Abstract
Given the tremendous growth in the application of titanium dioxide nanoparticles (TNPs), concerns about the potential health hazards of TNPs to humans have been raised. Poly(ADP-ribose) polymerase 1 (PARP-1), a highly conserved DNA-binding protein, is involved in many molecular and cellular processes. Limited data demonstrated that certain nanomaterials induced the aberrant hypermethylation of PARP-1. However, the mechanism involved in TNP-induced PARP-1 abnormal methylation has not been studied. A549 cells were incubated with anatase TNPs (22.1 nm) for 24 hours pretreatment with or without methyltransferase inhibitor 5-aza-2′-deoxycytidine and the reactive oxygen species (ROS) scavenger α-lipoic acid to assess the possible role of methylation and ROS in the toxic effect of TNPs. After TNPs characterization, a battery of assays was performed to evaluate the toxic effect of TNPs, PARP-1 methylation status, and oxidative damage. Results showed that TNPs decreased the cell viability in a dose-dependent manner, in accordance with the increase of lactate dehydrogenase activity, which indicated membrane damage of cells. Similar to the high level of PARP-1 methylation, the generation of ROS was significantly increased after exposure to TNPs for 24 hours. Furthermore, α-lipoic acid decreased TNP-induced ROS generation and then attenuated TNP-triggered PARP-1 hypermethylation. Meanwhile, 5-aza-2′-deoxycytidine simultaneously decreased the ROS generation induced by TNPs, resulting in the decline of PARP-1 methylation. In summary, TNPs triggered the aberrant hypermethylation of the PARP-1 promoter and there was a cross talk between oxidative stress and PARP-1 methylation in the toxic effect of TNPs.
Keywords: titanium dioxide nanoparticles, PARP-1, oxidative stress, DNA methylation
Introduction
Titanium dioxide (TiO2) is a natural mineral used broadly in domestic and cosmetic products, including antifouling paints, coatings, ceramics, and additives in pharmaceuticals, food colorants, and sunscreen owing to its typical characteristics such as surface adsorption, photo-catalysis, and ultraviolet (UV) absorption. Titanium, either pure or in alloys, is also extensively used for implanted medical devices, such as dental implants, joint replacements, cardiovascular stents, and spinal fixation devices. Titanium dioxide nanoparticles (TNPs) are absorbed through inhalation, ingestion, and dermal penetration into the body, and distributed in important organs such as lungs,1,2 lymph nodes,3 brain, liver, and kidneys.1
There are growing concerns about the possible influence of TNPs on human health. It has been shown that ultrafine TiO2 particles could induce impairment of macrophage function, persistently high inflammatory reactions, and increased pulmonary retention, compared with fine TiO2.4 Particularly, TNPs were reported to induce reactive oxygen species (ROS) generation in many cell types, including rat neuronal cells5–8 and human hepatoma cells.9 Indeed, the capacity of TNPs to produce ROS is frequently proposed to account for their toxic effects.5,10,11 Based on the evidences outlined earlier, it seems that TNPs can exert their toxic effects through the formation of ROS. Hence, TNPs were evaluated by the World Health Organization (WHO)/International Agency for Research on Cancer (IARC) as a Group 2B compound,12 which is possibly carcinogenic to humans. However, much still remains to be elucidated concerning the possible health effects of TNPs.
DNA methylation, as an important regulatory factor of gene expression,13 not only is crucial in the development of normal mammalian cells, but also plays a fundamental role in epigenetic silencing of cancer-related genes in tumorigenesis.14 In general, a low level of DNA methylation is considered an activation of gene, while hypermethylation can contribute to inactivation.15 The expression of several cancer-related genes has been reported to be silenced by DNA methylation of their promoter regions.
Poly(ADP-ribose) polymerase 1 (PARP-1), a highly conserved DNA-binding protein, is involved in many molecular and cellular processes, which include DNA repair, proliferation, and chromatin modification.16 PARP-1 was also found in a complex with the DNA methyltransferase DNMT1, the his-tone H3K9 methyltransferase G9a, and the histone ubiquitin ligase Np95, indicative of a link between poly(ADP-ribosyl) ation and the epigenome,17 and was described as a fundamental constituent of the transcription machinery that interacts with and modulates the activities of several transcription factors.18,19 Chemical materials such as benzene and silica have been reported to cause the methylation of PARP-1 promoters.20,21 A certain nanomaterial, nano-SiO2, was confirmed to induce the aberrant hypermethylation and inactivity of PARP-1.22 However, the mechanisms responsible for TNP-induced PARP-1 abnormal methylation have not been elucidated.
It was reported that oxidative stress influenced the occurrence of disease through regulating the gene methylation status.23 Tunc and Tremellen found that oxidative stress was associated with gene hypomethylation, on the contrary, and antioxidant supplements reduced oxidative damage and reverted the gene methylation level.24 Therefore, this study focused on the regulating mechanisms between oxidative stress and PARP-1 methylation involved in the toxic effect of anatase TNPs. In this study, we analyzed for the first time the epigenetic regulatory mechanism of PARP-1 in TNP-treated cells. After TNP characterization, batteries of assays were performed to evaluate the toxic effect, PARP-1 methylation status, and oxidative damage in TNP-exposed A549 cells. Results showed that TNPs triggered the generation of ROS and aberrant hypermethylation in the PARP-1 promoter region. We also used the inhibitor 5-aza-2′-deoxycytidine (5-aza) and α-lipoic acid (α-LA) to analyze the underlying mechanisms of the toxic effect of TNPs, suggesting that there was a cross talk between oxidative stress and PARP-1 methylation.
Materials and methods
Characterization of the TNPs
Commercial anatase TNPs, with a purity of at least 99.7% TiO2, were purchased from Sigma-Aldrich Co. (St Louis, MO, USA). The primary particle size was <25 nm with a specific surface area of 45–55 m2/g. The particle size and distribution of the TNPs were measured by transmission electron microscope (TEM) (JEOL, Tokyo, Japan) and Image J software. The hydrodynamic sizes and zeta potential of TNPs in ultrapure water as stock media and serum-free Roswell Park Memorial Institute 1640 (RPMI 1640) as culture media were examined by Zetasizer (Malvern Nano-ZS90; Malvern, UK). Suspensions of TNPs were dispersed by a sonicator (Bioruptor UCD-200; Diagenode, Liège, Belgium) at 300 W for 10 minutes before adding to dispersion media in order to minimize their aggregation.
Cell culture and the TNPs exposure
The human alveolar epithelial cells (A549) were obtained from the National Institute of Occupational Health and Poisons Control, Chinese Center for Diseases Control and Prevention. The use of this cell line was approved by the Ethical Committee of Capital Medical University, Beijing, People’s Republic of China. The cells were cultured in RPMI 1640 (HyClone, Logan, UT, USA) supplemented with 10% fetal bovine serum (Gibco, product line of Thermo Fisher Scientific, Waltham, MA, USA), 100 U/mL penicillin, and 100 μg/mL streptomycin at 37°C in a humidified incubator with 5% CO2. For experiments, 3×105 A549 cells were seeded in a six-well culture plate for 24 hours. The cells were then washed twice with phosphate buffered saline and treated with TNPs suspended in RPMI 1640 of certain concentrations (6.25 μg/mL, 12.5 μg/mL, 25 μg/mL, 50 μg/mL, and 100 μg/mL) for another 24 hours. The suspension of TNPs was sterilized by an autoclave and dispersed by a sonicator (300 W, 10 minutes) at room temperature to minimize their aggregation. Cells maintained in RPMI 1640 without TNPs were used as the control group.
Cell viability assay
The cytotoxicity of TNPs was determined using 3-(4,5-dimethyl-2-thiazolyl)-2,5-diphenyl-2-H-tetrazolium bromide (MTT). Briefly, 1×104 cells were seeded into a 96-well plate in a volume of 100 μL RPMI 1640 and incubated for 24 hours at 37°C. Cells were treated with varying concentrations of TNPs (6.25 μg/mL, 12.5 μg/mL, 25 μg/mL, 50 μg/mL, and 100 μg/mL). After 24 hours incubation, 10 μL MTT was added to each well at 5 mg/mL and further incubated for 4 hours. Next, 150 μL of dimethylsulfoxide was added in and mixed thoroughly for 10 minutes. Optical density was then measured with a microplate reader (Thermo Multiskan MK3; Thermo Fisher Scientific) at 492 nm according to the instructions.
Assessment of LDH activity
The lactate dehydrogenase (LDH) leakage assay, which is based on the measurement of LDH activity in the extracellular medium, was determined using a commercial LDH kit (Nanjing Jiancheng Bioengineering Institute, Nanjing, People’s Republic of China) according to the manufacturer’s protocols. The loss of intracellular LDH and its release into the culture medium is an indicator of irreversible cell death due to cell membrane damage. After cells were treated with different concentrations of TNPs for 24 hours, the supernatants were collected for LDH measurement. A cell medium of 100 μL was used for LDH activity analysis and the absorbance at 440 nm was measured by a UV-visible spectrophotometer (SpectraMax M5; Molecular Devices, Sunnyvale, CA, USA).
Intracellular ROS measurement
Intracellular ROS were measured by flow cytometry using 2,7-dichlorofluorescein diacetate (DCFH-DA) (Nanjing Jiancheng Bioengineering Institute) as a probe. Briefly, ~3×105 cells were seeded into a six-well plate for 24-hours pretreatment with or without methyltransferase inhibitor, 5-aza (5 μM, 10 μM, and 20 μM), or ROS scavenger α-LA (0.1 μM, 10 μM, and 100 μM) for 1 hour, and A549 cells were incubated with TNPs (6.25 μg/mL, 12.5 μg/mL, 25 μg/mL, 50 μg/mL, and 100 μg/mL) for 24 hours at 37°C. Then, cells were washed twice with phosphate buffered saline and co-incubated with serum-free RPMI 1640 containing 10 μM DCFH-DA for 30 minutes at 37°C in dark, according to the manufacturer’s instructions. The cells were trypsinized and intracellular ROS were measured using flow cytometry (BD LSRFortessa™, BD, Franklin Lakes, NJ, USA) at 488 nm excitation and 525 nm emission.
DNA isolation and methylation-specific PCR
After A549 cells were pretreated with or without methyltransferase inhibitor, 5-aza (5 μM, 10 μM, and 20 μM), or ROS scavenger α-LA (0.1 μM, 10 μM, and 100 μM) for 1 hour, the cells were incubated with TNPs for another 24 hours at 37°C. Genomic DNA was extracted following the protocols of the DNA kit (Solarbio, Beijing, People’s Republic of China). One microgram purified DNA was subjected to bisulfate modification, which was performed using CpGenome DNA Modification Kit (Merck Millipore, Billerica, MA, USA) according to the manufacturer’s instructions. polymerase chain reaction (PCR) Magic Mix 3.0 (TIANDZ, Beijing, People’s Republic of China) was used to detect the PARP-1 methylation under the following conditions: 94°C for 30 seconds; 35 cycles of 94°C for 20 seconds, 60°C for 45 seconds, and 72°C for 1 minute; 72°C for 3 minutes. These methylation-specific PCR (MSP) products were analyzed by 2% agarose gel electrophoresis staining with ethidium bromide and visualized under a UV illuminator. Distinct visible band of the amplicon with methylation-specific primers was considered positive. The density of each band was analyzed by image analysis software (Gel-Pro 4.5) for quantization. The sequences of primers were as follows:
For methylated DNA: MF-PARP-1 (5′-GAGATTAGT TTAGTTAATTTGGCGA-3′) and MR-PARP-1 (5′-ATCT AAAACTCCTAAACCCAACGAT-3′), a 139 bp fragment (−299 to −161 relative to transcription start site), was chosen to amplify by M primer; and for unmethylated DNA: UF-PARP-1 (5′-GAGATTAGTTTAGTTAATTTGGTGA-3′) and UR-PARP-1 (5′-ATCTAAAACTCCTAAACCCAACA AT-3′), a 139 bp fragment (−299 to −161 relative to transcription start site), was chosen to amplify by U primer.
Statistical analysis
Statistical analysis was performed using SPSS 16.0 software (SPSS Inc., Chicago, IL, USA). Results were expressed as mean ± standard deviation of independent experiments. The least significant difference test was used to compare means of two samples, while one-way analysis of variance was used for group means. In all cases, P<0.05 was considered to be statistically significant.
Results
Characterization of TNPs
We first characterized the TNPs by TEM. As shown in Figure 1A, a near-spherical particle shape, with the mean pristine particle diameter of 22.1±8.9 nm (Figure 1B) was obtained by measuring 500 random particles using ImageJ. Hydrodynamic measurements of both ultrasonicated TNPs in ultrapure water and serum-free RPMI 1640 at 1 hour and a concentration of 100 μg/mL by dynamic light scattering are shown in Table 1. TNPs in both dispersion mediums exhibited a larger hydrodynamic size than the original dimension, suggesting that TNPs formed aggregates in mediums due to the van der Waals force and hydrophobic interaction with surrounding media. The polydispersity index values acquired in our study further corroborate the observations made, with stable values being observed for particles dispersed in both ultrapure water and RPMI 1640. Zeta potentials provide quantitative information on the stability of the particles, confirming that the particles are more likely to remain dispersed if the absolute value of zeta potentials is higher than 30 mV.25 Consequently, zeta potential values in RPMI 1640 and ultrapure water varied between −29.8 mV and −11.9 mV. Nonetheless, the larger aggregates represent the actual scenario that the cells were exposed to.
Figure 1.
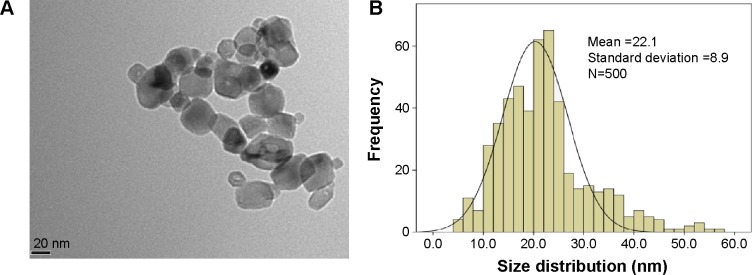
Characterization of TNPs by TEM.
Notes: Particle shape was analyzed by TEM (A) and the size distribution in the test media were evaluated by ImageJ software (B).
Abbreviations: TNPs, titanium dioxide nanoparticles; TEM, transmission electron microscope.
Table 1.
The hydrodynamic size and zeta potential of the TNPs in ultrapure water and RPMI 1640 culture media by DLS analysis
| Ultrapure water
|
RPMI 1640
|
|||||
|---|---|---|---|---|---|---|
| Hydrodynamic size (nm) | Zeta potential (mV) | PDI | Hydrodynamic size (nm) | Zeta potential (mV) | PDI | |
| 1 hour | 276.9±9.3 | −29.8±2.7 | 0.5±0.4 | 504.8±108.1 | −11.9±1.2 | 0.5±0.1 |
Abbreviations: TNPs, titanium dioxide nanoparticles; RPMI, Roswell Park Memorial Institute; DLS, dynamic light scattering; PDI, polydispersity index.
Cytotoxicity induced by the TNPs
To evaluate the possible toxicity of TNPs on A549 cells, cell viability was determined after exposing to TNPs (0 μg/mL, 6.25 μg/mL, 12.5 μg/mL, 25 μg/mL, 50 μg/mL, and 100 μg/mL) for 24 hours. As shown in Figure 2, with the dosages increasing, viability of A549 cells induced by TNPs was significantly decreased than control. In addition, the MTT results were strongly in accordance with the increased membrane damage measured by LDH release (Figure 3). Our results suggested that TNPs induced cytotoxicity in a dose-dependent manner.
Figure 2.
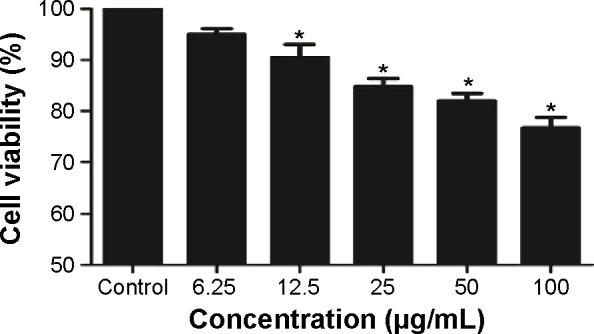
Cell viability of A549 cells was measured by MTT assay after 24 hours TNP exposure.
Notes: Cell viability was significantly decreased in a dose-dependent manner after TNP treatment. *P<0.05, compared with control group, n=5.
Abbreviations: MTT, 3-(4,5-dimethyl-2-thiazolyl)-2,5-diphenyl-2-H-tetrazolium bromide; TNP, titanium dioxide nanoparticle.
Figure 3.
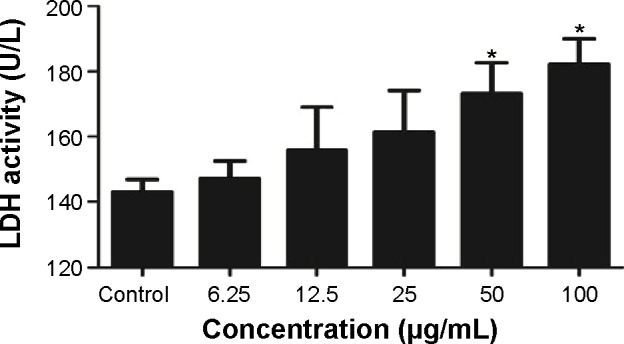
LDH activity was detected in TNP-treated A549 cells.
Notes: After cells were exposed to TNPs (0 μg/mL, 6.25 μg/mL, 12.5 μg/mL, 25 μg/mL, 50 μg/mL, and 100 μg/mL) for 24 hours, the supernatants were determined using a commercial LDH kit according to the manufacturer’s protocols. *P<0.05, compared with control group, n=5.
Abbreviations: LDH, lactate dehydrogenase; TNPs, titanium dioxide nanoparticles.
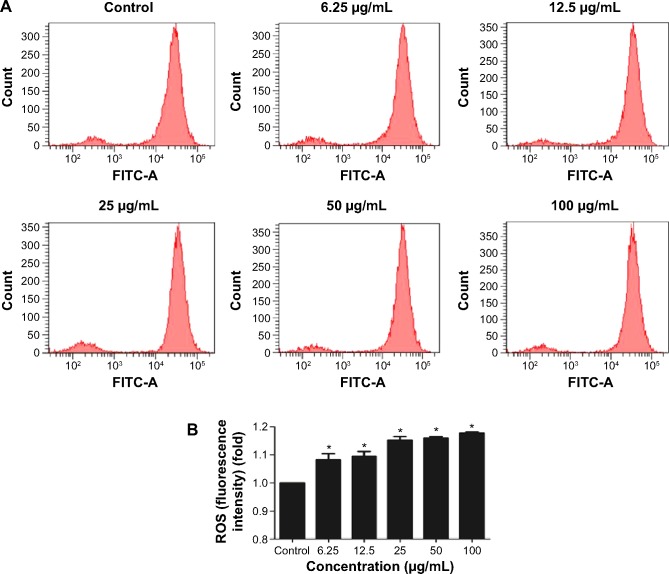
Intracellular ROS generation induced by TNPs
To get a closer insight into the possible mechanism of TNP-induced cellular toxicity, the intracellular ROS levels were determined by using the DCFH-DA probe. From Figure 4A and B, we observed the gradually elevated ROS levels in A549 cells in a dose-dependent manner. The generation of ROS in all TNP-treated groups was significantly different with control group. Compared to the group of treatment with TNPs only, the fluorescence intensity of ROS in the group of pretreatment with methyltransferase inhibitor 5-aza was relatively weak (Figure 5A), in line with the effect of ROS scavenger α-LA pretreatment (Figure 5B). It suggested that the methyltransferase inhibitor markedly decreased the generation of ROS induced by TNPs.
Figure 4.
Effects of TNPs on intracellular ROS generation in A549 cells.
Notes: The typical picture of ROS detected by flow cytometry (A); the intracellular ROS level significantly increased in a dose-dependent manner (B). Data are expressed as mean ± SD. n=5, *P<0.05, compared with control.
Abbreviations: FITC, fluorescein isothiocyanate; TNPs, titanium dioxide nanoparticles; ROS, reactive oxygen species; SD, standard deviation.
Figure 5.
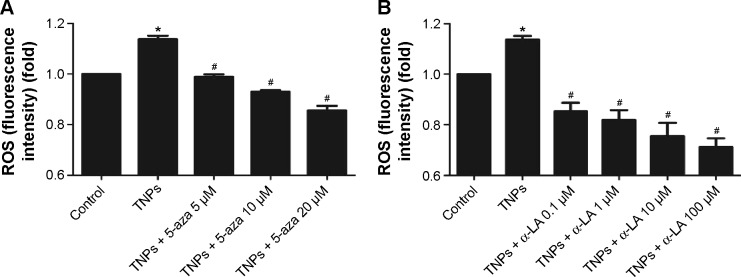
Effects of inhibitors, 5-aza or α-LA, on TNP-induced ROS generation in A549 cells.
Notes: The methyltransferase inhibitor, 5-aza decreased TNPs-induced ROS generation (A); the antioxidant agent, α-LA suppressed ROS generation triggered by TNPs (B). Data are expressed as mean ± SD; n=5, *P<0.05, compared with control; #P<0.05, compared with TNPs.
Abbreviations: 5-aza, 5-aza-2′-deoxycytidine; α-LA, α-lipoic acid; TNPs, titanium dioxide nanoparticles; ROS, reactive oxygen species; SD, standard deviation.
PARP-1 methylation alteration induced by TNPs
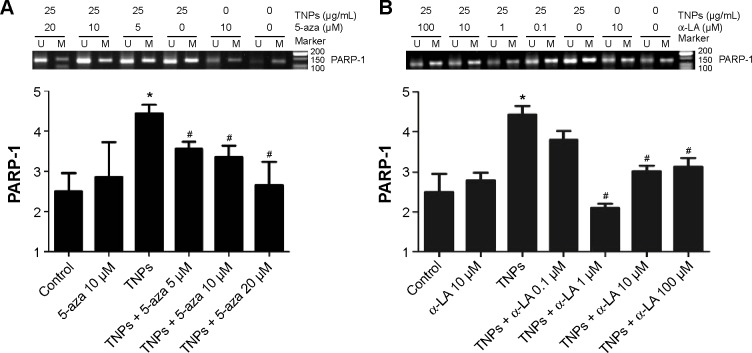
MSP, a common detection method of methylation, was performed to analyze the methylation status of PARP-1. Our results revealed that TNPs notably elevated the level of PARP-1 methylation in a dose-dependent way (Figure 6). However, pretreatment of both methyltransferase inhibitor 5-aza and ROS scavenger α-LA inversely altered the TNP-induced hypermethylation of PARP-1 promoter region (Figure 7A and B), implicating that the antioxidant agent α-LA decreased TNP-induced ROS generation and simultaneously attenuated TNP-triggered PARP-1 hypermethylation.
Figure 6.
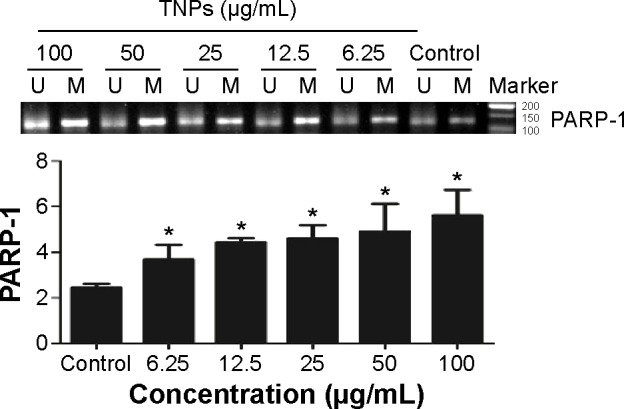
TNPs induced the alternation of PARP-1 methylation status.
Notes: A549 cells were exposed to various concentrations (6.25 μg/mL, 12.5 μg/mL, 25 μg/mL, 50 μg/mL, and 100 μg/mL) of TNPs for 24 hours, methylation-specific PCR was performed to examine the methylation status of PARP-1. U and M: primer sets specific to unmethylated (U) and methylated (M) DNA molecules. The data were typical examples of five independent experiments. *P<0.05, compared with control.
Abbreviations: TNPs, titanium dioxide nanoparticles; PARP-1, poly(ADP-ribose) polymerase 1; PCR, polymerase chain reaction.
Figure 7.
Effects of inhibitors, 5-aza or α-LA, on TNP-induced methylation of PARP-1 by MSP in A549 cells.
Notes: Treatment with methyltransferase inhibitor, 5-aza (A); treatment with antioxidant agent, α-LA (B). U and M: primer sets specific to unmethylated (U) and methylated (M) DNA molecules. The data were typical examples of five independent experiments. *P<0.05, compared with control; #P<0.05, compared with TNPs.
Abbreviations: 5-aza, 5-aza-2′-deoxycytidine; α-LA, α-lipoic acid; TNPs, titanium dioxide nanoparticles; PARP-1, poly(ADP-ribose) polymerase 1; MSP, methylation-specific PCR; PCR, polymerase chain reaction.
Discussion
With the increasing TNP-related products, the concern of governments and the public about the health and environmental risks are gradually growing. However, the effects induced by TNPs on human health or environment and the mechanisms of toxicity are still poorly investigated.
TEM provides the direct measurement of particle size, distribution, and morphology by image analysis, while dynamic light scattering, a common analysis for the size of nanoparticles and determining their aggregation in suspension,26 often supplies a larger value than Brunauer Emmett Teller or TEM diameter. The Brunauer Emmett Teller size of TNPs claimed by its commercial supplier was <25 nm, which was considered potentially toxic according to previous reports.27–29 In the TEM image, we obtained a near-spherical particle shape, with the mean pristine particle diameter at 22.1±8.9 nm. Furthermore, the mean hydrodynamic size of TNPs in RPMI 1640 was 504.8 nm, larger than that in ultrapure water (276.9 nm), suggesting that TNPs formed aggregates that were not thoroughly dispersed by ultrasonication. Therefore, the suspension was a mixture of nanoparticles and agglomerations/aggregations.
In our study, we evaluated the cytotoxicity of TNPs (6.25 μg/mL, 12.5 μg/mL, 25 μg/mL, 50 μg/mL, and 100 μg/mL) by MTT assays and LDH release. Results revealed that TNPs significantly decreased the viability of A549 cells, with the enhancement of LDH activity in a dose-dependent manner. In line with the LDH release, the generation of ROS was dramatically triggered by TNPs, consistent with several published studies.30,31 The cytotoxicity induced by TNPs has been demonstrated in various cell types such as WISH cells, human bronchial epithelial cell line, BEAS-2B, and mouse fibroblast cells.10,30,31 Available toxic assays confirmed that TNPs (5 nm) can inhibit A549 cell proliferation, cause DNA damage, and induce apoptosis via a mechanism primarily involving the activation of intrinsic mitochondrial pathway.32
Generally, DNA methylation is the first step in epigenetic phenomena that modulate gene expression via the recruitment of transcription factors. However, the underlying mechanisms involved in TNP-induced epigenetic toxicity have not been clearly clarified. Limited data demonstrated that certain nanomaterials such as nano-SiO2 caused aberrant PARP-1 hypermethylation and inactivity; yet the effects involved in TNP-related PARP-1 abnormal methylation have not been reported.
Our present studies focused for the first time on whether the epigenetic alteration of PARP-1 was responsible for TNP-induced cytotoxicity in A549 cells. Effects of the methyltransferase inhibitor 5-aza and antioxidant agent α-LA on TNP-induced PARP-1 methylation expression were investigated. The data showed that TNPs triggered aberrant hypermethylation of PARP-1 promoter, and there was a cross talk between oxidative stress and PARP-1 methylation in TNP-induced cellular toxicity.
Numerous studies have defined the crucial role of oxidative stress in the toxicity induced by TNPs.31–36 TNPs were demonstrated to have the potential to generate ROS and oxidative stress, leading to oxidative DNA damage and micronucleus formation in human epidermal cells (A431) subsequently.11 The biouptake of TiO2 in particulate form was the vital cause of ROS generation, which in turn was probably the cause of the DNA aberrations. Zhuang summarized that nano-SiO2 exposure induced the decrease of genomic DNA methylation and global hypoacetylation in HaCaT cells.37,38 Still, rare studies on nanomaterial triggered-epigenetic toxicity were published.39,40
Conclusion
In conclusion, to fully understand the effects and interaction mechanisms between ROS and PARP-1 methylation triggered by TNPs, we focused on whether the epigenetic alteration of PARP-1 was responsible for TNP-induced toxicity in A549 cells for the first time. Effects of methyltransferase inhibitor 5-aza and antioxidant agent α-LA respectively on TNP-induced ROS increasing and PARP-1 hypermethylation in promoter region suggested that there is a cross talk between oxidative stress and PARP-1 methylation in TNP-induced A549 cells toxicity. To our knowledge, this was the first study that showed TNPs can trigger PARP-1 hypermethylation and ROS generation may be involved in this process. These data further strengthened the notion that epigenetic alterations could play a significant role in TNP carcinogenesis.
This finding will not only be helpful to clarify the role of PARP-1 methylation in TNP-induced cytotoxicity but also offer a promising approach in prevention and therapy TNPs cytotoxicity using the methyltransferase inhibitor and antioxidant agent.
Acknowledgments
This work has been supported by grants from the Scientific Research Common Program of Beijing Municipal Commission of Education (KM201410025008), the National Natural Science Foundation of China (81472957, 81172639), the Beijing Natural Science Foundation (7142020), and the Importation and Development of High-Caliber Talents Project of Beijing Municipal Institutions (CIT&TCD201404187). We thank Jiajia Wang who helped us with English language editing.
Footnotes
Disclosure
The authors report no conflicts of interest in this work.
References
- 1.Wang J, Zhou G, Chen C, et al. Acute toxicity and biodistribution of different sized titanium dioxide particles in mice after oral administration. Toxicol Lett. 2007;168(2):176–185. doi: 10.1016/j.toxlet.2006.12.001. [DOI] [PubMed] [Google Scholar]
- 2.Warheit DB, Webb TR, Reed KL, Frerichs S, Sayes CM. Pulmonary toxicity study in rats with three forms of ultrafine-TiO2 particles: differential responses related to surface properties. Toxicology. 2007;230(1):90–104. doi: 10.1016/j.tox.2006.11.002. [DOI] [PubMed] [Google Scholar]
- 3.Bermudez E, Mangum JB, Wong BA, et al. Pulmonary responses of mice, rats, and hamsters to subchronic inhalation of ultrafine titanium dioxide particles. Toxicol Sci. 2004;77(2):347–357. doi: 10.1093/toxsci/kfh019. [DOI] [PubMed] [Google Scholar]
- 4.Baggs RB, Ferin J, Oberdorster G. Regression of pulmonary lesions produced by inhaled titanium dioxide in rats. Vet Pathol. 1997;34(6):592–597. doi: 10.1177/030098589703400607. [DOI] [PubMed] [Google Scholar]
- 5.Wu J, Sun J, Xue Y. Involvement of JNK and P53 activation in G2/M cell cycle arrest and apoptosis induced by titanium dioxide nanoparticles in neuron cells. Toxicol Lett. 2010;199(3):269–276. doi: 10.1016/j.toxlet.2010.09.009. [DOI] [PubMed] [Google Scholar]
- 6.Gurr JR, Wang ASS, Chen CH, Jan KY. Ultrafine titanium dioxide particles in the absence of photoactivation can induce oxidative damage to human bronchial epithelial cells. Toxicology. 2005;213(1–2):66–73. doi: 10.1016/j.tox.2005.05.007. [DOI] [PubMed] [Google Scholar]
- 7.Long TC, Saleh N, Tilton RD, Lowry GV, Veronesi B. Titanium dioxide (P25) produces reactive oxygen species in immortalized brain microglia (BV2): implications for nanoparticle neurotoxicity. Environ Sci Technol. 2006;40(14):4346–4352. doi: 10.1021/es060589n. [DOI] [PubMed] [Google Scholar]
- 8.Long TC, Tajuba J, Sama P, et al. Nanosize titanium dioxide stimulates reactive oxygen species in brain microglia and damages neurons in vitro. Environ Health Perspect. 2007;115(11):1631–1637. doi: 10.1289/ehp.10216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Petković J, Zegura B, Stevanović M, et al. DNA damage and alterations in expression of DNA damage responsive genes induced by TiO2 nanoparticles in human hepatoma HepG2 cells. Nanotoxicology. 2011;5(3):341–353. doi: 10.3109/17435390.2010.507316. [DOI] [PubMed] [Google Scholar]
- 10.Jin CY, Zhu BS, Wang XF, Lu QH. Cytotoxicity of titanium dioxide nanoparticles in mouse fibroblast cells. Chem Res Toxicol. 2008;21(9):1871–1877. doi: 10.1021/tx800179f. [DOI] [PubMed] [Google Scholar]
- 11.Shukla RK, Sharma V, Pandey AK, Singh S, Sultana S, Dhawan A. ROS-mediated genotoxicity induced by titanium dioxide nanoparticles in human epidermal cells. Toxicol In Vitro. 2011;25(1):231–241. doi: 10.1016/j.tiv.2010.11.008. [DOI] [PubMed] [Google Scholar]
- 12.Baan RA. Carcinogenic hazards from inhaled carbon black, titanium dioxide, and talc not containing asbestos or asbestiform fibers: recent evaluations by an IARC Monographs working group. Inhal Toxicol. 2007;19:213–228. doi: 10.1080/08958370701497903. [DOI] [PubMed] [Google Scholar]
- 13.Stirzaker C, Song JZ, Davidson B, Clark SJ. Transcriptional gene silencing promotes DNA hypermethylation through a sequential change in chromatin modifications in cancer cells. Cancer Res. 2004;64(11):3871–3877. doi: 10.1158/0008-5472.CAN-03-3690. [DOI] [PubMed] [Google Scholar]
- 14.Jones PA, Baylin SB. The epigenomics of cancer. Cell. 2007;128(4):683–692. doi: 10.1016/j.cell.2007.01.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jaenisch R, Bird A. Epigenetic regulation of gene expression: how the genome integrates intrinsic and environmental signals. Nat Genet. 2003;33:245–254. doi: 10.1038/ng1089. [DOI] [PubMed] [Google Scholar]
- 16.Chevanne M, Calia C, Zampieri M, et al. Oxidative DNA damage repair and parp 1 and parp 2 expression in Epstein-Barr virus-immortalized B lymphocyte cells from young subjects, old subjects, and centenarians. Rejuvenation Res. 2007;10(2):191–204. doi: 10.1089/rej.2006.0514. [DOI] [PubMed] [Google Scholar]
- 17.Sharif J, Muto M, Takebayashi S, et al. The SRA protein Np95 mediates epigenetic inheritance by recruiting Dnmt1 to methylated DNA. Nature. 2007;450(7171):U908–U925. doi: 10.1038/nature06397. [DOI] [PubMed] [Google Scholar]
- 18.Kraus WL. Transcriptional control by PARP-1: chromatin modulation, enhancer-binding, coregulation, and insulation. Curr Opin Cell Biol. 2008;20(3):294–302. doi: 10.1016/j.ceb.2008.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kraus WL, Lis JT. PARP goes transcription. Cell. 2003;113(6):677–683. doi: 10.1016/s0092-8674(03)00433-1. [DOI] [PubMed] [Google Scholar]
- 20.Gao A, Song S, Zuo X, Guo W, Niu P, Tian L. Epigenetic mediated transcriptional activation of PARP-1 participates in silica-associated malignant transformation of human bronchial epithelial cells. Toxicol Lett. 2010;193(3):236–241. doi: 10.1016/j.toxlet.2010.01.017. [DOI] [PubMed] [Google Scholar]
- 21.Gao A, Zuo X, Liu Q, Lu X, Guo W, Tian L. Methylation of PARP-1 promoter involved in the regulation of benzene-induced decrease of PARP-1 mRNA expression. Toxicol Lett. 2010;195(2–3):114–118. doi: 10.1016/j.toxlet.2010.03.005. [DOI] [PubMed] [Google Scholar]
- 22.Gong C, Tao G, Yang L, et al. Methylation of PARP-1 promoter involved in the regulation of nano-SiO2-induced decrease of PARP-1 mRNA expression. Toxicol Lett. 2012;209(3):264–269. doi: 10.1016/j.toxlet.2012.01.007. [DOI] [PubMed] [Google Scholar]
- 23.Franco R, Schoneveld O, Georgakilas AG, Panayiotidis MI. Oxidative stress, DNA methylation and carcinogenesis. Cancer Lett. 2008;266(1):6–11. doi: 10.1016/j.canlet.2008.02.026. [DOI] [PubMed] [Google Scholar]
- 24.Tunc O, Tremellen K. Oxidative DNA damage impairs global sperm DNA methylation in infertile men. J Assist Reprod Genet. 2009;26(9–10):537–544. doi: 10.1007/s10815-009-9346-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jiang JK, Oberdorster G, Biswas P. Characterization of size, surface charge, and agglomeration state of nanoparticle dispersions for toxicological studies. J Nanopart Res. 2009;11(1):77–89. [Google Scholar]
- 26.Rudell B, Sandstrom T, Hammarstrom U, Ledin ML, Horstedt P, Stjernberg N. Evaluation of an exposure setup for studying effects of diesel exhaust in humans. Int Arch Occup Environ Health. 1994;66(2):77–83. doi: 10.1007/BF00383361. [DOI] [PubMed] [Google Scholar]
- 27.Federici G, Shaw BJ, Handy RD. Toxicity of titanium dioxide nanoparticles to rainbow trout (Oncorhynchus mykiss): gill injury, oxidative stress, and other physiological effects. Aquat Toxicol. 2007;84(4):415–430. doi: 10.1016/j.aquatox.2007.07.009. [DOI] [PubMed] [Google Scholar]
- 28.Hao L, Wang Z, Xing B. Effect of sub-acute exposure to TiO2 nanoparticles on oxidative stress and histopathological changes in Juvenile Carp (Cyprinus carpio) J Environ Sci (China) 2009;21(10):1459–1466. doi: 10.1016/s1001-0742(08)62440-7. [DOI] [PubMed] [Google Scholar]
- 29.Vevers WF, Jha AN. Genotoxic and cytotoxic potential of titanium dioxide (TiO2) nanoparticles on fish cells in vitro. Ecotoxicology. 2008;17(5):410–420. doi: 10.1007/s10646-008-0226-9. [DOI] [PubMed] [Google Scholar]
- 30.Park EJ, Yi J, Chung KH, Ryu DY, Choi J, Park K. Oxidative stress and apoptosis induced by titanium dioxide nanoparticles in cultured BEAS-2B cells. Toxicol Lett. 2008;180(3):222–229. doi: 10.1016/j.toxlet.2008.06.869. [DOI] [PubMed] [Google Scholar]
- 31.Saquib Q, Al-Khedhairy AA, Siddiqui MA, Abou-Tarboush FM, Azam A, Musarrat J. Titanium dioxide nanoparticles induced cytotoxicity, oxidative stress and DNA damage in human amnion epithelial (WISH) cells. Toxicol In Vitro. 2012;26(2):351–361. doi: 10.1016/j.tiv.2011.12.011. [DOI] [PubMed] [Google Scholar]
- 32.Jugan ML, Barillet S, Simon-Deckers A, et al. Titanium dioxide nanoparticles exhibit genotoxicity and impair DNA repair activity in A549 cells. Nanotoxicology. 2012;6(5):501–513. doi: 10.3109/17435390.2011.587903. [DOI] [PubMed] [Google Scholar]
- 33.Wang Y, Cui H, Zhou J, et al. Cytotoxicity, DNA damage, and apoptosis induced by titanium dioxide nanoparticles in human non-small cell lung cancer A549 cells. Environ Sci Pollut Res Int. 2015;22(7):5519–5530. doi: 10.1007/s11356-014-3717-7. [DOI] [PubMed] [Google Scholar]
- 34.El-Said K, Ali E, Kanehira K, Taniguchi A. Molecular mechanism of DNA damage induced by titanium dioxide nanoparticles in toll-like receptor 3 or 4 expressing human hepatocarcinoma cell lines. J Nanobiotechnology. 2014;12(1):48. doi: 10.1186/s12951-014-0048-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gui S, Sang X, Zheng L, et al. Intragastric exposure to titanium dioxide nanoparticles induced nephrotoxicity in mice, assessed by physiological and gene expression modifications. Part Fibre Toxicol. 2013;10:4. doi: 10.1186/1743-8977-10-4. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 36.Huerta-García E, Pérez-Arizti JA, Márquez-Ramírez SG, et al. Titanium dioxide nanoparticles induce strong oxidative stress and mitochondrial damage in glial cells. Free Radic Biol Med. 2014;73:84–94. doi: 10.1016/j.freeradbiomed.2014.04.026. [DOI] [PubMed] [Google Scholar]
- 37.Gong C, Yang L, Tao G, Liu Q, Liu J, Zhuang Z. Genome DNA hypomethylation in HaCaT cells after short exposure to SiO2 nanoparticles. Wei Sheng Yan Jiu. 2013;42(2):179–184. Chinese. [PubMed] [Google Scholar]
- 38.Gong C, Tao G, Yang L, Liu J, Liu Q, Zhuang Z. SiO(2) nanoparticles induce global genomic hypomethylation in HaCaT cells. Biochem Biophys Res Commun. 2010;397(3):397–400. doi: 10.1016/j.bbrc.2010.05.076. [DOI] [PubMed] [Google Scholar]
- 39.Meng J, Xing J, Wang Y, et al. Epigenetic modulation of human breast cancer by metallofullerenol nanoparticles: in vivo treatment and in vitro analysis. Nanoscale. 2011;3(11):4713–4719. doi: 10.1039/c1nr10898k. [DOI] [PubMed] [Google Scholar]
- 40.Ng CT, Dheen ST, Yip WC, Ong CN, Bay BH, Lanry Yung LY. The induction of epigenetic regulation of PROS1 gene in lung fibroblasts by gold nanoparticles and implications for potential lung injury. Biomaterials. 2011;32(30):7609–7615. doi: 10.1016/j.biomaterials.2011.06.038. [DOI] [PubMed] [Google Scholar]