Abstract
Background
Prostate cancer (PCa) molecular subtypes have been defined by essentially mutually exclusive events, including ETS gene fusions (most commonly involving ERG) and SPINK1 over-expression. Clinical assessment may aid in disease stratification, complementing available prognostic tests.
Objective
To determine the analytical validity and clinicopatholgical associations of microarray-based molecular subtyping.
Design, Setting and Participants
We analyzed Affymetrix GeneChip expression profiles for 1,577 patients from eight radical prostatectomy (RP) cohorts, including 1,351 cases assessed using the Decipher prognostic assay (performed in a CLIA-certified laboratory). A microarray-based (m-) random forest ERG classification model was trained and validated. Outlier expression analysis was used to predict other mutually exclusive non-ERG ETS gene rearrangements (ETS+) or SPINK1 over-expression (SPINK1+).
Outcome Measurements
Associations with clinical features and outcomes by multivariable logistic regression analysis and receiver operating curves.
Results and Limitations
The m-ERG classifier showed 95% accuracy in an independent validation subset (n=155 samples). Across cohorts, 45%, 9%, 8% and 38% of PCa were classified as m-ERG+, m-ETS+, m-SPINK1+, and triple negative (m-ERG−/m-ETS−/m-SPINK1−), respectively. Gene expression profiling supports three underlying molecularly defined groups (m-ERG+, m-ETS+ and m-SPINK1+/triple negative). On multivariable analysis, m-ERG+ tumors were associated with lower preoperative serum PSA and Gleason scores, but enriched for extraprostatic extension (p<0.001). m-ETS+ tumors were associated with seminal vesicle invasion (p=0.01), while m-SPINK1+/triple negative tumors had higher Gleason scores and were more frequent in Black/African American patients (p<0.001). Clinical outcomes were not significantly different between subtypes.
Conclusions
A clinically available prognostic test (Decipher) can also assess PCa molecular subtypes, obviating the need for additional testing. Clinicopathological differences were found among subtypes based on global expression patterns.
Keywords: Prostate cancer, ERG, ETS, SPINK1, microarray, prognosis
INTRODUCTION
Prostate cancer (PCa) is clinically and molecularly heterogeneous. PCa genome and transcriptome characterization has identified molecular subtypes defined by essentially mutually exclusive genetic/transcriptomic events [1]. For example, approximately 50% of PCa foci from PSA-screened Caucasian cohorts harbor rearrangements between the 5′ untranslated region of androgen responsive genes (most commonly TMPRSS2) and members of the ETS transcription factor family [2,3]; fusions involving the ETS gene ERG are the most common (referred to as ERG+ (comprising ~90% of all ETS fusions), while mutually exclusive gene fusions involving non-ERG ETS genes, including ETV1, ETV4, ETV5 and FLI1, are infrequent (referred to as ERG−/ETS+ or ETS+; collectively comprising ~10% of all ETS fusions)[4]. Given the rarity of ETS+ PCa, it is unclear whether they are molecularly and clinicopathologically similar to ERG+ tumors. Approximately 10% of PCa, which are nearly exclusively negative for ERG or other ETS gene fusions (ETS−), harbor marked SPINK1 over-expression, consistent with a unique molecular subtype (SPINK1+)[3,5]. Although we and others have validated antibodies (against ERG and SPINK1) and fluorescence in situ hybridization (FISH) or RNA in situ hybridization (RISH) assays (against ETV1, ETV4, and ETV5)[4,6,7], routine comprehensive subtyping remains challenging and is cost prohibitive given the lack of current clinical indications.
High-throughput PCa transcriptome characterization has identified prognostic biomarkers, which have been translated to clinically available multi-gene prognostic tests compatible with routine formalin fixed paraffin embedded (FFPE) clinical biopsy or radical prostatectomy (RP) specimens[8–11]. Such tests must account for disease multifocality, as most men with PCa actually harbor multiple, genetically independent tumor clones that may have variable morphology (including Gleason score) and molecular alterations [12]. For example, 40-70% of RP samples harbor PCa foci with divergent TMPRSS2:ERG gene fusion status in distinct tumor foci, consistent with multiclonality [13–15]. Conflicting reports on associations of PCa molecular subtype defining lesions—such as TMPRSS2:ERG fusions and SPINK1 over-expression—with prognosis have been reported. Prognostic associations are confounded by cohort differences (i.e. PSA screened vs. unscreened, biopsy vs transurethral resection (TURP) detected, treatment modality and definition of “poor” outcome) as well as detection methodologies[16,17]. Nevertheless, inclusion of molecular subtyping in clinically available prognostic tests may provide additional information beyond routine prognosis in the post-RP setting, including assessment of multiclonality/multifocality [18–20] and predictive applications given clinical trials incorporating ETS status (NCT01576172). Additionally, it is unclear if prognostic signatures perform equally in different molecular PCa subtypes.
The goal of this study was to determine if PCa molecular subtyping could be performed from the “extra” data generated by a clinically available prognostic assay (Decipher), which utilizes genome-wide microarray expression profiling from formalin fixed paraffin embedded (FFPE) tissues to determine a prognostic score using the expression of 22 genes [9]. Hence, here we developed and validated computational tools for molecular subtyping using Decipher generated microarray expression data. We then determined clinicopathologic and prognostic associations from these microarray derived subtypes using 1,577 RP samples.
MATERIALS AND METHODS
Prostate cancer (PCa) samples
A total of 1,577 patient PCa expression profiles (1,351 from FFPE tissue) were analyzed from eight RP cohorts: Mayo Clinic (MCI & II) [9,21], Thomas Jefferson University (TJU)[22], Cleveland Clinic (CCF)[23], Johns Hopkins (JHMI), Memorial Sloan Kettering (MSKCC)[24], Erasmus MC (EMC)[25] and the German National Cancer Registry (DKFZ)[26] (Table S1). In each of the eight RP cohorts, a single tumor focus per patient was profiled (see Table S1 for selection criteria. The 1,351 FFPE samples were processed, assessed and analyzed using the Decipher clinical assay in the CLIA certified GenomeDx Biosciences Laboratory (San Diego, CA). The remaining 226 samples from three cohorts utilized RNA extracted in research laboratories from fresh-frozen or unfixed tissue preserved in RNAlater. These samples were profiled in microarray core facilities of major teaching hospitals and universities, although not to clinical grade standards. Data analysis was performed as for the Decipher clinical assay.
For development of microarray-based classifiers, MCI was used as a discovery cohort where 407/580 patients had ERG status (by fluorescence in situ hybridization [FISH]) as previously reported [27]; this cohort was split into training and validation sets of 252 and 155 patients, respectively, for training and validation of the microarray based ERG classifier (m-ERG). The other cohorts without FISH or IHC ERG status (or assessment of non-ERG ETS genes or SPINK1 expression) were used for classifier evaluation. See Appendix for additional details.
Microarray data processing
RNA extraction and microarray expression data generation using the Affymetrix Human Exon 1.0 ST arrays as part of the Decipher assay, including generation of the 22 gene prognostic score, were described previously[9,21,24–26]. See Appendix for additional details.
Development of ERG microarray-based classification models
We developed a Random Forrest (RF) supervised model (m-ERG) to predict FISH assessed ERG rearrangement status using the MCI cohort, which has available FISH-ERG information. The RF model was developed in a training subset of tumor patient profiles (n=252) combined with 29 benign prostate tissue profiles (from the MSKCC cohort) prior to assessment in the validation subset of MCI patient profiles (n=155) with known FISH-ERG status. The m-ERG model generated scores ranging from 0 to 1, with higher scores indicating increased likelihood of ERG rearrangement presence. Based on cut-off optimization methods[28], a m-ERG score above 0.6 was used to define m-ERG+ profiles from the training subset prior to application in the validation subset.
Development of ETV1, ETV4, ETV5, FLI1 and SPINK1 microarray-based classification models
FISH and/or IHC was not available for the other non-ERG ETS family members or SPINK1 in cohorts assessed herein, precluding gold standard validation of classifiers for these alterations. Hence, to develop classifiers, we performed unsupervised outlier analysis using the ‘extremevalues’ R package on expression of core probesets (those in canonical exons) for each gene using the entire MCI cohort (discovery) to define an expression threshold to classify each sample as an outlier (or not) for each gene. This outlier detection method estimates a model distribution for the discovery population (MCI) and using regression analysis identifies outliers as observations that are unlikely to be drawn from the same distribution. The minimum value of outliers in MCI was then set as the cutpoint to classify samples from the evaluation cohorts as outliers. Patients with outlier profiles defined as just described were annotated as m-ETV1+, m-ETV4+, m-ETV5+, m-FLI1+ or m-SPINK1+.
PCa Molecular subtyping
In this study, we initially classified patient profiles into four previously reported subtypes based on the results of the m-ERG, m-ETS and m-SPINK1 models. Tumor profiles with high m-ERG score (m-ERG+) and m-ETV1−, m-ETV4−, m-ETV5−, m-FLI1− and m-SPINK1− were classified as m-ERG+ subtype. Profiles that were m-ETV1+, m-ETV4+, m-ETV5+ or m-FLI1+ and m-ERG− were classified as m-ETS+ subtype, and those that were m-SPINK1+ and m-ERG− were classified as m-SPINK1+ subtype. Finally, patient profiles that are m-ERG−, m-ETV1−, m-ETV4−, m-ETV5−, m-FLI1− and m-SPINK1− were classified as ‘triple negative’. The four subtypes from this step were used to characterize the clinical and molecular characteristics of each subtype. m-ERG+/m-ETS+ or m-ERG+/m-SPINK1+ profiles were considered as “conflict cases” and were assessed separately.
Statistical analysis
Statistical analyses were performed in R v3.0. All statistical tests were two-sided using p<0.05 significance level. Univariable and multivariable logistic regression analysis were performed to evaluate the statistical associations between microarray defined molecular subtypes and clinical variables including age, race/ethnicity, pre-operative PSA, surgical margin status (SMS), extraprostatic extension (EPE), seminal vesicle invasion (SVI), lymph node involvement (LNI) and Gleason Score (GS). The multiple cohorts were considered as random effect in the MVA regression model to remove individual cohort bias.
RESULTS
Clinical Characteristics of the Study Cohorts
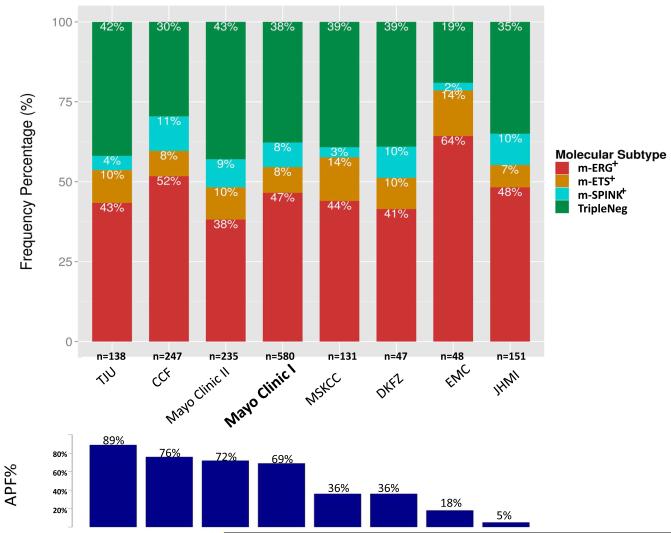
To develop and validate computational tools for basic PCa molecular subtyping by gene expression generated as part of the Decipher prognostic assay, we pooled RP samples from 1,577 patients from a total of 8 cohorts profiled using the Affymetrix Human Exon 1.0 ST arrays (Table S1). These cohorts represent the spectrum of RP treated PCa from low to high-risk localized disease. Overall, 61% of patients in the pooled cohort had one or more adverse pathology findings (APF: RP Gleason score ≥8, >pT2 or pN1); however, APF incidence of ranged from 5%-89% between individual cohorts (Table S1). The majority of patients were Caucasian (89%) and had aggressive PCa. Most of the patients received post-operative hormonal and/or radiation therapy. Patients with metastasis had a median follow up time of 62 months (range, 3-213), and patients with no metastasis had a median follow up of 129 months [1-280]. Detailed clinicopathological information for the pooled cohort provided in Table S2.
Microarray-based ERG (m-ERG) Model Development and Validation
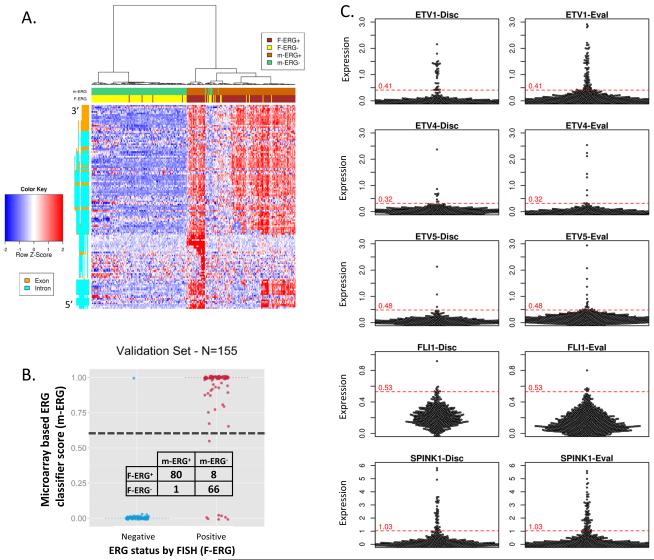
The most informative microarray probesets for the m-ERG model were identified through a multi-step procedure. First, in the training set (n=252), expression clustering of the 132 ERG locus probesets demonstrated that most are highly correlated and informative of FISH-ERG status (Figures 1A and S1A). Filtering of redundant and non-informative features (e.g., not expressed above background) was performed prior to training a random forests (RF) classifier for predicting FISH-ERG status. The final model used the expression values of 3 ERG locus probesets and 2 probesets associated with FISH-ERG+ but low ERG expression; AUC for predicting FISH-ERG status in the training set using the final m-ERG model was 0.98. In the validation subset (n=155 profiles, not used for training), the m-ERG model had an AUC of 0.94 and an overall accuracy of 95% (Figure 1B). Most misclassified patients had low ERG expression, consistent with previous reports that very small subsets of ERG fusion-positive tumors (as detected by FISH) do not overexpress ERG protein [29]. Validation in benign tissue, cell line controls and technical replicates are reported in the Appendix and Figure S1. When then applied to 1,170 patient profiles from the 7 cohorts not used for training or testing, 550 (47%) were classified as m-ERG+ (38-64% across cohorts) as shown in Figure 2.
Figure 1. Development and validation of microarray based prostate cancer (PCa) molecular subtyping using genome wide expression profiling data from the Decipher assay.
A. Development of a microarray based ERG rearrangement classifier (m-ERG) from genome wide expression profiling data from the Decipher assay. Unsupervised clustering of the training subset (n=252 samples) from the discovery cohort (Mayo Clinic I) was performed using gene expression from ERG exon (orange) and intron (green) probe sets (5′ on bottom). Expression of five summarized features was used to train a random forest (RF) classifier based on known fluorescence in situ hybridization (FISH) assessment of ERG rearrangement status (F-ERG) . m-ERG and F-ERG status for each profiled sample are indicated in the header according to the legend. B. m-ERG scores in the validation subset (n=155) of the discovery cohort are plotted with stratification by F-ERG status. The predefined m-ERG+/ERG− score cutoff is indicated by the black dashed line. Classification results are shown in the contingency table. C. Development of microarray based classifiers for other ETS gene rearrangements and SPINK1 over-expression using outlier analysis. Beeswarm plots show core-level expression of ETV1, ETV4, ETV5, FLI1 and SPINK1 in the discovery (n=580 samples, left panels) and evaluation cohorts (n=997 samples, right panels). Outlier analysis was used to define indicated cut-off scores for m-ETV1, m-ETV4, m-ETV5, m-FLI1, and m-SPINK1 classifiers in the discovery cohort and then applied to the evaluation cohort.
Figure 2. Distribution of molecular PCa subtypes across assessed cohorts.
In each cohort (total samples assessed given), the percentage of samples in each microarray defined subypte (m-ERG+, m-ETS1+, m-SPINK1+ and m-ERG−/m-ETS−/m-SPINK1− [TripleNeg]) are shown according to the legend. Cohorts are ordered based on frequency of Adverse Pathology Findings (APF%, bottom panel). Mayo Clinic I (bolded) was used as the discovery cohort, while the remaining cohorts were used as the evaluation cohort.
Development of ETS and SPINK1 microarray classifiers
We next sought to classify patients based on SPINK1 over-expression, or outlier expression of non-ERG ETS genes (ETV1, ETV4, ETV5 and FLI1) using over-expression as a surrogate for rearrangement of the respective ETS gene. Heatmaps of all probesets from the ETV1, ETV4, ETV5 and SPINK1 loci showed that a subset of patients over-expressed each gene, as expected (Figure S2). Outlier analysis was first performed in the MCI cohort to define outlier threshold cut-points for each gene (see Methods), which were then applied to the remaining evaluation cohorts, as no gold standard data was available for profiled datasets (Figure 1C). In MCI, microarray outlier analysis classified 5%, 1.7%, 0.5%, 1% and 7.7% as m-ETV1+, m-ETV4+, m-ETV5+, m-FLI+ and m-SPINK1+, respectively (Table S3). Performance of these classifiers in benign tissue, cell line controls and technical replicates are reported in the Appendix.
Molecular subtyping of 1,577 RP patient specimens using the microarray-based classifiers
Across the 1,577 profiles from 8 cohorts, microarray analysis classified 46%, 8%, 1.1%, 1.6%, 0.6% and 8% as m-ERG+, m-ETV1+, m-ETV4+, m-ETV5+, m-FLI+ and m-SPINK1+, respectively; 36% (n=575) lacked any outlier expression and were considered TripleNeg (Table S3). Additionally, 3% of patient profiles had outlier expression for two or more markers (m-ERG+/m-ETS+ or m-ERG+/m-SPINK1+ profiles), as shown in Table S3, which we consider as conflict cases (see Discussion). To focus on cases with clearly defined subtypes, we considered these conflict cases separately and collapsed the four ETS family members into one group (given the low numbers of individual m-ETV1+, m-ETV4+, m-ETV5+ and m-FLI1+ profiles even in this large cohort). This analysis resulted in in four molecular subtypes—m-ERG+, m-ETS+, m-SPINK1+ and TripleNeg— at 45%, 9%, 8% and 38% of samples, respectively (Figure 2), consistent with distributions in other predominantly Caucasian cohorts assessed by for these individual subtypes by gene expression, FISH and/or IHC[2,30].
Gene expression clustering of PCa molecular subtypes
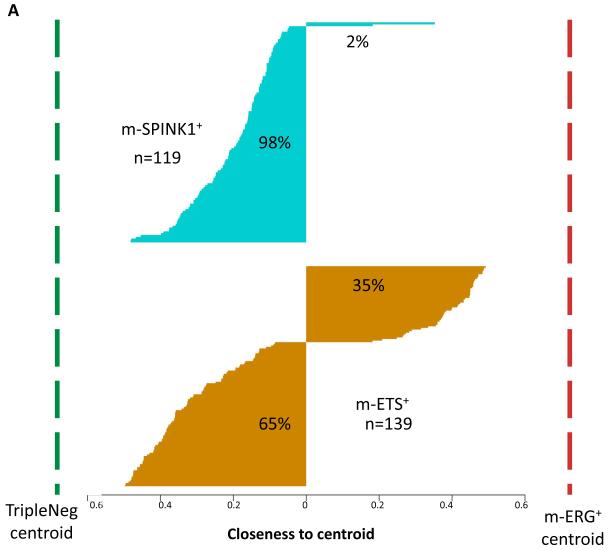
The low frequency of ETS+ and SPINK1+ subtypes has precluded comprehensive molecular and clinicopathological evaluation in large cohorts to determine whether these minor PCa subtypes represent distinct molecular subtypes, or are best classified as ERG+ and TripleNeg, respectively. Hence, we first assessed whether m-ETS+ tumors (or m-SPINK1+ tumors) show global transcriptional profiles more similar to m-ERG+ or TripleNeg tumors. Thus, we first defined expression centroids for m-ERG+ and TripleNeg tumors using transcriptome-wide differential expression analysis. To define m-ERG+ and TripleNeg expression centroids, we selected all probesets with AUC>0.75 for discriminating for these two subtypes (n=360 probesets). Calculating the distance between each m-ETS+ or m-SPINK1+ sample and the m-ERG+ and TripleNeg subtypes centroids demonstrated that 98% (117/119) of m-SPINK1+ tumors had cluster distances closer to the TripleNeg centroid. In contrast, 35% of m-ETS+ tumors (48/139) had cluster distances closer to the m-ERG+ centroid, while 65% of m-ETS+ tumors were closer to the TripleNeg centroid (Figure 3A). Defining m-ERG+ and TripleNeg expression centroids using other gene sets, as well as fuzzy c-means clustering supported these results as described in Table S4, the Appendix, and Figure S3. Together these analyses demonstrate that m-SPINK1+ tumors are highly similar to TripleNeg tumors while m-ETS+ tumors are distinct from m-ERG+ tumors.
Figure 3. Gene expression profiling support high similarity of m-SPINK1+ and TripleNeg subtypes, unlike m-ETS+ and m-ERG+ PCa.
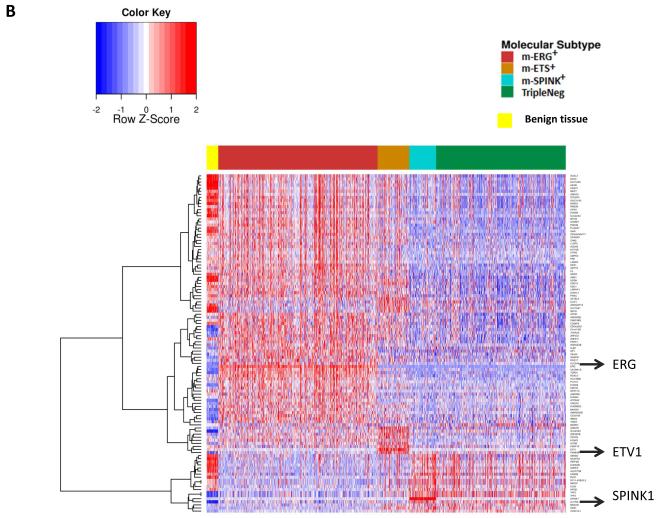
A. m-ERG+ and TripleNeg expression centroids were generated by identifying all probesets (n=360) with AUC>0.7 for discriminating m-ERG+ and TripleNeg samples across all cohorts (n=1,531 samples). To assess the relationship of m-SPINK1+ and m-ETS+ PCa to m-ERG+ and TripleNeg, the relative closeness of each m-SPINK1+ or m-ETS+ sample to the m-ERG+/TripleNeg centroids is plotted (larger value indicates more similarity). B. Clustering of subtype defining gene expression demonstrates overlap of m-SPINK1+ and TripleNeg PCa and unique profiles of m-ETS+ PCa. Expression of the most predictive genes for each subtype (AUC>0.70 for discrimination from all other subtypes, n=360) were used for clustering all profiled samples (n=1531). Benign specimens (yellow) from the DKFZ cohort clustered separately from all PCa samples.
To gain additional insight into subtype relationships, the most predictive genes for each subtype were defined based on AUC for discrimination of each subtype from the others. Seventy six, 15, 14 and 3 genes had an AUC>0.7 for m-ERG+, m-ETS+, m-SPINK1+, and TripleNeg, respectively (Table S5). Clustering expression of these discriminatory genes across all samples demonstrated two main dendrogram branches corresponding to m-ERG+ and Triple Negative predictive genes. While m-ETS+ tumors shared expression of m-ERG+ predictive genes and expressed a unique subset of genes, the expression pattern of m-SPINK1+ tumors was highly similar to TripleNeg PCa (Figure 3B). As expected, benign samples clustered separately from all tumor samples. Subtype specific genes are described in the Appendix.
Clinical associations of PCa molecular subtypes
On univariable analysis, race, preoperative PSA, Gleason score (GS), extraprostatic extension (EPE) and seminal vesicle invasion (SVI) status were non-uniformly distributed across microarray defined subtypes (Table S6). We used multinomial multivariable analysis to compare clinical and pathological characteristics between subtypes (Table 1). Compared to TripleNeg, m-ERG+ PCa is associated with lower pre-operative PSA (OR=0.47, p<0.001) and lower Gleason score (OR=0.43, p<0.001), but nearly twice as likely to have EPE (OR=1.80, p<0.001) and nearly five times more likely to occur in Caucasian men (p<0.001) (Table 1). The m-ETS+ subtype was more likely to have SVI compared to both TripleNeg (OR=2.27, p=0.004) or m-ERG+ PCa (OR=1.96, p=0.01) (Table 1). Both TripleNeg and m-SPINK1+ tumors had significantly higher preoperative PSA (OR=2.12, p<0.001 and OR=1.73, p=0.05, respectively) and Gleason scores (OR=2.3, p<0.001 and OR=3.0, p<0.001, respectively), and were more common in African American patients (OR=5.44, p=0.002 and OR=16.87, p<0.001, respectively), compared to m-ERG+ tumors. Interestingly, m-SPINK1+ is significantly associated with lack of SMS compared to m-ERG+ (OR=0.58, p=0.006). Together, these clinicopathologic associations are consistent with our transcriptome analysis demonstrating that m-SPINK1+ and TripleNeg are highly similar, while m-ERG+ and m-ETS+ have distinct features.
Table 1.
Multinomial multivariable logistic regression analysis between clinicopathological variables and molecular subtypes across 1531 profiled samples
| Variable | m-ERG+ OR Estimate (95% CI) |
MVA p value |
m-ETS+ OR Estimate (95% CI) |
MVA p value |
m-SPINK+ OR Estimate (95% CI) |
MVA p value |
ANOVA p - value |
|
|---|---|---|---|---|---|---|---|---|
| Reference: TripleNeg | Pre-Op PSA | 0.47 (0.33-0.68) | <0.001 | 0.48 (0.26-0.88) | 0.021 | 0.81 (0.44-1.51) | 0.42 | <0.001 |
| Race(Black/African American) | 0.18 (0.07-0.52) | 0.002 | 0.21 (0.03-1.6) | 0.12 | 3.10 (1.23-7.82) | 0.02 | <0.001 | |
| EPE | 1.80 (1.34-2.41) | <0.001 | 1.23 (0.75-2.01) | 0.34 | 0.76 (0.46-1.26) | 0.37 | <0.001 | |
| SVI | 1.16 (0.83-1.62) | 0.24 | 2.27 (1.35-3.82) | 0.004 | 0.84 (0.47-1.53) | 0.51 | 0.01 | |
| PathGS<7 | 0.96 (0.61-1.51) | 0.93 | 0.75 (0.31-1.81) | 0.58 | 0.89 (0.39-2.04) | 0.79 | <0.001 | |
| PathGS>7 | 0.43 (0.32-0.6) | <0.001 | 0.75 (0.45-1.26) | 0.46 | 1.31 (0.78-2.21) | 0.39 | <0.001 | |
| SMS | 1.18 (0.89-1.56) | 0.29 | 1.27 (0.79-2.04) | 0.53 | 0.69 (0.42-1.12) | 0.13 | 0.13 | |
| Age | 1.00 (0.98-1.02) | 0.71 | 0.97 (0.94-1.00) | 0.045 | 1.01 (0.97-1.05) | 0.62 | 0.24 | |
| LNI | 1.27 (0.77-2.11) | 0.42 | 1.6 (0.78-3.27) | 0.22 | 1.16 (0.49-2.76) | 0.73 | 0.60 |
| Variable | m-ETS+ OR Estimate (95% CI) |
MVA p value |
m-SPINK+ OR Estimate (95% CI) |
MVA p value |
TripleNeg OR Estimate (95% CI) |
MVA p value |
ANOVA p - value |
|
|---|---|---|---|---|---|---|---|---|
| Reference: m-ERG+ | Pre-Op PSA | 1.01 (0.54-1.88) | 0.92 | 1.73 (0.91-3.27) | 0.05 | 2.12 (1.47-3.06) | <0.001 | <0.001 |
| Race(Black/African American) | 1.12 (0.13-9.88) | 0.61 | 16.87 (5.13-55.48) | <0.001 | 5.44 (1.94-15.29) | 0.002 | <0.001 | |
| EPE | 0.68 (0.42-1.11) | 0.13 | 0.42 (0.25-0.7) | 0.001 | 0.56 (0.41-0.75) | <0.001 | <0.001 | |
| SVI | 1.96 (1.18-3.24) | 0.01 | 0.73 (0.4-1.32) | 0.37 | 0.86 (0.62-1.2) | 0.24 | 0.014 | |
| PathGS<7 | 0.78 (0.33-1.85) | 0.5 | 0.93 (0.41-2.13) | 0.96 | 1.04 (0.66-1.64) | 0.93 | <0.001 | |
| PathGS>7 | 1.74 (1.05-2.88) | 0.05 | 3.01 (1.77-5.13) | <0.001 | 2.30 (1.68-3.15) | <0.001 | <0.001 | |
| SMS | 1.08 (0.68-1.72) | 0.91 | 0.58 (0.36-0.95) | 0.006 | 0.85 (0.64-1.12) | 0.29 | 0.12 | |
| Age | 0.97 (0.94-1.00) | 0.08 | 1.01 (0.98-1.05) | 0.71 | 1.00 (0.98-1.02) | 0.71 | 0.23 | |
| LNI | 1.25 (0.62-2.53) | 0.59 | 0.91 (0.38-2.17) | 0.57 | 0.78 (0.47-1.3) | 0.42 | 0.60 |
Multinomial multivariable logistic regression analysis was performed for clinicopathological variables and molecular subtypes across all 1,531 profiled samples with reference to TripleNeg (upper panel) and m-ERG+ (lower panel) as the reference. OR = odds ratio; CI= confidence interval; Pre-OP PSA = pre-operative serum PSA (reference: <20ng/mL); Race (reference: Caucasian); EPE = extraprostatic extension; SVI = seminal vesicle invasion; PathGS = pathologic Gleason score at prostatectomy (reference: Gleason score 7); SMS = surgical margin status; LNI = lymph node involvement.
Impact of molecular subtypes on prognosis
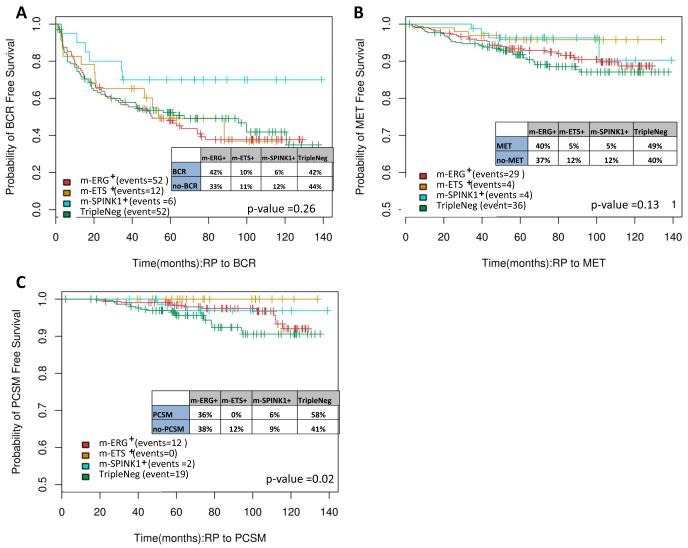
To evaluate the impact of molecular subtyping on prognosis, we assessed the ability of the subtypes to predict patient outcomes such as biochemical recurrence (BCR), metastasis (MET) and prostate cancer death (PCSM) after RP (Table S7). ROC analysis showed that the subtypes do not discriminate well for these endpoints (AUC ~0.5). Likewise, across all cohorts (excluding the MCI cohort used for development), Decipher [9] showed similar discrimination (as measured by AUC) for metastasis in all four subtypes (Figure 4). Other prognostic signatures such as CCP, GPS and the Penney et al. signature [10,31,32], which can be derived from our global gene expression data, also showed similar discrimination for metastasis in all subtypes except the microarray derived (md-)GPS signature, which was not discriminative in the m-SPINK+ subtype (Figure S4). Lastly, Kaplan-Meier analyses in the MCII cohort failed to show significant differences in time to events for BCR (Figure 5A) and metastasis (Figure 5B) endpoints between the subtypes. However, a trend toward significance was observed with the Triple Negative subtype patients having worse PCSM than the other subtypes (Figure 5C). Together, these results support limited prognostic utility for molecular subtypes in the post-RP setting where most patients receive post-operative hormonal and/or radiation therapy.
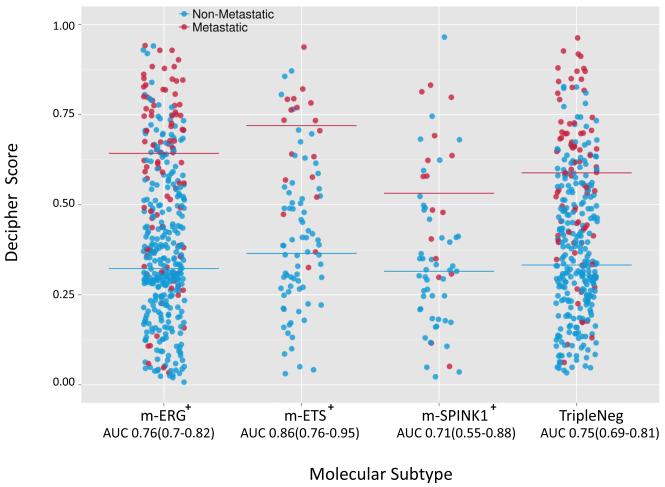
Figure 4. Performance of a multigene PCa prognostic predictor (Decipher) is similar across molecular subtypes.
The Decipher score (greater score predicts increased aggressiveness) for each profiled sample in the pooled cohorts (n= 997, excluding Mayo Clinic I as it was used for Decipher discovery) is plotted, stratified by assigned molecular subtypes. Patients who developed metastasis or not are indicated by blue and red points, respectively, and median scores per subtype are indicated by bars. The AUC for Decipher score prediction of metastasis development in each subtype (along with 95% CI) is given. AUCs in each subtype were significantly greater than expected by chance (p<0.0001 for all subtypes).
Figure 5. Kaplan Meier analysis demonstrates similar PCa outcome measures across molecular subtypes.
Kaplan Meier analysis was performed for all Mayo Clinic II cohort samples (case cohort, n=235 samples) stratified by assigned molecular subtype for A) biochemical recurrence (BCR), B) metastasis (MET) and C) prostate cancer specific mortality (PCSM) free survival. Log Rank p values are given, along with the percentage of each subtype experiencing each outcome.
DISCUSSION
High throughput technologies such as DNA microarrays and next generation sequencing have greatly increased our understanding of PCa molecular alterations, including defining molecular subtypes and the identification of prognostic gene expression signatures. Although well validated antibodies and FISH/RISH assays have been developed for research and clinical applications, comprehensive subtyping using these assays remains challenging and has not been applied to large translational research cohorts. Likewise, the lack of current clinical indications in PCa makes comprehensive molecular subtyping cost prohibitive in routine clinical practice. This lack of large, comprehensively subtyped clinical cohorts has hindered thorough evaluation of subtype specific clinicopathological associations and molecular features. However, the development and uptake of clinically available, FFPE compatible, gene expression based prognostic assays [8–11] suggests that expression profiling data will be available for tens of thousands of patients in the immediate future.
Hence, in this study, we sought to determine whether the “extra” gene expression data generated as part of the clinically available Decipher assay, which derives a 22 gene prognostic score from genome-wide microarray expression profiling, can be used to determine molecular subtypes. Thus, we built computational models to predict the most common PCa molecular subtypes defined by alterations resulting in marked transcript over-expression: ERG, ETV1, ETV4, ETV5 and FLI1(due to rearrangement) as well as SPINK1 (unknown mechanism). Our m-ERG classifier demonstrated 95% accuracy in predicting FISH-ERG status in an independent validation set, similar to the reported accuracy of ERG IHC [29,33,34] as used diagnostically in challenging cases [35]. In the pooled cohort, 45% of patients were predicted as m-ERG+, similar to 47% ERG rearrangement positive frequency reported from a meta-analysis of over 10,000 PCa samples [36]. We also demonstrated the robustness of this m-ERG classifier using PCa/normal prostate tissue pairs and technical replicates. Hence, using this validated classifier, the Decipher prognostic assay can also assess ERG status, without the cost or delay of separate IHC or FISH based evaluation.
We also developed microarray based classifiers for gene fusions involving other non-ERG ETS genes (m-ETS), as well as SPINK1 over-expression (m-SPINK1). As gold standard FISH/IHC data for assessing the performance of these classifiers was unavailable, findings from these analyses should be considered exploratory, representing a limitation of our study. We also identified a total of 3% of cases with m-ERG+/m-ETS+ or m-ERG+/m-SPINK1+ profiles. In our experience, ERG, non-ERG ETS and SPINK1 subtype defining alterations are nearly always mutually exclusive, and observed co-occurrence is most likely due to either misclassification (given the lack of gold standard training data for non-ERG classifiers) or profiling of collisions between genetically distinct tumor clones (which may appear morphologically indistinguishable), although exceptionally rare examples of focal SPINK1 expression in otherwise ERG+ tumor have been reported[7,37,38]. As shown by multivariate analysis (Table S8), conflict cases identified herein show similar clinicopathological associations as m-ERG+ PCa, consistent with an enrichment of m-ERG+ tumors in these conflict cases Thus, studies are ongoing to generate gold standard data for these non-ERG based classifiers. Importantly, however, for clinicopathological assessment of our microarray defined subtypes, only cases with clearly defined single subtypes were included, limiting the impact of these conflict cases on our findings.
Our combined cohort was comprised of over 1,500 PCa patient profiles, allowing us to explore clinicopathological and molecular correlates from these microarray defined subtypes which have not been addressed in smaller or less comprehensive studies. By multivariable analysis, m-ERG+ status was significantly associated with lower Gleason score, lower pre-operative serum PSA, and European American race. These findings were in keeping with other large RP cohorts [36] and further support the validity of our approach. Interestingly, although m-ETS+ PCa was associated with lower PSA when compared to TripleNeg PCa, this subtype was specifically associated with increased SVI when compared to both m-ERG+ and TripleNeg. On the other hand, m-SPINK1+ PCa was specifically associated with Black/African American race, in line with recent findings from an IHC based RP study assessing molecular subtypes and race [39].
At the molecular level, we also attempted to address whether m-ETS+ and m-SPINK1+ PCa are similar to m-ERG+ or TripleNeg tumors, respectively, or represent distinct subtypes. Results revealed that most m-SPINK1+ PCa cluster with TripleNeg based on global and supervised gene expression, unlike m-ETS+ PCa, which shared molecular overlap with both TripleNeg and m-ERG+ subtypes. Clinicopathologic associations similarly demonstrate the differences between m-ERG+ and other m-ETS+ PCa, as well as the similarity between m-SPINK1+ and TripleNeg PCa. Thus we believe PCa can be grouped into three clinically and molecularly distinct groups (m-ERG+, m-ETS+ and m-SPINK1+/TripleNeg), although the detection of SPINK1 may still be useful as a single gene marker, particularly given the frequency of over-expression in Black/African American patients (even compared to TripleNeg).
A limitation of our study is the lack of assessment of other relevant genomic lesions that occur across (e.g. PTEN deletion) or within specific molecular subtypes (e.g. SPOP mutations or CHD1 deletions/mutations in ERG− PCa). Efforts are ongoing to develop classifiers for these events, however they are more challenging to detect in gene expression data than outlier over-expression based events. Likewise, although our study suggests that ERG+ and non-ERG ETS+ PCa subtypes should not be combined in clinicopathological analyses, several thousand more samples will need to be profiled to inform on whether the non-ERG ETS+ subtype should be stratified by individual alterations (e.g. ETV1+ vs. ETV4+). Additional limitations include the lack of central histological review and the inclusion of a small subset of fresh frozen samples (n=226) assessed outside the CLIA laboratory. All cohorts derive from centers with expert genitourinary pathologists and we have shown high concordance of Decipher profiles from matched fresh frozen and FFPE samples assessed as herein ([40] and data not shown). Hence, although these factors may limit our ability to observe associations, our results are likely more generalizable and several individual clinicopathological associations observed herein (e.g. less frequent ERG+ in African American men) are consistent with prior studies.
The recognition of over-treatment has led to an enormous interest in the development of prognostic biomarkers, including several commercially available gene expression based prognostic assays applicable to routine biopsy or RP specimens [9,10,31]. Whether such assays are similarly prognostic across previously defined molecular subtypes had not been assessed. Our results herein, which show limited effect of subtyping on prognosis and prognostic assay performance, are consistent with large FISH/IHC based studies of ERG and SPINK1 status in RP cohorts that show a lack of prognostic ability for predicting post-surgical outcomes. Why subtypes lack prognostic ability despite strong associations with known prognostic pathological parameters (i.e. m-ETS+ associated with SVI), and the reasons for potential conflicting prognostic associations (i.e. m-ERG+ associated with Gleason score <7 and EPE), will require additional research. We and others have hypothesized that subtype defining lesions may play a more important role in tumor initiation and local growth characteristics, rather than in the factors that drive post-resection recurrence[36,41], suggesting prognostic/predictive applications in non-RP based cohorts (as discussed below).
Despite the lack of impact on post-RP prognosis, we anticipate that incorporating molecular subtyping into a clinically available prognostic assay has several areas of potential near term clinical utility. For example, ERG status has been reported as prognostic in several non-RP cohorts. Most notably, assessing a cohort of 217 active surveillance (AS) patients, Berg et al. reported that patients with any ERG+ cores at diagnosis (by IHC) were more than twice as likely to progress compared to ERG− patients; ERG+ was the most significant predictor of AS progression in multivariable Cox regression analysis[42]. These findings are in keeping with the hypothesis that ERG rearrangements may drive local growth as described above and hence molecular subtyping may be particularly relevant in the AS setting. Although Decipher has received Medicare coverage in the U.S. for post-RP prognosis, efforts to apply this assay to diagnostic biopsy specimen are ongoing. Given the use of other prognostic assays that assess biopsy specimens (such as Oncotype DX and Prolaris) largely in patients considering AS, application of Decipher in this setting will enable assessment of the impact of m-ERG, m-ETS, mSPINK1/TripleNeg subtypes on prognosis in this setting without the need for additional IHC/FISH/RISH based subtyping.
Likewise, incorporating ERG status into prognostic tests may have utility in evaluating multifocality/clonality. PCa is commonly multifocal, where a single prostate may harbor multiple genetically distinct tumor foci (as has been demonstrated through ERG status) that may be indistinguishable by routine histology. For example, patients on AS who have consecutive biopsies with discordant microarray subtypes would indicate that different tumor clones were sampled, as has been shown by IHC based ERG assessment in a patient who developed an aggressive interval cancer while on active surveillance [20]. Of critical importance, our approach can now be used to profile multiple foci at RP (or multiple involved prostate biopsy cores) and directly assess the impact of true multifocality (as indicated by discordant subtypes) on the Decipher prognostic score and other derived prognostic signatures. Although other prognostic assays have reported robustness to multifocality [10], molecular subtyping was not incorporating and hence it is unclear if separate areas of the same tumor focus or truly genetically independent tumors (as would be indicated by discordant subtypes) were profiled. Given the need for prognostic assays to be reflective of the most aggressive tumor focus, even if not sampled in the assessed biopsy specimen, our approach can be used to directly assess the robustness of prognostic assays to true multifocality.
Lastly, the ability to robustly detect molecular subtypes allows for pre-specified molecular subgroup analyses or enrichment of patient populations for clinical trials [20], which although common in precision medicine approaches in other cancers, are not routinely used in PCa. Importantly, ETS rearrangements are nearly always early, clonal alterations, suggesting that subtypes identified in diagnostic biopsy or RP samples will be maintained through advanced disease [1–3]. As an example of the potential predictive utility of PCa molecular subtypes, Galletti et al. demonstrated that in a pilot cohort of 34 men with metastatic castration resistant PCa (CRPC) treated by docetaxel chemotherapy, men with ERG+ primary tumors (by IHC) were nearly twice as likely to show resistance (by lack of PSA response) than men with ERG− tumors [43]. Likewise, pre-clinical data supports targeting of specific subtype defining alterations (e.g. targeting PARP in ERG+ or ETS+ and targeting EGFR in SPINK1+ PCa)[44,45], culminating in ongoing clinical trials are ongoing that require ERG and ETV1 status evaluation (NCT01576172). Importantly, several molecularly defined subtypes in other cancers, (i.e. KRAS/ALK/EGFR mutant lung cancer) show little to no association with prognosis but have distinct clincopathological features and became predictive and clinically useful only after the development of targeted therapies [46,47].
CONCLUSIONS
Several lines of evidence support PCa molecular subtypes, defined largely by mutually exclusive genomic/transcriptomic events. The most commons subtype defining lesion—ERG rearrangement—has been evaluated clinically by FISH and IHC. Herein, we validate the use of extra data from a clinically available gene expression based prognostic assay (Decipher) for assessing ERG rearrangement status. Additionally, although clinicopathological and molecular associations for m-ERG+ vs. m-ERG− PCa are well described, little is known about less frequent subtypes. Hence, we also developed classifiers for subtypes defined by rearrangements of other ETS genes or SPINK1 over-expression and explored associations in over 1,500 PCa. Importantly, gene expression profiles and clinicopathological associations support three general molecular subtypes (m-ERG+, m-ETS+, and m-SPINK1+/TripleNeg), providing the most comprehensive support for distinguishing ERG+ and ETS+ PCa. Of note, although IHC/FISH/RISH assays have been developed to assess these molecular subtypes, our microarray based classifiers are derived from “extra” data generated as part of the Decipher assay, a prognostic assay performed in a CLIA certified laboratory; hence a potential advantage of this assay compared to other assays is the inclusion of molecular subtyping information without the need for additional testing, delay or cost. Taken together, we demonstrate the validity of PCa molecular subtyping using extra data from a gene expression based prognostic assay and identify novel clinicopathological and molecular correlates to these subtypes. Although we demonstrate that molecular subtypes are not prognostic in the post-prostatectomy setting (nor impact the performance of currently available prognostic signatures), we anticipate that molecular subtyping will complement purely prognostic based tests in several areas of PCa management including non-RP cohorts.
Supplementary Material
PATIENT SUMMARY.
Prostate cancer molecular subtyping can be achieved using “extra” data generated from a clinical-grade genome wide expression profiling prognostic assay (Decipher). Transcriptomic and clinical analysis support three distinct molecular subtypes: 1) m-ERG+ 2) m-ETS+, and 3) m-SPINK1+/Triple Negative (m-ERG−/m-ETS−/m-SPINK1−). Incorporation of subtyping into a clinically available assay may enable additional applications beyond routine prognosis.
ACKNOWLEDGMENTS
S.A.T. and F.Y.F. are supported by the Prostate Cancer Foundation and the A. Alfred Taubman Medical Research Institute. GenomeDX funded specimen collection, performed the Decipher assay, participated in study and analysis design, and participated in manuscript revision, revision and the decision to submit for publication. S.A.T. and F.Y.F. had full access to all the data in the study and take responsibility for the integrity of the data and the accuracy of the data analysis.
Footnotes
FINANCIAL DISCLOSURE
The University of Michigan has been issued a patent on the detection of ETS gene fusions in prostate cancer, on which S.A.T. is listed as a co-inventor. The University of Michigan licensed the diagnostic field of use to Hologic/Gen-Probe, Inc, which sublicensed some rights to Ventana Medical Systems, Inc. The University of Michigan has filed a patent on SPINK1 in prostate cancer, on which S.A.T. is listed as a co-inventor. The University of Michigan licensed the diagnostic field of use to Hologic/Gen-Probe, Inc, which sublicensed some rights to Ventana Medical Systems, Inc. S.A.T. has received honoraria from, and served as a consultant to Ventana Medical Systems. M.A., E.D., N.E., K.Y., and Z.H. are employees of GenomeDX. The remaining authors declare no conflicts of interest.
REFERENCES
- [1].Svensson MA, LaFargue CJ, MacDonald TY, Pflueger D, Kitabayashi N, Santa-Cruz AM, et al. Testing mutual exclusivity of ETS rearranged prostate cancer. Lab Invest. 2011;91:404–12. doi: 10.1038/labinvest.2010.179. doi:10.1038/labinvest.2010.179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Rubin MA, Maher CA, Chinnaiyan AM. Common Gene Rearrangements in Prostate Cancer. J Clin Oncol. 2011;29:3659–68. doi: 10.1200/JCO.2011.35.1916. doi:10.1200/JCO.2011.35.1916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Rubin MA. ETS rearrangements in prostate cancer. Asian J Androl. 2012;14:393–9. doi: 10.1038/aja.2011.145. doi:10.1038/aja.2011.145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Barbieri CE, Tomlins SA. The prostate cancer genome: Perspectives and potential. Urol Oncol Semin Orig Investig. 2014;32 doi: 10.1016/j.urolonc.2013.08.025. doi:10.1016/j.urolonc.2013.08.025. [DOI] [PubMed] [Google Scholar]
- [5].Tomlins SA, Rhodes DR, Yu J, Varambally S, Mehra R, Perner S, et al. The Role of SPINK1 in ETS Rearrangement-Negative Prostate Cancers. Cancer Cell. 2008;13:519–28. doi: 10.1016/j.ccr.2008.04.016. doi:10.1016/j.ccr.2008.04.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Furusato B, Tan S-H, Young D, Dobi A, Sun C, Mohamed AA, et al. ERG oncoprotein expression in prostate cancer: clonal progression of ERG-positive tumor cells and potential for ERG-based stratification. Prostate Cancer Prostatic Dis. 2010;13:228–37. doi: 10.1038/pcan.2010.23. doi:10.1038/pcan.2010.23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Kunju LP, Carskadon S, Siddiqui J, Tomlins SA, Chinnaiyan AM, Palanisamy N. Novel RNA hybridization method for the in situ detection of ETV1, ETV4, and ETV5 gene fusions in prostate cancer. Appl Immunohistochem Mol Morphol. 2014;22:e32–40. doi: 10.1097/PAI.0000000000000095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Cuzick J, Swanson GP, Fisher G, Brothman AR, Berney DM, Reid JE, et al. Prognostic value of an RNA expression signature derived from cell cycle proliferation genes in patients with prostate cancer: A retrospective study. Lancet Oncol. 2011;12:245–55. doi: 10.1016/S1470-2045(10)70295-3. doi:10.1016/S1470-2045(10)70295-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Erho N, Crisan A, Vergara IA, Mitra AP, Ghadessi M, Buerki C, et al. Discovery and Validation of a Prostate Cancer Genomic Classifier that Predicts Early Metastasis Following Radical Prostatectomy. PLoS One. 2013;8:e66855. doi: 10.1371/journal.pone.0066855. doi:10.1371/journal.pone.0066855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Klein EA, Cooperberg MR, Magi-Galluzzi C, Simko JP, Falzarano SM, Maddala T, et al. A 17-gene assay to predict prostate cancer aggressiveness in the context of gleason grade heterogeneity, tumor multifocality, and biopsy undersampling. Eur Urol. 2014;66:550–60. doi: 10.1016/j.eururo.2014.05.004. doi:10.1016/j.eururo.2014.05.004. [DOI] [PubMed] [Google Scholar]
- [11].Davis J. Novel commercially available genomic tests for prostate cancer: a roadmap to understanding their clinical impact. BJU Int. 2014;114:320–2. doi: 10.1111/bju.12695. [DOI] [PubMed] [Google Scholar]
- [12].Arora R, Koch MO, Eble JN, Ulbright TM, Li L, Cheng L. Heterogeneity of Gleason grade in multifocal adenocarcinoma of the prostate. Cancer. 2004;100:2362–6. doi: 10.1002/cncr.20243. doi:10.1002/cncr.20243. [DOI] [PubMed] [Google Scholar]
- [13].Mehra R, Han B, Tomlins SA, Wang L, Menon A, Wasco MJ, et al. Heterogeneity of TMPRSS2 gene rearrangements in multifocal prostate adenocarcinoma: molecular evidence for an independent group of diseases. Cancer Res. 2007;67:7991–5. doi: 10.1158/0008-5472.CAN-07-2043. doi:10.1158/0008-5472.CAN-07-2043. [DOI] [PubMed] [Google Scholar]
- [14].Barry M, Perner S, Demichelis F, Rubin MA. TMPRSS2-ERG Fusion Heterogeneity in Multifocal Prostate Cancer: Clinical and Biologic Implications. Urology. 2007;70:630–3. doi: 10.1016/j.urology.2007.08.032. doi:10.1016/j.urology.2007.08.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Perner S, Demichelis F, Beroukhim R, Schmidt FH, Mosquera J-M, Setlur S, et al. TMPRSS2:ERG fusion-associated deletions provide insight into the heterogeneity of prostate cancer. Cancer Res. 2006;66:8337–41. doi: 10.1158/0008-5472.CAN-06-1482. doi:10.1158/0008-5472.CAN-06-1482. [DOI] [PubMed] [Google Scholar]
- [16].Demichelis F, Rubin MA. TMPRSS2-ETS fusion prostate cancer: biological and clinical implications. J Clin Pathol. 2007;60:1185–6. doi: 10.1136/jcp.2007.046557. doi:10.1136/jcp.2007.046557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Tomlins SA, Bjartell A, Chinnaiyan AM, Jenster G, Nam RK, Rubin MA, et al. ETS Gene Fusions in Prostate Cancer: From Discovery to Daily Clinical Practice. Eur Urol. 2009;56:275–86. doi: 10.1016/j.eururo.2009.04.036. doi:10.1016/j.eururo.2009.04.036. [DOI] [PubMed] [Google Scholar]
- [18].Minner S, Gärtner M, Freudenthaler F, Bauer M, Kluth M, Salomon G, et al. Marked heterogeneity of ERG expression in large primary prostate cancers. Mod Pathol. 2012 doi: 10.1038/modpathol.2012.130. doi:10.1038/modpathol.2012.130. [DOI] [PubMed] [Google Scholar]
- [19].Haffner MC, Mosbruger T, Esopi DM, Fedor H, Heaphy CM, Walker DA, et al. Tracking the clonal origin of lethal prostate cancer. J Clin Invest. 2013;123:4918–22. doi: 10.1172/JCI70354. doi:10.1172/JCI70354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Haffner MC, De Marzo AM, Yegnasubramanian S, Epstein JI, Carter HB. Diagnostic Challenges of Clonal Heterogeneity in Prostate Cancer. J Clin Oncol. 2014;32:2013–5. doi: 10.1200/JCO.2013.50.3540. doi:10.1200/JCO.2013.50.3540. [DOI] [PubMed] [Google Scholar]
- [21].Karnes RJ, Bergstralh EJ, Davicioni E, Ghadessi M, Buerki C, Mitra AP, et al. Validation of a Genomic Classifier that Predicts Metastasis Following Radical Prostatectomy in an At Risk Patient Population. J Urol. 2013 doi: 10.1016/j.juro.2013.06.017. doi:10.1016/j.juro.2013.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Den RB, Feng FY, Showalter TN, Mishra MV, Trabulsi EJ, Lallas CD, et al. Genomic Prostate Cancer Classifier Predicts Biochemical Failure and Metastases in Patients After Postoperative Radiation Therapy. Int J Radiat Oncol. 2014;89:1038–46. doi: 10.1016/j.ijrobp.2014.04.052. doi:10.1016/j.ijrobp.2014.04.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Klein EA, Yousefi K, Haddad Z, Choeurng V, Buerki C, Stephenson AJ, et al. A Genomic classifier improves prediction of metastatic disease within 5 years after surgery in node-negative high-risk prostate cancer patients managed by radical prostatectomy without adjuvant therapy. Eur Urol. 2014 doi: 10.1016/j.eururo.2014.10.036. [DOI] [PubMed] [Google Scholar]
- [24].Taylor BS, Schultz N, Hieronymus H, Gopalan A, Xiao Y, Carver BS, et al. Integrative Genomic Profiling of Human Prostate Cancer. Cancer Cell. 2010;18:11–22. doi: 10.1016/j.ccr.2010.05.026. doi:10.1016/j.ccr.2010.05.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Boormans JL, Korsten H, Ziel-Van Der Made AJC, Van Leenders GJLH, De Vos CV, Jenster G, et al. Identification of TDRD1 as a direct target gene of ERG in primary prostate cancer. Int J Cancer. 2013;133:335–45. doi: 10.1002/ijc.28025. doi:10.1002/ijc.28025. [DOI] [PubMed] [Google Scholar]
- [26].Brase JC, Johannes M, Mannsperger H, Falth M, Metzger J, Kacprzyk LA, et al. TMPRSS2-ERG -specific transcriptional modulation is associated with prostate cancer biomarkers and TGF-beta signaling. BMC Cancer. 2011;11:507. doi: 10.1186/1471-2407-11-507. doi:10.1186/1471-2407-11-507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Nakagawa T, Kollmeyer TM, Morlan BW, Anderson SK, Bergstralh EJ, Davis BJ, et al. A tissue biomarker panel predicting systemic progression after PSA recurrence post-definitive prostate cancer therapy. PLoS One. 2008;3:e2318. doi: 10.1371/journal.pone.0002318. doi:10.1371/journal.pone.0002318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Ruopp MD, Perkins NJ, Whitcomb BW, Schisterman EF. Youden Index and optimal cut- point estimated from observations affected by a lower limit of detection. Biometrical J. 2008;50:419–30. doi: 10.1002/bimj.200710415. doi:10.1002/bimj.200710415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Minner S, Enodien M, Sirma H, Luebke AM, Krohn A, Mayer PS, et al. ERG status is unrelated to PSA recurrence in radically operated prostate cancer in the absence of antihormonal therapy. Clin Cancer Res. 2011;17:5878–88. doi: 10.1158/1078-0432.CCR-11-1251. doi:10.1158/1078-0432.CCR-11-1251. [DOI] [PubMed] [Google Scholar]
- [30].Kumar-Sinha C, Tomlins SA, Chinnaiyan AM. Recurrent gene fusions in prostate cancer. Nat Rev Cancer. 2008;8:497–511. doi: 10.1038/nrc2402. doi:10.1038/nrc2402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Cuzick J, Swanson GP, Fisher G, Brothman AR, Berney DM, Reid JE, et al. Prognostic value of an RNA expression signature derived from cell cycle proliferation genes in patients with prostate cancer: a retrospective study. Lancet Oncol. 2011;12:245–55. doi: 10.1016/S1470-2045(10)70295-3. doi:10.1016/S1470-2045(10)70295-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Penney KL, Sinnott JA, Fall K, Pawitan Y, Hoshida Y, Kraft P, et al. mRNA expression signature of Gleason grade predicts lethal prostate cancer. J Clin Oncol. 2011;29:2391–6. doi: 10.1200/JCO.2010.32.6421. doi:10.1200/JCO.2010.32.6421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Braun M, Goltz D, Shaikhibrahim Z, Vogel W, Böhm D, Scheble V, et al. ERG protein expression and genomic rearrangement status in primary and metastatic prostate cancer—a comparative study of two monoclonal antibodies. Prostate Cancer Prostatic Dis. 2012;15:165–9. doi: 10.1038/pcan.2011.67. doi:10.1038/pcan.2011.67. [DOI] [PubMed] [Google Scholar]
- [34].Park K, Tomlins SA, Mudaliar KM, Chiu Y-L, Esgueva R, Mehra R, et al. Antibody- based detection of ERG rearrangement-positive prostate cancer. Neoplasia. 2010;12:590–8. doi: 10.1593/neo.10726. doi:10.1593/neo.10726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Tomlins SA, Palanisamy N, Siddiqui J, Chinnaiyan AM, Kunju LP. Antibody-based detection of erg rearrangements in prostate core biopsies, including diagnostically challenging cases: ERG staining in prostate core biopsies. Arch. Pathol. Lab. Med. 2012;136:935–46. doi: 10.5858/arpa.2011-0424-OA. doi:10.5858/arpa.2011-0424-OA. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Pettersson A, Graff RE, Bauer SR, Pitt MJ, Lis RT, Stack EC, et al. The TMPRSS2:ERG rearrangement, ERG expression, and prostate cancer outcomes: a cohort study and meta-analysis. Cancer Epidemiol Biomarkers Prev. 2012;21:1497–509. doi: 10.1158/1055-9965.EPI-12-0042. doi:10.1158/1055-9965.EPI-12-0042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Smith S, Tomlins SA. Prostate cancer SubtyPINg biomarKers and outcome: is clarity emERGing? Clin Cancer Res. 2014 doi: 10.1158/1078-0432.CCR-14-0818. doi:10.1158/1078-0432.CCR-14-0818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Flavin RJ, Pettersson A, Hendrickson WK, Fiorentino M, Finn SP, Kunz L, et al. SPINK1 Protein Expression and Prostate Cancer Progression. Clin Cancer Res. 2014 doi: 10.1158/1078-0432.CCR-13-1341. doi:10.1158/1078-0432.CCR-13-1341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Khani F, Mosquera JM, Park K, Blattner M, O’Reilly C, MacDonald TY, et al. Evidence for Molecular Differences in Prostate Cancer between African American and Caucasian Men. Clin Cancer Res. 2014:1078–0432. doi: 10.1158/1078-0432.CCR-13-2265. CCR – 13–2265 – . doi:10.1158/1078-0432.CCR-13-2265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Abdueva D, Wing M, Schaub B, Triche T, Davicioni E. Quantitative expression profiling in formalin-fixed paraffin-embedded samples by affymetrix microarrays. J Mol Diagn. 2010;12:409–17. doi: 10.2353/jmoldx.2010.090155. doi:10.2353/jmoldx.2010.090155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Schaefer G, Mosquera J-M, Ramoner R, Park K, Romanel a, Steiner E, et al. Distinct ERG rearrangement prevalence in prostate cancer: higher frequency in young age and in low PSA prostate cancer. Prostate Cancer Prostatic Dis. 2013;16:132–8. doi: 10.1038/pcan.2013.4. doi:10.1038/pcan.2013.4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Berg KD, Vainer B, Thomsen FB, Røder MA, Gerds TA, Toft BG, et al. ERG Protein Expression in Diagnostic Specimens Is Associated with Increased Risk of Progression During Active Surveillance for Prostate Cancer. Eur Urol. 2014 doi: 10.1016/j.eururo.2014.02.058. doi:10.1016/j.eururo.2014.02.058. [DOI] [PubMed] [Google Scholar]
- [43].Galletti G, Matov A, Beltran H, Fontugne J, Mosquera J-M, Cheung C, et al. ERG induces taxane resistance in castration-resistant prostate cancer. Nat Commun. 2014;25 doi: 10.1038/ncomms6548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Ateeq B, Tomlins SA, Laxman B, Asangani IA, Cao Q, Cao X, et al. Therapeutic targeting of SPINK1-positive prostate cancer. Sci Transl Med. 2011;3:72ra17. doi: 10.1126/scitranslmed.3001498. doi:10.1126/scitranslmed.3001498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Brenner JC, Ateeq B, Li Y, Yocum AK, Cao Q, Asangani IA, et al. Mechanistic Rationale for Inhibition of Poly(ADP-Ribose) Polymerase in ETS Gene Fusion-Positive Prostate Cancer. Cancer Cell. 2011;19:664–78. doi: 10.1016/j.ccr.2011.04.010. doi:10.1016/j.ccr.2011.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Shepherd FA, Domerg C, Hainaut P, Jänne PA, Pignon JP, Graziano S, et al. Pooled analysis of the prognostic and predictive effects of KRAS mutation status and KRAS mutation subtype in early-stage resected non-small-cell lung cancer in four trials of adjuvant chemotherapy. J Clin Oncol. 2013;31:2173–81. doi: 10.1200/JCO.2012.48.1390. doi:10.1200/JCO.2012.48.1390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Shaw AT, Yeap BY, Mino-Kenudson M, Digumarthy SR, Costa DB, Heist RS, et al. Clinical features and outcome of patients with non-small-cell lung cancer who harbor EML4-ALK. J Clin Oncol. 2009;27:4247–53. doi: 10.1200/JCO.2009.22.6993. doi:10.1200/JCO.2009.22.6993. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.