Abstract
The foodborne pathogen Listeria monocytogenes is able to survive and grow in ready-to-eat foods, in which it is likely to experience a number of environmental stresses due to refrigerated storage and the physicochemical properties of the food. Little is known about the specific molecular mechanisms underlying survival and growth of L. monocytogenes under different complex conditions on/in specific food matrices. Transcriptome sequencing (RNA-seq) was used to understand the transcriptional landscape of L. monocytogenes strain H7858 grown on cold smoked salmon (CSS; water phase salt, 4.65%; pH 6.1) relative to that in modified brain heart infusion broth (MBHIB; water phase salt, 4.65%; pH 6.1) at 7°C. Significant differential transcription of 149 genes was observed (false-discovery rate [FDR], <0.05; fold change, ≥2.5), and 88 and 61 genes were up- and downregulated, respectively, in H7858 grown on CSS relative to the genes in H7858 grown in MBHIB. In spite of these differences in transcriptomes under these two conditions, growth parameters for L. monocytogenes were not significantly different between CSS and MBHIB, indicating that the transcriptomic differences reflect how L. monocytogenes is able to facilitate growth under these different conditions. Differential expression analysis and Gene Ontology enrichment analysis indicated that genes encoding proteins involved in cobalamin biosynthesis as well as ethanolamine and 1,2-propanediol utilization have significantly higher transcript levels in H7858 grown on CSS than in that grown in MBHIB. Our data identify specific transcriptional profiles of L. monocytogenes growing on vacuum-packaged CSS, which may provide targets for the development of novel and improved strategies to control L. monocytogenes growth on this ready-to-eat food.
INTRODUCTION
Listeria monocytogenes is a psychrotolerant foodborne pathogen that causes a potentially severe disease, listeriosis. This pathogen is of particular concern to the ready-to-eat (RTE) meat and seafood industries due to its ability to grow at temperatures as low as −0.4°C and under conditions of salt content as high as 25% (at 4°C) (1–3). Cold smoked salmon (CSS), an RTE seafood, represents a typical food product that can support the growth of L. monocytogenes from low numbers to potentially hazardous levels (4–9). The heat treatment applied during processing of CSS is not sufficient to inactivate microbes present on the raw material, including L. monocytogenes (10, 11). In addition, RTE food products, including CSS, can be contaminated with L. monocytogenes from environmental sources in processing facilities (10–13). Importantly, typical product characteristics of CSS, including pH, water activity (aw), salt, and phenolic components, do not seem to be sufficient to control the growth of L. monocytogenes if it is present (8, 9).
With the aim of developing control strategies that prevent or reduce growth of this pathogen in RTE seafood products, there is a need for a better understanding of the mechanisms that L. monocytogenes uses to survive and grow under the complex conditions of specific food matrices. Characterization of bacterial gene expression patterns in different environments can be used to assess the physiological state of L. monocytogenes under different conditions and to help identify the metabolic pathways that are important for survival and growth of L. monocytogenes in food products. This will facilitate the identification of new compounds that could specifically interfere with these metabolic pathways and thereby control the growth of L. monocytogenes (14). Extensive studies on the transcriptome of L. monocytogenes have been conducted to assess how it responds to the physical, chemical, or biological stresses that it may encounter on/in food matrices (15–22). The majority of data from these experiments are based on exposure of L. monocytogenes to specific stresses in laboratory media, providing information about specific stress responses and transcriptional profiles in a controlled environment. This information may not provide the full extent of the bacterial transcriptional landscape in a more complex environment, such as a food matrix.
We characterized the transcriptome of late-log-phase L. monocytogenes strain H7858 (a serotype 4b lineage I strain) grown on CSS and the same strain grown to late log phase in modified brain heart infusion broth (MBHIB; water phase [w.p.] salt, 4.65%; pH 6.10). While the two conditions chosen here are distinct, they do facilitate characterization of the L. monocytogenes transcriptional landscape in a real food as well as comparisons against commonly used reference conditions. Our approach is similar to studies that provided significant insights into the pathogen transcriptional landscape in human or animal hosts, which also, by necessity, must choose reference conditions (e.g., growth in rich medium) that differ by a multitude of factors from complex host-associated environments (e.g., presence in intestinal lumen or human blood). For example, previous characterization of the L. monocytogenes transcriptome in BHI (designated the “reference condition” by the authors) and physiologically relevant conditions (e.g., stationary phase, low temperature) as well as in the intestinal lumen of infected mice and inoculated human blood provided crucial knowledge of the L. monocytogenes transcriptional landscape under various conditions that this pathogen may experience during transmission (23). In the present study, we demonstrate the use of transcriptome sequencing (RNA-seq) technology to study the global gene expression of L. monocytogenes in an RTE seafood product. We overcame the technical difficulties associated with isolating high-quality bacterial RNA from the seafood matrix and took advantage of the probe and annotation independence of RNA-seq technology to explore the genome-wide transcriptional landscape of L. monocytogenes grown under the complex conditions on this food matrix.
MATERIALS AND METHODS
Bacterial strain and inoculum preparation.
L. monocytogenes strain H7858 was used in this study (24). H7858 is a lineage I, serotype 4b strain (representing epidemic clone II) isolated from RTE meat and was linked to a multistate listeriosis outbreak from 1998 to 1999 (24, 25). We selected H7858 for this study because we had previously quantified the phenotypic and transcriptomic responses of this strain to a number of stresses relevant to food products, including organic acids and bactericidal additives, and know that it can grow to high levels on cold smoked salmon (26–28). L. monocytogenes H7858 was streaked from frozen BHI stock stored at −80°C in 15% glycerol onto a BHI agar plate, followed by incubation at 37°C for 24 h. A single colony was subsequently inoculated into 5 ml of BHIB (in 16-mm tubes), followed by incubation at 37°C with shaking (230 rpm) for 18 h (series 25 incubator; New Brunswick Scientific, Edison, NJ). After 18 h, 50 μl BHI culture was inoculated into 5 ml chemically defined medium (DM) (29) and grown to stationary phase in DM at 16°C statically, as described previously (30). This culture was used to inoculate CSS and MBHIB as detailed below. DM was used to approximate a nutrient-limited environment (e.g., food processing plants) that L. monocytogenes may encounter before contaminating food.
Growth conditions in BHI and on salmon.
BHIB was modified to have 4.65% w.p. NaCl and pH 6.1 to simulate the levels typically present in commercially processed CSS (30). The stationary-phase DM culture was used to inoculate 100 ml of MBHIB in 300-ml Erlenmeyer culture flasks with metal caps (Bellco Glass Co., Vineland, NJ), with an initial population of approximately 1 × 106 CFU/ml, followed by static incubation at 7°C.
Commercially produced wet-cured CSS fillets were stored at −20°C and thawed at 4°C overnight. A mixture of natural hardwood and fruitwood had been used to cold smoke the salmon. The background microbiota (mainly lactic acid bacteria) and physicochemical characteristics of the untreated salmon slices used in the present study have been described previously by Kang et al. (31). All CSS samples were from the same batch of product. The concentration of lactic acid bacteria on uninoculated CSS was ∼4 log CFU/g on day 0, ∼6 log CFU/g on day 5, and ∼7 log CFU/g on day 10 during incubation of the vacuum-packaged slices at 7°C. For the uninoculated CSS, the pH was ∼6.18, water activity (aw) was 0.96, moisture content was ∼63.58%, and fat content was ∼8.87% (31). Uninoculated salmon samples were plated onto Oxford agar and incubated at 30°C for 48 h to confirm the absence of L. monocytogenes.
Salmon slices were weighed (10 ± 0.5 g each) and transferred into sterile petri dishes. Both sides of the salmon slice were inoculated with 500 μl stationary-phase cultures from DM that were diluted in 0.1% sterile peptone water to a target population of approximately 1 × 106 CFU/g and spread with sterile plastic cell spreaders. Inoculated salmon slices were then placed in a biosafety cabinet for 15 min to dry the surface before being transferred into storage bags (oxygen permeability, 38.10 ml/m2 to 40.50 ml/m2 at 23°C dry/24 h) and packaged using a commercial vacuum sealer (FoodSaver model V2244). All samples were stored at 7°C. Incubator temperature was recorded every 20 min by an automated thermal recorder during the storage of both MBHIB cultures and CSS samples. The recorded incubation temperature was 7.0 ± 0.5°C.
Determination of exponential-phase sampling points.
To monitor L. monocytogenes growth, cell density was determined every day, starting from day 0, until log-phase cells were collected for RNA extraction on day 7; another three time points were taken on days 10, 11, and 12 after RNA extraction to determine the maximum cell density. For MBHIB samples, cultures were diluted with 0.1% sterile peptone water and spiral plated in duplicate onto BHI agar using an Autoplate 4000 (Spiral Biotech, Inc., Norwood, MA). BHI plates were incubated at 37°C for 24 h before colonies were counted with the Q-Count Colony Counter (Spiral Biotech). For salmon samples, 2 vacuum-packed samples were aseptically opened for each time point and stomached for 30 s at the high-speed setting (Stomacher 400; Seward, West Sussex, United Kingdom) with 40 ml of 0.1% sterile peptone water. CSS homogenates were spiral plated on Oxford agar (catalog no. 222530 from BD and catalog no. SR0140 from Oxoid, Ltd., Hampshire, United Kingdom) using the Autoplate 4000.
Measurements of L. monocytogenes cell density over time in MBHIB and on CSS were fitted with a three-phase linear model described by Buchanan et al. (32) using the NLStools package (v0.0 to 11) in R v 2.13.0. Four growth parameters including lag phase (λ, in days), maximum growth rate (μmax, in log CFU/milliliter or/gram per day), initial cell density (N0, in log CFU/milliliter or /gram), and maximum cell density (Nmax, in log CFU/milliliter or /gram) were calculated. Extracted RNA was considered to be qualified for downstream processing and analysis if it had an RNA integrity number (RIN) of ≥7 (33). Pilot experiments showed that L. monocytogenes cell density on CSS needed to be at least 8 log CFU/g to obtain total RNA with a RIN of ≥7; at this cell density, H7858 was still in log phase. To predict the time point for RNA isolation (TRNA-extraction) (when L. monocytogenes cell density reaches 8 log CFU/ml or /g), we obtained the growth parameters by fitting growth data collected from days 0 to 5 as described earlier and calculated TRNA-extraction using the equation derived from the Buchanan model (32): TRNA-extraction = λ + (1 × 108 CFU/g − N0)/μmax. The real Nmax of each growth experiment was confirmed by sampling on days 10, 11, and 12 after RNA extraction to ensure accuracy of the prediction. The time points for RNA extraction fell on day 7 for all four replicates of both CSS and BHI samples.
RNA isolation, integrity, and quality assessment.
Similar procedures were used to extract RNA from L. monocytogenes growing in MBHIB and on CSS. For MBHIB, a total of 7 ml of RNA Protect reagent (Qiagen, Valencia, CA) was added to 7-ml samples in 14-ml Sarstedt tubes (Sarstedt, Nümbrecht, Germany), followed by vigorous vortexing for 10 s to mix well; for CSS, 17 ml of RNA Protect reagent was added to 10-g samples, followed by massaging of the sample bag to mix well. The mixture was incubated at room temperature for 10 min to ensure that the bacterial RNA was stabilized. For CSS samples, the liquid part of the mixture was then filtered out with a 207-ml filter bag (catalog no. B01385WA, Whirl-Pak bag; Nasco, Fort Atkinson, WI) to remove salmon particles. Cells were pelleted by centrifugation (4,637 × g, 15 min) at 4°C and suspended in nuclease-free water with proteinase K (12.5 mg/ml) and lysozyme (25 mg/ml), followed by incubation at 37°C for 30 min. TRI reagent (Ambion, Austin, TX) was then added to each sample (in screw-cap tubes with 3 ml of 0.1-mm acid-washed zirconium beads), followed by mechanical lysis for 5 min in a Mini-Beadbeater-8 (BioSpec Products, Inc., Barlesville, OK) and subsequent RNA extraction according to the manufacturer's recommendations. Total RNA was incubated with RQ1 DNase (Promega, Madison, WI) in the presence of RNasin (Promega) to remove any remaining DNA. Subsequently, RNA was purified using two phenol-chloroform extractions and one chloroform extraction, followed by RNA precipitation and suspension of the RNA in RNase free Tris-EDTA (TE; 10 mM Tris, 1 mM EDTA [pH 8.0]; Ambion). UV spectrophotometry (Nanodrop, Wilmington, DE) was used to quantify and assess purity of the RNA. The efficacy of the DNase treatment was assessed by TaqMan quantitative PCR (qPCR) analysis of DNA levels for the housekeeping gene rpoB (34). qPCR was performed using Taq-Man One-Step RT-PCR Master Mix reagent and the ABI Prism 7000 sequence detection system (all from Applied Biosystems, Foster City, CA). All samples showed CT values of >35 for rpoB, indicating negligible levels of DNA contamination. As a final step, RNA integrity was assessed using the 2100 Bioanalyzer (Agilent, Foster City, CA). All experiments were performed in quadruplicate.
Preparation of cDNA fragment libraries and RNA-seq.
Preparation of cDNA fragment libraries was performed using the ScriptSeq Complete kit (Bacteria)-Low Input kit (Epicentre, Madison, WI). To remove 16S and 23S rRNA from total RNA and enrich for mRNA, 1 μg total RNA was treated with Ribo-Zero rRNA Removal Reagents (Bacteria)-Low Input and Magnetic Core kit-Low Input according to the manufacturer's protocol. Enriched mRNA samples were run on the 2100 Bioanalyzer (Agilent Technology, Santa Clara, CA) to confirm reduction of 16S and 23S rRNA and purified using Agencourt RNAClean XP kit (Beckman Coulter Inc., Brea, CA) prior to preparation of cDNA fragment libraries. The mRNA-enriched fraction was converted to indexed RNA-seq libraries with the ScriptSeq v2 RNA-seq Library Preparation kit. This protocol allows the identification of the specific strand from which each read was generated, resulting in a strand-specific analysis of the RNA-seq results. Indexed and purified libraries (8 libraries, including 4 replicates for each CSS and MBHIB) were loaded together onto a flow cell without any other samples; sequencing was carried out on a Hiseq 2000 (single end, 100 bp per read).
RNA-seq alignment and coverage.
As the H7858 genome has not been completely closed (GenBank accession number AADR00000000), the sequence reads were aligned to a H7858 pseudochromosome. The pseudochromosome was created through alignment of the contigs of the H7858 draft genome to the completely closed genome of the L. monocytogenes strain EGD-e (GenBank accession number NC_003210) and subsequent concatenation of these contigs into a pseudochromosome. Alignment of reads was carried out using the BWA mem algorithm in BWA version 0.7.3a (35). Default parameters were used for the alignment. Coverage at each base position along the chromosome was calculated by enumerating the number of reads that aligned to a given base for each DNA strand separately.
Differential expression analysis. Differential expression of genes under the conditions in MBHIB and on CSS was statistically assessed using the BaySeq method (36) implemented in the BaySeq 1.16.0 package available from Bioconductor. This package implements a full Bayesian model of negative binomial distributions to simultaneously assess the likelihood of various models, each representing a possible pattern of expression for a given gene. Library sizes were normalized using the approach described by Bullard et al. (37). To allow for quantitative comparisons among genes and treatments, we used the average normalized RNA-seq coverage (NRC) generated by BaySeq for each gene of the four replicates to identify the genes with the highest average NRC of L. monocytogenes grown on CSS. Genes were considered differentially expressed if they showed a false-discovery rate (FDR) of <0.05 and a fold change (FC) of ≥2.5 (for genes upregulated on CSS) or ≤0.4 (for genes downregulated on CSS), where FC is the average NRC for CSS divided by the average NRC for MBHIB. To confirm annotation of differentially expressed genes, the NCBI BLAST standalone program was used to search the H7858 amino acid sequence for each of these genes against the amino acid sequences of the CDS of L. monocytogenes strains EGD-e (GenBank accession number NC_003210), 10403S (GenBank accession number NC_017544), and F2365 (GenBank accession number NC_002973), as well as Salmonella enterica serovar Typhimurium LT2 (GenBank accession number NC_003197) and Escherichia coli K-12 MG1655 (GenBank accession number NC_000913).
GO enrichment analysis.
Rather than validating the upregulation of individual genes, we focused on identifying the metabolic pathways that contained multiple differentially expressed genes by using the results generated by Gene Ontology (GO) enrichment analysis. This method allowed us to statistically confirm the upregulation of specific metabolic pathways that may facilitate the survival and growth of L. monocytogenes on CSS. Enrichment of GO terms among genes upregulated on CSS was assessed using the GOseq 1.18.0 package (38) available from Bioconductor. GO term classification for each gene in H7858 was obtained using the blast2go program.
Microarray data accession number.
RNA-seq data from this study are available in the NCBI GEO Short Read Archives (GSE64353).
RESULTS
L. monocytogenes growth parameters are similar on CSS and in MBHIB.
L. monocytogenes growth parameters, including λ, μmax, N0, and Nmax, did not differ significantly between CSS and MBHIB (P > 0.05; t test) (Fig. 1). The average λ values were 1.78 ± 1.05 and 2.12 ± 0.35 days for CSS and MBHIB, respectively. The average μmax was 0.35 ± 0.06 log CFU/g/day and 0.40 ± 0.02 log CFU/ml/day for CSS and MBHIB, respectively. The average N0 and average Nmax were 6.48 ± 0.16 and 8.68 ± 0.14 log CFU/g for CSS, respectively, and 6.35 ± 0.06 and 8.82 ± 0.07 log CFU/ml for MBHIB, respectively.
FIG 1.
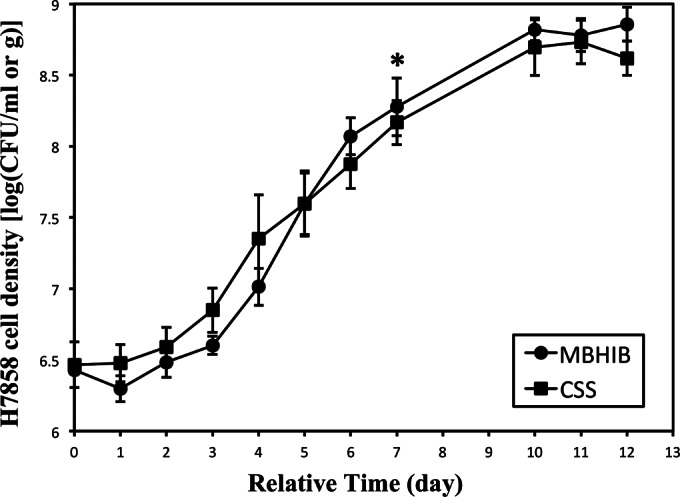
Growth of H7858 on cold smoked salmon (CSS; squares) and modified BHI broth (w.p. salt 4.65%, pH 6.10) (MBHIB; circles) at 7°C. RNA was extracted at the average H7858 cell density of 8.17 ± 0.16 log CFU/g on CSS and 8.28 ± 0.21 log CFU/ml in MBHIB (*).
Late-log-phase H7858 has 88 up- and 61 downregulated genes on CSS compared to cultures in MBHIB.
RNA-seq was performed on H7858 RNA samples representing four independent biological replicates of H7858 grown on CSS or in MBHIB. Samples for RNA isolation were collected when H7858 was grown to late log phase under the conditions of these two matrices; the average cell densities of collected L. monocytogenes samples were 8.17 ± 0.16 log CFU/ml for CSS and 8.28 ± 0.21 log CFU/g for MBHIB (Fig. 1). Since the growth parameters of H7858 were not significantly different on CSS and in MBHIB, the L. monocytogenes cells from both conditions at the time point for RNA isolation were expected to be at the same growth phase, indicating that observed differences in transcript levels were not likely to reflect different growth phases of H7858. Transcriptome sequencing generated 1.6 million to 10.9 million reads per sample (Table 1). For RNA samples from H7858 grown in MBHIB and on CSS, of the reads that mapped to the reference pseudochromosome, ∼80% and ∼82% mapped to protein-coding sequences, respectively. The remaining reads mapped to noncoding RNA, including rRNA and tRNAs. Among the unmapped reads from CSS, 83% on average were found to represent sequences that mapped to Atlantic Salmon genomic DNA (GenBank accession number AGKD00000000.3), suggesting contamination with residual salmon RNA, and 6% on average mapped to the genome of Carnobacterium maltaromaticum LMA28 (GenBank accession number NC_019425.2), which is representative of a genus of Gram-positive bacteria that are found in food products and grow anaerobically (39) and likely represent part of the resident microbiota of CSS.
TABLE 1.
Summary of RNA-seq coverage dataa
| Matrix and sample ID | No. of mapped reads | No. of reads mapped to CDS | % of reads mapped to CDS |
|---|---|---|---|
| MBHIB | |||
| MBHIB-1 | 1,555,154 | 1,293,750 | 83 |
| MBHIB-2 | 2,020,923 | 1,684,965 | 83 |
| MBHIB-3 | 6,182,126 | 4,828,777 | 78 |
| MBHIB-4 | 6,302,268 | 4,821,223 | 76 |
| Avg MBHIB | 4,015,118 | 3,157,179 | 80 |
| CSS | |||
| CSS-1 | 1,611,849 | 1,314,001 | 82 |
| CSS-2 | 2,303,793 | 1,946,859 | 85 |
| CSS-3 | 3,557,097 | 2,830,220 | 80 |
| CSS-4 | 3,204,674 | 2,684,028 | 84 |
| Avg CSS | 2,669,353 | 2,193,777 | 82 |
Abbreviations: MBHIB, modified brain heart infusion broth (water phase salt, 4.65%, pH 6.10); CSS, cold smoked salmon; CDS, coding DNA sequence for protein.
As RNA-seq allows for absolute quantification, our data allowed us to identify the genes that showed the highest transcript levels in H7858 on CSS (Table 2). The three genes with the highest average NRC were fusA, eno, and tuf, which encode translation elongation factor G, an enolase, and translation elongation factor Tu, respectively. Other genes with well-defined functions and high average NRC include gadT2D2 (40), which encode proteins involved in glutamate-dependent acid resistance, gap, which encodes a NAD-dependent glyceraldehyde-3-phosphate dehydrogenase involved in glycolysis, and cspLA, which encode cold shock proteins involved in adaptation to atypical conditions (Table 2).
TABLE 2.
Genes with highest average normalized RNA-seq coverage (NRC)
| Gene name in: |
H7858 gene product | Avg NRC rank for: |
Avg NRCb for: |
||||
|---|---|---|---|---|---|---|---|
| H7858 | EGD-ea | 10403Sa | CSS | MBHIB | CSS | MBHIB | |
| LMOh7858_2915 | lmo2654 (fusA) | LMRG_02199 | Translation elongation factor G | 1 | 1 | 3,643,359 | 3,404,382 |
| LMOh7858_2604 | lmo2455 (eno) | LMRG_01793 | Enolase (EC 4.2.1.11) | 2 | 3 | 3,348,509 | 2,505,110 |
| LMOh7858_2914 | lmo2653 (tuf) | LMRG_02198 | Translation elongation factor Tu | 3 | 4 | 3,296,021 | 2,329,659 |
| LMOh7858_2506 | lmo2363 (gadD2) | LMRG_01479 | Glutamate decarboxylase (EC 4.1.1.15) | 4 | 65 | 3,186,282 | 627,851 |
| LMOh7858_2608 | lmo2459 (gap) | LMRG_01789 | NAD-dependent glyceraldehyde-3-phosphate dehydrogenase (EC 1.2.1.12) | 5 | 15 | 2,650,840 | 1,618,077 |
| LMOh7858_2505 | lmo2362 | LMRG_01480 | Probable glutamate/gamma-aminobutyrate antiporter | 6 | 86 | 2,589,714 | 490,907 |
| LMOh7858_1751 | lmo1634 | LMRG_01332 | Alcohol dehydrogenase (EC 1.1.1.1); acetaldehyde dehydrogenase (EC 1.2.1.10) | 7 | 8 | 2,504,749 | 1,849,977 |
| LMOh7858_2605 | lmo2456 (gpmI) | LMRG_01792 | 2,3-Bisphosphoglycerate-independent phosphoglycerate mutase (EC 5.4.2.1) | 8 | 12 | 2,229,167 | 1,648,039 |
| LMOh7858_2609 | lmo2460 | LMRG_01788 | Central glycolytic genes regulator | 9 | 32 | 2,197,665 | 1,008,437 |
| LMOh7858_2607 | lmo2458 (pgk) | LMRG_01790 | Phosphoglycerate kinase (EC 2.7.2.3) | 10 | 15 | 2,158,721 | 1,592,390 |
| LMOh7858_2353 | lmo2219 (prsA2) | LMRG_01613 | Foldase protein PrsA precursor (EC 5.2.1.8); foldase clustered with pyrimidine conversion | 11 | 2 | 1,919,848 | 2,662,595 |
| LMOh7858_2899 | lmo2638 | LMRG_02183 | Pyridine nucleotide-disulfide oxidoreductase family protein | 12 | 665 | 1,913,850 | 1,275,847 |
| LMOh7858_2708 | lmo2556 (fbaA) | LMRG_01691 | Fructose-bisphosphate aldolase class II (EC 4.1.2.13) | 13 | 1,378 | 1,892,452 | 951,955 |
| LMOh7858_2043 | lmo1917 (pflA) | LMRG_01064 | Pyruvate formate-lyase (EC 2.3.1.54) | 14 | 19 | 1,886,299 | 1,435,588 |
| LMOh7858_1070 | lmo1003 (ptsI) | LMRG_02103 | Phosphoenolpyruvate-protein phosphotransferase of PTS (EC 2.7.3.9) | 15 | 6 | 1,810,680 | 1,933,477 |
| LMOh7858_1451 | lmo1364 (cspLA) | LMRG_00814 | Cold shock protein | 16 | 67 | 1,723,202 | 615,514 |
| LMOh7858_1331 | lmo1250 | LMRG_00696 | Transporter | 17 | 250 | 1,711,134 | 210,841 |
| LMOh7858_0642 | lmo0582 (iap) | LMRG_00264 | P60 extracellular protein, invasion-associated protein Iap | 18 | 5 | 1,675,726 | 2,318,702 |
| NGNc | lmo1388 (tcsA) | LMRG_00840 | Unspecified monosaccharide ABC transport system, substrate-binding compound | 19 | 9 | 1,607,769 | 1,818,968 |
| LMOh7858_2330 | lmo2196 (oppA) | LMRG_01636 | Oligopeptide ABC transporter, periplasmic oligopeptide-binding protein O | 20 | 10 | 1,603,950 | 1,694,246 |
Strains EGD-e (GenBank accession no. NC_003210) and 10403S (GenBank accession no. NC_017544) are L. monocytogenes.
NRC, normalized RNA-seq coverage; genes with average NRC of >1,500,000 are presented.
NGN, no gene name given; in the published version of the H7858 genome (27), this gene was not annotated and therefore did not receive a locus name; the gene was identified in our annotation.
Initial analysis of the RNA-seq data identified 88 and 61 genes that showed significantly higher and lower transcript levels, respectively, for H7858 grown on CSS than in MBHIB (see Tables S1 and S2 in the supplemental material for lists of up- and downregulated genes). The 88 upregulated genes included genes encoding proteins annotated as being involved in cobalamin biosynthesis (26 genes), ethanolamine utilization (8 genes), 1,2-propanediol utilization (7 genes), carbohydrate transport and utilization (14 genes), the nonoxidative branch of the pentose pathway (5 genes), and agmatine deiminase (4 genes), as well as genes regulated by PrfA (5 genes) (see Tables S3 to S7 in the supplemental material). The 61 downregulated genes included genes encoding proteins annotated as being involved in pyrimidine nucleotide biosynthesis (6 genes) and l-cystine ABC transporter (3 genes) (see Table S2 in the supplemental material). GO enrichment analysis identified 37 GO terms that were overrepresented among genes upregulated in L. monocytogenes grown on CSS compared to MBHIB (Table 3). No GO terms were found to be enriched among the downregulated genes.
TABLE 3.
GO terms enriched among genes upregulated in H7858 grown on CSS compared to genes in H7858 grown in MBHIB at 7°C
| GO ID | GO term | Pathway associated with GO term |
||
|---|---|---|---|---|
| Cobalamin biosynthesis/transport | Ethanolamine/1,2-propanediol utilization | Carbohydrate utilization/transport | ||
| GO:0005363 | Maltose transmembrane transporter activity | Ya | ||
| GO:0006580 | Ethanolamine metabolic process | Y | ||
| GO:0006766 | Vitamin metabolic process | Y | ||
| GO:0006767 | Water-soluble vitamin metabolic process | Y | ||
| GO:0006778 | Porphyrin-containing compound metabolic process | Y | ||
| GO:0006779 | Porphyrin-containing compound biosynthetic process | Y | ||
| GO:0006824 | Cobalt ion transport | Y | ||
| GO:0008757 | S-Adenosylmethionine-dependent methyltransferase activity | Y | ||
| GO:0009110 | Vitamin biosynthetic process | Y | ||
| GO:0009235 | Cobalamin metabolic process | Y | ||
| GO:0009236 | Cobalamin biosynthetic process | Y | ||
| GO:0015151 | Alpha-glucoside transmembrane transporter activity | Y | ||
| GO:0015157 | Oligosaccharide transmembrane transporter activity | |||
| GO:0015235 | Cobalamin transporter activity | Y | ||
| GO:0015420 | Cobalamin-transporting ATPase activity | Y | ||
| GO:0015422 | Oligosaccharide-transporting ATPase activity | Y | ||
| GO:0015423 | Maltose-transporting ATPase activity | Y | ||
| GO:0015768 | Maltose transport | Y | ||
| GO:0015889 | Cobalamin transport | Y | ||
| GO:0015994 | Chlorophyll metabolic process | Y | ||
| GO:0016628 | Oxidoreductase activity, acting on the CH-CH group of donors, NAD or NADP as acceptor | Y | ||
| GO:0022804 | Active transmembrane transporter activity | Y | Y | |
| GO:0033013 | Tetrapyrrole metabolic process | Y | ||
| GO:0033014 | Tetrapyrrole biosynthetic process | Y | ||
| GO:0034311 | Diol metabolic process | Y | ||
| GO:0034313 | Diol catabolic process | Y | ||
| GO:0042364 | Water-soluble vitamin biosynthetic process | Y | ||
| GO:0042439 | Ethanolamine-containing compound metabolic process | Y | ||
| GO:0042440 | Pigment metabolic process | Y | Y | |
| GO:0044237 | Cellular metabolic process | Y | Y | Y |
| GO:0046483 | Heterocycle metabolic process | Y | Y | |
| GO:0051143 | Propanediol metabolic process | Y | ||
| GO:0051144 | Propanediol catabolic process | Y | ||
| GO:0051180 | Vitamin transport | Y | ||
| GO:0051183 | Vitamin transporter activity | Y | ||
| GO:0051186 | Cofactor metabolic process | Y | ||
| GO:0051188 | Cofactor biosynthetic process | Y | ||
Y, yes (this gene ontology term is associated with the corresponding function).
Genes involved in cobalamin synthesis, ethanolamine utilization, and 1,2-propanediol utilization were upregulated in H7858 growing on CSS.
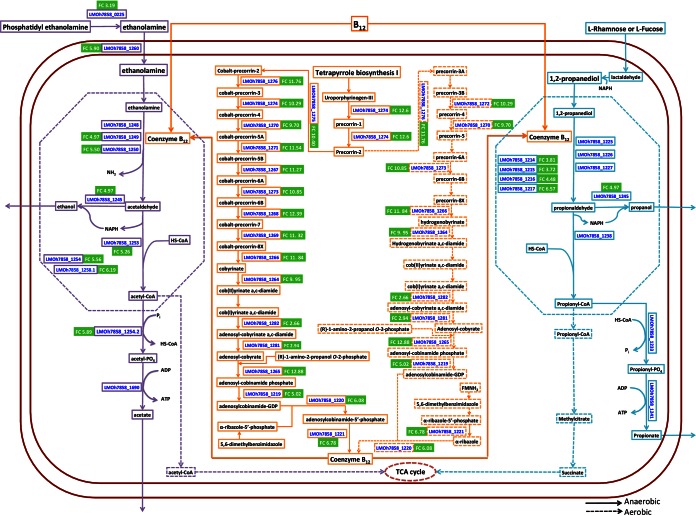
In addition to the identification of 26 upregulated genes with cobalamin metabolism-related annotations (see Table S3 in the supplemental material), we also found the GO terms “cobalamin biosynthetic process” and “cobalamin transport” to be overrepresented among upregulated genes of H7858 on CSS (Table 3). We mapped these genes to the overall cobalamin biosynthesis pathways (Fig. 2) constructed based on the metabolic pathway data of EGD-e and 10403S available in the BioCyc database (biocyc.org) (41). Among the 21 genes mapped to the cobalamin biosynthesis pathways (Fig. 2), 18 were found to be upregulated (FC, 2.76 to 12.88).
FIG 2.
Cobalamin biosynthesis and ethanolamine and 1,2-propanediol utilization pathways in H7858. Pathways were constructed based on information provided in the BioCyc database and previous studies (42–44). H7858 protein designations for enzymes involved in these pathways are shown in blue text. Enzymes encoded by genes upregulated in H7858 grown on cold smoked salmon (CSS) compared to genes in H7858 grown in modified BHI broth (MBHIB) are designated by display of the fold change (FC) in green boxes. Microcompartments are depicted by dotted lines. Purple, orange, and blue boxes mark molecules involved in ethanolamine, cobalamin, and 1,2-propanediol metabolism, respectively. Solid and dotted orange boxes mark reactions under anaerobic and aerobic conditions, respectively.
We also identified eight upregulated genes with ethanolamine utilization-related annotations (see Table S4 in the supplemental material) and the GO term “ethanolamine metabolic process”as overrepresented among upregulated genes in H7858 grown on CSS compared to in MBHIB (Table 3). Mapping of these genes to the ethanolamine utilization pathway, constructed based on previous studies for S. enterica (42–44) and information about the eut operon provided by Staib and Fuchs (44), showed that genes encoding 8 of the 11 key enzymes mapped to the ethanolamine degradation pathway were upregulated (FC, 4.97 to 6.19) (Fig. 2). Furthermore, we identified seven upregulated genes with 1,2-propanediol utilization-related annotations (see Table S4 in the supplemental material) and the GO term “propanediol catabolic process” as overrepresented among upregulated genes (Table 3). We mapped these genes to the 1,2-propanediol utilization pathway, which was constructed based on data from S. enterica (42–44) and information about the pdu operon provided by Staib and Fuchs (44). This analysis showed that 6 of the 13 genes encoding key enzymes mapped to the 1,2-propanediol degradation pathway (Fig. 2) were upregulated on CSS (FC, 2.88 to 6.57).
Genes involved in carbohydrate transport and utilization were upregulated in H7858 growing on CSS.
We identified 14 upregulated genes with carbohydrate and alcohol transport and utilization-related annotations (see Table S5 in the supplemental material), and the GO terms “alpha-glucoside transmembrane transporter activity” and “oligosaccharide-transporting ATPase activity” were overrepresented among upregulated genes (Table 3). As the 14 upregulated genes represented functions related to transport and metabolism of galactitol, mannose, and maltose, we diagrammed the galactitol- and mannose-specific phosphotransferase system (PTS), maltose-specific ATP-binding cassette (ABC) transporter system, as well as a few catabolism reactions for each of these three molecules (Fig. 3) to further assess expression of these pathways. For the mannose-specific PTS of L. monocytogenes, genes encoding all four components (PTS-IIAMan, PTS-IIBMan, PTS-IICMan, and PTS-IIDMan) were upregulated (Fig. 3). For the galactitol-specific PTS, which includes three components, genes encoding two components (PTS-IIBGat and PTS-IICGat) were upregulated. For the maltose-specific ABC transporter system of L. monocytogenes, genes encoding all three domains of this ABC transporter (MalE, MalF, and MalG) were upregulated.
FIG 3.
Schematic of galactitol- and mannose-specific phosphotransferase system (PTS), maltose-specific ATP-binding cassette (ABC) transporter system, catabolism reactions for each of the three molecules, and the nonoxidative branch of the pentose phosphate pathway in H7858. Pathways were constructed based on information provided in the BioCyc database. H7858 protein designations for enzymes involved in these reactions and pathways are shown in blue text. Enzymes encoded by genes upregulated in H7858 grown on cold smoked salmon (CSS) compared to genes in H7858 grown in modified BHI broth (MBHIB) are designated by display of the fold change (FC) in green boxes. Pink, red, and blue boxes mark molecules involved in galactitol- and mannose-specific PTS and catabolism reactions, maltose-specific ABC transporter system and catabolism reactions, and the nonoxidative branch of the pentose phosphate pathway, respectively.
Additionally, we found that 4 of the 16 genes involved in the nonoxidative branch of the L. monocytogenes pentose phosphate pathway were significantly upregulated in H7858 growing on CSS (see Table S5 in the supplemental material); these genes encode enzymes involved in four of five key reactions of this pathway branch based on the 10403S database on BioCyc (Fig. 3). Moreover, 6-phospho-d-gluconate, generated by maltose utilization reactions, and d-glyceraldehyde-3-phosphate, generated from galactitol utilization reactions, are also found to be the participants of the nonoxidative branch of the pentose phosphate pathway in L. monocytogenes (Fig. 3), which reflect a potential connection between these two carbohydrate utilization pathways and the nonoxidative branch of the pentose phosphate pathway.
Genes involved in the agmatine deiminase system were upregulated in H7858 growing on CSS.
We also identified four upregulated genes with agmatine deiminase-related annotations (see Table S6 in the supplemental material); the fold changes of these genes ranged from 13.67 to 31.36 and were the highest among all upregulated genes. To further explore the functions of these genes, we reconstructed the reactions involved in the L. monocytogenes agmatine deiminase system, using previous studies on L. monocytogenes and other Gram-positive bacteria (45–48). All four genes encoding the key enzymes involved in the breakdown of agmatine to CO2 and NH3 showed higher transcript levels in H7858 grown on CSS (Fig. 4), indicating upregulation of the overall agmatine deiminase pathway.
FIG 4.
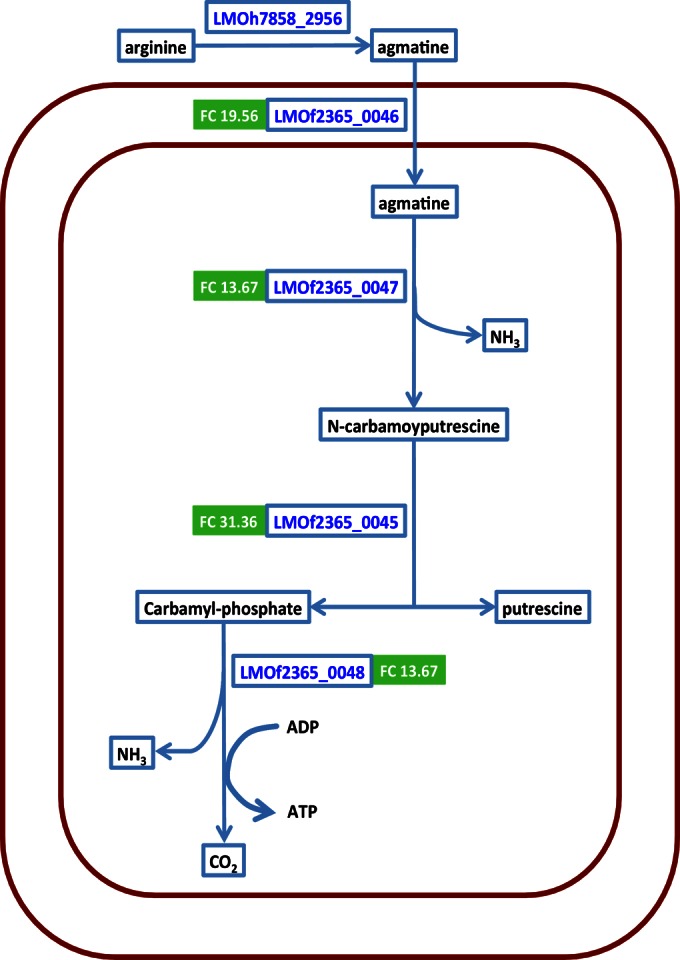
The agmatine deiminase system in H7858 was constructed by connecting reactions associated with agmatine deiminase based on previous studies (45–48). H7858 protein designations for enzymes involved in these reactions are shown in blue text. Enzymes encoded by genes upregulated in H7858 grown on cold smoked salmon (CSS) compared to genes in H7858 grown in modified BHI broth (MBHIB) are designated by display of the fold change (FC) in green boxes. Blue boxes mark molecules involved in the agmatine deiminase system.
L. monocytogenes grown on CSS shows higher transcript levels of PrfA-dependent genes.
Five genes known to be regulated by the master regulator of virulence genes, PrfA, were found to be upregulated in H7858 grown on CSS. These PrfA-dependent genes include inlB (FC = 2.71), plcA (FC = 2.83), hly (FC = 2.54), actA (FC = 2.86), and plcB (FC = 3.19) (see Table S7 in the supplemental material). Statistical analysis showed that PrfA-dependent genes as a group were significantly enriched among upregulated genes (P < 0.0001; Fisher's exact test).
DISCUSSION
In this study, we used RNA-seq to explore the transcriptional landscape of L. monocytogenes H7858 growing on CSS and in BHIB modified to reflect the pH and water phase salt concentration of CSS. Our data indicate that H7858 grown on vacuum-packaged CSS (i) upregulates cobalamin biosynthesis pathways as well as ethanolamine and 1,2-propanediol utilization pathways, (ii) differentially regulates carbohydrate transport functions, and (iii) upregulates arginine deiminase genes, likely facilitating adaptation to anaerobic conditions, utilization of nutrients available on CSS, and growth in the presence of the resident microbiota.
Limitations of using MBHIB as a reference condition to CSS for studying the gene expression profile of L. monocytogenes during growth.
A number of previous studies have analyzed the transcriptomes of foodborne pathogens under stress conditions commonly present on/in food matrices (including hyperosmotic stress, cold stress, hydrostatic pressure stress, antimicrobial stress, acid stress, and alkali stress), using laboratory media modified to simulate these conditions as a model (15–22, 49–63). These laboratory media include BHI broth (18, 20, 21, 59), BHI agar (19), Luria-Bertani broth (60, 61), tryptic soy broth (62), M9-glucose (63), and Listeria minimal medium (22) as reference conditions. The key differences between the reference condition that we used and CSS were that the CSS was vacuum packaged while the reference medium was not anaerobic (though MBHIB was incubated without shaking), L. monocytogenes was grown on the surface of CSS, while growth was planktonic in the liquid MBHIB, and CSS contains a number of unique components that cannot be easily added to BHIB at comparable concentrations (e.g., different phenolics, lipids, trace elements, etc.). In addition, a previous study in our lab on the same batch of CSS demonstrated that the resident lactic acid bacteria were able to grow to ∼7 log CFU/g by day 10 (with an initial density of ∼4 log CFU/g on day 0) under the same growth conditions as those used in the current study (31). Lactic acid bacteria are known to be one of the dominant microbes on CSS and may constitute a natural form of antimicrobial control via competition for particular nutrients or production of organic acids, hydrogen peroxide, and bacteriocins (64–66). Furthermore, Lactobacillus has been demonstrated to be able to reshape L. monocytogenes protein-coding genes and small RNA (sRNA) expression profiles (67). Even though we modified the laboratory medium to present the same major conditions as those found in CSS and L. monocytogenes had similar growth patterns under the conditions of food matrix and laboratory medium, the aforementioned differences between these two conditions add limitations to the comparison of the transcriptional profiles.
The inoculation level of L. monocytogenes in MBHIB and on CSS was high in the present study (∼1 × 106 CFU/g). L. monocytogenes contamination of cold smoked fish is typically at low levels (8, 68), and there is the potential for differences in responses of L. monocytogenes when inoculated at different levels. However, L. monocytogenes can grow in food products at refrigeration temperatures to high levels, and the infection dose in an immunocompetent individual can be high (up to ∼9 log CFU/g) (69). While our study does not encompass all possible variations in inoculum level, medium preparation, and competitive microbes found on some naturally contaminated products, data from previous studies indicate that what we found was not unusual. Consistent with our results, studies with lower inoculum levels also found similar growth rates (e.g., 0.37 log units/day) and similar final concentrations (e.g., ∼8 log CFU/ml) for L. monocytogenes on vacuum-packed CSS at refrigerated temperatures (e.g., 4°C) (7, 70). Our results do provide insights into the relative differences in transcriptomes of L. monocytogenes at the same growth phase under different complex conditions and reflect the gene expression profile of L. monocytogenes in food at high levels that could cause human disease.
L. monocytogenes grown on vacuum-packaged CSS upregulates cobalamin biosynthesis pathways as well as ethanolamine and 1,2-propanediol degradation pathways, likely facilitating adaptation to available nutrient sources.
Our data showed that L. monocytogenes significantly upregulated both cobalamin biosynthesis and transport systems, presumably to increase the availability of cobalamin in the bacterial cells. Cobalamin (coenzyme B12 [71]) is found in high levels in CSS. Smoked salmon has up to 18.10 μg B12 per 100 g according to the USDA National Nutrient Database for Standard Reference (Basic report no. 35190, salmon, red (sockeye), smoked). We found that transcript levels of genes involved in both aerobic and anaerobic pathways (72–74) were significantly higher in L. monocytogenes grown on CSS than in BHIB. Roth et al. (42) proposed that the primary function of cobalamin in many bacteria is to support fermentation of small molecules such as ethanolamine and 1,2-propanediol by catalyzing molecular rearrangements. We propose that one of the reasons L. monocytogenes uptakes or synthesizes cobalamin C is to facilitate growth under this condition by using cobalamin to support the utilization of ethanolamine and 1,2-propanediol.
To date, three foodborne bacterial pathogens have been shown to be able to use both ethanolamine and 1,2-propanediol as a carbon source (75) and to use ethanolamine as a nitrogen source: L. monocytogenes, S. enterica, and Clostridium perfringens (76). Ethanolamine is a major constituent of lipids in eukaryotic cells (77), including in salmon (78), and thus may become available for L. monocytogenes through the breakdown of salmon cells. Broad-range phospholipases such as PlcB (plcB was upregulated in CSS) of L. monocytogenes might serve to reduce phosphatidylethanolamine to ethanolamine (44). 1,2-Propanediol is produced during bacterial anaerobic catabolism of the common methylpentoses rhamnose and/or fucose (44, 79). 1,2-Propanediol may be available for L. monocytogenes through the breakdown of salmon mucosal glycoconjugates, which contain fucose and rhamnose (80, 81). Moreover, in L. monocytogenes, one cobalamin-binding riboswitch is located upstream of the first gene in the eut locus; this riboswitch regulates expression of eut in response to cobalamin availability (82). A second cobalamin-binding riboswitch is located upstream of the pdu locus; this riboswitch maximizes the expression of 1,2-propanediol utilization genes when both 1,2-propanediol and cobalamin are present (83). These findings further demonstrate the close relationship between ethanolamine and 1,2-propanediol utilization pathways and cobalamin biosynthesis/transport pathways.
Recent research has identified potential roles for ethanolamine and 1,2-propanediol utilization in L. monocytogenes and S. Typhimurium during growth in foods and/or in the host environment (84–87). Srikumar and Fuchs (88) found that nonpolar deletions of pocR (regulating pdu and cob-cbi) and eutR in S. enterica serovar Typhimurium led to significantly reduced proliferation in milk and egg yolk. Likewise, Goudeau et al. (84) reported that S. enterica mutants with deletion of pduD or cobS show decreased fitness in cilantro soft rot. Archambaud et al. (67) reported that L. monocytogenes shows higher transcript levels of genes encoding functions involved in ethanolamine and 1,2-propanediol utilization as well as cobalamin biosynthesis when present in the intestine of gnotobiotic mice that contained Lactobacillus spp. than in mice without Lactobacillus spp. This suggests that 1,2-propanediol and ethanolamine utilization may provide L. monocytogenes a mechanism to effectively coexist with the members of the resident microbiota, which are usually not able to utilize these organic compounds. Overall, our study, along with other studies, suggests that foodborne pathogens, including L. monocytogenes, may utilize ethanolamine and/or 1,2-propanediol to proliferate in food and host environments in which these molecules are available. Further studies will be needed to confirm this and to identify food matrices or growth conditions under which ethanolamine and/or 1,2-propanediol utilization by L. monocytogenes may occur.
Listerial physiological adaptation to the end products of ethanolamine and 1,2-propanediol utilization on CSS may provide targets for novel interventions.
Under anaerobic conditions, two of the major products of the ethanolamine and 1,2-propanediol utilization pathways are acetate (42–44, 89) and propionate (42–44, 90), respectively. We propose that L. monocytogenes utilizes the agmatine deiminase system to attenuate the acidification caused by these two acids. This is supported by the observation that L. monocytogenes grown in vacuum-packaged CSS upregulated genes encoding functions involved in the agmatine deiminase system. This system has been demonstrated to catalyze arginine and/or agmatine deamination, which generates two NH3 molecules, facilitating pH buffering (45–48, 91) and thus possibly buffering acid end products (acetate and propionate) created by the ethanolamine and 1,2-propanediol degradation. However, the upregulation of genes related to this system has not been found in studies on L. monocytogenes growing on/in food matrices such as turkey deli meat (19), skim milk (21), or cut cabbage (22), which may indicate that this transcriptional response is specific to the growth conditions tested here. The production of acetate and propionate as by-products of ethanolamine and 1,2-propanediol degradation suggests that growth inhibitors that include these two organic acids may be able to more effectively inhibit L. monocytogenes growth than currently used growth inhibitors. This is supported by a recent transcriptomic study in BHIB-grown L. monocytogenes (17), which showed that exposure of L. monocytogenes to acetate and lactate led to decreased expression of lactate- and acetate-creating energy pathways, shifting ATP production to a less efficient pathway with acetoin, a noncharged molecule, as an end product.
Higher transcript levels of genes encoding specific carbohydrate PTS components and ABC transporter domains indicate that L. monocytogenes may uptake and utilize a broad range of carbohydrates on CSS.
As L. monocytogenes upregulated genes encoding proteins involved in utilization and transport of galactitol, mannose, and maltose, we propose that L. monocytogenes growing on CSS broadens the range of carbohydrates that are utilized to compensate for the limited availability of glucose (relative to BHIB, which contains 2 g added glucose/liter). Galactitol is the reduction product of galactose, which together with mannose, may be available as components of mucin glycoconjugates at the mucosal tissue of fish (80, 81, 92, 93). While maltose is found in many processed products that have been sweetened (94), it is unclear whether there is a specific maltose source in CSS. Consistent with our findings, genes involved in carbohydrate metabolism were identified to be more expressed on cut cabbage (22) and on ready-to-eat turkey deli meat (19) than what was observed with the growth of L. monocytogenes in laboratory media. However, no genes involved in carbohydrate metabolism were found to be upregulated in skim milk in Liu and Ream's study (21), which may be attributed to the abundance of lactose as a carbon source in milk. In the host environment, Toledo-Arana et al. (23) and Chatterjee et al. (95) found increased transcript levels of specific PTSs in L. monocytogenes, which enabled it to utilize sugars, such as mannose and fructose, and/or their alcohols, including galactitol, during growth in the intestine of axenic mice (23) or upon entering epithelial cells (95). In sum, exploiting a wider range of carbohydrates appears to be a successful strategy of enteric pathogens to overcome nutrient limitations or adapt to specific nutrient compositions when proliferating under the complex conditions of food matrices such as CSS.
Our data suggest that L. monocytogenes growing on CSS may use the nonoxidative branch of the pentose phosphate pathway for gluconeogenesis, which is plausible since the pentose phosphate pathway is more efficient in anabolism than in glycolysis. In a result similar to our finding, Bae et al. (19) also found that the gene LMOf2365_1395 of L. monocytogenes F2365, involved in the pentose phosphate pathway, was induced on RTE turkey deli meat (19). Zhou et al. (96) found that several key enzymes involved in the pentose phosphate pathway were more prevalent in biofilms than in planktonic cells of L. monocytogenes, and Hefford et al. (97) reported that proteins involved in sugar metabolism were highly expressed in biofilms. These findings may indicate that upregulation of the aforementioned carbohydrate utilization genes, especially the pentose phosphate pathway-related genes, is due to growth of L. monocytogenes on the surface of foods, such as deli meat and CSS.
Differential regulation of PrfA-dependent genes in vacuum-packaged CSS supports that growth conditions can have multifaceted effects on gene expression and cell physiology.
Interestingly, PrfA-dependent genes showed higher transcript levels in vacuum-packaged CSS than in MBHIB, which may be triggered by (i) the low level of glucose on CSS, (ii) the oxygen restriction condition presented in vacuum-packaged CSS, and/or (iii) specific signals associated with salmon tissue or specific compounds in the BHIB that could suppress the expression/activity of PrfA. As the presence of glucose and fermentable carbohydrates can affect expression of the PrfA regulon (98–100), the apparent utilization of different carbohydrates by L. monocytogenes grown on CSS may affect the transcript level of PrfA-dependent genes in L. monocytogenes grown on CSS. The importance of the anaerobic environment is supported by data from Larsen et al. (101), who reported higher invasion of Caco-2 cells by L. monocytogenes grown for 2 and 4 weeks on modified atmosphere-packaged ham at 4°C than by bacteria grown in BHIB. Andersen et al. (102) reported that L. monocytogenes grown under oxygen-restricted conditions were more invasive to Caco-2 cells and yielded higher bacterial loads in organs after oral guinea pig challenge than did bacteria grown without oxygen restriction. In contrast to our findings, Olesen et al. (20) reported that L. monocytogenes strain O57 prfA transcript levels were significantly higher in bacteria grown in BHI than in those grown in liver pâté at 7°C. Possible explanations for these observed differences include the fact that L. monocytogenes was grown on liver pâtés under aerobic conditions and differences in the length of incubation.
Although PrfA-dependent genes represent bona fide virulence genes (103–105), differential regulation of these genes under the complex conditions of food matrices appears to occur, with oxygen-restricting conditions and carbohydrate availability possibly representing environmental cues affecting expression of the PrfA regulon. In addition, a role for PrfA in the survival and proliferation of L. monocytogenes outside the human host and on/in specific food matrices cannot be excluded, as supported by data that suggest that PrfA contributes to L. monocytogenes biofilm formation (106). Along with previously reported studies, our data suggest that growth conditions can have multifaceted effects on gene expression and cell physiology, which reach beyond specific adaptations to nutrient availability and stress conditions encountered.
Increasing evidence supports that food-related factors that are not easily simulated in laboratory medium likely play important roles in growth and survival of foodborne pathogens in different foods. Use of RNA-seq-based transcriptomic profiling allows for detailed assessment of the physiological state of pathogens present on/in food matrices and provides novel insights in adaptations of foodborne pathogens to the complex conditions on/in specific food matrices and environmental conditions. This type of detailed information will open up a number of new avenues to improve food safety. For example, the type of information presented here could pave the way for developing better detection methods (e.g., methods targeting highly expressed RNA molecules) and may even provide for improved risk assessments that account for the fact that the virulence of a given pathogen may be affected considerably by its physiological state, which clearly depends on a number of factors, including, but not limited to, food matrix, temperature, and packaging strategies. Most importantly, detailed data on pathogen adaptation to different complex conditions of food matrices may hold the key to the development of more-efficient control strategies and will move the development of control strategies from traditional trial and error approaches to rational-design type approaches for the development of new growth inhibitors.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by New York Sea Grant R/SHH-15, funded under award NA07OAR4170010 from the National Sea Grant College Program of the U.S. Department of Commerce's National Oceanic and Atmospheric Administration to the Research Foundation of State University of New York, and by Agriculture and Food Research Initiative grant 2010-65201-20575 from the U.S. Department of Agriculture, National Institute of Food and Agriculture, Food Safety Program.
We thank Veronica Guariglia-Oropeza and Sherry E. Roof for assistance with the RNA preparation for RNA-seq and Maureen Gunderson for assistance with preparation of media.
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AEM.01752-15.
REFERENCES
- 1.Wan Norhana MN, Poole SE, Deeth HC, Dykes GA. 2010. Prevalence, persistence and control of Salmonella and Listeria in shrimp and shrimp products: a review. Food Control 21:343–361. doi: 10.1016/j.foodcont.2009.06.020. [DOI] [Google Scholar]
- 2.Swaminathan B, Cabanes D, Zhang W, Cossart P. 2007. Listeria monocytogenes, p 457–492. In Doyle MP, Beuchat LR (ed), Food microbiology: fundamentals and frontiers, 3rd ed ASM Press, Washington, DC. [Google Scholar]
- 3.Walker S, Archer P, Banks JG. 1990. Growth of Listeria monocytogenes at refrigeration temperatures. J Appl Bacteriol 68:157–162. doi: 10.1111/j.1365-2672.1990.tb02561.x. [DOI] [PubMed] [Google Scholar]
- 4.Norton DM, McCamey MA, Gall KL, Scarlett JM, Boor KJ, Wiedmann M. 2001. Molecular studies on the ecology of Listeria monocytogenes in the smoked fish processing industry. Appl Environ Microbiol 67:198–205. doi: 10.1128/AEM.67.1.198-205.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sabanadesan S, Lammerding AM, Griffiths MW. 2000. Survival of Listeria innocua in salmon following cold-smoke application. J Food Prot 63:715–720. [DOI] [PubMed] [Google Scholar]
- 6.USDA. 2003. Interpretative summary: quantitative assessment of the relative risk to public health from foodborne Listeria monocytogenes among selected categories of ready-to-eat foods. Food Safety and Inspection Services, US Food and Drug Administration, Silver Spring, MD. [Google Scholar]
- 7.Yoon K, Burnette C, Abou-Zeid K, Whiting R. 2004. Control of growth and survival of Listeria monocytogenes on smoked salmon by combined potassium lactate and sodium diacetate and freezing stress during refrigeration and frozen storage. J Food Prot 67:2465–2471. [DOI] [PubMed] [Google Scholar]
- 8.Eklund MW, Poysky FT, Paranjpye RN, Lashbrook LC, Peterson ME, Pelroy GA. 1995. Incidence and sources of Listeria monocytogenes in cold-smoked fishery products and processing plants. J Food Prot 58:502–508. [DOI] [PubMed] [Google Scholar]
- 9.Mejlholm O, Dalgaard P. 2007. Modeling and predicting the growth boundary of Listeria monocytogenes in lightly preserved seafood. J Food Prot 70:70–84. [DOI] [PubMed] [Google Scholar]
- 10.Gombas DE, Chen Y, Clavero RS, Scott VN. 2003. Survey of Listeria monocytogenes in ready-to-eat foods. J Food Prot 66:559–569. [DOI] [PubMed] [Google Scholar]
- 11.Hoffman A, Gall K, Wiedmann M. 2003. The microbial safety of minimally processed seafood with respect to Listeria monocytogenes, p 53–75. In Novak J, Sapers G, Juneja V (ed), Microbial safety of minimally processed foods. CRC Press, Boca Raton, FL. [Google Scholar]
- 12.Lappi VR, Thimothe J, Nightingale KK, Gall K, Scott VN, Wiedmann M. 2004. Longitudinal studies on Listeria in smoked fish plants: impact of intervention strategies on contamination patterns. J Food Prot 67:2500–2514. [DOI] [PubMed] [Google Scholar]
- 13.Gram L. 2001. Potential hazards in cold-smoked fish: Listeria monocytogenes. J Food Sci 66:S1072–S1081. doi: 10.1111/j.1365-2621.2001.tb15526.x. [DOI] [Google Scholar]
- 14.Bergholz TM, Moreno Switt AI, Wiedmann M. 2014. Omics approaches in food safety: fulfilling the promise? Trends Microbiol 22:275–281. doi: 10.1016/j.tim.2014.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ivy RA, Wiedmann M, Boor KJ. 2012. Listeria monocytogenes grown at 7°C shows reduced acid survival and an altered transcriptional response to acid shock compared to L. monocytogenes grown at 37°C. Appl Environ Microbiol 78:3824–3836. doi: 10.1128/AEM.00051-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bergholz TM, Bowen B, Wiedmann M, Boor KJ. 2012. Listeria monocytogenes shows temperature-dependent and -independent responses to salt stress, including responses that induce cross-protection against other stresses. Appl Environ Microbiol 78:2602–2612. doi: 10.1128/AEM.07658-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Stasiewicz MJ, Wiedmann M, Bergholz TM. 2011. The transcriptional response of Listeria monocytogenes during adaptation to growth on lactate and diacetate includes synergistic changes that increase fermentative acetoin production. Appl Environ Microbiol 77:5294–5306. doi: 10.1128/AEM.02976-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rantsiou K, Greppi A, Garosi M, Acquadro A, Mataragas M, Cocolin L. 2012. Strain dependent expression of stress response and virulence genes of Listeria monocytogenes in meat juices as determined by microarray. Int J Food Microbiol 152:116–122. doi: 10.1016/j.ijfoodmicro.2011.08.009. [DOI] [PubMed] [Google Scholar]
- 19.Bae D, Crowley MR, Wang C. 2011. Transcriptome analysis of Listeria monocytogenes grown on a ready-to-eat meat matrix. J Food Prot 74:1104–1111. [DOI] [PubMed] [Google Scholar]
- 20.Olesen I, Thorsen L, Jespersen L. 2010. Relative transcription of Listeria monocytogenes virulence genes in liver pâtés with varying NaCl content. Int J Food Microbiol 141(Suppl):S60–S68. [DOI] [PubMed] [Google Scholar]
- 21.Liu Y, Ream A. 2008. Gene expression profiling of Listeria monocytogenes strain F2365 during growth in ultrahigh-temperature-processed skim milk. Appl Environ Microbiol 74:6859–6866. doi: 10.1128/AEM.00356-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Palumbo JD, Kaneko A, Nguyen KD, Gorski L. 2005. Identification of genes induced in Listeria monocytogenes during growth and attachment to cut cabbage, using differential display. Appl Environ Microbiol 71:5236–5243. doi: 10.1128/AEM.71.9.5236-5243.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Toledo-Arana A, Dussurget O, Nikitas G, Sesto N, Guet-Revillet H, Balestrino D, Loh E, Gripenland J, Tiensuu T, Vaitkevicius K. 2009. The Listeria transcriptional landscape from saprophytism to virulence. Nature 459:950–956. doi: 10.1038/nature08080. [DOI] [PubMed] [Google Scholar]
- 24.Nelson KE, Fouts DE, Mongodin EF, Ravel J, DeBoy RT, Kolonay JF, Rasko DA, Angiuoli SV, Gill SR, Paulsen IT. 2004. Whole genome comparisons of serotype 4b and 1/2a strains of the food borne pathogen Listeria monocytogenes reveal new insights into the core genome components of this species. Nucleic Acids Res 32:2386–2395. doi: 10.1093/nar/gkh562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.CDC. 1999. Update: multistate outbreak of listeriosis—United States, 1998-1999. MMWR Morb Mortal Wkly Rep 47:1117–1118. [PubMed] [Google Scholar]
- 26.Kang J, Wiedmann M, Boor KJ, Bergholz TM. 2015. VirR-mediated resistance of Listeria monocytogenes against food antimicrobials and cross-protection induced by exposure to organic acid salts. Appl Environ Microbiol 81:4553–4562. doi: 10.1128/AEM.00648-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tang S, Stasiewicz MJ, Wiedmann M, Boor KJ, Bergholz TM. 2013. Efficacy of different antimicrobials on inhibition of Listeria monocytogenes growth in laboratory medium and on cold-smoked salmon. Int J Food Microbiol 165:265–275. doi: 10.1016/j.ijfoodmicro.2013.05.018. [DOI] [PubMed] [Google Scholar]
- 28.Bergholz TM, Tang S, Wiedmann M, Boor KJ. 2013. Nisin resistance of Listeria monocytogenes is increased by exposure to salt stress and is mediated via LiaR. Appl Environ Microbiol 79:5682–5688. doi: 10.1128/AEM.01797-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Amezaga MR, Davidson I, McLaggan D, Verheul A, Abee T, Booth IR. 1995. The role of peptide metabolism in the growth of Listeria monocytogenes ATCC 23074 at high osmolarity. Microbiology 141:41–49. doi: 10.1099/00221287-141-1-41. [DOI] [PubMed] [Google Scholar]
- 30.Stasiewicz MJ, Wiedmann M, Bergholz TM. 2010. The combination of lactate and diacetate synergistically reduces cold growth in brain heart infusion broth across Listeria monocytogenes lineages. J Food Prot 73:631–640. [DOI] [PubMed] [Google Scholar]
- 31.Kang J, Tang S, Liu RH, Wiedmann M, Boor KJ, Bergholz TM, Wang S. 2012. Effect of curing method and freeze-thawing on subsequent growth of Listeria monocytogenes on cold-smoked salmon. J Food Prot 75:1619–1626. doi: 10.4315/0362-028X.JFP-11-561. [DOI] [PubMed] [Google Scholar]
- 32.Buchanan RL, Whiting RC, Damert WC. 1997. When is simple good enough: a comparison of the Gompertz, Baranyi, and three-phase linear models for fitting bacterial growth curves. Food Microbiol 14:313–326. doi: 10.1006/fmic.1997.0125. [DOI] [Google Scholar]
- 33.Schroeder A, Mueller O, Stocker S, Salowsky R, Leiber M, Gassmann M, Lightfoot S, Menzel W, Granzow M, Ragg T. 2006. The RIN: an RNA integrity number for assigning integrity values to RNA measurements. BMC Mol Biol 7:3. doi: 10.1186/1471-2199-7-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sue D, Boor KJ, Wiedmann M. 2003. σB-dependent expression patterns of compatible solute transporter genes opuCA and Imo1421 and the conjugated bile salt hydrolase gene bsh in Listeria monocytogenes. Microbiology 149:3247–3256. doi: 10.1099/mic.0.26526-0. [DOI] [PubMed] [Google Scholar]
- 35.Li H, Durbin R. 2010. Fast and accurate long-read alignment with Burrows–Wheeler transform. Bioinformatics 26:589–595. doi: 10.1093/bioinformatics/btp698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hardcastle TJ, Kelly KA. 2010. baySeq: empirical Bayesian methods for identifying differential expression in sequence count data. BMC Bioinformatics 11:422. doi: 10.1186/1471-2105-11-422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bullard J, Purdom E, Hansen K, Dudoit S. 2010. Evaluation of statistical methods for normalization and differential expression in mRNA-Seq experiments. BMC Bioinformatics 11:94. doi: 10.1186/1471-2105-11-94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Young MD, Wakefield MJ, Smyth GK, Oshlack A. 2010. Gene ontology analysis for RNA-seq: accounting for selection bias. Genome Biol 11:R14. doi: 10.1186/gb-2010-11-2-r14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Leisner JJ, Laursen BG, Prévost H, Drider D, Dalgaard P. 2007. Carnobacterium: positive and negative effects in the environment and in foods. FEMS Microbiol Rev 31:592–613. doi: 10.1111/j.1574-6976.2007.00080.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Cotter PD, Ryan S, Gahan CGM, Hill C. 2005. Presence of GadD1 glutamate decarboxylase in selected Listeria monocytogenes strains is associated with an ability to grow at low pH. Appl Environ Microbiol 71:2832–2839. doi: 10.1128/AEM.71.6.2832-2839.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Orsi RH, Bergholz TM, Wiedmann M, Boor KJ. 2015. The Listeria monocytogenes strain 10403S BioCyc database. Database (Oxford) 2015:pii: bav027. doi: 10.1093/database/bav027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Roth J, Lawrence J, Bobik T. 1996. Cobalamin (coenzyme B12): synthesis and biological significance. Annu Rev Microbiol 50:137–181. doi: 10.1146/annurev.micro.50.1.137. [DOI] [PubMed] [Google Scholar]
- 43.Price-Carter M, Tingey J, Bobik TA, Roth JR. 2001. The alternative electron acceptor tetrathionate supports B12-dependent anaerobic growth of Salmonella enterica serovar Typhimurium on ethanolamine or 1,2-propanediol. J Bacteriol 183:2463–2475. doi: 10.1128/JB.183.8.2463-2475.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Staib L, Fuchs TM. 2014. From food to cell: nutrient exploitation strategies of enteropathogens. Microbiology 160:1020–1039. doi: 10.1099/mic.0.078105-0. [DOI] [PubMed] [Google Scholar]
- 45.Griswold AR, Chen Y-YM, Burne RA. 2004. Analysis of an agmatine deiminase gene cluster in Streptococcus mutans UA159. J Bacteriol 186:1902–1904. doi: 10.1128/JB.186.6.1902-1904.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Chen J, Cheng C, Xia Y, Zhao H, Fang C, Shan Y, Wu B, Fang W. 2011. Lmo0036, an ornithine and putrescine carbamoyltransferase in Listeria monocytogenes, participates in arginine deiminase and agmatine deiminase pathways and mediates acid tolerance. Microbiology 157:3150–3161. doi: 10.1099/mic.0.049619-0. [DOI] [PubMed] [Google Scholar]
- 47.Llácer JL, Polo LM, Tavárez S, Alarcón B, Hilario R, Rubio V. 2007. The gene cluster for agmatine catabolism of Enterococcus faecalis: study of recombinant putrescine transcarbamylase and agmatine deiminase and a snapshot of agmatine deiminase catalyzing its reaction. J Bacteriol 189:1254–1265. doi: 10.1128/JB.01216-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lucas PM, Blancato VS, Claisse O, Magni C, Lolkema JS, Lonvaud-Funel A. 2007. Agmatine deiminase pathway genes in Lactobacillus brevis are linked to the tyrosine decarboxylation operon in a putative acid resistance locus. Microbiology 153:2221–2230. doi: 10.1099/mic.0.2007/006320-0. [DOI] [PubMed] [Google Scholar]
- 49.Feng SL, Eucker TP, Holly MK, Konkel ME, Lu XN, Wang S. 2014. Investigating the responses of Cronobacter sakazakii to garlic-derived organosulfur compounds: a systematic study of pathogenic-bacterium injury by use of high-throughput whole-transcriptome sequencing and confocal micro-Raman spectroscopy. Appl Environ Microbiol 80:959–971. doi: 10.1128/AEM.03460-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ribeiro VB, Mujahid S, Orsi RH, Bergholz TM, Wiedmann M, Boor KJ, Destro MT. 2014. Contributions of sigma(B) and PrfA to Listeria monocytogenes salt stress tinder food relevant conditions. Int J Food Microbiol 177:98–108. doi: 10.1016/j.ijfoodmicro.2014.02.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Cameron A, Frirdich E, Huynh S, Parker CT, Gaynor EC. 2012. Hyperosmotic stress response of Campylobacter jejuni. J Bacteriol 194:6116–6130. doi: 10.1128/JB.01409-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Tessema GT, Møretrø T, Snipen L, Heir E, Holck A, Naterstad K, Axelsson L. 2012. Microarray-based transcriptome of Listeria monocytogenes adapted to sublethal concentrations of acetic acid, lactic acid, and hydrochloric acid. Can J Microbiol 58:1112–1123. doi: 10.1139/w2012-091. [DOI] [PubMed] [Google Scholar]
- 53.Malone AS, Chung Y-K, Yousef AE. 2006. Genes of Escherichia coli O157:H7 that are involved in high-pressure resistance. Appl Environ Microbiol 72:2661–2671. doi: 10.1128/AEM.72.4.2661-2671.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Bowman JP, Bittencourt CR, Ross T. 2008. Differential gene expression of Listeria monocytogenes during high hydrostatic pressure processing. Microbiology 154:462–475. doi: 10.1099/mic.0.2007/010314-0. [DOI] [PubMed] [Google Scholar]
- 55.Giotis ES, Muthaiyan A, Natesan S, Wilkinson BJ, Blair IS, McDowell DA. 2010. Transcriptome analysis of alkali shock and alkali adaptation in Listeria monocytogenes 10403S. Foodborne Pathog Dis 7:1147–1157. doi: 10.1089/fpd.2009.0501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Du H, Wang M, Luo Z, Ni B, Wang F, Meng Y, Xu S, Huang X. 2011. Coregulation of gene expression by sigma factors RpoE and RpoS in Salmonella enterica serovar Typhi during hyperosmotic stress. Curr Microbiol 62:1483–1489. doi: 10.1007/s00284-011-9890-8. [DOI] [PubMed] [Google Scholar]
- 57.Liu YH, Morgan S, Ream A, Huang LH. 2013. Gene expression profiling of a nisin-sensitive Listeria monocytogenes Scott A ctsR deletion mutant. J Ind Microbiol Biotechnol 40:495–505. doi: 10.1007/s10295-013-1243-0. [DOI] [PubMed] [Google Scholar]
- 58.Oliver H, Orsi R, Ponnala L, Keich U, Wang W, Sun Q, Cartinhour S, Filiatrault M, Wiedmann M, Boor K. 2009. Deep RNA sequencing of L. monocytogenes reveals overlapping and extensive stationary phase and sigma B-dependent transcriptomes, including multiple highly transcribed noncoding RNAs. BMC Genomics 10:641. doi: 10.1186/1471-2164-10-641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Makhzami S, Quénée P, Akary E, Bach C, Aigle M, Delacroix-Buchet A, Ogier JC, Serror P. 2008. In situ gene expression in cheese matrices: application to a set of enterococcal genes. J Microbiol Methods 75:485–490. doi: 10.1016/j.mimet.2008.07.025. [DOI] [PubMed] [Google Scholar]
- 60.Fink RC, Black EP, Hou Z, Sugawara M, Sadowsky MJ, Diez-Gonzalez F. 2012. Transcriptional responses of Escherichia coli K-12 and O157:H7 associated with lettuce leaves. Appl Environ Microbiol 78:1752–1764. doi: 10.1128/AEM.07454-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Deng X, Li Z, Zhang W. 2012. Transcriptome sequencing of Salmonella enterica serovar Enteritidis under desiccation and starvation stress in peanut oil. Food Microbiol 30:311–315. doi: 10.1016/j.fm.2011.11.001. [DOI] [PubMed] [Google Scholar]
- 62.Fratamico PM, Wang S, Yan X, Zhang W, Li Y. 2011. Differential gene expression of E. coli O157:H7 in ground beef extract compared to tryptic soy broth. J Food Sci 76:M79–M87. doi: 10.1111/j.1750-3841.2010.01952.x. [DOI] [PubMed] [Google Scholar]
- 63.Kyle JL, Parker CT, Goudeau D, Brandl MT. 2010. Transcriptome analysis of Escherichia coli O157:H7 exposed to lysates of lettuce leaves. Appl Environ Microbiol 76:1375–1387. doi: 10.1128/AEM.02461-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Calo-Mata P, Arlindo S, Boehme K, de Miguel T, Pascoal A, Barros-Velazquez J. 2008. Current applications and future trends of lactic acid bacteria and their bacteriocins for the biopreservation of aquatic food products. Food Bioprocess Tech 1:43–63. doi: 10.1007/s11947-007-0021-2. [DOI] [Google Scholar]
- 65.Tomé E, Gibbs PA, Teixeira PC. 2007. Could modifications of processing parameters enhance the growth and selection of lactic acid bacteria in cold-smoked salmon to improve preservation by natural means? J Food Prot 70:1607–1614. [DOI] [PubMed] [Google Scholar]
- 66.Concha-Meyer A, Schobitz R, Brito C, Fuentes R. 2011. Lactic acid bacteria in an alginate film inhibit Listeria monocytogenes growth on smoked salmon. Food Control 22:485–489. doi: 10.1016/j.foodcont.2010.09.032. [DOI] [Google Scholar]
- 67.Archambaud C, Nahori M-A, Soubigou G, Bécavin C, Laval L, Lechat P, Smokvina T, Langella P, Lecuit M, Cossart P. 2012. Impact of lactobacilli on orally acquired listeriosis. Proc Natl Acad Sci U S A 109:16684–16689. doi: 10.1073/pnas.1212809109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Jørgensen LV, Huss HH. 1998. Prevalence and growth of Listeria monocytogenes in naturally contaminated seafood. Int J Food Microbiol 42:127–131. doi: 10.1016/S0168-1605(98)00071-3. [DOI] [PubMed] [Google Scholar]
- 69.Azadian B, Finnerty G, Pearson A. 1989. Cheese-borne Listeria meningitis in immunocompetent patient. Lancet 333:322–323. [DOI] [PubMed] [Google Scholar]
- 70.Cornu M, Beaufort A, Rudelle S, Laloux L, Bergis H, Miconnet N, Serot T, Delignette-Muller ML. 2006. Effect of temperature, water-phase salt and phenolic contents on Listeria monocytogenes growth rates on cold-smoked salmon and evaluation of secondary models. Int J Food Microbiol 106:159–168. doi: 10.1016/j.ijfoodmicro.2005.06.017. [DOI] [PubMed] [Google Scholar]
- 71.McDowell LR. 2008. Vitamins in animal and human nutrition, 2nd ed, p 523–564 Iowa State University Press, Ames, IA. [Google Scholar]
- 72.Raux E, Schubert H, Warren M. 2000. Biosynthesis of cobalamin (vitamin B12): a bacterial conundrum. Cell Mol Life Sci 57:1880–1893. doi: 10.1007/PL00000670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Martens JH, Barg H, Warren MJ, Jahn D. 2002. Microbial production of vitamin B12. Appl Microbiol Biotechnol 58:275–285. doi: 10.1007/s00253-001-0902-7. [DOI] [PubMed] [Google Scholar]
- 74.Rasetti V, Pfaltz A, Kratky C, Eschenmoser A. 1981. Ring contraction of hydroporphinoid to corrinoid complexes. Proc Natl Acad Sci U S A 78:16–19. doi: 10.1073/pnas.78.1.16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Salyers AA. 1979. Energy sources of major intestinal fermentative anaerobes. Am J Clin Nutr 32:158–163. [DOI] [PubMed] [Google Scholar]
- 76.Korbel JO, Doerks T, Jensen LJ, Perez-Iratxeta C, Kaczanowski S, Hooper SD, Andrade MA, Bork P. 2005. Systematic association of genes to phenotypes by genome and literature mining. PLoS Biol 3:e134. doi: 10.1371/journal.pbio.0030134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Bakovic M, Fullerton MD, Michel V. 2007. Metabolic and molecular aspects of ethanolamine phospholipid biosynthesis: the role of CTP: phosphoethanolamine cytidylyltransferase (Pcyt2). Biochem Cell Biol 85:283–300. doi: 10.1139/O07-006. [DOI] [PubMed] [Google Scholar]
- 78.Bell JG, McVicar AH, Park MT, Sargent JR. 1991. High dietary linoleic acid affects the fatty acid compositions of individual phospholipids from tissues of Atlantic salmon (Salmo salar): association with stress susceptibility and cardiac lesion. J Nutr 121:1163–1172. [DOI] [PubMed] [Google Scholar]
- 79.Lin E. 1996. Dissimilatory pathways for sugars, polyols, and carboxylates, p 307–342. Escherichia coli and Salmonella: cellular and molecular biology, 2nd ed ASM Press, Washington, DC. [Google Scholar]
- 80.Cummings J, Pomare E, Branch W, Naylor C, Macfarlane G. 1987. Short chain fatty acids in human large intestine, portal, hepatic and venous blood. Gut 28:1221–1227. doi: 10.1136/gut.28.10.1221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Micallef G, Bickerdike R, Reiff C, Fernandes JM, Bowman AS, Martin SA. 2012. Exploring the transcriptome of Atlantic salmon (Salmo salar) skin, a major defense organ. Mar Biotechnol 14:559–569. doi: 10.1007/s10126-012-9447-2. [DOI] [PubMed] [Google Scholar]
- 82.Mellin JR, Koutero M, Dar D, Nahori M-A, Sorek R, Cossart P. 2014. Sequestration of a two-component response regulator by a riboswitch-regulated noncoding RNA. Science 345:940–943. doi: 10.1126/science.1255083. [DOI] [PubMed] [Google Scholar]
- 83.Mellin JR, Tiensuu T, Becavin C, Gouin E, Johansson J, Cossart P. 2013. A riboswitch-regulated antisense RNA in Listeria monocytogenes. Proc Natl Acad Sci U S A 110:13132–13137. doi: 10.1073/pnas.1304795110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Goudeau DM, Parker CT, Zhou YG, Sela S, Kroupitski Y, Brandl MT. 2013. The Salmonella transcriptome in lettuce and cilantro soft rot reveals a niche overlap with the animal host intestine. Appl Environ Microbiol 79:250–262. doi: 10.1128/AEM.02290-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Joseph B, Przybilla K, Stühler C, Schauer K, Slaghuis J, Fuchs TM, Goebel W. 2006. Identification of Listeria monocytogenes genes contributing to intracellular replication by expression profiling and mutant screening. J Bacteriol 188:556–568. doi: 10.1128/JB.188.2.556-568.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Hautefort I, Thompson A, Eriksson-Ygberg S, Parker M, Lucchini S, Danino V, Bongaerts R, Ahmad N, Rhen M, Hinton J. 2008. During infection of epithelial cells Salmonella enterica serovar Typhimurium undergoes a time-dependent transcriptional adaptation that results in simultaneous expression of three type 3 secretion systems. Cell Microbiol 10:958–984. doi: 10.1111/j.1462-5822.2007.01099.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Harvey P, Watson M, Hulme S, Jones M, Lovell M, Berchieri A, Young J, Bumstead N, Barrow P. 2011. Salmonella enterica serovar Typhimurium colonizing the lumen of the chicken intestine grows slowly and upregulates a unique set of virulence and metabolism genes. Infect Immun 79:4105–4121. doi: 10.1128/IAI.01390-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Srikumar S, Fuchs TM. 2011. Ethanolamine utilization contributes to proliferation of Salmonella enterica serovar Typhimurium in food and in nematodes. Appl Environ Microbiol 77:281–290. doi: 10.1128/AEM.01403-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Tsoy O, Ravcheev D, Mushegian A. 2009. Comparative genomics of ethanolamine utilization. J Bacteriol 191:7157–7164. doi: 10.1128/JB.00838-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Van Dyk TK, LaRossa RA. 1987. Involvement of ack-pta operon products in α-ketobutyrate metabolism by Salmonella typhimurium. Mol Gen Genet 207:435–440. doi: 10.1007/BF00331612. [DOI] [PubMed] [Google Scholar]
- 91.Beenken KE, Dunman PM, McAleese F, Macapagal D, Murphy E, Projan SJ, Blevins JS, Smeltzer MS. 2004. Global gene expression in Staphylococcus aureus biofilms. J Bacteriol 186:4665–4684. doi: 10.1128/JB.186.14.4665-4684.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Cummings J, Macfarlane G. 1991. The control and consequences of bacterial fermentation in the human colon. J Appl Bacteriol 70:443–459. doi: 10.1111/j.1365-2672.1991.tb02739.x. [DOI] [PubMed] [Google Scholar]
- 93.Bhattacharyya SN, Kaufman B, Khorrami A, Enriquez JI, Manna B. 1988. Fibronectin: source of mannose in a highly purified respiratory mucin. Inflammation 12:433–446. doi: 10.1007/BF00919437. [DOI] [PubMed] [Google Scholar]
- 94.Guzmán-Maldonado H, Paredes-López O, Biliaderis CG. 1995. Amylolytic enzymes and products derived from starch: a review. Crit Rev Food Sci Nutr 35:373–403. doi: 10.1080/10408399509527706. [DOI] [PubMed] [Google Scholar]
- 95.Chatterjee SS, Hossain H, Otten S, Kuenne C, Kuchmina K, Machata S, Domann E, Chakraborty T, Hain T. 2006. Intracellular gene expression profile of Listeria monocytogenes. Infect Immun 74:1323–1338. doi: 10.1128/IAI.74.2.1323-1338.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Zhou Q, Feng X, Zhang Q, Feng F, Yin X, Shang J, Qu H, Luo Q. 2012. Carbon catabolite control is important for Listeria monocytogenes biofilm formation in response to nutrient availability. Curr Microbiol 65:35–43. doi: 10.1007/s00284-012-0125-4. [DOI] [PubMed] [Google Scholar]
- 97.Hefford M, D'Aoust S, Cyr T, Austin J, Sanders G, Kheradpir E, Kalmokoff M. 2005. Proteomic and microscopic analysis of biofilms formed by Listeria monocytogenes 568. Can J Microbiol 51:197–208. doi: 10.1139/w04-129. [DOI] [PubMed] [Google Scholar]
- 98.Joseph B, Mertins S, Stoll R, Schär J, Umesha KR, Luo Q, Müller-Altrock S, Goebel W. 2008. Glycerol metabolism and PrfA activity in Listeria monocytogenes. J Bacteriol 190:5412–5430. doi: 10.1128/JB.00259-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Behari J, Youngman P. 1998. Regulation of hly expression in Listeria monocytogenes by carbon sources and pH occurs through separate mechanisms mediated by PrfA. Infect Immun 66:3635–3642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Eylert E, Schär J, Mertins S, Stoll R, Bacher A, Goebel W, Eisenreich W. 2008. Carbon metabolism of Listeria monocytogenes growing inside macrophages. Mol Microbiol 69:1008–1017. doi: 10.1111/j.1365-2958.2008.06337.x. [DOI] [PubMed] [Google Scholar]
- 101.Larsen MH, Koch AG, Ingmer H. 2010. Listeria monocytogenes efficiently invades Caco-2 cells after low-temperature storage in broth and on deli meat. Foodborne Pathog Dis 7:1013–1018. doi: 10.1089/fpd.2009.0470. [DOI] [PubMed] [Google Scholar]
- 102.Andersen JB, Roldgaard BB, Christensen BB, Licht TR. 2007. Oxygen restriction increases the infective potential of Listeria monocytogenes in vitro in Caco-2 cells and in vivo in guinea pigs. BMC Microbiol 7:55. doi: 10.1186/1471-2180-7-55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.de las Heras A, Cain RJ, Bieleckal MK, Vazquez-Boland JA. 2011. Regulation of Listeria virulence: PrfA master and commander. Curr Opin Microbiol 14:118–127. doi: 10.1016/j.mib.2011.01.005. [DOI] [PubMed] [Google Scholar]
- 104.Freitag NE, Port GC, Miner MD. 2009. Listeria monocytogenes—from saprophyte to intracellular pathogen. Nat Rev Microbiol 7:623–628. doi: 10.1038/nrmicro2171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Gray MJ, Freitag NE, Boor KJ. 2006. How the bacterial pathogen Listeria monocytogenes mediates the switch from environmental Dr. Jekyll to pathogenic Mr. Hyde. Infect Immun 74:2505–2512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Lemon KP, Freitag NE, Kolter R. 2010. The virulence regulator PrfA promotes biofilm formation by Listeria monocytogenes. J Bacteriol 192:3969–3976. doi: 10.1128/JB.00179-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.