Abstract
Wnt signaling plays important roles in development and tumorigenesis. A central question about the Wnt pathway is the regulation of β-catenin. Phosphorylation of β-catenin by CK1α and GSK3 promotes β-catenin binding to β-TrCP, leading to β-catenin degradation through the proteasome. The phosphorylation and ubiquitination of β-catenin have been well characterized; however, it is unknown whether and how a deubiquitinase is involved. In this study, by screening RNA interference (RNAi) libraries, we identified USP47 as a deubiquitinase that prevents β-catenin ubiquitination. Inactivation of USP47 by RNAi increased β-catenin ubiquitination, attenuated Wnt signaling, and repressed cancer cell growth. Furthermore, USP47 deubiquitinates itself, whereas β-TrCP promotes USP47 ubiquitination through interaction with an atypical motif in USP47. Finally, in vivo studies in the Drosophila wing suggest that UBP64E, the USP47 counterpart in Drosophila, is required for Armadillo stabilization and plays a positive role in regulating Wnt target gene expression.
INTRODUCTION
Wnt/β-catenin signaling plays an essential role in animal development and tumorigenesis (1, 2). In normal cells, β-catenin is sequentially phosphorylated by CKIα and GSK-3 in a protein complex containing the tumor suppressor proteins axin and APC (3). Phosphorylated β-catenin is recognized by the ubiquitin (Ub) ligase β-TrCP, an F-box and WD40 repeat protein (4). The WD40 repeat domain of β-TrCP binds β-catenin. β-TrCP also binds, via its F-box, to components of the ubiquitination machinery, including Skp1, cullin 1, ring box protein 1 (Rbx1), and ubiquitin-conjugating enzyme (E2). Ubiquitinated β-catenin is degraded via the 26S proteasome (4–7). Upon Wnt stimulation, the Wnt protein binds its receptor, Frizzled, and coreceptor, LRP5/6; disrupts the axin complex; blocks the phosphorylation required for β-TrCP-mediated ubiquitination; and thus stabilizes β-catenin (8–10). Wnt further promotes the accumulated β-catenin entry into the nucleus; binding of TCF/LEF; and recruitment of transcriptional coactivators, such as Bcl9/legless, Pygopus, and CBP/p300, to activate downstream target genes (2, 11).
In human cancers, particularly in colorectal cancer, β-catenin is stabilized by mutations in APC or β-catenin (12, 13). β-Catenin mutations prevent CKIα or GSK-3 phosphorylation and β-TrCP recognition of β-catenin, resulting in abnormal β-catenin accumulation that ultimately leads to cancer (3, 4). APC truncations inhibit both phosphorylation and ubiquitination of β-catenin, also resulting in β-catenin accumulation (14–16). Understanding the molecular mechanisms of β-catenin ubiquitination/degradation is essential for developing therapeutic agents targeting this important pathway for cancer prevention and therapeutics (17).
The kinases and ubiquitin protein ligase for β-catenin phosphorylation and ubiquitination have been well studied. Phosphatase 2A (PP2A) and its regulatory subunit, PR55α, directly interact with and inhibit β-catenin phosphorylation (18). Whether β-catenin ubiquitination is also directly regulated by a deubiquitinase is not known. Several deubiquitinases have been shown to play positive or negative roles in the Wnt pathway. Trabid, a positive regulator of Wnt-mediated transcription, interacts with ubiquitinated APC and affects the activity of APC in transcription (19). USP8/UBPY regulates the cell surface levels of Frizzled by preventing its lysosomal trafficking and degradation (20). USP8/UBPY is required for sensory bristle formation in the Drosophila wing (20). Disheveled (Dvl) is also regulated by ubiquitination. Hyperubiquitination of polymerized Dvl enhances Wnt signaling (21). CYLD is a tumor suppressor; it regulates Dvl deubiquitination and plays a negative role in the Wnt pathway (21). In addition, USP4 was identified as a regulator of NLK and TCF4 and plays a negative role in Wnt signaling (22); USP15 stabilizes APC and also plays a negative role in Wnt signaling (23), and USP34 regulates axin stability (24). However, these deubiquitinases have not been characterized in vivo, and the deubiquitinase for β-catenin has not been identified. In this study, we screened the deubiquitinase RNA interference (RNAi) libraries and identified a deubiquitinase, USP47/UBP64E, that regulates β-catenin ubiquitination/degradation in vitro and in vivo.
MATERIALS AND METHODS
Screening of an siRNA library for human deubiquitinases.
The Dharmacon siGENOME RTF SMARTpool siRNA library for human deubiquitinating enzymes (H-004705; lot 08138) was used for screening for human deubiquitinases. HEK293T cells were stably transfected with a modified TOPFlash reporter, and the cells were selected with puromycin. The cells were plated in 96-well plates containing the small interfering RNA (siRNA) library. The siRNAs were reverse transfected into cells with DharmaFECT1 transfection reagent (Dharmacon) following the manufacture's protocol. After 48 h, the cells were treated with Wnt3A-conditioned medium for an additional 12 h. Cells were harvested, and the luciferase activities were analyzed with a luciferase assay system and the GloMax-Multi+Microplate multimode reader (Promega).
RNAi, wing disc immunostaining, and generating transgenic lines in Drosophila.
The RNAi lines that targeted each deubiquitinase in the Drosophila genome were obtained from the Vienna Drosophila RNAi Center (VDRC) (25). Wing-specific MS1096 Gal4 was used to assess for the induction of an adult wing phenotype. Upstream activation sequence (UAS)-Dicer was coexpressed with RNAi lines to enhance the RNAi effects. UBP64E RNAi lines (v26027 and v103743) were consistent in terms of the adult wing phenotypes and the effects on Armadillo (Arm) accumulation in wing discs. A standard protocol was used for the wing disc immunostaining. Briefly, wing discs from third-instar larvae with specific genotypes were dissected in phosphate-buffered saline (PBS) and then fixed with 4% formaldehyde in PBS for 20 min. After permeabilization with PBS supplemented with 1% Triton X-100 (PBT), the discs were incubated with the indicated primary antibodies for 3 h and the corresponding secondary antibodies for 1 h sequentially and then washed with PBT three times, for 20 min per wash, following the incubations. The antibodies used in this study were mouse anti-Arm (DSHB; 1:10), anti-Wg (Developmental Studies Hybridoma Bank [DSHB]; 1:50), anti-Ptc (DSHB; 1:10), rabbit anti-Flag (ABR; 1:150), anti-Dll (from Grace Boekhoff-Falk; 1:150), and guinea pig anti-Sens (from Hugo Bellen; 1:150). UAS-UBP64E and UAS-UBP64EC>S transgenic lines were generated by using integrase-mediated transgenesis in combination with the VK5 attP locus (26).
Quantitative RT-PCR.
Drosophila wing discs from third-instar larvae with specific genotypes were dissected, and total RNA was extracted using TRIzol reagent (Invitrogen). cDNA was synthesized, using random primer 6 (NEB; S1230S) and Moloney murine leukemia virus (M-MULV) reverse transcriptase (NEB; 0230908), from 1.0 μg total RNA according to the manufacturer's instructions. Quantitative reverse transcription (RT)-PCRs were carried out using SYBR green PCR master mix reagents (Thermo) on the ABI StepOnePlus real-time PCR system (Applied Biosystems). Thermal cycling was conducted at 95°C for 10 min, followed by 40 cycles of amplification at 95°C for 15 s and 60°C for 1 min, and then the following melting curve: 95°C for 15 s, 60°C for 1 min, and 95°C for 15 s. The relative quantification of gene expression for each sample was analyzed by the ΔCT method. Primer sequences were as follows: UBP64E-F, 5′-CAGAAACTGAGGCAGAGGC-3′; UBP64E-R, 5′-TCCCAGCGGTACAGAGCA-3′; Actin-F, 5′-GCGTCGGTCAATTCAATCTT-3′; and Actin-R, 5′-AAGCTGCAACCTCTTCGTCA-3′.
siRNA, shRNA, and plasmids.
Additional USP47 siRNA was ordered from Qiagen (reference sequence, NM_017944; catalog number SI00758716). USP47 short hairpin RNAs (shRNAs) were ordered from Sigma (reference sequence, NM_017944; CDS 764 to 4627). Plasmids are described in the figures. Site-specific mutagenesis was performed as previously described (27). The primer sequences for subcloning are available upon request. siRNA, shRNA, and plasmids were transfected into mammalian cells with Lipofectamine 2000 (Invitrogen) or calcium phosphate, as previously described (3).
Cell culture.
HEK293T and PC3 cells were grown in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% fetal bovine serum and 1% penicillin-streptomycin. A549 cells were grown in RPMI medium supplemented with 10% fetal bovine serum and 1% penicillin-streptomycin. For proliferation assays, cell lines were seeded at 0.04 × 106 cells/well in 12-well plates and counted using a cell viability analyzer (Beckman Coulter; Vi-Cell XR). For protein degradation assays, cells were treated with cycloheximide (CHX) (40 μM) and MG132 (25 μM) for 6 h before harvesting.
Detection of ubiquitin-conjugated proteins.
HEK293T cells were transfected with pCMV-His-Ub for 48 h and then treated with 2.5 μM MG132 for an additional 6 h. The cells were washed with cold PBS, lysed in PTU lysis buffer (100 mM NaH2PO4, 10 mM Tris, 8 M urea, pH 8.0) (28), sonicated 5 times for 3 s each time, and centrifuged for 10 min at 13,000 rpm. The supernatant was transferred to a new Eppendorf tube and incubated with 20 μl Ni-nitrilotriacetic acid (Ni-NTA) beads (Qiagen; catalog number 30250) at 4°C overnight. The beads were washed once with PTU lysis buffer and twice with PTU washing buffer (100 mM NaH2PO4, 10 mM Tris, 8 M urea, pH 6.8). The ubiquitin-conjugated proteins were analyzed by Western blotting.
Purification of ubiquitinated Myc-tagged β-catenin.
HEK293T cells in 10-cm plates were cotransfected with histidine-tagged ubiquitin (His-Ub) and Myc-tagged β-catenin (5 μg each) for 48 h and then treated with 2.5 μM MG132 for an additional 6 h. The cells were lysed in 1 ml PTU lysis buffer, and the lysates were incubated with Ni-NTA beads at 4°C overnight. The beads were washed with PTU washing buffer and then dialyzed against dialysis buffer (20 mM Tris, pH 7.5, 150 mM NaCl, 0.1% Triton X-100, 0.2% NP-40, 1 μg/ml pepstatin, 3 μg/ml aprotinin, 10 mM NaF, and 2.5 μM MG132).
In vitro deubiquitination assay.
The in vitro deubiquitination assay was performed according to published methods (29, 30). Flag-tagged USP47CD or USP47CDm (5 μg) was transfected into HEK293T cells in 10-cm plates for 48 h. Cell lysates were immunoprecipitated with M2 (anti-Flag antibody [Ab]-conjugated) beads and eluted with 100 μl glycine buffer, pH 3.5, and then neutralized with Tris-HCl buffer, pH 8.0. Purified ubiquitinated β-catenin (100 μl) was incubated with purified USP47CD or USP47CDm (10 μl) for 1 h at room temperature in dialysis solution supplemented with 5% glycerol, 5 mM MgCl2, 2 mM ATP, and 2 mM dithiothreitol (DTT). Ubiquitinated β-catenin on the Ni-NTA beads and deubiquitinated β-catenin in the supernatant were analyzed by Western blotting with an anti-Myc Ab.
Western blot and binding assays.
Bacterially expressed glutathione S-transferase (GST) fusion proteins were purified with Glutathione Sepharopore 4B beads as previously described (4). Cell fractionation, Western blotting, immunoprecipitation, and GST pulldown experiments were performed as previously described (16). Proteins were analyzed with the following antibodies: rabbit anti-USP47 (Bethyl; IHC-00235), rabbit anti-axin2 (Cell Signaling; 2151), rabbit anti-c-myc (Epitomics; 1472-1), rabbit anti-Sox9 (Abcam; ab185966), mouse anti-Flag (Sigma; F1804), rabbit antisurvivin (BioLegend; 614702), and mouse anti-glyceraldehyde-3-phosphate dehydrogenase (anti-GAPDH) (Gentex; GTX627408).
RESULTS
Identification of USP47 as a positive regulator of Wnt signaling.
The human genome has more than 100 deubiquitinase genes (31). In order to identify novel regulators of Wnt signaling, we generated a stable cell line that expresses the TOPFlash reporter. Using this cell line, we screened a human siRNA library and identified USP47 as a putative positive regulator of Wnt signaling.
To validate the function of USP47, we knocked down USP47 in mammalian cells with a different siRNA and analyzed Wnt signaling activity. USP47 siRNA strongly inhibited Wnt-mediated TOPFlash reporter activity (Fig. 1A). Two USP47 shRNAs that effectively knocked down USP47 expression in HEK293T cells also severely blocked the luciferase reporter activity (Fig. 1B). These data indicate that inactivation of USP47 by different approaches inhibits Wnt signaling, suggesting that USP47 is a novel positive regulator of Wnt signaling.
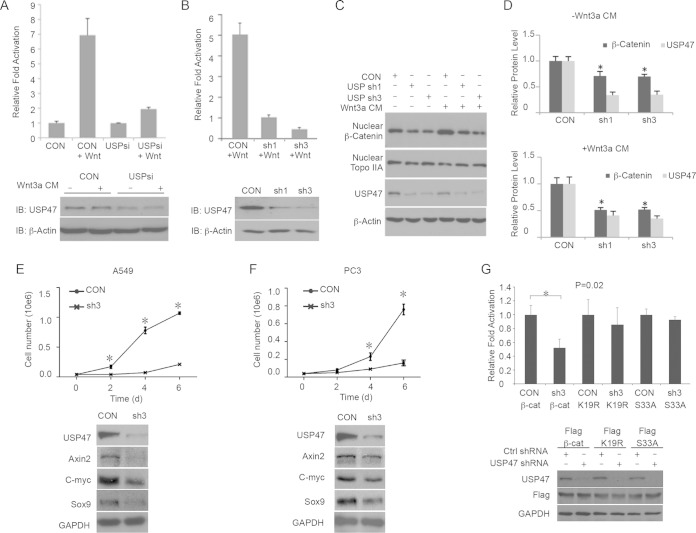
FIG 1.
USP47 regulates Wnt signaling by regulating β-catenin. (A) Effects of USP47 siRNA on the TOPFlash reporter. (Top) TOPFlash and Renilla reporters were cotransfected with USP47 siRNA (USPsi) or control siRNA (CON) into HEK293T cells. After 48 h, the cells were treated or not with Wnt-conditioned medium for an additional 12 h, and the luciferase activities were analyzed and normalized with that of Renilla. The relative luciferase activities compared with the control sample are presented. (Bottom) USP47 siRNA reduced USP47 protein levels in HEK293T cells. (B) Effects of USP47 shRNAs on the TOPFlash reporter. USP47 shRNAs were cotransfected with TOPFlash and Renilla into HEK293T cells. After 48 h, the cells were treated with Wnt-conditioned medium for an additional 12 h, and the luciferase activities were analyzed. (C and D) USP47 regulates β-catenin stability in the nucleus. HEK293T cells were transfected with USP47 shRNAs. The cells were treated or not with Wnt3A-conditioned medium for 6 h before harvesting. USP47 and nuclear β-catenin were analyzed by Western blotting, with β-actin and TopoIIA as loading controls. (D) Statistical analysis of protein levels in three independent experiments. (E and F) (Top) Stable cell lines that express control shRNA or USP47 shRNA were established. USP47 is required for the proliferation of A549 lung cancer cells and PC3 prostate cancer cells (*, P < 0.05). (Bottom) USP47 regulates endogenous Wnt target genes. The expression of Wnt target genes in A549 and PC3 cells was analyzed by Western blotting. USP47 shRNA reduced the protein levels of axin2, c-myc, and Sox-9. (G) Effects USP47 shRNA on wild-type and mutated β-catenin. (Top) TOPFlash and Renilla plasmids were cotransfected with Flag-β-catenin, Flag-β-catenin S33A, or Flag-β-catenin K19R, together with USP47 shRNA or control shRNA, into HEK293T cells. The luciferase activities were analyzed after 48 h. The relative luciferase activities compared with the control sample are presented. (Bottom) USP47 shRNA reduced USP47 protein levels in HEK293T cells. The error bars represent standard deviations.
We wondered whether USP47 regulates β-catenin stability. We knocked down USP47 in HEK293T cells using shRNAs and analyzed the levels of β-catenin. We found that USP47 shRNAs significantly decreased the protein levels of nuclear β-catenin in HEK293T cells with or without Wnt treatment (Fig. 1C and D). To understand the biological functions of USP47, we analyzed the effects of USP47 shRNAs on A549 lung cancer cells and PC3 prostate cancer cells. Knocking down USP47 inhibited the expression of Wnt target genes and significantly repressed the proliferation of these cancer cells (Fig. 1E and F), suggesting that USP47 is required for cancer cell growth. β-Catenin degradation is controlled by N-terminal S/T phosphorylation and K19/K49 ubiquitination; blocking phosphorylation or ubiquitination by mutating these residues may bypass its regulation by the deubiquitinase. Indeed, β-catenin with an S33 or K19 mutation is less sensitive to USP47 shRNA than wild-type β-catenin (Fig. 1G). These results suggest that USP47 regulates Wnt signaling through stabilizing β-catenin.
USP47 directly interacts with β-catenin and regulates β-catenin ubiquitination.
USP47 has a catalytic domain at the N terminus (Fig. 2A). Point mutations and truncations of USP47 were generated as indicated in Fig. 2A. We found that wild-type USP47, but not USP47m, a mutant form of USP47 that contains a C109S mutation at the enzyme activation site (Fig. 2A), rescued Wnt reporter inhibition by USP47 shRNA, suggesting that the catalytic activity of USP47 is required for Wnt signaling (Fig. 2B). To explore the mechanisms by which USP47 regulates β-catenin, we first determined whether USP47 acts in the same protein complex as β-catenin. Since HCT116 cells have high levels of β-catenin, we immunoprecipitated endogenous β-catenin from HCT116 cells and analyzed the physical interaction between β-catenin and USP47 with an anti-USP47 antibody. We found that endogenous USP47 bound endogenous β-catenin (Fig. 2C).
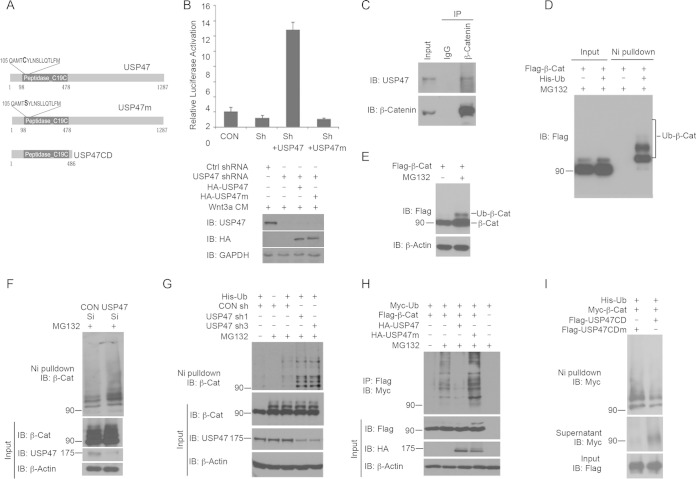
FIG 2.
USP47 regulates β-catenin ubiquitination. (A) Schematic overview of human USP47. The full-length protein and N-terminal catalytic domain and a C109S mutant that lost the catalytic activity are shown. (B) USP47 regulates Wnt signaling through it catalytic domain. TOPFlash and Renilla plasmids were cotransfected with USP47 or USP47m, together with USP47 shRNA or control shRNA, into HEK293T cells. After 48 h, the cells were treated with Wnt-conditioned medium for an additional 12 h, and the luciferase activities were analyzed. The relative luciferase activities compared with the control sample are presented. (C) Ubiquitination of β-catenin. Flag-tagged β-catenin was transfected into HEK293T cells. The cells were treated or not with MG132 for 6 h. Total and ubiquitinated β-catenin were analyzed by Western blotting with an anti-β-Flag Ab. IP, immunoprecipitation; IB, immunoblotting. (D) Detection of ubiquitinated β-catenin. Flag-tagged β-catenin was cotransfected with His-Ub or a control vector into HEK293T cells. The cells were treated with MG132 for 6 h before harvesting. Ubiquitin-conjugated proteins were pulled down with Ni-NTA resins under denaturing conditions. Total and ubiquitinated Flag-tagged β-catenin were analyzed by Western blotting with an anti-Flag Ab. (E) Endogenous USP47 interacts with endogenous β-catenin. β-Catenin in HCT116 cells was immunoprecipitated with an anti-β-catenin Ab or a control IgG. The immunoprecipitated β-catenin and USP47 were analyzed by Western blotting. (F) Knocking down USP47 with siRNA increased β-catenin ubiquitination. HEK293T cells were cotransfected with His-Ub and control siRNA or USP47 siRNA. The cells were treated with MG132 for 6 h before harvesting. The ubiquitinated proteins were pulled down with Ni-NTA beads under denaturing conditions, and β-catenin was analyzed by Western blotting with the anti-β-catenin Ab. (G) Knocking down USP47 with shRNA increased β-catenin ubiquitination. HEK293T cells were cotransfected with His-Ub and control shRNA or USP47 shRNA. Cells were treated or not with MG132 for 6 h before harvesting. The ubiquitinated proteins were pulled down with Ni-NTA beads under denaturing conditions, and β-catenin was analyzed by Western blotting with the anti-β-catenin Ab. (H) USP47 reduced β-catenin ubiquitination in mammalian cells. HEK293T cells were transfected with Flag-tagged β-catenin, together with Myc-Ub, HA-tagged USP47, or C109S-mutated USP47m. Cells were treated or not with MG132 for 6 h before harvesting. The cells were lysed and denatured at 100°C for 10 min. The samples were diluted 10 times and immunoprecipitated with an anti-Flag Ab. Ubiquitinated Flag-tagged β-catenin was detected by Western blotting with an anti-Myc Ab (top). The protein levels of overexpressed β-catenin and USP47 in cell lysates were analyzed by Western blotting with an anti-Flag Ab and an anti-HA Ab, respectively. (I) In vitro deubiquitination assay for β-catenin. (Bottom) A Flag-tagged USP47CD or USP47CDm construct was transfected into HEK293T cells. USP47CD and USP47CDm proteins were purified by immunoprecipitation with anti-Flag resin. (Top and middle) His-Ub and myc-tagged β-catenin were cotransfected into HEK293T cells; ubiquitinated β-catenin was purified by Ni-NTA resins under denaturing condition and then dialyzed overnight. Equal amounts of purified ubiquitinated β-catenin were incubated with purified USP47CD or control USP47CDm. The remaining ubiquitinated β-catenin on the Ni-NTA resins (top) and deubiquitinated β-catenin released from the beads (middle) were analyzed by Western blotting. The error bars represent standard deviations. Masses of protein standards are given in kilodaltons.
In the absence of Wnt, wild-type β-catenin is degraded through the ubiquitination/proteasome pathway. In our previous studies, when HEK293T cells were treated with MG132, a proteasome inhibitor, the levels of endogenous and overexpressed β-catenin, including ubiquitinated β-catenin, were increased (16). To precisely detect ubiquitinated β-catenin, His-Ub and Flag-tagged β-catenin (Flag-β-Cat) were cotransfected into HEK293T cells. The cells were treated with MG132 to stabilize the ubiquitinated β-catenin. Ubiquitinated proteins were pulled down by Ni-NTA beads under denaturing conditions and analyzed by Western blotting (28). The mobility shift indicates the specific ubiquitination of the protein (Fig. 2D and E).
To determine if USP47 controls β-catenin ubiquitination, we examined the levels of β-catenin ubiquitination when USP47 was inactivated. We knocked down USP47 in HEK293T cells using siRNA and found that the levels of ubiquitinated β-catenin were increased compared with the sample treated with control siRNA (Fig. 2F). Similarly, knocking down USP47 with shRNAs also increased the levels of β-catenin ubiquitination (Fig. 2G). Expression of wild-type USP47 decreased β-catenin ubiquitination (Fig. 2H). However, USP47m increased β-catenin ubiquitination (Fig. 2H). These results suggest that the enzymatic activity of USP47 is required for its function in regulating β-catenin ubiquitination. USP47m may act as a dominant-negative mutant. To further determine if USP47 directly catalyzes β-catenin deubiquitination, we performed an in vitro deubiquitination assay. Ubiquitinated β-catenin purified from HEK293T cells overexpressing Myc-β-catenin was incubated with the Flag-tagged USP47 catalytic domain (Flag-USP47CD) or its mutant (Flag-USP47CDm) purified separately from HEK293T cells. Compared with the control, USP47CDm, USP47CD decreased the levels of ubiquitinated β-catenin (Fig. 2I, top) and increased the levels of deubiquitinated β-catenin released into the supernatant from the Ni-NTA beads (Fig. 2I, middle), suggesting that USP47 is an enzyme that directly catalyzes β-catenin deubiquitination.
β-TrCP/USP47 regulate USP47 ubiquitination.
β-Catenin ubiquitination is mediated by β-TrCP; it has been indicated that USP47 also interacts with β-TrCP (32), but the function of this interaction in the Wnt pathway is not clear. We found that the catalytic domain of USP47 was sufficient for β-TrCP binding in an immunoprecipitation assay carried out with HEK293T cells cotransfected with Flag-USP47CD and Myc-β-TrCP (Fig. 3A). We noted that the protein levels of USP47m were lower than the levels of wild-type USP47 (Fig. 2E). Also, in HEK293T cells transfected with USP47m and His-Ub, the protein levels of USP47m were decreased by ubiquitin overexpression and increased by MG132 treatment (Fig. 3B). As a control, the protein levels of green fluorescent protein (GFP) were not affected (Fig. 3B). These results indicate that USP47m is less stable than the wild-type USP47 and that the USP47m protein could be regulated by proteasome-mediated degradation.
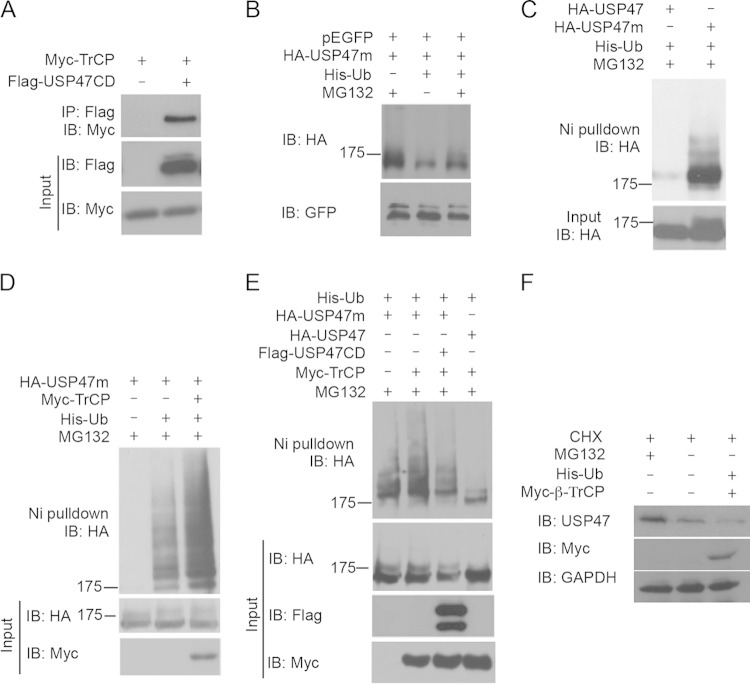
FIG 3.
β-TrCP regulates USP47 ubiquitination. (A) USP47 interacts with β-TrCP. HEK293T cells were transfected with Myc-tagged β-TrCP, together with Flag-tagged USP47CD or a control vector. β-TrCP was immunoprecipitated from cell lysates with an anti-Flag Ab. β-TrCP and USP47CD were analyzed by Western blotting with anti-Myc and anti-Flag Abs. (B) Stability of USP47m. HEK293T cells were transfected with HA-tagged USP47m and pEGFP, with or without His-Ub. Cells were treated or not with MG132 for 6 h before harvesting. The protein levels of USP47m were analyzed by Western blotting, with GFP as a control. (C) Ubiquitination of USP47. HEK293T cells were transfected with His-Ub, together with HA-tagged USP47 or USP47m constructs. Cells were treated with MG132 for 6 h before harvesting and then lysed in PTU buffer. Ubiquitin-conjugated proteins were pulled down by Ni-NTA. Ubiquitinated USP47 (top) and total USP47 (bottom) proteins were analyzed by Western blotting with an anti-HA Ab. (D) β-TrCP regulates USP47 ubiquitination. HEK293T cells were transfected with HA-tagged USP47m, together with or without His-Ub or Myc-tagged β-TrCP. The cells were treated with MG132 for 6 h before harvesting. The cells were lysed in PTU buffer, and ubiquitin-conjugated proteins were pulled down with Ni-NTA. Ubiquitinated USP47 (top) and total USP47 (middle) proteins were analyzed by Western blotting with an anti-HA Ab. (Bottom) β-TrCP was analyzed with an anti-Myc Ab. (E) The catalytic domain of USP47 is sufficient for deubiquitinating USP47. HEK293T cells were transfected with His-Ub. Myc-tagged β-TrCP, HA-tagged USP47, USP47m, and Flag-tagged USP47CD were transfected as indicated. The cells were treated with MG132 for 6 h and lysed in PTU buffer. Ubiquitin-conjugated proteins were pulled down by Ni-NTA. (Top) Ubiquitinated USP47 and total USP47 proteins were analyzed by Western blotting with an anti-HA Ab. (Bottom) β-TrCP proteins were analyzed with an anti-Myc Ab. USP47CD was analyzed with an anti-Flag Ab. (F) β-TrCP regulates USP47 degradation. HEK293T cells were transfected with control vector or Myc-β-TrCP plus His-Ub. After 36 h, cells were treated with CHX (40 μM) with or without MG132 (25 μM) for 6 h before harvesting. The protein levels of USP47 and myc-tagged β-TrCP were analyzed by Western blotting, with GAPDH as a loading control.
Since USP47m contains a C109S mutation at the catalytic active site, we hypothesized that the wild-type USP47 could decrease its own ubiquitination and resist ubiquitination/degradation while USP47m lost the deubiquitinase activity and was more sensitive to proteasome degradation. To test this hypothesis, His-Ub and USP47m were cotransfected into HEK293T cells. The ubiquitinated proteins were pulled down with Ni-NTA beads, and USP47 was analyzed by Western blotting. We found that USP47m exhibited a mobility shift, indicative of ubiquitination (Fig. 3C). In contrast, wild-type USP47 never showed such a mobility shift (Fig. 3C). These data suggest that USP47 regulates it own ubiquitination.
To test if the ubiquitination of USP47 could be regulated by β-TrCP, we transfected His-Ub, hemagglutinin-tagged USP47m (HA-USP47m), and Myc-β-TrCP into HEK293T cells and performed the Ni-NTA pulldown assay. We found that the overexpression of β-TrCP increased USP47m ubiquitination (Fig. 3D), indicating that β-TrCP is the ubiquitin ligase for USP47. To further test the enzymatic activity of USP47 in regulating its own ubiquitination, USP47CD was cotransfected with USP47m, His-Ub, and β-TrCP into HEK293T cells. We found that both USP47 and USP47CD decreased the ubiquitination of USP47m (Fig. 3E), suggesting that the catalytic domain of USP47 is sufficient to catalyze the deubiquitination of USP47m. These results further indicate that USP47 could regulate its own ubiquitination and that USP47m lost the deubiquitinase activity with the consequent elevation of ubiquitination. To further validate the role of ubiquitination in USP47 degradation, we treated HEK293T cells with MG132 and cycloheximide. MG132 stabilized endogenous USP47 and expression of β-TrCP, and His-Ub further enhanced USP47 degradation (Fig. 3F), suggesting that USP47 is degraded by the β-TrCP-mediated ubiquitination/proteasome pathway.
Identification of an atypical β-TrCP-binding motif in USP47.
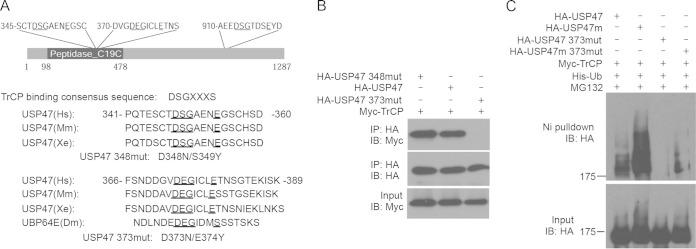
The consensus β-TrCP-binding site for β-catenin is DSGXXS (conserved amino acids are in boldface), where S is a phosphoserine or phosphothreonine (3). There is a consensus β-TrCP-binding site in the C terminus of USP47 (DSGTDS; amino acids [aa] 913 to 918) (Fig. 3F). However, deletion of the C terminus, which contains amino acids 913 to 918, did not affect β-TrCP binding (Fig. 4A). Since glutamic acid (E) can mimic phosphoserine/phosphothreonine, we identified two potential β-TrCP-binding sites in the catalytic domain of USP47 (DSGNEAE at amino acids 348 to 354 and DEGICLE at amino acids 373 to 379) (Fig. 4A). The latter site is conserved in UBP64E, the Drosophila homolog of USP47. We mutated these sites and analyzed the interaction between each mutant and β-TrCP using an immunoprecipitation assay. We found that USP47CD with 348-349 mutations can still bind β-TrCP (Fig. 4B), whereas 373-374 mutations abolished the binding (Fig. 4B), indicating that 373-374 in USP47 is responsible for its interaction with β-TrCP. These findings suggest that β-TrCP mediates the ubiquitination of USP47 through optimized motifs.
FIG 4.
Identification of the β-TrCP-binding motif in USP47. (A) Schematic overview of the potential β-TrCP-binding motifs in USP47. Mutations at two potential β-TrCP-binding motifs in the catalytic domain of USP47 are underlined. (B) The 373-to-379 motif, but not the 348-to 354 motif, of USP47 is required for β-TrCP binding. HEK293T cells were transfected with Myc-tagged β-TrCP, together with HA-tagged USP47 constructs with or without mutations. (Top) The USP47 proteins were immunoprecipitated with an anti-HA Ab, and the interacting β-TrCP was analyzed by Western blotting with an anti-Myc Ab. (Middle and bottom) The expression levels of USP47 (middle) and β-TrCP (bottom) were analyzed with anti-HA and anti-Myc Abs. (C) The β-TrCP-binding motif is required for USP47 ubiquitination. HEK293T cells were transfected with His-Ub, together with different HA-tagged USP47 constructs. The cells were treated with MG132 for 6 h before harvesting and lysed in PTU. Ubiquitin-conjugated proteins were pulled down by Ni-NTA. Ubiquitinated USP47 (top) and total USP47 (bottom) proteins were analyzed by Western blotting with an anti-HA Ab.
To further characterize the role of these potential β-TrCP-binding sites in USP47 ubiquitination, we transfected wild-type or mutant forms of USP47 with His-Ub and β-TrCP into HEK293T cells. USP47 ubiquitination was analyzed by Ni-NTA pulldown and Western blotting. Wild-type USP47 had a low level of ubiquitination even in the presence of β-TrCP (Fig. 4C). USP47m had a much higher level of ubiquitination (Fig. 4C). USP47 with 373-374 mutations had no detectable ubiquitination (Fig. 4C). These results suggest that the interaction between aa 373 to 379 of USP47 and β-TrCP is essential for β-TrCP-mediated ubiquitination of USP47.
Interactions of USP47 with β-catenin and β-TrCP.
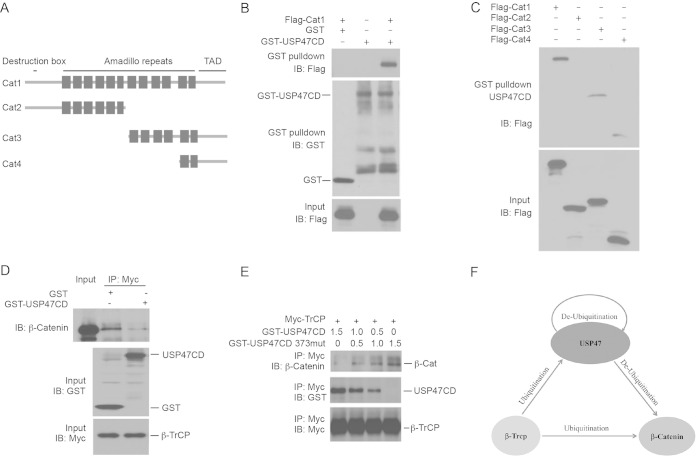
β-Catenin interacts with multiple proteins through its N-terminal destruction domain, central armadillo domain, and C-terminal transcription activation domain (Fig. 5A). USP47CD contains the N-terminal catalytic domain and is sufficient to interact with β-catenin (Fig. 2A and 5B). To determine which domain of β-catenin interacts with USP47, we generated several deletion mutants of β-catenin (Fig. 5A). We analyzed the binding between GST-USP47CD and the β-catenin mutants and found that the C terminus of β-catenin containing two armadillo repeats (Fig. 5A, Cat4) was sufficient for USP47 binding (Fig. 5C).
FIG 5.
Functional interactions of USP47, β-catenin, and β-TrCP. (A) Schematic overview of wild-type and truncated β-catenin constructs. TAD, transcription activation domain. (B) GST-USP47CD interacts with β-catenin. HEK293T cells were transfected with Flag-tagged full-length β-catenin. The cell lysates were incubated with purified GST-USP47CD proteins. GST47CD was pulled down by glutathione-Sepharose 4B beads (GST). The interacting β-catenin was analyzed by Western blotting with an anti-Flag Ab. (C) USP47 interacts with the C terminus of β-catenin. HEK293T cells were transfected with Flag-tagged Cat1, -2, -3, and -4. The cell lysates were incubated with GST-USP47CD fusion proteins purified from bacteria. The interacting β-catenin proteins were analyzed by Western blotting with an anti-Flag Ab. (D) USP47 inhibits the interaction between β-TrCP and β-catenin. The lysates of HEK 293T cells transfected with Myc-tagged β-TrCP were mixed with the lysates of SW480 cells containing high levels of β-catenin. Purified GST or GST-USP47CD protein was added to the mixture. The β-TrCP complex was immunoprecipitated with an anti-Myc Ab, and β-catenin was analyzed by Western blotting with an anti-β-catenin Ab. Purified GST fusion proteins were analyzed by Western blotting with an anti-GST Ab. The expression of β-TrCP was analyzed with an anti-Myc Ab. (E) Wild-type USP47CD, but not the 373-to-379 mutant, inhibits binding between β-TrCP and β-catenin. The lysates of HEK 293T cells transfected with Myc-tagged β-TrCP were mixed with the lysates of SW480 cells. Different amounts of purified wild-type or mutant GST-USP47CD were added to the mixture. (Top) The β-TrCP complex was immunoprecipitated with an anti-Myc Ab, and β-catenin was analyzed by Western blotting with an anti-β-catenin Ab. (Middle) GST-USP47CD proteins in the β-TrCP complex were analyzed with an anti-GST Ab. (Bottom) β-TrCP proteins were analyzed with an anti-Myc Ab. (F) Interactions of USP47, β-TrCP, and β-catenin.
Since both β-catenin and USP47 bind β-TrCP, the proteins may compete with each other for the binding. To test this possibility, we carried out experiments to examine the interactions among the proteins. Myc-tagged β-TrCP was transfected into HEK293T cells. The cell lysates were incubated with purified GST or GST-USP47CD protein from bacteria. The β-TrCP complex was immunoprecipitated with the anti-Myc Ab, and β-catenin was analyzed by Western blotting. We found that USP47CD inhibited the binding between β-TrCP and β-catenin (Fig. 5D). To further evaluate our findings, we performed the assay with different concentrations of purified GST-USP47 and a GST–USP47–373-374 mutant. We found that wild-type USP47CD, which interacts with β-TrCP, inhibited the interaction between β-catenin and β-TrCP (Fig. 5E). In contrast, the mutant USP47CD, which cannot bind β-TrCP, did not block the binding between β-catenin and β-TrCP (Fig. 5E). These results suggest that USP47 may inhibit β-catenin ubiquitination by multiple mechanisms: deubiquitinating β-catenin and interfering with the interactions between β-catenin and β-TrCP (Fig. 5F).
USP47/UBP64E regulates Wnt signaling in Drosophila.
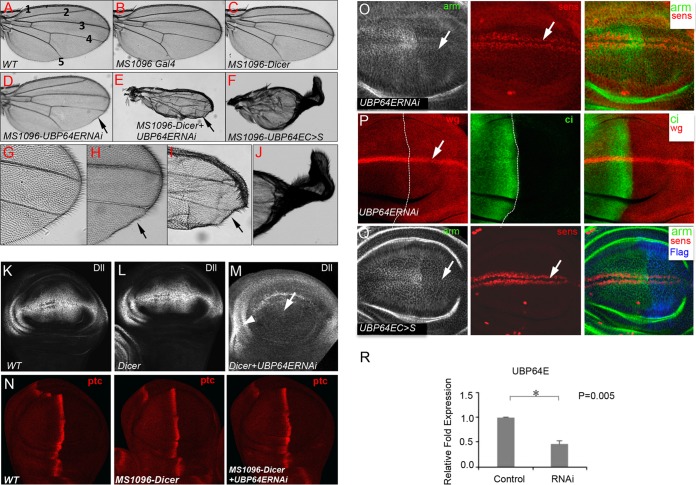
To examine the physiological function of USP47 in vivo, we turned to the Drosophila wing model. We performed both loss- and gain-of-function studies in Drosophila wings to examine the regulation of β-catenin/Arm by USP47/UBP64E. We found that the expression of UBP64E RNAi by MS1096 Gal4 induced a wing margin bristle phenotype, which is a partial loss of the Wingless/Wnt phenotype (Fig. 6, compare panel D with wild-type wings in panels A to C). The coexpression of Dicer with UBP64E RNAi dramatically enhanced this phenotype and severely disrupted the wing morphology (Fig. 6, compare panels E and F with wild-type wings in panels A and G, respectively). The wing margin phenotype induced by UBP64E RNAi is unlikely to be due to an off-target effect because another transgenic RNAi line (v103743) targeting a nonoverlapping region of the UBP64E coding sequence produced a similar wing margin phenotype in the adult wing. Consistently, the overexpression of a UBP64EC >S mutant by MS1096 Gal4 caused severe morphology change of the wings (Fig. 6F and J), suggesting that, like other deubiquitinases, mutating the catalytic Cys in the catalytic domain of UBP64E leads to a dominant-negative form.
FIG 6.
UBP64E positively regulates Wnt/Wg by stabilizing β-catenin/Arm. (A) Wild-type adult wing showing interveins 1 to 5. (B and C) Wings from flies expressing either MS1096 Gal4 alone or along with UAS-Dicer exhibit wild-type structures. (D) Wing from flies expressing UAS-UBP64E RNAi by MS1096 Gal4 showing a wing margin bristle phenotype, indicated by the arrow. (E) Wing from a fly coexpressing UAS-UBP64E RNAi (VDRC stock no. v26027) with UAS-Dicer by MS1096 Gal4 showing a severe wing phenotype with the loss of margin bristles, indicated by the arrow. (F) Wing from a fly expressing UAS-UBP64EC>S by MS1096 Gal4 exhibiting a severe phenotype. (G) High magnification of wild-type wing showing normal margin bristles. (H) High magnification of panel D showing wing margin bristle phenotype. (I) High magnification of panel E showing the loss of bristles on the wing margin. (J) High magnification of panel F. (K to M) Wing discs with the indicated genotypes were stained for Dll. (M) Coexpression of UAS-UBP64E RNAi with UAS-Dicer blocked Dll expression in the wing pouch (arrow). Dll staining outside the wing pouch served as a control (arrowhead). (N) Wing discs with the indicated genotypes were immunostained with the anti-Ptc antibody. Inactivation of UBP64E by RNAi does not regulate Ptc expression. (O) A wing disc expressing UBP64ERNAi by en-Gal4 was immunostained with the indicated antibodies. Inactivation of UBP64E by RNAi blocked the accumulation of Arm (left, arrow) and attenuated the expression of Sens (middle, arrow). (P) A wing disc expressing UBP64ERNAi by en-Gal4 was immunostained with the indicated antibodies. Inactivation of UBP64E by RNAi attenuated the expression of Wingless (Wg) (left, arrow, and right). (Q) A wing disc expressing Flag-UBP64EC >S by en-Gal4 was immunostained with the indicated antibodies. UBP64EC >S has dominant-negative activity and blocks the accumulation of Arm (left, arrow) and attenuated the expression of Sens (middle, arrow). (Right) The Flag staining marks the en-Gal4 expression domain. (R) The effects of UBP64E RNAi on UBP64E mRNA levels were analyzed by qPCR. UBP64E mRNA levels were significantly decreased in RNAi-treated discs. The error bars represent standard deviations.
To examine the role of UBP64E in regulating Wnt target genes, UBP64E RNAi was coexpressed with Dicer in wing discs by MS1096 Gal4. We found that the inactivation of UBP64E abolished the expression of the Wnt Distalless (Dll) target gene (Fig. 6, compare panel M with panels K and L) but did not affect the expression of Patched (Ptc), which is a target gene in Hedgehog signaling (Fig. 6N), suggesting the pathway specificity of UBP64E.
We next examined whether UBP64E regulates the stability of Arm in wing discs. As shown in Fig. 6O, the expression of UBP64E RNAi by the posterior cell-specific Engrailed (en)-Gal4 inhibited Arm accumulation in these cells (Fig. 6O, left), leading to the attenuated expression of Senseless (Sens) (Fig. 6O, middle), and Wingless (Fig. 6P, left). We also found that the overexpression of UBP64EC >S blocked the accumulation of Arm and attenuated the expression of Sens (Fig. 6Q). The efficiency of UBP64E RNAi was analyzed by quantitative PCR (qPCR) (Fig. 6R). These data suggest that UBP64E is required for Wingless/Wnt signal transduction in Drosophila wing discs. The levels of Arm accumulation in wing discs correlated with the adult wing phenotypes when UBP64E RNAi or UBP64EC >S was expressed, suggesting that the adult wing phenotypes were caused, at least in part, by UBP64E-mediated regulation of Arm.
DISCUSSION
β-Catenin degradation is tightly regulated by Wnt signaling in normal cells. Mutations in the Wnt pathway are associated with human cancers and many other diseases. β-Catenin is degraded by a ubiquitin/proteasome pathway that is mediated by the ubiquitin protein ligase β-TrCP. Our collaborative efforts led to the identification of a new regulator for β-catenin ubiquitination. We found that the deubiquitinase USP47/UBP64E interacts with and regulates β-catenin ubiquitination and degradation. The ubiquitination of USP47 is also regulated by β-TrCP/USP47. In vivo studies in the Drosophila wing disc demonstrated that USP47/UBP64E regulates the accumulation of β-catenin and the expression of Wnt target genes. Taken together, the data show that USP47/UBP64E directly deubiquitinates β-catenin and is a novel positive regulator for β-catenin stabilization.
USP47 was first identified as a β-TrCP-binding protein that regulates Cdc25A expression and cell survival (32). The effects of USP47 on other β-TrCP substrates, such as IκBα and β-catenin, are not clear. Since the majority of β-catenin proteins are localized on the cell membrane and only cytoplasmic and nuclear β-catenins are sensitive to degradation, it is difficult to examine the change in total β-catenin levels. We found that USP47 RNAi decreased the levels of nuclear β-catenin. More importantly, USP47/UBP64E RNAi decreased Arm levels in Drosophila, suggesting that USP47 regulates β-catenin stability both in vitro and in vivo.
Previous studies did not detect the effects of β-TrCP on USP47 (32). In our study, HEK293T cells were transfected with His-Ub, followed by immunoprecipitation with Ni-NTA beads, which was likely a more sensitive and reliable approach. Our results clearly demonstrated that β-TrCP regulates USP47 ubiquitination, which may lead to USP47 degradation. Since wild-type USP47 has deubiquitinase activity, it is difficult to detect the ubiquitination of wild-type USP47. It is much easier to detect the ubiquitination of mutant USP47, which loses the deubiquitinase activity.
In some cases, deubiquitinase binds its substrate through a ubiquitin ligase. For example, Mdm2 mediates the binding of HAUSP and P53 (33). The binding between USP47 and β-catenin is not β-TrCP dependent, because β-TrCP binds the N terminus of β-catenin and USP47 binds the C terminus of β-catenin. We found that USP47 has a novel β-TrCP recognition motif (DEGXXXE). This is not a typical β-TrCP-binding motif (DSGXXS) found previously (3). Several nontypical β-TrCP-binding motifs have been reported. For example, β-TrCP binds a DDGXXD motif in Cdc25A and Cdc25B (34), suggesting that acidic residues can replace phosphoserine/phosphothreonine in the consensus β-TrCP-binding site. The β-TrCP-binding motif is located in a predicted loop of USP47 (35) and is thus likely to be surface exposed and poised to mediate protein interactions.
The Wnt pathway is conserved from invertebrates to vertebrates (2). The role of Wnt/Wingless signaling is well established in Drosophila. Many key components of the Wnt pathway, such as Wingless (Wg), Armadillo (β-catenin), Zester-white 3 (GSK-3), and Slimb (β-TrCP), were identified or characterized in Drosophila (36–40). In Drosophila, Wingless promotes the accumulation and nuclear translocation of Armadillo, leading to the expression of its target genes, such as wg, dll, and sens (41, 42). Our previous work has demonstrated that CKIα regulates Drosophila embryonic development by regulating β-catenin phosphorylation and degradation (3). We have also demonstrated that PP2A (Twins) regulates Drosophila wing development by regulating β-catenin dephosphorylation (18). The Drosophila model provides a powerful tool to study Wnt/Wg signaling in vivo. In this study, in vivo screening identified several RNAi lines, including USP8, causing wing phenotypes, such as small wing, sick wing, or abnormal margin bristles. However, only UBP64E RNAi gave the most obvious Wnt phenotype. It has been reported that the expression of UBP64E-RNAi under the ubiquitous Gal4 or panneuronal Gal4 caused a drooping-wing phenotype (43). In our study, to examine the wingless phenotype, we used the wing-specific MS1096 Gal4. We analyzed the levels of Arm and the expression of Wnt target genes, and only UBP64E RNAi decreased the levels of both Arm and its targets (Fig. 6).
In conclusion, we have identified a β-catenin deubiquitinase that regulates Wnt signaling in vitro and in vivo. Recently, it has been reported that the RNF220/USP7 complex can also deubiquitinate β-catenin (44). USP7 is closely related to USP47; it will be interesting to investigate whether they play redundant roles in β-catenin regulation. Although our studies suggest that USP47 regulates β-catenin deubiquitination, we cannot rule out the possibility that USP47 also regulates other components in the Wnt pathway, and the precise mechanism of β-catenin regulation by USP47 needs further study. In addition, USP47 has been reported to be a deubiquitinating enzyme for DNA polymerase β that regulates DNA repair and genome integrity (45). In Drosophila, UBP64E also controls cell fate by regulating the transcriptional repressor tramtrack (46). As demonstrated in Fig. 1E and F, knockdown of USP47 inhibited cancer cell proliferation, probably by affecting multiple targets of USP47. Given the roles of USP47 in Wnt signaling, cell survival, and DNA repair, USP47 may play an important role in cancer biology and may become a novel therapeutic target for anticancer agents.
ACKNOWLEDGMENTS
We are grateful to Randy Moon for the 8× TOPFlash plasmid, Hugo Bellen for the anti-Sens antibody, and Grace Boekhoff-Falk for the anti-Dll antibody. We thank Ruohan Xia and Xi Chen for help with initiation of the project and generation of some of the constructs and transgenic fly lines. We thank the VDRC for RNAi flies and DSHB for antibodies.
J.J. was supported by R01GM079684 and C.L. was supported by R01CA172379 from the National Institutes of Health.
REFERENCES
- 1.Giles RH, van Es JH, Clevers H. 2003. Caught up in a Wnt storm: Wnt signaling in cancer. Biochim Biophys Acta 1653:1–24. [DOI] [PubMed] [Google Scholar]
- 2.van Amerongen R, Nusse R. 2009. Towards an integrated view of Wnt signaling in development. Development 136:3205–3214. doi: 10.1242/dev.033910. [DOI] [PubMed] [Google Scholar]
- 3.Liu C, Li Y, Semenov M, Han C, Baeg GH, Tan Y, Zhang Z, Lin X, He X. 2002. Control of beta-catenin phosphorylation/degradation by a dual-kinase mechanism. Cell 108:837–847. doi: 10.1016/S0092-8674(02)00685-2. [DOI] [PubMed] [Google Scholar]
- 4.Liu C, Kato Y, Zhang Z, Do VM, Yankner BA, He X. 1999. Beta-Trcp couples beta-catenin phosphorylation-degradation and regulates Xenopus axis formation. Proc Natl Acad Sci U S A 96:6273–6278. doi: 10.1073/pnas.96.11.6273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hart M, Concordet JP, Lassot I, Albert I, del los Santos R, Durand H, Perret C, Rubinfeld B, Margottin F, Benarous R, Polakis P. 1999. The F-box protein beta-TrCP associates with phosphorylated beta-catenin and regulates its activity in the cell. Curr Biol 9:207–210. doi: 10.1016/S0960-9822(99)80091-8. [DOI] [PubMed] [Google Scholar]
- 6.Spencer E, Jiang J, Chen ZJ. 1999. Signal-induced ubiquitination of IkappaBalpha by the F-box protein Slimb/beta-TrCP. Genes Dev 13:284–294. doi: 10.1101/gad.13.3.284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Winston JT, Strack P, Beer-Romero P, Chu CY, Elledge SJ, Harper JW. 1999. The SCFbeta-TRCP-ubiquitin ligase complex associates specifically with phosphorylated destruction motifs in IkappaBalpha and beta-catenin and stimulates IkappaBalpha ubiquitination in vitro. Genes Dev 13:270–283. doi: 10.1101/gad.13.3.270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tamai K, Semenov M, Kato Y, Spokony R, Liu C, Katsuyama Y, Hess F, Saint-Jeannet JP, He X. 2000. LDL-receptor-related proteins in Wnt signal transduction. Nature 407:530–535. doi: 10.1038/35035117. [DOI] [PubMed] [Google Scholar]
- 9.Tamai K, Zeng X, Liu C, Zhang X, Harada Y, Chang Z, He X. 2004. A mechanism for Wnt coreceptor activation. Mol Cell 13:149–156. doi: 10.1016/S1097-2765(03)00484-2. [DOI] [PubMed] [Google Scholar]
- 10.Zeng X, Tamai K, Doble B, Li S, Huang H, Habas R, Okamura H, Woodgett J, He X. 2005. A dual-kinase mechanism for Wnt co-receptor phosphorylation and activation. Nature 438:873–877. doi: 10.1038/nature04185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kramps T, Peter O, Brunner E, Nellen D, Froesch B, Chatterjee S, Murone M, Zullig S, Basler K. 2002. Wnt/wingless signaling requires BCL9/legless-mediated recruitment of pygopus to the nuclear beta-catenin-TCF complex. Cell 109:47–60. doi: 10.1016/S0092-8674(02)00679-7. [DOI] [PubMed] [Google Scholar]
- 12.Kinzler KW, Vogelstein B. 1996. Lessons from hereditary colorectal cancer. Cell 87:159–170. doi: 10.1016/S0092-8674(00)81333-1. [DOI] [PubMed] [Google Scholar]
- 13.Polakis P. 2000. Wnt signaling and cancer. Genes Dev 14:1837–1851. [PubMed] [Google Scholar]
- 14.Ha NC, Tonozuka T, Stamos JL, Choi HJ, Weis WI. 2004. Mechanism of phosphorylation-dependent binding of APC to beta-catenin and its role in beta-catenin degradation. Mol Cell 15:511–521. doi: 10.1016/j.molcel.2004.08.010. [DOI] [PubMed] [Google Scholar]
- 15.Xing Y, Clements WK, Le Trong I, Hinds TR, Stenkamp R, Kimelman D, Xu W. 2004. Crystal structure of a beta-catenin/APC complex reveals a critical role for APC phosphorylation in APC function. Mol Cell 15:523–533. doi: 10.1016/j.molcel.2004.08.001. [DOI] [PubMed] [Google Scholar]
- 16.Yang J, Zhang W, Evans PM, Chen X, He X, Liu C. 2006. Adenomatous polyposis coli (APC) differentially regulates beta-catenin phosphorylation and ubiquitination in colon cancer cells. J Biol Chem 281:17751–17757. doi: 10.1074/jbc.M600831200. [DOI] [PubMed] [Google Scholar]
- 17.Barker N, Clevers H. 2006. Mining the Wnt pathway for cancer therapeutics. Nat Rev Drug Discov 5:997–1014. doi: 10.1038/nrd2154. [DOI] [PubMed] [Google Scholar]
- 18.Zhang W, Yang J, Liu Y, Chen X, Yu T, Jia J, Liu C. 2009. PR55 alpha, a regulatory subunit of PP2A, specifically regulates PP2A-mediated beta-catenin dephosphorylation. J Biol Chem 284:22649–22656. doi: 10.1074/jbc.M109.013698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tran H, Hamada F, Schwarz-Romond T, Bienz M. 2008. Trabid, a new positive regulator of Wnt-induced transcription with preference for binding and cleaving K63-linked ubiquitin chains. Genes Dev 22:528–542. doi: 10.1101/gad.463208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mukai A, Yamamoto-Hino M, Awano W, Watanabe W, Komada M, Goto S. 2010. Balanced ubiquitylation and deubiquitylation of Frizzled regulate cellular responsiveness to Wg/Wnt. EMBO J 29:2114–2125. doi: 10.1038/emboj.2010.100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tauriello DV, Haegebarth A, Kuper I, Edelmann MJ, Henraat M, Canninga-van Dijk MR, Kessler BM, Clevers H, Maurice MM. 2010. Loss of the tumor suppressor CYLD enhances Wnt/beta-catenin signaling through K63-linked ubiquitination of Dvl. Mol Cell 37:607–619. doi: 10.1016/j.molcel.2010.01.035. [DOI] [PubMed] [Google Scholar]
- 22.Zhao B, Schlesiger C, Masucci MG, Lindsten K. 2009. The ubiquitin specific protease 4 (USP4) is a new player in the Wnt signalling pathway. J Cell Mol Med 13:1886–1895. doi: 10.1111/j.1582-4934.2008.00682.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Huang X, Langelotz C, Hetfeld-Pechoc BK, Schwenk W, Dubiel W. 2009. The COP9 signalosome mediates beta-catenin degradation by deneddylation and blocks adenomatous polyposis coli destruction via USP15. J Mol Biol 391:691–702. doi: 10.1016/j.jmb.2009.06.066. [DOI] [PubMed] [Google Scholar]
- 24.Lui TT, Lacroix C, Ahmed SM, Goldenberg SJ, Leach CA, Daulat AM, Angers S. 2011. The ubiquitin-specific protease USP34 regulates axin stability and Wnt/beta-catenin signaling. Mol Cell Biol 31:2053–2065. doi: 10.1128/MCB.01094-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dietzl G, Chen D, Schnorrer F, Su KC, Barinova Y, Fellner M, Gasser B, Kinsey K, Oppel S, Scheiblauer S, Couto A, Marra V, Keleman K, Dickson BJ. 2007. A genome-wide transgenic RNAi library for conditional gene inactivation in Drosophila. Nature 448:151–156. doi: 10.1038/nature05954. [DOI] [PubMed] [Google Scholar]
- 26.Jia H, Liu Y, Yan W, Jia J. 2009. PP4 and PP2A regulate Hedgehog signaling by controlling Smo and Ci phosphorylation. Development 136:307–316. doi: 10.1242/dev.030015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Evans PM, Liu C. 2005. SiteFind: a software tool for introducing a restriction site as a marker for successful site-directed mutagenesis. BMC Mol Biol 6:22. doi: 10.1186/1471-2199-6-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ling R, Colon E, Dahmus ME, Callis J. 2000. Histidine-tagged ubiquitin substitutes for wild-type ubiquitin in Saccharomyces cerevisiae and facilitates isolation and identification of in vivo substrates of the ubiquitin pathway. Anal Biochem 282:54–64. doi: 10.1006/abio.2000.4586. [DOI] [PubMed] [Google Scholar]
- 29.Dupont S, Mamidi A, Cordenonsi M, Montagner M, Zacchigna L, Adorno M, Martello G, Stinchfield MJ, Soligo S, Morsut L, Inui M, Moro S, Modena N, Argenton F, Newfeld SJ, Piccolo S. 2009. FAM/USP9x, a deubiquitinating enzyme essential for TGFbeta signaling, controls Smad4 monoubiquitination. Cell 136:123–135. doi: 10.1016/j.cell.2008.10.051. [DOI] [PubMed] [Google Scholar]
- 30.Strayhorn WD, Wadzinski BE. 2002. A novel in vitro assay for deubiquitination of I kappa B alpha. Arch Biochem Biophys 400:76–84. doi: 10.1006/abbi.2002.2760. [DOI] [PubMed] [Google Scholar]
- 31.Sowa ME, Bennett EJ, Gygi SP, Harper JW. 2009. Defining the human deubiquitinating enzyme interaction landscape. Cell 138:389–403. doi: 10.1016/j.cell.2009.04.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Peschiaroli A, Skaar JR, Pagano M, Melino G. 2010. The ubiquitin-specific protease USP47 is a novel beta-TRCP interactor regulating cell survival. Oncogene 29:1384–1393. doi: 10.1038/onc.2009.430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Brooks CL, Li M, Hu M, Shi Y, Gu W. 2007. The p53-Mdm2-HAUSP complex is involved in p53 stabilization by HAUSP. Oncogene 26:7262–7266. doi: 10.1038/sj.onc.1210531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kanemori Y, Uto K, Sagata N. 2005. Beta-TrCP recognizes a previously undescribed nonphosphorylated destruction motif in Cdc25A and Cdc25B phosphatases. Proc Natl Acad Sci U S A 102:6279–6284. doi: 10.1073/pnas.0501873102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ye Y, Scheel H, Hofmann K, Komander D. 2009. Dissection of USP catalytic domains reveals five common insertion points. Mol Biosyst 5:1797–1808. doi: 10.1039/b907669g. [DOI] [PubMed] [Google Scholar]
- 36.Bejsovec A, Martinez Arias A. 1991. Roles of wingless in patterning the larval epidermis of Drosophila. Development 113:471–485. [DOI] [PubMed] [Google Scholar]
- 37.Jiang J, Struhl G. 1998. Regulation of the Hedgehog and Wingless signalling pathways by the F-box/WD40-repeat protein Slimb. Nature 391:493–496. doi: 10.1038/35154. [DOI] [PubMed] [Google Scholar]
- 38.Peifer M, Pai LM, Casey M. 1994. Phosphorylation of the Drosophila adherens junction protein Armadillo: roles for wingless signal and zeste-white 3 kinase. Dev Biol 166:543–556. doi: 10.1006/dbio.1994.1336. [DOI] [PubMed] [Google Scholar]
- 39.Peifer M, Rauskolb C, Williams M, Riggleman B, Wieschaus E. 1991. The segment polarity gene armadillo interacts with the wingless signaling pathway in both embryonic and adult pattern formation. Development 111:1029–1043. [DOI] [PubMed] [Google Scholar]
- 40.Peifer M, Sweeton D, Casey M, Wieschaus E. 1994. wingless signal and Zeste-white 3 kinase trigger opposing changes in the intracellular distribution of Armadillo. Development 120:369–380. [DOI] [PubMed] [Google Scholar]
- 41.Cadigan KM, Nusse R. 1997. Wnt signaling: a common theme in animal development. Genes Dev 11:3286–3305. doi: 10.1101/gad.11.24.3286. [DOI] [PubMed] [Google Scholar]
- 42.Nusse R. 1997. A versatile transcriptional effector of Wingless signaling. Cell 89:321–323. doi: 10.1016/S0092-8674(00)80210-X. [DOI] [PubMed] [Google Scholar]
- 43.Tsou WL, Sheedlo MJ, Morrow ME, Blount JR, McGregor KM, Das C, Todi SV. 2012. Systematic analysis of the physiological importance of deubiquitinating enzymes. PLoS One 7:e43112. doi: 10.1371/journal.pone.0043112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ma P, Yang X, Kong Q, Li C, Yang S, Li Y, Mao B. 2014. The ubiquitin ligase RNF220 enhances canonical Wnt signaling through USP7-mediated deubiquitination of beta-catenin. Mol Cell Biol 34:4355–4366. doi: 10.1128/MCB.00731-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Parsons JL, Dianova II, Khoronenkova SV, Edelmann MJ, Kessler BM, Dianov GL. 2011. USP47 is a deubiquitylating enzyme that regulates base excision repair by controlling steady-state levels of DNA polymerase beta. Mol Cell 41:609–615. doi: 10.1016/j.molcel.2011.02.016. [DOI] [PubMed] [Google Scholar]
- 46.Bajpe PK, van der Knaap JA, Demmers JA, Bezstarosti K, Bassett A, van Beusekom HM, Travers AA, Verrijzer CP. 2008. Deubiquitylating enzyme UBP64 controls cell fate through stabilization of the transcriptional repressor tramtrack. Mol Cell Biol 28:1606–1615. doi: 10.1128/MCB.01567-07. [DOI] [PMC free article] [PubMed] [Google Scholar]