Abstract
The transmembrane protein CRB3A controls epithelial cell polarization. Elucidating the molecular mechanisms of CRB3A function is essential as this protein prevents the epithelial-to-mesenchymal transition (EMT), which contributes to tumor progression. To investigate the functional impact of altered CRB3A expression in cancer cells, we expressed CRB3A in HeLa cells, which are devoid of endogenous CRB3A. While control HeLa cells display a patchy F-actin distribution, CRB3A-expressing cells form a circumferential actomyosin belt. This reorganization of the cytoskeleton is accompanied by a transition from an ameboid cell shape to an epithelial-cell-like morphology. In addition, CRB3A increases the cohesion of HeLa cells. To perform these functions, CRB3A recruits p114RhoGEF and its activator Ehm2 to the cell periphery using both functional motifs of its cytoplasmic tail and increases RhoA activation levels. ROCK1 and ROCK2 (ROCK1/2), which are critical effectors of RhoA, are also essential to modulate the cytoskeleton and cell shape downstream of CRB3A. Overall, our study highlights novel roles for CRB3A and deciphers the signaling pathway conferring to CRB3A the ability to fulfill these functions. Thereby, our data will facilitate further investigation of CRB3A functions and increase our understanding of the cellular defects associated with the loss of CRB3A expression in cancer cells.
INTRODUCTION
The physiology of epithelial cells relies on the asymmetric distribution of specific cellular constituents—a structural organization referred to as epithelial polarity (1). Epithelial cell polarization results in the regionalization of the plasma membrane into apical, lateral, and basal domains. In vertebrate epithelial cells, the apical and lateral domains are segregated by tight junctions (TJ), which seal the intercellular space to prevent passive diffusion across the tissue (2). Different groups of apical and lateral proteins cooperate within their respective domains to elaborate membrane territories with specific compositions and functions (3). In addition, the mutual antagonism between apical and lateral protein complexes defines a sharp apicolateral boundary (3). Pioneer studies in Drosophila melanogaster have established that one of these protein complexes is articulated around the transmembrane apical protein Crumbs (Crb) (4–7). The mammalian genome encodes three Crb orthologs, namely, CRB1, CRB2, and CRB3 (8). CRB1 is expressed mainly in the brain, cornea, and retina (9, 10). Mutations in the human CRB1 or mouse Crb1 gene cause retinal dystrophies (11–14). CRB2 distribution overlaps that of CRB1, but CRB2 is also found in other organs such as kidneys (15). CRB2 is required for retinal integrity and for gastrulation of mouse embryos (16, 17). CRB3 is widely expressed in epithelial tissues and exists as two isoforms, namely, CRB3A and CRB3B (18, 19). The latter associates with spindle poles during mitosis or is found in the primary cilium of epithelial cells to control cytokinesis and ciliogenesis (19). CRB3A is apically localized and is required for the formation of tight junctions in cultured epithelial cells (18, 20–22). Moreover, CRB3A is required for apical-basal polarity and promotes apical membrane growth (23, 24). Knockout of mouse Crb3 is associated with epithelial tissue morphogenesis defects and perinatal lethality (25). The CRB3A isoform shares a conserved cytoplasmic tail with Crb, CRB1, and CRB2, but the sequence of its extracellular domain diverged from the sequences of other CRB proteins (8, 18).
The extracellular domain of CRB3 is N-glycosylated (18), but the function of this posttranslational modification remains unknown. The last four amino acids of the cytoplasmic tail of CRB3A (ERLI) define a PDZ domain-binding motif (PBM), which interacts with the PDZ domain protein PALS1 that recruits PATJ into the CRB3A complex (26). PALS1 and PATJ act as critical downstream effectors of CRB3A and contribute to epithelial cell polarity and tight junction formation (24, 27–29). The cytoplasmic tail of CRB3A also contains a FERM (4.1, ezrin, radixin, and moesin) domain-binding motif (FBM), which has poorly defined functional roles. However, it was shown that the intracellular domain of CRB3A associates with FERM domain proteins, including Ehm2 (also referred to as Lulu2) (25, 30, 31). Ehm2 enhances the activity of p114RhoGEF, which is recruited to cell-cell contacts by PATJ (32). p114RhoGEF is a guanine nucleotide exchange factor (GEF) activating the small GTPase RhoA at cell-cell contacts (33). The Ehm2/p114RhoGEF module organizes the circumferential actomyosin belt by activating RhoA and its effector kinases ROCK1 and ROCK2 (ROCK1/2) (31–34). These kinases modulate the contractility of the actomyosin ring via phosphorylation of the myosin regulatory light chain (MRLC), thereby activating myosin II activity (35, 36). Mechanical forces generated by the actomyosin ring are required for cell-cell adhesion and cell morphology in epithelial tissues (37–42). In addition, ROCK1/2 phosphorylate and activate ERM proteins (43–45). The ERM family is composed of the closely related proteins ezrin, radixin, and moesin. These proteins contain a FERM domain at their N termini and an F-actin binding site at their C-terminal ends (46, 47). ERM proteins link the plasma membrane or membrane-associated proteins to actin filaments. Thereby, ERM family members contribute to actin organization and stabilize epithelial cell architecture (46, 47). Although some actin regulators interact with components of the CRB3A complex, it remains to be determined whether CRB3A contributes to the organization and regulation of the actin-myosin meshwork.
The importance of the polarized architecture of epithelial cells is emphasized by the fact that most human cancers are associated with epithelial polarity defects (48, 49). Loss of CRB3 expression correlates with the increased tumorigenic potential of mouse kidney epithelial cells transformed by repeated cycles of injection in nude mice and in in vitro culture (50). Reexpression of CRB3A in these cells restored cell polarization while limiting cell proliferation, cell motility, and metastasis (50). In addition, CRB3 expression is directly repressed by Snail and ZEB1, which promote the epithelial-to-mesenchymal transition (EMT) (51–53). EMT is a hallmark of cancer progression characterized by loss of epithelial polarity, alteration of cell-cell adhesion, and increased cell invasion (54). Importantly, expression of exogenous CRB3A partially blocks Snail-induced EMT (53). Altogether, these studies suggest that CRB3A plays a crucial role in maintaining the epithelial phenotype and in restricting tumor progression. However, the functional impact of the loss of CRB3A expression in human cancer cells has not been thoroughly investigated so far. In addition, the molecular mechanisms conferring to CRB3A its putative ability to repress tumor growth and progression remain elusive. Here, we expressed CRB3A in cervical carcinoma HeLa cells, which do not express endogenous CRB3A (19). We found that CRB3A organizes a cortical actin-myosin meshwork to improve cell-cell cohesion and restore an epithelial-cell-like morphology through activation of an Ehm2-p114RhoGEF-RhoA-ROCK1/2 pathway. Using a structure-function analysis, we also established that these roles of CRB3A require both functional motifs of its cytoplasmic tail but are independent of glycosylation of its extracellular domain.
MATERIALS AND METHODS
Plasmid constructs and mutagenesis.
The cDNA encoding Myc-tagged CRB3A was obtained from B. Margolis (University of Michigan, Ann Harbor, MI). Deletion of the PBM (last four amino acids of the cytoplasmic tail [ERLI]; referred to as CRB3AΔERLI) or point mutations in the FBM (Y93A, P95A, and E99A; here, CRB3AMutFBM) or in the N-glycosylation site of CRB3A (N36A; here, CRB3AN36A) were generated by PCR and cloned into an MFGi puromycin expression vector (gift from M. Caruzo, Laval University, Canada) using an In-Fusion cloning kit (Clontech Laboratories, Mountain View, CA). The cDNA encoding Flag-tagged Ehm2 was synthesized at GenScript (Piscataway, NJ). Positive clones were fully sequenced and purified using a HiPure Plasmid MidiPrep kit (Life Technologies, Burlington, Canada).
Cell culture and transfection.
HeLa, HEK 293T, MDCK, MDA-MB-231, and Caco-2 cells were cultured at 37°C in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% fetal bovine serum (Wisent, Inc., Saint-Bruno, Canada), 2 mM glutamine, 10 mM HEPES, 50 units/ml penicillin, and 50 μg/ml streptomycin (Life Technologies) under a humidified atmosphere containing 5% CO2. Minimal essential medium (MEM; 1×) with nonessential amino acids (Life Technologies) was also added to the culture medium of MDCK and MDA-MB-231 cells. MCF7 cells were grown in the alpha modification of Eagle's medium (AMEM; Wisent, Inc.) supplemented with 10% fetal bovine serum, 2 mM glutamine, 10 mM HEPES, 50 units/ml penicillin, and 50 μg/ml streptomycin. Cells were transfected using Fugene HD transfection reagent according to the manufacturer's instructions (Promega, Madison, WI). Transfected cells were selected with 2 μg/ml puromycin (Bioshop, Burlington, Canada), which was maintained in the culture medium. For immunostaining experiments, cells were seeded and grown on glass coverslips (VWR, Radnor, Germany). Where indicated, chemical inhibitors were added to cell cultures at 48 h postseeding (6 h at 37°C) at the following concentrations: Y-27632 (Santa Cruz Biotechnology, Dallas, TX), 50 μM; blebbistatin (Santa Cruz Biotechnology), 50 μM. Dimethyl sulfoxide (DMSO; Bioshop), which was used to dissolve blebbistatin, was added to control cells.
Immunofluorescence microscopy.
For all stainings excepting p114RhoGEF immunolabeling, cells were washed in phosphate-buffered saline (PBS) and fixed in 4% paraformaldehyde (diluted in PBS) for 20 min at room temperature (RT). Fixed cells were permeabilized with a solution of 0.1% Triton X-100 in PBS (PBT) for 15 min and saturated with PBT containing 2% bovine serum albumin (BSA) for 30 min at RT. Primary antibodies were diluted in PBT containing 0.2% BSA and incubated for 1 h at RT. For p114RhoGEF staining, cells were fixed in 1% paraformaldehyde for 10 min at RT, permeabilized in PBT for 15 min, and blocked in PBT supplemented with 2% BSA for 1 h at 37°C. Incubation with the p114RhoGEF antibody (diluted in PBT–0.2% BSA) was performed at 37°C for 90 min. In all cases, cells were then washed three times with PBT and incubated with secondary antibodies diluted in PBT–0.2% BSA for 30 min at RT. F-actin was stained with Cy3-coupled phalloidin (1 U/ml; Invitrogen/Life Technologies), which was coincubated with secondary antibodies. Nuclei were stained with 1 μg/ml 4′,6′-diamidino-2-phenylindole (DAPI; Roche, Mississauga, Canada) for 5 min at RT. Finally, cells were washed three times with PBT and once with distilled water, and coverslips were mounted using Vectashield (Vector Laboratories, Inc., Burlington, Ontario, Canada). Confocal images were acquired using a confocal microscope (FV1000; Olympus) using a 40× Apochromat lens with a numerical aperture of 0.90. Images were analyzed and processed uniformly using ImageJ (National Institutes of Health) or Photoshop (Adobe). Primary antibodies used were rabbit anti-Myc (1/100; Sigma-Aldrich, Oakville, Canada), rabbit anti-PALS1 (1/50; Santa Cruz Biotechnology), mouse anti-phospho-myosin regulatory light chain (Ser19) (1/50; New England Biolabs, Whitby, Canada), rabbit anti-p114RhoGEF (1/50; GeneTex, Irvine, CA), and goat anti-Ehm2 (1/50; GeneTex). Secondary antibodies were conjugated to Cy3 (Jackson ImmunoResearch Laboratories, West Grove, PA), Alexa Fluor 488 (Molecular Probes/Life Technologies), or Alexa Fluor 647 (Jackson ImmunoResearch Laboratories) and used at a dilution of 1/400.
Western blotting.
Cells were rinsed with PBS, homogenized in ice-cold lysis buffer (40 mM Tris-HCl, pH 7.6, 1% Triton X-100, 40 mM β-glycero-phosphate, 100 mM NaCl, 1 mM EDTA, 5% glycerol, 50 mM NaF, 0.1 mM sodium orthovanadate, 0.1 mM phenylmethylsulfonyl fluoride, 10 μg/ml aprotinin, 10 μg/ml leupeptin, and 0.7 μg/ml pepstatin), and processed for SDS-PAGE and Western blotting as previously described (55). Primary antibodies used were rabbit anti-Myc (1/10,000; Sigma-Aldrich), mouse anti-RhoA (1/500) (26C4; Santa Cruz Biotechnology), rabbit anti-ROCK2 (1/2,000) (H85; Santa Cruz Biotechnology), rabbit anti-ROCK1 (1/2,000; Abcam), rabbit anti-p114RhoGEF (1/1,000; GeneTex), goat anti-Ehm2 (1/1,000; GeneTex), rabbit anti-glutathione S-transferase (anti-GST; 1:10,000) (56), rabbit antiezrin (1/2,000; New England Biolab), rabbit antimoesin (1/2,000) (Q480; New England Biolabs), mouse antiactin (1/10,000; Millipore, Etobicoke, Canada), rat anti-CRB3A (1/1,000; Medimabs, Montreal, Canada), and rat anti-CRB3 (1/5,000; kindly provided by A. Le Bivic). Horseradish peroxidase (HRP)-conjugated secondary antibodies were from GE Healthcare and were used at a 1/1,000 dilution.
Knockdown.
Small interfering RNA (siRNA)-mediated knockdowns were performed as previously described (57). Briefly, 24 h after cell plating, HeLa cells were transfected with siRNAs by calcium phosphate precipitation, whereas MDCK cells were transfected with JetPrime transfection reagent (Polyplus Transfection, France). At 16 h posttransfection, the culture medium was changed, and cells were further incubated in standard growth medium for 32 h. Cells were then harvested for Western blotting or processed for immunofluorescence. Ehm2 siRNA 1 was purchased from Qiagen (FlexiTube gene solution for EPB41L4B, GS54566; Qiagen, Toronto, Canada). All other siRNAs were Ambion siRNA duplexes obtained from Life Technologies/Applied Biosystems (Carlsbad, CA). These siRNAs contain a DNA TT dinecleotide overhang at the 3′ end. The following specific sequences were used (tt indicates the dinucleotide overhang): human Ehm2 number 2 (EPB41L4B, validated siRNA AM16708) sense, 5′-CCUGCUUAUGCUUUACACUtt-3′ human p114RhoGEF number 1 (predesigned siRNA) sense, 5′-GGACGCAACUCGGACCAAUtt-3′; human p114RhoGEF number 2 (predesigned siRNA) sense, 5′-UCAGGGCGCUUGAAAGAUAtt-3′; human RhoA number 1 (predesigned siRNA) sense, 5′-AUGGAAAGCAGGUAGAGUUtt-3′; human RhoA number 2 (predesigned siRNA) sense, 5′-GAACUAUGUGGCAGAUAUCtt-3′; human ROCK1 (predesigned siRNA) sense, 5′-GAAGAAACAUUCCCUAUUCtt-3′; human ROCK2 (predesigned siRNA) sense, 5′-GCAAAUCUGUUAAUACUCGtt-3′; human ezrin (predesigned siRNA) sense, 5′-CAAGAAGGCACCUGACUUUtt-3′; human moesin (predesigned siRNA) sense, 5′-AUAAGGAAGUGCAUAAGUCtt-3′; canine CRB3 number 1 (predesigned siRNA) sense, 5′-GGAGAUGUGCUGCUCACAUtt-3′; and canine CRB3 number 2 (predesigned siRNA) sense, 5′-GCCAUCACUGCCAUCAUUGtt-3′. Ambion Silencer Negative Control 2 siRNA (Life Technologies/Applied Biosystems) was used as a control.
RT-PCR analysis.
Cells were lysed using TRIzol reagent, and RNA was extracted according to the manufacturer's instructions (Life Technologies). Reverse transcription-PCR (RT-PCR) was performed on 2 μg of total RNA using oligo(dT) (Invitrogen/Life Technologies) and a SuperScript II reverse transcriptase kit (Invitrogen/Life Technologies) according to the manufacturer's protocol. A portion of the CRB3 cDNA was then PCR amplified using the following primers: 5′-CCTTCATCCACCAGCTCC-3′ (forward; part of exon 3) and 5′-GCTTCTCCCGAAGCTTCC-3′ (reverse; part of exon 4). A portion of actin cDNA was amplified as a control using the following primers: 5′-TGGGACGACATGGAGAAAATCT-3′ (forward; part of exon 3) and 5′-GTGTTGAAGGTCTCAAACATGA-3′ (reverse; part of exon 4).
RhoA pulldown assay.
The level of active RhoA was determined using a RhoA pulldown activation assay kit (Cytoskeleton, Inc., Denver, CO) according to the manufacturer's protocol. Briefly, cleared cell lysates were incubated with beads coupled with GST-rhotekin-RBD (where RBD is Rho binding domain) to precipitate GTP-loaded RhoA. Active, GTP-bound RhoA as well as total RhoA was detected by Western blotting.
Phase-contrast microscopy and CI.
Cells were observed with a 20× lens mounted on a CKX41 microscope (Olympus), and phase-contrast images were acquired using a DP20 camera (Olympus) coupled with MicroSuite FIVE software (Olympus). Surface area and perimeter of colonies were measured using ImageJ. The colony circularity index (CI) was then calculated using the following formula: CI = 4π(surface area/perimeter2) (58, 59). A perfect circle has a CI of 1. For each experiment, the CI was expressed as a mean of a minimum of three independent experiments in which at least 10 colonies were measured.
Immunoprecipitation.
HeLa cells were rinsed with PBS and homogenized in ice-cold lysis buffer (20 mM Tris-HCl, pH 7.4, 150 mM NaCl, 0.5% NP-40, 1.5 mM MgCl2, 1.5 mM EGTA, 10% glycerol, 1 mM phenylmethylsulfonyl fluoride, 0.1 mM sodium orthovanadate, 10 μg/ml aprotinin, 10 μg/ml leupeptin, and 0.7 μg/ml pepstatin). Two micrograms of cleared lysate was incubated with 20 μl of anti-Flag magnetic beads (Sigma-Aldrich) for 2 h at 4°C. Beads were washed five times with lysis buffer and further processed for SDS-PAGE and Western blotting.
Wound-healing assay.
Confluent control or CRB3A-expressing HeLa cells were scratched with a 200-μl micropipette tip. A picture of the resulting cell-free area was taken immediately after wounding (0 h) and then 24 h later. The percentage of the wound covered at 24 h was calculated and used as the cell migration index. Results represent the means of at least six independent experiments.
Tumor growth assay in nude mice.
Xenografting in nude mice was performed as previously described (50). Briefly, cells were trypsinized and resuspended in PBS at a concentration of 4 × 106 cells/ml. Cells were injected subcutaneously in two nude mice (1 × 106 cells/injection site; four injection sites/mouse). Three independent experiments were performed for each cell type for a total of 24 injection sites. Tumors were harvested 21 days postinjection and weighed.
Statistical analysis.
Statistical significance was determined using one-way analysis of variance (StatPlus software; Analysoft, Inc.). Results were considered significant if they reached a 95% confidence level.
RESULTS
CRB3 promotes cortical F-actin organization and establishes an epithelial-cell-like morphology.
The functional impact of altered CRB3 expression in human cancer cells remains elusive. To shed light on this issue, we expressed Myc-tagged CRB3A in HeLa cells, which are devoid of endogenous CRB3A (Fig. 1A and B) (19). As expected, Myc-CRB3A showed membrane localization and was able to recruit its binding partner PALS1 (Fig. 1C to F). Consistent with the smooth and uninterrupted distribution of Myc-CRB3A, cells expressing this protein formed tightly packed colonies of cohesive cells and adopted an epithelial-cell-like morphology. In contrast, control HeLa cells displayed an ameboid morphology and poorly adhered to each other (Fig. 1G to J). This striking change in cell morphology and cell-cell organization was quantified using the colony circularity index (58, 59), which was significantly increased by the expression of Myc-CRB3A (Fig. 1K). Importantly, this effect was not specific to HeLa cells as expression of Myc-CRB3A caused a similar phenotype in the breast cancer cell lines MCF7 and MDA-MB-231, as well as in transformed HEK 293T cells (see Fig. S1A to G in the supplemental material).
FIG 1.
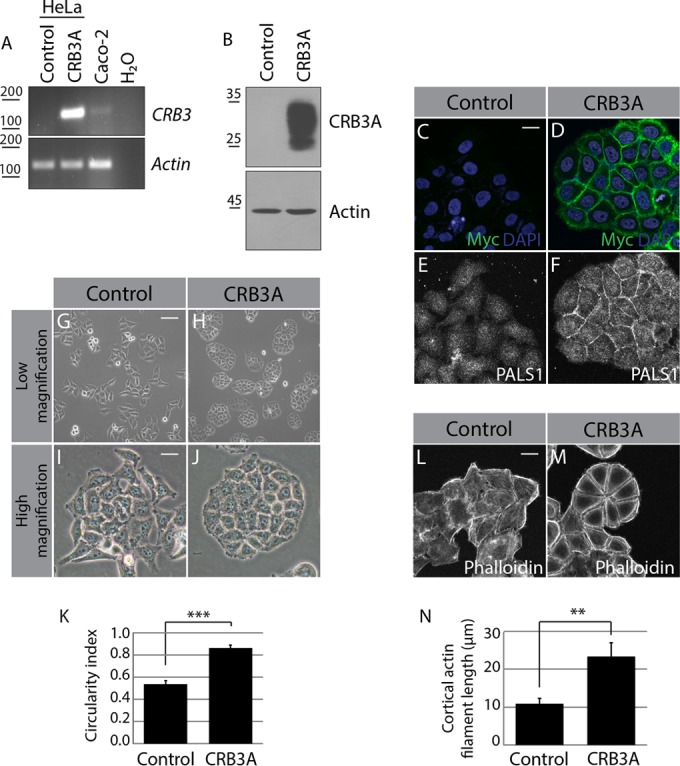
CRB3A modifies cell morphology and actin network organization. (A) RT-PCR/PCR analysis of CRB3 expression in control HeLa cells (transfected with the empty MFGi vector) or Myc-CRB3A-expressing cells (CRB3A). Caco-2 cells were used as a positive control for endogenous CRB3 expression, whereas a reaction without cDNA was used as a negative control (H2O). (B) Western blot analysis of CRB3A expression in control HeLa cells or Myc-CRB3A-expressing cells. (C and D) Immunofluorescence performed on control HeLa cells or Myc-CRB3A-expressing cells using an anti-Myc tag antibody (green). Nuclei were stained with DAPI (blue). (E and F) Immunostaining of PALS1 in control HeLa cells or Myc-CRB3A-expressing cells. (G to J) Panels show low-magnification phase-contrast images of control HeLa cells or Myc-CRB3A-expressing cells or high-magnification pictures of these cells. (K) Histogram showing the circularity indexes of the colonies formed by control HeLa cells or Myc-CRB3A-expressing cells. Bars represent means ± standard deviations. (L and M) F-actin labeling using phalloidin in control HeLa cells or Myc-CRB3A-expressing cells. (N) Histogram showing the average length of cortical actin filaments in control and Myc-CRB3A-expressing HeLa cells. Bars represent means ± standard deviations (**, P < 0.01; ***, P < 0.001). Scale bars, 20 μm (C and L), 100 μm (G), 30 μm (I).
The actin cytoskeleton plays crucial roles in coordinating cell morphology and cell-cell adhesion (42, 60). F-actin primarily appeared as scattered fibers as well as cytoplasmic and membrane-associated patches in control HeLa cells (Fig. 1L). In contrast, actin filaments formed thick and regular subcortical bundles in cells expressing Myc-CRB3A (Fig. 1M; see also Fig. S1H to M in the supplemental material). Of note, cortical actin filaments were significantly longer upon expression of Myc-CRB3A (Fig. 1N). This CRB3A-induced phenotype is reminiscent of the actin network found in normal adherent epithelial cells (42). This suggests that CRB3A usually contributes to the organization of the cytoskeleton and cell architecture in epithelial tissue. To support this premise, we knocked down endogenous CRB3 in MDCK cells, a common model for normal epithelial cells. Knockdown of CRB3 in MDCK cells decreased the amount of cortical F-actin and PALS1 (see Fig. S2A to D). Moreover, CRB3-depleted cells showed partially impaired cell-cell cohesion and formed loose colonies compared to cells transfected with a control siRNA (see Fig. S2B to D, arrows). Of note, these results were confirmed using another siRNA targeting CRB3 (data not shown). Together, these data show that CRB3A promotes the organization of cortical microfilaments, supports cell-cell cohesion, and sustains epithelial cell morphology.
To further explore the consequences of CRB3A expression in cancer cells, we examined the behavior of control and Myc-CRB3A-expressing HeLa cells in a wound-healing assay and in a xenograft experiment in nude mice. The former evaluates directional migration, whereas the latter reflects cellular growth and survival in an in vivo environment. These cellular processes are crucial for tumorigenesis and tumor progression. We found that Myc-CRB3A expression decreased the ability of HeLa cells to migrate and cover the wounded area (see Fig. S3A to E in the supplemental material). Myc-CRB3A expression also significantly reduced the efficiency of tumor formation in nude mice (see Fig. S3F). In addition, Myc-CRB3A-expressing HeLa cells produced smaller tumors than control cells (see Fig. S3G). These observations support the notion that loss of CRB3A expression contributes to the tumorigenic potential of epithelial cells.
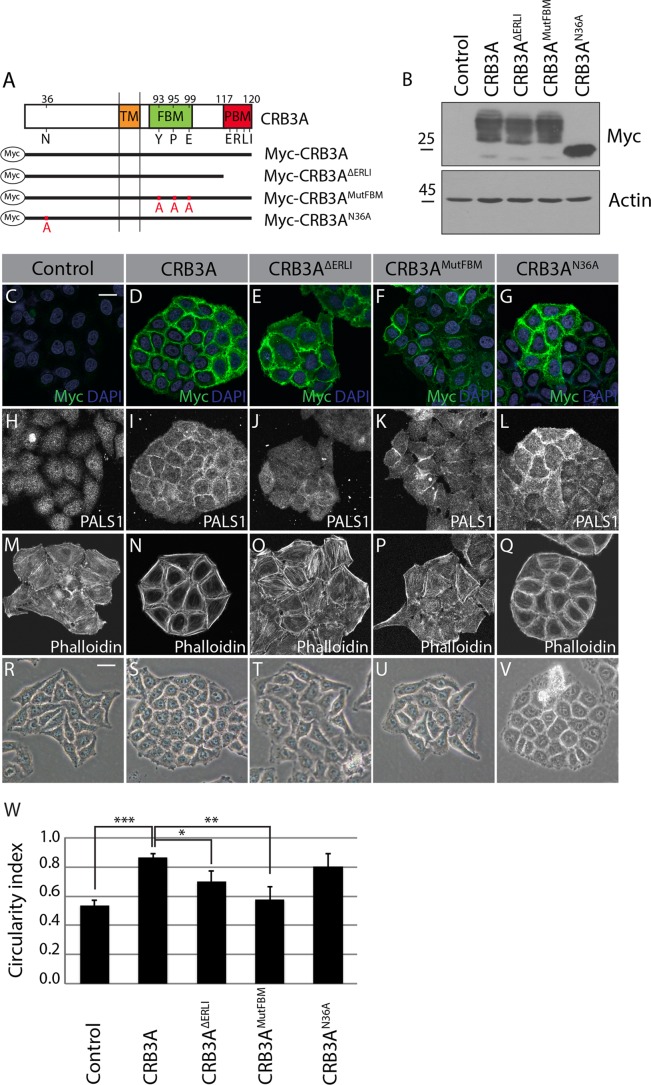
CRB3A-dependent actin organization and changes in cell shape require both the FBM and PBM.
To explore the molecular basis sustaining CRB3A functions in actin organization and epithelial cell morphology, we generated truncated or mutated Myc-CRB3A proteins and performed a structure-function analysis (Fig. 2A). Mutant proteins were expressed at levels comparable to the wild-type Myc-CRB3A level and were mainly localized at the plasma membrane (Fig. 2B to G). In agreement with previous findings (22, 24), truncation of the PBM (Myc-CRB3AΔERLI) abolished recruitment of PALS1 to the membrane, whereas mutation of the FBM (Myc-CRB3AMutFBM) allowed normal PALS1 distribution (Fig. 2H to K). In addition, we found that mutation of the N-glycosylation site (Myc-CRB3AN36A) had a limited impact on PALS1 localization (Fig. 2L). Phalloidin staining revealed that the CRB3A-dependent actin reorganization requires the FBM and PBM. Indeed, the actin cytoskeleton failed to form cortical bundles in cells expressing Myc-CRB3AΔERLI or Myc-CRB3AMutFBM, which showed instead an actin organization similar to that of control HeLa cells (Fig. 2M to P). In contrast, cells expressing Myc-CRB3AN36A produced a prominent continuous circumferential actin belt analogous to that of cells expressing wild-type Myc-CRB3A (Fig. 2N and Q). Phase-contrast images illustrated that the ability of CRB3A mutant proteins to reorganize the cytoskeleton paralleled their capacity to induce morphological changes. Specifically, Myc-CRB3A- and Myc-CRB3AN36A-expressing cells adopted an epithelial-cell-like morphology and formed tightly packed colonies with a higher circularity index than control HeLa cells (Fig. 2S, V, and W). In contrast, the circularity index of Myc-CRB3AΔERLI- or Myc-CRB3AMutFBM-expressing cells was reduced compared to the CIs of cells expressing wild-type Myc-CRB3A (Fig. 2S to U and W). Collectively, these results establish that CRB3A uses both functional motifs of its cytoplasmic tail to organize a cortical actin network and to support an epithelial-cell-like morphology.
FIG 2.
The CRB3A-dependent reorganization of the cytoskeleton requires both the FBM and PBM. (A) Schematics of the truncated or mutated Myc-tagged CRB3A proteins used for the structure-function analysis. Myc-CRB3AΔERLI lacks the PBM, Myc-CRB3AMutFBM contains a mutated FBM (Y93A, P95A, and E99A), and Myc-CRB3AN36A contains a mutated N-glycosylation site (N36A). Numbers indicate amino acid positions in CRB3A sequence. TM, transmembrane domain. (B) Western blotting using an anti-Myc antibody showing the relative expression of Myc-CRB3A mutant proteins in HeLa cells. Control cells were transfected with the empty MFGi vector. (C to G) Myc-tag immunostaining performed on control HeLa cells (empty vector) or HeLa cells expressing wild-type Myc-CRB3A or one of the mutant Myc-CRB3A proteins depicted in panel A. (H to L) Immunofluorescence of PALS1 in control HeLa cells (empty vector) or HeLa cells expressing Myc-CRB3A, Myc-CRB3AΔERLI, Myc-CRB3AMutFBM, or Myc-CRB3AN36A. (M to Q) F-actin staining using phalloidin in control HeLa cells (empty vector) or HeLa cells expressing Myc-CRB3A, Myc-CRB3AΔERLI, Myc-CRB3AMutFBM, or Myc-CRB3AN36A. (R to V) Phase-contrast images of control HeLa cells (empty vector) or HeLa cells expressing Myc-CRB3A, Myc-CRB3AΔERLI, Myc-CRB3AMutFBM, or Myc-CRB3AN36A. (W) Histogram showing the circularity indexes of the colonies formed by the cell lines shown in panels R to V. Bars represent means ± standard deviations (*, P < 0.05; **, P < 0.01; ***, P < 0.001). Scale bars, 20 μm (C) and 30 μm (R).
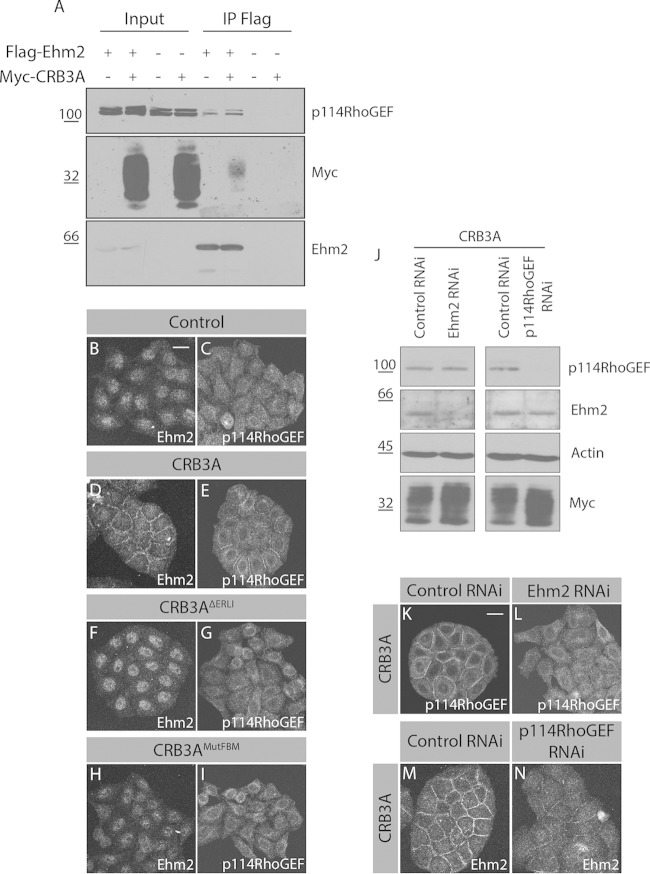
CRB3A-induced cortical actin remodeling requires Ehm2 and p114RhoGEF.
The FBM of CRB1 binds to Ehm2, which also interacts with the cytoplasmic tail of CRB3A (30). The Ehm2-associated protein p114RhoGEF interacts with PATJ (34). The latter is recruited into the CRB3A complex in a PBM-dependent manner (7). These observations led us to propose that the PBM and FBM of CRB3A cooperate to recruit the Ehm2/p114RhoGEF signaling module and to organize actin at the cell cortex. In support of this hypothesis, we found that CRB3A and p114RhoGEF were coprecipitated with Ehm2 (Fig. 3A). In addition, Ehm2 and p114RhoGEF accumulated at cell-cell boundaries in Myc-CRB3A-expressing cells, whereas they showed a diffuse distribution in control cells (Fig. 3B to E). Moreover, Ehm2 and p114RhoGEF relocalization was abolished by mutation of the FBM or truncation of the PBM (Fig. 3F to I). To investigate the functional role of Ehm2 and p114RhoGEF downstream of CRB3A, we used RNA interference (RNAi) to knock down their expression in HeLa cells expressing Myc-CRB3A. Depletion of Ehm2 had no impact on p114RhoGEF expression levels and vice versa, but p114RhoGEF remained cytoplasmic in the absence of Ehm2 (Fig. 3J to L). Reciprocally, Ehm2 cortical localization was reduced in the absence of p114RhoGEF (Fig. 3M and N). Thus, the localization of these proteins is interdependent.
FIG 3.
CRB3A recruits the Ehm2/p114RhoGEF signaling module to cell-cell contacts. (A) Flag-Ehm2 was immunoprecipitated (IP) from control or CRB3A-expressing HeLa cells. Western blotting detected immunopurified Flag-Ehm2, as well as p114RhoGEF and CRB3A that were coimmunoprecipitated. (B to I) Immunofluorescence of Ehm2 or p114RhoGEF in control HeLa cells (empty vector) or HeLa cells expressing Myc-CRB3A, Myc-CRB3AΔERLI, or Myc-CRB3AMutFBM. (J) Western blot analysis validating Ehm2 or p114RhoGEF knockdown in Myc-CRB3A-expressing HeLa cells. Actin was used as a loading control, whereas Western blotting using an anti-Myc antibody shows that Myc-CRB3A expression is maintained in knocked down cells. (K and L) Immunofluorescence of p114RhoGEF in Myc-CRB3A-expressing HeLa cells transfected with a scrambled siRNA (control RNAi) or knockdown for Ehm2 (Ehm2 RNAi). (M and N) Immunostaining of Ehm2 in Myc-CRB3A-expressing HeLa cells knocked down for p114RhoGEF (p114RhoGEF RNAi). A scrambled siRNA was used as a control (control RNAi). Scale bar, 20 μm.
Knockdown of either Ehm2 or p114RhoGEF suppressed CRB3A-induced actin reorganization (Fig. 4A to C). Loss of Ehm2 or p114RhoGEF also altered cell morphology and significantly decreased the circularity index of Myc-CRB3A-expressing cells (Fig. 4D to G). These results were confirmed using another siRNA targeting Ehm2 or p114RhoGEF (data not shown). In addition, the ability of CRB3A to limit cell migration in a wound-healing assay required Ehm2 and p114RhoGEF. Indeed, while expression of CRB3A repressed cell migration, Myc-CRB3A HeLa cells knocked down for either Ehm2 or p114RhoGEF showed a comparable migration ability to that of control HeLa cells (see Fig. S3A to E and S4 in the supplemental material). Collectively, these data show that CRB3A uses both the FBM and PBM motifs to gather a membrane-associated Ehm2/p114RhoGEF module, which is required for the formation of subcortical actin bundles and acquisition of an epithelial-cell-like morphology, and to control cell migration downstream of CRB3A.
FIG 4.
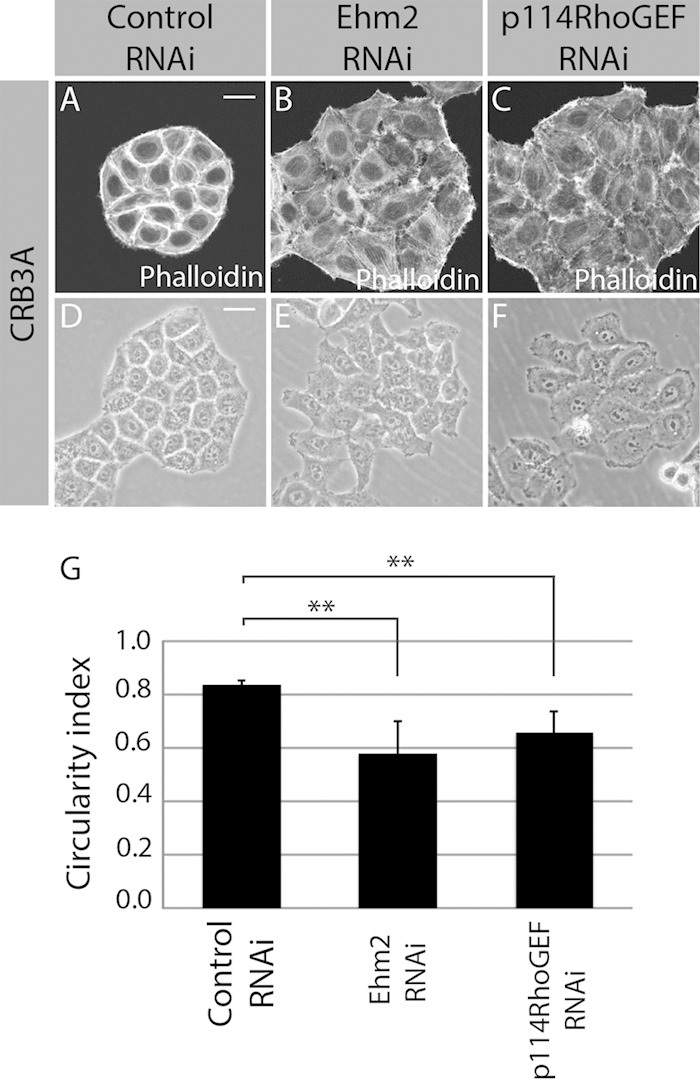
Ehm2 and p114RhoGEF mediate CRB3A functions. (A to C) F-actin staining using phalloidin in Myc-CRB3A-expressing cells transfected with a scrambled siRNA (control RNAi), an siRNA targeting Ehm2 (Ehm2 RNAi), or an siRNA targeting p114RhoGEF (p114RhoGEF RNAi). (D to F) Phase-contrast images of Myc-CRB3A-expressing cells knocked down for Ehm2 (Ehm2 RNAi) or p114RhoGEF (p114RhoGEF RNAi). A scrambled siRNA was used as a control (control RNAi). The circularity indexes of the colonies formed by these cells were measured and plotted in the histogram shown in panel G. Bars represent means ± standard deviations (**, P < 0.01). Scale bars, 20 μm (A) and 30 μm (D).
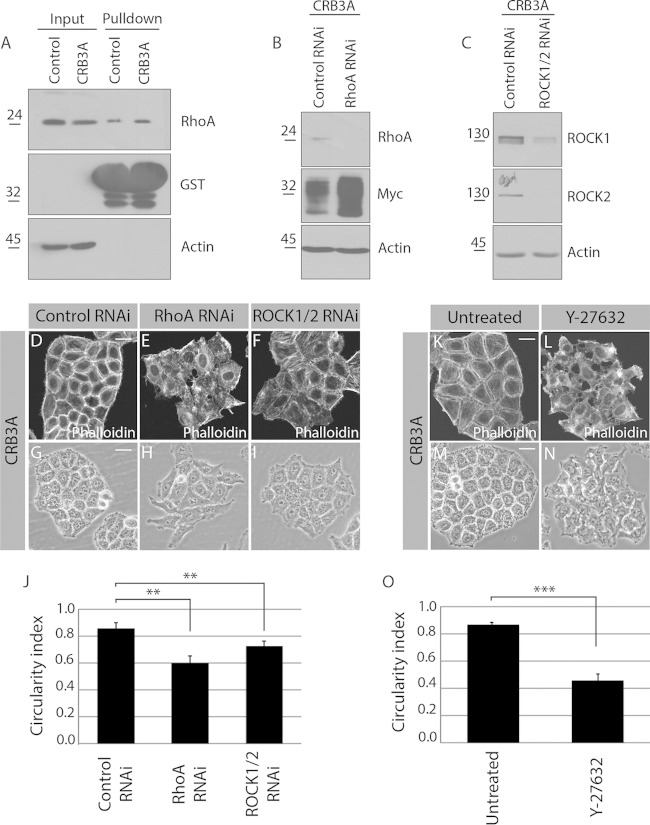
RhoA and ROCK1/2 act downstream of CRB3A to support actin organization.
The primary function of p114RhoGEF is to activate the small GTPase RhoA (33). Thus, the CRB3A-dependent recruitment of p114RhoGEF likely results in a local activation of RhoA and of its effector kinases ROCK1/2, which may be responsible for actin reorganization downstream of CRB3A. Accordingly, Myc-CRB3A-expressing cells showed an increased RhoA activation level compared to that of control cells (Fig. 5A). In addition, knockdown of RhoA or ROCK1/2 suppressed the formation of cortical actin bundles associated with the expression of Myc-CRB3A (Fig. 5B to F). Depletion of RhoA or ROCK1 and ROCK2 also decreased the circularity index of colonies formed by Myc-CRB3A-expressing cells (Fig. 5G to J). We obtained similar results with two independent siRNAs targeting RhoA (data not shown). To further support the role of ROCK1/2 downstream of CRB3A, we blocked their activity using the chemical inhibitor Y-27632. This inhibitor disrupted the actin ring in Myc-CRB3A cells, which then showed actin organization similar to that of control HeLa cells (Fig. 1L and 5K and L). The morphology of Myc-CRB3A-expressing cells also changed drastically upon inhibition of ROCK1/2. Indeed, Y-27632-treated Myc-CRB3A cells formed long protrusions in contrast to the morphology of untreated cells. Moreover, Myc-CRB3A-expressing cells in which the activity of ROCK1/2 was inhibited showed poor cell-cell contacts and a low circularity index, as observed for control HeLa cells (Fig. 1G, I, and K and 5M to O). Together, these data establish that CRB3A signals through the RhoA-ROCK1/2 module to generate a cortical actin network and to modify cell shape and intercellular interactions.
FIG 5.
RhoA and ROCK1/2 are required for the CRB3A-induced reorganization of the cytoskeleton. (A) Control HeLa cells and Myc-CRB3A-expressing HeLa cells were homogenized. A portion of each homogenate was kept to monitor RhoA expression levels (input), and GST-rhotekin-RBD was used to pull down active RhoA, which was detected by Western blotting. Western blotting also controlled the amount of GST-rhotekin-RBD used in each experiment. Expression of Myc-CRB3A is associated with a 25% increase in RhoA activation (n = 3). (B and C) Western blots validating the knockdown of RhoA (RhoA RNAi) or ROCK1 and ROCK2 (ROCK1/2 RNAi). Actin was used as a loading control. (D to F) Phalloidin was used to stain F-actin in Myc-CRB3A-expressing HeLa cells knocked down for RhoA (RhoA RNAi) or ROCK1 and ROCK2 (ROCK1/2 RNAi). A scrambled siRNA was used as a control (control RNAi). (G to I) Phase-contrast images of Myc-CRB3A-expressing HeLa cells transfected with a scrambled siRNA (control RNAi), an siRNA targeting RhoA (RhoA RNAi), or siRNAs targeting ROCK1 and ROCK2 (ROCK1/2 RNAi). The circularity indexes of the colonies formed by these cells are shown in the histogram depicted in panel J. Bars represent means ± standard deviations (**, P < 0.01). (K and L) Staining of F-actin using phalloidin in Myc-CRB3A-expressing HeLa cells incubated with or without the ROCK1/2 inhibitor Y-27632. (M and N) Phase-contrast images of Myc-CRB3A-expressing HeLa cells treated or not with the ROCK1/2 inhibitor Y-27632. (O) Histogram showing the circularity indexes of Myc-CRB3A-expressing HeLa cells incubated with or without the ROCK1/2 inhibitor Y-27632. Bars represent means ± standard deviations (***, P < 0.001). Scale bars, 20 μm (D and K) and 30 μm (G and M).
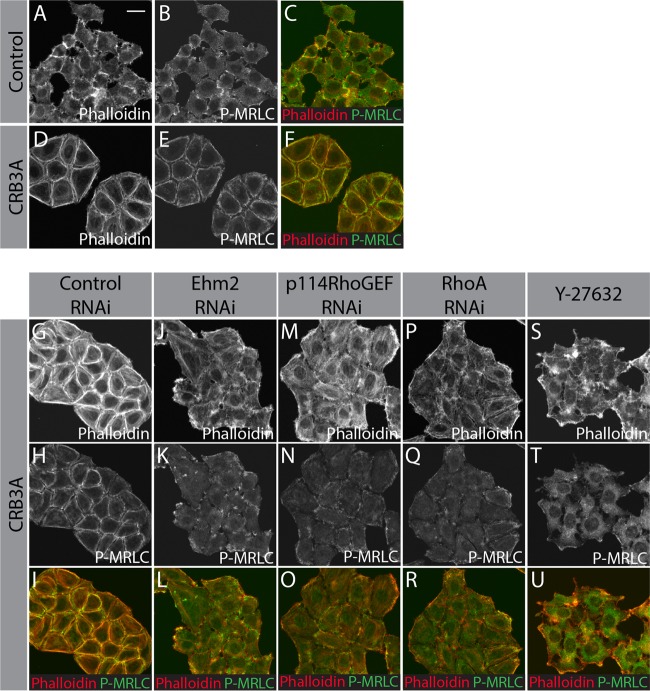
Myosin II and ERM proteins contribute to the CRB3A-dependent reorganization of cell architecture.
ROCK1/2 phosphorylate myosin regulatory light chain (MRLC), thereby stimulating myosin II activity (35). The contractile properties of the actomyosin network are fundamental for cell-cell cohesion and cell architecture (38). Strikingly, phosphorylated MRLC formed a continuous ring colocalizing with F-actin at the cell periphery in Myc-CRB3A-expressing cells, whereas control HeLa cells showed scattered patches of phospho-MRLC (Fig. 6A to F). Knockdown of Ehm2, p114RhoGEF, or RhoA or inhibition of ROCK1/2 activity fully suppressed the CRB3A-induced phospho-MRLC relocalization (Fig. 6G to U). These results suggest that CRB3A organizes a contractile circumferential actomyosin belt through the Ehm2-p114RhoGEF-RhoA-ROCK1/2 pathway.
FIG 6.
CRB3A promotes relocalization of phospho-MRLC to cell-cell contacts. F-actin staining (phalloidin; red in the merged picture) and phospho-MRLC immunostaining (P-MRLC; green) in control HeLa cells (A to C), Myc-CRB3A-expressing cells (D to F), Myc-CRB3A-expressing cells transfected with a scrambled siRNA (control RNAi; G to I), Myc-CRB3A-expressing cells knocked down for Ehm2 (Ehm2 RNAi; J to L), Myc-CRB3A-expressing cells knocked down for p114RhoGEF (p114RhoGEF RNAi; M to O), Myc-CRB3A-expressing cells knocked down for RhoA (RhoA RNAi; P to R), or Myc-CRB3A-expressing cells treated with the ROCK1/2 inhibitor Y-27632 (S to U). Scale bar, 20 μm.
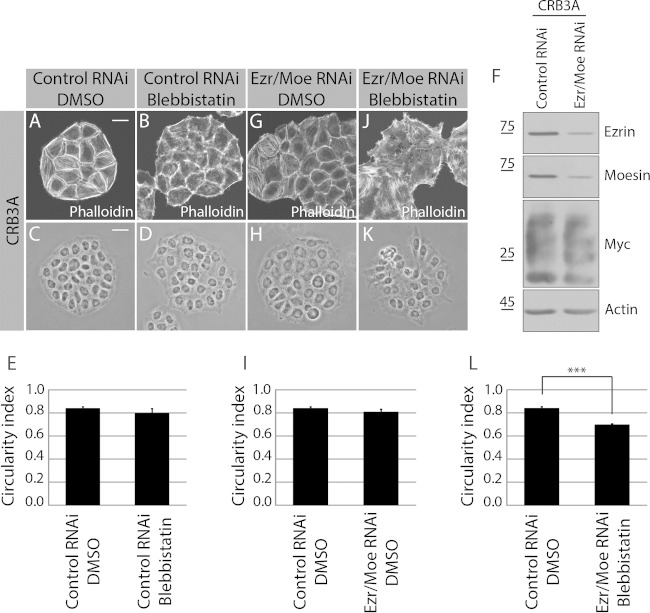
Inhibition of myosin II with blebbistatin in Myc-CRB3A-expressing cells altered cell-cell contacts, which were less strained than in untreated cells (Fig. 7A and B). This establishes that the tension built by myosin II is required downstream of CRB3A to form smooth and regular cell-cell boundaries. However, blebbistatin had a weak impact on cell morphology and the circularity index in comparison to the effect of ROCK1/2 inhibition (Fig. 5K to O and 7C to E). This suggests that in addition to myosin II, other pathways are activated downstream of ROCK1/2 to mediate the CRB3A-induced actin organization and generation of an epithelial-cell-like morphology. ROCK1/2 also phosphorylates and activates ERM proteins, which play important roles in organizing cortical actin and cell shape (46). Combined knockdown of ezrin and moesin, two ERM proteins, did not modify significantly the phenotype of Myc-CRB3A-expressing cells (Fig. 7F to I). However, concomitant inhibition of myosin II and depletion of ezrin and moesin disrupted the CRB3A-dependent cortical actin network and reduced the colony circularity index (Fig. 7J to L). Together, these data establish that both myosin II and ERM proteins contribute to establish a circumferential actin belt and an epithelial-cell-like morphology downstream of CRB3A.
FIG 7.
Combined inhibition of myosin II and depletion of ezrin and moesin suppress the CRB3A-induced phenotypes. (A and B) Phalloidin was used to stain F-actin in Myc-CRB3A-expressing HeLa cells transfected with a control RNAi and incubated with DMSO or with the myosin II inhibitor blebbistatin. (C and D) Phase-contrast images of Myc-CRB3A-expressing HeLa cells transfected with a scrambled siRNA (control RNAi) and treated or not with the myosin II inhibitor blebbistatin. The circularity indexes of the colonies formed by these cells are reported in the histogram shown in panel E. (F) Western blot validating the knockdown of ezrin and moesin. (G) F-actin staining using phalloidin in Myc-CRB3A-expressing cells depleted of ezrin and moesin (Ezr/Moe RNAi). (H) Phase-contrast images of Myc-CRB3A-expressing HeLa cells knocked down for ezrin and moesin (Ezr/Moe RNAi). (I) Histogram showing the circularity index of Myc-CRB3A-expressing HeLa cells transfected with a scrambled siRNA (control RNAi) or an siRNA targeting ezrin and an siRNA targeting moesin (Ezr/Moe RNAi). (J) F-actin staining (phalloidin) in Myc-CRB3A-expressing HeLa cells treated with blebbistatin and depleted of ezrin and moesin. (K) Phase-contrast image of Myc-CRB3A-expressing HeLa cells treated with blebbistatin and knocked down for ezrin and moesin. The circularity indexes of colonies formed by these cells are shown in panel L. Bars represent means ± standard deviations (***, P < 0.001). Scale bars, 20 μm (A) and 30 μm (C).
DISCUSSION
In this study, we highlighted a novel role for CRB3A in epithelial cell architecture and cell-cell cohesion. The cytoplasmic tail of CRB3A is similar to the intracellular domain of Drosophila Crb, which is essential for the coalescence of adherens junction material into a zonula adherens (ZA) (61). This E-cadherin-based adhesion belt maintains the cohesion of epithelial tissues. However, HeLa cells are devoid of E-cadherin (62), and Myc-CRB3A does not induce E-cadherin expression in these cells (E. Loie and P. Laprise, unpublished data). In addition, the localization of β-catenin, which is a cytoplasmic binding partner of classical cadherins, was similar in control and CRB3A-expressing cells (Loie and Laprise, unpublished data). Although these observations do not exclude the possibility that CRB3A normally contributes to ZA integrity, as reported for its Drosophila ortholog (61), they strongly argue that CRB3A-induced cell-cell cohesion is independent of cadherin-based adherens junction in HeLa cells. It was recently suggested that the extracellular domain of Drosophila Crb and zebrafish Crb2a and Crb2b self-interact in trans to support cell-cell contacts (63, 64). Interestingly, it was also shown that CRB3A acts as a sensor of cell density (65), thus implying that CRB3A is involved in cell-cell interactions. Together with our data, these reports raise the intriguing possibility that CRB3A has conserved the ability to promote cell-cell adhesion through homotypic connections although the sequence of its extracellular domain has diverged from sequences of other CRB proteins (8).
We observed that the ability of CRB3A to induce cell-cell adhesion and to sustain an epithelial-cell-like cell shape intimately correlates with the organization of a subcortical circumferential actomyosin belt. This belt strengthens cell-cell adhesion and organizes and maintains cell morphology (37–41). We reported for the first time that CRB3A controls RhoA signaling, which is required for CRB3A-dependent reorganization of the actin cytoskeleton according to our data. The level of RhoA activation downstream of CRB3A is modest, thus suggesting that CRB3A acts to properly localize RhoA signaling rather than stimulating general and strong activation of this small GTPase. Accordingly, we found that CRB3A promotes the membrane localization of p114RhoGEF, which specifically increases RhoA signaling at cell-cell contacts (33). Knockdown of either Ehm2 or p114RhoGEF mimics RhoA depletion. Similarly, inhibition of ROCK1/2, which are critical effectors of RhoA (35), totally abrogates actin reorganization and cell shape changes associated with CRB3A expression. Thus, our data suggest that CRB3A locally activates an Ehm2-p114RhoGEF-RhoA-ROCK1/2 signaling pathway, thereby allowing formation of a cortical actin ring that supports epithelial cell morphology and cohesiveness. ROCK1/2 phosphorylate MRLC to activate myosin II activity, thereby modulating the contractility of the actomyosin circumferential belt (35). We observed that CRB3A expression induces relocation of phosphorylated MRLC to the cell periphery where it colocalizes with F-actin. Inhibition of myosin II with blebbistatin limits the apical tension resulting from Ehm2 overexpression in MDCK cells (34). Similarly, we observed that blebbistatin perturbs the linearity of cell-cell contacts in CRB3A-expressing cells. However, blebbistatin has a milder effect than the ROCK1/2 inhibitor, suggesting that these kinases target other critical effectors to fully support CRB3A-mediated reorganization of the cytoskeleton. ROCK1/2 also activate ERM proteins, such as ezrin and moesin (43–45). We found that knockdown of ezrin and moesin has a mild effect on the cytoskeleton and cell shape but that combined inhibition of myosin II and depletion of ezrin and moesin suppress the CRB3A-induced phenotype. This suggests that ROCK1/2 act on both myosin II activity and functions of ERM proteins downstream of CRB3A. Our data establishing that ERM proteins are mediators of CRB3A functions are consistent with studies showing that the knockout of mouse Crb3 or Ezrin causes similar phenotypes, including abnormal brush borders in enterocytes and fusion of intestinal villi (25, 66, 67). Moreover, it was shown that CRB3A binds to ezrin (25) and that Drosophila Crb associates with the sole ERM protein expressed in flies, namely, moesin (68). Future work is required to establish whether ERM protein activation by ROCK1/2 favors their association with CRB proteins.
We also established that both the PBM and the FBM contained in the cytoplasmic tail of CRB3A are required to recruit Ehm2 and p114RhoGEF to the membrane and to support actin organization, thus providing the first evidence of cooperativity between the PBM and FBM. The configuration of the CRB3A-Ehm2-p114RhoGEF signaling module has not yet been fully revealed, but several pieces of evidence suggest that these proteins are part of a common macromolecular complex. Indeed, we showed that CRB3A and p114RhoGEF coprecipitate with Ehm2. In addition, Ehm2 and p114RhoGEF directly bind together (34). This interaction may explain why the recruitment of these proteins to the cell periphery is interdependent to a large extent. The recruitment of the Ehm2/p114RhoGEF module could result from the interaction of the FERM domain of Ehm2 with the FBM of CRB3A (30). Alternatively, the binding of p114RhoGEF to PATJ (34), which is recruited into the CRB3A complex by PALS1 in a PBM-dependent manner (8, 32), could account for the membrane localization of Ehm2 and p114RhoGEF. Our structure-function analysis suggests that both of these interactions could be required to form a stable complex as mutation of either the FBM or the PBM contained in the cytoplasmic tail of CRB3A compromises the recruitment of Ehm2 and p114RhoGEF.
Although CRB3 shows wide distribution in different types of epithelia (18, 69), the role of this protein was mostly studied in models of columnar or cuboidal epithelial cells, which are polarized along the apical-basal axis (3). As HeLa cells originate from the stratified epithelium of the cervix that expresses CRB3 (69), our data provide evidence that CRB3A also sustains the morphology of squamous epithelial cells. In addition, we established that CRB3A promotes intercellular cohesion of HeLa cells, likely through its action on the cytoskeleton. Interestingly, our data show that CRB3A also promotes cell-cell cohesion and cytoskeleton organization in cell lines originating from the breast and kidney epithelia, which are both composed of polarized cells. It remains to be determined whether CRB3A assumes these newly described functions in parallel to its role in establishing polarity in these epithelia or whether the roles of CRB3A that we described here contribute to the organization of the apical-basal axis.
Several pieces of evidence suggest that CRB3A restricts tumor growth and progression. On the one hand, CRB3A restricts cell growth and contributes to contact-mediated inhibition of cell proliferation (50, 65). Similarly, we show in this study that CRB3A limits tumor growth in nude mice. On the other hand, the CRB3A promoter is targeted by transcription factors promoting EMT (51–53), and loss of CRB3A alters the epithelial phenotype and favors metastasis in nude mice (50). Recent studies have highlighted signaling pathways acting downstream of CRB proteins that may provide a molecular basis explaining how CRB3A restricts cell proliferation. Indeed, it was shown that CRB3A limits cell proliferation of a confluent epithelial cell culture by activating the Hippo pathway (65), a function shared by its Drosophila ortholog (70, 71). Moreover, it was shown that Crb and vertebrate CRB proteins limit the activity of the Notch signaling pathway (72–74), which is well known for its contribution to tumor cell growth (75). However, much less is known about the molecular mechanisms used by CRB3A to maintain the epithelial phenotype and how the loss of CRB3A contributes to EMT. Our data suggest that loss of CRB3A causes a transition from an epithelial-cell-like morphology to an ameboid shape, which is associated with tumor cell migration and invasion. In line with this premise, we show that CRB3A limits migration in a wound-healing assay. Moreover, our results show that CRB3A favors cell-cell cohesion, thus suggesting that CRB3A may limit cell scattering through this function. Importantly, we decipher a complex RhoA-dependent signaling pathway conferring to CRB3A its ability to organize the cytoskeleton and cell shape and to favor cell-cell interactions. Of note, Ehm2 is an essential component of this signaling pathway. Interestingly, Ehm2 is overexpressed in breast, prostate, and skin cancer (76–78). Ehm2 overexpression correlates with a high metastatic potential and poor prognosis (78). These studies may seem in contradiction with our data showing that Ehm2 is important for epithelial cell morphology and cohesion and acts downstream of CRB3A to limit migration. However, we showed that Ehm2 is localized in the cytoplasm in the absence of CRB3A, whereas it is recruited to the cell periphery in CRB3A-expressing cells. This suggests that Ehm2 fills different functions depending on its subcellular distribution. Thus, when Ehm2 is overabundant compared to CRB3A levels, it likely ectopically accumulates in the cytoplasm and serves promigratory functions. One critical function of CRB3A in limiting tumor cell invasion could thus be to localize Ehm2 to the membrane to favor its function in cell-cell adhesion rather than cell migration. Overall, our study highlighted novel functions of CRB3A and a complex signaling pathway acting downstream of this protein. Thereby, our study has a broad interest in epithelial cell biology and oncology.
Supplementary Material
ACKNOWLEDGMENTS
We are grateful to M. Caruso, A. Le Bivic, and B. Margolis for reagents. We also thank A. Turcotte for technical support. Confocal microscopy was performed at the CRCHU-Hôtel-Dieu imaging facility. DNA sequencing was carried out at the Genome Sequencing and Genotyping Platform of the CHU de Québec Research Centre.
This work was supported by an operating grant from the Cancer Research Society-Société de Recherche sur le Cancer (SRC-CRS) to P.L., who is a Fonds de Recherche du Québec-Santé junior 2 investigator.
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/MCB.00673-15.
REFERENCES
- 1.Tepass U, Tanentzapf G, Ward R, Fehon R. 2001. Epithelial cell polarity and cell junctions in Drosophila. Annu Rev Genet 35:747–784. doi: 10.1146/annurev.genet.35.102401.091415. [DOI] [PubMed] [Google Scholar]
- 2.Van Itallie CM, Anderson JM. 2014. Architecture of tight junctions and principles of molecular composition. Semin Cell Dev Biol 36:157–165. doi: 10.1016/j.semcdb.2014.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Laprise P, Tepass U. 2011. Novel insights into epithelial polarity proteins in Drosophila. Trends Cell Biol 21:401–408. doi: 10.1016/j.tcb.2011.03.005. [DOI] [PubMed] [Google Scholar]
- 4.Tepass U, Theres C, Knust E. 1990. crumbs encodes an EGF-like protein expressed on apical membranes of Drosophila epithelial cells and required for organization of epithelia. Cell 61:787–799. doi: 10.1016/0092-8674(90)90189-L. [DOI] [PubMed] [Google Scholar]
- 5.Tepass U, Knust E. 1993. Crumbs and stardust act in a genetic pathway that controls the organization of epithelia in Drosophila melanogaster. Dev Biol 159:311–326. doi: 10.1006/dbio.1993.1243. [DOI] [PubMed] [Google Scholar]
- 6.Bachmann A, Schneider M, Theilenberg E, Grawe F, Knust E. 2001. Drosophila Stardust is a partner of Crumbs in the control of epithelial cell polarity. Nature 414:638–643. doi: 10.1038/414638a. [DOI] [PubMed] [Google Scholar]
- 7.Bulgakova NA, Knust E. 2009. The Crumbs complex: from epithelial-cell polarity to retinal degeneration. J Cell Sci 122:2587–2596. doi: 10.1242/jcs.023648. [DOI] [PubMed] [Google Scholar]
- 8.Bazellieres E, Assemat E, Arsanto JP, Le Bivic A, Massey-Harroche D. 2009. Crumbs proteins in epithelial morphogenesis. Front Biosci 14:2149–2169. [DOI] [PubMed] [Google Scholar]
- 9.den Hollander AI, Ghiani M, de Kok YJ, Wijnholds J, Ballabio A, Cremers FP, Broccoli V. 2002. Isolation of Crb1, a mouse homologue of Drosophila crumbs, and analysis of its expression pattern in eye and brain. Mech Dev 110:203–207. doi: 10.1016/S0925-4773(01)00568-8. [DOI] [PubMed] [Google Scholar]
- 10.Beyer J, Zhao XC, Yee R, Khaliq S, McMahon TT, Ying H, Yue BY, Malicki JJ. 2010. The role of crumbs genes in the vertebrate cornea. Invest Ophthalmol Vis Sci 51:4549–4556. doi: 10.1167/iovs.09-4549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.den Hollander AI, Davis J, van der Velde-Visser SD, Zonneveld MN, Pierrottet CO, Koenekoop RK, Kellner U, van den Born LI, Heckenlively JR, Hoyng CB, Handford PA, Roepman R, Cremers FP. 2004. CRB1 mutation spectrum in inherited retinal dystrophies. Hum Mutat 24:355–369. doi: 10.1002/humu.20093. [DOI] [PubMed] [Google Scholar]
- 12.Mehalow AK, Kameya S, Smith RS, Hawes NL, Denegre JM, Young JA, Bechtold L, Haider NB, Tepass U, Heckenlively JR, Chang B, Naggert JK, Nishina PM. 2003. CRB1 is essential for external limiting membrane integrity and photoreceptor morphogenesis in the mammalian retina. Hum Mol Genet 12:2179–2189. doi: 10.1093/hmg/ddg232. [DOI] [PubMed] [Google Scholar]
- 13.van de Pavert SA, Meuleman J, Malysheva A, Aartsen WM, Versteeg I, Tonagel F, Kamphuis W, McCabe CJ, Seeliger MW, Wijnholds J. 2007. A single amino acid substitution (Cys249Trp) in Crb1 causes retinal degeneration and deregulates expression of pituitary tumor transforming gene Pttg1. J Neurosci 27:564–573. doi: 10.1523/JNEUROSCI.3496-06.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Richard M, Roepman R, Aartsen WM, van Rossum AG, den Hollander AI, Knust E, Wijnholds J, Cremers FP. 2006. Towards understanding CRUMBS function in retinal dystrophies. Hum Mol Genet 15:R235–R243. doi: 10.1093/hmg/ddl195. [DOI] [PubMed] [Google Scholar]
- 15.van den Hurk JA, Rashbass P, Roepman R, Davis J, Voesenek KE, Arends ML, Zonneveld MN, van Roekel MH, Cameron K, Rohrschneider K, Heckenlively JR, Koenekoop RK, Hoyng CB, Cremers FP, den Hollander AI. 2005. Characterization of the Crumbs homolog 2 (CRB2) gene and analysis of its role in retinitis pigmentosa and Leber congenital amaurosis. Mol Vis 11:263–273. [PubMed] [Google Scholar]
- 16.Xiao Z, Patrakka J, Nukui M, Chi L, Niu D, Betsholtz C, Pikkarainen T, Vainio S, Tryggvason K. 2011. Deficiency in Crumbs homolog 2 (Crb2) affects gastrulation and results in embryonic lethality in mice. Dev Dyn 240:2646–2656. doi: 10.1002/dvdy.22778. [DOI] [PubMed] [Google Scholar]
- 17.Alves CH, Sanz AS, Park B, Pellissier LP, Tanimoto N, Beck SC, Huber G, Murtaza M, Richard F, Gurubaran IS, Garrido MG, Levelt CN, Rashbass P, Le Bivic A, Seeliger MW, Wijnholds J. 2013. Loss of CRB2 in the mouse retina mimics human retinitis pigmentosa due to mutations in the CRB1 gene. Hum Mol Genet 22:35–50. doi: 10.1093/hmg/dds398. [DOI] [PubMed] [Google Scholar]
- 18.Makarova O, Roh MH, Liu CJ, Laurinec S, Margolis B. 2003. Mammalian Crumbs3 is a small transmembrane protein linked to protein associated with Lin-7 (Pals1). Gene 302:21–29. doi: 10.1016/S0378111902010843. [DOI] [PubMed] [Google Scholar]
- 19.Fan S, Fogg V, Wang Q, Chen XW, Liu CJ, Margolis B. 2007. A novel Crumbs3 isoform regulates cell division and ciliogenesis via importin beta interactions. J Cell Biol 178:387–398. doi: 10.1083/jcb.200609096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lemmers C, Michel D, Lane-Guermonprez L, Delgrossi MH, Medina E, Arsanto JP, Le Bivic A. 2004. CRB3 binds directly to Par6 and regulates the morphogenesis of the tight junctions in mammalian epithelial cells. Mol Biol Cell 15:1324–1333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hurd TW, Gao L, Roh MH, Macara IG, Margolis B. 2003. Direct interaction of two polarity complexes implicated in epithelial tight junction assembly. Nat Cell Biol 5:137–142. doi: 10.1038/ncb923. [DOI] [PubMed] [Google Scholar]
- 22.Fogg VC, Liu CJ, Margolis B. 2005. Multiple regions of Crumbs3 are required for tight junction formation in MCF10A cells. J Cell Sci 118:2859–2869. doi: 10.1242/jcs.02412. [DOI] [PubMed] [Google Scholar]
- 23.Schluter MA, Pfarr CS, Pieczynski J, Whiteman EL, Hurd TW, Fan S, Liu CJ, Margolis B. 2009. Trafficking of Crumbs3 during cytokinesis is crucial for lumen formation. Mol Biol Cell 20:4652–4663. doi: 10.1091/mbc.E09-02-0137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Roh MH, Fan S, Liu CJ, Margolis B. 2003. The Crumbs3-Pals1 complex participates in the establishment of polarity in mammalian epithelial cells. J Cell Sci 116:2895–2906. doi: 10.1242/jcs.00500. [DOI] [PubMed] [Google Scholar]
- 25.Whiteman EL, Fan S, Harder JL, Walton KD, Liu CJ, Soofi A, Fogg VC, Hershenson MB, Dressler GR, Deutsch GH, Gumucio DL, Margolis B. 2014. Crumbs3 is essential for proper epithelial development and viability. Mol Cell Biol 34:43–56. doi: 10.1128/MCB.00999-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Roh MH, Makarova O, Liu CJ, Shin K, Lee S, Laurinec S, Goyal M, Wiggins R, Margolis B. 2002. The Maguk protein, Pals1, functions as an adapter, linking mammalian homologues of Crumbs and Discs Lost. J Cell Biol 157:161–172. doi: 10.1083/jcb.200109010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Straight SW, Shin K, Fogg VC, Fan S, Liu CJ, Roh M, Margolis B. 2004. Loss of PALS1 expression leads to tight junction and polarity defects. Mol Biol Cell 15:1981–1990. doi: 10.1091/mbc.E03-08-0620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Michel D, Arsanto JP, Massey-Harroche D, Beclin C, Wijnholds J, Le Bivic A. 2005. PATJ connects and stabilizes apical and lateral components of tight junctions in human intestinal cells. J Cell Sci 118:4049–4057. doi: 10.1242/jcs.02528. [DOI] [PubMed] [Google Scholar]
- 29.Shin K, Straight S, Margolis B. 2005. PATJ regulates tight junction formation and polarity in mammalian epithelial cells. J Cell Biol 168:705–711. doi: 10.1083/jcb.200408064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Laprise P, Beronja S, Silva-Gagliardi NF, Pellikka M, Jensen AM, McGlade CJ, Tepass U. 2006. The FERM protein Yurt is a negative regulatory component of the Crumbs complex that controls epithelial polarity and apical membrane size. Dev Cell 11:363–374. doi: 10.1016/j.devcel.2006.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nakajima H, Tanoue T. 2010. Epithelial cell shape is regulated by Lulu proteins via myosin-II. J Cell Sci 123:555–566. doi: 10.1242/jcs.057752. [DOI] [PubMed] [Google Scholar]
- 32.Nakajima H, Tanoue T. 2012. The circumferential actomyosin belt in epithelial cells is regulated by the Lulu2-p114RhoGEF system. Small GTPases 3:91–96. doi: 10.4161/sgtp.19112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Terry SJ, Zihni C, Elbediwy A, Vitiello E, Leefa Chong San IV, Balda MS, Matter K. 2011. Spatially restricted activation of RhoA signalling at epithelial junctions by p114RhoGEF drives junction formation and morphogenesis. Nat Cell Biol 13:159–166. doi: 10.1038/ncb2156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nakajima H, Tanoue T. 2011. Lulu2 regulates the circumferential actomyosin tensile system in epithelial cells through p114RhoGEF. J Cell Biol 195:245–261. doi: 10.1083/jcb.201104118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Julian L, Olson MF. 2014. Rho-associated coiled-coil containing kinases (ROCK): structure, regulation, and functions. Small GTPases 5:e29846. doi: 10.4161/sgtp.29846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Matsumura F, Hartshorne DJ. 2008. Myosin phosphatase target subunit: Many roles in cell function. Biochem Biophys Res Commun 369:149–156. doi: 10.1016/j.bbrc.2007.12.090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ivanov AI, Bachar M, Babbin BA, Adelstein RS, Nusrat A, Parkos CA. 2007. A unique role for nonmuscle myosin heavy chain IIA in regulation of epithelial apical junctions. PLoS One 2:e658. doi: 10.1371/journal.pone.0000658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lecuit T, Lenne PF. 2007. Cell surface mechanics and the control of cell shape, tissue patterns and morphogenesis. Nat Rev Mol Cell Biol 8:633–644. doi: 10.1038/nrm2222. [DOI] [PubMed] [Google Scholar]
- 39.Miyake Y, Inoue N, Nishimura K, Kinoshita N, Hosoya H, Yonemura S. 2006. Actomyosin tension is required for correct recruitment of adherens junction components and zonula occludens formation. Exp Cell Res 312:1637–1650. doi: 10.1016/j.yexcr.2006.01.031. [DOI] [PubMed] [Google Scholar]
- 40.Shewan AM, Maddugoda M, Kraemer A, Stehbens SJ, Verma S, Kovacs EM, Yap AS. 2005. Myosin 2 is a key Rho kinase target necessary for the local concentration of E-cadherin at cell-cell contacts. Mol Biol Cell 16:4531–4542. doi: 10.1091/mbc.E05-04-0330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Smutny M, Cox HL, Leerberg JM, Kovacs EM, Conti MA, Ferguson C, Hamilton NA, Parton RG, Adelstein RS, Yap AS. 2010. Myosin II isoforms identify distinct functional modules that support integrity of the epithelial zonula adherens. Nat Cell Biol 12:696–702. doi: 10.1038/ncb2072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Harris TJ, Tepass U. 2010. Adherens junctions: from molecules to morphogenesis. Nat Rev Mol Cell Biol 11:502–514. doi: 10.1038/nrm2927. [DOI] [PubMed] [Google Scholar]
- 43.Fukata Y, Kimura K, Oshiro N, Saya H, Matsuura Y, Kaibuchi K. 1998. Association of the myosin-binding subunit of myosin phosphatase and moesin: dual regulation of moesin phosphorylation by Rho-associated kinase and myosin phosphatase. J Cell Biol 141:409–418. doi: 10.1083/jcb.141.2.409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Matsui T, Maeda M, Doi Y, Yonemura S, Amano M, Kaibuchi K, Tsukita S, Tsukita S. 1998. Rho-kinase phosphorylates COOH-terminal threonines of ezrin/radixin/moesin (ERM) proteins and regulates their head-to-tail association. J Cell Biol 140:647–657. doi: 10.1083/jcb.140.3.647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Oshiro N, Fukata Y, Kaibuchi K. 1998. Phosphorylation of moesin by rho-associated kinase (Rho-kinase) plays a crucial role in the formation of microvilli-like structures. J Biol Chem 273:34663–34666. doi: 10.1074/jbc.273.52.34663. [DOI] [PubMed] [Google Scholar]
- 46.Fehon RG, McClatchey AI, Bretscher A. 2010. Organizing the cell cortex: the role of ERM proteins. Nat Rev Mol Cell Biol 11:276–287. doi: 10.1038/nrm2866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Neisch AL, Fehon RG. 2011. Ezrin, radixin and moesin: key regulators of membrane-cortex interactions and signaling. Curr Opin Cell Biol 23:377–382. doi: 10.1016/j.ceb.2011.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ellenbroek SI, Iden S, Collard JG. 2012. Cell polarity proteins and cancer. Semin Cancer Biol 22:208–215. doi: 10.1016/j.semcancer.2012.02.012. [DOI] [PubMed] [Google Scholar]
- 49.Dow LE, Humbert PO. 2007. Polarity regulators and the control of epithelial architecture, cell migration, and tumorigenesis. Int Rev Cytol 262:253–302. doi: 10.1016/S0074-7696(07)62006-3. [DOI] [PubMed] [Google Scholar]
- 50.Karp CM, Tan TT, Mathew R, Nelson D, Mukherjee C, Degenhardt K, Karantza-Wadsworth V, White E. 2008. Role of the polarity determinant crumbs in suppressing mammalian epithelial tumor progression. Cancer Res 68:4105–4115. doi: 10.1158/0008-5472.CAN-07-6814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Aigner K, Dampier B, Descovich L, Mikula M, Sultan A, Schreiber M, Mikulits W, Brabletz T, Strand D, Obrist P, Sommergruber W, Schweifer N, Wernitznig A, Beug H, Foisner R, Eger A. 2007. The transcription factor ZEB1 (δEF1) promotes tumour cell dedifferentiation by repressing master regulators of epithelial polarity. Oncogene 26:6979–6988. doi: 10.1038/sj.onc.1210508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Spaderna S, Schmalhofer O, Wahlbuhl M, Dimmler A, Bauer K, Sultan A, Hlubek F, Jung A, Strand D, Eger A, Kirchner T, Behrens J, Brabletz T. 2008. The transcriptional repressor ZEB1 promotes metastasis and loss of cell polarity in cancer. Cancer Res 68:537–544. doi: 10.1158/0008-5472.CAN-07-5682. [DOI] [PubMed] [Google Scholar]
- 53.Whiteman EL, Liu CJ, Fearon ER, Margolis B. 2008. The transcription factor snail represses Crumbs3 expression and disrupts apico-basal polarity complexes. Oncogene 27:3875–3879. doi: 10.1038/onc.2008.9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Thiery JP, Acloque H, Huang RY, Nieto MA. 2009. Epithelial-mesenchymal transitions in development and disease. Cell 139:871–890. doi: 10.1016/j.cell.2009.11.007. [DOI] [PubMed] [Google Scholar]
- 55.Laprise P, Chailler P, Houde M, Beaulieu JF, Boucher MJ, Rivard N. 2002. Phosphatidylinositol 3-kinase controls human intestinal epithelial cell differentiation by promoting adherens junction assembly and p38 MAPK activation. J Biol Chem 277:8226–8234. doi: 10.1074/jbc.M110235200. [DOI] [PubMed] [Google Scholar]
- 56.Gamblin CL, Hardy EJ, Chartier FJ, Bisson N, Laprise P. 2014. A bidirectional antagonism between aPKC and Yurt regulates epithelial cell polarity. J Cell Biol 204:487–495. doi: 10.1083/jcb.201308032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Lafleur VN, Richard S, Richard DE. 2014. Transcriptional repression of hypoxia-inducible factor-1 (HIF-1) by the protein arginine methyltransferase PRMT1. Mol Biol Cell 25:925–935. doi: 10.1091/mbc.E13-07-0423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Poh YC, Chen J, Hong Y, Yi H, Zhang S, Chen J, Wu DC, Wang L, Jia Q, Singh R, Yao W, Tan Y, Tajik A, Tanaka TS, Wang N. 2014. Generation of organized germ layers from a single mouse embryonic stem cell. Nat Commun 5:4000. doi: 10.1038/ncomms5000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Caslavsky J, Klimova Z, Vomastek T. 2013. ERK and RSK regulate distinct steps of a cellular program that induces transition from multicellular epithelium to single cell phenotype. Cell Signal 25:2743–2751. doi: 10.1016/j.cellsig.2013.08.024. [DOI] [PubMed] [Google Scholar]
- 60.Martin AC, Goldstein B. 2014. Apical constriction: themes and variations on a cellular mechanism driving morphogenesis. Development 141:1987–1998. doi: 10.1242/dev.102228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Tepass U. 1996. Crumbs, a component of the apical membrane, is required for zonula adherens formation in primary epithelia of Drosophila. Dev Biol 177:217–225. doi: 10.1006/dbio.1996.0157. [DOI] [PubMed] [Google Scholar]
- 62.Vessey CJ, Wilding J, Folarin N, Hirano S, Takeichi M, Soutter P, Stamp GW, Pignatelli M. 1995. Altered expression and function of E-cadherin in cervical intraepithelial neoplasia and invasive squamous cell carcinoma. J Pathol 176:151–159. doi: 10.1002/path.1711760208. [DOI] [PubMed] [Google Scholar]
- 63.Roper K. 2012. Anisotropy of Crumbs and aPKC drives myosin cable assembly during tube formation. Dev Cell 23:939–953. doi: 10.1016/j.devcel.2012.09.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Zou J, Wang X, Wei X. 2012. Crb apical polarity proteins maintain zebrafish retinal cone mosaics via intercellular binding of their extracellular domains. Dev Cell 22:1261–1274. doi: 10.1016/j.devcel.2012.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Varelas X, Samavarchi-Tehrani P, Narimatsu M, Weiss A, Cockburn K, Larsen BG, Rossant J, Wrana JL. 2010. The Crumbs complex couples cell density sensing to Hippo-dependent control of the TGF-beta-SMAD pathway. Dev Cell 19:831–844. doi: 10.1016/j.devcel.2010.11.012. [DOI] [PubMed] [Google Scholar]
- 66.Casaletto JB, Saotome I, Curto M, McClatchey AI. 2011. Ezrin-mediated apical integrity is required for intestinal homeostasis. Proc Natl Acad Sci U S A 108:11924–11929. doi: 10.1073/pnas.1103418108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Saotome I, Curto M, McClatchey AI. 2004. Ezrin is essential for epithelial organization and villus morphogenesis in the developing intestine. Dev Cell 6:855–864. doi: 10.1016/j.devcel.2004.05.007. [DOI] [PubMed] [Google Scholar]
- 68.Medina E, Williams J, Klipfell E, Zarnescu D, Thomas G, Le Bivic A. 2002. Crumbs interacts with moesin and βHeavy-spectrin in the apical membrane skeleton of Drosophila. J Cell Biol 158:941–951. doi: 10.1083/jcb.200203080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.FANTOM Consortium and the RIKEN PMI and CLST (DGT), Forrest AR, Kawaji H, Rehli M, Baillie JK, de Hoon MJ, Haberle V, Lassman T, Kulakovskiy IV, Lizio M, Itoh M, Andersson R, Mungall CJ, Meehan TF, Schmeier S, Bertin N, Jorgensen M, Dimont E, Arner E, Schmidl C, Schaefer U, Medvedeva YA, Plessy C, Vitezic M, Severin J, Semple C, Ishizu Y, Young RS, Francescatto M, Alam I, Albanese D, Altschuler GM, Arakawa T, Archer JA, Arner P, Babina M, Rennie S, Balwierz PJ, Beckhouse AG, Pradhan-Bhatt S, Blake JA, Blumenthal A, Bodega B, Bonetti A, Briggs J, Brombacher F, Burroughs AM, Califano A, Cannistraci CV, Chen Y, Chierici M, et al. 2014. A promoter-level mammalian expression atlas. Nature 507:462–470. doi: 10.1038/nature13182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Robinson BS, Huang J, Hong Y, Moberg KH. 2010. Crumbs regulates Salvador/Warts/Hippo signaling in Drosophila via the FERM-domain protein Expanded. Curr Biol 20:582–590. doi: 10.1016/j.cub.2010.03.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Laprise P. 2011. Emerging role for epithelial polarity proteins of the crumbs family as potential tumor suppressors. J Biomed Biotechnol 2011:868217. doi: 10.1155/2011/868217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Richardson EC, Pichaud F. 2010. Crumbs is required to achieve proper organ size control during Drosophila head development. Development 137:641–650. doi: 10.1242/dev.041913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Mitsuishi Y, Hasegawa H, Matsuo A, Araki W, Suzuki T, Tagami S, Okochi M, Takeda M, Roepman R, Nishimura M. 2010. Human CRB2 inhibits gamma-secretase cleavage of amyloid precursor protein by binding to the presenilin complex. J Biol Chem 285:14920–14931. doi: 10.1074/jbc.M109.038760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Ohata S, Aoki R, Kinoshita S, Yamaguchi M, Tsuruoka-Kinoshita S, Tanaka H, Wada H, Watabe S, Tsuboi T, Masai I, Okamoto H. 2011. Dual roles of Notch in regulation of apically restricted mitosis and apicobasal polarity of neuroepithelial cells. Neuron 69:215–230. doi: 10.1016/j.neuron.2010.12.026. [DOI] [PubMed] [Google Scholar]
- 75.Guo S, Liu M, Gonzalez-Perez RR. 2011. Role of Notch and its oncogenic signaling crosstalk in breast cancer. Biochim Biophys Acta 1815:197–213. doi: 10.1016/j.bbcan.2010.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Shimizu K, Nagamachi Y, Tani M, Kimura K, Shiroishi T, Wakana S, Yokota J. 2000. Molecular cloning of a novel NF2/ERM/4.1 superfamily gene, ehm2, that is expressed in high-metastatic K1735 murine melanoma cells. Genomics 65:113–120. doi: 10.1006/geno.2000.6154. [DOI] [PubMed] [Google Scholar]
- 77.Wang J, Cai Y, Penland R, Chauhan S, Miesfeld RL, Ittmann M. 2006. Increased expression of the metastasis-associated gene Ehm2 in prostate cancer. Prostate 66:1641–1652. doi: 10.1002/pros.20474. [DOI] [PubMed] [Google Scholar]
- 78.Yu H, Ye L, Mansel RE, Zhang Y, Jiang WG. 2010. Clinical implications of the influence of Ehm2 on the aggressiveness of breast cancer cells through regulation of matrix metalloproteinase-9 expression. Mol Cancer Res 8:1501–1512. doi: 10.1158/1541-7786.MCR-10-0186. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.