Abstract
BACKGROUND & AIMS
The tumor suppressors retinoblastoma (RB) and p53 are important regulators of the cell cycle. Although human cancer cells inactivate RB and p53 by many mechanisms, the cooperative roles of these proteins in tumorigenesis are complex and tissue-specific. We analyzed the cooperation of RB and p53 in liver development and pathogenesis of hepatocellular carcinoma (HCC).
METHODS
Spontaneous and carcinogen-induced (diethylnitrosamine) tumorigenesis were studied in mice with liver-specific deletions of Rb and/or p53 (Rbf/f;albcre+, p53f/f;albcre+ and Rbf/f; p53f/f;albcre+ mice). Genotype, histologic, immunohistochemical, microarray, quantitative PCR, immunoblot, and comparative genomic hybridization analyses were performed using normal and tumor samples. Comparative microarray analyses were performed against publicly available human microarray datasets.
RESULTS
Deletion of RB and p53 from livers of mice deregulated the transcriptional programs associated with human disease; these changes were not sufficient for spontaneous tumorigenesis—potent quiescence mechanisms compensated for loss of these tumor suppressors. In response to hepatocarcinogen-induced damage, distinct and cooperative roles of RB and p53 were revealed; their loss affected cell cycle control, checkpoint response, and genome stability. In damaged tissue, combined loss of RB and p53 resulted in early lesion formation, aggressive tumor progression, and gene expression signatures and histological characteristics of advanced human HCC.
CONCLUSIONS
This study demonstrates that the impact of RB and p53 loss is modified by tissue environment, and critical functions of these tumor suppressors are revealed specifically in response to cellular stresses that promote aggressive disease.
Keywords: Liver cancer, mouse model, genetics, tumor environment, DEN
Introduction
Hepatocellular carcinoma (HCC) is the most prevalent form of primary liver cancer and results in greater than 500,000 deaths annually 1. Like many epithelial cancers, HCC is a complex disease characterized by numerous molecular abnormalities and chromosomal aberrations 2, 3. However, HCC is unique in that the majority of cases can be traced to specific environmental stresses that include viral infection (e.g. Hepatitis C Virus), exposure to hepatotoxins (e.g. Aflatoxin B1), or metabolic stresses (e.g. Nonalcoholic Steatohepatitis) 4. These etiological exposures are believed to influence cell cycle machinery by impinging on critical tumor suppressor pathways via discrete mechanisms, ultimately playing an important role in driving the genesis of HCC 5, 6. Amongst the pathways frequently disrupted in concert in HCC are those modulated by the retinoblastoma (RB) and p53 tumor suppressors 2, 7.
In the case of RB, loss of heterozygosity at 13q14 is a relatively common event in HCC and is associated with a particularly aggressive form of disease 2, 8. Physiologically, RB regulates cell cycle progression through control of the E2F family of transcription factors 9. Transcriptional targets of RB/E2F include genes involved in diverse processes including DNA replication, cell cycle progression, and DNA damage response 10, 11. Loss of RB results in the deregulation of this gene expression program, which is associated with human cancer and poor disease outcome 12. Interestingly, liver-specific deletion of the Rb gene in mice does not result in spontaneous tumor development, presumably a reflection of compensatory tumor suppressor pathways that regulate E2F activity 13.
One pathway likely to mediate cell cycle control in the absence of RB is the p53 pathway. In this context, many tumors exhibit coordinate loss of RB and p53 14, and DNA tumor viruses have evolved mechanisms to inactivate both pathways 15. The p53 tumor suppressor binds directly to target promoters, functioning as either a transcriptional activator or repressor for specific genes 16. In response to cellular stresses, p53 is activated and modulates a transcriptional program that influences cell cycle checkpoints, DNA damage response, and apoptosis 17, 18. These functions of p53 are selected against during tumorigenesis, and multiple mechanisms have been discovered that result in the disruption of the p53 pathway in HCC 2, 5.
Although combined loss of RB and p53 is a frequent occurrence in human cancer, the basis of cooperation between these tumor suppressor pathways has proven to be complex and tissue-specific. For example, in mouse models of medulloblastoma, loss of RB and p53 promotes tumorigenesis by delaying normal differentiation and apoptotic responses, which results in genomic instability 19. In models of mammary tumorigenesis, RB/p53-deficiency not only promotes aggressive tumor formation, but also influences the molecular characteristics (i.e. subtype) of the tumor 20. The fact that many human cancers exhibit inactivation of both tumor suppressor pathways suggests an intrinsic cooperation in tumorigenesis. However, while there is evidence that RB and p53 play partially overlapping roles in regulating cell cycle checkpoints, the mechanism by which combined inactivation of these tumor suppressors ultimately promotes tumor development is unknown. Here, we used a mouse model of liver tumorigenesis in which the impact of RB/p53 loss is dominated by extrinsic stresses. In the context of liver carcinogenesis, distinct functions of RB and p53 were revealed specifically in response to genotoxic damage, which lead to cooperative bypass of critical cell cycle checkpoints that ultimately promoted genomic instability and tumorigenesis.
Materials and Methods
Animals and Treatment
Rbf/f and Rbf/f;albcre+ mice, and p53f/f mice on a mixed FVB;129 background have been previously described 13, 21. Rbf/f;albcre+ mice were crossed with p53f/f mice to generate p53f/f;albcre+ and Rbf/f; p53f/f;albcre+ mice. 14-day old mice were given a single interperitoneal injection (20mg/kg) of diethylnitrosamine (DEN) (Sigma-Aldrich, St. Louis, MO). Experiments were conducted in accordance with the NIH Guide for Care and Use of Laboratory Animals and approved by Thomas Jefferson University Institutional Animal Care and Use Committee.
Tissue Genotyping
Genomic DNA isolation, primer sequences, and polymerase chain reaction (PCR) specifications have been previously described 13, 21.
Histology and Immunohistochemistry
Liver tissue was embedded in paraffin and sectioned at 4μM. Slide preparation and tissue staining procedures are described in detail in the Supplementary Materials and Methods.
Tumor Analyses
Surface lesions and dissected liver tumors were scored, and histology was analyzed by microscopic evaluation in a blinded fashion by a surgical pathologist (A.W.).
Microarray Analyses
Microarray experiments were preformed using the Affymetrix GeneChip Mouse Gene 1.0 ST array (Affymetrix, Inc., Santa Clara, CA). Detailed descriptions of RNA purification, sample preparation, microarray data processing, normalization, and differential expression analysis are provided in Supplementary Materials and Methods. All microarray data are MIAME compliant and have been deposited in Gene Expression Omnibus (GEO) (accession number GSE24347). Additional human HCC microarray datasets (GSE4108, GSE6764, and GSE9843) were obtained from GEO.
qPCR Analyses
RNA was harvested from frozen livers using TRIzol (Invitrogen, Carlsbad, CA) according to manufacturer’s instructions. Superscript reverse transcriptase (Invitrogen) was used to generate cDNA with random hexamer primers. Primers and PCR conditions available upon request.
Immunoblot Analyses
Frozen liver tissue was processed and immunoblotted as previously described 13, 21. Antibody details are provided in the Supplementary Materials and Methods.
Ploidy Determination
Liver nuclei were prepared as previously described 13, 21. Fluorescence-activated cell sorting was performed using a Coulter XL Flow Cytometer (Beckman Coulter, Inc., Brea, CA).
Comparative Genomic Hybridization (CGH)
DNA isolation and labeling, and Agilent 244A array processing was performed according to manufacturer’s instructions (Oligonucleotide Array-Based CGH for Genomic DNA Analysis, Agilent Technologies, Palo Alto, CA). Detailed analyses are described in the Supplementary Materials and Methods. Raw data is accessible through GEO accession number GSE24347.
Statistical Analyses
With the exception of microarray and CGH analyses, all statistical analyses were performed with GraphPad Prism (GraphPad Prism Software, Inc.). P values were calculated by performing Student’s T-Tests. P values <0.05 were considered significant.
Results
Loss of RB/p53 in the liver promotes deregulated proliferation and transcriptional programs similar to human disease
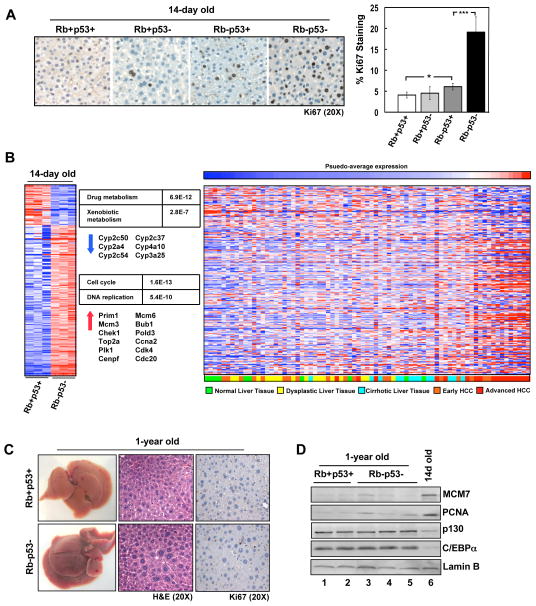
To investigate the cooperation of RB/p53 in the liver, a model for tissue-specific inactivation of Rb and p53 was employed. Mice transgenic for Cre recombinase under the albumin promoter (Alb-cre) were crossed to mice harboring loxP sites within the Rb and/or p53 gene 13, 21. Gene recombination and expression were documented by PCR (Supplementary Figure 1A). To examine the influence of RB/p53 loss in developing livers, 14-day old mice were utilized. In these mice, p53 deletion had no effect on hepatocyte proliferation as determined by Ki67 staining (Figure 1A). RB loss allowed for a modest yet significant increase in proliferation; however, this effect was miniscule compared to the proliferative advantage afforded by combined RB/p53 loss (Figure 1A). Furthermore, livers harboring combined RB/p53 loss displayed increased levels of DNA replication proteins, MCM7 and PCNA, relative to loss of a single tumor suppressor (Supplementary Figure 1B).
Figure 1. RB/p53 loss in the liver promotes cell cycle deregulation but not spontaneous tumor formation.
(A) Left, Ki67 staining in 14-day old livers. Right, percentage Ki67-positive hepatocytes (*p=0.03; ***p<0.0001). (B) Gene expression data for 14-day old, RB/p53-deficient mouse livers compared to wild-type livers. Left, changes in gene expression displayed as a heat map. Center, KEGG Pathway analysis with corresponding P-values and representative genes. Right, human liver samples (GSE6764) clustered based on average intensities of deregulated genes in RB/p53-deficient mouse livers. (C) Dissected 1-year old mouse livers, H&E and Ki67-stained sections. (D) Total protein lysates from 1-year old and 14-day old livers immunoblotted for the indicated proteins.
As RB and p53 are known to regulate transcriptional programs involved in proliferation, the transcriptional profile of livers harboring deletion of both genes compared to wild-type livers was determined via microarray analyses. These analyses identified 937 genes that were significantly altered in RB/p53-deficient livers, most of which were up-regulated genes (718 genes) significantly associated with cell cycle control (p=1.6E-13) and DNA replication (p=5.4E-10) as determined by KEGG Pathway analysis (Figure 1B; Supplementary Figure 1C). Interestingly, genes that were down-regulated in response to RB/p53 loss were associated with metabolic pathways (e.g. drug and xenobiotic metabolism), but surprisingly were absent of genes activated by p53 (e.g. Cdkn1a, Apaf1, Bax). To delineate the significance of this transcriptional profile to human disease, average intensities of deregulated genes were used to cluster human tissue samples of varying disease stage (p=0.0127) (Figure 1B). These analyses demonstrated that the transcriptional deregulation observed in RB/p53-deficient mouse livers was most similar to that observed with progression to advanced HCC.
Combined loss of RB/p53 does not promote spontaneous tumor formation in the liver
To determine whether RB/p53 loss in the liver was sufficient for spontaneous tumorigenesis, wild-type and RB/p53-deficient mice were aged to one year. Surprisingly, RB/p53-deficient livers displayed no evidence of gross abnormality, with the exception of increased liver weight (Figure 1C; Supplementary Figure 2A). Histological examination revealed clusters of hepatocytes with large nuclei (Figure 1C), indicating elevated DNA ploidy. Flow cytometric analysis displayed a modest yet significant increase in the number of hepatocytes with 4N DNA content, but no difference in the number of hepatocytes with >4N DNA content (Supplementary Figure 2B).
Livers of adult mice are maintained in a quiescent state until stimulated to proliferate. To determine whether RB/p53 loss influences the quiescent state of adult tissue, 1-year old livers were stained for Ki67. Unlike neonatal livers, 1-year old livers displayed virtually no cell proliferation (Figure 1C), and DNA replication factors (MCM7 and PCNA) were nearly undetectable (Figure 1D), regardless of RB/p53-status. Thus, despite enhanced proliferation observed in neonatal livers in response to RB/p53 loss, adult livers displayed a quiescent phenotype identical to wild-type tissue. Accordingly, protein levels of p130 and C/EBPα, two factors involved in establishing and maintaining quiescence 22, 23, were significantly elevated in both wild-type and RB/p53-deficient adult livers as compared to 14-day old livers (Figure 1D). Examination of these proliferation and quiescence markers was also performed at the onset of liver quiescence (i.e. 5-weeks old). Reduced proliferation (along with decreased levels of MCM7 and PCNA) and increased levels of p130 and C/EBPα were observed in both wild-type and RB/p53-deficient, 5-week old livers (Supplementary Figure 2C). Combined, these data indicate that there are additional mechanisms that serve to protect the liver from tumorigenesis independent of RB and p53.
RB/p53 loss results in bypass of hepatocarcinogen-induced damage response in the liver
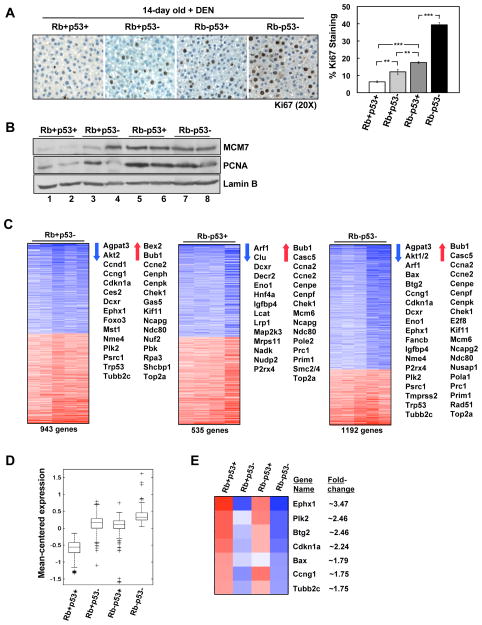
HCC is associated with multiple extrinsic risk factors, many of which are directly genotoxic (e.g. Aflatoxin B1) or induce mutation through chronic tissue damage (e.g. HCV) 5, 6. While RB/p53 loss was not sufficient for spontaneous tumorigenesis in the liver, the role of these tumor suppressors in mediating cellular response to genotoxic agents may be important in preventing tumor development. As such, 14-day old mice were exposed to a single dose of the hepatocarcinogen diethylnitrosamine (DEN), and the acute response of the liver (24h post-treatment) was examined. Loss of either RB or p53 alone resulted in significantly deregulated hepatocyte proliferation in response to DEN (Figure 2A). RB-deficient livers displayed a greater proliferative fraction of hepatocytes relative to p53-deficient livers, suggesting that RB has greater impact on cell cycle entry in the presence of genotoxic stress than p53. Combined RB/p53 loss resulted in a significant increase in proliferation above that of RB or p53 loss alone, indicating a cooperative damage response by these tumor suppressors (Figure 2A). Increased levels of DNA replication proteins (MCM7 and PCNA) were observed in all tumor suppressor-deficient livers compared to wild-type livers (Figure 2B).
Figure 2. RB/p53 loss promotes deregulation of transcriptional programs that cooperate in proliferation and bypass of hepatocarcinogen-induced checkpoints.
(A) Left, Ki67 staining in 14-day old, DEN-treated livers (24h post-treatment). Right, percentage Ki67-positive hepatocytes (**p<0.04; ***p<0.0001). (B) Total protein lysates from 14-day old, DEN-treated livers immunoblotted for the indicated proteins. (C) Gene expression data for 14-day old, DEN-treated mouse livers compared to wild-type livers. Representative gene lists are displayed. (D) Expression data for genes specifically up-regulated in RB/p53-deficient livers were clustered and compared. (E) Expression data for genes specifically altered by p53 loss displayed as a heat map with fold-change values for RB/p53-deficient livers.
To determine the impact of RB/p53-mediated transcriptional control on the response to hepatocarcinogen, gene expression profiling was performed on DEN-treated livers. With p53 loss alone, 943 genes were significantly altered compared to wild-type livers (Figure 2C, left panel). These gene changes consisted of down-regulated genes encompassing known p53 targets (e.g. Cdkn1a, Ccng1), as well as a subset of genes previously shown to be involved in response to hepatocarcinogens (e.g. Ephx1, Plk2, Ccng1, and Tubb2c)24. Additionally, an equivalent number of up-regulated genes was observed, most significantly associated with cell cycle progression (e.g. Bub1, Chek1, Top2α) (Figure 2C, left panel). RB deletion alone resulted in the deregulation of 535 genes, most notably including enhanced expression of a large number of genes involved in cell cycle progression and DNA replication (e.g. Bub1, Ccna2, Mcm6, Prim1, Top2α) (Figure 2C, center panel). Thus, inactivation of either p53 or RB results in elevated expression of genes involved in cell cycle progression, which would account for the enhanced Ki67 incorporation observed upon loss of either tumor suppressor.
In the context of combined RB/p53 loss, 1192 genes were significantly deregulated (Figure 2C, right panel). Interestingly, this analysis revealed a gene expression profile that was reflective of both additive gene deregulation via loss of RB and p53 individually, and gene deregulation specifically dependent on loss of one particular tumor suppressor. For example, while genes involved in cell cycle progression and DNA replication (e.g. Ccna2, Ndc80, Top2α) were similarly up-regulated with single tumor suppressor loss, additive enhancement of these genes was observed with combined loss of RB/p53 (Figure 2D). Additionally, a striking pattern of gene deregulation was observed that was strictly dependent on p53 loss and included genes previously implicated in response to hepatocarcinogens (e.g. Bax, Btg2, Ccng1, Cdkn1a, Ephx1, Plk2, and Tubb2c)24. These genes were observed to be significantly down-regulated in RB/p53-deficient livers (Figure 2E), and several genes (e.g. Ccng1, Cdkn1a, Plk2) are specifically involved in mediating a G2/M checkpoint and dependent on p53 status 25–28. To validate the described microarray results, qPCR was performed on select genes (Supplementary Figure 3). Combined, these data suggest that the overall enhanced proliferation observed in RB/p53-deficient mouse livers represents both discrete and cooperative roles for the RB and p53 pathways.
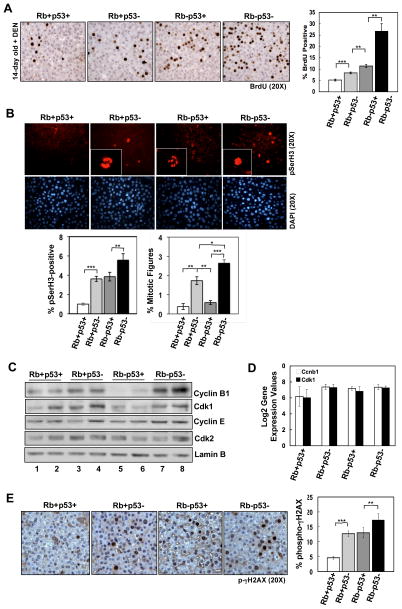
Mitotic bypass of RB/p53-deficient hepatocytes in response to genotoxic damage is dependent on p53 status
Microarray analyses suggested that combined loss of RB/p53 in the presence of hepatocarcinogen resulted in a cooperative transcriptional response to promote proliferation and bypass of cell cycle checkpoints. To examine this concept functionally, we assessed the impact of RB/p53 loss on DNA replication and mitotic progression in response to DEN. In the context of DNA replication, analysis of BrdU incorporation indicated that RB/p53 loss cooperates to promote S-phase progression (Figure 3A). In contrast, analysis of mitotic progression via phospho-histone H3 Serine10 (pSerH3) staining revealed a distinct p53-dependent G2/M-checkpoint (Figure 3B). Few pSerH3-positive cells were detected in wild-type livers, with most exhibiting interphase staining consistent with mitotic arrest (Figure 3B). Interestingly, while RB-deficient livers displayed increased pSerH3 staining, these hepatocytes also exhibited interphase staining. Thus, while more RB-deficient cells were entering G2/M-phase, they did not demonstrate productive mitoses. In contrast, increased levels of mitotic figures were observed in livers deficient in either p53 alone or RB/p53 combined (Figure 3B), indicating that loss of p53 is sufficient to promote mitotic progression.
Figure 3. p53 loss is required for mitotic progression in response to hepatocarcinogen.
(A) Left, BrdU staining in 14-day old, DEN-treated livers. Right, percentage BrdU-positive hepatocytes (**p<0.002, ***p=0.0006). (B) Top, pSerH3 staining in 14-day old, DEN-treated livers. Insets display representative mitotic figures (anaphase and prophase for Rb+p53− and Rb-p53−, respectively) or staining indicative of G2-arrest (Rb-p53+). Bottom, percentage pSerH3-positive hepatocytes (**p=0.0067, ***p=0.0008) and mitotic figures (*p=0.02; **p=0.003, ***p<0.0001). (C) Total protein lysates from 14-day old, DEN-treated livers immunoblotted for the indicated proteins. (D) Normalized Log2 gene expression values for Ccnb1 and Cdk1 from microarray analyses. (E) Left, p-γH2AX staining in 14-day old, DEN-treated livers. Right, percentage p-γH2AX-positive hepatocytes (**p=0.0069; ***p<0.0001).
To examine the mechanism associated with the observed mitotic phenotypes, expression levels of proteins involved in mitotic progression (Cyclin B1 and Cdk1) were monitored. In wild-type livers, basal levels of Cyclin B1 and Cdk1 were present (Figure 3C, lanes 1–2), consistent with the low level of hepatocytes observed in G2/M-phase. While loss of p53 resulted in elevated levels of Cyclin B1 and Cdk1 (Figure 3C, lanes 3–4), these proteins were significantly decreased in RB-deficient livers (Figure 3C, lanes 5–6), suggesting a G2/M-checkpoint. This decrease is likely indicative of checkpoint-dependent protein degradation, as RNA levels of Cyclin B1 (Ccnb1) and Cdk1 were not altered according to microarray analyses (Figure 3D). Strikingly, Cyclin B1 and Cdk1 protein levels were elevated in livers deficient for both RB/p53 (Figure 3C, lanes 7–8), corresponding with the highest percentage of hepatocytes exhibiting mitotic progression. Cyclin E and Cdk2 protein levels were unaltered, demonstrating a specific G2/M-checkpoint. Combined, these data indicate that while either RB or p53 loss alone can promote deregulated cell cycle entry in response to genotoxic damage, only loss of p53 can promote mitotic progression.
To examine the impact of the described cell cycle deregulation on DNA damage generated in response to DEN, phospho-γH2AX (p-γH2AX) was examined. Livers deficient in either RB or p53 displayed a similar and significant increase in p-γH2AX compared to wild-type livers, and tissues deficient in both RB/p53 displayed a modest yet significant increase in pγH2AX above that observed with single tumor suppressor loss (Figure 3E). Thus, deregulation of cell cycle control via loss of either tumor suppressor promotes elevated levels of DNA damage in response to hepatocarcinogen, and deregulation of both cell cycle checkpoints via combined RB/p53 loss ultimately enhances this damage. No difference was observed in the levels of apoptosis in this model regardless of RB//p53-status (data not shown).
RB/p53 loss in the liver promotes early lesion formation and aggressive tumor development similar to human HCC
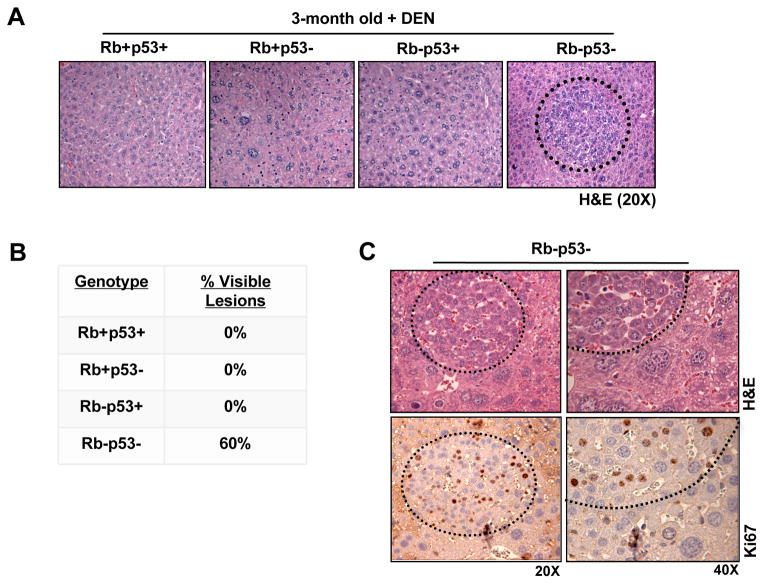
To determine the relative impact of RB and/or p53 loss on tumor development, mice treated with DEN were aged. At three months post-treatment, gross examination of the liver revealed little effect of tumor suppressor loss on liver size or visible lesions (Supplementary Figure 4). However, histological analyses revealed the presence of neoplastic foci only in livers deficient for both RB/p53 (Figure 4A), which were observed in 60% of sections analyzed (Figure 4B). Ki67 staining of RB/p53-deficient livers demonstrated that the majority of the liver is quiescent, while the foci are hyperproliferative (Figure 4C), indicating that combined RB/p53 loss contributes to the age of onset of liver tumor development but is restricted to focal lesions.
Figure 4. Combined RB/p53 loss promotes early lesion formation in response to hepatocarcinogen.
(A) 3-month old, DEN-treated livers stained with H&E. (B) Percentage of 3-month old, DEN-treated livers with visible lesions. (C) Top, RB/p53-deficient lesions stained with H&E. Bottom, Ki67-stained lesions.
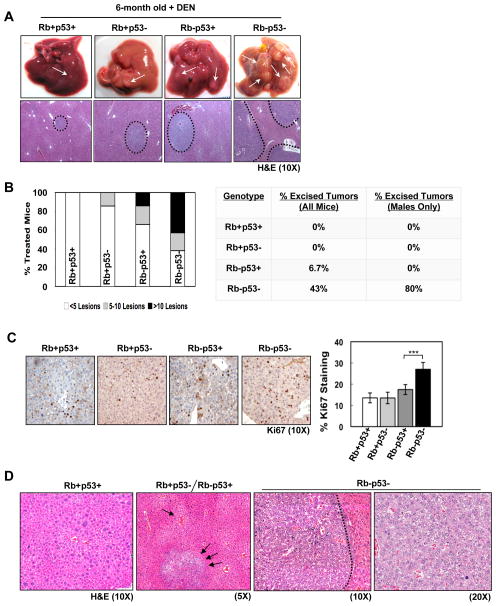
To interrogate disease progression, mice were aged to 6 months post-DEN. At this age, wild-type livers, and livers deficient in either RB or p53, displayed small surface lesions (Figure 5A). Interestingly, RB-deficient livers displayed a greater number of individual lesions than wild-type or p53-deficient livers (Figure 5B); however, the level of proliferation within the lesions was not significantly different (Figure 5C). These data indicate that RB loss is a more potent determinant of lesion multiplicity, and perhaps suggest a more significant role for RB in tumor initiation. Livers harboring combined RB/p53-deficiency exhibited dramatically enhanced tumor burden with 60% of mice displaying >10 lesions, and 43% of mice (80% of males) displaying large tumors that were easily dissected (Figure 5B). Furthermore, RB/p53-deficient lesions displayed enhanced cell proliferation compared to lesions harboring single tumor suppressor loss (Figure 5C).
Figure 5. Combined RB/p53 loss drives aggressive tumor progression in response to hepatocarcinogen.
(A) Top, dissected 6-month old, DEN-treated livers. Bottom, liver sections stained with H&E. (B) Left, number of surface lesions per liver. Right, percentage of mice with dissected tumors. (C) Left, Ki67 staining in lesions. Right, percentage Ki67-positive hepatocytes (***p<0.0001). (D) Representative images of disease stages observed in mouse liver tissue 6-months post-treatment.
The enhanced tumor burden and elevated proliferation observed in RB/p53-deficient livers suggested a potential increase in disease progression specifically associated with combined tumor suppressor loss. To address this hypothesis, histological examination of lesions from all liver genotypes was performed in a blinded fashion by a surgical pathologist (A.W.). While wild-type livers were characterized by diffuse, panlobular atypia with few small dysplastic foci (Figure 5D, left), livers harboring single tumor suppressor loss displayed a significant increase in centrilobular atypia and multiple larger dysplastic foci (Figure 5D, middle). Livers deficient for both RB/p53 displayed histological characteristics similar to human HCC, including thickened trabeculae and a focal adenoid pattern, as well as enhanced cytologic atypia and mitotic figures (Figure 5D, right). Thus, not only does combined loss of RB/p53 promote early lesion formation, but these lesions exhibit histological phenotypes indicative of aggressive tumor progression similar to human disease.
RB/p53-deficient liver tumors display chromosomal aberrations and gene deregulation indicative of advanced HCC
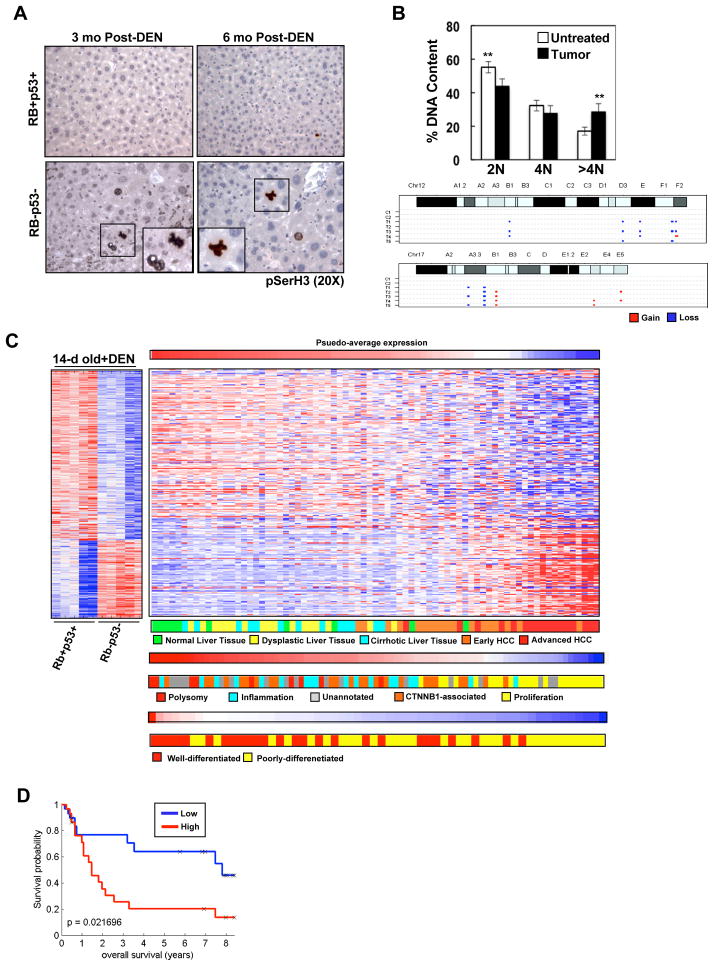
A key feature of human HCC is chromosome instability 3, 29. Since RB/p53-deficient liver tumors were histologically similar to human HCC, these tissues were analyzed for the presence of chromosomal abnormalities. Initially, histological examination and pSerH3 staining was utilized to assess potential nuclear and mitotic abnormalities in wild-type versus RB/p53-deficient, DEN-treated livers (Figure 6A). At an acute stage (24h post-treatment), RB/p53-deficient livers were histologically identical to wild-type livers and displayed seemingly normal chromosome condensation and segregation (Figure 3B). In contrast, at 3- and 6-months post-treatment, RB/p53-deficient tissues displayed hepatocytes with dramatically larger nuclei compared to wild-type livers (Figure 6A). Furthermore, whereas wild-type adult livers are quiescent and mitotic figures are virtually undetectable, RB/p53-deficent livers harbor pSerH3-positive hepatocytes and mitotic figures displaying abnormal chromosome condensation.
Figure 6. Liver tumors harboring combined RB/p53 loss display chromosomal aberrations and gene deregulation indicative of advanced HCC.
(A) Wild-type and RB/p53-deficient livers stained for pSerH3. (B) Top, percentage of hepatocytes harboring 2N, 4N and >4N DNA content (**p=0.0018, 0.0015). Bottom, selected chromosome gain/loss images from CGH analyses. C1-2 indicates non-tumor liver tissue and T1-5 indicates liver tumor samples from mice 6-months post-DEN. (C) Gene expression data comparing 14-day old, RB/p53-deficient and wild-type DEN-treated livers. Left, changes in gene expression displayed as a heat map. Right, human liver samples (GSE6764, GSE9843, and GSE4108) clustered based on average intensities of deregulated genes in RB/p53-deficient, DEN-treated mouse livers. (D) Kaplan Meier analysis demonstrating recurrence-free survival in human tissue samples (GSE4108) clustered based on average intensities of up-regulated genes (i.e. additive gene deregulation) in RB/p53-deficient, DEN-treated mouse livers. Samples were divided into high expression (Red) or low expression (Blue) of the gene profile.
In keeping with the aberrant nuclear morphology demonstrated in RB/p53-deficient livers, analyses of dissected tumors demonstrated a significant increase in hepatocytes with >4N DNA content (Figure 6B, top), indicating the possibility of genomic aberrations. CGH analyses of RB/p53-deficient tumors revealed a significant increase in overall genomic instability compared to normal livers (Figure 6B, bottom; Supplementary Figure 5). Furthermore, RB/p53-deficient tumors displayed a significant increase in genomic instability compared to tumors harboring RB loss alone (data not shown). These data indicate that combined loss of RB/p53 yields deregulation of multiple checkpoints that facilitate tumor development via acquisition of additional mutations capable of driving disease pathogenesis.
These functional data demonstrate that combined RB/p53 loss in the presence of hepatocarinogen promotes aggressive tumor development in the mouse liver, and could be particularly reflective of conditions associated with advanced human disease. To interrogate this concept, the gene expression profile for RB/p53-deficient, DEN-treated livers was employed to cluster independent human gene expression datasets (Figure 6C). These data sets are annotated for disease stage, enhanced proliferation, differentiation status, and patient survival. As shown, the gene expression signature associated with RB/p53-pathway inactivation in the presence of heptocarcinogen is significantly associated with advanced human disease (p=1.19E-7) (Figure 6C). Furthermore, the additive gene deregulation associated with aberrant cell cycle control observed with combined RB/p53 loss was indicative of poor clinical outcome (Figure 6D). Thus, the RB/p53-deficient mouse model of liver tumorigenesis described herein demonstrates both transcriptional deregulation and chromosomal instability indicative of aggressive human disease.
Discussion
Hepatocellular carcinoma is complex disease that involves multiple oncogenic and tumor suppressive pathways. In this context, loss of RB and p53 has been observed via cytogenetic or CGH approaches in many cancers 2, 3, 29. Additionally, in the case of the RB-pathway, loss of p16ink4a or overexpression of Cyclin D1 is also observed in human cancers, including HCC 30, 31. Similarly, in the context of the p53-pathway, expression of MDM2 is associated with liver tumor development 32. These findings suggest that, like in other human cancers, there is a selection for loss of these tumor suppressor pathways in the development of HCC. In the present study, we used an in vivo model of liver-specific inactivation of Rb and p53 to determine how loss of these tumor suppressive pathways cooperates in liver tumor development.
Initial observations revealed that while RB/p53 loss influenced molecular and biological events in developing mouse livers, spontaneous tumor formation did not occur. In fact, RB/p53-deficient livers demonstrated strikingly normal tissue development with the exception of enhanced liver size, which was not related to deregulated proliferation and likely a reflection of larger hepatocytes. While in many tissues RB loss alone is not sufficient for tumor development, coordinate loss of RB/p53 has been shown to induce tumors in all tissues analyzed to date 33–35. Here, RB/p53 loss did not significantly impact the establishment of quiescence in the liver, indicating that there are potent mechanisms to suppress neoplastic transformation in this tissue. Specifically, our data suggests that quiescence mechanisms regulated by p130 and C/EBPα were not altered in response to RB/p53 loss, likely representing an important mechanism of tumor suppression in the liver. As such, a recent study in mice demonstrated a critical role for C/EBPα phosphorylation in preventing both spontaneous and carcinogen-induced liver cancer 36. Combined, these studies demonstrate that the liver harbors potent tumor suppressive mechanisms capable of maintaining normal proliferative control in the absence of functional RB/p53 pathways.
Interestingly, while combined RB/p53 loss was not sufficient to drive spontaneous tumor formation, the transcriptional profile of RB/p53-deficient neonatal livers was strikingly similar to the transcriptional profile of human HCC. These results suggest that while transcriptional deregulation as a result of tumor suppressor loss is seemingly predictive of disease, gene deregulation alone is not sufficient to promote disease development under otherwise unchallenged conditions, and additional aberrations are required to promote tumorigenesis. A key etiological aspect of HCC is genetic damage to hepatocytes, which occurs as the result of proliferation under chronic stress conditions (i.e. inflammation) or the presence of genotoxic agents 2, 3, 29. Presumably, such environmental stresses exploit the loss of tumor suppressor pathways to drive transformation. Consistent with this concept, loss of either RB or p53 had a significant impact on response to hepatocarcinogen. Through both transcriptional profiling and functional analyses, we demonstrate that the influence of RB and p53 on DNA damage signaling is quite distinct and represents additive pathways in coordinating stress response. While RB loss alone deregulated cell cycle genes coordinated by E2F (e.g. Top2α, Ccna2, Bub1), loss of p53 augmented this response. Conversely, p53 loss prevented the induction of genes associated with DNA damage checkpoints (e.g. Ccng1, Cdkn1a, Plk2), which subsequently demonstrated a requisite role for p53-dependent signaling in the G2/M transition. Together, these data reveal a key molecular basis of cooperation between RB and p53 in DNA damage response. Ultimately, the critical nature of RB/p53 loss in response to genotoxic challenge was manifest through genomic instability, and aberrant loci observed in this model encompassed regions frequently disrupted in human HCC (e.g. 14q31-32, 6p21-23, 11q13) 2, 29, 37. Importantly, analyses of human liver samples revealed that the signature of RB/p53 loss in the presence of challenge is reflected in advanced disease, suggesting that continued hepatocarcinogenic stresses are present in the progression of disease and modulated by perturbations in the RB/p53 pathways. Consistent with this concept, genes that are most reflective of the common effect of RB/p53 loss in response to stress (i.e. genes coordinated in a synonymous fashion) were associated with human HCC exhibiting poor overall prognosis. Thus, these data demonstrate a model of HCC in which coordinate loss of RB/p53 leads to both transcriptional deregulation and chromosomal aberrations consistent with human disease.
The model described herein provided an opportunity to decipher which facets of DNA damage response and cell cycle control are particularly relevant to tumor development in the liver. These studies demonstrate a tissue in which loss of key tumor suppressor pathways are ultimately compensated for in an “unchallenged” setting, but are critical for preventing disease development in response to genotoxic stress. Furthermore, these data provide insight into the molecular mechanisms that promote tumorigenesis and the unique environmental factors that are associated with disease development in the liver.
Supplementary Material
Acknowledgments
Grant Support: This study was funded through NIH Grant 5R01CA127387
Abbreviations
- CDK
cyclin dependent kinase
- CGH
Comparative Genomic Hybridization
- DEN
diethylnitrosamine
- GEO
Gene Expression Omnibus
- H&E
hematoxylin and eosin
- HCC
hepatocellular carcinoma
- KEGG
Kyoto Encyclopedia of Genes and Genomes
- MIAME
Minimum Information About a Microarray Experiment
- MCM7
minichromosmoe maintenance 7
- MDM2
murine double minute 2
- PCNA
proliferating cell nuclear antigen
- PCR
polymerase chain reaction
- RB
retinoblastoma
Footnotes
Disclosures: There are no financial conflicts to disclose.
Microarray/aCGH Repository Information: Raw data is accessible through GEO Series accession number GSE24347
Reviewer URL: http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?token=ttytrcymmckuixm&acc=GSE24347
Writing Assistance: No writing assistance was provided.
Author Contributions:
A. Kathleen McClendon: study concept/design, acquisition of data, analysis/interpretation of data, drafting of manuscript
Jeffry L. Dean: acquisition of data, analysis/interpretation of data, drafting of manuscript
Adam Ertel: technical support, analysis/interpretation of data, statistical analysis, drafting of manuscript
Zhiyan Fu: technical/material support, analysis/interpretation of data, statistical analysis
Dayana B. Rivadeneira: acquisition of data, analysis/interpretation of data, critical reading of manuscript
Christopher A. Reed: technical support, analysis/interpretation of data, critical reading of manuscript
Ryan J. Bourgo: acquisition of data, technical/material support
Agnieszka Witkiewicz: technical/material support, analysis/interpretation of data
Sankar Addya: technical/material support, analysis/interpretation of data, statistical analysis
Christopher N. Mayhew: study concept/design, critical reading of manuscript
H. Leighton Grimes: study concept/design, material support
Paolo Fortina: technical/material support, analysis/interpretation of data
Erik S. Knudsen: study concept/design, analysis/interpretation of data, drafting of manuscript, obtained funding, study supervision
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.El-Serag HB, Rudolph KL. Hepatocellular carcinoma: epidemiology and molecular carcinogenesis. Gastroenterology. 2007;132:2557–76. doi: 10.1053/j.gastro.2007.04.061. [DOI] [PubMed] [Google Scholar]
- 2.Teufel A, Staib F, Kanzler S, Weinmann A, Schulze-Bergkamen H, Galle PR. Genetics of hepatocellular carcinoma. World J Gastroenterol. 2007;13:2271–82. doi: 10.3748/wjg.v13.i16.2271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wong CM, Ng IO. Molecular pathogenesis of hepatocellular carcinoma. Liver Int. 2008;28:160–74. doi: 10.1111/j.1478-3231.2007.01637.x. [DOI] [PubMed] [Google Scholar]
- 4.Yu MC, Yuan JM. Environmental factors and risk for hepatocellular carcinoma. Gastroenterology. 2004;127:S72–8. doi: 10.1016/j.gastro.2004.09.018. [DOI] [PubMed] [Google Scholar]
- 5.McGivern DR, Lemon SM. Tumor Suppressors, Chromosomal Instability, and Hepatitis C Virus-Associated Liver Cancer. Annu Rev Pathol. 2008 doi: 10.1146/annurev.pathol.4.110807.092202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Azam F, Koulaouzidis A. Hepatitis B virus and hepatocarcinogenesis. Ann Hepatol. 2008;7:125–9. [PubMed] [Google Scholar]
- 7.Iakova P, Timchenko L, Timchenko NA. Intracellular signaling and hepatocellular carcinoma. Semin Cancer Biol. 2010;21:28–34. doi: 10.1016/j.semcancer.2010.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lau SH, Guan XY. Cytogenetic and molecular genetic alterations in hepatocellular carcinoma. Acta Pharmacol Sin. 2005;26:659–65. doi: 10.1111/j.1745-7254.2005.00126.x. [DOI] [PubMed] [Google Scholar]
- 9.Nevins JR. The Rb/E2F pathway and cancer. Hum Mol Genet. 2001;10:699–703. doi: 10.1093/hmg/10.7.699. [DOI] [PubMed] [Google Scholar]
- 10.Markey MP, Angus SP, Strobeck MW, Williams SL, Gunawardena RW, Aronow BJ, Knudsen ES. Unbiased analysis of RB-mediated transcriptional repression identifies novel targets and distinctions from E2F action. Cancer Res. 2002;62:6587–97. [PubMed] [Google Scholar]
- 11.Vernell R, Helin K, Muller H. Identification of target genes of the p16INK4A-pRB-E2F pathway. J Biol Chem. 2003;278:46124–37. doi: 10.1074/jbc.M304930200. [DOI] [PubMed] [Google Scholar]
- 12.Knudsen ES, Knudsen KE. Tailoring to RB: tumour suppressor status and therapeutic response. Nat Rev Cancer. 2008 doi: 10.1038/nrc2401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mayhew CN, Carter SL, Fox SR, Sexton CR, Reed CA, Srinivasan SV, Liu X, Wikenheiser-Brokamp K, Boivin GP, Lee JS, Aronow BJ, Thorgeirsson SS, Knudsen ES. RB loss abrogates cell cycle control and genome integrity to promote liver tumorigenesis. Gastroenterology. 2007;133:976–84. doi: 10.1053/j.gastro.2007.06.025. [DOI] [PubMed] [Google Scholar]
- 14.Sherr CJ, McCormick F. The RB and p53 pathways in cancer. Cancer Cell. 2002;2:103–12. doi: 10.1016/s1535-6108(02)00102-2. [DOI] [PubMed] [Google Scholar]
- 15.Wise-Draper TM, Wells SI. Papillomavirus E6 and E7 proteins and their cellular targets. Front Biosci. 2008;13:1003–17. doi: 10.2741/2739. [DOI] [PubMed] [Google Scholar]
- 16.Menendez D, Inga A, Resnick MA. The expanding universe of p53 targets. Nat Rev Cancer. 2009;9:724–37. doi: 10.1038/nrc2730. [DOI] [PubMed] [Google Scholar]
- 17.Boehme KA, Blattner C. Regulation of p53--insights into a complex process. Crit Rev Biochem Mol Biol. 2009;44:367–92. doi: 10.3109/10409230903401507. [DOI] [PubMed] [Google Scholar]
- 18.Meek DW. Tumour suppression by p53: a role for the DNA damage response? Nat Rev Cancer. 2009;9:714–23. doi: 10.1038/nrc2716. [DOI] [PubMed] [Google Scholar]
- 19.Shakhova O, Leung C, van Montfort E, Berns A, Marino S. Lack of Rb and p53 delays cerebellar development and predisposes to large cell anaplastic medulloblastoma through amplification of N-Myc and Ptch2. Cancer Res. 2006;66:5190–200. doi: 10.1158/0008-5472.CAN-05-3545. [DOI] [PubMed] [Google Scholar]
- 20.Jiang Z, Deng T, Jones R, Li H, Herschkowitz JI, Liu JC, Weigman VJ, Tsao MS, Lane TF, Perou CM, Zacksenhaus E. Rb deletion in mouse mammary progenitors induces luminal-B or basal-like/EMT tumor subtypes depending on p53 status. J Clin Invest. 2010;120:3296–309. doi: 10.1172/JCI41490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jonkers J, Meuwissen R, van der Gulden H, Peterse H, van der Valk M, Berns A. Synergistic tumor suppressor activity of BRCA2 and p53 in a conditional mouse model for breast cancer. Nat Genet. 2001;29:418–25. doi: 10.1038/ng747. [DOI] [PubMed] [Google Scholar]
- 22.Litovchick L, Sadasivam S, Florens L, Zhu X, Swanson SK, Velmurugan S, Chen R, Washburn MP, Liu XS, DeCaprio JA. Evolutionarily conserved multisubunit RBL2/p130 and E2F4 protein complex represses human cell cycle-dependent genes in quiescence. Mol Cell. 2007;26:539–51. doi: 10.1016/j.molcel.2007.04.015. [DOI] [PubMed] [Google Scholar]
- 23.Wang GL, Salisbury E, Shi X, Timchenko L, Medrano EE, Timchenko NA. HDAC1 cooperates with C/EBPalpha in the inhibition of liver proliferation in old mice. J Biol Chem. 2008;283:26169–78. doi: 10.1074/jbc.M803544200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Watanabe T, Tanaka G, Hamada S, Namiki C, Suzuki T, Nakajima M, Furihata C. Dose-dependent alterations in gene expression in mouse liver induced by diethylnitrosamine and ethylnitrosourea and determined by quantitative real-time PCR. Mutat Res. 2009;673:9–20. doi: 10.1016/j.mrgentox.2008.11.004. [DOI] [PubMed] [Google Scholar]
- 25.Taylor WR, Stark GR. Regulation of the G2/M transition by p53. Oncogene. 2001;20:1803–15. doi: 10.1038/sj.onc.1204252. [DOI] [PubMed] [Google Scholar]
- 26.Kimura SH, Ikawa M, Ito A, Okabe M, Nojima H. Cyclin G1 is involved in G2/M arrest in response to DNA damage and in growth control after damage recovery. Oncogene. 2001;20:3290–300. doi: 10.1038/sj.onc.1204270. [DOI] [PubMed] [Google Scholar]
- 27.Burns TF, Fei P, Scata KA, Dicker DT, El-Deiry WS. Silencing of the novel p53 target gene Snk/Plk2 leads to mitotic catastrophe in paclitaxel (taxol)-exposed cells. Mol Cell Biol. 2003;23:5556–71. doi: 10.1128/MCB.23.16.5556-5571.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lee J, Kim JA, Barbier V, Fotedar A, Fotedar R. DNA damage triggers p21WAF1-dependent Emi1 down-regulation that maintains G2 arrest. Mol Biol Cell. 2009;20:1891–902. doi: 10.1091/mbc.E08-08-0818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Herath NI, Leggett BA, MacDonald GA. Review of genetic and epigenetic alterations in hepatocarcinogenesis. J Gastroenterol Hepatol. 2006;21:15–21. doi: 10.1111/j.1440-1746.2005.04043.x. [DOI] [PubMed] [Google Scholar]
- 30.Matsuda Y. Molecular mechanism underlying the functional loss of cyclindependent kinase inhibitors p16 and p27 in hepatocellular carcinoma. World J Gastroenterol. 2008;14:1734–40. doi: 10.3748/wjg.14.1734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhang YJ, Jiang W, Chen CJ, Lee CS, Kahn SM, Santella RM, Weinstein IB. Amplification and overexpression of cyclin D1 in human hepatocellular carcinoma. Biochem Biophys Res Commun. 1993;196:1010–6. doi: 10.1006/bbrc.1993.2350. [DOI] [PubMed] [Google Scholar]
- 32.Endo K, Ueda T, Ohta T, Terada T. Protein expression of MDM2 and its clinicopathological relationships in human hepatocellular carcinoma. Liver. 2000;20:209–15. doi: 10.1034/j.1600-0676.2000.020003209.x. [DOI] [PubMed] [Google Scholar]
- 33.Martinez-Cruz AB, Santos M, Lara MF, Segrelles C, Ruiz S, Moral M, Lorz C, Garcia-Escudero R, Paramio JM. Spontaneous squamous cell carcinoma induced by the somatic inactivation of retinoblastoma and Trp53 tumor suppressors. Cancer Res. 2008;68:683–92. doi: 10.1158/0008-5472.CAN-07-3049. [DOI] [PubMed] [Google Scholar]
- 34.Zhou Z, Flesken-Nikitin A, Corney DC, Wang W, Goodrich DW, Roy-Burman P, Nikitin AY. Synergy of p53 and Rb deficiency in a conditional mouse model for metastatic prostate cancer. Cancer Res. 2006;66:7889–98. doi: 10.1158/0008-5472.CAN-06-0486. [DOI] [PubMed] [Google Scholar]
- 35.Berman SD, Calo E, Landman AS, Danielian PS, Miller ES, West JC, Fonhoue BD, Caron A, Bronson R, Bouxsein ML, Mukherjee S, Lees JA. Metastatic osteosarcoma induced by inactivation of Rb and p53 in the osteoblast lineage. Proc Natl Acad Sci U S A. 2008;105:11851–6. doi: 10.1073/pnas.0805462105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wang GL, Shi X, Haefliger S, Jin J, Major A, Iakova P, Finegold M, Timchenko NA. Elimination of C/EBPalpha through the ubiquitin-proteasome system promotes the development of liver cancer in mice. J Clin Invest. 2010;120:2549–62. doi: 10.1172/JCI41933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Nishida N, Nishimura T, Ito T, Komeda T, Fukuda Y, Nakao K. Chromosomal instability and human hepatocarcinogenesis. Histol Histopathol. 2003;18:897–909. doi: 10.14670/HH-18.897. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.