Abstract
8,5’-Cyclopurine-2’-deoxynucleotides represent a class of oxidative DNA lesions that are specifically repaired by nucleotide excision repair but not by base excision repair or direct enzymatic reversion. 32P-postlabeling assay is an ultrasensitive method that has been extensively used for the detection of carcinogen-DNA adducts in laboratory animal and epidemiological studies. This assay under modified chromatographic conditions is also a suitable and sensitive method for the detection of 8,5'-cyclo-2'-deoxyadenosine (cA). After enzymatic digestion of DNA, and enrichment of the oxidative products from the DNA digest, four dinucleotides containing cA, i.e. Ap-cAp, Cp-cAp, Gp-cAp, and Tp-cAp, are 5’-labeled with 32P-orthophosphate form [γ-32P]ATP mediated by polynucleotide kinase (PNK). The 32P-labeled cA products are separated by two-dimensional thin-layer chromatography (TLC) and quantified by Instant Imager or by a scintillation counter. The assay only requires 1–10 µg of DNA sample and is capable of detecting cA lesions as low as 1 in 1010 normal nucleotides.
Keywords: 8,5’-Cyclopurine-2’-deoxynucleotides; 32P-postlabeling assay; Oxidative DNA damage; Thin-layer chromatography
INTRODUCTION
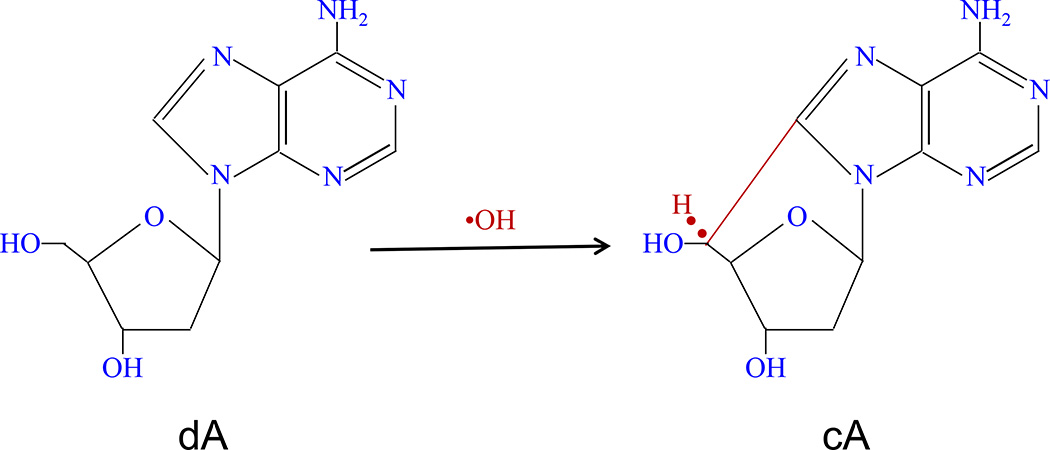
Endogenous and exogenous DNA oxidative damage play a major role in mutagenesis, carcinogenesis and ageing (De Bont and van Larebeke, 2004). Reactive oxygen species (ROS), including the highly reactive hydroxyl radical, are chemically-reactive molecules produced during a wide range of physiological processes (oxygen metabolism) (Chatgilialoglu et al., 2011). Excessive exposure to short wavelength UV radiation in sunlight results in premature aging of the skin, including an increased incidence of skin cancer. 8,5'-Cyclo-2'-deoxyadenosine (cA) and 8,5'-cyclo-2'-deoxyguanosine (cG) are among the major lesions formed in DNA by hydroxyl radical attack on 2'-deoxyadenosine and 2'-deoxyguanosine, respectively, followed by intramolecular cyclization between C5' and C8 (Jaruga and Dizdaroglu, 2008). Structure and formation of cA are displayed in Figure 1. These cA and cG oxidative lesions can only be repaired by nucleotide excision repair (NER) system, but not by base excision repair because of the presence of the C5′-C8 covalent bond between the nitrogen base and the sugar. Indeed, no glycosylase has been found to repair these oxidative lesions (Dizdaroglu, 1992; Randerath et al., 2001a). They may play a role in diseases including cancers with defective NER. Their biological effects include blocking DNA polymerases, inhibition of gene expression, transcriptional mutagenesis among others (Jaruga and Dizdaroglu, 2008).
Figure 1.
Structure and formation of 8,5’-cycloadenosine-2’-deoxynucleotide (cA), that is induced by hydroxyl radical attack on 2’-deoxyadenosine (dA).
32P-postlabeling analysis is a highly sensitive method for the detection and measurement of covalent carcinogen-adducts (Reddy and Randerath, 1986), some oxidative DNA lesions (Zhou et al., 2004), and other DNA modifications (Randerath et al., 1993; Zhou et al., 2005). This assay only requires less than 10 µg of DNA sample. Therefore, it is suitable for detection of DNA adducts and oxidative lesions in epidemiological studies (Everson et al., 1986; Tang et al., 2001) and animal experiments (Zhou et al., 2004). The property of 8,5'-Cyclo-2'-deoxypurine including cA and cG is that C8 of a purine base becomes covalently bound to the C5 of its own deoxyribose through oxidation by the highly reactive •OH (Marietta et al., 2002). Formation of this covalent bond causes an unusual puckering of the deoxyribose, as well as local distortion of the DNA helix (Miaskiewicz et al., 1995). This cyclic linkage between a purine base and its own deoxyribose also cause the formation of dinucleotide such as pAp-cAp, pCp-cAp, pGp-cAp, and pTp-cAp, which is not easily chopped off by enzymatic digestion (Randerath et al., 2001a). This dinucleotide, which is similar to bulky carcinogen-DNA adducts, is likely to induce mutations during replication. It is also suitable to be detected by 32P-postlabeling assay (Randerath et al., 2001a).
BASIC PROTOCOL 1: ENZYMATIC DIGESTION OF DNA AND ENRICHMENT OF DINUCLEOTIDES CONTAINING CYCLOADENOSINE
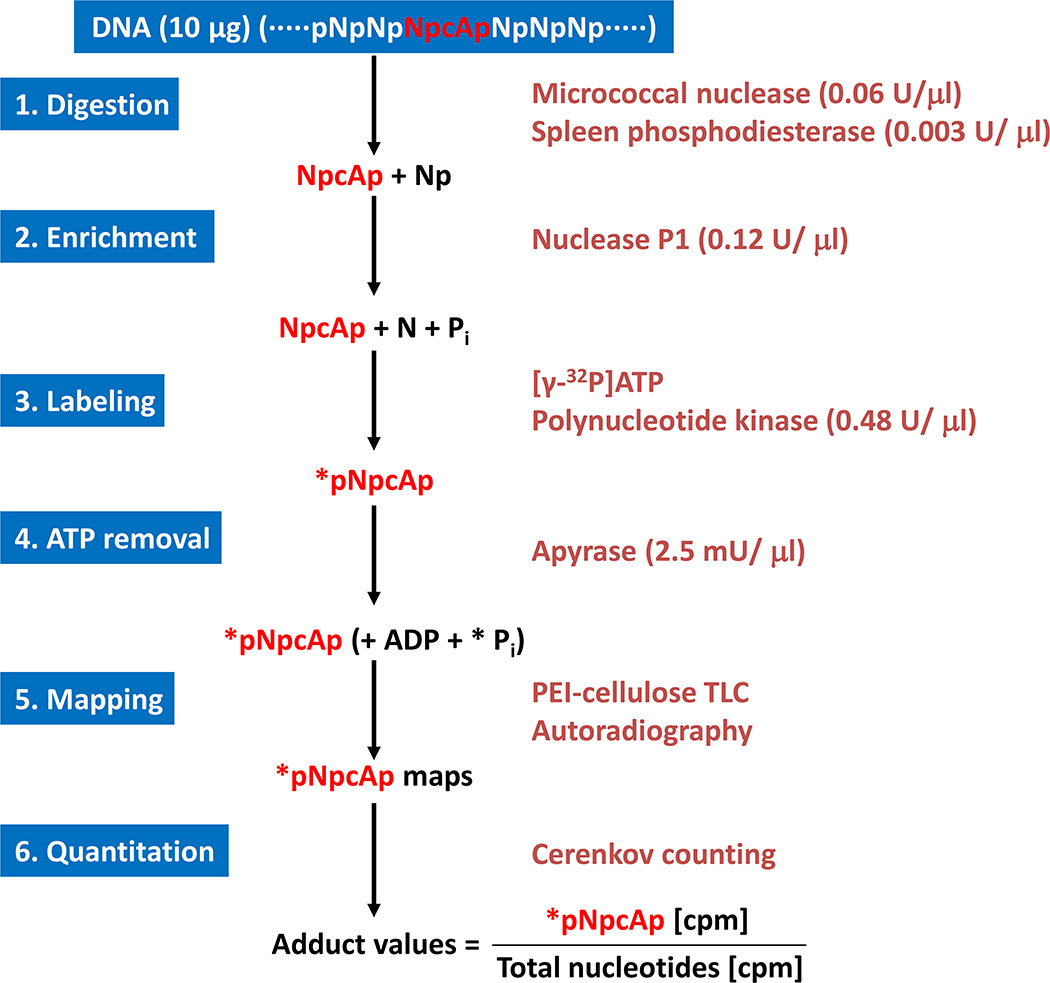
This protocol describes how the DNA is enzymatically degraded to normal (Np) and modified deoxyribonucleoside 3’-monophosphates containing cycloadenosine (Np-cAp). The first step, DNA (…pNpNpNp-cApNpNpNp…) is degraded to Np-cAp and Np by enzymes micrococcal nuclease (MN) and spleen phosphodiesterase (SPD). The modified dinucleotides containing cycloadenosine (Np-cAp) are further enriched by nuclease P1, which degrade the normal nucleotides (Np) to deoxyribonucleosides (N), but leave the Np-cAps intact. Only Np-cAp, but not N, is substrates for subsequent 32P-labeling. Nuclease P1 enrichment enhances the sensitivity of the assay significantly and is also reduces the amount of isotope for the assay. Figure 2 shows the methods and principals of 32P-postlabeling assay. The following protocol provides details for enzymatic digestion of DNA and enrichment of dinucleotides containing cycloadenosine.
Figure 2.
Procedures of 32P-postlabeling assay. cA, cycloadenosine; N, normal nucleotides; p, phosphate; *P, [32P]phosphate; *pNpcAp, 32P-labeled dinucleotide containing cycloadenosine.
Materials
Micrococcal nuclease (MN, 0.4 units/µl, Sigma-Aldrich, #M3755)
Spleen phosphodiesterase (SPD, 0.02 units/µl, Sigma-Aldrich, #P9041)
Calcium chloride (CaCl2, 100 mM)
Sodium succinate (300 mM, pH 6.0)
Nuclease P1 (NP1, 1 unit/µl, Sigma-Aldrich, #N8630)
Sodium acetate (1 M, pH 5.0)
Zinc chloride (ZnCl2, 1 mM)
1.5 ml plastic micro tubes
Enzymatic digestion of DNA
Transfer 1–10 µg (depending on extent of oxidative DNA lesions) of DNA into 1.5 ml micro-tubes (e.g., Eppendorf tubes). Routinely, 10 µg of DNA are needed for the detection of endogenous oxidative lesions in vivo, as the levels of adduction are very low. However, if animals are exposed to high concentrations of oxygen (hyperoxia), or animals are subjected to oxidative stress [e.g., by treatment with Ni(II))]l, then only 1–3 µg of DNA is required for the assay.]
Adjust the volume to 5.4 µl. If the volume is larger than 5.4 µl, evaporate DNA samples to dryness and then add 5.4 µl distilled H2O.
Pre-mix 1.5 µl MN, 1.5 µl SPD, 1.0 µl CaCl2, and 1.0 µl Sodium succinate. If you have 20 DNA samples for the experiments, you should prepare at least for 22 samples.
Add 5 µl mixed enzymes and buffers into each micro tube. Total volume of digest is 10.4 µl.
Vortex and centrifuge for a few seconds with microcentrifuge (8,000–10,000 RPM).
Incubate at 37°C for 3.5 hours.
Enrichment of dinucleotides containing cycloadenosine
Pre-mix 1.8 µl NP1, 1.6 µl ZnCl2, and 0.9 µl Sodium acetate.
Add 4.3 µl above mixed enzymes and buffers into each micro tube. Total volume of the digest after this step is 14.7 µl.
Vortex and centrifuge for a few seconds with microcentrifuge (8,000–10,000 RPM).
Incubate at 37°C for 40 minutes.
BASIC PROTOCOL 2: 5’-32P-LABELING OF DINUCLEOTIDES CONTAINING CYCLOADENOSINE
After nuclease enrichment, pH of DNA digest must be adjusted to alkaline. Nuclease PI cleaves deoxyribonucleoside 3'- monophosphates of normal nucleotides to deoxyribonucleosides (N) which do not serve as substrates for polynucleotide kinase, while most modified dinucleotides containing cycloadenosine (Np-cAp) are totally or partially resistant to the 3-dephosphorylating action of nuclease P1. Np-cAps are substrates for subsequent 32P-labeling, leading to their conversion to 5’-32P-labeled deoxyribodinucleotides 3’-monophosphates containing cycloadenosine (*pNp-cAp, where * = 32P-label) in the presence [γ-32P]ATP and polynucleotide kinase. The labeled products (*pNp-cAp) are separated by different conditions of multidirectional thin layer chromatography (TLC) and quantified by Instant Imager.
Materials
2-(Cyclohexylamino)ethanesulfonic acid (CHES, 500 mM, pH 9.5, Sigma-Aldrich, #C2885)
[γ-32P]Adenosine 5’-triphosphate ([γ-32P]ATP, MP Biomedicals, #35020)
T4 Polynucleotide kinase (PNK, 30 units/µl, Affymetrix/USB, #70031X)
T4 PNK reaction buffer (10×, 500mM Tris-HCl, pH 7.6, 100 mM MgCl2, 100 mM β-ME, included in T4 PNK package. Affymetrix/USB, #PN70083)
T4 PNT dilution buffer (50 mM Tris-HCl, pH 8.0, included in T4 PNK package. Affymetrix/USB, #PN71079)
Four dinucleotides containing cycloadenosine (5’-dAp-cdAp-3’, 5’-dCp-cdAp-3’, 5’-dGp-cdAp-3’, and 5’-Tp-cdAp-3’. These standards of cycloadenosines are only available from research laboratories).
Apyrase (40 mU/µl, Sigma-Aldrich, #A7646)
Preparation of standards of cycloadenosines
Add 0.1 µg of each dinucleotide containing cycloadenosine into micro tube.
Bring the volume to 14.7 µl with distilled H2O.
Adjust pH of DNA digest
Add 3 µl CHES into each micro tube containing DNA samples to stop nuclease P1 reaction or containing standards of cycloadenosine to bring pH to 9. Total volume of digest is 17.7 µl.
Labeling of dinucleotides containing cycloadenosine
-
Pre-mix 60 µCi [γ-32P]ATP (in 1.5 µl 20 mM CHES), 0.35 µl PNK, and 2.0 µl 1× PNK reaction buffer.
This labeling mixture is also used for determining specific activity. Therefore, one needs to prepare at least 6 samples more than actual sample size. For example, if there are 20 DNA samples, you need prepare at least 26 samples (1,560 µCi [γ-32P]ATP in 39 µl 20 mM CHES, 9.1 µl PNK, and 52 µl 1× PNK reaction buffer).
Label DNA digests and standards of cycloadenosines by adding 3.85 µl above labeling mixture. Total volume after this step is 21.55 µl.
Incubate at 37°C for 40 minutes.
Note: [γ-32P]ATP is a high-energy β-particle emitter. Exposure to 32P should be as little as possible. Attenuation of exposure time and effective shielding (glass or plexiglass) are most important measurements. Geiger counter should be used to check contamination, and personal dosimeters (ring and body badge) should be worn to monitor exposure dose.
BASIC PROTOCOL 3: DETERMINATION OF SPECIFIC ACTIVITY OF [γ-32P]ATP
The specific activity of [γ-32P]ATP must be determined in each labeling experiment. The data are very important for calculation of oxidative DNA lesions. 2′-Deoxyadenosine 3′-monophosphate (dAp) is converted to 5’ 32P-labeled deoxyadenosine 3’,5’-bisphosphates (d*pAp) using the above labeling mixture (see BASIC PROTOCOL 2).
Materials
2′-Deoxyadenosine 3′-monophosphate (dAp, 2.0 µM, Sigma-Aldrich, #D3139)
CHES (50 mM, pH 9.5)
T4 PNK
[γ-32P]ATP
Labeling of 2′-Deoxyadenosine 3′-monophosphate
Take 5 µl dAp (total 10 µg) each into tubes A and B (duplicates).
Add 2.5 µl CHES into each tube.
Add 2.5 µl above labeling mixture (see BASIC PROTOCOL 2).
Incubate at 37°C for 40 minutes.
Take 4 µl from each tube to a fresh micro tube with 996 µl 20 mM CHES. These samples are ready for chromatography.
BASIC PROTOCOL 4: PREPARATION OF THIN LAYER CHROMATOGRAPHY (TLC) PLATES
Thin-layer chromatography is used to separate cycloadenosine oxidative products. Polyethyleneimine (PEI)-impregnated cellulose TLC plates are available from Macherey-Nagel (Bethlehem, PA).
Materials
Polyethyleneimine (PEI)-impregnated cellulose TLC (Thickness: 0.5 mm, 20 × 20 cm, Macherey-Nagel, #801053)
Whatman filter paper (46 × 57 cm, Fisher Scientific, #1001917)
Pencil (EX-SOFT)
Preparation of D1 chromatographic plate
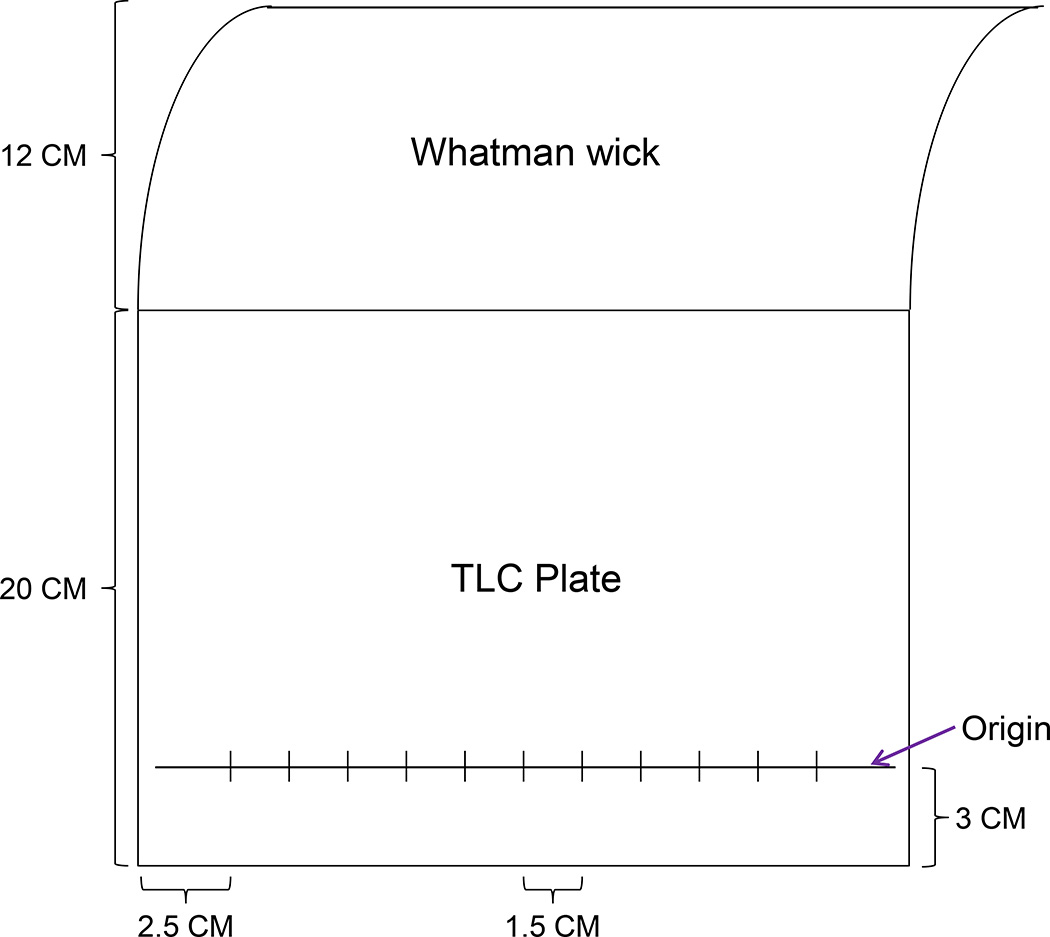
Take a PEI-cellulose TLC plate (20 × 20 cm). Draw an original line at 3 cm from bottom.
Draw sample spotting lines on original line with 1.5 cm distance between two samples (Figure 3).
Attach a Whatman filter paper wick (12 cm high) on top of the plate at cellulose side using staples.
Figure 3.
Template of D1 chromatogram (TLC plate).
Preparation of 2D chromatographic plate
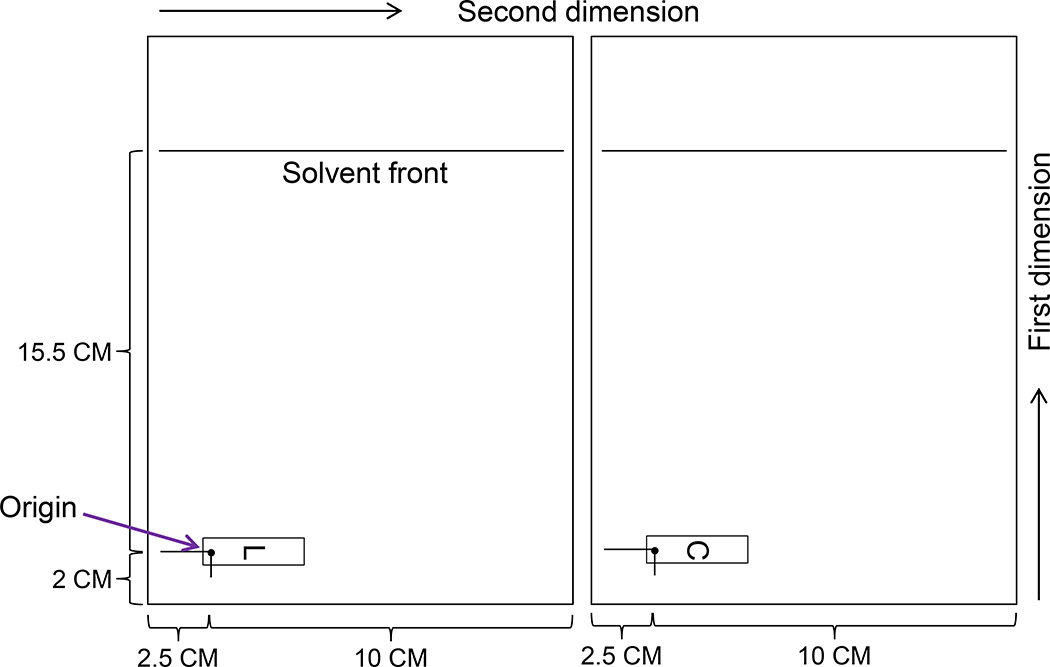
Cut PEI-cellulose TLC plate to 20 × 12.5 cm.
Draw a spot (2 cm from bottom and 2.5 cm from left margin) for cutouts obtained from D1 chromatogram transfer (Figure 4).
Draw a line to top (15.5 cm from cutout transfer spot) for solvent developing front (Figure 4).
Figure 4.
Template of 2-D chromatograms (TLC plates). L, lower cutout (donor) from D 1 TLC plate; C, central cutout (donor) from D 1 TLC plate.
Preparation of chromatogram for Specific Activity
Cut PEI-cellulose TLC plate to 20 × 12.5 cm.
Draw a spotting line at 2 cm from bottom (Figure 5).
Draw a line to top (13 cm from spotting line) for solvent developing front (Figure 5).
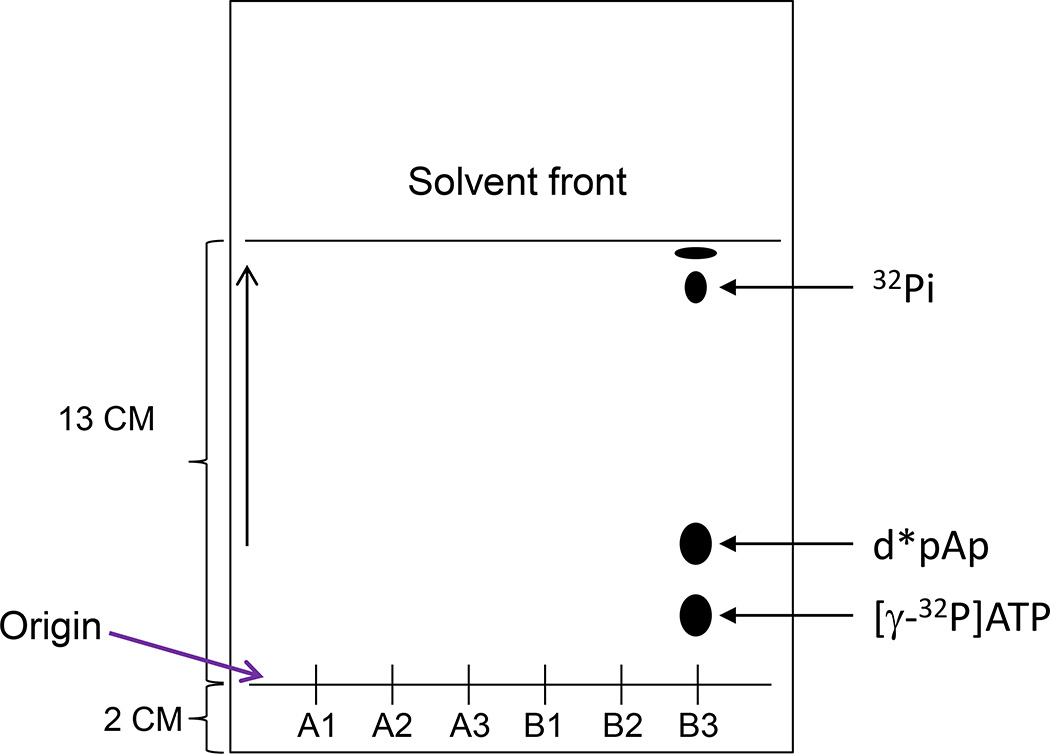
Figure 5.
Template of chromatogram for [γ-32P]ATP specific activity and locations of major labeled products after development in 0.28 M NH4(SO4)2 + 50 mM NaH2PO4, pH 6.7.
BASIC PROTOCOL 5: THIN LAYER CHROMATOGRAPHY (TLC)
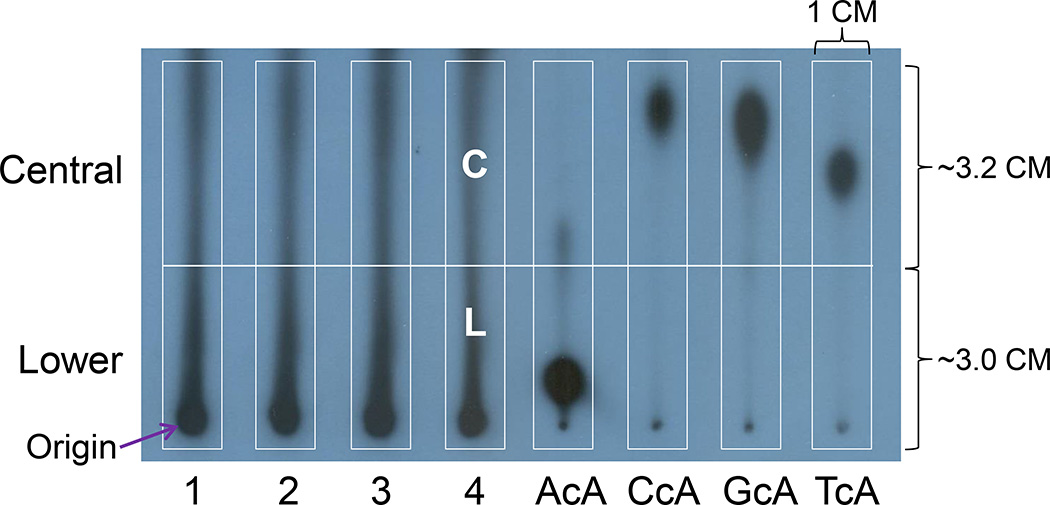
The labeled digests are applied to the origin of PEI-cellulose thin-layer plastic sheet (D1) and develop overnight (16 h) in 2.8 M sodium phosphate, pH 5.2 in order to remove 32P-labeled Pi and ATP and to purify dinucleotide containing cycloadenosine. Labeled dinucleotide containing cycloadenosine will retain in the lower (L, 2.8–3.0 × 1.0 cm) and central (C, 2.8–3.2 × 1.0 cm) parts of the D1 chromatogram (Figure 6). These cutouts are each contact-transferred to individual acceptor sheets (Figure 4) and resolved by two-dimensional TLC (Figure 7).
Figure 6.
Profiles of DNA samples (1–4) after D1 chromatography and chromatographic mobilities of the four possible dinucleotides containing A, C, G, or T, 5’ to cA. L, lower cutout; C, central cutout. These two cutouts are used to contact-transfer to 2D TLC plates.
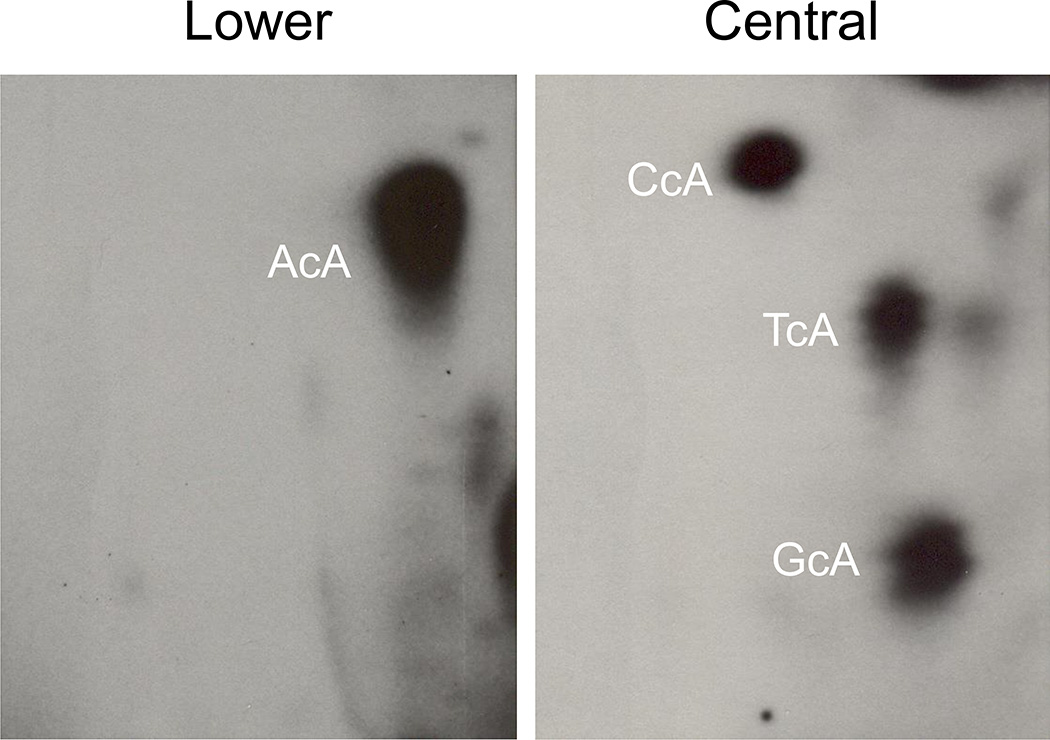
Figure 7.
Typical patterns of 32P-postlabeled cycloadenosins. AcA, CcA, GcA, and TcA represent different dinucleotides containing 8,5'-cycloadenosine-2'-deoxynucleotides.
Materials
Polyethyleneimine (PEI)-impregnated cellulose TLC (Thickness: 0.5 mm, 20 × 20 cm, Macherey-Nagel, #801053) for D1
Polyethyleneimine (PEI)-impregnated cellulose TLC (Thickness: 0.5 mm, 20 × 12.5 cm, Macherey-Nagel, #801053) for 2D
D1 solvent (NaH2PO4, 2.8 M, pH 5.2, Fisher Scientific, #S369)
Rectangular TLC developing tank with lid (Fisher Scientific, #K416180)
Ink with 99Tc (99Tc 100 µl = 56 µCi, India ink 800 µl, water 400 µl)
Kodak X-Omat LS Film (Fisher Scientific, #864 6770)
Magnet button (General Hardware MFG CO., INC, #372A)
2D solvent, first dimension (Lithium Formate, 2.12 M, Urea, 3.75 M, pH 3.35)
2D solvent, second dimension for L cutout (NaH2PO4, 0.4 M, Tris 0.25 M, urea 4.25 M, pH 8.2)
2D solvent, second dimension for C cutout (NaH2PO4, 1.0 M)
Solvent for specific activity (0.28 M NH4(SO4)2 + 50 mM NaH2PO4, pH 6.7)
Chromatography in D1
Mark the origins lightly with an extra soft pencil on D1 TLC plate (Figure 3). Attach Whatman wick (12 cm) on top of TLC plate by stapling.
Apply labeled digest with 20 µl pipette very slowly. Spot only 2 µl of digest for standards of dinucleotide containing cycloadenosine.
Set up TLC developing under the hood in advance with 70–80 ml D1 solvent. The tank and lid are draped with Saran wrap for prevention from 32P contamination.
Place the TLC plate with samples or standards in the tank and fold the wick on top of the tank. Develop in D1 solvent for 16 hours (overnight).
Note: The tank should be kept behind a plastic shielding to prevent unnecessary isotope exposure.
Transfer samples from D1 to 2D TLC Plate
After development, cut off the wick plus the top portion of TLC plate behind plastic shielding over a piece of aluminum foil. The cutting line is at 8–10 cm above the origin.
Soak the chromatogram in 1 L deionized water for 5 min with frequent shaking. Repeat this step one more time.
Dry the chromatogram in a stream of air (hair dryer can be used).
Mark the chromatogram with 99Tc ink on the edge area for the overlap autoradiogram and chromatogram.
Expose the chromatogram with Kodak X-OMAT LS Film for 20–30 minutes at room temperature.
Draw L and C cutouts on the film based on locations of cycloadenosine standards as showed on Figure 6.
Superimpose the film and chromatogram on a light box and copy L and C cutouts to chromatogram with a pencil on back of the chromatogram.
Sample number and the direction of D1 development should be indicated at the back of the cutouts. Then all origin areas (rectangles) are cut out by a pair of scissors.
Chromatography L and C cutouts in 2D TLC plates
2D (acceptor) sheets are 12.5 cm wide and 20 cm long as showed on figure 4. Mark L or C with sample number on right top corner.
Place the donor cutout from D1 chromatogram onto the origin area of the acceptor sheet (2D) with layer facing each other and the point of application on the cutout being superimposed on the origin mark of the 2D sheet (Figure 4).
The 2D sheets of both L and C are predeveloped with water to origin line (2 cm above the bottom edge). Then the sheet is developed in glass tank with prefilled 60 ml first dimension solvent (Lithium Formate, 2.12 M, Urea, 3.75 M, pH 3.35) till 15.5 cm above the bottom edge.
Cut off top portion of the 2D sheet (cut line is 1 cm below solvent front, i.e. 14.5 cm above bottom edge). Then soak the 2D sheet in 1 L deionized water for 4 min with frequent shaking. Repeat this step once more.
Dry the sheet in a stream of air (hair dryer can be used). Attach Whatman wick (1.2 cm) on top of second dimension (short dimension) of sheet by stapling.
Pre-develop the L sheet in 0.4 M NaH2PO4, pH 8.2 to 0.5 cm above origin corner. Then develop the sheet in a glass tank with prefilled 60 ml of second dimension solvent (NaH2PO4, 0.4 M, Tris 0.25 M, urea 4.25 M, pH 8.2) till top of the wick.
Cut off the wick (cut below the staplers). Then soak the 2D sheet in 1 L deionized water for 3 min with frequent shaking. Dry the sheet in a stream of air.
Predevelop the C sheet in water to origin corner. Then develop the C sheet in 1.0 M NaH2PO4, pH 6.0 till top of the wick.
Cut off the wick (cut below the staplers). Then dry the sheet in a stream of air without soaking.
Chromatography for specific activity (d*pAp)
Spot 5 µl of d*pAp solution (from BASIC PROTOCOL 3) to the chromatogram. Spot three times from each tube A and B to make triplicates for each tube.
Develop the sheet in 0.28 M NH4(SO4)2 + 50 mM NaH2PO4, pH 6.7 till 13 cm from spotting line (Figure 5).
Dry the sheet in a stream of air. Mark the chromatogram with 99Tc ink on the edge area to recognize samples.
BASIC PROTOCOL 6: AUTORADIOGRAPHY AND QUANTITATION OF CYCLOADENOSINE
32P-labeled products are visualized by screen-enhanced autoradiography using Kodak XAR-5 film and by imaging with an Instant Imager (Packard Instruments, Meriden, CT). Film exposure is necessary for observing the profiles of cycloadenosine oxidative lesions. It is also important for further study such as structure identification. Basically, there are three methods to quantify the levels of 32P-labeled oxidative DNA products, i.e. Instant Imager, Phosporimager, and Scintillation Analyzer.
Materials
Ink with 14C (14C 20 µl = 20 µCi, India ink 1,300 µl, water 700 µl)
Kodak XAR Film (Fisher Scientific, #165 1454)
Wolf Cassettes (8” × 10”, Medical Solution, #11118)
BioMax MS Intensifying Screen (8” × 10”, Fisher Scientific, #856 8706)
Autoradiography
Mark 2D sheet with 3 dots and sample number in four corners using ink with 14C.
Place 2–3 2D sheets (layer face up) into one cassette. Then put a Kodak XAR Film on top of 2D sheet. Finally, place an intensifying screen over the film.
Keep cassette in −80°C deep freezer if radioactive counts are lower.
Exposure time is depending on the levels of 32P-labeled products. For example, exposure time for standards of cycloadenosine is about 1 hour. However, 16–20 hours may be needed for in vivo samples.
Typical profiles of cycloadenosines are obtained in L and C 2D sheets as displayed in Figure 7.
Quantitation of cycloadenosine
Various instruments can be used to detect and quantify 32P-labeled products including Instant Imager (Zhou et al., 1999), Phosphorimager (Longo et al., 1997), and Scintillation Analyzer (Randerath et al., 1985). Please follow the operating manual of the instruments.
Obtain counts per minute (CPM) for every labeled product of each sample including d*pAp (specific activity).
BASIC PROTOCOL 7: CALCULATION OF OXIDATIVE DNA LESIONS
In order to evaluate the effects of oxidative DNA lesions on human or animals, CPM needs to be converted to biological parameters. Basically, we need obtain two sets of data, i.e. total nucleotides (normal + modified) and total nucleotides containing cycloadenosine lesions.
The extent of cycloadenosine DNA modification is estimated by calculating relative adduct labeling (RAL) values from corrected sample count rates, the amount of DNA assayed (expressed as pmol DNA monomer units or DNA-P), and the specific activity of [γ-32P]ATP according to
Quantitative data represented minimum estimates because 100% adduct recovery presumably was not achieved.
COMMENTARY
Background Information
The oxidative DNA lesions, 8,5′-cyclo-2′-deoxyadenosine (cA) and 8,5′-cyclo-2′-deoxyguanosine (cG) diastereomers, have been detected in DNA derived from various cells and organisms (Jaruga and Dizdaroglu, 2008). The hydroxyl radicals are known for their reactivity and ability to cause chemical modifications to DNA through the formation of strand breaks and nucleobase modifications (Chatgilialoglu et al., 2011). These oxidative products were first detected by 32P-postlabeling assay in mice treated with Ni(II) (Chang et al., 1994), rats exposed to ferric nitrilotriacetate (Fe-NTA) (Randerath et al., 1995), and untreated new born mice (Randerath et al., 1997a). These 32P-labeled products were named as type II I-compounds (Randerath et al., 1999; Zhou et al., 2000) because their structures were not identified. Current protocol of 32P-postlabeling assay to detect cA products was developed using Standards of cA (Randerath et al., 2001a). Randerath et al reported (Randerath et al., 2001b) that levels of cA oxidative lesions were significantly higher in normal new born mice compared to those in fetus. The possible mechanisms could be sudden increase in partial oxygen pressure at birth in blood and tissues, implying inadequate antioxidant defenses in the affected neonatal organs (Randerath et al., 1997a). Similar trends of oxidative DNA lesions were observed in newborn liver between cA and 8-hydroxy-2’-deoxyguanosine, a major mutagenic DNA oxidation product (Randerath et al., 1997b). The analytical techniques applied herein should provide helpful tools in experiments directed at the prevention of DNA oxidative damage in both model systems and humans.
Critical Parameters
Technical variation should be minimized by using standards of cA dinucleotide including AcA, CcA, GcA, and TcA. Each D1 sheet should include these four standards to get accurate location of AcA, CcA, GcA, and TcA for L and C cutouts (Figure 6). If space is limited, AcA and GcA, as well as CcA and TcA can be combined as one sample. These standards will be also used to adjust loss of cA during wash after chromatography. Determination of specific activity of [γ-32P]ATP each experiment can also attenuate experimental variation. If data from two experiments need to be combined, 3–6 DNA samples should be labeled in both experiments for normalization.
Troubleshooting
No cA spots of standards displayed on L and C cutout area of D1 chromatogram. Wrong D1 solvent could have been used. Correct D1 solvent is 2.8 M NaH2PO4, pH 5.2.
No cA spots and background radioactivity detectable on 2D sheets. Contact transfer from donor cutout of D1 chromatogram onto 2D sheet must be layered facing each other.
Lower labeling intensity displays on D1 sheets. Adjust pH to alkaline (pH 9.5) by CHES before labeling is very important. Forgetting to add 3 µl CHES before labeling could result in lower labeling efficiency.
Lower levels of cA products show on 2D map. Unusual lower cA products on 2D map could be caused by incomplete digestion of MN/SPD or nuclease P1. If normal nucleotides are not digested to nucleosides, then [γ-32P]ATP will be used up to label the Nps but not NpcAps.
Higher levels of cA products show on 2D map. Routinely, 10 µg of DNA are needed for the detection of endogenous oxidative lesions in vivo, as the levels of adduction are very low. However, if animals are exposed to high concentrations of oxygen (hyperoxia), or animals are subjected to oxidative stress [e.g., by treatment with Ni(II)], then only 1–3 µg of DNA is required for the assay.
Dark radioactive background on 2D map. Check DNA quality. The ratio of 260/280 should be between 1.6–1.8.
Anticipated Results
Typical patterns of cA standards including AcA, CcA, GcA and TcA as well as in vivo DNA samples should be observed in D1 chromatogram as shown in Figure 6. Location of AcA on D1 sheet is very stable. However, locations of CcA, GcA, and TcA may vary depending on different batch of TLC plates and solvent. Therefore, cA standards are very important and will help locate L and C cutouts, especially C cutout. After contact transfer and 2D chromatography, typical patterns of 32P-postlabeled dinucleotides containing 8,5'-cycloadenosine-2'-deoxynucleotides, AcA, CcA, GcA, and TcA, should be obtained as displayed in Figure 7 with AcA being shown in lower and CcA, GcA, and TcA being in Central. Similar pattern of cA adducts from in vivo or in vitro DNA samples should be observed.
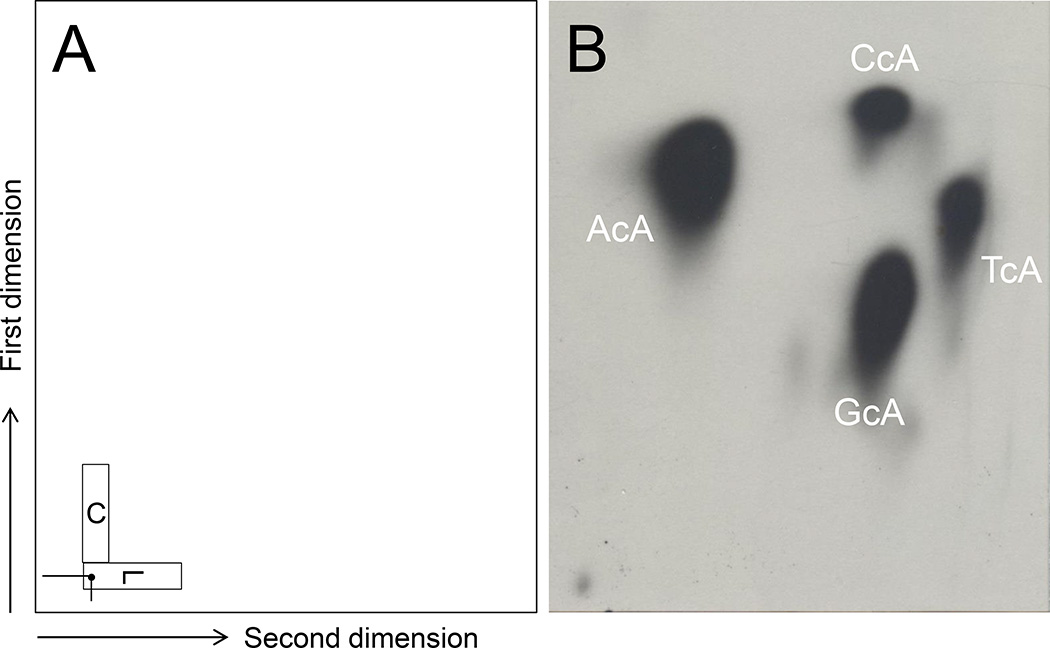
In order to improve low-throughput and to get better profiles of cA products, an alternate protocol of 2D chromatography was developed. L and C donor cutouts are placed on the same 2D chromatogram as showed in Figure 8 (left panel). Then this TLC plate is developed in first dimension solvent (Lithium Formate, 2.12 M, Urea, 3.75 M, pH 3.35). After soaking in 1 L deionized water for 4 min with frequent shaking, the plate is developed in 1 M NaH2PO4, pH 6.0. No soaking with water is needed after development of second dimension. Four cA products are displayed in Figure 8 (right panel). Note: this new chromatographic protocol was only used with cA standards but not with real DNA samples.
Figure 8.
A new chromatographic method combining L and C cutouts in a single 2D plate. Contact-transfer of L and C is displayed in panel A. Typical patterns of 32P-postlabeled AcA, CcA, GcA, and TcA are shown in panel B.
Time Consideration
If DNA samples are ready, the 32P-postlabeling assay for 20–24 samples will take about 5–8 days.
| Day 1: | Digest DNA samples with MN/SPD enzymes. |
| Prepare D1 and 2D chromatograms. | |
| Day 2: | Digest DNA samples with nuclease P1 followed by 5’-32P-labeling. Apply labeled digest on D1 sheet and develop in D1 solvent for 16 hours (overnight). |
| Day 3: | Get L and C cutouts from D1 sheet. Transfer cutouts to 2D sheet. Run first dimension of L sheets. |
| Day 4: | Run second dimension for L sheets and run first and second dimensions of C sheets. |
| Day 5: | Quantitation and imaging. |
| Day 6–8: | Data analysis. |
Acknowledgments
This work was in part supported by funds from RO1 grants from National Institutes of Health [ES-009132, HL-112516, HL-087174, and ES-019689] to B.M, and the Center for Translational Environmental Health Research (CTEHR), P30ES023512.
Literature Cited
- Chang J, Jaeschke H, Randerath K. Effect of Ni(II) on tissue hydrogen peroxide content in mice as inferred from glutathione and glutathione disulfide measurements. Life Sci. 1994;55:1789–1796. doi: 10.1016/0024-3205(94)90089-2. [DOI] [PubMed] [Google Scholar]
- Chatgilialoglu C, Ferreri C, Terzidis MA. Purine 5',8-cyclonucleoside lesions: chemistry and biology. Chemical Society reviews. 2011;40:1368–1382. doi: 10.1039/c0cs00061b. [DOI] [PubMed] [Google Scholar]
- De Bont R, van Larebeke N. Endogenous DNA damage in humans: a review of quantitative data. Mutagenesis. 2004;19:169–185. doi: 10.1093/mutage/geh025. [DOI] [PubMed] [Google Scholar]
- Dizdaroglu M. Oxidative damage to DNA in mammalian chromatin. Mutation research. 1992;275:331–342. doi: 10.1016/0921-8734(92)90036-o. [DOI] [PubMed] [Google Scholar]
- Everson RB, Randerath E, Santella RM, Cefalo RC, Avitts TA, Randerath K. Detection of smoking-related covalent DNA adducts in human placenta. Science (New York, N.Y. 1986;231:54–57. doi: 10.1126/science.3941892. [DOI] [PubMed] [Google Scholar]
- Jaruga P, Dizdaroglu M. 8,5'-Cyclopurine-2'-deoxynucleosides in DNA: mechanisms of formation, measurement, repair and biological effects. DNA repair. 2008;7:1413–1425. doi: 10.1016/j.dnarep.2008.06.005. [DOI] [PubMed] [Google Scholar]
- Longo JA, Nevaldine B, Longo SL, Winfield JA, Hahn PJ. An assay for quantifying DNA double-strand break repair that is suitable for small numbers of unlabeled cells. Radiation research. 1997;147:35–40. [PubMed] [Google Scholar]
- Marietta C, Gulam H, Brooks PJ. A single 8,5'-cyclo-2'-deoxyadenosine lesion in a TATA box prevents binding of the TATA binding protein and strongly reduces transcription in vivo. DNA repair. 2002;1:967–975. doi: 10.1016/s1568-7864(02)00148-9. [DOI] [PubMed] [Google Scholar]
- Miaskiewicz K, Miller JH, Fuciarelli AF. Theoretical analysis of DNA intrastrand cross linking by formation of 8,5'-cyclodeoxyadenosine. Nucleic acids research. 1995;23:515–521. doi: 10.1093/nar/23.3.515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Randerath E, Watson WP, Zhou GD, Chang J, Randerath K. Intensification and depletion of specific bulky renal DNA adducts (I-compounds) following exposure of male F344 rats to the renal carcinogen ferric nitrilotriacetate (Fe-NTA) Mutation research. 1995;341:265–279. doi: 10.1016/0165-1218(95)90098-5. [DOI] [PubMed] [Google Scholar]
- Randerath E, Zhou GD, Randerath K. Organ-specific oxidative DNA damage associated with normal birth in rats. Carcinogenesis. 1997a;18:859–866. doi: 10.1093/carcin/18.4.859. [DOI] [PubMed] [Google Scholar]
- Randerath K, Randerath E, Agrawal HP, Gupta RC, Schurdak ME, Reddy MV. Postlabeling methods for carcinogen-DNA adduct analysis. Environmental health perspectives. 1985;62:57–65. doi: 10.1289/ehp.856257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Randerath K, Randerath E, Zhou GD, Li D. Bulky endogenous DNA modifications (I-compounds) -possible structural origins and functional implications. Mutation research. 1999;424:183–194. doi: 10.1016/s0027-5107(99)00018-4. [DOI] [PubMed] [Google Scholar]
- Randerath K, Zhou GD, Hart RW, Turturro A, Randerath E. Biomarkers of aging: correlation of DNA I-compound levels with median lifespan of calorically restricted and ad libitum fed rats and mice. Mutation research. 1993;295:247–263. doi: 10.1016/0921-8734(93)90024-w. [DOI] [PubMed] [Google Scholar]
- Randerath K, Zhou GD, Monk SA, Randerath E. Enhanced levels in neonatal rat liver of 7,8-dihydro-8-oxo-2'-deoxyguanosine (8-hydroxydeoxyguanosine), a major mutagenic oxidative DNA lesion. Carcinogenesis. 1997b;18:1419–1421. doi: 10.1093/carcin/18.7.1419. [DOI] [PubMed] [Google Scholar]
- Randerath K, Zhou GD, Somers RL, Robbins JH, Brooks PJ. A 32P-postlabeling assay for the oxidative DNA lesion 8,5'-cyclo-2'-deoxyadenosine in mammalian tissues: evidence that four type II I-compounds are dinucleotides containing the lesion in the 3' nucleotide. The Journal of biological chemistry. 2001a;276:36051–36057. doi: 10.1074/jbc.M105472200. [DOI] [PubMed] [Google Scholar]
- Randerath K, Zhou GD, Somers RL, Robbins JH, Brooks PJ. A 32P-postlabeling assay for the oxidative DNA lesion 8,5'-cyclo-2'-deoxyadenosine in mammalian tissues: evidence that four type II I-compounds are dinucleotides containing the lesion in the 3' nucleotide. J Biol Chem. 2001b;276:36051–36057. doi: 10.1074/jbc.M105472200. [DOI] [PubMed] [Google Scholar]
- Reddy MV, Randerath K. Nuclease P1-mediated enhancement of sensitivity of 32P-postlabeling test for structurally diverse DNA adducts. Carcinogenesis. 1986;7:1543–1551. doi: 10.1093/carcin/7.9.1543. [DOI] [PubMed] [Google Scholar]
- Tang D, Phillips DH, Stampfer M, Mooney LA, Hsu Y, Cho S, Tsai WY, Ma J, Cole KJ, She MN, Perera FP. Association between carcinogen-DNA adducts in white blood cells and lung cancer risk in the physicians health study. Cancer research. 2001;61:6708–6712. [PubMed] [Google Scholar]
- Zhou G, Hernandez NS, Randerath E, Randerath K. Effects of different diets and dietary restriction on perinatal endogenous DNA adducts. Time dependence of oxidative and presumptive nonoxidative lesions. Mutation research. 2000;447:137–147. doi: 10.1016/s0027-5107(99)00211-0. [DOI] [PubMed] [Google Scholar]
- Zhou GD, Hernandez NS, Randerath E, Randerath K. Acute elevation by short-term dietary restriction or food deprivation of type I I-compound levels in rat liver DNA. Nutrition and cancer. 1999;35:87–95. doi: 10.1207/S1532791487-95. [DOI] [PubMed] [Google Scholar]
- Zhou GD, Popovic N, Lupton JR, Turner ND, Chapkin RS, Donnelly KC. Tissue-specific attenuation of endogenous DNA I-compounds in rats by carcinogen azoxymethane: possible role of dietary fish oil in colon cancer prevention. Cancer Epidemiol Biomarkers Prev. 2005;14:1230–1235. doi: 10.1158/1055-9965.EPI-04-0759. [DOI] [PubMed] [Google Scholar]
- Zhou GD, Randerath K, Donnelly KC, Jaiswal AK. Effects of NQO1 deficiency on levels of cyclopurines and other oxidative DNA lesions in liver and kidney of young mice. International journal of cancer. 2004;112:877–883. doi: 10.1002/ijc.20375. [DOI] [PubMed] [Google Scholar]