Abstract
To trace evolution of canine parvovirus-2 (CPV-2), a total of 201 stool samples were collected from dogs with diarrhea in Heilongjiang province of northeast China from May 2014 to April 2015. The presence of CPV-2 in the samples was determined by PCR amplification of the VP2 gene (568 bp) of CPV-2. The results revealed that 95 samples (47.26%) were positive for CPV-2, and they showed 98.8%–100% nucleotide identity and 97.6%–100% amino acid identity. Of 95 CPV-2-positive samples, types new2a (Ser297Ala), new2b (Ser297Ala), and 2c accounted for 64.21%, 21.05%, and 14.74%, respectively. The positive rate of CPV-2 and the distribution of the new2a, new2b and 2c types exhibited differences among regions, seasons, and ages. Immunized dogs accounted for 48.42% of 95 CPV-2-positive samples. Coinfections with canine coronavirus, canine kobuvirus, and canine bocavirus were identified. Phylogenetic analysis revealed that the identified new2a, new2b, and CPV-2c strains in our study exhibited a close relationship with most of the CPV-2 strains from China; type new2a strains exhibited high variability, forming three subgroups; type new2b and CPV-2c strains formed one group with reference strains from China. Of 95 CPV-2 strains, Tyr324Ile and Thr440Ala substitutions accounted for 100% and 64.21%, respectively; all type new2b strains exhibited the Thr440Ala substitution, while the unique Gln370Arg substitution was found in all type 2c strains. Recombination analysis using entire VP2 gene indicated possible recombination events between the identified CPV-2 strains and reference strains from China. Our data revealed the co-circulation of new CPV-2a, new CPV-2b, and rare CPV-2c, as well as potential recombination events among Chinese CPV-2 strains.
Introduction
Canine parvovirus-2 (CPV-2) causes a highly contagious and often fatal disease characterized by vomiting and hemorrhagic gastroenteritis in dogs of all ages, and by myocarditis in pups of less than three months of age [1]. The vaccine could provide protective immunity against the classic CPV-2 infection in canine populations, reducing the mortality of animals and spreading of the virus. However, the long-term interaction of the virus and its host may have led to the emergence of CPV-2 antigenic variants. During the past decades, new antigenic types of the CPV-2a, CPV-2b, CPV-2c, new CPV-2a, and new CPV-2b were frequently reported in the dog population, and they replaced the original CPV-2, which has caused widespread concern [2–9]. Therefore, molecular epidemiological investigation of CPV-2 is an undergoing hotspot issue worldwide.
The VP2 protein of CPV-2 is the major capsid protein, and it is highly antigenic, playing important roles in determining viral host ranges and tissue tropisms [10, 11]. At present, VP2 is widely used for the identification and monitoring of the types of CPV-2 circulating in the canine population based on the amino acid residues of the VP2 protein at positions 426Asp/Glu/Asn and 297Ser/Ala [10, 12–15]. In the current study, the molecular epidemiology of the CPV-2 strains circulating in northeast China was investigated by polymerase chain reaction (PCR) targeting the partial VP2 gene. Moreover, potential recombination events of the identified CPV-2 strains were analyzed using the selected entire VP2 gene. Our aim was to provide insights into the epidemiology and genetic diversity of the CPV-2 strains circulating in northeast China, which can be useful for preventing and controlling CPV-2 infections.
Materials and Methods
Ethics statement
All animal experiments were conducted according to the Guide for the Care and Use of Laboratory Animals of Harbin Veterinary Research Institute (HVRI), Chinese Academy of Agricultural Sciences, China. Sampling and data publication also were approved by animal’s owners. The field study did not involve endangered or protected species. No specific permissions were required for locations of samples because the samples were collected from public areas or non-protection areas.
Sampling
Commercial virus sampling tubes (YOCON Biological Technology Co. Ltd., Beijing, China) containing 3.5 mL of Hank’s balanced salt solution, streptomycin, and penicillin, were used to collect fecal samples. A total of 201 fecal samples were collected in the form of rectal swabs from animal hospitals of the Harbin, Daqing, and Mudanjiang districts of Heilongjiang province in northeast China from May 2014 to April 2015. These animals, which were aged between 1 month and 1 year, included animals with symptoms of different degrees of diarrhea, animals with or without a vaccination history, and animals from different breeds and both genders. Of the 201 samples, 141 were collected in Harbin, 20 were collected in Daqing, and 40 were collected in Mudanjiang. All rectal swab samples were stored at −80°C, and they were also used for etiological investigations in our other studies.
DNA extraction
After 1 mL of fecal samples was centrifuged at 1,500 × g for 10 min at 4°C, the supernatant of each sample was transferred to a 1.5 mL EP tube. Genomic DNA was extracted from the supernatant using a commercial TIANamp Stool DNA Kit (Tiangen Biotech Co., Ltd., Beijing, China) according to the manufacturer’s instructions. The extracted genomic DNAs were stored at −40°C. A commercial vaccine was used as a positive control, and distilled water was used as a negative control.
Sequencing of the VP2 fragment
For all samples, 568 bp of the VP2 gene was amplified using the primers CPV-FP (5’−TGATTGTAAACCATGTAGACTAAC−3’) and CPV-RP (5’−TAATGCAGTTAAAGGACCATAAG−3’) as described by Mittal et al. (2014). PCR amplifications were conducted in a 50 μL reaction volume containing 0.1 μM of forward primer, 0.1 μM of reverse primer, 4 μL of genomic DNA, 25μL of EmeraldAmp PCR Master Mix (2 × Premix) (TaKaRa Biotechnology Co. Ltd., Dalian, China), and an appropriate volume of double-distilled (dd) H2O. The mixtures were amplified by 35 cycles of 94°C for 30 s, 55°C for 30 s, and 72°C for 1 min, with a final extension at 72°C for 10 min using an automated Applied Biosystems GeneAmp PCR System 9700 thermal cycler (Thermo Fisher Scientific, Waltham, MA, USA). After the amplification, products were purified using the AxyPrep DNA Gel Extraction kit (A Corning Brand, Suzhou, China), and they were directly subjected to Sanger sequencing. Each sample was sequenced three times. Searches for nucleotide similarities were conducted using the BLAST algorithm at the National Center for Biotechnology Information (NCBI) (http://www.ncbi.nlm.nih.gov/blast). Sequence analysis was performed using the EditSeq program in the Lasergene DNASTAR version 5.06 software (DNASTAR Inc., Madison, WI, USA). Multiple sequence alignments were performed using the Multiple Sequence Alignment program of the DNAMAN version 6.0 software (Lynnon BioSoft, Point-Claire, Quebec, Canada). A total of 95 sequences (507 bp in length) were submitted to the NCBI Genbank (http://www.ncbi.nlm.nih.gov) under the accession numbers KT074258–KT074352.
Phylogenetic analysis
For the phylogenetic analysis, the VP2 gene sequences of CPV-2, CPV-2a, CPV-2b, CPV-2c, new CPV-2a, and new CPV-2b strains from different geographical locations within China and the rest of the world were retrieved from the NCBI nucleotide database. A nucleotide phylogenetic tree of the VP2 fragments (507 bp) of CPV-2 was generated from the ClustalX-generated alignments by MEGA6.06 software using the neighbor-joining method [16]. Neighbor-joining phylogenetic tree was built with the p-distance model and 1000 replicates of interior-branch test; otherwise, the default parameters in MEGA 6 were used. The support of reliability (above 50%) was shown in the obtained phylogenetic tree.
Sequencing of the entire selected VP2 genes
A total of 13 entire VP2 genes identified in our study, representing different genetic diversity in the phylogenetic tree of the 507 bp VP2 fragments of CPV-2, were chosen for recombination analysis. The entire VP2 gene of CPV-2 was amplified using the primers CPVvp2-F (5'-AACTAAAAGAAGTAAACCAC-3') and CPVvp2-R (5'-TGTAATAAACATAAAAACAT-3'), which resulted in a 1882 bp fragment covering the open reading frame of the VP2 gene. PCR amplifications were conducted in a 50 μL reaction volume containing 0.1 μM of forward primer, 0.1 μM of reverse primer, 4 μL of genomic DNA, 5 μL of 10 × buffer, 0.5 U of ExTaq DNA polymerase (TaKaRa), and an appropriate volume of ddH2O. The mixtures were amplified by 35 cycles of 94°C for 1 min, 50°C for 1 min, and 72°C for 2 min, with a final extension at 72°C for 10 min using an automated Applied Biosystems GeneAmp PCR System 9700 thermal cycler. The VP2 PCR products were purified using the AxyPrep DNA Gel Extraction kit (Corning), and then purified products were directly subjected to Sanger sequencing. Each sample was sequenced three times. All sequences were submitted to the NCBI Genbank (http://www.ncbi.nlm.nih.gov) under the accession numbers KT156825–KT156837.
Recombination analysis
Thirteen entire VP2 sequences in this study and 25 VP2 gene reference sequences from different areas in China were used for the recombination analysis (Table 1). The 38 VP2 genes were aligned in the ClustalX (1.83) program, and then were screened for possible recombination by using the Recombination Detection Program (RDP) [17], GENECONV [18], BOOTSCAN [19], MaxChi [20], CHIMAERA [21], and SISCAN [22] methods embedded in RDP4. Only potential recombination events detected by two or more of the programs, coupled with phylogenetic evidence of recombination, were considered significant using the highest acceptable P-value cutoff of 0.05.
Table 1. List of the entire VP2 gene for recombination analysis.
| No. | Strains | Accession no. | Genotyping | Location in China | Year |
|---|---|---|---|---|---|
| 1 | MDJ-1 | KT156825 | New2a | Northeast (Mudanjiang) | 2014 |
| 2 | MDJ-7 | KT156826 | New2a | Northeast (Mudanjiang) | 2014 |
| 3 | MDJ-9 | KT156827 | New2a | Northeast (Mudanjiang) | 2014 |
| 4 | MDJ-15 | KT156828 | New2a | Northeast (Mudanjiang) | 2014 |
| 5 | MDJ-20 | KT156829 | New2a | Northeast (Mudanjiang) | 2015 |
| 6 | MDJ-22 | KT156830 | New2a | Northeast (Mudanjiang) | 2015 |
| 7 | HRB-A4 | KT156831 | New2b | Northeast (Harbin) | 2014 |
| 8 | HRB-A6 | KT156832 | New2c | Northeast (Harbin) | 2014 |
| 9 | HRB-b2 | KT156833 | New2b | Northeast (Harbin) | 2014 |
| 10 | HRB-e2 | KT156834 | New2a | Northeast (Harbin) | 2015 |
| 11 | HRB-ee7 | KT156835 | New2a | Northeast (Harbin) | 2014 |
| 12 | HRB-F8 | KT156836 | New2a | Northeast (Harbin) | 2014 |
| 13 | HRB-J8 | KT156837 | New2b | Northeast (Harbin) | 2014 |
| 14 | bj-5 | GQ169549 | New2a | North (Beijing) | 2010 |
| 15 | Wu | KJ674820 | New2a | South-central (Wuhan) | 2013 |
| 16 | CPV/WH02/06 | EU377537 | New2a | South-central (Wuhan) | 2006 |
| 17 | 06–11-NJ | FJ432716 | New2a | East (Nanjing) | 2006 |
| 18 | 08–5-WH | FJ432717 | New2a | South-central (Wuhan) | 2008 |
| 19 | Wh-3 | GQ169539 | New2a | South-central (Wuhan) | 2009 |
| 20 | 1-nj | GQ169550 | New2a | East (Nanjing) | 2009 |
| 21 | 09/09 | GU452713 | New2a | NA | 2009 |
| 22 | 11/09 | GU452715 | New2a | NA | 2009 |
| 23 | CPV-6(10) | JQ743904 | New2a | NA | 2010 |
| 24 | CPVSH-3/2011 | JX121627 | New2a | East (Shanghai) | 2011 |
| 25 | GZ0202 | GU569937 | New2b | Southwest (Guizhou) | 2002 |
| 26 | YN0201 | GU569938 | New2b | Southwest (Yunnan) | 2002 |
| 27 | YN0203 | GU569940 | New2b | Southwest (Yunnan) | 2002 |
| 28 | GZ0201 | GU569944 | New2b | Southwest (Guizhou) | 2002 |
| 29 | CPV-4(10) | JQ743890 | New2b | NA | 2010 |
| 30 | CPV-10(10) | JQ743891 | New2b | NA | 2010 |
| 31 | CPV-BG(11) | JQ743892 | New2b | NA | 2011 |
| 32 | CPV-HS(11) | JQ743893 | New2b | NA | 2011 |
| 33 | CPV-BM(11) | JQ743894 | New2b | NA | 2011 |
| 34 | CPV-2b-wuhan2 | KC881278 | New2b | South-central (Wuhan) | 2010 |
| 35 | 06/09 | GU380303 | 2c | NA | 2009 |
| 36 | 08/09 | GU380305 | 2c | NA | 2009 |
| 37 | G1 | KF482468 | 2c | South | 2009 |
| 38 | G15 | KF482471 | 2c | South | 2009 |
Screening for canine enteric pathogens
Fecal samples that tested positive for CPV-2 were also screened for canine bocavirus (CBoV), canine coronavirus (CCoV), canine astrovirus (CaAstV), canine norovirus (CNoV), canine kobuvirus (CaKV), and group A-rotavirus (RV-A), by either PCR or reverse transcription-PCR (RT-PCR) and sequencing, as previously described [23–28].
Results
Investigation and genotyping of CPV-2
A total of 201 samples from the Harbin, Daqing, and Mudanjiang districts of Heilongjiang province in northeast China were detected via PCR amplification of 568 bp of the VP2 gene of CPV-2. Nucleotide sequences of the partial VP2 of CPV-2 strains identified in our study were shown in Supporting Information (S1 Fig). The characteristics of the CPV-2-positive dogs, the genotypes of CPV-2 strains, and the amino acid substitutions of the VP2 protein are shown in Table 2, and a further analysis of the CPV-2-positive samples is shown in Table 3. Of the 201 samples, 47.26% (95/201) were positive for CPV-2, and three types, new2a (Ser297Ala), new2b (Ser297Ala), and 2c, were found, accounting for 64.21%, 21.05%, and 14.74%, respectively. The new2a type was identified in Harbin, Daqing, and Mudanjiang; the new2b type was identified in Harbin and Daqing; and the 2c type was identified in Harbin and Mudanjiang.
Table 2. Characteristics of the CPV-2 positive dogs, the genotypes of CPV-2 strains, and amino acid substitution of the VP2 protein in northeast China.
| No. | Strain | Accession No. | Collection date | Location | Breed | Gender | Age | Vaccine | Other enteric viruses (Accession No.) | CPV type | Substitution of amino acid residues in VP2 protein of CPV | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 297 | 324 | 370 | 419 | 426 | 440 | ||||||||||||
| Reference strains | 1 | CPV-b | M38245 | 1996 | USA | NA | NA | NA | NA | NA | 2 | Ser | Tyr | Gln | Asn | Asn | Thr |
| 2 | YB8301 | GU569943 | 1983 | China | NA | NA | NA | NA | NA | 2 | Ser | Try | Gln | Asn | Asn | Thr | |
| 3 | CPV-15 | M24003 | 1984 | USA | NA | NA | NA | NA | NA | 2a | Ser | Tyr | Gln | Asn | Asn | Thr | |
| 4 | CC8601 | GU569948 | 1986 | China | NA | NA | NA | NA | NA | 2a | Ser | Tyr | Gln | Asn | Asn | Thr | |
| 5 | CPV-39 | M74849 | 1984 | USA | NA | NA | NA | NA | NA | 2b | Ser | Tyr | Gln | Asn | Asp | Thr | |
| 6 | 56/00 | FJ222821 | 2009 | Italy | NA | NA | NA | NA | NA | 2c | Ala | Tyr | Gln | Asn | Glu | Thr | |
| 7 | 06/09 | GU380303 | 2009 | China | NA | NA | NA | NA | NA | 2c | Ala | Ile | Gln | Asn | Glu | Thr | |
| 8 | G1 | KF482468 | 2009 | China | NA | NA | NA | NA | NA | 2c | Ala | Tyr | Gln | Asn | Glu | Thr | |
| 9 | CPV-435 | AY742953 | 2005 | USA | NA | NA | NA | NA | NA | New2a | Ala | Tyr | Gln | Asn | Asn | Thr | |
| 10 | Wh-6 | GQ169542 | 2009 | China | NA | NA | NA | NA | NA | New2a | Ala | Ile | Gln | Asn | Asn | Thr | |
| 11 | CPVSH-2/2011 | JX121626 | 2011 | China | NA | NA | NA | NA | NA | New2a | Ala | Ile | Gln | Asn | Asn | Ala | |
| 12 | CPV-436 | AY742955 | 2005 | USA | NA | NA | NA | NA | NA | New2b | Ala | Tyr | Gln | Asn | Asp | Thr | |
| 13 | GZ0201 | GU569944 | 2002 | China | NA | NA | NA | NA | NA | New2b | Ala | Tyr | Gln | Asn | Asp | Thr | |
| 14 | CPV-2b-wuhan2 | KC881278 | 2010 | China | NA | NA | NA | NA | NA | New2b | Ala | Ile | Gln | Asn | Asp | Ala | |
| Strains in this study | 1 | MDJ-1 | KT074258 | Dec-2014 | MDJ | SH | F | 2M | Yes | CCoV (KT192642) | New2a | Ala | Ile | Gln | Asn | Asn | Ala |
| 2 | MDJ-22 | KT074265 | Oct-2014 | MDJ | GM | F | 6M | Yes | — | New2a | Ala | Ile | Gln | Asn | Asn | Ala | |
| 3 | MDJ-28 | KT074267 | Dec-2014 | MDJ | JS | M | 3M | Yes | — | New2a | Ala | Ile | Gln | Asn | Asn | Ala | |
| 4 | MDJ-29 | KT074268 | May-2015 | MDJ | MX | M | 5M | No | — | New2a | Ala | Ile | Gln | Asn | Asn | Ala | |
| 5 | MDJ-32 | KT074269 | Jan-2015 | MDJ | NA | NA | NA | Yes | — | New2a | Ala | Ile | Gln | Asn | Asn | Ala | |
| 6 | MDJ-40 | KT074272 | May-2015 | MDJ | PD | F | NA | Yes | — | New2a | Ala | Ile | Gln | Asn | Asn | Ala | |
| 7 | DQ-alpha1 | KT074273 | Dec-2014 | DQ | NA | F | 3M | No | CCoV (KT192660) | New2a | Ala | Ile | Gln | Asn | Asn | Ala | |
| 8 | DQ-beta1 | KT074274 | May-2015 | DQ | PD | NA | 3M | Yes | — | New2a | Ala | Ile | Gln | Asn | Asn | Ala | |
| 9 | HRB-A5 | KT074278 | Sep-2014 | HRB | MX | M | NA | NA | — | New2a | Ala | Ile | Gln | Asn | Asn | Ala | |
| 10 | HRB-A8 | KT074280 | Oct-2014 | HRB | BF | M | 4M | NA | CCoV (KT192655), CaKV (KT210398) | New2a | Ala | Ile | Gln | Asn | Asn | Ala | |
| 11 | HRB-B0 | KT074279 | Oct-2014 | HRB | PM | F | 3M | Yes | CCoV (KT192665), CaKV (KT210404) | New2a | Ala | Ile | Gln | Asn | Asn | Ala | |
| 12 | HRB-B7 | KT074283 | Oct-2014 | HRB | MI | F | 3M | Yes | CCoV (KT192668) | New2a | Ala | Ile | Gln | Asn | Asn | Ala | |
| 13 | HRB-b9 | KT074318 | Sep-2014 | HRB | NA | M | 3M | Yes | — | New2a | Ala | Ile | Gln | Asn | Asn | Ala | |
| 14 | HRB-C2 | KT074285 | Oct-2014 | HRB | TM | F | NA | No | CCoV (KT192678), CaKV (KT210410) | New2a | Ala | Ile | Gln | Asn | Asn | Ala | |
| 15 | HRB-C5 | KT074286 | Oct-2014 | HRB | YT | M | NA | No | — | New2a | Ala | Ile | Gln | Asn | Asn | Ala | |
| 16 | HRB-C6 | KT074287 | Oct-2014 | HRB | BF | F | 4M | Yes | — | New2a | Ala | Ile | Gln | Asn | Asn | Ala | |
| 17 | HRB-C9 | KT074289 | Oct-2014 | HRB | MX | F | 3M | Yes | — | New2a | Ala | Ile | Gln | Asn | Asn | Ala | |
| 18 | HRB-b5 | KT074290 | Oct-2014 | HRB | NA | NA | 3M | No | — | New2a | Ala | Ile | Gln | Asn | Asn | Ala | |
| 19 | HRB-bb7 | KT074281 | Oct-2014 | HRB | GM | NA | 2M | NA | CCoV (KT192671), | New2a | Ala | Ile | Gln | Asn | Asn | Ala | |
| 20 | HRB-bb8 | KT074284 | Oct-2014 | HRB | SM | F | 2M | No | CCoV (KT192672), CaKV (KT210406) | New2a | Ala | Ile | Gln | Asn | Asn | Ala | |
| 21 | HRB-dd2 | KT074291 | Nov-2014 | HRB | BD | M | 5M | NA | — | New2a | Ala | Ile | Gln | Asn | Asn | Ala | |
| 22 | HRB-D4 | KT074294 | Oct-2014 | HRB | YT | F | 4M | NA | CCoV (KT192683) | New2a | Ala | Ile | Gln | Asn | Asn | Ala | |
| 23 | HRB-D8 | KT074295 | Nov-2014 | HRB | YT | M | 3.5M | No | — | New2a | Ala | Ile | Gln | Asn | Asn | Ala | |
| 24 | HRB-D9 | KT074296 | Nov-2014 | HRB | NA | NA | 3.5M | Yes | CaKV (KT210414) | New2a | Ala | Ile | Gln | Asn | Asn | Ala | |
| 25 | HRB-E0 | KT074297 | Nov-2014 | HRB | NA | F | 3.5M | No | — | New2a | Ala | Ile | Gln | Asn | Asn | Ala | |
| 26 | HRB-E1 | KT074298 | Nov-2014 | HRB | NA | M | 3.5M | Yes | CCoV (KT192686) | New2a | Ala | Ile | Gln | Asn | Asn | Ala | |
| 27 | HRB-e2 | KT074299 | Apr-2015 | HRB | NA | M | 3M | No | New2a | Ala | Ile | Gln | Asn | Asn | Ala | ||
| 28 | HRB-E8 | KT074300 | Nov-2014 | HRB | GM | M | 3M | Yes | CCoV (KT192687), CaKV (KT210415) | New2a | Ala | Ile | Gln | Asn | Asn | Ala | |
| 29 | HRB-F0 | KT074301 | Feb-2015 | HRB | SM | F | 4M | Yes | — | New2a | Ala | Ile | Gln | Asn | Asn | Ala | |
| 30 | HRB-F4 | KT074302 | Nov-2014 | HRB | NA | M | NA | No | — | New2a | Ala | Ile | Gln | Asn | Asn | Ala | |
| 31 | HRB-G4 | KT074305 | Mar-2015 | HRB | YT | F | 4M | Yes | — | New2a | Ala | Ile | Gln | Asn | Asn | Ala | |
| 32 | HRB-H2 | KT074307 | Dec-2014 | HRB | PD | F | 3M | NA | CaKV (KT210423) | New2a | Ala | Ile | Gln | Asn | Asn | Ala | |
| 33 | HRB-I1 | KT074308 | Sep-2014 | HRB | NA | F | 4M | NA | CCoV (KT192697) | New2a | Ala | Ile | Gln | Asn | Asn | Ala | |
| 34 | HRB-I3 | KT074309 | Dec-2014 | HRB | NA | F | 3M | Yes | New2a | Ala | Ile | Gln | Asn | Asn | Ala | ||
| 35 | HRB-I4 | KT074310 | Sep-2014 | HRB | SN | M | 3.5M | NA | — | New2a | Ala | Ile | Gln | Asn | Asn | Ala | |
| 36 | HRB-I8 | KT074312 | Feb-2015 | HRB | MI | M | 3M | No | — | New2a | Ala | Ile | Gln | Asn | Asn | Ala | |
| 37 | HRB-J1 | KT074313 | Sep-2014 | HRB | NA | M | 3M | Yes | — | New2a | Ala | Ile | Gln | Asn | Asn | Ala | |
| 38 | HRB-K1 | KT074316 | Mar-2015 | HRB | NA | M | 3.5M | Yes | — | New2a | Ala | Ile | Gln | Asn | Asn | Ala | |
| 39 | HRB-K3 | KT074317 | Jan-2015 | HRB | NA | F | 3M | Yes | CaKV (KT210425) | New2a | Ala | Ile | Gln | Ser | Asn | Ala | |
| 40 | HRB-G0 | KT074304 | Dec-2014 | HRB | PK | M | 4M | Yes | — | New2a | Ala | Ile | Gln | Ser | Asn | Ala | |
| 41 | HRB-F8 | KT074303 | Nov-2014 | HRB | PD | M | 3M | No | CCoV (KT192691), CaKV (KT210418) | New2a | Ala | Ile | Gln | Ser | Asn | Ala | |
| 42 | MDJ-7 | KT074259 | Nov-2014 | MDJ | NA | M | 6M | Yes | — | New2a | Ala | Ile | Gln | Asn | Asn | Thr | |
| 43 | MDJ-9 | KT074260 | Dec-2014 | MDJ | RC | M | 3M | No | — | New2a | Ala | Ile | Gln | Asn | Asn | Thr | |
| 44 | MDJ-15 | KT074261 | Nov-2014 | MDJ | RC | F | 1.5M | Yes | CBoV (KR998482) | New2a | Ala | Ile | Gln | Asn | Asn | Thr | |
| 45 | MDJ-18 | KT074262 | Mar-2015 | MDJ | MX | M | 3M | Yes | — | New2a | Ala | Ile | Gln | Asn | Asn | Thr | |
| 46 | MDJ-20 | KT074263 | Mar-2015 | MDJ | NA | F | 3M | Yes | — | New2a | Ala | Ile | Gln | Asn | Asn | Thr | |
| 47 | MDJ-21 | KT074264 | Dec-2014 | MDJ | RC | M | 3M | No | CBoV (KR998484) | New2a | Ala | Ile | Gln | Asn | Asn | Thr | |
| 48 | MDJ-27 | KT074266 | Apr-2015 | MDJ | NA | F | NA | NA | CBoV (KR998487), CaKV (KT210392) | New2a | Ala | Ile | Gln | Asn | Asn | Thr | |
| 49 | MDJ-33 | KT074270 | Jan-2015 | MDJ | MX | NA | 3M | No | — | New2a | Ala | Ile | Gln | Asn | Asn | Thr | |
| 50 | MDJ-37 | KT074271 | Apr-2015 | MDJ | NA | M | NA | Yes | — | New2a | Ala | Ile | Gln | Asn | Asn | Thr | |
| 51 | DQ-beta7 | KT074275 | Feb-2015 | DQ | NA | M | 1M | No | CBoV (KR998491) | New2a | Ala | Ile | Gln | Asn | Asn | Thr | |
| 52 | HRB-A2 | KT074277 | Sep-2014 | HRB | CH | M | 4M | No | — | New2a | Ala | Ile | Gln | Asn | Asn | Thr | |
| 53 | HRB-B8 | KT074282 | Oct-2014 | HRB | GM | NA | NA | NA | — | New2a | Ala | Ile | Gln | Asn | Asn | Thr | |
| 54 | HRB-C7 | KT074288 | Oct-2014 | HRB | NA | M | 2M | NA | CBoV (KR998493) | New2a | Ala | Ile | Gln | Asn | Asn | Thr | |
| 55 | HRB-a1 | KT074276 | Sep-2014 | HRB | NA | F | 3.5M | Yes | CCoV (KT192656), CBoV (KR998488), CaKV (KT210394) | New2a | Ala | Ile | Gln | Asn | Asn | Thr | |
| 56 | HRB-ee7 | KT074292 | Dec-2014 | HRB | NA | M | 3.5M | Yes | — | New2a | Ala | Ile | Gln | Asn | Asn | Thr | |
| 57 | HRB-D2 | KT074293 | Oct-2014 | HRB | PD | F | 2M | No | — | New2a | Ala | Ile | Gln | Asn | Asn | Thr | |
| 58 | HRB-G6 | KT074306 | Dec-2014 | HRB | GM | F | 3.5M | Yes | — | New2a | Ala | Ile | Gln | Asn | Asn | Thr | |
| 59 | HRB-I7 | KT074311 | Nov-2014 | HRB | NA | F | NA | Yes | — | New2a | Ala | Ile | Gln | Asn | Asn | Thr | |
| 60 | HRB-J4 | KT074314 | Oct-2014 | HRB | NA | M | 4M | No | — | New2a | Ala | Ile | Gln | Asn | Asn | Thr | |
| 61 | HRB-J7 | KT074315 | Nov-2014 | HRB | NA | F | 3.5M | Yes | — | New2a | Ala | Ile | Gln | Asn | Asn | Thr | |
| 62 | DQ-beta0 | KT074336 | Dec-2014 | DQ | PM | M | 1.5M | No | CCoV (KT192673) | New2b | Ala | Ile | Gln | Asn | Asp | Ala | |
| 63 | DQ-beta4 | KT074337 | Dec-2014 | DQ | SN | F | 5M | Yes | — | New2b | Ala | Ile | Gln | Asn | Asp | Ala | |
| 64 | DQ-beta9 | KT074338 | Mar-2015 | DQ | CO | M | NA | Yes | — | New2b | Ala | Ile | Gln | Asn | Asp | Ala | |
| 65 | HRB-A4 | KT074319 | Sep-2014 | HRB | NA | F | 3M | NA | CCoV (KT192652), CaKV (KT210396) | New2b | Ala | Ile | Gln | Asn | Asp | Ala | |
| 66 | HRB-A9 | KT074320 | Sep-2014 | HRB | PM | NA | 3M | NA | — | New2b | Ala | Ile | Gln | Asn | Asp | Ala | |
| 67 | HRB-B6 | KT074321 | Oct-2014 | HRB | CH | F | 2M | No | — | New2b | Ala | Ile | Gln | Asn | Asp | Ala | |
| 68 | HRB-aa8 | KT074322 | Oct-2014 | HRB | BF | M | 4M | NA | — | New2b | Ala | Ile | Gln | Asn | Asp | Ala | |
| 69 | HRB-b2 | KT074323 | Oct-2014 | HRB | GM | NA | 3M | NA | CBoV (KR998490) | New2b | Ala | Ile | Gln | Asn | Asp | Ala | |
| 70 | HRB-d1 | KT074324 | Nov-2014 | HRB | LR | F | 7M | Yes | — | New2b | Ala | Ile | Gln | Asn | Asp | Ala | |
| 71 | HRB-d3 | KT074325 | Oct-2014 | HRB | RC | NA | NA | NA | — | New2b | Ala | Ile | Gln | Asn | Asp | Ala | |
| 72 | HRB-d5 | KT074326 | Sep-2014 | HRB | NA | M | 3M | No | — | New2b | Ala | Ile | Gln | Asn | Asp | Ala | |
| 73 | HRB-E7 | KT074327 | Nov-2014 | HRB | SM | F | 3M | Yes | — | New2b | Ala | Ile | Gln | Asn | Asp | Ala | |
| 74 | HRB-F5 | KT074328 | Nov-2014 | HRB | YT | F | 4M | Yes | CCoV (KT192690) | New2b | Ala | Ile | Gln | Asn | Asp | Ala | |
| 75 | HRB-F6 | KT074329 | Nov-2014 | HRB | NA | M | 3M | No | — | New2b | Ala | Ile | Gln | Asn | Asp | Ala | |
| 76 | HRB-F7 | KT074330 | Nov-2014 | HRB | MI | F | 3M | Yes | — | New2b | Ala | Ile | Gln | Asn | Asp | Ala | |
| 77 | HRB-H3 | KT074331 | Jan-2015 | HRB | SM | M | 3M | No | CCoV (KT192693) | New2b | Ala | Ile | Gln | Asn | Asp | Ala | |
| 78 | HRB-H4 | KT074332 | Dec-2014 | HRB | NA | F | 3.5M | No | — | New2b | Ala | Ile | Gln | Asn | Asp | Ala | |
| 79 | HRB-H8 | KT074333 | Dec-2014 | HRB | BF | M | 3.5M | No | New2b | Ala | Ile | Gln | Asn | Asp | Ala | ||
| 80 | HRB-I5 | KT074334 | Oct-2014 | HRB | BD | M | 3.5M | Yes | — | New2b | Ala | Ile | Gln | Asn | Asp | Ala | |
| 81 | HRB-J8 | KT074335 | Dec-2014 | HRB | SM | NA | 3.5M | Yes | — | New2b | Ala | Ile | Gln | Asn | Asp | Ala | |
| 82 | MDJ-4 | KT074348 | Apr-2015 | MDJ | NA | M | 4M | No | — | 2c | Ala | Ile | Arg | Asn | Glu | Thr | |
| 83 | MDJ-6 | KT074349 | Dec-2014 | MDJ | NA | F | NA | Yes | — | 2c | Ala | Ile | Arg | Asn | Glu | Thr | |
| 84 | MDJ-11 | KT074350 | Oct-2014 | MDJ | PD | F | 4M | No | CCoV (KT192644) | 2c | Ala | Ile | Arg | Asn | Glu | Thr | |
| 85 | MDJ-24 | KT074351 | Nov-2014 | MDJ | RC | F | 1.5M | Yes | — | 2c | Ala | Ile | Arg | Asn | Glu | Thr | |
| 86 | MDJ-39 | KT074352 | Mar-2015 | MDJ | NA | F | 3M | No | — | 2c | Ala | Ile | Arg | Asn | Glu | Thr | |
| 87 | HRB-A6 | KT074339 | Sep-2014 | HRB | NA | F | 3M | Yes | CCoV (KT192653) | 2c | Ala | Ile | Arg | Asn | Glu | Thr | |
| 88 | HRB-aa9 | KT074340 | Oct-2014 | HRB | CH | M | 3.5M | No | 2c | Ala | Ile | Arg | Asn | Glu | Thr | ||
| 89 | HRB-b3 | KT074341 | Oct-2014 | HRB | PM | NA | 2M | No | CCoV (KT192670), CaKV (KT210409) | 2c | Ala | Ile | Arg | Asn | Glu | Thr | |
| 90 | HRB-E5 | KT074342 | Nov-2014 | HRB | NA | M | 3M | Yes | — | 2c | Ala | Ile | Arg | Asn | Glu | Thr | |
| 91 | HRB-e6 | KT074343 | Mar-2015 | HRB | PD | F | 3.5M | No | — | 2c | Ala | Ile | Arg | Asn | Glu | Thr | |
| 92 | HRB-E9 | KT074344 | Nov-2014 | HRB | CH | NA | NA | Yes | — | 2c | Ala | Ile | Arg | Asn | Glu | Thr | |
| 93 | HRB-F9 | KT074345 | Feb-2015 | HRB | NA | M | NA | NA | — | 2c | Ala | Ile | Arg | Asn | Glu | Thr | |
| 94 | HRB-G8 | KT074346 | Nov-2014 | HRB | PM | NA | 4M | Yes | CaKV (KT210421) | 2c | Ala | Ile | Arg | Asn | Glu | Thr | |
| 95 | HRB-J6 | KT074347 | Nov-2014 | HRB | PD | NA | NA | Yes | — | 2c | Ala | Ile | Arg | Asn | Glu | Thr | |
Note. For breed, GM = Golden Malinois, LR = Labrador Retriever, RC = Rough Collie, CO = Caucasian Owtcharka, TM = Tibetan Mastiff, PD = Poodle, CH = Chihuahua, and JS = Japanese Spitz; For gender, F = female, and M = male; for age, M = month; for location, MDJ = Mudanjiang, HRB = Harbin, DQ = Daqing.
Table 3. Further analysis of the CPV-2 positive samples.
| Harbin | Daqing | Mudanjiang | Total | ||||
|---|---|---|---|---|---|---|---|
| Numbers of sample | 141 | 20 | 40 | 201 | |||
| Positive rate for CPV-2 | 48.94% (69/141) | 30% (6/20) | 50% (20/40) | 47.26% (95/201) | |||
| Genotyping of CPV-2 | New2a | 62.32% (43/69) | 50% (3/6) | 75% (15/20) | 64.21% (61/95) | ||
| New2b | 24.64% (17/69) | 50% (3/6) | — | 21.05% (20/95) | |||
| 2c | 13.04% (9/69) | — | 25% (5/20) | 14.74% (14/95) | |||
| Substitution of amino acid residues in VP2 protein of CPV-2 | 297Ser/Ala | Ser | — | — | — | — | |
| Ala | 100% (69/69) | 100% (6/6) | 100% (20/20) | 100% (95/95) | |||
| 324Tyr/Ile | Tyr | — | — | — | — | ||
| Ile | 100% (69/69) | 100% (6/6) | 100% (20/20) | 100% (95/95) | |||
| 370Gln/Arg | Gln | 86.96% (60/69) | 100% (6/6) | 75% (15/20) | 85.26% (81/95) | ||
| Arg | 13.04% (9/69) | — | 25% (5/20) | 14.74% (14/95) | |||
| 419Asn/Ser | Asn | 95.65% (66/69) | 100% (6/6) | 100% (20/20) | 96.84% (92/95) | ||
| Ser | 4.35% (3/69) | — | — | 3.16% (3/95) | |||
| 440Thr/Ala | Thr | 27.54% (19/69) | 16.67% (1/6) | 70% (14/20) | 35.79% (34/95) | ||
| Ala | 72.46% (50/69) | 83.33% (5/6) | 30% (6/20) | 64.21% (61/95) | |||
| Vaccined | Yes | 44.93% (31/69) | 50% (3/6) | 60% (12/20) | 48.42% (46/95) | ||
| No | 30.43% (21/69) | 50% (3/6) | 35% (7/20) | 32.63% (31/95) | |||
| Collection data | Jan. to Mar. | 11.59% (8/69) | 33.33% (2/6) | 25% (5/20) | 15.78% (15/95) | ||
| Apr. to Jun. | 1.45% (1/69) | 16.67% (1/6) | 25% (5/20) | 7.37% (7/95) | |||
| Jul. to Sep. | 15.94% (11/69) | — | — | 11.58% (11/95) | |||
| Oct. to Dec. | 71.01% (49/69) | 50% (3/6) | 50% (10/20) | 65.26% (62/95) | |||
| Age | 0< Age ≤2M | 8.70% (6/69) | 33.33% (2/6) | 15% (3/20) | 11.58% (11/95) | ||
| 2M< Age ≤4M | 73.91% (51/69) | 33.33% (2/6) | 45% (9/20) | 65.26% (62/95) | |||
| 4M< Age ≤6M | 1.45% (1/69) | 16.67% (1/6) | 15% (3/20) | 5.26% (5/95) | |||
| >6M | 1.45% (1/69) | — | — | 1.05% (1/95) | |||
| Other enteric viruses in the CPV positive samples | CCoV | 24.64% (17/69) | 33.33% (2/6) | 10% (2/20) | 22.11% (21/95) | ||
| CaKV | 18.84% (13/69) | — | 5% (1/20) | 14.74% (14/95) | |||
| CBoV | 4.35% (3/69) | 16.67% (1/6) | 15% (3/20) | 7.37% (7/95) | |||
| CCoV+CaKV | 13.04% (9/69) | — | 5% (1/20) | 10.53% (10/95) | |||
| CBoV+CaKV | 1.45% (1/69) | — | 5% (1/20) | 2.11% (2/95) | |||
| CCoV+CBoV+CaKV | 1.45% (1/69) | — | — | 1.05% (1/95) | |||
| Identity | Nucleotides | New2a | 99.2%–100% | 99.2%–100% | 99.2%–100% | 99.0%–100% | |
| New2b | 99.6%–100% | 100% | — | 99.8%–100% | |||
| 2c | 100% | — | 100% | 100% | |||
| New2a+ New2b+2c | 98.8%–100% | 99.0%–100% | 98.6%–100% | 98.8%–100% | |||
| Amino acids | New2a | 98.8%–100% | 99.4%–100% | 99.4%–100% | 98.8%–100% | ||
| New2b | 100% | 100% | — | 100% | |||
| 2c | 100% | — | 100% | 100% | |||
| New2a+ New2b+2c | 97.6%–100% | 98.8%–100% | 98.2%–100% | 97.6%–100% | |||
Sequence comparisons revealed 99.0%–100%, 99.8%–100%, and 100% nucleotide identities within the new2a, new2b, and 2c strains, respectively, and 98.8%–100% nucleotide identities between the new2a, new2b, and 2c types, respectively. At the amino acid level, 98.8%–100%, 100%, 100%, 97.6%–100% identities were revealed within the new2a, new2b, and 2c strains, and between the new2a, new2b, and 2c types, respectively. A total of five substitutions of amino acid residues occurred in all 95 CPV-2-positive samples based on an analysis of partial VP2 sequences. Residue 324Ile was present in all 95 CPV-2-positive samples when compared with the reference strains, and residue 370Arg was specific for type CPV-2c when compared with CPV-2c reference strains. Only three new CPV-2a strains, HRB-K3, HRB-G0, and HRB-F8, showed amino acids substitutions (Asn→Ser) at position 419. At position 440, all new2b strains and most of the new2a strains identified in our study showed amino acids substitutions (Thr→Ala) when compared with the reference strains.
For the CPV-2-positive animals, 65.26% (62/95) were aged from 2–4 months, 48.42% (46/95) had a vaccination history, and 32.63% (31/95) had no vaccination history, while 65.26% (62/95) were collected from October to December. Coinfections with CCoV, CaKV, and CBoV were found in the 95 CPV-2-positive samples, of which 22.11% (21/95) were positive for CCoV, 14.74% (14/95) were positive for CaKV, 7.37% (7/95) were positive for CBoV, 10.53% (10/95) were positive for CCoV and CaKV, 2.11% (2/95) were positive for CBoV and CaKV, and 1.05% (1/95) were positive for CCoV, CBoV, and CaKV.
Phylogenetic analysis
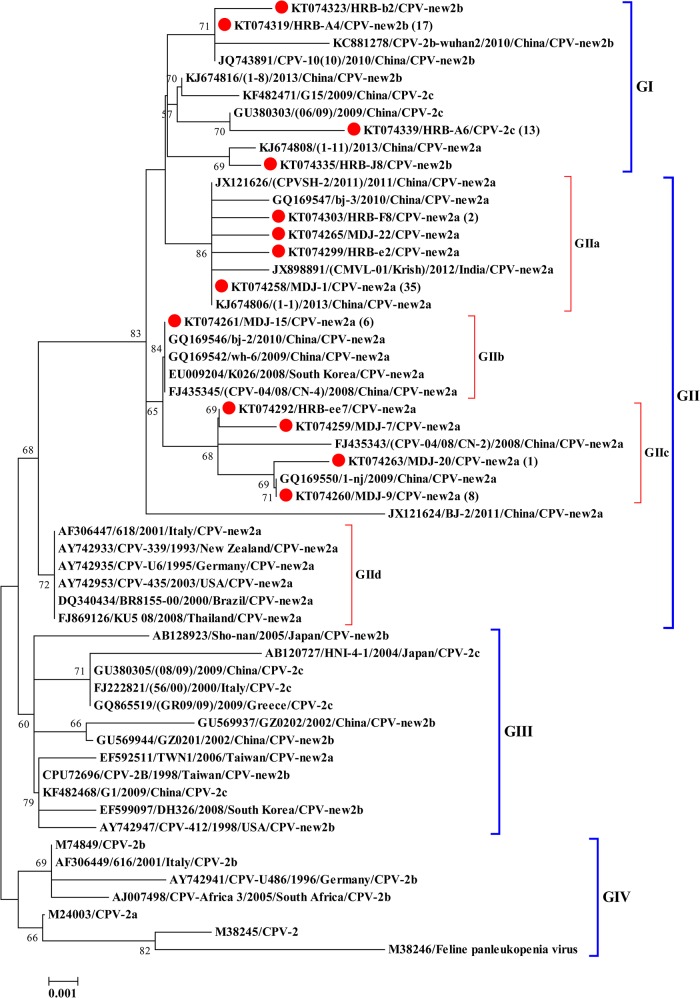
To analyze the genetic diversity of the new2a, new2b, and 2c strains identified in northeast China, partial VP2 gene sequences of CPV-2, CPV-2a, CPV-2b, CPV-2c, new CPV-2a, and new CPV-2b strains from different geographical locations within China and the rest of the world were used to construct a phylogenetic tree. Nucleotide sequences of the partial VP2 used for construction of phylogenetic tree was shown in Supporting Information (S2 Fig). The generated phylogenetic tree was composed of four groups: GI, GII, GIII, and GIV. The 95 CPV-2 strains from northeast China were divided into two different groups: GI and GII (Fig 1). All 14 CPV-2c strains and 20 new CPV-2b strains identified in our study were divided into the GI group. Meanwhile, the GI group included the new2a, new2b and 2c reference strains from China. The GII group, consisting of four subgroups, GIIa, GIIb, GIIc, and GIId, was specific for the new CPV-2a strains, except for one new CPV-2b strain (HRB-J8). The new CPV-2a strains identified in northeast China were divided into the three subgroups GIIa, GIIb, and GIIc, and the new CPV-2a reference strains from other countries were divided into the subgroup GIId. The GIV group consisted of different genotypes of CPV-2 reference strains from China and other countries.
Fig 1. Phylogenetic analysis of CPV-2 strains on the basis of the partial VP2 gene (507 bp).
The red spot diagram represents CPV-2 strains identified in this study. The values in parentheses indicate the number of strains with identical sequences to that included in the tree. (1): KT074275 from Daqing; (2): KT074317 and KT074304 from Harbin; (6): KT074266, KT074270 from Mudanjiang; KT074276, KT074288, KT074293, and KT074306 from Harbin; (8): KT074262, KT074264, and KT074271 from Mudanjiang; KT074277, KT074282, KT074311, KT074314, and KT074315 from Harbin. (13): KT074340-KT074347 from Harbin; KT074348-KT074352 from Mudanjiang. (17): KT074320-KT074322 and KT074324-KT074334 from Harbin; KT074336-KT074338 from Daqing; (35): KT074267-KT074269 and KT074272 from Mudanjiang; KT074273 and KT074274 from Daqing; KT074278, KT074279, KT074280-KT074287, KT074289- KT074291, KT074294- KT074298, KT074300- KT074302, KT074305, KT074307- KT074310, KT074312, KT074313, KT074316, and KT074318 from Harbin.
Recombination analysis
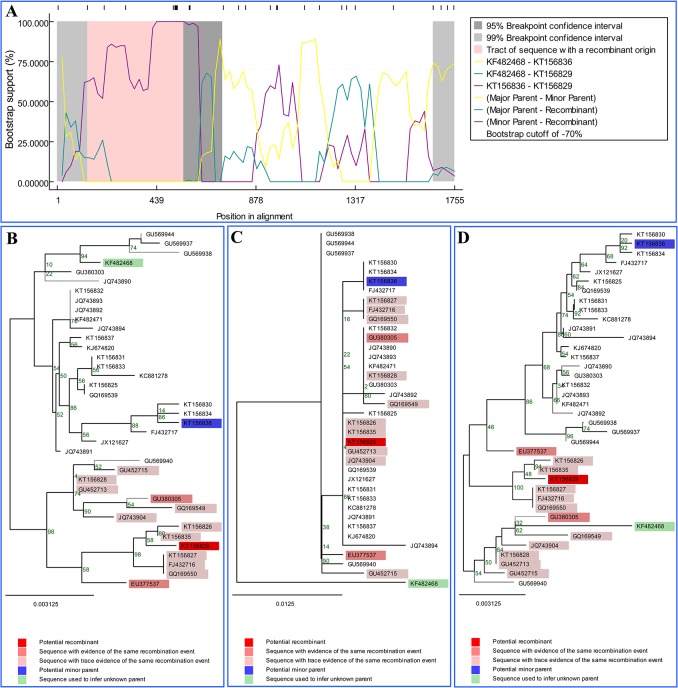
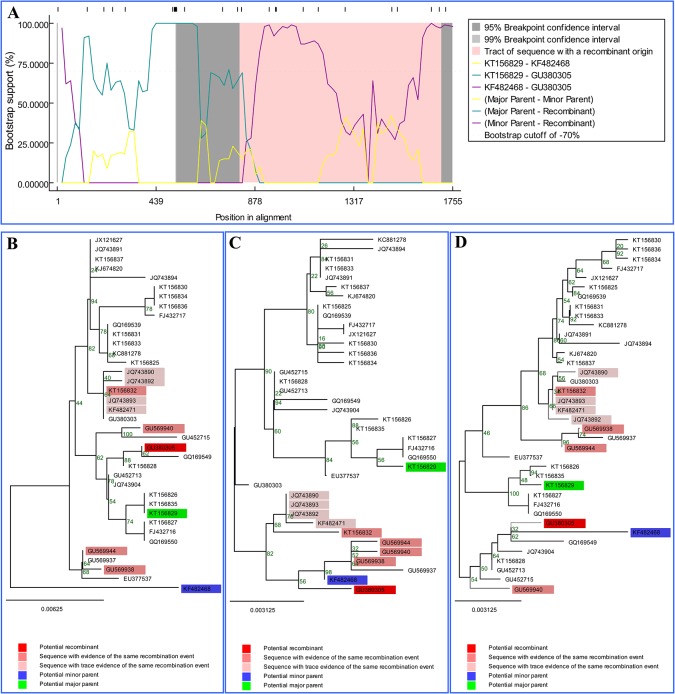
The recombination analysis based on the VP2 gene revealed that two potential recombination events occurred between the selected 13 strains in this study and the 25 reference strains from China, but no potential recombination events were found within the selected 13 strains in this study. Nucleotide sequences of the entire VP2 of CPV-2 strains identified in our study were shown in Supporting Information (S3 Fig), and the breakpoint positions of recombination events were shown in Supporting Information (S4 Fig). Fig 2 shows the recombination event that occurred between the HRB-F8 strain (accession no. KT156836) and a G1 strain (accession no. KF482468), which led to the recombinant MDJ-20 strain (accession no. KT156829). The bootscan plot of this recombination event is shown in Fig 2A, which used HRB-F8 and G1 as minor parent strain, and major parent strain, respectively. The recombination event was confirmed by fast neighbor-joining trees that were constructed using the regions derived from minor parent strain (1–133 and 558–1755) (Fig 2B), the recombinant region (134–557) (Fig 2C), and the non-recombinant region (Fig 2D). Fig 3 shows the recombination event that occurred between MDJ-20 strain (KT156829) and a G1 strain (KF482468), which led to the recombinant 08/09 strain (GU380305). The bootscan plot of this recombination event is shown in Fig 3A, which used MDJ-20 and G1 as major parent strain, and minor parent strain, respectively. The recombination event was confirmed by fast neighbor-joining trees that were constructed using the regions derived from minor parent strain (1–809 and 1708–1755) (Fig 3B), the recombinant region (810–1707) (Fig 3C), and the non-recombinant region (Fig 3D). The two recombinant strains MDJ-20 and 80/09 were divided into different clusters in the recombination region-based phylogenetic trees when compared with the non-recombination region-based phylogenetic trees.
Fig 2. Identification of the recombination event between the minor parent strain HRB-F8 (KT156836) (blue) and the major parent strain G1 (KF482468) (green), which led to the recombinant MDJ-20 strain (KT156829) (red).
(A) Bootscan evidence for the recombination origin on the basis of pairwise distance, modelled with a window size 200, step size 20 and 100 bootstrap replicates. (B, C, and D) Fast neighbour-joining (NJ) tree (1000 replicates, Kimura two-parameter distance) constructed using the regions derived from minor parent strain (1–133, 558–1755) (B), the recombination region (134–557) (C), and non-recombinant region (D). Note. The potential recombination event was detected in Maxchi (P = 3.74E-02) and SiSscan (P = 1.42E-04) methods.
Fig 3. Identification of the recombination event between the major parent strain MDJ-20 (KT156829) (green) and the minor parent strain G1 (KF482468) (blue), which led to the recombinant 08/09 strain (GU380305) (red).
(A) Bootscan evidence for the recombination origin on the basis of pairwise distance, modelled with a window size 200, step size 20 and 100 bootstrap replicates. (B, C, and D) Fast neighbour-joining (NJ) tree (1000 replicates, Kimura two-parameter distance) constructed using the regions derived from minor parent strain (1–809, 1708–1755) (B), the recombination region (810–1707) (C), and non-recombinant region (D). Note. The potential recombination event was detected in Maxchi (P = 8.51E-04), SiSscan (P = 1.05E-02), and Chimaera (P = 7.25E-03) methods.
Discussion
It has been reported that a high proportion (29.2%–84%) of fecal samples from diarrhea-affected dogs tested positive for CPV-2 [11, 29]. In our study, 201 samples were collected from diarrhea-affected dogs in three districts in Heilongjiang province in northeast China, and 95 CPV-2-positive samples were identified, accounting for 47.26% of all samples. The results were in line with recent reports in China [15, 30]. The CPV-2-positive rate differed among the three districts (Harbin, 48.94%; Daqing, 30%; Mudanjiang, 50%). In our study, mixed infections of CCoV (22.11%), CaKV (14.74%), and CBoV (7.37%) were found in the CPV-2-positive samples. A coinfection of CCoV, CBoV, and CaKV with CPV-2 was also found in one sample. Cavalli et al. (2014) reported that mixed infections of CPV-2 and CCoV reached 49.12% in diarrheic dogs in Albania [31]. CaKV and CBoV had been reported to be associated with severe enteritis in a litter of puppies [28, 32]. The high prevalence of co-infections of CPV-2 with three enteric viruses may be associated with viral diarrhea in dogs in northeast China.
In our study, CPV-2-positive rates showed clear differences among seasons and ages. 65.26% of CPV-2-positive samples were collected from October to December. The high prevalence of CPV-2 between October and December may be indicative of a seasonal variation in northeast China. Additionally, a high CPV-2-positive rate was found in animals aged from 2–4 months, which is similar to the report described by Cavalli et al. (2014) [31]. At present, most dogs in China are vaccinated for CPV-2. However, in the CPV-2-positive samples identified in this study, 48.42% came from dogs with a vaccination history. This data demonstrated that the CPV-2 vaccines used in northeast China may fail to generate protective antibody titers against heterogeneous CPV antigenic types.
The Ser297Ala mutation may have had a remarkable influence on the process of host adaptation [33]. This mutation was used as a marker of the new CPV-2a/2b variant. This study indicated that the new CPV-2a (Ser297Ala) variant, accounting for 64.21% of CPV-2 infections, was the predominating strain circulating in the three districts of Heilongjiang province in northeast China. The new CPV-2b (Ser297Ala) variant, accounting for 21.05% of CPV-2 infections, was the second most predominant strain circulating in the Harbin and Daqing districts of Heilongjiang province. In our study, the prevalence of the new CPV-2a is in agreement with most of the reports that were recently conducted in China [8, 15, 30, 34, 35], African countries [36], other Asian countries, including Korea, Thailand, Taiwan, and Turkey, and Australia [6, 37–39]. However, the prevalence of the new CPV-2b variant reported here in CPV-2-positive samples was higher than those in other reports in China. Interestingly, the presence of the rare CPV-2c, accounting for 14.74% of the CPV-2-positive samples, was confirmed in the Harbin and Mudanjiang of Heilongjiang province. The co-circulation of the CPV-2c, new CPV-2a, and new CPV-2b has previously not been reported in China. It has been reported that the CPV-2c variant is widely distributed and co-exists with other CPV types in Europe and North and South America [40–45]. Compared with an earlier report that detected the CPV-2, new CPV-2a, and CPV-2b variants in China [46], the emergence of the CPV-2c in northeast China suggests that CPV-2 strains circulating in China exhibit genetic variations. Additionally, our results are similar to those reported for the genotypes of CPV-2 strains circulating in India, where the new CPV-2a and new CPV-2b were the predominant strains [14], and the co-circulation of the CPV-2c was also found [47].
In the VP2 protein of CPV-2, the Tyr324Ile mutation is adjacent to residue 323, which affects binding to canine transferrin receptors, resulting in changes in the host range of canine parvovirus [13]. Presumably, this mutation may result in stronger receptor binding. In our study, all 95 identified CPV-2 strains, including the new2a, new2b, and 2c types, contained the unique Tyr324Ile mutation. The Tyr324Ile mutation in CPV-2a strains has been reported in China [8, 15, 30, 46], South Korea [38], Thailand [48], Uruguay [49], Japan [50], Taiwan [6], and India [51]. These data support the view that the Try324Ile substitution in CPV-2 strains may be a common amino acid alteration in Asia countries, especially in China. This conclusion needs to be validated by an extensive epidemiological survey in a future study. In addition, further studies should be conducted to understand the relationship between the Try324Ile substitution and the severity of clinical symptoms.
In the VP2 protein of CPV-2, the amino acid residue at position 440 is located in the GH loop. It has been reported that the high levels of the Thr440Ala substitution in VP2 are associated with the evolution of antigenic variants in circulating parvovirus types [52, 53]. In our study, the Thr440Ala substitution was found in 64.21% of the 95 CPV-2-positive samples, of which 100% of the new2b strains had the Thr440Ala substitution, 67.22% of the new2a strains exhibited the Thr440Ala substitution, while the type 2c strains did not have the Thr440Ala substitution. At present, the Thr440Ala substitution has been frequently reported in China and other countries [8, 14, 15, 36]. Therefore, we suggest further studies to better understand the relationship between this mutation and the severity of clinical symptoms.
In this study, both new CPV-2b strains and CPV-2c strains exhibited 100% amino acid identity on the basis of the deduced sequence of the partial VP2 protein among different districts. These data demonstrated that type new2b and 2c strains of CPV-2 circulating in northeast China exhibited genetic stability. However, compared with CPV-2 reference strains, the type new2b strains showed a unique Thr440Ala substitution (100%), and the type 2c strains exhibited a unique Gln370Arg substitution (100%). The amino acid substitutions resulted in phylogenetic changes in the new2b and 2c strains identified in our study. The new2b and 2c strains (except one strain) differed genetically from the reference strains of other countries, forming one group (GI) with Chinese reference strains of the new2b and 2c types. In our study, the close relationship between the new 2b and 2c types suggests that these two types share a similar evolutionary history in China. For the identified type 2c strains, the unique Gln370Arg substitution, which was previously not described, was highlighted as evidence of a potential CPV-2c variant or new CPV-2c. Further studies regarding potential variant CPV-2c strains should be conducted to understand the relationship between the Gln370Arg substitution and viral pathogenicity.
The phylogenetic analysis revealed that the type new2a strains identified in our study exhibited high variability, forming three subgroups. Of the 61 new2a strains, a total of two amino acid substitution sites were revealed, Asn419Ser and Thr440Ala. The two amino acid substitutions, as well as some non-synonymous substitutions, accounted for the genetic diversity of the new2a strains in the phylogenetic tree. In our study, the Asn419Ser substitution was detected in only three of the 61 new2a strains. We suggest that the Asn419Ser substitution, first described here, should be the focus of further epidemiological investigations of CPV-2. Accumulating reports have indicated that the new CPV-2a strains, accounting for high positive rates among CPV-2 types, are the predominating strains circulating in China. The high variability of the new CPV-2a strains in our study may be attributed to long-term interactions between the new CPV-2a strains and their hosts. It is suggested that vaccinations against CPV-2 should be updated on the basis of these new CPV-2a strains.
Although point mutations are considered the main mechanism for generating genomic diversity in CPV, co-infection and recombination have also been explored as variability-inducing mechanisms [54]. Pérez et al. (2014) revealed that a recombinant strain arose from inter-genotypic recombination between CPV-2c and CPV-2a strains within the VP1/VP2 gene boundary [55]. In our study, two inter-genotypic recombination events between the 13 CPV-2 strains in this study and the Chinese reference strains were identified in the VP2 gene. The two recombination events occurred between type new2a and 2c strains, resulting in two potential recombinant type new2a strains. Our finding is in line with the occurrence of a possible recombination event in the CPV-2 strain 364-rec, as described by Pérez et al. (2014) [55]. Although the evidence of naturally occurring recombination events was not obtained in our study, possible recombination events among Chinese CPV-2 strains suggest that co-infections with CPV-2 strains with different genotypes, especially types 2a and 2c, as well as potential recombinant strains of CPV-2, should be monitored in China by extensive molecular epidemiology investigations in the future.
Since 1983, the epidemiological information of CPV-2 strains circulating in China has been frequently reported [8, 15, 30, 34, 35, 46]. Accumulating reports indicated that circulation of CPV-2, CPV-2a, CPV-2b, new CPV-2a, and new CPV-2b strains was identified in different districts of China, of which new CPV-2a was predominant strains in China, followed by new CPV-2b. Compared with the previous epidemiological investigation of CPV-2 in China, the detailed information of samples used in our study, including collection date, location, animal breed, animal gender, animal age, animal immunization, and coinfection with other enteric viruses, resulted in the generation of more precise and valuable epidemiological data which not only enriches basic epidemiological data of Chinese CPV-2 strains, but also increases the understanding of epidemic characteristics of CPV-2 strains circulating in northeast China. In addition, our findings revealed novel epidemiological information of CPV-2 in China, involving in the co-circulation of new CPV-2a with high variation, new CPV-2b, and rare CPV-2c in Heilongjiang province of northeast China, the unique amino acids substitutions including Gln370Arg substitution in identified CPV-2c strains, and Thr440Ala substitution in identified new CPV-2b strains, and the potential recombination events which could happen among type new2a and 2c CPV-2 field strains circulating in China.
Conclusions
Taken together, in the current study, the resulting data increase our understanding of the genetic evolution of CVP-2 strains in China, and they also provide valuable information for further studies of CPV-2 in other countries. However, some speculations resulting from our data still need to be validated by further and extensive epidemiological investigation in the future.
Supporting Information
(TXT)
(TXT)
(TXT)
(XLS)
Data Availability
All sequence files are available from the GenBank database (accession numbers KT074258–KT074352 and KT156825–KT156837).
Funding Statement
This work was supported by the Program for New Century Excellent Talents in Heilongjiang Provincial University (grant no. 1252-NCET-016).
References
- 1. Kelly WR. An enteric disease of dogs resembling feline panleukopenia. Aust. Vet J. 1978; 54:593 [DOI] [PubMed] [Google Scholar]
- 2. Parrish CR, Aquadro CF, Strassheim ML, Evermann JF, Sgro JY, Mohammed HO. Rapid antigenic-type replacement and dna sequence evolution of canine parvovirus. J Virol. 1991; 65:6544–6552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Parrish CR, O’Connell PH, Evermann JF, Carmichael LE. Natural variation of canine parvovirus. Science 1985; 230:1046–1048. [DOI] [PubMed] [Google Scholar]
- 4. Buonavoglia C, Martella V, Pratelli A, Tempesta M, Cavalli A, Buonavoglia D, et al. Evidence for evolution of canine parvovirus type 2 in Italy. J Gen Virol. 2001; 82:3021–3025. [DOI] [PubMed] [Google Scholar]
- 5. Ohshima T, Hisaka M, Kawakami K, Kishi M, Tohya Y, Mochizuki M. Chronological analysis of canine parvovirus type 2 isolates in Japan. J Vet Med Sci. 2008; 70:769–775. [DOI] [PubMed] [Google Scholar]
- 6. Lin CN, Chien CH, Chiou MT, Chueh LL, Hung MY, Hsu HS. Genetic characterization of type 2a canine parvoviruses from Taiwan reveals the emergence of an Ile324 mutation in VP2. Virol J. 2014; 11:39 10.1186/1743-422X-11-39 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Mohan Raj J, Mukhopadhyay HK, Thanislass J, Antony PX, Pillai RM. Isolation, molecular characterization and phylogenetic analysis of canine parvovirus. Infect Genet Evol. 2010; 10:1237–1241. 10.1016/j.meegid.2010.08.005 [DOI] [PubMed] [Google Scholar]
- 8. Han SC, Guo HC, Sun SQ, Shu L, Wei YQ, Sun DH, et al. Full-length genomic characterizations of two canine parvoviruses prevalent in Northwest China. Arch Microbiol. 2015; 197:621–626. 10.1007/s00203-015-1093-4 [DOI] [PubMed] [Google Scholar]
- 9. Pedroza-Roldán C, Páez-Magallan V, Charles-Niño C, Elizondo-Quiroga D, Leonel De Cervantes-Mireles R, López-Amezcua MA. Genotyping of Canine parvovirus in western Mexico. J Vet Diagn Invest. 2015; 27:107–111. 10.1177/1040638714559969 [DOI] [PubMed] [Google Scholar]
- 10. Nelson CDS, Palermo LM, Hafenstein SL, Parrish CR. Different mechanisms of antibody-mediated neutralization of parvoviruses revealed using the Fab fragments of monoclonal antibodies. Virology 2007; 361:283–293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Pinto LD, Streck AF, Gonçalves KR, Souza CK, Corbellini ÂO, Corbellini LG, et al. Typing of canine parvovirus strains circulating in Brazil between 2008 and 2010. Virus Res. 2012; 165:29–33. 10.1016/j.virusres.2012.01.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Decaro N, Buonavoglia C. Canine parvovirus—a review of epidemiological and diagnostic aspects, with emphasis on type 2c. Vet Microbiol. 2012; 155:1–12. 10.1016/j.vetmic.2011.09.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Hueffer K, Parker JS, Weichert WS, Geisel RE, Sgro JY, Parrish CR. The natural host range shift and subsequent evolution of canine parvovirus resulted from virus-specific binding to the canine transferrin receptor. J Virol. 2003; 77:1718–1726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Mittal M, Chakravarti S, Mohapatra JK, Chug PK, Dubey R, Upmanuyu V, et al. Molecular typing of canine parvovirus strains circulating from 2008 to 2012 in an organized kennel in India reveals the possibility of vaccination failure. Infect Genet Evol. 2014; 23:1–6. 10.1016/j.meegid.2014.01.015 [DOI] [PubMed] [Google Scholar]
- 15. Yi L, Tong M, Cheng Y, Song W, Cheng S. Phylogenetic Analysis of Canine Parvovirus VP2 Gene in China. Transbound Emerg Dis. 2014. September 11 10.1111/tbed.12268 [DOI] [PubMed] [Google Scholar]
- 16. Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. MEGA6: Molecular Evolutionary Genetics Analysis version 6.0. Mol Biol Evol. 2013; 30:2725–2729. 10.1093/molbev/mst197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Martin D, Rybicki E. RDP: detection of recombination amongst aligned sequences. Bioinformatics 2000; 16:562–563. [DOI] [PubMed] [Google Scholar]
- 18. Padidam M, Sawyer S, Fauquet CM. Possible emergence of new geminiviruses by frequent recombination. Virology 1999; 265:218–225. [DOI] [PubMed] [Google Scholar]
- 19. Martin DP, Posada D, Crandall KA, Williamson C. A modified bootscan algorithm for automated identification of recombinant sequences and recombination breakpoints. AIDS Res Hum Retroviruses 2005; 21:98–102. [DOI] [PubMed] [Google Scholar]
- 20. Smith JM. Analyzing the mosaic structure of genes. J Mol Evol. 1992; 34:126–129. [DOI] [PubMed] [Google Scholar]
- 21. Posada D, Crandall KA. Evaluation of methods for detecting recombination from DNA sequences: computer simulations. Proc Natl Acad Sci USA. 2001; 98:13757–13762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Gibbs MJ, Armstrong JS, Gibbs AJ. Sister-scanning: a Monte Carlo procedure for assessing signals in recombinant sequences. Bioinformatics 2000; 16:573–582. [DOI] [PubMed] [Google Scholar]
- 23. Costa EM, de Castro TX, Bottino Fde O, Garcia Rde C. Molecular characterization of canine coronavirus strains circulating in Brazil. Vet Microbiol. 2014; 68:8–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Gómez MM, de Mendonça MC, Volotão Ede M, Tort LF, da Silva MF, Cristina J, et al. Rotavirus A genotype P[4]G2: genetic diversity and reassortment events among strains circulating in Brazil between 2005 and 2009. J Med Virol. 2011. 83:1093–1106. 10.1002/jmv.22071 [DOI] [PubMed] [Google Scholar]
- 25. Grellet A, De Battisti C, Feugier A, Pantile M, Marciano S, Grandjean D, et al. Prevalence and risk factors of astrovirus infection in puppies from French breeding kennels. Vet Microbiol. 2012; 157:214–219. 10.1016/j.vetmic.2011.11.012 [DOI] [PubMed] [Google Scholar]
- 26. Lau SK, Woo PC, Yeung HC, Teng JL, Wu Y, Bai R, et al. Identification and characterization of bocaviruses in cats and dogs reveals a novel feline bocavirus and a novel genetic group of canine bocavirus. J Gen Virol. 2012; 93:1573–1582. 10.1099/vir.0.042531-0 [DOI] [PubMed] [Google Scholar]
- 27. Mesquita JR, Nascimento MS. Molecular epidemiology of canine norovirus in dogs from Portugal, 2007–2011. BMC Vet Res. 2012; 8:107 10.1186/1746-6148-8-107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Di Martino B, Di Felice E, Ceci C, Di Profio F, Marsilio F. Canine kobuviruses in diarrhoeic dogs in Italy. Vet Microbiol. 2013; 166:246–249. 10.1016/j.vetmic.2013.05.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Cságola A, Varga S, Lőrincz M, Tuboly T. Analysis of the full-length VP2 protein of canine parvoviruses circulating in Hungary. Arch Virol. 2014; 159:2441–2444. 10.1007/s00705-014-2068-5 [DOI] [PubMed] [Google Scholar]
- 30. Xu J, Guo HC, Wei YQ, Shu L, Wang J, Li JS, et al. Phylogenetic analysis of canine parvovirus isolates from sichuan and gansu provinces of China in 2011. Transbound Emerg Dis. 2015; 62:91–95. 10.1111/tbed.12078 [DOI] [PubMed] [Google Scholar]
- 31. Cavalli A, Desario C, Kusi I, Mari V, Lorusso E, Cirone F, et al. Detection and genetic characterization of Canine parvovirus and Canine coronavirus strains circulating in district of Tirana in Albania. J Vet Diagn Invest. 2014; 26:563–566. [DOI] [PubMed] [Google Scholar]
- 32. Bodewes R, Lapp S, Hahn K, Habierski A, Förster C, König M, et al. Novel canine bocavirus strain associated with severe enteritis in a dog litter. Vet Microbiol. 2014; 174:1–8. 10.1016/j.vetmic.2014.08.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Pereira CA, Leal ES, Durigon EL. Selective regimen shift and demographic growth increase associated with the emergence of high-fitness variants of canine parvovirus. Infect Genet Evol. 2007; 7:399–409. [DOI] [PubMed] [Google Scholar]
- 34. Zhao Y, Lin Y, Zeng X, Lu C, Hou J. Genotyping and pathobiologic characterization of canine parvovirus circulating in Nanjing, China. Virol J. 2013; 10:272 10.1186/1743-422X-10-272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Zhong Z, Liang L, Zhao J, Xu X, Cao X, Liu X, et al. First isolation of new canine parvovirus 2a from Tibetan mastiff and global analysis of the full-length VP2 gene of canine parvoviruses 2 in China. Int J Mol Sci. 2014; 15:12166–12187. 10.3390/ijms150712166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Dogonyaro BB, Bosman AM, Sibeko KP, Venter EH, van Vuuren M. Genetic analysis of the VP2-encoding gene of canine parvovirus strains from Africa. Vet Microbiol. 2013; 165:460–465. 10.1016/j.vetmic.2013.04.022 [DOI] [PubMed] [Google Scholar]
- 37. Meers J, Kyaw-Tanner M, Bensink Z, Zwijnenberg R. Genetic analysis of canine parvovirus from dogs in Australia. Aust Vet J. 2007; 85:392–396. [DOI] [PubMed] [Google Scholar]
- 38. Jeoung SY, Ahn SJ, Kim D. Genetic analysis of VP2 gene of canine parvovirus isolates in Korea. J Vet Med Sci. 2008; 70:719–722. [DOI] [PubMed] [Google Scholar]
- 39. Timurkan M, Oğuzoğlu T. Molecular characterization of canine parvovirus (CPV) infection in dogs in Turkey. Vet Ital. 2015; 51:39–44. 10.12834/VetIt.263.908.3 [DOI] [PubMed] [Google Scholar]
- 40. Decaro N, Desario C, Addie DD, Martella V, Vieira MJ, Elia G, et al. The study molecular epidemiology of canine parvovirus, Europe. Emerg Infect Dis. 2007; 13:1222–1224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Decaro N, Desario C, Billi M, Mari V, Elia G, Cavalli A, et al. Western European epidemiological survey for parvovirus and coronavirus infections in dogs. Vet J. 2011; 187:195–199. 10.1016/j.tvjl.2009.10.027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Kapil S, Cooper E, Lamm C, Murray B, Rezabek G, Johnston L 3rd, et al. Canine parvovirus types 2c and 2b circulating in North American dogs in 2006 and 2007. J Clin Microbiol. 2007; 45:4044–4047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Hong C, Decaro N, Desario C, Tanner P, Pardo MC, Sanchez S, et al. Occurrence of canine parvovirus type 2c in the United States. J Vet Diagn Invest. 2007; 19:535–539. [DOI] [PubMed] [Google Scholar]
- 44. Calderon MG, Mattion N, Bucafusco D, Fogel F, Remorini P, La Torre J. Molecular characterization of canine parvovirus strains in Argentina: Detection of the pathogenic variant CPV2c in vaccinated dogs. J Virol Methods 2009; 159:141–145. 10.1016/j.jviromet.2009.03.013 [DOI] [PubMed] [Google Scholar]
- 45. Pérez R, Francia L, Romero V, Maya L, López I, Hernández M. First detection of canine parvovirus type 2c in South America. Vet Microbiol. 2007; 124:147–152. [DOI] [PubMed] [Google Scholar]
- 46. Zhang R, Yang S, Zhang W, Zhang T, Xie Z, Feng H, et al. Phylogenetic analysis of the VP2 gene of canine parvoviruses circulating in China. Virus Genes 2010; 40:397–402. 10.1007/s11262-010-0466-7 [DOI] [PubMed] [Google Scholar]
- 47. Nandi S, Chidri S, Kumar M, Chauhan RS. Occurrence of canine parvovirus type 2c in the dogs with haemorrhagic enteritis in India. Res Vet Sci. 2010; 88:169–171. 10.1016/j.rvsc.2009.05.018 [DOI] [PubMed] [Google Scholar]
- 48. Phromnoi S, Sirinarumitr K, Sirinarumitr T. Sequence analysis of VP2 gene of canine parvovirus isolates in Thailand. Virus Genes 2010; 41:23–29. 10.1007/s11262-010-0475-6 [DOI] [PubMed] [Google Scholar]
- 49. Maya L, Calleros L, Francia L, Hernández M, Iraola G, Panzera Y, et al. Phylodynamics analysis of canine parvovirus in Uruguay: evidence of two successive invasions by different variants. Arch Virol. 2013; 158:1133–1141. 10.1007/s00705-012-1591-5 [DOI] [PubMed] [Google Scholar]
- 50. Soma T, Taharaguchi S, Ohinata T, Ishii H, Hara M. Analysis of the VP2 protein gene of canine parvovirus strains from affected dogs in Japan. Res Vet Sci. 2013; 94:368–371. 10.1016/j.rvsc.2012.09.013 [DOI] [PubMed] [Google Scholar]
- 51. Mukhopadhyay HK, Matta SL, Amsaveni S, Antony PX, Thanislass J, Pillai RM. Phylogenetic analysis of canine parvovirus partial VP2 gene in India. Virus Genes 2014; 48:89–95. 10.1007/s11262-013-1000-5 [DOI] [PubMed] [Google Scholar]
- 52. Ikeda Y, Mochizuki M, Naito R, Nakamura K, Miyazawa T, Mikami T, et al. Predominance of canine parvovirus (CPV) in unvaccinated cat populations and emergence of new antigenic types of CPVs in cats. Virology 2000; 278:13–19. [DOI] [PubMed] [Google Scholar]
- 53. Decaro N, Desario C, Parisi A, Martella V, Lorusso A, Miccolupo A, et al. Genetic analysis of canine parvovirus type 2c. Virology 2009; 385:5–10. 10.1016/j.virol.2008.12.016 [DOI] [PubMed] [Google Scholar]
- 54. Hoelzer K, Shackelton LA, Parrish CR, Holmes EC. Phylogenetic analysis reveals the emergence, evolution and dispersal of carnivore parvoviruses. J Gen Virol. 2008; 89:2280–2289. 10.1099/vir.0.2008/002055-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Pérez R, Calleros L, Marandino A, Sarute N, Iraola G, Grecco S, et al. Phylogenetic and genome-wide deep-sequencing analyses of canine parvovirus reveal co-infection with field variants and emergence of a recent recombinant strain. PLoS One 2014; 9:e111779 10.1371/journal.pone.0111779 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(TXT)
(TXT)
(TXT)
(XLS)
Data Availability Statement
All sequence files are available from the GenBank database (accession numbers KT074258–KT074352 and KT156825–KT156837).