Abstract
Peptides were immobilized onto superparamagnetic beads via photocleavable linkers. This enabled simple, rapid, and label-free protein kinase assays via MALDI-TOF MS detection of substrate peptide phosphorylation. Abltide, a model substrate for the Abl protein tyrosine kinase model, was coupled onto amine-terminated beads, incubated with ATP and recombinant c-Abl kinase, and released and further detected to determine phosphorylation. Abltide phosphorylation was found to depend significantly on the length and composition of linkers to the bead surface. Inserting a diblock spacer of poly(glycine) and poly(ethylene glycol) segments markedly enhanced phosphorylation. To validate the assay, the activity of two small-molecule kinase inhibitors, imatinib and dasatinib, which target the oncogenic mutant tyrosine kinase Bcr-Abl to treat chronic myeloid leukemia (CML), was tested. Examining inhibition of the purified c-Abl or Bcr-Abl in K562 CML cell extracts, IC50 values were determined to be consistent with the literature. This simple, label-free, MALDI-based protein kinase assay can be readily adapted to allow multiplexed assays of multiple peptide substrates and/or analysis of alternative post-translational modifications as a tool for drug discovery and clinical testing.
INTRODUCTION
Protein kinases play critical roles in cellular signal transduction by regulating intracellular processes such as cell growth, proliferation, differentiation, and apotosis (1–3). Deregulation of protein kinase activities and resulting perturbations of downstream targets in phosphorylation cascades have been implicated as underlying causes of a number of diseases, including cancer, diabetes, and chronic inflammation (4–7). Thus, protein kinases have emerged as a major class of drug targets, leading to several small-molecule kinase inhibitors already in use, and many more in clinical trials (8–12). Multiple technologies have been developed for analyzing protein kinase activities and screening of small molecules for inhibition of protein kinases (13–23).
A key challenge in protein kinase assays lies in the detection of the formation of the phosphorylated product by transfer of the ATP γ-phosphate to the unphosphorylated substrate. A reasonable goal is to detect the activity and inhibition of a kinase in its cellular context, but this requires extraordinary sensitivity and specificity, given the ubiquitous nature of protein phosphorylation and the expression of hundreds of different kinases in many cells. Heterogeneous kinase assays, where kinase-specific protein or peptide substrates are tethered to the surface of solid supports, have unique advantages over homogeneous assays for this purpose. The added steps of substrate immobilization and separation and/or washing are offset by enhanced sensitivity and specificity offered by ready discrimination of phosphorylated products from the many phosphorylated proteins in a cell extract. Diverse strategies are described for immobilization of peptides or proteins on solid supports, including in situ peptide synthesis (24, 25), noncovalent attachment via streptavidin-biotin interaction, and covalent chemistries such as NHS-amine reaction (26), Diel-Alder reaction (13), native chemical ligation (27), and Michael addition (28–30). However, direct attachment of peptide substrates to the support surface may limit accessibility of kinases to their substrates. Indeed, inserting a spacer between the peptide substrate and the support surface can significantly improve phosphorylation efficiency and reduce background (31–34).
Most heterogeneous kinase assays adapted to screening exploit indirect detection of phosphorylation via labels (35–37). Traditional kinase assays based on transfer of radioactive (thio)phosphate from γ-labeled ATP to peptide or protein substrates are sensitive and quantitative but not well-suited to analysis of cell extracts. Antibodies that recognize substrate phosphopeptides offer sufficient sensitivity and specificity but require additional binding and washing steps and offer limited quantitation. An attractive label-free approach is mass spectrometry (MS), which offers independent, quantitative detection of both phosphorylated and unphosphorylated substrate, and offers potential for multiplexed assays with high sensitivity, selectivity, and speed (38, 39). Direct MS analysis of kinase reaction mixtures suffers from signal suppression due to salt and detergent interference and is confounded by the complexity of cell extracts. Direct matrix-assisted laser desorption ionization (MALDI) analysis of substrates immobilized via tethering to self-assembled monolayers (40) or onto gold nanoparticles coated onto glass slides (32) has demonstrated the potential of mass spectrometry for heterogeneous kinase assays.
Photocleavable linkers offer a number of advantages, because light is easy to apply, relatively harmless to living organisms, and importantly controllable both spatially and temporally (41). o-Nitrobenzyl ether derivatives are especially convenient photocleavable moieties and have been used in a variety of fields including live cell culture and imaging due to unique properties that include stability under ambient light, clean cleavage via two-photon photolysis mechanism upon exposure to UV irradiation, and fast fragmentation reactions (within nanoseconds) upon photoexcitation (42–44). Previously, we reported a chip-based kinase assay using substrate peptides copolymerized into a hydrogel via a UV-photocleavable linker (45). After release by 365 nm light, peptide phosphorylation can be readily detected by MALDI-TOF MS. Although hydrogel arrays offer a robust and inert surface that can support a high peptide density, adaptation to high-throughput screening is not straightforward. As an alternative strategy to decrease background after phosphorylation, biotin-tagged peptide substrates can be captured onto streptavidin-coated magnetic beads (46) prior to MALDI-TOF analysis. We recently extended this work to create a practical kinase activity screening assay and demonstrated rapid determination of IC50 for small-molecule kinase inhibitors (47). In this study, we have used Fmoc solid-phase peptide synthesis to install diblock linkers terminated by a photocleavable group onto paramagnetic particles, tethered substrate peptides, and validated these conjugates as reporters of protein kinase activity and inhibition using a MALDI-TOF readout (Figure 1). Our data suggest that this approach may be readily adapted to high-throughput, highly multiplexed analysis of kinases in whole cell lysates.
Figure 1.

Schematic representation of the new label-free MALDI-based protein kinase assay technique using magnetic beads as a solid-phase analysis platform to facilitate separation and photocleavable linkers as a trigger to release peptide substrates under UV light for MALDI detection. Both phosphorylated and unphosphorylated substrate peptides can be detected by MALDI with a characteristic 80 Da difference in mass between the two peaks.
EXPERIMENTAL PROCEDURES
Materials
Fmoc-aminoethyl photolinker (4-[4-(1-(Fmoc-amino)ethyl)-2-methoxy-5-nitrophenoxy)] butanoic acid, ≥96%, Novabiochem), Fmoc-Gly-OH (N-α-Fmoc-glycine, Peptides International), HCTU coupling reagent (1-[bis(dimethylamino)methylene]-5-chlorobenzotriazolium-3-oxide hexafluorophosphate, Peptides International), 4-methylmorpholine (NMM, 99.5+%, Sigma-Aldrich), piperidine (99%, Sigma-Aldrich), N,N-dimethylformamide (DMF, anhydrous, 99.8%, Acros Organics), N-[p-maleimidophe-nyl]isocyanate (PMPI, Pierce), N-Fmoc-amido-dPEG4-acid, N-Fmoc-amido-dPEG8-acid, N-Fmoc-amido-dPEG12-acid (Quanta Biodesign), BcMag amine-terminated magnetic beads (1 μm, Bioclone Inc.), BupH phosphate buffered saline (0.1 M sodium phosphate, 0.15 M NaCl, pH 7.2) (Pierce), and recombinant c-Abl (Upstate) were used as received. Imatinib mesylate (Novartis) was provided by Wendy Stock (The University of Chicago). Dasatinib (free base) was purchased from LC Laboratories (Woburn, MA). Cysteineterminated G4SG3 Abltide substrate (CGGGGSGGGKGEAIYAAPFAKKKG) and K562 cell lysate (human chronic myelogenous leukemia cell extract) were prepared as reported previously (28).
Surface Modification of Magnetic Beads
BcMag particles are uniform superparamagnetic beads consisting of silica-coated iron oxide, presenting surface primary amino groups (MNP@NH2). The beads were washed in turn with water and DMF, and diluted to a concentration of 5 mg/mL in DMF. 500 μL of washed beads (equivalent to 0.625 μmol NH2 groups) was added to a mixture of 100 μL of 240 mM Fmoc-Gly-OH and 900 μL of coupling reagent solution containing 0.76 M HCTU, 1.60 M NMM in DMF. The coupling reaction mixture was rotated at room temperature for 40 min; then, the beads were separated and washed with DMF three times to remove excess reactants. The Fmoc protection group was removed by treatment with 20% piperidine in DMF for 30 min. After deprotection, beads were washed with DMF three times. Four additional cycles of coupling and deprotection reactions were performed in the same manner. As a result, beads with surface poly(glycine) spacers (MNP@Gly5-NH2) were obtained and further washed with DMF.
300 μL of surface poly(glycine) spacer-modified beads (MNP@ Gly5-NH2) in DMF (equivalent 0.375 μmol of free NH2 groups) was added to a mixture of 20 μL of 400 mM N-Fmoc-amido-dPEG4-acid and 780 μL of coupling reagent solution containing 0.76 M HCTU, 1.60 M NMM in DMF. The coupling reaction was again carried out at room temperature for 40 min, and deprotection for 30 min, as above. After washing with DMF, beads with surface spacers consisting of both poly(glycine) and poly-(ethylene glycol) segments (MNP@Gly5-PEG4-NH2) were produced.
Superparamagnetic beads with poly(ethylene glycol) spacers of different length (MNP@PEGn-NH2, n = 4, 8, 12) were prepared using three different PEG linkers (N-Fmoc-amido-dPEG4-acid, N-Fmoc-amido-dPEG8-acid, and N-Fmoc-amido-dPEG12-acid) according to the above-described procedure.
Preparation of Photocleavable Abltide-Conjugated Superparamagnetic Beads
Beads with different surface spacer structures (MNP@NH2, MNP@Gly5-NH2, MNP@Gly5-PEG4-NH2, and MNP@PEGn-NH2, n = 4, 8, 12) were used for preparation of photocleavable Abltide-conjugated beads with different spacer linkers. 300 μL of each kind of bead with amino groups (equivalent to 0.375 μmol of free NH2 groups) were added to a mixture of 60 μL of 180 mM Fmoc-aminoethyl photolinker in DMF and 540 μL of coupling reagent solution containing 0.76 M HCTU, 1.60 M NMM in DMF. The coupling reaction was carried out at room temperature for 40 min. Excess reactants were removed by washing with DMF. The removal of the Fmoc group was completed by treating the beads with 20% piperidine in DMF for 30 min. The deprotected beads were washed in turn with DMF and anhydrous DMF, and reacted with the heterobifunctional cross-linker PMPI in a 1:30 ratio of amino group to isocyanate group over 13 h. After completion of the reaction, beads were washed successively with DMF and BupH PBS buffer (pH 7.2). The maleimide-functionalized beads were reacted with the cysteine-terminated G4SG3 Abltide (3.33 equiv to maleimide groups) in BupH PBS buffer medium for 2.5 h. The resulting photocleavable Abltide-conjugated superparamagnetic beads were washed with water and stored in water at 4 °C.
In Vitro Kinase Assay
On-bead phosphorylation reactions by c-Abl kinase and K562 cell lysates were performed. Kinase assays using c-Abl were as reported (47). For kinase assays using K562 cell lyaste, a typical 50 μL kinase reaction contained 50 mM Tris-Cl (pH 7.5), 10 mM MgCl2, 1 mM EGTA, 2 mM DTT, 0.01% Brij-35, 50 μM ATP, 0.50 mg/mL BSA-blocked photocleavable Abltide beads with 100 μg/mL K562 cell lysate, and 1× complete protease inhibitor cocktail (Roche) and was incubated at 37 °C for 1 h. After the kinase reactions, beads were separated from the reaction buffer, washed twice with water, and subjected to MALDI-TOF MS analysis.
In Vitro Inhibition Assay
Dasatinib and imatinib were assayed in vitro using both recombinant c-Abl and K562 cell lysate. Assays with c-Abl or K562 cell lysate were as above with the addition of varying concentrations of inhibitors. A series of kinase reactions with 50 μM ATP and 0, 1, 10, 100, 500, 103, 104, or 105 nM Imatinib and 10 μM ATP and 0, 0.2, 0.5, 1, 2, 5, 10, 100, or 1000 nM Dasatinib were performed for 1 h using c-Abl at 30 °C and K562 cell lysate at 37 °C, respectively. After the kinase reactions, the beads were separated and washed with water twice, and stored in 5 μL of water at 4 °C. The degree of phosphorylation for each sample was determined by MALDI-TOF MS.
MALDI-TOF MS Analysis
Sample preparation for MALDI-TOF analysis was as described (47), except that the dry spotted samples were irradiated by UV light of 366 nm for 20 min prior to addition of R-cyano-4-hydroxycinnamic acid (CHCA) matrix solution onto each sample spot and analysis on a Voyager DE. The phosphorylation ratio of Abltide immobilized on magnetic beads was determined according to the previously described computational method (47).
RESULTS
We used solid-phase peptide synthesis to immobilize a tyrosine kinase peptide substrate with a photocleavable linker onto silica-coated superparamagnetic beads to enable simple and straightforward kinase assays (Figure 1). After the kinase reaction, both phosphorylated and unphosphorylated substrate peptide are released from the beads by UV photocleavage and then detected by MALDI-TOF MS. The phosphorylated and unphosphorylated peptide peaks are offset by 80 Da (48, 49) and the degree of phosphorylation can be estimated by the relative intensities of the two peaks.
To test the new protein kinase assay technique, we chose the model system of the oncogenic protein kinase, Bcr-Abl, and a synthetic substrate peptide, Abltide. Bcr-Abl is a fusion gene product resulting from the Ph1 reciprocal 9:22 translocation (50, 51), and its constitutive activity is responsible for cancer onset and progression in CML (52). Bcr-Abl kinase from cell extracts of the K562 cell line derived from human chronic myeloid leukemia and its counterpart recombinant c-Abl kinase were both used for this study. With this model system, Abltide was readily immobilized on magnetic beads and phosphorylated by kinases. After the reaction, MALDI-TOF MS could detect both phosphorylated and unphosphorylated peptides cleaved from the magnetic beads upon UV light exposure (λ = 366 nm).
Currently, two kinds of spacer linkers have been extensively used to distance peptides from solid supports: (i) poly(ethylene glycol) (PEG) linkers, well-known for their ability to prevent nonspecific interactions, also act as hydrophilic spacers minimizing any detrimental interaction between the attached peptide and solid surface (53–55); and (ii) glycine spacers, generally inserted into the peptide sequence, can improve the accessibility of enzymes to peptide substrates (56). To optimize the kinase reactions, we investigated the effect of two different linkers on phosphorylation of peptide substrates. First, the surface of amino-functionalized silica-coated superparamagnetic beads (MNP@NH2) was modified by introducing a five glycine spacer (Gly5) by a five-step standard solid-phase peptide synthesis (SPPS) procedure shown in Figure 2A. Glycine spacer-modified beads (MNP@Gly5-NH2) were further modified by insertion of a PEG linker using N-Fmoc-amino-dPEG4 acid according to the standard SPPS protocol (Figure 2B). As a result, dual spacer-modified beads (MNP@Gly5-PEG4-NH2) were produced. For comparison, the beads were also modified by single PEG spacers of varying lengths (Figure 2C). The beads with different surface spacers, MNP@NH2, MNP@Gly5-NH2, MNP@PEGn-NH2, and MNP@Gly5-PEG4-NH2, were then coupled to an Fmocphotolinker and deprotected and the amino groups reacted with N-[p-maleimidophenyl]isocyanate to form surface maleimide-functionalized beads. The maleimide-functionalized beads were then reacted with the amino-terminal cysteine on CG4SG3-Abltide (CGGGGSGGGKEAIYAAPFAKKKG) in PBS buffer to produce photocleavable Abltide-conjugated superparamagnetic beads with different surface spacer linkers (Figure 3). These beads were examined in kinase assays.
Figure 2.

Structural design and synthesis of magnetic beads bearing different surface spacers via the solid-phase peptide synthesis (SPPS) technique. (A) Pentaglycine spacers. (B) Diblock spacers consisting of PEG and pentaglycine segments. (C) Single PEG spacers with different lengths.
Figure 3.

General synthetic route for photocleavable Abltide-conjugated magnetic beads with different surface spacer structures (pentaglycine spacer, single PEG spacers with different lengths, and diblock spacer consisting of PEG and polyglycine) through successive coupling-deprotection, amino-isocyanate, and highly selective thiol-maleimide reactions starting from magnetic beads bearing different surface spacers (denoted by the green bar).
Abltide conjugated to superparamagnetic beads was phosphorylated in kinase buffer for 1 h with c-Abl at 30 °C or K562 cell lysate at 37 °C. The degree of phosphorylation of peptide substrates conjugated to beads via different linkers was determined by the relative intensity of phosphorylated and unphosphorylated peptide peaks in the MALDI spectrum, as summarized in Figure 4. It is clear that both single glycine and PEG spacers are able to improve the phosphorylation efficiency, most likely due to the increase in molecular mobility of the substrate peptide and kinase accessibility to peptide. The number of PEG units in the spacer had a measurable effect on phosphorylation, with longer spacers yielding a slightly higher phosphorylation ratio. We found that insertion of PEG12 between the peptide and solid support yielded optimal phosphorylation efficiency, consistent with Katayama et al. (33). In particular, the diblock spacer consisting of poly(glycine) and PEG markedly increased the phosphorylation efficiency of peptide substrates, perhaps due to the complementary features of the two spacers. Therefore, the photocleavable Abltide-conjugated magnetic beads with a diblock spacer of poly(glycine) and PEG were used for the kinase inhibition assays.
Figure 4.

Effect of the surface spacer between the Abltide substrate and magnetic bead support on phosphorylation of Abltide by both c-Abl at 30 °C and K562 cell lysate at 37 °C for 1 h, respectively. The general structure of photocleavable abltide-conjugated magnetic beads was given in the upper panel, and the green bar represents different surface spacer structures shown in the lower left panel. The phosphoryaltion ratios corresponding to each surface spacer by both c-Abl and K562 cell lysate were determined by MALDI according to the intensity of phosphorylated and unphosphorylated peaks. Each experiment represented in the lower right panel was performed in triplicate.
A common application for assays reporting the phosphorylation of peptides is to evaluate the inhibitory ability of different drugs on protein kinases. Two clinically useful Abl kinase inhibitors, imatinib and dasatinib, have been used in the treatment of CML via targeting the oncogenic Bcr-Abl kinase. Imatinib and dasatinib were evaluated using our magnetic bead-based kinase assays. The Abltide-conjugated beads were incubated for 1 h with c-Abl kinase at 30 °C or K562 cell lysate at 37 °C with 50 μM ATP + 0–100 μM for imatinib, and with 10 μM ATP + 0–1 μM for dasatinib. The peptides were then released and analyzed by MALDI-TOF MS. Figure 5A shows a representative set of MALDI spectra of Abltide released from beads after phosphorylation by c-Abl kinase at different imatinib concentrations. The expected 80 Da increase in mass relative to the unmodified peptide was observed after peptides were phosphorylated, yielding two singly charged peaks at m/z = 2478.22 and 2398.35. With no addition of inhibitor, the Abltide substrate was 86% phosphorylated, calculated according to the relative intensity of the phosphorylated and unphosphorylated peptide ions. The c-Abl kinase activity was gradually inhibited with the increase of imatinib concentration, and fully blocked at 10 μM imatinib (Figure 5A). There is a minor peak (denoted by a star) at m/z = 2355.70 attributed to fragmentation at a site other than the site predicted for photocleavage (Figure 5B). We found that minor peak intensity depends on the completion of photocleavage prior to analysis, as well as the amount of matrix spotted and laser intensity during MALDI analysis. We found that the relative intensity ratio of the minor peaks formed from phosphorylated and unphosphorylated peptides was equal to the major ones. Therefore, the phosphorylation ratio of Abltide substrate was calculated according to the relative intensities of the major phosphorylated and unphosphorylated peaks. A dose-dependent effect on phosphorylation of Abltide by c-Abl or K562 cell lysate was observed for each inhibitor (Figure 6), and IC50 values were calculated by fitting to sigmoidal dose–response curves (Table 1). The IC50 values of imatinib against c-Abl and K562 cell lysate were 0.53 and 1.10 μM, respectively. These values are higher than the literature IC50 values, perhaps reflecting the use of a higher ATP concentration, 50 μM, in our experiments. However, for dasatinib, where we used 10 μM ATP, the IC50 values against c-Abl and K562 cell lysate of 12.0 and 2.3 nM are close to reported values.
Figure 5.
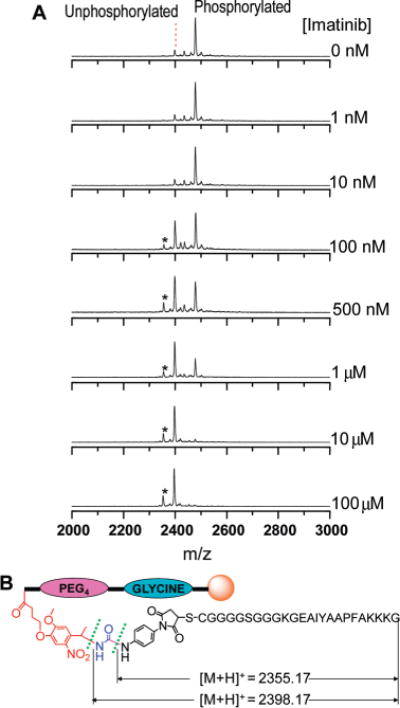
(A) A representative set of MALDI-TOF MS spectra of the photocleavable Abltide-conjugated magnetic beads after 1 h phosphorylation by c-Abl at different imatinib concentrations. The peaks of phosphorylated and unphorylated peptide substrates were observed at m/z = 2478.22 and 2398.35, respectively. The minor peak (denoted by a star) at m/z = 2355.70 is attributed to fragmentation at a site other than that predicted for photocleavage. (B) Predicted photocleavage and minor cleavage sites and theoretical exact mass of their corresponding generated peptide substrate fragments.
Figure 6.

Inhibition assays using imatinib and dasatinib against c-Abl kinase and K562 cell lysate using the photocleavable substrate on magnetic beads, and MALDI-TOF MS analysis. The degree of phosphorylation was calculated from the ratio of phosphorylated and unphosphorylated peak intensities. Each data point is the average of three spectra. ATP concentrations for imatinib and dasatinib inhibition assays were 50 μM and 10 μM, respectively.
Table 1.
IC50 of Inhibitors
DISCUSSION
Although much progress has been made in developing protein kinase assay techniques for understanding biological regulatory pathways, many challenges still exist (57). Only a fraction of techniques are compatible with cell lysates or within cells (57). It has become clear that MALDI-based label-free heterogeneous kinase assays may have a significant application value in this field. In this study, we explored the use of magnetic beads for heterogeneous kinase assays. We chose magnetic beads as solid supports, because they can greatly facilitate on-bead preparation of peptide conjugates and rapid separation of phosphorylated products as well as being amenable to automated processing. In addition, they can promote the ionization of phosphorylated products during the process of MALDI analysis and further increase assay sensitivity (58). However, the immobilization of peptide substrates on the solid support plays a key role in the overall heterogeneous kinase assays. There are many strategies for immobilization of peptide substrates onto solid supports (30, 59). However, our magnetic bead-based SPPS strategy has shown distinct advantages over the published approaches, because it conveniently inserts different spacers between the peptide substrate and magnetic beads.
We performed a systematic study by introducing different spacers between peptide substrates and magnetic beads to understand their influence on the kinase assays. For this purpose, poly(glycine), PEG spacers, and their combinations were employed. Without spacers, the phosphorylation ratios of peptide substrates were quite low. Both the single pentaglycine spacer and PEG spacer were able to enhance the phosphorylation ratios of peptide substrates. Notably, a diblock spacer consisting of polyglycine and PEG prepared in situ by the magnetic bead-based SPPS strategy greatly improved the phosphorylation ratios as compared with the other spacers due to the synergic effects of the two types of spacers. The introduction of the diblock spacer made the MALDI-based kinase assays work well with both recombinant kinases and cell lysates. As this new kinase assay technique can rapidly quantify protein kinase activities directly from complex cellular mixtures, it provides a reasonable strategy for the development of cell-based screens for drug discovery. Unlike traditional methods, MALDI can differentiate multiple peptides and their modified forms. Thus, the use of the label-free MALDI detection tool provides this new kinase assay with the potential for quantitative evaluation of the relative potencies of a drug to different kinases.
CONCLUSIONS
We have developed a new label-free protein kinase assay technique in which magnetic beads were used as an analytical platform to immobilize peptide substrates via photocleavable linkers and released by UV light for MALDI detection. Both poly(glycine) and PEG spacers are able to improve phosphorylation efficiency and a diblock poly(glycine) and PEG spacer was found to be even more efficient than any single spacers. This simple and efficient assay technique may be adapted to high-throughput screening and generalized to a wide range of multiple post-translational modification analysis.
Acknowledgments
We thank Wendy Stock for kindly providing imatinib and Jonathan Goya for his helpful comments on the manuscript. This work was supported by National Institutes of Health grants HG003864, GM074691, and CA126764.
LITERATURE CITED
- 1.Lu ZM, Hunter T. Degradation of activated protein kinases by ubiquitination. Annu Rev Biochem. 2009;78:435–475. doi: 10.1146/annurev.biochem.013008.092711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hunter T. Signaling - 2000 and beyond. Cell. 2000;100:113–127. doi: 10.1016/s0092-8674(00)81688-8. [DOI] [PubMed] [Google Scholar]
- 3.Manning G, Whyte DB, Martinez R, Hunter T, Sudarsanam S. The protein kinase complement of the human genome. Science. 2002;298:1912–1934. doi: 10.1126/science.1075762. [DOI] [PubMed] [Google Scholar]
- 4.Noble MEM, Endicott JA, Johnson LN. Protein kinase inhibitors: insights into drug design from structure. Science. 2004;303:1800–1805. doi: 10.1126/science.1095920. [DOI] [PubMed] [Google Scholar]
- 5.Alonso A, Sasin J, Bottini N, Friedberg I, Osterman A, Godzik A, Hunter T, Dixon J, Mustelin T. Protein tyrosine phosphatases in the human genome. Cell. 2004;117:699–711. doi: 10.1016/j.cell.2004.05.018. [DOI] [PubMed] [Google Scholar]
- 6.Cohen P. Protein kinases - the major drug targets of the twenty-first century? Nat Rev Drug Discovery. 2002;1:309–315. doi: 10.1038/nrd773. [DOI] [PubMed] [Google Scholar]
- 7.Swanson CD, Paniagua RT, Lindstrom TM, Robinson WH. Tyrosine kinases as targets for the treatment of rheumatoid arthritis. Nat Rev Rheumatol. 2009;5:317–324. doi: 10.1038/nrrheum.2009.82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhang JM, Yang PL, Gray NS. Targeting cancer with small molecule kinase inhibitors. Nat Rev Cancer. 2009;9:28–39. doi: 10.1038/nrc2559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Druker BJ, Tamura S, Buchdunger E, Ohno S, Segal GM, Fanning S, Zimmermann J, Lydon NB. Effects of a selective inhibitor of the Abl tyrosine kinase on the growth of Bcr-Abl positive cells. Nat Med. 1996;2:561–566. doi: 10.1038/nm0596-561. [DOI] [PubMed] [Google Scholar]
- 10.Grant S. Therapeutic protein kinase inhibitors. Cell Mol Life Sci. 2009;66:1163–1177. doi: 10.1007/s00018-008-8539-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Johnson LN. Protein kinase inhibitors: contributions from structure to clinical compounds. Q Rev Biophys. 2009;42:1–40. doi: 10.1017/S0033583508004745. [DOI] [PubMed] [Google Scholar]
- 12.Sebolt-Leopold JS, English JM. Mechanisms of drug inhibition of signalling molecules. Nature. 2006;441:457–462. doi: 10.1038/nature04874. [DOI] [PubMed] [Google Scholar]
- 13.Houseman BT, Huh JH, Kron SJ, Mrksich M. Peptide chips for the quantitative evaluation of protein kinase activity. Nat Biotechnol. 2002;20:270–274. doi: 10.1038/nbt0302-270. [DOI] [PubMed] [Google Scholar]
- 14.Rathore R, Corr J, Scott G, Vollmerhaus P, Greis KD. Development of an inhibitor screening platform via mass spectrometry. J Biomol Screen. 2008;13:1007–1013. doi: 10.1177/1087057108326143. [DOI] [PubMed] [Google Scholar]
- 15.Kondo N, Nishimura SI. MALDI-TOF mass-spectrometry-based versatile method for the characterization of protein kinases. Chem—Eur J. 2009;15:1413–1421. doi: 10.1002/chem.200801650. [DOI] [PubMed] [Google Scholar]
- 16.von Ahsen O, Bomer U. High-throughput screening for kinase inhibitors. ChemBioChem. 2005;6:481–490. doi: 10.1002/cbic.200400211. [DOI] [PubMed] [Google Scholar]
- 17.Shults MD, Janes KA, Lauffenburger DA, Imperiali B. A multiplexed homogeneous fluorescence-based assay for protein kinase activity in cell lysates. Nat Methods. 2005;2:277–283. doi: 10.1038/nmeth747. [DOI] [PubMed] [Google Scholar]
- 18.Zondlo SC, Gao F, Zondlo NJ. Design of an encodable tyrosine kinase-inducible domain: detection of tyrosine kinase activity by terbium luminescence. J Am Chem Soc. 2010;132:5619–5621. doi: 10.1021/ja100862u. [DOI] [PubMed] [Google Scholar]
- 19.Placzek EA, Plebanek MP, Lipchik AM, Kidd SR, Parker LL. A peptide biosensor for detecting intracellular Abl kinase activity using matrix-assisted laser desorption/ionization time-of-flight mass spectrometry. Anal Biochem. 2010;397:73–78. doi: 10.1016/j.ab.2009.09.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wu D, Sylvester JE, Parker LL, Zhou G, Kron SJ. Peptide reporters of kinase activity in whole cell lysates. Biopolym (Pept Sci) 2010;94:475–486. doi: 10.1002/bip.21401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mand MR, Wu D, Veach DR, Kron SJ. Cell treatment and lysis in 96-well filter-bottom plates for screening Bcr-Abl activity and inhibition in whole-cell extracts. J Biomol Screen. 2010;15:434–440. doi: 10.1177/1087057110363307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sylvester JE, Kron SJ. A bead-based activity screen for small-molecule inhibitors of signal transduction in chronic myelogenous leukemia cells. Mol Cancer Ther. 2010;9:1469–1481. doi: 10.1158/1535-7163.MCT-10-0157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kim M, Shin DS, Kim J, Yoon-Sik Lee. Substrate screening of protein kinases: Detection methods and combinatorial peptide libraries. Pept Sci. 2010 doi: 10.1002/bip.21506. [DOI] [PubMed] [Google Scholar]
- 24.Reineke U, Volkmer-Engert R, Schneider-Mergener J. Applications of peptide arrays prepared by the SPOT-technology. Curr Opin Biotechnol. 2001;12:59–64. doi: 10.1016/s0958-1669(00)00178-6. [DOI] [PubMed] [Google Scholar]
- 25.Fodor SPA, Read JL, Pirrung MC, Stryer L, Lu AT, Solas D. Light-directed, spatially addressable parallel chemical synthesis. Science. 1991;251:767–773. doi: 10.1126/science.1990438. [DOI] [PubMed] [Google Scholar]
- 26.MacBeath G, Schreiber SL. Printing proteins as microarrays for high-throughput function determination. Science. 2000;289:1760–1763. doi: 10.1126/science.289.5485.1760. [DOI] [PubMed] [Google Scholar]
- 27.Lesaicherre ML, Uttamchandani M, Chen GYJ, Yao SQ. Developing site-specific immobilization strategies of peptides in a microarray. Bioorg Med Chem Lett. 2002;12:2079–2083. doi: 10.1016/s0960-894x(02)00379-7. [DOI] [PubMed] [Google Scholar]
- 28.Wu D, Mand MR, Veach DR, Parker LL, Clarkson B, Kron SJ. A solid-phase Bcr-Abl kinase assay in 96-well hydrogel plates. Anal Biochem. 2008;375:18–26. doi: 10.1016/j.ab.2007.12.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Uttamchandani M, Yao SQ. Peptide microarrays: Next generation biochips for detection, diagnostics and high-throughput screening. Curr Pharm Des. 2008;14:2428–2438. doi: 10.2174/138161208785777450. [DOI] [PubMed] [Google Scholar]
- 30.Kohn M. Immobilization strategies for small molecule, peptide and protein microarrays. J Pept Sci. 2009;15:393–397. doi: 10.1002/psc.1130. [DOI] [PubMed] [Google Scholar]
- 31.Ding H, Prodinger WM, Kopecek J. Identification of CD21-binding peptides with phage display and investigation of binding properties of HPMA copolymer-peptide conjugates. Bioconjugate Chem. 2006;17:514–523. doi: 10.1021/bc0503162. [DOI] [PubMed] [Google Scholar]
- 32.Kim YP, Oh E, Oh YH, Moon DW, Lee TG, Kim HS. Protein kinase assay on peptide-conjugated gold nanoparticles by using secondary-ion mass spectrometric imaging. Angew Chem, Int Ed. 2007;46:6816–6819. doi: 10.1002/anie.200701418. [DOI] [PubMed] [Google Scholar]
- 33.Inamori K, Kyo M, Matsukawa K, Inoue Y, Sonoda T, Tatematsu K, Tanizawa K, Mori T, Katayama Y. Optimal surface chemistry for peptide immobilization in on-chip phosphorylation analysis. Anal Chem. 2008;80:643–650. doi: 10.1021/ac701667g. [DOI] [PubMed] [Google Scholar]
- 34.Ding H, Prodinger WM, Kopecek J. Two-step fluorescence screening of CD21-binding peptides with one-bead one-compound library and investigation of binding properties of N-(2-hydroxypropyl)methacrylamide copolymer-peptide conjugates. Biomacromolecules. 2006;7:3037–3046. doi: 10.1021/bm060508f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Li YJ, Xie WH, Fang GJ. Fluorescence detection techniques for protein kinase assay. Anal Bioanal Chem. 2008;90:2049–2057. doi: 10.1007/s00216-008-1986-z. [DOI] [PubMed] [Google Scholar]
- 36.Olive DM. Quantitative methods for the analysis of protein phosphorylation in drug development. Expert Rev Proteomics. 2004;1:327–341. doi: 10.1586/14789450.1.3.327. [DOI] [PubMed] [Google Scholar]
- 37.Ma HC, Deacon S, Horiuchi K. The challenge of selecting protein kinase assays for lead discovery optimization. Expert Opin Drug Discovery. 2008;3:607–621. doi: 10.1517/17460441.3.6.607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.McLachlin DT, Chait BT. Analysis of phosphorylated proteins and peptides by mass spectrometry. Curr Opin Chem Biol. 2001;5:591–602. doi: 10.1016/s1367-5931(00)00250-7. [DOI] [PubMed] [Google Scholar]
- 39.Reinders J, Sickmann A. State-of-the-art in phosphoproteomics. Proteomics. 2005;5:4052–4061. doi: 10.1002/pmic.200401289. [DOI] [PubMed] [Google Scholar]
- 40.Min DH, Tang WJ, Mrksich M. Chemical screening by mass spectrometry to identify inhibitors of anthrax lethal factor. Nat Biotechnol. 2004;22:717–723. doi: 10.1038/nbt973. [DOI] [PubMed] [Google Scholar]
- 41.Kostiainen MA, Smith DK, Ikkala O. Optically triggered release of DNA from multivalent dendrons by degrading and charge-switching multivalency. Angew Chem, Int Ed. 2007;46:7600–7604. doi: 10.1002/anie.200701200. [DOI] [PubMed] [Google Scholar]
- 42.Yan FN, Chen LH, Tang QL, Rong W. Synthesis and characterization of a photocleavable cross-linker and its application on tunable surface modification and protein photodelivery. Bioconjugate Chem. 2004;15:1030–1036. doi: 10.1021/bc049901d. [DOI] [PubMed] [Google Scholar]
- 43.Saran D, Burke DH. A versatile photocleavable bifunctional linker for facile synthesis of substrate-DNA conjugates for the selection of nucleic acid catalysts. Bioconjugate Chem. 2007;18:275–279. doi: 10.1021/bc060221f. [DOI] [PubMed] [Google Scholar]
- 44.Kloxin AM, Kasko AM, Salinas CN, Anseth KS. Photodegradable hydrogels for dynamic tuning of physical and chemical properties. Science. 2009;324:59–63. doi: 10.1126/science.1169494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Parker LL, Brueggemeier SB, Rhee WJ, Wu D, Kent SBH, Kron SJ, Palecek SP. Photocleavable peptide hydrogel arrays for MALDI-TOF analysis of kinase activity. Analyst. 2006;131:1097–1104. doi: 10.1039/b607180e. [DOI] [PubMed] [Google Scholar]
- 46.Kinumi T, Niki E, Shigeri Y, Matsumoto H. Affinity-tagged phosphorylation assay by matrix-assisted laser desorption/ionization time-of-flight mass spectrometry (ATPA-MALDI): Application to calcium/calmodulin-dependent protein kinase. J Biochem. 2005;138:791–796. doi: 10.1093/jb/mvi178. [DOI] [PubMed] [Google Scholar]
- 47.Zhou G, Sylvester JE, Wu D, Veach DR, Kron SJ. A magnetic bead-based protein kinase assay with dual detection techniques. Anal Biochem. 2010 doi: 10.1016/j.ab.2010.08.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Parker L, Engel-Hall A, Drew K, Steinhardt G, Helseth DL, Jabon D, McMurry T, Angulo DS, Kron SJ. Investigating quantitation of phosphorylation using MALDI-TOF mass spectrometry. J Mass Spectrom. 2008;43:518–527. doi: 10.1002/jms.1342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Annan RS, Carr SA. Phosphopeptide analysis by matrix-assisted laser desorption time-of-flight mass spectrometry. Anal Chem. 1996;68:3413–3421. doi: 10.1021/ac960221g. [DOI] [PubMed] [Google Scholar]
- 50.Rowley JD. New consistent chromosomal abnormality in chronic myelogenous leukemia identified by quinacrine fluorescence and giemsa staining. Nature. 1973;243:290–293. doi: 10.1038/243290a0. [DOI] [PubMed] [Google Scholar]
- 51.Nagar B, Hantschel O, Young MA, Scheffzek K, Veach D, Bornmann V, Clarkson B, Superti-Furga G, Kuriyan J. Structural basis for the autoinhibition of c-Abl tyrosine kinase. Cell. 2003;112:859–871. doi: 10.1016/s0092-8674(03)00194-6. [DOI] [PubMed] [Google Scholar]
- 52.Warmuth M, Danhauser-Riedl S, Hallek M. Molecular pathogenesis of chronic myeloid leukemia: implications for new therapeutic strategies. Ann Hematol. 1999;78:49–64. doi: 10.1007/s002770050473. [DOI] [PubMed] [Google Scholar]
- 53.Cheung CL, Camarero JA, Woods BW, Lin TW, Johnson JE, De Yoreo JJ. Fabrication of assembled virus nanostructures on templates of chemoselective linkers formed by scanning probe nanolithography. J Am Chem Soc. 2003;125:6848–6849. doi: 10.1021/ja034479h. [DOI] [PubMed] [Google Scholar]
- 54.Camarero JA, Kwon Y, Coleman MA. Chemoselective attachment of biologically active proteins to surfaces by expressed protein ligation and its application for “protein chip” fabrication. J Am Chem Soc. 2004;126:14730–14731. doi: 10.1021/ja0456611. [DOI] [PubMed] [Google Scholar]
- 55.Niculescu-Duvaz D, Getaz J, Springer CJ. Long functionalized poly(ethylene glycol)s of defined molecular weight: Synthesis and application in solid-phase synthesis of conjugates. Bioconjugate Chem. 2008;19:973–981. doi: 10.1021/bc060242+. [DOI] [PubMed] [Google Scholar]
- 56.Wegner GJ, Lee HJ, Corn RM. Characterization and optimization of peptide arrays for the study of epitope-antibody interactions using surface plasmon resonance imaging. Anal Chem. 2002;74:5161–5168. doi: 10.1021/ac025922u. [DOI] [PubMed] [Google Scholar]
- 57.Jia Y, Gu XJ, Brinker A, Warmuth M. Measuring the tyrosine kinase activity: a review of biochemical and cellular assay technologies. Expert Opin Drug Discovery. 2008;3:959–978. doi: 10.1517/17460441.3.8.959. [DOI] [PubMed] [Google Scholar]
- 58.Ficarro SB, Adelmant G, Tomar MN, Zhang Y, Cheng VJ, Marto JA. Magnetic bead processor for rapid evaluation and optimization of parameters for phosphopeptide enrichment. Anal Chem. 2009;81:4566–4575. doi: 10.1021/ac9004452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Rusmini F, Zhong ZY, Feijen J. Protein immobilization strategies for protein biochips. Biomacromolecules. 2007;8:1775–1789. doi: 10.1021/bm061197b. [DOI] [PubMed] [Google Scholar]
- 60.Brasher BB, Van Etten RA. c-Ab1 has high intrinsic tyrosine kinase activity that is stimulated by mutation of the Src homology 3 domain and by autophosphorylation at two distinct regulatory tyrosines. J Biol Chem. 2000;275:35631–35637. doi: 10.1074/jbc.M005401200. [DOI] [PubMed] [Google Scholar]
- 61.Radujkovic A, Schad M, Topaly J, Veldwijk MR, Laufs S, Schultheis BS, Jauch A, Melo JV, Fruehauf S, Zeller WJ. Synergistic activity of imatinib and 17-AAG in imatinib-resistant CML cells overexpressing BCR-ABL -Inhibition of P-glycoprotein function by 17-AAG. Leukemia. 2005;19:1198–1206. doi: 10.1038/sj.leu.2403764. [DOI] [PubMed] [Google Scholar]
- 62.De Keersmaecker K, Versele M, Cools J, Superti-Furga G, Hantschel O. Intrinsic differences between the catalytic properties of the oncogenic NUP214-ABL1 and BCR-ABL1 fusion protein kinases. Leukemia. 2008;22:2208–2216. doi: 10.1038/leu.2008.242. [DOI] [PubMed] [Google Scholar]
- 63.Rix U, Hantschel O, Duernberger G, Rix LLR, Planyavsky M, Fernbach NV, Kaupe I, Bennett KL, Valent P, Colinge J, Kocher T, Superti-Furga G. Chemical proteomic profiles of the BCR-ABL inhibitors imatinib, nilotinib, and dasatinib, reveal novel kinase and nonkinase targets. Blood. 2007;110:4055–4063. doi: 10.1182/blood-2007-07-102061. [DOI] [PubMed] [Google Scholar]
- 64.Hantschel O, Rix U, Schmidt U, Burckstummer T, Kneidinger M, Schutze G, Colinge J, Bennett KL, Ellmeier WR, Valent P, Superti-Furga G. The Btk tyrosine kinase is a major target of the Bcr-Abi inhibitor dasatinib. Proc Natl Acad Sci USA. 2007;104:13283–13288. doi: 10.1073/pnas.0702654104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Okabe S, Tauchi T, Ohyashiki K. Characteristics of dasatinib- and imatinib-resistant chronic myelogenous leukemia cells. Clin Cancer Res. 2008;14:6181–6186. doi: 10.1158/1078-0432.CCR-08-0461. [DOI] [PubMed] [Google Scholar]


