Abstract
Viral diversity is an important predictor of hepatitis C virus (HCV) treatment response and may influence viral pathogenesis. HIV influences HCV variability in the plasma; however, limited data on viral variability are available from distinct tissue/cell compartments in patients co-infected with HIV and HCV. Thus, this exploratory study evaluated diversity of the hypervariable region 1 (HVR1) of HCV in the plasma and liver for 14 patients co-infected with HIV and HCV. Median intra-patient genetic distances and entropy values were similar in the plasma and liver compartments. Positive immune selection pressure was observed in the plasma for five individuals and in the liver for three individuals. Statistical evidence supporting viral compartmentalization was found in five individuals. Linear regression identified ALT (P = 0.0104) and AST (P = 0.0130) as predictors of viral compartmentalization. A total of 12 signature amino acids that distinguish liver from plasma E1/HVR1 were identified. One signature amino acid was shared by at least two individuals. These findings suggest that HCV compartmentalization is relatively common among patients co-infected with HIV and HCV. These data also imply that evaluating viral diversity, including drug resistance patterns, in the serum/plasma only may not adequately represent viruses replicating with in the liver and, thus, deserves careful consideration in future studies.
Keywords: HIV, HCV, co-infection, diversity, quasispecies, compartmentalization
INTRODUCTION
Similar to other RNA viruses, variability is a hallmark of HCV infection. Within an infected individual, HCV exists as a population of distinct but closely related viral variants, termed the viral quasispecies. Quasispecies diversity has important biologic implications for viral persistence, host cell tropism, antiviral drug resistance, and the development of an effective HCV vaccine (reviewed in [Farci, 2011]). The associations between HCV heterogeneity and liver transplantation outcome, disease progression, and chronicity of infection have been explored previously [Martell et al., 1994; Gretch et al., 1996; Brambilla et al., 1998; Manzin et al., 1998; Sullivan et al., 1998; Ray et al., 1999; Farci et al., 2000; Hayashi et al., 2000; Curran et al., 2002]. An association between HCV variability and treatment outcome has been reported as well [Pawlotsky, 2000; Layden-Almer et al., 2005; Sherman et al., 2010]. To date, most studies of HCV diversity have focused on the hypervariable region 1 (HVR1). HVR1 is subject to selective immune pressure [Weiner et al., 1992], and subsequent studies have demonstrated that HVR1 diversity may correlate with viral clearance [Chen and Wang, 2005; Liu et al., 2012].
Due to shared routes of transmission, co-infection with HCV is increasingly recognized as a major cause of morbidity and mortality among HIV-positive persons [Bica et al., 2001; Tedaldi et al., 2003; Salmon-Ceron et al., 2000]. In the US, 150,000–300,000 persons are infected with HIV and HCV, representing 15–30% of all HIV-infected persons and 5–10% of all HCV-infected persons [Alter et al., 1999; Sherman et al., 2002]. It is well established that HIV negatively impacts the clinical course of HCV disease. While the precise mechanism(s) remain unclear, HCV RNA levels are significantly elevated during HIV/HCV co-infection as compared to HCV mono-infection [Yokozaki et al., 2000; Tedaldi et al., 2003]. Furthermore, HIV seroconversion and antiretroviral therapy (ART) initiation [Beld et al., 1998; John et al., 1998; Rutschmann et al., 1998; Ragni and Bontempo, 1999; Chung et al., 2002; Cooper and Cameron, 2002; Pett et al., 2002] are both associated with sustained increases in HCV RNA. The clinical course of HCV infection is also accelerated in patients co-infected with HIV and HCV, as they experience more advanced liver fibrosis, cirrhosis, and death than patients with HCV mono-infected [Soto et al., 1997; Lesens et al., 1999; Yee et al., 2000; Bica et al., 2001; Puoti et al., 2001; Ragni and Belle, 2001; Martinez-Sierra et al., 2003; Mohsen et al., 2003; Poynard et al., 2003; Tedaldi et al., 2003]. Further compounding the clinical management of HCV, HIV co-infection results in decreased response rates to HCV treatment compared to HCV mono-infection [DiMartino et al., 2001; Perez-Olmeda et al., 2003; Chung et al., 2004]. Previous studies have focused largely on HCV mono-infected individuals and reported a non-random distribution of viral variants in various cell types and tissues (reviewed in [Farci, 2011]). This implies that the requirements for HCV replication are cell type-dependent and/or that there are distinct selection pressures acting upon viral variants depending upon their major site(s) of replication.
Relatively, few studies have assessed HCV diversity during HIV/HCV co-infection. Two cross-sectional studies found increased diversity in HVR1 in patients co-infected with HIV and HCV compared to patients with HCV mono-infection [Sherman et al., 1996] or to HIV/HCV co-infected subjects with higher CD4 cell counts [Toyoda et al., 1997]. Others have demonstrated that HCV was more diverse and under less selection pressure in patients co-infected with HIV and HCV compared to the patients with HCV mono-infection [Tanaka et al., 2007]; however, this was not observed in all studies [Mao et al., 2001; Shuhart et al., 2006]. Genomic regions other than the viral envelope proteins also exhibit intra-patient diversity, but the selection pressures on these regions may be distinct. Higher quasispecies diversity was observed in HVR1 compared to core or the 5′ untranslated region from the same patients co-infected with HIV and HCV [Blackard et al., 2004]. Similarly, higher E1/HVR1 genetic distances were observed compared to NS5B sequences from the same patients [Blackard et al., 2012]. This is particularly interesting given that HCV titers are elevated during HIV co-infection [Beld et al., 1998; Yokozaki et al., 2000; Tedaldi et al., 2003] and that co-infection is associated with lower HCV treatment response [DiMartino et al., 2001; Perez-Olmeda et al., 2003; Chung et al., 2004]. However, direct comparisons of HCV diversity in non-structural genes between mono-infected and patients co-infected with HIV and HCV are rare. Similarly, others have shown that p7 diversity was higher during HCV mono-infection compared to HIV/HCV co-infection, and liver fibrosis was inversely correlated with p7 synonymous substitutions [Li et al., 2012]. Therefore, viral diversity and compartmentalization was characterized in a well-characterized cohort of patients co-infected with HIV and HCV.
METHODS
Study Patients
For this pilot study, informed consent was obtained from 14 patients co-infected with HIV and HCV genotype 1 enrolled at the University of Cincinnati. Liver biopsy and plasma samples were obtained prior to the initiation of antiretroviral therapy (ART).
PCR Amplifications
RNA was extracted from 140 ul of plasma using the QIAamp Viral RNA Kit or from homogenized liver biopsies (typically 1–2 mm in length) using the RNeasy Mini kit. cDNA corresponding to E1/HVR1 was generated using the antisense primer 5′-AGG CTT TCA TTG CAG TTC AAG GCC TTG CTA TTG ATG TGC C-3′ (nucleotides 1639–1600) as described elsewhere [Blackard et al., 2004, 2007]. First round PCR amplification using the sense primer 5′-GCG TCC GGG TTC TGG AAG ACG GCG TGA ACT ATG CAA CAG G-3′ (nucleotides 802–841) was performed as follows: 94°C for 2 min, followed by 35 cycles at 94°C for 1 min, 65°C for 2 min, and 72°C for 2 min, with a final elongation step at 72°C for 8 min. Second round PCR conditions were the same as the first round and utilized the antisense primer 5′-AGT TCA AGG CCG TGC TAT TGA TGT GCC AAC TGC CGT TGG T-3′ (nucleotides 1626–1587) and the sense primer 5′-GGC ATG GGA TAT GAT GAT GAA CTG GTC CCC TAC-3′ (nucleotides 1295–1327). These primers have been utilized extensively and amplify HCV genotypes 1–4. All RT-PCR amplifications included one reaction containing no reverse transcriptase and another containing no template as negative controls. PCR products were gel purified and ligated into the pGEM-T Easy Vector. An average of 11 plasmids per sample type per individual were sequenced using dye terminator chemistry and edited using CodonCode Aligner. Previous reports have shown that a similar approach accurately and reproducibly represents the overall HCV quasispecies diversity within an individual and that multiple amplifications of the same template yielded similar estimates of quasispecies diversity [Torres-Puente et al., 2003; Blackard et al., 2004].
Phylogenetic Analysis and Quantification of Diversity
All alignments were performed using the neighbor-joining method implemented in Clustal X [Thompson et al., 1997]. GenBank references used to confirm genotype included D10749 (1a), M62321 (1a), AF511950 (1a), AF177040 (H77; 1a), D90208 (1b), AY460204 (1b), D14853 (1c), AY051292 (1c), AF238481 (2a), AB047639 (2a), AB030907 (2b), AY232746 (2b), D50409 (2c), AF046866 (3a), D17763 (3a), D28917 (3a), D49374 (3b), Y11604 (4a), DQ516084 (4a), FJ025854 (4b), Y13184 (5a), AF064490 (5a), Y12083 (6a), and D84262 (6b). The statistical robustness and reliability of the branching order within each phylogenetic tree were confirmed by bootstrap analysis using 1,000 replicates [Felsenstein, 1985]. Phylogenetic inference was performed using a Bayesian Markov chain Monte Carlo (MCMC) approach as implemented in the BEAST v1.7.2 program [Drummond et al., 2012] under an uncorrelated log-normal relaxed molecular clock and the general time-reversible model with nucleotide site heterogeneity estimated using a gamma distribution. The MCMC analysis was run for a chain length of 50,000,000 with sampling every 5,000th generation. Results were visualized in Tracer v1.5 to confirm chain convergence, and the effective sample size (ESS) was calculated for each parameter. All ESS values were >500 indicating sufficient sampling. The maximum clade credibility tree was selected from the posterior tree distribution after a 10% burn-in using TreeAnnotator v1.7.2. Clonal sequences have been submitted to GenBank and given the accession numbers KC192048–KC192373.
Intra-patient genetic distances were calculated by pairwise comparison of nucleotide sequences using the Kimura method of MegAlign (DNASTAR; Madison, WI). Shannon entropy , where pi is the frequency of each distinct amino acid sequence, and N is the total number of sequences analyzed, was also calculated. Non-synonymous (dN)-synonymous (dS) values were calculated via the Nei-Gojobori method in MEGA v5.1 [Kumar et al., 2008].
Evaluation of Compartmentalization
Compartmentalization of HCV variants was assessed using Mantel's test as described previously using PASSaGE version 2 with 10,000 permutations and the Slatkin-Maddison (S-M) test for population gene flow as implemented in the HyPhy program using 10,000 permutations [Slatkin and Maddison, 1989; Afonso et al., 1999; Ducoulombier et al., 2004; Pond et al., 2005; Roque-Afonso et al., 2005; Zehender et al., 2005; Sobesky et al., 2007; Schramm et al., 2008; Ramirez et al., 2009; Rosenberg and Anderson, 2011]. However, because there is no agreed upon gold standard for evaluating viral compartmentalization, multiple methods were utilized. P values <0.05 were considered statistically significant evidence that viral sequences from the liver were compartmentalized as compared to those from the corresponding plasma. Signature amino acid patterns that distinguish liver viral variants from those in the corresponding plasma were identified using the Viral Epidemiology Signature Pattern Analysis program with a threshold of 70% [Korber and Myers, 1992].
Statistical Analyses
Paired-sample t tests were used to compare intra-patient variability in the plasma versus liver. All analyses were performed using SAS software. Best subset regression models were used to determine which predictor variables—alanine aminotransferase (ALT), aspartate aminotransferase (AST), age, CD4 cell count, log10 HCV viral load, log10 HIV viral load, race, and gender—to include in a multiple regression model. Least squares linear regression was then used to model the relationship between these clinical and/or demographic predictors (with P values ≤ 0.10) and the various methods utilized to assess compartmentalization.
RESULTS
Patient Characteristics
The mean age for study participants was 50.5 years and included 11 males and 3 females (Table I). Twelve participants were African American. The median CD4 cell count was 342 cells/mm3. Median ALT and AST levels were 56.0 and 85.5 IU/ml, respectively. Median baseline HIV and HCV RNA levels were 4.6 log10 copies/ml and 6.3 log10 IU/ml, respectively. All participants were ART-naïve and were not receiving interferon or direct-acting HCV agents at the time of sample collection.
TABLE I.
Demographic and Clinical Characteristics of Patients Co-infected With HIV and HCV
| Characteristic | |
| Age at enrollment (years) ± SD | 50.5 ± 9.2 |
| Race | |
| African American | 12 (85.7%) |
| Caucasian | 2 (14.3%) |
| Gender (% male) | 11 (78.6%) |
| Log10 HIV viral load (copies/ml) ± SD | 4.6 ± 0.6 |
| Log10 HCV viral load (IU/ml) ± SD | 6.3 ± 0.6 |
| Median CD4 cell count (cells/mm3) ± SD | 342.0 ± 119.5 |
| AST (IU/ml) ± SD | 85.5 ± 89.2 |
| ALT (IU/ml) ± SD | 56.0 ± 80.6 |
IU, international units; ALT, alanine aminotransferase; AST, aspartate aminotransferase; SD, standard deviation.
Intra-patient Variability
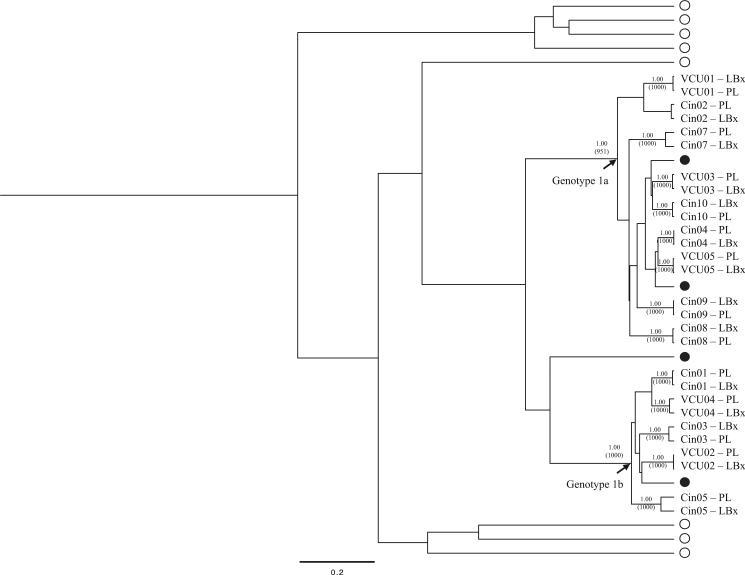
All consensus E1/HVR1 nucleotide sequences belonged to HCV genotype 1, and plasma and liver sequences clustered by patient in all instances (Fig. 1). Intra-patient variability was similar between plasma and liver samples with respect to median genetic distance (0.98% in plasma versus 0.78% in liver; P = 0.65), entropy (0.89 vs. 0.80; P = 0.21), and dN-dS (−0.0004 vs. −0.0011; P = 30). Positive dN-dS values—indicating positive immune selection pressure—were observed in the plasma and liver for six and six individuals, respectively.
Fig. 1.
Consensus E1/HVR1 nucleotide sequences from 14 pairs of matched plasma and liver biopsies analyzed using a Bayesian inference approach. Relevant posterior probabilities greater than 90% are shown. Relevant bootstrap values >700 out of 1,000 from a Neighbor-Joining approach are shown in parentheses. Closed circles denote HCV genotype 1 references, while open circles denote HCV genotype 2–6 references.
Analysis of HCV Compartmentalization
By phylogenetic comparison of patient-specific viral variants from the plasma and liver, three subjects—Cin03, Cin05, and VCU01–had plasma-specific E1/HVR1 variants (Supplementary Fig. S1). HCV compartmentalization was explored further using the Slatkin-Maddison (S-M) test and Mantel's test, which do not rely on interpretation of a phylogenetic tree (Table II). When comparing patient-specific plasma- and liver-derived E1/HVR1 variants, the S-M test demonstrated statistically significant quasispecies compartmentalization for Cin05 (P = 0.002) and VCU05 (P = 0.0004) and trends towards compartmentalization for Cin03 (P = 0.088) and Cin10 (P = 0.071). Similarly, Mantel's test demonstrated statistically significant evidence of compartmentalization for Cin05 (P = 0.001) and Cin10 (P = 0.017). Thus, five patients showed evidence of viral compartmentalization using at least one measurement.
TABLE II.
Compartmentalization of HCV E1/HVR1 Sequences in the Liver Versus the Plasma
| Patient ID | Phylogenetic compartmentalizationa | Slatkin-Maddison test | Mantel's test |
|---|---|---|---|
| Cin01 | No | NS | NS |
| Cin02 | No | NS | NS |
| Cin03 | Yes (4) | 0.088 | NS |
| Cin04 | No | NS | NS |
| Cin05 | Yes (6)* | 0.002 | 0.001 |
| Cin07 | No | NS | NS |
| Cin08 | No | NS | NS |
| Cin09 | No | NS | NS |
| Cin10 | No | 0.071 | 0.017 |
| VCU01 | Yes (2) | NS | NS |
| VCU02 | No | NS | NS |
| VCU03 | No | NS | NS |
| VCU04 | No | NS | NS |
| VCU05 | No | 0.0004 | NS |
Phylogenetic compartmentalization indicates the presence of clustered sequences from the same location separated by a posterior probability of at least 0.9. Numbers in parentheses indicate the number of clones within a cluster of sequences.
The asterisk indicates an additional cluster of three sequences. Bold values indicate statistically significant evidence of compartmentalization.
NS, not significant (P-value > 0.10).
Next, potential predictors of viral compartmentalization were evaluated (Table III). Using the phylo-genetic test, best subset regression identified CD4 cell count and gender as factors possibly associated with HCV compartmentalization (adjusted R2 for the model = 0.2162). With Mantel's test, ALT, AST, log10 HCV, log10 HIV, race, and gender were identified as factors possibly associated with compartmentalization (adjusted R2 = 0.5273). Using the S-M test, ALT, AST, and age were identified as factors possibly associated with compartmentalization (adjusted R2 = 0.1536). Using a combination of phylogenetic, Mantel's, and/or SM tests, age, race, and gender were identified as factors possibly associated with compartmentalization (adjusted R2 = 0.2668). The Mantel's test model had the highest R2 value and was deemed the best predictive model overall. Linear regression identified ALT (P = 0.0104) and AST (P = 0.0130) as predictors of viral compartmentalization (R2 value = 0.5365 and P value for the model = 0.0453).
TABLE III.
Results of Best Subset Regression for Four Models of HCV Compartmentalization
| Model | Adjusted R2 | Model variables |
|---|---|---|
| 1 = Phylogeny | 0.2162 | CD4, gender |
| 2 = Mantel's testa | 0.5273 | ALT, AST, log10 HCV, log10 HIV, race, gender |
| 3 = Slatkin-Maddison test | 0.1536 | ALT, AST, age |
| 4 = Models 1, 2, and/or 3 | 0.2668 | Age, race, gender |
Best predictive model based on highest adjusted R2.
Signature Sequence Analysis of Compartment-Specific Amino Acids
To identify specific amino acids associated with compartmentalization in patients co-infected with HIV and HCV, signature sequence analysis was performed as shown in Figure 2. A total of 12 (range: 1–7) signature amino acids were detected in E1/HVR1 for Cin05, Cin07, and Cin10. One signature amino acid—position 401–was shared by two individuals.
Fig. 2.
Signature sequence analysis showing amino acid positions at which the distributions of liver- and plasma-specific E1/HVR1 variants are significantly different. Shown are the consensus amino acids at each position relative to the reference strain H77. Numbers in parentheses indicate the number of amino acid signatures between the plasma-liver sequences. The underlined amino acids represent signatures, while the signature amino acid denoted by an asterisk (*) is shared by two individuals.
CONCLUSIONS
During HIV infection, viral compartmentalization has been documented in the brain, cerebrospinal fluid, and genital tract, and there are emerging data for HIV compartmentalization within the gut, lung, liver, kidney, and breast milk (reviewed in [Blackard, 2012]). HIV alters HCV diversity [Sherman et al., 1996; Toyoda et al., 1997; Mao et al., 2001; Shuhart et al., 2006; Tanaka et al., 2007; Li et al., 2012]. Extrahepatic complications of HCV infection include glomerulonephritis, mixed cryoglobulinemia, porphyria cutanea tarda, arthralgias, diabetes, autoimmune disorders, and thyroid disease. Extrahepatic replication of HCV has been reported in peripheral blood mononuclear cells (PBMCs)—including granulocytes, monocytes/macrophages, dendritic cells, and B lymphocytes—as well as extrahepatic tissues [Laskus et al., 1997, 1998a,b, 2000; Maggi et al., 1997, 1999; Shimizu et al., 1997; Lerat et al., 1998; Navas et al., 1998; Okuda et al., 1999; Roque-Afonso et al., 1999; Caussin-Schwemling et al., 2001; Radkowski et al., 2002; Goutagny et al., 2003; Radkowski et al., 2004; Bartolomé et al., 2008]. Extrahepatic replication of HCV has important implications for transmission, disease progression, and treatment [Bare, 2009]. For instance, several studies detected HCV RNA in PBMCs without detection in the corresponding serum or liver [Saleh et al., 1994; Taliani et al., 1995; Mazur et al., 2001]. In addition, patients with negative-strand HCV RNA in PBMCs had lower IFN responses compared to those with no negative-strand RNA in PBMCs [Gong et al., 2003]. Studying HCV diversity provides further evidence of extrahepatic replication by demonstrating a non-random distribution of viral variants in hepatic and/or extrahepatic compartments (reviewed in [Farci, 2011]). By definition, viral compartments are characterized by a restriction of viral gene flow between cells or tissues [Nickle et al., 2003] and occur due to physical isolation of a particular tissue/cell-type, selective migration of susceptible cells, different determinants of virulence or tropism depending upon the cell-type (i.e., distinct levels of entry factors or co-factors required for replication), and/or localized selection pressures. Significant diversity exists among hypervariable region 1 (HVR1) variants in the serum/plasma, liver, and/or PBMC variants [Maggi et al., 1997, 1999; Shimizu et al., 1997; Navas et al., 1998; Okuda et al., 1999; Zehender et al., 2005; Ramirez et al., 2009] due to variation in the viral envelope determinants of HCV entry and/or the varied capacity of the host immune response to interact with and remove certain viral variants. Diversity in the 5′ untranslated region (UTR) and/or internal ribosome entry site (IRES) also exists in PBMCs [Di Liberto et al., 2006; Vera-Otarola et al., 2009]. The precise mechanisms leading to the development of compartmentalization have not been adequately examined but likely include (i) differential immune selection pressures, (ii) cell-type specific differences in viral replication, (iii) local concentrations of antiviral drugs and/or drug resistance, and (iv) co-infections that alter the cellular microenvironment. Functional studies further demonstrate that HCV variation also affects translational efficiency and implies that HCV adapts for efficient replication in a cell type-dependent manner [Laporte et al., 2000, 2003; Lerat et al., 2000; Forton et al., 2004].
In the current analysis, both phylogenetic and statistical evidence of viral compartmentalization were observed between the liver and the plasma for a subset of patients co-infected with HIV and HCV. These findings are supported by other studies demonstrating that the circulating quasispecies population does not always reflect the composition of the intrahepatic quasispecies population. For example, when examining quasispecies diversity in the brain, lymph node, liver, and serum, 50% of E2/HVR1 variants in the liver were not observed in the serum [Forton et al., 2004]. When evaluating UTR diversity in the serum and liver by a single-strand conformational polymorphism assay, 4 of 6 liver samples contained variants not present in the corresponding sera [Jang et al., 1999]. Others reported that E2/NS1 variants present in the liver and/or PBMCs were not detected in the corresponding plasma, while other variants were present only in the plasma [Maggi et al., 1997]. Similarly, at least one HVR1 sequence unique to the serum was detected in 12 of 13 patients analyzed [Okuda et al., 1999]. Thus, HCV compartmentalization is relatively frequent in vivo, although only a minority of studies have included patients co-infected with HIV and HCV or evaluated the clinical factors associated with compartmentalization.
To our knowledge, only one other study has evaluated factors associated with HCV compartmentalization. Using a single-strand conformational polymorphism assay, others reported that compartmentalization was more frequent in former injection drug users and less frequent with genotype 1 [Di Liberto et al., 2006]. In this study, ALT and AST were identified as variables significantly associated with viral compartmentalization, suggesting that higher transaminase levels may be associated with higher rates of hepatocyte cell death and turnover. However, the current study only included individuals with HCV genotype 1; therefore, the role of HCV genotype could not be examined further. Others have examined the association between hepatic injury—as measured by ALT/AST levels—and viral diversity [Nagayama et al., 1999; Bozdayi et al., 2000; Farci et al., 2006; Hanada et al., 2007]. In a pilot study of five patients, reduced transaminase levels were associated with decreased immune pressure targeting the E2 region, suggesting that the vigor of the immune response may drive both ALT elevations and viral diversity [Hanada et al., 2007]. HVR1 diversity also correlates with ALT levels in perinatally infected children [Farci et al., 2006]. Unfortunately, these studies did not evaluate compartmentalization or patients co-infected with HIV and HCV as included in the current study. In theory, the development of viral compartmentalization may serve as a defense mechanism by which the virus hides for certain immunologic responses and/or impacts the degree of immune-mediated liver injury, although this hypothesis has not been adequately explored to date.
Limitations to the current study include the modest number of study patients evaluated in this pilot study, the possible lack of generalizability across HCV genotypes, and an inability to evaluate compartment-specific HCV variants over time. As well, by sequencing a limited number of variants and a small portion of the HCV genome, the true magnitude of viral diversity and compartmentalization was likely underestimated. As discussed elsewhere, there are several methods for evaluating viral compartmentalization; however, there is no consensus as to which are most accurate. Nonetheless, the utilization of several complementary methods—as performed here —provides the most reliable assessment of viral compartmentalization [Zárate et al., 2007].
The current study focused on the viral envelope as a critical determinant of HCV replication, cell tropism, and compartmentalization. However, non-structural genomic regions are also involved in viral replication, tropism, and/or compartmentalization. For instance, diversity in the 5′ UTR/IRES affects translational efficiency, implying that HCV has adapted for replication in a cell type-dependent manner [Laporte et al., 2000, 2003; Lerat et al., 2000; Forton et al., 2004; Di Liberto et al., 2006; Vera-Otarola et al., 2009]. Our group also reported that UTR diversity was associated with plasma HCV RNA [Blackard et al., 2004], although this has not yet been examined in extrahepatic sites. As well, NS5A variability modifies its transcriptional activity and regulates virus-host interactions in a cell type-dependent manner [Pellerin et al., 2004]. Similarly, NS5B variability impacts HCV RNA levels and treatment outcome in vivo [Lohmann et al., 2000; Lévêque and Wang, 2002; Hamano et al., 2005; Itakura et al., 2005; Le Pogam et al., 2008; Blackard et al., 2010]. We identified previously NS5B compartmentalization in PBMCs even when E1/E2 compartmentalization was absent [Blackard et al., 2012].
Multiple explanations may support the presence of divergent viruses in the serum and liver. First, different viral variants may possess distinct replication kinetics. For variants that replicate at low levels, this could provide a means of avoiding immunologic surveillance and maintaining persistent infection. Alternatively, liver-specific variants could represent defective genomes that cannot be packaged and released into the peripheral circulation or newly emerged variants that have not yet entered the circulation. Finally, it is possible that rapid elimination of major viral variants by neutralizing antibodies leads to over-representation of minor variants over time; however, longitudinal analysis of viral variants will be necessary to explore such a possibility in the future. Importantly, the association between compartmentalization and treatment outcome has not been examined to date in the context of direct-acting antiviral agents. These findings imply that evaluating viral diversity—including drug resistance patterns—in the serum/plasma only may not adequately represent viruses replicating within the liver. This potential divergence has implications for the effective management of chronic HCV infection and deserves careful consideration in future studies.
Supplementary Material
ACKNOWLEDGMENTS
This work has been supported in part by NIAID R01 (AI 065256) to KES. This work was presented at the 6th International HIV and Hepatitis Co-Infection Workshop held in Tel Aviv, Israel from May 31–June 2, 2010.
Grant sponsor: NIAID R01; Grant number: AI 065256.
Footnotes
SUPPORTING INFORMATION
Additional supporting information may be found in the online version of this article at the publisher's web-site.
REFERENCES
- Afonso A, Jian J, Penin F, Tareau C, Samuel D, Petit M, Bismuth H, Dussaix E, Feray C. Nonrandom distribution of hepatitis C virus quasispecies in plasma and peripheral blood mononuclear cell subsets. J Virol. 1999;73:9213–9221. doi: 10.1128/jvi.73.11.9213-9221.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alter M, Druszon-Moran D, Nainan O, McQuillan G, Gao F, Moyer L, Kaslow R, Margolis H. The prevalence of hepatitis C virus infection in the United States, 1988 through 1994. N Engl J Med. 1999;341:556–562. doi: 10.1056/NEJM199908193410802. [DOI] [PubMed] [Google Scholar]
- Bare P. Hepatitis C virus and peripheral blood mononuclear cell reservoirs. World J Hepatol. 2009;1:67–71. doi: 10.4254/wjh.v1.i1.67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartolomé J, Rodríguez-Iñigo E, Quadros P, Vidal S, Pascual-Miguelañez I, Rodríguez-Montes JA, García-Sancho L, Carreño V. Detection of hepatitis C virus in thyroid tissue from patients with chronic HCV infection. J Med Virol. 2008;80:1588–1594. doi: 10.1002/jmv.21269. [DOI] [PubMed] [Google Scholar]
- Beld M, Penning M, Lukashov V, McMorrow M, Roos M, Pakker N, van den Hoek A, Goudsmit J. Evidence that both HIV and HIV-induced immunodeficiency enhance HCV replication among HCV seroconverters. Virology. 1998;244:504–512. doi: 10.1006/viro.1998.9130. [DOI] [PubMed] [Google Scholar]
- Bica I, McGovern B, Dhar R, Stone D, McGowan K, Scheib R, Snydman D. Increasing mortality due to end-stage liver disease in patients with human immunodeficiency virus infection. Clin Infect Dis. 2001;32:492–497. doi: 10.1086/318501. [DOI] [PubMed] [Google Scholar]
- Blackard J. HIV compartmentalization: A review on a clinically important phenomenon. Current HIV Res. 2012;10:133–142. doi: 10.2174/157016212799937245. [DOI] [PubMed] [Google Scholar]
- Blackard JT, Yang Y, Bordoni P, Sherman KE, Chung R, for the AIDS Clinical Trials Group 383 Study Team Hepatitis C virus (HCV) diversity in HIV-HCV coinfected subjects initiating highly active antiretroviral therapy. J Infect Dis. 2004;189:1472–1481. doi: 10.1086/382959. [DOI] [PubMed] [Google Scholar]
- Blackard JT, Hiasa Y, Smeaton L, Jamieson DJ, Rodriguez I, Mayer KH, Chung R. Compartmentalization of hepatitis C virus (HCV) during HCV/HIV coinfection. J Infect Dis. 2007;195:1765–1773. doi: 10.1086/518251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blackard JT, Ma G, Limketkai BN, Welge JA, Dryer PD, Martin CM, Hiasa Y, Taylor LE, Mayer KH, Jamieson DJ, Sherman K. Variability of the polymerase gene (NS5B) in hepatitis C virus-infected women. J Clin Microbiol. 2010;48:4256–4259. doi: 10.1128/JCM.01613-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blackard JT, Ma G, Welge JA, Martin CM, Sherman KE, Taylor LE, Mayer KH, Jamieson D. Analysis of a non-structural gene reveals evidence of possible hepatitis C virus (HCV) compartmentalization. J Med Virol. 2012;84:242–252. doi: 10.1002/jmv.22269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bozdayi AM, Uzunalimoglu O, Aslan N, Bozkaya H, Türkyilmaz AR, Kayhan B, Sahin T, Sivri B, Uygun A, Altiok A, Cetinkaya H, Karayalçin S, Yurdaydin C. Influence of viral load and alanine aminotransferase on viral genetic heterogeneity in patients with chronic hepatitis C virus infection. Intervirology. 2000;43:61–66. doi: 10.1159/000025024. [DOI] [PubMed] [Google Scholar]
- Brambilla S, Bellati G, Asti M, Lisa A, Candusso M, D'Amico M, Grassi G, Giacca M, Franchini A, Bruno S, Ideo G, Mondelli M, Silini E. Dynamics of hypervariable region 1 variation in hepatitis C virus infection and correlation with clinical and virological features of liver disease. Hepatology. 1998;27:1678–1686. doi: 10.1002/hep.510270629. [DOI] [PubMed] [Google Scholar]
- Caussin-Schwemling C, Schmitt C, Stoll-Keller F. Study of the infection of human blood derived monocyte/macrophages with hepatitis C virus in vitro. J Med Virol. 2001;65:14–22. doi: 10.1002/jmv.1095. [DOI] [PubMed] [Google Scholar]
- Chen S, Wang Y. Multigene tracking of quasispecies in viral persistence and clearance of hepatitis C virus. World J Gastroenterol. 2005;11:2874–2884. doi: 10.3748/wjg.v11.i19.2874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung R, Evans S, Yang Y, Theodore D, Valdez H, Clark R, Shikuma C, Nevin T, Sherman K, for the AIDS Clinical Trials Group 383 Study Team Immune recovery is associated with persistent rise in hepatitis C RNA, infrequent liver test flares, and is not impaired by hepatitis C in co-infected subjects. AIDS. 2002;16:1915–1923. doi: 10.1097/00002030-200209270-00008. [DOI] [PubMed] [Google Scholar]
- Chung RT, Andersen J, Volberding P, Robbins GK, Liu T, Sherman KE, Peters MG, Koziel MJ, Bhan AK, Alston B, Colquhoun D, Nevin T, Harb G, van der Horst C, for the AIDS Clinical Trials Group A5071 Study Team Peginterferon alpha-2a plus ribavirin versus interferon alpha-2a plus ribavirin for chronic hepatitis C in HIV-coinfected persons. N Engl J Med. 2004;351:451–459. doi: 10.1056/NEJMoa032653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper C, Cameron D. Review of the effect of highly active antiretroviral therapy on hepatitis C virus (HCV) RNA levels in human immunodeficiency virus and HCV coinfection. Clin Infect Dis. 2002;35:873–879. doi: 10.1086/342388. [DOI] [PubMed] [Google Scholar]
- Curran R, Jameson C, Craggs J, Grabowska A, Thomson B, Robins A, Irving W, Ball J. Evolutionary trends of the first hypervariable region of the hepatitis C virus E2 protein in individuals with differing liver disease severity. J Gen Virol. 2002;83:11–23. doi: 10.1099/0022-1317-83-1-11. [DOI] [PubMed] [Google Scholar]
- Di Liberto G, Roque-Afonso AM, Kara R, Ducoulombier D, Fallot G, Samuel D, Feray C. Clinical and therapeutic implications of hepatitis C virus compartmentalization. Gastroenterology. 2006;131:76–84. doi: 10.1053/j.gastro.2006.04.016. [DOI] [PubMed] [Google Scholar]
- DiMartino V, Rufat P, Boyer N, Renard P, Degos F, Martinot-Peignoux M, Matheron S, Le Moing V, Vachon F, Degott C, Valla D, Marcellin P. The influence of human immunodeficiency virus coinfection on chronic hepatitis C in injection drug users: A long-term retrospective cohort study. Hepatology. 2001;34:1193–1199. doi: 10.1053/jhep.2001.29201. [DOI] [PubMed] [Google Scholar]
- Drummond AJ, Suchard MA, Xie D, Rambaut A. Bayesian phylogenetics with BEAUti and the BEAST 1.7. Mol Biol Evol. 2012;29:1969–1973. doi: 10.1093/molbev/mss075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ducoulombier D, Roque-Afonso AM, Di Liberto G, Penin F, Kara R, Richard Y, Dussaix E, Feray C. Frequent compartmentalization of hepatitis C virus variants in circulating B cells and monocytes. Hepatology. 2004;39:817–825. doi: 10.1002/hep.20087. [DOI] [PubMed] [Google Scholar]
- Farci P. New insights into the HCV quasispecies and compartmentalization. Semin Liver Dis. 2011;31:356–374. doi: 10.1055/s-0031-1297925. [DOI] [PubMed] [Google Scholar]
- Farci P, Shimoda A, Coiana A, Diaz G, Peddis G, Melpolder J, Strazzera A, Chien D, Munoz S, Balestrieri A, Purcell R, Alter H. The outcome of acute hepatitis C predicted by the evolution of the viral quasispecies. Science. 2000;288:339–344. doi: 10.1126/science.288.5464.339. [DOI] [PubMed] [Google Scholar]
- Farci P, Quinti I, Farci S, Alter HJ, Strazzera R, Palomba E, Coiana A, Cao D, Casadei AM, Ledda R, Iorio R, Vegnente A, Diaz G, Tovo P. Evolution of hepatitis C viral quasispecies and hepatic injury in perinatally infected children followed prospectively. Proc Natl Acad Sci U S A. 2006;103:8475–8848. doi: 10.1073/pnas.0602546103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Felsenstein J. Confidence limits on phylogenies: An approach using the bootstrap. Evolution. 1985;39:783–791. doi: 10.1111/j.1558-5646.1985.tb00420.x. [DOI] [PubMed] [Google Scholar]
- Forton DM, Karayiannis P, Mahmud N, Taylor-Robinson SD, Thomas H. Identification of unique hepatitis C virus quasispecies in the central nervous system and comparative analysis of internal translational efficiency of brain, liver, and serum variants. J Virol. 2004;78:5170–5183. doi: 10.1128/JVI.78.10.5170-5183.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gong G, Lai L, Jiang Y, He Y, Su X. HCV replication in PBMC and its influence on interferon therapy. World J Gastroenterol. 2003;9:291–294. doi: 10.3748/wjg.v9.i2.291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goutagny N, Fatmi A, De Ledinghen V, Penin F, Couzigou P, Ichauspe G, Bain C. Evidence of viral replication in circulating dendritic cells during hepatitis C virus infection. J Infect Dis. 2003;187:1951–1958. doi: 10.1086/375350. [DOI] [PubMed] [Google Scholar]
- Gretch D, Polyak S, Wilson J, Carithers R, Perkins J, Corey L. Tracking hepatitis C virus quasispecies major and minor variants in symptomatic and asymptomatic liver transplant recipients. J Virol. 1996;70:7622–7631. doi: 10.1128/jvi.70.11.7622-7631.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamano K, Sakamoto N, Enomoto N, Izumi N, Asahina Y, Kurosaki M, Ueda E, Tanabe Y, Maekawa S, Itakura J, Watanabe H, Kakinuma S, Watanabe M. Mutations in the NS5B region of the hepatitis C virus genome correlate with clinical outcomes of interferon-alpha plus ribavirin combination therapy. J Gastroenterol Hepatol. 2005;20:1401–1409. doi: 10.1111/j.1440-1746.2005.04024.x. [DOI] [PubMed] [Google Scholar]
- Hanada K, Tanaka Y, Mizokami M, Gojobori T, Alter H. A reduction in selective immune pressure during the course of chronic hepatitis C correlates with diminished biochemical evidence of hepatic inflammation. Virology. 2007;361:27–33. doi: 10.1016/j.virol.2006.10.041. [DOI] [PubMed] [Google Scholar]
- Hayashi J, Furusyo N, Ariyama I, Sawayama Y, Etoh Y, Kashiwagi S. A relationship between the evolution of hepatitis C virus variants, liver damage, and hepatocellular carcinoma in patients with hepatitis C viremia. J Infect Dis. 2000;181:1523–1527. doi: 10.1086/315431. [DOI] [PubMed] [Google Scholar]
- Itakura J, Nagayama K, Enomoto N, Hamano K, Sakamoto N, Fanning LJ, Kenny-Walsh E, Shanahan F, Watanabe M. Viral load change and sequential evolution of entire hepatitis C virus genome in Irish recipients of single source-contaminated anti-D immunoglobulin. J Viral Hepat. 2005;12:594–603. doi: 10.1111/j.1365-2893.2005.00645.x. [DOI] [PubMed] [Google Scholar]
- Jang SJ, Wang LJ, Radkowski M, Rakela J, Laskus T. Differences between hepatitis C virus 5’ untranslated region quasispecies in serum and liver. J Gen Virol. 1999;80:711–716. doi: 10.1099/0022-1317-80-3-711. [DOI] [PubMed] [Google Scholar]
- John M, Flexman J, French M. Hepatitis C virus-associated hepatitis following treatment of HIV-infected patients with HIV protease inhibitors: An immune response restoration disease? AIDS. 1998;12:2289–2293. doi: 10.1097/00002030-199817000-00010. [DOI] [PubMed] [Google Scholar]
- Korber B, Myers G. Signature pattern analysis: A method for assessing viral sequence relatedness. AIDS Res Hum Retroviruses. 1992;8:1549–1560. doi: 10.1089/aid.1992.8.1549. [DOI] [PubMed] [Google Scholar]
- Kumar S, Dudley J, Nei M, Tamura K. MEGA: A biologist-centric software for evolutionary analysis of DNA and protein sequences. Brief Bioinform. 2008;9:299–306. doi: 10.1093/bib/bbn017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laporte J, Malet I, Andrieu T, Thibault V, Toulme J, Wychowski C, Pawlotsky J, Huraux J, Agut H, Cahour A. Comparative analysis of translation efficiencies of hepatitis C virus 5’ untranslated regions among intraindividual quasispecies present in chronic infection: Opposite behaviors depending on cell type. J Virol. 2000;74:10827–10833. doi: 10.1128/jvi.74.22.10827-10833.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laporte J, Bain C, Maurel P, Inchauspe G, Agut H, Cahour A. Differential distribution and internal translation efficiency of hepatitis C virus quasispecies present in dendritic and liver cells. Blood. 2003;101:52–57. doi: 10.1182/blood-2002-03-0818. [DOI] [PubMed] [Google Scholar]
- Laskus T, Radkowski M, Wang L, Cianciara J, Vargas H, Rakela J. Hepatitis C virus negative strand RNA is not detected in peripheral blood mononuclear cells and viral sequences are identical to those in serum: A case against extrahepatic replication. J Gen Virol. 1997;78:2747–2750. doi: 10.1099/0022-1317-78-11-2747. [DOI] [PubMed] [Google Scholar]
- Laskus T, Radkowski M, Wang L, Jang S, Vargas H, Rakela J. Hepatitis C virus quasispecies in patients infected with HIV-1: Correlation with extrahepatic replication. Virology. 1998a;248:164–171. doi: 10.1006/viro.1998.9269. [DOI] [PubMed] [Google Scholar]
- Laskus T, Radkowski M, Wang L, Vargas H, Rakela J. Search for hepatitis C virus extrahepatic replication sites in patients with acquired immunodeficiency syndrome: Specific detection of negative-strand viral RNA in various tissues. Hepatology. 1998b;28:1398–1401. doi: 10.1002/hep.510280531. [DOI] [PubMed] [Google Scholar]
- Laskus T, Radkowski M, Wang L, Nowicki M, Rakela J. Uneven distribution of hepatitis C virus quasispecies in tissues from subjects with end-stage liver disease: Confounding effect of viral adsorption and mounting evidence for the presence of low-level extrahepatic replication. J Virol. 2000;74:1014–1017. doi: 10.1128/jvi.74.2.1014-1017.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Layden-Almer JE, Kuiken C, Ribeiro RM, Kunstman KJ, Perelson AS, Layden TJ, Wolinsky SM. Hepatitis C virus genotype 1a NS5A pretreatment sequence variation and viral kinetics in African American and white patients. J Infect Dis. 2005;192:1078–1087. doi: 10.1086/432760. [DOI] [PubMed] [Google Scholar]
- Le Pogam S, Seshaadri A, Kosaka A, Chiu S, Kang H, Hu S, Rajyaguru S, Symons J, Cammack N, Nájera I. Existence of hepatitis C virus NS5B variants naturally resistant to nonnucleoside, but not to nucleoside, polymerase inhibitors among untreated patients. J Antimicrob Chemother. 2008;61:1205–1216. doi: 10.1093/jac/dkn085. [DOI] [PubMed] [Google Scholar]
- Lerat H, Rumin S, Habersetzer F, Berby F, Trabaud M, Trepo C, Inchauspe G. In vivo tropism of hepatitis C virus genomic sequences in hematopoietic cells: Influence of viral load, viral genotype, and cell phenotype. Blood. 1998;91:3841–3849. [PubMed] [Google Scholar]
- Lerat H, Shimizu Y, Lemon S. Cell type-specific enhancement of hepatitis C virus internal ribosome entry site-directed translation due to 5’ nontranslated region substitutions selected during passage of virus in lymphoblastoid cells. J Virol. 2000;74:7024–7031. doi: 10.1128/jvi.74.15.7024-7031.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lesens O, Deschenes M, Steben M, Belanger G, Tsoukas G. Hepatitis C virus is related to progressive liver disease in human immunodeficiency virus-positive hemophiliacs and should be treated as an opportunistic infection. J Infect Dis. 1999;179:1254–1258. doi: 10.1086/314720. [DOI] [PubMed] [Google Scholar]
- Lévêque VJ, Wang Q. RNA-dependent RNA polymerase encoded by hepatitis C virus: Biomedical applications. Cell Mol Life Sci. 2002;59:909–919. doi: 10.1007/s00018-002-8478-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Atkins E, Bruckner J, McArdle S, Qiu WC, Thomassen LV, Scott J, Shuhart MC, Livingston S, Townshend-Bulson L, McMahon BJ, Harris M, Griffin S, Gretch D. Genetic and functional heterogeneity of the hepatitis C virus p7 ion channel during natural chronic infection. Virology. 2012;423:30–37. doi: 10.1016/j.virol.2011.11.011. [DOI] [PubMed] [Google Scholar]
- Liu L, Fisher BE, Thomas DL, Cox AL, Ray S. Spontaneous clearance of primary acute hepatitis C virus infection correlated with high initial viral RNA level and rapid HVR1 evolution. Hepatology. 2012;55:1684–1691. doi: 10.1002/hep.25575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lohmann V, Roos A, Körner F, Koch JO, Bartenschlager B. Biochemical and structural analysis of the NS5B RNA-dependent RNA polymerase of the hepatitis C virus. J Viral Hepat. 2000;7:167–174. doi: 10.1046/j.1365-2893.2000.00218.x. [DOI] [PubMed] [Google Scholar]
- Maggi F, Fornai C, Vatteroni ML, Giorgi M, Morrica A, Pistello M, Cammarota G, Marchi S, Ciccorossi P, Bionda A, Bendinelli M. Differences in hepatitis C virus quasispecies composition between liver, peripheral blood mononuclear cells and plasma. J Gen Virol. 1997;78:1521–1525. doi: 10.1099/0022-1317-78-7-1521. [DOI] [PubMed] [Google Scholar]
- Maggi F, Fornai C, Morrica A, Vatteroni M, Giorgi M, Marchi S, Ciccorossi P, Bendinelli M, Pistello M. Divergent evolution of hepatitis C virus in liver and peripheral blood mononuclear cells of infected patients. J Med Virol. 1999;57:57–63. [PubMed] [Google Scholar]
- Manzin A, Solforosi L, Petrelli E, Macarri G, Tosone G, Piazza M, Clementi M. Evolution of hypervariable region 1 of hepatitis C virus in primary infection. J Virol. 1998;72:6271–6276. doi: 10.1128/jvi.72.7.6271-6276.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mao Q, Ray SC, Laeyendecker O, Ticehurst JR, Strathdee SA, Vlahov D, Thomas D. Human immunodeficiency virus seroconversion and evolution of the hepatitis C virus quasispecies. J Virol. 2001;75:3259–3267. doi: 10.1128/JVI.75.7.3259-3267.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martell M, Esteban J, Quer J, Vargas V, Esteban R, Guardia J, Gomez J. Dynamic behavior of hepatitis C virus quasispecies in patients undergoing orthotopic liver transplantation. J Virol. 1994;68:3425–3436. doi: 10.1128/jvi.68.5.3425-3436.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez-Sierra C, Arizcorreta A, Diaz F, Roldan R, Martin-Herrera L, Perez-Guzman E, Giron-Gonzalez J. Progression of chronic hepatitis C to liver fibrosis and cirrhosis in patients coinfected with hepatitis C virus and human immunodeficiency virus. Clin Infect Dis. 2003;36:491–498. doi: 10.1086/367643. [DOI] [PubMed] [Google Scholar]
- Mazur W, Mazurek U, Jurzak M, Wilczok T, Bulanowski Z, Gonciarz Z. Positive and negative strands of HCV-RNA in sera and peripheral blood mononuclear cells of chronically hemodialyzed patients. Med Sci Monit. 2001;7:108–115. [PubMed] [Google Scholar]
- Mohsen A, Easterbrook P, Taylor C, Portmann B, Kulasegaram R, Murad S, Wiselka M, Norris S. Impact of human immunodeficiency virus (HIV) infection on the progression of liver fibrosis in hepatitis C virus infected patients. Gut. 2003;52:1035–1040. doi: 10.1136/gut.52.7.1035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagayama K, Kurosaki M, Enomoto N, Maekawa SY, Miyasaka Y, Tazawa J, Izumi N, Marumo F, Sato C. Time-related changes in full-length hepatitis C virus sequences and hepatitis activity. Virology. 1999;263:244–253. doi: 10.1006/viro.1999.9924. [DOI] [PubMed] [Google Scholar]
- Navas S, Martin J, Quiroga JA, Castillo I, Carreno V. Genetic diversity and tissue compartmentalization of the hepatitis C virus genome in blood mononuclear cells, liver, and serum from chronic hepatitis C patients. J Virol. 1998;72:1640–1646. doi: 10.1128/jvi.72.2.1640-1646.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nickle DC, Shriner D, Mittler JE, Frenkel LM, Mullins J. Importance and detection of virus reservoirs and compartments of HIV infection. Curr Opin Microbiol. 2003;6:410–416. doi: 10.1016/s1369-5274(03)00096-1. [DOI] [PubMed] [Google Scholar]
- Okuda M, Hino K, Korenaga M, Yamaguchi Y, Katoh Y, Okita K. Differences in hypervariable region 1 quasispecies of hepatitis C virus in human serum, peripheral blood mononuclear cells, and liver. Hepatology. 1999;29:217–222. doi: 10.1002/hep.510290117. [DOI] [PubMed] [Google Scholar]
- Pawlotsky J. Hepatitis C virus resistance to antiviral therapy. Hepatology. 2000;32:889–896. doi: 10.1053/jhep.2000.19150. [DOI] [PubMed] [Google Scholar]
- Pellerin M, Lopez-Aguirre Y, Penin F, Dhumeaux D, Pawlotsky J. Hepatitis C virus quasispecies variability modulates nonstructural protein 5A transcriptional activation, pointing to cellular compartmentalization of virus-host interactions. J Virol. 2004;78:4617–4627. doi: 10.1128/JVI.78.9.4617-4627.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perez-Olmeda M, Nunez M, Romero M, Gonzalez J, Castro A, Arribas JR, Pedreira J, Barreiro P, Garcia-Samaniego J, Martin-Carbonero L, Jimenez-Nacher I, Soriano V. Pegylated IFN-alpha2b plus ribavirin as therapy for chronic hepatitis C in HIV-infected patients. AIDS. 2003;17:1023–1028. doi: 10.1097/00002030-200305020-00011. [DOI] [PubMed] [Google Scholar]
- Pett S, Dore G, Fielden R, Cooper D. Cyclical hepatitis and early liver cirrhosis after hepatitis C seroconversion during pulsed antiretroviral therapy for primary HIV-1. AIDS. 2002;16:2364–2365. doi: 10.1097/00002030-200211220-00029. [DOI] [PubMed] [Google Scholar]
- Pond SL, Frost SD, Muse S. HyPhy: Hypothesis testing using phylogenetics. Bioinformatics. 2005;21:676–679. doi: 10.1093/bioinformatics/bti079. [DOI] [PubMed] [Google Scholar]
- Poynard T, Mathurin P, Lai C, Guyader D, Poupon R, Tainturier M, Myers R, Muntenau M, Ratziu V, Manns M, Vogel A, Capron F, Chedid A, Bedossa P. A comparison of fibrosis progression in chronic liver diseases. J Hepatol. 2003;38:257–265. doi: 10.1016/s0168-8278(02)00413-0. [DOI] [PubMed] [Google Scholar]
- Puoti M, Bonancini M, Spinetti A, Putzolu V, Govindarajan S, Zaltron S, Favret M, Callea F, Gargiulo F, Donato F, Carosi G, for the HIV-HCV Coinfection Study Group Liver fibrosis progression is related to CD4 cell depletion in patients coin-fected with hepatitis C virus and human immunodeficiency virus. J Infect Dis. 2001;183:134–137. doi: 10.1086/317644. [DOI] [PubMed] [Google Scholar]
- Radkowski M, Wilkinson J, Nowicki M, Adair D, Vargas H, Ingui C, Rakela J, Laskus T. Search for hepatitis C virus negative-strand RNA sequences and analysis of viral sequences in the central nervous system: Evidence of replication. J Virol. 2002;76:600–608. doi: 10.1128/JVI.76.2.600-608.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radkowski M, Bednarska A, Horban A, Stanczak J, Wilkinson J, Adair DM, Nowicki M, Rakela J, Laskus T. Infection of primary human macrophages with hepatitis C virus in vitro: Induction of tumour necrosis factor-a and interleukin 8. J Gen Virol. 2004;85:47–59. doi: 10.1099/vir.0.19491-0. [DOI] [PubMed] [Google Scholar]
- Ragni M, Belle S. Impact of human immunodeficiency virus infection on progress to end-stage liver disease in individuals with hemophilia and hepatitis C virus infection. J Infect Dis. 2001;183:1112–1115. doi: 10.1086/319273. [DOI] [PubMed] [Google Scholar]
- Ragni M, Bontempo F. Increase in hepatitis C viral load in hemophiliacs during treatment with highly active antiretroviral therapy. J Infect Dis. 1999;180:2027–2029. doi: 10.1086/315143. [DOI] [PubMed] [Google Scholar]
- Ramirez S, Perez-Del-Pulgar S, Carrion JA, Costa J, Gonzalez P, Massaguer A, Fondevila C, Garcia-Valdecasas JC, Navasa M, Forns X. Hepatitis C virus compartmentalization and infection recurrence after liver transplantation. Am J Transplant. 2009;9:1591–1601. doi: 10.1111/j.1600-6143.2009.02666.x. [DOI] [PubMed] [Google Scholar]
- Ray S, Wang Y, Laeyendecker O, Ticehurst J, Villano S, Thomas D. Acute hepatitis C virus structural gene sequences as predictors of persistent viremia: Hypervariable region 1 as a decoy. J Virol. 1999;73:2938–2946. doi: 10.1128/jvi.73.4.2938-2946.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roque-Afonso A, Jiang J, Penin F, Tareau C, Samuel D, Petit M, Bismuth H, Dussaix E, Feray C. Nonrandom distribution of hepatitis C virus quasispecies in plasma and peripheral blood mononuclear cell subsets. J Virol. 1999;73:9213–9221. doi: 10.1128/jvi.73.11.9213-9221.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roque-Afonso A, Ducoulombier D, Di Liberto G, Kara R, Gigou M, Dussaix E, Samuel D, Feray C. Compartmentalization of hepatitis C virus genotypes between plasma and peripheral blood mononuclear cells. J Virol. 2005;79:6349–6357. doi: 10.1128/JVI.79.10.6349-6357.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenberg MS, Anderson C. PASSaGE: Pattern analysis, spatial statistics and geographic exegesis. Version 2. Methods Ecol Evol. 2011;2:229–232. [Google Scholar]
- Rutschmann O, Negro F, Hirschel B, Hadengue A, Anwar D, Perrin L. Impact of treatment with human immunodeficiency virus (HIV) protease inhibitors on hepatitis C viremia in patients coinfected with HIV. J Infect Dis. 1998;177:783–785. doi: 10.1086/517808. [DOI] [PubMed] [Google Scholar]
- Saleh M, Tibbs C, Koskinas J, Pereira L, Bomford A, Portmann B, McFarlane I, Williams R. Hepatic and extrahepatic hepatitis C virus replication in relation to response to interferon therapy. Hepatology. 1994;20:1399–1404. doi: 10.1002/hep.1840200604. [DOI] [PubMed] [Google Scholar]
- Salmon-Ceron D, Lewden C, Morlat P, Bévilacqua S, Jougla E, Bonnet F, Héripret L, Costagliola D, May T, Chêne G, for the Mortality Group 2000 Liver disease as a major cause of death among HIV infected patients: Role of hepatitis C and B viruses and alcohol. J Hepatol. 2005;42:799–805. doi: 10.1016/j.jhep.2005.01.022. [DOI] [PubMed] [Google Scholar]
- Schramm F, Soulier E, Royer C, Weitten T, Fafi-Kremer S, Brignon N, Meyer N, Ellero B, Woehl-Jaegle ML, Meyer C, Wolf P, Doffoël M, Baumert TF, Stoll-Keller F, Schvoerer E. Frequent compartmentalization of hepatitis C virus with leukocyte-related amino acids in the setting of liver transplantation. J Infect Dis. 2008;198:1656–1666. doi: 10.1086/592986. [DOI] [PubMed] [Google Scholar]
- Sherman KE, Andreatta C, O'Brien J, Gutierrez A, Harris R. Hepatitis C in human immunodeficiency virus-coinfected patients: Increased variability in the hypervariable envelope coding domain. Hepatology. 1996;23:688–694. doi: 10.1002/hep.510230405. [DOI] [PubMed] [Google Scholar]
- Sherman K, Rouster S, Chung R, Rajicic N. Hepatitis C virus prevalence among patients infected with human immunodeficiency virus: A cross-sectional analysis of the US Adult AIDS Clinical Trials Groups. Clin Infect Dis. 2002;34:831–837. doi: 10.1086/339042. [DOI] [PubMed] [Google Scholar]
- Sherman KE, Rouster SD, Stanford S, Blackard JT, Shire N, Koziel M, Peters M, Chung RT, for the AIDS Clinical Trials Group 5071 Study Team Hepatitis C virus (HCV) quasispecies complexity and selection in HCV/HIV coinfected subjects treated with interferon-based regimens. J Infect Dis. 2010;201:712–719. doi: 10.1086/650490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimizu Y, Igarashi H, Kanematu T, Fujiwara K, Wong D, Purcell R, Yoshikura H. Sequence analysis of the hepatitis C virus genome recovered from serum, liver, and peripheral blood mononuclear cells of infected chimpanzees. J Virol. 1997;71:5769–5773. doi: 10.1128/jvi.71.8.5769-5773.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shuhart MC, Sullivan DG, Bekele K, Harrington RD, Kitahata MM, Mathisen TL, Thomassen LV, Emerson SS, Gretch D. HIV infection and antiretroviral therapy: Effect on hepatitis C virus quasispecies variability. J Infect Dis. 2006;193:1211–1218. doi: 10.1086/502974. [DOI] [PubMed] [Google Scholar]
- Slatkin M, Maddison W. A cladistic measure of gene flow inferred from the phylogenies of alleles. Genetics. 1989;123:603–613. doi: 10.1093/genetics/123.3.603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sobesky R, Feray C, Rimlinger F, Derian N, Dos Santos A, Roque-Afonso AM, Samuel D, Bréchot C, Thiers V. Distinct hepatitis C virus core and F protein quasispecies in tumoral and nontumoral hepatocytes isolated via microdissection. Hepatology. 2007;46:1704–1712. doi: 10.1002/hep.21898. [DOI] [PubMed] [Google Scholar]
- Soto B, Sanchez-Quijano A, Rodrigo L, del Olmo J, Garcia-Bengoechea M, Hernandez-Quero J, Rey C, Abad M, Rodriquez M, Gilabert M, Gonzalez F, Miron P, Caruz A, Relimpio F, Torronteras F, Leal M, Lissen E. Human immunodeficiency virus infection modifies the natural history of chronic parenterally-acquired hepatitis C with an unusually rapid progression to cirrhosis. J Hepatol. 1997;26:1–5. doi: 10.1016/s0168-8278(97)80001-3. [DOI] [PubMed] [Google Scholar]
- Sullivan D, Wilson J, Carithers R, Perkins J, Gretch D. Multigene tracking of hepatitis C virus quasispecies after liver transplantation: Correlation of genetic diversification in the envelope region with asymptomatic or mild disease patterns. J Virol. 1998;72:10036–10043. doi: 10.1128/jvi.72.12.10036-10043.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taliani G, Badolato M, Lecce R, Poliandri G, Bozza A, Duca F, Pasquazzi C, Clementi C, Furlan C, Bac CD. Hepatitis C virus RNA in peripheral blood mononuclear cells: Relation with response to interferon treatment. J Med Virol. 1995;47:16–22. doi: 10.1002/jmv.1890470105. [DOI] [PubMed] [Google Scholar]
- Tanaka Y, Hanada K, Hanabusa H, Kurbanov F, Gojobori T, Mizokami M. Increasing genetic diversity of hepatitis C virus in haemophiliacs with human immunodeficiency virus coinfection. J Gen Virol. 2007;88:2513–2519. doi: 10.1099/vir.0.82974-0. [DOI] [PubMed] [Google Scholar]
- Tedaldi EM, Baker RK, Moorman AC, Alzola CF, Furhrer J, McCabe RE, Wood KC, Holmberg SD, for the HIV Outpatient Study (HOPS) Investigators Influence of coinfection with hepatitis C virus on morbidity and mortality due to human immunodeficiency virus infection in the era of highly active antiretroviral therapy. Clin Infect Dis. 2003;36:363–367. doi: 10.1086/345953. [DOI] [PubMed] [Google Scholar]
- Thompson J, Gibson T, Plewniak F, Jeanmougin F, Higgins D. The CLUSTAL X windows interface: Flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 1997;25:4876–4882. doi: 10.1093/nar/25.24.4876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torres-Puente M, Bracho M, Jiminez N, Garcia-Robles I, Moya A, Gonzalez-Candelas F. Sampling and repeatability in the evaluation of hepatitis C virus genetic variability. J Gen Virol. 2003;84:2343–2350. doi: 10.1099/vir.0.19273-0. [DOI] [PubMed] [Google Scholar]
- Toyoda H, Fukuda Y, Koyama Y, Takamatsu J, Saito H, Hayakawa T. Effect of immunosuppression on composition of quasispecies population of hepatitis C virus in patients with chronic hepatitis C coinfected with human immunodeficiency virus. J Hepatol. 1997;26:975–982. doi: 10.1016/s0168-8278(97)80105-5. [DOI] [PubMed] [Google Scholar]
- Vera-Otarola J, Barría MI, León U, Marsac D, Carvallo P, Soza A, López-Lastra M. Hepatitis C virus quasispecies in plasma and peripheral blood mononuclear cells of treatment naïve chronically infected patients. J Viral Hepat. 2009;16:633–643. doi: 10.1111/j.1365-2893.2009.01112.x. [DOI] [PubMed] [Google Scholar]
- Weiner A, Geysen H, Christopherson C, Hall J, Mason T, Saracco G, Bonino F, Crawford K, Marion C, Crawford K, Brunetto M, Barr P, Miyamura T, McHutchinson J, Houghton M. Evidence for immune selection of hepatitis C virus (HCV) putative envelope glycoprotein variants: Potential role in chronic HCV infections. Proc Natl Acad Sci U S A. 1992;89:3468–3472. doi: 10.1073/pnas.89.8.3468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yee T, Griffioen A, Sabin C, Dusheiko G, Lee C. The natural history of HCV in a cohort of haemophilic patients infected between 1961 and 1985. Gut. 2000;47:845–851. doi: 10.1136/gut.47.6.845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yokozaki S, Takamatsu J, Nakano I, Katano Y, Toyoda H, Hayashi K, Hayakawa T, Fukuda Y. Immunological dynamics in hemophiliac patients infected with hepatitis C virus and human immunodeficiency virus: Influence of antiretroviral therapy. Blood. 2000;96:4293–4299. [PubMed] [Google Scholar]
- Zárate S, Pond SL, Shapshak P, Frost S. Comparative study of methods for detecting sequence compartmentalization in human immunodeficiency virus type 1. J Virol. 2007;81:6643–6651. doi: 10.1128/JVI.02268-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zehender G, De Maddalena C, Bernini F, Ebranati E, Monti G, Pioltelli P, Galli M. Compartmentalization of hepatitis C virus quasispecies in blood mononuclear cells of patients with mixed cryoglobulinemic syndrome. J Virol. 2005;79:9145–9156. doi: 10.1128/JVI.79.14.9145-9156.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.