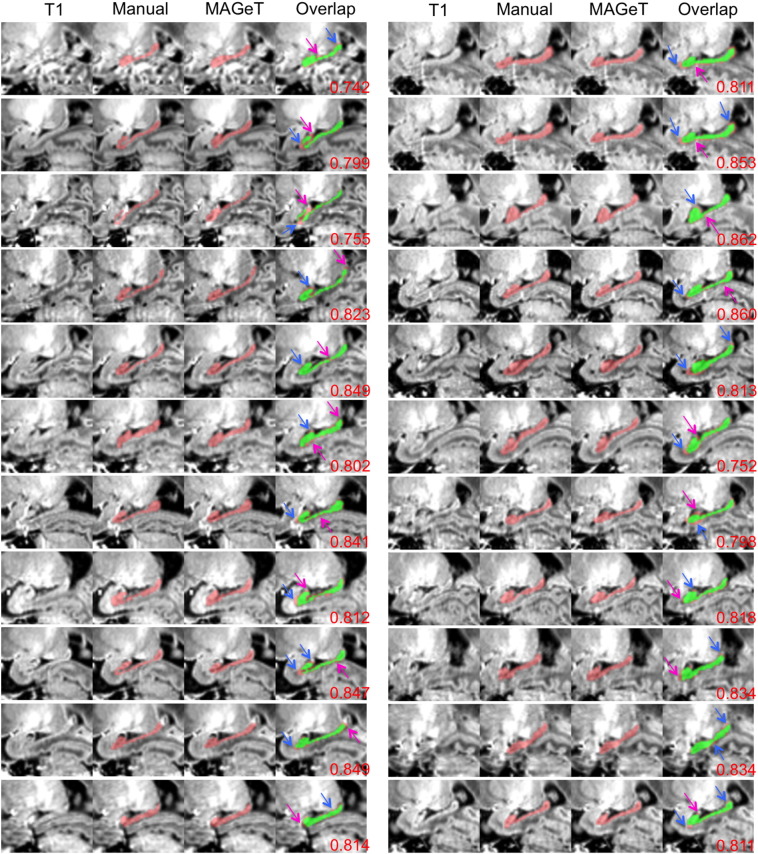
Figs. 8 and 9 display the visual analysis results of the manual and MAGeT-Brain hippocampal segmentations on the sagittal view in the 22 early-in-life and 22 term images. Excellent agreement between the hippocampi segmented manually and those by MAGeT-Brain can be found in these two figures. Discrepancy is only in peripheral areas.
Fig. 8.
Comparison of manual hippocampal segmentations with MAGeT-Brain-based hippocampal segmentations on the 22 early-in-life images of very preterm-born infants. Red: hippocampal regions segmented either manually or by MAGeT-Brain; green: the common regions of manual and MAGeT-Brain segmentations. Magenta arrows point to areas over-estimated by MAGeT-Brain; blue arrows point to areas under-estimated by MAGeT-Brain. Kappa values are at bottom right corners.
Fig. 9.
Comparison of manual hippocampal segmentations with MAGeT-Brain-based hippocampal segmentations on the 22 term images of very preterm-born infants (sagittal view). Red: hippocampal regions segmented either manually or by MAGeT-Brain; green: the common regions of manual and MAGeT-Brain segmentations. Magenta arrows point to areas over-estimated by MAGeT-Brain; blue arrows point to areas under-estimated by MAGeT-Brain. Kappa values are at bottom right corners.
5.3. Experiment 3: association of hippocampal volume with prematurity
5.3.1. Experiment 3: methods
We calculated the average volume of the left and right hippocampi for each of the 125 neonates who had both early-in-life and term images from their MAGeT-Brain segmentations. The effect of prematurity on the volume of the hippocampi was assessed using linear regression analysis. The average of the right and left hippocampal volumes at early-in-life and those at term-equivalent age scans were used as the dependent variable and GA at birth was used as the independent variable. The days of life (DOL) at the time of the MRI scans was considered as a covariate in the analysis. Statistical analyses were performed using Matlab (MATLAB and Statistics Toolbox, The MathWorks, Inc., Natick, MA, United States).
5.3.2. Experiment 3: Results
Linear regression analysis to assess the impact of premature birth on the volume of the hippocampi shows a positive significant effect (t = 17.9, df = 247, p < 0.001; Fig. 10). Results indicate that hippocampi volume assessed at the time of early-in-life and term-equivalent age is linearly associated with GA at birth, whereby smaller volumes are associated with earlier birth (~24 weeks) and the volumes are progressively larger in infants born later (25–32 weeks).
Fig. 10.
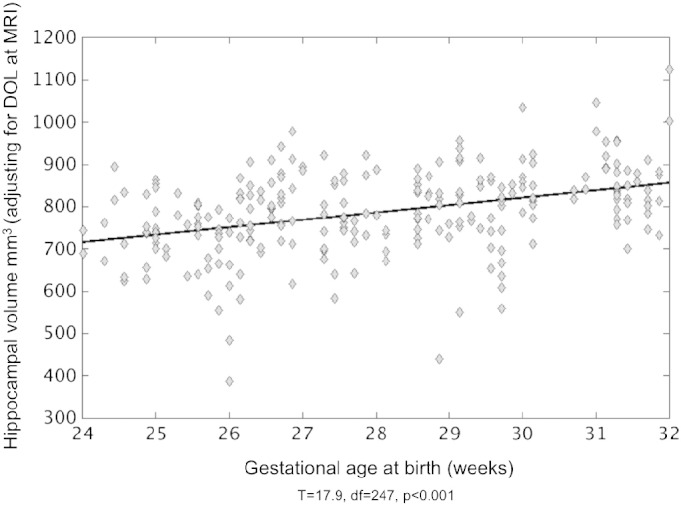
Impact of premature birth on hippocampal volume assessed within the first weeks of life and at term-equivalent age in very preterm born neonates. Data represent the average left and right hippocampal volumes (mm3) acquired at early-in-life and those at term-equivalent age for each neonate. Smaller hippocampal volume is associated with an earlier GA at birth (T = 17.9, p < 0.0001, adjusting for days of life [DOL] at MRI).
6. Discussion and conclusions
This manuscript presents a segmentation methodology that can effectively delineate the hippocampi of very preterm neonates (born 24–32 weeks GA) on both their early-in-life images (mean PMA 32 weeks) and term images (mean PMA 40 weeks). The three-step manual segmentation protocol described in this manuscript is a novel contribution that addresses the rules for the segmentation of hippocampus in the rapidly changing neuroanatomy of preterm-born infants from early-in-life to term-equivalent age. Particular care was taken to be sure that the segmentation rules that were developed to delineate the early-in-life images of very preterm neonates were also applicable to the neuroanatomy of neonates at term-equivalent age in order to maintain consistency amongst our neuroanatomical definitions. Given the 6-month intra-rater reliability interval, we also demonstrate that our manual segmentation protocol is consistent and easy to follow. These hippocampal segmentations are thus reliable definitions that are able to serve as the input atlases in our validation experiments of the automatically generated segmentations.
Although the expert rater strictly followed the 3-step manual segmentation protocol for both the early-in-life and term images, the intra-rater reliability of manual hippocampal segmentation is slightly lower for term images than that for early-in-life images. This could be caused by the fact that at term-equivalent age a higher proportion of images may suffer from motion artifacts and the shape of the hippocampus is much more convoluted. In this study the images were acquired without sedation and the babies at term-equivalent age have developed better muscle control over their neck and can move their heads much more than they could at early-in-life stage. Although we took exceptional care to pre-select MRIs that suffered from motion artifacts, it is indeed possible that some images with visually less obvious motion artifacts were included in this manuscript since the artifacts were undetected. Quantifying the effects of movement in this neonatal population, much in the same way that has been done in pediatric and adolescent populations would be of exceptional use to the field (Satterthwaite et al., 2012; Fair et al., 2012). Similarly, other methods have been developed to automatically detect MRI data suffering from large motion artifacts (Gedamu et al., 2008; Trampel et al., 2011), although it is unclear how well this method would work in neonates. Firstly, the contrast profile of these images varies greatly as a function of age. Therefore using a standard set of regions of interest employed in Gedamu et al. (2008) and Trampel et al. (2011) may not be feasible in this setting. The regions of interest used to define the signal-to-noise ratio are exclusively based on our knowledge of the contrast profile of the fully developed children or the adult human brain. It is unlikely that measuring signal-to-noise ratios using the same ROIs would work in this application. Moreover, due to the changing size and intensity/contrast-profiles of the brains, it is also unlikely that any pre-defined set of ROIs would work well here and that a set of regions that can be dynamically generated are what is required. Generating a novel algorithm that is able to automatically detect outliers due to excessive motion is of course of great interest to the neonatal imaging community. The development and validation of such an algorithm, are outside the scope of this manuscript.
In addition, rapid brain growth and maturation occur from early-in-life to term-equivalent age (Fig. 1a, b) and the more convoluted shape of the hippocampus at term significantly increases the difficulty in manual segmentation. Moreover, it is possible that design choices that were made for the manual segmentation protocol contributed to potential confounders. For example, although the images were in the original space during the segmentation process, a tricubic interpolant was used for the purposes of visualization but was never applied directly to the data that was used. As a result, the segmentations might be skewed to include voxels suffering from partial-volume effects. Nonetheless, the segmentations were verified without the effects of the interpolant and once more using the surface-based representation of the hippocampus. Therefore, our multi-stage manual segmentation technique should buffer against these confounders.
The rigorous 10-fold Monte Carlo cross-validation (MCCV) conducted in Experiment 1 on the early-in-life and the term groups reveals that the segmentation accuracy improves with the increased number of atlases and templates. In our previous experiments in the segmentation of the adult hippocampus in the context of Alzheimer's disease and first episode psychosis (Pipitone et al., 2014), the segmentation accuracy appears to plateau when the number of input atlases and templates exceeds 5 and 15 respectively. In the first and second validation studies, when we employ early-in-life atlases and templates to segment early-in-life images and term-equivalent atlases and templates to segment term-equivalent images, we are able to obtain consistently reliable and accurate segmentations. Whereas, in the third validation study, we employ randomly mixed atlases and templates to segment target images that are not age matched to the atlases and templates. The age difference between the input atlases/templates and the target images can be as far as 15 weeks. As we can see in Fig. 1, the size and shape of the neonatal brains even for the same subject 11 weeks apart can be dramatically different and the morphological differences can be too large to be accommodated by any high performance registration or segmentation algorithm. It should be noted that even though randomly selected age-mismatched atlases and templates are used in this study, MAGeT-Brain is still able to achieve accurate hippocampal segmentations in 98.3% of the 82,280 cases in this group and only 1.7% of the MAGeT-Brain segmentations have Kappa values lower than 0.6 when compared to the gold standard manual segmentations. Given these circumstances, it is reasonable to consider that MAGeT-Brain performs robustly to neuroanatomical variation that is present through the developmental period. When we combine the input atlases acquired at different post-menstrual ages, the segmentation accuracy (in Kappa) is slightly decreased. These results suggest that age appropriate atlases and templates are required to obtain better segmentation accuracy. Given the rapid growth of the brain and the heterogeneous morphology at these early points in the human life-span, it is encouraging to note that the robust segmentations are still possible even if the neuroanatomy in the atlas library is not homologous to the anatomy of the subjects to be labeled.
As demonstrated by the Dice's Kappa and Euclidean distance between the gold standard manual segmentations and the MAGeT-Brain-based segmentations that were generated in Experiment 2, the hippocampi on the early-in-life images and those on term-equivalent images can be accurately delineated using MAGeT-Brain (Table 1A). We evaluated the hippocampal segmentation accuracy of MAGeT-Brain employing strictly defined MR-PMA appropriate atlases and templates in delineating the hippocampi on images scanned at similar MR-PMA. However, no statistically significant enhancement was found for images in either the early-preterm or the term group defined by their MR-PMA (Table 1A). This further demonstrates that MAGeT-Brain performs robustly in hippocampal segmentation with different selections of atlases and templates. Accurate segmentation of the hippocampi allows us to evaluate whether younger birth GA is related to the sizes of hippocampal volumes. The high segmentation accuracy can be achieved by MAGeT-Brain in both early-in-life and term-equivalent images, suggesting its applicability in studying the volumetric and morphometric changes of hippocampus of preterm neonates.
Another quality metric that was used in this study was bias estimation using the Bland–Altman plot. While metrics such as the Dice similarity coefficient and the analysis of the centroids provide information regarding the accuracy and precision of the final segmentation, the analysis of the Bland–Altman plots provides more information regarding any fixed or proportional biases that may exist within the hippocampal segmentations. The plots of the volume demonstrate a slight bias where larger hippocampi are under-estimated while smaller hippocampi are over-estimated, regardless of the choice of the initial atlas library composition (see Fig. 6). This is consistent with the proportional bias reported in our previous work (Pipitone et al., 2014). However, these plots also demonstrate a slight underestimation of the overall hippocampal volume (~50–150 mm3). This may be due to the majority voting methodology used in the label fusion stage, which may be potentially filtering too many uncertain labels using a conservative threshold. In recent work from our group (Bhagwat et al., 2015), we did evaluate MAGeT Brain performance using STAPLE (Warfield et al., 2004) (an algorithm commonly used for label-fusion), and surprisingly, found no improvements in performance over majority vote. While this does not explicitly suggest that the performance of MAGeT Brain would not be improved through the inclusion of a more sophisticated label-fusion technique, the exploration of the performance of this technique using more state-of-the art label fusion methods (Coupe et al., 2011; Hao et al., 2014; Jorge Cardoso et al., 2013; Khan et al., 2011; Wang et al., 2011; Warfield et al., 2004; Wu et al., 2014; Yushkevich et al., 2010) is necessary and is an area of active investigation. More work is required to properly identify a label fusion method that improves the final segmentations but that can fully account for the changing intensity and contrast characteristics in the MRI data in this population through the course of development and maturation.
There are multi-atlas segmentation techniques that employ sophisticated atlas selection (Aljabar et al., 2009; Collins and Pruessner, 2010; Heckemann et al., 2006; Heckemann et al., 2010; Tong et al., 2013; Wolz et al., 2010) and label fusion methods (Coupe et al., 2011; Hao et al., 2014; Jorge Cardoso et al., 2013; Khan et al., 2011; Wang et al., 2011; Warfield et al., 2004; Wu et al., 2014; Yushkevich et al., 2010). It is indeed likely that employing such a strategy for the segmentation of the neonatal brain could be beneficial. It should be noted that in previous work, we did not see significant benefit of using cross-correlation weighted and normalized mutual information weighted voting over the simple majority voting fusion method in improving the segmentation accuracy of MAGeT-Brain from our previously published studies (Pipitone et al., 2014; Chakravarty et al., 2013). These studies also demonstrate that when given the same number of atlases, MAGeT-Brain is more accurate than multi-atlas segmentation and is more reliable than the probabilistic atlas approach (Diedrichsen et al., 2009). Further, in previous manuscripts (Pipitone et al., 2014) we also validated hippocampal segmentations against commonly used methods in the field, including: FreeSurfer (Fischl et al., 2002), FSL FIRST (Patenaude et al., 2011), MAPER (Heckemann et al., 2006; Heckemann et al., 2010), and SNT (Medtronic Surgical Navigation Technologies, Louisville, CO, USA). In all cases we found that MAGeT Brain outperformed many of these methods in terms of consistency and quality of segmentation. Nonetheless, improving the label fusion stage is an active area of research for improving this algorithm.
Although the average volume of the right hippocampus is only marginally larger than that of the left hippocampus for the 22 manually segmented subjects in Table 1, asymmetrical hippocampal volumes are noted in the MAGeT-Brain segmentation results for the 168 early-in-life images and the 154 term-equivalent images in Experiment 2 with the right side being larger than the left side. We should note that we did not observe these asymmetries in the originally manually segmented data. This is likely to be a product of the differences in the sample sizes analyzed. The finding of the hippocampal asymmetry (right hippocampus > left hippocampus) in the term images of preterm-born neonates is in concordance with what was reported in an earlier study by Thompson et al. (2009). Hippocampal asymmetry was also observed in children (Giedd et al., 1996; Pfluger et al., 1999; Utsunomiya et al., 1999) and adults (Jack et al., 2000; Watson et al., 1992), however the underlying mechanisms for this phenomenon still remain unknown. To our knowledge, this is the earliest demonstration of hippocampal asymmetry well prior to term in preterm neonates (mean MR-PMA: 32 weeks), in what would have been the third-trimester of gestation. These data suggest that the hippocampal asymmetry previously described at term-age and in older children and adults has its onset in the 3rd trimester of gestation or earlier. Given the size of the sample analyzed here, it is unlikely that this finding is artifactual. Nonetheless, it is possible that it may be the product of some sort of systematic bias causing data quality to be lower on the right side of the image (due to field inhomogeneities or even noise).
We note that the absolute error was larger in the y direction (anterior–posterior) than that in the other two directions when we calculated the Euclidean distances between the centroids of the manually and automatically segmented hippocampi (Table 2A). This was very likely due to the fact that the longest section of the hippocampus runs almost parallel to the y-axis. The error measured with the average Euclidean distance in the term group was larger than that in the early-in-life group, because of the brain growth and the hippocampal growth from preterm to term ages. Although good agreement is found between the MAGeT-Brain based segmentations and the gold standard manual segmentations, the automatically segmented hippocampi obtained with MAGeT-Brain tend to be smaller than the manually segmented hippocampal volumes.
Contrary to what we expected, employing a pre-processing step (N3) to correct the image intensity non-uniformity (Sled et al., 1998) did not improve the hippocampal segmentation accuracy of MAGeT-Brain for neither the early-in-life nor the term images. The hippocampal volumes that were segmented from pre-processed images were at the correct positions, but significantly smaller in size when they were compared with the manual segmentations; these problems were persistent regardless of our choice of parameter settings. We were surprised that the benefits that were seen in previous studies for the adult brain segmentations (Zheng et al., 2009) were not achieved in segmenting the hippocampi on the images of preterm infants. Since the average size of an infant brain is only one-third that of an adult brain (Ponce de León et al., 2008), acquiring infant brain images requires much smaller field of view (FOV) than acquiring adult brain images, and the artifacts caused by magnetic field inhomogeneity may be less pronounced for the neonatal brain images. As a result, N3 may over-correct the non-uniformity on neonatal images thereby affecting our segmentation results negatively, making this an important area for further investigation.
High-resolution T2-weighted images usually require much longer acquisition time than T1-weighted images in the same resolution. Due to the limited available scanner time, it is often not feasible to acquire high-resolution 3D T2-weighted images. T2-weighted images have usually been acquired with high in-plane resolution but large inter-slide distance. This kind of T2 weighted images alone is not suitable for direct volumetric analysis of small brain structures, such as the hippocampus. While we have not assessed this directly here, it is possible that post-processing techniques such as simple addition of the T2 and PD images can improve contrast to noise (CNR) and facilitate the definition of hippocampal boundaries on images acquired at term-equivalent age (Thompson et al., 2012). The manual segmentation results obtained by two independent raters on the combined T2 and PD images were better than those only on T2 images. Utilizing multi-spectral data may provide more detailed information on the hippocampal borders and allow more accurate delineation of this convoluted structure. Furthermore, MR image quality, and subsequent segmentation quality, can be improved with better accessibility to higher field strength MR scanners, custom-made multi-channel coils (Panigrahy et al., 2012; Reiss-Zimmermann et al., 2013), and more advanced acquisition techniques (Wargo et al., 2013).
Only a few semi-automatic and automatic segmentation approaches are available to delineate the hippocampus on neonatal brain images of preterm infants (Nishida et al., 2006; Gousias et al., 2013; Makropoulos et al., 2014). Nishida's semi-automatic method is the first that was applied to segment hippocampus on images scanned before term though quantitative validations of their segmentation results were not presented. The work presented by Gousias and that by Makropoulos is more closely related to our hippocampal segmentation technique. Gousias et al. performed two types of automatic segmentation on the MR images of preterm infants scanned at term (Gousias et al., 2013). The first study used a multi-atlas based approach which generated the final segmentation through fusion of the atlas labels from multiple neonatal atlases, while the second study employed a model-based technique which was achieved by propagating labels from one of the eight maximum probability neonatal ALBERT (MPNA) to the target brain. It is extremely difficult to compare segmentation results between laboratories given the differences in neuroanatomical definitions and the underlying MRI dataset itself, it is, nonetheless, important to compare the results from our two groups against one another. In Makropoulos et al. (2014), the authors report maximum hippocampus Dice similarity coefficients (0.797 and 0.783; left and right sides respectively) that are comparable with what are reported here (0.783 and 0.808; left and right sides respectively). The main difference in the evaluation of the segmentation lies in our 10-fold Monte Carlo cross-validation (MCCV) and their leave-one-out validation. In addition, we have taken particular care to report a robust and reliable manual segmentation protocol, something that we have yet to see in the literature.
Different from multi-atlas based segmentation methods, MAGeT-brain does not require an exhaustive manually defined atlas library (typically only five input atlases is required to achieve a consistent and accurate labeling). However, it is important to note that there are other works that have explicitly addressed this issue. Both the label fusion techniques of Wolz et al. (2010) and Coupé et al. (2012) demonstrate that accurate segmentation can be achieved through the use of 20 and 30 manually derived atlases as input. To the best of our knowledge, however, neither of these methods has been validated for the unique neuroanatomy of the neonate brain. Further, neither performed their validations using a multi-fold analysis to demonstrate that the final segmentations are robust to the choice of input parameters.
The intermediate template images can be chosen according to specific selection strategies to best suit the segmentation of the targeted subject images. The nonlinear registration algorithm, ANTs (Avants et al., 2008; Avants et al., 2011), employed to accommodate the inter-subject anatomical variability in this study uses more than twenty million degrees of freedom of the deformation and was ranked amongst the highest in a comprehensive validation study of 14 popularly used registration methods (Klein et al., 2009). MAGeT-Brain also has the flexibility that it can be implemented using any validated nonlinear registration algorithm. Although this technique requires considerable computational resources, given rapidly improved accessibility of modern super computing infrastructure, this limitation can be readily overcome. MAGeT-Brain's minimal demand on labor-intense and time consuming manual labeling makes it a desirable approach to analyze the changes in hippocampus and/or other brain structures for cohorts with a large number of subjects.
One of the disadvantages of the MAGeT-Brain algorithm is its computational complexity. While it would be possible to run this on a single computer, the computation time would scale directly with the size of the atlas and template libraries. In our case, all implementations were done using a high-performance computing infrastructure in a grossly parallel fashion in order to generate all of the segmentations described in this manuscript. Given the increasing availability of this type of infrastructure, we would recommend that this algorithm be implemented in the same way.
In conclusion, we have developed a reliable and efficient segmentation protocol to outline the boundaries of hippocampal volumes in preterm neonates, even early-in-life. Strong agreement is found between automatic MAGeT-Brain segmentations and the gold-standard manual hippocampal segmentations in size and location. No significant difference is observed when MR-PMA appropriate atlases and templates are applied to segment hippocampi of images acquired at similar MR-PMA. Hippocampal asymmetry is demonstrated in both the early-in-life and term images segmented with MAGeT-Brain. Using MAGeT-Brain segmentation pipeline, we are able to analyze the hippocampal growth rates of neonates who have both early-in-life and term images, and assess the effect of preterm birth on the volumes of hippocampi. Smaller hippocampal volumes are associated with earlier gestational age at birth. Although a number of methods were published for the segmentation of hippocampus in images of preterm-born infants that were acquired at term-equivalent age (Ball et al., 2012; Gousias et al., 2012; Gousias et al., 2013; Makropoulos et al., 2014; Nishida et al., 2006; Thompson et al., 2008; Thompson et al., 2012; Thompson et al., 2013), to the best of our knowledge, this is the first segmentation approach that is specifically designed to delineate the hippocampus from the first weeks of life in very preterm-born neonates. This segmentation pipeline also shows promise in segmenting fetal images in the third trimester.
Acknowledgments
Computations were performed on the SciNet supercomputer at the SciNet HPC Consortium. SciNet is funded by the Canada Foundation for Innovation under the auspices of Compute Canada, the Government of Ontario, Ontario Research Fund — Research Excellence and the University of Toronto. Supported by the Canadian Institutes of Health Research (CIHR) operating grants MOP-79262 (S.P.M.) and MOP-86489 (R.E.G.). SPM is currently the Bloorview Children's Hospital Chair in Pediatric Neuroscience and was supported by a Tier 2 Canada Research Chair in Neonatal Neuroscience, and Michael Smith Foundation for Health Research Scholar Award. MMC receives salary support from the Fonds de Recherche Québec Santé and is supported by CIHR, Natural Sciences and Engineering Research Council of Canada, Weston Brain Institute, the Alzheimer's Society, Michael J Fox Foundation for Parkinson's Research, and Brain, Canada. REG holds a Senior Scientist salary award from the Child and Family Research Institute. We thank Dr. Margot Taylor, The Hospital for Sick Children (Toronto, Ontario), for the longitudinal MRI images presented in Fig. 1.
Contributor Information
Ting Guo, Email: jessie.guo@sickkids.ca.
M. Mallar Chakravarty, Email: mallar@cobralab.ca.
Appendix A
Table 1A.
The Dice's Kappa measurements of similarity between the manually segmented hippocampi and the MAGeT-Brain segmented hippocampal volumes. In Experiment 2, 15 atlases and 21 templates were used and in MR-PMA appropriate study, 17 atlases and 21 templates that were acquired at similar MR-PMA with each target image were used.
| Mean Dice's kappa | Left | Right | Both | |
|---|---|---|---|---|
| Early-in-life | Experiment 2 | 0.784 ± 0.053 | 0.804 ± 0.036 | 0.794 ± 0.036 |
| MR-PMA appropriate | 0.782 ± 0.049 | 0.800 ± 0.033 | 0.791 ± 0.033 | |
| Term | Experiment 2 | 0.781 ± 0.048 | 0.812 ± 0.030 | 0.797 ± 0.031 |
| MR-PMA appropriate | 0.780 ± 0.048 | 0.814 ± 0.031 | 0.805 ± 0.032 |
Table 2A.
The absolute displacements in x, y, and z directions and the Euclidean distances between the centroids of the manually segmented hippocampi and those of the MAGeT-Brain segmented hippocampi in early-in-life and term groups of Experiment 1.
| Mean of absolute displacement | x (mm) | y (mm) | z (mm) | Euclidean distance (mm) |
|---|---|---|---|---|
| Early in life — left | 0.45 ± 0.30 | 0.47 ± 0.31 | 0.41 ± 0.32 | 0.90 ± 0.35 |
| Early in life — right | 0.34 ± 0.28 | 0.61 ± 0.50 | 0.45 ± 0.35 | 0.97 ± 0.49 |
| Term — left | 0.46 ± 0.39 | 0.86 ± 0.60 | 0.43 ± 0.37 | 1.20 ± 0.65 |
| Term — right | 0.49 ± 0.30 | 0.82 ± 0.48 | 0.46 ± 0.37 | 1.20 ± 0.45 |
References
- Aarnoudse-Moens C.S., Duivenvoorden H.J., Weisglas-Kuperus N., Van Goudoever J.B., Oosterlaan J. The profile of executive function in very preterm children at 4 to 12 years. Dev. Med. Child Neurol. 2012;54(3):247–253. doi: 10.1111/j.1469-8749.2011.04150.x. 22126188 [DOI] [PubMed] [Google Scholar]
- Aarnoudse-Moens C.S., Smidts D.P., Oosterlaan J., Duivenvoorden H.J., Weisglas-Kuperus N. Executive function in very preterm children at early school age. J. Abnorm. Child Psychol. 2009;37(7):981–993. doi: 10.1007/s10802-009-9327-z. 19488851 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akhondi-Asl A., Jafari-Khouzani K., Elisevich K., Soltanian-Zadeh H. Hippocampal volumetry for lateralization of temporal lobe epilepsy: automated versus manual methods. Neuroimage. 2011;54(Suppl. 1):S218–S226. doi: 10.1016/j.neuroimage.2010.03.066. 20353827 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aljabar P., Heckemann R.A., Hammers A., Hajnal J.V., Rueckert D. Multi-atlas based segmentation of brain images: atlas selection and its effect on accuracy. Neuroimage. 2009;46(3):726–738. doi: 10.1016/j.neuroimage.2009.02.018. 19245840 [DOI] [PubMed] [Google Scholar]
- Anbeek P., Išgum I., van Kooij B.J.M., Mol C.P., Kersbergen K.J., Groenendaal F., Viergever M.A., de Vries L.S., Benders M.J.N.L. Automatic segmentation of eight tissue classes in neonatal brain MRI. PLOS One. 2013;8(12):e81895. doi: 10.1371/journal.pone.0081895. 24358132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Avants B.B., Epstein C.L., Grossman M., Gee J.C. Symmetric diffeomorphic image registration with cross-correlation: evaluating automated labeling of elderly and neurodegenerative brain. Med. Image Anal. 2008;12(1):26–41. doi: 10.1016/j.media.2007.06.004. 17659998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Avants B.B., Tustison N.J., Song G., Cook P.A., Klein A., Gee J.C. A reproducible evaluation of ANTs similarity metric performance in brain image registration. Neuroimage. 2011;54(3):2033–2044. doi: 10.1016/j.neuroimage.2010.09.025. 20851191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Back S.A., Miller S.P. Brain injury in premature neonates: a primary cerebral dysmaturation disorder? Ann. Neurol. 2014;75(4):469–486. doi: 10.1002/ana.24132. 24615937 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ball G., Boardman J.P., Rueckert D., Aljabar P., Arichi T., Merchant N., Counsell S.J. The effect of preterm birth on thalamic and cortical development. Cereb. Cortex. 2012;22(5):1016–1024. doi: 10.1093/cercor/bhr176. 21772018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beauchamp M.H., Thompson D.K., Howard K., Doyle L.W., Egan G.F., Inder T.E., Anderson P.J. Preterm infant hippocampal volumes correlate with later working memory deficits. Brain. 2008;131(11):2986–2994. doi: 10.1093/brain/awn227. 18799516 [DOI] [PubMed] [Google Scholar]
- Bhagwat N., Pipitone J., Voineskos A., Pruessner J., Chakravarty M.M. Improving multi-atlas segmentation accuracy by leveraging local neighborhood information during label-fusion. International Symposium on Biomedical Imaging (ISBI) 2015; 2015. [Google Scholar]
- Bland J.M., Altman D.G. Statistical methods for assessing agreement between two methods of clinical measurement. Lancet. 1986;1(8476):307–310. 2868172 [PubMed] [Google Scholar]
- Boccardi M., Bocchetta M., Ganzola R., Robitaille N., Redolfi A., Duchesne S., Jack C.R., Frisoni G.B., EADC-ADNI Working Group on The Harmonized Protocol for Manual Hippocampal Segmentation and for the Alzheimer's Disease Neuroimaging Initiative Operationalizing protocol differences for EADC-ADNI manual hippocampal segmentation. Alzheimers Dement. 2015;11(2):184–194. doi: 10.1016/j.jalz.2013.03.001. 23706515 [DOI] [PubMed] [Google Scholar]
- Cardoso M.J., Melbourne A., Kendall G.S., Modat M., Robertson N.J., Marlow N., Ourselin S. AdaPT: an adaptive preterm segmentation algorithm for neonatal brain MRI. Neuroimage. 2013;65:97–108. doi: 10.1016/j.neuroimage.2012.08.009. 22906793 [DOI] [PubMed] [Google Scholar]
- Chakravarty M.M., Steadman P., van Eede M.C., Calcott R.D., Gu V., Shaw P., Raznahan A., Collins D.L., Lerch J.P. Performing label-fusion-based segmentation using multiple automatically generated templates. Hum. Brain Mapp. 2013;34(10):2635–2654. doi: 10.1002/hbm.22092. 22611030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang H.H., Larson J., Blencowe H., Spong C.Y., Howson C.P., Cairns-Smith S., Lackritz E.M., Lee S.K., Mason E., Serazin A.C., Walani S., Simpson J.L., Lawn J.E. Preventing preterm births: analysis of trends and potential reductions with interventions in 39 countries with very high human development index. Lancet. 2013;381(9862):223–234. doi: 10.1016/S0140-6736(12)61856-X. 23158883 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chau V., Synnes A., Grunau R.E., Poskitt K.J., Brant R., Miller S.P. Abnormal brain maturation in preterm neonates associated with adverse developmental outcomes. Neurology. 2013;81(24):2082–2089. doi: 10.1212/01.wnl.0000437298.43688.b9. 24212394 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheong J.L.Y., Anderson P.J., Roberts G., Burnett A.C., Lee K.J., Thompson D.K., Molloy C., Wilson-Ching M., Connelly A., Seal M.L., Wood S.J., Doyle L.W. Contribution of brain size to IQ and educational underperformance in extremely preterm adolescents. PLOS One. 2013;8(10):e77475. doi: 10.1371/journal.pone.0077475. 24130887 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choe M.-S., Ortiz-Mantilla S., Makris N., Gregas M., Bacic J., Haehn D., Kennedy D., Pienaar R., Caviness V.S., Jr, Benasich A.A., Grant P.E. Regional infant brain development: an MRI-based morphometric analysis in 3 to 13 month olds. Cereb. Cortex. 2013;23(9):2100–2117. doi: 10.1093/cercor/bhs197. 22772652 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chupin M., Gérardin E., Cuingnet R., Boutet C., Lemieux L., Lehéricy S., Benali H., Garnero L., Colliot O. Fully automatic hippocampus segmentation and classification in Alzheimer's disease and mild cognitive impairment applied on data from ADNI. Hippocampus. 2009;19(6):579–587. doi: 10.1002/hipo.20626. 19437497 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chupin M., Hammers A., Liu R.S.N., Colliot O., Burdett J., Bardinet E., Duncan J.S., Garnero L., Lemieux L. Automatic segmentation of the hippocampus and the amygdala driven by hybrid constraints: method and validation. Neuroimage. 2009;46(3):749–761. doi: 10.1016/j.neuroimage.2009.02.013. 19236922 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins D.L., Pruessner J.C. Towards accurate, automatic segmentation of the hippocampus and amygdala from MRI by augmenting ANIMAL with a template library and label fusion. Neuroimage. 2010;52(4):1355–1366. doi: 10.1016/j.neuroimage.2010.04.193. 20441794 [DOI] [PubMed] [Google Scholar]
- Coupé P., Eskildsen S.F., Manjón J.V., Fonov V.S., Collins D.L., Alzheimer’s Disease Neuroimaging Initiative Simultaneous segmentation and grading of anatomical structures for patient's classification: application to Alzheimer's disease. Neuroimage. 2012;59(4):3736–3747. doi: 10.1016/j.neuroimage.2011.10.080. 22094645 [DOI] [PubMed] [Google Scholar]
- Coupé P., Manjón J.V., Fonov V., Pruessner J., Robles M., Collins D.L. Patch-based segmentation using expert priors: application to hippocampus and ventricle segmentation. Neuroimage. 2011;54(2):940–954. doi: 10.1016/j.neuroimage.2010.09.018. 20851199 [DOI] [PubMed] [Google Scholar]
- Csernansky J.G., Joshi S., Wang L., Haller J.W., Gado M., Miller J.P., Grenander U., Miller M.I. Hippocampal morphometry in schizophrenia by high dimensional brain mapping. Proc. Natl. Acad. Sci. U. S. A. 1998;95(19):11406–11411. doi: 10.1073/pnas.95.19.11406. 9736749 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Kieviet J.F., Zoetebier L., van Elburg R.M., Vermeulen R.J., Oosterlaan J. Brain development of very preterm and very low-birthweight children in childhood and adolescence: a meta-analysis. Dev. Med. Child Neurol. 2012;54(4):313–323. doi: 10.1111/j.1469-8749.2011.04216.x. 22283622 [DOI] [PubMed] [Google Scholar]
- Diedrichsen J., Balsters J.H., Flavell J., Cussans E., Ramnani N. A probabilistic MR atlas of the human cerebellum. Neuroimage. 2009;46(1):39–46. doi: 10.1016/j.neuroimage.2009.01.045. 19457380 [DOI] [PubMed] [Google Scholar]
- Duvernoy H., Cattin F., Naidich T., Fatterpekar G., Raybaud C., Risold P.Y., Salvolini U., Scarabino T. Springer-Verlag; Berlin, Germany: 2005. The Human Hippocampus: Functional Anatomy, Vascularization and Serial Sections with MRI. [Google Scholar]
- Fair D.A., Nigg J.T., Iyer S., Bathula D., Mills K.L., Dosenbach N.U.F., Schlaggar B.L., Mennes M., Gutman D., Bangaru S., Buitelaar J.K., Dickstein D.P., Di Martino A., Kennedy D.N., Kelly C., Luna B., Schweitzer J.B., Velanova K., Wang Y.-F., Mostofsky S., Castellanos F.X., Milham M.P. Distinct neural signatures detected for ADHD subtypes after controlling for micro-movements in resting state functional connectivity MRI data. Front. Syst. Neurosci. 2012;6:80. doi: 10.3389/fnsys.2012.00080. 23382713 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischl B., Salat D.H., Busa E., Albert M., Dieterich M., Haselgrove C., van der Kouwe A., Killiany R., Kennedy D., Klaveness S., Montillo A., Makris N., Rosen B., Dale A.M. Whole brain segmentation: automated labeling of neuroanatomical structures in the human brain. Neuron. 2002;33(3):341–355. doi: 10.1016/s0896-6273(02)00569-x. 11832223 [DOI] [PubMed] [Google Scholar]
- Ford R.M., Neulinger K., O’Callaghan M., Mohay H., Gray P., Shum D. Executive function in 7–9-year-old children born extremely preterm or with extremely low birth weight: effects of biomedical history, age at assessment, and socioeconomic status. Arch. Clin. Neuropsychol. 2011;26(7):632–644. doi: 10.1093/arclin/acr061. 21816952 [DOI] [PubMed] [Google Scholar]
- Gao Y., Corn B., Schifter D., Tannenbaum A. Multiscale 3D shape representation and segmentation with applications to hippocampal/caudate extraction from brain MRI. Med. Image Anal. 2012;16(2):374–385. doi: 10.1016/j.media.2011.10.002. 22119491 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gedamu E.L., Collins D.L., Arnold D.L. Automated quality control of brain MR images. J. Magn. Reson. Imaging. 2008;28(2):308–319. doi: 10.1002/jmri.21434. [DOI] [PubMed] [Google Scholar]
- Giedd J.N., Vaituzis A.C., Hamburger S.D., Lange N., Rajapakse J.C., Kaysen D., Vauss Y.C., Rapoport J.L. Quantitative MRI of the temporal lobe, amygdala, and hippocampus in normal human development: ages 4–18 years. J. Comp. Neurol. 1996;366(2):223–230. doi: 10.1002/(SICI)1096-9861(19960304)366:2<223::AID-CNE3>3.0.CO;2-7. 8698883 [DOI] [PubMed] [Google Scholar]
- Gimenez M., Soria-Pastor S., Junque C., Caldu X., Narberhaus A., Botet F., Bargallo N., Falcon C., Mercader J.M. Proton magnetic resonance spectroscopy reveals medial temporal metabolic abnormalities in adolescents with history of preterm birth. Pediatr. Res. 2008;64(5):572–577. doi: 10.1203/PDR.0b013e3181841eab. 18596571 [DOI] [PubMed] [Google Scholar]
- Gousias I.S., Edwards A.D., Rutherford M.A., Counsell S.J., Hajnal J.V., Rueckert D., Hammers A. Magnetic resonance imaging of the newborn brain: manual segmentation of labelled atlases in term-born and preterm infants. Neuroimage. 2012;62(3):1499–1509. doi: 10.1016/j.neuroimage.2012.05.083. 22713673 [DOI] [PubMed] [Google Scholar]
- Gousias I.S., Hammers A., Counsell S.J., Srinivasan L., Rutherford M.A., Heckemann R.A., Hajnal J.V., Rueckert D., Edwards A.D. Magnetic resonance imaging of the newborn brain: automatic segmentation of brain images into 50 anatomical regions. PLOS One. 2013;8(4):e59990. doi: 10.1371/journal.pone.0059990. 23565180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gui L., Lisowski R., Faundez T., Hüppi P.S., Lazeyras F., Kocher M. Morphology-driven automatic segmentation of MR images of the neonatal brain. Med. Image Anal. 2012;16(8):1565–1579. doi: 10.1016/j.media.2012.07.006. 22921305 [DOI] [PubMed] [Google Scholar]
- Haller J.W., Banerjee A., Christensen G.E., Gado M., Joshi S., Miller M.I., Sheline Y., Vannier M.W., Csernansky J.G. Three-dimensional hippocampal MR morphometry with high-dimensional transformation of a neuroanatomic atlas. Radiology. 1997;202(2):504–510. doi: 10.1148/radiology.202.2.9015081. 9015081 [DOI] [PubMed] [Google Scholar]
- Hao Y., Wang T., Zhang X., Duan Y., Yu C., Jiang T., Fan Y. Local label learning (LLL) for subcortical structure segmentation: application to hippocampus segmentation. Hum. Brain Mapp. 2014;35(6):2674–2697. doi: 10.1002/hbm.22359. 24151008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heckemann R.A., Hajnal J.V., Aljabar P., Rueckert D., Hammers A. Automatic anatomical brain MRI segmentation combining label propagation and decision fusion. Neuroimage. 2006;33(1):115–126. doi: 10.1016/j.neuroimage.2006.05.061. 16860573 [DOI] [PubMed] [Google Scholar]
- Heckemann R.A., Keihaninejad S., Aljabar P., Rueckert D., Hajnal J.V., Hammers A. Improving intersubject image registration using tissue-class information benefits robustness and accuracy of multi-atlas based anatomical segmentation. Neuroimage. 2010;51(1):221–227. doi: 10.1016/j.neuroimage.2010.01.072. 20114079 [DOI] [PubMed] [Google Scholar]
- Holsti L., Grunau R.V.E., Whitfield M.F. Developmental coordination disorder in extremely low birth weight children at nine years. J. Dev. Behav. Pediatr. 2002;23(1):9–15. doi: 10.1097/00004703-200202000-00002. 11889346 [DOI] [PubMed] [Google Scholar]
- Hu S., Coupé P., Pruessner J.C., Collins D.L. Appearance-based modeling for segmentation of hippocampus and amygdala using multi-contrast MR imaging. Neuroimage. 2011;58(2):549–559. doi: 10.1016/j.neuroimage.2011.06.054. 21741485 [DOI] [PubMed] [Google Scholar]
- Insausti R., Cebada-Sánchez S., Marcos P. Postnatal development of the human hippocampal formation. Adv. Anat. Embryol. Cell Biol. 2010;206:1–86. 20329667 [PubMed] [Google Scholar]
- Jack C.R., Petersen R.C., Xu Y., O’Brien P.C., Smith G.E., Ivnik R.J., Boeve B.F., Tangalos E.G., Kokmen E. Rates of hippocampal atrophy correlate with change in clinical status in aging and AD. Neurology. 2000;55(4):484–489. doi: 10.1212/wnl.55.4.484. 10953178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jorge Cardoso M.J., Leung K., Modat M., Keihaninejad S., Cash D., Barnes J. STEPS: similarity and truth estimation for propagated segmentations and its application to hippocampal segmentation and brain parcelation. Med. Image Anal. 2013;17(6):671–684. doi: 10.1016/j.media.2013.02.006. 23510558 [DOI] [PubMed] [Google Scholar]
- Khan A.R., Cherbuin N., Wen W., Anstey K.J., Sachdev P., Beg M.F. Optimal weights for local multi-atlas fusion using supervised learning and dynamic information (SuperDyn): validation on hippocampus segmentation. Neuroimage. 2011;56(1):126–139. doi: 10.1016/j.neuroimage.2011.01.078. 21296166 [DOI] [PubMed] [Google Scholar]
- Klein A., Andersson J., Ardekani B.A., Ashburner J., Avants B., Chiang M.-C., Parsey R.V. Evaluation of 14 nonlinear deformation algorithms applied to human brain MRI registration. Neuroimage. 2009;46(3):786–802. doi: 10.1016/j.neuroimage.2008.12.037. 19195496 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konrad C., Ukas T., Nebel C., Arolt V., Toga A.W., Narr K.L. Defining the human hippocampus in cerebral magnetic resonance images — an overview of current segmentation protocols. Neuroimage. 2009;47(4):1185–1195. doi: 10.1016/j.neuroimage.2009.05.019. 19447182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Limperopoulos C., Tworetzky W., McElhinney D.B., Newburger J.W., Brown D.W., Robertson R.L., Jr., Guizard N., McGrath E., Geva J., Annese D., Dunbar-Masterson C., Trainor B., Laussen P.C., du Plessis A.J. Brain volume and metabolism in fetuses with congenital heart disease: evaluation with quantitative magnetic resonance imaging and spectroscopy. Circulation. 2010;121(1):26–33. doi: 10.1161/CIRCULATIONAHA.109.865568. 20026783 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lodygensky G.A., Rademaker K., Zimine S., Gex-Fabry M., Lieftink A.F., Lazeyras F., Groenendaal F., de Vries L.S., Huppi P.S. Structural and functional brain development after hydrocortisone treatment for neonatal chronic lung disease. Pediatrics. 2005;116(1):1–7. doi: 10.1542/peds.2004-1275. 15995023 [DOI] [PubMed] [Google Scholar]
- Lodygensky G.A., Seghier M.L., Warfield S.K., Tolsa C.B., Sizonenko S., Lazeyras F., Hüppi P.S. Intrauterine growth restriction affects the preterm infant's hippocampus. Pediatr. Res. 2008;63(4):438–443. doi: 10.1203/PDR.0b013e318165c005. 18356754 [DOI] [PubMed] [Google Scholar]
- Lorensen W.E., Cline H.E. Marching cubes: a high resolution 3D surface construction algorithm. A.C.M. Computer Graphics. 1987;21(4):163–169. [Google Scholar]
- Lötjönen J.M., Wolz R., Koikkalainen J.R., Thurfjell L., Waldemar G., Soininen H. Fast and robust multi-atlas segmentation of brain magnetic resonance images. Neuroimage. 2010;49(3):2352–2365. doi: 10.1016/j.neuroimage.2009.10.026. 19857578 [DOI] [PubMed] [Google Scholar]
- Makropoulos A., Gousias I.S., Ledig C., Aljabar P., Serag A., Hajnal J.V., Edwards A.D., Counsell S.J., Rueckert D. Automatic whole brain MRI segmentation of the developing neonatal brain. I.E.E.E. Trans. Med. Imaging. 2014;33(9):1818–1831. doi: 10.1109/TMI.2014.2322280. 24816548 [DOI] [PubMed] [Google Scholar]
- Mathur A., Inder T. Magnetic resonance imaging — insights into brain injury and outcomes in premature infants. J. Commun. Disord. 2009;42(4):248–255. doi: 10.1016/j.jcomdis.2009.03.007. 19406431 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morey R.A., Petty C.M., Xu Y., Hayes J.P., Wagner H.R., Lewis D.V., LaBar K.S., Styner M., McCarthy G. A comparison of automated segmentation and manual tracing for quantifying hippocampal and amygdala volumes. Neuroimage. 2009;45(3):855–866. doi: 10.1016/j.neuroimage.2008.12.033. 19162198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moster D., Lie R.T., Markestad T. Long-term medical and social consequences of preterm birth. N. Engl. J. Med. 2008;359(3):262–273. doi: 10.1056/NEJMoa0706475. 18635431 [DOI] [PubMed] [Google Scholar]
- Mulder H., Pitchford N.J., Marlow N. Processing speed and working memory underlie academic attainment in very preterm children. Arch. Dis. Child. Fetal Neonatal. Ed. 2010;95(4):F267–F272. doi: 10.1136/adc.2009.167965. 20488865 [DOI] [PubMed] [Google Scholar]
- Nishida M., Makris N., Kennedy D.N., Vangel M., Fischl B., Krishnamoorthy K.S., Caviness V.S., Grant P.E. Detailed semiautomated MRI based morphometry of the neonatal brain: preliminary results. Neuroimage. 2006;32(3):1041–1049. doi: 10.1016/j.neuroimage.2006.05.020. 16857388 [DOI] [PubMed] [Google Scholar]
- Nosarti C., Al-Asady M.H.S., Frangou S., Stewart A.L., Rifkin L., Murray R.M. Adolescents who were born very preterm have decreased brain volumes. Brain. 2002;125(7):1616–1623. doi: 10.1093/brain/awf157. 12077010 [DOI] [PubMed] [Google Scholar]
- Panigrahy A., Wisnowski J.L., Furtado A., Lepore N., Paquette L., Bluml S. Neuroimaging biomarkers of preterm brain injury: toward developing the preterm connectome. Pediatr. Radiol. 2012;42(Suppl. 1):S33–S61. doi: 10.1007/s00247-011-2239-4. 22395719 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papile L.A., Burstein J., Burstein R., Koffler H. Incidence and evolution of subependymal and intraventricular hemorrhage: a study of infants with birth weights less than 1,500 gm. J. Pediatr. 1978;92(4):529–534. doi: 10.1016/s0022-3476(78)80282-0. 305471 [DOI] [PubMed] [Google Scholar]
- Park M.T., Pipitone J., Baer L.H., Winterburn J.L., Shah Y., Chavez S., Schira M.M., Lobaugh N.J., Lerch J.P., Voineskos A.N., Chakravarty M.M. Derivation of high-resolution MRI atlases of the human cerebellum at 3 T and segmentation using multiple automatically generated templates. Neuroimage. 2014;95:217–231. doi: 10.1016/j.neuroimage.2014.03.037. 24657354 [DOI] [PubMed] [Google Scholar]
- Patenaude B., Smith S.M., Kennedy D.N., Jenkinson M.A. Bayesian model of shape and appearance for subcortical brain. NeuroImage. 2011;56(3):907–922. doi: 10.1016/j.neuroimage.2011.02.046. 21352927 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peterson B.S., Vohr B., Staib L.H., Cannistraci C.J., Dolberg A., Schneider K.C., Katz K.H., Westerveld M., Sparrow S., Anderson A.W., Duncan C.C., Makuch R.W., Gore J.C., Ment L.R. Regional brain volume abnormalities and long-term cognitive outcome in preterm infants. JAMA. 2000;284(15):1939–1947. doi: 10.1001/jama.284.15.1939. 11035890 [DOI] [PubMed] [Google Scholar]
- Pfluger T., Weil S., Weis S., Vollmar C., Heiss D., Egger J., Scheck R., Hahn K. Normative volumetric data of the developing hippocampus in children based on magnetic resonance imaging. Epilepsia. 1999;40(4):414–423. doi: 10.1111/j.1528-1157.1999.tb00735.x. 10219266 [DOI] [PubMed] [Google Scholar]
- Pipitone J., Park M.T.M., Winterburn J., Lett T.A., Lerch J.P., Pruessner J.C., Lepage M., Voineskos A.N., Chakravarty M.M., Alzheimer’s Disease Neuroimaging Initiative Multi-atlas segmentation of the whole hippocampus and subfields using multiple automatically generated templates. Neuroimage. 2014;101(101):494–512. doi: 10.1016/j.neuroimage.2014.04.054. 24784800 [DOI] [PubMed] [Google Scholar]
- Ponce de León M.S., Golovanova L., Doronichev V., Romanova G., Akazawa T., Kondo O., Ishida H., Zollikofer C.P.E. Neanderthal brain size at birth provides insights into the evolution of human life history. Proc. Natl. Acad. Sci. U. S. A. 2008;105(37):13764–13768. doi: 10.1073/pnas.0803917105. 18779579 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prastawa M., Gilmore J.H., Lin W., Gerig G. Automatic segmentation of MR images of the developing newborn brain. Med. Image Anal. 2005;9(5):457–466. doi: 10.1016/j.media.2005.05.007. 16019252 [DOI] [PubMed] [Google Scholar]
- Pruessner J.C., Li L.M., Serles W., Pruessner M., Collins D.L., Kabani N., Lupien S., Evans A.C. Volumetry of hippocampus and amygdala with high-resolution MRI and three-dimensional analysis software: minimizing the discrepancies between laboratories. Cereb. Cortex. 2000;10(4):433–442. doi: 10.1093/cercor/10.4.433. 10769253 [DOI] [PubMed] [Google Scholar]
- Raznahan A., Shaw P.W., Lerch J.P., Clasen L.S., Greenstein D., Berman R., Pipitone J., Chakravarty M.M., Giedd J.N. Longitudinal four-dimensional mapping of subcortical anatomy in human development. Proc. Natl. Acad. Sci. U. S. A. 2014;111(4):1592–1597. doi: 10.1073/pnas.1316911111. 24474784 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reiss-Zimmermann M., Gutberlet M., Köstler H., Fritzsch D., Hoffmann K.T. Improvement of SNR and acquisition acceleration using a 32-channel head coil compared to a 12-channel head coil at 3T. Acta Radiol. 2013;54(6):702–708. doi: 10.1177/0284185113479051. 23474767 [DOI] [PubMed] [Google Scholar]
- Rogers C.E., Anderson P.J., Thompson D.K., Kidokoro H., Wallendorf M., Treyvaud K., Roberts G., Doyle L.W., Neil J.J., Inder T.E. Regional cerebral development at term relates to school-age social–emotional development in very preterm children. J. Am. Acad. Child Adolesc. Psychiatry. 2012;51(2):181–191. doi: 10.1016/j.jaac.2011.11.009. 22265364 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saigal S., Doyle L.W. An overview of mortality and sequelae of preterm birth from infancy to adulthood. Lancet. 2008;371(9608):261–269. doi: 10.1016/S0140-6736(08)60136-1. 18207020 [DOI] [PubMed] [Google Scholar]
- Satterthwaite T.D., Wolf D.H., Loughead J., Ruparel K., Elliott M.A., Hakonarson H. Impact of in-scanner head motion on multiple measures of functional connectivity: relevance for studies of neurodevelopment in youth. Neuroimage. 2012;60(1):623–632. doi: 10.1016/j.neuroimage.2011.12.063. 22233733 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shao J. Linear model selection by cross-validation. J. Am. Stat. Assoc. 1993;88(422):486–494. [Google Scholar]
- Shaw P., De Rossi P., Watson B., Wharton A., Greenstein D., Raznahan A., Sharp W., Lerch J.P., Chakravarty M.M. Mapping the development of the basal ganglia in children with attention-deficit/hyperactivity disorder. J. Am. Acad. Child Adolesc. Psychiatry. 2014;53(7):780–789. doi: 10.1016/j.jaac.2014.05.003. 24954827 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen D., Davatzikos C. HAMMER: hierarchical attribute matching mechanism for elastic registration. I.E.E.E. Trans. Med. Imaging. 2002;21(11):1421–1439. doi: 10.1109/TMI.2002.803111. 12575879 [DOI] [PubMed] [Google Scholar]
- Shen D., Moffat S., Resnick S.M., Davatzikos C. Measuring size and shape of the hippocampus in MR images using a deformable shape model. Neuroimage. 2002;15(2):422–434. doi: 10.1006/nimg.2001.0987. 11798276 [DOI] [PubMed] [Google Scholar]
- Shi F., Shen D., Yap P.-T., Fan Y., Cheng J.-Z., An H., Wald L.L., Gerig G., Gilmore J.H., Lin W. CENTS: cortical enhanced neonatal tissue segmentation. Hum. Brain Mapp. 2011;32(3):382–396. doi: 10.1002/hbm.21023. 20690143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sled J.G., Zijdenbos A.P., Evans A.C. A nonparametric method for automatic correction of intensity nonuniformity in MRI data. I.E.E.E. Trans. Med. Imaging. 1998;17(1):87–97. doi: 10.1109/42.668698. 9617910 [DOI] [PubMed] [Google Scholar]
- Taylor H.G., Minich N.M., Klein N., Hack M. Longitudinal outcomes of very low birth weight: neuropsychological findings. J. Int. Neuropsychol. Soc. 2004;10(2):149–163. doi: 10.1017/S1355617704102038. 15012835 [DOI] [PubMed] [Google Scholar]
- Thompson D.K., Adamson C., Roberts G., Faggian N., Wood S.J., Warfield S.K., Doyle L.W., Anderson P.J., Egan G.F., Inder T.E. Hippocampal shape variations at term equivalent age in very preterm infants compared with term controls: perinatal predictors and functional significance at age 7. Neuroimage. 2013;7:70. doi: 10.1016/j.neuroimage.2012.12.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson D.K., Ahmadzai Z.M., Wood S.J., Inder T.E., Warfield S.K., Doyle L.W., Egan G.F. Optimizing hippocampal segmentation in infants utilizing MRI post-acquisition processing. Neuroinformatics. 2012;10(2):173–180. doi: 10.1007/s12021-011-9137-7. 22194186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson D.K., Omizzolo C., Adamson C., Lee K.J., Stargatt R., Egan G.F., Doyle L.W., Inder T.E., Anderson P.J. Longitudinal growth and morphology of the hippocampus through childhood: impact of prematurity and implications for memory and learning. Hum. Brain Mapp. 2014;35(8):4129–4139. doi: 10.1002/hbm.22464. 24523026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson D.K., Wood S.J., Doyle L.W., Warfield S.K., Egan G.F., Inder T.E. MR-determined hippocampal asymmetry in full-term and preterm neonates. Hippocampus. 2009;19(2):118–123. doi: 10.1002/hipo.20492. 18767066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson D.K., Wood S.J., Doyle L.W., Warfield S.K., Lodygensky G.A., Anderson P.J., Egan G.F., Inder T.E. Neonate hippocampal volumes: prematurity, perinatal predictors, and 2-year outcome. Ann. Neurol. 2008;63(5):642–651. doi: 10.1002/ana.21367. 18384167 [DOI] [PubMed] [Google Scholar]
- Tong T., Wolz R., Coupé P., Hajnal J.V., Rueckert D. Segmentation of MR images via discriminative dictionary learning and sparse coding: application to hippocampus labeling. Neuroimage. 2013;76:11–23. doi: 10.1016/j.neuroimage.2013.02.069. 23523774 [DOI] [PubMed] [Google Scholar]
- Trampel R., Ott D.V.M., Turner R. Do the congenitally blind have a stria of Gennari? First intracortical insights in vivo. Cereb. Cortex. 2011;21(9):2075–2081. doi: 10.1093/cercor/bhq282. [DOI] [PubMed] [Google Scholar]
- Utsunomiya H., Takano K., Okazaki M., Mitsudome A. Development of the temporal lobe in infants and children: analysis by MR-based volumetry. A.J.N.R. Am. J. Neuroradiol. 1999;20(4):717–723. 10319988 [PMC free article] [PubMed] [Google Scholar]
- Van Leemput K., Bakkour A., Benner T., Wiggins G., Wald L.L., Augustinack J., Dickerson B.C., Golland P., Fischl B. Automated segmentation of hippocampal subfields from ultra-high resolution in vivo MRI. Hippocampus. 2009;19(6):549–557. doi: 10.1002/hipo.20615. 19405131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vohr B.R., Allan W.C., Westerveld M., Schneider K.C., Katz K.H., Makuch R.W., Ment L.R. School-age outcomes of very low birth weight infants in the indomethacin intraventricular hemorrhage prevention trial. Pediatrics. 2003;111(4 Pt 1):e340–e346. doi: 10.1542/peds.111.4.e340. 12671149 [DOI] [PubMed] [Google Scholar]
- Wang H., Suh J.W., Pluta J., Altinay M., Yushkevich P. Optimal weights for multi-atlas label fusion. Inf Proc. Med Imaging. 2011;22:73–84. doi: 10.1007/978-3-642-22092-0_7. 21761647 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L., Shi F., Li G., Gao Y., Lin W., Gilmore J.H., Shen D. Segmentation of neonatal brain MR images using patch-driven level sets. Neuroimage. 2014;84:141–158. doi: 10.1016/j.neuroimage.2013.08.008. 23968736 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warfield S.K., Zou K.H., Wells W.M. Simultaneous truth and performance level estimation (STAPLE): an algorithm for the validation of image segmentation. I.E.E.E. Trans. Med. Imaging. 2004;23(7):903–921. doi: 10.1109/TMI.2004.828354. 15250643 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wargo C.J., Moore J., Gore J.C. A comparison and evaluation of reduced-FOV methods for multi-slice 7 T human imaging. Magn. Reson. Imaging. 2013;31(8):1349–1359. doi: 10.1016/j.mri.2013.05.003. 23891434 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watson C., Andermann F., Gloor P., Jones-Gotman M., Peters T., Evans A., Olivier A., Melanson D., Leroux G. Anatomic basis of amygdaloid and hippocampal volume measurement by magnetic resonance imaging. Neurology. 1992;42(9):1743–1750. doi: 10.1212/wnl.42.9.1743. 1513464 [DOI] [PubMed] [Google Scholar]
- Weisenfeld N.I., Warfield S.K. Automatic segmentation of newborn brain MRI. Neuroimage. 2009;47(2):564–572. doi: 10.1016/j.neuroimage.2009.04.068. 19409502 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winterburn J.L., Pruessner J.C., Chavez S., Schira M.M., Lobaugh N.J., Voineskos A.N., Chakravarty M.M. A novel in vivo atlas of human hippocampal subfields using high-resolution 3 T magnetic resonance imaging. Neuroimage. 2013;74:254–265. doi: 10.1016/j.neuroimage.2013.02.003. 23415948 [DOI] [PubMed] [Google Scholar]
- Wolz R., Aljabar P., Hajnal J.V., Hammers A., Rueckert D. LEAP: learning embeddings for atlas propagation. Neuroimage. 2010;49(2):1316–1325. doi: 10.1016/j.neuroimage.2009.09.069. 19815080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong A.P., Pipitone J., Park M.T., Dickie E.W., Leonard G., Perron M., Pike B.G., Richer L., Veillette S., Chakravarty M.M., Pausova Z., Paus T. Estimating volumes of the pituitary gland from T1-weighted magnetic-resonance images: effects of age, puberty, testosterone, and estradiol. Neuroimage. 2014;94:216–221. doi: 10.1016/j.neuroimage.2014.02.030. 24632090 [DOI] [PubMed] [Google Scholar]
- Wu G., Wang Q., Zhang D., Nie F., Huang H., Shen D. A generative probability model of joint label fusion for multi-atlas based brain segmentation. Med. Image Anal. 2014;18(6):881–890. doi: 10.1016/j.media.2013.10.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue H., Srinivasan L., Jiang S., Rutherford M., Edwards A.D., Rueckert D., Hajnal J.V. Automatic segmentation and reconstruction of the cortex from neonatal MRI. Neuroimage. 2007;38(3):461–477. doi: 10.1016/j.neuroimage.2007.07.030. 17888685 [DOI] [PubMed] [Google Scholar]
- Yu X., Zhang Y., Lasky R.E., Datta S., Parikh N.A., Narayana P.A. Comprehensive brain MRI segmentation in high risk preterm newborns. PLOS One. 2010;5(11):e13874. doi: 10.1371/journal.pone.0013874. 21079730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yushkevich P.A., Wang H., Pluta J., Das S.R., Craige C., Avants B.B., Weiner M.W., Mueller S. Nearly automatic segmentation of hippocampal subfields in in vivo focal T2-weighted MRI. Neuroimage. 2010;53(4):1208–1224. doi: 10.1016/j.neuroimage.2010.06.040. 20600984 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng W., Chee M.W.L., Zagorodnov V. Improvement of brain segmentation accuracy by optimizing non-uniformity correction using N3. Neuroimage. 2009;48(1):73–83. doi: 10.1016/j.neuroimage.2009.06.039. 19559796 [DOI] [PubMed] [Google Scholar]