Abstract
Pristinamycin production in Streptomyces pristinaespiralis Pr11 is tightly regulated by an interplay between different repressors and activators. A γ-butyrolactone receptor gene (spbR), two TetR repressor genes (papR3 and papR5), three SARP (Streptomyces antibiotic regulatory protein) genes (papR1, papR2, and papR4), and a response regulator gene (papR6) are carried on the large 210-kb pristinamycin biosynthetic gene region of Streptomyces pristinaespiralis Pr11. A detailed investigation of all pristinamycin regulators revealed insight into a complex signaling cascade, which is responsible for the fine-tuned regulation of pristinamycin production in S. pristinaespiralis.
INTRODUCTION
Streptomycetes are filamentous, Gram-positive soil bacteria that are well known for their ability to produce varieties of bioactive secondary metabolites, including more than 70% of the commercially important antibiotics (1). The production of antibiotics is controlled by a vast array of physiological and nutritional conditions, communicated by extracellular and intracellular signaling molecules (2). The beginning of antibiotic biosynthesis is often coordinated with processes of morphological differentiation. The characteristic Streptomyces life cycle involves the formation of a feeding substrate mycelium and subsequent development of aerial hyphae, which finally septate into spores (3). Generally, antibiotic production begins as the culture enters stationary growth in liquid culture and coincidences with the onset of morphological differentiation in agar-grown cultures (reviewed in reference 4). In many Streptomyces strains, antibiotic production is regulated by low-molecular-weight compounds, called γ-butyrolactone autoregulators (GBLs) (5, 6). GBLs are small diffusible signaling molecules that are synthesized and gradually accumulated in a growth-dependent manner, at or near the middle of the exponential phase of Streptomyces growth, when they trigger the onset of antibiotic biosynthesis and/or morphological differentiation at nanomolar concentrations (7). Often, the GBL signal is transmitted via a hierarchical signaling cascade including pleiotropic and pathway-specific regulators, which all together control the antibiotic production: when the GBL concentration reaches a critical level, the signal is transmitted into the cells by binding to specific cytoplasmic receptor proteins, the GBL receptors (7). GBL receptors belong to the TetR family of transcriptional regulators (8). In the absence of the corresponding ligand, the GBL receptor binds to conserved AT-rich, partially palindromic sequences (9), the so-called “ARE” sequences (autoregulatory element) (10), within the promoter regions of its target genes and thereby represses the transcription of these genes. By binding of the GBLs to their receptors, the latter undergo a conformational change and dissociate from the target DNA, allowing expression of the derepressed genes (11). Predominantly, targets of GBL receptors are transcriptional regulatory genes, such as TetR and SARP (Streptomyces antibiotic regulatory protein) genes. The different families of regulatory proteins together control secondary metabolite production in a complex cascade. Such cascades can consist of several levels of regulation, which can have either a pleiotropic mode of action, by affecting a broad range of morphological and physiological processes, or a pathway-specific activity that affects only a single antibiotic biosynthetic pathway (4). TetR regulators prevalently have a repressive function on the transcription of their target genes and act on a higher level within the regulatory signaling cascade (12), whereas SARP-type regulators are a family of pathway-specific transcriptional activators that directly control the expression of the respective antibiotic biosynthetic gene cluster (13). The best-understood model for a targeted coordination of antibiotic biosynthesis is the A-factor regulatory cascade of Streptomyces griseus, which controls streptomycin biosynthesis (14). Further examples have been reported, e.g., for the regulation of the pristinamycin-related antibiotic virginiamycin of Streptomyces virginiae (15, 16), tylosin production in Streptomyces fradiae (17), lankacidin/lankamycin biosynthesis in Streptomyces rochei (18), or auricin biosynthesis in Streptomyces aureofaciens (19).
Streptomyces pristinaespiralis Pr11 produces the streptogramin-type antibiotic pristinamycin, which consists of two chemically diverse antibiotics: the cyclohexadepsipeptide pristinamycin I (PI) and the polyunsaturated macrolactone pristinamycin II (PII) (Fig. 1). PI and PII are coproduced in a 30:70 ratio (20). Each compound alone displays only a little bacteriostatic activity by binding to the 50S subunit of the bacterial ribosome and thereby blocking protein synthesis (21). In combination, the pristinamycins exhibit a strong synergistic antibacterial activity against a wide range of Gram-positive and some Gram-negative bacteria, including methicillin- and vancomycin-resistant strains (22). The genes that code for PI and PII biosynthesis, regulation, and resistance are organized together with a cryptic type II polyketide synthase (PKS) gene cluster (cpp) in a large supercluster (prs) (23). This gene region contains a variety of diverse regulatory genes, including a GBL receptor-like gene (spbR), two TetR-type regulatory genes (papR3 and papR5), and four SARP-type genes (papR1, papR2, papR4, and cpp1), as well as one response regulator gene (papR6) (20, 23). SpbR has already been characterized as an autoregulator receptor protein, which acts as a pleiotropic regulator and hereby influences growth, morphological differentiation and pristinamycin biosynthesis of S. pristinaespiralis (10). Production of pristinamycin is induced by nanomolar concentrations of A-factor-like quorum-sensing molecules, which are secreted by the strain 3 h prior to the initiation of antibiotic production (24). However, the chemical structure of the S. pristinaespiralis-specific GBL receptor ligand(s) is still not elucidated. papR1 encodes the SARP-type regulator PapR1 and itself is under transcriptional control of SpbR (10). As a papR1 deletion mutant still produces 30% pristinamycin, it was assumed that there is another SARP-type regulator (PapR2), which is involved in the control of pristinamycin biosynthesis (10). The function of the other papR genes has not been investigated before. The SARP gene cpp1, which is located within the cpp subcluster, is not involved in pristinamycin biosynthesis, as deletion of cpp1 had no influence on pristinamycin production (23). Altogether, the multitude of different regulatory genes within the pristinamycin supercluster suggests a complex network for the regulation of the antibiotic's synthesis.
FIG 1.
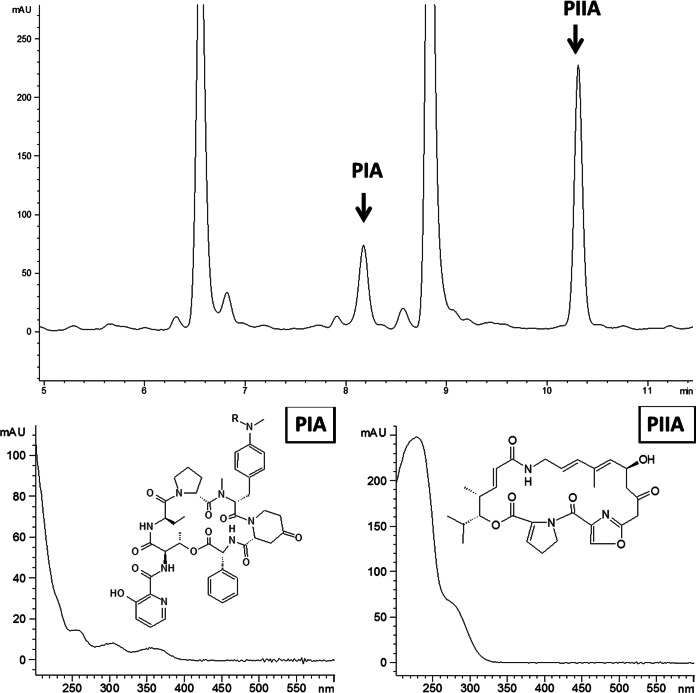
(Upper panel) HPLC spectrum of the culture extract from S. pristinaespiralis Pr11 wild-type strain (48-h sample). Wavelength monitoring was performed at 210 nm. PIA-specific (Rt = 8.2 min) and PIIA-specific (Rt = 10.3 min) peaks are indicated by arrows. (Lower panels) Corresponding UV-visible (UV-Vis) spectra of PIA (left) and PIIA (right) with their respective chemical structures; R (in PIA structure) represents CH3.
In this study, we describe the characterization of the regulatory function of PapR1 to PapR6 by mutational as well as overexpression, electrophoretic mobility shift assay (EMSA), and RT-PCR analyses. These regulators constitute a hierarchical signaling cascade governing the fine-tuned biosynthesis of pristinamycin in S. pristinaespiralis, which is summarized in a comprehensive regulatory model.
MATERIALS AND METHODS
Bacterial strains, plasmids, and cultivation conditions.
The bacterial strains and plasmids used in this study are listed in Table 1. For routine cloning strategies, Escherichia coli XL1-Blue (25) was used. E. coli strains were grown in Luria-Bertani (LB) medium at 37°C (26) supplemented with kanamycin, apramycin, or ampicillin (50, 100, or 150 μg/ml, respectively) when appropriate. S. pristinaespiralis Pr11 (Aventis Pharma) was used for the generation of gene insertion mutants and overexpression strains. Streptomyces lividans T7 (27) was used for protein expression experiments. Streptomyces strains were grown at 28°C on yeast malt (YM), R5, R2YE, or mannitol soy flour (MS) solid medium for isolation of spores (28). For cultivation and harvesting of genomic DNA, Streptomyces strains were grown in 100 ml of S-medium (28) in 500-ml Erlenmeyer flasks (with steel springs) on an orbital shaker (180 rpm) at 28°C. Kanamycin and apramycin (50 and 100 μg/ml, respectively) were added to liquid cultures when required. For production analysis, S. pristinaespiralis strains were cultivated as reported previously (23).
TABLE 1.
Bacterial strains and plasmids
| Bacterial strain or plasmid | Description | Source or reference |
|---|---|---|
| E. coli XL1 Blue | recA1 endA1 gyrA96 thi-1 hsdR17 supE44 relA1 lac [F′ proAB lac1q ZΔM15 Tn10 (Tetr)] | 25 |
| S. pristinaespiralis | ||
| Pr11 | Pristinamycin-producing strain/wild type; natural isolate of S. pristinaespiralis ATCC 25486 | Aventis Pharma |
| papR1::apr mutant | Gene interruption of papR1, aac(3)IV | This work |
| papR2::apr mutant | Gene interruption of papR2, aac(3)IV | This work |
| papR3::apr mutant | Gene interruption of papR3, aac(3)IV | This work |
| papR4::apr mutant | Gene interruption of papR4, aac(3)IV | This work |
| papR5::apr mutant | Gene interruption of papR5, aac(3)IV | This work |
| papR6::apr mutant | Gene interruption of papR6, aac(3)IV | This work |
| ΔpapR1′ mutant | papR1 deletion mutant of S. pristinaespiralis NRRL2958 | Aventis Pharma |
| ΔpapR1 ΔpapR4 mutant | Gene interruption of papR4, aac(3)IV in ΔpapR1′ mutant | This work |
| SPpGM190 | S. pristinaespiralis/pGM190 | This work |
| SPpapR1-OE | S. pristinaespiralis/pYM17 | This work |
| SPpapR2-OE | S. pristinaespiralis/pYM18 | This work |
| SPpapR3-OE | S. pristinaespiralis/pYM19 | This work |
| SPpapR4-OE | S. pristinaespiralis/pYM20 | This work |
| SPpapR5-OE | S. pristinaespiralis/pYM21 | This work |
| papR5::apr/pYM21 | Gene interruption of papR5, aac(3)IV, pYM21 | This work |
| ΔpapR2::apr/papR1-OE | papR2::apr/pYM18 | This work |
| ΔpapR2::apr/papR4-OE | papR2::apr/pYM20 | This work |
| S. lividans | ||
| T7 | tsr, T7 RNA polymerase gene | 27 |
| SLpGM190 | S lividans/pGM190 | This work |
| SLpapR1-OE | S lividans/pYM17 | This work |
| SLpapR2-OE | S lividans/pYM18 | This work |
| SLpapR3-OE | S lividans/pYM19 | This work |
| SLpapR4-OE | S lividans/pYM20 | This work |
| SLpapR5-OE | S lividans/pYM21 | This work |
| Plasmids | ||
| pDRIVE | lacZ′ complementation system, ampicillin and kanamycin resistance, multiple cloning site | Qiagen |
| pK18 | pUC derivative, aphII, lacZ′ complementation system | 29 |
| pEH13 | pUC21 derivative carrying the 1.8-kb apramycin resistance cassette (Aprr) | 30 |
| pRSETB | bla, pT7, pUC derivative | Invitrogen |
| pGM190 | Streptomyces-E. coli shuttle vector, tsr aphII, pSG5 derivative, tipA promoter shuttle vector | 31 |
| pUC18C | bla cya-T18 | 32 |
| pKT25 | aph cya-T25 | 32 |
| Plasmids for papR mutant construction | ||
| pYM11 | pk18 derivative, aphII Aprr lacZ′α papR1ab | This work |
| pYM12 | pk18 derivative, aphII Aprr lacZ′α papR2ab | This work |
| pYM13 | pk18 derivative, aphII Aprr lacZ′α papR3ab | This work |
| pYM14 | pk18 derivative, aphII Aprr lacZ′α papR4ab | This work |
| pYM15 | pk18 derivative, aphII Aprr lacZ′α papR5ab | This work |
| pYM16 | pk18 derivative, aphII Aprr lacZ′α papR6ab | This work |
| Plasmids for papR overexpression | ||
| pYM17 | pGM190 derivative, PtipA tsr aphII his papR1 | This work |
| pYM18 | pGM190 derivative, PtipA tsr aphII his papR2 | This work |
| pYM19 | pGM190 derivative, PtipA tsr aphII his papR3 | This work |
| pYM20 | pGM190 derivative, PtipA tsr aphII his papR4 | This work |
| pYM21 | pGM190 derivative, PtipA tsr aphII his papR5 | This work |
Molecular cloning.
The basic procedures for DNA manipulation were performed as described by Sambrook et al. (28) for E. coli and for Streptomyces. The primers used for PCR were obtained from MWG Biotech AG (Ebersberg, Germany) and Integrated DNA Technologies (IDT; Coralville, IA, USA) and are listed in Table 2.
TABLE 2.
Primers used in this study
| Primer | Primer sequence (5′–3′)a | Temp (°C) |
|---|---|---|
| For papR mutant construction | ||
| papR1m1 | GCGTAGAGGTGGTCGGTGAT | 59 |
| papR1m2 | ATGATATCACGGCCGGCTGACCGG | 65 |
| papR1m3 | ATGATATCATGGCGTTTCGTCTTC | 55 |
| papR1m4 | TACGCCGCCGACCACGGCAT | 67 |
| papR2m1 | GTATCTGCCCGCTCCT | 59 |
| papR2m2 | ATGATATCACTAGGCCCTGCCCCG | 66 |
| papR2m3 | TAGATATCCATTGTCTTCCTCGCA | 55 |
| papR2m4 | CGGGCACTTCTACTTCC | 58 |
| papR3m1 | GGCGAGCGCGTTGTG | 52 |
| papR3m2 | ATGATATCCACGCCGCCTGAACCC | 65 |
| papR3m3 | ATGATATCGGTACGCCCGTGCGGATCG | 71 |
| papR3m4 | GGGAGACGGCGTGGACATCG | 62 |
| papR4m1 | ATCTATAGCAGCGCTCGATCCTGATG | 61 |
| papR4m2 | ATGATATCCAGCGCTCGATCCTGATG | 63 |
| papR4m3 | ATGATATCCAGGGCCGCCGAGATCAG | 67 |
| papR4m4 | CCGCCGTCCGTCAGTGAG | 57 |
| papR5m1 | CGATCTGGCCTGCATCCCGGTTTC | 68 |
| papR5m2 | ATGATATCTAGCAGACCACCCGCCCTGTTTC | 68 |
| papR5m3 | ATGATATCACGCTCCTGCTTGAC | 55 |
| papR5m4 | ATGATCTCGACCCTGAAC | 44 |
| papR6m1 | AACAGGGTGTGCAGCGCGGG | 72 |
| papR6m2 | ATGATATCTGCGCTGACGCCCGCA | 70 |
| papR6m3 | ATGATATCTTCCGTCATGACCCGC | 61 |
| papR6m4 | TGCGCGTCGACCCCGAGACC | 73 |
| For papR overexpression | ||
| papR1ex1 | ATGGATCCAGACATCGACATACTCGGCGC | 70 |
| papR1ex2 | ATAAGCTTTCAGCCGGCCGTGGCGGCG | 73 |
| papR2ex1 | TGGATCCAAGTTCCGCATTCTCGGTCCGGTG | 75 |
| papR2ex2 | TAAGCTTAGTGGCCCGAGGCCGGGTTG | 75 |
| papR3ex1 | ATGGATCCCGGCACGCGGCACGCGATCC | 81 |
| papR3ex2 | TAAGCTTTCAGGCGGCGTGGGCGGGGC | 77 |
| papR4ex1 | ATGGATCCGACATCGATGTGCTGGGGGAG | 73 |
| papR4ex2 | TAAGCTTTCAGCCGGCCCGGCTCAGCCG | 76 |
| papR5ex1 | ATGGATCCTGCGGTCCGCCGTCCGTCAGTG | 78 |
| papR5ex2 | TAAGCTTCTATTGGGGGGTGGGGGTGC | 68 |
| For amplification of promoter regions | ||
| propapR1fw | AGCCAGTGGCGATAAGAACGACGGCTGCCTGACCGC | 77 |
| propapR1rev | AGCCAGTGGCGATAAGTATGTCGATGTCCATGGCGT | 73 |
| propapR2fw | AGCCAGTGGCGATAAGATCGGCCGGCCCGGGCGCGA | 81 |
| propapR2rev | AGCCAGTGGCGATAAGGCAGGGCGTTGGTCGCGTTC | 77 |
| propapR3fw | AGCCAGTGGCGATAAGAGCGGCTGAGCCGGGCCGGC | 81 |
| propapR3rev | AGCCAGTGGCGATAAGTTGCCCCTGGTGACCCCTGG | 77 |
| propapR4fw | AGCCAGTGGCGATAAGACCCCCTTACGCCCCGTTTT | 75 |
| propapR4rev | AGCCAGTGGCGATAAGAGCTCCCCCAGCACATCGAT | 75 |
| propapR5fw | AGCCAGTGGCGATAAGTCTGTCCCGGTTCCCGGCCC | 78 |
| propapR5rev | AGCCAGTGGCGATAAGATCGCAGCGATCCCCTCACT | 75 |
| propapR6fw | AGCCAGTGGCGATAAGAATGTCTCCATGATCGTCCC | 73 |
| propapR6rev | AGCCAGTGGCGATAAGACGGAGGCCTTGCCGCCGTG | 78 |
| prospbRfw1 | AGCCAGTGGCGATAAGAGCGTTCGTGCCGGCTCCGG | 78 |
| prospbRrev1 | AGCCAGTGGCGATAAGTCCCTTTCAAGCGGAATGAT | 72 |
| prospbRfw2 | AGCCAGTGGCGATAAGATCATTCCGCTTGAAAGGGA | 72 |
| prospbRrev2 | AGCCAGTGGCGATAAGTCGGCCGCCGCCACCAAAAT | 76 |
| prosnaBfw | AGCCAGTGGCGATAAGAGCCGCCTGCTCGTCCGTGG | 78 |
| prosnaBrev | AGCCAGTGGCGATAAGTGTCGAGGGTGGCGACGAGG | 77 |
| prosnaDfw | AGCCAGTGGCGATAAGAAGGGCGGGGAACGGCTGCC | 78 |
| prosnaDrev | AGCCAGTGGCGATAAGTCCGGCGGTCGCGGGGTTCT | 78 |
| prosnaE3fw | AGCCAGTGGCGATAAGCGCGGCGGCCTCGGCCCGGTC77 | |
| prosnaE3rev | AGCCAGTGGCGATAAGTCCTGCGCGCGGCAGCGGAG | 76 |
| prosnaFfw | AGCCAGTGGCGATAAGAACTGGGCCTGGAAC | 60 |
| prosnaFrev | AGCCAGTGGCGATAAGCGCGGTGGAAACATC | 60 |
| procpp1-snaRfw | AGCCAGTGGCGATAAGGGTTCCTCCGTA | 74 |
| procpp1-snaRrev | AGCCAGTGGCGATAAGGGCGCCCGAAAGTA | 77 |
| propipA-snbAfw | AGCCAGTGGCGATAAGAACGCATCCGTCCAGCATCG | 75 |
| propipA-snbArev | AGCCAGTGGCGATAAGTCGCGCCGGCCCAGGACCCA | 79 |
| prosnbCfw | AGCCAGTGGCGATAAGGCAGAACCTGCTGAACAAG | 60 |
| prosnbCrev | AGCCAGTGGCGATAAGTGTTGAAGACGGGACTGTG | 60 |
| propapB-papCfw | AGCCAGTGGCGATAAGACGTCCAGCCAGGTCACCGC | 77 |
| propapB-papCrev | AGCCAGTGGCGATAAGAGGGCGGCGTCCGCGGCGTC | 81 |
| prosnbRfw | AGCCAGTGGCGATAAGGGATCCCCTCGCCCAGGGCC | 79 |
| prosnbRrev | AGCCAGTGGCGATAAGGTTGTCGAGCAGGACGACGA | 75 |
| Cy5 | AGCCAGTGGCGATAAG | 60 |
| For amplification of cDNA (RT-PCR) | ||
| papR1int1 | ACCGTGCAGACCTACATCC | 62 |
| papR1int2 | TCAGTTCGGCGAGCAGTTC | 62 |
| papR2int1 | GCGGGAACGTTTCTACGACCTG | 66 |
| papR2int2 | TTCGAGGGAGAGGTGCTCGATG | 66 |
| papR3int1 | ACCTCGGTGATCCAGGTCTG | 65 |
| papR3int2 | TCCTCGCGGGCGCCCAGATG | 71 |
| papR4int1 | AACTGGCCGTGCAGGTTCTC | 65 |
| papR4int2 | AAGGACGTGCTGGTGACCTC | 65 |
| papR5int1 | AGAAGCCGGTGATCTTGC | 60 |
| papR5int2 | GCACTTCCACTTCGAGAAC | 60 |
| papR6int1 | GTGTCATAGGGGAGGACGAG | 60 |
| papR6int2 | CTGGGAGGTGGTGGAGTG | 60 |
| spbRint1 | AGATCCTGCGTCTGCTCCATCC | 66 |
| spbRint2 | GGTGTTCGACGAGGTCGGTTAC | 66 |
| snaBint1 | ATCACCGCCCCGCTCCCGGC | 73 |
| snaBint2 | ATCTTGGCGTGCGGTGCCTG | 67 |
| snaE3int1 | CTGTCCTACCTGCTGGACCT | 60 |
| snaE3int2 | GAGGGGTGCAGGTAGAGGTT | 60 |
| snbAint1 | GGAGGTGAAGGTGACGTGTT | 60 |
| snbAint2 | AGTGTCTATCTGGCGGTGCT | 60 |
| snbCint1 | CCTACGTCCAGTGGCTGAC | 60 |
| snbCint2 | CCTCCCTCGTAGGGGTAGC | 60 |
| hrdBfw | CCGGTCAAGGACTACCTGAA | 60 |
| hrdBrv | GTGGCGTACGTGGAGAACTT | 60 |
Restriction sites are underlined; Cy5 homologous base pairs are in bold.
Targeted disruption of pristinamycin regulatory genes.
For the construction of gene insertion mutants ∼1- to 1.5-kb fragments up- and downstream of the regulatory genes papR1, papR2, papR3, papR4, papR5, and papR6 were amplified by PCR using S. pristinaespiralis Pr11 genomic DNA and the primers listed in Table 2. For amplification of the downstream fragments, primer pairs labeled m1/m2, which added an artificial EcoRV restriction site to the 5′ end, were used, resulting in amplificates papR1a (1.1 kb), papR2a (1.0 kb), papR3a (1.0 kb), papR4a (1.2 kb), papR5a (1.3 kb), and papR6a (1.5 kb), respectively. For the amplification of the upstream fragments, primer pairs labeled m3/m4, which added an artificial EcoRV restriction site to the 3′ end, were used, resulting in amplificates papR1b (1.2 kb), papR2b (1.3 kb), papR3b (1.2 kb), papR4b (1.1 kb), papR5b (1.5 kb), and papR6b (1.5 kb), respectively. All fragments were subcloned into the EcoRV-restricted E. coli vector pDRIVE, resulting in the constructs pDRIVE/papR1a and -b; pDRIVE/papR2a and -b; pDRIVE/papR3a and -b; pDRIVE/papR4a and -b; pDRIVE/papR5a and -b; and pDRIVE/papR6a and -b, respectively. All “a” fragments were isolated from the respective pDRIVE/papRa constructs as XbaI/BamHI fragments and ligated into the XbaI/BamHI-restricted E. coli vector pK18 (29), resulting in the constructs pK18/papR1a, pK18/papR2a, pK18/papR3a, pK18/papR4a, pK18/papR5a, and pK18/papR6a, respectively. The respective “b” fragments were excised as HindIII/EcoRV fragments from the respective pDRIVE/papRb constructs and were ligated into the HindIII/EcoRV site of the respective pK18/papRa plasmids, which resulted in the constructs pK18/papR1′, pK18/papR2′, pK18/papR3′, pK18/papR4′, pK18/papR5′, and pK18/papR6′, respectively. A 1.5-kb aac(3)IV cassette (Aprr) was isolated as an EcoRV/SmaI fragment from pEH13 (30) and was cloned into the EcoRV restriction site between fragments “a” and “b” of the respective pK18/papR′ derivatives, resulting in the mutational constructs pYM11, pYM12, pYM13, pYM14, pYM15, and pYM16, in which the regulatory genes were inactivated by the insertion of the Aprr cassette. The pYM11 to -16 plasmids were each transferred into S. pristinaespiralis Pr11 by protoplast transformation, followed by selection for apramycin-resistant and kanamycin-sensitive transformants, resulting in the papR1::apr, papR2::apr, papR3::apr, papR4::apr, papR5::apr, and papR6::apr mutants. Transformants were confirmed by PCR and Southern hybridization (data not shown). The ΔpapR1 ΔpapR4 double mutant was constructed on the basis of an S. pristinaespiralis NRRL2958 ΔpapR1′ deletion mutant, kindly provided by Sanofi-Aventis Pharma GmbH. This mutant shows the same pristinamycin production profile as the Pr11 papR1::apr mutant (data not shown). Plasmid pYM14 was introduced into the ΔpapR1′ mutant. Transformants were selected for apramycin-resistant and kanamycin-sensitive clones, resulting in the ΔpapR1 ΔpapR4 strain.
Construction of papR overexpression strains.
For overexpression experiments, the genes papR1, papR2, papR3, papR4, and papR5 were amplified by PCR using S. pristinaespiralis Pr11 genomic DNA and the primers listed in Table 2. The primers were designed in such a way that a BamHI restriction site at the 5′ end and a HindIII restriction site at the 3′ end were added to the papR coding sequence. This resulted in the fragments papR1ex (1.0 kb), papR2ex (1.0 kb), papR3ex (0.8 kb), papR4ex (0.9 kb), and papR5ex (0.7 kb), respectively. All “ex” fragments were restricted by BamHI/HindIII and subcloned into the BglII/HindIII restriction site of the E. coli expression plasmid pRSETB. With this cloning procedure, each papR gene is fused to a His tag-encoding sequence, which is localized behind the IPTG (isopropyl-β-d-thiogalactopyranoside)-inducible T7 promoter. From the resulting constructs pRSETB/papR1, pRSETB/papR2, pRSETB/papR3, pRSETB/papR4, and pRSETB/papR5, respectively, the hispapR fragments were excised with NdeI/HindIII and ligated into the NdeI/HindIII-restricted E. coli/Streptomyces shuttle plasmid pGM190 (31), where they are under the control of the thiostrepton-inducible PtipA promoter. The targeting plasmids pYM17, pYM18, pYM19, pYM20, and pYM21, respectively, were each transferred to S. pristinaespiralis Pr11 for pristinamycin production analyses, resulting in the strains SPpapR1-OE, SPpapR2-OE, SPpapR3-OE, SPpapR4-OE, and SPpapR5-OE, respectively, and into S. lividans T7 for protein purification experiments, resulting in the strains SLpapR1-OE, SLpapR2-OE, SLpapR3-OE, SLpapR4-OE, and SLpapR5-OE, respectively.
Fermentation and pristinamycin production analysis.
For pristinamycin production analyses, the S. pristinaespiralis Pr11 wild-type strain, the papR mutant strains, and the SPpapR-OE strains (see above) were cultivated as described previously (23). Strains harboring the pGM190 plasmid were induced for gene expression by adding 25 μg/ml thiostrepton.
Expression and purification of the His-tagged pristinamycin regulators.
For protein purification, the SLpapR-OE strains were grown in 100 ml of yeast extract-malt extract (YEME) medium with 50 μg/ml kanamycin in 500-ml Erlenmeyer flasks (with steel springs) on an orbital shaker (180 rpm) at 28°C. After 2 to 3 days, 17 ml of the YEME preculture was inoculated into 200 ml of fresh YEME medium with 25 μg/ml thiostrepton for gene expression and then incubated for a further 3 to 4 days in 1-liter Erlenmeyer flasks with steel springs on an orbital shaker (180 rpm) at 28°C. Cells were harvested by centrifugation at 5,000 rpm for 10 min at 4°C. Cell pellets were resuspended in ice-cold lysis buffer (50 mM NaH2PO4, 300 mM NaCl, 10 mM imidazole, pH 8), and then cells were disrupted by a French press (10,000 lb/in2; American Instruments) with two consecutive passages. Cell debris and the insoluble protein fraction were harvested by centrifugation at 10,000 rpm for 20 min and 4°C. The His-tagged proteins were purified from the soluble crude extract by metal chelate affinity chromatography using Ni-nitrilotriacetic acid resins according to the standard protocol by Qiagen. The collected fractions were analyzed by standard SDS-polyacrylamide gel electrophoresis (PAGE) in 12% gels (26) and Western blot analysis.
EMSAs.
DNA fragments (100 to 250 bp) of the upstream regions of the various genes were amplified by PCR from genomic DNA of S. pristinaespiralis Pr11 with the primers listed in Table 2. For Cy5 labeling, the DNA amplificates were used as the templates in a second PCR approach together with the Cy5 primer (Table 2). DNA binding reactions were performed at room temperature in 20 μl of 100 mM HEPES (pH 7.6), 5 mM EDTA, 50 mM (NH4)2SO4, 5 mM dithiothreitol, 1% (wt/vol) Tween 20, 150 mM KCl, 5 mM MgCl2, 2 ng of Cy5-labeled DNA, and 8 μl of crude extract. After a 10-min incubation, 2 μl loading buffer (0.25× Tris-borate EDTA [TBE] buffer, 60% glycerol) was added, and the samples were loaded onto a 2% agarose gel. To verify the specificity of the regulator-DNA binding, an excess of unlabeled, specific, or nonspecific DNA was added to the EMSA mixture, separately. DNA bands were visualized by fluorescence imaging using a Typhoon Trio Variable Mode Imager (GE Healthcare).
Transcriptional analysis by RT-PCR experiments.
S. pristinaespiralis Pr11 strains were grown in 100 ml of preculture medium (see above) and incubated at 28°C in 500-ml Erlenmeyer flasks (with steel springs) on an orbital shaker (180 rpm). Samples were taken after 24, 36, 48, 72, and 96 h. Cell disruption was carried out with glass beads (150 to 212 μm; Sigma) using a Precellys Homogenizer (6,500 rpm, once for 20 to 30 s; Peqlab). Total RNA was extracted from S. pristinaespiralis and used as the template for RT-PCR in accordance with the instructions from the RNeasy minikit (Qiagen). DNA that bound to the RNA purification column was digested with RNase-free DNase (Fermentas) once on the column for 30 min at 24°C and a second time for 1.5 h at 24°C before elution of the RNA. RNA concentrations and quality were checked using a NanoDrop ND-1000 spectrophotometer (Thermo Fisher Scientific). cDNA from 3 mg RNA was generated with random nonamer primers (Sigma), reverse transcriptase, and cofactors (Fermentas). For PCRs, primers that amplify cDNA of 200 to 300 bp from internal gene sequences were used (Table 2). PCR conditions were 98°C for 5 min, followed by 35 cycles of 95°C for 30 s, 55°C for 30 s and 72°C for 40 s, and finally 72°C for 5 min. As a positive control, cDNA was amplified from the major vegetative sigma factor (hrdB) transcript, which is expressed constitutively. To exclude DNA contamination, negative controls were carried out by using total RNA as a template for each RT-PCR.
BTH assays.
Bacterial two-hybrid (BTH) complementation assays were carried out with the nonreverting adenylate cyclase-deficient (cya) E. coli strain BTH101 (32). For the construction of recombinant plasmids used for BTH assays, the genes of interest were amplified by PCR using Taq polymerase (Qiagen) and the oligonucleotide pairs described in Table 2. PCR products were cloned as XbaI/BamHI- or XbaI/KpnI-digested DNA fragments in frame with either the T18 or the T25 fragment of the catalytic domain of the Bordetella pertussis adenylate cyclase (cyaA) into the equally restricted pUT18C and pKT25 vectors (BACTH System kit; Euromedex). For BTH complementation assays, recombinant pKT25 and pUT18C plasmids carrying the genes of interest (Table 1) were used in various combinations to cotransform E. coli BTH101/pRARE2 cells. The transformants were plated onto M63-X-Gal-IPTG (where X-Gal is 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside) or MacConkey medium with ampicillin (75 mg/ml), kanamycin (25 mg/ml), and chloramphenicol (25 mg/ml) supplemented with lactose. E. coli BTH101/pRARE2 with the empty pUT18C and pKT25 vectors was used as a negative control. E. coli BTH101/pRARE2 with the plasmids pUT18-zip and pKT25-zip was used as a positive control. Protein-protein interaction is observed when cells stain blue on M63-X-Gal-IPTG and red on MacConkey agar.
Phylogenetic analysis.
SARP sequences from different antibiotic producers were identified with the BLAST software and aligned using the Clustal Omega software. A phylogenetic analysis was performed with the aligned sequence data using the Clustal W2 program.
RESULTS
In silico analysis of the regulatory gene products.
In the course of sequence analysis of the pristinamycin gene region, several regulatory genes were identified. The respective genes were designated spbR, papR1, papR2, papR3, papR4, papR5, and papR6 (23). The spbR gene is localized at the right border of the pristinamycin gene region (see Fig. S1A in the supplemental material) and encodes the autoregulator receptor SpbR, with high similarity to TylP and BarA of S. fradiae and S. virginiae, respectively (Table 3). papR1, papR2, and papR4 code for SARP-type regulators belonging to the “small SARP” group (2). The deduced proteins PapR1 and PapR4 both show high similarity to the SARP TylS of S. fradiae, whereas PapR2 is similar to VmsS of S. virginiae and TylT of S. fradiae (Table 3; see also Fig. S2A in the supplemental material). Two TetR-type regulatory genes are present in the pristinamycin gene region, papR3 and papR5, both of which are part of a “regulatory island,” consisting of papR3-papR4-papR5, in the proximity of the cluster border (see Fig. S1A in the supplemental material). The deduced PapR3 protein is similar to BarB of S. virginiae, whereas the predicted PapR5 protein is more similar to TylQ of S. fradiae (Table 3). PapR3 and PapR5, as well as the autoregulator receptor SpbR, consist of the typical TetR-like protein structure (see Fig. S2B in the supplemental material). The gene papR6 encodes a predicted response regulator of bacterial two-component transduction systems that shows similarity to VmsT of S. virginiae (Table 3). However, no cognate sensor kinase gene is present in the pristinamycin gene cluster. Thus, PapR6 represents an orphan response regulator, like Aur1P in Streptomyces aureofaciens (activator of auricin biosynthesis) (33) or JadR1 in Streptomyces venezuelae (activator of jadomycin biosynthesis) (34).
TABLE 3.
Characteristics of genes, predicted functions, and protein matches from other Streptomyces species
| Gene | Size (bp) | No. of amino acids | pI | Predicted function | ID/SMa (%) | Match | Origin reference | GenBank accession no. |
|---|---|---|---|---|---|---|---|---|
| spbR | 1,050 | 228 | 5.72 | GBL receptor | 63/77 | TylP | S. fradiae | AAD40801 |
| 46/64 | SrrA | S. rochei | BAC76540 | |||||
| 46/64 | BarA | S. virginiae | BAA06981 | |||||
| papR1 | 857 | 285 | 10.58 | SARP-type regulator | 72/81 | TylS | S. fradiae | AAD40804 |
| 71/83 | SrrW | S. rochei | BAC76513 | |||||
| papR2 | 995 | 331 | 7.03 | SARP-type regulator | 63/72 | VmsS | S. virginiae | BAF50715 |
| 60/69 | TylT | S. fradiae | AAD40805 | |||||
| 44/55 | RedD | S. lividans TK24 | EFD66212 | |||||
| papR3 | 824 | 275 | 9.92 | TetR-type regulator | 39/55 | BarB | S. virginiae | BAA23612 |
| 53/65 | SrrC | S. rochei | BAC76532 | |||||
| papR4 | 902 | 300 | 10.05 | SARP-type regulator | 73/83 | TylS | S. fradiae | AAD40804 |
| 72/78 | SrrY | S. rochei | BAC76533 | |||||
| papR5 | 662 | 220 | 6.08 | TetR-type regulator | 57/68 | TylQ | S. fradiae | AAD40803 |
| 56/69 | SrrB | S. rochei | BAC76537 | |||||
| papR6 | 749 | 249 | 9.74 | Response regulator | 52/65 | VmsT | S. virginiae | BAF50712 |
ID/SM, % identity/similarity of amino acid sequences.
Regulatory influence on S. pristinaespiralis morphological development.
To analyze the functions of papR1, papR2, papR3, papR4, papR5, and papR6, the genes were inactivated by insertion of an apramycin resistance cassette (Aprr) and the morphologies of the respective mutants, i.e., papR1::apr, papR2::apr, papR3::apr, papR4::apr, papR5::apr, and papR6::apr mutants, were analyzed on MS solid medium. An S. pristinaespiralis NRRL2958 spbR apramycin insertion mutant (spbR25) has been constructed before and was described to have severe defects in growth and morphological differentiation and not produce any pristinamycin (10). The papR1::apr, papR2::apr, papR3::apr, papR4::apr, and papR6::apr mutants grew as the wild-type strain, which forms the aerial mycelium after ∼3 days and starts producing gray spores after around 7 days. The papR5::apr mutant failed to form any aerial mycelium or spores on solid medium (Fig. 2). This phenotype was observed on different media, such as LB, R5, R2YE, YM, or MS agar (see Fig. S3 in the supplemental material), and was restored after complementation with the native papR5 gene (see Fig. S4A in the supplemental material). Altogether, these data show that papR1, papR2, papR3, papR4, and papR6 do not exert any effect on morphological development, whereas papR5 strongly influences differentiation in S. pristinaespiralis.
FIG 2.
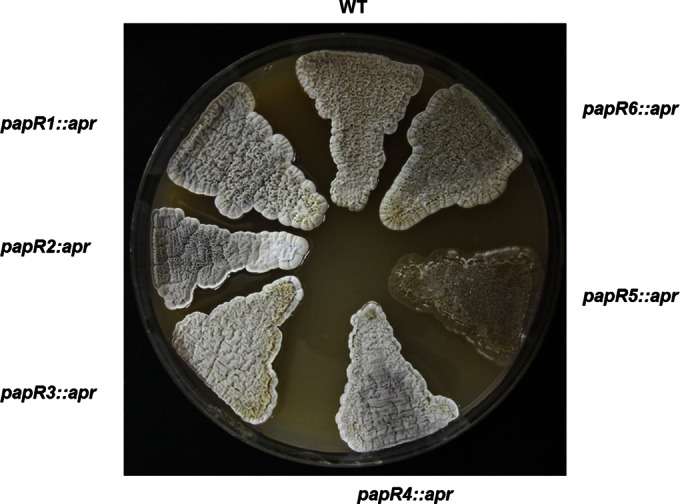
Morphological phenotype of S. pristinaespiralis wild-type strain and the papR1::apr, papR2::apr, papR3::apr, papR4::apr, papR5::apr, and papR6::apr mutants on MS solid agar after 5 days.
Pristinamycin production of papR mutants and overexpression strains.
To investigate the regulatory functions of papR1, papR2, papR3, papR4, papR5, and papR6, the pristinamycin production of the respective regulatory mutants (papR1::apr, papR2::apr, papR3::apr, papR4::apr, papR5::apr, and papR6::apr mutants) and that of overexpression strains were analyzed by HPLC and compared to the antibiotic production of the S. pristinaespiralis wild-type strain. For the overexpression experiments, additional copies of papR1, papR2, papR3, papR4, and papR5 were each cloned into the medium-copy-number plasmid pGM190 under the control of the thiostrepton-inducible promoter PtipA and then transferred into S. pristinaespiralis, resulting in the overexpression strains SPpapR1-OE, SPpapR2-OE, SPpapR3-OE, SPpapR4-OE, and SPpapR5-OE, respectively. S. pristinaespiralis with the empty pGM190 plasmid served as a control. All strains were cultivated in pristinamycin production medium. Samples were taken at different time points (24 h, 48 h, 72 h, and 96 h) and then analyzed for pristinamycin production by high-performance liquid chromatography (HPLC).
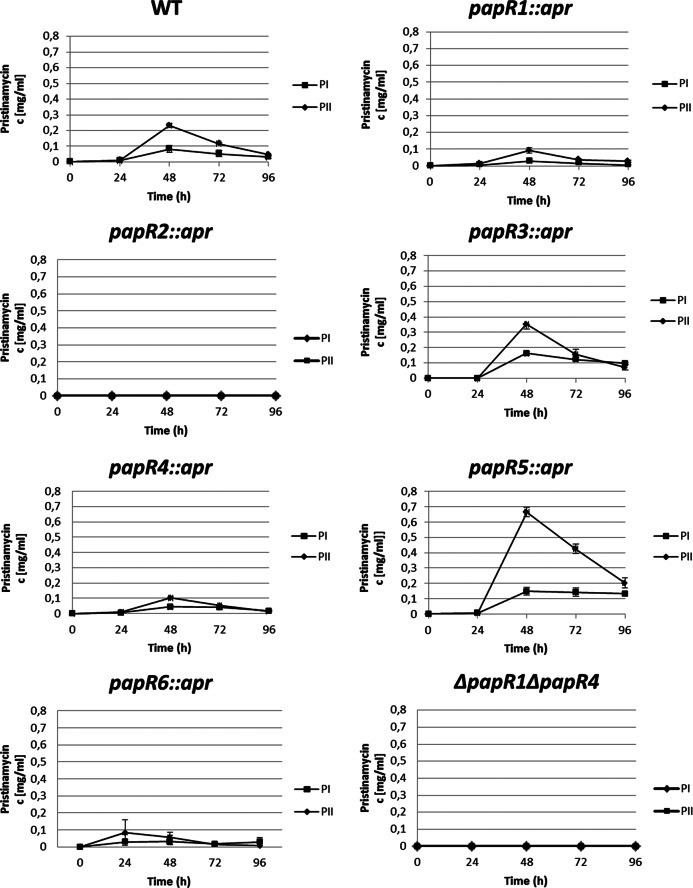
The papR1::apr and papR4::apr mutant strains showed similar production profiles and produced less pristinamycin than the wild-type strain. The production performance was around 30% to 50% that of the wild-type strain. The deletion of papR2 led to a complete loss of antibiotic production (Fig. 3). In contrast, the overexpression of the SARP genes papR1 and papR2 each led to an earlier and greater (increased by up to 100%) pristinamycin production, whereas the papR4 overexpression did not have any significant effect on the starting time or amount of production (see Fig. S5 in the supplemental material). Altogether, these data show that PapR1, PapR2, and PapR4 act as activators of pristinamycin biosynthesis. Thereby, papR2 is essential for pristinamycin biosynthesis, whereas papR1 and papR4 are not.
FIG 3.
Pristinamycin production of the S. pristinaespiralis wild-type strain (WT) and the papR mutant papR1::apr, papR2::apr, papR3::apr, papR4::apr, papR5::apr, papR6::apr, and ΔpapR1 ΔpapR4 mutant strains.
The deletion of the TetR-like papR genes led to an increase of pristinamycin production: the papR3::apr mutant produced up to 150% and the papR5::apr mutant even 300% more pristinamycin than did the wild-type strain (Fig. 3). Here, deletion of papR5 had a more dramatic effect on PII than on PI production. In contrast, the overexpression of papR3 resulted in a decreased pristinamycin production, which was around 40% that of the wild-type strain (see Fig. S5 in the supplemental material). The overexpression of papR5 in S. pristinaespiralis caused a complete loss of antibiotic production. This was observed when papR5 was overexpressed in S. pristinaespiralis Pr11 (strain SPpapR5-OE) (see Fig. S5 in the supplemental material) but also in the pYM21-complemented papR5::apr mutant strain (see Fig. S4B in the supplemental material). These data show that both PapR3 and PapR5 are repressors of pristinamycin biosynthesis but papR5 has a more dramatic effect on production than papR3.
The deletion of the response regulator gene papR6 led to a decrease of pristinamycin production, which was around 40% that of the wild-type level (Fig. 3), whereas papR6 overexpression resulted in a slightly increased pristinamycin production (data not shown), suggesting that PapR6 is an activator of pristinamycin biosynthesis.
The pristinamycin production profiles of all mutant strains were restored, at least partially, to wild-type levels after complementation with the respective papR genes using the pGM190 derivatives described above (data not shown).
Identification of TetR- and SARP-binding sequences within the pristinamycin gene cluster.
To identify binding sequences in the upstream regions of the pristinamycin gene cluster that are potential targets of the diverse PapR regulators, we performed bioinformatical analyses using the software PatScan (35). TetR-like regulators are known to bind to conserved partially palindromic sequence motifs, the so-called ARE sequences (10). Three ARE motifs have already been identified in the upstream region of spbR (PspbR-ARE1 and PspbR-ARE2) and papR1 (at bp −19 to −42; PpapR1) (10). Our analysis identified two further ARE motifs, upstream of papR4 and papR5, as well as another less conserved one in front of papR2 (Fig. 4A). No ARE sequences were found in the upstream region of papR3 and papR6 or any of the pristinamycin biosynthetic genes. Thus, except for the promoter region of the regulatory genes papR3 and papR6, all other pristinamycin regulatory promoters contain ARE binding motifs and therefore are suggested to be regulated by TetR-like proteins.
FIG 4.

(A) ARE sequences and their respective conformities in front of the genes spbR, papR1, papR2, papR4, and papR5. The sequences were compared to the consensus “IUPAC string” mentioned by Folcher et al. The S. pristinaespiralis-specific ARE consensus sequence (prist) is shown in the lowest row. The underlined sequences represent half sites of the palindrome, supposed to be bound by the TetR-like monomers. (B) SARP binding sequences of pristinamycin-related genes. Heptameric repeats, which are supposed to be bound by two SARP monomers and RNA polymerase, are shown in bold. The S. pristinaespiralis-specific SARP consensus sequence is shown in the lowest row.
In the same manner, the cluster was screened for putative SARP binding sites. SARP regulators are known to bind to specific direct heptameric sequence motifs, of which the 3′ repeat is located 8 bp from the −10 promoter element of the target gene (for example, ActII-ORF4, 5′-TCGAGCC/G-3′). It is suggested that two SARP monomers cooperatively bind to a direct repeat at the same face of the DNA, whereas the RNA polymerase is recruited to the opposite face of the DNA (36). Putative SARP binding motifs were detected upstream of the PI-specific genes snbC and snbA-pipA and the PII-specific genes snaB, snaD, snaE3, and snaR-cpp1, where cpp1 is not related to pristinamycin biosynthesis, as well in front of the regulatory gene papR1 and the predicted ABC transporter gene snbR (Fig. 4B). Thus, nearly all predicted pristinamycin operons (37) contain a SARP binding motif in their cognate promoter regions, except for the intergenic regions in front of snaF and papB-papC.
PapR2 is the central SARP activator of pristinamycin biosynthesis.
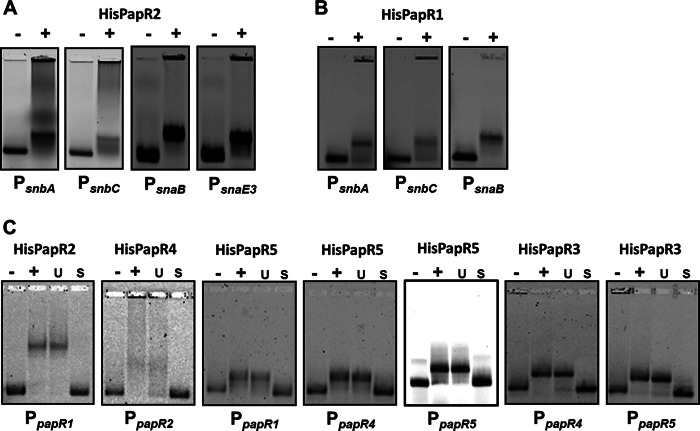
From the pristinamycin production analysis reported above, we know that PapR2 is essential for pristinamycin biosynthesis. To identify the targets of the PapR2 regulation, we performed EMSAs with the purified HisPapR2 protein and the SARP-motif containing intergenic regions of all regulatory and structural genes (see above). In these analyses, HisPapR2 specifically bound to the upstream regions of the SARP regulator gene papR1 (Fig. 5C), the PI structural genes snbA-pipA and snbC, and the PII structural genes snaB and snaE3, which all contain conserved SARP-binding motifs (Fig. 4A). To confirm an effect on transcription, reverse transcription-PCR (RT-PCR) experiments using RNA isolated from the papR2::apr mutant and the wild-type strain were performed. The isolated RNA was used as the template in RT-PCRs with primers annealing to internal parts of the various genes. The transcriptional analysis showed that there is no or almost no transcription of papR1 and the pristinamycin structural genes snbA, snbC, and snaE3 in the papR2::apr mutant (Fig. 6A and B). Furthermore, transcripts of the PII structural genes snaB and snaD or the transporter gene snbR were absent in the papR2::apr mutant samples, whereas all these genes were transcribed in the wild-type strain (data not shown). Thus, even if not all SARP motif-containing samples shifted in our EMSAs (snaD and snbR), we have supporting evidence from RT-PCR data that these genes are also regulated by PapR2. Altogether, these data indicate that PapR2 is a superior SARP-type regulator that directly activates the transcription of the SARP regulatory gene papR1 as well as the transcription of the pristinamycin structural and putative resistance gene(s).
FIG 5.
(A) EMSAs with His PapR2 and Cy5-labeled promoter regions of the pristinamycin structural genes snbA-pipA, snbC, snaB, and snaE3. −, negative control without protein; +, addition of purified His-tagged PapR protein. (B) EMSAs with His PapR1 and Cy5-labeled promoter regions of the pristinamycin structural genes snbA-pipA, snbC, and snaB. (C) EMSAs with the His PapR2, His PapR4, His PapR5, and His PapR3 and Cy5-labeled promoter regions of different papR genes. The specificity of the reaction was checked by the addition of 500-fold specific (S) and unspecific (U) unlabeled DNA.
FIG 6.
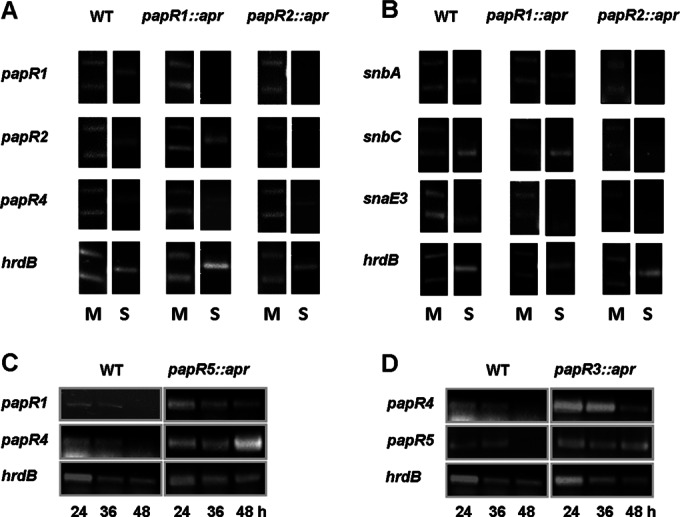
Transcriptional analysis of the S. pristinaespiralis wild-type and different papR::apr mutant strains. (A and B) A 5-μl volume of the GeneRuler 1-kb ladder (Fermentas) was loaded onto each gel as an internal control. The first picture in a row shows the 250-bp and 500-bp bands (lower and upper bands, respectively) of the 1-kb ladder (M); the second picture in a row shows the RT-PCR sample (S). RT-PCR analysis of genes papR1, papR2, and papR4 (A) and of genes snbA, snbC, and snaE3 (B) in the WT, papR1::apr, and papR2::apr strains at 36 h. (C and D) RT-PCR analysis of the genes papR1 and papR4 in the WT and papR5::apr strains (C) and of the genes papR4 and papR5 in the WT and papR3::apr strains (D). hrdB was used as a control.
PapR1 and PapR4 are accessory regulators.
The production analyses of the papR1::apr and papR4::apr mutant showed that the two SARP regulators are dispensable for pristinamycin biosynthesis, as they still produced low levels of the antibiotics (Fig. 3). In EMSAs, HisPapR1 specifically bound to the promoter regions of the PI structural genes snbA-pipA and snbC and the PII structural genes snaB and snaE3 (Fig. 5B), which indicates that PapR1 directly targets the pristinamycin biosynthesis genes. HisPapR4 bound to the promoter region of papR2, suggesting that PapR4 has an activating function in papR2 gene transcription (Fig. 5C). Due to the fact that PapR1 and PapR4 are nonessential for pristinamycin biosynthesis, we concluded that both SARPs play only a subordinate role in the activation of pristinamycin biosynthesis.
To examine if papR4 (similar to papR1) is a target of PapR2, we analyzed the papR4 transcription profile in the papR2::apr mutant by RT-PCR. Here, we found that papR4 is transcribed in papR2::apr, which indicates that the papR4 expression is not under the control of PapR2 (Fig. 6A). To investigate if the loss of pristinamycin production in papR2::apr is due to a failure of inducing another SARP gene's expression, we overexpressed papR1 and papR4 each in papR2::apr, using plasmids pYM17 and pYM20, respectively, and measured pristinamycin production by HPLC. Here, we did not detect any pristinamycin production (data not shown), which showed that PapR1 and PapR4 cannot compensate for the loss of PapR2 activity and alone cannot directly activate pristinamycin biosynthesis. To investigate the significance of PapR1 and PapR4 for antibiotic production in more detail, a ΔpapR1 ΔpapR4 double mutant was constructed and analyzed for pristinamycin production by HPLC. Interestingly, the ΔpapR1 ΔpapR4 double mutant did not produce any pristinamycin at all (Fig. 3), which shows that even if PapR1 and PapR4 each alone is dispensable for pristinamycin production, the existence of both regulators together is a prerequisite for production. To analyze if PapR2 alone, in sufficient amounts, can activate pristinamycin biosynthesis, we overexpressed PapR2 in the ΔpapR1 ΔpapR4 double mutant. Here we found that production was restored to a low level (see Fig. S6 in the supplemental material), which was around 10% that of the wild-type production level. These data show that PapR2 can activate pristinamycin production without its helping partners and underline its important role in pristinamycin regulation. In summary, we conclude that each of the SARPs has a specific function in pristinamycin regulation, which cannot be compensated by another SARP. Furthermore, we suggest that PapR1 and PapR4 both contribute to the activating function of PapR2 as assisting regulatory proteins.
PapR5 is an important pleiotropic regulator that represses papR1 and papR4 transcription and itself is under the control of PapR3 repression.
The papR5::apr mutant is the S. pristinaespiralis strain that produces the largest amounts of pristinamycin (up to 300% more than the wild-type strain) and also shows a morphological defect on solid agar plates. Thus, the TetR-like protein PapR5 is a pleiotropic regulator of morphological development and pristinamycin biosynthesis (see above). To investigate the regulatory role of PapR5 in pristinamycin biosynthesis, EMSAs were performed with the HisPapR5 protein and the promoter regions of the pristinamycin regulatory and structural genes. Here it was found that HisPapR5 binds to the promoter region of its own gene as well as to the promoters of papR1 and papR4 (Fig. 5C). No shifted band was observed with HisPapR5 and any promoter regions of the pristinamycin structural genes. RT-PCR experiments supported these data, as papR1 and papR4 transcription was prolonged in the papR5::apr mutant compared to the wild-type strain (Fig. 6C). Thus, we conclude that PapR5 acts as an autoregulatory protein, which regulates the transcription of the two SARP genes papR1 and papR4. Interestingly, we observed in BTH analysis that PapR5 interacts on the protein level with the response regulator PapR6, which suggests that the two proteins may represent a new type of two-component system in bacteria (Fig. 7). However, this interaction will need to be verified and investigated in depth by prospective biochemical analysis.
FIG 7.

BTH assays. E. coli BTH101/pRARE2 with the empty pUT18C and pKT25 vector was used as a negative control (NC). E. coli BTH101/pRARE2 with the plasmids pUT18-zip and pKT25-zip was used as a positive control (PC). Protein-protein interactions between the different pristinamycin regulators were studied. The first item (before the slash [/]) represents the pUT18C derivative; the second item (after the slash) represents the pKT25 derivative; e.g., R1/R1 is E. coli BTH101/pRARE2 harboring the pUT18/papR1 and pKT25/papR1 plasmid; R1/R2 is E. coli BTH101/pRARE2 harboring the pUT18/papR1 and pKT25/papR2 plasmid, etc. Abbreviations: R1, PapR1; R2, PapR2; R3, PapR3; R4, PapR4; R5, PapR5; R6, PapR6; SR, SpbR. Protein-protein interaction is observed when cells stain red on MacConkey agar. Agar plates were incubated for 2 days at 30°C.
In a similar way, the regulatory function of the TetR-like protein PapR3 was investigated. EMSA analysis showed that HisPapR3 specifically binds to the promoter regions of papR4 and papR5 (Fig. 5D). Furthermore, RT-PCR analyses demonstrated that papR4 and papR5 transcription is prolonged in the papR3::apr mutant compared to the wild type (Fig. 6D). Thus, we propose that PapR3 represses the transcription of the SARP gene papR4 and the TetR-like gene papR5. The role of PapR3 could be to fine-tune the expression amount of these PapR regulators. As we showed above that PapR5 has a significant influence on the production rate of pristinamycin, there might be the need to control the amount of protein PapR5, which could be the function of PapR3.
Altogether, our analyses show that PapR2 and PapR5 are two important set screws in the pristinamycin regulatory signaling cascade, with PapR2 being the essential activator for biosynthesis and PapR5 exerting a major influence on the production rate.
DISCUSSION
Regulatory influence on the synergistic production ration.
From our regulatory studies, we know that at least seven different regulators are involved in controlling the pristinamycin biosynthesis. The question is why S. pristinaespiralis needs such a multitude of control elements. We speculated that this might be needed to ensure the cosynthesis of the two different streptogramin-type antibiotics in the synergistic active 30:70 ratio (PI/PII). However, whenever we inactivated or overexpressed our regulators in S. pristinaespiralis, we did not observe any influence on the PI/PII production ratio (except for PapR6). Thus, the synergistic production ratio is not the result of a fine-tuned regulation but rather is based on the antibiotic synthesis efficiency: The polyketide-type antibiotic PII is composed mainly of malonyl-coenzyme A (-CoA) units and three proteinogenic amino acids, whereas the peptide antibiotic PI consists of two proteinogenic and five aproteinogenic amino acids (23). Overall, the PI biosynthesis may be more laborious, since all the aproteinogenic amino acid precursors have to be synthesized beforehand, whereas in terms of PII synthesis, malonyl-CoA and the proteinogenic amino acids are available immediately. This assumption is supported by more-detailed HPLC analyses (data not shown) and the production curves presented in reference 37, in which PI production is seen to increase during the later growth phase.
Effector synthesis and autoregulator receptor SpbR.
In a previous study, it has been shown that the autoregulator receptor SpbR is the major regulator of pristinamycin biosynthesis. It was suggested that SpbR senses an A-factor-like effector molecule (24); however, the SpbR-interactive ligand has not been characterized so far. In S. griseus, the GBL synthase AfsA is responsible for the biosynthesis of the A-factor signaling molecule (38). However, no putative orthologous gene is present in the pristinamycin biosynthetic gene region. Instead, at the right border of the cluster, between the autoregulator receptor gene spbR and the TetR-like gene papR5, a putative P450 monooxygenase-encoding gene, snbU, is localized. The predicted SnbU protein shows similarity to Orf16 of S. fradiae (64% identity, 75% similarity). Orf16 is a deduced cytochrome P450 and together with Orf18 (a deduced acyl-CoA oxidase) has been suggested to play a role in the synthesis of the GBL receptor (TylP)-interacting ligand in S. fradiae (12). Actually, no orf18 orthologous gene is present in the pristinamycin gene cluster. However, due to the sequence homology to Orf16, we suggest that SnbU is involved in the pristinamycin effector synthesis. The respective autoregulator receptor SpbR has previously been shown to bind to the promoter region of its own gene, as well as to the papR1 promoter (10). Our SpbR EMSA analyses additionally showed a binding to the papR2, papR4, and papR5 promoter regions (data not shown), meaning that SpbR is an autoregulator that binds to all promoters harboring an ARE element and thereby controls the transcription of nearly all papR genes (except for papR3 and papR6).
TetR-like regulators PapR3 and PapR5 and their putative receptor role.
A protein alignment of the TetR-like proteins from S. pristinaespiralis and those from other antibiotic-producing streptomycetes revealed that PapR3 (275 amino acids) has a 60-amino-acid-longer N-terminal sequence residue (see Fig. S2B in the supplemental material). Within the respective gene region, a less conserved ARE motif (57.7%) was identified. Thus, it may be possible that the papR3 gene has been annotated incorrectly and the actual PapR3 protein is shorter (215 amino acids). However, we are quite sure that the data generated with the longer PapR3 version are authentic, for several reasons: overexpression analyses with the larger PapR3 version clearly showed a repressive function on pristinamycin biosynthesis, and EMSAs exhibited a specific binding performance property (see above); furthermore, from BTH analysis we know that the longer PapR3 protein is capable of forming functional dimers (Fig. 7), whereas the shorter version is not (see Fig. S7 in the supplemental material).
In silico analyses of the amino acid composition of the TetR-like regulators PapR5 and PapR3 revealed that the two proteins considerably differ from each other in respect to their deduced isoelectric points (pI values), which is a rather acidic one for PapR5 (pI = 6.08) and a more basic one for PapR3 (pI = 9.92) (Table 3). Such a difference in the pI values of antibiotic cluster-originated TetR-like regulators has been observed before. Here, it was stated that the basic TetR-like proteins are supposed to be “pseudo-GBL receptors,” whereas the acidic ones act as “real” GBL receptors (19, 39, 40). Given this classification, PapR3 would be expected to be the pseudo-GBL receptor whereas PapR5 should act as the real GBL receptor protein. However, previously published phylogenetic analyses suggest a pseudo-GBL receptor function for PapR5 (19, 41). At present, the receptor role of PapR3 and PapR5 is unclear and cannot be predicted from phylogenetic trees; instead, it should be resolved experimentally in future studies, e.g., by effector-dependent EMSAs. Our data showed that PapR5 has the strongest effect on pristinamycin production amount, which theoretically makes it a good candidate for a sensor protein that may detect pristinamycin and/or its intermediates as effector(s) and in a feed-forward mechanism drives the antibiotic biosynthesis in a way similar to that described for the JadR2-mediated regulation of jadomycin biosynthesis in Streptomyces venezuelae (42, 43).
Different SARPs exert discrete regulatory effects.
As outlined above, PapR2 is the essential activator for pristinamycin biosynthesis, which controls papR1- and pristinamycin structural gene transcription, whereas PapR1 and PapR4 play only a subordinate role, as they are dispensable for pristinamycin production. PapR1, like PapR2, directly bound to the promoter regions of the pristinamycin structural genes. Thus, we suggest that PapR1 has an assisting function as a helper protein for PapR2. A similar “SARP helper” activity has already been proposed for TylU of S. fradiae, which is a nonessential, non-SARP-type regulator that together with the essential SARP TylS activates the expression of the pathway-specific activator TylR of tylosin biosynthesis (44). In EMSAs, PapR4 bound to the papR2 promoter region, which suggests that it may have an activating function in papR2 gene transcription. A comparable SARP-SARP system has been described for the lankamycin/lankacidin producer S. rochei, whereby the SARP-type regulator SrrY activates the transcription of the SARP gene srrZ (45). In summary, we can state that each of the pristinamycin SARPs exerts its individual role during pristinamycin biosynthesis.
Correlation between pI characteristics and functionalization of SARPs.
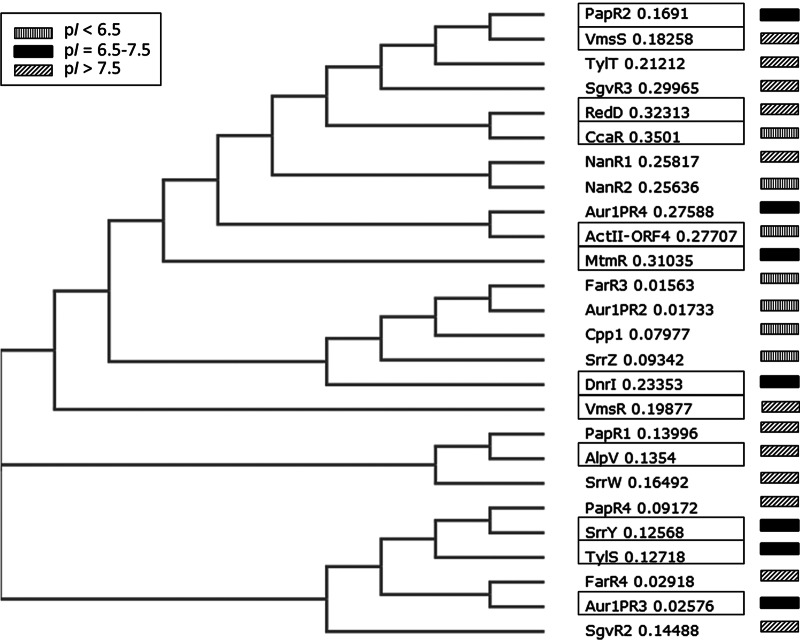
Interestingly, in silico analyses of the amino acid composition of the SARP-type regulators PapR1, PapR2, and PapR4 also revealed striking differences in pI values: PapR2, the essential regulator of pristinamycin biosynthesis, has a neutral character (pI = 7.03), whereas PapR1 and PapR4 are very basic proteins (pI = 10.58 and 10.05, respectively) (Fig. 8; Table 3). To investigate if there is a relationship between the essentiality of different SARPs, such as TylS (activator of tylosin biosynthesis) from S. fradiae, SrrY from S. rochei (activator of lankacidin and lankamycin biosynthesis), DnrI from Streptomyces peucetius (activator of daunorubicin biosynthesis), RedD and ActII-Orf4 from Streptomyces coelicolor A3(2) (activator of undecylprodigiosin and actinorhodin biosynthesis, respectively), and Aur1PR3 from S. aureofaciens (activator of auricin biosynthesis) (46, 47, 48, 49, 50, 51) in antibiotic biosynthesis and different pI values, a phylogenetic analysis was performed with the known SARP-type regulators from various antibiotic-producing streptomycetes (Fig. 8). From this analysis, we found that, with a few exceptions, acidic and basic SARPs are part of discrete clades. Essential SARPs have a rather neutral or basic character. Interestingly, we observed that, except for the NanR regulators of Streptomyces nanchangensis (52), the SARPs from different strains are more similar to each other than the different SARPs from a specific host strain, meaning that the different SARPs from one strain most likely did not evolve from duplication events in the host strain but rather were acquired separately by horizontal gene transfer. In conclusion, the difference in protein acidity seems to be a more unitary tool that nature uses to develop functionally different regulators from the same protein family. Thus, the importance of protein acidity in the differential functionalization of regulators definitely needs further attention.
FIG 8.
Phylogenetic tree of different SARPs. Sources: S. pristinaespiralis (PapR1, CBW45751; PapR2, CBW45736; PapR4, CBW45766; Cpp1, CBW45731); S. fradiae (TylS, AAD40804; TylT, AAD40805); S. lavendulae (FarR3, BAG74713; FarR4, BAG74714); S. aureofaciens (Aur1PR2, ADM72850; Aur1PR3, ADM72849; Aur1PR4, ACK77758); S. rochei (SrrY, BAC76533; SrrZ, BAC76529; SrrW, BAC76513); S. virginiae (VmsR, BAA96296; VmsS, BAF50715); S. griseoviridis (SgvR3, AGN74872; SgvR2, AGN74902); S. nanchangensis (NanR1, AAP42853; NanR2, AAP42854); S. coelicolor A3(2) (RedD, AAA88556; ActII-ORF4, CAC44198); S. peucetius (DnrI, AAA26736); S. clavuligerus ATCC 27064 (CcaR, AAC32494); S. argillaceus (MtmR, CAK50770); S. ambofaciens ATCC 23877 (AlpV, CAJ87890). Phylogenetic distances are given as numbers. Essential SARP regulators are framed. Proteins with an acidic (pI < 6.5), neutral (pI = 6.5 to 7.5), or basic (pI > 7.5) character are indicated by boxes with vertical lines, black boxes, or boxes with diagonal lines, respectively.
Putative interaction partners of response regulator PapR6.
Our analysis showed that nearly all predicted pristinamycin operons (37) contain a SARP binding motif in their cognate promoter regions, except snaF and papB-papC. Thus, the expression of the SARP motif-containing genes is suggested to be controlled by SARP-type regulators, whereas the snaF and papB-papC operon expression may be governed by a non-SARP-type regulator. Such a regulatory function most likely is exerted by the response regulator PapR6. This is supported by data from reference 37, which showed by EMSAs that PapR6 binds to the snaF promoter region. PapR6 is an orphan response regulator with no cognate sensor kinase gene in the pristinamycin cluster. A sensor kinase, Spy1, of which the encoding gene is located outside the pristinamycin gene cluster, has previously been reported to positively influence PI biosynthesis but has a negative effect on PII biosynthesis (53). The Dun et al. study (37) showed that a papR6 deletion or overexpression had a more dramatic effect on PII production than on PI biosynthesis. Even if this is not obvious from the graph in Fig. 3, such a differential influence on PI and PII production has also been observed for several individual growth curves of papR6 deletion and overexpression strains in the present study (data not shown). Thus, we concluded that PapR6 is an activator of PII but not PI biosynthesis. Whether Spy1 interacts with PapR6 is doubtful, as the two regulators have opposite effects on PI and PII production.
With BTH analysis, we found that PapR6 interacts on the protein level with the TetR-like regulator PapR5 (Fig. 7). The fact that PapR6 interacts with PapR5 fits more with the data of jadomycin regulation in S. venezuelae and auricin regulation in S. aureofaciens. Here, it was hypothesized that the response regulator JadR1 forms a novel two-component system with the pseudo-GBL receptor JadR2, and together they govern jadomycin biosynthesis (53). A similar protein pair, Aur1P (response regulator)/Aur1R (TetR-like regulator), controls auricin production in S. aureofaciens (33). However, in these analyses the regulation has always been reported to take place on the protein-DNA level. Our data show an additional regulatory level, which occurs on the protein-protein stage and may disclose a completely novel regulatory interaction pair.
Pristinamycin regulation model.
To comprehend our data obtained from the regulatory studies of the PapR regulators into a broader context, we derived a hierarchical organized regulatory model that governs pristinamycin biosynthesis in S. pristinaespiralis (Fig. 9): in the absence of ligands, the autoregulator receptor SpbR represses its own transcription, as well as that of papR1, papR2, papR4, and papR5. As this repression prohibits the expression of all SARPs, which are the pathway-specific activators, no (or only basal) pristinamycin biosynthesis can occur. In the presence of a critical concentration of ligands, the repressive function of SpbR is relieved and the signaling cascade switches on. PapR2, as the major activator of pristinamycin biosynthesis, together with PapR1, activates the transcription of the pristinamycin structural genes.
FIG 9.
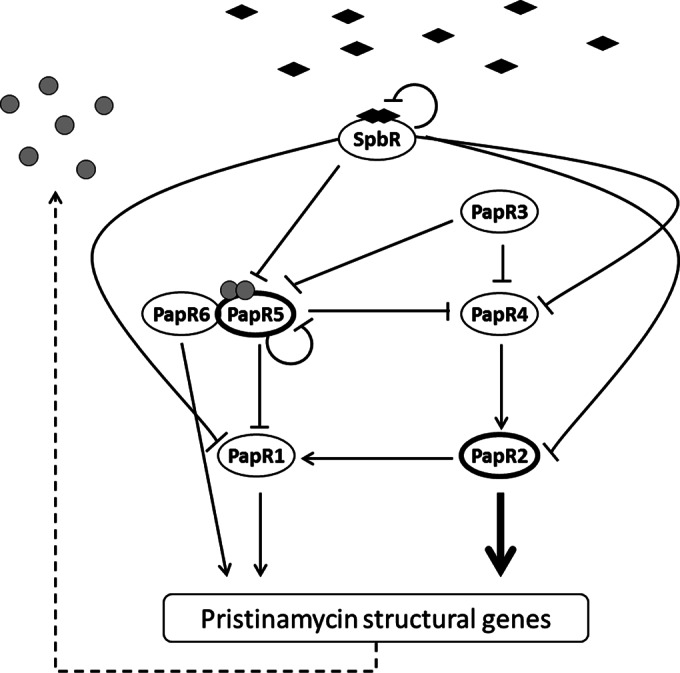
Model of the regulation of pristinamycin biosynthesis in Streptomyces pristinaespiralis. SpbR ligands are indicated as black-filled diamonds. Pristinamycins and/or intermediates are shown as gray-filled circles. Regulators are represented by ellipses. The major pristinamycin regulators PapR2 and PapR5 are highlighted by thick lines. Arrows indicate transcriptional activation, and perpendicular lines represent transcriptional repression. The dashed line illustrates pristinamycin (intermediate) production.
PapR2 expression is indirectly regulated by PapR5, as PapR5 represses the transcription of papR4, the product of which activates papR2 transcription. Furthermore, PapR5 interacts on the protein level with the PII activator PapR6. The function of PapR6 in the regulatory cascade remains unclear. PapR5 represents the major control element for the pristinamycin production amount. As a GBL receptor-like protein, PapR5 may sense pristinamycin or intermediates(s) of the pathway. During the early growth phase, PapR5 directly represses the transcription of the SARP genes papR1 and papR4 and indirectly that of papR2 (via PapR4), which results in a tight inhibition of pristinamycin production. At the beginning of the stationary phase, when the ligand-induced derepression of SpbR is initiated, the protein concentration of PapR5 is decreased by the function of PapR3, which inhibits the transcription of papR4 and papR5 and in turn allows low-level pristinamycin production. Initially, such a timely controlled drug homeostasis may be necessary for the strain to develop self-resistance and thereby prevents self-toxicity from the compound. The low-level pristinamycin synthesis finally drives the feed-forward mechanism and by inactivating the PapR5 repression leads to full pristinamycin production. Alternatively, PapR5 may act as a late repressor that allows to switch off pristinamycin production. In such a scenario, the ligand-induced derepression of SpbR allows papR5 transcription, which leads to an accumulation of PapR5 protein in the cell. When the PapR5 concentration is high enough, it represses the SARP gene transcription, resulting in a shutdown of pristinamycin production.
Regulation in related streptogramin producers.
A comparison of the two other characterized streptogramin antibiotic gene clusters—the virginiamycin cluster (vir) from S. virginiae (54) and the griseoviridin/viridogrisein (GV/VG) cluster (sgv) from Streptomyces griseoviridis (55)—revealed that the amount of pathway-specific regulatory genes, as well as their overall localization within the gene cluster, is quite conserved (see Fig. S1A in the supplemental material). However, a closer look at the abundance of different regulator types and their function, which has been partially shown for virginiamycin regulation (15, 16, 56, 57, 58) but only deduced for GV/VG regulation (55), suggests that there exist different regulatory networks in all streptogramin producers (see Fig. S1B in the supplemental material): e.g., the virginiamycin cascade has no equivalent for PapR1 and PapR5 in abundance and function. The autoregulator ligand synthesis seems to be different in all three streptogramin producers: in S. virginiae, BarX, BarS1, and BarS2 are involved in virginiae butanolide synthesis (58), and in S. pristinaespiralis a P450 monooxygenase is suggested to be involved in the biosynthesis of an A-factor-like GBL (see above), whereas in S. griseoviridis a barX homologous gene is present, in addition to two other putative GBL synthesis genes, of which the products are not homologous to BarS1 and BarS2. In conclusion, it seems that even in strains with a similar—or in the case of virginiamycin, nearly identical—gene cluster, the regulatory network governing the respective antibiotic biosynthesis is unique for every strain.
So far, there are only few reports on antibiotic signaling cascades consisting of multiple regulators. However, regulator-based strain engineering is an efficient tool, and knowledge of regulatory effects is a prerequisite for improving antibiotic production and/or activating a silent gene cluster. In this regard, our data contribute to a better understanding of antibiotic regulation processes by disclosing a comprehensive new regulatory cascade that governs the fine-tuned biosynthesis of pristinamycin in S. pristinaespiralis.
Supplementary Material
ACKNOWLEDGMENTS
This study was supported by The German Center for Infection Research (DZIF) grant number TTU 09.802. J.G. acknowledges a grant from the Promotionsverbund Antibakterielle Wirkstoffe of the University of Tübingen. Y.M. was supported by scholarships funded by the Landesgraduiertenförderungsgesetz des Landes Baden-Württemberg and the DFG (Graduiertenkolleg Infektionsbiologie), as well as by a grant from the Athene-Programm für Nachwuchswissenschaftlerinnen of the University of Tübingen. Sanofi-Aventis partially financed this work.
We thank H.-P. Fiedler and Andreas Kulik (Universität Tübingen) for help in HPLC measurements and Regina Ort-Winklbauer for excellent technical assistance.
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AEM.00728-15.
REFERENCES
- 1.Tanaka Y, Omura S. 1990. Metabolism and products of actinomycetes: an introduction. Actinomycetologica 4:13–14. doi: 10.3209/saj.4_13. [DOI] [Google Scholar]
- 2.Liu G, Chater KF, Chandra G, Niu G, Tan H. 2013. Molecular regulation of antibiotic biosynthesis in Streptomyces. Microbiol Mol Biol Rev 77:112–143. doi: 10.1128/MMBR.00054-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Claessen D, de Jong W, Dijjkhuizen L, Wosten HA. 2006. Regulation of Streptomyces development: reach for the sky! Trends Microbiol 14:313–319. [DOI] [PubMed] [Google Scholar]
- 4.Bibb MJ. 2005. Regulation of secondary metabolism in streptomycetes. Curr Opin Microbiol 8:208–215. doi: 10.1016/j.mib.2005.02.016. [DOI] [PubMed] [Google Scholar]
- 5.Nihira T, Shimizu Y, Kim HS, Yamada Y. 1988. Structure-activity relationships of virginiae butanolide C, an inducer of virginiamycin production in Streptomyces virginiae. J Antibiot 41:1828–1837. doi: 10.7164/antibiotics.41.1828. [DOI] [PubMed] [Google Scholar]
- 6.Sato K, Nihira T, Sakuda S, Yanagimoto M, Yamada Y. 1989. Isolation and structure of a new butyrolactone autoregulator from Streptomyces sp. FRI-5. J Ferment Biotechnol 68:170–173. doi: 10.1016/0922-338X(89)90131-1. [DOI] [Google Scholar]
- 7.Nishida H, Ohnishi Y, Beppu T, Horinouchi S. 2007. Evolution of γ-butyrolactone synthases and receptors in Streptomyces. Environ Microbiol 9:1986–1994. doi: 10.1111/j.1462-2920.2007.01314.x. [DOI] [PubMed] [Google Scholar]
- 8.Natsume R, Ohnishi Y, Senda T, Horinouchi S. 2004. Crystal structure of a γ-butyrolactone autoregulator receptor protein in Streptomyces coelicolor A3(2). J Mol Biol 336:409–419. doi: 10.1016/j.jmb.2003.12.040. [DOI] [PubMed] [Google Scholar]
- 9.Kinoshita H, Tsuji T, Ipposhi H, Nihira T, Yamada Y. 1999. Characterization of binding sequences for butyrolactone autoregulator receptors in streptomycetes. J Bacteriol 181:5075–5080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Folcher M, Gaillard H, Nguyen LT, Nguyen KT, Lacroix P, Bamas-Jacques N, Rinkel M, Thompson CJ. 2001. Pleiotropic functions of a Streptomyces pristinaespiralis autoregulator receptor in development, antibiotic biosynthesis, and expression of a superoxide dismutase. J Biol Chem 276:44297–22306. doi: 10.1074/jbc.M101109200. [DOI] [PubMed] [Google Scholar]
- 11.Ramos JL, Martínez-Bueno M, Molina-Henares AJ, Terán W, Watanabe K, Zhang X, Gallegos MT, Brennan R, Tobes R. 2005. The TetR family of transcriptional repressors. Microbiol Mol Biol Rev 69:326–356. doi: 10.1128/MMBR.69.2.326-356.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bignell DR, Bate N, Cundliffe E. 2007. Regulation of tylosin production: role of a TylP-interactive ligand. Mol Microbiol 63:838–847. [DOI] [PubMed] [Google Scholar]
- 13.Wietzorrek A, Bibb M. 1997. A novel family of proteins that regulates antibiotic production in streptomycetes appears to contain an OmpR-like DNA-binding fold. Mol Microbiol 25:1181–1184. doi: 10.1046/j.1365-2958.1997.5421903.x. [DOI] [PubMed] [Google Scholar]
- 14.Horinouchi S. 2002. A microbial hormone, A-factor, as a master switch for morphological differentiation and secondary metabolism in Streptomyces griseus. Front Biosci 7:2045–2057. [DOI] [PubMed] [Google Scholar]
- 15.Pulsawat N, Kitani S, Fukushima E, Nihira T. 2009. Hierarchical control of virginiamycin production in Streptomyces virginiae by three pathway-specific regulators: VmsS, VmsT, and VmsR. Microbiology 155:1250–1259. doi: 10.1099/mic.0.022467-0. [DOI] [PubMed] [Google Scholar]
- 16.Matsuno K, Yamada Y, Lee CK, Nihira T. 2004. Identification by gene deletion analysis of barB as a negative regulator controlling an early process of virginiamycin biosynthesis in Streptomyces virginiae. Arch Microbiol 181:52–59. doi: 10.1007/s00203-003-0625-5. [DOI] [PubMed] [Google Scholar]
- 17.Cundliffe E. 2006. Antibiotic production by actinomycetes: the Janus face of regulation. J Ind Microbiol Biotechnol 33:500–506. doi: 10.1007/s10295-006-0083-6. [DOI] [PubMed] [Google Scholar]
- 18.Arakawa K, Mochizuki S, Yamada K, Noma T, Kinashi H. 2007. γ–butyrolactone autoregulator-receptor system involved in lankacidin and lankamycin production and morphological differentiation in Streptomyces rochei. Microbiology 153:1817–1827. doi: 10.1099/mic.0.2006/002170-0. [DOI] [PubMed] [Google Scholar]
- 19.Kormanec J, Novakova R, Mingyar E, Feckova L. 2014. Intriguing properties of the angucycline antibiotic auricin and complex regulation of its biosynthesis. Appl Microbiol Biotechnol 98:45–60. doi: 10.1007/s00253-013-5373-0. [DOI] [PubMed] [Google Scholar]
- 20.Mast Y, Wohlleben W. 2014. Streptogramins—two are better than one! Int J Med Microbiol 304:44–50. doi: 10.1016/j.ijmm.2013.08.008. [DOI] [PubMed] [Google Scholar]
- 21.Cocito CG. 1979. Antibiotics of the virginiamycin family, inhibitors which contain synergistic components. Microbiol Rev 43:145–198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Qadri SM, Ueno Y, Abu Mostafa FM, Halim M. 1997. In vitro activity of quinupristin/dalfopristin, RP59500, against gram-positive clinical isolates. Chemotherapy 43:94–99. doi: 10.1159/000239542. [DOI] [PubMed] [Google Scholar]
- 23.Mast Y, Weber T, Gölz M, Ort-Winklbauer R, Gondran A, Wohlleben W, Schinko E. 2011. Characterization of the ‘pristinamycin supercluster’ of Streptomyces pristinaespiralis. Microb Biotechnol 4:192–206. doi: 10.1111/j.1751-7915.2010.00213.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Paquet V, Goma G, Soucaille P. 1992. Induction of pristinamycins production in Streptomyces pristinaespiralis. Biotechnol Lett 14:1065–1070. doi: 10.1007/BF01021060. [DOI] [Google Scholar]
- 25.Bullock WO, Fernandez JM, Short JM. 1987. Xl1-Blue, a high efficiency plasmid transforming recA Escherichia coli strain with β-galactosidase selection. Biotechniques 5:376–378. [Google Scholar]
- 26.Sambrook J, Fritsch T, Maniatis T. 1989. Molecular cloning: a laboratory manual, 2nd ed Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. [Google Scholar]
- 27.Fischer J. 1996. Entwicklung eines regulierbaren Expressionssystems zur effizienten Synthese rekombinanter Proteine in Streptomyces lividans. Ph.D. thesis University of Stuttgart, Stuttgart, Germany. [Google Scholar]
- 28.Kieser T, Bibb MJ, Buttner MJ, Chater KF, Hopwood DA. 2000. Practical Streptomyces genetics. The John Innes Foundation, Norwich, England. [Google Scholar]
- 29.Pridmore RD. 1987. New and versatile cloning vectors with kanamycin-resistance marker. Gene 56:309–312. doi: 10.1016/0378-1119(87)90149-1. [DOI] [PubMed] [Google Scholar]
- 30.Heinzelmann E, Kienzlen G, Kaspar S, Recktenwald J, Wohlleben W, Schwartz D. 2001. The phosphinomethylmalate isomerase gene pmi, encoding an aconitase-like enzyme, is involved in the synthesis of phosphinothricin tripeptide in Streptomyces viridochromogenes. Appl Environ Microbiol 67:3603–3609. doi: 10.1128/AEM.67.8.3603-3609.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wohlleben W, Stegmann E, Süßmuth RD. 2009. Molecular genetic approaches to analyze glycopeptide biosynthesis. Methods Enzymol 458:459–486. doi: 10.1016/S0076-6879(09)04818-6. [DOI] [PubMed] [Google Scholar]
- 32.Karimova G, Pidoux J, Ullmann A, Ladant D. 1998. A bacterial two-hybrid system based on a reconstituted signal transduction pathway. Proc Natl Acad Sci U S A 95:5752–5756. doi: 10.1073/pnas.95.10.5752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Novakova R, Homerova D, Feckova L, Kormanec J. 2005. Characterization of a regulatory gene essential for the production of the angucycline-like polyketide antibiotic auricin in Streptomyces aureofaciens CCM 3239. Microbiology 151:2693–2706. doi: 10.1099/mic.0.28019-0. [DOI] [PubMed] [Google Scholar]
- 34.Wang L, Tian X, Wang J, Yang H, Fan K, Xu G, Yang K, Tan H. 2009. Autoregulation of antibiotic biosynthesis by binding of the end product to an atypical response regulator. Proc Natl Acad Sci U S A 106:8617–8622. doi: 10.1073/pnas.0900592106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Dsouza M, Larsen N, Overbeek R. 1997. Searching for patterns in genomic data. Trends Genet 13:497–498. [DOI] [PubMed] [Google Scholar]
- 36.Tanaka A, Takano Y, Ohnishi Y, Horinouchi S. 2007. AfsR recruits RNA polymerase to the afsS promoter: a model for transcriptional activation by SARPs. J Mol Biol 369:322–333. doi: 10.1016/j.jmb.2007.02.096. [DOI] [PubMed] [Google Scholar]
- 37.Dun J, Zhao Y, Zheng G, Zhu H, Ruan L, Wang W, Ge M, Jiang W, Lu Y. 2015. PapR6, a putative atypical response regulator, functions as a pathway-specific activator of pristinamycin II biosynthesis in Streptomyces pristinaespiralis. J Bacteriol 197:441–450. doi: 10.1128/JB.02312-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Horinouchi S, Kumada Y, Beppu T. 1984. Unstable genetic determinant of A-factor biosynthesis in streptomycin producing organisms: cloning and characterization. J Bacteriol 158:481–487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Salehi-Najafabadi Z, Barreiro C, Rodríguez-García A, Cruz A, López GE, Martín JF. 2014. The gamma-butyrolactone receptors BulR1 and BulR2 of Streptomyces tsukubaensis: tacrolimus (FK506) and butyrolactone synthetases production control. Appl Microbiol Biotechnol 98:4919–4936. doi: 10.1007/s00253-014-5595-9. [DOI] [PubMed] [Google Scholar]
- 40.Wang J, Wang W, Wang L, Zhang G, Fan K, Tan H, Yang K. 2011. A novel role of pseudo γ-butyrolactone receptors in controlling γ-butyrolactone biosynthesis in Streptomyces. Mol Microbiol 82:236–250. doi: 10.1111/j.1365-2958.2011.07811.x. [DOI] [PubMed] [Google Scholar]
- 41.Cuthbertson L, Nodwell JR. 2013. The TetR family of regulators. Microbiol Mol Biol Rev 77:440–475. doi: 10.1128/MMBR.00018-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhang Y, Pan G, Zou Z, Fan K, Yang K, Tan H. 2013. JadR*-mediated feed-forward regulation of cofactor supply in jadomycin biosynthesis. Mol Microbiol 90:884–897. doi: 10.1111/mmi.12406. [DOI] [PubMed] [Google Scholar]
- 43.Wang W, Ji J, Li X, Wang J, Li S, Pan G, Fan K, Yang K. 2014. Angucyclines as signals modulate the behaviors of Streptomyces coelicolor. Proc Natl Acad Sci U S A 111:5688–5693. doi: 10.1073/pnas.1324253111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bate N, Bignell DRD, Cundliffe E. 2006. Regulation of tylosin biosynthesis involving ‘SARP-helper’ activity. Mol Microbiol 62:148–156. doi: 10.1111/j.1365-2958.2006.05338.x. [DOI] [PubMed] [Google Scholar]
- 45.Suzuki T, Mochizuki S, Yamamoto S, Arakawa K, Kinashi H. 2010. Regulation of lankamycin biosynthesis in Streptomyces rochei by two SARP genes, srrY and srrZ. Biosci Biotechnol Biochem 74:819–827. doi: 10.1271/bbb.90927. [DOI] [PubMed] [Google Scholar]
- 46.Bate N, Stratigopoulos G, Cundliffe E. 2002. Differential roles of two SARP-encoding regulatory genes during tylosin biosynthesis. Mol Microbiol 43:449–458. doi: 10.1046/j.1365-2958.2002.02756.x. [DOI] [PubMed] [Google Scholar]
- 47.Yamamoto S, He Y, Arakawa K, Kinashi H. 2008. Gamma-butyrolactone-dependent expression of the Streptomyces antibiotic regulatory protein gene srrY plays a central role in the regulatory cascade leading to lankacidin and lankamycin production in Streptomyces rochei. J Bacteriol 190:1308–1316. doi: 10.1128/JB.01383-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Tang L, Grimm A, Zhang YX, Hutchingson CR. 1996. Purification and characterization of the DNA-binding protein DnrI, a transcriptional factor of daunorubicin biosynthesis in Streptomyces peucetius. Mol Microbiol 22:801–813. doi: 10.1046/j.1365-2958.1996.01528.x. [DOI] [PubMed] [Google Scholar]
- 49.Narva KE, Feitelson JS. 1990. Nucleotide sequence and transcriptional analysis of the red locus of Streptomyces coelicolor A3(2). J Bacteriol 172:326–333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Fernández-Moreno MA, Caballero JL, Hopwood DA, Malpartida F. 1991. The act cluster contains regulatory and antibiotic export genes, direct targets for translational control by the bldA tRNA gene of Streptomyces. Cell 66:769–780. doi: 10.1016/0092-8674(91)90120-N. [DOI] [PubMed] [Google Scholar]
- 51.Novakova R, Rehakova A, Kutas P, Feckova L, Kormanec J. 2011. The role of two SARP family transcriptional regulators in regulation of the auricin gene cluster in Streptomyces aureofaciens CCM 3239. Microbiology 157:1629–1639. doi: 10.1099/mic.0.047795-0. [DOI] [PubMed] [Google Scholar]
- 52.Yu Q, Du A, Liu T, Deng Z, He X. 2012. The biosynthesis of the polyether antibiotic nanchangmycin is controlled by two pathway-specific transcriptional activators. Arch Microbiol 194:415–426. doi: 10.1007/s00203-011-0768-8. [DOI] [PubMed] [Google Scholar]
- 53.Jin Q, Yin H, Hong X, Jin Z. 2012. Isolation and functional analysis of spy1 responsible for pristinamycin yield in Streptomyces pristinaespiralis. J Microbiol Biotechnol 22:793–799. doi: 10.4014/jmb.1111.11031. [DOI] [PubMed] [Google Scholar]
- 54.Yang K, Han L, He J, Wang L, Vining LC. 2001. A repressor-response regulator gene pair controlling jadomycin B production in Streptomyces venezuelae ISP5230. Gene 279:165–173. doi: 10.1016/S0378-1119(01)00723-5. [DOI] [PubMed] [Google Scholar]
- 55.Xie Y, Wang B, Liu J, Zhou J, Ma J, Huang H, Ju J. 2012. Identification of the biosynthetic gene cluster and regulatory cascade for the synergistic antibacterial antibiotics griseoviridin and viridogrisein in Streptomyces griseoviridis. Chembiochem 13:2745–2757. doi: 10.1002/cbic.201200584. [DOI] [PubMed] [Google Scholar]
- 56.Kinoshita H, Ipposhi H, Okamoto S, Nakano H, Nihira T, Yamada Y. 1997. Butyrolactone autoregulator receptor protein (BarA) as a transcriptional regulator in Streptomyces virginiae. J Bacteriol 179:6986–6993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kawachi R, Wangchaisoonthorn U, Nihira T, Yamada Y. 2000. Identification by gene deletion analysis of a regulator, VmsR, that controls virginiamycin biosynthesis in Streptomyces virginiae. J Bacteriol 182:6259–6263. doi: 10.1128/JB.182.21.6259-6263.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Lee YJ, Kitani S, Nihira T. 2010. Null mutation analysis of an afsA-family gene, barX, that is involved in biosynthesis of the {gamma}-butyrolactone autoregulator in Streptomyces virginiae. Microbiology 256:206–210. doi: 10.1099/mic.0.032003-0. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.