Abstract
Distinction of hydatidiform moles (HMs) from non-molar specimens (NMs) and subclassification of HMs as complete hydatidiform moles (CHMs) and partial hydatidiform moles (PHMs) are important for clinical practice and investigational studies; yet, diagnosis based solely on morphology is affected by interobserver variability. Molecular genotyping can distinguish these entities by discerning androgenetic diploidy, diandric triploidy, and biparental diploidy to diagnose CHMs, PHMs, and NMs, respectively. Eighty genotyped cases (27 CHMs, 27 PHMs, and 26 NMs) were selected from a series of 200 potentially molar specimens previously diagnosed using p57 immunostaining and genotyping. Cases were classified by 3 gynecologic pathologists on the basis of H&E slides (masked to p57 immunostaining and genotyping results) into 1 of 3 categories (CHM, PHM, or NM) during 2 diagnostic rounds; a third round incorporating p57 immunostaining results was also conducted. Consensus diagnoses (those rendered by 2 of 3 pathologists) were determined. Genotyping results were used as the gold standard for assessing diagnostic performance. Sensitivity of a diagnosis of CHM ranged from 59% to 100% for individual pathologists and from 70% to 81% by consensus; specificity ranged from 91% to 96% for individuals and from 94% to 98% by consensus. Sensitivity of a diagnosis of PHM ranged from 56% to 93% for individual pathologists and from 70% to 78% by consensus; specificity ranged from 58% to 92% for individuals and from 74% to 85% by consensus. The percentage of correct classification of all cases by morphology ranged from 55% to 75% for individual pathologists and from 70% to 75% by consensus. The κ values for interobserver agreement ranged from 0.59 to 0.73 (moderate to good) for a diagnosis of CHM, from 0.15 to 0.43 (poor to moderate) for PHM, and from 0.13 to 0.42 (poor to moderate) for NM. The κ values for intraobserver agreement ranged from 0.44 to 0.67 (moderate to good). Addition of the p57 immunostain improved sensitivity of a diagnosis of CHM to a range of 93% to 96% for individual pathologists and 96% by consensus; specificity was improved from a range of 96% to 98% for individual pathologists and 96% by consensus; there was no substantial impact on diagnosis of PHMs and NMs. Interobserver agreement for interpretation of the p57 immunostain was 0.96 (almost perfect). Even with morphologic assessment by gyneco-logic pathologists and p57 immunohistochemistry, 20% to 30% of cases will be misclassified, and, in particular, distinction of PHMs and NMs will remain problematic.
Keywords: hydatidiform mole, reproducibility, p57, molecular genotyping
Distinction of hydatidiform moles (HMs) from non-molar specimens (NMs) and subclassification of HMs as complete hydatidiform moles (CHMs) and partial hydatidiform moles (PHMs) are important for clinical management and ascertainment of the actual risk of persistent/subsequent gestational trophoblastic disease (GTD) for investigational studies. CHMs have higher risks for persistent GTD than do PHMs: 15% to 20% versus 0.2% to 4%, respectively.15,53,60 Despite this lower risk associated with PHMs, choriocarcinoma arising in a PHM, PHM with synchronous metastatic GTD, and development of chorio-carcinoma and placental site trophoblastic tumor after a diagnosis of PHM have been reported.5,41,47,54 Furthermore, the correct diagnosis of PHMs, particularly their distinction from NMs, is important for appropriate clinical management, as a diagnosis of PHM generates follow-up with serum β-human chorionic gonadotropin levels and contraception, which would be unnecessary for a diagnosis of NM and undesirable for infertility patients.
Microscopically, CHMs are characterized by enlarged hydropic chorionic villi with notable circumferential trophoblastic hyperplasia, cistern formation, trophoblastic inclusions, cytologic atypia, and apoptotic bodies within the villous stroma. Also, an early form of CHM has been described showing less well-developed yet characteristic features.31 In contrast, PHMs usually exhibit 2 populations of villi (large hydropic villi and small fibrotic ones) with irregular contours (scalloping), trophoblastic inclusions, and less trophoblastic hyperplasia than in CHMs. Despite these differences, a substantial degree of morphologic overlap exists between these 2 types of HMs. In addition, distinction of HMs from NMs can be problematic in several situations, including: (a) products of conception specimens (POCs) with abnormal villous morphology (a nonmolar type of villous abnormality having some morphologic features suggestive of a PHM but lacking the diandric triploidy required for a definitive diagnosis of PHM, sometimes attributable to other genetic abnormalities such as trisomy)7,46,49,51; (b) early nonmolar abortuses with prominent trophoblastic hyperplasia; (c) hydropic abortuses; and (d) mosaic/chimeric conceptions.25,26,30,51,57 These problems are partially because of the fact that histologic criteria for diagnosis of HMs are imperfect, not all pathologists apply criteria similarly, and because the histologic appearance can vary depending on gestational age. With the widespread use of routine first-trimester ultrasonography, the latter factor has become significant as most POC specimens, including molar and nonmolar ones, are encountered at much earlier gestational ages,32 when microscopic features are less well developed, than in the past. Indeed, previously conducted studies have demonstrated poor interobserver and intraobserver reproducibility of the diagnosis of HMs when based only on assessment of H&E slides.11,22,27,28,43
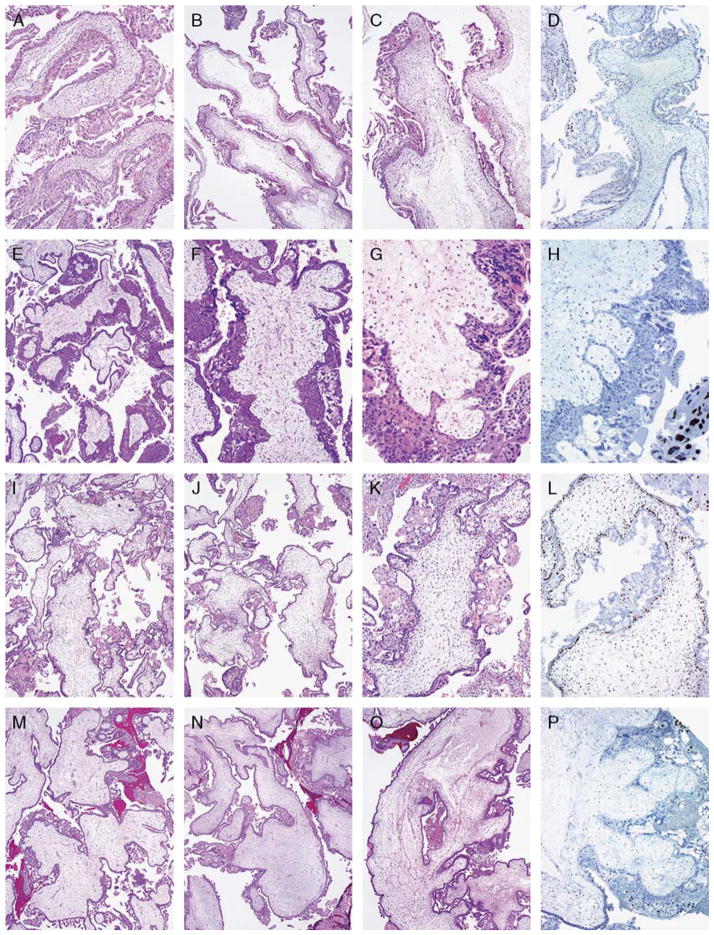
FIGURE 2.
Details are provided in Table 7. A to D, An androgenetic diploid CHM with a negative p57 stain. This example was recognized as such in all 3 rounds. E to H, An androgenetic diploid CHM with features of the early form, with a negative p57 stain. This example was recognized as such in all 3 rounds. I to L, A diandric triploid PHM with a positive p57 stain. This example was recognized as such in all 3 rounds. M to P, An androgenetic diploid CHM with a negative p57 stain. This example was recognized as molar but subclassification was problematic, being diagnosed variably as PHM versus CHM. All reviewers recognized this as a CHM with the p57 stain.
Numerous ancillary techniques to aid the diagnosis of HMs have been described and include conventional cytogenetics (karyotype), DNA ploidy determination [by flow cytometry, image analysis, and fluorescence in situ hybridization (FISH)],6–8,12,19,23,25,26,33,35,37,38,56,62 and immunohistochemistry for p57.3,4,9,12,20,21,25,26,29,39,42,48,50,52 Although previous studies have shown that the supplementation of histologic evaluation with techniques that determine DNA content (ploidy) improves the accuracy and reproducibility of a diagnosis,11,22 these techniques have the limitation of not being able to determine the maternal and paternal contributions of chromosome complements; thus, they cannot distinguish androgenetic diploidy (CHM) from biparental diploidy (NM) or diandric triploidy (PHM) from digynic triploidy (NM).
One ancillary technique with the capability of specifically distinguishing CHMs, PHMs, and NMs from one another is molecular genotyping using polymerase chain reaction amplification of multiple microsatellite loci (short tandem repeats) from multiple different chromosomes.2,3,39,42,44,45,48,51 This technique allows for determination of both ploidy and the maternal/paternal contributions of chromosome complements. Previously, we developed a working algorithm for the diagnosis of HMs using p57 immunostaining coupled with molecular genotyping and have found this approach to be quite useful in the evaluation of potentially molar specimens.39,45,51
Unlike other ancillary techniques, genotyping has the ability to establish diagnostic truth. As previous studies have not used a definitive gold standard to determine the true accuracy/reproducibility of diagnosing HMs and their distinction from NMs, the goals of this study were to establish the accuracy and interobserver and intraobserver reproducibility of diagnosing HMs by gynecologic pathologists on the basis of morphologic assessment and with the aid of p57 immunohistochemistry, using genotyping as the true gold standard against which performance is assessed.
MATERIALS AND METHODS
Study Cases, Histologic Assessment, p57 Immunohistochemistry, and Molecular Genotyping
A large prospective and consecutive series of potentially molar specimens on the Gynecologic Pathology Service of the Johns Hopkins Hospital, Baltimore, Maryland (comprising ~90% consultation cases), previously diagnosed using p57 immunostaining (monoclonal antibody from Neomarkers; Fremont, CA) and molecular genotyping (n = 200) served as the pool of material from which cases were obtained. Molecular genotyping consisted of microdissection of separate foci of villous and decidual tissue in each case. DNA was subjected to polymerase chain reaction amplification of 9 polymorphic markers (microsatellites) from multiple (8) different chromosomes and the amelogenin locus (AmpFlSTR Profiler kit; Applied Biosystems; Foster City, CA). The alleles at each locus were identified for both villous and decidual tissues, and the patterns were compared. Additional details of immunohistochemical and molecular analysis are provided elsewhere.39,44,51
Eighty genotyped cases (1 H&E slide per case), including 27 CHMs, 27 PHMs, and 26 NMs, were selected by 1 of the authors (B.M.R.) familiar with the cases to ensure the quality of the H&E-stained section and p57 immunostain. Three faculty pathologists practicing in an active gynecologic pathology service, with varying experience in gynecologic pathology (~5 to >30 y), participated in the study (A.V.Y., R.V., and R.J.K., respectively). Cases were classified by the reviewers on the basis of H&E slides, masked to p57 immunostaining and genotyping results, into 1 of 3 categories (CHM, PHM, or NM) during 2 diagnostic rounds without training sessions, separated by 8 to 12 weeks. All observers used those histologic criteria used in routine practice; these criteria are essentially similar to those described elsewhere.51 A third diagnostic round incorporating p57 immunostaining results was conducted after an interval of at least 16 weeks from the second round, with a brief explanation of interpretation of p57 immunostaining conducted using a multiheaded microscope with all reviewers.
Briefly, to interpret immunohistochemical stains for p57, the presence or absence of nuclear positivity was assessed in villous stromal cells, cytotrophoblast, intermediate trophoblast, and maternal decidua. Reviewers were instructed that the p57 immunostain is interpreted as “negative” and satisfactory when maternal decidua and/or intermediate trophoblastic cells exhibit nuclear expression of p57 (serving as internal positive control in all cases, including CHMs) but villous stromal cells and cytotrophoblast are either entirely negative or demonstrate only limited expression (nuclear staining in <10% of these cell types). The p57 immunostain is interpreted as “positive” when the extent of staining is extensive or diffuse in these cell types. In addition to the typical diffusely positive result, 2 variants of positive staining can be encountered occasionally (<10% of cases in our large series): equivocal and discordant. The p57 immunostain is interpreted as “equivocal” when nuclear expression in both villous stromal cells and cytotrophoblast is in the focally positive range (≥10% but <50% of the villi in the stained section). The p57 immunostain is interpreted as “discordant” when there is any combination/admixture of negative and positive results for villous stromal cells and cytotrophoblast within individual villi, including positive staining in cytotrophoblast and negative staining in villous stromal cells or vice versa. Further details of immunohistochemical patterns of p57 expression have been provided elsewhere.51
Statistical Analysis
The diagnoses of individual pathologists in each round were tabulated, and consensus diagnoses (those rendered by 2 of 3 pathologists) were determined. Genotyping results were used as the gold standard (true) diagnosis for assessing diagnostic performance. Sensitivity, specificity, and positive and negative predictive values for the various diagnostic categories were calculated as well.
Interobserver and intraobserver agreement for diagnoses (rounds 1 and 2) and interobserver agreement for interpretation of the p57 stain (round 3) were tested using Cohen kappa statistics.36 Statistical analyses were performed with the STATA software package (version 11.1; StataCorp LP).
RESULTS
Results are presented in Tables 1 to 6 and Figure 1. Representative examples of cases are illustrated in Figures 2 and 3, with details provided in Table 7.
TABLE 1.
Performance of Morphologic Assessment and p57 Immunostaining for Predicting a Genotyping-Confirmed Diagnosis of any Kind of HM
| Diagnosis of HM (CHM+PHM vs. NM) | Sensitivity (%) | Specificity (%) | Positive Predictive Value (%) | Negative Predictive Value (%) |
|---|---|---|---|---|
| Pathologist 1 | ||||
| Round 1 | 80 | 42 | 74 | 50 |
| Round 2 | 83 | 69 | 85 | 67 |
| Round 3 (p57) | 93 | 39 | 76 | 71 |
| Pathologist 2 | ||||
| Round 1 | 100 | 31 | 75 | 100 |
| Round 2 | 93 | 50 | 79 | 76 |
| Round 3 (p57) | 93 | 58 | 82 | 79 |
| Pathologist 3 | ||||
| Round 1 | 76 | 92 | 95 | 65 |
| Round 2 | 74 | 88 | 93 | 62 |
| Round 3 (p57) | 78 | 92 | 96 | 67 |
| Consensus diagnosis | ||||
| Round 1 | 83 | 62 | 82 | 64 |
| Round 2 | 87 | 73 | 87 | 73 |
| Round 3 (p57) | 93 | 69 | 86 | 82 |
CHM indicates complete hydatidiform mole (either classic or early type); HM, hydatidiform mole; NM, non-molar specimen; PHM, partial hydatidiform mole.
TABLE 6.
Overall Intraobserver Agreement (Comparison of Rounds 1 and 2)
| Pathologist | κ (Interpretation) | Agreement |
|---|---|---|
| 1 | 0.44 (moderate) | 64% (51/80) |
| 2 | 0.66 (good) | 79% (63/80) |
| 3 | 0.67 (good) | 79% (63/80) |
FIGURE 1.
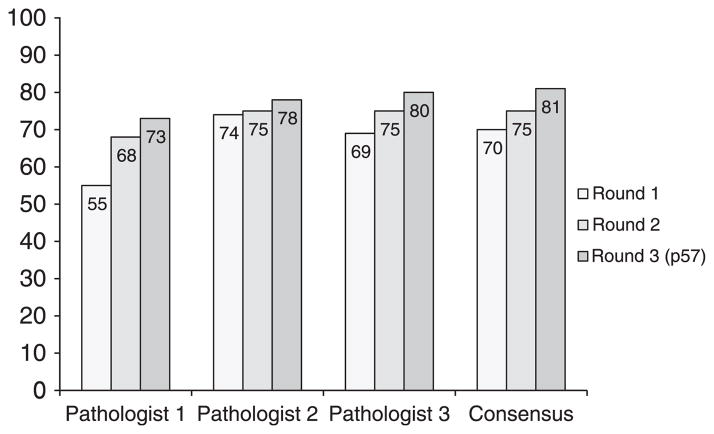
Percentage of correct classification of all cases. Percentages are listed on the y axis.
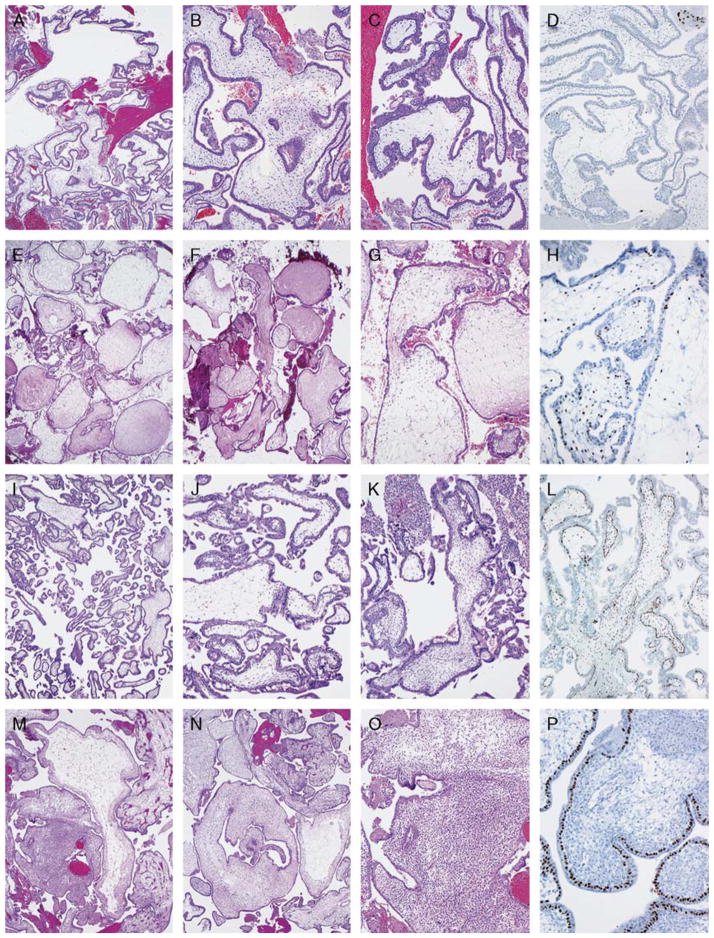
FIGURE 3.
Details are provided in Table 7. A to D, An androgenetic diploid CHM with a negative p57 stain. This example received no consensus diagnosis in the first round and a consensus of NM in the second round. All reviewers recognized this as a CHM with the p57 stain. E to H, A diandric triploid PHM with positive p57 stain. This subtle example was most often misinterpreted as NM. I to L, A biparental diploid (trisomy 16 and 21) NM with abnormal villous morphology and positive p57 stain. This example was often misinterpreted as a PHM. M to P, An androgenetic/biparental mosaic/chimeric conception with a discordant positive p57 stain. This example was often misinterpreted as a PHM; 2 of 3 reviewers recognized the discordant p57 pattern that characterizes mosaic/chimeric conceptions.
TABLE 7.
Details for Figures
| Figure | Consensus Diagnoses/Interpretations
|
Final Diagnosis (Genotyping Result) | |||
|---|---|---|---|---|---|
| Round 1* | Round 2* | Round 3 (With p57)* | p57* w | ||
| 2A-D | CHM (3/3) | CHM (3/3) | CHM (3/3) | Negative (3/3) | CHM (androgenetic diploidy) |
| 2E-H | CHM (3/3) | CHM (3/3) | CHM (3/3) | Negative (3/3) | CHM (androgenetic diploidy) |
| 2I-L | PHM (3/3) | PHM (3/3) | PHM (3/3) | Positive (3/3) | PHM (diandric triploidy) |
| 2M-P | PHM (2/3) | PHM (2/3) | CHM (3/3) | Negative (3/3) | CHM (androgenetic diploidy) |
| 3A-D | None | NM (2/3) | CHM (3/3) | Negative (3/3) | CHM (androgenetic diploidy) |
| 3E-H | NM (2/3) | NM (3/3) | NM (2/3) | Positive (3/3) | PHM (diandric triploidy) |
| 3I-L | PHM (2/3) | NM (3/3) | PHM (2/3) | Positive (3/3) | NM (abnormal villous morphology) (biparental diploidy, trisomy 16 and 21) |
| 3M-P | PHM (2/3) | PHM (2/3) | NM (2/3) | Positive (3/3)zy | NM (mosaic/chimeric) (androgenetic/biparental diploidy) |
Result (no. pathologists rendering the diagnosis/interpretation).
The p57 result for all observers for each case was concordant with the officially reported p57 interpretation.
Two of 3 pathologists recognized a discordant pattern.
Mosaic/chimeric conception with p57 expression in villous cytotrophoblast but not in villous stromal cells (see Ronnett et al51 for additional details of p57 expression patterns in mosaic/chimeric conceptions).
CHM, complete hydatidiform mole (either classic or early type); NM, non-molar specimen; PHM, partial hydatidiform mole.
Sensitivity of a diagnosis of any kind of HM by morphologic assessment ranged from 74% to 100% for individual pathologists and from 83% to 87% by consensus; specificity ranged from 31% to 92% for individual pathologists and from 62% to 73% by consensus (rounds 1 and 2; Table 1). Sensitivity of a diagnosis of CHM ranged from 59% to 100% for individual pathologists and from 70% to 81% by consensus; specificity ranged from 91% to 96% for individual pathologists and from 94% to 98% by consensus (rounds 1 and 2; Table 2). Sensitivity of a diagnosis of PHM ranged from 56% to 93% for individual pathologists and from 70% to 78% by consensus; specificity ranged from 58% to 92% for individual pathologists and from 74% to 85% by consensus (rounds 1 and 2; Table 3).
TABLE 2.
Performance of Morphologic Assessment and p57 Immunostaining for Predicting a Genotyping-Confirmed Diagnosis of CHM
| Diagnosis of CHM (CHM vs. PHM+NM) | Sensitivity (%) | Specificity (%) | Positive Predictive Value (%) | Negative Predictive Value (%) |
|---|---|---|---|---|
| Pathologist 1 | ||||
| Round 1 | 63 | 94 | 85 | 83 |
| Round 2 | 63 | 94 | 85 | 83 |
| Round 3 (p57) | 93 | 96 | 93 | 96 |
| Pathologist 2 | ||||
| Round 1 | 96 | 92 | 87 | 98 |
| Round 2 | 100 | 91 | 84 | 100 |
| Round 3 (p57) | 96 | 96 | 93 | 98 |
| Pathologist 3 | ||||
| Round 1 | 59 | 96 | 89 | 82 |
| Round 2 | 78 | 96 | 91 | 89 |
| Round 3 (p57) | 93 | 98 | 96 | 96 |
| Consensus diagnosis | ||||
| Round 1 | 70 | 98 | 95 | 87 |
| Round 2 | 81 | 94 | 88 | 91 |
| Round 3 (p57) | 96 | 96 | 93 | 98 |
CHM indicates complete hydatidiform mole (either classic or early type); NM, non-molar specimen; PHM, partial hydatidiform mole.
TABLE 3.
Performance of Morphologic Assessment and p57 Immunostaining for Predicting a Genotyping-Confirmed Diagnosis of PHM
| Diagnosis of PHM (PHM vs. CHM+NM) | Sensitivity (%) | Specificity (%) | Positive Predictive Value (%) | Negative Predictive Value (%) |
|---|---|---|---|---|
| Pathologist 1 | ||||
| Round 1 | 59 | 58 | 42 | 74 |
| Round 2 | 70 | 74 | 58 | 83 |
| Round 3 (p57) | 85 | 70 | 59 | 90 |
| Pathologist 2 | ||||
| Round 1 | 93 | 68 | 60 | 95 |
| Round 2 | 74 | 79 | 65 | 86 |
| Round 3 (p57) | 78 | 77 | 64 | 87 |
| Pathologist 3 | ||||
| Round 1 | 56 | 81 | 60 | 78 |
| Round 2 | 59 | 92 | 80 | 82 |
| Round 3 (p57) | 56 | 94 | 83 | 81 |
| Consensus diagnosis | ||||
| Round 1 | 78 | 74 | 60 | 87 |
| Round 2 | 70 | 85 | 70 | 85 |
| Round 3 (p57) | 78 | 83 | 70 | 88 |
CHM indicates complete hydatidiform mole (either classic or early type); NM, non-molar specimen; PHM, partial hydatidiform mole.
The percentage of correct classification of all cases ranged from 55% to 80% for individual pathologists and from 70% to 81% by consensus (rounds 1 to 3; Fig. 1). Details of diagnostic concordance between genotyping and consensus diagnoses are provided in Table 4. The most common problem in misclassification of cases by morphology was distinction of PHMs from NMs (13 cases in round 1 and 12 cases in round 2). This was followed by difficulty in distinction of CHMs from PHMs (5 cases in round 1 and 4 cases in round 2). Problems with distinction of CHMs from NMs were uncommon but did occur (no cases in round 1 and 2 cases in round 2). Of note, some cases did not achieve a consensus diagnosis (6 cases in round 1 and 2 cases in round 2).
TABLE 4.
Diagnostic Concordance Between Morphologic Assessment and Genotyping-Confirmed Diagnoses (Rounds 1 and 2)
| Genotyping Diagnosis | Consensus Diagnosis*
|
Total | |||
|---|---|---|---|---|---|
| CHM | PHM | NM | None | ||
| Round 1 | |||||
| CHM | 19 | 4 | 0 | 4 | 27 |
| PHM | 1 | 21 | 3 | 2 | 27 |
| NM | 0 | 10 | 16 | 0 | 26 |
| Total | 20 | 35 | 19 | 6 | 80 |
| Round 2 | |||||
| CHM | 22 | 2 | 1 | 2 | 27 |
| PHM | 2 | 19 | 6 | 0 | 27 |
| NM | 1 | 6 | 19 | 0 | 26 |
| Total | 25 | 27 | 26 | 2 | 80 |
Diagnosis rendered by 2 of 3 reviewers.
CHM indicates complete hydatidiform mole (either classic or early type); NM, non-molar specimen; PHM, partial hydatidiform mole.
Addition of the p57 immunostain improved sensitivity of a diagnosis of CHM to a range of 93% to 96% for individual pathologists and 96% by consensus; specificity was improved from a range of 96% to 98% for individual pathologists and 96% by consensus; there was no substantial impact on diagnosis of PHMs and NMs (Tables 1–3). The CHM that was not recognized as such in round 3 incorporating the p57 stain was a p57-positive androgenetic diploid CHM with a retained maternal copy of chromosome 11 (location of p57 gene).40 This case was diagnosed by all 3 reviewers as a CHM in both rounds 1 and 2 but was interpreted as a PHM by 2 reviewers and as NM by 1 reviewer in round 3 based on the diffusely positive p57 stain. Of the cases lacking consensus in the H&E rounds, the one that failed to achieve consensus in both rounds was a p57-negative androgenetic diploid CHM that was recognized as such by consensus with the assistance of the p57 stain.
The κ values for interobserver agreement for morphologic assessment ranged from 0.59 to 0.73 (moderate to good) for a diagnosis of CHM, from 0.15 to 0.43 (poor to moderate) for a diagnosis of PHM, and from 0.13 to 0.42 (poor to moderate) for a diagnosis of NM (rounds 1 to 2; Table 5). Intraobserver agreement for each pathologist between rounds 1 and 2 ranged from 0.44 to 0.67 (moderate to good), corresponding to raw agreement of 64% to 79% (Table 6). Interobserver agreement for interpretation of the p57 immunostain [including all variants of a positive interpretation (equivocal and discordant) as simply positive] was 0.96 (almost perfect). Positive cases were most often interpreted as diffusely positive, with only a minority of cases assessed as equivocal or discordant positive results. The 3 reviewers subclassified the positive stains as equivocally positive in 0, 5, and 3 cases and as discordant in 0, 5, and 6 cases. Of 3 androgenetic/biparental diploid mosaic/chimeric cases, the discordant staining pattern was recognized by 2 of 3 reviewers for each of these.
TABLE 5.
Interobserver Agreement
| Diagnostic Category | Round 1: κ (Interpretation) | Round 2: κ (Interpretation) |
|---|---|---|
| CHM | 0.59 (moderate) | 0.73 (good) |
| PHM | 0.15 (poor) | 0.43 (moderate) |
| NM | 0.13 (poor) | 0.42 (moderate) |
CHM indicates complete hydatidiform mole (either classic or early type); NM, non-molar specimen; PHM, partial hydatidiform mole.
DISCUSSION
The literature has demonstrated that there is marked variability in the diagnosis of HMs based only on H&E slides, even among experienced pathologists with specialized training, and that the reproducibility for CHMs is higher than that for PHMs.11,22,27,28,43 Depending on the number of pathologists and distribution of cases included in previous studies, the level of inter-observer agreement for diagnosis of HMs has been shown to be 55% to 80% (percentage of consensus agreement)22,27,28,43 and poor (κ statistics),11 and corresponding figures for intraobserver agreement are 50% to 90%22,27 and poor to excellent.11 The current study demonstrates similar findings. Although intraobserver reproducibility was moderate to good, our findings show that, even among experienced gynecologic pathologists, only 70% to 81% of all POCs are correctly classified solely on the basis of H&E slides, even when using p57 immunohistochemistry. Our study confirms that the main diagnostic problem relates to distinguishing PHMs from NMs (interobserver reproducibility of poor to moderate); distinction of CHMs from PHMs (primarily) and NMs (occasionally) is also problematic, and failure to recognize all CHMs by morphology persists even for experienced gynecologic pathologists. As expected, addition of p57 immunohistochemistry, which is a highly reproducibly interpreted stain, to the H&E evaluation improved the sensitivity of diagnosis of CHMs (from 70% to 81% to 96%) but not of PHMs or NMs.
It has been suggested that the addition of ploidy analysis improves both the interbserver and intraobserver reproducibility of diagnosis of HMs.11,22 However, it should be noted that previous studies assessing the accuracy/reproducibility of a diagnosis of HM with ancillary techniques have not included any type of definitive gold standard to validate the diagnoses; therefore, it has not been possible to determine true diagnostic accuracy using only H&E stains. Most ancillary techniques, including conventional cytogenetics (karyotype), DNA ploidy (flow cytometry, image analysis, and FISH),6–8,11,12,19,22,23,25,26,33,35,37,38,56,62 and p57 immunohistochemistry (the product of a paternally imprinted, maternally expressed gene),3,4,9,12,20,21,25,26,29,39,42,48,50,52 have the limitation of not being able to establish maternal/parental contributions of chromosome complements. For example, a diploid result by karyotyping, flow cytometry/image analysis, or FISH cannot distinguish a CHM (androgenetic diploidy) from a diploid NM (biparental diploidy), and a triploid result cannot discern a PHM (diandric triploidy) from a triploid NM (digynic triploidy). Similarly, although a p57 immunostain can distinguish a CHM (negative p57 because of lack of maternal DNA) from an NM (positive p57 because of the presence of a maternal chromosomal complement), it cannot discern a PHM from a diploid (biparental) or triploid (digynic) NM (all of these share the same pattern of p57 expression because of the presence of a maternal chromosomal complement). These problems can be overcome with molecular genotyping using short tandem repeat markers because this technique allows for determination of both ploidy and the maternal/paternal contributions of chromosome complements.39,44,45,51
As all of the cases in this study were derived from our gynecologic pathology consultation service, we acknowledge some degree of bias; however, this set of cases included a wide morphologic spectrum of cases, including typical HMs with characteristic features (as illustrated in some of the figures). Interestingly, this prospectively collected set has had, since its beginning, both a substantial number of HMs and a greater proportion of CHMs than PHMs. The 200 case set from which these 80 study cases were extracted comprised 76 CHMs, 48 PHMs, 69 NMs, and 7 mosaic/chimeric conceptions. This has been a surprise to us, given that diagnosis of PHMs and NMs is more problematic than that of CHMs. Although early forms are expected to constitute a notable subset of CHMs, given that nowadays a substantial number of HMs present at relatively early gestational ages, they did not constitute the exclusive form of CHM represented in this set (as judged by subjective morphologic assessment, as genotyping is identical for all forms of CHMs). Thus, as a set of consultation cases derived from the community and comprising sufficient numbers of CHMs, we believe that the cases in this study are not unduly biased toward unusually difficult cases.
The findings in this study support the use of the algorithm that we previously developed for improving the diagnosis of HMs, which combines p57 immunohistochemistry and molecular genotyping.39,45,51 Briefly, immunohistochemical staining for p57 is first performed. In the context of appropriate morphologic features, a negative p57 result establishes a diagnosis of CHM, and no further ancillary evaluation is required. If the p57 result is positive, then a CHM has been excluded, but molecular genotyping is required to distinguish a PHM from an NM. When the molecular analysis reveals a diandric triploid or biparental diploid result, then a diagnosis of PHM or NM, respectively, can be rendered. We advocate use of this algorithm for all POCs with any suspicion for an HM, which includes either a clinical concern for an HM (eg, abnormally elevated β-human chorionic gonadotropin level, abnormal ultrasound findings, clinical diagnosis of “rule-out mole,” etc.) or pathologic concern because of some morphologic abnormality of chorionic villi.
The algorithm yields a definitive diagnosis in essentially all cases, with the exception of a few situations, some rare, in which diagnostic evaluation may be limited or confusing. CHMs with a maternally derived trisomy of chromosome 11 will produce a positive p57 result (p57 is the product of the CDKN1C gene, which is on chromosome 11), which can lead to misinterpretation as a PHM or NM (as in the current study).18,40 Conversely, PHMs with loss of the maternal chromosome 11 will yield a negative p57 result, leading to an incorrect diagnosis of CHM.13 However, both of these situations, which are quite rare (1 example of each encountered in our prospective analysis of nearly 400 cases to date), can be resolved with molecular genotyping. CHMs that arise in the setting of a multiple gestation pregnancy will have more than 1 population of chorionic villi: the CHM component, which is p57 negative, and the normal NM components, which are p57 positive (overall divergent p57 staining pattern in different populations of villi).10,51,58 Failure to recognize these morphologically and immunohistochemically distinct populations can lead to incorrect interpretation as a PHM (“2” populations of villi, with some being p57 positive) and false assurance that a CHM has been excluded because of p57 positivity in at least some villi. In addition, mosaic/chimeric conceptions, particularly those with focal features of a CHM, also can have >1 morphologically and immunohistochemically distinct population of villi, leading to misclassification as a typical HM (as occurred in the current study).25,26,30,51,57 In such cases, discordant p57 staining patterns (eg, positive cytotrophoblast with negative villous stromal cells) can cause diagnostic confusion when pathologists are not familiar with this pattern or this entity. Recognition of these discordant and divergent staining patterns, respectively, in mosaic/chimeric conceptions and in the CHM and NM components of a multiple gestation is the key to correct interpretation of these complex specimens and is necessary for specific microdissection of the different components to ensure accurate molecular genotyping. Yet another rare problematic situation is the rare familial/biparental form of CHM, which is a result of an inherited homozygous mutation of the NLRP7 gene on chromosome 19 (rather than the usual uniparental androgenetic diploid origin of most CHMs).1,14,16,17,24,34,55,59,61 The molecular genotyping in this situation produces a biparental diploid result, which can erroneously lead to interpretation as NM. However, the microscopic features and loss of p57 expression are similar to that of uniparental androgenetic diploid CHMs (this is the one rare scenario in which the algorithm actually prevents confusion that might be introduced by genotyping if one is unaware of this form of CHM). Occasionally, POCs can have complex genotypes that preclude molecular interpretation, but these are uncommon in our experience (~1% of our prospectively analyzed cases). Finally, technical problems can limit molecular evaluation. Cases that include the presence of only villous tissue without decidua for comparison of allele patterns to determine the maternal and nonmaternal (paternal) allele contributions at each locus cannot be definitively assessed. Both contaminations due to a sufficient admixture of both villous and decidual tissue within the same microdissected focus and poor DNA amplification yield uninterpretable molecular results.
Comments regarding any cost-benefit analysis of this algorithmic approach to evaluation of potentially molar specimens are beyond the scope of this study. It is worth pointing out, however, that this approach represents a compromise between simple traditional microscopic evaluation, which leads to incorrect classification of at least 20% of cases in the hands of gynecologic/placental pathologists, even when a consensus diagnosis is used, and genotyping of all cases. The p57 component of the algorithm should essentially capture all CHMs, the most important group to be readily identified for clinical management purposes; this assessment is highly reproducible and is a technique that can be performed in most immunohistochemistry laboratories without the need for highly specialized equipment and expertise, such as that required for genotyping. As PHMs have a low but real risk of persistent GTD,5,41,47,54 and distinction from NMs has implications for infertility patients, genotyping of all p57-positive cases as per the algorithm provides for definitive diagnosis of all these specimens. The alternative approach of performing molecular genotyping in all potentially molar cases without utilizing p57 immunohistochemistry as the initial triage step would allow for a definitive diagnosis in virtually all cases but would clearly be more costly. This approach would potentially fail to identify the rare familial/biparental form of CHM, particularly in its early stage when morphologic features might be sufficiently subtle to not trigger recognition of a discrepancy between morphology and genotyping. Genotyping alone also can preclude specific recognition of CHMs arising in the setting of multiple gestation pregnancies and mosaic/chimeric conceptions, as the distinctive p57 immunostaining patterns that are a clue to these diagnoses and are critical to guiding micro-dissection would not be identified.25,51,57 Therefore, the most ideal method for identifying all HMs is a combined approach including correlation of morphologic features, p57 immunohistochemistry, and molecular genotyping.
In conclusion, even at the hands of experienced gynecologic pathologists, diagnosis of HMs and their distinction from NMs is imperfect. Ancillary techniques can substantially improve the diagnosis of HMs, with p57 enabling recognition of virtually all CHMs and molecular genotyping providing a definitive diagnosis for the ~20% to 30% of specimens that are misclassified by morphology.
Footnotes
Conflicts of Interest and Source of Funding: The authors have disclosed that they have no significant relationships with, or financial interest in, any commercial companies pertaining to this article.
References
- 1.Ambani LM, Vaidya RA, Rao CS, et al. Familial occurrence of trophoblastic disease—report of recurrent molar pregnancies in sisters in three families. Clin Genet. 1980;18:27–29. doi: 10.1111/j.1399-0004.1980.tb01360.x. [DOI] [PubMed] [Google Scholar]
- 2.Bell KA, Van Deerlin V, Addya K, et al. Molecular genetic testing from paraffin-embedded tissue distinguishes nonmolar hydropic abortion from hydatidiform mole. Mol Diagn. 1999;4:11–19. doi: 10.1016/s1084-8592(99)80045-9. [DOI] [PubMed] [Google Scholar]
- 3.Bifulco C, Johnson C, Hao L, et al. Genotypic analysis of hydatidiform mole: an accurate and practical method of diagnosis. Am J Surg Pathol. 2008;32:445–451. doi: 10.1097/PAS.0b013e3181520034. [DOI] [PubMed] [Google Scholar]
- 4.Castrillon DH, Sun D, Weremowicz S, et al. Discrimination of complete hydatidiform mole from its mimics by immunohistochemistry of the paternally imprinted gene product p57KIP2. Am J Surg Pathol. 2001;25:1225–1230. doi: 10.1097/00000478-200110000-00001. [DOI] [PubMed] [Google Scholar]
- 5.Cheung AN, Khoo US, Lai CY, et al. Metastatic trophoblastic disease after an initial diagnosis of partial hydatidiform mole: genotyping and chromosome in situ hybridization analysis. Cancer. 2004;100:1411–1417. doi: 10.1002/cncr.20107. [DOI] [PubMed] [Google Scholar]
- 6.Cheville JC, Greiner T, Robinson RA, et al. Ploidy analysis by flow cytometry and fluorescence in situ hybridization in hydropic placentas and gestational trophoblastic disease. Hum Pathol. 1995;26:753–757. doi: 10.1016/0046-8177(95)90223-6. [DOI] [PubMed] [Google Scholar]
- 7.Chew SH, Perlman EJ, Williams R, et al. Morphology and DNA content analysis in the evaluation of first trimester placentas for partial hydatidiform mole (PHM) Hum Pathol. 2000;31:914–924. doi: 10.1053/hupa.2000.9085. [DOI] [PubMed] [Google Scholar]
- 8.Chiang S, Fazlollahi L, Nguyen A, et al. Diagnosis of hydatidiform moles by polymorphic deletion probe fluorescence in situ hybridization. J Mol Diagn. 2011;13:406–415. doi: 10.1016/j.jmoldx.2011.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chilosi M, Piazzola E, Lestani M, et al. Differential expression of p57kip2, a maternally imprinted cdk inhibitor, in normal human placenta and gestational trophoblastic disease. Lab Invest. 1998;78:269–276. [PubMed] [Google Scholar]
- 10.Choi-Hong SR, Genest DR, Crum CP, et al. Twin pregnancies with complete hydatidiform mole and coexisting fetus: use of fluorescent in situ hybridization to evaluate placental X- and Y-chromosomal content. Hum Pathol. 1995;26:1175–1180. doi: 10.1016/0046-8177(95)90189-2. [DOI] [PubMed] [Google Scholar]
- 11.Conran RM, Hitchcock CL, Popek EJ, et al. Diagnostic considerations in molar gestations. Hum Pathol. 1993;24:41–48. doi: 10.1016/0046-8177(93)90061-k. [DOI] [PubMed] [Google Scholar]
- 12.Crisp H, Burton JL, Stewart R, et al. Refining the diagnosis of hydatidiform mole: image ploidy analysis and p57KIP2 immunohistochemistry. Histopathology. 2003;43:363–373. doi: 10.1046/j.1365-2559.2003.01716.x. [DOI] [PubMed] [Google Scholar]
- 13.DeScipio C, Haley L, Beierl K, et al. Diandric triploid hydatidiform mole with loss of maternal chromosome 11. Am J Surg Pathol. 2011;35:1586–1591. doi: 10.1097/PAS.0b013e31822d5cff. [DOI] [PubMed] [Google Scholar]
- 14.El-Maarri O, Seoud M, Riviere JB, et al. Patients with familial biparental hydatidiform moles have normal methylation at imprinted genes. Eur J Hum Genet. 2005;13:486–490. doi: 10.1038/sj.ejhg.5201353. [DOI] [PubMed] [Google Scholar]
- 15.Feltmate CM, Growdon WB, Wolfberg AJ, et al. Clinical characteristics of persistent gestational trophoblastic neoplasia after partial hydatidiform molar pregnancy. J Reprod Med. 2006;51:902–906. [PubMed] [Google Scholar]
- 16.Fisher RA, Hodges MD, Newlands ES. Familial recurrent hydatidiform mole: a review. J Reprod Med. 2004;49:595–601. [PubMed] [Google Scholar]
- 17.Fisher RA, Khatoon R, Paradinas FJ, et al. Repetitive complete hydatidiform mole can be biparental in origin and either male or female. Hum Reprod. 2000;15:594–598. doi: 10.1093/humrep/15.3.594. [DOI] [PubMed] [Google Scholar]
- 18.Fisher RA, Nucci MR, Thaker HM, et al. Complete hydatidiform mole retaining a chromosome 11 of maternal origin: molecular genetic analysis of a case. Mod Pathol. 2004;17:1155–1160. doi: 10.1038/modpathol.3800175. [DOI] [PubMed] [Google Scholar]
- 19.Fukunaga M. Flow cytometric and clinicopathologic study of complete hydatidiform moles with special reference to the significance of cytometric aneuploidy. Gynecol Oncol. 2001;81:67–70. doi: 10.1006/gyno.2000.6100. [DOI] [PubMed] [Google Scholar]
- 20.Fukunaga M. Immunohistochemical characterization of p57(KIP2) expression in early hydatidiform moles. Hum Pathol. 2002;33:1188–1192. doi: 10.1053/hupa.2002.129421. [DOI] [PubMed] [Google Scholar]
- 21.Fukunaga M. Immunohistochemical characterization of p57Kip2 expression in tetraploid hydropic placentas. Arch Pathol Lab Med. 2004;128:897–900. doi: 10.5858/2004-128-897-ICOPEI. [DOI] [PubMed] [Google Scholar]
- 22.Fukunaga M, Katabuchi H, Nagasaka T, et al. Interobserver and intraobserver variability in the diagnosis of hydatidiform mole. Am J Surg Pathol. 2005;29:942–947. doi: 10.1097/01.pas.0000157996.23059.c1. [DOI] [PubMed] [Google Scholar]
- 23.Gschwendtner A, Neher A, Kreczy A, et al. DNA ploidy determination of early molar pregnancies by image analysis: comparison to histologic classification. Arch Pathol Lab Med. 1998;122:1000–1004. [PubMed] [Google Scholar]
- 24.Hayward BE, De Vos M, Talati N, et al. Genetic and epigenetic analysis of recurrent hydatidiform mole. Hum Mutat. 2009;30:E629–E639. doi: 10.1002/humu.20993. [DOI] [PubMed] [Google Scholar]
- 25.Hoffner L, Dunn J, Esposito N, et al. P57KIP2 immunostaining and molecular cytogenetics: combined approach aids in diagnosis of morphologically challenging cases with molar phenotype and in detecting androgenetic cell lines in mosaic/chimeric conceptions. Hum Pathol. 2008;39:63–72. doi: 10.1016/j.humpath.2007.05.010. [DOI] [PubMed] [Google Scholar]
- 26.Hoffner L, Parks WT, Swerdlow SH, et al. Simultaneous detection of imprinted gene expression (p57(KIP2)) and molecular cytogenetics (FICTION) in the evaluation of molar pregnancies. J Reprod Med. 2010;55:219–228. [PubMed] [Google Scholar]
- 27.Howat AJ, Beck S, Fox H, et al. Can histopathologists reliably diagnose molar pregnancy? J Clin Pathol. 1993;46:599–602. doi: 10.1136/jcp.46.7.599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Javey H, Borazjani G, Behmard S, et al. Discrepancies in the histological diagnosis of hydatidiform mole. Br J Obstet Gynaecol. 1979;86:480–483. doi: 10.1111/j.1471-0528.1979.tb10793.x. [DOI] [PubMed] [Google Scholar]
- 29.Jun SY, Ro JY, Kim KR. p57kip2 is useful in the classification and differential diagnosis of complete and partial hydatidiform moles. Histopathology. 2003;43:17–25. doi: 10.1046/j.1365-2559.2003.01667.x. [DOI] [PubMed] [Google Scholar]
- 30.Kaiser-Rogers KA, McFadden DE, Livasy CA, et al. Androgenetic/biparental mosaicism causes placental mesenchymal dysplasia. J Med Genet. 2006;43:187–192. doi: 10.1136/jmg.2005.033571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Keep D, Zaragoza MV, Hassold T, et al. Very early complete hydatidiform mole. Hum Pathol. 1996;27:708–713. doi: 10.1016/s0046-8177(96)90402-5. [DOI] [PubMed] [Google Scholar]
- 32.Kerkmeijer LG, Massuger LF, Ten Kate-Booij MJ, et al. Earlier diagnosis and serum human chorionic gonadotropin regression in complete hydatidiform moles. Obstet Gynecol. 2009;113:326–331. doi: 10.1097/AOG.0b013e3181945a4f. [DOI] [PubMed] [Google Scholar]
- 33.Kipp BR, Ketterling RP, Oberg TN, et al. Comparison of fluorescence in situ hybridization, p57 immunostaining, flow cytometry, and digital image analysis for diagnosing molar and nonmolar products of conception. Am J Clin Pathol. 2010;133:196–204. doi: 10.1309/AJCPV7BRDUCX0WAQ. [DOI] [PubMed] [Google Scholar]
- 34.Kou YC, Shao L, Peng HH, et al. A recurrent intragenic genomic duplication, other novel mutations in NLRP7 and imprinting defects in recurrent biparental hydatidiform moles. Mol Hum Reprod. 2008;14:33–40. doi: 10.1093/molehr/gam079. [DOI] [PubMed] [Google Scholar]
- 35.Lage JM, Mark SD, Roberts DJ, et al. A flow cytometric study of 137 fresh hydropic placentas: correlation between types of hydatidiform moles and nuclear DNA ploidy. Obstet Gynecol. 1992;79:403–410. doi: 10.1097/00006250-199203000-00016. [DOI] [PubMed] [Google Scholar]
- 36.Lee J, Fung KP. Confidence interval of the kappa coefficient by bootstrap resampling. Psychiatry Res. 1993;49:97–98. doi: 10.1016/0165-1781(93)90033-d. [DOI] [PubMed] [Google Scholar]
- 37.LeGallo RD, Stelow EB, Ramirez NC, et al. Diagnosis of hydatidiform moles using p57 immunohistochemistry and HER2 fluorescent in situ hybridization. Am J Clin Pathol. 2008;129:749–755. doi: 10.1309/7XRL378C22W7APBT. [DOI] [PubMed] [Google Scholar]
- 38.Lescoat D, Jouan H, Loeuillet-Olivo L, et al. Fluorescent in situ hybridization (FISH) on paraffin-embedded placental tissues as an adjunct for understanding the etiology of early spontaneous abortion. Prenat Diagn. 2005;25:314–317. doi: 10.1002/pd.1132. [DOI] [PubMed] [Google Scholar]
- 39.McConnell TG, Murphy KM, Hafez M, et al. Diagnosis and subclassification of hydatidiform moles using p57 immunohistochemistry and molecular genotyping: validation and prospective analysis in routine and consultation practice settings with development of an algorithmic approach. Am J Surg Pathol. 2009;33:805–817. doi: 10.1097/PAS.0b013e318191f309. [DOI] [PubMed] [Google Scholar]
- 40.McConnell TG, Norris-Kirby A, Hagenkord JM, et al. Complete hydatidiform mole with retained maternal chromosomes 6 and 11. Am J Surg Pathol. 2009;33:1409–1415. doi: 10.1097/PAS.0b013e3181a90e01. [DOI] [PubMed] [Google Scholar]
- 41.Medeiros F, Callahan MJ, Elvin JA, et al. Intraplacental choriocarcinoma arising in a second trimester placenta with partial hydatidiform mole. Int J Gynecol Pathol. 2008;27:247–251. doi: 10.1097/PGP.0b013e3181577dc8. [DOI] [PubMed] [Google Scholar]
- 42.Merchant SH, Amin MB, Viswanatha DS, et al. p57KIP2 immunohistochemistry in early molar pregnancies: emphasis on its complementary role in the differential diagnosis of hydropic abortuses. Hum Pathol. 2005;36:180–186. doi: 10.1016/j.humpath.2004.12.007. [DOI] [PubMed] [Google Scholar]
- 43.Messerli ML, Parmley T, Woodruff JD, et al. Inter- and intrapathologist variability in the diagnosis of gestational trophoblastic neoplasia. Obstet Gynecol. 1987;69:622–626. [PubMed] [Google Scholar]
- 44.Murphy KM, McConnell TG, Hafez MJ, et al. Molecular genotyping of hydatidiform moles: analytic validation of a multiplex short tandem repeat assay. J Mol Diagn. 2009;11:598–605. doi: 10.2353/jmoldx.2009.090039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Murphy KM, Ronnett BM. Molecular analysis of hydatidiform moles: utilizing p57 immunohistochemistry and molecular genotyping to refine morphologic diagnosis. Pathol Case Rev. 2010;15:126–134. [Google Scholar]
- 46.Norris-Kirby A, Hagenkord JM, Kshirsagar MP, et al. Abnormal villous morphology associated with triple trisomy of paternal origin. J Mol Diagn. 2010;12:525–529. doi: 10.2353/jmoldx.2010.090184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Palmieri C, Fisher RA, Sebire NJ, et al. Placental site trophoblastic tumour arising from a partial hydatidiform mole. Lancet. 2005;366:688. doi: 10.1016/S0140-6736(05)67143-7. [DOI] [PubMed] [Google Scholar]
- 48.Popiolek DA, Yee H, Mittal K, et al. Multiplex short tandem repeat DNA analysis confirms the accuracy of p57(KIP2) immunostaining in the diagnosis of complete hydatidiform mole. Hum Pathol. 2006;37:1426–1434. doi: 10.1016/j.humpath.2006.04.030. [DOI] [PubMed] [Google Scholar]
- 49.Redline RW, Hassold T, Zaragoza M. Determinants of villous trophoblastic hyperplasia in spontaneous abortions. Mod Pathol. 1998;11:762–768. [PubMed] [Google Scholar]
- 50.Romaguera RL, Rodriguez MM, Bruce JH, et al. Molar gestations and hydropic abortions differentiated by p57 immunostaining. Fetal Pediatr Pathol. 2004;23:181–190. doi: 10.1080/15227950490890351. [DOI] [PubMed] [Google Scholar]
- 51.Ronnett BM, DeScipio C, Murphy KM. Hydatidiform moles: ancillary techniques to refine diagnosis. Int J Gynecol Pathol. 2011;30:101–116. doi: 10.1097/PGP.0b013e3181f4de77. [DOI] [PubMed] [Google Scholar]
- 52.Sarmadi S, Izadi-Mood N, Abbasi A, et al. p57KIP2 immunohistochemical expression: a useful diagnostic tool in discrimination between complete hydatidiform mole and its mimics. Arch Gynecol Obstet. 2011;283:743–748. doi: 10.1007/s00404-010-1433-1. [DOI] [PubMed] [Google Scholar]
- 53.Sebire NJ, Fisher RA, Foskett M, et al. Risk of recurrent hydatidiform mole and subsequent pregnancy outcome following complete or partial hydatidiform molar pregnancy. BJOG. 2003;110:22–26. [PubMed] [Google Scholar]
- 54.Seckl MJ, Fisher RA, Salerno G, et al. Choriocarcinoma and partial hydatidiform moles. Lancet. 2000;356:36–39. doi: 10.1016/S0140-6736(00)02432-6. [DOI] [PubMed] [Google Scholar]
- 55.Slim R, Mehio A. The genetics of hydatidiform moles: new lights on an ancient disease. Clin Genet. 2007;71:25–34. doi: 10.1111/j.1399-0004.2006.00697.x. [DOI] [PubMed] [Google Scholar]
- 56.Sumithran E, Cheah PL, Susil BJ, et al. Problems in the histological assessment of hydatidiform moles: a study on consensus diagnosis and ploidy status by fluorescent in situ hybridisation. Pathology. 1996;28:311–315. doi: 10.1080/00313029600169254. [DOI] [PubMed] [Google Scholar]
- 57.Surti U, Hoffner L, Kolthoff M, et al. Persistent gestational trophoblastic disease after an androgenetic/biparental fetal chimera: a case report and review. Int J Gynecol Pathol. 2006;25:366–372. doi: 10.1097/01.pgp.0000215295.45738.ed. [DOI] [PubMed] [Google Scholar]
- 58.Van de Kaa CA, Robben JC, Hopman AH, et al. Complete hydatidiform mole in twin pregnancy: differentiation from partial mole with interphase cytogenetic and DNA cytometric analyses on paraffin embedded tissues. Histopathology. 1995;26:123–129. doi: 10.1111/j.1365-2559.1995.tb00641.x. [DOI] [PubMed] [Google Scholar]
- 59.Van den Veyver I, Al-Hussaini TK. Biparental hydatidiform moles: a maternal effect mutation affecting imprinting in the offspring. Hum Reprod Update. 2006;12:233–242. doi: 10.1093/humupd/dmk005. [DOI] [PubMed] [Google Scholar]
- 60.Wielsma S, Kerkmeijer L, Bekkers R, et al. Persistent trophoblast disease following partial molar pregnancy. Aust N Z J Obstet Gynaecol. 2006;46:119–123. doi: 10.1111/j.1479-828X.2006.00539.x. [DOI] [PubMed] [Google Scholar]
- 61.Williams D, Hodgetts V, Gupta J. Recurrent hydatidiform moles. Eur J Obstet Gynecol Reprod Biol. 2010;150:3–7. doi: 10.1016/j.ejogrb.2010.01.003. [DOI] [PubMed] [Google Scholar]
- 62.Yver M, Carles D, Bloch B, et al. Determination of DNA ploidy by fluorescence in situ hybridization (FISH) in hydatidiform moles: evaluation of FISH on isolated nuclei. Hum Pathol. 2004;35:752–758. doi: 10.1016/j.humpath.2004.01.020. [DOI] [PubMed] [Google Scholar]