SUMMARY
The probabilistic expression of cytokine genes in differentiated T helper (Th) cell populations remains ill defined. By single-cell analyses and mathematical modeling, we show that one stimulation featured stable cytokine nonproducers as well as stable producers with wide cell-to-cell variability in the magnitude of expression. Focusing on interferon-γ (IFN-γ) expression by Th1 cells, mathematical modeling predicted that this behavior reflected different cell-intrinsic capacities and not mere gene-expression noise. In vivo, Th1 cells sort purified by secreted IFN-γ amounts preserved a quantitative memory for both probability and magnitude of IFN-γ re-expression for at least 1 month. Mechanistically, this memory resulted from quantitatively distinct transcription of individual alleles and was controlled by stable expression differences of the Th1 cell lineage-specifying transcription factor T-bet. Functionally, Th1 cells with graded IFN-γ production competence differentially activated Salmonella-infected macrophages for bacterial killing. Thus, individual Th cells commit to produce distinct amounts of a given cytokine, thereby generating functional intrapopulation heterogeneity.
INTRODUCTION
Cytokines are key regulators of immune responses. Differentiated T helper (Th) cells rapidly secrete specific cytokines upon antigen challenge (Löhning et al., 2002; Zhu et al., 2010). The lineage-specifying transcription factors T-bet, GATA-3, and RORγt program the expression of Th1 (interferon-γ [IFN-γ]), Th2 (interleukin-4 [IL-4], IL-5, and IL-13), and Th17 (IL-17) cell-associated cytokines, respectively (Zhu et al., 2010). However, only a fraction of activated Th cells expressing such a “master regulator” transcription factor produces the associated cytokines (Bucy et al., 1994; Openshaw et al., 1995; Peine et al., 2013). Such intrapopulation heterogeneity has been attributed to a stochastic “choice” of the cells (Apostolou and Thanos, 2008; Guo et al., 2005; Rand et al., 2012). However, mammalian gene transcription occurs in brief bursts, separated by random intervals of up to several hours (Harper et al., 2011; Suter et al., 2011). Thus, antigen-stimulated T cells might rapidly switch between cytokine-producing and silent states, implying that all cells in a population are producers—but at different time points. Alternatively, the decision to express a cytokine could be made only once, resulting in stable producing and nonproducing subpopulations.
A rapid-switching model based on transcriptional bursting implies that the amounts of a given cytokine produced by an individual cell fluctuate over time. Such rapid fluctuations have been observed for constitutively expressed genes in human cell lines (Sigal et al., 2006), suggesting that each individual cell recapitulates the entire variability in the population. By contrast, individual Th cells might have different inherent capacities to express cytokine genes. This capacity might be influenced by response thresholds caused by heterogeneous expression of receptors, signaling proteins, and key transcription factors (Feinerman et al., 2008; Peine et al., 2013). Intrapopulation heterogeneity might result in functional diversification of Th cell responses (O’Garra et al., 2011) and—presumably—of T-cell-mediated immunological memory.
Previous studies on cytokine expression are based on conventional “snapshot” flow cytometry that would have missed a rapid switching between cytokine-producing and -nonproducing states. Here, we have developed an experimental method to track the expression of endogenous cytokine genes in individual Th cells over time without resorting to genetic alterations. Our approach combined the fluorescent labeling of viable cytokine producers by a cytokine capture matrix on the cell surface (“secretion assay”) (Assenmacher et al., 1998; Löhning et al., 2003) with time-delayed intracellular staining. We show that in a given stimulation, T cells made a stable decision whether to produce a given cytokine or not. In addition, the producers committed to individual magnitudes of expression. Mathematical modeling predicted different cell-intrinsic capacities to express the respective cytokine genes. Using a prototypical example, we found that the amount of IFN-γ production was a stable feature of individual Th1 cells that was memorized for at least 1 month in vivo, even upon immunological challenge. This memory was based on quantitatively distinct transcription at single alleles, controlled by different quantities of T-bet protein, and associated with graded DNA methylation at the Ifng and Tbx21 loci. In functional terms, the produced IFN-γ amount defined a cell’s capacity to stimulate macrophages to kill bacteria. Thus, individual T cells can stably maintain and inherit distinct expression rates of a given cytokine, thereby regulating their potential to stimulate immune responses.
RESULTS
Differentiated Th Cells Segregate into Stable Cytokine-Producing and -Nonproducing Subsets during One Stimulation
We analyzed the cytokine production behavior of Th1, Th2, and Th17 cells in a kinetic fashion. To obtain homogeneous populations, we derived them from naive precursors. Cytokine-producing cells reached their maximal frequency within ~3 hr after stimulation (Figure S1A available online). An interruption of stimulation led to the rapid termination of cytokine production, and resumption of stimulation caused rapid reinitiation (Figure S1B). At every time point, a fraction of the cells did not produce cytokines. However, this behavior did not reflect heterogeneous differentiation or activation, because all cells underwent multiple cell divisions (data not shown), upregulated the activation marker CD44 (Figure S1C), and homogeneously expressed the lineage-specifying transcription factors T-bet, GATA-3, or RORγt, respectively. Thus, cytokine expression by Th cell populations appeared heterogeneous and required recent and continuous stimulation, consistent with previous studies on CD8+ T cells (Corbin and Harty, 2005; Slifka et al., 1999).
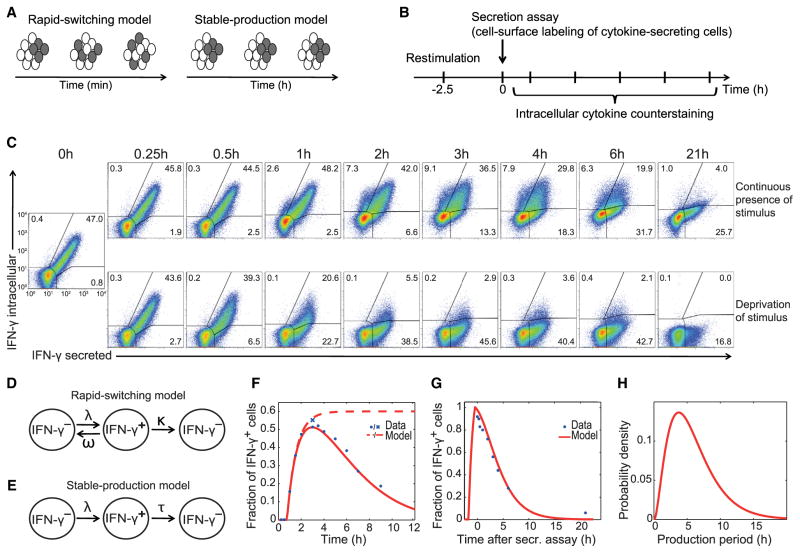
To distinguish whether all stimulated Th cells transiently produce cytokines but rapidly cycle between producing and nonproducing states or whether there are stable producing sub-populations (Figure 1A), we tracked the behavior of individual cells over time. We surface labeled viable cytokine producers via the cytokine secretion assay technology (Assenmacher et al., 1998; Löhning et al., 2003) and counterstained for the same cytokine intracellularly at various time points (Figure 1B). The vast majority of Th1 cells that initiated IFN-γ production maintained it for several hours in the presence of the stimulus (Figure 1C; top row, upper right quadrants). Controls without cell permeabilization confirmed the accurate detection of intra-cellular versus surface-captured IFN-γ (Figure S2A). Similarly, individual Th17 cells continuously produced IL-17 (Figure S2B). A small fraction of cells switched on cytokine production with some delay (Figure 1C, upper left quadrants), consistent with the gradual culmination of cytokine production (cf. Figures S1A and S1B). Virtually all cytokine-producing cells switched off cytokine production within 21 hr (Figure 1C, lower right quadrants). A substantial fraction of cells did not produce cytokine throughout the experiment (Figure 1C, lower left quadrants). These results show that upon stimulation, fully differentiated Th cells segregate into stable cytokine-producing and -nonproducing subpopulations.
Figure 1. Individual Th Cells Maintain Their Specific Rate of Cytokine Production.
(A) Alternative models of cytokine production by T cell populations.
(B) Experimental setup.
(C) Th1 cells were restimulated with PMA and ionomycin. Live IFN-γ+ cells were labeled by secretion assay and cultured with or without stimulus. Intracellular IFN-γ counterstainings were performed at the indicated time points. Percentages of IFN-γ+ cells are indicated.
(D) Scheme of the “rapid-switching model” that allows cells to cycle between producing and nonproducing states at rates λ and ω before production is irreversibly ceased at a rate κ.
(E) Scheme of the “stable-production model” used to extract the IFN-γ production length distribution τ in the population. Cells become IFN-γ+ at a rate λ.
(F) Fit of the stable-production model (solid line) to the time series data of IFN-γ production (intracellular staining, dots). The brefeldin A control (cross) is modeled by leaving out the second step in the model (dashed line).
(G) Model fit (simultaneously with the data from F) to the kinetics of the secreted-IFN-γ+ cells from (C) (IFN-γ measured by intracellular staining).
(H) The resulting production period is 5.9 ± 3.6 hr (mean production period and variability within the population).
Data are representative of three independent experiments. See also Figures S1 and S2.
Individual Th Cells Maintain Their Specific Rate of Cytokine Production
To assess how long an individual cell produced a given cytokine, we fitted mathematical models of cytokine production to the data in Figure 1C. The models describe IFN-γ− cells becoming IFN-γ+ upon stimulation and expressing the cytokine for a certain time. In the first model, we allowed for rapid switching between on and off states, e.g., by transcriptional bursting (Figure 1D). We fitted the model to the fraction of IFN-γ+ cells over time, gating on either all cells or only those that had initially been surface labeled (Figures 1C; 0 hr, upper right gate). This fit constrained the backward rate from the transcriptionally active state, ω, to be less than 0.09/hr (upper bound of the 95% confidence interval, Table S1), implying a half-life of this state of 7.7 hr or longer. Hence, repeated on-off switching of IFN-γ expression could be neglected. Instead, we considered a stable-production model where switching off cytokine expression is irreversible after a gamma-distributed production period τ (Figure 1E). The model accurately fitted the time courses of intracellular IFN-γ+ cells among both total and surface-labeled cells simultaneously (Figures 1F and 1G). The best-fit parameters implied that, on average, after an initial delay of 40 min, the cells start IFN-γ expression within the following 1 hr and continue production for 5.9 ± 3.6 hr (Figure 1H, Table S1). Thus, to describe the data, the stable-production model was required where individual cells switch on continuous cytokine production once (cf. Figures 1A and 1E). Switching on was more synchronous than switching off, explaining that the decline of the IFN-γ+ fraction was slower than the initial increase (Figure 1F). Stable production rather than rapid switching was also observed for IL-17 expression by Th17 cells (Figure S2B) and thus appeared to be a common mode of effector cytokine expression.
Mathematical Modeling Predicts Inherently Distinct Cytokine Expression Capacities of Individual Cells
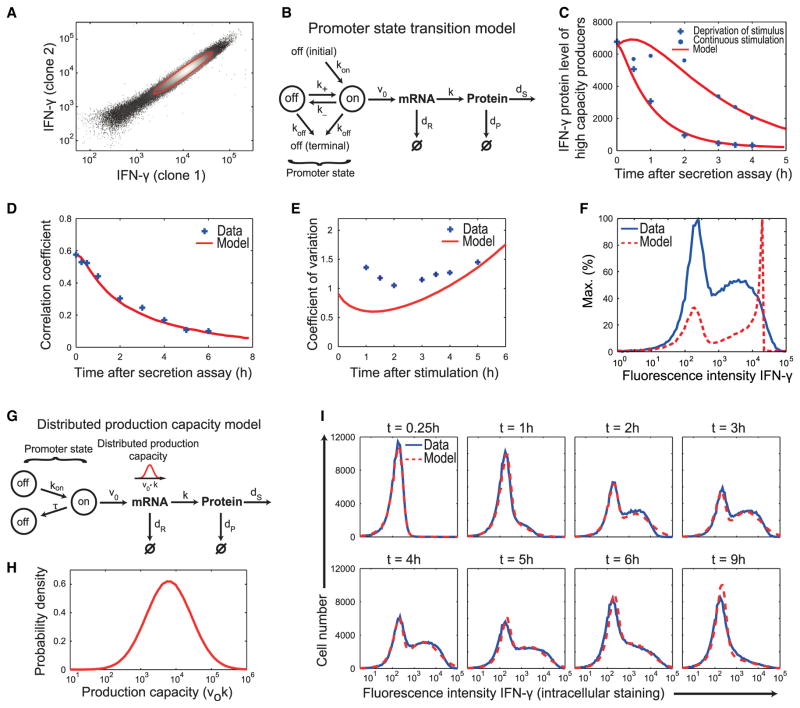
Among IFN-γ+ cells, the IFN-γ amount per cell varied by more than one order of magnitude. Our detection method introduced only a marginal experimental error (relative error 17%, Figure 2A), so this was primarily due to true cell-to-cell variability that could result from stochastic fluctuations in IFN-γ expression (e.g., in transcription rate) or intrinsically different IFN-γ expression capacities of individual cells, or both. Addressing this question, we asked whether a standard stochastic gene-expression model based on transcriptional bursting could describe the data (Raj et al., 2006). To account for the transient nature of cytokine production, we extended the standard model by including initial and terminal off states (Figure 2B). This promoter state transition model produces cell-to-cell heterogeneity in IFN-γ expression due to switching between inactive and active promoter states (with rates rates k±; [Friedman et al., 2006; Mariani et al., 2010; Miller-Jensen et al., 2011]) as well as asynchronous induction and terminal switching off of cytokine transcription (with rates kon and koff, respectively).
Figure 2. Mathematical Modeling Predicts Inherently Distinct Cytokine Expression Capacities of Individual Cells.
(A) Intracellular IFN-γ detection with two different antibodies. The red line represents the one standard deviation error ellipse of the IFN-γ+ cells corresponding to a relative measurement error of 17%.
(B) Scheme of the “promoter state transition model:” Cytokine transcription becomes active with rate kon after stimulation and is terminally inactivated with rate koff; intermittently, the promoter can switch between transcriptional on and off states (transcriptional bursting, k±). Transcription, translation, degradation of IFN-γ mRNA and protein, and protein secretion are described with rate constants v0, k, dR, dP, and dS.
(C) Decline of the mean protein amount in IFN-γ+ cells with persistent stimulus (dots) and after removal of the stimulus (crosses) (cf. Figure 1C) together with fits of the model in (B) (solid lines).
(D) Correlation coefficients (crosses) calculated for the secreted-IFN-γ+ cells and fit of the model (solid line).
(E) Coefficients of variation of the distributions of IFN-γ+ cells (crosses) and simulation of the model substantially deviating from the data (solid line).
The experimental data in (C)–(E) were used simultaneously for fitting the model parameters.
(F) Intracellular IFN-γ staining of Th1 cells 3 hr after restimulation (blue) and simulation of the promoter state transition model (red).
(G) Scheme of the “distributed production capacity model:” As in (B) but allowing only for a single switch-on and switch-off event: The promoter switches to an on-state at a rate kon and switches back to a nonproductive state after a gamma-distributed production period τ. The IFN-γ expression capacity, defined as the product of transcription and translation rates, v0 k, is assumed to be lognormally distributed within the cell population.
(H) Lognormal distribution of the IFN-γ production capacity (v0 k) resulting from the fit in (I).
(I) Fit of the model (dashed line) to the time evolution of the distribution of intracellular IFN-γ amounts within the total Th1 cell population (solid line).
Data are representative of three independent experiments. See also Figure S3.
Given the short half-life of IFN-γ protein in the cells (~1 hr; Figure 2C, blue crosses), transcription fluctuations would manifest themselves at the protein level. However, the correlation of IFN-γ protein amounts at two different time points in the same cell (autocorrelation) persisted for longer than the IFN-γ protein half-life (Figure 2D). We asked whether the model in Figure 2B could explain this autocorrelation and the observed cell-to-cell variability in IFN-γ expression, as quantified by the coefficients of variation of the IFN-γ+ cells (Figure 2E, blue crosses). Systematic parameter estimation (Table S1) revealed that the model accounted for the kinetics of IFN-γ+ cells (Figure 2C, red curves) as well as the temporal correlations of IFN-γ quantities in individual cells (Figure 2D, red curve) but failed to reproduce the cell-to-cell variability of IFN-γ expression. The model accounted neither for the width (Figure 2E) nor the shape (Figure 2F) of the distribution. Thus, intrinsic noise in gene expression alone could not explain the observed cell-to-cell variability of IFN-γ expression.
Therefore, we extended the model by cell-to-cell differences in the IFN-γ expression capacity, defined as the product of transcription and translation rates (Figures 2G and 2H). These differences would result from the variability in regulators of transcription and/or translation between the cells, including epigenetic mechanisms. Moreover, we found that transcriptional bursting (rates k±) made a negligible contribution to the protein variability, explaining <7% of the CV of the IFN-γ+ cells (Table S1), so we neglected it. The resulting distributed production capacity model accurately described the dynamics of IFN-γ expression upon stimulation (Figure 2I). In the beginning, the broad IFN-γ distribution was due to the distributed expression capacities whereas the further increase until t = 3 hr resulted from the different switching-off times of individual cells. We also fitted the model to the distribution of initially surface-labeled cells, achieving a good fit with the same parameter values (Figure S3). To conclude, the observed cell-to-cell variability in IFN-γ protein amounts is consistent with a model in which individual cells have inherently distinct capacities for IFN-γ expression.
Individual Th1 Cells Exhibit a Stable Quantitative Memory for IFN-γ and T-bet Expression
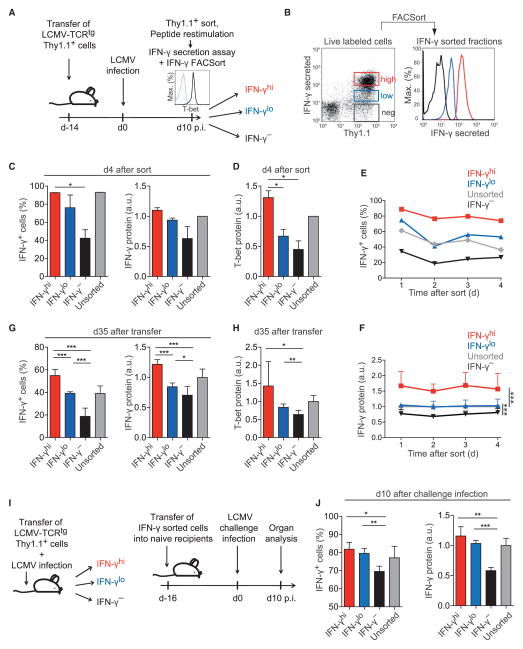
According to our data-driven modeling, a given antigen stimulation of Th1 cells featured stable IFN-γ high producers, low producers, and nonproducers. We therefore hypothesized that the stability of these qualitative (decision to express) and quantitative (expression magnitude) characteristics might persist in subsequent stimulations. To generate IFN-γ-producing cells in vivo, we adoptively transferred naive lymphocytic choriomeningitis virus (LCMV)-T cell receptor (TCR)-transgenic (tg) Th cells into wild-type (WT) mice and infected the recipients with LCMV, a strongly Th1-cell-polarizing pathogen (Hegazy et al., 2010). At the peak of infection, we isolated the transferred cells, which all expressed T-bet (Figure 3A, histogram), and sorted them by secreted amounts of IFN-γ upon antigen-specific restimulation (Figure 3B). In a second restimulation 4 days later, those cells that had initially produced the highest amounts of IFN-γ showed a higher probability to reexpress it than sorted IFN-γlo or IFN-γ− cells, and they again produced more IFN-γ per cell (Figure 3C). IFN-γhi cells also expressed the highest amounts of T-bet directly after sorting (data not shown), and this correlation was stable for at least 4 days (Figure 3D). Thus, individual Th1 cells generated during a viral infection in vivo had a quantitative memory for IFN-γ production that correlated with their degree of T-bet expression. Moreover, kinetic analyses revealed that the graded IFN-γ production capacity of IFN-γ-sorted Th1 cells was a stable property that could be observed at every time point in daily restimulations (Figures 3E, 3F, and S4).
Figure 3. Individual Th1 Cells Exhibit a Stable Quantitative Memory for IFN-γ and T-bet Expression.
(A) Experimental set up of (B)–(D). WT recipients of 2 × 105 LCMV-TCRtg CD4+Thy1.1+ T cells were infected with LCMV. Thy1.1+ cells were reisolated on day 10 and analyzed for T-bet expression (histogram inset; black, staining; gray, isotype control).
(B) Cells were restimulated with LCMV-GP64–80, sorted by secreted IFN-γ amounts, and cultured.
(C) Frequency of IFN-γ+ cells and normalized IFN-γ amount per cell in the sorted fractions on day 4 after sort.
(D) Normalized T-bet amounts per cell on day 4 after sort.
Representative results (B) and means + SD (C, D) of two experiments are shown.
(E and F) In-vitro-differentiated Th1 cells were sorted by secreted IFN-γ amounts and cultured with IL-2.
(E) Frequency of IFN-γ+ cells in the sorted fractions.
(F) Normalized IFN-γ amount per cell in the sorted fractions.
Representative results of three (E) and means + SD of two (F) independent experiments are shown.
(G and H) In-vitro-differentiated Th1 cells were sorted by secreted IFN-γ amounts, transferred into WT mice (1.5 × 106 cells/mouse), and reisolated on day 35. Means + SD of two independent experiments are shown.
(G) Frequency of IFN-γ+ cells and normalized IFN-γ amount per cell in the sorted fractions.
(H) Normalized T-bet amounts per cell in the sorted fractions.
(I) Experimental setup of (J). WT recipients of 2 × 105 LCMV-TCRtg CD4+Thy1.1+ T cells were infected with LCMV. Thy1.1+ cells were reisolated on day 10, restimulated with LCMV-GP64–80, sorted by secreted IFN-γ amounts, and transferred into naive WT mice (5 × 104 cells/mouse). After 16 days, secondary recipients were infected with LCMV. On day 10 after challenge infection, Thy1.1+ cells were reisolated.
(J) Frequency of IFN-γ+ cells and normalized IFN-γ amount per cell in the sorted fractions are shown (means + SD of n = 3–4 mice).
See also Figures S4 and S5.
Even among Th1 cells that were strongly polarized in LCMV infections, some did not produce IFN-γ in every restimulation (cf. Figure 3B). To formally show that fully differentiated Th1 cells had a quantitative cytokine memory, we performed a similar sort-and-track experiment starting with purified IFN-γ producers. Again, individual cells memorized both probability and per-cell amount of IFN-γ production, and this correlated with their degree of T-bet expression (Figure S5). Thus, the probability and amount of IFN-γ expression are stable properties of bona fide Th1 cells.
The Quantitative Memory for IFN-γ Expression Persists upon Viral Challenge Infection In Vivo
To analyze whether quantitative differences in IFN-γ expression were stable in the long term in memory Th1 cells in vivo, we adoptively transferred purified IFN-γhi, IFN-γlo, or IFN-γ− Th1 cells into WT mice. After more than 1 month, we analyzed the capacity of the resting cells to reexpress IFN-γ. Both probability and per-cell expression still recapitulated the IFN-γ expression capacity which the cells had been sorted by (Figure 3G). In addition, T-bet expression was still positively correlated with the amount of IFN-γ production (Figure 3H). Thus, the magnitude of expression of both T-bet (a constitutively expressed transcription factor) and IFN-γ (a stimulation-induced cytokine) are stable cell-intrinsic features.
We then examined whether individual Th1 cells maintain their quantitative cytokine memory after a strongly Th1-cell-polarizing challenge. We isolated in-vivo-differentiated Th1 cells from LCMV-infected mice, sorted them by secreted quantities of IFN-γ, and transferred the sorted fractions into naive recipients (Figure 3I). After at least 2 weeks of resting, the recipient mice were infected with LCMV. Upon reisolation at the peak of the secondary infection, the cells still recapitulated their initial graded differences in IFN-γ expression probability and amount (Figure 3J). Notably, the stably constrained IFN-γ production of the sorted IFN-γ− cells was not due to impaired proliferation, because these cells expanded at least 50-fold upon LCMV challenge. Thus, differentiated Th cells can remain committed to produce distinct quantities of effector cytokines while participating in sequential immune reactions in vivo.
Quantitative Memory for IFN-γ Production Is Regulated at the Level of Transcription at Individual Alleles
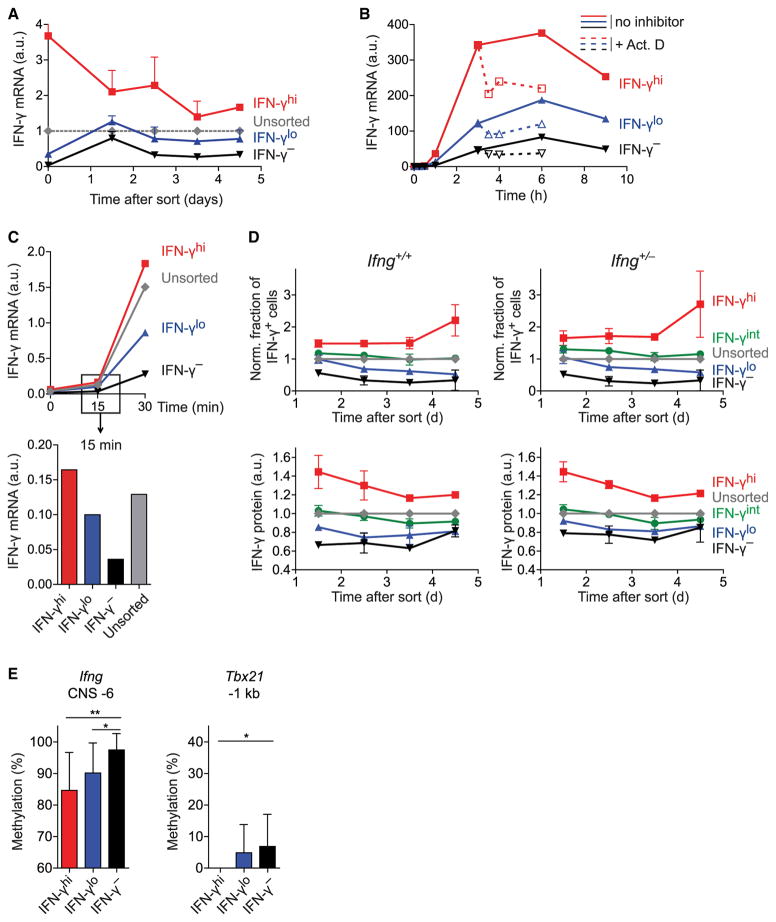
Next, we asked whether the quantitative cytokine memory was regulated at the RNA or protein level. Th1 cells sorted by their amount of IFN-γ secretion continuously featured graded IFN-γ mRNA quantities in subsequent restimulations (Figure 4A), matching their stably graded IFN-γ protein amounts and probabilities to produce IFN-γ (cf. Figures 3E, 3F, and S4). Thus, the secretion of distinct amounts of IFN-γ by sub-populations of Th1 cells does not reflect different translation rates but different mRNA amounts. To distinguish the possibility of enhanced transcription at the Ifng locus in IFN-γhi cells from that of enhanced IFN-γ mRNA degradation in IFN-γlo cells, we used the transcription inhibitor actinomycin D. Blocking transcription 3 hr after stimulation onset reduced IFN-γ mRNA (Figure 4B). However, this reduction was most profound in IFN-γhi cells, showing that degradation was at least as efficient in IFN-γhi cells as in IFN-γlo cells. Moreover, at just 15 min after stimulation onset, when mRNA amounts reflect transcription rather than degradation rates, IFN-γ transcripts were graded among the sorted subsets (Figure 4C). Taken together, these results indicate that the specific amounts of IFN-γ secreted by individual Th1 cells resulted from differences in IFN-γ transcription rates and not from differences in mRNA degradation or translation.
Figure 4. The Quantitative Memory for IFN-γ Production Is Regulated at the Level of Transcription at Individual Alleles.
(A–C) Th1 cells were sorted by secreted IFN-γ amounts and tracked.
(A) IFN-γ mRNA upon restimulation was normalized to HPRT (means + SD).
(B) On day 3 after sort, cells were restimulated. 3 hr after onset, transcription was inhibited in some cells (dotted line, open symbols). IFN-γ mRNA normalized to HPRT over the course of stimulation is shown.
(C) Data as in (B) with a focus on early time points after stimulation onset.
Data are pooled from (A) or are representative of (B, C) two independent experiments.
(D) Ifng+/+ or Ifng+/− Th1 cells were sorted by secreted IFN-γ amounts and cultured. Top, frequency of IFN-γ+ cells in sorted fractions normalized to that in unsorted cells. Bottom, normalized IFN-γ amount per cell in the sorted fractions. Means ± SD of two independent experiments are shown.
(E) Degree of DNA methylation (means + SD) is depicted in Th1 cells sorted by graded IFN-γ secretion and analyzed by bisulfite sequencing at a CpG island corresponding to CNS −6 at the Ifng locus (left) and at a CpG island approximately 1 kb upstream of the Tbx21 promoter (right).
See also Figure S4.
One possible mechanism underlying Ifng expression differences could be stable usage of either one or two alleles, such that IFN-γhi cells would always express biallelically and IFN-γlo cells would only express monoallelically. To test this hypothesis, we differentiated Ifng+/+ and Ifng+/− Th1 cells in a coculture and sorted them for differential IFN-γ secretion. We found that the graded differences in IFN-γ expression were similarly stable in WT and Ifng+/− cells (Figure 4D). Thus, it is not differential allelic usage but constant transcription rate differences at individual alleles that regulate the quantitative memory for IFN-γ. These cell-specific transcription rates could result from different chromatin states allowing distinct degrees of transcription factor binding at regulatory sites. One mechanism assumed to stably suppress transcription is DNA methylation. We found less methylation in IFN-γhi and IFN-γlo cells than in IFN-γ− Th1 cells at the key regulatory conserved noncoding sequence (CNS) −6 (Balasubramani et al., 2010b; Shnyreva et al., 2004) at the Ifng locus (Figure 4E). We also found graded DNA methylation upstream of the Tbx21 promoter (Figure 4E), matching the higher T-bet expression in IFN-γhi cells compared with that in their IFN-γlo/− counterparts (cf. Figures 3D and 3F). In summary, our findings imply a model where DNA methylation differences contribute to quantitatively distinct IFN-γ transcription rates at individual alleles.
T-bet Quantitatively Controls IFN-γ Expression in Fully Differentiated Th1 Cells
Cells sorted for a high amount of IFN-γ secretion had more T-bet mRNA and protein than their IFN-γlo and IFN-γ− counterparts (Figures S6, 3D, and 3H). This corresponds with the more pronounced DNA methylation of IFN-γlo and IFN-γ− cells at the Tbx21 locus (cf. Figure 4E). Next, we analyzed the quantitative relationship between T-bet and IFN-γ expression in fully committed Th1 cells at the single-cell level by coexpression analysis. We found that the more T-bet protein was expressed by a cell, the higher was its probability to produce IFN-γ and the produced IFN-γ amount (Figure 5A). To test whether T-bet amounts are predictive of IFN-γ expression in subsequent re-stimulations, we sorted Th1 cells from T-bet-ZsGreen reporter (TBGR) mice (Zhu et al., 2012) by their intensity of ZsGreen (i.e., T-bet) expression (Figure 5B) and analyzed their capacity to express IFN-γ. The initial T-bet expression predicted the production of IFN-γ in terms of probability and amount per cell immediately after the sort and also several days later (Figures 5C and 5D). Upon adoptive transfer into WT mice, Th1 cells sorted by T-bet amounts preserved their differential T-bet and IFN-γ expression for at least 1 month in vivo (Figures 5E and 5F). Distinct T-bet protein amounts were stably maintained by these resting memory cells independent of restimulation (Figure S6C). Moreover, the same functional relationship between T-bet and IFN-γ expression described the data both immediately after the sort and 4 weeks later (Figure 5G), suggesting that T-bet quantitatively controlled IFN-γ expression in the same manner in activated effector and in memory cells.
Figure 5. T-bet and IFN-γ Expression Are Quantitatively Correlated.
(A) Th1 cells were stained intracellularly for IFN-γ, combined with either T-bet staining (left plot, colored dots) or isotype control staining (left plot, gray dots). IFN-γ expression in subpopulations with different T-bet expression is shown. Frequencies of IFN-γ+ cells and geometric mean of IFN-γ in IFN-γ+ cells (bold numbers) are indicated. Data are representative of three independent experiments.
(B) TBGR Th1 cells were sorted by ZsGreen expression and cultured.
(C) Frequency of IFN-γ+ cells in the sorted fractions and IFN-γ amount per cell in the sorted fractions, both normalized to those in unsorted cells, are shown as means + SD on day 4 after sort.
(D) Kinetic analysis of the frequency of IFN-γ+ cells in the sorted fractions (means ± SD).
Data are pooled from three (C) or two (D) independent experiments.
(E–G) TBGR LCMV-TCRtg Thy1.1+ Th1 cells were sorted by ZsGreen expression into T-bethi or T-betlo fractions and transferred into WT mice (2 × 106 cells/mouse).
(E) T-bet expression and frequency of IFN-γ+ cells directly before transfer, both normalized to those in unsorted cells.
(F) T-bet expression and frequencies of IFN-γ+ cells, both normalized to those in unsorted controls, are shown in cells reisolated from spleens on day 29 after transfer (n = 3 mice/group).
Data in (E) and (F) represent means + SD from four independent experiments.
(G) Correlation of IFN-γ+ frequency with ZsGreen expression directly after sort (purple dots) and on day 29 after transfer (blue dots). Each blue dot represents transferred cells recovered from one recipient. Data are representative of two independent experiments. The purple line shows the best fit to the data obtained directly after sort via a two-parameter model (cf. Supplemental Experimental Procedures). The shaded region indicates the 95% confidence prediction bands. The predicted functional relationship captures the measured data on day 29.
See also Figure S6.
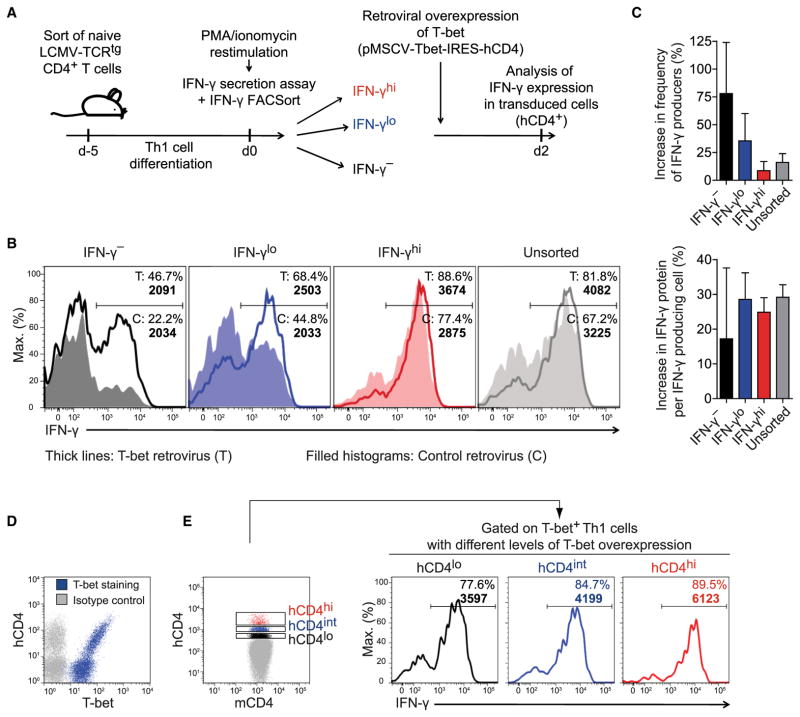
T-bet had been identified as the master regulator of the Th1 cell lineage because of its capacity to instruct non-Th1 cells to acquire IFN-γ production competence (Szabo et al., 2000). To address whether a causal relationship dictated the quantitative correlation between T-bet protein and IFN-γ production in fully committed, already IFN-γ-competent Th1 cells, we sorted Th1 cells for different amounts of IFN-γ secretion (and thus indirectly also for different T-bet expression). We then further increased their respective T-bet amount by retroviral overexpression (Figure 6A). Notably, all of these Th1 cells stained positive for T-bet protein already before the transduction (cf. Figure S1C and hCD4− cells in Figure 6D). Upon T-bet overexpression, sorted IFN-γhi, IFN-γlo, and IFN-γ− Th1 cells exhibited a strong increase in both probability and per-cell amount of IFN-γ production compared with their counterparts that were transduced with a control retrovirus (Figures 6B and 6C). Both quantitative parameters were also graded among unsorted T-bet-overexpressing cells, correlating with the degree of ectopic T-bet expression (Figures 6D and 6E). Taken together, an increase in T-bet amount in already T-bet+ Th1 cells can overcome an otherwise stably restrained cellular capacity to produce IFN-γ. This result identifies T-bet as a quantitative regulator of IFN-γ expression in fully differentiated Th1 cells.
Figure 6. T-bet Quantitatively Controls IFN-γ Expression in Fully Differentiated Th1 Cells.
(A) Experimental setup. Th1 cells were sorted by secreted IFN-γ amounts and transduced with a T-bet-encoding or control retrovirus. IFN-γ expression was analyzed in transduced (hCD4+) cells 2 days later.
(B) Frequencies of IFN-γ+ cells and geometric mean of IFN-γ in IFN-γ+ cells (bold numbers).
(C) Relative increase in IFN-γ expression probability and per-cell amount upon T-bet overexpression (means + SD).
(D and E) Unsorted Th1 cells were transduced with a T-bet-encoding retrovirus and analyzed 2 days later.
(D) Counterstaining of T-bet and hCD4.
(E) Frequencies of IFN-γ+ cells and geometric mean of IFN-γ in IFN-γ+ cells (bold numbers) in cells overexpressing different amounts of hCD4, i.e., T-bet.
Representative results of (B, D, E) or pooled data from (C) two independent experiments are shown.
Graded IFN-γ Production by Th1 Cells Regulates Bacterial Killing by Macrophages
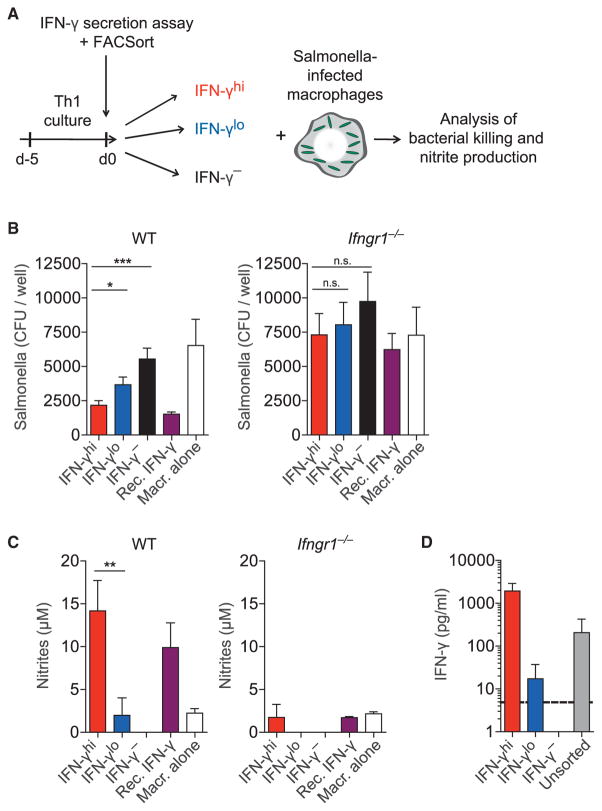
To test the functional capacity of Th1 cells with distinct IFN-γ production, we analyzed their ability to activate macrophages for bacterial killing. We infected macrophages with the facultative intracellular pathogen Salmonella enterica serovar Typhimurium (S. Typhimurium) and cocultured them with sorted IFN-γhi, IFN-γlo, or IFN-γ− Th1 cells (Figure 7A). IFN-γhi Th1 cells were most efficient in inducing bacterial killing, followed by IFN-γlo and finally IFN-γ− Th1 cells (Figure 7B). Graded bacterial killing was associated with different amounts of nitric oxide (NO) production by the macrophages (Figure 7C). Ifngr1−/− macrophages could not kill the bacteria nor produce NO when cocultured with either Th1 cell population (Figures 7B and 7C, right graphs), demonstrating that the effects were IFN-γ dependent. The IFN-γ amount secreted by IFN-γhi and IFN-γlo Th1 cells differed by two orders of magnitude (Figure 7D). Thus, in addition to the frequency of cytokine producers in a population, the per-cell amount of cytokine production critically influences the functional capacity of Th cells to control intracellular bacterial infections.
Figure 7. Graded IFN-γ Production by Th1 Cells Regulates Bacterial Killing by Macrophages.
(A) Th1 cells were sorted by secreted IFN-γ amounts. WT or Ifngr1−/− BM-derived macrophages were infected with S. Typhimurium. IFN-γ-sorted fractions were cocultured for 36 hr with infected macrophages at a 1:5 ratio, or recombinant IFN-γ (10 ng/ml) was added as a control.
(B) Bacterial colonies were counted after plating macrophage lysates for 24 hr (means + SEM).
(C) Nitrite accumulation in the culture medium (means + SD).
(D) Sorted fractions from (A) were cultured without macrophages for 36 hr. IFN-γ concentrations in the supernatants of 4 × 105 cells/ml are shown (means + SD; dotted line, detection limit).
Data are pooled from (B, D) or are representative of (C) three independent experiments.
DISCUSSION
Although cytokines have been recognized for more than three decades as key effector molecules of T cells, quantitative aspects of their expression and underlying regulatory mechanisms are poorly understood. Here, we have shown that the well-known phenomenon of a heterogeneous cytokine production within a cell population is caused by stable cellular decisions and not governed by short-term transcription noise (Suter et al., 2011). We found that during an antigenic challenge, individual Th cells expressed their effector cytokines in stable amounts that varied widely between cells. Focusing on IFN-γ as a prototypical cytokine, we demonstrated that the magnitude of its expression per cell was an intrinsic feature of Th1 cells. In vivo, this magnitude was maintained by the cells and their progeny for weeks, even in the face of a strongly Th1-cell-polarizing challenge infection that was expected to reduce cell-to-cell differences. Moreover, the expression magnitude of the Th1-cell-specific, Ifng-transactivating transcription factor T-bet was also a stable and heritable quantitative feature of individual Th1 cells. It predicted the magnitude of IFN-γ expression according to a dose-response function. Thus, T cells can quantitatively control a key effector function in a stable manner by quantitatively regulating a “master regulator” transcription factor.
How is such intrapopulation heterogeneity established? A TCR repertoire with diverse antigen affinities is likely to contribute (Constant and Bottomly, 1997) but is not required, given that we found similar heterogeneity within TCR-transgenic T cell populations. As Th1 cell differentiation proceeds, cooperative actions of STAT4 as well as T-bet together with the transcription factors Hlx and Runx3 induce permissive chromatin remodeling at the Ifng locus (Balasubramani et al., 2010a). The cell-specific fine tuning of this process might be achieved by the regulation of cytokine signaling. This can occur through either the control of the expression of cytokine receptors or the availability and/or activity of downstream signal transduction molecules and transcription factors. Indeed, quantitative regulation of IL-12Rβ2 and STAT4 (Szabo et al., 1997; Usui et al., 2003) as well as of IFN-γR expression (de Weerd and Nguyen, 2012) during Th cell differentiation have been described, e.g., due to asymmetric cell division (Chang et al., 2007). Such kinds of adjustment might generate Th1 effector cells with distinct IFN-γ production probabilities at the population level and distinct IFN-γ as well as T-bet expression rates in individual cells. We found that the probability to express IFN-γ and its amount produced per cell were heterogeneous in Th1 cell populations, and both features were stably memorized by individual cells. We did not detect a correlation between the IFN-γ expression of sorted cell populations and their survival in vivo. In extension of our previous study (Löhning et al., 2008), this finding indicated that IFN-γhi cells did not represent short-lived effectors but could efficiently form a memory compartment.
Upon lineage commitment, the loci of signature cytokines exhibit stable lineage-specific epigenetic marks (Wei et al., 2009), allowing the rapid reexpression of the appropriate effector cytokines. However, key transcription factors continuously serve important regulatory functions. In fully differentiated Th2 cells, GATA-3 remains crucial for IL-13 and IL-5 production, although it appears largely dispensable for IL-4 production (Zhu et al., 2004). Overexpression of a dominant-negative T-bet mutant is most detrimental during early Th1 cell differentiation, but still results in a significant decrease of IFN-γ production per cell when introduced after sequential polarizations with IL-12 (Martins et al., 2005). We found that although all Th1 cells expressed T-bet, its protein amounts varied in the effector population, and these differences were stably maintained in memory cells in vivo. Consistent with a continuous requirement of T-bet for efficient Ifng expression, we showed that IFN-γ production increased even in fully committed Th1 cells as a direct consequence of a retrovirus-induced gradual T-bet over-expression. Thus, T-bet not only orchestrates the commitment of naive T cells to the Th1 cell differentiation program but continuously serves as a quantitative regulator of Th1 cell functions.
How does T-bet quantitatively control IFN-γ expression in memory Th1 cells? Recent studies indicate that epigenetic marks are subjected to a certain turnover and have to be actively maintained (Barth and Imhof, 2010; Dalton and Bellacosa, 2012). Here, T-bet seems a likely candidate because it contributes to the opening of the Ifng locus during primary Th1 cell differentiation (Mullen et al., 2001; Szabo et al., 2000). We found that stable IFN-γ expression differences were associated with corresponding DNA methylation patterns at both the Ifng and Tbx21 loci. In addition to DNA methylation, various histone modifications are thought to orchestrate gene expression activity (Barth and Imhof, 2010). The graded DNA methylation we observed at CNS −6 of the Ifng gene and at the Tbx21 promoter might partially contribute to a stable quantitative cytokine memory. However, we hypothesize that a quantitative cytokine memory is rather based on a combination of multiple permissive and repressive epigenetic modifications at several regulatory sites. They might act together with distinct T-bet expression rates retained by the cells through transcriptional autoactivation (Afkarian et al., 2002; Mullen et al., 2001). Moreover, T-bet might cooperate with NF-κB family members to facilitate Ifng expression upon antigen-driven restimulation—in analogy to STAT4 enabling NF-κB binding to the IFN-γ locus (Balasubramani et al., 2010b) in the scenario of IL-12- and IL-18-driven antigen-independent IFN-γ expression by Th1 cells (Robinson et al., 1997).
Furthermore, cytokine expression might generally include a stochastic element (Zhao et al., 2012). Then, changes of the expression probability would require a regulator, and the maintenance of a cell’s individual production magnitude of this regulator would constitute a quantitative cytokine memory. Here, distinct amounts of transcriptional repressors such as twist1 (Niesner et al., 2008; Pham et al., 2012) could contribute to the stable differences in IFN-γ expression between individual Th1 cells. We found a major regulatory step to generate these differences already at the level of transcription, controlled by the positive regulator T-bet. Thus, we suggest that posttranscriptional mechanisms such as different mRNA decay rates or modulation by microRNAs are unlikely to be key mechanisms.
Cytokine genes can be expressed either mono- or biallelically (Guo et al., 2005; Hu-Li et al., 2001). Therefore, graded cytokine expression rates might be the result of transcription from either one or two alleles. However, we found that even cells with only one functional Ifng allele maintained quantitative expression differences. This proves that not allelic usage but different transcription at individual alleles constitutes the decisive mechanism underlying a cell’s quantitative cytokine memory.
Although the frequency of cytokine-producing cells within a population and the expression per cell both matter for the local cytokine concentration, most studies focus exclusively on the former. Yet, we found that Th1 cells exhibiting a mere 3-to 5-fold difference in their IFN-γ secretion accumulated to a 100-fold difference over time. Thus, relatively small per-cell differences in the production magnitude of a given cytokine probably have a great impact on immune responses.
IFN-γ is crucial for the control of Salmonella infections (Eckmann and Kagnoff, 2001). We observed that distinct IFN-γ expression rates of Th1 cells translated into graded activation of infected macrophages to kill intracellular bacteria. Hence, the amount of IFN-γ produced by individual Th1 cells was decisive for the functional outcome of the T cell-macrophage interaction. Thus, a population of seemingly homogeneously differentiated T cells indeed features stable functional diversity that could quantitatively regulate various immune reactions. This mechanism might also provide a possibility to limit immunopathology. Under changing environmental challenges, plasticity of Th cell programs can be beneficial (Hegazy et al., 2010). IFN-γlo Th1 cells express T-bet only modestly and thus might retain certain plasticity, e.g., to adjust to a second pathogen that shares an epitope with the first but requires a different type of immune response.
Cytokine production must be controlled tightly, because a misbalance can induce pathology. Here, we have demonstrated that individual T cells stably commit to express distinct amounts of a given cytokine. This fine tuning of cytokine production could contribute to the regulation of immune responses and the prevention of excessive inflammation. The persistent memory for individual IFN-γ expression rates shown here could result from regulation at several levels. Yet the specific amount of T-bet produced by a Th1 cell is decisive for its IFN-γ expression magnitude. With regard to potential clinical application, individual rates of cytokine and/or transcription factor expression could serve as predictive markers for the quantitative functional behavior of T cells and their progeny in later antigenic challenges. These findings could lead to therapeutic strategies to improve the protective capacity of T cell responses and dampen associated immunopathology.
EXPERIMENTAL PROCEDURES
Mice
DO11.10 ovalbumin-TCRtg mice, Ifngr1−/− mice, LCMV-TCRtg (SMARTA1) Thy1.1+ mice, or TBGR mice (Zhu et al., 2012) crossed to SMARTA1 Thy1.1+ mice were used as organ donors. C57BL/6 mice or TBGR Thy1.2+ mice were used as recipients for cell transfers. Mice were bred under SPF conditions at the Charité animal facility, Berlin. All mouse experiments were performed in accordance with the German law for animal protection and with permission from the local veterinary offices. For details, see Supplemental Information.
Viruses and Bacteria
For LCMV, mice were infected intravenously with 200 plaque-forming units. For S. Typhimurium, macrophages were infected with an MOI of 1:10 and cocultured with IFN-γ-sorted Th1 cells for 36 hr. Macrophage lysate was plated onto LB agar plates. Bacterial colonies were counted after 24 hr. For details, see Supplemental Information.
Primary T Cell Cultures
Naive CD4+CD62LhiCD44lo T cells were differentiated into Th1 cells via 3 ng/ml IL-12 and 10 μg/ml anti-IL-4 (11B11), into Th2 cells via 30 ng/ml IL-4, 10 μg/ml anti-IL-12 (C17.8), and 10 μg/ml anti-IFN-γ (AN18.17.24), or into Th17 cells via 20 ng/ml IL-6, 1 ng/ml TGF-β, 10 ng/ml IL-23, 10 μg/ml anti-IL-4, and 10 μg/ml anti-IFN-γ. Cells were analyzed on day 5. For details, see Supplemental Information.
Flow Cytometry
Cells were stained as described (Hegazy et al., 2010). Antibodies and buffers were purchased from eBioscience and BD Biosciences. For detailed protocols and antibody clones, see Supplemental Information.
For cytokine production analysis, cells were restimulated with PMA and ionomycin. To normalize the per-cell cytokine protein amount of sorted cell populations, the geometric mean (GM) of cytokine-positive cells in a sorted subset was divided by the GM of the respective cytokine-positive cells from an unsorted population.
For transcription factor protein quantification, GM indices were calculated as the GM of stained cells divided by the GM of isotype control-stained cells. Unless indicated otherwise, GM indices of sorted cell subsets were normalized to those of unsorted cells.
Bone-Marrow-Derived Macrophages
BM from WT or Ifngr1−/− mice was cultured via standard macrophage differentiation protocols. For details, see Supplemental Information.
Cytokine Secretion Assay
The cytometric cytokine secretion assay was performed as described (Assenmacher et al., 1998; Löhning et al., 2003) upon PMA and ionomycin restimulation unless indicated otherwise. For details, see Supplemental Information.
Retroviral Transduction
Ecotrophic retroviruses (encoding pMSCV-Tbet-I-hCD4 or pMSCV-I-hCD4) were generated by transfection of Phoenix cells. Retrovirus supernatants were used to spin-infect T cells in the presence of 8 μg/ml polybrene (Sigma).
RNA Isolation and Real-Time PCR
RNA isolation and qPCR were performed by standard protocols. For details, see Supplemental Information.
Cytokine Analysis in Culture Supernatants
IFN-γ concentrations in cell culture supernatants were determined by cytometric Bead Array (BD Biosciences) according to the manufacturer’s instructions.
Bisulfite Sequencing
DNA was isolated with the NucleoSpin Blood Kit (Macherey-Nagel). Amplicon design and bisulfite sequencing was performed by Epiontis GmbH.
Statistical Analysis
Two groups were compared with two-tailed unpaired Student’s t test; n.s., not significant; *p < 0.05; **p < 0.01; ***p < 0.001.
Mathematical Modeling
For detailed description of all mathematical models, see Supplemental Information.
Supplementary Material
Acknowledgments
We thank V. Holecska, S. Ebel, I. Panse, and U. Zedler for expert technical assistance and the FCCF at DRFZ for cell sorting. This work was supported by the German Federal Ministry of Education and Research (FORSYS and e:Bio/T-Sys), the German Research Foundation (SFB618, TPC3; SFB650, TP28; LO1542/3-1, and HO02050/4-1), and the Volkswagen Foundation (Lichtenberg program to M.L.). C.H., M.P., and M.A.D.-C. were fellows of the International Max Planck Research School for Infectious Diseases and Immunology. A.K. was supported by the National Health and Medical Research Council of Australia through a CJ Martin Biomedical Early Career Fellowship (APP1052764). A.N.H. is supported by an EMBO long-term fellowship (ALTF 116-2012) and a Marie Curie fellowship (FP7-PEOPLE-2012-IEF, Proposal 330621). J.Z. and W.E.P. are supported by the Division of Intramural Research, NIAID, NIH, USA. T.H. and M.F. are supported in part by the US National Science Foundation (NSF PHY11-25915). T.H. is a member of CellNetworks.
Footnotes
SUPPLEMENTAL INFORMATION
Supplemental Information includes six figures, one table, and Supplemental Experimental Procedures and can be found with this article online at http://dx.doi.org/10.1016/j.immuni.2014.12.018.
AUTHOR CONTRIBUTIONS
C.H. and M.P. designed the study, performed most experiments, analyzed data, and wrote the manuscript. M.F. and T.H. conceptualized, performed, and described mathematical modeling. A.K., A.N.H., M.A.D.-C., Q.Z., and Y.V. performed experiments. J.Z., A.R., S.H.K., and W.E.P. provided tools and advice. T.H. and M.L. designed and directed the study and wrote the manuscript.
References
- Afkarian M, Sedy JR, Yang J, Jacobson NG, Cereb N, Yang SY, Murphy TL, Murphy KM. T-bet is a STAT1-induced regulator of IL-12R expression in naïve CD4+ T cells. Nat Immunol. 2002;3:549–557. doi: 10.1038/ni794. [DOI] [PubMed] [Google Scholar]
- Apostolou E, Thanos D. Virus infection induces NF-kappaB-dependent interchromosomal associations mediating monoallelic IFN-beta gene expression. Cell. 2008;134:85–96. doi: 10.1016/j.cell.2008.05.052. [DOI] [PubMed] [Google Scholar]
- Assenmacher M, Löhning M, Scheffold A, Manz RA, Schmitz J, Radbruch A. Sequential production of IL-2, IFN-gamma and IL-10 by individual staphylococcal enterotoxin B-activated T helper lymphocytes. Eur J Immunol. 1998;28:1534–1543. doi: 10.1002/(SICI)1521-4141(199805)28:05<1534::AID-IMMU1534>3.0.CO;2-R. [DOI] [PubMed] [Google Scholar]
- Balasubramani A, Mukasa R, Hatton RD, Weaver CT. Regulation of the Ifng locus in the context of T-lineage specification and plasticity. Immunol Rev. 2010a;238:216–232. doi: 10.1111/j.1600-065X.2010.00961.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balasubramani A, Shibata Y, Crawford GE, Baldwin AS, Hatton RD, Weaver CT. Modular utilization of distal cis-regulatory elements controls Ifng gene expression in T cells activated by distinct stimuli. Immunity. 2010b;33:35–47. doi: 10.1016/j.immuni.2010.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barth TK, Imhof A. Fast signals and slow marks: the dynamics of histone modifications. Trends Biochem Sci. 2010;35:618–626. doi: 10.1016/j.tibs.2010.05.006. [DOI] [PubMed] [Google Scholar]
- Bucy RP, Panoskaltsis-Mortari A, Huang GQ, Li J, Karr L, Ross M, Russell JH, Murphy KM, Weaver CT. Heterogeneity of single cell cytokine gene expression in clonal T cell populations. J Exp Med. 1994;180:1251–1262. doi: 10.1084/jem.180.4.1251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang JT, Palanivel VR, Kinjyo I, Schambach F, Intlekofer AM, Banerjee A, Longworth SA, Vinup KE, Mrass P, Oliaro J, et al. Asymmetric T lymphocyte division in the initiation of adaptive immune responses. Science. 2007;315:1687–1691. doi: 10.1126/science.1139393. [DOI] [PubMed] [Google Scholar]
- Constant SL, Bottomly K. Induction of Th1 and Th2 CD4+ T cell responses: the alternative approaches. Annu Rev Immunol. 1997;15:297–322. doi: 10.1146/annurev.immunol.15.1.297. [DOI] [PubMed] [Google Scholar]
- Corbin GA, Harty JT. T cells undergo rapid ON/OFF but not ON/OFF/ON cycling of cytokine production in response to antigen. J Immunol. 2005;174:718–726. doi: 10.4049/jimmunol.174.2.718. [DOI] [PubMed] [Google Scholar]
- Dalton SR, Bellacosa A. DNA demethylation by TDG. Epigenomics. 2012;4:459–467. doi: 10.2217/epi.12.36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Weerd NA, Nguyen T. The interferons and their receptors—distribution and regulation. Immunol Cell Biol. 2012;90:483–491. doi: 10.1038/icb.2012.9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eckmann L, Kagnoff MF. Cytokines in host defense against Salmonella. Microbes Infect. 2001;3:1191–1200. doi: 10.1016/s1286-4579(01)01479-4. [DOI] [PubMed] [Google Scholar]
- Feinerman O, Veiga J, Dorfman JR, Germain RN, Altan-Bonnet G. Variability and robustness in T cell activation from regulated heterogeneity in protein levels. Science. 2008;321:1081–1084. doi: 10.1126/science.1158013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedman N, Cai L, Xie XS. Linking stochastic dynamics to population distribution: an analytical framework of gene expression. Phys Rev Lett. 2006;97:168302. doi: 10.1103/PhysRevLett.97.168302. [DOI] [PubMed] [Google Scholar]
- Guo L, Hu-Li J, Paul WE. Probabilistic regulation of IL-4 production. J Clin Immunol. 2005;25:573–581. doi: 10.1007/s10875-005-8218-5. [DOI] [PubMed] [Google Scholar]
- Harper CV, Finkenstädt B, Woodcock DJ, Friedrichsen S, Semprini S, Ashall L, Spiller DG, Mullins JJ, Rand DA, Davis JR, White MR. Dynamic analysis of stochastic transcription cycles. PLoS Biol. 2011;9:e1000607. doi: 10.1371/journal.pbio.1000607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hegazy AN, Peine M, Helmstetter C, Panse I, Fröhlich A, Bergthaler A, Flatz L, Pinschewer DD, Radbruch A, Löhning M. Interferons direct Th2 cell reprogramming to generate a stable GATA-3(+) T-bet(+) cell subset with combined Th2 and Th1 cell functions. Immunity. 2010;32:116–128. doi: 10.1016/j.immuni.2009.12.004. [DOI] [PubMed] [Google Scholar]
- Hu-Li J, Pannetier C, Guo L, Löhning M, Gu H, Watson C, Assenmacher M, Radbruch A, Paul WE. Regulation of expression of IL-4 alleles: analysis using a chimeric GFP/IL-4 gene. Immunity. 2001;14:1–11. doi: 10.1016/s1074-7613(01)00084-x. [DOI] [PubMed] [Google Scholar]
- Löhning M, Richter A, Radbruch A. Cytokine memory of T helper lymphocytes. Adv Immunol. 2002;80:115–181. doi: 10.1016/s0065-2776(02)80014-1. [DOI] [PubMed] [Google Scholar]
- Löhning M, Richter A, Stamm T, Hu-Li J, Assenmacher M, Paul WE, Radbruch A. Establishment of memory for IL-10 expression in developing T helper 2 cells requires repetitive IL-4 costimulation and does not impair proliferation. Proc Natl Acad Sci USA. 2003;100:12307–12312. doi: 10.1073/pnas.2035254100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Löhning M, Hegazy AN, Pinschewer DD, Busse D, Lang KS, Höfer T, Radbruch A, Zinkernagel RM, Hengartner H. Long-lived virus-reactive memory T cells generated from purified cytokine-secreting T helper type 1 and type 2 effectors. J Exp Med. 2008;205:53–61. doi: 10.1084/jem.20071855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mariani L, Schulz EG, Lexberg MH, Helmstetter C, Radbruch A, Löhning M, Höfer T. Short-term memory in gene induction reveals the regulatory principle behind stochastic IL-4 expression. Mol Syst Biol. 2010;6:359. doi: 10.1038/msb.2010.13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martins GA, Hutchins AS, Reiner SL. Transcriptional activators of helper T cell fate are required for establishment but not maintenance of signature cytokine expression. J Immunol. 2005;175:5981–5985. doi: 10.4049/jimmunol.175.9.5981. [DOI] [PubMed] [Google Scholar]
- Miller-Jensen K, Dey SS, Schaffer DV, Arkin AP. Varying virulence: epigenetic control of expression noise and disease processes. Trends Biotechnol. 2011;29:517–525. doi: 10.1016/j.tibtech.2011.05.004. [DOI] [PubMed] [Google Scholar]
- Mullen AC, High FA, Hutchins AS, Lee HW, Villarino AV, Livingston DM, Kung AL, Cereb N, Yao TP, Yang SY, Reiner SL. Role of T-bet in commitment of TH1 cells before IL-12-dependent selection. Science. 2001;292:1907–1910. doi: 10.1126/science.1059835. [DOI] [PubMed] [Google Scholar]
- Niesner U, Albrecht I, Janke M, Doebis C, Loddenkemper C, Lexberg MH, Eulenburg K, Kreher S, Koeck J, Baumgrass R, et al. Autoregulation of Th1-mediated inflammation by twist1. J Exp Med. 2008;205:1889–1901. doi: 10.1084/jem.20072468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Garra A, Gabryšová L, Spits H. Quantitative events determine the differentiation and function of helper T cells. Nat Immunol. 2011;12:288–294. doi: 10.1038/ni.2003. [DOI] [PubMed] [Google Scholar]
- Openshaw P, Murphy EE, Hosken NA, Maino V, Davis K, Murphy K, O’Garra A. Heterogeneity of intracellular cytokine synthesis at the single-cell level in polarized T helper 1 and T helper 2 populations. J Exp Med. 1995;182:1357–1367. doi: 10.1084/jem.182.5.1357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peine M, Rausch S, Helmstetter C, Fröhlich A, Hegazy AN, Kühl AA, Grevelding CG, Höfer T, Hartmann S, Löhning M. Stable T-bet(+)GATA-3(+) Th1/Th2 hybrid cells arise in vivo, can develop directly from naive precursors, and limit immunopathologic inflammation. PLoS Biol. 2013;11:e1001633. doi: 10.1371/journal.pbio.1001633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pham D, Vincentz JW, Firulli AB, Kaplan MH. Twist1 regulates Ifng expression in Th1 cells by interfering with Runx3 function. J Immunol. 2012;189:832–840. doi: 10.4049/jimmunol.1200854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raj A, Peskin CS, Tranchina D, Vargas DY, Tyagi S. Stochastic mRNA synthesis in mammalian cells. PLoS Biol. 2006;4:e309. doi: 10.1371/journal.pbio.0040309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rand U, Rinas M, Schwerk J, Nöhren G, Linnes M, Kröger A, Flossdorf M, Kály-Kullai K, Hauser H, Höfer T, Köster M. Multi-layered stochasticity and paracrine signal propagation shape the type-I interferon response. Mol Syst Biol. 2012;8:584. doi: 10.1038/msb.2012.17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson D, Shibuya K, Mui A, Zonin F, Murphy E, Sana T, Hartley SB, Menon S, Kastelein R, Bazan F, O’Garra A. IGIF does not drive Th1 development but synergizes with IL-12 for interferon-gamma production and activates IRAK and NFkappaB. Immunity. 1997;7:571–581. doi: 10.1016/s1074-7613(00)80378-7. [DOI] [PubMed] [Google Scholar]
- Shnyreva M, Weaver WM, Blanchette M, Taylor SL, Tompa M, Fitzpatrick DR, Wilson CB. Evolutionarily conserved sequence elements that positively regulate IFN-gamma expression in T cells. Proc Natl Acad Sci USA. 2004;101:12622–12627. doi: 10.1073/pnas.0400849101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sigal A, Milo R, Cohen A, Geva-Zatorsky N, Klein Y, Liron Y, Rosenfeld N, Danon T, Perzov N, Alon U. Variability and memory of protein levels in human cells. Nature. 2006;444:643–646. doi: 10.1038/nature05316. [DOI] [PubMed] [Google Scholar]
- Slifka MK, Rodriguez F, Whitton JL. Rapid on/off cycling of cytokine production by virus-specific CD8+ T cells. Nature. 1999;401:76–79. doi: 10.1038/43454. [DOI] [PubMed] [Google Scholar]
- Suter DM, Molina N, Gatfield D, Schneider K, Schibler U, Naef F. Mammalian genes are transcribed with widely different bursting kinetics. Science. 2011;332:472–474. doi: 10.1126/science.1198817. [DOI] [PubMed] [Google Scholar]
- Szabo SJ, Dighe AS, Gubler U, Murphy KM. Regulation of the interleukin (IL)-12R beta 2 subunit expression in developing T helper 1 (Th1) and Th2 cells. J Exp Med. 1997;185:817–824. doi: 10.1084/jem.185.5.817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szabo SJ, Kim ST, Costa GL, Zhang X, Fathman CG, Glimcher LH. A novel transcription factor, T-bet, directs Th1 lineage commitment. Cell. 2000;100:655–669. doi: 10.1016/s0092-8674(00)80702-3. [DOI] [PubMed] [Google Scholar]
- Usui T, Nishikomori R, Kitani A, Strober W. GATA-3 suppresses Th1 development by downregulation of Stat4 and not through effects on IL-12Rbeta2 chain or T-bet. Immunity. 2003;18:415–428. doi: 10.1016/s1074-7613(03)00057-8. [DOI] [PubMed] [Google Scholar]
- Wei G, Wei L, Zhu J, Zang C, Hu-Li J, Yao Z, Cui K, Kanno Y, Roh TY, Watford WT, et al. Global mapping of H3K4me3 and H3K27me3 reveals specificity and plasticity in lineage fate determination of differentiating CD4+ T cells. Immunity. 2009;30:155–167. doi: 10.1016/j.immuni.2008.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao M, Zhang J, Phatnani H, Scheu S, Maniatis T. Stochastic expression of the interferon-β gene. PLoS Biol. 2012;10:e1001249. doi: 10.1371/journal.pbio.1001249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu J, Min B, Hu-Li J, Watson CJ, Grinberg A, Wang Q, Killeen N, Urban JF, Jr, Guo L, Paul WE. Conditional deletion of Gata3 shows its essential function in T(H)1-T(H)2 responses. Nat Immunol. 2004;5:1157–1165. doi: 10.1038/ni1128. [DOI] [PubMed] [Google Scholar]
- Zhu J, Yamane H, Paul WE. Differentiation of effector CD4 T cell populations (*) Annu Rev Immunol. 2010;28:445–489. doi: 10.1146/annurev-immunol-030409-101212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu J, Jankovic D, Oler AJ, Wei G, Sharma S, Hu G, Guo L, Yagi R, Yamane H, Punkosdy G, et al. The transcription factor T-bet is induced by multiple pathways and prevents an endogenous Th2 cell program during Th1 cell responses. Immunity. 2012;37:660–673. doi: 10.1016/j.immuni.2012.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.