Summary
At every cell cycle, faithful inheritance of metazoan genomes requires the concerted activation of thousands of DNA replication origins. However, the genetic and chromatin features defining metazoan replication start sites remain largely unknown. Here, we delineate the origin repertoire of the Drosophila genome at high resolution. We address the role of origin-proximal G-quadruplexes and suggest that they transiently stall replication forks in vivo. We dissect the chromatin configuration of replication origins and identify a rich spatial organization of chromatin features at initiation sites. DNA shape and chromatin configurations, not strict sequence motifs, mark and predict origins in higher eukaryotes. We further examine the link between transcription and origin firing and reveal that modulation of origin activity across cell types is intimately linked to cell-type-specific transcriptional programs. Our study unravels conserved origin features and provides unique insights into the relationship between DNA topology, chromatin, transcription and replication initiation across metazoa.
Graphical Abstract
Introduction
Maintenance of cellular identity critically relies on the faithful transmission of the parental genome through DNA replication and a reestablishment of the epigenome (Alabert and Groth, 2012). Perturbation of this finely orchestrated process poses a major threat to genome stability, thus linking aberrant DNA replication to several human diseases (Zeman and Cimprich, 2014).
In the circular chromosome of bacteria and archea, DNA replication starts from a single locus termed replication origin (Mott and Berger, 2007). In contrast, eukaryotic DNA replication requires the concerted activation of thousands of replication origins (Leonard and Méchali, 2013). The firing of a eukaryotic origin is preceded by the orderly recruitment of protein factors to potential initiation sites. In G1 phase, the origin recognition complex (ORC) binds replication origins and along with the help of Cdc6 and Cdt1 nucleates the pre-replication complex (pre-RC) through the loading of an inactive form of the mini-chromosome maintenance (MCM) helicase. At the onset of S-phase, Dbf4-dependent kinase (DDK) and cyclin-dependent kinases (CDKs) catalyze sequential phosphorylation events, which recruit initiation factors. These in turn stimulate MCM activity, complete replisome assembly and trigger the initiation of DNA synthesis, a process referred to as origin firing (Masai et al., 2010). Whereas sixty years of genetic and biochemical dissection have elucidated much of the activation cascade underlying origin firing, the mechanisms that target replisomes to replication origins remain poorly understood, raising the question of which sequence and chromatin features define origins in vivo. A comprehensive answer to this question is missing, partly because prior to the genomic era only a handful of origins were precisely mapped (Leonard and Méchali, 2013) and partly because isolation and characterization of transient replication intermediates is experimentally challenging (reviewed in Gilbert, 2010). The isolation of small nascent leading-strands (SNS, Bielinsky and Gerbi, 1998) is currently considered the most reliable method to map replication origins (Leonard and Méchali, 2013).
Recent high-throughput approaches coupled SNS purification with tiling arrays (SNS-chip) (Sequeira-Mendes et al., 2009; Cayrou et al., 2011) or next-generation sequencing (NGS, SNS-Seq) (Besnard et al., 2012; Picard et al., 2014), enabling systematic origin mapping genome-wide. Particularly, origin profiling in four human cell types (Besnard et al., 2012) identified thousands of active origins and suggested that cell-type specific origin-usage signatures are responsible for the observed plasticity of replication programs. However, despite considerable efforts, these works fell short in identifying a eukaryotic consensus sequence and converged to associate G-rich elements and G-quadruplexes (Maizels and Grey, 2013) to a variable fraction of initiation sites, suggesting that features beyond nucleotide sequence define metazoan replication origins. Cooperation between genetic elements and epigenetic features is therefore likely to be key for origin function, but chromatin configurations of replication start sites remain largely unexplored.
In this study, we delineate the origin repertoire of the Drosophila melanogaster genome at an unprecedented resolution. We examine the role of origin-proximal G-quadruplexes and provide evidence that these DNA secondary structures act as replication fork barriers in vivo. We carefully dissect the chromatin configuration of replication start sites and demonstrate that specific DNA shape and chromatin configurations, as opposed to strict sequence specificity, mark and accurately predict replication origins in higher eukaryotes. Finally, our study reveals that differential origin usage across cell types is tightly connected to cell-type-specific transcriptional programs, thus providing a means to couple chromatin processes crucial for maintenance of cellular identity.
Results
High-resolution mapping of Drosophila replication origins
Upon origin firing, two nascent leading strands extend from a short RNA primer and emanate bidirectionally from the origin. SNS-Seq aims at selectively isolating these covalent RNA-DNA hybrids, whose 5′ ends define the site of replication initiation. In the SNS purification protocol, origin-proximal SNS are first size-separated from Okazaki fragments and then enriched by lambda-exonuclease (Lexo) digestion of non-RNA-primed DNA. As this 5′ to 3′ processive nuclease exhibits very weak activity on ribonucleotides, SNS are protected from digestion while contaminating DNA species are degraded. However, even in rapidly dividing cells, SNS account for only ~0.002% of total genomic DNA (Gilbert, 2012). Accuracy and resolution of origin detection, therefore, critically depends on efficient degradation of contaminant, unreplicated DNA. Moreover, in the absence of other DNA species, the relative abundance of SNS from all origins firing throughout S-phase is expected to reflect their firing efficiency within a cell population, thus allowing estimates of aggregated firing probabilities (Gilbert, 2010). Treatment with Lexo has proven essential in eliminating contamination and previous work enriched for SNS through two or three rounds of Lexo digestion (Cayrou et al., 2011; Besnard et al., 2012; Picard et al., 2014).
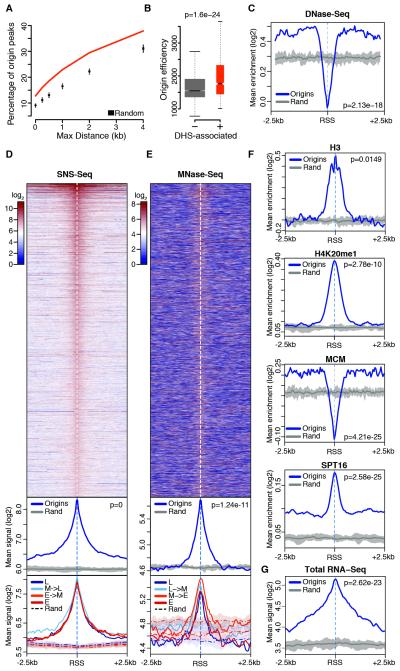
Here, we adopted an enhanced sensitivity SNS purification protocol (Cayrou et al., 2011; see Experimental Procedures) to map active replication origins genome-wide in two Drosophila cell lines, the late embryo-derived S2 and the neuronal-derived Bg3 cells, whose epigenomes have been extensively profiled by the modENCODE project (Celniker et al., 2009). For each cell type, we obtained highly pure SNS preparations from two biological replicates by subjecting size-selected genomic DNA to up to five rounds of Lexo digestion (Figure S1B, inset). High-coverage, saturating deep sequencing of these SNS yielded a total of 119 and 251 million reads aligning to the Drosophila genome for S2 and Bg3 cells, respectively (Figure S1A). This led us to identify 7268 and 8212 high-confidence replication origins in S2 and Bg3 cells (Figure 1A), respectively, whose replication start sites (RSSs) were defined as the summit of highly-resolved origin peaks (Figure S1G-I). Furthermore, position and firing efficiency of a subset of origins was confirmed by quantitative PCR (qPCR;Figure S1B and Table S1).
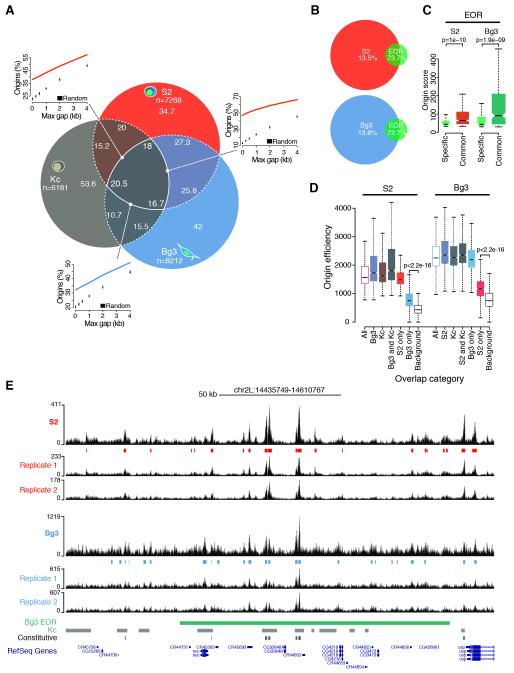
Figure 1. High-resolution mapping of the Drosophila origin repertoire.
(A) Percentage overlap of origin peaks identified in S2, Bg3 and Kc (Cayrou et al., 2011) Drosophila cells and comparison of observed pairwise overlaps (lines) with random expectations (boxplots). n, total number of origin peaks. (B) Percentage overlap of S2 and Bg3 origin peaks and modENCODE early origin regions (EOR). (C) Origin score of EOR overlapping with S2 and Bg3 origins (common) or solely identified by modENCODE (specific). (D) Efficiency of S2 and Bg3 origins partitioned and color-coded according to (A). Background estimates are shown. (E) A representative snapshot of the SNS-Seq coverage in S2 and Bg3 cells from two biological replicates and detected origin peaks. A single EOR (green) spans most of this 175 kb genomic region. Kc (gray) and constitutive (black) origins are also shown. p-values are from Wilcoxon rank-sum test. See also Figure S1.
Notably, 73-81% and 69-75% of origin peaks were independently detected within S2 and Bg3 biological replicates, respectively (Figure S1D-E). These values not only compare favorably to previous studies, but also exceed the technical reproducibility of recently published SNS-Seq data in human K562 cells (Picard et al., 2014). Moreover, SNS-Seq signals exhibited a nearly perfect correlation (r=0.98) across biological replicates when computed in the union of all origin peaks from a given cell type (Figure S1F). To verify the accuracy of our origin mapping, we compared our results with previously published data sets. S2 and Bg3 origins covered a total of 6.9 and 6.4 Mb, respectively, a considerably smaller fraction of the Drosophila genome than the 27.3 Mb spanned by 6184 origins previously identified in Drosophila Kc cells (Cayrou et al., 2011) (Figures 1A,E and S1G). In addition, <14% of S2 and Bg3 origin peaks sufficed to recall >72% of modENCODE early origin regions (Eaton et al., 2010) (Figure 1B), which were mapped by BrdU immunoprecipitation from G1/S synchronized cells and resulted in broad initiation zones. Our data finely resolved the composition of initiation zones exhibiting significantly higher origin scores than regions exclusively identified by modENCODE (Figure 1C; 1E for an example), suggesting that the remaining low-confidence events contain dormant origins that do not fire in an unperturbed S-phase (Zeman and Cimprich, 2014). Taken together, these results demonstrate the high quality and unmatched resolution of our origin mapping, which markedly expands the origin repertoire of the Drosophila genome to 14005 distinct genomic loci.
Modulation of origin activity, not site selection, defines cell-type specific replication programs
Next, we analyzed the overlap between S2, Bg3 and Kc origins. We found 16-20% of origin peaks common to all three cell types (constitutive origins) and 35-45% of origin sites activated by at least two cell types. This overlap is significantly larger than expected by chance (p<0.001, Figure 1A) and indicates a preferred origin localization across cell types. Previous work reported that cell-type specific origins were on average poorly used (Besnard et al., 2012). However, whether firing of these sites was restricted to the cell type of detection or whether these origins are also marginally used in other cell types remained unclear. To address this question, we estimated firing efficiencies by integrating SNS-Seq signals within all detected origin peaks and in ten matched background sets (see Experimental Procedures). In line with previous reports (Besnard et al., 2012), we found that constitutive origins exhibited on average the highest efficiency values across the entire origin repertoire (Figure 1D). However, cell type-specific origin peaks were not only characterized by low efficiency in the cell type of detection, but they also yielded SNS-Seq signals well above background in other cell types (Figure 1D). This result indicates that virtually all origins we identified fire in each Drosophila cell type but with characteristic frequencies. Cell-type specific origins, therefore, could rather be termed cell-type preferred origins, thus reflecting cell type-specific preferences for low efficiency origins. Our results strongly suggest that modulation of origin activity, not the selection of origin sites, is likely to define cell-type specific replication programs in Drosophila.
Origin proximal G-quadruplexes act as transient replication fork barriers in vivo
Recent work investigated the role of G-quadruplexes (G4) in DNA replication (Valton et al., 2014; Castillo Bosch et al., 2014), yet the potential contribution of G4 structures to replication initiation remained controversial. Here, we examined the association between G4 and origins genome-wide.
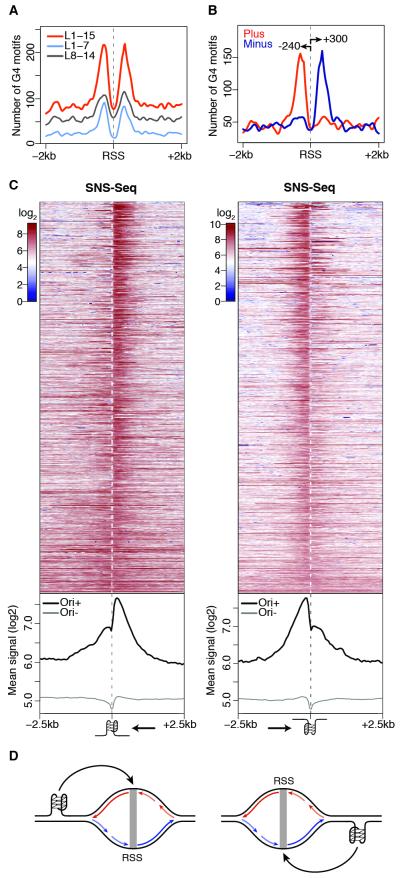
We predicted G4 occurrences in the Drosophila genome and used the loop size (L) to define nested classes of G4 motifs. We found a significant association between origins and predicted G4 as compared to random expectation (p<0.001, Figure S2A), with 9% and 22% of S2 origins overlapping 7% and 5% of L1-7 and L1-15 G4 motifs, respectively. G4-associated origins were more efficient than G4-negative origins (Figure S2B). This result is not mediated by colocalization with transcription start sites (TSSs, Figure S2C), which do not significantly associate with origins in Drosophila (p=0.81, Figure S2D; Cayrou et al., 2011). We then examined the position and orientation of G4 motifs with respect to S2 origins. The alignment of G4-associated origins relative to their RSSs revealed a strong positional preference (Figure 2A), with strand-specific occurrences of G4 peaking at 240-300 bp from RSSs and largely restricted to their flanking regions (Figures 2B and S2E-F).
Figure 2. A G-quadruplex signature at S2 replication origins.
(A) Spatial distribution of G4 motifs within ± 2 kb of S2 RSSs. (B) Same as (A) for strand-specific annotation of G4 L1-15 motifs. Arrows indicate peak distances (bp) from the RSS. (C) S2 SNS-Seq signals within ± 2.5 kb of origin-associated G4 L1-15 motifs occurring on the plus (left) and minus (right) strands, ranked by coefficient of variation. Bottom panels show the average of the signals above (Ori+) and at origin-negative (Ori−) G4 motifs. Arrows indicate the direction of the leading strand facing the G4. (D) Model describing how origin-proximal G4 motifs could orient (black arrows) replication forks. Leading strands (long arrows) and Okazaki fragments (short) replicating the plus (red) and minus (blue) strands are indicated. See also Figure S2.
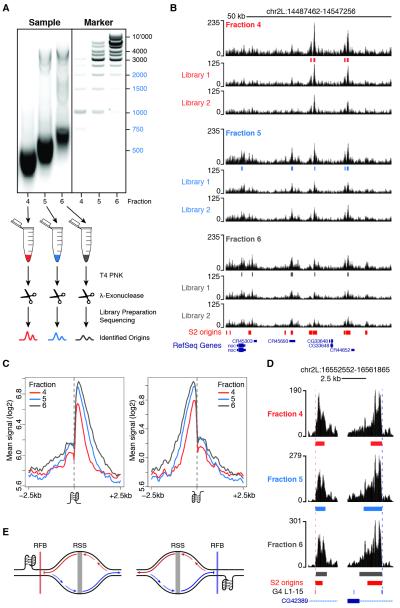
Next, we quantified the spatial distribution of SNS-Seq signals within a 5kb region centered on each G4 L1-15 motif and partitioned G4 occurrences by strand and association with S2 origins. We observed a skewed distribution of SNS at origin-proximal G4, with most of the signal contributed by RSSs located downstream and upstream of G4 motifs mapping to the plus and the minus strand, respectively (Figure 2C). However, we noted that the SNS signal was not only asymmetrically distributed at these G4 sites, but it also sharply dropped exactly at the G4 position (Figure 2C). Thus, we reasoned that our data might capture G4-proximal replication fork stalling events and that, if this is the case, only synthesis of the leading strand replicating the G4 template should be affected. To test this hypothesis, we set out to indirectly monitor the progression of replication forks emanating from origin-associated G4 by purifying, barcoding and deep sequencing SNS of increasingly larger sizes (Figure 3A). Two sequencing libraries were prepared for each of three gradient fractions, for a total of 298 million reads aligning to the Drosophila genome (Figure S3A). Fractions 4 (shortest DNA molecules), 5 (intermediate) and 6 (largest) independently identified 4505, 6448 and 5814 origin peaks, respectively, most of which (>81%) overlapped in two or more fractions (Figures S3B and 3B). The SNS enrichment of a subset of origin peaks was further confirmed by qPCR and negatively correlated with SNS sizes (Figure S1C and Table S1). SNS-Seq signals in the union of origin peaks were highly correlated across fractions (r=0.94-0.98, Figure S3C). Moreover, >70% of origins detected by all fractions (n=3246) overlapped S2 origins previously identified in separate biological replicates (Figure S1B), thus demonstrating the high sensitivity of our approach. These data allowed us to quantify the relative contribution of individual fractions to the SNS profile previously observed at G4 motifs (Figure 2C). Strikingly, we found that while leading strands traveling away from the G4 motif extend normally, DNA synthesis on the strand replicating the G4 template is blocked at the G4 site (Figures 3C; 3D for an example). This finding suggests that most origin-proximal G4 are folded at the time of origin activation and function as replication fork barriers in vivo (Figure 3E).
Figure 3. Origin-proximal G-quadruplexes stall replication forks in vivo.
(A) An outline of the experimental strategy used to indirectly monitor replication fork progression at origin-associated G4. Fractions 4-6, corresponding to marker lanes 4-6, were individually purified and subjected to two sequential rounds of T4 PNK phosphorylation and Lexo digestion. Two sequencing libraries were prepared for each sample and origin peaks were called on their union. (B) A representative snapshot of the single-fraction SNS-Seq coverage. Origin peaks identified in each fraction and S2 origin peaks from standard SNS-Seq experiments are shown. (C) Average single-fraction SNS-Seq signal within ± 2.5 kb of origin-associated G4 L1-15 motifs occurring on the plus (left) and minus (right) strands. (D) Two representative G4 motifs occurring on opposite strands are shown. (E) Model describing how origin-proximal G4 motifs could act as replication fork barriers. Origin-proximal G4 pause the synthesis of the nascent leading strands replicating the G4 template. See also Figure S3.
A DNA shape signature of metazoan replication origins
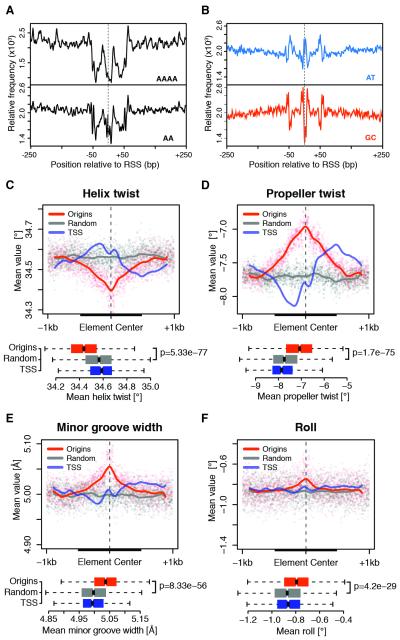
With the high resolution of our data, it was possible to revisit the sequence characteristics of replication origins that have been elusive in lower resolution studies. We started by examining the local DNA sequence composition at RSSs. Interestingly, nucleosome-repelling AAAA polynucleotides and AA dinucleotides (Kaplan et al., 2009; Tillo and Hughes, 2009) were symmetrically distributed around RSSs, with depletion of these elements marking both the RSS and two proximal sites localized within 50 bp (Figure 4A). Moreover, depletion of poly(A) stretches at flanking regions was accompanied by features characteristic of nucleosome container sites such as a central core of GC-rich sequences and a moderate decrease in AT content (Figure 4B; Tillo and Hughes, 2009). In contrast, RSSs reside at the global minimum in GC content and at a local maximum in AT content (Figure 4B), suggesting an enrichment of TpA base pair steps, which are characterized by the weakest base stacking interactions among all possible dinucleotides (Rohs et al., 2009). Interestingly, nucleosome containers similarly marked RSSs of human HeLa replication origins (Figure S4A-B), thus indicating that this feature is conserved across higher eukaryotes. Taken together, these results strongly suggest an increase in conformational flexibility of RSSs and immediately adjacent regions.
Figure 4. Specific DNA shape features mark metazoan replication origins.
(A) Relative frequency of AAAA polynucleotides and AA dinucleotides within ± 250 bp of S2 RSSs. (B) Same as (A) for AT and GC dinucleotides. (C-F) Average of DNA shape features within ± 1 kb of RSSs for constitutive Drosophila origins, background regions and TSSs. The latter were extended while preserving orientation. Solid lines are Loess fitted curves from single-nucleotide resolution shape predictions (dots). Boxplots of average feature values within 500 bp windows (thick black lines) are shown (bottom panels). p-values are from Wilcoxon rank-sum test. See also Figure S4.
To further test this hypothesis, we generated high-throughput predictions of DNA shape features at origins (Zhou et al., 2013). Strikingly, a specific DNA shape signature common to Drosophila (Figure 4C-F) and human (Figure S4C-F) origins emerged, characterized by reduced helix twist and increased propeller twist, minor groove width and roll. A decrease in helix twist (Figure 4C) indicates helical unwinding, which renders bending and other DNA deformations energetically more favorable (Chen et al., 2013). The increase in propeller twist (Figure 4D) suggests a reduction in inter-base pair hydrogen bonds in the major groove, which is the main stabilizing force for the formation of rigid poly(A) elements (Rohs et al., 2009). This, in turn, has been previously correlated with widening of the minor groove at the corresponding positions (Figure 4E; Hancock et al., 2013). Moreover, the local increase in roll (Figure 4F) suggests an enrichment in pyrimidine-purine base pair steps, such as TpA dinucleotides, thus generating weak stacking interactions that enhance local flexibility of RSSs (Rohs et al., 2010). Together, these data provide compelling evidence that degenerate sequence features dictate a conserved DNA structure that is likely to play a key role in origin function.
The chromatin composition of Drosophila replication origins
Previous studies noted increased chromatin accessibility at sites of early replication in Drosophila (Bell et al., 2010; MacAlpine et al., 2010) and identified DNase I hypersensitive sites (DHSs) as a determinant of replication initiation in human cells (Gindin et al., 2014). In line with these studies, S2 origins were significantly associated with DHSs (p<0.001, Figure 5A), and their firing efficiency positively correlated with local chromatin accessibility (Figure 5B). Moreover, averaging of DNase-Seq signals across 5 kb windows centered on RSSs revealed a strong enrichment for DNase I digested fragments at origins as compared to randomized genomic regions (Figure 5C). At first glance, these results are incompatible with a nucleosome container signature at initiation sites, and this apparent contradiction prompted us to examine the spatial distribution of DNase-Seq signals across RSSs. To our surprise, we found a striking difference between chromatin accessibility of RSSs and their flanking regions. Indeed, while the latter exhibit clear features of open chromatin, a sharp reduction in DNase I digested fragments was seen at the RSS (Figure 5C). These results reconcile our observations and led us to posit that a rich, spatially organized chromatin configuration marks eukaryotic origins.
Figure 5. The chromatin composition of Drosophila replication origins.
(A) Percentage overlap of S2 origin peaks with DHSs and random expectation. (B) Efficiency of S2 origins localizing within (+) or outside (−) DHSs. (C) Average DNase-Seq enrichment within ± 2.5 kb of S2 RSSs and within ten sets of randomized genomic regions (Rand). The thick gray line traces average background values. (D) Spatial distribution of SNS-Seq signal within ± 2.5 kb of S2 RSSs (top), metaprofiles comparing origins with ten sets of randomized genomic regions (middle), and further partitioning of the signal above in four timing classes (L: late S-phase; M: mid; E: early) based on replication timing quartiles (bottom). (E) Same as (D) for MNase-Seq. (F-G) Same as (C) for the indicated features. p-values are from Wilcoxon rank-sum test. See also Figure S5.
To test this hypothesis, we set out to survey the chromatin landscape of replication origins at an unprecedented resolution. First, we compiled a comprehensive representation of the chromatin landscape of S2 cells comprising 85 chromatin features profiled by the modENCODE project (Celniker et al., 2009) or independent studies (Table S2). Second, we analyzed the spatial distribution of each feature within 5 kb windows centered on inferred RSSs or in ten sets of matched control regions at 50 bp resolution. The potentially confounding contribution of TSS-associated chromatin features was limited by excluding origin-TSSs from the analysis. Third, as chromatin features do not uniformly distribute across replication timing compartments, we partitioned origins and control regions in four timing classes (from early to late replicating) based on replication timing quartiles. This allowed us to probe for evidence of timing-specific chromatin signatures by directly comparing matched timing classes.
As expected, origins were strongly enriched for SNS compared to control regions, with SNS-Seq signals sharply peaking at the inferred RSS positions (Figure 5D). Notably, nearly no difference in the average SNS-Seq signal was observed across timing classes indicating that firing efficiency does not correlate with replication timing at the single origin level. Next, we focused on direct and indirect measurements of nucleosome occupancy. Interestingly, while the genomic regions flanking the RSSs exhibit background MNase-Seq signals (Figure 5E) and histone H3/H4 enrichments (Figures 5F and S5A), RSSs correspond to positions of high nucleosome occupancy, a feature that is shared across timing classes. Moreover, we found that nucleosomes at RSSs are decorated by histone modifications such as lysine mono-methylation and acetylation. H3K9me1, H3K23me1 and H4K20me1, a PRSet7-dependent histone modification that promotes loading of the pre-RC at origins (Tardat et al., 2010), sharply peaked at RSSs (Figures 5F and S5B). Intriguingly, enrichment of these marks was not limited to early replicating regions (Figure S5B), suggesting that initiation sites could be invariably bookmarked by these modifications. In contrast, pre-RC binding was restricted to the accessible chromatin regions flanking RSSs (Figures 5F and S5C), in line with the concept that ORC-mediated pre-RC nucleation requires direct contact with the DNA template (Masai et al., 2010) and occurs adjacent to RSSs (Lombraña et al., 2013).
Rapid nucleosome turnover has emerged as a distinguishing feature of both promoters and origins (Deal et al., 2010) and binding of chromatin remodelers was previously correlated with early replication timing in Drosophila (Eaton et al., 2011; Comoglio and Paro, 2014). In agreement, members of different remodeling complexes and the histone chaperone Spt16, a core component of the facilitates chromatin transcription (FACT) complex, were markedly enriched at origins (Figures 5F and S5D) with the highest values throughout early replicating, active chromatin. However, while the overall enrichment of these proteins largely varied across timing classes, RSSs were invariably marked by the highest occupancy and exhibited similar enrichments relative to flanking regions in each timing class (Figure S5E). Finally, as the correlation between transcription and origin firing remained largely unexplored, we leveraged our high-resolution data to examine local transcriptional outputs at origins. Strikingly, analysis of the spatial distribution of both total and poly(A)+ RNA-Seq signals at origins revealed conspicuous transcription at RSSs, once again, irrespective of their replication timing (Figures 5G and S5F).
Chromatin landscape and transcriptional output predict origin activity of CG-rich regions
Several CpG-islands (CGIs) in mammals and CG-rich regions (CGRs) in Drosophila share the potential to initiate DNA replication (Cayrou et al., 2012a; Besnard et al., 2012). Indeed, 18% of S2 and 22% of Bg3 origin peaks were significantly associated with CGRs (p<0.001, Figure 6A). However, only 6-37% of origin-CGRs are highly efficient, constitutive origins (Figure 6A-B), raising the question of which features might favor or prevent replication initiation at these sites. Here, we asked whether the local chromatin context at CGRs correlates with their firing potential.
Figure 6. Origin activity of CG-rich regions is predicted by chromatin landscape and transcriptional output.
(A) Percentage overlap of origin peaks associated with CGRs in S2, Bg3 and Kc (Cayrou et al., 2011) cells, and comparison of the observed overlap between S2 and Bg3 origin peaks and CGRs (lines) with random expectation (boxplots). n, total number of CGRs. (B) Two representative snapshots of the S2 SNS-Seq coverage from two biological replicates, origin peaks and poly(A)+ RNA-Seq coverage across several CGRs. Constitutive origins (black) are also shown. (C) Spatial distribution of SNS-Seq signal within ± 2.5 kb of S2 origin-CGR midpoints (top) and metaprofiles (bottom) comparing origin-CGRs (Ori+) with origin-negative CGRs (Ori−). (D) Same as (C) for MNase-Seq. (E) Same as bottom panel of (C) for the indicated features. (F) An outline of the modeling strategy used to classify CGRs. (G) ROC curves and AUC values for lasso models trained on the indicated sets of features. The inset shows selection probabilities of the top-ranked features selected by bootstrap-lasso. Bars are color-coded according to coefficient signs (positive, red; negative, blue) and absolute value of coefficient z-scores. (H) S2/Bg3 RNA-Seq fold change for the indicated classes of origin-CGRs. p-values are from Wilcoxon rank-sum test. See also Figure S6.
We started by contrasting the chromatin landscape of origin-CGRs active in S2 cells (Figure 6C), with that of origin-negative CGRs. Intriguingly, contrary to our expectation, we found that chromatin was overall similarly configured across CGRs, irrespective of origin activity (Figures 6D-E and S6A). A few noteworthy features, therefore, likely render the chromatin configuration of origin-CGRs compatible with efficient origin firing: i) higher chromatin accessibility and higher pre-RC loading proximal to the CGR-center (a proxy for the RSS); ii) higher nucleosome occupancy; and iii) markedly higher levels of Spt16 throughout the entire origin region that sharply peaked at the CGR-center (Figure 6E). Moreover, while origin-negative CGRs were on average poorly transcribed, origin-CGRs were strongly enriched for RNA-Seq reads (Figure 6E).
These findings led us to test whether chromatin configurations and transcriptional outputs could predict the firing potential of CGRs. To this purpose, we trained binary classifiers based on lasso logistic regression (Tibshirani, 1996), using DNA sequence content (k-mers, k ≤ 4), chromatin feature enrichments and RNA-Seq signal at CGRs as predictors. A test set of CGRs that was not previously seen by the models was used to evaluate performances (Figure 6F, see Experimental Procedures). Interestingly, while the DNA sequence content of CGRs was a poor predictor of origin activity (area under the receiver operating characteristic curve, AUC = 0.59), chromatin features and transcription were able to accurately classify CGRs (AUC = 0.78). Moreover, a more complex model combining genetic and epigenetic features did not perform better than epigenetic features alone (AUC = 0.78), indicating that these two sets of features are highly redundant. Next, we unbiasedly assessed the importance of individual predictors by estimating feature selection probabilities with bootstrap-lasso (Comoglio and Paro, 2014). Intuitively, the more a feature is required for accurate predictions, the higher its selection probability. Our analysis identified RNA-Seq, H3K36me1 and RNA polymerase II (Pol II) as top-ranked, positive predictors of origin firing at CGRs. In contrast, local GC content and H4K16ac were stably selected, negative predictors of origin activity (Figure 6G, inset). Further analysis of the H4K16ac distribution at CGRs revealed a striking contrast between early and late replicating origin-CGRs, which were depleted and enriched for this mark, respectively (Figure S6B).
As the RNA-Seq signal was stably selected by all models, we investigated whether differential expression of origin-CGRs could explain differential usage of these sites across cell types. To this end, we computed the S2/Bg3 RNA-Seq fold change of origin-CGRs that efficiently fired in both cell types or that were efficiently activated only in one of the two. Intriguingly, while the former were on average similarly transcribed in S2 and Bg3 cells, transcription of the latter was significantly upregulated in the cell type of efficient activation (Figure 6H). Taken together, these results establish a tight coupling between transcription and origin firing at origin-CGRs.
Differential origin activity mirrors differences in cell-type-specific transcriptional programs
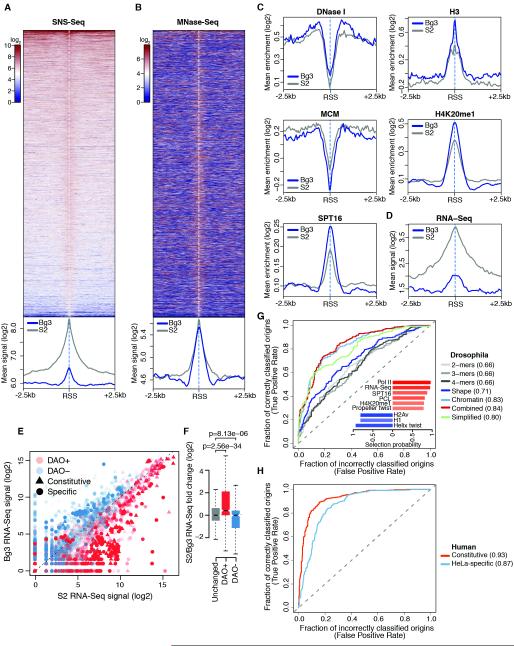
Bg3-preferred origins exhibited poor, yet highly significant, firing efficiency in S2 cells (Figure 1B). Indeed, analysis of S2 SNS-Seq signals at these sites indicated that virtually all Bg3-preferred origins also fire in S2 cells (Figure 7A). Therefore, we reasoned that a systematic comparison between these low-efficiency sites and S2 origin peaks could shed light on poorly understood epigenetic determinants of origin firing.
Figure 7. Differential origin activity mirrors differences in cell-type-specific transcriptional programs.
(A) Spatial distribution of S2 SNS-Seq signal within ± 2.5 kb of RSSs of origin peaks solely identified in Bg3 cells (top) and metaprofiles (bottom) comparing all S2 origin peaks with these sites. (B) Same as (A) for MNase-Seq. (C-D) Same as bottom panel of (A) for the indicated features. (E) Scatter plot of S2 and Bg3 RNA-Seq signals at differentially activated origins (DAOs) that were more efficiently used by S2 (DAO+) or Bg3 (DAO−) cells. Triangles, constitutive origins; circles, origin peaks solely detected in one cell type. Opacity reflects the statistical significance of differential origin activity and is proportional to -log10-transformed adjusted p-values. (F) S2/Bg3 RNA-Seq fold change of equally activated origins (unchanged) and of differentially activated ones. p-values are from Wilcoxon rank-sum test. (G) ROC curves and AUC values for origin-classifiers trained on the indicated set of features in Drosophila. The inset shows selection probabilities of the top-ranked features selected by bootstrap-lasso and used to train the simplified model. Bars are color-coded according to coefficient signs (positive, red; negative, blue) and absolute values of coefficient z-scores. (H) Same as (G) for constitutive and HeLa-specific human origins. See also Figure S7.
Dissection of the chromatin configuration of these two origin sets revealed an enrichment of H3K9me3, Su(var)3-9 and increased binding of insulator proteins (Figure S7A) at Bg3-preferred origins in S2 cells compared to S2 origin peaks, suggesting that these sites are preferentially embedded within constitutive heterochromatin in S2 cells. However, despite their heterochromatic localization, the distinctive chromatin signature previously identified at S2 origin peaks invariably marked Bg3-preferred origins in the S2 epigenome (Figure 7B-C). In fact, Bg3-preferred origins shared several features with efficient origins including accessible flanking regions as well as high H4K20me1, Spt16 and chromatin remodeler levels (Figure 7C and Figure S7B). Conversely, a markedly lower transcriptional output distinguished Bg3-preferred origins from S2 origin peaks (Figure 7D), suggesting that a chromatin environment less permissive to transcription might suffice to prevent efficient origin firing irrespective of local chromatin cues. These results, along with convergence of transcription and replication programs at CGRs, prompted us to test whether differential origin activity across cell types could similarly mirror differences in the cell-type-specific transcriptional outputs genome-wide. To this end, we identified 5917 differentially activated origins (DAOs, see Experimental Procedures) between S2 and Bg3 cells. Notably, DAOs encompassed 38% of constitutive origins and >49% of origins efficiently firing only in one cell type at a very stringent significance threshold (adjusted p-value <=1e-5). When S2 and Bg3 RNA-Seq signals were compared at DAOs, a clear-cut correlation emerged between differential origin activity and local transcription (Figure 7E). Indeed, while equally activated origins were similarly transcribed in S2 and Bg3 cells (Figures 7F and S7C), DAOs that were more efficiently used by S2 cells were in turn significantly more transcribed in this cell type than in Bg3, with the opposite trend being observed at DAOs more efficiently used by Bg3 cells (Figure 7F).
DNA shape and epigenetic features accurately predict active origins in the Drosophila and human genomes
Next, we asked whether the identified genetic and epigenetic characteristics of replication origins could discriminate active replication initiation sites from the rest of the Drosophila genome. To this end, we trained lasso origin-classifiers on DNA kmers and shape features, chromatin feature enrichments and transcriptional output, using the same learning scheme of CGR-classifiers (Figure 6F).
Intriguingly, DNA shape features alone not only exhibited a moderately high predictive power (AUC=0.71), but they also outperformed DNA sequence content (AUC=0.66) in classifying constitutive origins (Figure 7G). Moreover, chromatin feature enrichments and transcription were able to generate accurate origin predictions (AUC=0.83). Further inclusion of k-mers only marginally improved model performances (AUC=0.84) (Figure 7G). Indeed, no k-mer was consistently selected in this more complex model. Conversely, a simplified origin-classifier solely based on the nine most stably selected features (selection probability ≥ 0.8), including RNA-Seq signal, six chromatin features and two DNA shape features (Figure 7G, inset), generated remarkably accurate predictions (AUC=0.80, Figure 7G). These results led us to examine whether local DNA shape and transcription at origins could similarly predict active origins in the human genome. Strikingly, a classifier solely based on helix twist, propeller twist, and RNA-Seq signal was not only able to discriminate active origins from the rest of the human genome, but it also outperformed the Drosophila origin-classifier at both constitutive (AUC = 0.93) and HeLa-specific (AUC = 0.87) origin peaks (Figure 7H). Interestingly, model performances correlated with the DNA shape profiles observed within these origin sets, with poorly efficient origins exhibiting a less pronounced DNA shape signature at RSSs (Figure S4).
Discussion
Origin specification in higher eukaryotes involves mechanisms other than simple replicator-initiator interactions (Leonard and Méchali, 2013), yet the genetic and epigenetic features that specify replication origins in vivo remain enigmatic. ORC is the first replication factor to bind origins but it lacks a sequence-specific binding motif (Gilbert, 2010). Interestingly, studies have suggested that DNA topology rather than sequence motifs might mediate nucleation of the pre-RC at origins. Mainly, Drosophila ORC exhibits much higher affinity for negatively supercoiled DNA than for linear or relaxed DNA in vitro (Remus et al., 2004) and human ORC binds preferentially to G4-forming RNA and single-stranded DNA (Hoshina et al., 2013). Moreover, these findings are reminiscent of observations in bacteria where negatively supercoiled DNA coordinates replication initiation (Mott and Berger, 2007).
By high-throughput predictions of DNA shape, our study demonstrates that a specific DNA topology, characterized by increased conformational flexibility of RSSs, marks both Drosophila and human replication origins. This relaxed DNA conformation likely serves two important functions. First, positioning of nucleosomes at the RSS, previously observed at efficient mammalian origin-CGIs (Lombraña et al., 2013), is likely to be weaker in this region, thus favoring nucleosome displacement. Formation of atypical nucleosomes at RSSs could also contribute to this process. Second, melting of the double helix at the RSS is energetically assisted, further facilitating initiation. Interestingly, while enhanced DNA flexibility similarly marks Drosophila(Figure 4) and human (Figure S4) origins, the sequence composition of RSSs is remarkably different between these two organisms. Indeed, while Drosophila RSSs are locally AT-rich, high GC content is found at human RSSs (Figure S4B). Intriguingly, the increased GC content of human RSSs might reflect a role for cytosine methylation of CpG dinucleotides, which is widespread in mammals but not Drosophila (Takayama et al. 2014), in granting conformational flexibility to human origin sequences, thus functionally replacing TpA base pair steps (Lazarovici et al., 2013). Together, these findings indicate that by integrating over degenerate sequence features, DNA shape appears to represent a universally conserved origin bookmark.
A confounding feature of metazoan origins, however, is that mechanisms of origin specification are likely not sufficient in defining origin activity. Origin firing is intrinsically stochastic at the single-cell level (Bechhoefer and Rhind, 2012; Cayrou et al., 2011), yet aggregated firing probabilities within a replicon could be modulated by extrinsic factors. A major question in the field is which cis- and trans-acting factors influence these probabilities. G-quadruplexes and noncoding RNAs (Ge and Lin, 2014) are likely candidates. G4 have been proposed to orient replication forks and enhance the efficiency of origin firing at the chicken βA promoter (Valton et al., 2014) but they are not sufficient for origin activation. Our data suggest that origin-associated G4 determine the precise position of replication initiation at a subset of origins (Figure 2D). However, single-fraction SNS-Seq experiments indicate that synthesis of the leading strand pauses at origin-associated G4 motifs in vivo (Figure 3) despite the presence of Pif1 and other G4-unwinding helicases (Maizels and Grey, 2013). Replication fork stalling might therefore be responsible for the observed accumulation of SNS at G4-associated origins (Valton et al., 2014). Moreover, it provides an alternative explanation to the repeatedly observed, yet enigmatic, higher firing efficiency of these sites (Figure S2B; Besnard et al., 2012; Valton et al., 2014). Our finding is unlikely to be a technical artifact. First, SNS purifications underwent up to five rounds of Lexo digestion. Second, SNS were absent from non-dividing cells and degraded upon RNase or alkali treatment (Cayrou et al., 2011; Cayrou et al., 2012b). Third, recent work based on independent assays revealed transient replication fork stalling at exogenous G4 sequences in vitro (Castillo Bosch et al., 2014).
Whereas most studies have focused on the relation between chromatin features and replication timing (Bell et al., 2010; Eaton et al., 2010; Gindin et al., 2014), little is known about the chromatin configuration of metazoan origins. Moreover, conflicting evidence has been reported on the role of transcription in origin firing (Sequeira-Mendes et al., 2009; Martin et al., 2011; Lubelsky et al, 2014). An answer to this question likely depends on the genomic scale at which conclusions are drawn. Here we show that a specific chromatin configuration similarly marks efficient and poorly used origins, irrespective of replication timing (Figures 5, 6 and 7). However, differential transcription at RSSs discriminates active from inactive origin-CGRs, explaining differential usage of these sites (Figure 6), and reflects differential origin activity across cell types (Figure 7). These findings reinforce the convergence of transcriptional and replication programs at replication origins and support a model in which DNA shape and chromatin features primarily define origin localization. Transcription at RSSs, in contrast, likely contributes to modulate aggregated firing probabilities, which could be directly reflected in the number of MCM molecules recruited at the origin site (Bechhoefer and Rhind, 2012).
While our study does not establish a causal link between transcription and origin firing, we anticipate that the genome-wide data sets reported here will facilitate mechanistic studies of the interplay between transcription and origin function in higher eukaryotes.
Experimental procedures
Cells and cell culture
Drosophila S2-DRSC cells and ML-DmBG3-c2 cells were cultured at 25 °C in 145 mm plates (Greiner) at a density of 1.5 × 10^6 cells/ml in Schneider’s Insect Cell Medium (Sigma S0146) and Shields and Sang M3 insect medium (Sigma S3652) with10 μg/ml insulin (Sigma), respectively, both supplemented with 10 % FBS (Pansera ES).
SNS Purification
SNS fragments were isolated essentially as described in (Cayrou et al. 2011) with the following modifications. Adjustments were made according to SW-41 Ti rotor specifications (Beckman-Coulter) and centrifugation was carried out at 4 °C and 26700 rpm for 21 hours. DNA fragments of 0.5-2.5 kb were collected, purified and subjected to four/five rounds of T4 PNK (Fermentas) phosphorylation and lambda Exonuclease (Fermentas) digestion. SNS were then prepared for sequencing by digesting the RNA-primer, second strand synthesis (NEBNext mRNA Second Strand Synthesis) and purified with AMPure XP beads (Beckman-Coulter). This procedure ensures that RNA species do not contribute to SNS-Seq signals. For a complete description of the experimental procedures, see Supplemental Experimental Procedures.
Data analysis and statistical learning
S2 and Bg3 SNS-Seq reads were aligned to the dm3 Drosophila reference genome using Bowtie2 (Langmead et al., 2012). Human SNS-Seq data (Besnard et al., 2012) were mapped to the hg19 human reference genome. Origin peaks were called using MACS (Zhang et al., 2008). The coverage function was used to define the position of the RSS within each origin peak and read counts normalized to the peak length in kb were used as a proxy for origin efficiency. G4 motif occurrences in the Drosophila genome were predicted with QuadParser (Huppert and Balasubramanian, 2005). DNA shape features were obtained from high-throughput predictions (Zhou et al., 2013). CG-rich regions were predicted according to (Gardiner-Garden and Frommer, 1987). Feature scoring was performed essentially as described (Comoglio and Paro, 2014). Lasso logistic regression models were trained with ten-fold cross validation. Feature selection probabilities were computed with bootstrap-Lasso (Comoglio and Paro 2014). Differential origin activity analysis was carried out using DESeq2 (Love et al., 2014). For a complete description of the algorithms and analysis procedures, see the Supplemental Experimental Procedures.
Supplementary Material
Acknowledgements
We thank Katja Eschbach and Ina Nissen of the Quantitative Genomics Facility, DBSSE, ETH Zurich, for excellent technical assistance in DNA library preparation and Illumina sequencing and Radostina Pirovska for performing preliminary experimental work. We are grateful to Mauro Giacca and Alessandro Carrer for support with the SNS protocol, Sarah Geisler and Allwyn Pereira for critical reading of the manuscript, and to Dirk Schübeler, Kenji Shimada, Niko Beerenwinkel and Maurizio Rinaldi for insightful discussion. F.C. is a member of the Life Science Zurich Graduate School, PhD program in Systems Biology. This research was supported by the Swiss National Science Foundation, by Epigenesys and the ETH Zurich to R.P. and the National Institutes of Health (grants R01GM106056 and U01GM103804 to R.R.).
Footnotes
Accession numbers
The Gene Expression Omnibus (GEO) accession number for the SNS-Seq data reported in this paper is GSE65692.
Author Contributions
F.C., T.S., C.B. and R.P. designed the study. F.C. carried out most of the experimental work with the help of T.S. and V.S. F.C. carried out computational and statistical analyses and wrote the manuscript with the help of T.S, R.P. and R.R., who also contributed to the DNA shape analysis.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Alabert C, Groth A. Chromatin replication and epigenome maintenance. Nat. Rev. Mol. Cell Biol. 2012;13:153–167. doi: 10.1038/nrm3288. [DOI] [PubMed] [Google Scholar]
- Bechhoefer J, Rhind N. Replication timing and its emergence from stochastic processes. Trends Genet. 2012;28:374–381. doi: 10.1016/j.tig.2012.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bell O, Schwaiger M, Oakeley EJ, Lienert F, Beisel C, Stadler MB, Schübeler D. Accessibility of the Drosophila genome discriminates PcG repression, H4K16 acetylation and replication timing. Nat. Struct. Mol. Biol. 2010;17:894–900. doi: 10.1038/nsmb.1825. [DOI] [PubMed] [Google Scholar]
- Besnard E, Babled A, Lapasset L, Milhavet O, Parrinello H, Dantec C, Marin JM, Lemaitre JM. Unraveling cell type-specific and reprogrammable human replication origin signatures associated with G-quadruplex consensus motifs. Nat. Struct. Mol. Biol. 2012;19:837–844. doi: 10.1038/nsmb.2339. [DOI] [PubMed] [Google Scholar]
- Bielinsky AK, Gerbi SA. Discrete start sites for DNA synthesis in the yeast ARS1 origin. Science. 1998;279:95–98. doi: 10.1126/science.279.5347.95. [DOI] [PubMed] [Google Scholar]
- Castillo Bosch P, Segura-Bayona S, Koole W, van Heteren JT, Dewar JM, Tijsterman M, Knipscheer P. FANCJ promotes DNA synthesis through G-quadruplex structures. EMBO J. 2014;33:2521–2533. doi: 10.15252/embj.201488663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cayrou C, Coulombe P, Vigneron A, Stanojcic S, Ganier O, Peiffer I, Rivals E, Puy A, Laurent-Chabalier S, Desprat R, Méchali M. Genome-scale analysis of metazoan replication origins reveals their organization in specific but flexible sites defined by conserved features. Genome Res. 2011;21:1438–1449. doi: 10.1101/gr.121830.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cayrou C, Coulombe P, Puy A, Rialle S, Kaplan N, Segal E, Méchali M. New insights into replication origin characteristics in metazoans. Cell Cycle. 2012a;11:658–667. doi: 10.4161/cc.11.4.19097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cayrou C, Grégoire D, Coulombe P, Danis E, Méchali M. Genome-scale identification of active DNA replication origins. Methods. 2012b;57:158–164. doi: 10.1016/j.ymeth.2012.06.015. [DOI] [PubMed] [Google Scholar]
- Celniker SE, Dillon LA, Gerstein MB, Gunsalus KC, Henikoff S, Karpen GH, Kellis M, Lai EC, Lieb JD, MacAlpine DM, Micklem G, Piano F, Snyder M, Stein L, White KP, Waterston RH, modENCODE Consortium Unlocking the secrets of the genome. Nature. 2009;459:927–930. doi: 10.1038/459927a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y, Zhang X, Dantas Machado AC, Ding Y, Chen Z, Qin PZ, Rohs R, Chen L. Structure of p53 binding to the BAX response element reveals DNA unwinding and compression to accommodate base-pair insertion. Nucleic Acids Res. 2013;41:8368–8376. doi: 10.1093/nar/gkt584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Comoglio F, Paro R. Combinatorial modeling of chromatin features quantitatively predicts DNA replication timing in Drosophila. PLoS Comput. Biol. 2014;10:e1003419. doi: 10.1371/journal.pcbi.1003419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deal RB, Henikoff JG, Henikoff S. Genome-wide kinetics of nucleosome turnover determined by metabolic labeling of histones. Science. 2010;328:1161–1164. doi: 10.1126/science.1186777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eaton ML, Prinz JA, MacAlpine HK, Tretyakov G, Kharchenko PV, MacAlpine DM. Chromatin signatures of the Drosophila replication program. Genome Res. 2011;21:164–174. doi: 10.1101/gr.116038.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gardiner-Garden M, Frommer M. CpG islands in vertebrate genomes. J. Mol. Biol. 1987;196:261–282. doi: 10.1016/0022-2836(87)90689-9. [DOI] [PubMed] [Google Scholar]
- Ge XQ, Lin H. Noncoding RNAs in the regulation of DNA replication. Trends Biochem. Sci. 2014;39:341–343. doi: 10.1016/j.tibs.2014.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilbert DM. Evaluating genome-scale approaches to eukaryotic DNA replication. Nat. Rev. Genet. 2010;11:673–684. doi: 10.1038/nrg2830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilbert DM. Replication origins run (ultra) deep. Nat. Struct. Mol. Biol. 2012;19:740–742. doi: 10.1038/nsmb.2352. [DOI] [PubMed] [Google Scholar]
- Gindin Y, Valenzuela MS, Aladjem MI, Meltzer PS, Bilke S. A chromatin structure-based model accurately predicts DNA replication timing in human cells. Mol. Syst. Biol. 2014;10:722. doi: 10.1002/msb.134859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hancock SP, Ghane T, Cascio D, Rohs R, Di Felice R, Johnson RC. Control of DNA minor groove width and Fis protein binding by the purine 2-amino group. Nucleic Acids Res. 2013;41:6750–6760. doi: 10.1093/nar/gkt357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoshina S, Yura K, Teranishi H, Kiyasu N, Tominaga A, Kadoma H, Nakatsuka A, Kunichika T, Obuse C, Waga S. Human origin recognition complex binds preferentially to G-quadruplex-preferable RNA and single-stranded DNA. J. Biol. Chem. 2013;288:30161–30171. doi: 10.1074/jbc.M113.492504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huppert JL, Balasubramanian S. Prevalence of quadruplexes in the human genome. Nucleic Acids Res. 2005;33:2908–2916. doi: 10.1093/nar/gki609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaplan N, Moore IK, Fondufe-Mittendorf Y, Gossett AJ, Tillo D, Field Y, LeProust EM, Hughes TR, Lieb JD, Widom J, Segal E. The DNA-encoded nucleosome organization of a eukaryotic genome. Nature. 2009;458:362–366. doi: 10.1038/nature07667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B, Salzberg SL. Fast gapped-read alignment with Bowtie 2. Nat. Methods. 2012;9:357–359. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lazarovici A, Zhou T, Shafer A, Dantas Machado AC, Riley TR, Sandstrom R, Sabo PJ, Lu Y, Rohs R, Stamatoyannopoulos JA, Bussemaker HJ. Probing DNA shape and methylation state on a genomic scale with DNase I. Proc. Natl. Acad. Sci. U S A. 2013;110:6376–6381. doi: 10.1073/pnas.1216822110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leonard AC, Méchali M. DNA replication origins. Cold Spring Harb. Perspect. Biol. 2013;5:a010116. doi: 10.1101/cshperspect.a010116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15:550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lombraña R, Almeida R, Revuelta I, Madeira S, Herranz G, Saiz N, Bastolla U, Gómez M. High-resolution analysis of DNA synthesis start sites and nucleosome architecture at efficient mammalian replication origins. EMBO J. 2013;32:2631–2644. doi: 10.1038/emboj.2013.195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lubelsky Y, Prinz JA, DeNapoli L, Li Y, Belsky JA, MacAlpine DM. DNA replication and transcription programs respond to the same chromatin cues. Genome Res. 2014;24:1102–1114. doi: 10.1101/gr.160010.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacAlpine HK, Gordân R, Powell SK, Hartemink AJ, MacAlpine DM. Drosophila ORC localizes to open chromatin and marks sites of cohesin complex loading. Genome Res. 2010;20:201–211. doi: 10.1101/gr.097873.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maizels N, Gray LT. The G4 genome. PLoS Genet. 2013;9:e1003468. doi: 10.1371/journal.pgen.1003468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin MM, Ryan M, Kim R, Zakas AL, Fu H, Lin CM, Reinhold WC, Davis SR, Bilke S, Liu H, Doroshow JH, Reimers MA, Valenzuela MS, Pommier Y, Meltzer PS, Aladjem MI. Genome-wide depletion of replication initiation events in highly transcribed regions. Genome Res. 2011;21:1822–1832. doi: 10.1101/gr.124644.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masai H, Matsumoto S, You Z, Yoshizawa-Sugata N, Oda M. Eukaryotic chromosome DNA replication: where, when, and how? Annu. Rev. Biochem. 2010;79:89–130. doi: 10.1146/annurev.biochem.052308.103205. [DOI] [PubMed] [Google Scholar]
- Mott ML, Berger JM. DNA replication initiation: mechanisms and regulation in bacteria. Nat. Rev. Microbiol. 2007;5:343–354. doi: 10.1038/nrmicro1640. [DOI] [PubMed] [Google Scholar]
- Picard F, Cadoret J-C, Audit B, Arneodo A, Alberti A, Battail C, Duret L, Prioleau M-N. The spatiotemporal program of DNA replication is associated with specific combinations of chromatin marks in human cells. PLoS Genet. 2014;10:e1004282. doi: 10.1371/journal.pgen.1004282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Remus D, Beall EL, Botchan MR. DNA topology, not DNA sequence, is a critical determinant for Drosophila ORC-DNA binding. EMBO J. 2004;23:897–907. doi: 10.1038/sj.emboj.7600077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rohs R, Jin X, West SM, Joshi R, Honig B, Mann RS. Origins of specificity in protein-DNA recognition. Annu. Rev. Biochem. 2010;79:233–269. doi: 10.1146/annurev-biochem-060408-091030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rohs R, West SM, Sosinsky A, Liu P, Mann RS, Honig B. The role of DNA shape in protein-DNA recognition. Nature. 2009;461:1248–1253. doi: 10.1038/nature08473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sequeira-Mendes J, Díaz-Uriarte R, Apedaile A, Huntley D, Brockdorff N, Gómez M. Transcription initiation activity sets replication origin efficiency in mammalian cells. PLoS Genet. 2009;5:e1000446. doi: 10.1371/journal.pgen.1000446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takayama S, Dhahbi J, Roberts A, Mao G, Heo SJ, Pachter L, Martin DI, Boffelli D. Genome methylation in D. melanogaster is found at specific short motifs and is independent of DNMT2 activity. Genome Res. 2014;24:821–830. doi: 10.1101/gr.162412.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tardat M, Brustel J, Kirsh O, Lefevbre C, Callanan M, Sardet C, Julien E. The histone H4 Lys 20 methyltransferase PR-Set7 regulates replication origins in mammalian cells. Nat. Cell Biol. 2010;12:1086–1093. doi: 10.1038/ncb2113. [DOI] [PubMed] [Google Scholar]
- Tibshirani R. Regression shrinkage and selection via the lasso. J. R. Stat. Soc. B. 1996;58:267–288. [Google Scholar]
- Tillo D, Hughes TR. G+C content dominates intrinsic nucleosome occupancy. BMC Bioinformatics. 2009;10:442. doi: 10.1186/1471-2105-10-442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valton AL, Hassan-Zadeh V, Lema I, Boggetto N, Alberti P, Saintomé C, Riou JF, Prioleau M-N. G4 motifs affect origin positioning and efficiency in two vertebrate replicators. EMBO J. 2014;33:732–746. doi: 10.1002/embj.201387506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeman MK, Cimprich KA. Causes and consequences of replication stress. Nat. Cell Biol. 2014;16:2–9. doi: 10.1038/ncb2897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Liu T, Meyer CA, Eeckhoute J, Johnson DS, Bernstein BE, Nusbaum C, Myers RM, Brown M, Li W, Liu XS. Model-based analysis of ChIP-Seq (MACS) Genome Biol. 2008;9:R137. doi: 10.1186/gb-2008-9-9-r137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou T, Yang L, Lu Y, Dror I, Dantas Machado AC, Ghane T, Di Felice R, Rohs R. DNAshape: a method for the high-throughput prediction of DNA structural features on a genomic scale. Nucleic Acids Res. 2013;41:W56–62. doi: 10.1093/nar/gkt437. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.