Abstract
Eukaryotic gene expression is often controlled by distant regulatory elements. In developing B lymphocytes, transcription is associated with V(D)J recombination at immunoglobulin loci. This process is regulated by remote cis-acting elements. At the immunoglobulin heavy chain (IgH) locus, the 3′ regulatory region (3′RR) promotes transcription in mature B cells. This led to the notion that the 3′RR orchestrates the IgH locus activity at late stages of B cell maturation only. However, long-range interactions involving the 3′RR were detected in early B cells, but the functional consequences of these interactions were unknown. Here we show that not only does the 3′RR affect transcription at distant sites within the IgH variable region but also it conveys a transcriptional silencing activity on both sense and antisense transcription. The 3′RR-mediated silencing activity is switched off upon completion of VH-DJH recombination. Our findings reveal a developmentally controlled, stage-dependent shift in the transcriptional activity of a master regulatory element.
INTRODUCTION
The spatial and temporal control of gene expression in metazoans is effected by regulatory elements that are often located far from gene promoters (1). This pattern of gene expression regulation is a hallmark of antigen receptor loci, whose expression involves both transcription and recombination. The mouse IgH locus contains ∼195 variable (VH) genes subdivided into domain-organized gene families, including the distal VH family, by far the largest, and the proximal VH family. The VH genes are followed by a dozen diversity (D) segments, 4 joining (JH) segments, and 8 constant (CH) genes (2, 3) (Fig. 1A, top scheme). The assembly of an IgH variable region exon through V(D)J recombination occurs in early developing B cells in an ordered manner, first D to JH and then VH to DJH. While the first recombination step (D-JH) can also be detected in developing T cells, VH-DJH recombination is strictly B cell specific (4).
FIG 1.
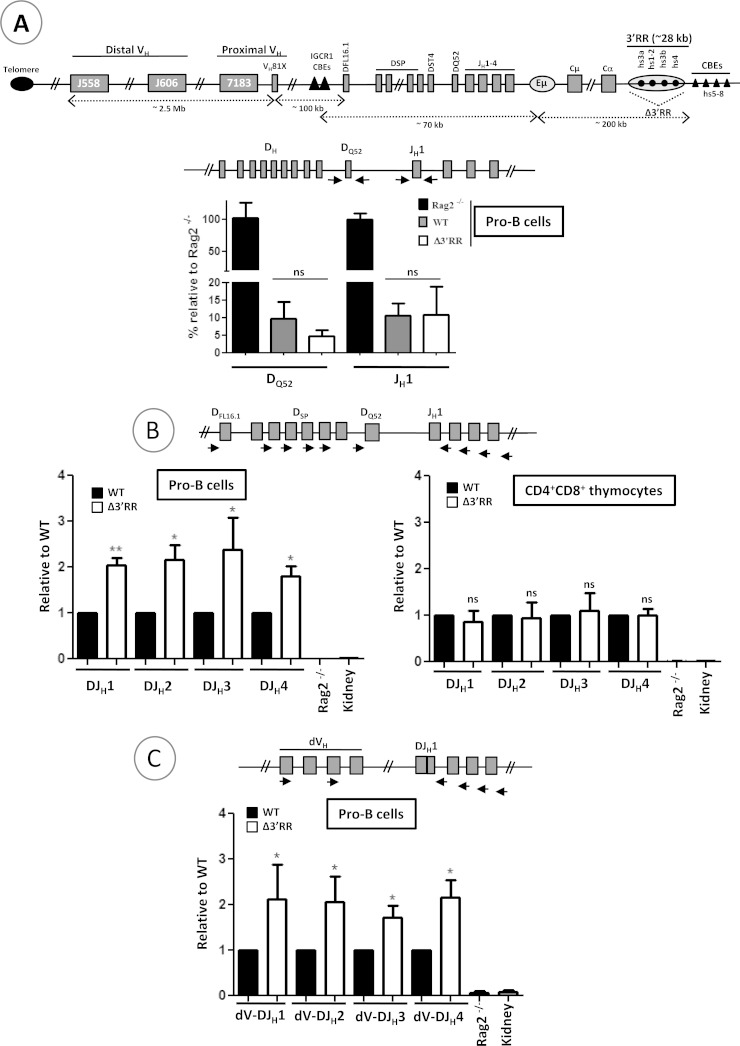
V(D)J recombination in mutant mice. (A) The schematic at the top shows the mouse IgH locus (not to scale). CBEs, CTCF-binding elements. Not all CBEs are shown. Genomic DNAs were prepared from sorted WT and Δ3′RR pro-B cells and subjected to qPCR to amplify unrearranged DQ52 and JH1 gene segments. The relative positions of the primers are indicated in the schematic above the graph. Genomic DNA from Rag2−/− mice was used as a control. The hs5 sequence was used for normalization (n = 4). (B) Genomic DNAs from sorted pro-B cells and double-positive thymocytes were subjected to qPCR to quantify D-JH1, D-JH2, D-JH3, and D-JH4 recombination events. Rag2−/− and kidney DNAs were used as negative controls (n ≥ 4). (C) Quantification of distal (dVH) VH-DJH recombination events in pro-B cells by qPCR (n ≥ 4). **, P < 0.01; *, P < 0.05; ns, not significant. Error bars indicate SEM.
In addition to its B cell lineage specificity, VH-DJH rearrangement is regulated by allelic exclusion, which enables monoallelic expression of only one IgH locus by a given B cell (4, 5). In this process, a productive V(D)J rearrangement on one allele ultimately leads to surface expression of a μ heavy chain which signals arrest of VH-DJH recombination on the second allele. If the first rearrangement is not productive (i.e., no μ heavy chain production), then the second allele can undergo VH-DJH recombination (4, 5). There is considerable evidence to support the notion that VH-DJH rearrangement is the regulated step in IgH allelic exclusion and its maintenance through a feedback mechanism (4, 5), although the molecular mechanisms through which feedback signaling inhibits VH-DJH recombination remain unclear.
Another level of regulation of VH-DJH recombination relates to the physical location of VH gene segments within the variable domain. Indeed, several gene-targeted studies showed that recombinations of the distal and proximal domains are regulated very differently (4, 6, 7). Additionally, allelic exclusion of the distal VH genes is more stringent than that of the most proximal VH genes (4).
In developing B lymphocytes, sense and antisense transcription is associated with V(D)J recombination in a cell type- and developmental stage-specific manner (7). The process is regulated by distant cis-acting elements, including enhancers, promoters, and insulators (6, 7). Three long-range regulatory elements were identified within the IgH locus and were shown by targeted deletion studies to regulate the locus activity. The Eμ enhancer, located between the variable and constant regions, plays a critical role in V(D)J recombination and associated germ line transcription (8–12). Additionally, CTCF-binding elements (CBEs) with insulator activity were identified in the VH-D intergenic region (13–15). This regulatory region (called IGCR1) is important for the order and lineage specificity of V(D)J rearrangements and for allelic exclusion of proximal VH genes (16). A locus control region called the 3′ regulatory region (3′RR) contains four enhancers (hs3a, hs1-2, hs3b, and hs4) (17) and was shown to synergistically promote germ line transcription of CH genes during class switch recombination in mature B cells and also to promote IgH expression in plasma cells (18, 19). Previous targeted deletion studies showed that the 3′RR affected μ heavy chain gene expression in resting B cells (18, 19) but was dispensable for the repertoire diversity in pre-B cells (20). In contrast, its role in allelic exclusion is still unknown.
The established role of the 3′RR in IgH locus expression in late B cells and the lack of an effect on repertoire diversity led to the notion that 3′RR activity is restricted to the late stages of B cell maturation (19, 20). However, various studies described long-range interactions between the 3′RR and various upstream sequences, including the Eμ enhancer (15, 16, 21, 22), though their functional significance was unclear. Strikingly, deletion of either Eμ or the 3′RR had no effect on long-range interactions mediating locus contraction of the IgH locus, leading to the proposal that the activity of these elements may be restricted to the ∼270-kb D-CH region (22).
In this study, we used a mouse line devoid of the 3′RR (19) (here called Δ3′RR) to explore the role of the 3′RR in V(D)J recombination and associated germ line transcription, which occurs at distances ∼220 kb to megabases away from the 3′RR. Here we report the striking finding that the 3′RR mediates a transcriptional silencing activity which is switched off after completion of VH-DJH recombination.
MATERIALS AND METHODS
Mice.
The generation of 3′RR-deleted mice was described previously (19). Throughout the study, the RAG2-deficient mice used as controls were of the 129Sv genetic background. Experiments on mice were carried out according to the CNRS ethical guidelines and were approved by the Regional Ethical Committee.
Antibodies.
Allophycocyanin (APC)-conjugated anti-B220 and fluorescein isothiocyanate (FITC)-conjugated anti-IgM were purchased from BioLegend. Phycoerythrin (PE)-conjugated anti-CD43, FITC-conjugated anti-Igκ, PE-conjugated CD4, and FITC-conjugated CD8 were obtained from BD-Pharmingen.
FACS analyses.
Bone marrows from 6- to 8-week-old mice were prepared by standard techniques. A total of 5 × 105 cells/assay were stained with anti-B220, anti-CD43, and anti-IgM or anti-Igκ and gated on either the IgM− or Igκ− population. Data were obtained for 2.0 × 104 viable cells by using a BD FACSCalibur flow cytometer. Fluorescence-activated cell sorter (FACS) acquisitions included isotype controls and exclusion of dead cells by labeling with propidium iodide.
V(D)J rearrangement assays.
B cells from bone marrows were sorted by using CD19− magnetic microbeads and LS columns (Miltenyi) and were labeled with anti-B220, anti-CD43, and either anti-IgM or (as a cross-check) anti-Igκ. The purity of the sorted pro-B cell populations was checked by FACS analysis (>95%) and by the rearrangement status of the Igκ locus. The CD4+ CD8+ thymocytes were sorted as described previously (16). Genomic DNAs from the sorted pro-B cells (B220+ Igκ− CD43high or B220+ IgM− CD43high) and CD4+ CD8+ thymocytes were prepared by standard techniques and diluted for the (q)PCR assays (23). Controls included genomic DNAs from kidney and RAG-2-deficient pro-B cells. The hs5 sequence, located downstream of the 3′RR (24), was used for normalization. The primers are listed in Table S1 in the supplemental material.
Reverse transcription-quantitative PCR (RT-qPCR).
RAG-2-deficient pro-B cells were sorted using CD19 magnetic microbeads (Miltenyi). RAG-2-deficient thymuses were prepared as described previously (25). B220− CD4+ CD8+ thymocytes were sorted as described previously (16). Pro-B and pre-B cells (B220+ Igκ− CD43low or B220+ IgM− CD43low) were sorted as described above. Unstimulated splenic B cells were sorted by using CD43 magnetic microbeads (Miltenyi) and activated by culturing for 48 h in the presence of 20 μg/ml lipopolysaccharide (Sigma) and 2 ng/ml anti-IgD–dextran (Fina Biosolutions). Total RNAs were reverse transcribed (Fermentas) and subjected to semiquantitative PCR, using SYBR green I (Invitrogen) and ImageQuant software as described previously (26), or to qPCR, using Sso Fast Eva Green (Bio-Rad). The relative transcription levels were normalized using β-actin and Gapdh RNAs as controls.
Statistical analysis.
Results are expressed as means ± standard errors of the means (SEM) (GraphPad Prism), and overall differences between values for wild-type (WT) and mutant mice were evaluated by the Student t test. The difference between means is considered significant if the P value is <0.05, very significant if the P value is <0.01, and extremely significant if the P value is <0.001.
RESULTS
The 3′RR downmodulates V(D)J recombination.
To analyze the effect of the 3′RR on V(D)J recombination, we performed sensitive qPCR-based V(D)J recombination assays (23) on genomic DNAs from sorted WT and Δ3′RR pro-B cells and CD4+ CD8+ thymocytes.
We first quantified the proportions of the DQ52 and JH1 segments that had retained their germ line configuration in purified pro-B cells (Fig. 1A). The total number of alleles with unrearranged DQ52 and JH1 segments on the mutant alleles was comparable to that for WT controls (Fig. 1A), clearly indicating that there was no obvious delay in the onset of D-JH recombination. Thus, any potential effect of the 3′RR on V(D)J recombination is likely to occur after the onset of the process.
We also quantified recombined DJΗ segments and fully rearranged VHDJH segments. We used forward degenerate primers that recognize most D segments (Fig. 1B, schematic) or distal VH genes (Fig. 1C, schematic) and specific backward primers located downstream of each JH segment (Fig. 1B and C, schematics). We did not analyze the recombination of proximal VH genes because the mutant allele is derived from 129Ola ES cells, which bear an ∼120-kb internal deletion in the proximal VH domain (22).
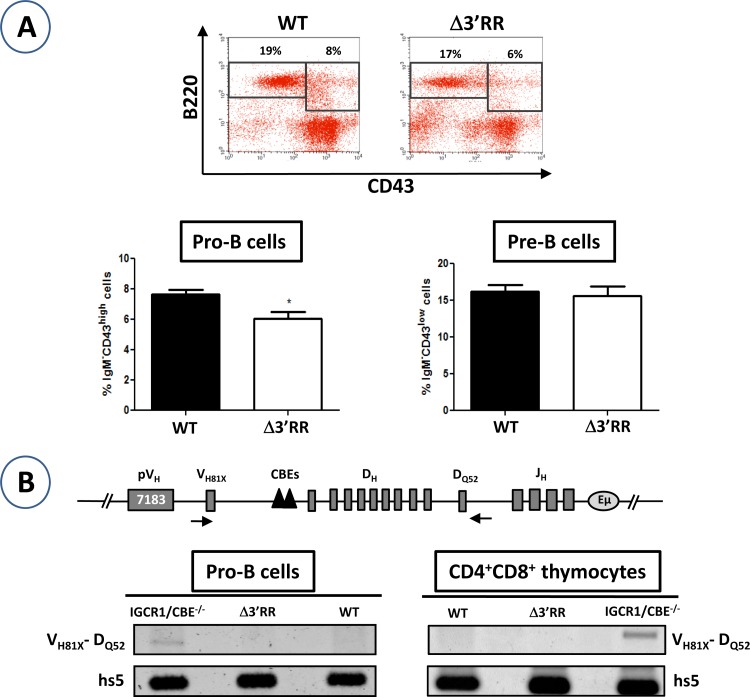
Surprisingly, a mild increase in DJH alleles was detected in mutant pro-B cells but not in mutant CD4+ CD8+ thymocytes (Fig. 1B). Interestingly, a similar increase was also detected for distal VHDJH alleles (Fig. 1C), which could be due, at least in part, to accumulated DJH substrates. Inspection of D-JH and VH-D-JH junction sequences in pro-B cells showed no evidence of an overrepresentation of a D gene segment family or an anomaly regarding the number of inserted or deleted nucleotides (Table 1). The increase seen for VHDJH alleles was not due to a block at the pro-B cell stage either, as the pro-B compartment was rather slightly reduced (Fig. 2A). The data suggest an enhancement of V(D)J recombination (see Discussion).
TABLE 1.
D segment usagea
| Junction | No. of sequences |
|||
|---|---|---|---|---|
| Total | DFL16 | DSP | DQ52 | |
| D-JH (WT) | 39 | 9 | 29 | 1 |
| D-JH (Δ3′RR) | 42 | 10 | 31 | 1 |
| VH-D-JH (WT) | 22 | 9 | 11 | 2 |
| VH-D-JH (Δ3′RR) | 23 | 8 | 14 | 1 |
Genomic DNAs were purified from sorted WT and Δ3′RR pro-B cells and subjected to PCR using degenerate primers that amplify DJH or VHDJH segments. Amplicons were cloned and sequenced. The junctional diversity was used to check for the clonality of the sequences. Thus, sequences with identical insertions/deletions were considered to be one. Within the limits of our data set, there was no obvious anomaly with regard to D gene segment usage or to the number of inserted or deleted nucleotides.
FIG 2.
Early B cell development and order of rearrangements. (A) To determine the distribution of pro-B and pre-B cell populations, single-cell suspensions from the bone marrows of WT and Δ3′RR mice were stained with anti-B220, anti-CD43, and anti-IgM and gated on the IgM− population (n = 11). *, P < 0.05. Data are presented as means and SEM. (B) Genomic DNAs were prepared from sorted pro-B cells (B220+ IgM− CD43high) and CD4+ CD8+ thymocytes and were assayed for VH81X-DQ52 rearrangement (n = 2).
The 3′RR does not affect the order of rearrangements.
As mentioned previously (see the introduction), VH-DJH recombination is strictly B cell specific, and Eμ deletion impairs V(D)J recombination (8, 9, 12); additionally, mutation of IGCR1 CBEs affects the order of V(D)J rearrangements (16). Given the reported CTCF-mediated loop formation between IGCR1 CBEs and CBEs downstream of the 3′RR (15, 16, 21, 22), we asked whether deletion of the 3′RR, which would disrupt the stable Eμ-3′RR interaction (16), and potentially the architecture of the larger CTCF-mediated domain, would somehow deregulate the order of V(D)J rearrangements.
To this end, we attempted to detect VHD amplicons, which result from a direct VH-D recombination. Genomic DNAs were extracted from purified pro-B cells and CD4+ CD8+ thymocytes and assayed for VH-D recombination by using a forward primer pairing with the VH81X gene and a backward primer downstream of the DQ52 segment (Fig. 2B). This sequence is deleted following any D-JH rearrangement, but not if the VH81X segment directly recombines with the DQ52 segment. As a positive control, we used genomic DNAs from IGCR1 CBE−/− pro-B cells and CD4+ CD8+ thymocytes, which undergo VH81X-DQ52 recombination (16). We found no evidence of VHD amplicons in Δ3′RR pro-B cells or in CD4+ CD8+ thymocytes (Fig. 2B). Thus, the 3′RR does not affect the order of rearrangements.
The 3′RR downregulates sense and antisense transcription in the distal variable region.
Germ line transcription in the variable region precedes VH-DJH recombination (27, 28). To investigate the effect of the 3′RR on germ line transcription of unrearranged genes, we first introduced the Δ3′RR mutation into the RAG-2-deficient background, which precludes V(D)J recombination. We mainly focused on the D-Cμ and distal VH domains, for which high levels of transcription are detected (7, 16).
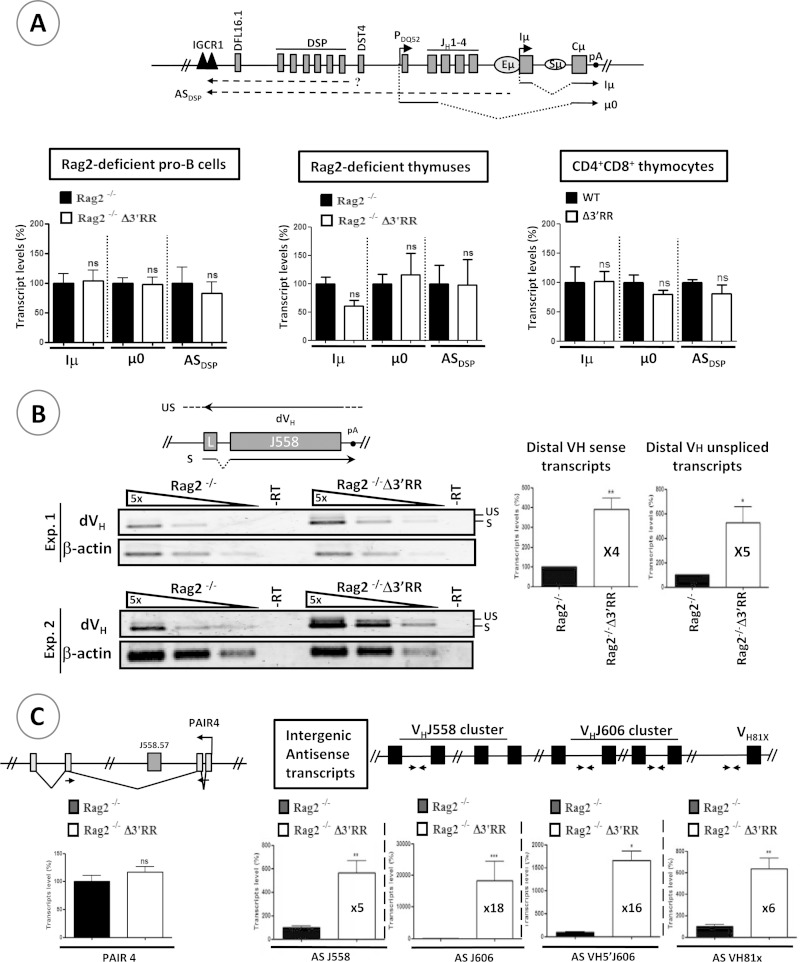
Germ line transcription within the D-Cμ domain, but not that within the distal VH domain, is regulated by the Eμ enhancer (8–12, 29). In contrast, the effect of the 3′RR is unknown. We found no obvious effect on Iμ or μ0 sense transcripts (derived from the Eμ enhancer and the DQ52 promoter, respectively) or on DSP antisense transcripts (derived from the Eμ region and/or an ill-known promoter upstream of the DST4 segment [30]) (Fig. 3A). Concordantly, normal levels of Iμ, μ0, and DSP GL transcripts were found in RAG-2-deficient thymuses and RAG-2-proficient CD4+ CD8+ thymocytes (Fig. 3A). Thus, within the D-Cμ domain, sense and antisense transcription was not altered in the absence of the 3′RR, indicating that the Eμ-mediated control of germ line transcription in the D-Cμ domain does not require the 3′RR.
FIG 3.
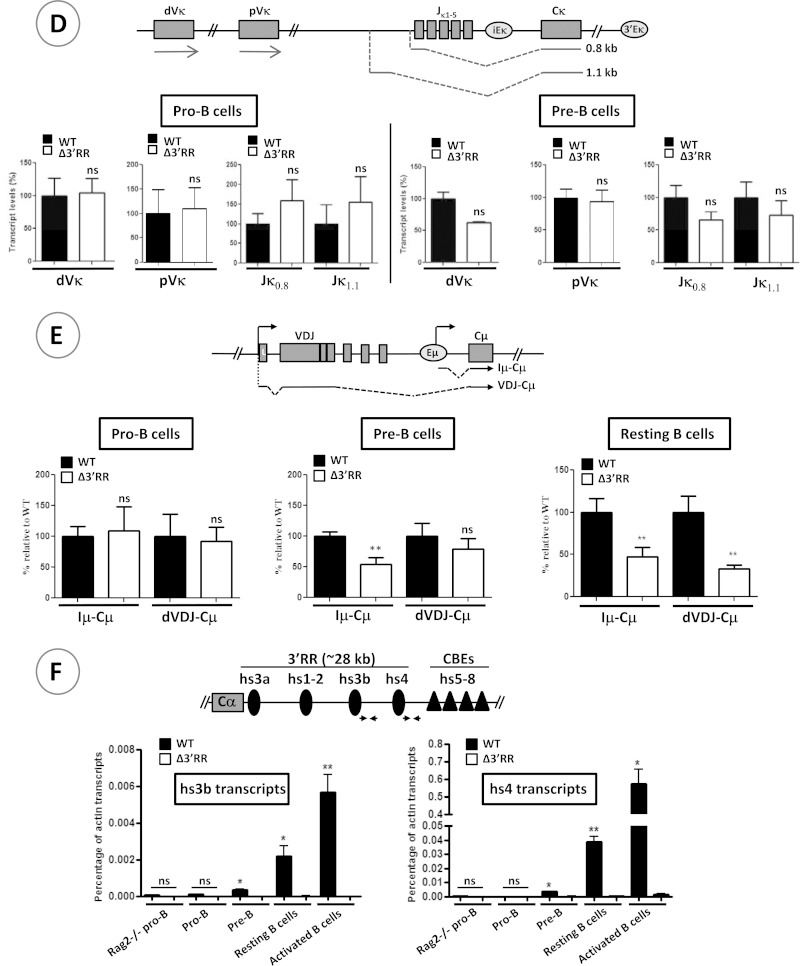
Sense and antisense transcription in the IgH variable locus. (A) The schematic at the top shows the germ line transcripts analyzed in the D-Cμ domain. The Iμ and μ0 sense transcripts are derived from Eμ and the DQ52 promoter, respectively, while antisense transcripts originate from the Eμ region and an ill-defined promoter around the DST4 segment (30). Dots indicate that the initiation and termination sites of the indicated transcripts were not precisely mapped. pA, polyadenylation site. Germ line transcripts in RAG-2-deficient pro-B cells (left; n ≥ 6) and thymuses (middle; n ≥ 3) and RAG2-proficient CD4+ CD8+ thymocytes (right; n ≥ 3) were quantified by RT-qPCR. (B) (Left) Analysis of distal (dVH) germ line transcripts by semiquantitative RT-PCR. Results of two independent experiments are shown (n = 4). S, spliced transcripts (sense); US, unspliced (antisense/primary sense) transcripts. Quantification of the bands is displayed in the histograms on the right. (C) The schematics at top show the relative positions of the primers used along the IgH variable domain. The histograms display the antisense transcript levels as measured by RT-qPCR (n ≥ 6). AS, antisense. (D) The schematic at top indicates the relative positions of the analyzed germ line transcripts along the Igκ locus. The histograms display the transcript levels in pro-B and pre-B cells (n = 3). (E) RT-qPCR analysis of μ (VHDJHCμ) and Iμ transcripts in pro-B, pre-B, and unstimulated splenic B cells. Forward primers that bind the distal VH (dVH) genes or the Iμ exon and a reverse primer that pairs with the Cμ1 exon were used to quantify μ and Iμ cDNAs (n ≥ 6). (F) RT-qPCR analysis of hs3b and hs4 transcripts at various stages of B cell development (n = 3). ***, P < 0.001; **, P < 0.01; *, P < 0.05; ns, not significant. Error bars indicate SEM.
Remarkably, the distal VH region yielded increased levels of both spliced, sense transcripts and unspliced, antisense transcripts in Δ3′RR mice (Fig. 3B). To exclude any bias potentially introduced by the increased level of primary VH sense transcripts, we quantified intergenic, antisense germ line transcript levels within the VHJ558 and VHJ606 clusters. The levels of these exclusively antisense transcripts were also increased (Fig. 3C). In contrast, the Pax5-activated intergenic repeat 4 (PAIR4) antisense germ line transcripts (31) were unaltered (Fig. 3C), suggesting that regulation of these transcripts, derived from the PAIR4 promoter/enhancer element (31), is 3′RR independent. Thus, within the distal VH domain (except for PAIR elements), the 3′RR affects both sense and antisense transcription.
Within the proximal VH domain, we also found increased antisense transcription upstream of VH81X (the most 3′ functional VH gene segment, which is not encompassed by the ∼120-kb deletion in Δ3′RR mice [22]) (Fig. 3C), suggesting a variable region-wide effect of the 3′RR.
Overall, and within the limits of the transcripts analyzed, the data show that the 3′RR downregulates sense and antisense germ line transcription along the remote IgH variable domain.
Transcription of some loci could be regulated by elements located on a different chromosome (32). Specifically, it was suggested that the Igκ locus and its 3′ enhancer (on chromosome 6) are involved in directing the IgH locus (on chromosome 12) to a repressive nuclear compartment and inducing IgH locus decontraction (33). To investigate whether the 3′RR can act in trans, we analyzed germ line transcription along the Igκ locus and found that it was unchanged (Fig. 3D), excluding, at least with regard to the Igκ locus, any trans effect of the 3′RR.
Switching off the 3′RR-mediated silencing activity coincides with the completion of V(D)J recombination.
To elucidate precisely at which step the 3′RR-mediated silencing activity is turned off, we quantified the transcript levels of the fully rearranged μ gene at various developmental stages. To avoid potential bias introduced by cellular selection and/or selective use of distal versus proximal VH genes, we also measured Iμ transcript levels. We found normal levels of the distal VH exon-containing μ transcripts (dVHDJHCμ) in pro-B cells (Fig. 3E). These transcript levels were unchanged in pre-B cells but were clearly decreased in unstimulated splenic B cells (Fig. 3E). Downregulation of Iμ transcripts was clearly detectable in pre-B cells and was more pronounced in unstimulated cells (Fig. 3E). Interestingly, the shift from a silencer (in pro-B cells) to an enhancer (in pre-B cells) activity correlated with the appearance of 3′RR enhancer transcripts (Fig. 3F) (34).
Therefore, the 3′RR-mediated silencing effect appears to be switched off upon completion of V(D)J recombination at the pro-B cell stage, and it correlates with the onset of 3′RR transcription.
DISCUSSION
We report here the first demonstration of a stepwise shift in the transcriptional activity of a long-range regulatory element in higher eukaryotes. The downregulating activity of the 3′RR targets multiple upstream sense and antisense promoters in the remote IgH variable region, but it spares known enhancers/promoters (Eμ and PAIR4). Specifically, the 3′RR does not affect sense and antisense transcription within the D-Cμ domain, consistent with the notion that transcription within this domain is controlled mainly by the Eμ enhancer (8, 10).
As mentioned previously, the Δ3′RR mouse line is derived from 129Ola ES cells, which have a 120-kb internal deletion in the proximal VH domain; this is not the case for RAG-2-deficient mice, which are derived from strain 129Sv. Thus, although we cannot formally exclude the possibility that the 120-kb deletion within the proximal VH domain affected distal VH germ line transcription and V(D)J recombination, we think that it is unlikely, for various reasons. Multiple studies clearly showed that the proximal and distal VH domains are differentially regulated. Thus, recombination of distal but not proximal VH genes is inhibited in mice deficient in the histone-modifying enzyme EZH2 and in different transcription factors involved in V(D)J recombination, such as PAX5, YY1, and Ikaros (35–38). Mutations targeting various cis-acting elements at the IgH locus similarly showed a differential effect on germ line transcription and recombination of proximal versus distal VH genes (12, 16, 39–42). Importantly, deletion of the 3′RR in the context of the 120-kb deletion had no effect on long-range interactions across the IgH locus in RAG2-deficient pro-B cells (22). Additionally, within the IgH variable region, the viewpoints that were found by chromosome conformation capture-derived (4C-seq) analyses to strongly or minimally interact with the 3′RR correlated well with our transcriptional analyses. For instance, antisense transcription upstream of the VH81X gene (which is intact in the 129Ola background) was enhanced in the absence of the 3′RR (present study), and this gene was efficiently contacted by the hs3b enhancer of the 3′RR (22), whereas PAIR4, whose expression was not affected by the 3′RR deletion (present study), did not significantly interact with the 3′RR (21, 22). Moreover, antisense transcription within the J606 family was increased (present study), which correlated well with an interaction of this region with the 3′RR-Eμ loop (21, 22; reviewed in reference 7). Finally, it is difficult to figure out how the 120-kb deletion would affect the 3′RR-mediated effect on D-JH recombination, which takes place in the stable IGCR1-IgH 3′CBE chromatin domain (15, 16, 21, 22).
Determining whether the long-range 3′RR-mediated silencing activity is due to an unidentified, developmentally regulated silencer within the 3′RR itself or is mediated by interacting partners requires further investigations involving combined mutations. The strong correlation between the extinction of the 3′RR-mediated silencing activity and the completion of V(D)J recombination suggests that the interacting partner(s) should be deleted upon VH-DJH recombination. Likely candidates could be the IGCR1 (16, 21, 22) and/or the newly identified interaction site upstream of IGCR1 (22). This could be a means through which recombination regulates the 3′RR transcriptional activity. Alternatively, though not mutually exclusive, the correlation between the triggering of 3′RR transcription and its enhancer activity suggests that the long-range activity of the 3′RR during B cell development may be modulated by its enhancers' transcripts.
The B cell-specific downmodulating effect of the 3′RR on D-JH recombination suggests that 3′RR-Eμ interaction may affect Eμ-mediated control of recombination rather than transcription. Various studies have found that transcription and V(D)J recombination can be mediated by distinct activities of accessibility control elements, including the Eμ enhancer (43; reviewed in reference 44), and there is some evidence that recombination can be recapitulated in vitro in the absence of transcription (45).
By quantifying the proportions of the DQ52 and JH1 segments in their unrearranged configuration, we found no obvious delay in the onset of D-JH recombination, clearly indicating that the effect of the 3′RR occurs after the initiation of V(D)J recombination. In contrast, there was an accumulation of DJH intermediates and fully recombined VHDJH alleles, with no obvious block at the pro-B cell stage, at which V(D)J recombination at the IgH locus occurs. Thus, it appears that we are dealing with a specific phenomenon that is restricted to pro-B cells, i.e., an increased number of recombination events in a “fixed” time window. One simple explanation is that the process runs faster in the absence of the 3′RR. Recent evidence highlighted the importance of spatial confinement for the kinetics of V(D)J recombination and the time of encounter with regulatory elements (46). It is tempting to speculate that 3′RR interactions with its partners are a critical component of the mechanisms that regulate the kinetics of V(D)J recombination. Within a newly generated topological domain that forms upon DJH recombination, the 3′RR may, for instance, contribute to the control of the kinetics of VH-DJH recombination by affecting germ line transcription within the VH domain, while Eμ is focused on DJH transcription (40).
Why does the 3′RR mediate transcriptional silencing within the VH domain? It should be noted that VH-DJH recombination is the regulated step in allelic exclusion (4, 5) and that the control of germ line transcription is likely the primary event during allelic exclusion (42). In the absence of the 3′RR, we found increases of germ line transcription within the distal VH domain (with the exception of PAIR promoter/enhancer-derived transcripts) and of the proportion of distal VHDJH alleles, with no obvious block at the pro-B cell stage. This increase could be due, at least in part, to accumulated DJH substrates. However, it may also indicate a disruption of allelic exclusion. Thus, our findings of enhanced VH-DJH recombination and germ line transcription may be explained by a speculative model (Fig. 4) in which a productive rearrangement on one allele instructs the 3′RR on the second allele to downregulate antisense transcription, leading to the inhibition of VH-DJH recombination. In the absence of the 3′RR, a productive VHDJH rearrangement on the first allele (and subsequent surface expression of the μ heavy chain) would not block VH-DJH recombination on the second allele, leading to an overall accumulation of VHDJH alleles.
FIG 4.
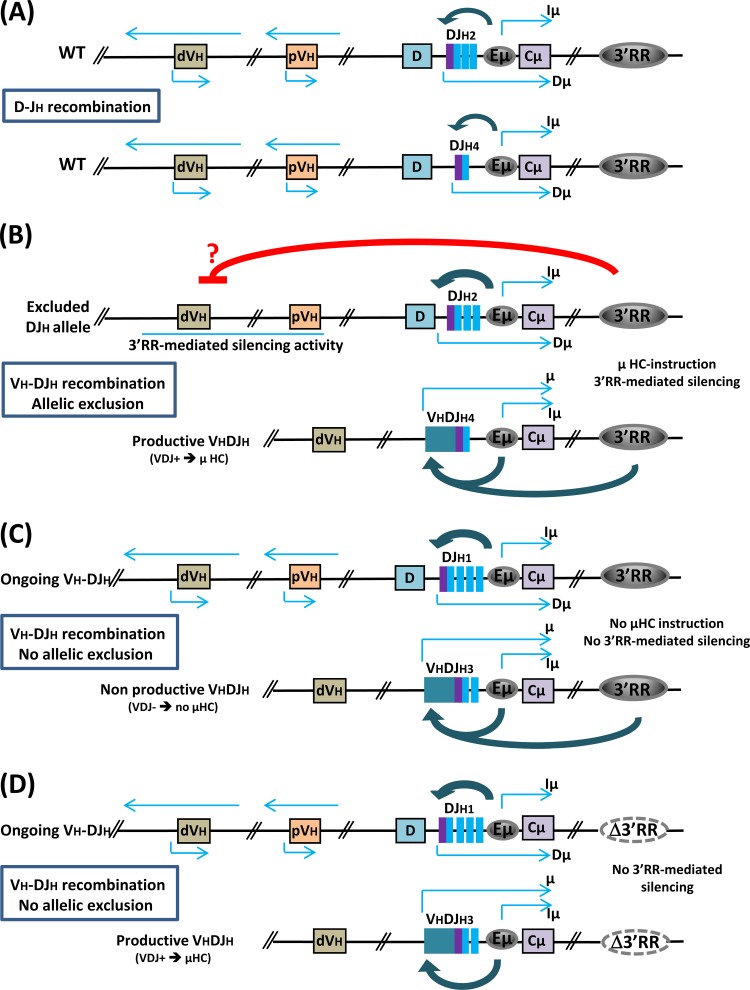
Speculative model linking 3′RR-mediated silencing activity to allelic exclusion. This model stipulates an interplay between cis-acting elements and μ heavy chain (HC) signaling. Among the cis-regulatory elements which play a role in allelic exclusion, only the interactions between the Eμ enhancer and the 3′RR are shown. (A) Upon D-JH recombination, the Eμ enhancer upregulates DJH transcription (Dμ transcripts), and sense and antisense germ line transcripts are detected at the IgH variable region. (B) A productive rearrangement on one allele will lead to μ HC surface expression, in association with VpreB and λ5 surrogate light chains and the Igα/Igβ heterodimer, which will signal to the 3′RR on the second allele to mediate a transcriptional silencing activity within the VH region, leading to downregulation of sense and antisense transcription and VH-DJH recombination. Cooperation between Eμ and the 3′RR on the productive allele upregulates rearranged μ HC gene expression, leading to the enforcement and maintenance of allelic exclusion. (C) If the first rearrangement is not productive (not in frame and therefore no μ HC production), the 3′RR receives no signal to mediate its silencing activity, and VH-DJH recombination can therefore occur on the second allele. (D) In the absence of the 3′RR, the link between μ HC instruction and germ line transcription in the IgH variable region is lost, and allelic exclusion is disrupted. pVH, proximal VH cluster; dVH, distal VH cluster.
In this regard, our model may fill a gap in the regulated/feedback inhibition model of allelic exclusion. Indeed, how can we explain that a productive rearrangement on the first allele inhibits VH-DJH recombination on the second allele? Our interpretation is that surface expression of the μ heavy chain instructs the 3′RR to inhibit germ line transcription within the variable domain, and therefore VH-DJH recombination. Thus, 3′RR-mediated inhibition of VH germ line transcription could be the missing link between surface expression of the heavy chain and effective allelic exclusion. One prediction of this model is that deletion of the 3′RR will result in increased VH germ line transcription and VH-DJH frequency, and we found this result in the present study. Another prediction is that if a heavy chain is expressed prematurely [that is, prior to V(D)J recombination], the 3′RR will be instructed to inhibit VH germ line transcription, and the outcome will be an impairment of VH-DJH recombination; this is indeed the case (42).
The wide impact of the 3′RR on sense and antisense germ line transcription within the variable region raises the question of whether the 3′RR targets sense and antisense promoters simultaneously. We favor the view that the 3′RR targets primarily antisense promoters and that the silencing of sense transcripts may be a downstream consequence of this primary effect. It should be noted that antisense transcripts are long and extend through multiple VH genes (28). Thus, the control of a limited number of antisense promoters would be sufficient for a wide transcriptional impact. Nonetheless, the 3′RR must somehow reach its distant target promoters. The possibility of a long-range effect mediated by 3′RR transcripts was ruled out because such transcripts were undetectable in pro-B cells. This implies that the 3′RR-mediated silencing activity correlates with a lack of 3′RR transcription. A likely explanation is that the 3′RR exploits developmentally regulated, 3′RR-independent (22) mechanisms that allow compaction of the IgH locus. In particular, the large-scale reorganization of the distal variable region into rosette-like structures following D-JH recombination and the compaction of the IgH locus in pro-B cells (47) may bring the 3′RR and its target promoters into close proximity.
How the 3′RR can mediate its silencing activity is presently unknown and may involve a developmental stage-dependent interplay between the topological reorganization of the IgH locus, which may be modulated by CTCF insulators and transcription factor-mediated loops, posttranslational modifications of factors such as CTCF, and the 3′RR epigenetic modifications and recruitment of transcription factors and corepressors (48–51). Interestingly, there is some evidence that the human β-globin locus control region can in some contexts repress gene expression through transcriptional interference, potentially involving transcripts initiating in flanking repetitive sequences and running through the β-globin locus (52). This suggests that besides their established role in gene expression activation, locus control regions also have the potential to mediate transcriptional silencing activity, which may depend on the developmental stage, lineage specificity, interacting partners, and chromatin context.
In conclusion, our study reveals a hitherto unsuspected function of the 3′RR during early B cell development. The 3′RR emerges as a master regulatory element that mediates a transcriptional silencing activity along the distant and large IgH variable region, leading to the inhibition of VH-DJH recombination, likely to promote allelic exclusion.
Supplementary Material
ACKNOWLEDGMENTS
We thank F. W. Alt for providing genomic DNA from IGCR1-deficient mice and for advice. We also thank the animal facility staff at the IPBS and F. L'Faqihi, V. Duplan-Eche, and A.-L. Iscache at the Purpan CPTP platform for their excellent assistance.
This work was supported by the Fondation ARC (grant PJA 20141201647), Agence Nationale de la Recherche, Institut National du Cancer (PLBIO15-134), Ligue Contre le Cancer-Comité de Haute-Garonne, and Cancéropôle Grand-Sud-Ouest.
F.-Z.B. and C.C. performed research and analyzed data; M.M., Y.D., and M.C. contributed new reagents or analytic tools; and A.A.K. designed research, analyzed data, and wrote the paper.
We declare that we have no conflicts of interest.
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/MCB.00509-15.
REFERENCES
- 1.Bulger M, Groudine M. 2011. Functional and mechanistic diversity of distal transcription enhancers. Cell 144:327–339. doi: 10.1016/j.cell.2011.01.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Johnston CM, Wood AL, Bolland DJ, Corcoran AE. 2006. Complete sequence assembly and characterization of the C57BL/6 mouse Ig heavy chain V region. J Immunol 176:4221–4234. doi: 10.4049/jimmunol.176.7.4221. [DOI] [PubMed] [Google Scholar]
- 3.Retter I, Chevillard C, Scharfe M, Conrad A, Hafner M, Im TH, Ludewig M, Nordsiek G, Severitt S, Thies S, Mauhar A, Blocker H, Muller W, Riblet R. 2007. Sequence and characterization of the Ig heavy chain constant and partial variable region of the mouse strain 129S1. J Immunol 179:2419–2427. doi: 10.4049/jimmunol.179.4.2419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Jung D, Giallourakis C, Mostoslavsky R, Alt FW. 2006. Mechanism and control of V(D)J recombination at the immunoglobulin heavy chain locus. Annu Rev Immunol 24:541–570. doi: 10.1146/annurev.immunol.23.021704.115830. [DOI] [PubMed] [Google Scholar]
- 5.Vettermann C, Schlissel MS. 2010. Allelic exclusion of immunoglobulin genes: models and mechanisms. Immunol Rev 237:22–42. doi: 10.1111/j.1600-065X.2010.00935.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Subrahmanyam R, Sen R. 2012. Epigenetic features that regulate IgH locus recombination and expression. Curr Top Microbiol Immunol 356:39–63. doi: 10.1007/82_2011_153. [DOI] [PubMed] [Google Scholar]
- 7.Stubbington MJ, Corcoran AE. 2013. Non-coding transcription and large-scale nuclear organisation of immunoglobulin recombination. Curr Opin Genet Dev 23:81–88. doi: 10.1016/j.gde.2013.01.001. [DOI] [PubMed] [Google Scholar]
- 8.Perlot T, Alt FW, Bassing CH, Suh H, Pinaud E. 2005. Elucidation of IgH intronic enhancer functions via germ-line deletion. Proc Natl Acad Sci U S A 102:14362–14367. doi: 10.1073/pnas.0507090102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Afshar R, Pierce S, Bolland DJ, Corcoran A, Oltz EM. 2006. Regulation of IgH gene assembly: role of the intronic enhancer and 5′DQ52 region in targeting DHJH recombination. J Immunol 176:2439–2447. doi: 10.4049/jimmunol.176.4.2439. [DOI] [PubMed] [Google Scholar]
- 10.Bolland DJ, Wood AL, Afshar R, Featherstone K, Oltz EM, Corcoran AE. 2007. Antisense intergenic transcription precedes Igh D-to-J recombination and is controlled by the intronic enhancer Emu. Mol Cell Biol 27:5523–5533. doi: 10.1128/MCB.02407-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Perlot T, Li G, Alt FW. 2008. Antisense transcripts from immunoglobulin heavy-chain locus V(D)J and switch regions. Proc Natl Acad Sci U S A 105:3843–3848. doi: 10.1073/pnas.0712291105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chakraborty T, Perlot T, Subrahmanyam R, Jani A, Goff PH, Zhang Y, Ivanova I, Alt FW, Sen R. 2009. A 220-nucleotide deletion of the intronic enhancer reveals an epigenetic hierarchy in immunoglobulin heavy chain locus activation. J Exp Med 206:1019–1027. doi: 10.1084/jem.20081621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Degner SC, Wong TP, Jankevicius G, Feeney AJ. 2009. Cutting edge: developmental stage-specific recruitment of cohesin to CTCF sites throughout immunoglobulin loci during B lymphocyte development. J Immunol 182:44–48. doi: 10.4049/jimmunol.182.1.44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Featherstone K, Wood AL, Bowen AJ, Corcoran AE. 2010. The mouse immunoglobulin heavy chain V-D intergenic sequence contains insulators that may regulate ordered V(D)J recombination. J Biol Chem 285:9327–9338. doi: 10.1074/jbc.M109.098251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Degner SC, Verma-Gaur J, Wong TP, Bossen C, Iverson GM, Torkamani A, Vettermann C, Lin YC, Ju Z, Schulz D, Murre CS, Birshtein BK, Schork NJ, Schlissel MS, Riblet R, Murre C, Feeney AJ. 2011. CCCTC-binding factor (CTCF) and cohesin influence the genomic architecture of the Igh locus and antisense transcription in pro-B cells. Proc Natl Acad Sci U S A 108:9566–9571. doi: 10.1073/pnas.1019391108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Guo C, Yoon HS, Franklin A, Jain S, Ebert A, Cheng HL, Hansen E, Despo O, Bossen C, Vettermann C, Bates JG, Richards N, Myers D, Patel H, Gallagher M, Schlissel MS, Murre C, Busslinger M, Giallourakis CC, Alt FW. 2011. CTCF-binding elements mediate control of V(D)J recombination. Nature 477:424–430. doi: 10.1038/nature10495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Khamlichi AA, Pinaud E, Decourt C, Chauveau C, Cogné M. 2000. The 3′ IgH regulatory region: a complex structure in a search for a function. Adv Immunol 75:317–345. doi: 10.1016/S0065-2776(00)75008-5. [DOI] [PubMed] [Google Scholar]
- 18.Pinaud E, Khamlichi AA, Le Morvan C, Drouet M, Nalesso V, Le Bert M, Cogné M. 2001. Localization of the 3′ IgH locus elements that effect long-distance regulation of class switch recombination. Immunity 15:187–199. doi: 10.1016/S1074-7613(01)00181-9. [DOI] [PubMed] [Google Scholar]
- 19.Vincent-Fabert C, Fiancette R, Pinaud E, Truffinet V, Cogné N, Cogné M, Denizot Y. 2010. Genomic deletion of the whole IgH 3′ regulatory region (hs3a, hs1,2, hs3b, and hs4) dramatically affects class switch recombination and Ig secretion to all isotypes. Blood 116:1895–1898. doi: 10.1182/blood-2010-01-264689. [DOI] [PubMed] [Google Scholar]
- 20.Rouaud P, Vincent-Fabert C, Fiancette R, Cogné M, Pinaud E, Denizot Y. 2012. Enhancers located in heavy chain regulatory region (hs3a, hs1,2, hs3b, and hs4) are dispensable for diversity of VDJ recombination. J Biol Chem 287:8356–8360. doi: 10.1074/jbc.M112.341024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Guo C, Gerasimova T, Hao H, Ivanova I, Chakraborty T, Selimyan R, Oltz EM, Sen R. 2011. Two forms of loops generate the chromatin conformation of the immunoglobulin heavy-chain gene locus. Cell 147:332–343. doi: 10.1016/j.cell.2011.08.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Medvedovic J, Ebert A, Tagoh H, Tamir IM, Schwickert TA, Novatchkova M, Sun Q, Huis In't Veld PJ, Guo C, Yoon HS, Denizot Y, Holwerda SJ, de Laat W, Cogne M, Shi Y, Alt FW, Busslinger M. 2013. Flexible long-range loops in the VH gene region of the Igh locus facilitate the generation of a diverse antibody repertoire. Immunity 39:229–244. doi: 10.1016/j.immuni.2013.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Braikia F-Z, Chemin G, Moutahir M, Khamlichi AA. 2014. Quantification of V(D)J recombination by real-time quantitative PCR. Immunol Lett 162:119–123. doi: 10.1016/j.imlet.2014.08.002. [DOI] [PubMed] [Google Scholar]
- 24.Garrett FE, Emelyanov AV, Sepulveda MA, Flanagan P, Volpi S, Li F, Loukinov D, Eckhardt LA, Lobanenkov VV, Birshtein BK. 2005. Chromatin architecture near a potential 3′ end of the IgH locus involves modular regulation of histone modifications during B-cell development and in vivo occupancy at CTCF sites. Mol Cell Biol 25:1511–1525. doi: 10.1128/MCB.25.4.1511-1525.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Shinkai Y, Rathbun G, Lam K-P, Oltz EM, Stewart V, Mendelsohn M, Charron J, Datta M, Young F, Stall AM, Alt FW. 1992. RAG-2-deficient mice lack mature lymphocytes owing to inability to initiate V(D)J recombination. Cell 68:855–867. doi: 10.1016/0092-8674(92)90029-C. [DOI] [PubMed] [Google Scholar]
- 26.Haddad D, Oruc Z, Puget N, Laviolette-Malirat N, Philippe M, Carrion C, Le Bert M, Khamlichi AA. 2011. Sense transcription through the S region is essential for immunoglobulin class switch recombination. EMBO J 30:1608–1620. doi: 10.1038/emboj.2011.56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yancopoulos GD, Alt FW. 1985. Developmentally controlled and tissue-specific expression of unrearranged VH gene segments. Cell 40:271–281. doi: 10.1016/0092-8674(85)90141-2. [DOI] [PubMed] [Google Scholar]
- 28.Bolland DJ, Wood AL, Johnston CM, Bunting SF, Morgan G, Chakalova L, Fraser PJ, Corcoran AE. 2004. Antisense intergenic transcription in V(D)J recombination. Nat Immunol 5:630–637. doi: 10.1038/ni1068. [DOI] [PubMed] [Google Scholar]
- 29.Verma-Gaur J, Torkamani A, Schaffer L, Head SR, Schork NJ, Feeney AJ. 2012. Noncoding transcription within the Igh distal V(H) region at PAIR elements affects the 3D structure of the Igh locus in pro-B cells. Proc Natl Acad Sci U S A 109:17004–17009. doi: 10.1073/pnas.1208398109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Giallourakis CC, Franklin A, Guo C, Cheng HL, Yoon HS, Gallagher M, Perlot T, Andzelm M, Murphy AJ, Macdonald LE, Yancopoulos GD, Alt FW. 2010. Elements between the IgH variable (V) and diversity (D) clusters influence antisense transcription and lineage-specific V(D)J recombination. Proc Natl Acad Sci U S A 107:22207–22212. doi: 10.1073/pnas.1015954107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ebert A, McManus S, Tagoh H, Medvedovic J, Salvagiotto G, Novatchkova M, Tamir I, Sommer A, Jaritz M, Busslinger M. 2011. The distal V(H) gene cluster of the Igh locus contains distinct regulatory elements with Pax5 transcription factor-dependent activity in pro-B cells. Immunity 34:175–187. doi: 10.1016/j.immuni.2011.02.005. [DOI] [PubMed] [Google Scholar]
- 32.Williams A, Spilianakis CG, Flavell RA. 2010. Interchromosomal association and gene regulation in trans. Trends Genet 26:188–197. doi: 10.1016/j.tig.2010.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hewitt SL, Farmer D, Marszalek K, Cadera E, Liang HE, Xu Y, Schlissel MS, Skok JA. 2008. Association between the Igk and Igh immunoglobulin loci mediated by the 3′ Igk enhancer induces ‘decontraction’ of the Igh locus in pre-B cells. Nat Immunol 9:396–404. doi: 10.1038/ni1567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Péron S, Laffleur B, Denis-Lagache N, Cook-Moreau J, Tinguely A, Delpy L, Denizot Y, Pinaud E, Cogné M. 2012. AID-driven deletion causes immunoglobulin heavy chain locus suicide recombination in B cells. Science 336:931–934. doi: 10.1126/science.1218692. [DOI] [PubMed] [Google Scholar]
- 35.Su IH, Basavaraj A, Krutchinsky AN, Hobert O, Ullrich A, Chait BT, Tarakhovsky A. 2003. Ezh2 controls B cell development through histone H3 methylation and Igh rearrangement. Nat Immunol 4:124–131. doi: 10.1038/ni876. [DOI] [PubMed] [Google Scholar]
- 36.Fuxa M, Skok J, Souabni A, Salvagiotto G, Roldan E, Busslinger M. 2004. Pax5 induces V-to-DJ rearrangements and locus contraction of the immunoglobulin heavy-chain gene. Genes Dev 18:411–422. doi: 10.1101/gad.291504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Liu H, Schmidt-Supprian M, Shi Y, Hobeika E, Barteneva N, Jumaa H, Pelanda R, Reth M, Skok J, Rajewsky K. 2007. Yin Yang 1 is a critical regulator of B-cell development. Genes Dev 21:1179–1189. doi: 10.1101/gad.1529307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Reynaud D, Demarco IA, Reddy KL, Schjerven H, Bertolino E, Chen Z, Smale ST, Winandy S, Singh H. 2008. Regulation of B cell fate commitment and immunoglobulin heavy-chain gene rearrangements by Ikaros. Nat Immunol 9:927–936. doi: 10.1038/ni.1626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Volpi SA, Verma-Gaur J, Hassan R, Ju Z, Roa S, Chatterjee S, Werling U, Hou H Jr, Will B, Steidl U, Scharff M, Edelman W, Feeney AJ, Birshtein BK. 2012. Germline deletion of Igh 3′ regulatory region elements hs 5, 6, 7 (hs5-7) affects B cell-specific regulation, rearrangement, and insulation of the Igh locus. J Immunol 188:2556–2566. doi: 10.4049/jimmunol.1102763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Puget N, Hirasawa R, Nguyen Hu NS, Laviolette-Malirat N, Feil R, Khamlichi AA. 2015. Insertion of an imprinted insulator into the IgH locus reveals developmentally regulated, transcription-dependent control of V(D)J recombination. Mol Cell Biol 35:529–543. doi: 10.1128/MCB.00235-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lin SG, Guo C, Su A, Zhang Y, Alt FW. 2015. CTCF-binding elements 1 and 2 in the Igh intergenic control region cooperatively regulate V(D)J recombination. Proc Natl Acad Sci U S A 112:1815–1820. doi: 10.1073/pnas.1424936112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Puget N, Leduc C, Oruc Z, Moutahir M, Le Bert M, Khamlichi AA. 2015. Complete cis exclusion upon duplication of Eμ enhancer at the immunoglobulin heavy chain locus. Mol Cell Biol 35:2231–2241. doi: 10.1128/MCB.00294-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Fernex C, Capone M, Ferrier P. 1995. The V(D)J recombinational and transcriptional activities of the immunoglobulin heavy-chain intronic enhancer can be mediated through distinct protein-binding sites in a transgenic substrate. Mol Cell Biol 15:3217–3226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Cobb RM, Oestreich KJ, Osipovich OA, Oltz EM. 2006. Accessibility control of V(D)J recombination. Adv Immunol 91:45–109. doi: 10.1016/S0065-2776(06)91002-5. [DOI] [PubMed] [Google Scholar]
- 45.Du H, Ishii H, Pazin MJ, Sen R. 2008. Activation of 12/23-RSS-dependent RAG cleavage by hSWI/SNF complex in the absence of transcription. Mol Cell 31:641–649. doi: 10.1016/j.molcel.2008.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lucas JS, Zhang Y, Dudko OK, Murre C. 2014. 3D trajectories adopted by coding and regulatory DNA elements: first-passage times for genomic interactions. Cell 158:339–352. doi: 10.1016/j.cell.2014.05.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Jhunjhunwala S, van Zelm MC, Peak MM, Cutchin S, Riblet R, van Dongen JJ, Grosveld FG, Knoch TA, Murre C. 2008. The 3D structure of the immunoglobulin heavy-chain locus: implications for long-range genomic interactions. Cell 133:265–279. doi: 10.1016/j.cell.2008.03.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Giambra V, Volpi S, Emelyanov AV, Pflugh D, Bothwell AL, Norio P, Fan Y, Ju Z, Skoultchi AI, Hardy RR, Frezza D, Birshtein BK. 2008. Pax5 and linker histone H1 coordinate DNA methylation and histone modifications in the 3′ regulatory region of the immunoglobulin heavy chain locus. Mol Cell Biol 28:6123–6133. doi: 10.1128/MCB.00233-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Cobaleda C, Schebesta A, Delogu A, Busslinger M. 2007. Pax5: the guardian of B cell identity and function. Nat Immunol 8:463–470. doi: 10.1038/ni1454. [DOI] [PubMed] [Google Scholar]
- 50.Atchison ML. 2014. Function of YY1 in long-distance DNA interactions. Front Immunol 5:45. doi: 10.3389/fimmu.2014.00045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Birshtein BK. 2014. Epigenetic regulation of individual modules of the immunoglobulin heavy chain locus 3′ regulatory region. Front Immunol 5:163. doi: 10.3389/fimmu.2014.00163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Feng YQ, Warin R, Li T, Olivier E, Besse A, Lobell A, Fu H, Lin CM, Aladjem MI, Bouhassira EE. 2005. The human beta-globin locus control region can silence as well as activate gene expression. Mol Cell Biol 25:3864–3874. doi: 10.1128/MCB.25.10.3864-3874.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.