Abstract
APOL1 genetic variants account for much of the excess risk of chronic and end stage kidney disease, which results in a significant global health disparity for persons of African ancestry. We estimate the lifetime risk of kidney disease in APOL1 dual-risk allele individuals to be at least 15%. Experimental evidence suggests a direct role of APOL1 in pore formation, cellular injury, and programmed cell death in renal injury. The APOL1 BH3 motif, often associated with cell death, is unlikely to play a role in APOL1-induced cytotoxicity as it is not conserved within the APOL family and is dispensable for cell death in vitro. We discuss two models for APOL1 trypanolytic activity: one involving lysosome permeabilization and another colloid-osmotic swelling of the cell body, as well as their relevance to human pathophysiology. Experimental evidence from human cell culture models suggests that both mechanisms may be operative. A systems biology approach whereby APOL1-associated perturbations in gene and protein expression in affected individuals are correlated with molecular pathways may be productive to elucidate APOL1 function in vivo.
Keywords: health disparities, chronic kidney disease, focal segmental glomerulosclerosis, innate immunity, APOL1
Introduction
End-stage kidney disease (ESKD) constitutes a major health disparity in African Americans. While ESKD incidence rates increased steadily in all American populations between 1980 and 2000 and stabilized since, the rate in African Americans rose more rapidly than any other ethnic/racial group. ESKD incidence rates are approximately 3.5 fold higher in African Americans compared to European Americans, affecting almost 0.1% of the African American population in 2010 compared to 0.03% in European Americans [1]. ESKD incidence rates are higher in African Americans compared to European Americans for hypertension-attributed ESKD (HA-ESKD), focal segmental glomerulosclerosis (FSGS) and HIV-associated nephropathy (HIVAN) [1]. Overall, the cumulative lifetime risk of reaching ESKD in African Americans is ~7.5%, compared to ~2% in European Americans; in African Americans, ESKD is responsible for nearly as much loss of life-years as breast cancer in women and more loss of life-years than colorectal cancer in men [2].
Much of the excess risk is attributable to two common protein-changing alleles in the APOL1 gene, encoding apolipoprotein L1 (APOL1) [3]. The APOL1 G1 allele comprises two missense variants (p.S342G:I384M) and the G2 variant allele is a 6 base pair deletion that removes two amino acids (N388Y389); these alleles are on opposing chromosomes and, owing to their close proximity, have not undergone recombination. These alleles for the most part are equivalent in their effect sizes and strongly recessive [4]: APOL1 high-risk genotypes are defined as two risk alleles in any combination (homozygous G1/G1, homozygous G2/G2, or compound heterozygous G1/G2). The APOL1 renal risk variants are common in African Americans (>50% carry at least one risk allele) and throughout sub-Saharan African populations (from 5% up to 50% in sub-Saharan Africa)[4–6]. High-risk genotypes are greatly enriched in African Americans with FSGS and HIVAN (72%) and HA-ESKD (44%) compared to 12–14% in healthy controls. Compared to individuals carrying APOL1 low-risk genotypes (0 or 1 risk allele), the odds ratio for these diseases for carriers of high-risk genotypes is 17 for FSGS, 29 for HIVAN and 7 for HA-ESKD [3,4].
Approximately 13% of African Americans (~five million individuals) carry high-risk genotypes; a substantial fraction will develop APOL1-associated chronic kidney disease (Table 1). As shown, the lifetime risk for HIV-associated nephropathy has been estimated at 50% among HIV positive African Americans who have two APOL1 risk alleles and do not receive anti-retroviral therapy and the lifetime risk for focal segmental glomerulosclerosis (FSGS) has estimated as 4.25% among African Americans who have two APOL1 risk alleles [reference pending]. Using data from the United States Renal Data System on the incidence of HA-ESKD, and the odds ratio of 7 for those with two APOL1 risk alleles [3], we have calculated the lifetime risk for HA-ESKD as 11% in these subjects. Thus, taken together, the lifetime risk for these two APOL1 nephropathies is estimated as 15%. The table also shows the explained variance (the proportion of a disease that is explained by the factor, among all contributing factors) and the attributable risk (the fraction of the disease that would be eliminated if the factor were absent). These estimates are subject to confounding by other contributing factors; they may also understate APOL1 nephropathy incidence because other renal conditions, notably clinically-diagnosed diabetic nephropathy, may in fact be driven by APOL1 variants.
Table 1. Lifetime risk estimates for APOL1 nephropathies.
The frequency of APOL1 risk alleles among the African American population is shown, as well as lifetime risks for three common APOL1 nephropathies among individuals with two APOL1 risk alleles. For individuals with HIV who are not receiving effective anti-retroviral therapy, the lifetime risk for HIV-associated nephropathy (HIVAN) is estimated at 50%. In the general US population, lifetime risk for those with two APOL1 risk alleles is estimated at ~4% for focal segmental glomerulosclerosis (FSGS) and 12% for those with hypertension attributed end-stage kidney disease (ESKD). Shown are the explained variance and population attributable risk for two APOL1 risk alleles.
| Disease lifetime frequency |
Low risk genotypes | High risk genotype |
High risk genotypes | |||
|---|---|---|---|---|---|---|
| 0 risk alleles |
1 risk allele | 2 risk alleles |
Explained variance |
Population attributable risk |
||
| African American population genotype frequencies | ~42% | ~45% | ~13% | N/A | N/A | |
| HIVAN | 10% HIV+ 1:10 |
2.5% 1:40 |
4% 1:25 |
50% 1:2 |
37% | 68% |
| FSGS | 0.8% 1:125 |
0.2% 1:500 |
0.3% 1:333 |
4.25% 1:24 |
18% | 68% |
| Hypertension attributed ESKD | 2.75% 1:36 |
1.5% 1:65 |
11% 1:9 |
7% | 52% | |
The evolution of APOL family genes
APOL1 is a member of the APOL gene family, which comprises six genes on human chromosome 22, all of which are presumed to play a role in innate immunity [7,8]. The APOL family has evolved rapidly in primates by multiple events of gene duplication, gene loss and pseudogenization [7]. This dynamic evolution has led to both a variable number of APOL genes among primate species and variable exon content among the different genes (Figures 1 and 2). The complete loss (e.g. chimpanzee) or pseudogenization (e.g. macaque) of APOL1 from most primates suggests that the gene is not critical for normal physiologic function. The account of an APOL1-null individual with apparently normal renal function supports this conclusion [9]. Moreover, both APOL1 and APOL6 are toxic to human cells when overexpressed [10–13], introducing the possibility that its retention comes at a fitness cost that often outweighs its benefits. An understanding of the mechanism of toxicity might provide valuable clues to the mechanisms of APOL1 nephropathy.
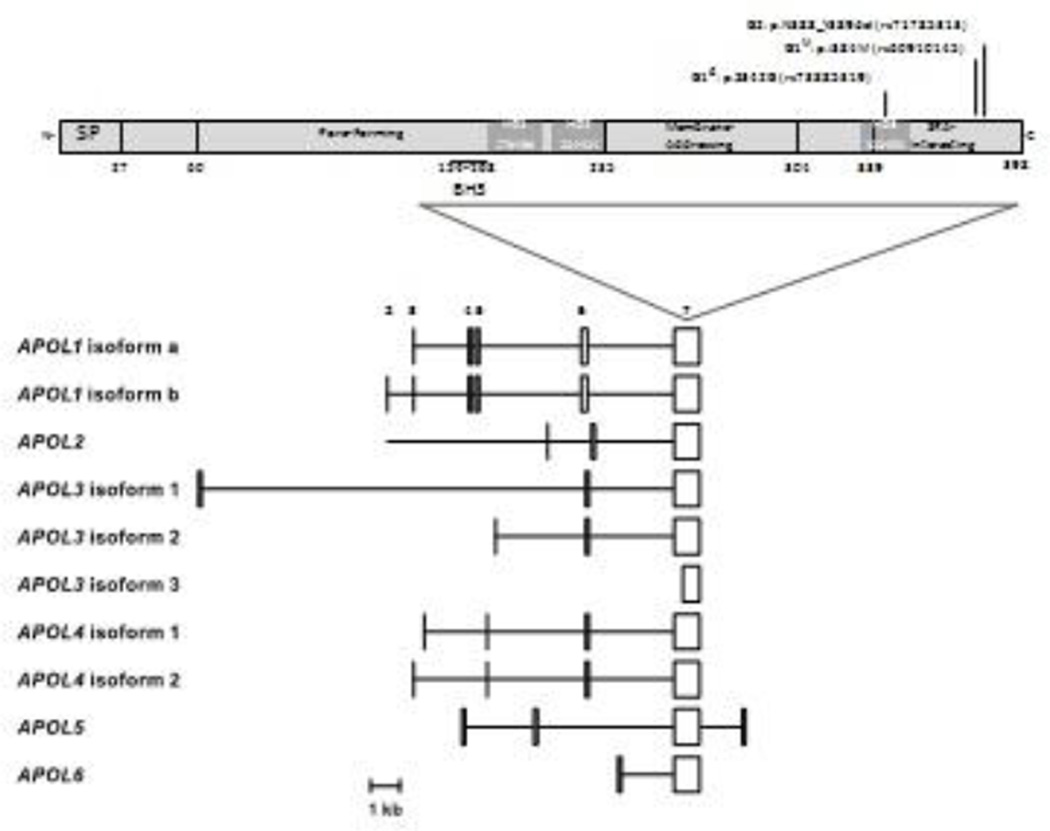
Figure 1. APOL1 predicted protein domains and human predicted APOL1 isoform transcripts.
The upper panel shows APOL1 domain organization as proposed by Pays and colleagues, with residues numbered according to APOL1 isoform A. The BH3 domain proposed by Hu and colleagues is also shown [10]. MB, predicted membrane binding domains; SP, signal peptide. The locations of mutations that comprise the G1 and G2 alleles are shown. The lower panel shows the RNA transcript structures for the six human APOL family members, limited to protein-coding exons or portions of exons. Exon numbering is according to APOL1. Exon 7 encodes amino acid residues 106 to 398 of APOL1. APOL1 has three transcripts; only a and b are shown. [Permission pending from Smith and Malik, Genome Res. 2009. 19: 850–858 - PMID: 19299565]
Figure 2. APOL gene family in selected primate species and mouse.
Genes, lost genes, and pseudogenes are represented by plain black box, empty box, and light gray box with Ψ, respectively. The hashed boxes represent mouse homologous, but not orthologous, genes. (1) Macaque also exhibits an APOL2.1 gene and an APOL7 pseudogene.
(2) Baboon also exhibits two APOL2.1 pseudogenes and an APOL7 pseudogene. The trypanolytic potential against T. brucei is also depicted. [Permission pending from Capewell et al., Parasitology, 2015 - PMID:25656360]
All APOL genes show more missense mutations than would be predicted by chance, suggestive of positive selective pressure in response to environmental stressors during primate evolution [7]. In primates [7] and human beings [14], the strongest selection pattern in APOL1 is noted in the C-terminal serum resistance associated (SRA)-interacting domain, suggesting the importance of this domain in regulating APOL1 function. APOL1 is the only secreted member of the APOL family, having acquired a signal peptide from a gain-of-function mutation occurring after the APOL1/APOL2 divergence [7]. For this reason, extracellular pathogens are thought to have shaped the evolution of APOL1, whereas intracellular pathogens would have driven evolution of the other APOL genes [7]. The up-regulation of APOL genes by pro-inflammatory cytokines (e.g., interferon-γ [IFNγ] and tumor necrosis factor [TNF]) and their involvement in autophagy and apoptosis suggest that most APOL genes may be involved in innate immune defense[10,12,13,15].
A co-evolutionary arms race
APOL1 is the circulating factor that confers human resistance to Trypanosoma brucei brucei (T. b. brucei), the parasite responsible for trypanosomiasis [16]. While humans are resistant to T. b. brucei, two subspecies of trypanosome, T. b. rhodesiense and T. b. gambiense, have evolved to strategies to avoid APOL1-mediated lysis. These sub-species are responsible for human African sleeping sickness across sub-Saharan Africa.
APOL1 circulates systemically on high-density lipoprotein (HDL) particles, the primary nutrient source for trypanosomes. Following endocytosis by the trypanosome and trafficking to the lysosome, the acidic environment induces a transition of APOL1 from the HDL particle to the inner leaflet of the lysosome, followed by membrane insertion, pore formation, and trypanolysis [17–19]. T. b. gambiense and T. b. rhodesiense have each evolved different mechanisms to preempt APOL1 pore formation: T. b. rhodensiense encodes the serum resistance-associated (SRA) protein [8,9,37] and T .b. gambiense expresses T. b. gambiense-specific glycoprotein (TgsGP) [38,40]. SRA preempts APOL1 pore formation directly through binding of the α- helical SRA-interacting domain at the C terminus of APOL1 [5,16,17]. The association occurs in a pH-dependent manner, with optimal affinity at acidic pH within the trypanosome lysosome [9,11]. By contrast, TgsGP indirectly preempts APOL1 pore formation by stiffening the lysosomal membrane and blocking APOL1 membrane insertion [38]. By inactivating APOL1, SRA and TgsGP enable T. b. rhodesiense and T. b. gambiense proliferation, resulting in acute and chronic African sleeping sickness, respectively. Both the G1 and G2 renal risk variants are located in the SRA-interacting domain of APOL1 (Figure 1A). Genovese and colleagues showed that the G1 and G2 variant protein isoforms are able to bypass SRA inhibition, thereby extending APOL1-mediated immunity to T. b. rhodesiense at the cost of increasing risk for renal diseases [3][5,11,41]. The G1 and G2 variants were ineffective against T .b. gambiense [3].
The G1 and to a lesser extent G2 alleles exhibit signatures of a recent selective sweep in West Africa, which is attributed to protection against T. b. rhodesiense [3]. Although both G1 and G2 alleles show the highest frequency in West Africa, the G2 allele is more evenly distributed across sub-Saharan Africa [20]; it remains an enigma why the only signals of positive selection occur in West Africa where T. b. rhodesiense is not endemic [3,5]. The widespread distribution of the G2 allele, the effective trypanolytic activity of the G2 variant in vitro and in mice, and its lower affinity for SRA relative to the G1 variant, both compared to the G0 variant, support the hypothesis that the G2 allele is older than the G1 allele and might have evolved in response to T. b. rhodesiense SRA-mediated resistance mechanism [5]. Although we cannot rule out the possibility of a shift in the geographic distribution of trypanosomes during evolution, the more recent emergence of the G1 allele and its very high prevalence in West Africa suggests that the evolutionary histories of these two trypanolytic variants might be different. Indeed, pathogens other than Tb Rhodesiense may have exerted selective pressure on the APOL1 risk alleles. Capewell et al [23] showed differences in susceptibility to APOL1 trypanolysis within subgroups of T. b. gambiense, and unlike Tb rhodesiense, these strains of Tb gambiense are endemic to West Africa. It is possible that the APOL1 risk variants reduce susceptibility to infection of certain strains of T. b. gambiense but not others, thereby conferring a selective advantage in these regions. The APOL1 ortholog APOL2, which shares considerable sequence homology with APOL1, is retained in the cell and nevertheless shows a pattern of positive selection in the SRA-interacting domain. [7] This introduces the possibility that not only extracellular, but also intracellular pathogens have driven the selection of APOL1. In addition to its antitrypanosomal role, evidence suggests that APOL1 has broad immune properties because it can ameliorate Leishmania infection and restrict HIV-1 replication in macrophages in vitro [21,22].
Possible pathomechanisms: APOL1 toxicity
Multiple mechanisms have been proposed for how APOL1 might contribute to glomerulopathies, including lysosomal membrane permeabilization [24,25], autophagic cell death [10,12,26], apoptosis [7,11], and necrosis [24], but clearly much emphasis has been placed on cell death. Non-risk APOL1 (G0) induces autophagic cell death in p53-null human colorectal cancer cells [10], but this pathway of programmed cell death is not well understood and remains controversial [27–29]. Both the G1 and G2 risk variants display increased toxicity compared to G0 in HEK-293 cells and human podocytes in vitro [15,24]. It is unclear whether this gain of toxicity occurs through the same or separate pathways, and a better understanding of mechanisms of APOL1-induced toxicity may elucidate the pathomechanisms of APOL1-associated disorders.
Proposed mechanisms of APOL1 toxicity in human cells: to BH3 or not to BH3?
Early studies investigating the function of APOL1 in human cells suggest that APOL1 induces autophagic cell death through Bcl-2 homology 3 (BH3) domain-mediated pathways. All APOL proteins contain a putative BH3 domain (Figure 3A), a motif commonly found in pro-death BH3-only proteins. Deletion of the BH3 domain in APOL1 and APOL6 ablated the associated cytotoxicity, suggesting that both are BH3-only pro-death proteins [10–13,19]. BH3-only proteins augment cytotoxicity indirectly by competing with Bcl-2 family prodeath effector proteins for binding to pro-survival Bcl-2 family members, such as Bcl-2, Bcl-xL, and Mcl-1. Once displaced from a pro-survival Bcl-2 partner, Bcl-2 family effector proteins engage death pathways directly through pore formation, as is the case with Bax, which induces apoptosis via mitochondrial outer membrane permeabilization [30–33].
Figure 3. APOL1 domain organization and homology of the BH3 domain.
Panel A shows APOL1 domain organization as in figure 1. Also shown are the reported cleavage sites at amino acid residues 27 (signal peptidase) and 54 (unknown), and the Nglycosylation site (□) at N264. Panel B presents sequence analysis of residues that comprise the proposed BH3 domain of APOL family members, compared to the canonical BH3 domain motif. The middle panel shows an alignment of the amino residues for the six human APOL family members. The upper and lower panels show alignment logos generated with Skyligne software using a hidden Markov model (HMM), where the height of the amino acid code letter reflects residue frequency among the analyzed protein sequences. The upper panel shows an alignment logo for APOL family proteins found in the Pfam database, while the lower panel shows that of BH3 domain-containing proteins found in the InterPro database. For both HMM logos, sequences were filtered to remove redundancy. Each logo was generated from at least 300 aligned sequences, using sequences from all species included in the respective databases.
The BH3 domain of APOL1 is required to induce autophagic cell death [10,13], but the mechanism is not well understood. Typically, BH3-only proteins such as BCL2/adenovirus E1B 19kDa interacting protein 3 (BNIP3) induce autophagy by displacing the BH3-only protein Beclin 1[32–35]. Unlike pro-apoptotic Bax however, displaced Beclin 1 does not activate cell death pathways. Instead, Beclin 1 associates with Vps34, Atg14, and UV-radiation resistance associated gene protein (UVRAG) to form the class III phosphoinositide 3-kinase (PI3K) complex. This complex phosphorylates phosphoinositol (PI) at the 3 position to form PI(3)P, leading to autophagosome membrane nucleation and autophagy initiation [35–37]. Induction of autophagy through Beclin 1 is a pro-survival pathway that allows cells to maintain protein synthesis and energy homeostasis during nutrient deprivation and cell stress [27,33]. While APOL1 may induce autophagy through this pathway, it is difficult to comprehend how this BH3-mediated pro-survival pathway might be involved in APOL1-induced toxicity.
As shown in Figure 3B, the consensus sequence corresponding to the putative BH3 domain in APOL proteins, while well conserved, differs considerably from the BH3 consensus motif. This analysis suggests that although residues within this region are important for APOL function, a BH3 motif per se is not required. In support of this hypothesis, a recent article by Galindo-Moreno and colleagues showed that despite containing a BH3 consensus sequence and binding Bcl-2, APOL2 does not function as a classical BH3-only protein [38]. Our unpublished data indicates that mutating L158 and D163 does not inhibit APOL1 toxicity, although these residues are critical for cytotoxicity in canonical BH3-only pro-death proteins [39–42]. Moreover, Heneghan et al, working in Xenopus oocytes, showed that while deletion of the proposed BH3 domain abolished APOL1 toxicity (as previously shown by Wan et al. [10]), toxicity remained after replacing the same residues with a string of alanines [43]. Taken together, these data indicate that APOL1 does not require a BH3 motif to induce toxcity, and therefore initiates cell death through BH3-independent mechanisms.
Lessons from trypanosomal cell death: APOL1 pore formation
The earlier-described data throw into question the model by which APOL1 induces toxicity in human cells. If APOL1 is not a BH3-only prodeath protein, then how does it induce toxicity? Although less is known about APOL1 function in human cells, considerable work has been performed to elucidate the mechanisms by which APOL1 induces toxicity in the trypanosome. A more thorough understanding of APOL1 trypanolytic activity may point toward analogous functions in human beings.
The trypanolytic activity of APOL1 depends upon its ability to form a pore in the trypanosome membrane [5,17,19,44–46]. The leading model for APOL1 function, as proposed by Pays and colleagues (Figure 4A), suggests that APOL1 is both structurally and functionally similar to bacterial colicins which are pore-forming toxins released by certain bacteria and active at acidic pH [17,18]. APOL1 is proposed to consist of a colicin-like pore-forming domain adjacent to a pH-sensitive, membrane-addressing domain, followed by the SRA-interacting domain. APOL1 is modeled as associating with the HDL particle through the hairpin-like membrane-addressing domain. According to this model, following endocytosis and trafficking to the lysosome, the acidic environment promotes a pH-dependent opening of the hairpin-like membrane-addressing domain [17,18] (Figure 4B). This decreases affinity for the neutral HDL lipids, enabling APOL1 to transition from the HDL particle to the inner leaflet of the lysosomal membrane. APOL1 inserts into the membrane via the membrane-addressing domain, which forms an anion-selective channel resulting in chloride entry and lysosomal membrane depolarization. Chloride influx is followed by irreversible osmotic swelling of the lysosome, and trypanosome death. This model is supported by experiments using E. coli and trypanosomes using recombinant full-length and truncated forms of APOL1, and in vitro liposome assays and electrophysiology studies using only the pore-forming domain of APOL1 [17]. It is unclear what role the SRA-interacting domain plays in this process, though interaction with SRA inactivates APOL1 by preventing membrane insertion.
Figure 4. APOL1 domain organization and proposed mechanisms of APOL1 trypanolysis.
Panel A shows the domain structure suggested by Pays and colleagues, identical that shown in figure 1. Panel B shows the model of APOL1 trypanolysis proposed by Pays and colleagues. In this model the HDL-bound APOL1 is endocytosed by the trypanosome and trafficked to the trypanosome lysosome. Once in the lysosome, the acidic environment leads to opening of the hairpin-like membrane-addressing domain, followed by dissociation of APOL1 from the HDL particle and insertion membrane-addressing domain into the lysosomal membrane. The pore-forming domain of APOL1 forms a chloride channel that depolarizes the lysosomal membrane, leading to irreversible swelling of the lysosome, followed by trypanolysis. SRA inhibits APOL1 within the lysosome by binding the SRA-interacting domain on APOL1, though the mechanism of inhibition is unclear. Panel C depicts the model of trypanolysis proposed by Thomson and Finkelstein. In this model, following endocytosis and trafficking to the lysosome, the acidic environment leads to irreversible insertion of APOL1 into the lysosomal membrane, a process mediated by the SRA-interacting domain. The APOL1 channel is inactive until recycled to the trypanosome cell surface, where neutral pH leads to APOL1 channel activation. Cation influx through APOL1 depolarizes the plasma membrane and is coupled to potassium efflux and anion and water influx, followed by loss of osmoregulation, cytoplasmic vacuolization, and lysis. SRA binding to APOL1 is speculated to inhibit insertion of the SRA-interacting domain into the lysosomal membrane, thereby preempting APOL1 channel formation and lysis. Panel D depicts a revised model for APOL1 domain organization. The pore-forming domain has been removed, as the residues responsible for channel formation remain to be identified. The membrane-addressing domain has been replaced by a proposed membrane insertion domain at the C terminus, within the SRA-interacting domain. PF, pore-forming domain. MA, membrane-addressing domain. SI,SRA-interacting domain. Other abbreviations as in Figure 1. [Permission pending from Pays et al., Nat Rev Microbiol. 2006, PMID: 16710327; Thomson and Finkelstein, Proc Natl Acad Sci U S A. 2015PMID: 25730870]
An alternate model of APOL1 pore formation, proposed by Raper and colleagues, suggests that APOL1 forms cation-selective pores in the trypanosome plasma membrane, leading to a loss of osmoregulation that results in cytoplasmic vacuolization, colloid-osmotic swelling of the cell body, and ultimately trypanolysis [46]. This model was recently supported and further developed by Thomson and Finkelstein, who showed that the APOL1 channel is not only cation-selective, but also pH-gated, with optimal activity at neutral pH (pKa ~7.1) [19]. The investigators suggest that the acidic pH of the lysosome induces irreversible insertion of APOL1 into the lysosomal membrane (a process mediated by the SRA-interacting domain, not the membrane-addressing domain) but that channel activity is not induced until endosomes containing APOL1 are recycled to the cell surface, where the neutral pH leads to channel opening (Figure 4C). In support of this model, the investigators used electrophysiology to show that, while a small but noticeable conductance is present when APOL1 is at acidic pH, this signal can be amplified up to ~3000-fold upon return to neutral pH. Importantly, preincubation with SRA inhibited signal amplification in non-risk G0 APOL1, which binds SRA, but not G2 APOL1, which does not bind SRA. Moreover, channel activity was not present when APOL1 lacked the SRA-interacting domain, supporting a critical role for the SRA-interacting domain in APOL1 channel activity. This study redefines APOL1 as a pH-gated cation channel and suggests that the SRA-interacting domain is required for channel activity, although its precise role in this process remains to be defined.
APOL1 channel formation and initiation is clearly a multi-stage process. Although Thomson and Finkelstein [19] speculated that the SRA-interacting domain mediated membrane insertion, it is not clear which regions of APOL1 are involved in other stages of channel assembly and membrane permeabilization, calling into question the previously-proposed domain organization (Fig. 4D). The study also sheds light on mechanisms of APOL1 inhibition: similar to TgsGP, SRA may inhibit APOL1 pore formation by preempting pH-dependent insertion of APOL1 into the lysosomal membrane. The investigators did not investigate whether APOL1 channel properties, such as pKa or maximum conductance, are altered by the risk mutations. If APOL1 pore formation is relevant to human physiology, this data may shed light on whether the increased toxicity of the risk variants in human cells is due to changes in APOL1 pore activity itself, or to altered regulation by endogenous inhibitors.
The relevance of trypanocentric models to human pathophysiology
While the work by Pays and colleagues and Thomson and Finkelstein illuminates the molecular function of APOL1 and introduce novel pathways for APOL1-induced toxicity, it is unclear whether lysosome permeabilization and osmotic swelling of the cell body are mutually exclusive mechanisms of cell death. In support of both models, Singhal and colleagues showed that overexpression of APOL1 induced both podocyte swelling and lysosomal membrane permeability [24], with both phenotypes increased with the G1 and G2 risk variants. These data suggest that the trypanocentric models of APOL1 function translate at least in part to human physiology and may be important pathomechanisms in APOL1-associated diseases.
There may be limits to this parallelism, however. According to these trypanocentric models, one might hypothesize that systemically circulating APOL1 is responsible for the pathogenesis of glomerular injury, but there is evidence to the contrary. Specifically, transplanted kidneys from high-risk donors fail more rapidly when compared to those of donors bearing low-risk genotypes [47,48] while recipient genotype has no effect on allograft survival five years post-transplant [49]. Although these studies did not include data on corresponding recipient or donor genotype, taken in combination they suggest that APOL1-associated renal diseases result from APOL1 generated within the kidney rather than systemically circulating APOL1. These data do not rule out an autocrine or paracrine function of APOL1 within the kidney, however. Moreover, human cells are exposed to and endocytose circulating APOL1 [50,51], but cells that are most exposed, such as immune cells and cells of the vasculature, are apparently healthy. These data suggest that APOL1 toxicity may be prevented in such cell types by alternate trafficking, endogenous APOL1 inhibitors, or an increased threshold for APOL1 channel activity. Though there may be physiologically relevant circumstances when APOL1 is trafficked to the plasma membrane, endogenous APOL1 has a primarily vesicular subcellular localization, and does not appear at considerable levels on the cell surface [52].
There has been much speculation about a human analog to SRA that would act as a trigger lock to inhibit APOL1 activity, with the G1 and G2 risk variants having a decreased affinity to the postulated host protein, accounting for the increased cytotoxicity of the risk variants [53]. O'Toole and colleagues Use the following citation:[20] used a bioinformatics-based structural homolgy search to identify human orthologs of trypanosomal SRA, and identified VAMP8, an R-SNARE involved in autophagosome-lysosome fusion. Co-immunoprecipitation demonstrated an interaction between non-risk G0 APOL1 but not with G1 or G2, suggesting an altered affinity similar to SRA. While VAMP8 may indeed be an important inhibitor of APOL1 function, according to the trypanocentric models, endocytosed APOL1 would be in the lumen of the lysosome while VAMP8 is for the most part cytoplasmic [54]. The presence of the lysosomal membrane therefore would preclude an interaction between these two proteins. While these considerations are based on endocytosed APOL1, it cannot be ruled out that endogenously expressed APOL1 may be on the cytoplasmic side of the membrane, thereby enabling the interaction. While the search for human analogs of trypanosomal APOL1 inhibitors has focused primarily on SRA, less attention has been paid to human TgsGP analogs. If the principle of APOL1 inhibition by TgsGP and SRA are truly the same (preempting channel formation by inhibiting APOL1 membrane insertion), the search should not be limited to SRA homologs alone, as there may be numerous proteins that can fulfill this role.
While most work to illuminate APOL1 intracellular function has used overexpression models, it remains to be seen whether APOL1-induced toxicity occurs at physiologic levels in the podocyte. The low penetrance, chronic presentation, and relatively late onset of APOL1-associated diseases suggest that APOL1-induced toxicity may not be the whole story. Indeed, the role of APOL1 in human physiology (and pathophysiology) is very likely multifactorial, and subtler experimental approaches may be required to uncover these functions.
Circulating APOL1 and lipid biology
APOL1, as a lipoprotein, binds lipids; whether it has role in regulatory lipid metabolism remains to be determined. Circulating APOL1 is found primarily in the HDL3b and c subfractions[55], where it associates with two distinct complexes termed trypanolytic factors (TLF) 1 and 2 [56–59]. TLF1 is an appfoximately 500-kDa lipid-rich complex found in the densest HDL3c subfraction and containing APOL1, APOA1, and haptoglobin-related protein (Hpr), and lower levels of paraoxonase, hemoglobin, and APOA2. TLF2 is an approximately 1000-kDa lipid-poor complex that is more dense that normal HDL particles [56,60]. Along with APOL1, APOA1, and Hpr, TLF2 also contains IgM molecules [56,58]. It is not known whether TLF1 and 2 have functions beyond trypanolysis, and a better understanding of their role in lipid biology may help to elucidate APOL1 function. Subclass HDL3 attenuates low-density lipoprotein oxidation, and APOL1 levels in HDL3 are highly correlated with HDL anti-oxidative function and with presence of paraoxonases 1 and 3 [55], important anti-oxidant and anti-inflammatory proteins [61]. Thus far, all that can be said is that APOL1 is a fellow traveler with anti-oxidant passengers in an anti-inflammatory vehicle; whether APOL1 contributes in any way to these activities remains to be determined.
Several studies have investigated associations between APOL1 risk variants and circulating lipids. Plasma APOL1 levels correlate with plasma triglycerides, total cholesterol, and low-density lipoprotein cholesterol [62–64], but no differences were found by APOL1 genotype [63]. Bentley and colleagues found no difference in mean plasma HDL cholesterol, an approximation of total HDL number, by APOL1 genotype [65]. They showed an unexpected inverse relationship between plasma HDL cholesterol and estimated glomerular filtration rate (eGFR) in G1/G1 African Americans (i.e., higher HDL associated with lower eGFR), while no correlation was witnessed in other subjects. This pattern did not repeat in a West African cohort, although the investigators noted considerable metabolic differences between the populations. The G2 risk allele was not genotyped, however, and therefore the controls almost certainly included individuals homozygous for G2 allele, an important limitation in these data. Freedman and colleagues found an inverse correlation between HDL2c/3a concentration and risk allele number, while the concentration of HDL3b/c (the subfractions reported to contain APOL1) was not different [66]. Bruggeman et al. found no correlation between APOL1 genotype and levels of any lipid class, though HDL subfractions were not studied. Moreover, they saw no correlation between plasma APOL1 levels and CKD, and no change in circulating APOL1 levels by genotype [63]. Despite the largely negative findings of these studies, it still would be premature to conclude that the risk variants have no effect on HDL function.
APOL1 tissue and cellular expression
APOL1 RNA and protein expression have been documented in many tissues, with high expression in pancreas, prostate, spleen, liver, kidney, and placenta and highest in lung [8,67–69]. This would suggest either ubiquitous expression, possibly pan-cellular, or expression restricted to a cell-type that is common to the various tissues. APOL1 expression first was noted in TNF-stimulated endothelial cells [67]. Subsequent work demonstrated broader cellular expression. In particular two reports elaborated APOL1 distribution in normal and diseased kidneys and explored the impact of APOL1 risk allele status on APOL1 expression and distribution. In normal kidney parenchyma, APOL1 mRNA and protein expression is present in glomerular endothelium, podocytes, proximal tubular epithelium, arteriolar endothelium and in HIVAN and FSGS was noted in arteriolar media [51]. Mesangial APOL1 expression was not observed [51]. Comparisons between APOL1 expression levels in podocytes in tissue versus the lower levels observed in cultured podocytes were interpreted to suggest uptake of APOL1 by podocytes, a process supported by experimental evidence, with a preference for uptake by podocytes compared to other renal parenchymal cells (mesangial, endothelial and tubular epithelial cells)[51].
As an innate immune protein, APOL1 RNA and protein expression is driven by cytokines such as TNF and interferons in innate immune cells such as macrophages. Furthermore, IFN-α induced expression of multiple APOL1 transcript variants in cultured endothelial cells and podocytes [15]. All APOL genes are induced by interferon and TNF [10,12,13,15,70], and binding sites for the interferon-associated transcription factors Interferon Regulatory Factors 1 and 2 (IRF1 and IRF2), and Signal Transducer and Activator of Transcription 2 (STAT2) are present in the promoter region of the APOL1 gene [15]. In support of its role in innate immune function, Divers and colleagues showed an interaction between APOL1 genotype and JC polyoma virus infection in non-diabetic kidney disease [71], and Nichols and colleagues found that all individuals who developed interferon-associated FSGS within a small cohort had two APOL1 risk alleles [15].
Beyond innate immunity, there may be role for APOL1 in the brain, and possibly in schizophrenia. Studies have shown expression in relevant brain regions such as the frontal cortex [72] [73], altered expression in schizophrenia [72–75], and genetic variants in APOL1 are associated with increased rates of the disorder [76–78], the physiological role of ApoL1 in schizophrenia is largely speculative [79]. Interestingly, the podocyte, the central target in several APOL1 nephropathies, shares characteristics with neurons. Certain proteins that are expressed by podocytes and implicated in glomerulopathies, such as podocin, nephrin, podocalxyin, and synaptopodin, are also important neuronal markers, indicating some degree of transcriptome similarity [80,81]. Furthermore, podocyte foot processes display some similarities to neuronal dendrites [82].
Despite these efforts to elucidate the expression pattern of APOL1 and the role of the risk alleles, the pathomechanism of APOL1 risk allele-related disease at the tissue and cell level is unclear. What is the role for APOL1 in arteriopathy? Does its presence reflect non-specific trapping of circulating APOL1 or local production by smooth muscle cells? [Among commercial APOL1 antibodies, we have witnessed heterogeneity in staining patterns, including within arterial/arteriolar media across disease states (unpublished)]
A spectrum of APOL1-related nephropathies
Pollak and colleagues determined that substantial risk for FSGS and HIVAN can be attributed to the G1 and G2 APOL1 risk alleles [3], and subsequent studies have extended the renal phenotype to additional clinical settings, connecting distinct histopathologies through the APOL1 genetic variants [83]. The spectrum of APOL1-related nephropathies consists of HIVAN [4,84–88], FSGS [4,89,90], sickle-cell nephropathy [91], arterionephrosclerosis [92], lupus nephritis [93,94], microalbuminuria [95], CKD [96,97], and ESKD [98,99]. There is also increased deceased donor kidney transplant rejection [47], and greater decline in glomerular number and greater increase in glomerular volume over lifespan [84], as summarized in Table 1. The strongest association is with collapsing glomerulopathy in many settings that include HIVAN [86], primary collapsing glomerulopathy [4,90], and collapsing glomerulopathy seen in the setting of in systemic lupus erythematosus [94,100] and PLA2R-positive membranous glomerulopathy [101]. Furthermore, APOL1 two-risk allele status predisposes patients receiving interferon to collapsing glomerulopathy, suggesting a interferon-related pathomechanism [15].
Less is known about the role of APOL1 in other histopathologic entities. For example further work will be needed to elaborate the impact of APOL1 risk allele status on renal allograft outcomes where donor (but not recipient) APOL1 risk allele status correlates with accelerated loss of renal allografts [47,49] but did not impact HIV reinfection of allograft in HIV-positive recipients [102]. Importantly, we need data on the long-term outcomes of two-risk allele kidney donors, to establish whether it is safe for these individuals to serve as donors. Finally, APOL1 risk alleles have been associated with faster progression of clinically-diagnosed type 2 diabetes-associated ESKD [103]; it remains to be determined whether this effect is due to compound effects of diabetes and APOL1 risk alleles on the glomerulus and the vasculature, or whether some of these cases have only APOL1 nephropathy.
APOL1 kidney risk variants may have effects on phenotypes beyond the kidney, in addition to their role in innate immunity. There may be a role in cardiovascular disease [104], but this area remains controversial [48,105]. Additional cohorts with genome-wide data will be needed to better understand the role of APOL1 risk allele status in more complex genetic states. That such a diverse array of pathologies are associated with variants in a single gene suggests that disease etiology is in part mediated by either environmental factors or genegene interactions that place excess strain on biological processes already susceptible to dysregulation, followed by varied compensatory mechanisms to achieve homeostasis.
APOL1 and systems biology
Ongoing work from human renal disease cohorts, including particularly those that include renal tissue obtained from individuals of African descent and non-African descent, offer the promise to integrate epidemiologic, genetic, genomic, phenotypic, and histomorphologic data. This multi-level integrated analysis may offer insights into how an interplay between genes and environment, leading to protein expression in a one or more cell types (particularly podocyte and vascular cells), results in chronic kidney disease. The NEPTUNE study provides just such an opportunity to carry out deep multi-level analysis and begin to answer these questions.
Conclusions and future questions
The spectrum of APOL1-asociated diseases has expanded considerably since the initial publication by Genovese et al. [3]. That APOL1 genotype is a susceptibility factor for such a diverse array of complex diseases suggests that it has a physiological function at the nexus of biological pathways important in multiple renal diseases. A better understanding of APOL1 biology will not only suggest targeted treatments for APOL1-associated diseases and expand possibilities for individualized medicine, but may also shed light on pathways dysregulated in these diseases generally.
Our understanding of APOL1 biology and potential pathomechansims has also been enhanced. The molecular function of APOL1 has been clarified, as a recent report suggests that APOL1 functions as a pH-gated cation channel. The SRA-interacting domain is required for APOL1 channel activity and is speculated to play a role in membrane insertion. It is unclear whether this domain plays a role in channel activity beyond this initial stage, or whether the risk mutations affect fundamental properties of APOL1 channel activity. These data suggest that recycling of APOL1-containing endosomes is important for APOL1 channel activation in the trypanosome.
We look forward to seeing whether the trypanocentric models are applicable to human physiology, as a report by Singhal and colleagues suggests. A better understanding of APOL1 intracellular trafficking and protein binding partners will provide important information for the pathophysiological mechanism(s) that targets the podocyte. While endogenous binding partners analogous to trypanosomal SRA, including VAMP8, have been discovered, their physiologic relevance remains unknown. Data suggests that APOL1 is unlikely to function as a canonical BH3-only protein to induce cell death. The mechanism(s) by which APOL1 induces cell death require clarification, but plausible pathways include necrosis, apoptosis, autophagy, lysosomal permeabilization, and colloid osmotic swelling.
Transplant studies suggest that APOL1-associated diseases are due to locally expressed, rather than systemically circulating APOL1. Studies investigating the localization of endogenous APOL1 support expression in glomerular podocytes. Whether APOL1 secretion and uptake are involved in pathology is unclear, however. These findings have important implications for not only the disease model, but also treatments and donor screening.
There are many barriers to progress that are worth noting. While a number polyclonal and monoclonal antibodies are available, specificity for APOL1 rather than other APOL family members or other proteins sharing sequence homology has not often been rigorously characterized. Stably-transfected cells are difficult to generate, particularly for the G1 and G2 alleles, likely due to toxicity, and stable cell lines that have been generated may have genetic mutations in APOL1 that have promoted survival (our unpublished data). Relevant animal models are critical to assess the in vivo consequences of APOL1 variant isoforms and will be important for pre-clinical testing of potential therapeutic agents. APOL1 is only present in a few primates, however, and the utility of introducing APOL1 into mice, rats, fish and flies may be limited if critical interacting proteins are also missing. Studies using human tissues and patient samples may shed light on pathogenesis, but will not provide a suitable system to safely test therapeutics in a pre-clinical setting. While barriers to progress exist, integrating data on APOL1 biology across multiple experimental and clinical platforms will enable clinicians and researchers to side-step the limitations of any individual system. An integrated approach will illuminate disease mechanisms and reveal new and unexpected routes to treatment, thereby providing hope for millions worldwide.
Areas for which clarity is needed include:
Are there other infectious agents, beyond T. brucei, that have driven APOL1 evolution? Could one of these other agents better explain the high prevalence of APOL1 risk alleles in West African populations?
How is APOL1 trafficked within cells and between intracellular compartments?
Does the SRA domain have a distinct function in the host cell, as suggested by its binding to VAMP8?
Does APOL1 have other protein partners beyond VAMP8? APOL1 is a toxic protein, as the difficulty of obtaining stable cell lines attests. Are there endogenous inhibitors or regulators of APOL1 activity? Do the risk mutations exert toxicity by subverting these control mechanisms?
In the context of widespread APOL1 expression across tissues, why is renal disease most impacted by the risk alleles?
What is the mechanism of APOL1 risk variant cytoxicity?
What are the most effective therapeutic paradigms to manage APOL1 related diseases?
What additional evidence is needed to support a role for genetic screening for APOL1 risk alleles to drive implementation of prevention strategies for individuals with two APOL1 risk alleles?
How should we counsel prospective kidney donors with two APOL1 risk alleles and will this help clinical management?
Table 2. APOL1 risk allele kidney phenotypes.
Displayed are the range of kidney diseases and conditions for which APOL1 risk variants have been shown to increase risk or alter phenotype.
| Kidney disease | Phenotype | Geographic region, or age |
Odds ratio | Reference |
|---|---|---|---|---|
| HIV-associated nephropathy | N/A | USA | 29 | [4] |
| N/A | South Africa | 89 | [88] | |
| Focal segmental glomerulosclerosis and collapsing glomerulopathy | N/A | Mostly adults | 17 | [4, 89] |
| N/A | Mostly children | N/A | [90] | |
| Sickle cell nephropathy | Proteinuria | Mostly adults | 3.4 | [91] |
| Diabetic nephropathy | Faster progression to ESKD | USA | N/A | [106] |
| Arterionephro-sclerosis | All CKD stages | USA | 2.7 | [92] |
| ESKD | USA | 7 | [3] | |
| Lupus nephritis | ESKD | USA | 2.7 | [93] |
| Collapsing glomerulopathy | USA | 5.4 | [94] | |
| Microalbuminuria | N/A | USA | N/A | [107] |
| Chronic kidney disease | Not otherwise specified | USA | 1.5 | [96] |
| Nigeria | N/A | [97] | ||
| ESKD | Younger age at dialysis initiation | USA | N/A | [98][99] |
| Not otherwise specified | USA | 2.2 | [96] | |
| Deceased donor allograft loss | N/A | USA | 3.8 | [47] |
| Kidney number and size | Glomerular loss and hypertrophy | Mississippi | N/A | [84] |
Acknowledgement
This work was supported in part the Intramural Research Programs of the NIDDK, NIH. This project has been funded in whole or in part with federal funds from the National Cancer Institute, National Institutes of Health, under contract HHSN26120080001E. The content of this publication does not necessarily reflect the views or policies of the Department of Health and Human Services, nor does mention of trade names, commercial products, or organizations imply endorsement by the U.S. Government. This Research was supported [in part] by the Intramural Research Program of the NIH, National Cancer Institute, Center for Cancer Research.
Financial support: funded by NIH.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
conflict of interest: No conflicts of interest.
REFERENCES
- 1.United States Renal Data System. Bethesda, MD: National Institutes of Health, National Institute of Diabetes and Digestive and Kidney Diseases; 2014. Annual Data Report: Epidemiology of Kidney Disease in the United States. [Google Scholar]
- 2.Kiberd BA. Cumulative Risk for Developing End-Stage Renal Disease in the US Population. Journal of the American Society of Nephrology. 2002;13:1635–1644. doi: 10.1097/01.asn.0000014251.87778.01. [DOI] [PubMed] [Google Scholar]
- 3.Genovese G, Friedman DJ, Ross MD, Lecordier L, Uzureau P, et al. Association of trypanolytic ApoL1 variants with kidney disease in African Americans. Science. 2010;329:841–845. doi: 10.1126/science.1193032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kopp JB, Nelson GW, Sampath K, Johnson RC, Genovese G, et al. APOL1 genetic variants in focal segmental glomerulosclerosis and HIV-associated nephropathy. J Am Soc Nephrol. 2011;22:2129–2137. doi: 10.1681/ASN.2011040388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Thomson R, Genovese G, Canon C, Kovacsics D, Higgins MK, et al. Evolution of the primate trypanolytic factor APOL1. Proc Natl Acad Sci U S A. 2014;111:E2130–E2139. doi: 10.1073/pnas.1400699111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Limou S, Nelson GW, Kopp JB, Winkler CA. APOL1 Kidney Risk Alleles: Population Genetics and Disease Associations. Adv Chronic Kidney Dis. 2014;21:426–433. doi: 10.1053/j.ackd.2014.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Smith EE, Malik HS. The apolipoprotein L family of programmed cell death and immunity genes rapidly evolved in primates at discrete sites of host-pathogen interactions. Genome Res. 2009;19:850–858. doi: 10.1101/gr.085647.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Page NM, Butlin DJ, Lomthaisong K, Lowry PJ. The human apolipoprotein L gene cluster: identification, classification, and sites of distribution. Genomics. 2001;74:71–78. doi: 10.1006/geno.2001.6534. [DOI] [PubMed] [Google Scholar]
- 9.Johnstone DB, Shegokar V, Nihalani D, Rathore YS, Mallik L, et al. APOL1 null alleles from a rural village in India do not correlate with glomerulosclerosis. PLoS One. 2012;7:e51546. doi: 10.1371/journal.pone.0051546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wan G, Zhaorigetu S, Liu Z, Kaini R, Jiang Z, et al. Apolipoprotein L1, a novel Bcl-2 homology domain 3-only lipid-binding protein, induces autophagic cell death. J Biol Chem. 2008;283:21540–21549. doi: 10.1074/jbc.M800214200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Liu Z, Lu H, Jiang Z, Pastuszyn A, Hu CA. Apolipoprotein l6, a novel proapoptotic Bcl-2 homology 3-only protein, induces mitochondria-mediated apoptosis in cancer cells. Mol Cancer Res. 2005;3:21–31. [PubMed] [Google Scholar]
- 12.Zhaorigetu S, Yang Z, Toma I, McCaffrey TA, Hu CA. Apolipoprotein L6, induced in atherosclerotic lesions, promotes apoptosis and blocks Beclin 1-dependent autophagy in atherosclerotic cells. J Biol Chem. 2011;286:27389–27398. doi: 10.1074/jbc.M110.210245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhaorigetu S, Wan G, Kaini R, Jiang Z, Hu CA. ApoL1, a BH3-only lipid-binding protein, induces autophagic cell death. Autophagy. 2008;4:1079–1082. doi: 10.4161/auto.7066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ko WY, Rajan P, Gomez F, Scheinfeldt L, An P, et al. Identifying Darwinian Selection Acting on Different Human APOL1 Variants among Diverse African Populations. Am J Hum Genet. 2013;93:54–66. doi: 10.1016/j.ajhg.2013.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nichols B, Jog P, Lee JH, Blackler D, Wilmot M, et al. Innate immunity pathways regulate the nephropathy gene Apolipoprotein L1. Kidney Int. 2014 doi: 10.1038/ki.2014.270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Vanhamme L, Paturiaux-Hanocq F, Poelvoorde P, Nolan DP, Lins L, et al. Apolipoprotein L-I is the trypanosome lytic factor of human serum. Nature. 2003;422:83–87. doi: 10.1038/nature01461. [DOI] [PubMed] [Google Scholar]
- 17.Perez-Morga D, Vanhollebeke B, Paturiaux-Hanocq F, Nolan DP, Lins L, et al. Apolipoprotein L-I promotes trypanosome lysis by forming pores in lysosomal membranes. Science. 2005;309:469–472. doi: 10.1126/science.1114566. [DOI] [PubMed] [Google Scholar]
- 18.Pays E, Vanhollebeke B, Vanhamme L, Paturiaux-Hanocq F, Nolan DP, et al. The trypanolytic factor of human serum. Nat Rev Microbiol. 2006;4:477–486. doi: 10.1038/nrmicro1428. [DOI] [PubMed] [Google Scholar]
- 19.Thomson R, Finkelstein A. Human trypanolytic factor APOL1 forms pH-gated cation-selective channels in planar lipid bilayers: Relevance to trypanosome lysis. Proc Natl Acad Sci U S A. 2015;112:2894–2899. doi: 10.1073/pnas.1421953112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.O’Toole JFMS, Thomas DB, Barisoni L, Bruggeman LA, Sedor JR. APOL1 Is an innate immunity effector that induces stress autophagy by interacting with the SNARE Protein,VAMP8 [Abstract] ASN Annual Meeting. Atlanta, GA J Am Soc Nephrol. 2013;24:93A. [Google Scholar]
- 21.Samanovic M, Molina-Portela MP, Chessler AD, Burleigh BA, Raper J. Trypanosome lytic factor, an antimicrobial high-density lipoprotein, ameliorates Leishmania infection. PLoS Pathog. 2009;5:e1000276. doi: 10.1371/journal.ppat.1000276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Taylor HE, Khatua AK, Popik W. The innate immune factor apolipoprotein L1 restricts HIV-1 infection. J Virol. 2014;88:592–603. doi: 10.1128/JVI.02828-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Capewell P, Veitch NJ, Turner CM, Raper J, Berriman M, et al. Differences between Trypanosoma brucei gambiense groups 1 and 2 in their resistance to killing by trypanolytic factor 1. PLoS Negl Trop Dis. 2011;5:e1287. doi: 10.1371/journal.pntd.0001287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lan X, Jhaveri A, Cheng K, Wen H, Saleem MA, et al. APOL1 risk variants enhance podocyte necrosis through compromising lysosomal membrane permeability. Am J Physiol Renal Physiol. 2014;307:F326–F336. doi: 10.1152/ajprenal.00647.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Vanhollebeke B, Pays E. The function of apolipoproteins L. Cell Mol Life Sci. 2006;63:1937–1944. doi: 10.1007/s00018-006-6091-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hu CA, Klopfer EI, Ray PE. Human apolipoprotein L1 (ApoL1) in cancer and chronic kidney disease. FEBS Lett. 2012;586:947–955. doi: 10.1016/j.febslet.2012.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Codogno P, Meijer AJ. Autophagy and signaling: their role in cell survival and cell death. Cell Death Differ. 2005;12(Suppl 2):1509–1518. doi: 10.1038/sj.cdd.4401751. [DOI] [PubMed] [Google Scholar]
- 28.Fougeray S, Pallet N. Mechanisms and biological functions of autophagy in diseased and ageing kidneys. Nat Rev Nephrol. 2015;11:34–45. doi: 10.1038/nrneph.2014.201. [DOI] [PubMed] [Google Scholar]
- 29.Kroemer G, Jaattela M. Lysosomes and autophagy in cell death control. Nat Rev Cancer. 2005;5:886–897. doi: 10.1038/nrc1738. [DOI] [PubMed] [Google Scholar]
- 30.Wei MC, Zong WX, Cheng EH, Lindsten T, Panoutsakopoulou V, et al. Proapoptotic BAX and BAK: a requisite gateway to mitochondrial dysfunction and death. Science. 2001;292:727–730. doi: 10.1126/science.1059108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lovell JF, Billen LP, Bindner S, Shamas-Din A, Fradin C, et al. Membrane binding by tBid initiates an ordered series of events culminating in membrane permeabilization by Bax. Cell. 2008;135:1074–1084. doi: 10.1016/j.cell.2008.11.010. [DOI] [PubMed] [Google Scholar]
- 32.Lomonosova E, Chinnadurai G. BH3-only proteins in apoptosis and beyond: an overview. Oncogene. 2008;27(Suppl 1):S2–S19. doi: 10.1038/onc.2009.39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sinha S, Levine B. The autophagy effector Beclin 1: a novel BH3-only protein. Oncogene. 2008;27(Suppl 1):S137–S148. doi: 10.1038/onc.2009.51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. (!!! INVALID CITATION !!!). [Google Scholar]
- 35.Kang R, Zeh HJ, Lotze MT, Tang D. The Beclin 1 network regulates autophagy and apoptosis. Cell Death Differ. 2011;18:571–580. doi: 10.1038/cdd.2010.191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Axe EL, Walker SA, Manifava M, Chandra P, Roderick HL, et al. Autophagosome formation from membrane compartments enriched in phosphatidylinositol 3-phosphate and dynamically connected to the endoplasmic reticulum. The Journal of Cell Biology. 2008;182:685–701. doi: 10.1083/jcb.200803137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hamasaki M, Furuta N, Matsuda A, Nezu A, Yamamoto A, et al. Autophagosomes form at ER-mitochondria contact sites. Nature. 2013;495:389–393. doi: 10.1038/nature11910. [DOI] [PubMed] [Google Scholar]
- 38.Galindo-Moreno J, Iurlaro R, El Mjiyad N, Diez-Perez J, Gabaldon T, et al. Apolipoprotein L2 contains a BH3-like domain but it does not behave as a BH3-only protein. Cell Death Dis. 2014;5:e1275. doi: 10.1038/cddis.2014.237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zha J, Harada H, Osipov K, Jockel J, Waksman G, et al. BH3 domain of BAD is required for heterodimerization with BCL-XL and pro-apoptotic activity. J Biol Chem. 1997;272:24101–24104. doi: 10.1074/jbc.272.39.24101. [DOI] [PubMed] [Google Scholar]
- 40.Oda E, Ohki R, Murasawa H, Nemoto J, Shibue T, et al. Noxa, a BH3-only member of the Bcl-2 family and candidate mediator of p53-induced apoptosis. Science. 2000;288:1053–1058. doi: 10.1126/science.288.5468.1053. [DOI] [PubMed] [Google Scholar]
- 41.Puthalakath H, Villunger A, O'Reilly LA, Beaumont JG, Coultas L, et al. Bmf: a proapoptotic BH3-only protein regulated by interaction with the myosin V actin motor complex, activated by anoikis. Science. 2001;293:1829–1832. doi: 10.1126/science.1062257. [DOI] [PubMed] [Google Scholar]
- 42.Fleischer A, Ayllon V, Dumoutier L, Renauld JC, Rebollo A. Proapoptotic activity of ITM2B(s), a BH3-only protein induced upon IL-2-deprivation which interacts with Bcl-2. Oncogene. 2002;21:3181–3189. doi: 10.1038/sj.onc.1205464. [DOI] [PubMed] [Google Scholar]
- 43.Heneghan J, Alper S, Pollak M. BH3 domain-independent apolipoprotein L1 toxicity rescued by Bcl2 pro-survival proteins [abstract] J Am Soc Nephrol. 2014;25:452A. doi: 10.1152/ajpcell.00142.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lecordier L, Vanhollebeke B, Poelvoorde P, Tebabi P, Paturiaux-Hanocq F, et al. C-terminal mutants of apolipoprotein L-I efficiently kill both Trypanosoma brucei brucei and Trypanosoma brucei rhodesiense. PLoS Pathog. 2009;5:e1000685. doi: 10.1371/journal.ppat.1000685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Uzureau P, Uzureau S, Lecordier L, Fontaine F, Tebabi P, et al. Mechanism of Trypanosoma brucei gambiense resistance to human serum. Nature. 2013;501:430–434. doi: 10.1038/nature12516. [DOI] [PubMed] [Google Scholar]
- 46.Molina-Portela Mdel P, Lugli EB, Recio-Pinto E, Raper J. Trypanosome lytic factor, a subclass of high-density lipoprotein, forms cation-selective pores in membranes. Mol Biochem Parasitol. 2005;144:218–226. doi: 10.1016/j.molbiopara.2005.08.018. [DOI] [PubMed] [Google Scholar]
- 47.Reeves-Daniel AM, DePalma JA, Bleyer AJ, Rocco MV, Murea M, et al. The APOL1 gene and allograft survival after kidney transplantation. Am J Transplant. 2011;11:1025–1030. doi: 10.1111/j.1600-6143.2011.03513.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Freedman BI, Langefeld CD, Lu L, Palmer ND, Smith SC, et al. APOL1 associations with nephropathy, atherosclerosis, and all-cause mortality in African Americans with type 2 diabetes. Kidney Int. 2015;87:176–181. doi: 10.1038/ki.2014.255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lee BT, Kumar V, Williams TA, Abdi R, Bernhardy A, et al. The APOL1 genotype of African American kidney transplant recipients does not impact 5-year allograft survival. Am J Transplant. 2012;12:1924–1928. doi: 10.1111/j.1600-6143.2012.04033.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Molina-Portela MP, Samanovic M, Raper J. Distinct roles of apolipoprotein components within the trypanosome lytic factor complex revealed in a novel transgenic mouse model. J Exp Med. 2008;205:1721–1728. doi: 10.1084/jem.20071463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ma L, Shelness GS, Snipes JA, Murea M, Antinozzi PA, et al. Localization of APOL1 Protein and mRNA in the Human Kidney: Nondiseased Tissue, Primary Cells, and Immortalized Cell Lines. J Am Soc Nephrol. 2015;26:339–348. doi: 10.1681/ASN.2013091017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Human Protein Atlas [Google Scholar]
- 53.Limou S, Dummer P, Nelson G, Kopp J, Winkler C. APOL1 multimer model and kidney function. Kidney Int. (Accepted) [Google Scholar]
- 54.Rathore SS, Ghosh N, Ouyang Y, Shen J. Topological arrangement of the intracellular membrane fusion machinery. Mol Biol Cell. 2011;22:2612–2619. doi: 10.1091/mbc.E11-03-0190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Davidson WS, Silva RA, Chantepie S, Lagor WR, Chapman MJ, et al. Proteomic analysis of defined HDL subpopulations reveals particle-specific protein clusters: relevance to antioxidative function. Arterioscler Thromb Vasc Biol. 2009;29:870–876. doi: 10.1161/ATVBAHA.109.186031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Raper J, Fung R, Ghiso J, Nussenzweig V, Tomlinson S. Characterization of a novel trypanosome lytic factor from human serum. Infect Immun. 1999;67:1910–1916. doi: 10.1128/iai.67.4.1910-1916.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Hajduk SL, Moore DR, Vasudevacharya J, Siqueira H, Torri AF, et al. Lysis of Trypanosoma-Brucei by a Toxic Subspecies of Human High-Density Lipoprotein. Journal of Biological Chemistry. 1989;264:5210–5217. [PubMed] [Google Scholar]
- 58.Pays E, Vanhollebeke B, Uzureau P, Lecordier L, Perez-Morga D. The molecular arms race between African trypanosomes and humans. Nat Rev Microbiol. 2014;12:575–584. doi: 10.1038/nrmicro3298. [DOI] [PubMed] [Google Scholar]
- 59.Capewell P, Cooper A, Clucas C, Weir W, Macleod A. A co-evolutionary arms race: trypanosomes shaping the human genome, humans shaping the trypanosome genome. Parasitology. 2015;142(Suppl 1):S108–S119. doi: 10.1017/S0031182014000602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wheeler RJ. The trypanolytic factor-mechanism, impacts and applications. Trends Parasitol. 2010;26:457–464. doi: 10.1016/j.pt.2010.05.005. [DOI] [PubMed] [Google Scholar]
- 61.Litvinov D, Mahini H, Garelnabi M. Antioxidant and anti-inflammatory role of paraoxonase 1: implication in arteriosclerosis diseases. N Am J Med Sci. 2012;4:523–532. doi: 10.4103/1947-2714.103310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Albert TS, Duchateau PN, Deeb SS, Pullinger CR, Cho MH, et al. Apolipoprotein L-I is positively associated with hyperglycemia and plasma triglycerides in CAD patients with low HDL. J Lipid Res. 2005;46:469–474. doi: 10.1194/jlr.M400304-JLR200. [DOI] [PubMed] [Google Scholar]
- 63.Bruggeman LA, O'Toole JF, Ross MD, Madhavan SM, Smurzynski M, et al. Plasma apolipoprotein L1 levels do not correlate with CKD. J Am Soc Nephrol. 2014;25:634–644. doi: 10.1681/ASN.2013070700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Duchateau PN, Movsesyan I, Yamashita S, Sakai N, Hirano K, et al. Plasma apolipoprotein L concentrations correlate with plasma triglycerides and cholesterol levels in normolipidemic, hyperlipidemic, and diabetic subjects. J Lipid Res. 2000;41:1231–1236. [PubMed] [Google Scholar]
- 65.Bentley AR, Doumatey AP, Chen G, Huang H, Zhou J, et al. Variation in APOL1 Contributes to Ancestry-Level Differences in HDLc-Kidney Function Association. Int J Nephrol. 2012;2012:748984. doi: 10.1155/2012/748984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Freedman BI, Langefeld CD, Murea M, Ma L, Otvos JD, et al. Apolipoprotein L1 nephropathy risk variants associate with HDL subfraction concentration in African Americans. Nephrol Dial Transplant. 2011;26:3805–3810. doi: 10.1093/ndt/gfr542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Monajemi H, Fontijn RD, Pannekoek H, Horrevoets AJ. The apolipoprotein L gene cluster has emerged recently in evolution and is expressed in human vascular tissue. Genomics. 2002;79:539–546. doi: 10.1006/geno.2002.6729. [DOI] [PubMed] [Google Scholar]
- 68.Duchateau PN, Pullinger CR, Cho MH, Eng C, Kane JP. Apolipoprotein L gene family: tissue-specific expression, splicing, promoter regions; discovery of a new gene. J Lipid Res. 2001;42:620–630. [PubMed] [Google Scholar]
- 69.Uhlen M, Fagerberg L, Hallstrom BM, Lindskog C, Oksvold P, et al. Proteomics. Tissue-based map of the human proteome. Science. 2015;347:1260419. doi: 10.1126/science.1260419. [DOI] [PubMed] [Google Scholar]
- 70.Liao W, Goh FY, Betts RJ, Kemeny DM, Tam J, et al. A novel anti-apoptotic role for apolipoprotein L2 in IFN-gamma-induced cytotoxicity in human bronchial epithelial cells. J Cell Physiol. 2011;226:397–406. doi: 10.1002/jcp.22345. [DOI] [PubMed] [Google Scholar]
- 71.Divers J, Nunez M, High KP, Murea M, Rocco MV, et al. JC polyoma virus interacts with APOL1 in African Americans with nondiabetic nephropathy. Kidney Int. 2013;84:1207–1213. doi: 10.1038/ki.2013.173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Mimmack ML, Ryan M, Baba H, Navarro-Ruiz J, Iritani S, et al. Gene expression analysis in schizophrenia: reproducible up-regulation of several members of the apolipoprotein L family located in a high-susceptibility locus for schizophrenia on chromosome 22. Proc Natl Acad Sci U S A. 2002;99:4680–4685. doi: 10.1073/pnas.032069099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Hwang Y, Kim J, Shin JY, Kim JI, Seo JS, et al. Gene expression profiling by mRNA sequencing reveals increased expression of immune/inflammation-related genes in the hippocampus of individuals with schizophrenia. Transl Psychiatry. 2013;3:e321. doi: 10.1038/tp.2013.94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Tomita Y, Ikeda M, Mutoh H, Inada T, Iwata N, et al. Association study between Apolipoprotein L and schizophrenia by exhaustive and rule-based combination analysis for identification of multilocus interactions. J Biosci Bioeng. 2007;103:303–310. doi: 10.1263/jbb.103.303. [DOI] [PubMed] [Google Scholar]
- 75.Venkatasubramanian G, Arasappa R, Christopher R, Gangadhar BN. Neuropharmacology of schizophrenia: is there a role for leptin? Clin Chem Lab Med. 2010;48:895–896. doi: 10.1515/CCLM.2010.158. [DOI] [PubMed] [Google Scholar]
- 76.Carrera N, Arrojo M, Paz E, Ramos-Rios R, Agra S, et al. Testing the antagonistic pleiotropy model of schizophrenia susceptibility by analysis of DAOA, PPP1R1B, and APOL1 genes. Psychiatry Res. 2010;179:126–129. doi: 10.1016/j.psychres.2009.05.014. [DOI] [PubMed] [Google Scholar]
- 77.McGhee KA, Morris DW, Schwaiger S, Nangle JM, Donohoe G, et al. Investigation of the apolipoprotein-L (APOL) gene family and schizophrenia using a novel DNA pooling strategy for public database SNPs. Schizophr Res. 2005;76:231–238. doi: 10.1016/j.schres.2005.01.006. [DOI] [PubMed] [Google Scholar]
- 78.Takahashi S, Cui YH, Han YH, Fagerness JA, Galloway B, et al. Association of SNPs and haplotypes in APOL1–2 and 4 with schizophrenia. Schizophr Res. 2008;104:153–164. doi: 10.1016/j.schres.2008.05.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Sutcliffe JG, Thomas EA. The neurobiology of apolipoproteins in psychiatric disorders. Mol Neurobiol. 2002;26:369–388. doi: 10.1385/mn:26:2-3:369. [DOI] [PubMed] [Google Scholar]
- 80.Rastaldi MP, Armelloni S, Berra S, Calvaresi N, Corbelli A, et al. Glomerular podocytes contain neuron-like functional synaptic vesicles. FASEB J. 2006;20:976–978. doi: 10.1096/fj.05-4962fje. [DOI] [PubMed] [Google Scholar]
- 81.Sistani L, Rodriguez PQ, Hultenby K, Uhlen M, Betsholtz C, et al. Neuronal proteins are novel components of podocyte major processes and their expression in glomerular crescents supports their role in crescent formation. Kidney Int. 2013;83:63–71. doi: 10.1038/ki.2012.321. [DOI] [PubMed] [Google Scholar]
- 82.Kobayashi N. Mechanism of the process formation; podocytes vs. neurons. Microsc Res Tech. 2002;57:217–223. doi: 10.1002/jemt.10077. [DOI] [PubMed] [Google Scholar]
- 83.Freedman BI, Langefeld CD. The new era of APOL1-associated glomerulosclerosis. Nephrol Dial Transplant. 2012;27:1288–1291. doi: 10.1093/ndt/gfr812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Hoy WE, Hughson MD, Kopp JB, Mott SA, Bertram JF, et al. APOL1 risk alleles are associated with exaggerated age-related changes in glomerular number and volume in African American adults: an autopsy study. J Am Soc Nephrol. doi: 10.1681/ASN.2014080768. (in press) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Atta MG, Estrella MM, Kuperman M, Foy MC, Fine DM, et al. HIV-associated nephropathy patients with and without apolipoprotein L1 gene variants have similar clinical and pathological characteristics. Kidney Int. 2012;82:338–343. doi: 10.1038/ki.2012.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Fine DM, Wasser WG, Estrella MM, Atta MG, Kuperman M, et al. APOL1 risk variants predict histopathology and progression to ESRD in HIV-related kidney disease. J Am Soc Nephrol. 2012;23:343–350. doi: 10.1681/ASN.2011060562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Estrella MM, Wyatt CM, Pearce CL, Li M, Shlipak MG, et al. Host APOL1 genotype is independently associated with proteinuria in HIV infection. Kidney Int. 2013;84:834–840. doi: 10.1038/ki.2013.203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Kasembeli AN, Duarte R, Ramsay M, Mosiane P, Dickens C, et al. APOL1 Risk Variants Are Strongly Associated with HIV-Associated Nephropathy in Black South Africans. Journal of the American Society of Nephrology. 2015 doi: 10.1681/ASN.2014050469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Papeta N, Kiryluk K, Patel A, Sterken R, Kacak N, et al. APOL1 variants increase risk for FSGS and HIVAN but not IgA nephropathy. J Am Soc Nephrol. 2011;22:1991–1996. doi: 10.1681/ASN.2011040434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Kopp JB, Winkler CA, Zhao X, Radeva MK, Gassman JJ, et al. Clinical Features and Histology of Apolipoprotein L1-Associated Nephropathy in the FSGS Clinical Trial. J Am Soc Nephrol. 2015 doi: 10.1681/ASN.2013111242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Ashley-Koch AE, Okocha EC, Garrett ME, Soldano K, De Castro LM, et al. MYH9 and APOL1 are both associated with sickle cell disease nephropathy. Br J Haematol. 2011;155:386–394. doi: 10.1111/j.1365-2141.2011.08832.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Lipkowitz MS, Freedman BI, Langefeld CD, Comeau ME, Bowden DW, et al. Apolipoprotein L1 gene variants associate with hypertension-attributed nephropathy and the rate of kidney function decline in African Americans. Kidney Int. 2013;83:114–120. doi: 10.1038/ki.2012.263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Freedman BI, Langefeld CD, Andringa KK, Croker JA, Williams AH, et al. End-stage renal disease in African Americans with lupus nephritis is associated with APOL1. Arthritis Rheumatol. 2014;66:390–396. doi: 10.1002/art.38220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Larsen CP, Beggs ML, Saeed M, Walker PD. Apolipoprotein L1 risk variants associate with systemic lupus erythematosus-associated collapsing glomerulopathy. J Am Soc Nephrol. 2013;24:722–725. doi: 10.1681/ASN.2012121180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Friedman JR, Dibenedetto JR, West M, Rowland AA, Voeltz GK. Endoplasmic reticulum-endosome contact increases as endosomes traffic and mature. Mol Biol Cell. 2013;24:1030–1040. doi: 10.1091/mbc.E12-10-0733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Foster MC, Coresh J, Fornage M, Astor BC, Grams M, et al. APOL1 variants associate with increased risk of CKD among African Americans. J Am Soc Nephrol. 2013;24:1484–1491. doi: 10.1681/ASN.2013010113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Ulasi II, Tzur S, Wasser WG, Shemer R, Kruzel E, et al. High population frequencies of APOL1 risk variants are associated with increased prevalence of non-diabetic chronic kidney disease in the Igbo people from south-eastern Nigeria. Nephron Clin Pract. 2013;123:123–128. doi: 10.1159/000353223. [DOI] [PubMed] [Google Scholar]
- 98.Kanji Z, Powe CE, Wenger JB, Huang C, Ankers E, et al. Genetic variation in APOL1 associates with younger age at hemodialysis initiation. J Am Soc Nephrol. 2011;22:2091–2097. doi: 10.1681/ASN.2010121234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Tzur S, Rosset S, Skorecki K, Wasser WG. APOL1 allelic variants are associated with lower age of dialysis initiation and thereby increased dialysis vintage in African and Hispanic Americans with non-diabetic end-stage kidney disease. Nephrol Dial Transplant. 2012;27:1498–1505. doi: 10.1093/ndt/gfr796. [DOI] [PubMed] [Google Scholar]
- 100.Kofman T, Narjoz C, Raimbourg Q, Loriot MA, Karras A, et al. Collapsing glomerulopathy associated lupus in a black female with homozygous APOL1 mutation. Lupus. 2012;21:1459–1462. doi: 10.1177/0961203312460114. [DOI] [PubMed] [Google Scholar]
- 101.Larsen CP, Beggs ML, Walker PD, Saeed M, Ambruzs JM, et al. Histopathologic effect of APOL1 risk alleles in PLA2R-associated membranous glomerulopathy. Am J Kidney Dis. 2014;64:161–163. doi: 10.1053/j.ajkd.2014.02.024. [DOI] [PubMed] [Google Scholar]
- 102.Canaud G, Dejucq-Rainsford N, Avettand-Fenoel V, Viard JP, Anglicheau D, et al. The kidney as a reservoir for HIV-1 after renal transplantation. J Am Soc Nephrol. 2014;25:407–419. doi: 10.1681/ASN.2013050564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Palmer ND, Ng MC, Hicks PJ, Mudgal P, Langefeld CD, et al. Evaluation of candidate nephropathy susceptibility genes in a genome-wide association study of African American diabetic kidney disease. PLoS One. 2014;9:e88273. doi: 10.1371/journal.pone.0088273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Ito K, Bick AG, Flannick J, Friedman DJ, Genovese G, et al. Increased burden of cardiovascular disease in carriers of APOL1 genetic variants. Circ Res. 2014;114:845–850. doi: 10.1161/CIRCRESAHA.114.302347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Langefeld CD, Divers J, Pajewski NM, Hawfield AT, Reboussin DM, et al. Apolipoprotein L1 gene variants associate with prevalent kidney but not prevalent cardiovascular disease in the Systolic Blood Pressure Intervention Trial. Kidney Int. 2015;87:169–175. doi: 10.1038/ki.2014.254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Parsa A, Kao WH, Xie D, Astor BC, Li M, et al. APOL1 risk variants, race, and progression of chronic kidney disease. N Engl J Med. 2013;369:2183–2196. doi: 10.1056/NEJMoa1310345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Friedman DJ, Kozlitina J, Genovese G, Jog P, Pollak MR. Population-based risk assessment of APOL1 on renal disease. J Am Soc Nephrol. 2011;22:2098–2105. doi: 10.1681/ASN.2011050519. [DOI] [PMC free article] [PubMed] [Google Scholar]