Abstract
β-2-microglobulin (β2m) self-aggregates to form amyloid fibril in renal patients taking long-term dialysis treatment. Despite the extensive structural and mutation studies carried out so far, the molecular details on the factors that dictate amyloidogenic potential of β2m remain elusive. Here we report molecular dynamics simulations followed by the solvation thermodynamic analyses on the wild-type β2m and D76N, D59P, and W60C mutants at the native (N) and so-called aggregation-prone intermediate (IT) states, which are distinguished by the native cis- and non-native trans-Pro32 backbone conformations. Three major structural and thermodynamic characteristics of the IT-state relative to the N-state in β2m protein are detected that contribute to the increased amyloidogenic potential: (i) the disruption of the edge D-strand, (ii) the increased solvent-exposed hydrophobic interface, and (iii) the increased solvation free energy (less affinity toward solvent water). Mutation effects on these three factors are shown to exhibit a good correlation with the experimentally observed distinct amyloidogenic propensity of the D76N (+), D59P (+), and W60C (−) mutants (+/− for enhanced/decreased). Our analyses thus identify the structural and thermodynamic characteristics of the amyloidogenic intermediates, which will serve to uncover molecular mechanisms and driving forces in β2m amyloid fibril formation.
Amyloid fibril formation in the osteoarticular tissues resulting from the aggregation of β-2-microglobulin (β2m) is the hallmark of dialysis-related amyloidosis (DRA)1. With the full renal activity, β2m released from the major histocompatibility complex class I is filtered and degraded in the kidney to retain its low plasma concentration2. Upon renal failure, on the other hand, a 60-fold increase in the β2m concentration is observed in the serum2. The resulting high concentration has been implicated to be the cause of the fibrillogenesis associated with DRA. However, a sensible correlation has not been detected between the fibril load in the osteoarticular tissues and the β2m concentration in the serum3, implying that additional factors and/or events must be involved in initiating the β2m aggregation in vivo. Furthermore, the monomeric form of β2m is highly stable under physiological conditions and does not exhibit a tendency to aggregate even at elevated concentrations in vitro4,5.
It is now widely recognized that partial unfolding, also called misfolding, of monomeric species is a necessary step to initiate aggregation6. In this regard, a long-lived folding intermediate has been identified in the kinetics study of β2m folding that adopts a non-native trans-Pro32 backbone conformation7,8,9,10,11. This partially unfolded intermediate, termed as IT-state, has been recognized as a crucial amyloidogenic precursor since enhanced fibrillogenesis has been observed in variants of β2m in which the non-native trans-Pro32 isomer is stabilized5,11,12,13. However, it has also been reported that some mutants of β2m, which predominantly adopt a trans conformation at the proline residue 32, cannot form amyloid fibrils spontaneously13, indicating that the trans peptide conformation at this position alone is insufficient to endow amyloidogenic potential to β2m. It therefore remains to investigate in more details the changes in structural and possibly thermodynamic features induced by the cis to trans Pro32 backbone isomerization in order to unveil the characteristics of more direct relevance to the amyloidogenic propensity of β2m.
In this paper, we report in silico studies on the structural and thermodynamic features of the native (N) and intermediate (IT) states of the wild-type β2m and its D76N, D59P, and W60C mutants. Molecular dynamics (MD) simulations were conducted to analyze the atomic-level structural changes caused by the cis-to-trans Pro32 isomerization. Solvation free energy calculations using the molecular theory of solvation were also carried out to analyze thermodynamic consequences of the structural changes upon the Pro32 isomerization. Mutation effects were also studied to examine whether the structural and thermodynamic traits that we observe at the IT-state are indeed responsible for the amyloidogenic potential. The three mutants, D76N, D59P, and W60C, were chosen because of their distinct aggregation propensity14,15; the pathogenic familial mutant D76N and synthetic mutant D59P are known to exhibit more enhanced amyloidogenic propensity than the wild-type β2m, whereas the synthetic mutant W60C is less aggregation-prone. Thereby, we aim to identify the structural and thermodynamic characteristics of the amyloidogenic intermediates of β2m.
Results
Protein structures at the N-state are highly stable
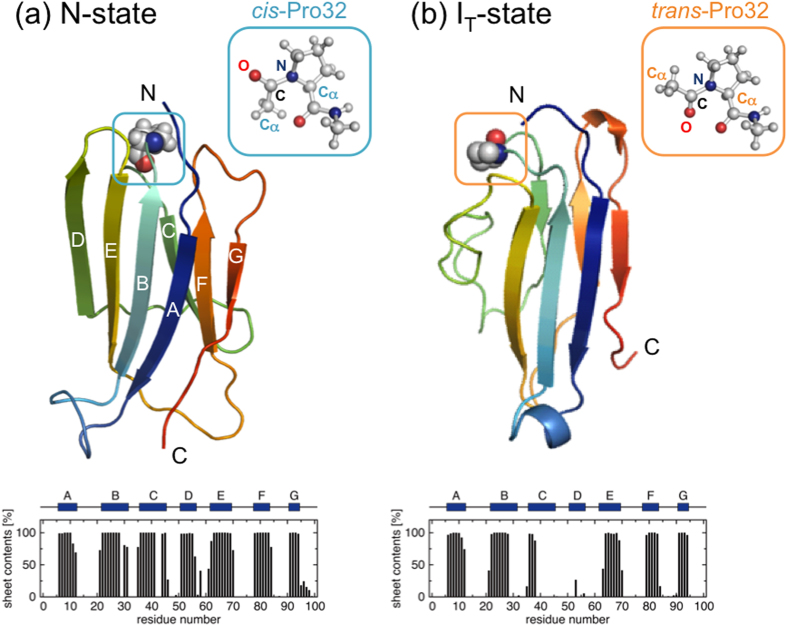
The wild-type β2m (99 residues) determined by X-ray (PDB entry 2YXF16), in which Pro32 adopts a native cis-conformation, shows an immunoglobulin fold comprising seven β-strands (termed as A to G) organized into two β-sheets (ABED and CFG) connected with a disulfide bridge (Cys25-Cys80) (see Fig. 1a). The X-ray structures of the D76N (PDB entry 4FXL14), D59P (3DHM15), and W60C (3DHJ15) mutants are quite similar to the wild-type structure. Indeed, Cα root-mean-square deviation (RMSD) values are within 0.8 Å between all of these X-ray structures.
Figure 1. Representative wild-type structures at N-state (a) and IT-state (b) containing, respectively, the native cis- and a non-native trans-Pro32 residue.
The average β-sheet contents versus amino acid residue are also displayed.
We performed the N-state simulations of the wild-type β2m and its D76N, D59P, and W60C mutants starting from the respective X-ray structures. The protein conformations during the N-state simulations were found to remain close to the X-ray structures (Table 1, Figs 1a and 2a–c): the average Cα RMSD values from the respective X-ray structures are within a few Å, the radii of gyration are nearly the same as those of the X-ray structures, the fraction of the native hydrophobic contacts are ~90%, and the β-sheets as found in the X-ray structures are well conserved. Thus, the native structures of the wild-type β2m as well as the D76N, D59P, and W60C mutants are highly stable at the simulated condition.
Table 1. Structural characteristics (average ± standard deviation).
| Cα RMSD(Å)a | Rg (Å)b | number ofhydrophobiccontacts | fraction of nativehydrophobic contacts(%)c | |
|---|---|---|---|---|
| X-ray structure | ||||
| wild type | 0 | 14.3 | 34 | 100 |
| D76N | 0 | 14.0 | 35 | 100 |
| D59P | 0 | 14.5 | 36 | 100 |
| W60C | 0 | 14.3 | 34 | 100 |
| N-state | ||||
| wild type | 2.4 ± 0.5 | 14.7 ± 0.1 | 32.8 ± 1.5 | 89.3 ± 3.6 |
| D76N | 2.6 ± 0.5 | 14.7 ± 0.1 | 32.8 ± 1.8 | 86.8 ± 3.9 |
| D59P | 2.1 ± 0.4 | 14.6 ± 0.1 | 33.4 ± 1.6 | 86.7 ± 3.3 |
| W60C | 2.0 ± 0.4 | 14.6 ± 0.1 | 33.2 ± 1.8 | 91.1 ± 3.3 |
| IT-state | ||||
| wild type | 4.9 ± 0.3 | 14.4 ± 0.2 | 25.3 ± 2.3 | 54.0 ± 3.9 |
| D76N | 5.8 ± 0.4 | 14.3 ± 0.2 | 26.7 ± 3.0 | 51.3 ± 3.8 |
| D59P | 6.2 ± 0.6 | 14.4 ± 0.2 | 25.2 ± 2.2 | 51.6 ± 3.9 |
| W60C | 6.2 ± 0.4 | 14.4 ± 0.2 | 23.0 ± 2.0 | 49.5 ± 3.4 |
aCα RMSD relative to the respective X-ray structure.
bRadius of gyration.
cFraction of the native hydrophobic contacts defined by the respective X-ray structure.
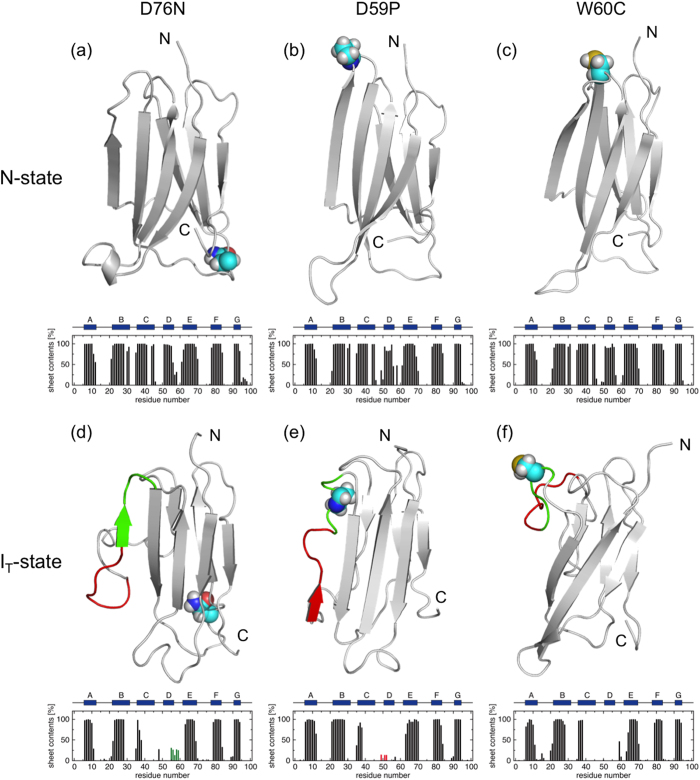
Figure 2. Representative N-state structures of D76N (a), D59P (b), and W60C (c) and IT-state structures of these proteins in this order (d–f).
The mutation sites are represented with the space-filling model. The average β-sheet contents versus amino acid residue are also displayed. In (d–f), the D-strand and DE-loop regions are colored with red and green, respectively.
Structural characteristics of the wild-type β2m at the IT-state
We carried out the IT-state simulations of the wild-type β2m, in which Pro32 takes a non-native trans conformation (Fig. 1b), to investigate structural changes induced by the Pro32 isomerization. The simulations were initiated from the partially unfolded structure obtained at a high temperature to enhance the otherwise slow cis to trans Pro32 isomerization (see Methods for details).
The global structure and the overall topology of the wild-type β2m are well conserved between the N- and IT-states as can be inferred from the representative structures and the average β-sheet contents displayed in Fig. 1a,b, with the most notable exception being the unstructured edge D-strand at the IT-state. The disordered D-strand is accompanied by the disruption and solvent-exposure of the hydrophobic core residues surrounding this region (Fig. 3a). Indeed, the fraction of the native hydrophobic contacts decreases to ~50% (Table 1) and the solvent accessible surface area (SASA) of hydrophobic residues increases (Table 2) at the IT-state. However, the number of hydrophobic contacts (25.3) of the wild-type β2m at the IT-state is not decreased much as compared to the one (32.8) at the N-state (Table 1), indicating that a reorganized, or repacked, hydrophobic core is formed at IT-state.
Figure 3. Surface representation of the hydrophobic residues located in the C-, D-, E-strands and CD- and DE-loop regions (residues 36 to 69) of the wild-type β2m (a), D76N (b), D59P (c), and W60C (d) in their representative IT-state conformations.
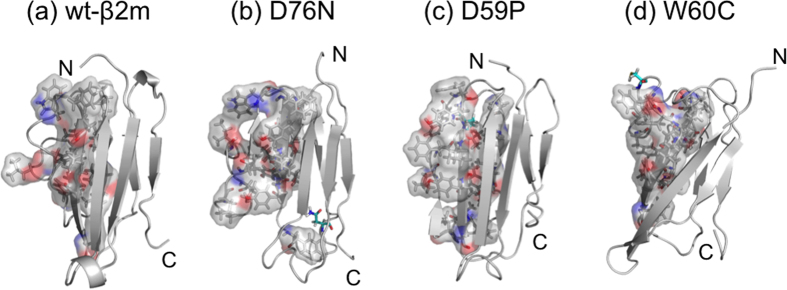
The mutation sites are indicated with cyan stick representation.
Table 2. Solvent accessible surface area (SASA) of hydrophobic side chains.
| hydrophobic SASA (Å2)a | |
|---|---|
| N-state | |
| wild type | 911.1 ± 58.3 |
| D76N | 882.3 ± 77.8 |
| D59P | 848.2 ± 59.5 |
| W60C | 770.3 ± 60.9 |
| IT-state | |
| wild type | 1037.2 ± 87.1 |
| D76N | 1057.7 ± 95.6 |
| D59P | 1142.3 ± 76.5 |
| W60C | 996.3 ± 104.7 |
aAverage ± standard deviation.
Mutation effects on the structural characteristics at the IT-state
The IT-state simulations for the three mutants, D76N, D59P, and W60C, were performed to investigate the mutation effects on the IT-state structural characteristics. The overall structural features are conserved between N- and IT-states also for these mutants as demonstrated in Fig. 2a–f. On the other hand, we find the increased β-sheet forming propensity around the edge D-strand region of the D76N and D59P mutants at the IT-state (Fig. 2d,e), whereas the D-strand remains unstructured in the W60C mutant (Fig. 2f).
The hydrophobic cores are also disrupted and reorganized in all the mutants at the IT-state compared to the N-state (Table 1). However, the degrees of the disruption and the resulting solvent exposure of the hydrophobic-core residues exhibit distinct behavior depending on the site of mutation. We find that the hydrophobic SASA at the IT-state is larger in the D76N and D59P mutants than in the wild-type β2m, whereas it is smaller in the W60C mutant (Table 2). This is exemplified in Fig. 3a–d showing the hydrophobic residues located in the C- to E-strand regions of the representative IT-state structures. It is observed that the solvent-exposure of the hydrophobic residues is substantial in the D76N and D59P mutants, whereas the solvent-exposed hydrophobic residues are rather compact in the W60C mutant.
Solvation thermodynamic characteristics of the IT-state
While the structural features in the IT-state caused by the Pro32 isomerization presented so far are candidates of the protein intrinsic factors relevant to understanding amyloidogenic potential, it is also necessary to pay attention to environmental factors such as the role of solvent water. Indeed, two negatively charged β2m proteins (the total charge of the wild-type β2m at neutral pH is −2) would not approach each other if there were no water-mediated driving force to overcome the electrostatic repulsion. (This is true also for proteins in which positively and negatively charged residues are unevenly distributed on the protein surface; See Supplementary Discussion and Fig. S1 on this point.) The key quantity that quantifies the water-mediated interaction is the solvation free energy ΔGsolv measuring an overall affinity of a protein to water17. We computed ΔGsolv of the wild-type β2m both at the N- and IT-states by using the molecular theory of solvation (see Methods), and found that ΔGsolv(IT) is larger than ΔGsolv(N) by +47.9 kcal/mol. Since a larger ΔGsolv value is associated with a less affinity toward solvent water, this indicates that the IT-state protein conformations are more “hydrophobic” and possess a more tendency to self-associate in aqueous environments than the N-state conformations.
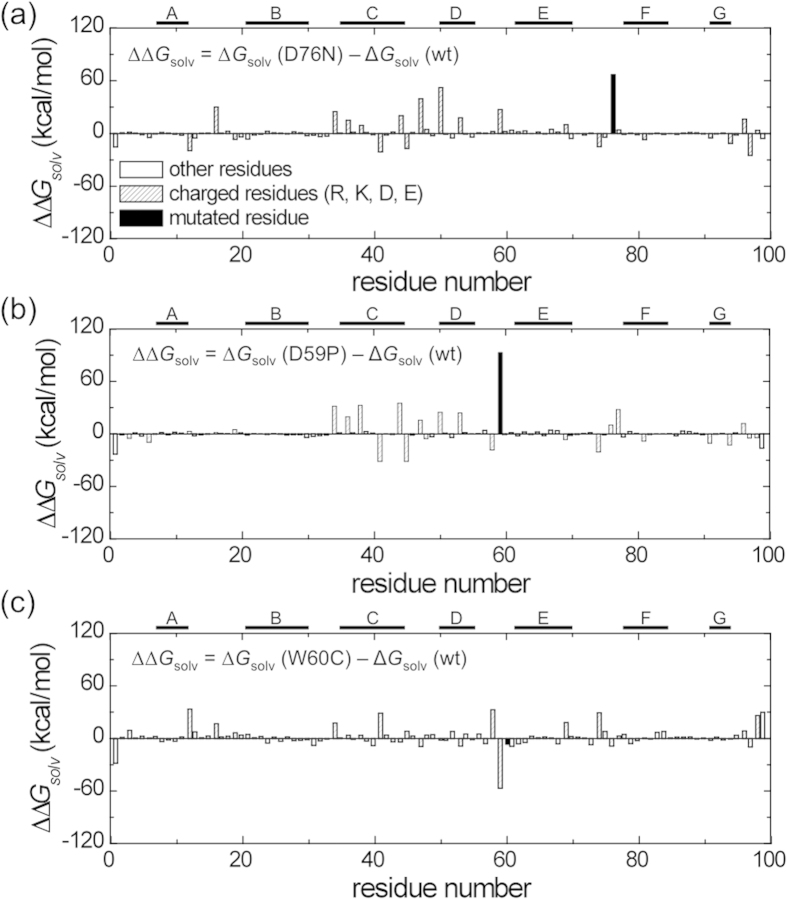
Solvation free energies for the D76N, D59P, and W60C mutants at the IT-state were also computed to examine the mutation effects, and the results relative to the wild-type β2m at the IT-state, ΔGsolv(mutant) − ΔGsolv(wild type), are presented in Table 3. The solvation free energies for D76N and D59P mutants are found to be significantly larger than the one for the wild-type, whereas the one for W60C is lower. These results indicate that the water-induced attraction is stronger for the D76N and D59P mutants than the one for the wild type, while it is weaker for the W60C mutant. We also note that the mutation site is not the sole cause of the solvation free energy change upon mutation. Indeed, the residue-decomposed solvation free energy values illustrate the significant contributions from the amino acid residues other than the mutation site (Fig. 4). In particular, the C- through E-strand regions contribute to total solvation free energy changes by adapting the conformational changes upon mutation. It is noted that the large contributions from the charged residues reflect the fact that the electrostatic component dominates the change in solvation free energy upon mutation18,19.
Table 3. Solvation free energy change upon mutation at IT-state.
| ΔΔGsolv (kcal/mol)a | |
|---|---|
| D76N | +99.0 ± 26.2 |
| D59P | +135.8 ± 9.9 |
| W60C | −32.0 ± 13.1 |
aDifference in the solvation free energy, ΔΔGsolv = ΔGsolv(mutant) − ΔGsolv(wild type), from the wild type ± standard error.
Figure 4. Contribution from each amino acid residue to the solvation free energy change, ΔΔGsolv = ΔGsolv(mutant) − ΔGsolv(wild type), upon mutation at IT-state for D76N (a), D59P (b), and W60C (c).
Discussion
Crucial to uncovering molecular mechanisms behind amyloid fibril formation is to elucidate factors that confer amyloidogenic potential to otherwise an inert soluble protein. Structural changes induced by the native cis to a non-native trans Pro32 isomerization has therefore been a subject of intense experimental investigations12,20,21,22,23,24. While detailed structural analysis of amyloidogenic intermediates has been difficult to investigate because of their transient nature, it has recently been demonstrated that the D-strand at IT-state exhibits less populated β-sheet conformation compared to N-state23 and that a repacking and exposure of the hydrophobic residues occur to accommodate the non-native trans-Pro32 conformation12,20. We find the disruption of the D-strand (Fig. 1) and the repacked (Table 1) as well as solvent-exposed (Table 2) hydrophobic residues at IT-state relative to N-state, and these structural features observed in our IT-state simulations are fully consistent with the experimental measurements.
We analyzed the D76N, D59P, and W60C mutants to examine whether the structural features observed in the simulated IT-state are relevant to experimentally observed amyloidogenic propensity. In this regard, the larger β-sheet forming propensity and the exposure of hydrophobic residues have been suggested as major factors dictating the protein aggregation propensity25,26. In our simulated IT-state structures, the enhanced β-sheet forming propensity was observed in the disordered D-strand region of the D76N and D59P mutants (Fig. 2d,e), while no such enhancement was detected in the W60C mutant (Fig. 2f). Furthermore, we find that D76N and D59P exhibit larger hydrophobic SASA than the wild-type β2m, while W60C shows smaller hydrophobic SASA (Table 2). These mutation-dependent structural features correlate quite well with an enhanced aggregation propensity of D76N14 and D59P15 and a reduced propensity of W60C15, indicating that these structural features are the characteristics associated with the amyloidogenic potential. Notably, the region where these structural changes are observed upon the Pro32 isomerization is the edge D-strand region whose potential role in assembling into amyloid fibrils has been proposed27,28,29.
Interaction with surrounding water of a protein also plays a critical role in dictating the protein aggregation propensity30. Indeed, it has been demonstrated for a substantial number of mutants that the change in protein aggregation propensity upon mutation has a significant correlation with the change in solvation free energy30. Furthermore, the decrease in solvation free energy has been shown to be the major driving force for proteins of the same charge to approach from a long distance to a short contact31,32. This indicates that a protein of a larger solvation free energy has a more tendency to self-associate. In this regard, we find at the IT-state that the D76N and D59P mutants exhibit a larger solvation free energy than the wild type, while the W60C mutant shows a smaller solvation free energy (Table 3). These trends are also in accord with the experimentally observed amyloidogenic propensity of these mutants. Such changes in the solvation free energy upon mutation are the results of structural changes induced by the Pro32 isomerization (Fig. 4). Thus, our structural and solvation thermodynamic analyses suggest that the β-sheet forming propensity of the disordered edge D-strand region and the solvent exposure of the hydrophobic residues, together with a concomitant solvation free energy change, are responsible for the amyloidogenic potential of the IT-state of β2m.
In summary, we report de novo computational studies for characterizing aggregation-prone intermediate (IT) state of the wild type and three mutants (D76N, D59P, and W60C) of β2m to investigate the molecular determinants of their distinct amyloidogenic potential. We focus on the changes in the structural and thermodynamic features caused by the native cis (N-state) to a non-native trans (IT-state) isomerization of Pro32, which has been recognized as a trigger to initiate the β2m aggregation. We find for the wild-type β2m that the edge D-strand becomes unstructured, the hydrophobic core is disrupted and solvent-exposed, and the solvation free energy is consequently increased upon the Pro32 cis-trans isomerization. These features are mutation-dependent, and through the comparison with the experimentally-measured aggregation propensity, it is suggested that (i) more enhanced β-sheet forming propensity of the disordered D-strand region, (ii) more solvent-exposed hydrophobic residues, and (iii) the larger value of the solvation free energy, are the structural and thermodynamic characteristics at the IT-state that impart more amyloidogenic potential. We believe that our work contributes to uncovering the structural motif and molecular mechanisms of β2m amyloid fibril formation.
Methods
MD simulations at N-state
Fully atomistic MD simulations with explicit water were performed at 330 K and 1 bar under neutral pH for the wild type as well as three D76N, D59P, and W60C mutants of β2m with the AMBER12 simulation suite33. The ff99SB force field34 was used for protein and the TIP3P model35 for water. The initial structures of the N-state simulations, in which Pro32 adopts a cis-conformation, were taken from X-ray: PDB entry 2YXF16 was used for the wild-type β2m, 4FXL14 for D76N, 3DHM15 for D59P, and 3DHJ15 for W60C. Each protein was solvated with ~10,000 water molecules. The control of temperature and pressure was done with Berendsen’s method36. The SHAKE algorithm37 was employed for bonds including hydrogen atoms, and a 2.0 fs time step was used.
We used the following common procedures for all MD simulations. After the energy minimization, a 20 ps equilibration simulation under constant volume was performed in which temperature was set to 330 K. (The choice of this temperature, which is higher than the room/body temperature, was intended to enhance the conformational changes at IT-state induced by the Pro32 isomerization, and for consistency, this temperature was also adopted for the N-state simulations.) We then carried out a 200 ps equilibration simulation with a constant pressure of 1 bar. Finally, production run at 330 K and 1 bar was conducted for 200 ns. Three independent production runs with differing initial random velocities were conducted for each system to compute statistical averages of various observables. No cis to trans Pro32 isomerization was observed during all N-state simulations.
MD simulations at IT-state
We conducted partial unfolding simulations at a high temperature to obtain initial structures for the IT-state simulations in which Pro32 displays a non-native trans conformation. The high-temperature partial unfolding simulation was necessary since no atomic-level structure for the IT-state is available from experiments. It was also done because the cis to trans proline isomerization is one of the slowest protein conformational changes that is difficult to capture without an enhanced sampling method. There are other possible ways to obtain the IT-state from simulations, such as replica exchange simulations. In any case, high-temperature simulations or replicas are necessary to facilitate the Pro32 isomerization. At the same time, we consider it necessary to keep the high-temperature simulations short, in order to suppress high-temperature-induced conformational changes as much as possible. For these two reasons, we have chosen to simply perform short simulations at a high temperature, which are to be terminated as soon as the occurrence of the cis-to-trans Pro32 isomerization.
For the wild-type β2m, we carried out several short simulations at 600 K with a constant volume, starting from the N-state conformation equilibrated at 330 K and 1 bar. In one of the high-temperature trajectories, a cis to trans isomerization of Pro32 was observed at ~6 ns, and the protein structure just after the proline isomerization was taken as the starting structure of the IT-state simulations for the wild-type β2m. The initial structures of the IT-state simulations for the mutants (D76N, D59P, and W60C) were generated based on the one for the wild-type β2m by mutating the corresponding residues D76, D59, and W60 to N76, P59, and C60, respectively, using Swiss PDB Viewer38. Our method for generating the initial IT-state structures for the mutants is reasonable in light of the experimental observations that the cis-to-trans Pro32 isomerization is the trigger step to initiate the aggregation and that, before the occurrence of the Pro32 isomerization (i.e., in N-state), the structures of the D76N, D59P, and W60C mutants are quite similar to the wild-type structure (Cα RMSD values are within 0.8 Å between all of their X-ray structures). We then carried out three independent 200-ns IT-state production runs for each protein system. Pro32 maintained the trans conformation during the IT-state simulations in all cases.
To support our IT-state simulations initiated from the partial unfolding at a high temperature, we additionally performed 200 ns MD simulations for the wild-type β2m as well as its three mutants at the normal body temperature (310 K) and 1 bar starting from the protein structures containing a non-native trans-Pro32, which were obtained by locally changing the backbone torsions from the respective X-ray structures of cis-Pro32. We also observe from the supporting simulations that the most distinctive structural characteristic caused by the non-native trans-Pro32 is the disruption of the edge D-strand region, including the subtle mutation effect on the β-sheet forming propensity, which is consistent with our IT-state simulations (Supplementary Fig. S2). This indicates that the IT-state structural characteristics that we discuss in the main text are not due to the disruptive impact from the high temperature, but reflect genuine structural features caused by the trans-Pro32 conformation.
Structural and solvation free energy analyses
Root-mean-square deviation of Cα atoms (Cα RMSD), radius of gyration (Rg), secondary structure contents, number of hydrophobic contacts, and solvent accessible surface area (SASA) were computed to analyze the simulated protein structures. The DSSP39 was used to calculate the secondary-structure contents. A hydrophobic contact between a pair of hydrophobic residues is considered formed when the distance between the heavy side-chain atoms is less than 5.4 Å. Representative protein structures were obtained by performing the RMSD-based K-clustering analysis (3.5-Å cutoff).
We used the three-dimensional reference interaction site model (3D-RISM) theory40 to compute the solvation free energy ΔGsolv of the simulated protein conformations. This theory provides the equilibrium water distribution function around a given protein structure, from which ΔGsolv can be computed (see Supplementary Methods for details on the ΔGsolv calculations). For each protein system, we took 2,400 protein structures with a 250 ps interval from the three independent trajectories of 200 ns length to compute the average ΔGsolv. An exact decomposition method is also available that divides ΔGsolv of a protein into contributions from the constituent amino acid residues41, which was used to analyze residue-specific contributions to the solvation free energy change upon mutation (see also Supplementary Methods for details).
Amyloid fibril formation involves a number of steps, including the initial partial unfolding of monomers to form the aggregation-prone (IT-state) structures, the oligomer formation from those monomers, and the final fibril growth through the attachment of monomers/oligomers. Overall, the structural and thermodynamic features of monomers at IT-state would play a crucial role. Therefore, although there is a huge difference between the timescale of our simulations (sub-microseconds) and that of the fibril formation measured experimentally (hours)14,15, our computational analyses revealing the IT-state characteristics will be relevant to uncovering the molecular determinants of the amyloidogenic potential.
Additional Information
How to cite this article: Chong, S.-H. et al. Structural and Thermodynamic Characteristics of Amyloidogenic Intermediates of β-2-Microglobulin. Sci. Rep. 5, 13631; doi: 10.1038/srep13631 (2015).
Supplementary Material
Acknowledgments
This work was supported by Samsung Science and Technology Foundation under Project Number SSTF-BA1401-13.
Footnotes
Author Contributions S.H. designed the research. S.-H.C. and J.H. performed the calculations. S.L., S.C. and J.L. contributed to analyze the data. All authors wrote and reviewed the manuscript.
References
- Heegaard N. H. H. β2-microglobulin: From physiology to amyloidosis. Amyloid 16, 151–173 (2009). [DOI] [PubMed] [Google Scholar]
- Floege J. & Ehlerding G. Beta-2-microglobulin-associated amyloidosis. Nephron 72, 9–26 (1996). [DOI] [PubMed] [Google Scholar]
- Gejyo F. et al. β2-microglobulin: A new form of amyloid protein associated with chronic hemodialysis. Kidney Int. 30, 385–390 (1986). [DOI] [PubMed] [Google Scholar]
- Myers S. L. et al. A systematic study of the effect of physiological factors on β2-microglobulin amyloid formation at neutral pH. Biochemistry 45, 2311–2321 (2006). [DOI] [PubMed] [Google Scholar]
- Eichner T. & Radford S. E. A generic mechanism of β2-microglobulin amyloid assembly at neutral pH involving a specific proline switch. J. Mol. Biol. 386, 1312–1326 (2009). [DOI] [PubMed] [Google Scholar]
- Dobson C. M. Protein folding and misfolding. Nature 426, 884–890 (2003). [DOI] [PubMed] [Google Scholar]
- Chiti F. et al. Detection of two partially structured species in the folding process of the amyloidogenic protein β2-microglobulin. J. Mol. Biol. 307, 379–391 (2007). [DOI] [PubMed] [Google Scholar]
- Kameda A. et al. Nuclear magnetic resonance characterization of the refolding intermediate of β2-microglobulin trapped by non-native prolyl peptide bond. J. Mol. Biol. 348, 383–397 (2005). [DOI] [PubMed] [Google Scholar]
- Chiti F. et al. A partially structured species of β2-microglobulin is significantly populated under physiological conditions and involved in fibrillogenesis. J. Biol. Chem. 276, 46714–46721 (2001). [DOI] [PubMed] [Google Scholar]
- Sakata M. et al. Kinetic coupling of folding and prolyl isomerization of β2-microglobulin studied by mutational analysis. J. Mol. Biol. 382, 1242–1255 (2008). [DOI] [PubMed] [Google Scholar]
- Jahn T. R., Parker M. J., Homans S. W. & Radford S. E. Amyloid formation under physiological conditions proceeds via a native-like folding intermediate. Nat. Struct. Mol. Biol. 13, 195–201 (2006). [DOI] [PubMed] [Google Scholar]
- Eichner T., Kalverda A. P., Thompson G. S., Homans S. W. & Radford S. E. Conformational conversion during amyloid formation at atomic resolution. Mol. Cell 41, 161–172 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Platt G. W. & Radford S. E. Glimpses of the molecular mechanisms of β2-microglobulin fibril formation in vitro: Aggregation on a complex energy landscape. FEBS Lett. 583, 2623–2629 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valleix S. et al. Hereditary systemic amyloidosis due to Asp76Asn variant β2-microglobulin. N. Engl. J. Med. 366, 2276–2283 (2012). [DOI] [PubMed] [Google Scholar]
- Ricagno S. et al. DE loop mutations affect β2-microglobulin stability and amyloid aggregation. Biochem. Biophys. Res. Commun. 377, 146–150 (2008). [DOI] [PubMed] [Google Scholar]
- Iwata K., Matsuura T., Sakurai K., Nakagawa A. & Goto Y. High-resolution crystal structure of β2-microglobulin formed at pH 7.0. J. Biochem. 142, 413–419 (2007). [DOI] [PubMed] [Google Scholar]
- Ben-Naim A. Hydrophobic Interactions (Plenum Press, New York, 1980). [Google Scholar]
- Chong S.-H., Lee C., Kang G., Park M. & Ham S. Structural and thermodynamic investigations on the aggregation and folding of acylphosphatase by molecular dynamics simulations and solvation free energy analysis. J. Am. Chem. Soc. 133, 7075–7083 (2011). [DOI] [PubMed] [Google Scholar]
- Chong S.-H. & Ham S. Site-directed analysis on protein hydrophobicity. J. Comput. Chem. 35, 1364–1370 (2014). [DOI] [PubMed] [Google Scholar]
- Eakin C. M., Berman A. J. & Miranker A. D. A native to amyloidogenic transition regulated by a backbone trigger. Nat. Struct. Mol. Biol. 13, 202–208 (2006). [DOI] [PubMed] [Google Scholar]
- Corazza A. et al. Native-unlike long-lived intermediates along the folding pathway of the amyloidogenic protein β2-microglobulin revealed by real-time two-dimensional NMR. J. Biol. Chem. 285, 5827–5835 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eichner T. & Radford S. E. Understanding the complex mechanisms of β2-microglobulin amyloid assembly. FEBS J. 278, 3868–3883 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rennella E. et al. Real-time NMR characterization of structure and dynamics in a transiently populated protein folding intermediate. J. Am. Chem. Soc. 134, 8066–8069 (2012). [DOI] [PubMed] [Google Scholar]
- Rennella E. et al. Oligomeric states along the folding pathways of β2-microglobulin: Kinetics, thermodynamics, and structure. J. Mol. Biol. 425, 2722–2736 (2013). [DOI] [PubMed] [Google Scholar]
- Chiti F., Stefani M., Taddei N., Ramponi G. & Dobson C. M. Rationalization of the effects of mutations on peptide and protein aggregation rates. Nature 424, 805–808 (2003). [DOI] [PubMed] [Google Scholar]
- Ahmed A. B. & Kajava A. V. Breaking the amyloidogenecity code: Methods to predict amyloids from amino acid sequence. FEBS Lett. 587, 1089–1095 (2013). [DOI] [PubMed] [Google Scholar]
- Trinh C. H., Smith D. P., Kalverda A. P., Phillips S. E. & Radford S. E. Crystal structure of monomeric human β-2-microglobulin reveals clues to its amyloidogenic properties. Proc. Natl. Acad. Sci. USA 99, 9771–9776 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rennella E. et al. Equilibrium unfolding thermodynamics of β2-microglobulin analyzed through native-state H/D exchange. Biophys. J. 96, 169–179 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su Y. et al. Secondary structure in the core of amyloid fibrils formed from human β2m and its truncated variant ΔN6. J. Am. Chem. Soc. 136, 6313–6325 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chong S.-H. & Ham S. Interaction with the surrounding water plays a key role in determining the aggregation propensity of proteins. Angew. Chem. Int. Ed. 53, 3961–3964 (2014). [DOI] [PubMed] [Google Scholar]
- Chong S.-H. & Ham S. Impact of chemical heterogeneity on protein self-assembly in water. Proc. Natl. Acad. Sci. USA 109, 7636–7641 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chong S.-H. & Ham S. Atomic-level investigations on the amyloid-β dimerization process and its driving forces in water. Phys. Chem. Chem. Phys. 14, 1573–1575 (2012). [DOI] [PubMed] [Google Scholar]
- Case D. A. et al. AMBER 12 (University of California, San Francisco, 2012). [Google Scholar]
- Hornak V. et al. Comparison of multiple amber force fields and development of improved protein backbone parameters. Proteins 65, 712–725 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jorgensen W. L., Chandrasekhar J., Madura J. D., Impey R. W. & Klein M. L. Comparison of simple potential functions for simulating liquid water. J. Chem. Phys. 79, 926–935 (1983). [Google Scholar]
- Berendsen H. J. C., Postma J. P. M., van Gunsteren W. F., DiNola A. & Haak J. R. Molecular dynamics with coupling to an external bath. J. Chem. Phys. 81, 3684–3690 (1984). [Google Scholar]
- Ryckaert J.-P., Ciccotti G. & Berendsen H. J. C. Numerical integration of the Cartesian equations of motion of a system with constraints: Molecular dynamics of n-Alkanes. J. Comput. Phys. 23, 327–341 (1977). [Google Scholar]
- Guex N. & Peitsch M. C. SWISS-MODEL and the Swiss-PDB Viewer: An environment for comparative protein modeling. Electrophoresis 18, 2714–2723 (1997). [DOI] [PubMed] [Google Scholar]
- Kabsch W. & Sander C. Dictionary of protein secondary structure: Pattern recognition of hydrogen-bonded and geometrical features. Biopolymers 22, 2577–2637 (1983). [DOI] [PubMed] [Google Scholar]
- Imai T., Harano Y., Kinoshita M., Kovalenko A. & Hirata F. A theoretical analysis on hydration thermodynamics of proteins. J. Chem. Phys. 125, 024911 (2006). [DOI] [PubMed] [Google Scholar]
- Chong S.-H. & Ham S. Atomic decomposition of the protein solvation free energy and its application to amyloid-beta protein in water. J. Chem. Phys. 135, 034506 (2011). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.