Abstract
The bacterium Microbacterium aurum strain B8.A, originally isolated from a potato plant wastewater facility, is able to degrade different types of starch granules. Here we report the characterization of an unusually large, multidomain M. aurum B8.A α-amylase enzyme (MaAmyA). MaAmyA is a 1,417-amino-acid (aa) protein with a predicted molecular mass of 148 kDa. Sequence analysis of MaAmyA showed that its catalytic core is a family GH13_32 α-amylase with the typical ABC domain structure, followed by a fibronectin (FNIII) domain, two carbohydrate binding modules (CBM25), and another three FNIII domains. Recombinant expression and purification yielded an enzyme with the ability to degrade wheat and potato starch granules by introducing pores. Characterization of various truncated mutants of MaAmyA revealed a direct relationship between the presence of CBM25 domains and the ability of MaAmyA to form pores in starch granules, while the FNIII domains most likely function as stable linkers. At the C terminus, MaAmyA carries a 300-aa domain which is uniquely associated with large multidomain amylases; its function remains to be elucidated. We concluded that M. aurum B8.A employs a multidomain enzyme system to initiate degradation of starch granules via pore formation.
INTRODUCTION
Starch is an excellent carbon and energy source for many microorganisms, which employ a dedicated set of proteins for extracellular hydrolysis of this polysaccharide, uptake of shorter oligosaccharides into the cell, and further degradation into glucose. Most studies on degradation of starch by microbial enzymes have focused on soluble starch. This has resulted in the identification and characterization of a large variety of enzymes cleaving either α(1→4) or α(1→6) linkages in amylose and amylopectin. Most of these enzymes belong to the glycoside hydrolase 13 (GH13) family (1). Sequence diversity is such that, at the moment, the GH13 family contains a total of 40 subfamilies (1). Most of the new members in subfamilies are identified in DNA sequencing projects, and biochemical information about the activity and specificity of these potentially new enzymes is highly lagging.
Many plants produce starch in a granular form for the storage of carbohydrates. The crystallinity of such granules varies with the plant source. Potato starch granules have a relatively high degree of crystallinity, making them notoriously resistant to bacterial and fungal degradation (2–4). Nevertheless, some microorganisms have been reported to employ enzymes that are able to digest granular starch (5, 6).
Amylases found to be involved in granular starch degradation are often multidomain enzymes that include one or more carbohydrate binding modules (CBMs), which aid in the binding of the enzyme to the granular substrate (7–10).
In previous work, various bacteria able to grow on potato starch granules as a carbon source were isolated, and their enzymatic degradation mechanism was evaluated. Initially, this resulted in the identification of an enzyme mechanism involving peeling off layer after layer of the starch granules in Paenibacillus granivorans (11).
In this paper, we focus on the bacterium Microbacterium aurum strain B8.A, which was originally isolated from a potato plant wastewater facility. This strain is able to degrade different types of starch granules as a carbon and energy source. We recently reported that it attacks and degrades starch by an alternative mechanism initially involving pore formation in wheat, tapioca, and potato starch granules (12).
This paper reports the characterization of an unusually large M. aurum B8.A α-amylase enzyme (MaAmyA) that is able to form pores in starch granules in vitro and that belongs to GH13 subfamily 32 (GH13_32). MaAmyA is about 2 times larger than the other GH13_32 members that have a single catalytic domain. Next to the catalytic domain, MaAmyA also contains two CBM25 domains, and it is the only known GH13_32 member with FNIII domains. Multiple deletion constructs of MaAmyA were expressed and characterized to study the roles of the different domains in the degradation of both soluble and granular starches. A direct relationship between the presence of CBM25 domains in MaAmyA and its ability to form pores in starch granules was observed.
MATERIALS AND METHODS
Bacterial strains, media, and plasmids.
Microbacterium aurum strain B8.A was isolated from a wastewater treatment plant of a potato starch processing factory. Isolation and growth conditions have been described previously (12). Escherichia coli Top10 and BL21(DE3) were cultivated at 37°C overnight in LB with orbital shaking (220 rpm). When required, ampicillin or kanamycin was added to a final concentration of 100 or 50 μg ml−1, respectively. The vector pZERO-1 (Invitrogen) was used to construct a genomic library of M. aurum B8.A in E. coli Top10. The pCR-XL-TOPO vector (Sigma) was used for sequencing of the MaAmyA-encoding gene; pET-15b (Novagen) was used as an expression vector for the amyA gene constructs in E. coli BL21(DE3).
Bioinformatic tools.
All BLAST searches were performed with NCBI BLASTP, using standard settings. To find all sequences related to the catalytic domain of MaAmyA, amino acids (aa) 57 to 504 were used as queries in BLAST searches using standard settings, but with the maximum number of target sequences increased to 1,000 (instead of the default of 100). Conserved domains were detected using both the NCBI conserved domain finder (13) with forced live search, without the low-complexity filter, using the Conserved Domain Database (CDD), and dbCAN (14), using standard settings. The signal sequence was predicted by use of SignalP4.1 (15), using standard settings. Catalytic AB regions of the GH13 domain sequences were extracted from the full-length enzymes based on dbCAN domain information. CBM25 domain sequences were extracted from the full-length protein sequences based on CDD information. Alignments of the extracted domain sequences were made with Mega6.0 (16), using the built-in muscle alignment with standard settings. Alignments were checked for the correct positions of conserved residues and manually tuned when needed. Phylogenetic trees were made with Mega6.0, using the maximum likelihood method, with gap/missing data treatment set on “partial deletion” instead of “full deletion.” Trees were visualized with Interactive Tree Of Life v2 (17). The protein domain annotations shown in the trees are based on combined data from CDD and dbCAN. Information about GH13 subfamilies was obtained from the CAZy database (1). An initial tree with a diverse selection of all GH13 subfamilies was constructed to select the closest related subfamilies for display in the final tree. The selection included multiple amylases for each of the 40 defined subfamilies, which varied in domain organization and origin. The final tree, based on catalytic domain region AB, included MaAmyA, all GH13_32 members present in the CAZy database, and a selection of the most closely related proteins based on the initial tree. The tree based on CBM25 domain sequences included the CBM25 domains from MaAmyA as well as 153 CBM25 domains listed in CAZy with a GenBank link.
Genomic DNA library and gene identification.
Chromosomal DNA isolated from M. aurum B8.A was partially digested with the Sau3A restriction enzyme (NEB), cloned into the pZERO-1 vector (Invitrogen), and transformed into E. coli Top10. Transformants were grown on LB agar plates containing red amylopectin to screen for α-amylase activity (18, 19). Colonies producing the largest halo sizes were isolated and the plasmid inserts sequenced.
Expression constructs for MaAmyA in E. coli.
To easily create C-terminal truncations of MaAmyA, pBAD-VV was constructed by insertion of a synthesized multiple-cloning site (containing removable N- and C-terminal His tags, a stop codon after the C-terminal His tag, and various restriction sites [NdeI, SpeI, NotI, PstI, and EcoO109I]) into pBAD/Myc-His B, using NcoI and EcoRI. The existing NdeI site in pBAD/Myc-His B was disrupted by site-directed mutagenesis. The full-length amyA gene was obtained by PCR with the forward primer GATGCATGATATCATATGTATCCGAAAGGAACAGGCGCA and the reverse primer GCTACTCTAGAGGATCCTTAACACCTTGGGGTGGGTGTGTGGACTA (restriction sites are underlined). The resulting PCR product was ligated into the pCR-XL-TOPO vector and then inserted into pBAD-VV by using NdeI and SpeI restriction, resulting in the pamyApre plasmid (see Fig. S1 in the supplemental material). The final constructs were made through single restriction and self-ligation of pamyApre and subsequently transferred to pET15b by using NdeI and EcoRI. The amyA construct was obtained through PstI restriction, the amyA2 construct through NotI restriction, and the amyA4 construct through EcoO109I restriction (see Fig. S1). For the amyA7 construct, an additional PstI site was inserted into the pamyApre construct through site-directed mutagenesis, and the construct was then cut with PstI, self-ligated, and transferred to pET15b (see Fig. S1). All products were confirmed by sequencing (GATC-Biotech). Final constructs all had an N-terminal His tag from the pET15b vector and a C-terminal His tag from pamyApre. The terminator present in pET15b was removed. All constructs were prepared with both N- and C-terminal His tags.
Protein expression.
Recombinant E. coli strains were grown as 500-ml cultures in 3-liter flasks with 100 μg ml−1 ampicillin and 50 μM (final concentration) isopropyl-β-d-thiogalactopyranoside (IPTG) for 6 h at 30°C (220 rpm) and then for 40 h at 18°C (220 rpm). Cells were collected by centrifugation at 4,250 × g for 20 min at 4°C (Thermo Lynx 4000 centrifuge). Pellets were resuspended in 50 mM Tris-HCl buffer, pH 6.8, containing 10 mM CaCl2. Protease inhibitors (Mini EDTA-free protease inhibitor; Roche) were added, and cells were broken by sonication (15 s at 10,000 Ω, with 30 s of cooling, performed 7 times). Cell debris and intact cells were removed by centrifugation at 15,000 × g for 20 min at 4°C. Resulting cell extracts were immediately used for His tag purification of recombinant proteins.
His tag purification.
The pH of E. coli cell lysates was adjusted to pH 7.5, and after being mixed with Ni-nitrilotriacetic acid (Ni-NTA; Sigma-Aldrich), the samples were left to bind for 1 h at 4°C. The column was washed 5 times with 2 column volumes of 50 mM Tris buffer, pH 8.0, containing 10 mM CaCl2, 250 mM NaCl, and 20 mM imidazole. The sample was then eluted 3 times with 2 column volumes of 50 mM Tris buffer, pH 8.0, containing 10 mM CaCl2, 250 mM NaCl, and 500 mM imidazole. Elution fractions were desalted using a 5-ml desalting column (Amersham Pharmacia Biotech) and stored at 4°C in standard assay buffer (50 mM Tris-HCl buffer, pH 6.8, containing 10 mM CaCl2).
M. aurum B8.A culture fluid production.
M. aurum B8.A was grown as a preculture overnight at 37°C and 220 rpm in LB medium. MMTV medium (12) containing 1% (wt/vol) granular potato starch (200 ml in a 1-liter flask) was inoculated with 200 μl preculture and grown for 48 h at 30°C and 220 rpm. The golden yellow cultures were harvested by centrifugation at 4,250 × g for 20 min at 4°C (Thermo Lynx 4000 centrifuge). The resulting cell-free culture fluid containing the extracellular enzymes was collected, 100 μg ml−1 ampicillin and 100 μg ml−1 kanamycin were added, and the culture fluid was stored at 4°C.
Standard assay buffer.
A 50 mM Tris-HCl buffer, pH 6.8, containing 10 mM CaCl2 was used as the standard assay buffer for enzymatic incubations. Unless indicated otherwise, all incubations were performed at 37°C.
Activity staining of SDS-PAGE gels.
SDS-PAGE analysis was used to determine protein masses. The locations of starch-acting enzymes in the gels were visualized on the basis of their activity. SDS-PAGE was performed as described by Laemmli (20), using precast THX gels (Bio-Rad). Use of THX gels prevented the appearance of additional bands for protein fragments which were observed previously (12) (see Discussion). After loading and running, gels were washed 3 times for 5 min each in MilliQ water to remove SDS and then incubated for 2 h in standard assay buffer containing 0.5% soluble potato starch (Sigma-Aldrich) (7). After incubation, the gel was stained with Lugol's iodine (2.5% I2, 5% KI) to visualize active protein bands. After imaging, gels were partly destained by washing in MilliQ water and then stained with Bio-Safe Coomassie stain (Bio-Rad) to visualize proteins. The Fermentas PageRuler prestained marker was included in each gel.
Activity assay and activity unit determination using CNPG3 as the substrate.
The CNPG3 compound, with a 2-chloro-4-nitrophenol (CNP) group coupled to maltotriose (G3), is a suitable substrate for determining α-amylase activity (21). The assay is based on the detection of the released CNP group from the CNPG3 substrate by α-amylase activity. The enzyme solution (10 μl) to be tested was prepared in a 96-well microtiter plate. Prewarmed substrate (100 μl 2 mM CNPG3) was added, and the reaction was monitored by reading the absorbance at 405 nm for 10 min in a microtiter plate reader (Spectramax Plus; Molecular Devices, Sunnyvale, CA) set at 37°C. A calibration curve was prepared using 0.03 to 0.15 mM CNP (Sigma) in assay buffer. The activity was calculated as the micromoles of CNP released per minute per microliter of enzyme solution. One unit was defined as the amount of enzyme needed to release 1 mmol CNP min−1 under standard assay conditions.
Enzymatic activity on soluble potato starch.
Hydrolysis of soluble potato starch was determined with the 2,4-dinitrosalicylic acid (DNS) method by measuring the increase of reducing ends over time (22). A 0.4% soluble potato starch (Sigma-Aldrich) solution in assay buffer was preheated at 37°C in a heating block (VWR). An enzyme preparation (2.6 U ml−1 in assay buffer) (CNPG3 assay) was then added at 0 min, in a total volume of 500 μl. Samples of 50 μl were taken at 0, 7, 14, 21, and 28 min. The reaction was stopped by addition of 50 μl DNS reagent (22) and immediate incubation at 100°C for 7 min in a heating block. Afterwards, the samples were left to cool down, and 400 μl of MilliQ water was added. The absorbance at 540 nm was measured in a spectrophotometer (Spectramax Plus). A standard curve of 833 to 8,333 μM glucose, as well as a blank sample to which no enzyme was added, was included.
Granular starch degradation.
The degradation of granular starch was followed by measuring the release of soluble carbohydrates from the granules, as determined by the anthrone method (23). Two different granular starches were used: milled potato starch (Avebe) and wheat starch (Sigma-Aldrich). Granules were sterilized by gamma irradiation (Synergy Health). The moisture content of the starches was determined with a dry-matter balance (Sartorius). For calculations of the percentage of starch degradation, the dry-matter content of the starches was considered to be 100% carbohydrate, since other components, such as protein, form a minor part of the contents (<2%). Ampicillin and kanamycin (100 μg ml−1 [final concentration]) were added to prevent bacterial growth. A His-tag-purified E. coli cell extract blank (empty vector) was included as a negative control.
The granular starch substrate (2.3 ml; 3.3% [wt/vol]) was prepared in a 15-ml tube including 300 U (CNPG3 assay) of the enzyme to be tested. Samples were incubated at 37°C in a rotating wheel and gently mixed. At 0, 1, 2, 3, 4, 5, 6, 14, 24, and 48 h, 200-μl samples were taken from the suspension and centrifuged for 20 min at 14,000 × g. Supernatants (50 μl) were transferred to clean glass test tubes. Samples containing more than 8 mg ml−1 carbohydrate were diluted with assay buffer before analysis. MilliQ water (200 μl) and 2 ml anthrone reagent (23) were added and mixed. Samples were incubated in a boiling water bath for 10 min and then cooled in a cold water bath, and the absorbance at 620 nm was measured in a spectrophotometer (Spectramax Plus). A calibration curve of 1.66 to 8.33 mg ml−1 glucose was included. Determination of activity on granular starch (Table 1) was based on the amount of hydrolysis during the first 6 h of incubation.
TABLE 1.
Relative activities of MaAmyA and truncated derivatives on 0.4% soluble potato starch and 3.3% granular wheat starcha
| Protein or sample | Relative activity on soluble starch (%) | Relative activity on wheat starch granules (%) |
|---|---|---|
| MaAmyA2 | 79 ± 3 | 9 ± 1 |
| MaAmyA4 | 99 ± 2 | 94 ± 2 |
| MaAmyA7 | 97 ± 4 | 83 ± 10 |
| MaAmyA | 100 ± 3 | 100 ± 10 |
| M. aurum B8.A culture fluid | 123 ± 5 | 122 ± 16 |
| Negative control | <2 | <5 |
Incubations with soluble starch were performed in triplicate, and those with granular starch were performed in duplicate. An empty vector construct was used as the negative control. Data are means ± standard deviations.
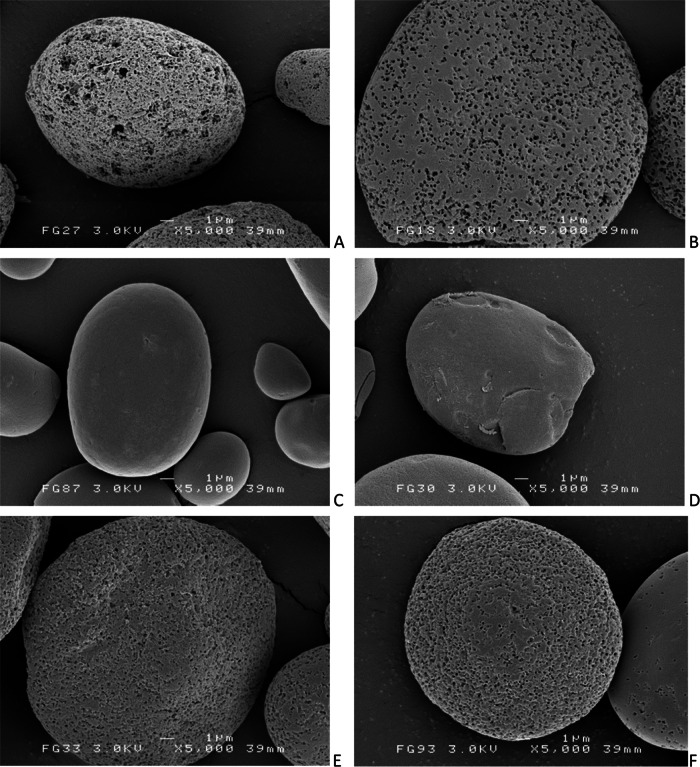
Pelleted starch granules were dried for 48 h at 48°C and stored in a desiccator for at least 1 day. The dried starch granules were transferred to scanning electron micrograph (SEM) stubs with double-sided adhesive tape and coated with gold. SEMs were taken using a JSM-6301F scanning electron microscope (JEOL, Groningen, The Netherlands) at the UMCG Microscopy and Imaging Center (UMIC). The accelerating voltage (3.0 kV) and the magnification are shown on the micrographs. For each wheat starch sample, three images of the same area were recorded, at magnifications of ×750, ×2,000, and ×5,000. For each potato starch sample, two images of the same area were recorded, at magnifications of ×2,500 and ×25,000.
Determination of pore surface sizes was performed using the digitally recorded SEM images with the lowest magnification. Pictures were loaded into Adobe Photoshop (version 6.0) in grayscale picture modus, zoomed to true pixels. Pores were selected with the “magic wand” selection tool, using the following settings: size, 1 pixel; tolerance, 40; contiguous; and no Anti-Alias. A total of 10 pores were randomly chosen, and the number of pixels selected by the “magic wand” was recorded for each pore. The averages and standard deviations were calculated. The scale bar was used to determine the conversion of pixel numbers to square micrometers.
Nucleotide sequence accession number.
The sequence for the amylase gene amyA isolated from M. aurum B8.A has been deposited in the GenBank database under accession no. KP901246.
RESULTS
Gene identification.
In previous work (12), we reported that culture fluid of M. aurum B8.A degraded granular starches initially by introducing pores. To identify the starch-acting enzyme(s) involved, an M. aurum B8.A DNA library of 70,000 clones was screened for α-amylase activity. The clone with the highest activity carried a vector with an insert of 11.6 kb. DNA sequencing revealed a complete open reading frame (ORF) of 4,227 nucleotides (nt), encoding a large, multidomain (putative) α-amylase enzyme of 1,409 aa (with a predicted mass of 148 kDa), including a signal sequence of 34 aa. The sequence of this unusually large α-amylase, designated MaAmyA, was confirmed when the full genome of M. aurum B8.A was sequenced recently (V. Valk , R. M. van der Kaaij, and L. Dijkhuizen, unpublished data).
The catalytic domain of MaAmyA shows similarity to those of the members of glycoside hydrolase family 13, subfamily 32 (GH13_32), which currently includes 109 members and is found mainly in bacteria (1). The catalytic domain contains the A, B, and C regions typical for the α-amylase superfamily (24), as well as all catalytic residues and regions generally conserved in α-amylases (25). The characterization of MaAmyA revealed that it was enzymatically active with soluble starch but also degraded starch granules by introducing pores (see below).
Domain organization.
A CDD (13) search revealed a total of 7 domains in MaAmyA: the N-terminal GH13 catalytic domain is followed by 1 FNIII domain, 2 CBM25 domains, and 3 highly similar FNIII domains (>95% identity) (Fig. 1). The C-terminal tail of MaAmyA (331 aa) does not match any known domain. A BLAST search nevertheless returned 26 hits for similar, 300-aa-long fragments (34 to 60% identity; 48 to 72% similarity). Of the 26 hits, 21 are part of predicted multidomain α-amylases. An additional four hits are genomically linked to a nearby ORF encoding an α-amylase, further indicating a relationship between this domain and α-amylases. None of these putative amylases belong to subfamily GH13_32; instead, 21 belong to GH13_28. The 300-aa fragment is usually located C terminal to the catalytic domain, either at the C terminus or between other domains. None of these multidomain α-amylases have been characterized biochemically.
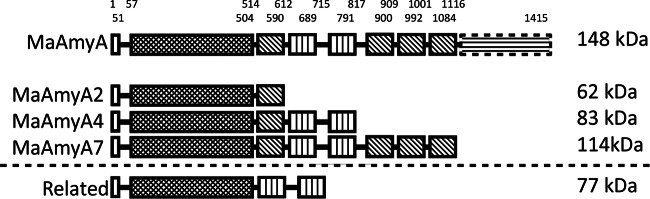
FIG 1.
Schematic representation of the domain organization of MaAmyA, the truncated derivatives MaAmyA2, MaAmyA4, and MaAmyA7, and the two related GH13_32 amylases of Bacillus sp. strain 195 and K. varians ATCC 21971. Domains are indicated as follows: white box, signal sequence; dark box, GH13 catalytic domain; hatched box, FNIII domain; vertically lined box, CBM25 domain; dashed box with horizontal lines, unknown domain. Predicted molecular masses are indicated.
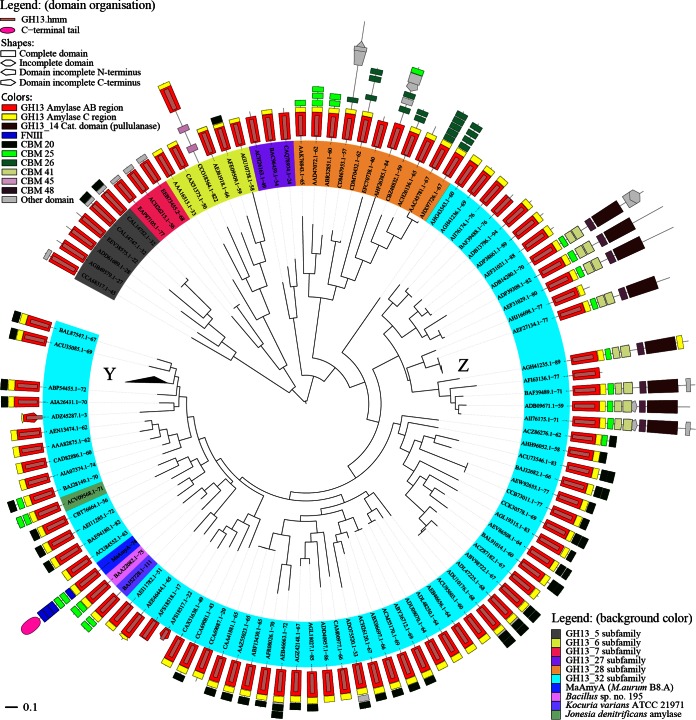
Phylogenetic tree of subfamily GH13_32.
In a BLAST search using full-length MaAmyA (26), the GH13_32 α-amylases of Bacillus sp. strain 195 (GenBank accession no. BAA22082) and Kocuria varians ATCC 21971 (GenBank accession no. BAJ52728) were the hits with the highest similarities. The latter enzymes consist of a catalytic domain and two CBM25 domains (Fig. 1). With a predicted mass of 77 kDa, they are much smaller than the 148-kDa MaAmyA protein. Both enzymes have been described as granular starch-degrading GH13_32 amylases, but their mechanism of action on starch granules has not been investigated (7, 8, 27–30). A phylogenetic tree based on the catalytic AB regions of MaAmyA and all other GH13_32 members (1) shows the diversity within this subfamily (Fig. 2). MaAmyA closely clusters with 5 other members, in a subgroup with a total of 52 GH13_32 members. Surprisingly, MaAmyA is the only protein in this subfamily with FNIII domains and the C-terminal tail of 300 aa. Interestingly, one blade of the tree contains GH13_32 enzymes with a second GH13_14 catalytic domain (pullulanase) and several binding domains. Only one of these large enzymes has been described in the literature (31).
FIG 2.
Phylogenetic tree of all 109 GH13_32 members in CAZy, Jonesia denitrificans amylase (GenBank accession no. ACV09568.1), and MaAmyA. The tree is based on the alignment of α-amylase catalytic domains (regions AB) obtained from DbCAN (aa 74 to 362 for MaAmyA). The domain organization was based on combined CDD and DbCAN data. For comparison, mixed selections of the related subfamilies GH13_5, GH13_6, GH13_7, GH13_27, and GH13_28 are also included. At Y and Z, 32 and 5 sequences similar to others in the respective branch were collapsed to improve the readability of the tree. The GH13 subfamilies are indicated by the background colors of the protein names. Domain colors and shapes are explained in the legend.
Members of GH13_32 are richly decorated with diverse starch binding domains (Fig. 2). Approximately 66% of all GH13_32 subfamily members contain one or two CBM20 domains. CBM25 domains are relatively rare among the enzymes with a single catalytic domain (present in 7 members of GH13_32, including MaAmyA). However, none of the other currently known GH13_32 enzymes in CAZy or in the NCBI database containing nonredundant protein sequences possesses FNIII domains or the 300-aa C-terminal tail (data not shown). This makes MaAmyA an exceptional member of GH13_32.
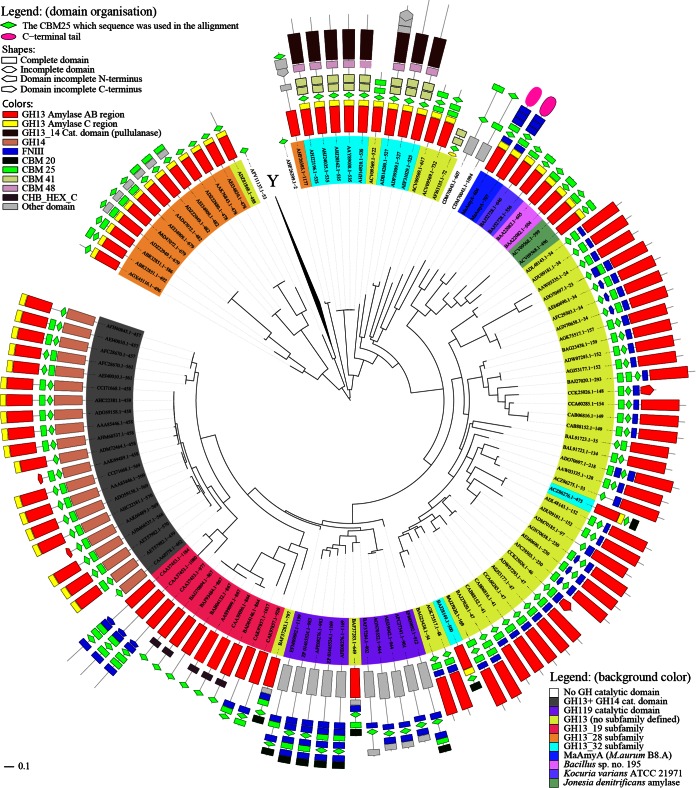
To obtain more insight into the evolutionary origin of the CBM25 domains in MaAmyA, a phylogenetic analysis was performed (Fig. 3). The phylogenetic tree shows clustering of the CBM25 domains present as tandems in MaAmyA, the two closely related GH13_32 enzymes of Bacillus and K. varians, and a third enzyme, from Jonesia denitrificans DSM 20603 (GenBank accession no. ACV09568.1), with a similar global domain organization. The latter enzyme has not been allocated to a specific GH13 subfamily in CAZy, but it clearly clusters with other GH13_32 amylases (Fig. 2).
FIG 3.
Phylogenetic tree of all 153 CBM25 domains in CAZy with sequences in GenBank and of those in MaAmyA. The tree is based on the CBM25 domains obtained from the CDD (aa 604 to 681 and 707 to 783 for MaAmyA); therefore, proteins with multiple CBM25 domains also have multiple entries in the tree. The domain organization was based on CDD data. At Y, a branch containing 34 sequences of single CBM25 domains was collapsed to improve the readability of the tree. The GH (sub)families are indicated by the background colors of the protein names; proteins without a catalytic domain are not colored. Domain colors and shapes are explained in the legend.
Expression and His tag purification of MaAmyA.
MaAmyA was successfully expressed in E. coli as a full-length protein and in truncated forms, MaAmyA2, MaAmyA4, and MaAmyA7 (Fig. 1), with His tags at both the N and C termini of all constructs. Studies with proteins carrying a single His tag showed that only the C-terminal His tags were functional in metal-affinity chromatography purification; the presence of the N-terminal His tag resulted in higher expression levels (data not shown). Removal of the predicted N-terminal signal sequence did not improve protein yields (data not shown), and expression levels were low for all forms. Different expression strains, vectors, and conditions, including pBAD vector-based constructs, different E. coli strains, and a Rhodococcus expression system, were tested as ways to improve expression, without noticeable improvement (data not shown). The amounts of protein obtained after purification of MaAmyA and the truncated derivatives were relatively low, and no protein bands were visible after SDS-PAGE analysis with silver staining. In-gel activity staining (after SDS-PAGE and washing steps) of the MaAmyA protein and the truncated versions by use of soluble potato starch confirmed the presence of all MaAmyA protein derivatives, with the expected masses (Fig. 4). The full-length MaAmyA protein of 148 kDa and the MaAmyA7 truncation of 114 kDa, lacking the C-terminal tail, both showed a minor additional activity band between 200 and 300 kDa, which may be the result of protein dimerization. MaAmyA also showed a band at the same height as that for the MaAmyA7 truncation, which may indicate early expression termination in E. coli and a (partial) loss of the C-terminal tail. A similar effect was seen with the MaAmyA4 truncation of 83 kDa, which showed a lower band of approximately 73 kDa, corresponding to the expected mass of MaAmyA4 without the second CBM25 domain. Storage of the enzymes for 5 months at 4°C did not affect these results, indicating that the observed fragments were not formed due to enzyme instability (data not shown). M. aurum culture fluid showed a single activity band at the same height as that for MaAmyA. No background E. coli amylase activity was detected (empty vector construct).
FIG 4.
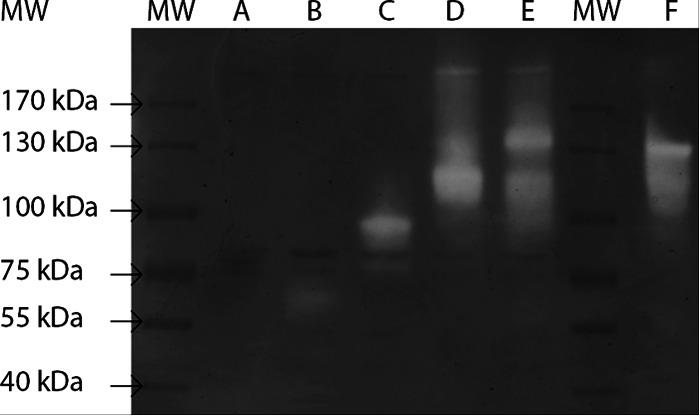
Activity staining of heterologously expressed, His-tag-purified M. aurum B8.A proteins on SDS-PAGE with soluble starch. Lanes: MW, size marker; A, empty vector; B, MaAmyA2 (62 kDa); C, MaAmyA4 (83 kDa); D, MaAmyA7 (114 kDa); E, MaAmyA (148 kDa); F, M. aurum B8.A culture fluid. Marker masses are indicated. The main activity bands correspond to the expected masses of the expressed proteins.
Enzyme activity.
The activities of the purified MaAmyA protein and the truncated enzyme derivatives were determined on both soluble potato starch and granular wheat and potato starch (Table 1). The MaAmyA activity was set at 100% for each substrate. The activity on granular potato starch was too low to be determined. MaAmyA4, MaAmyA7, and MaAmyA had similar activity ratios for both soluble and granular starch (Table 1). The truncated enzyme MaAmyA2 clearly acted on soluble starch (79%), but its activity on granular starch was strongly reduced compared to that of the full enzyme. In particular, the two CBM25 domains thus play a crucial role in the activity of MaAmyA on granular starch (Fig. 1).
Granular starch degradation.
To study the roles of the different MaAmyA domains in pore formation in granular starch, wheat starch granules were incubated with MaAmyA and the truncated MaAmyA2, MaAmyA4, and MaAmyA7 enzymes.
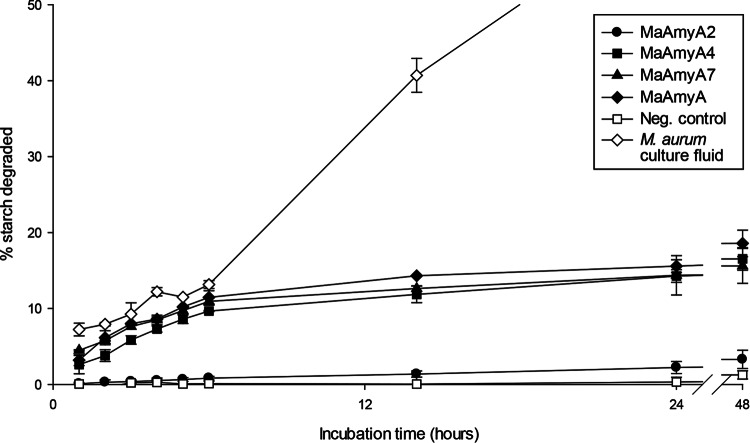
During the first 6 h of incubation, MaAmyA, MaAmyA7, and MaAmyA4, as well as the M. aurum B8.A culture fluid, showed similar rates of degradation of granular starch, whereas MaAmyA2 and the negative control showed much lower rates (Fig. 5). To exclude a possible effect of the FNIII domain present in MaAmyA2, the experiment was also performed with this domain removed, which gave similar results (data not shown). After longer incubation times, MaAmyA, MaAmyA7, and MaAmyA4 continued to show similar curves, but their degradation rates decreased over time, and their activities reached a threshold at about 20%. The M. aurum culture fluid was most active in degrading granular starches, reaching about 80% degradation of wheat starch after 72 h.
FIG 5.
Degradation of granular wheat starch over time by M. aurum B8.A culture fluid, MaAmyA, and truncated MaAmyA enzymes. To each reaction mixture, 300 U of enzyme (CNPG3 assay) was added. His-tag-purified empty vector E. coli extract was added as a negative control. The total carbohydrate concentration in the supernatant was determined by the anthrone method, and the percentage of granular starch degraded was calculated.
When a fresh MaAmyA4 enzyme sample (300 U; CNPG3 assay) was added at 24 h and 48 h to MaAmyA4 incubations with wheat starch granules, the degradation rate increased again, reaching 40% degradation after 72 h (data not shown). This indicated that the rate of degradation decreased at least partly due to a loss of enzyme activity within the first 48 h. The granular substrate itself was thus further degradable by MaAmyA4.
A selection of samples with similar degradation percentages was used for SEM imaging analysis (Fig. 5 and 6). After 48 h of incubation, the wheat starch granules treated with M. aurum B8.A culture fluid had lost their granular structure, and only debris remained (data not shown).
FIG 6.
SEM images of wheat starch granules following incubation with the different M. aurum B8.A enzyme samples (300 U; CNPG3 assay). Magnification, ×5,000. All images show granules that were incubated for 48 h, except the culture fluid samples, which were incubated for 6 h. M. aurum culture fluid (A), MaAmyA (B), the negative control (C), MaAmyA2 (D), MaAmyA4 (E), or MaAmyA7 (F).
The SEM images were used for analysis of pore sizes by using imaging software (Fig. 7). Incubations with MaAmyA clearly resulted in homogeneous pore formation (Fig. 6B). Wheat starch granules degraded with M. aurum B8.A culture fluid (Fig. 6A) additionally showed a set of large pores (LP) that were approximately 20 times larger and were absent in the MaAmyA-treated granules (Fig. 7). The truncated proteins MaAmyA7 and MaAmyA4 also introduced pores with homogeneous sizes, although these pores were approximately 3 times smaller than those formed by MaAmyA (Fig. 7). This indicated a possible role for the C-terminal protein tail in the formation of larger pores. MaAmyA2 (Fig. 6D) did not show pore formation, and the granules looked similar to those of the negative-control samples (Fig. 6C). Apparently, the presence of the CBM25 domains in MaAmyA4 is sufficient for pore formation in granular starches. The incubations and subsequent image analysis were also performed with granular potato starch, which gave similar results: pore formation in granules was observed for the truncated MaAmyA constructs that included the CBM25 domains but was absent in granules incubated with MaAmyA2 and the negative control (data not shown).
FIG 7.
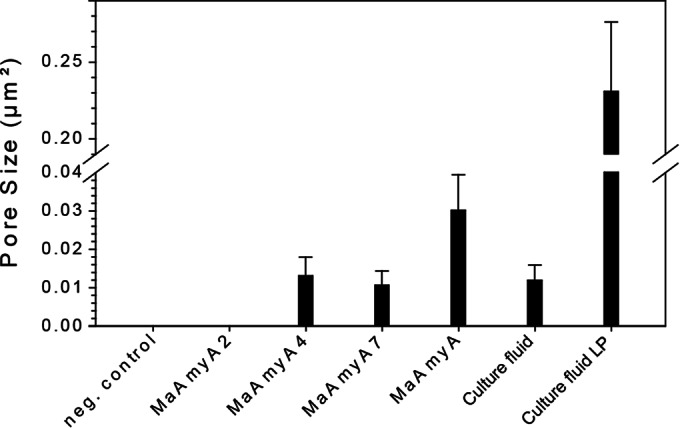
Pore sizes in wheat starch granules after incubation with different MaAmyA enzymes or M. aurum culture fluid. The images used to determine the pore sizes were lower-magnification images (×750) taken in the same region as the images shown in Fig. 6. All samples had been incubated for 48 h, except the culture fluid samples, which were incubated for 6 h. Culture fluid-treated samples also showed large pores (LP), which were included as a separate group.
DISCUSSION
This paper reports the characterization of MaAmyA, a multidomain α-amylase enzyme of Microbacterium aurum B8.A carrying four FNIII and two CBM25 domains plus a C-terminal domain of 300 aa. The MaAmyA enzyme degrades granular starches by initially introducing pores. Deletion of the additional domains did not affect the activity on soluble starch. The data clearly show that the CBM25 domains are essential for activity on granular starch and for pore formation. CBM25 domains have previously been shown to be involved in granular starch degradation (7, 8), but a role specifically in pore formation has not been reported before.
The MaAmyA enzyme thus allows M. aurum B8.A to degrade granular starches directly, without the need for starch gelatinization. This may be a more general situation in the natural environment. The occurrence of extracellular bacterial α-amylases degrading starch granules by peeling off layer after layer (11) or by introducing pores into starch granules (12, 32, 33; this paper) may be more widespread than currently described in the literature.
On the basis of its catalytic domain, MaAmyA belongs to the CAZy GH13_32 subfamily, but its length and the presence of FNIII domains and a C-terminal tail make it an exceptional member of this subfamily (Fig. 2). To study the roles of the different domains, MaAmyA and four C-terminally truncated proteins were successfully expressed in E. coli, purified, and characterized. Low levels of expression were achieved; therefore, the activity with the CNPG3 substrate was used to standardize the amount of protein used in further experiments. We assumed that the presence or absence of additional domains had no effect on the activity with CNPG3, in view of the small size of this substrate, and that a direct relationship exists between CNPG3 activity and the enzyme concentration. However, these assumptions can be made only for MaAmyA and its truncated versions, and it may not hold for M. aurum culture fluid, which may contain additional enzymes. M. aurum culture fluid showed a very high ratio of granular starch activity to CNPG3 activity (Fig. 5) compared to that for MaAmyA. It is therefore likely that the culture fluid contains one or more additional enzymes with low activity on CNPG3 but high activity on starch granules.
FNIII domains.
MaAmyA has four FNIII domains, which is an unusually large number compared to the case for other GH13 enzymes. α-Amylases generally lack FNIII domains (none are found in the other 109 GH13_32 members), and when these are present, there are rarely more than two. The three adjacent FNIII domains in MaAmyA are almost identical and may be the result of recent duplications, suggesting that they provide a strong evolutionary advantage. In eukaryotes, FNIII domains are widespread and well known for their protein-protein interactions (34). In prokaryotes, FNIII domains were initially found only in carbohydrate-acting enzymes (35), but more recently they have been identified in a wide variety of bacterial proteins (36, 37). Little is known about the function(s) of FNIII domains in amylases. Published reports about FNIII domains in carbohydrate-acting enzymes do not show a clear common function, with results pointing at possible roles in substrate binding or enzymatic activity (38–43). So far, there are no reports of FNIII domains in prokaryotes with a function in protein-protein interactions.
In the present study, no direct effect of the FNIII domains on substrate binding or enzyme activity was observed (Fig. 5 to 7; Table 1). Previously, FNIII domains were suggested to function as stable linkers between separate domains in bacterial cellobiohydrolase (42) and chitinase (41) enzymes. Also, the FNIII domains of MaAmyA may function as stable linkers between the catalytic domain, the CBM25 domains, and the C-terminal tail.
CBM25 domains.
Enzymes acting on insoluble carbohydrates, such as granular starches, often possess carbohydrate binding modules (CBMs) (5). When they function in starch binding, these are known as starch binding domains (SBDs). At present, SBDs have been identified in CBM families 20, 21, 25, 26, 34, 41, 45, 48, 53, 58, and 69 (1). The functions of CBM25 domains have been described previously, and their three-dimensional (3D) structures have been resolved (30, 44–46). It has also been demonstrated that the CBM25-26 tandem in the maltohexaose-forming amylase from Bacillus halodurans C-125 has a 50-fold stronger binding affinity for granular corn starch than each of the single domains (44). A similar effect, though much smaller, was observed for the CBM25 tandem found in the two homologs closely related to MaAmyA: in the halophilic K. varians α-amylase, the binding affinity of the CBM25 tandem was only 20% higher than that of a single CBM25 domain (28). Despite this small increase in binding affinity, the homologous enzyme in Bacillus sp. strain 195 showed 2- to 4-fold increases in the degradation rates of different granular starches compared to those with a single CBM25 domain attached to the C terminus (8). This work shows that CBM25 domains greatly enhance the ability of MaAmyA to degrade granular starch through the formation of pores in the granules (Fig. 5 to 7), while they have little effect on the activity on soluble substrates (Table 1). Phylogenetic analysis revealed that the CBM25 domains of MaAmyA are most closely related to each other, which suggests that a duplication of this CBM25 domain occurred. The same is true for the other GH13_32 amylases with two CBM25 domains (Fig. 2 and 3). The CBM25 domain tree (Fig. 3) suggests that an ancestral GH13_32 amylase acquired a single CBM25 domain, which duplicated only recently in the different enzymes, independently, resulting in a tandem of CBM25 domains. These duplication events may have improved the binding capability and the granular starch degradation rates of the enzymes (8, 28).
Another remarkable feature of MaAmyA is the presence of a single FNIII domain between the catalytic core and the 2 CBM25 domains, even though both the CBM25 domains (Fig. 3) and the catalytic domain (Fig. 2) cluster with two related GH13_32 amylases lacking FNIII domains. In view of the recent duplications of the CBM25 and FNIII domains, we conclude that M. aurum MaAmyA is able to easily acquire and include other domains and then duplicate them.
C-terminal tail.
The presence of three FNIII domains between the preceding CBM25 domains and the C-terminal tail, potentially functioning as stable linkers, suggests that the C-terminal tail of MaAmyA has a specific functional role. Homologs were found in 21 other large, multidomain α-amylases. The 300-aa C-terminal tail thus appears to represent a domain that is often part of a large, multidomain α-amylase. The pores formed by full-length MaAmyA on granular wheat starch were approximately 3 times larger than the pores formed by AmyA4 and AmaA7 lacking the tail (Fig. 6 and 7). At present, the mechanism by which the C-terminal protein tail has an effect on granule pore size is unknown; deletion had no apparent effect on enzyme activity (Table 1; Fig. 5). Further research is needed to elucidate the precise role of this domain.
Comparison of MaAmyA with M. aurum B8.A culture fluid.
M. aurum B8.A culture fluid degraded wheat starch up to 60% more than MaAmyA only did. Part of this difference can be explained by the instability of MaAmyA. Still, the sharp increase in the degradation rate (from 6 to 12 h) (Fig. 5) could not be achieved with MaAmyA alone. Furthermore, the granules degraded by culture fluid showed a different pore pattern from that for those degraded by MaAmyA, indicating the presence of additional enzymes in the culture fluid. The recently obtained genome sequence of M. aurum B8.A (V. Valk, R. M. van der Kaaij, and L. Dijkhuizen, unpublished data) revealed genes for 14 other GH13 family enzymes, one of them in close proximity to the MaAmyA gene. In previous work with M. aurum B8.A culture fluids, multiple activity bands were visible in gels with soluble starch (12). The current study shows that most of the sizes of these activity bands correspond to the sizes of the four truncated enzymes. Therefore, it is likely that the SDS-PAGE gel analysis procedure resulted in the loss of one or more domains from MaAmyA. When precast THX gels were used, these artifacts were strongly reduced (Fig. 4). However, the culture fluid sample showed a second weak activity band, of approximately 130 kDa, in the smear below the main band. This band does not correspond to any of the truncated MaAmyA masses. Also, in view of the difference between CNPG3 activity and granular starch activity of the culture fluid, it appears likely that M. aurum B8.A harbors one or more additional enzymes involved in granular starch degradation.
Supplementary Material
ACKNOWLEDGMENTS
This study was partly funded by the Top Institute of Food & Nutrition (project B1003) and the University of Groningen. The SEM studies were performed at the UMCG Microscopy and Imaging Center (UMIC), Groningen, The Netherlands, which is sponsored by NWO grants 40-00506-98-9021 and 175-010-2009-023.
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AEM.01029-15.
REFERENCES
- 1.Lombard V, Golaconda Ramulu H, Drula E, Coutinho PM, Henrissat B. 2014. The carbohydrate-active enzymes database (CAZy) in 2013. Nucleic Acids Res 42:D490–D495. doi: 10.1093/nar/gkt1178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Williamson G, Belshaw NJ, Self DJ, Noel TR, Ring SG, Cairns P, Morris VJ, Clark SA, Parker ML. 1992. Hydrolysis of A- and B-type crystalline polymorphs of starch by α-amylase, β-amylase and glucoamylase 1. Carbohydr Polym 18:179–187. doi: 10.1016/0144-8617(92)90062-U. [DOI] [Google Scholar]
- 3.Gallant D, Mercier C, Lbot A. 1972. Electron microscopy of starch granules modified by bacterial or α-amylase. Cereal Chem 49:354–365. [Google Scholar]
- 4.Cone JW, Wolters MGE. 1990. Some properties and degradability of isolated starch granules. Starch 42:298–301. doi: 10.1002/star.19900420804. [DOI] [Google Scholar]
- 5.Sun H, Zhao P, Ge X, Xia Y, Hao Z, Liu J, Peng M. 2010. Recent advances in microbial raw starch degrading enzymes. Appl Biochem Biotechnol 160:988–1003. doi: 10.1007/s12010-009-8579-y. [DOI] [PubMed] [Google Scholar]
- 6.Lei Y, Peng H, Wang Y, Liu Y, Han F, Xiao Y, Gao Y. 2012. Preferential and rapid degradation of raw rice starch by an α-amylase of glycoside hydrolase subfamily GH13_37. Appl Microbiol Biotechnol 94:1577–1584. doi: 10.1007/s00253-012-4114-0. [DOI] [PubMed] [Google Scholar]
- 7.Yamaguchi R, Tokunaga H, Ishibashi M, Arakawa T, Tokunaga M. 2011. Salt-dependent thermo-reversible α-amylase: cloning and characterization of halophilic α-amylase from moderately halophilic bacterium, Kocuria varians. Appl Microbiol Biotechnol 89:673–684. doi: 10.1007/s00253-010-2882-y. [DOI] [PubMed] [Google Scholar]
- 8.Sumitani J, Tottori T, Kawaguchi T, Arai M. 2000. New type of starch-binding domain: the direct repeat motif in the C-terminal region of Bacillus sp. no. 195 α-amylase contributes to starch binding and raw starch degrading. Biochem J 350:477–484. doi: 10.1042/0264-6021:3500477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Christiansen C, Hachem MA. 2009. The carbohydrate-binding module family 20—diversity, structure, and function. FEBS J 276:5006–5029. doi: 10.1111/j.1742-4658.2009.07221.x. [DOI] [PubMed] [Google Scholar]
- 10.Majzlová K, Janeček Š. 2014. Two structurally related starch-binding domain families CBM25 and CBM26. Biologia (Bratisl) 69:1087–1096. [Google Scholar]
- 11.Wijbenga DD-J, Beldman G, Veen A, Binnema D. 1991. Production of native-starch-degrading enzymes by a Bacillus firmus/lentus strain. Appl Microbiol Biotechnol 35:180–184. [Google Scholar]
- 12.Sarian F, van der Kaaij R, Kralj S, Wijbenga D-J, Binnema D, van der Maarel M, Dijkhuizen L. 2012. Enzymatic degradation of granular potato starch by Microbacterium aurum strain B8.A. Appl Microbiol Biotechnol 93:645–654. doi: 10.1007/s00253-011-3436-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Marchler-Bauer A, Lu S, Anderson JB, Chitsaz F, Derbyshire MK, DeWeese-Scott C, Fong JH, Geer LY, Geer RC, Gonzales NR, Gwadz M, Hurwitz DI, Jackson JD, Ke Z, Lanczycki CJ, Lu F, Marchler GH, Mullokandov M, Omelchenko MV, Robertson CL, Song JS, Thanki N, Yamashita RA, Zhang D, Zhang N, Zheng C, Bryant SH. 2011. CDD: a conserved domain database for the functional annotation of proteins. Nucleic Acids Res 39:D225–D229. doi: 10.1093/nar/gkq1189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yin Y, Mao X, Yang J, Chen X, Mao F, Xu Y. 2012. dbCAN: a web resource for automated carbohydrate-active enzyme annotation. Nucleic Acids Res 40:W445–W451. doi: 10.1093/nar/gks479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Petersen TN, Brunak S, von Heijne G, Nielsen H. 2011. SignalP 4.0: discriminating signal peptides from transmembrane regions. Nat Methods 8:785–786. doi: 10.1038/nmeth.1701. [DOI] [PubMed] [Google Scholar]
- 16.Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. 2013. MEGA6: Molecular Evolutionary Genetics Analysis version 6.0. Mol Biol Evol 30:2725–2729. doi: 10.1093/molbev/mst197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Letunic I, Bork P. 2011. Interactive Tree Of Life v2: online annotation and display of phylogenetic trees made easy. Nucleic Acids Res 39:W475–W478. doi: 10.1093/nar/gkr201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sax SM, Bridgwater AB, Moore JJ. 1971. Determination of serum and urine amylase with use of procion brilliant red M-2BS amylopectin. Clin Chem 17:311–315. [PubMed] [Google Scholar]
- 19.Groudieva T, Kambourova M, Yusef H, Royter M, Grote R, Trinks H, Antranikian G. 2004. Diversity and cold-active hydrolytic enzymes of culturable bacteria associated with Arctic sea ice, Spitzbergen. Extremophiles 8:475–488. doi: 10.1007/s00792-004-0409-0. [DOI] [PubMed] [Google Scholar]
- 20.Laemmli UK. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 21.Foo AY, Bais R. 1998. Amylase measurement with 2-chloro-4-nitrophenyl maltotrioside as substrate. Clin Chim Acta 272:137–147. doi: 10.1016/S0009-8981(98)00009-6. [DOI] [PubMed] [Google Scholar]
- 22.Miller GL. 1959. Use of dinitrosalicylic acid reagent for determination of reducing sugar. Anal Chem 31:426–428. doi: 10.1021/ac60147a030. [DOI] [Google Scholar]
- 23.Fales FW. 1951. The assimilation and degradation of carbohydrates by yeast cells. J Biol Chem 193:113–124. [PubMed] [Google Scholar]
- 24.MacGregor EA, Janeček Š, Svensson B. 2001. Relationship of sequence and structure to specificity in the α-amylase family of enzymes. Biochim Biophys Acta 1546:1–20. doi: 10.1016/S0167-4838(00)00302-2. [DOI] [PubMed] [Google Scholar]
- 25.Van der Maarel MJEC, van der Veen B, Uitdehaag JCM, Leemhuis H, Dijkhuizen L. 2002. Properties and applications of starch-converting enzymes of the α-amylase family. J Biotechnol 94:137–155. doi: 10.1016/S0168-1656(01)00407-2. [DOI] [PubMed] [Google Scholar]
- 26.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. 1990. Basic local alignment search tool. J Mol Biol 215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 27.Stam MR, Danchin EGJ, Rancurel C, Coutinho PM, Henrissat B. 2006. Dividing the large glycoside hydrolase family 13 into subfamilies: towards improved functional annotations of α-amylase-related proteins. Protein Eng Des Sel 19:555–562. doi: 10.1093/protein/gzl044. [DOI] [PubMed] [Google Scholar]
- 28.Yamaguchi R, Arakawa T, Tokunaga H, Ishibashi M, Tokunaga M. 2012. Distinct characteristics of single starch-binding domain SBD1 derived from tandem domains SBD1-SBD2 of halophilic Kocuria varians alpha-amylase. Protein J 31:250–258. doi: 10.1007/s10930-012-9400-2. [DOI] [PubMed] [Google Scholar]
- 29.Yamaguchi R, Arakawa T, Tokunaga H, Ishibashi M, Tokunaga M. 2012. Effects of salt and ligand concentrations on the thermal unfolding and refolding of halophilic starch-binding domain from Kocuria varians α-amylase. Protein Pept Lett 19:326–332. doi: 10.2174/092986612799363082. [DOI] [PubMed] [Google Scholar]
- 30.Yamaguchi R, Ishibashi M, Tokunaga H, Arakawa T, Tokunaga M. 2013. Structure of starch binding domains of halophilic alpha-amylase at low pH. Protein Pept Lett 20:755–760. doi: 10.2174/0929866511320070004. [DOI] [PubMed] [Google Scholar]
- 31.O'Connell Motherway M, Fitzgerald GF, Neirynck S, Ryan S, Steidler L, van Sinderen D. 2008. Characterization of ApuB, an extracellular type II amylopullulanase from Bifidobacterium breve UCC2003. Appl Environ Microbiol 74:6271–6279. doi: 10.1128/AEM.01169-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Roy JK, Borah A, Mahanta CL, Mukherjee AK. 2013. Cloning and overexpression of raw starch digesting α-amylase gene from Bacillus subtilis strain AS01a in Escherichia coli and application of the purified recombinant α-amylase (AmyBS-I) in raw starch digestion and baking industry. J Mol Catal B Enzym 97:118–129. doi: 10.1016/j.molcatb.2013.07.019. [DOI] [Google Scholar]
- 33.Giraud E, Champailler A, Raimbault M. 1994. Degradation of raw starch by a wild amylolytic strain of Lactobacillus plantarum. Appl Environ Microbiol 60:4319–4323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Watanabe T, Suzuki K, Oyanagi W, Ohnishi K, Tanaka H. 1990. Gene cloning of chitinase A1 from Bacillus circulans WL-12 revealed its evolutionary relationship to Serratia chitinase and to the type III homology units of fibronectin. J Biol Chem 265:15659–15665. [PubMed] [Google Scholar]
- 35.Hansen CK. 1992. Fibronectin type III-like sequences and a new domain type in prokaryotic depolymerases with insoluble substrates. FEBS Lett 305:91–96. doi: 10.1016/0014-5793(92)80871-D. [DOI] [PubMed] [Google Scholar]
- 36.Henderson B, Nair S, Pallas J, Williams MA. 2011. Fibronectin: a multidomain host adhesin targeted by bacterial fibronectin-binding proteins. FEMS Microbiol Rev 35:147–200. doi: 10.1111/j.1574-6976.2010.00243.x. [DOI] [PubMed] [Google Scholar]
- 37.Konkel ME, Larson CL, Flanagan RC. 2010. Campylobacter jejuni FlpA binds fibronectin and is required for maximal host cell adherence. J Bacteriol 192:68–76. doi: 10.1128/JB.00969-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lin F-P, Ma H-Y, Lin H-J, Liu S-M, Tzou W-S. 2011. Biochemical characterization of two truncated forms of amylopullulanase from Thermoanaerobacterium saccharolyticum NTOU1 to identify its enzymatically active region. Appl Biochem Biotechnol 165:1047–1056. doi: 10.1007/s12010-011-9319-7. [DOI] [PubMed] [Google Scholar]
- 39.Lin H-Y, Chuang H-H, Lin F-P. 2008. Biochemical characterization of engineered amylopullulanase from Thermoanaerobacter ethanolicus 39E—implicating the non-necessity of its 100 C-terminal amino acid residues. Extremophiles 12:641–650. doi: 10.1007/s00792-008-0168-4. [DOI] [PubMed] [Google Scholar]
- 40.Watanabe T, Ito Y, Yamada T, Hashimoto M, Sekine S, Tanaka H. 1994. The roles of the C-terminal domain and type III domains of chitinase A1 from Bacillus circulans WL-12 in chitin degradation. J Bacteriol 176:4465–4472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chuang H-H, Lin H-Y, Lin F-P. 2008. Biochemical characteristics of C-terminal region of recombinant chitinase from Bacillus licheniformis: implication of necessity for enzyme properties. FEBS J 275:2240–2254. doi: 10.1111/j.1742-4658.2008.06376.x. [DOI] [PubMed] [Google Scholar]
- 42.Kataeva IA, Seidel RD, Shah A, West LT, Li X-L, Ljungdahl LG. 2002. The fibronectin type 3-like repeat from the Clostridium thermocellum cellobiohydrolase CbhA promotes hydrolysis of cellulose by modifying its surface. Appl Environ Microbiol 68:4292–4300. doi: 10.1128/AEM.68.9.4292-4300.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kim DY, Han MK, Park D-S, Lee JS, Oh H-W, Shin D-H, Jeong T-S, Kim SU, Bae KS, Son K-H, Park H-Y. 2009. Novel GH10 xylanase, with a fibronectin type 3 domain, from Cellulosimicrobium sp. strain HY-13, a bacterium in the gut of Eisenia fetida. Appl Environ Microbiol 75:7275–7279. doi: 10.1128/AEM.01075-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Boraston AB, Healey M, Klassen J, Ficko-Blean E, Lammerts van Bueren A, Law V. 2006. A structural and functional analysis of α-glucan recognition by family 25 and 26 carbohydrate-binding modules reveals a conserved mode of starch recognition. J Biol Chem 281:587–598. doi: 10.1074/jbc.M509958200. [DOI] [PubMed] [Google Scholar]
- 45.Machovič M, Janeček Š. 2006. Starch-binding domains in the post-genome era. Cell Mol Life Sci 63:2710–2724. doi: 10.1007/s00018-006-6246-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Mehta D, Satyanarayana T. 2014. Domain C of thermostable α-amylase of Geobacillus thermoleovorans mediates raw starch adsorption. Appl Microbiol Biotechnol 98:4503–4519. doi: 10.1007/s00253-013-5459-8. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.