Abstract
DNA polymerase eta (POLH), a target of p53 tumor suppressor, plays a key role in translesion DNA synthesis (TLS). Loss of POLH is responsible for human cancer prone syndrome, Xeroderma Pigmentosum Variant (XPV). Due to its critical role in DNA repair and genome stability, POLH expression and activity are regulated by multiple pathways. In this study, we found that the levels of both POLH transcript and protein were decreased upon knockdown of the transcript encoding poly(rC)-binding protein 1 (PCBP1). We also found that the half-life of POLH mRNA was markedly decreased upon knockdown of PCBP1. Moreover, we found that PCBP1 directly bound to POLH 3′UTR and the PCBP1-binding site in POLH mRNA is an atypical AU-rich element. Finally, we showed that the AU-rich element in POLH 3′UTR was responsive to PCBP1 and sufficient for PCBP1 to regulate POLH expression. Altogether, we uncovered a novel mechanism by which POLH expression is controlled by PCBP1 via mRNA stability.
Keywords: POLH, p53, RNA binding protein, PCBP1, mRNA decay, AU-rich element
INTRODUCTION
DNA polymerase eta (POLH), a member of the Y-family DNA polymerases, is necessary for repair of DNA lesions induced by ultraviolet irradiation and carcinogens via translesion DNA synthesis (TLS) [1–6]. POLH can accurately repair cyclobutane pyrimidine dimers (CPDs), pyrimidine (6-4) pyrimidone photoadducts, 8,5′-cyclopurine-2′-deoxynucleosides (cPus) and 7,8-dihydro-8-oxoguanine (8-oxoG) caused by UV-irradiation or oxidative stress [7–11]. Upon DNA damage, POLH can be recruited to the sites of replication fork stalling by interacting with FANCD2 and PCNA [12–14]. Mutation of the POLH gene is associated with human syndrome, Xeroderma Pigmentosum Variant (XPV) [15–17]. XPV patients are prone to skin cancer [18–20]. Consistently, repression of POLH expression is observed in various types of skin cancer [18]. In addition to its role in TLS, POLH is necessary for hypermutation of immunoglobulin genes [21, 22] and for maintenance of genome stability [23–26].
POLH expression is found to be regulated by multiple mechanisms, including transcriptional regulation by DNA damage in a p53-dependent manner [25] and protein stability by Pirh2 and Mdm2 E3 ligases [27, 28]. C. elegans POLH is targeted for proteasomal degradation upon SUMOylation by the Cul4-Ddb1-Cdt2 pathway [29]. Additionally, the enzymatic activity of POLH is regulated by posttranslational modifications, such as SUMOylation and monoubiquitination [30, 31]. In this study, we found that POLH expression is regulated by poly(rC)-binding protein 1 (PCBP1, also called heterogeneous nuclear ribonucleoprotein E1 (hnRNP E1) or α-CP1) via mRNA stability. We also found that PCBP1 directly binds to POLH 3′UTR. Interestingly, we found that an AU-rich element in POLH mRNA is recognized by and responsive to PCBP1 although several PCBP1-binding sites are CU-rich elements or oligo(rC) elements [32–35]. Together, we uncovered a novel mechanism by which POLH expression is regulated by PCBP1 via mRNA stability.
EXPERIMENTAL PROCEDURE
Cell culture
Human pancreatic cancer cell line MIA-PaCa2, human colon cancer cell line p53−/− HCT116, human cervical carcinoma cell line ME180 and human breast cancer cell line MCF7 were cultured in DMEM (Invitrogen) with 10% fetal bovine serum (Hyclone) and maintained at 37°C in 5% CO2 incubator.
Plasmid
Lentiviral vectors (pLKO.1-puro) expressing shRNA targeting luciferase and PCBP1 were purchased from Sigma Inc. The targeting sequences are 5′-CGCTGAGTACTTCGAAATGTC-3′ for control luciferase shRNA and 5′-CCCATGATCCAACTGTGTAAT-3′ (shPCBP1) or GCTCCTCTGGTAGGCAGGTTACT (shPCBP1*) for PCBP1 shRNA. pGEX-4T-3 plasmid was used to express GST and GST-fused PCBP1 proteins as previously described [36]. To generate mutant p53(R175H) reporter vector, the DNA fragments amplified from POLH 3′UTR were digested with XhoI and NheI and then ligated into pcDNA3-p53(R175H) vector [25] cut by XhoI and XbaI. The primers used for amplification of POLH 3′ UTR are listed in Table 1. Plasmid RP11-22I24 (BACPAC Resources, Children’s Hospital and Research Center at Oakland, CA), which carries the POLH locus, was used as a template to amplify POLH 3′UTR.
Table 1.
Primers used in this study.
| Primer name | Sequence |
|---|---|
| Primers for RT-PCR | |
| POLH-exo4-F | 5′-tcgagccattgaaataagcc-3′ |
| POLH-exo5-R | 5′-acaaggtcagcctatctcgg-3′ |
| actin-exo3-F | 5′-ctgaagtaccccatcgagcacggca-3′ |
| actin-exo4-R | 5′-ggatagcacagcctggatagcaacg-3′ |
| PCBP1-F | 5′-ggcgggtgtaagatcaaaga-3′ |
| PCBP1-R | 5′-gagcggagaaatggtgtgtt-3′ |
| p63-1751-F | 5′-gaggttgggctgttcatcat-3′ |
| p63-2023-R | 5′-gtgaatcgcacagcatcaat-3′ |
| ΔNp63-153-F | 5′-ggaaaacaatgcccagactc-3′ |
| ΔNp63-513-R | 5′-tggggtcatcaccttgatct-3′ |
| Primers for generation of reporter vectors | |
| POLH-XhoI-2447-F | 5′-atcgctcgagtgctgccctcaggcttgcctgtaggattta-3′ |
| POLH-NheI-8412-R | 5′-atcggctagctattgtacagaataaaaatgttttattgaatac-3′ |
| POLH-NheI-3607-R | 5′-atcggctagccctgacgacagagggaga-3′ |
| POLH-XhoI-3515-F | 5′-atgcctcgagaatgtaatgagacttgcatagtt-3′ |
| POLH-NheI-4619-R | 5′-atcggctagctgccctagttaccatatcactt-3′ |
| POLH-XhoI-4580-F | 5′-atcgctcgaggaagccttgaaaccctaaa-3′ |
| POLH-NheI-5879-R | 5′-atcggctagccacctggtcattagtatcttttag-3′ |
| POLH-XhoI-5843-F | 5′-atcgctcgaggagaaatgctgatctaaaaga-3′ |
| POLH-NheI-7177-R | 5′-atcggctagcgattcaggtgatcctccc-3′ |
| POLH-XhoI-7032-F | 5′-atgcctcgaggaggtgggtggactactgga-3′ |
| POLH-NheI-2723-R | 5′-atcggctagccaaggcccacacacttttta-3′ |
| POLH-XhoI2447-2471- | 5′-atcgctcgagtgctgccctcaggcttgcctgtagg-3′ |
| Primers for generation of REMSA RNA probes | |
| POLH-2447-T7-F | 5′-ggatcctaatacgactcactatagggagtgctgccctcaggcttgcctg-3′ |
| POLH-3607-R | 5′-cctgacgacagagggaga-3′ |
| POLH-3515-T7-F | 5′-ggatcctaatacgactcactatagggagaatgtaatgagacttgcatagtt-3′ |
| POLH-4619-R | 5′-tgccctagttaccatatcactt-3′ |
| POLH-4580-T7-F | 5′-ggatcctaatacgactcactatagggaggaagccttgaaaccctaaa-3′ |
| POLH-5879-R | 5′-cacctggtcattagtatcttttag-3′ |
| POLH-5843-T7-F | 5′-ggatcctaatacgactcactatagggaggagaaatgctgatctaaaaga-3′ |
| POLH-7177-R | 5′-gattcaggtgatcctccc-3′ |
| POLH-7032-T7-F | 5′-ggatcctaatacgactcactatagggaggaggtgggtggactactgga-3′ |
| POLH-8412-R | 5′-tattgtacagaataaaaatgtt-3′ |
| POLH-2964-R | 5′-ggctggtctcaaactcctga-3′ |
| POLH-2945-T7-F | 5′-ggatcctaatacgactcactatagggagtcaggagtttgagaccagcc-3′ |
| POLH-2723-R | 5′-caaggcccacacacttttta-3′ |
| POLH-2704-T7-F | 5′-ggatcctaatacgactcactatagggagtaaaaagtgtgtgggccttg-3′ |
| POLH-2587-R | 5′-tcagcacctaaatggattattttt-3′ |
| POLH-2564-T7-F | 5′-ggatcctaatacgactcactatagggagaaaaataatccatttaggtgctga-3′ |
| T7-deltaARE-A-F | 5′-ggatcctaatacgactcactatagggagtgctgccctcaggcttgcctgtaggcagatctttatctttaatat-3′ |
| T7-deltaARE-B-F | 5′-ggatcctaatacgactcactatagggagtgctgccctcaggcttgcctgtaggatttaatattttttatctttacagatctcagatttccctgagaaag-3′ |
| T7-deltaARE-AB-F | 5′-ggatcctaatacgactcactatagggagtgctgccctcaggcttgcctgtaggcagatctcagatttccctgagaaagggaat-3′ |
| POLH-ARE-A-t2a-F | 5′-tgctgccctcaggcttgcctgtaggaaaaaaaaaaaaaaaacaaaacagatctttatctttaatattttatctttacagatttccctgagaaag-3′ |
| POLH-ARE-B-t2a-F | 5′-tgctgccctcaggcttgcctgtaggatttaatattttttatctttacagatctaaaacaaaaaaaaaaaaacaaaacagatttccctgagaaag-3′ |
| T7-POLH-2447-2467-F | 5′-ggatcctaatacgactcactatagggagtgctgccctcaggcttgcctg-3′ |
F, forward; R, reverse.
RNA interference
For lentivirus preparation, shRNA-expressing vector (10 μg) and packaging plasmids (pMDL g/p RRE (5 μg), pCMV-VSVG (5 μg) and pRSV-REV (5 μg)) were co-transfected into HEK 293T cells (6×106) using Expressfect™ transfection reagent (Denville Scientific). Lentiviral particles were collected from the medium every 24 h for 2 days and then filtered and concentrated by ultracentrifugation at 107,000 g in a Beckman SW41TI rotor for 2 h at 4°C. Cells were transduced with concentrated lentiviral particles and then treated with puromycin for 3 days to eliminate untransduced cells. For MCF7 and p53−/− HCT116 cells, 1 μ g/ml of puromycin was used whereas 0.5 μg/ml of puromycin was used for MIA-PaCa2 and ME180 cells.
Antibodies and western blot analysis
Mouse anti-PCBP1 (E-2), mouse anti-p63 (4A4) and rabbit anti-POLH (H-300) purchased from Santa Cruz were used for western blots. Rabbit anti-PCBP1 (catalog#8534) from cell signaling was used for immunoprecipitation. Mouse anti-HA antibody was purchased from Covance. Rabbit anti-actin was purchased from Sigma.
Whole cell lysates were prepared with 2×SDS sample buffer and separated in 8–10% SDS-PAGE, transferred to nitrocellulose membrane and then probed with primary and secondary antibodies, followed by chemiluminescent detection.
RNA isolation and RT-PCR
Total RNAs were extracted from cells using Trizol reagent (Invitrogen) according to the manufacturer’s protocol. cDNA was synthesized using M-MLV reverse transcriptase (Promega) according to the manufacturer’s manual. Semi-quantitative PCR was performed with GoTaq DNA polymerase (Promega). RT-qPCR was performed with Maxima SYBR Green qPCR Master Mix (Thermo) and the relative expression level was calculated upon normalization to the level of actin transcript. The sequences of primers used for PCR are listed in Table 1.
RNA immunoprecipitation assay (RNA-IP)
RNA-IP was carried out as previously described [37]. Briefly, ~3 ×106 cells were lysed with 1 mL lysis buffer (10 mM HEPES, pH 7.0, 100 mM KCl, 100 mM NaCl, 10 mM MgCl2, 0.5% NP-40, 1 mM DTT) supplemented with RiboLock™ ribonuclease inhibitor (Thermo Scientific) and protease inhibitor cocktails (Sigma). The cell lysates were centrifuged for 10 min at 13,000 rpm at 4°C, followed by imunoprecipitation with 2 μg of rabbit anti-PCBP1 antibody or isotype control IgG at 4°C for 6 hours. The RNA-protein immunocomplexes were brought down by protein A-agrose beads (Sigma), followed by RT-PCR analysis.
RNA electrophoretic mobility shift assay (REMSA)
The probes used for REMSA were labeled during in vitro transcription of a DNA fragment containing the T7 promoter and a part or all of POLH 3′UTR. Briefly, 250 ng of purified DNA fragments was incubated with 20 μCi of [α-32P]-UTP (800 Ci/mL, PerkinElmer), 0.5 mM each of rNTP (A, G and C), 10 U of T7 RNA polymerase (Ambion) and 20 U of RNase inhibitor (Thermo) in 10 μL of reaction mixture at 37°C for 1 h. One unit of DNase I (Promega) was added to the reaction mixture to remove the DNA template. The labeled RNA probes were purified by Sephadex G-50 column to remove unlabeled free nucleotides. The radioactivity of probes was measured by a liquid scintillation counter. REMSA was performed as previously described [36]. Briefly, 50,000 CPM of [α-32P]-labeled RNA probe, 250 nM GST or GST-PCBP1, and 100 ng/μL of yeast tRNA were mixed in 20 μL of binding buffer (10 mM Tris-Cl, pH 8.0, 25 mM KCl, 10 mM MgCl2 and 1 mM DTT) at room temperature for 20 min, followed by treatment with 100 U of RNase T1 (Ambion) for 15 min at 37°C to digest unprotected RNA fragments. The RNA-protein complexes were then separated in 7% native polyacrylamide gel and visualized by autoradiography.
RESULTS
POLH expression is decreased by knockdown of PCBP1
A transcript with a long 3′UTR is often subject to posttranscriptional regulation, including mRNA stability. Since POLH has a long 3′UTR (~6,000 nt) along with several CU- and AU-rich elements, we examined whether PCBP1, a poly(rC)-binding protein, regulates POLH expression. To test this, PCBP1 was knocked down in MCF7 cells transduced with lentivirus expressing PCBP1 shRNAs for 3 days. A lentivirus expressing shRNA targeting luciferase mRNA was used as a negative control. We found that the level of POLH protein in MCF7 cells, which carries wild-type p53, was decreased by knockdown of PCBP1, but not control shRNA (Fig. 1A). To rule out potential effects of wild-type p53 on expression of POLH, a target of p53 [25], PCBP1 was knocked down in p53−/− HCT116 cells, MIA-PaCa2 cells which carry a mutant p53 [38], and ME180 cells which express undetectable wild-type p53 [39]. We showed that knockdown of PCBP1 led to decreased expression of POLH regardless of the status of the p53 gene (Fig. 1B–D). As a control, ΔNp63 in ME180 cells and TAp63 in MIA-PaCa2 cells were decreased by knockdown of PCBP1, consistent with a previous report [36]. Next, we examined whether the decreased levels of POLH protein are due to decreased levels of POLH transcript. Indeed, we found that the levels of POLH transcript were decreased in cells by knockdown of PCBP1 regardless of the status of the p53 gene (Fig. 2A–D). Together, these results suggest that PCBP1 is necessary for appropriate expression of POLH.
Figure 1. POLH expression is decreased upon knockdown of PCBP1.
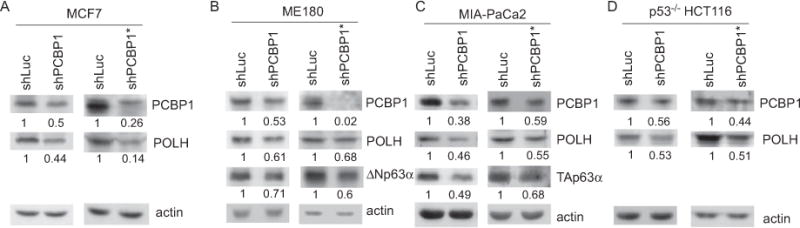
MCF7 (A), ME180 (B), MIA-PaCa2 (C) and p53−/− HCT116 (D) cells were transduced with a lentivirus expressing luciferase shRNA (shLuc) or one of the two PCBP1 shRNAs (shPCBP1 or shPCBP1*), followed by puromycin selection for 3 days. Whole cell lysates were collected and used for western blotting to measure the levels of PCBP1, POLH, actin, ΔNp63α, and TAp63α. The level of actin protein was used as a loading control. Western blots shown in the figure were representative of three independent experiments. The values below the strips were the relative intensities normalized to actin.
Figure 2. POLH mRNA is decreased upon knockdown of PCBP1.
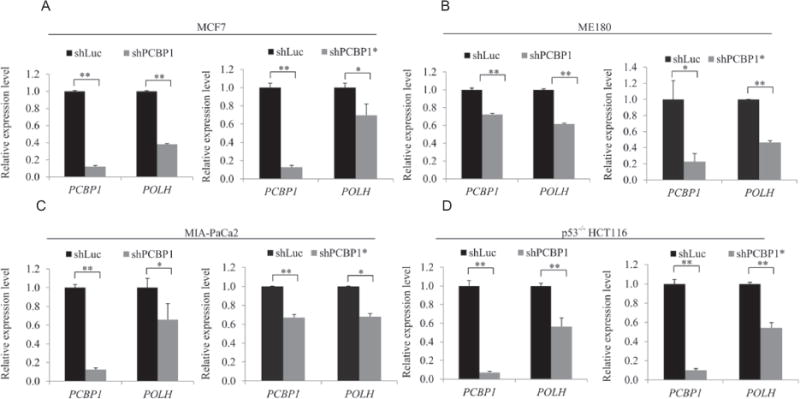
MCF7 (A), ME180 (B), MIA-PaCa2 (C) and p53−/− HCT116 (D) cells were transduced with a lentivirus expressing a control luciferase shRNA (shLuc) or one of the two PCBP1 shRNAs (shPCBP1 or shPCBP1*), followed by puromycin selection for 3 days. Total RNA was purified and RT-qPCR was performed to determine the levels of POLH, PCBP1 and actin transcripts. Error bars indicate S.D. and significance was calculated using t-test (**P<0.01, * P<0.05).
POLH mRNA stability is regulated by PCBP1
As an RNA-binding protein, PCBP1 may bind to its target and then regulate the target’s mRNA stability. To test this, the half-life of POLH mRNA was measured in p53−/− HCT116 cells transduced with a lentivirus expressing luciferase shRNA or PCBP1 shRNA (shPCBP1) for 3 days. The cells were then treated with 100 μ M 5, 6-Dichloro-1-β-D-ribofuranosylbenzimidazole (DRB), a transcription inhibitor, to block de novo RNA synthesis. We found that the half-life of POLH mRNA was decreased from ~2.57 h in control cells to ~1.44 h in PCBP1-knockdown cells (Fig. 3).
Figure 3. POLH mRNA stability is regulated by PCBP1.
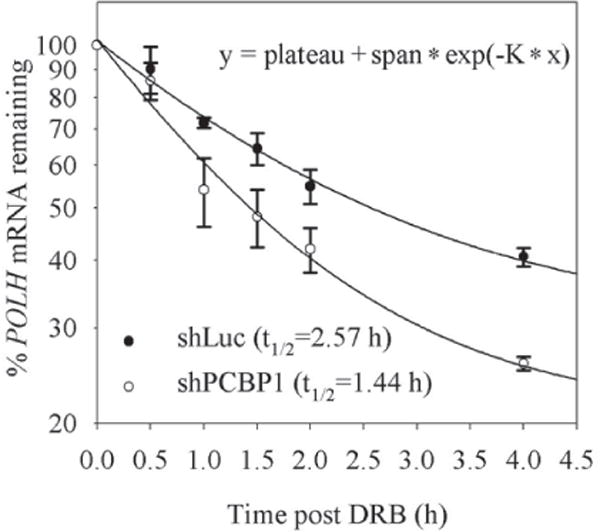
p53−/− HCT116 cells were transduced with a lentivirus expressing shLuc or shPCBP1 and selected with puromycin for 3 days, followed by treatment with 5, 6-Dichloro-1-β-D-ribofuranosylbenzimidazole (DRB) for the indicated times. The levels of POLH and actin transcripts were determined by RT-qPCR. The relative levels of POLH transcript were normalized with the levels of actin, which were then plotted along with the times following DRB treatment to determine the relative half-life of POLH mRNA. One of the three independent experiments was shown here.
To examine whether PCBP1 physically associates with POLH mRNA in vivo, RNA immunoprecipitation assay followed by RT-PCR was performed with MIA-PaCa2, p53−/− HCT116 and ME180 cells. We found that the level of POLH mRNA was highly enriched in anti-PCBP1 immunocomplexes (Fig. 4A–C). The levels of TAp63 α and ΔNp63 α transcripts were also examined as positive controls and found to be enriched in anti-PCBP1 immunocomplexes (Fig. 4A–C), consistent with a previous report [36]. In contrast, no interaction was found between actin transcript and PCBP1 (Fig. 4).
Figure 4. PCBP1 physically interacts with POLH transcript in vivo.
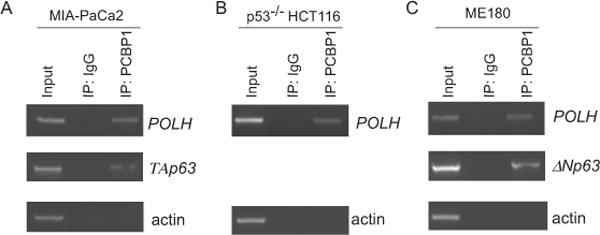
MIA-PaCa2 (A), p53−/− HCT116 (B) and ME180 (C) cells were lysed by immunoprecipitation buffer and incubated with rabbit anti-PCBP1 antibody or control IgG, followed by washing and RNA extraction. RT-PCR was performed to examine the level of POLH in the control IgG and anti-PCBP1 immunocomplexes. The levels of ΔNp63 transcript in ME180 cells, and TAp63 transcript in MIA-PaCa2 cells, were measured as a positive control. The levels of actin transcript were used as a negative control. The RT-PCR result is representative of at least three independent experiments.
POLH 3′UTR is recognized by and responsive to PCBP1
To identify a region in POLH 3′UTR that is responsive to PCBP1, we generated seven reporter (mutant p53 R175H) plasmids which carry none or a segment from POLH 3′UTR (Fig. 5A). Mutant p53 R175H was chosen as a reporter since mutant p53 is highly stable and can be easily detected. These reporters were expressed in MIA-PaCa2 cells transduced with a lentivirus expressing shRNA (shPCBP1) against luciferase or PCBP1. As expected, knockdown of PCBP1 led to decreased expression of endogenous POLH (Fig. 5B). We also found that knockdown of PCBP1 was capable of decreasing the level of mutant p53 protein from a reporter vector that contains the full-length or fragment A of POLH 3′UTR (Fig. 5B, 3′UTR and A panels). In contrast, knockdown of PCBP1 had no obvious effect on the expression of mutant p53 from the control vector that does not carry POLH 3′UTR and vectors that contain fragments B–E of POLH 3′UTR (Fig. 5B, Ctrl and B–E panels). Thus, fragment A (nt 2447–3607) of POLH 3′UTR carries a PCBP1-responsive element.
Figure 5. POLH 3′UTR is responsive to PCBP1.
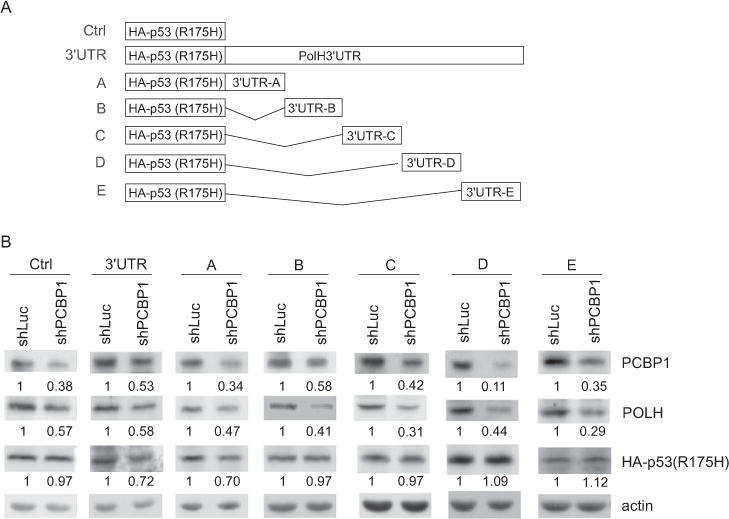
(A) Schematic presentation of mutant p53 (R175H) reporter vectors that carry none, the full-length, or a part of POLH 3′UTR. Ctrl: no POLH 3′UTR; 3′UTR: the full-length POLH 3′UTR from nt 2447–8412; A: nt 2447–3607 from POLH mRNA; B: nt 3515–4619 from POLH mRNA; C: nt 4580–5879 from POLH mRNA; D: nt 5843–7177 from POLH mRNA; E: 7032–8412 from POLH mRNA. (B) The reporter vectors were transfected into MIA-PaCa2 cells transduced with a lentivirus expressing luciferase shRNA or PCBP1 shRNA (shPCBP1) for 3 days. Western blots were performed to determine the levels of the reporter (HA-tagged mutant p53 R175H), endogenous POLH, PCBP1, and actin. Western blots shown in the figure were representative of three independent experiments. The values below the strips were the relative intensities normalized to actin.
To define the PCBP1-responsive element in POLH 3′UTR, REMSA was performed with five RNA probes (A–E), which span the entire POLH 3′UTR, to map the binding site of PCBP1 in POLH transcript (Fig 6A). p63 3′UTR was used as a positive control [36]. We showed that GST-fused PCBP1 bound strongly to fragment A and p63 probe, weakly to fragment C, but little if any to fragments B, D, and E (Fig. 6B). As expected, GST alone didn’t bind to these probes (Fig. 6B). The specificity was confirmed by competition assay. As indicated in Fig. 6C, the binding of PCBP1 to fragment A was inhibited by addition of an excess amount of un-labeled fragment A or p63 RNA probe. Next, we prepared six sub-fragments from fragment A: A1 (nt 2447–2964); A2 (nt 2945–3607); A11 (nt 2447–2723); A12 (nt 2704–2964); A111 (nt 2447–2587); A112 (nt 2564–2723) (Fig. 6A). We showed that GST-fused PCBP1 bound to A1, A11, and A111, but not to A2, A12, and A112 (Fig. 6D–F). These results suggest that sub-fragment A111 (nt 2447–2587) contains the PCBP1-binding site.
Figure 6. PCBP1 directly binds to POLH 3′UTR.
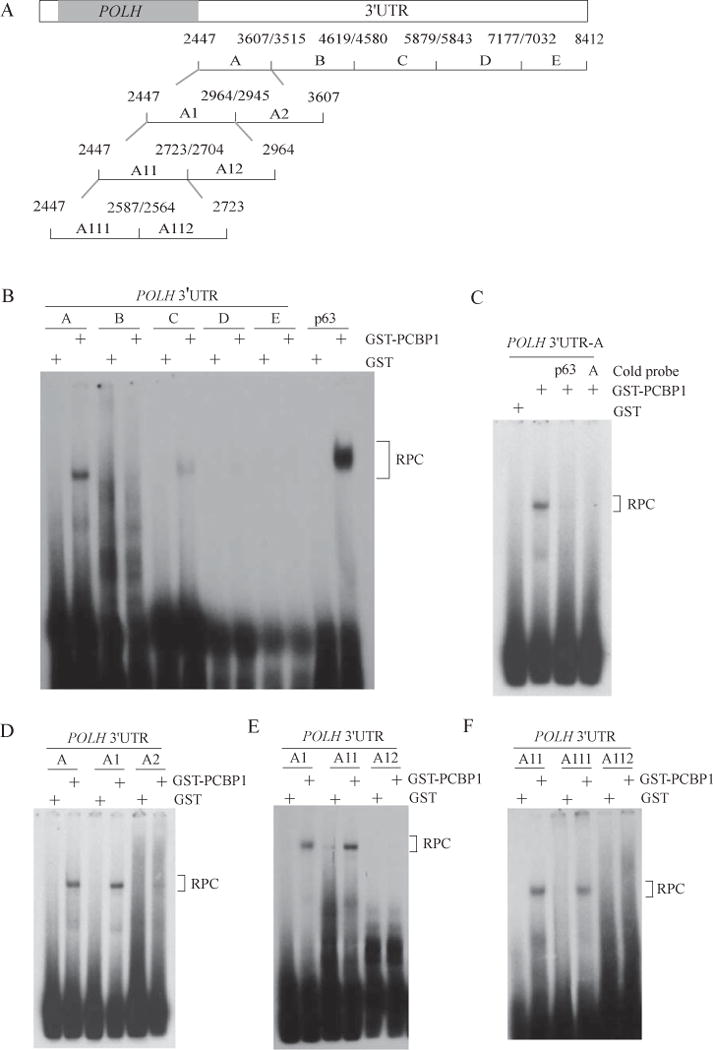
(A) Schematic presentation of the transcript of POLH and the locations of the probes used for REMSA. (B) PCBP1 binds to fragment A of POLH 3′UTR (nt 2447–3607). REMSA was performed with GST or GST-fused PCBP1 along with P32-labeled RNA probes (A, B, C, D, E and p63). p63 probe was used as positive control. RPC, RNA-protein complex. (C) Competition assay was performed by mixing P32-labeled RNA probe A along with or without 50-fold of unlabeled p63 probe or probe A. (D) REMSA was performed by mixing P32-labeled RNA probes (A, A1 and A2) with GST or GST-PCBP1. (E) REMSA was performed by mixing P32-labeled RNA probes (A1, A11 and A12) with GST or GST-PCBP1. (F) REMSA was performed by mixing P32-labeled RNA probes (A11, A111 and A112) with GST or GST-PCBP1. One of the two independent experiments was shown here.
PCBP1 directly binds to an AU-rich element in POLH 3′UTR
PCBP proteins are shown to preferentially recognize CU/C-rich elements [33]. Thus, we searched for such elements in A11 region (nt 2447–2723) of POLH 3′UTR. We found two well-conserved AU-rich elements, ARE-A (nt 2472–2492) and ARE-B (nt 2500–2522), but no CU/C-rich elements (Fig. 7A). Since PCBP1 is also found to recognize a poly(rU) element [40], we examined whether one or both ARE elements are recognized by PCBP1. To test this, we generated three RNA probes in which one or both ARE elements were deleted: ΔARE-A, ΔARE-B and ΔARE-AB (Fig. 7A). We found that ΔARE-A was still recognized by PCBP1 whereas the binding of PCBP1 to ΔARE-B and ΔARE-AB was markedly deceased (Fig. 7B), suggesting that ARE-B is the primary PCBP1-binding site. To further define the PCBP1 responsive element in POLH mRNA, we tested whether the poly(rU) sequence in ARE-B is required for PCBP1 binding. To address this, two RNA probes were generated: ARE-A-U2A and ARE-B-U2A in which U to A substitutions were made (Fig. 7C). REMSA was performed and showed that the binding of PCBP1 to probe ARE-B-U2A was nearly abolished (Fig. 7D). In contrast, the binding of PCBP1 to probe ARE-A-U2A was not decreased, but instead increased (Fig. 7D). These results suggest that the poly (rU) sequence in ARE-B is essential for PCBP1 binding.
Figure 7. An ARE element in POLH 3′UTR is recognized by and responsive to PCBP1.
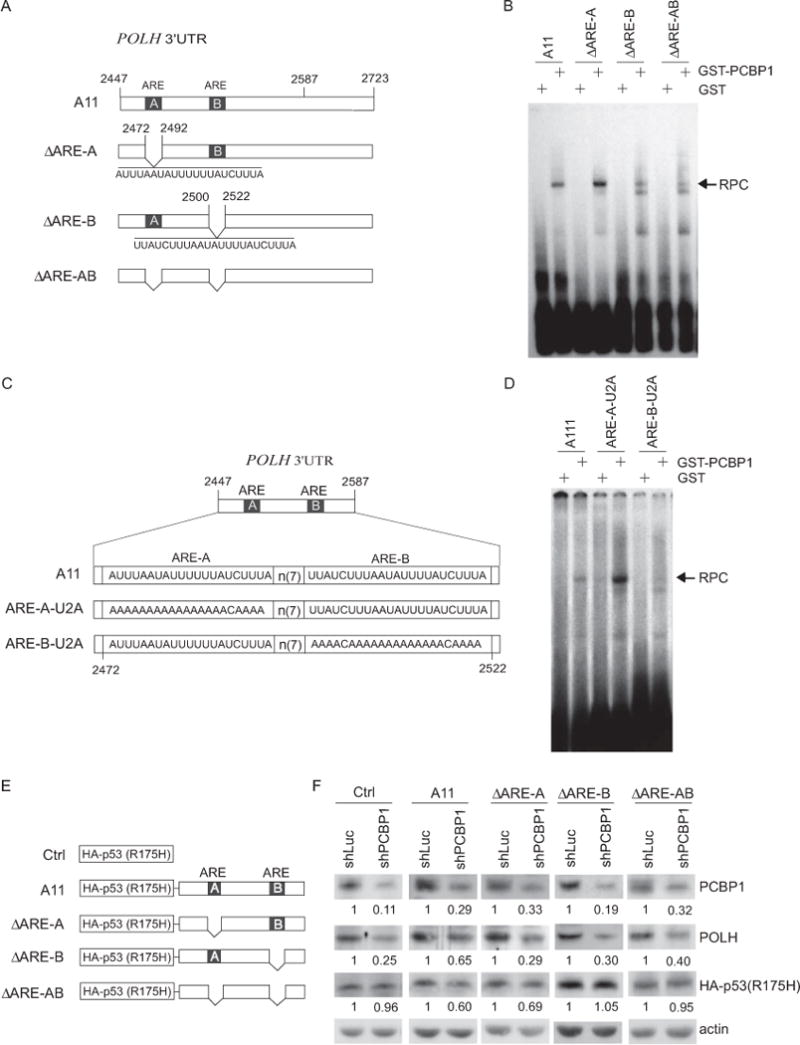
(A) Schematic presentation of the wild-type and deletion mutant probes used for REMSA. (B) REMSA was performed by mixing P32-labeled probes (A11, ΔARE-A, ΔARE-B and ΔARE-AB) with GST or GST-PCBP1 to identify the PCBP1-binding site in POLH 3′UTR. RPC, RNA-protein complex. (C) Schematic presentation of wild-type and mutant probes used for REMSA. (D) Poly(rU) nucleotides in ARE-B of POLH 3′UTR are crucial for PCBP1 binding. REMSA was performed by mixing P32-labeled RNA probes (A111, ARE-A-U2A and ARE-B-U2A) with GST and GST-PCBP1. One of the two independent experiments was shown here. (E) Schematic presentation of the reporter vectors for identification of the PCBP1-binding site in POLH 3′UTR. (F) ARE-B in POLH 3′UTR is responsive to PCBP1. The reporter vectors were transfected into MIA-PaCa2 cells transduced with a lentivirus expressing luciferase shRNA or PCBP1 shRNA (shPCBP1) for 3 days. The levels of the reporter (HA-tagged mutant p53 R175H), endogenous POLH, PCBP1, and actin were determined by western blotting. Western blots shown here were representative of three independent experiments. The values below the strips were the relative intensities normalized to actin.
To determine whether ARE-B is responsive to PCBP1 in vivo, we generated four additional reporter plasmids carrying mutant p53 R175H and a portion of POLH 3′UTR (nt 2447–nt 2723) with or without ARE-A, ARE-B, or both (Fig. 7E). These reporter plasmids were expressed in MIA-PaCa2 cells transduced with a lentivirus expressing shRNA against luciferase or PCBP1. As a positive control, the level of endogenous POLH was measured and found to be decreased upon knockdown of PCBP1 (Fig. 7F). As a negative control, we found that knockdown of PCBP1 had no obvious effect on the expression of mutant p53 (R175H) from a reporter vector that carries no sequence from POLH 3′UTR (Fig. 7F, Ctrl panel). Interestingly, we found that the level of mutant p53 was decreased by knockdown of PCBP1 for the reporter vectors that carry an intact ARE-B (A11 and ΔARE-A) (Fig. 7F, A11 and ΔARE-A panels). In contrast, knockdown of PCBP1 had no effect on mutant p53 expression for reporter vectors that carry ΔARE-B and ΔARE-AB region, respectively (Fig. 7F, ΔARE-B and ΔARE-AB panels). These data suggest that PCBP1 binds to ARE-B and the poly(rU) nucleotides in ARE-B are crucial for the binding of PCBP1 to POLH mRNA.
DISCUSSION
Previous studies have shown that POLH expression can be regulated at the transcriptional level by p53 and at post-translational levels by Pirh2 and MDM2 [25, 27, 28]. It is not clear, however, whether POLH expression is regulated by other mechanisms. In this study, we found that PCBP1 regulates POLH expression via mRNA stability. Thus, an obvious question would be: is there a functional connection between PCBP1 and POLH? Indeed, PCBP1 is known to inhibit tumor invasiveness and metastasis by repressing PRL3 [41] and CD44 [42]. Additionally, dephosphorylated PCBP1 is capable of repressing epithelial-mesenchymal transition (EMT) via decreased translation of disabled-2 (Dab2) and interleukin-like EMT inducer (ILEI) [43]. Consistently, PCBP1 is found to be downregulated in cervical tumor tissues [44], in breast cancer cell lines [45], and during malignant transformation of hydatidiform moles [46]. Since POLH is necessary for the maintenance of genome stability [23, 24], downregulation of PCBP1 in cancer cells would reduce POLH mRNA stability, leading to genome instability. Additionally, since POLH plays a role in p53 activation upon DNA damage [25], downregulation of PCBP1 in cancer cells would reduce the level of POLH, thus weakening p53 activation. Thus, further studies are warranted to address the relationship between PCBP1 and POLH in normal and cancer cells, which may provide an insight into the possibility that PCBP1 might be explored as a marker or target for anticancer therapeutic strategies.
PCBP1 is known to recognize poly(rC)- or CU-rich elements in its targets, including tyrosine hydroxylase [47], β-globin [48], androgen receptor [49], collagens [50], erythropoietin [47], and 15-lipoxygenase [51]. PCBP1 also recognizes to non-poly(rC) elements in other targets, including human papilloma virus L2 transcript [52] and Dab2 and ILEI transcripts [43]. However, PCBP1 consensus element has not been defined in these transcripts. Here, we found that PCBP1 regulates POLH expression via binding to an AU-rich element located in the proximal POLH 3′UTR. Although POLH mRNA harbors at least two AREs, our data suggest that only one ARE element is recognized by and responsive to PCBP1. Thus, future studies are needed to define how POLH mRNA stability is regulated by PCBP1 and other RNA-binding proteins via the AU-rich element.
SUMMARY.
DNA polymerase eta (POLH) is mutated in Xeroderma Pigmentosum Variant patients and required for genome stability. However, it is still uncertain how POLH expression is regulated at mRNA level. Here we found that POLH expression is regulated by poly(rC)-binding protein PCBP1 via mRNA stability.
Acknowledgments
FUNDING: This work was supported in part by NIH grant CA123227, CA076069, and CA081237.
Abbreviations
- PCBP1
poly(rC)-binding protein 1
- POLH
DNA polymerase eta
- ARE
AU-rich element
- TLS
translesion DNA synthesis
- REMSA
RNA electrophoretic mobility shift assay
- UTR
untranslated region
- shRNA
short hairpin RNA
- GST
glutathione S-transferase
Footnotes
AUTHOR CONTRIBUTION
R.C. and X.C. designed the research, analyzed the data and wrote the manuscript; R.C. performed experiments; S.-J. C. and Y.-S. J. contributed reagents and analyzed the data.
References
- 1.Cleaver JE, Lam ET, Revet I. Disorders of nucleotide excision repair: the genetic and molecular basis of heterogeneity. Nat Rev Genet. 2009;10:756–768. doi: 10.1038/nrg2663. [DOI] [PubMed] [Google Scholar]
- 2.Friedberg EC, Wagner R, Radman M. Specialized DNA polymerases, cellular survival, and the genesis of mutations. Science. 2002;296:1627–1630. doi: 10.1126/science.1070236. [DOI] [PubMed] [Google Scholar]
- 3.McCulloch SD, Kunkel TA. The fidelity of DNA synthesis by eukaryotic replicative and translesion synthesis polymerases. Cell Res. 2008;18:148–161. doi: 10.1038/cr.2008.4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Prakash S, Johnson RE, Prakash L. Eukaryotic translesion synthesis DNA polymerases: specificity of structure and function. Annu Rev Biochem. 2005;74:317–353. doi: 10.1146/annurev.biochem.74.082803.133250. [DOI] [PubMed] [Google Scholar]
- 5.Fischhaber PL, Friedberg EC. How are specialized (low-fidelity) eukaryotic polymerases selected and switched with high-fidelity polymerases during translesion DNA synthesis? DNA Repair (Amst) 2005;4:279–283. doi: 10.1016/j.dnarep.2004.08.011. [DOI] [PubMed] [Google Scholar]
- 6.Lange SS, Takata K, Wood RD. DNA polymerases and cancer. Nat Rev Cancer. 2011;11:96–110. doi: 10.1038/nrc2998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Maga G, Villani G, Crespan E, Wimmer U, Ferrari E, Bertocci B, Hubscher U. 8-oxo-guanine bypass by human DNA polymerases in the presence of auxiliary proteins. Nature. 2007;447:606–608. doi: 10.1038/nature05843. [DOI] [PubMed] [Google Scholar]
- 8.Haracska L, Yu SL, Johnson RE, Prakash L, Prakash S. Efficient and accurate replication in the presence of 7,8-dihydro-8-oxoguanine by DNA polymerase eta. Nature genetics. 2000;25:458–461. doi: 10.1038/78169. [DOI] [PubMed] [Google Scholar]
- 9.You C, Swanson AL, Dai X, Yuan B, Wang J, Wang Y. Translesion synthesis of 8,5′-cyclopurine-2′-deoxynucleosides by DNA polymerases eta, iota, and zeta. J Biol Chem. 2013;288:28548–28556. doi: 10.1074/jbc.M113.480459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Albertella MR, Green CM, Lehmann AR, O’Connor MJ. A role for polymerase eta in the cellular tolerance to cisplatin-induced damage. Cancer Res. 2005;65:9799–9806. doi: 10.1158/0008-5472.CAN-05-1095. [DOI] [PubMed] [Google Scholar]
- 11.Cruet-Hennequart S, Gallagher K, Sokol AM, Villalan S, Prendergast AM, Carty MP. DNA polymerase eta, a key protein in translesion synthesis in human cells. Subcell Biochem. 2010;50:189–209. doi: 10.1007/978-90-481-3471-7_10. [DOI] [PubMed] [Google Scholar]
- 12.Fu D, Dudimah FD, Zhang J, Pickering A, Paneerselvam J, Palrasu M, Wang H, Fei P. Recruitment of DNA polymerase eta by FANCD2 in the early response to DNA damage. Cell Cycle. 2013;12:803–809. doi: 10.4161/cc.23755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kannouche PL, Wing J, Lehmann AR. Interaction of human DNA polymerase eta with monoubiquitinated PCNA: a possible mechanism for the polymerase switch in response to DNA damage. Mol Cell. 2004;14:491–500. doi: 10.1016/s1097-2765(04)00259-x. [DOI] [PubMed] [Google Scholar]
- 14.Acharya N, Yoon JH, Gali H, Unk I, Haracska L, Johnson RE, Hurwitz J, Prakash L, Prakash S. Roles of PCNA-binding and ubiquitin-binding domains in human DNA polymerase eta in translesion DNA synthesis. Proc Natl Acad Sci U S A. 2008;105:17724–17729. doi: 10.1073/pnas.0809844105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Di Lucca J, Guedj M, Lacapere JJ, Fargnoli MC, Bourillon A, Dieude P, Dupin N, Wolkenstein P, Aegerter P, Saiag P, Descamps V, Lebbe C, Basset-Seguin N, Peris K, Grandchamp B, Soufir N. Variants of the xeroderm a pigmentosum variant gene (POLH) are associated with melanoma risk. Eur J Cancer. 2009;45:3228–3236. doi: 10.1016/j.ejca.2009.04.034. [DOI] [PubMed] [Google Scholar]
- 16.Guo J, Zhou G, Zhang W, Song Y, Bian Z. A novel mutation causes XP-V disease and XP-V tumor proneness may involve imbalance of numerous DNA polymerases. Oncol Lett. 2013;6:1583–1590. doi: 10.3892/ol.2013.1604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Liu X, Zhang X, Qiao J, Fang H. Identification of a novel nonsense mutation in POLH in a Chinese pedigree with xeroderma pigmentosum, variant type. Int J Med Sci. 2013;10:766–770. doi: 10.7150/ijms.6095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Flanagan AM, Rafferty G, O’Neill A, Rynne L, Kelly J, McCann J, Carty MP. The human POLH gene is not mutated, and is expressed in a cohort of patients with basal or squamous cell carcinoma of the skin. Int J Mol Med. 2007;19:589–596. [PubMed] [Google Scholar]
- 19.Glick E, White LM, Elliott NA, Berg D, Kiviat NB, Loeb LA. Mutations in DNA polymerase eta are not detected in squamous cell carcinoma of the skin. Int J Cancer. 2006;119:2225–2227. doi: 10.1002/ijc.22099. [DOI] [PubMed] [Google Scholar]
- 20.Broughton BC, Cordonnier A, Kleijer WJ, Jaspers NG, Fawcett H, Raams A, Garritsen VH, Stary A, Avril MF, Boudsocq F, Masutani C, Hanaoka F, Fuchs RP, Sarasin A, Lehmann AR. Molecular analysis of mutations in DNA polymerase eta in xeroderma pigmentosum-variant patients. Proc Natl Acad Sci U S A. 2002;99:815–820. doi: 10.1073/pnas.022473899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Delbos F, Aoufouchi S, Faili A, Weill JC, Reynaud CA. DNA polymerase eta is the sole contributor of A/T modifications during immunoglobulin gene hypermutation in the mouse. J Exp Med. 2007;204:17–23. doi: 10.1084/jem.20062131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zeng X, Winter DB, Kasmer C, Kraemer KH, Lehmann AR, Gearhart PJ. DNA polymerase eta is an A-T mutator in somatic hypermutation of immunoglobulin variable genes. Nat Immunol. 2001;2:537–541. doi: 10.1038/88740. [DOI] [PubMed] [Google Scholar]
- 23.Rey L, Sidorova JM, Puget N, Boudsocq F, Biard DS, Monnat RJ, Jr, Cazaux C, Hoffmann JS. Human DNA polymerase eta is required for common fragile site stability during unperturbed DNA replication. Mol Cell Biol. 2009;29:3344–3354. doi: 10.1128/MCB.00115-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bergoglio V, Boyer AS, Walsh E, Naim V, Legube G, Lee MY, Rey L, Rosselli F, Cazaux C, Eckert KA, Hoffmann JS. DNA synthesis by Pol eta promotes fragile site stability by preventing under-replicated DNA in mitosis. J Cell Biol. 2013;201:395–408. doi: 10.1083/jcb.201207066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Liu G, Chen X. DNA polymerase eta, the product of the xeroderma pigmentosum variant gene and a target of p53, modulates the DNA damage checkpoint and p53 activation. Mol Cell Biol. 2006;26:1398–1413. doi: 10.1128/MCB.26.4.1398-1413.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Durando M, Tateishi S, Vaziri C. A non-catalytic role of DNA polymerase eta in recruiting Rad18 and promoting PCNA monoubiquitination at stalled replication forks. Nucleic Acids Res. 2013;41:3079–3093. doi: 10.1093/nar/gkt016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jung YS, Liu G, Chen X. Pirh2 E3 ubiquitin ligase targets DNA polymerase eta for 20S proteasomal degradation. Mol Cell Biol. 2010;30:1041–1048. doi: 10.1128/MCB.01198-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jung YS, Qian Y, Chen X. DNA polymerase eta is targeted by Mdm2 for polyubiquitination and proteasomal degradation in response to ultraviolet irradiation. DNA Repair (Amst) 2012;11:177–184. doi: 10.1016/j.dnarep.2011.10.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kim SH, Michael WM. Regulated proteolysis of DNA polymerase eta during the DNA-damage response in C. elegans. Mol Cell. 2008;32:757–766. doi: 10.1016/j.molcel.2008.11.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bienko M, Green CM, Sabbioneda S, Crosetto N, Matic I, Hibbert RG, Begovic T, Niimi A, Mann M, Lehmann AR, Dikic I. Regulation of translesion synthesis DNA polymerase eta by monoubiquitination. Mol Cell. 2010;37:396–407. doi: 10.1016/j.molcel.2009.12.039. [DOI] [PubMed] [Google Scholar]
- 31.Jung YS, Hakem A, Hakem R, Chen X. Pirh2 E3 ubiquitin ligase monoubiquitinates DNA polymerase eta to suppress translesion DNA synthesis. Mol Cell Biol. 2011;31:3997–4006. doi: 10.1128/MCB.05808-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chaudhury A, Chander P, Howe PH. Heterogeneous nuclear ribonucleoproteins (hnRNPs) in cellular processes: Focus on hnRNP E1’s multifunctional regulatory roles. RNA. 2010;16:1449–1462. doi: 10.1261/rna.2254110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ostareck-Lederer A, Ostareck DH, Hentze MW. Cytoplasmic regulatory functions of the KH-domain proteins hnRNPs K and E1/E2. Trends in biochemical sciences. 1998;23:409–411. doi: 10.1016/s0968-0004(98)01301-2. [DOI] [PubMed] [Google Scholar]
- 34.Waggoner SA, Johannes GJ, Liebhaber SA. Depletion of the poly(C)-binding proteins alphaCP1 and alphaCP2 from K562 cells leads to p53-independent induction of cyclin-dependent kinase inhibitor (CDKN1A) and G1 arrest. J Biol Chem. 2009;284:9039–9049. doi: 10.1074/jbc.M806986200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ostareck-Lederer A, Ostareck DH. Control of mRNA translation and stability in haematopoietic cells: the function of hnRNPs K and E1/E2. Biology of the cell / under the auspices of the European Cell Biology Organization. 2004;96:407–411. doi: 10.1016/j.biolcel.2004.03.010. [DOI] [PubMed] [Google Scholar]
- 36.Cho SJ, Jung YS, Chen X. Poly (C)-binding protein 1 regulates p63 expression through mRNA stability. PLoS One. 2013;8:e71724. doi: 10.1371/journal.pone.0071724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Peritz T, Zeng F, Kannanayakal TJ, Kilk K, Eiriksdottir E, Langel U, Eberwine J. Immunoprecipitation of mRNA-protein complexes. Nat Protoc. 2006;1:577–580. doi: 10.1038/nprot.2006.82. [DOI] [PubMed] [Google Scholar]
- 38.Srivastava S, Tong YA, Devadas K, Zou ZQ, Chen Y, Pirollo KF, Chang EH. The status of the p53 gene in human papilloma virus positive or negative cervical carcinoma cell lines. Carcinogenesis. 1992;13:1273–1275. doi: 10.1093/carcin/13.7.1273. [DOI] [PubMed] [Google Scholar]
- 39.Thomas M, Pim D, Banks L. The role of the E6-p53 interaction in the molecular pathogenesis of HPV. Oncogene. 1999;18:7690–7700. doi: 10.1038/sj.onc.1202953. [DOI] [PubMed] [Google Scholar]
- 40.Leffers H, Dejgaard K, Celis JE. Characterisation of two major cellular poly(rC)-binding human proteins, each containing three K-homologous (KH) domains. Eur J Biochem. 1995;230:447–453. [PubMed] [Google Scholar]
- 41.Wang H, Vardy LA, Tan CP, Loo JM, Guo K, Li J, Lim SG, Zhou J, Chng WJ, Ng SB, Li HX, Zeng Q. PCBP1 suppresses the translation of metastasis-associated PRL-3 phosphatase. Cancer cell. 2010;18:52–62. doi: 10.1016/j.ccr.2010.04.028. [DOI] [PubMed] [Google Scholar]
- 42.Lian WX, Yin RH, Kong XZ, Zhang T, Huang XH, Zheng WW, Yang Y, Zhan YQ, Xu WX, Yu M, Ge CH, Guo JT, Li CY, Yang XM. THAP11, a novel binding protein of PCBP1, negatively regulates CD44 alternative splicing and cell invasion in a human hepatoma cell line. FEBS letters. 2012;586:1431–1438. doi: 10.1016/j.febslet.2012.04.016. [DOI] [PubMed] [Google Scholar]
- 43.Chaudhury A, Hussey GS, Ray PS, Jin G, Fox PL, Howe PH. TGF-beta-mediated phosphorylation of hnRNP E1 induces EMT via transcript-selective translational induction of Dab2 and ILEI. Nature cell biology. 2010;12:286–293. doi: 10.1038/ncb2029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Pillai MR, Chacko P, Kesari LA, Jayaprakash PG, Jayaram HN, Antony AC. Expression of folate receptors and heterogeneous nuclear ribonucleoprotein E1 in women with human papillomavirus mediated transformation of cervical tissue to cancer. Journal of clinical pathology. 2003;56:569–574. doi: 10.1136/jcp.56.8.569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Thakur S, Nakamura T, Calin G, Russo A, Tamburrino JF, Shimizu M, Baldassarre G, Battista S, Fusco A, Wassell RP, Dubois G, Alder H, Croce CM. Regulation of BRCA1 transcription by specific single-stranded DNA binding factors. Molecular and cellular biology. 2003;23:3774–3787. doi: 10.1128/MCB.23.11.3774-3787.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Shi Z, Zhang T, Long W, Wang X, Zhang X, Ling X, Ding H. Down-regulation of poly(rC)-binding protein 1 correlates with the malignant transformation of hydatidiform moles. International journal of gynecological cancer: official journal of the International Gynecological Cancer Society. 2012;22:1125–1129. doi: 10.1097/IGC.0b013e3182606ac3. [DOI] [PubMed] [Google Scholar]
- 47.Czyzyk-Krzeska MF, Beresh JE. Characterization of the hypoxia-inducible protein binding site within the pyrimidine-rich tract in the 3′-untranslated region of the tyrosine hydroxylase mRNA. J Biol Chem. 1996;271:3293–3299. doi: 10.1074/jbc.271.6.3293. [DOI] [PubMed] [Google Scholar]
- 48.Yu J, Russell JE. Structural and functional analysis of an mRNP complex that mediates the high stability of human beta-globin mRNA. Mol Cell Biol. 2001;21:5879–5888. doi: 10.1128/MCB.21.17.5879-5888.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Yeap BB, Voon DC, Vivian JP, McCulloch RK, Thomson AM, Giles KM, Czyzyk-Krzeska MF, Furneaux H, Wilce MC, Wilce JA, Leedman PJ. Novel binding of HuR and poly(C)-binding protein to a conserved UC-rich motif within the 3′-untranslated region of the androgen receptor messenger RNA. J Biol Chem. 2002;277:27183–27192. doi: 10.1074/jbc.M202883200. [DOI] [PubMed] [Google Scholar]
- 50.Thiele BJ, Doller A, Kahne T, Pregla R, Hetzer R, Regitz-Zagrosek V. RNA-binding proteins heterogeneous nuclear ribonucleoprotein A1, E1, and K are involved in post-transcriptional control of collagen I and III synthesis. Circulation research. 2004;95:1058–1066. doi: 10.1161/01.RES.0000149166.33833.08. [DOI] [PubMed] [Google Scholar]
- 51.Reimann I, Huth A, Thiele H, Thiele BJ. Suppression of 15-lipoxygenase synthesis by hnRNP E1 is dependent on repetitive nature of LOX mRNA 3′-UTR control element DICE. J Mol Biol. 2002;315:965–974. doi: 10.1006/jmbi.2001.5315. [DOI] [PubMed] [Google Scholar]
- 52.Collier B, Goobar-Larsson L, Sokolowski M, Schwartz S. Translational inhibition in vitro of human papillomavirus type 16 L2 mRNA mediated through interaction with heterogenous ribonucleoprotein K and poly(rC)-binding proteins 1 and 2. J Biol Chem. 1998;273:22648–22656. doi: 10.1074/jbc.273.35.22648. [DOI] [PubMed] [Google Scholar]


