Abstract
This unit describes a method for the separation of a mixture of quadruplex conformations formed from the same parent sequence via reversed-phase chromatography (RPC). Polymorphism is inherent to quadruplex formation and even relatively simple quadruplex-forming sequences can fold into a cornucopia of possible conformations and topologies. Isolation of a specific conformation for study can be problematic. This is especially true for conformations of the human telomere sequence d(GGG(TTAGGG)3), High Performance Liquid Chromatography (HPLC), especially reversed-phase chromatography, has been a mainstay of nucleic acids research and purification for many decades. We have successfully applied this method to the problem of separating individual quadruplex species in the ensemble from the same parent sequence.
Keywords: RPC, quadruplex, reversed-phase chromatography, telomere
INTRODUCTION
The polymorphic characteristic of G-quadruplex formation has been an ongoing stumbling block to efforts in studying, understanding, and exploiting these complex structures. There are few methods for the isolation of a single quadruplex species from the ensemble of topologies formed from the same sequence of DNA. Many separation strategies that have been employed, such as gel electrophoresis and centrifugation, either lack the necessary resolution or are impractical for recovery of samples for further study. The successful separation strategies that have overcome these obstacles, such as Size Exclusion Chromatography (SEC), have not been particularly useful for isolation of individual topologies of variations of the human telomere sequence (Miller et al, 2011, Miller and Trent, 2011, Largy and Mergny, 2014). This lack of separation methodology has led to a trend to modify quadruplex-forming sequences to achieve a single significantly enriched species for study, with the assumption that the product of the modification will in some way embody the diverse range of species that may actually be present in solution or in-vivo. In reality, the products of such modifications may not represent the exact biologically relevant conformation. Further, a single isolated conformation and its properties may not be sufficiently representative of the original diversity present. The study of several topologies formed from a single unmodified parent sequence may be necessary to fully explore the biophysical characteristics and the potential biological roles of these complex structures. Reversed-phase chromatography has proven to be useful for separating the heretofore un-separable conformations resulting from quadruplex formation by DNA sequences derived from the human telomere. With the possibility of separation comes the opportunity to study these complex systems in greater detail and to gain better insight into the factors that influence quadruplex structure and stability, the polymorphism and dynamics of quadruplex formation, and the equilibrium among various conformations formed from the same parent sequence. Separation may also afford the opportunity to separate more than one configuration from a single sequence and set of annealing conditions, which is especially useful as quadruplexes are increasingly attractive for drug targets and biotechnology applications (Yogesh S. Sanghvi, 2011(UNIT 4.1) and Sannohe and Sugiyama, 2010 (UNIT 17.2)).
This unit describes a facile RPC method for separating quadruplex conformations formed from the same sequence. This method has proven to be successful in dealing with the particularly difficult problem of isolation of conformations formed by variations of the human telomere sequence. The unit details a general method for separating quadruplex DNA (see Basic Protocol), outlines useful procedures for handling and preparing quadruplex samples (see Basic Protocol and Support Protocol), and presents and discusses results for separation of several quadruplex-forming sequences.
Strategic Planning
This protocol describes a general method for separating quadruplex species formed from the same parent DNA sequence using RPC. Each step of the method is explained in detail and followed by a set of critical points. This method can easily be modified to accommodate individual sample requirements, procedures, and equipment. The results presented in this method are representative of the general conditions described and have not been altered optimize separation for any of the sequences used. This method can be modified as needed to provide better separation for specific sequences.
It is important to plan the procedure thoroughly, especially if the user is relatively unfamiliar with HPLC methods and equipment. Materials must be gathered and prepared ahead of time. For example, putative quadruplex DNA must be annealed and HPLC mobile phases must be prepared and degassed well in advance. Several key steps, such as equilibration of the RPC column, cannot be accomplished on the fly and can be very time consuming. The separation step itself takes approximately 1 hour. The method may need to be modified based on constraints imposed by the handling and preparation protocols of individual samples or to adapt to specific laboratory equipment.
HPLC System
The HPLC system selected should be capable of reliable flow rates from 0.1 to 1.0 ml/minute, be capable of mixing at least two mobile phases in a gradient manner, be equipped with either a manual injection port or an auto sampler, and be outfitted with a UV/fluorescence or UV-vis detector. Other features such as computer control, a photodiode array detector, a fraction collector, or an auto-sampler are optional. The HPLC system used in this protocol is a Waters 600 pump equipped with a Waters 2998 photodiode array detector, a Waters 2707 Auto-sampler, and a Waters Fraction collector III. The autosampler was equipped with a 250 μl sample loop. The system was operated via computer equipped with Empower Software (Waters).
Reverse phase column selection
In general, most reverse phase columns are similar in construction and function. For this method a μRPC C2/C18 ST 4.6/100 column (Catalog No. 17-5057-01) from GE Healthcare was used. Other reverse phase columns may be used. Various column matrixes may have slightly different characteristics such as particle size, pore size, and hydrodynamic properties so results may vary. Experimentation to reveal the best column material and column configuration for your specific application is recommended.
HPLC mobile phase
The mobile phases and gradient described in this procedure have been tested for purification of quadruplex DNA. If this procedure is modified then experimentation to assess the suitability of the mobile phase is highly recommended. This protocol may also be adapted to use alternate buffers and solvents. Experimentation to evaluate the suitability of alternate solvents before use with this procedure is highly recommended. Great care and consideration should be used when choosing mobile phase conditions for quadruplex separation. Quadruplex formation and conformational equilibrium have been shown to be sensitive to variations in buffer composition and especially to the presence of organic solvents such as polyethylene glycol (PEG) or acetonitrile. Any variation between mobile phase and annealing buffer may cause variation in the stability of the various quadruplex species in solution.
Filtration
All Buffers, solvents, and samples should be filtered using 0.22 μm filter. Alternatively, samples may be microcenrtrifuged at max speed for 10 minutes. Solvents labeled HPLC grade can be assumed to be free of contamination within the limits of their certification and may not need additional filtering.
Buffer compatibility
Each component of all buffers (for quadruplex sample preparation or mobile phase) should be checked for compatibility with the selected RPC column. Incompatible buffer components may damage the column or degrade the DNA sequence and lead to unsatisfactory results.
BASIC PROTOCOL: Reversed-phase CHROMATOGRAPHY OF G-QUADRUPLEXES
This protocol describes a general method for separating quadruplex species formed from the same parent sequence using RPC. Each step of the method is explained in detail. Each step is then followed up with a set of critical points. While this protocol attempts to be thorough, common aspects of HPLC should also be considered as should the following general guidelines for preparation and handling of quadruplex DNA. This method can easily be modified to accommodate individual sample requirements and procedures.
Materials
Distilled, deionized water (18 MΩ or HPLC grade)
HPLC mobile phases:
Buffer A: water with 5% (v/v) acetonitrile, 50 mM triethylamine, 50 mM Acetic Acid, 35 mM KCl, pH 7.0
Buffer B: water with 85% (v/v) acetonitrile, 50 mM triethylamine, 50 mM Acetic Acid, 35 mM KCl, pH 7.0
Quadruplex sample (see Support Protocol)
HPLC system equipped with:
Waters 600 pump and controller
Waters 2998 UV-vis photodiode array detector
Waters 2707 autosampler with 250-μl sample loop
Waters fraction collector III (optional)
Computer with Waters Empower Software (optional)
μRPC C2/C18 ST 4.6/100 column (GE Healthcare Catalog No. 17-5057-01)
Microcentrifuge (optional)
0.22 μm Syringe filter (optional)
1.5-mL microcentrifuge tubes (optional)
Set up HPLC and prepare column
Install μRPC C2/C18 ST 4.6/100 column to the HPLC pump output. Be sure to follow all manufacturer’s recommendations and procedures for individual column and HPLC equipment.
-
Set up HPLC with buffers A and B in the correct reservoirs.
Any buffer or solvent used should be completely degassed before introducing it into the system. Vacuum degassing is best, degassing with helium is acceptable; some HPLC systems are capable of degassing solvents during sample analysis.
-
Pump ~2 column volumes of buffer B through the column followed by at least 8 ml of buffer A. At this point, switch the column to buffer A with a flow rate of 0.5 ml/min.
An initial solvent/buffer wash to wet the column matrix is critical. Reversed-phase columns are, in general, shipped in storage solution consisting of water with an alcohol preservative. The μRPC C2/C18 ST 4.6/100 column is shipped in 70% methanol/30% water. This storage solution must be completely flushed out, the column matrix wetted with the solvent/buffer of choice, and the column equilibrated in the running buffer before use. Flushing the column with distilled, deionized water before buffer equilibration is recommended. Periodic cleaning is critical to maintain proper separation. Cleaning of the column should be performed per manufacturer’s directions. After cleaning, the column should then be re-equilibrated as described in steps 1 through 3.
Separate Quadruplex Sample
4. Make sure that the HPLC system is on and working properly. Ensure that the column has been properly installed and equilibrated, and is running buffer A. Set flow rate to 0.5 ml/min. Typical UV-vis detectors should be turned on ~20 min in advance of use to allow time for the instrument to stabilize.
-
5. Set UV-vis detector to A260 or A280.
Typically DNA has an absorbance maxima at ≈ 260 nm. However, some instruments that might be utilized for this work may not have this capacity. A photodiode array detector is not necessary to accomplish this method. Many of the instruments available for separation of biomolecules are primarily focused on protein separation and may only have one or two wavelengths available for use. Most instruments will at least be capable of monitoring the elution at 280 nm and as such can still be used for this method.
Additionally, there may be advantages to monitoring the elution of DNA at wavelengths other than 260 nm. For example, for DNA the absorbance at 280 nm is typically only about 50% of the absorbance at 260 nm. Monitoring at 280 nm might be useful for monitoring concentrated samples without overloading the detector. Further, monitoring the elution at multiple wavelengths may provide valuable information. Purification strategies often result in small molecule contamination of DNA. For example, strong absorbance at 230 nm may be indicative of contamination by phenolate, thiocyanates, or other organic molecules. Phenol absorbs strongly at 270 nm. Monitoring elution at 330 nm may reveal the presence of particulates due to aggregation. Conversely, the A260/A280 ratio, often used for determination of DNA contamination of protein samples, is not useful for monitoring protein contamination of DNA. The molar absorbance of DNA is typically so much greater than the molar absorbance of protein that even when a sample contains more protein than DNA the A260/A280 ratio will change only a little with respect to the DNA. For example, the typical A260/A280 ratio for pure protein is 0.6, and for pure DNA the A260/A280 ratio is 1.8. At 70% protein, 30% DNA the A260/A280 ratio is approximately 1.75.
-
6. Program HPLC gradient upon injection to run Buffer A for 5 minutes, followed by a 40 minute linear gradient to 100% Buffer B, followed by 5 minutes of 100% Buffer B, and then returning to buffer A for an additional 5 minutes.
Total run time should be approximately 55 minutes. This gradient profile ensures that the instrument will be immediately ready for another injection once a run is completed.
-
7. Inject 10 to 100 μl of crude quadruplex sample manually or using an autosampler.
Elution of quadruplex DNA can vary greatly depending on sequence, preparation, and other factors. The best results will have well isolated peaks. (Figure 1.)
For the data presented in this method, injections of 20 μl of a 100 μM sample were used. Injection volumes will vary depending on the concentration of DNA in the sample and on the specifications of the column and detector. Concentrated sample may be injected at lower volumes. Dilute samples will require injection of greater volumes. This method may also be used to concentrate dilute samples. A few initial trial runs may be useful for determining the appropriate injection volume to use.
The μRPC C2/C18 ST 4.6/100 column used in this protocol is capable of handling sample volumes of up to 100 μl and approximately 5 to 10 mg of sample. These limits are reasonable for a protein sample; however, the typical quadruplex forming DNA sequence tends to have molar absorptivity on the order of 100 times the molar absorptivity of a typical protein. Therefore a DNA sample will saturate the detector of the typical UV-vis detector long before the maximum sample load is achieved. Several initial trial runs may be required to determine the appropriate injection volume and sample concentration to use for individual applications.
The sample should be passed through a 0.22-μm filter prior to injection on the HPLC. Alternately, the sample may be centrifuged at max speed for ≈10 minutes. A microcentrifuge and 1.5 ml microcentrifuge tube are ideal for this operation.
Figure 1.
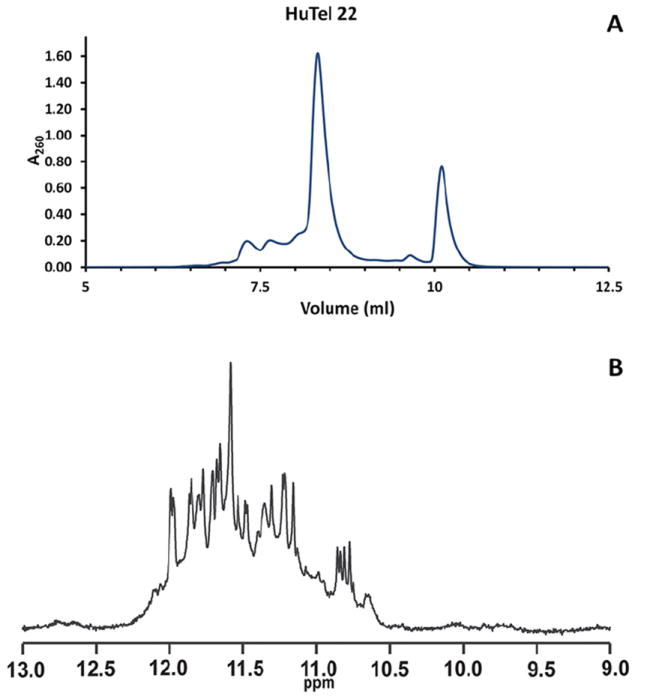
A. Typical elution profile for HuTel 22. The elution profile reveals the presence of two major conformations and a host of less abundant conformations and is consistent with the available data on this sequence. (Miller et al., 2010, Gray et al., 2012) The two major peaks at ≈8.3 ml and ≈10.2 ml have CD spectra typical of Hybrid and Parallel conformations respectively. B. 1H NMR of imino regions of the HuTel 22 sequence. The spectrum exhibits ≈24 strong GN1H signals, indicative of the presence of at least two quadruplex species. The spectrum also exhibits many weaker signals, evidence of the presence of several less abundant topographies. NMR samples were freshly annealed DNA samples which had been diluted into fresh buffer as described above and D2O added to 10%. Sample concentrations were approximately 200 to 500 μM. Samples without acetonitrile were loaded into 5 mm Shigemi NMR tubes. NMR spectra were recorded on a Varian Inova spectrometers at 293 K and 800 MHz. All spectra were referenced to internal DSS-d6.
Collect fractions (optional)
-
8. During sample elution, collect fractions in 1.5-ml microcentrifuge tubes using any commercial fraction collector or collect desired fractions by hand.
If the fraction collection method does not adequately match fractions with chromatogram then orientation of chromatogram to fraction can usually be accomplished via standard UV-vis analysis with low volume cuvettes or via low volume devices such as the Nanodrop (Thermo Scientific).
Since UV-vis detectors generally have much smaller path lengths than desktop UV-vis instruments, the actual optical density of the collected fractions may be much higher than indicated by the detector. For the Waters 2998 UV-vis photodiode array detector, the path length can range from 0.5 mm to 10 mm depending on flow cell configuration, so an A260 of 1.0 may result in a fraction with an actual A260 of up to 10.0. If one assumes that a typical optical density for quadruplex forming sequences is approximately 250,000 L/(M·cm), then a sample with an A260 of 10.0 has a concentration of 40 μM. In many cases the collected fractions will be concentrated enough that further dilution may be necessary for UV-vis measurements or other downstream applications.
-
9. Use or freeze the desired fractions immediately.
Quadruplex formation and conformational equilibrium has been shown to be sensitive to variations in buffer composition and especially to the presence of organic solvents such as PEG or acetonitrile. If used immediately fractions should be diluted with buffers that do not contain organic additives to reduce the impact of solvent interaction. For applications that require samples of relatively low concentration, such as circular dichroism and analytical ultra-centrifugation which generally utilize samples with A260 of approximately 1.0 or less (less than 5 μM for most samples), fractions with high optical densities can be diluted into the desired buffer and used immediately.
In the case of a downstream application requiring a fairly concentrated sample, such as NMR or calorimetry, it may not be possible to use individual fractions as samples without further processing. If more concentrated samples are needed, samples may be lyophilized and rehydrated at lower volumes to concentrate them.
Fractions not used when they are collected should be frozen immediately. Fractions that are not frozen or diluted immediately may re-equilibrate into an ensemble of conformations. Once frozen, fractions can be stored indefinitely at −80°C for future use. Once individual fractions have been collected, they may be combined with succeeding sets of identical fractions.
Samples that are prone to re-equilibration or degradation of the quadruplex species should be frozen at −80°C, and can be stored indefinitely at this temperature. Very stable quadruplex conformations may be refrigerated. A high-resolution technique such as NMR should be used to ensure that individual samples are stable in the conditions they will be exposed to (Dailey et al., 2010).
Concentrate final sample (optional)
-
10. Further concentrate fractions by lyophilization.
Once lyophilized, samples may be rehydrated at higher concentrations. Do not attempt to concentrate samples by Centrivap concentrator. Quadruplex formation and conformational equilibrium has been shown to be sensitive to variations in buffer composition and especially to the presence of organic solvents such as PEG or acetonitrile. Do not attempt to concentrate samples by using devices such as centrifugal concentrators. Samples should be concentrated by lyophilization followed by rehydration at lower volumes. Lyophilization has the added advantage of effective removal of the volatile buffer components. Lyophilized fractions may also be rehydrated in any buffer desired. A high-resolution technique such as NMR should be used to ensure that individual samples are stable in the conditions they will be exposed to.
SUPPORT PROTOCOL: PREPARATION OF QUADRUPLEX SAMPLES
Quadruplex DNA samples should be prepared as required for individual lab protocols or experimental needs. Any quadruplex-forming sequence is sufficient. Results for several sequences are shown in Table 1. Any oligonucleotide sample used for this procedure should be dissolved and dialyzed into water before dilution/dialysis into annealing buffer to ensure purity. Samples are then equilibrated for 10 min in a 100°C water bath, cooled gradually to room temperature overnight, and stored at 4°C until use.
Table 1.
Quadruplex-forming sequences used in this study.
| Sequence Name: | Type: | DNA Sequence: | Reference: |
|---|---|---|---|
| HuTel 22 | HT | AGGGTTAGGGTTAGGGTTAGGG | Wand and Patel 1993 |
| 2GKU | HT | TTGGGTTAGGGTTAGGGTTAGGGA | Luu et al., 2006 |
| 2HY9 | HT | AAAGGGTTAGGGTTAGGGTTAGGGAA | Dai et al., 2006a |
| 2JSM | HT | TAGGGTTAGGGTTAGGGTTAGGG | Phan et al., 2007a |
| 2JPZ | HT | TTAGGGTTAGGGTTAGGGTTAGGGTT | Dai et al., 2007a |
| 2JSL | HT | TAGGGTTAGGGTTAGGGTTAGGGTT | Phan et al., 2007a |
| 2KF8 | HT | GGGTTAGGGTTAGGGTTAGGGT | Lim et al., 2009 |
| 2KKA | HT | AGGGTTAGGGTTAGGGTTAGGGT | Zhang et al., 2010 |
| 2KJ2 | PR | CGGGCGGGCGCGAGGGAGGGT | Hsu et al., 2009, Kuryavyi et al., 2010 |
| 3O2M | PR | AGGGAGGGCGCTGGGAGGAGGG | Phan et al., 2007b |
| RB | PR | CGGGGGGTTTTGGGCGGC | Xu and Sugiyama 2006 |
| Her2 | PR | AGGAGAAGGAGGAGGTGGAGGAGGAGGGC | Ziemba et al., 2005 |
| 2F8U | PR | AGGGGCGGGCGCGGGAGGAAGGGGGCGGGAGCGGGGC | Dai et al., 2006a |
| VEGF | PR | GGGCGGGCCGGGGGCGGGGTCCCGGCGGGGCGGGAG | Sun et al., 2005 |
| KRAS | PR | GGGAAGAGGGAAGAGGGGGAGG | Cogoi and Xodo 2006 |
| HIF-1α | PR | GCGCGGGGAGGGGAGAGGGGGCGGGAGCGCG | De Armand et al., 2005 |
| c-myc | PR | TGGGGAGGGTGGGGAGGGTGGGGAAGG | Simonsson et al., 1998 |
| 1XAV | PR | TGAGGGTGGGTAGGGTGGGTAA | Ambrus et al., 2005 |
HT denotes sequences that are derived from the human telomere sequence. PR denotes sequences that are derived from oncogene promoter sequences.
Samples should be diluted to final concentrations ranging from 200 μM to 1 mM before injection into the HPLC. Do not depend on the quantities/yields of DNA as stated by the manufacturer. Measure DNA quantities and check purity directly whenever possible. All oligonucleotides used to produce data in this protocol (Table 1) were obtained as 1 μMole preps from Integrated DNA Technologies as purified, dried, and desalted pellets.
Although many different annealing buffers and profiles are acceptable, phosphate and cacodylate buffers are generally best for this application. Buffers such as Tris, Tricine, HEPES, MOPS, TAPS, MES, BES, TES, and acetate are not recommended. Phosphate buffers should be avoided if working with some divalent ions such as Mg2+ and Ca2+. Due to the sensitivity of quadruplex formation to ion type and concentration, care should be taken not to introduce inappropriate ions as counter ions to buffer constituents.
Samples used in this protocol were purchased from Integrated DNA Technologies as 1 μmole samples and used as delivered. The samples were diluted to 500 μM, filtered, and dialyzed as described below. Final samples were diluted to 100 μM in folding buffer and annealed as described below.
Materials
DNA sample
Annealing buffer: 100 mM KCl, 25 mM K2HPO4, pH = 7.0
Distilled, deionized water
Dialysis unit
1- to 2-liter beakers
100°C hotplate or water bath
Aluminum foil
-
Dissolve DNA sample in distilled, deionized water.
In general, it is better to dissolve the sample at a concentration slightly higher than the final desired concentration (e.g., 1.5×).
-
Filter sample through a 0.22-μm filter.
Small volumes of DNA solution can be effectively filtered with small gauge syringe filters. Alternately, the sample can be filtered or centrifuged at max speed for ≈10 minutes prior to injection on the HPLC.
-
Place DNA sample in the dialysis unit of choice and dialyze first against distilled, deionized water and then into annealing buffer. For samples up to 1 mL in volume, dialyze into 1000 mL with at least two changes of buffer. Gentle stirring is recommended.
The dialysis unit should be of the appropriate size and molecular weight cut off (MWCO) for the sequence being prepared. Most quadruplex-forming sequences studied herein are no more than 30 bases in length, with an average molecular weight of ~7,000 to 8,000 Da. Slide-A-Lyzer Dialysis Cassettes with 0.2-0.5 mL total volume and 2,000 Da MWCO (Thermo Sci. Cat. No. 66205) will work well for this step. Other systems can be used.
Any desired annealing buffer can be used for this step. A basic annealing buffer that yields good results in simply 100 mM KCl, 25 mM K2HPO4, pH=7.0. Other formulations and additives may be used. For example, lower potassium or total salt levels may be employed. Care should be taken to insure that the individual components selected for an annealing buffer will not adversely affect quadruplex formation or select for a specific conformation. For example, reagents that introduce alternate cations should be avoided. Organic additives should be evaluated thoroughly to assess their suitability for the desired application.
Transfer the dialyzed sample to 1.5- or 2.0-ml microcentrifuge tube with a screw cap and rubber gasket.
-
Immerse quadruplex samples for 10 min in a 100°C water bath using a commercially available float. To reduce the possibility for contamination, do not allow the top of the tube and cap to be immersed.
Alternatively, a laboratory beaker with 1 to 2 liters of boiling water may be used.
-
Turn off water bath and allow sample to cool overnight in the water bath.
If using a beaker of boiling water, remove beaker from heat source and cover it with aluminum foil to allow slower, more even cooling.
-
7. Check samples to verify concentration and confirm quadruplex formation.
Typically, quadruplex formation can be verified by spectroscopic methods such as CD or NMR or by biophysical testing such as AUC or calorimetric methods such as DSC.
COMMENTARY
Background Information
Guanine-rich DNA with at least four runs of two or more guanines can form three-dimensional structures called G-quadruplexes in the presence of certain monovalent cations such as Na+ or K+. In general, quadruplex-forming sequences are of the type d(G3+N1-7G3+N1-7G3+N1-7G3+), where N is any base (Todd et al., 2005). G-quadruplexes are made up of stacks of two or more square planer arrays of four guanines (a G-quartet) stabilized by Hoogsteen hydrogen bonds with coordination of a monovalent cation to the O6 of the guanines (Figure 2.) (Yang et al., 2010). Although most often associated with telomeres, potential quadruplex-forming sequences have been found throughout the genome (Ambrus et al., 2005, Dai et al., 2006a, De Armond et al., 2005, Huppert et al., 2005, Hsu et al., 2009, Patel et al., 2007, Phan et al., 2007, Sun et al., 2005). Approximately 380,000 putative quadruplex forming sequences have been identified in the human genome (Huppert et al., 2005). Quadruplex DNA is of interest because of putative roles in biological regulation, potential bio/nanotechnology applications, as potential targets for developing therapeutic drugs or as therapeutic agents themselves (Cogoi et al., 2004, Huppert et al., 2007a, Huppert et al., 2007b, Ireson et al., 2006, Maizels et al., 2006, Neidle 2010, Neidle and Parkinson 2002).
Figure 2.
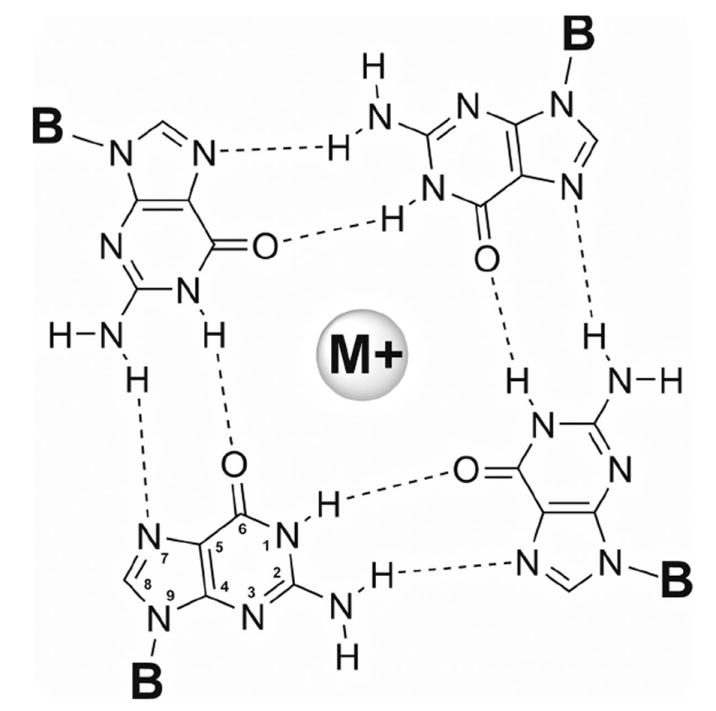
A Guanine quartet. G-quartets are stabilized by Hoogsteen hydrogen bonding between O6 and N1, between N7 and N2, and by coordination of the O6 to a monovalent cation. Typically the cation is potassium or sodium. Each guanine is attached to the sugar backbone at N9-position.
G-quadruplex formation can be highly polymorphic. When factors such as strand orientation, loop type and arrangement, glycosyl torsion angles, intramolecular versus intermolecular formation, ion type and concentration, DNA concentration, and annealing profile are considered, even sequences based on the relatively simple human telomeric sequence, d(GGGTTA)n, have the potential to fold into more than 200 intramolecular and intermolecular conformations (Figure 3.) (Lane et al., 2008, Li et al., 2005, Miller and Trent, 2011, Webba da Silva, 2007). This inherent polymorphism has been an ongoing stumbling block to efforts to study, understand, and exploit these complex structures. Structural and biophysical studies of quadruplex DNA have been hindered by our inability to isolate and study the individual quadruplex conformations. Low-resolution methods such as circular dichroism, UV-vis spectrophotometry, and analytical ultracentrifugation are not able to distinguish between conformations with similar physical properties. High-resolution techniques such as NMR spectroscopy are equally unsuited for examining these complex mixtures (Dailey et al., 2010).
Figure 3.
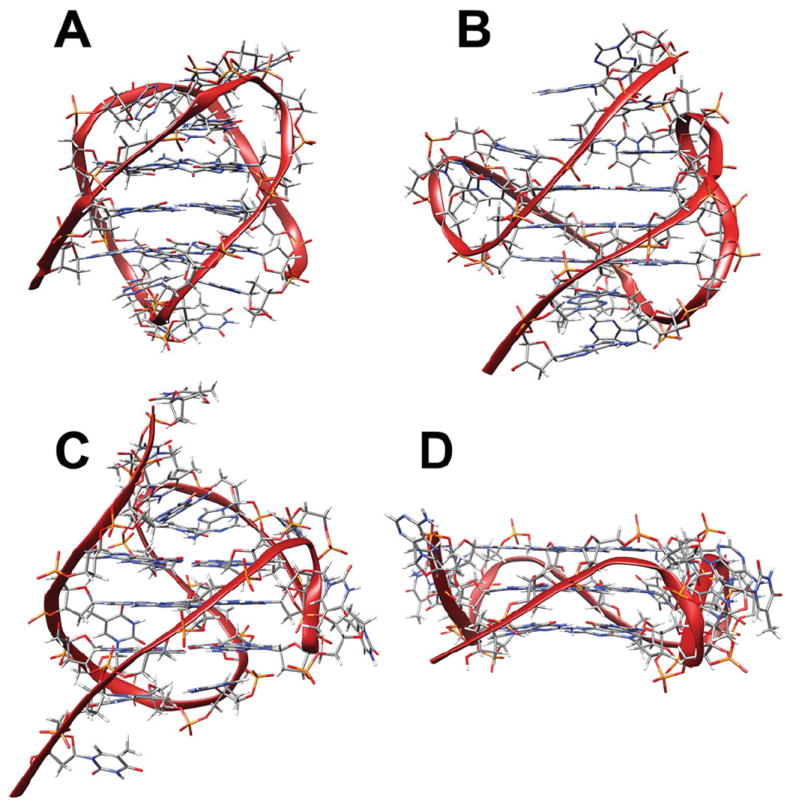
Possible topologies of the human telomere sequence include (A) basket topology (Protein Database code: 143D), (B) hybrid-1 topology (2HY9), (C) hybrid-2 topology (2JPZ), and (D) propeller type all parallel topology (1KF1).
Several methods have been used to reduce or eliminate this inherent polymorphism. The primary means of dealing with the polymorphism of quadruplex formation has been sequence modification. This generally takes the form of adding or deleting bases at the 5’- or 3’- ends of a sequence, lengthening or shortening of guanine runs, or eliminating guanines from putative loop regions to ensure that a single conformation is achieved (Dai et al., 2008, Yang et al., 2010). Putative quadruplex-forming sequences often undergo a series of modifications and truncations before producing a sequence suitable for study. Extreme examples of this can be seen with the sequences used to produce the solution structures of c-kit, bcl-2, and c-myc. In each case the parent quadruplex forming sequence underwent a series of modifications, up to 12 separate modification steps, before producing an enriched conformation of sufficient purity for study (Ambrus et al., 2005, Dai et al., 2006a, Dai et al., 2006b, Dexheimer et al., 2006, Kuryavyi et al., 2010, Masiero et al., 2010, Phan et al., 2004).
There are many other sequence modifications that can be used to influence the envelope of potential quadruplex conformations. Substitution of 8-aminoguanine promotes formation of tetramolecular parallel quadruplexes such as those formed by TG4T (Gros et al., 2008). Incorporation of 8-methylguanine, 8-bromoguanine, or use of RNA and LNA force adoption of a syn glycosidic guanosine conformation (Bonifacio et al., 2008, Esposito et al., 2004, Kumar et al., 2007, Qi et al., 2007, Sannohe and Sugiyama, 2010 (UNIT 17.2), Tang et al., 2006, Virgilio et al., 2005a, Virgilio et al., 2005b). Alterations can also include modification of the sugar phosphate backbone by insertion of a 5’-5’ or a 3’-3’ polarity inversion has also been shown to have a remarkable effect on quadruplex formation and stability (Esposito et al., 2005). While the use of inosine, O6-methylguanine, or 6-thioguanine has been shown to destabilize quadruplex formation (Marathias et al., 1999, Mekmaysy et al., 2008, Petrovic et al., 2008, Spackova et al., 2004).
Quadruplex polymorphism can be reduced by using methods other than sequence modification. Annealing buffer conditions can greatly affect the range of conformations formed. The type and concentration of monovalent cation, primarily K+ or Na+, or inclusion of divalent cations, such as Ca2+, Mg2+, Mn2+, Co2+, Ni2+, Pb2+, or Sr2+, may either stabilize or destabilize quadruplex structures. For example, cation concentration has been shown to be essential in selecting which quadruplex conformation is formed by the human telomere sequence (Gray et al., 2009, Gray et al., 2010). Inclusion of divalent cations has been shown to induce a transition from antiparallel to parallel quadruplex conformation for the G4T4G4 sequence (Blume et al., 1997, Miyoshi et al., 2001). Addition of organic co-solvents such as PEG and acetonitrile, or biological molecules such as proteins or polysaccharides, can also greatly affect quadruplex formation, conformation, and stability (Buscaglia et al., 2013, Miller et al., 2010, Xue et al., 2007).
Several recent advances have been made in separation of quadruplex conformations formed from the same parent sequence. The method that has proven to be most useful for this purpose is Size Exclusion Chromatography (Dailey et al., 2010, Largy and Mergney 2014, Miller et al, 2011, Miller and Trent, 2011). While SEC has proven fairly effective for separation of quadruplex conformations formed by sequences derived from oncogene promoter sequences, the method has not been useful for separation of quadruplex conformations derived from variations of the human telomere sequence (Miller and Trent, 2011). This unit describes a facile RPC method for separating quadruplex conformations formed from the same sequence. This method has been successful in dealing with the particularly difficult problem of isolation of conformations formed by variations of the human telomere sequence.
Critical Parameters and Troubleshooting
The reversed-phase chromatography method described in this unit is straightforward and easy to duplicate. However, there are several key points that may make a difference when executing this method. First, many reversed-phase columns and HPLC systems are available for use; therefore, the conditions outlined in this procedure may not be optimal for the specific combination of instrument and column available for use. Conditions should be tested thoroughly for suitability. Any changes made to the protocol described in this unit should be investigated thoroughly before implementation. For example, variables such as buffer composition, attempts to speed up the procedure, or increase separation resolution by changing the flow rate or gradient should be examined thoroughly. In some cases, such as the addition of alternate organic constituents, a change in ion type or concentration, or differences between buffer and mobile phase, changes to this method may result in complete rearrangement of quadruplex configuration resulting in a misleading chromatographic outcome. Second, any buffer or solvent used should be 0.22 μm filtered before use. In addition, buffers need to be completely degassed before introducing them into the system. Unless the HPLC system used is capable of degassing solvents/buffers during operation vacuum degassing should be employed. Degassing with helium is acceptable.
If the resolution provided by this method is insufficient then modification of the elution buffers may prove useful. Increasing and/or decreasing the amount of acetonitrile in Buffer A and Buffer B, respectively, may result in greater separation between potentially isolatable quadruplex conformations. For example, if the quadruplex conformations of interest elute before the gradient reaches 50% Buffer B, reducing the amount of acetonitrile in Buffer B to 45% should result improved separation. Alternatively, the 40 minute linear gradient to 100% Buffer B can be altered to a 40 minute linear gradient to 50% Buffer B. In either case, each HPLC run should end with 5 minutes of 100% Buffer B at 85% acetonitrile, and then returning to buffer A for an additional 5 minutes. This ensures that all DNA is eluted from the column and the column is ready for the next injection.
Finally, while quadruplex structures are known for their stability and resistance to nucleases, some quadruplex samples, once isolated, have been shown to re-equilibrate. Therefore any fractions that are collected for downstream applications should be frozen at once to prevent degradation. If a specific quadruplex structure re-equilibrates too quickly to isolate under the conditions of this method, good results might be obtained by refrigeration of the column, buffers, and fraction collecting system. The elution profile can be altered to reduce the elution time. If the quadruplex conformation of interest elutes before the gradient reaches 50% Buffer B, the gradient can be altered to a 20 minute gradient to 50% Buffer B. This change can be used to reduce the time that a conformation is exposed to inhospitable conditions. Each HPLC run should end with 5 minutes of 100% Buffer B at 85% acetonitrile, and then returning to buffer A for an additional 5 minutes to ensure that all DNA is eluted from the column and the column is ready for the next injection. Alternatively, higher potassium, or other suitable cation, concentration may also stabilize some quadruplex conformations. If this option is exercised then care should be taken to ensure that the increased salt concentration does not cause Buffer B to separate into discrete organic and aqueous layers. If this occurs, the increased cation concentration may necessitate a lower percentage of acetonitrile for Buffer B which will shift the anticipated elution profile.
Again, any changes to the method as outlined should be investigated thoroughly before implementation. General laboratory safety is also a concern when hazardous materials, such as organic solvents, are involved. Some procedures for preparation and handling of quadruplex DNA call for the use of cacodylic acid or thallium compounds (Gill et al., 2006, Parkinson et al., 2002). Arsenic and thallium are acutely toxic. The utmost care and adherence to the appropriate safety procedures should be of priority at all times.
Anticipated Results
Several examples of the application of RPC to the separation of biologically relevant quadruplex-forming sequences are shown in Figure 4 and Figure 5. These results were obtained at 20 C and with the method as described above. No optimization to enhance separation was performed. These results are especially promising as reversed-phase chromatography and SEC may serve as excellent orthogonal methods for separation of quadruplex conformations formed from a single parent sequence. Unlike SEC, reversed-phase chromatography shows good separation of the conformations formed from derivatives of the human telomere sequence. (Figure 4.) Each of the sequences derived from the human telomere displays relatively few resolvable peaks. This result is in good agreement with the number of observed quadruplex conformations for these sequences (Lane et al., 2008, Le et al., 2013, Le et al., 2014). (Figure 1.)
Figure 4.
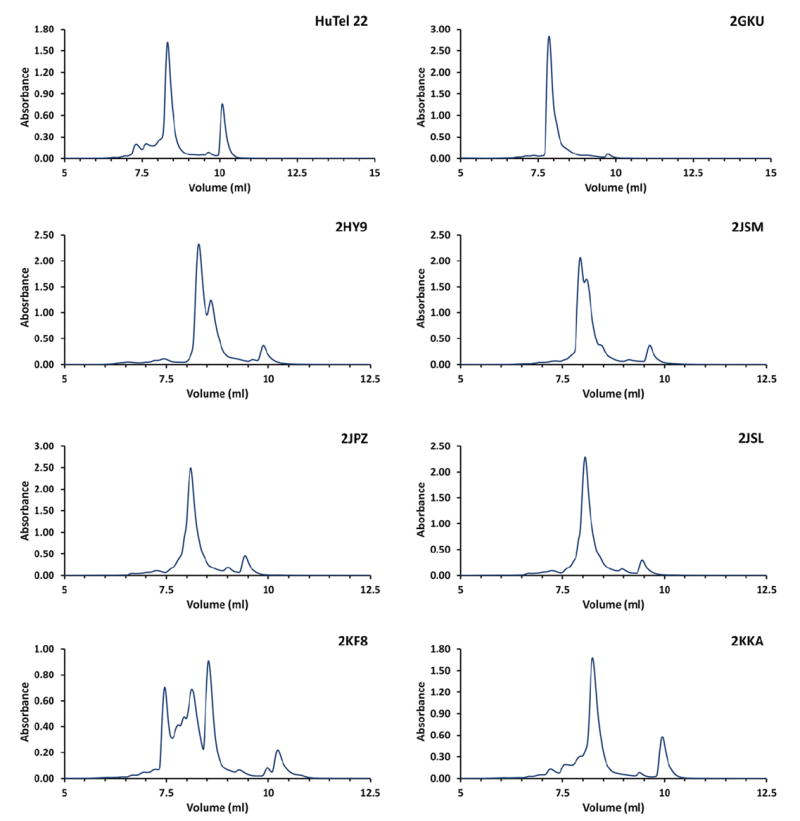
Reversed-phase HPLC results for sequences derived from the human telomere, d(GGGTTA)n. All samples were prepared in buffer consisting of 50 mM KCl and 25 mM K2HPO4, pH = 7.0 and annealed as outlined in the procedure above. These results were obtained at 20 C and with the method as described above. No optimization to enhance separation was performed.
Figure 5.
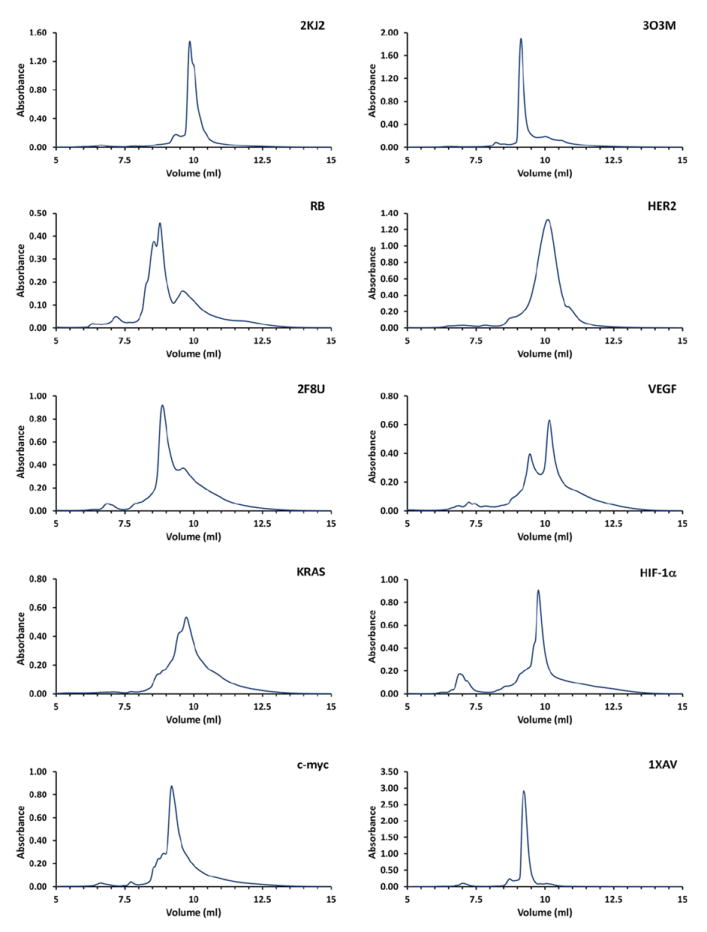
Reversed-phase HPLC results for sequences derived from oncogene promoters. All samples were prepared in buffer consisting of 50 mM KCl and 25 mM K2HPO4, pH = 7.0 and annealed as outlined in the procedure above. These results were obtained at 20 C and with the method as described above. No optimization to enhance separation was performed.
The results from the sequences derived from oncogene promoter sequences shows a broader envelope of separation and even greater number of potentially resolvable conformations. (Figure 5.) This is likely due to the presence of a larger variation in conformation for these sequences than is possible in the telomere derived sequences. Further, the conformations that are present may display a larger variation in physical properties. This is likely due to the fact that many of the promoter sequences possess more than four runs of guanine. Some sequences such as VEGF, bcl-2, and Her2 have 6 or more runs of guanine. Additionally, each individual run of guanines may contain up to five guanines. The increased complexity of these sequences provides the opportunity for quadruplex formation involving alternate runs of guanines and even alternate guanines with a single run resulting in a wide range of differences in loop length and arrangement, more diverse G-quartet stacking, and formation of alternate G-quartets. (Le et al., 2012)
In contrast, telomere derived sequences are limited to four runs of guanine each with exactly three guanines per run. This sequence homology results in very similar loop arrangements and G-quartet stacking. No matter the resulting quadruplex topology, the human telomere sequence is limited to three different loop arrangements an equal number of (Webba da Silva, 2007). (Figure 3.) While the telomere sequences can potentially form over 200 conformations, the number of observed conformations is actually quite limited. The telomere sequences have also been observed to have low thermodynamic and kinetic barriers to equilibration from one conformation to another (Gray et al., 2009, Gray et al., 2010, Gray et al., 2014).
Since quadruplex formation and conformational equilibrium has been demonstrated to be sensitive to variations in buffer composition and especially to the presence of organic solvents such as PEG, alcohols, or acetonitrile there is some concern that the presence of several resolvable conformations could be the result of re-equilibration due to exposure to either the organic buffer components or acetonitrile. Conformations formed by the human telomere sequence may be especially sensitive to re-equilibration because of relatively low thermodynamic and kinetic barriers to refolding. The 2GKU sequence provides a reasonable check for this problem. Under the annealing conditions outlined in this method, 2GKU forms a single predominant conformation. Comparison of the elution profile for 2GKU with high-resolution 1H NMR of imino regions of the sequence (Figure 6.) reveals that for this sequence the elution process and exposure to the buffer components do not seem to effect re-equilibration. In contrast the elution profile of a sequence known to have multiple components, HuTel22, (Figure 1.) points to the various conformations remaining stable throughout the elution process. These results are consistent with the established time frame required for conversion to the all parallel topology by dehydration or binding or PEG (Buscaglia et al., 2013, Miller et al., 2010).
Figure 6.
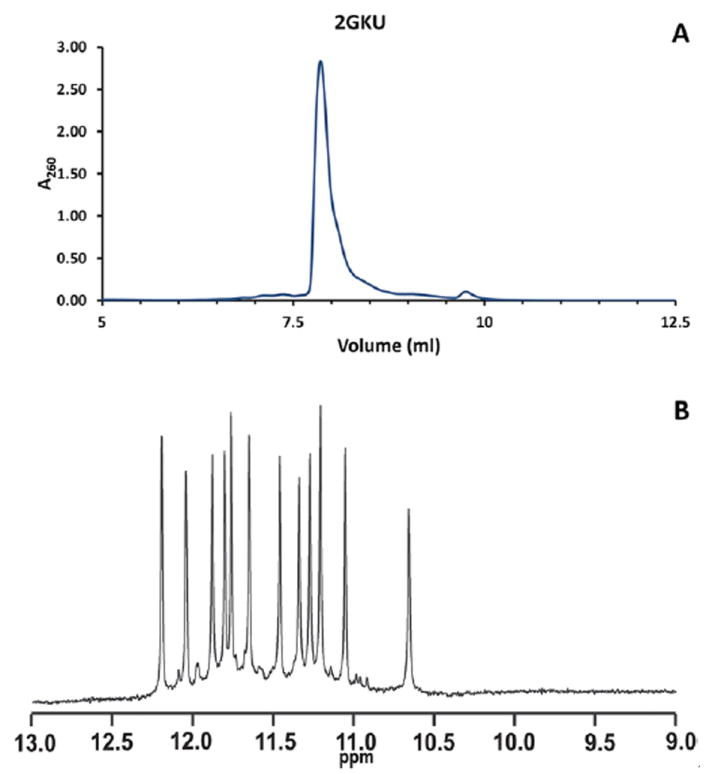
A. Typical elution profile for 2GKU sequence. The elution profile reveals the presence of only one major conformation and several minimally abundant conformations. This is consistent with the NMR data for this sequence under these annealing conditions. B. 1H NMR of imino regions of the 2GKU sequence. The spectrum exhibits 12 strong GN1H signals, indicative of the presence of a single predominant quadruplex species. The spectrum also exhibits many weaker signals, evidence of the presence of several minimally abundant species. NMR samples were freshly annealed DNA samples which had been diluted into fresh buffer as described above and D2O added to 10%. Sample concentrations were approximately 200 to 500 μM. Samples without acetonitrile were loaded into 5 mm Shigemi NMR tubes. NMR spectra were recorded on a Varian Inova spectrometers at 293 K and 800 MHz. All spectra were referenced to internal DSS-d6.
Time Considerations
For most quadruplex samples the entire procedure outlined in this method, including preparing HPLC buffers, column installation and equilibration, sample extraction, and multiple sample concentration steps can easily be accomplished in a single day. The user can expect to be able to run several repeats of this HPLC separation over the course of a single day. However, several steps of this procedure can be time-consuming, and any strategic planning should take these steps into account. Repetition of this method offers some economies of scale since some steps only need to be repeated periodically.
Once the HPLC system is prepared and equilibrated the actual HPLC run time for each sample drops to about one hour. Unless stability is proven by NMR or another high-resolution method, any fractions collected should be frozen at once to prevent degradation, especially if fractions from several different HPLC runs will be combined for a single use. Because the fractions should be frozen immediately for future use, the opportunity to use automation to collect samples is limited. Depending on the equipment available and specific user protocols, some steps, such as degassing of HPLC solvents, preparation of samples, or lyophilization, may require an overnight procedure.
Acknowledgments
This work was supported by National Institutes of Health (NIH) Grant GM077422 (J.O.T.). This work was also supported by the Department of Chemistry and Biochemistry at the University of North Georgia, the William N. Cannon Fund, and the University of North Georgia Foundation (C.M. and C.O.).
Literature Cited
- Ambrus A, Chen D, Dai J, Jones RA, Yang D. Solution structure of the biologically relevant G-quadruplex element in the human c-MYC promoter. Implications for G-quadruplex stabilization. Biochemistry. 2005;44:2048–2058. doi: 10.1021/bi048242p. [DOI] [PubMed] [Google Scholar]
- Blume SW, Guarcello V, Zacharias W, Miller DM. Divalent transition metal cations counteract potassium-induced quadruplex assembly of oligo(dG) sequences. Nucleic Acids Res. 1997;25:617–625. doi: 10.1093/nar/25.3.617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonifacio L, Church FC, Jarstfer MB. Effect of locked-nucleic acid on a biologically active G-quadruplex. A structure-activity relationship of the thrombin aptamer. Int J Mol Sci. 2008;9:422–433. doi: 10.3390/ijms9030422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buscaglia R, Miller MC, Dean WL, Gray RD, Lane AN, Trent JO, Chaires JB. Polyethylene glycol binding alters human telomere G-quadruplex structure by conformational selection. Nucleic Acids Res. 2013;41:7934–7946. doi: 10.1093/nar/gkt440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cogoi S, Quadrifoglio F, Xodo LE. G-rich oligonucleotide inhibits the binding of a nuclear protein to the Ki-ras promoter and strongly reduces cell growth in human carcinoma pancreatic cells. Biochemistry. 2004;43:2512–2523. doi: 10.1021/bi035754f. [DOI] [PubMed] [Google Scholar]
- Cogoi S, Xodo LE. G-quadruplex formation within the promoter of the KRAS proto-oncogene and its effect on transcription. Nucleic Acids Res. 2006;34:2536–2549. doi: 10.1093/nar/gkl286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai J, Chen D, Jones RA, Hurley LH, Yang D. NMR solution structure of the major G-quadruplex structure formed in the human BCL2 promoter region. Nucleic Acids Res. 2006a;34:5133–5144. doi: 10.1093/nar/gkl610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai J, Dexheimer TS, Chen D, Carver M, Ambrus A, Jones RA, Yang D. An intramolecular G-quadruplex structure with mixed parallel/antiparallel G-strands formed in the human BCL-2 promoter region in solution. J Am Chem Soc. 2006b;128:1096–1098. doi: 10.1021/ja055636a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai J, Carver M, Yang D. Polymorphism of human telomeric quadruplex structures. Biochimie. 2008;90:1172–1183. doi: 10.1016/j.biochi.2008.02.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dailey M, Miller MC, Bates PJ, Lane AN, Trent JO. Resolution and characterization of the structural polymorphism of a single quadruplex-forming sequence. Nucleic Acids Res. 2010;38:4877–4888. doi: 10.1093/nar/gkq166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Armond R, Wood S, Sun D, Hurley LH, Ebbinghaus SW. Evidence for the presence of a guanine quadruplex forming region within a polypurine tract of the hypoxia inducible factor 1 alpha promoter. Biochemistry. 2005;44:16341–16350. doi: 10.1021/bi051618u. [DOI] [PubMed] [Google Scholar]
- Dexheimer TS, Sun D, Hurley LH. Deconvoluting the structural and drug-recognition complexity of the G-quadruplex-forming region upstream of the bcl-2 P1 promoter. J Am Chem Soc. 2006;128:5404–5415. doi: 10.1021/ja0563861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esposito V, Randazzo A, Piccialli G, Petraccone L, Giancola C, Mayol L. Effects of an 8-bromodeoxyguanosine incorporation on the parallel quadruplex structure [d(TGGGT)](4) Org Biomol Chem. 2004;2:313–318. doi: 10.1039/b314672c. [DOI] [PubMed] [Google Scholar]
- Esposito V, Virgilio A, Randazzo A, Galeone A, Mayol L. A new class of DNA quadruplexes formed by oligodeoxyribonucleotides containing a 3’-3’ or 5’-5’ inversion of polarity site. Chem Commun (Camb) 2005;31:3953–3955. doi: 10.1039/b504455c. [DOI] [PubMed] [Google Scholar]
- Gill ML, Strobel SA, Loria JP. Crystallization and characterization of the thallium form of the Oxytricha nova G-quadruplex. Nucleic Acids Res. 2006;34(16):4506–4514. doi: 10.1093/nar/gkl616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray RD, Li J, Chaires JB. Energetics and kinetics of a conformational switch in G-quadruplex DNA. J Phys Chem B. 2009;113:2676–2683. doi: 10.1021/jp809578f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray RD, Petraccone L, Trent JO, Chaires JB. Characterization of a K+-induced conformational switch in a human telomeric DNA oligonucleotide using 2-aminopurine fluorescence. Biochemistry. 2010;49:179–194. doi: 10.1021/bi901357r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray RD, Buscaglia R, Chaires JB. Populated intermediates in the thermal unfolding of the human telomeric quadruplex. J Am Chem Soc. 2012;134(40):16834–16844. doi: 10.1021/ja307543z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray RD, Trent JO, Chaires JB. Folding and unfolding pathways of the Human telomeric G-quadruplex. J Mol Biol. 2014;426(8):1629–1650. doi: 10.1016/j.jmb.2014.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gros J, Avino A, Lopez de la Osa J, Gonzalez C, Lacroix L, Perez A, Orozco M, Eritja R, Mergny JL. 8-Amino guanine accelerates tetramolecular G-quadruplex formation. Chem Commun. 2008;25:2926–2928. doi: 10.1039/b801221k. [DOI] [PubMed] [Google Scholar]
- Hsu STD, Varnai P, Bugaut A, Reszka AP, Neidle S, Balasubramanian S. A G-rich sequence within the c-kit oncogene promoter forms a parallel G-quadruplex having asymmetric G-tetrad dynamics. J Am Chem Soc. 2009;131:13399–13409. doi: 10.1021/ja904007p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huppert JL, Balasubramanian S. Prevalence of quadruplexes in the human genome. Nucleic Acids Res. 2005;33:2908–2916. doi: 10.1093/nar/gki609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huppert JL, Balasubramanian S. G-quadruplexes in promoters throughout the human genome. Nucleic Acids Res. 2007a;35:406–413. doi: 10.1093/nar/gkl1057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huppert JL. Four-stranded DNA: Cancer, gene regulation and drug development. Philos Transact A Math Phys Eng Sci. 2007b;365:2969–2984. doi: 10.1098/rsta.2007.0011. [DOI] [PubMed] [Google Scholar]
- Ireson CR, Kelland LR. Discovery and development of anticancer aptamers. Mol Cancer Therapeut. 2006;5:2957–2962. doi: 10.1158/1535-7163.MCT-06-0172. [DOI] [PubMed] [Google Scholar]
- Kumar N, Maiti S. Role of locked nucleic acid modified complementary strand in quadruplex/Watson-Crick duplex equilibrium. J Phys Chem B. 2007;111:12328–12337. doi: 10.1021/jp072705u. [DOI] [PubMed] [Google Scholar]
- Kuryavyi V, Phan AT, Patel DJ. Solution structures of all parallel-stranded monomeric and dimeric G-quadruplex scaffolds of the human c-kit2 promoter. Nucleic Acids Res. 2010;38:6757–6773. doi: 10.1093/nar/gkq558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lane AN, Chaires JB, Gray RD, Trent JO. Stability and kinetics of G-quadruplex structures. Nucleic Acids Res. 2008;36:5482–5515. doi: 10.1093/nar/gkn517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Largy E, Mergny JL. Shape matters: size-exclusion HPLC for the study of nucleic acid structural polymorphism. Nucleic Acids Res. 2014;42:e149. doi: 10.1093/nar/gku751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le HT, Miller MC, Buscaglia R, Dean WL, Holt PA, Chaires JB, Trent JO. Not all G-quadruplexes are created equally: an investigation of the structural polymorphism of the c-Myc G-quadruplex-forming sequence and its interaction with the porphyrin TMPyP4. Org Biomol Chem. 2012;10:9393–9404. doi: 10.1039/c2ob26504d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le HT, Buscaglia R, Dean WL, Chaires JB, Trent JO. Calculation of hydrodynamic properties for G-quadruplex nucleic acid structures from in silico bead models. Top Curr Chem. 2013;330:179–210. doi: 10.1007/128_2012_351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le HT, Dean WL, Buscaglia R, Chaires JB, Trent JO. An investigation of G-quadruplex structural polymorphism in the human telomere using a combined approach of hydrodynamic bead modeling and molecular dynamics simulation. J Phys Chem B. 2014;118:5390–5405. doi: 10.1021/jp502213y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Correia JJ, Wang L, Trent JO, Chaires JB. Not so crystal clear: The structure of the human telomere G-quadruplex in solution differs from that present in a crystal. Nucleic Acids Res. 2005;33:4649–4659. doi: 10.1093/nar/gki782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim KW, Amrane S, Bouaziz S, Xu W, Mu Y, Patel DJ, Luu KN, Phan AT. Structure of the human telomere in K+ solution: a stable basket-type G-quadruplex with only two G-tetrad layers. J Am Chem Soc. 2009;131:4301–4309. doi: 10.1021/ja807503g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luu KN, Phan AT, Kuryavyi V, Lacroix L, Patel DJ. Structure of the human telomere in K+ solution: an intramolecular (3 + 1) G-quadruplex scaffold. J Am Chem Soc. 2006;128:9963–9970. doi: 10.1021/ja062791w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maizels N. Dynamic roles for G4 DNA in the biology of eukaryotic cells. Nat Struct Mol Biol. 2006;13:1055–1059. doi: 10.1038/nsmb1171. [DOI] [PubMed] [Google Scholar]
- Marathias VM, Sawicki MJ, Bolton PH. 6-Thioguanine alters the structure and stability of duplex DNA and inhibits quadruplex DNA formation. Nucleic Acids Res. 1999;27:2860–2867. doi: 10.1093/nar/27.14.2860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masiero S, Trotta R, Pieraccini S, De Tito S, Perone R, Randazzo A, Spada GP. A non-empirical chromophoric interpretation of CD spectra of DNA G-quadruplex structures. Org Biomol Chem. 2010:82683–82692. doi: 10.1039/c003428b. [DOI] [PubMed] [Google Scholar]
- Mekmaysy CS, Petraccone L, Garbett NC, Ragazzon PA, Gray R, Trent JO, Chaires JB. Effect of O6-methylguanine on the stability of G-quadruplex DNA. J Am Chem Soc. 2008;130:6710–6711. doi: 10.1021/ja801976h. [DOI] [PubMed] [Google Scholar]
- Miller MC, Buscaglia R, Chaires JB, Lane AN, Trent JO. Hydration is a major determinant of the G-quadruplex stability and conformation of the human telomere 3’ sequence of d(AG(3)(TTAG(3))(3)) J Am Chem Soc. 2010;132:1705–1707. doi: 10.1021/ja105259m. [DOI] [PubMed] [Google Scholar]
- Miller MC, Le HT, Dean WL, Holt PA, Chaires JB, Trent JO. Polymorphism and resolution of promoter quadruplex-forming sequences. Organic and Biomolecular Chemistry. 2011;9:7633–7637. doi: 10.1039/c1ob05891f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller MC, Trent JO. Size Exclusion Chromatography of G-Quadruplexes. Current Protocols in Nucleic Acids Chemistry. 2011;45:17.3.1–17.3.18. doi: 10.1002/0471142700.nc1703s45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyoshi D, Nakao A, Sugimoto N. Structural transition of d(G4T4G4) from antiparallel to parallel G-quartet induced by divalent cations. Nucleic Acids Res. 2001;1:259–260. doi: 10.1093/nass/1.1.259. [DOI] [PubMed] [Google Scholar]
- Neidle S. Human telomeric G-quadruplex: The current status of telomeric G-quadruplexes as therapeutic targets in human cancer. FEBS J. 2010;277:1118–1125. doi: 10.1111/j.1742-4658.2009.07463.x. [DOI] [PubMed] [Google Scholar]
- Neidle S, Parkinson G. Telomere maintenance as a target for anticancer drug discovery. Nat Rev Drug Discov. 2002;1:383–393. doi: 10.1038/nrd793. [DOI] [PubMed] [Google Scholar]
- Parkinson GN, Lee MP, Neidle S. Crystal structure of parallel quadruplexes from human telomeric DNA. Nature. 2002;417:876–880. doi: 10.1038/nature755. [DOI] [PubMed] [Google Scholar]
- Patel DJ, Phan AT, Kuryavyi V. Human telomere, oncogenic promoter and 5 -UTR G-quadruplexes: Diverse higher order DNA and RNA targets for cancer therapeutics. Nucleic Acids Res. 2007;35:7429–7455. doi: 10.1093/nar/gkm711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petrovic AG, Polavarapu PL. Quadruplex structure of polyriboinosinic acid: Dependence on alkali metal ion concentration, pH and temperature. J Phys Chem B. 2008;112:2255–2260. doi: 10.1021/jp075873v. [DOI] [PubMed] [Google Scholar]
- Phan AT, Modi YS, Patel DJ. Propeller-type parallel-stranded G-quadruplexes in the human c-myc promoter. J Am Chem Soc. 2004;126:8710–8716. doi: 10.1021/ja048805k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phan AT, Kuryavyi V, Luu KN, Patel DJ. Structure of two intramolecular G-quadruplexes formed by natural human telomere sequences in K+ solution. Nucleic Acids Res. 2007a;35:6517–6525. doi: 10.1093/nar/gkm706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phan AT, Kuryavyi V, Burge S, Neidle S, Patel DJ. Structure of an unprecedented G-quadruplex scaffold in the human c-kit promoter. J Am Chem Soc. 2007b;129:4386–4392. doi: 10.1021/ja068739h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi J, Shafer RH. Human telomere quadruplex: Refolding and selection of individual conformers via RNA/DNA chimeric editing. Biochemistry. 2007;46:7599–7606. doi: 10.1021/bi602392u. [DOI] [PubMed] [Google Scholar]
- Sanghvi YS. A Status Update of Modified Oligonucleotides for Chemotherapeutics Applications. Current Protocols in Nucleic Acid Chemistry. 2011;46:4.1:4.1.1–4.1.22. doi: 10.1002/0471142700.nc0401s46. [DOI] [PubMed] [Google Scholar]
- Sannohe Y, Sugiyama H. Overview of Formation of G-Quadruplex Structures. Current Protocols in Nucleic Acid Chemistry. 2010;40:17.2:17.2.1–17.2.17. doi: 10.1002/0471142700.nc1702s40. [DOI] [PubMed] [Google Scholar]
- Simonsson T, Pecinka P, Kubista M. DNA tetraplex formation in the control region of c-myc. Nucleic Acids Res. 1998;26:1167–1172. doi: 10.1093/nar/26.5.1167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spackova N, Cubero E, Sponer J, Orozco M. Theoretical study of the guanine→6-thioguanine substitution in duplexes, triplexes, and tetraplexes. J Am Chem Soc. 2004;126:14642–14650. doi: 10.1021/ja0468628. [DOI] [PubMed] [Google Scholar]
- Sun DY, Guo KX, Rusche JJ, Hurley LH. Facilitation of a structural transition in the polypurine/polypyrimidine tract within the proximal promoter region of the human VEGF gene by the presence of potassium and G-quadruplex-interactive agents. Nucleic Acids Res. 2005;33:6070–6080. doi: 10.1093/nar/gki917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang CF, Shafer RH. Engineering the quadruplex fold: Nucleoside conformation determines both folding topology and molecularity in guanine quadruplexes. J Am Chem Soc. 2006;128:5966–5973. doi: 10.1021/ja0603958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Todd AK, Johnston M, Neidle S. Highly prevalent putative quadruplex sequence motifs in human DNA. Nucleic Acids Research. 2005;33:2901–2907. doi: 10.1093/nar/gki553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Virgilio A, Esposito V, Randazzo A, Mayol L, Galeone A. 8-Methyl-2’-deoxyguanosine incorporation into parallel DNA quadruplex structures. Nucleic Acids Res. 2005a;33:6188–6195. doi: 10.1093/nar/gki924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Virgilio A, Esposito V, Randazzo A, Mayol L, Galeone A. Effects of 8-methyl-2’-deoxyadenosine incorporation into quadruplex forming oligodeoxyribonucleotides. Bioorg Med Chem. 2005b;13:1037–1044. doi: 10.1016/j.bmc.2004.11.037. [DOI] [PubMed] [Google Scholar]
- Webba da Silva M. Geometric formalism for DNA quadruplex folding. Chemistry. 2007;13:9738–9745. doi: 10.1002/chem.200701255. [DOI] [PubMed] [Google Scholar]
- Wang Y, Patel DJ. Solution structure of the human telomeric repeat d[AG3(T2AG3)3] G-tetraplex. Structure. 1993;1:263–282. doi: 10.1016/0969-2126(93)90015-9. [DOI] [PubMed] [Google Scholar]
- Xu Y, Sugiyama H. Formation of the G-quadruplex and i-motif structures in retinoblastoma susceptibility genes (Rb) Nucleic Acids Res. 2006;34:949–954. doi: 10.1093/nar/gkj485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue Y, Kan ZY, Wang Q, Yao Y, Liu J, Hao YH, Tan Z. Human telomeric DNA forms parallel-stranded intramolecular G-quadruplex in K+ solution under molecular crowding condition. J Am Chem Soc. 2007;129:11185–11191. doi: 10.1021/ja0730462. [DOI] [PubMed] [Google Scholar]
- Yang D, Okamoto K. Structural insights into G-quadruplexes: Towards new anticancer drugs. Future Med Chem. 2010;2:619–646. doi: 10.4155/fmc.09.172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z, Dai J, Veliath E, Jones RA, Yang D. Structure of a two-G-tetrad intramolecular G-quadruplex formed by a variant human telomeric sequence in K+ solution: insights into the interconversion of human telomeric G-quadruplex structures. Nucleic Acids Res. 2010;38:1009–1021. doi: 10.1093/nar/gkp1029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ziemba AJ, Zhilina ZV, Krotova-Khan Y, Stankova L, Ebbinghaus SW. Targeting and regulation of the HER-2/neu oncogene promoter with bis-peptide nucleic acids. Oligonucleotides. 2005;15:36–50. doi: 10.1089/oli.2005.15.36. [DOI] [PubMed] [Google Scholar]


