ZFIN (http://zfin.org) is the model organism database for Danio rerio, the zebrafish. As such, ZFIN provides a centralized resource for zebrafish genomic, genetic, phenotypic, and developmental data. The ZFIN database contains highly integrated, manually curated information about genes, gene expression, mutant phenotypes, antibodies, anatomical structures, publications and the zebrafish community (Sprague et al., 2008). Web-based search interfaces and tools allow viewing and analysis of the data. The integrated representation of data allows a search for one type of data to be viewed together with related data, thus facilitating the understanding of gene function and regulation, and promoting scientific discovery. A broad range of data types, including text, images, and graphical representations support the data. Links are also provided to related data at other species-specific and bioinformatics databases.
Data captured in ZFIN come from three primary sources. ZFIN scientific curators extract and attribute pertinent data from current literature. Zebrafish laboratories conducting large-scale screens submit data directly to ZFIN. Collaborations with other bioinformatics organizations, such as NCBI, the Sanger Institute, and UniProt, provide valuable cross-site integration via reciprocal links. In addition, some unpublished data are submitted by individual laboratories. Data are updated daily through ongoing curation by ZFIN staff. Software updates are frequent.
Protocols in this unit, describing methods for searching ZFIN major data types, include the following: navigating the home page and utilizing search functions (Basic Protocol 1); retrieving information about zebrafish genes (Basic Protocol 2); finding and viewing comprehensive gene expression data (Basic Protocol 3); finding antibodies (Basic Protocol 4); finding phenotype data (Basic Protocol 5); browsing the zebrafish anatomical ontology (Basic Protocol 6); using ZFIN BLAST to search for zebrafish sequences (Basic Protocol 7); exploring the zebrafish genome using GBrowse (Basic Protocol 8); contributing and collaborating via the community wiki (Basic Protocol 9).
NAVIGATING THE HOME PAGE AND UTILIZING SEARCH FUNCTIONS (Basic Protocol 1)
This protocol introduces the ZFIN home page, including its layout, features, navigation and search functions. The ZFIN home page serves as the main gateway to the zebrafish model organism database, where data, news, and information for the research community are centrally located. The home page also acts as the starting point for general and specialized searches of the ZFIN database.
Necessary Resources
Hardware
Computer with Internet access.
Software
Current or recent version of a web browser (e.g., Firefox, http://www.mozilla.com/firefox, Safari, http://www.apple.com/safari/, or Internet Explorer, http://www.microsoft.com/windows/internet-explorer), with JavaScript, applets, and cookies enabled for best performance.
Viewing the ZFIN home page
-
1. To view the ZFIN home page, type zfin.org into the address bar at the top of the web browser window and press the Enter key (Fig. 1).
The home page is divided into two main sections, with navigation tabs and a navigation tool bar at the top and bottom, respectively. In the upper left of the home page is the official ZFIN logo. The navigation bar at the bottom of the page includes links to help documents, tips for optimizing use of the site, a glossary of terms, contact information, and additional information about ZFIN. For ease of navigation, the ZFIN logo, upper navigation tabs and lower navigation bar are found on all ZFIN pages. Clicking the ZFIN logo always directs the user back to the home page.
-
2. On the left side of the main section, mouse over the top title in bold, Genes/Markers/Clones to view a description of the linked page in a tooltip.
The main body of the home page is divided into two sections, with links to ZFIN’s advanced search forms, tools, and community resources on the left side and links to external resources for the zebrafish research community on the right side. The ZFIN tools and search forms are described in more detail in the following protocols; however, placing the cursor over the links on the home page will provide a brief description of the linked pages in a tooltip. The home page links are duplicated in the navigation tabs to allow quick linking from any ZFIN page.
Figure 1.

The Zebrafish Model Organism Database (ZFIN) home page layout features three navigation tabs at the top of the page (Research, General Information, and ZIRC). The left side of the main section is dedicated to ZFIN resources and advanced search forms, and the right side is primarily external resource links (e.g., Zebrafish International Resource Center, external genome browsers, and genomics resources).
Using the navigation tabs
Located at the top of all ZFIN pages are three navigation tabs (Fig. 2). The first two tabs represent broad categories (Research and General Information) and provide links to pages within ZFIN. The third tab provides links to the Zebrafish International Resource Center (ZIRC).
3. From the ZFIN home page (http://zfin.org), click on the Research tab at the top of the page.
-
4. Mouse over the navigation bar subheading BLAST to read a brief description of the linked page.
Clicking on each tab will reveal a list of related links in the navigation bar below. A brief description of each page is displayed by placing the cursor over the linked page name. These links duplicate links found on the home page, allowing for ease of navigation from any ZFIN page.
Figure 2.
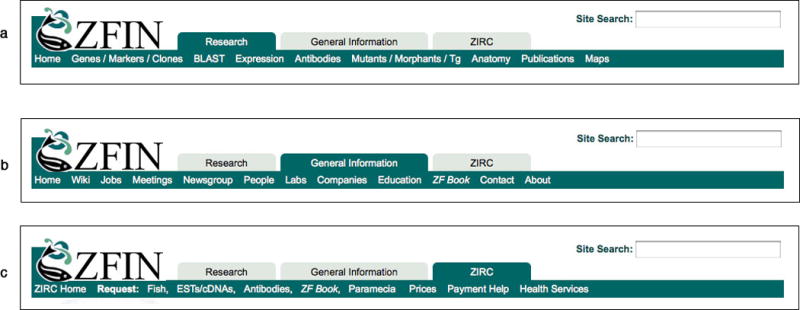
Appearing at the top of every ZFIN page is the Research navigation tab (a), General Information navigation tab (b) and ZIRC navigation tab (c). When a tab is selected by clicking, a list of related links is opened and displayed horizontally below the tab.
Performing a Site Search
From any page in ZFIN, a Site Search can be performed using the search box at the top right of the page. Site Search provides a simple method for searching the entire ZFIN website.
-
5. From the ZFIN home page (http://zfin.org), or any page in ZFIN, type the search term cadherin into the Site Search text field in the upper right corner of the page and press the Enter key (Fig. 3).
A typical query might be a gene symbol or a partial protein name. A wildcard is not necessary for Site Search queries. Wildcard matches are done automatically. For example, the search term hox will return data on all members of the hox gene family (e.g., hoxa1a, hoxa2b, hoxa3a). Search results are organized by categories that reflect the various data types in ZFIN (Fig. 3). With cadherin as the search query, results are returned in the following categories: Genes/Markers/Clones, Features, Mutants/Transgenics, Expression/Phenotype, Sequence Information, Antibody, Gene Product, Gene Ontology, People, Community Wiki: Antibodies, and Other. The number of matches for each category is displayed in parentheses next to the category header. A ZFIN record or page may be represented in more than one category.
-
6. To narrow the result set to the Genes/Markers/Clones category, click on the hyperlinked category header at the top of the second column in the results by category table.
The search results that appear (Fig. 4) will now be limited to results in the Genes/Markers/Clones category. An arrowhead is placed to the right of the selected category and the descriptive search results header in bold at the top of the page also displays the category selection and current search term (Genes/Marker/Clones search results for cadherin).
-
7. Click on the first item returned in the Genes/Markers/Clones category for the cadherin search, the hyperlinked result Gene: ctnna2.
Each of the items in the displayed result list links directly to the data for that category type. Clicking on the Gene: ctnna2 hyperlink in the cadherin result list will display the ctnna2 Gene Detail page in ZFIN (see Basic Protocol 2). To view the search results for a different category, click on the new hyperlinked category header. To return to the complete list of individual results in all categories using the current search term, click the All category header. For additional tips on performing a search of ZFIN, click the Tips hyperlink at the top of the results page (Fig. 4).
Figure 3.
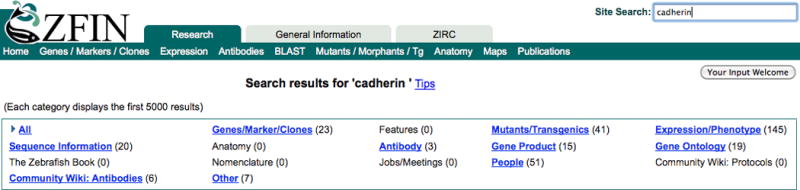
Site Search offers a quick search of ZFIN when a term is typed in the search field at the upper right of any ZFIN page. Results are displayed in a table sorted by category, with the number of results in each category listed in parentheses. Clicking the hyperlinked category name provides further filtering of results.
Figure 4.
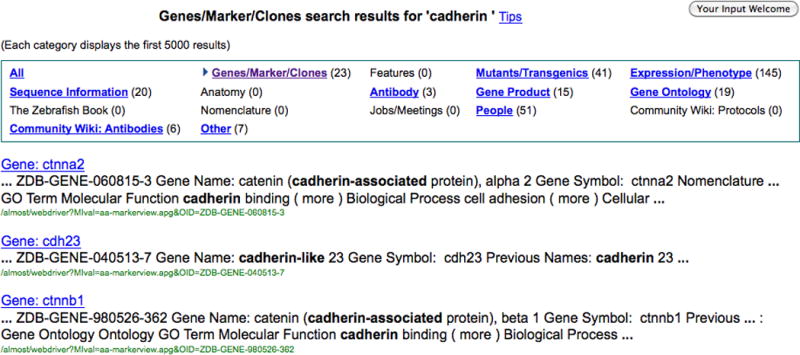
A view of the list of categories from Site Search with the individual results below. Clicking the headings in the individual results below the category list will direct the user to the selected detail page in ZFIN. The heading in bold above the table indicates the search performed. In this example, choosing the Genes/Markers/Clones category narrowed the search result set for cadherin. Note the arrow to the left of the selected category in the table.
Using the advanced ZFIN search forms
The advanced ZFIN search forms allow a customizable search to be performed on various categories of data in the ZFIN database. Each form is unique, depending on the data type being searched, but most forms share common features. A simple search can be performed by filling in a single text field and using the default settings, or a more complex search can be performed by using pulldown menus to customize the search, radio buttons to select filters, and the Affected Anatomy search field to focus the search based on anatomical structures. There are multiple advanced search forms for specific data types, including Genes/Markers/Clones (see Basic Protocol 2), Gene Expression (see Basic Protocol 3), Antibodies (see Basic Protocol 4), Mutants/Morphants/Transgenics (see Basic Protocol 5), Anatomy (see Basic Protocol 6), Publications, People, Labs, and Companies. Links to these advanced search forms are available from the home page and from the navigation tabs on any ZFIN page.
8. Click the ZFIN logo at the top left of the page to return to the home page.
9. Click the Publications link mid-way down the left side of the home page to open the Publication Search form (Fig. 5).
-
10. Click on the Abstract pulldown menu to view alternate search categories.
A search can be initiated using words found in the Abstract, keywords, journal abbreviation, PubMed IDs, and ZFIN publication IDs. Selecting a search category from the pulldown menu and entering a search term will limit the search to that category.
11. Click the Type pulldown menu to view options for limiting the search based on publication type.
-
12. To search by author’s name, enter the search term Wester in the first text field on the top left next to the Author Contains pulldown menu and click the Search button on the lower right.
As with the Site Search, it is not necessary to use wildcards in the text fields because the wildcard search results are displayed automatically (e.g., the search term Wester will return publications by authors Wester, Westerman, and Westerfield). Additionally, more than one author name can be included in the search field with a space as the delimiter.
Figure 5.
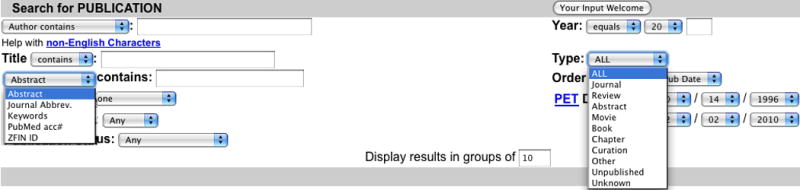
A view of the advanced search form for Publications. The form offers several options for searching publications in ZFIN, including the use of author name, publication date, keywords, or search terms from the title or abstract. The pulldown menu expanded on the left provides a mechanism for searching based on unique publication IDs, and the menu expanded on the right allows filtering based on publication type. An option is also provided to select the number of results to display per page.
Viewing and modifying advanced search results
Once an advanced search has been performed, the results are displayed in a list with each item linking the user to the detail page of interest. In our current example of a Publications search, the search results for the query on Wester would return the page seen in Fig. 6. At the top of the page, the number of records found in the search is listed in parentheses. Below this is a list of the first page of results, ordered and displayed based on the parameters selected from the search form (e.g., the default parameters order by publication date and display 10 results per page).
10. To review the search results, click on the first hyperlinked publication to view the abstract.
11. Click the browser Back button to return to the page of publication results.
12. Scroll to the bottom of the publications list and use the buttons on the pagination bar to page through additional results.
-
13. To modify the search parameters, click the Modify Search link at the upper right of the results page or scroll to the bottom of the page and view the search modification interface.
The search form is reproduced at the bottom of all advanced search results pages to expedite the modification of a search, to view the parameters of the current query, or to allow the user to begin a new search. Rather than a simple search on author name only, the search can be refined by any of the following methods: enter a keyword or the complete title in the Title field, enter a keyword or ID in the second text field on the left and select a category to filter from the pulldown menu, limit the search based on the year of the publication, select the publication Type using the pulldown menu, or use any combination of these search parameters. A pulldown menu on the lower right allows users to select the format in which the results will be displayed, either by the publication date or alphabetically by the first author’s name.
-
17. Once the combination of search parameters has been selected, click the Search button at the bottom right of the form to perform the search.
Clicking the Reset button resets the form to the default settings and removes all terms from the text fields. Clicking the List All Publications button ignores selections on the form and returns a complete list of all publications in ZFIN.
Figure 6.
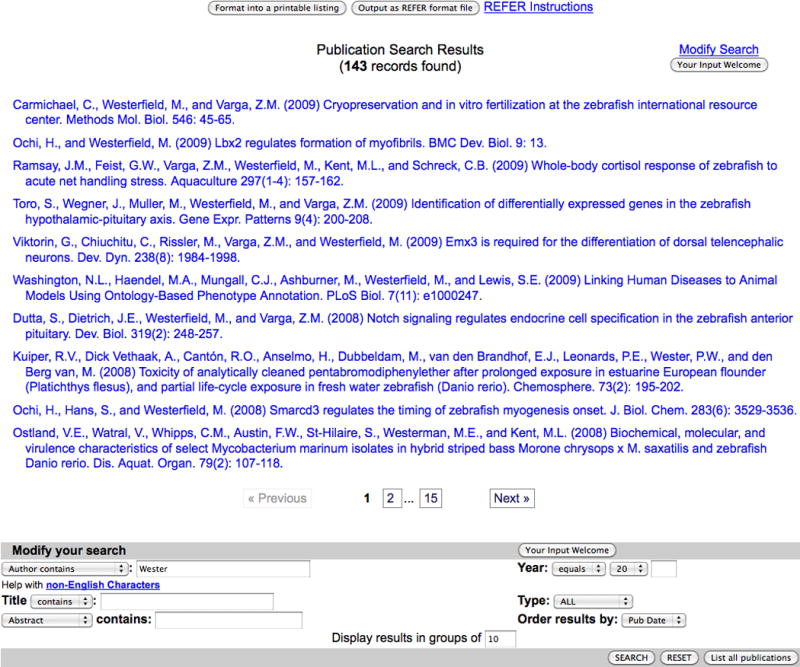
The Publication Search Results page features a list of records matching the search parameters, each hyperlinked to the publication abstract in ZFIN. Buttons at the top provide formatting and output options, and the pagination bar below the list provides the means to page through the results. The Modify Search hyperlink at the upper right redirects to the bottom of the page where the search form is reproduced, allowing further modification, filtering or initiation of a new search.
RETRIEVING INFORMATION ABOUT ZEBRAFISH GENES (Basic Protocol 2)
This protocol describes how to search for information about a zebrafish gene or set of genes.
Necessary Resources
See Basic Protocol 1.
Searching for information about a gene or set of genes using the Genes/Markers/Clones search
1. Go to the ZFIN home page (http://zfin.org). Click on the Genes/Markers/Clones link in the upper left side of the page to go the Genes/Markers/Clones search page.
2. Type myosin in the Name/Symbol field.
-
3. Click the Search button at the bottom of the page to initiate the search. The default settings are used for all search parameters in this example query. In this instance, the query retrieves all Genes, Markers and Clones that include the term “myosin” and have been mapped to any Linkage Group.
The Name/Symbol search option in the pulldown menu can be set to either Contains or Begins With. The Types option can be used to limit the search to specific marker types or clones in ZFIN. An alternative to name or symbol search is a sequence Accession Number search, where a GenBank, GenPept, RefSeq or UniProt sequence accession number can be used to search for the gene it is associated with in ZFIN.
4. All the markers containing a match to myosin are displayed on the Search Results page (Fig. 7).
Figure 7.
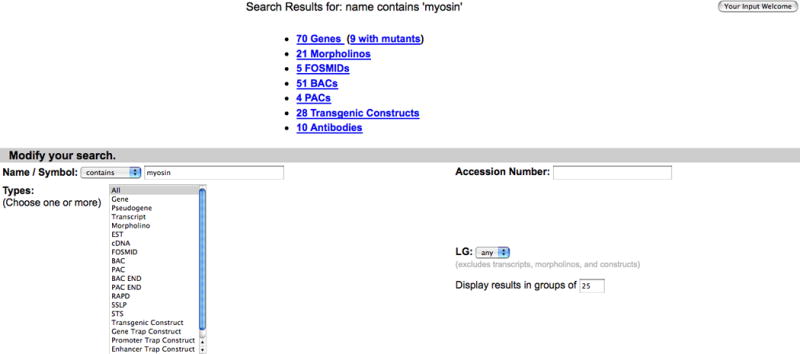
Genes/Markers/Clones Search Results page displaying the search results for myosin, with matches to genes, morpholinos, BACs, transgenic constructs and antibodies. Click on hyperlinks to view results for each category.
Reviewing the results from the Genes/Markers/Clones search
Search results from a Genes/Markers/Clones search are organized by the different types that are listed in the selection box. The search for myosin returns matches to genes, morpholinos, BACs, PACs, transgenic constructs, and antibodies (Fig. 7).
-
5. Click on the 70 Genes hyperlink (Fig. 7) to see the Gene Search Results page with the list of all genes that have a name, symbol, previous name, or an ortholog name match to the search string myosin (Fig. 8).
The Matching Text column indicates why a particular gene was returned in the search.
Results can be narrowed or broadened by changing the search string or by selecting or deselecting specific marker types or Linkage Groups (LG).
6. Click on the myh6 gene hyperlink (Fig. 8) to view the Gene Detail Page (Fig. 9).
Figure 8.
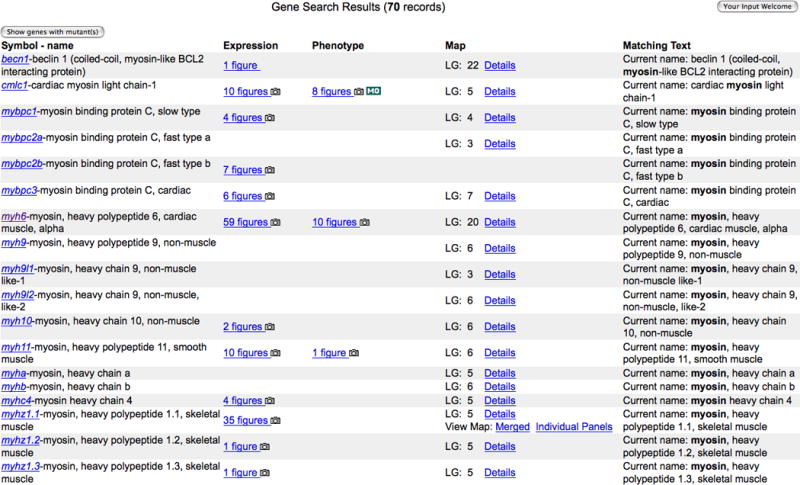
Gene Search Results page showing matches for myosin in Genes result category.
Figure 9.
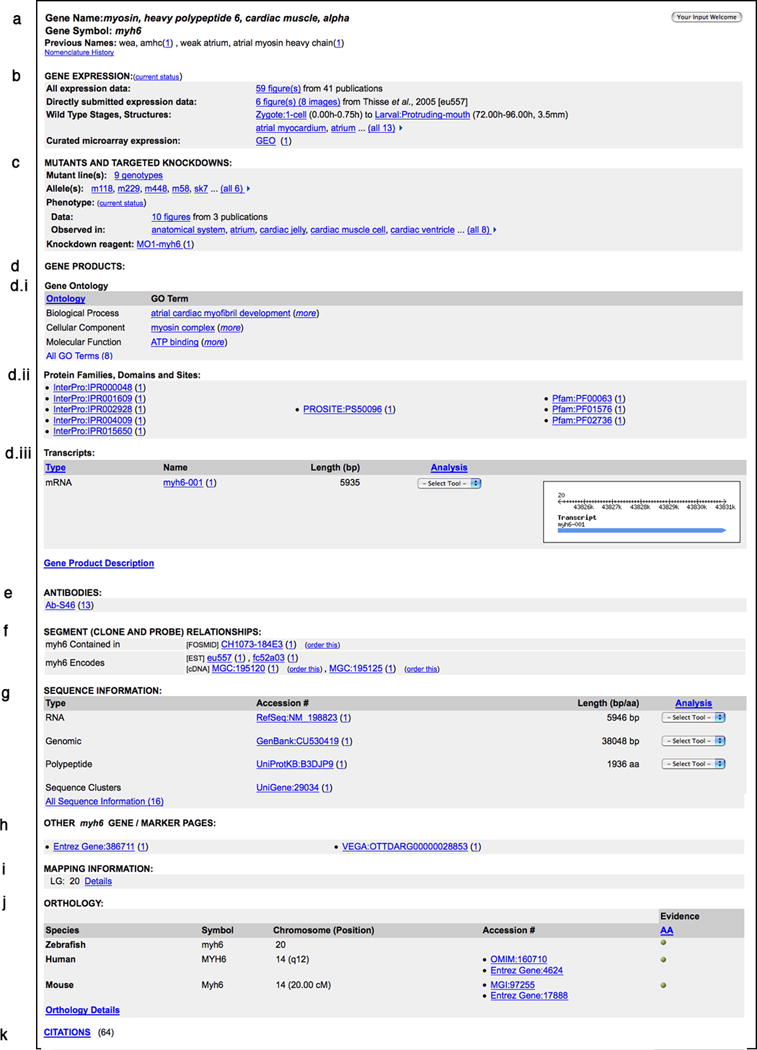
Gene Detail Page for myh6. This figure shows the various sections of the Gene page in an alternating color scheme. The different types of data can be grouped into the following categories: (a) Gene nomenclature, (b) Gene expression, (c) Mutants and Targeted knockdowns, (d) Gene product information, (e) Antibodies, (f) Clones and Probes, (g) Sequence Information, (h) Links to Gene pages at other resources, (i) Mapping data, (j) Orthology, and (k) Citations. A brief description of each section is provided in the text of the protocol.
Examining the Gene Detail page
The gene page in ZFIN presents the comprehensive set of information that has been curated for a specific gene. This information is presented in separate sections in an alternating color scheme (Fig. 9). The page can include information regarding nomenclature, expression, mutant phenotypes, Gene Ontology (GO) classifications, gene products, reagents, mapping information, orthology, and associated zebrafish literature. The primary purpose of the specific sections is to provide the user with a concise summary of the information available for specific data types. These sections have hypertext links that allow the user to access specific details and source information for the data.
A brief description of the various data types provided on the gene page is presented below.
Gene Nomenclature: The official name and symbol are listed on the gene page. The naming follows the guidelines approved by the Zebrafish Nomenclature Committee (ZNC). The previous names include all alternative variants of the gene name and symbol. The Nomenclature History link provides users with details regarding name changes such as gene merges and renames.
Gene Expression: Gene expression data in ZFIN come from two data sources: direct submission and figures curated from journal articles. These data are presented as a summary. The total number of figures annotated for gene expression and the associated publications are provided with a link to the details. Expression data that are submitted directly to ZFIN are available as a separate subset. The stage range and anatomical structures for which expression has been detected in wild-type fish is provided next to the Wild-Type Stages and Structures heading. Microarray expression data can be accessed by following the Gene Expression Omnibus (GEO) link in this section.
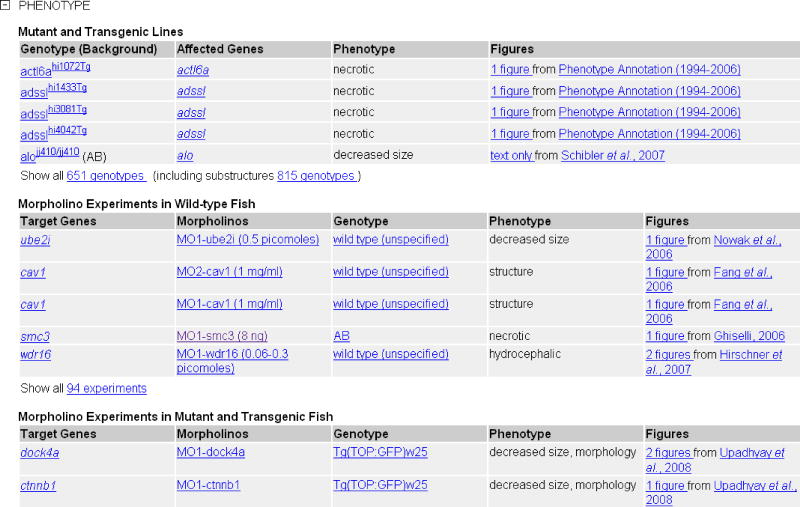
Mutants and Targeted Knockdowns: This section contains phenotypic data curated by ZFIN for mutants, morphants, and transgenic lines. Phenotype data in ZFIN are annotated using the Entity Quality Ontology (PATO, formerly known as Phenotype and Trait Ontology; Washington et al., 2009). Phenotype details can be accessed from the Genotype, Figure, Anatomy, and Morpholino pages. Phenotype data relevant to the specific data type are available at each location. In addition, specific information for Genotypes and Alleles, such as Background, Affected Genes, Zygosity and Mutation Type, are available at the respective Detail pages. Morpholino oligonucleotides are the most common anti-sense “knockdown” technique used in zebrafish (Bill et al., 2009). Additional details, including sequence information, are presented on the Morpholino Detail page.
- Gene Products: The Gene Products section provides information on Gene Ontology (GO) annotations, protein domains, and transcripts.
- The Gene Ontology is a set of structured vocabularies for describing the molecular function, biological role, and subcellular localization of the gene product (UNIT 7.2). These terms can be applied to gene products in many species, allowing effective cross-species comparison and querying. The Gene Ontology subsection on the gene page presents a representative set of terms from each of the vocabularies. The comprehensive set of annotations for a gene can be obtained by clicking on the All GO Terms link. The GO Details page provides complete details on the annotation, including the evidence supporting the annotation and the source of the annotation.
- Protein Domains: InterPro, PFAM, and PROSITE domain links are presented to users through data exchange with UniProt (UniProt Consortium, 2010).
- Transcripts: The transcripts subsection contains annotated transcripts from the zebrafish genome mapping and sequencing project at the Sanger Institute (Jekosch, 2004), as well as non-coding microRNA transcripts curated from literature at ZFIN. This section includes information on the transcript type, name, and length, and access to sequence Analysis Tools. Click on the transcript name to view additional information about transcripts such as Annotation Status, Sequence, Supporting Sequence data, and links to External database resources.
Antibodies: This section contains antibodies that recognize a protein product of the gene. Antibodies are used extensively to probe gene expression and to label anatomical structures. ZFIN curates antibodies from publications and detailed information about the antibody is presented on the Antibody Detail page (see Basic Protocol 4).
Segments (Clone and Probe) Relationships: Genomic clones (BACs, PACs and fosmids), cDNA clones (full length cDNAs, ESTs) and polymorphisms (SSLPs, SNPs) are included in this section. Order This links are provided next to clones that are available for purchase. Click the clone name to view additional details about the clone, including the relationship to other markers in ZFIN and Sequence Accession IDs.
Sequences: The Sequence Information section of the gene page provides a summary view of the longest mRNA, genomic, and protein sequences that have been identified and associated with the gene. A link to the RefSeq record for RNA and a UniProt record for the protein are provided as the representative sequences for the gene when they are available. Links to the sequence record for the longest genomic clone and the UniGene cluster are also available on the gene page. A comprehensive set of nucleotide and protein sequences that have been annotated for the gene can be accessed by clicking the All Sequence Information link at the bottom of this section. This will take the user to the full set of sequence accessions and the related clones that are annotated for the gene. This set of sequences is derived from curation of publications as well as from data exchanges with resources such as NCBI (Sayers et al., 2010), UniProt, Zebrafish Gene Collection (ZGC; http://zgc.nci.nih.gov/Info/Summary), and the genome annotation team at the Sanger Institute.
Links to other Resources: ZFIN exchanges links with the corresponding gene pages at Entrez Gene in NCBI and with the gene pages at the Ensembl (Hubbard et al., 2009) and Vega Genome Browsers (Wilming et al., 2008) at the Wellcome Trust Sanger Institute. This provides the user with access to as much information as possible. Other links to different data resources for sequences, polymorphisms, clones, and functional data are included in the relevant sections.
-
Mapping: The Mapping Information section displays the Linkage Group (chromosome) location for the gene. Further information about the experimental details and source of the mapping data can be obtained by selecting the Details link in this section. In addition, the location of the gene on a genetic map can be accessed through the View Map hyperlinks in this section.
In some instances, two or more map locations could be displayed for a Gene or Marker as a result of mapping discrepancies. ZFIN curates and displays all published and submitted data along with the details and sources. In such cases, it is useful to look at the map locations attributed to the latest version of the whole genome assembly at the Sanger Institute.
Orthology: The orthology section of the gene page primarily provides information regarding the mouse and human orthologs for the zebrafish gene of interest. The human and mouse records are linked to the relevant databases, such as Entrez Gene, OMIM, and MGI. In addition, the evidence used to assert orthology, such as sequence similarity or conserved synteny, is provided along with the citations.
Citations: The list of zebrafish publications associated with the gene is available by clicking on the Citations link at the bottom of the gene page. Citations include published literature imported from PubMed and other databases, as well as references to details of data exchanges with large-scale data producers.
FINDING AND VIEWING COMPREHENSIVE GENE EXPRESSION DATA (Basic Protocol 3)
Analysis of gene expression patterns promotes an understanding of gene function. ZFIN’s curation of gene expression data facilitates this analysis. Gene expression is curated at the figure level by associating genes, fish, stages, and anatomical terms from the zebrafish anatomical ontology (AO, see Basic Protocol 6). Experimental conditions (morpholinos, chemicals, and changes in light or temperature, etc.) and assay type are also reported. Gene expression data in ZFIN represent:
Annotated figures from the current literature
Annotated images that have been directly submitted to ZFIN by researchers
An index of gene expression data from older publications
Links to curated microarray data at Gene Expression Omnibus (GEO; Barrett et al., 2009)
All gene expression data are searched and displayed in the same figure-based format with the exception of microarray data, which are not stored at ZFIN and are not searchable.
Necessary Resources
See Basic Protocol 1.
1. From the ZFIN Home page (http://zfin.org), click on the Gene Expression link on the left side of the main page.
Searching for fibroblast growth factor 8a, fgf8a, gene expression patterns in wild-type fish
2. Enter fgf8a in the Gene/EST name text box. (Fig. 10)
-
3. Select Equals from the pulldown menu associated with the Gene/EST name text box.
Pulldown menu options (Contains, Begins with, Equals) allow you to specify an exact or inexact name match.
-
4. Click the Show Only WT Expression check box.
The selection of one check box may render others unavailable. Unavailable fields will be grayed out.
-
5. Leave other settings unchanged and click the Search button at the bottom of the form to initiate the search. Default values are used for all other search form options in this example.
Search results can be narrowed or broadened by adding or removing search specifications. Additional search options and their default values are as follows:- An entry in the Genotype or Background field will restrict the search to gene expression in a particular fish. An associated pulldown menu provides options to specify an exact or inexact match (Contains, Begins With, and Equals).
- The MO Knockdown: Gene name field is used to locate gene expression for a specific morpholino target gene. An associated pulldown menu provides options to specify an exact match or inexact match (Contains, Begins With, and Equals).
- Enter the last name of an author in the Author name field to find gene expression data published by a particular researcher. An associated pulldown menu provides options to specify an exact match or inexact match (Contains, Begins With, and Equals).
- Search for gene expression in specified anatomical structures using anatomy terms from the zebrafish anatomical ontology (AO). One or more structures may be entered. Basic Protocol 6 describes the Anatomy Terms autocomplete feature. The selection of a stage range and an anatomical term that is not present in the specified range will result in no results found.
- Gene expression may be examined for a given developmental stage or range by selecting stages from the Between Stages pulldown menu. Values are derived from Kimmel et al., 1995. Zygote: 1-cell to Adult is the default stage range.
- Searches may be restricted by assay type by selecting an assay from the Assay pulldown menu. By default, data from any assay type are included.
- Show Only WT Expression, Show Only Reporter Genes, and Show Only Figures With Images check boxes offer additional ways to constrain data. All data are included if no check boxes are selected.
- Show Only Direct Submission Data, Show Only Published Literature, and Show All radio buttons constrain searches by data source. By default, data from all data sources is included.
Figure 10.
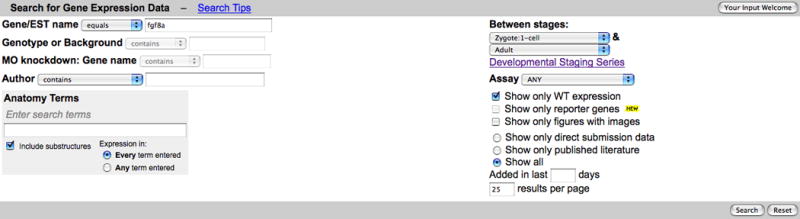
Gene Expression Search form showing a search for fgf8a expression data in wild-type fish only. Note that some options are mutually exclusive. Selection of the Show Only WT Expression invalidates the Genotype or Background and MO Knockdown: Gene name options. They are grayed out to indicate they are no longer valid options.
Reviewing results
The Gene Expression Search Results page (Fig. 11) summarizes patterns with fgf8a gene expression in wild-type fish only. Search results are presented as images and as text. A thumbnail strip is found at the top of the search results page. Each thumbnail in the strip represents a figure image from your expression search result. A typical search returns many thumbnail strips. A textual representation of search results follows the Figure Gallery. Several types of data are represented.
6. Navigate the Figure Gallery thumbnail strips using the controls immediately above the strip. There are ten thumbnails in each strip. Advance the strip by typing a number in the counter box or by clicking on the arrows. The Figure Gallery in Figure 11 shows 165 images returned for the example search. The Figure Gallery is advanced to strip 14 of 17.
-
7. Hover the mouse over a thumbnail to view a larger popup image. For this example, the mouse is placed over the fourth image in the strip.
Fig. 6 on the popup image links to a full-sized version of the image. dlx2a, fgf8a, and pax2a link to their representative Gene Detail pages. See Basic Protocol 2 for a discussion of Gene Detail pages.
-
8. Release the mouse and scroll down the page to view the results summary list immediately below the Figure Gallery.
Gene name searches, as depicted in this fgf8a example, provide publication, figure image thumbnails, genotype or background, developmental stage range, and anatomical structures. Summary lists for other searches provide an alphabetical list of genes, links to figures, and the stage range represented by the data. Details of each object type can be learned by clicking on the links
A camera icon indicates images are available. Copyright permissions may prohibit display of some images. An MO icon indicates experiments were performed using morpholinos.
Figure 11.
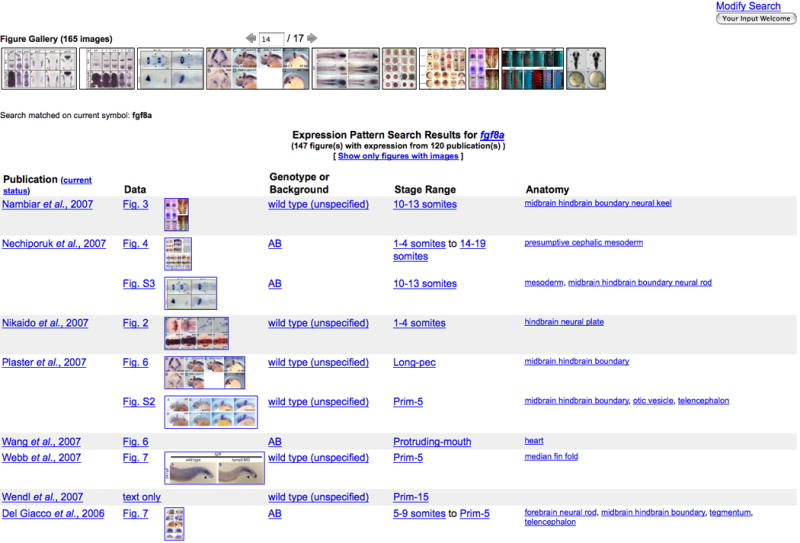
Gene Expression Search Results page showing the Figure Gallery; a collection of all images relevant to the query, depicting 165 images for fgf8a in wild-type fish. This figure depicts the summary list portion of search results page for fgf8a in wild-type fish. Matching entries are summarized by publication, figure (including thumbnail image when available), genotype or background, stage range, and anatomy terms. All items are hyperlinked to data detail pages.
Examining details
9. Click on the Plaster et al., 2007 Fig. 6 (Fig. 12) link to see details about this figure (Fig. 13).
-
10. View the four major sections of the Figure Detail page. The first section provides links to additional publication data. Click on the Author or Full text link to read the publication abstract or complete article, respectively. Follow the Additional Figures link to view all figures with gene expression data in the publication. The Expression/Labeling section summarizes experimental details, including gene, antibodies, anatomical term, and stage range. Follow these links for additional information. The figure image and figure legend follow. Click on the image to view a larger representation. Finally, view a tabular representation of the gene expression details for the publication.
ZFIN began to include published figures in 2004. Figures from older publications are added on an ad hoc basis. Images and legends are displayed when copyright permissions are available.
Figure 12.
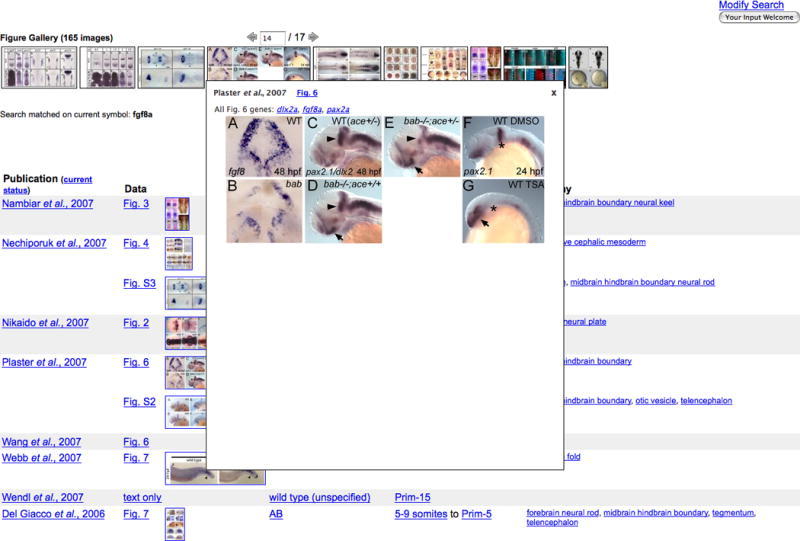
Gene Expression Search Results page. A larger image is shown when mousing over a thumbnail image. The text displayed on the image hyperlinks to detail pages. Plaster et al., 2007 links to the Publication Abstract page. Fig. 6 hyperlinks to the Figure Detail page for figure 6 from this publication. Expression patterns for dlx2a, fgf8a, and pax2a are depicted in this figure. Click on the gene symbols to view their respective Gene Detail pages.
Figure 13.
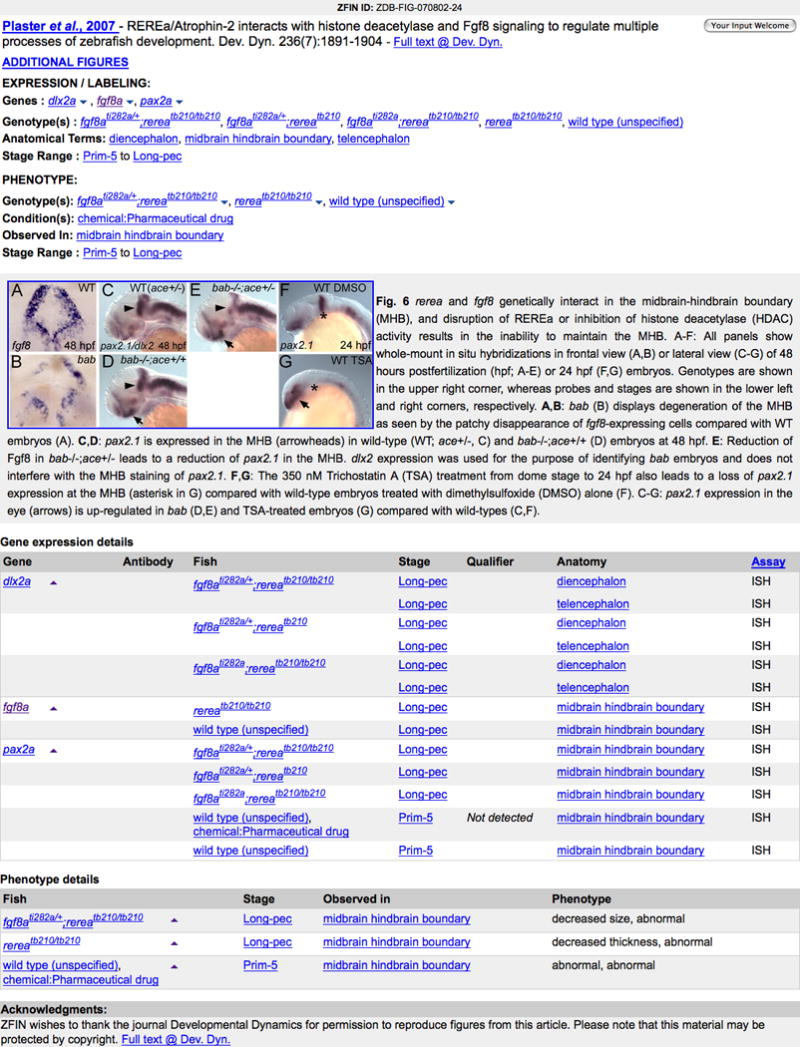
An example of a Figure Detail page, including figure 6 from Plaster et al., 2007 and the corresponding gene expression annotations. Follow hyperlinked text to corresponding data detail pages. Click the image to open a larger image. Click on the down arrows associated with gene symbols to advance to the Gene Expression Details summary of data annotated from this image. Note that the Not Detected qualifier indicates the author stated pax2a was not expressed in the midbrain hindbrain boundary at Prim-5. The Fish column data for the same annotation indicates that the wild-type fish were treated with a chemical.
FINDING ANTIBODIES (Basic Protocol 4)
ZFIN records antibodies that are used as reagents for labeling specific cellular or anatomical structures, or gene products in zebrafish. Gene products labeled by an antibody are reported when provided. Antibodies are commonly used as reagents to assay gene expression and mutant phenotypes. Researchers can use the Antibody Search Form to look for antibodies that target a specific structure or product of a gene. In addition to this information about antibodies, ZFIN captures external comments about usage in experiments that might be valuable for researchers to know when searching for a potential antibody.
Necessary Resources
See Basic Protocol 1.
Searching for Antibody Data
1. Follow the Antibodies link on the home page (http://zfin.org) to invoke the Antibody Search page (Fig. 14).
- 2. Type alcam in the Antigen Gene search field and head in the Labeled Anatomy search field. Basic Protocol 6 describes the Anatomy Terms autocomplete feature. This search will return antibodies that target proteins produced by genes containing the name alcam and labeling structures in the head of the zebrafish.
- Antibodies can be searched using the name of the antibody or the name of the gene producing proteins that are targeted by the antibodies. The associated pulldown menu provides the user with the choice of using either Begins With or Contains to restrict or broaden the search.
- One or more anatomy structures can be entered in the Labeled Anatomy field to limit the search for antibodies known to label specific structures.
- Additional search parameters include selecting the host organism where the antibody was created, choosing an assay type where the antibody was utilized and the ability to specify monoclonal or polyclonal antibodies. The default option for the antibody search is set to include both types of antibodies from any host organism used in any assay.
-
3. Click on the Search button to initiate the search.
To learn more about searching antibodies in ZFIN, click on the Search Tips link. ZFIN matches the Antigen Gene field against gene name, gene symbol and previous names.
Figure 14.
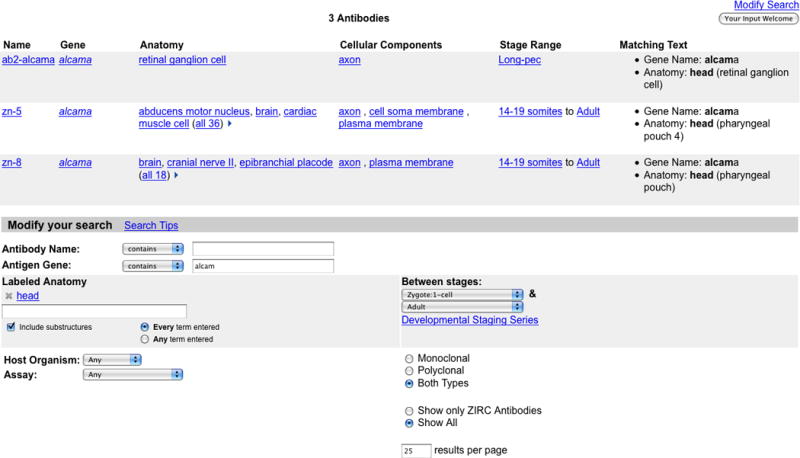
The Antibody Search results page. Shown here is a search for antibodies that label the head of the zebrafish and match the antigen gene alcam. The Antibody Search page with the specified search criteria can be viewed below the Search results.
Reviewing Search Results
4. Scroll down the list of antibodies (Fig. 14), ordered alphabetically by antibody name, that match the specified criteria. The targeted genes, labeled structures, and labeled cellular components are displayed.
-
5. Click on the zn-8 link to view all details about the antibody.
The Matching Text column indicates why the antibody was returned.
Viewing Antibody Details
-
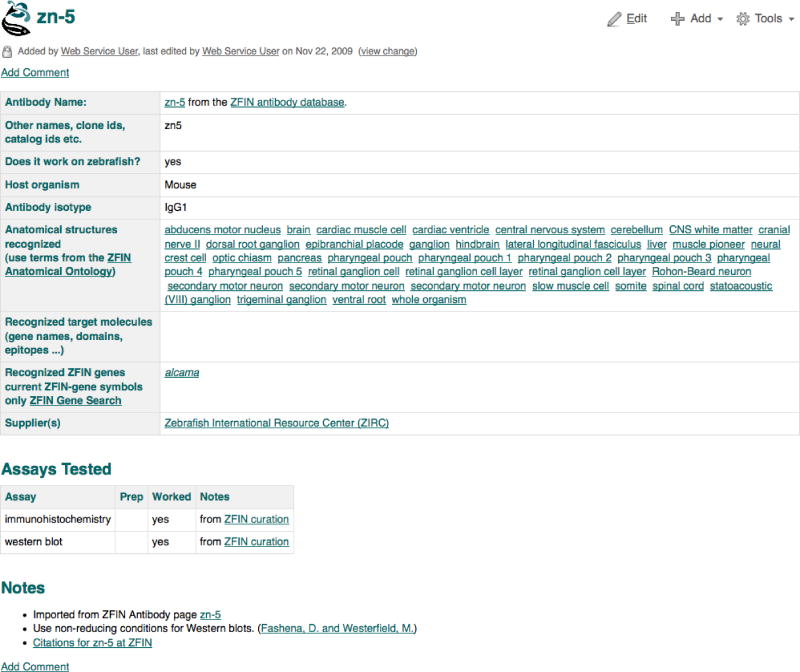
6. Examine the summary of the major characteristics of the zn-8 antibody on the Antibody Detail page (Fig. 15). Details include the host organism, isotype, clonal type and a list of antigen genes whose protein products are recognized by the antibody. In addition, the Anatomical Labeling section details the structures that are labeled by the antibody with links to the detailed annotations (experiments) in which the antibody was used.
The Notes section provides information about the usage of the antibody and a link is also provided to the Community Wiki antibody entry. The Source section lists suppliers of the antibody.
Most “zn-” named antibodies can be ordered at ZIRC.
Figure 15.
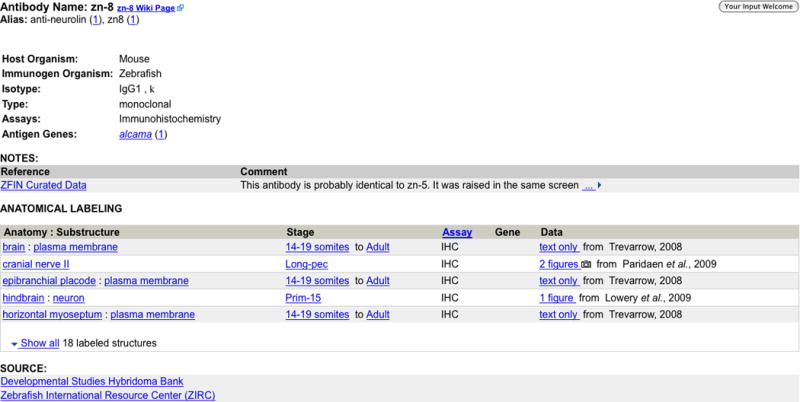
The Antibody Detail Page for the antibody zn-8. Click on Show all to view all labeled structures. The link next to the antibody name links to the antibody page at the Antibody Community Wiki.
FINDING PHENOTYPE DATA (Basic Protocol 5)
ZFIN curates phenotype data for mutants, morphants, and transgenic lines. Phenotype descriptions are found on the Figure View page. Phenotype data are annotated with terms from the Entity Quality Ontology (PATO, formerly known as Phenotype and Trait Ontology), indicating normal or abnormal, and the cellular or anatomical structure (see Basic Protocol 6) in which the phenotype is observed. Annotations use terms from the Zebrafish Anatomical Ontology (see Basic Protocol 6) and the Gene Ontology (see Unit 7.2) specific to a given fish and stage range. ZFIN displays the following types of phenotype data:
Annotated figures from the current literature
Annotated images that have been directly submitted to ZFIN by researchers
Necessary Resources
See Basic Protocol 1.
Searching for Phenotype Data
There are two ways to find phenotype data (Fig. 16), either through the Mutant/Morpholinos/Transgenics form or using the Anatomy Detail page (see Basic Protocol 6).
1. Navigate to the Mutant/Morpholinos/Transgenics search page by clicking on the Mutants/Morphants/Transgenics link on the home page (http://zfin.org). A variety of attributes can be set to help find specific mutant fish (Fig. 17). This example will search for point mutations affecting the gene bmp2b.
2. Enter bmp2b in the Gene/Allele Name field.
-
3. Select Equals from the pulldown menu associated with the Gene/Allele name text box.
Pulldown menu options (Contains, Begins with, Equals) allow you to specify an exact or inexact name match.
4. Select Point Mutation from the Mutation Type pulldown menu.
-
5. Leave other settings unchanged and click the Search button at the bottom of the form to initiate the search. Default values are used for all other search form options in this example.
Search results can be narrowed or broadened by adding or removing search specifications. Other search options and their default values are as follows:- One or more anatomy structures may be specified in the Affected Anatomy section (see Basic Protocol 6). Anatomical structures are not specified by default.
- The LG pulldown menu specifies the linkage group (chromosome) where a mutant gene is mapped. Any of the 25 linkage groups or the mitochondrion can be selected. The default value is Any.
- Select the type of mutagen used to create a mutation by selecting from the Type of Mutation pulldown menu. By default, any mutagen type will be included.
- Only Morphant, Morphants in WT/reporter, Characterized Mutant, Transgenic, Exclude morphants, and Show All radio buttons filter search results. By default, Show All is selected allowing all types of mutants, morphants, and transgenics to be included in the search results.
Figure 16.
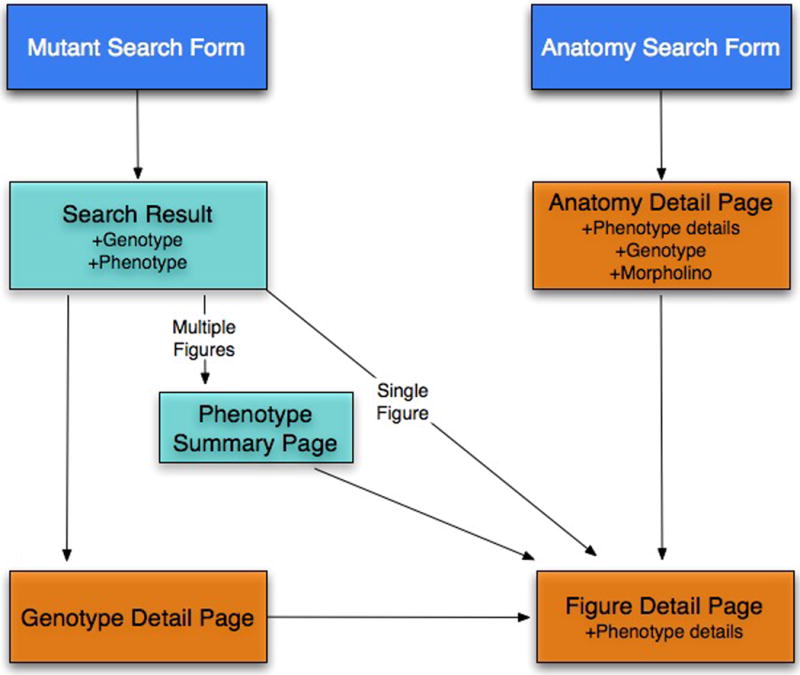
Site map that displays how to find genotype and phenotype details in ZFIN.
Figure 17.
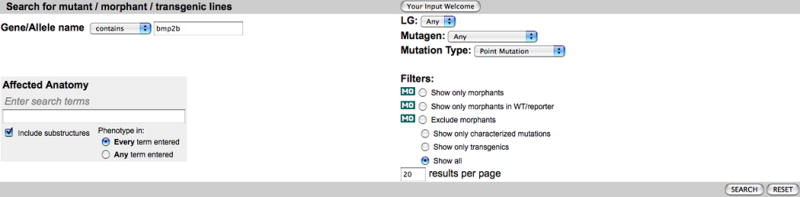
The Mutant Search form provides a variety of parameters that can be used to search for specific mutant phenotypes. This figure depicts a search for mutants containing a point mutation in the gene bmp2b.
Reviewing Search Results
-
6. Browse the search result list of all genotypes with point mutations in the gene bmp2b (Fig. 18). Each result includes the following information when possible: number of Figure View pages that show phenotypic data, genomic feature, mutation type, affected gene, LG (chromosome), and matching text that resulted in the selection of each mutant.
In the case of a wild-type morphant, i.e., a wild-type fish (genotype is the background) with a morpholino, the Allele and the Affected Genes may be blank. Multiple-mutants (double or higher) may have more than one allele and affected gene listed. If the background of the mutant fish is known, it is displayed in parentheses after the genotype name.
-
7. Click on the 2 figures link in the Phenotype data column for bmp2btc300a/tc300a (TU) to advance to its Phenotype Summary page.
A camera icon behind the figure number indicates that there are images available on the Figure View page. An MO icon indicates that data for morphant fish are available.
If there is a single figure available, the link navigates directly to the detail page.
8. Review the Phenotype Summary page (Fig. 19) for bmp2btc300a/tc300a (TU). Links are provided to the publication with matching data, the figures containing phenotype data, the genotype, any morpholinos, and affected anatomical structures and GO terms. All links lead to detailed information for each type of data.
Figure 18.
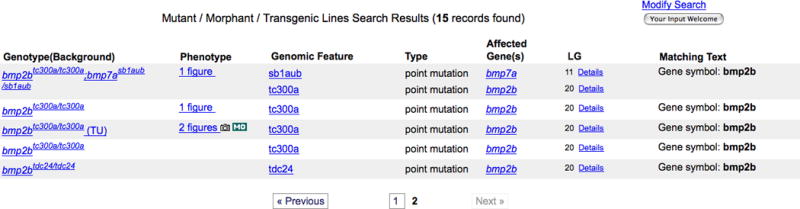
A portion of the Mutant/Morphant/Transgenics Search Results page for bmp2b point mutations.
Figure 19.

The Phenotype Summary page lists all individual figures grouped by publication for the genotype bmp2btc300a/tc300a(TU). To see the complete list of morpholinos associated with for Fig. 1 for Ramel et al., click on the all 4 link.
Viewing Phenotype Details
-
9. Click on Fig. 1 or the image thumbnail associated with Ramel et al., 2005 to see the phenotype details for Fig. 1 (Fig. 20).
Details are displayed on the Figure View page in 4 major sections. The first section provides links to additional publication data. Click on the Author or Full text link to read the publication abstract or complete article, respectively. Follow the Additional Figures link to view all figures with phenotype data from Ramel et al., 2005. The phenotype data section follows providing an alphabetized list of genotypes, experimental conditions, observed in anatomy structure or GO term, and stage. The image and legend for this figure are presented next. The links for the observed structures lead either to the Anatomy Detail page (Basic Protocol 6) or to EBI’s GO Detail page.
Figure 20.
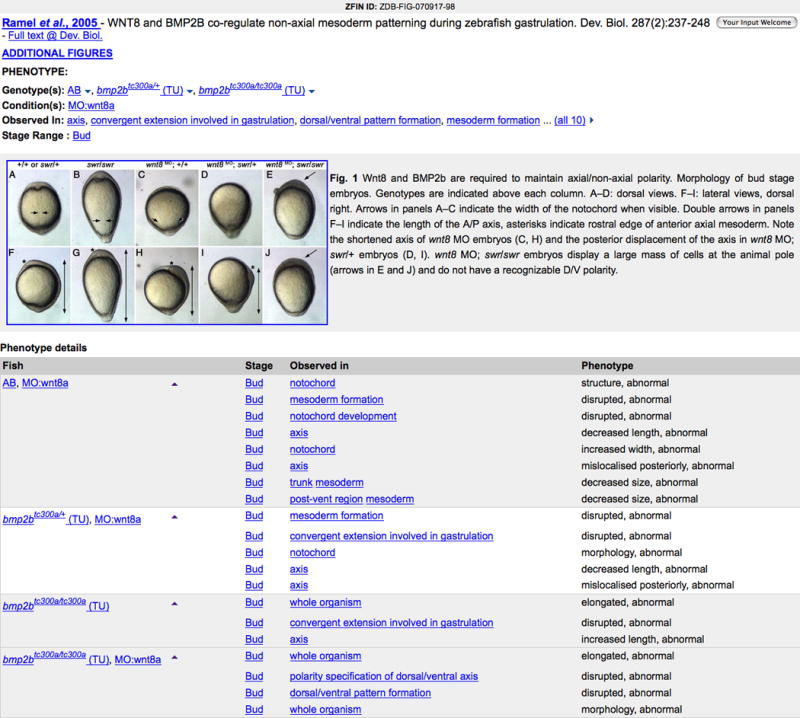
Phenotype Data on the Figure View page for Figure 1 from Ramel et al., 2005.
Viewing Genotypes
10. Click on the bmp2btc300a/tc300a(TU) Genotype link.
-
11. View mutant and transgenic line information on the Genotype Detail page (Fig. 21).
Details include the official genotype name, the wild-type fish background, and the affected genes. The parental genotype is displayed for each allele when known. Current Sources lists suppliers that carry all or one of the alleles.
All wild-type lines can be found at (http://zfin.org/cgi-bin/webdriver?MIval=aa-wtlist.apg). The Genotype Page for wild-type genotypes differs from the above because it lists only suppliers of the wild-type line, the lab of origin, and a note about the wild type.
Figure 21.
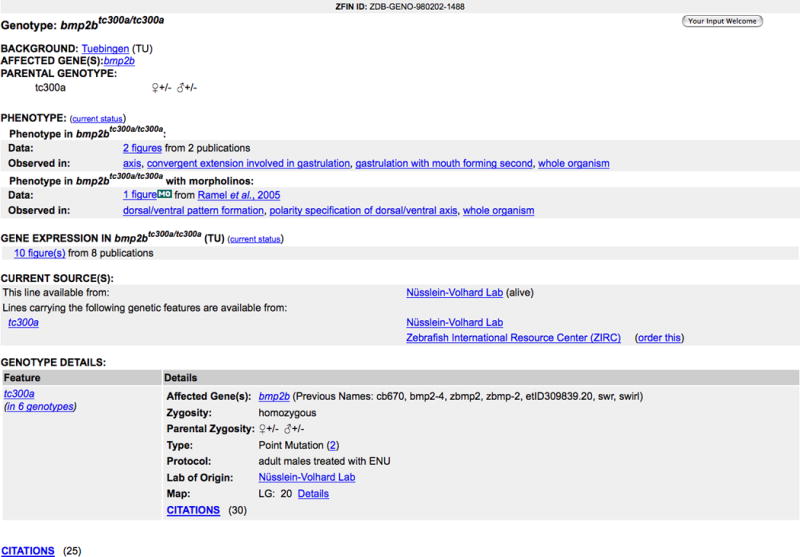
Genotype Detail page for bmp2btc300a/tc300a(TU).
BROWSING THE ZEBRAFISH ANATOMICAL ONTOLOGY (Basic Protocol 6)
ZFIN maintains the official Zebrafish Anatomical Ontology (AO), which contains about 2,400 terms as of April 2010. The AO is used for gene expression, antibody and phenotype annotations (see Basic Protocols 3, 4 and 5). The ontology consists of anatomical structures specific to zebrafish, as well as cell types present in zebrafish. Table 1 describes the three relationship types used by the AO to relate two terms to one another. The forward name is used in cases where the first term is a child term of the second term, and the reverse name is used in cases where the first term is the parent term.
Table 1.
Zebrafish Anatomical Ontology Relationships.
| Relationship Name | Example | |
|---|---|---|
| Forward | Reverse | |
| is a type of | has subtype |
white matter
is a type of
multi-tissue structure multi-tissue structure has subtype white matter |
| is part of | has parts |
hindbrain
is part of
brain brain has parts hindbrain |
| develops from | develops into |
posterior kidney
develops from
pronephros pronephros develops into posterior kidney |
In contrast to most other ontologies in biology, the anatomical ontology has a time component. Each structure exists within a specific stage range in the life cycle of the zebrafish. For instance, the pronephros appears at Segmentation stage and lasts until Day 5. The stages form an ontology themselves and are defined in The Zebrafish Book (http://zfin.org/zf_info/zfbook/stages/index.html). Each stage is defined by a time range, with a start and end time stated in hours or days. Stages do not overlap, although one stage term named Unknown is used as a start or end stage when the exact stage is yet to be determined. The Anatomy Detail page displays the definition, stage range, and relationships to other terms. Links to expression and phenotype data for the given anatomical structure are also provided.
The zebrafish anatomical ontology plays a key role in ZFIN gene expression, mutant, and antibody searches (see Basic Protocols 3, 4 and 5; for a more general discussion of ontologies, see the Commentary at the end of this unit).
Necessary Resources
See Basic Protocol 1.
Searching for an anatomical structure
-
1. View the Anatomical Ontology Browser by clicking on the Anatomy link on the home page (http://zfin.org). The AO may be browsed by structure or developmental stage.
The Request new Anatomical Term link allows users to submit a new anatomical structure for inclusion in the AO. ZFIN curators review and act on all requests.
-
2. To browse by structure, specifiy brain in the Anatomical Term field (Fig. 22).
Autocomplete provides possible AO matches after three or more characters are entered into the search field (Fig. 23b). The list contains structures or their synonyms that match the search term. Synonyms are displayed in brackets.
An alternative method of searching the AO is to browse by stage. Selecting a particular stage from the Development Stage pulldown menu will return a list of terms present at this stage. The terms are displayed ontologically, where child terms follow their parent term, indented by a small amount. The hyperlinked structure of your choice can be clicked to view details.
-
3. Select brain from the autocomplete list to view the brain Anatomy Detail page. Alternatively, press the Enter key when brain is entered.
This tool is also provided on the gene expression, mutant, and antibody search forms to specify Affected Anatomy (Fig. 23). The Enter Search Terms field is used to specify one or more terms from the zebrafish anatomical ontology (AO). Terms are entered one at a time and are added to the query by either pressing the enter key, if the complete search term has been typed, or selecting the term from the autocomplete list. Selected terms are displayed above the entry field (Fig. 23b). To remove a term from the list of those selected, click the X to the left of the term.
If more than one anatomical structure is selected, the radio buttons provide the option to search for annotations that are observed in all structures (Every term entered) or any of the structures (Any term entered).
The Include Substructures check box (Fig. 23a) ensures that the search finds all matches for the structure and its substructures. Searching for brain will find annotations to the brain and any structure that is part of the brain (e.g., the cerebellum which is part of the hindbrain which is part of the brain). The search also finds annotations to terms that are subtypes of an anatomical structure (e.g., cavitated compound organ has subtype brain). Deselecting the check box will limit the search to exact matches. In most cases, the default selection (Include substructures) should be used, because it ensures that all annotations are found even if the annotation is more specific than the selected structure.
Figure 22.

Anatomical Ontology Browser includes an Anatomical Term search field and a Developmental Stage pulldown menu. These two options can be used individually, or combined to narrow the search results. The Request New Anatomical Term hyperlink allows users to request new anatomical terms for addition to the ontology.
Figure 23.
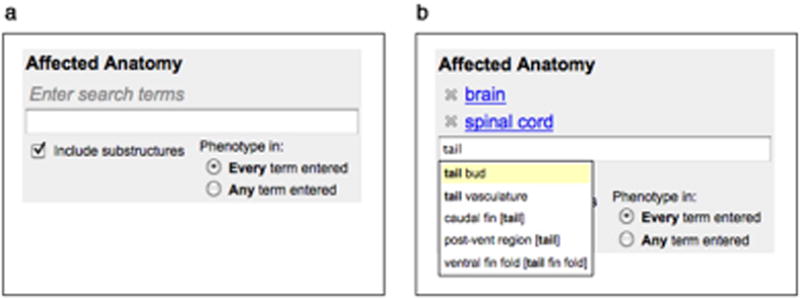
A view of the Affected Anatomy search interface before (a) and after entry of terms (b). One or several terms can be entered in the Enter Search Terms field (a). The Include Substructures check box is selected by default but can be deselected if an exact match is preferred. Selected terms are displayed above the search term field (b). Each term has an X at the left, which can be clicked to remove the term from the list. The search term field has an autocomplete feature, which provides a popup menu with a list of matching terms once three or more letters have been entered. The radio button selections on the lower right (b) allow a choice between finding phenotypes observed in all selected structures or any of the structures.
Viewing anatomical structure details
4. View the Anatomy Detail page (Fig. 24) for brain, including all synonyms, a definition, and the developmental stages during which the brain is present in wild-type fish. Links to the full developmental staging series description that defines each stage are provided. In addition, all parent and child structures are listed and grouped by relationship type.
- 5. Click on the plus (+) to the left of EXPRESSION to view brain-specific expression (Fig. 25). By default, the Expression and Phenotype sub-sections are collapsed.
- Gene Expression: Displayed are the first 5 genes for which ZFIN annotated gene expression data in wild-type fish in a standard environment (mutants, morphants, and transgenic lines are excluded). The genes are sorted by the number of figures available. Click on the figure number to navigate to the gene expression result page (see Basic Protocol 3) for the given gene name and anatomical structure. Below the table there is a link to see the complete list of all genes excluding and including substructures, again linking to the gene expression search (see Basic Protocol 3).
- In Situ Probes: In situ hybridization probes are cDNA or EST from the Thisse laboratory high-throughput expression data studies. The first five most highly ranked genes (see http://zfin.org/zf_info/stars.html) and the clones that encode them are displayed, and the link below the table navigates to pages that list all probes and their encoding genes. The entries are sorted by gene name.
- Antibodies: In this table you can find antibodies that are known to label the given structure. Targeted genes are provided in the second column. The links with the figure numbers can be used to navigate to the expression details on the Figure View Page (see Basic Protocol 3). To see all antibodies for this structure, click on the link below the table. This will bring you to the Antibody Search Result page (see Basic Protocol 4) with either only this structure or all substructures included.
- 6. Click on the plus (+) to the left of PHENOTYPE to see phenotypic data for the brain (Fig. 26).
- Mutant and Transgenic Lines: Mutants and transgenic lines for which a mutant phenotype is observed are listed alphabetically, sorted by genotype. Only non-wild-type fish treated under standard conditions are included. The affected genes and a detailed phenotype description using PATO are specified (for more about ontologies see the Commentary section). Follow the figure links to the Figure View page where the full phenotype annotation is provided (see Basic Protocol 5). To view all genotypes, click on the link below the table to navigate to the mutant, morphant, and transgenic line search (see Basic Protocol 5) including or excluding substructures.
- Morpholino Experiments in Wild-type Fish: A list of morphants for which a phenotype is observed in the specified structure. The morphant is described by the target genes, the morpholino experiment, the genotype, and the phenotype.
- Morpholino Experiments in Mutant and Transgenic Fish: The same data as for the wild-type fish in the previous table but for mutant and transgenic fish (under standard conditions).
Figure 24.
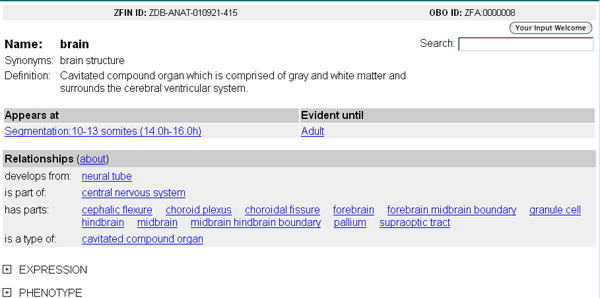
The Anatomy Detail Page for the structure brain.
Figure 25.
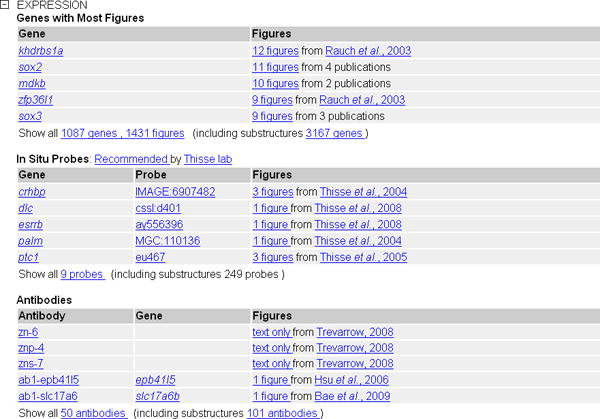
The Expression section of the Anatomy Detail page expanded for the structure brain.
Figure 26.
The Phenotype section of the Anatomy Detail page expanded for the structure brain.
USING ZFIN BLAST TO SEARCH FOR ZEBRAFISH SEQUENCES (Basic Protocol 7)
This protocol describes how to use Basic Local Alignment Search Tool (BLAST) to search for matching or similar sequences in ZFIN. Additional information regarding the BLAST algorithm and sequence similarity searching can be found in UNITS 3.1, 3.3, and 3.4. ZFIN BLAST is used to search for zebrafish genes, clones, and morpholinos using sequence. BLAST searches can reveal orthologies, paralogies, and gene family relationships that may be used to reveal information about gene function and evolution.
Necessary Resources
See Basic Protocol 1.
Searching zebrafish sequences at ZFIN
-
1. Click the BLAST at ZFIN link at the upper left of the home page (http://zfin.org). This will lead to the BLAST page (http://zfin.org/action/blast/blast; Fig. 27).
The Program pulldown menu can be used to select the desired sequence similarity search algorithm. The Database pulldown menu can be used to select from a variety of sequence datasets to compare with the search sequence. Click on the Database or Program link to get a description of the various datasets or BLAST programs.
-
2. Type the sequence accession ID BC050482 in the Sequence ID field (Fig. 27). The default sequence search setting performs a nucleotide sequence similarity search against a database of all zebrafish RNA/cDNA sequences at ZFIN.
Nucleotide or protein sequences can be entered in the Query sequence field in either FASTA or free-text format. Alternatively, a query sequence file can be uploaded from the user’s computer. Although default search parameters are provided for various datasets, the BLAST parameters and sequence filters can readily be adjusted in the options box by the user according to their requirements.
3. Click the Begin Search button to start the sequence similarity search.
Figure 27.
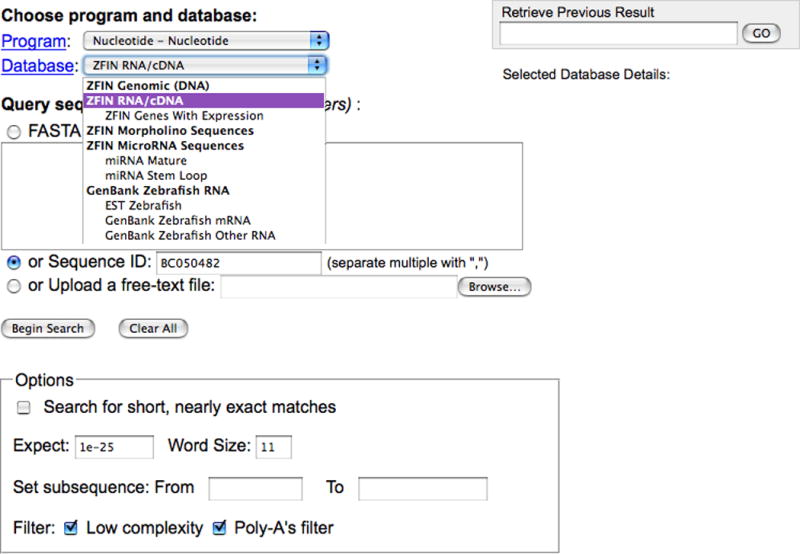
ZFIN BLAST Query page displaying programs and available datasets at the top, sequence entry choices, and BLAST search options at the bottom. The database menu is expanded in this view. Accession ID BC050482 is entered in the Sequence ID field.
Viewing the BLAST search results
-
4. To view the actual alignment between the query sequence BC050582 and a specific BLAST hit, click on the appropriate alignment in the graphic or on the score in the tabulated BLAST output (Fig. 28). Scrolling down the Result page past the tabulated output will also bring up the pairwise sequence alignments. Integration of sequence datasets within the ZFIN database, allows direct navigation from the BLAST results to ZFIN gene, transcript, and clone pages.
Genes with related Expression, Gene Ontology (GO), and Phenotype data are indicated by the E, G and P, icons, respectively. The camera icons indicate instances where ZFIN has been able to provide a figure containing expression or phenotype data. In the case of the BC050482 sequence, it is associated with the cDNA clone MGC:55230 and the gene alcama in ZFIN as displayed in the tabulated BLAST output.
The results from the BLAST output can be accessed for a week. The Result ID is displayed under the BLAST results (Fig. 28) and can be entered in the Retrieve Previous Result box in the BLAST page at ZFIN (Fig. 27).
Figure 28.
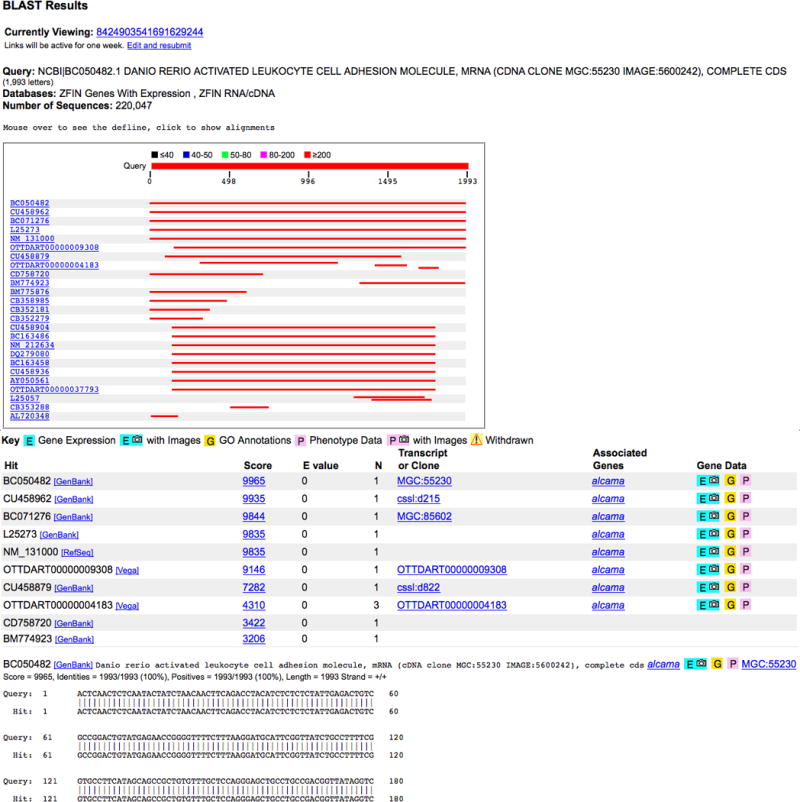
ZFIN BLAST Result page displays the search results for BC050482, an mRNA sequence accession ID. The Results page displays a graphical view of the sequence alignments, a tabulated view of the matches and relevant marker associations in ZFIN and the alignment of the query sequence with each hit.
EXPLORING THE ZEBRAFISH GENOME USING GBROWSE (Basic Protocol 8)
The zebrafish genomic sequence and annotated data can be explored at ZFIN using the Generic Genome Browser, or GBrowse (Stein et al., 2002), a web application developed for viewing genome annotations (see Unit 9.9). GBrowse provides an interactive web interface with the benefits of a customizable display, links to ZFIN detail pages, and the ability to launch sequence analysis tools. GBrowse at ZFIN hosts the Sanger Institute’s genome assembly, based on fully sequenced BAC and PAC clones from the Tübingen strain with Vega gene and transcript annotation (Jekosch, 2004; Ashurst, 2005). It should be noted that, because no whole genome shotgun (WGS) sequence was used to complete the assembly, many genes and markers are not yet represented in this version of the genome assembly. ZFIN will continue to add tracks to GBrowse incrementally, based on availability of data and community interest.
Necessary Resources
See Basic Protocol 1.
Displaying a landmark or region
-
1. To launch the zebrafish genome browser, click the GBrowse link at the upper left of the ZFIN home page (http://zfin.org), or go directly to GBrowse at http://zfin.org/genome-browser.
The page displayed is the zebrafish GBrowse home page (Fig. 29), which is divided into several main sections, each with a red plus/minus button that allows for collapse or expansion of the section, depending on the user’s needs. The Overview section is a schematic of the entire linkage group, with a highlighted section indicating the region being displayed in the Details section below. First time users may benefit from viewing the Help documentation or the OpenHelix tutorial (http://www.openhelix.com/gbrowse), both linked from the Instructions section.
An alternative way to launch GBrowse is directly from the detail page for many gene, transcript or BAC clones used in the assembly. After searching Genes/Markers/Clones and displaying the detail page of interest (see Basic Protocol 2), check to see whether there is a GBrowse graphic on the page. The image will be found in the Transcripts section on the Gene and Transcript Detail pages or the GBrowse section on the BAC Detail page. Click the image to launch GBrowse and view the selected region.
-
2. Go to the Landmark or Region search field (Fig. 29) and enter the search term 13:105101..105200, which represents a region on linkage group 13 with designated coordinates 105101–105200.
The search field in GBrowse requires a landmark or region, which can be in one of several formats. See the Examples in the Instructions section at the top of the page. Various gene IDs and sequence accessions can be used, in addition to the chromosome, or linkage group, with a colon followed by specific or approximate coordinates. A gene symbol, or a partial symbol followed by an asterisk as a wildcard, can also be used as a search term.
-
3. Click the Search button to view the displayed region.
If the search term used was followed by the wildcard (*) then, rather than going directly to a region of the genome, the search results will be displayed. A wildcard Example is shown in the Instructions section at the top of the page (i.e., hox*). The schematic results display includes the gene or marker name and the linkage group with coordinates. Each is a hyperlink to the genome display.
Figure 29.
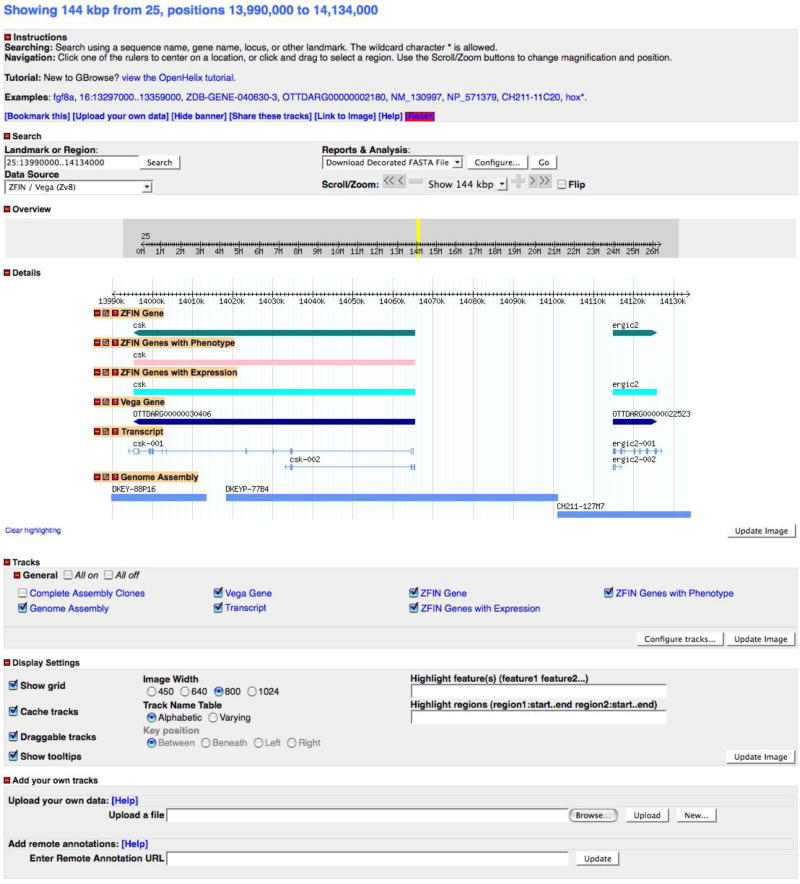
An expanded view of a selected region of the zebrafish genome as displayed in GBrowse. The search performed is shown in the Landmark or Region text field. The GBrowse page is divided into several main sections, including Instructions, Search, Overview, Details, Tracks, and Display Settings. The view is customizable by modifying the size of the region displayed and selecting and moving the tracks within the Details section. The data in the tracks show genome annotation, including the position of genes on the linkage groups, alternatively spliced transcripts, BACs used for assembly, and genes that have expression and/or phenotype data.
Navigating and modifying the displayed region
-
4. To reverse the orientation of the displayed region, click on the Flip check box located at the lower right of the Search section, and then click the Update Image button in the lower right corner of the Details section (Fig. 30).
The default genome display is from the smaller to the larger coordinate in the selected range. When the orientation is flipped, the text in the Landmark or Region search field remains the same but the coordinates in the Details section are flipped in orientation, i.e., 105200-105101. The coordinates shown in the Overview section are unaltered.
-
5. Click the Flip check box to deselect, and then click the Update Image button to refresh and return to the default orientation of increasing coordinates in the Details display.
The nucleotide sequence will be displayed in the Genome Assembly track in the Details section when the region displayed is less than 134 bp (Fig. 30).
6. In the Overview or Details section, click on the ruler to center on a location and refresh the display.
-
7. On the right side of the Search section, click on the pulldown menu and select Show 50 kbp to zoom out and expand the displayed region, centered on the coordinates of your last position.
The controls in the Search section allow the position and magnification to be changed within the Details display. To zoom in or out by 10%, click the minus (−) and plus (+) buttons. Click the left or right single or double arrows to scroll along the linkage group. As the coordinate range and position change, the highlighted region in the Overview section will reflect the new location.
8. In the Landmark or Region text field, type the symbol egr2b and click the Search button (Fig. 31).
-
9. Click and drag across the ruler in either the Overview or Details section (Fig. 31), as an alternate method to select a new region to display. Once the selection is made, release the mouse button, and then click on the hyperlink in the popup menu either to Zoom In or Recenter on the selected region.
As the mouse is dragged across the region, the coordinates are highlighted and the size of the selected region is displayed. If the selection is incorrect, it can be canceled from the popup menu.
Figure 30.
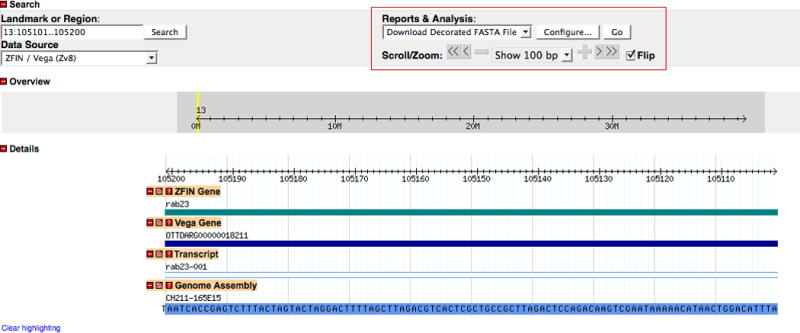
Navigating the genome in the Details section. Using the pulldown menu on the lower right side of the Search section (red boxed area), a region spanning 100 bp has been selected and displayed. The nucleotide sequence can be seen at this magnification in the blue schematic representation of the BAC component in the Genome Assembly track. The orientation of the rab23 gene in this view has been flipped. The Flip check box has been selected and the reversed coordinates can be confirmed in the Details section. Note that the region in the Landmark or Region search field is still displayed in the default orientation.
Figure 31.
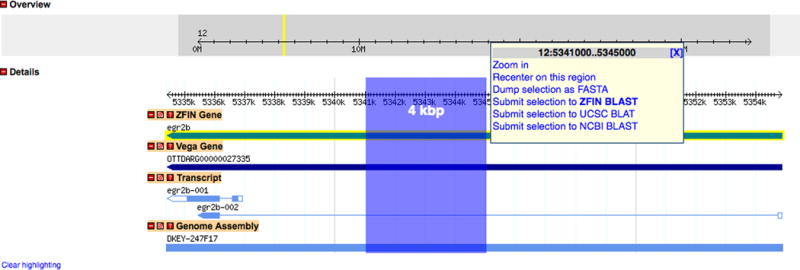
Capturing and analyzing sequence in GBrowse. A 4 kbp region has been selected in the Details section by clicking on the ruler and dragging the mouse. The selection is highlighted as the mouse is dragged across the ruler and the region size is displayed. When the selection is complete, a popup window appears offering hyperlinked choices for changing the magnification and location of the Details display, as well as tools for capturing the sequence as FASTA or performing BLAST analysis.
Capturing and analyzing sequence
10. Click on the ruler in the Details section and drag the mouse to select the region of interest (Fig. 31). Release the mouse button when the selection is complete.
11. Click on the Dump Selection as FASTA hyperlink to launch a new window containing the selected sequence in FASTA format.
-
12. Return to the GBrowse window, repeat Step 10, and click on the Submit Selection To ZFIN BLAST hyperlink to launch a new window for ZFIN BLAST analysis.
The selected sequence will be displayed in the ZFIN BLAST submission form as the Query Sequence. Once on this page, the Program, Database, and Options can be selected to set the parameters before clicking the Begin Search button to perform the BLAST analysis (see Basic Protocol 7).
-
13. Return to the GBrowse window, repeat Step 10, and click on the Submit Selection To UCSC BLAT (Kent, 2002) hyperlink or the Submit Selection To NCBI BLAST (Altschul et al., 1990) hyperlink.
A submission confirmation will launch in a new window. To analyze the selected sequence at either UCSC or NCBI, click the Confirm button to submit the sequence. Alternatively, click Cancel to close the window and return to GBrowse.
Exploring and modifying the ZFIN-specific tracks
14. Scroll down to the Tracks section of the page.
15. Click the All On check box in the Tracks section (Fig. 32) and click the Update Image button in the lower right to update the Details view and display all available tracks.
-
16. Mouse over ZFIN Genes with Phenotype in the Tracks section to view a tooltip with a brief description of the data displayed in the track.
Click on any track name to launch a new window with the complete list of tracks and descriptions.
17. Click the All Off check box in the Tracks section to deselect all available tracks.
-
18. Click the individual check boxes to select tracks of interest, and then click Update Image to refresh the Details display.
The ZFIN Genes with Expression track displays genes with expression data stored in ZFIN. The ZFIN Genes with Phenotype track displays genes with phenotype data in the ZFIN database.
19. In the Details section, mouse over the glyphs in each track to view a tooltip with a brief description of the annotated data and the displayed genomic coordinates.
20. Click on the track name in the Details section, then drag the track up or down and release to move the track to a new position, rearranging the tracks to create a customized Details view.
-
21. Click on a glyph in the ZFIN Gene, Transcript, or Assembly tracks to view its detail page at ZFIN.
The data viewed on marker detail pages is described in Basic Protocol 2. Nearly all the tracks in the zebrafish GBrowse viewer have glyphs that link to ZFIN pages. One exception is the glyph in the Vega Gene track, which links to the corresponding gene page at the Wellcome Trust Sanger Institute’s Vega site (http://vega.sanger.ac.uk/Danio_rerio/).
-
22. Click on a glyph in the ZFIN Genes with Expression or Phenotype track to view the Expression Pattern Search Results or Phenotype Summary for the gene of interest.
The data viewed on the Expression Search Results and Phenotype Summary pages, and their format, is described in Basic Protocols 3 and 5.
Figure 32.
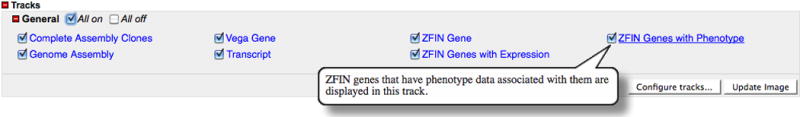
GBrowse tracks available for the zebrafish genome. The Assembly tracks show the components used in the Vega assembly. The Gene and Transcript tracks show schematic representations of the gene and the exon structure of the transcripts, with arrows indicating their orientation in the genome. The Expression and Phenotype tracks are ZFIN-specific tracks indicating the presence of associated data in the ZFIN database. Selecting and de-selecting the check boxes and updating the image allow customization of the Details view.
CONTRIBUTING AND COLLABORATING VIA THE COMMUNITY WIKI (Basic Protocol 9)
To facilitate collaboration in the zebrafish research community, ZFIN hosts a Community Wiki. The wiki is designed to be a repository for information, where contributions and comments from the community will encourage a dynamic exchange of information and expand the knowledge base. The goal of the wiki is to foster sharing and participation by providing a forum that responds quickly to changes and additions, thereby remaining current and supplementing the ZFIN database.
Necessary Resources
See Basic Protocol 1.
Launching the Community Wiki
1. Click on the Wiki hyperlink in the Community section on the left side of the ZFIN home page (http://zfin.org) or go directly to the wiki URL http://wiki.zfin.org. This will invoke the Community Wiki home page (Fig. 33).
Figure 33.
The ZFIN Community Wiki home uses Confluence software from Atlassian (http://www.atlassian.com/software/confluence/). The welcome page for the Community Wiki has links to the Protocol Wiki and the Antibody Wiki on the left side. Links to Additional Resources and Help documentation are available on the right. The search field at the upper right can be used to perform a search of all wiki content. The Log In and Sign Up links at the upper right are for registered users to log in or for new members to sign up as registered users, respectively.
Searching and navigating the Community Wiki
The main landing page for the Community Wiki (http://wiki.zfin.org) is formatted to provide quick tips and direction for getting started. On the left side of the page are the Welcome message and a table listing the main sections of the wiki (Protocols and Antibodies), with brief descriptions and hyperlinks to those areas. On the right side of the page are Resources, which direct users to pages on web sites outside of the wiki, and Help documentation, which will assist in familiarizing users with the wiki. There are also links at the bottom of every wiki page that direct users to the Help page and the User’s Guide. In the upper right corner of every page, there are links to allow users to Log In, or Sign Up if not already registered, and pulldown menus for adding Comments to the page and using Tools on the wiki.
-
2. To perform a general search of the wiki, click on the Search field at the upper right of the Community Wiki home page, enter the search term breeding and press the enter key.
The wiki pages found via the search term will be listed with their page titles as headers and a brief selection of text from the page below. Text matching the search term is shown in bold (Fig. 34). The titles are hyperlinked to the page of interest and below each result is listed the section of the wiki in which it is found (e.g., Protocols).
3. From the results page (Fig. 34), click on the Where, What, When pulldown menus on the right side to select additional search parameters as filters. Enter a name in the Who text field to narrow the results further. Click the Filter button to filter the results and refresh the list on the left.
Figure 34.
A view of a wiki search results page using the word breeding shows the term in the search field at the top followed by a list of results down the left side of the page. The search term is shown in bold within the individual titles and page excerpts in the list. On the right side of the page are several pulldown menus and a text field that provide filtering of the result set.
Registering for the Community Wiki
The Community Wiki is a resource for the zebrafish research community and will be most valuable when there is widespread participation. Sharing information and commenting on the wiki pages is encouraged. Pages on the wiki can be viewed by anyone but to add to the wiki, or comment on the current content, users must log in. Everyone is encouraged to register for an account and begin participating.
4. Click on the Log In link at the top right of any page on the Community Wiki (Fig. 34).
5. Enter your Username and Password, and then click the Log In button.
6. Click the Sign Up link at the top right of any wiki page, if you are not yet a member.
7. Enter Full Name and Email in the appropriate fields, and then select a Username and Password. After confirming the Password, enter the text from the challenge-response image and click the Sign Up button to register.
Launching and navigating the Protocol Wiki
The Protocol Wiki contains experimental protocols imported from The Zebrafish Book, 5th edition (Westerfield, 2007), in addition to those shared by zebrafish researchers through direct submission to the wiki. The protocols are organized into sections corresponding to the chapters of The Zebrafish Book. On the left side of the Protocol Wiki home page (Fig. 35), users can browse through the protocols by Section or use the search interface to query by keyword. On the right side of the page, Recent Comments and Recently Updated Pages are listed with hyperlinks to the most recently created and updated wiki pages.
8. From the ZFIN home page (http://zfin.org), click the Protocols hyperlink next to Wiki under the Community header on the left side of the page. Alternatively, click the Protocols link on the left side of the Community Wiki main landing page (http://wiki.zfin.org). This will invoke the ZFIN Protocol Wiki home page (Fig. 35).
-
9. On the lower left of the Protocol Wiki home page under Sections, click on the + (plus) button next to the chapter heading for Breeding. Continue to click on the + buttons to expand the chapter and reveal additional protocols.
To collapse the list, click the − (minus) button next to the chapter or subheading. Clicking on a chapter heading opens a wiki page where the protocols for that chapter are listed as hyperlinks.
Figure 35.
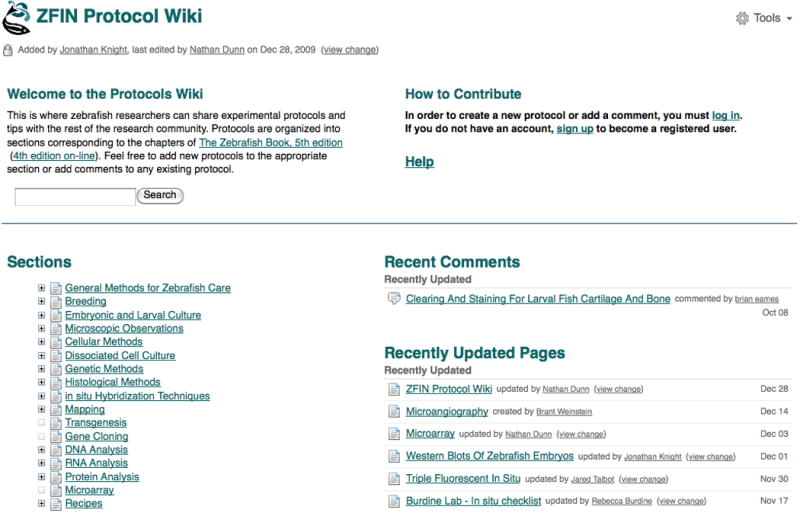
A view of the ZFIN Protocol Wiki home page. The protocols are organized into Sections corresponding to the chapters in The Zebrafish Book, 5th edition, which is the source material used to seed the Protocol Wiki. The headings under Sections can be expanded or collapsed by clicking on the + and – buttons. Section headers and protocol titles are hyperlinked to their respective pages on the wiki. The right side of the page provides links to recent wiki content, including added comments and updated pages.
Viewing a protocol and using the Tools menu
-
10. From the Breeding chapter under Sections, click on the hyperlinked protocol title for Embryo Collection. The wiki page for Embryo Collection (Westerfield, 2007) will be displayed (Fig. 36).
If the protocol is not found by browsing through the chapters and protocol titles, enter a keyword into the text field on the left side of the Protocol Wiki home page and click the Search button. Pages containing the search term in either their title or content will appear in the list of results. Protocols that reference solutions and recipes include a link to the Recipe chapter (Fig. 36).
11. Mouse over the Tools menu on the right side of the page to see a list of options.
-
12. Select Attachments from the menu to view a list of attachments associated with the protocol. The list includes the attachment name, size, creation date, and a comment field.
The attachments include any images included with the protocol. In the case of the Embryo Collection protocol, the image for Figure 1 is the only attachment. Other protocols may have supplementary documents or spreadsheets. From the attachment list page, click View in the upper right or the browser Back button to return to the protocol.
13. From the Tools menu, select Page History to see the history of edits to the page and to compare versions. Click View in the upper right to return to the protocol.
14. From the protocol Tools menu, select Export to PDF or Export to Word. An option is provided to open or save the protocol in the selected format.
Figure 36.
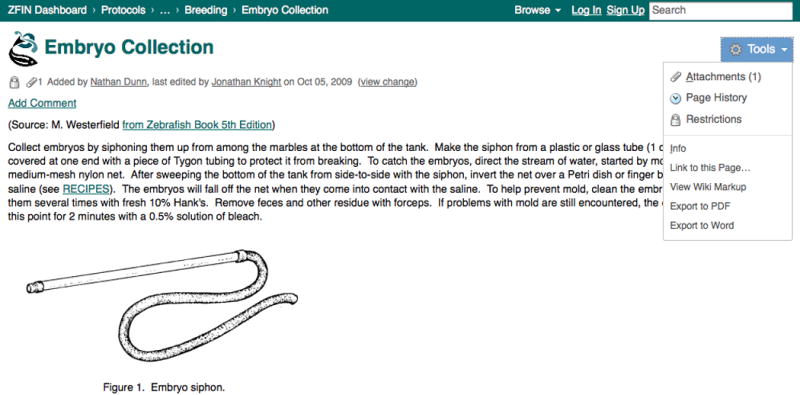
A view of a protocol page in the Protocol Wiki. Below the protocol title is a status line, indicating the registered user who added the protocol, as well as information about the most recent instance of editing. Each protocol can include figures and links to other wiki pages, as in this example where a recipe is hyperlinked to another section of the wiki. Registered users can use the Add Comment hyperlink to comment on the protocol and share their experience and tips. The Tools menu on the right provides links for tracking the edit history of the page and for exporting the protocol.
Launching and navigating the Antibody Wiki
The Antibody Wiki is a place where zebrafish researchers can help each other by sharing antibody information, including antibodies that work well on zebrafish, labeling protocols and tips for antibody usage. The antibodies that were used to seed the wiki include the full set of antibodies available from ZIRC and those curated from publications by ZFIN staff. The number of antibodies on the Antibody Wiki will continue to grow as the ZFIN antibody database grows through curation from publications and as registered users contribute to the wiki by creating new community submitted antibody records.
15. From the ZFIN home page, click the Antibodies hyperlink next to Wiki under the Community header on the left side of the page. Alternatively, click the Antibodies link on the left side of the Community Wiki home page. This will invoke the ZFIN Antibody Wiki home page (Fig. 37).
Figure 37.
A view of the ZFIN Antibody Wiki home page. To find an antibody, users can browse a list of antibodies by clicking the appropriate hyperlink on the lower left side of the page. Alternatively, a search can be performed using the text field at the upper left, a method that does not require knowledge of the official name on the antibody record. The lower right side of the Antibody Wiki home page provides hyperlinks to recently added or updated antibody pages and comments.
There are several ways to find an antibody from the Antibody Wiki home page. Users can take advantage of sections listing Community Submitted Antibodies and ZFIN Antibodies or, if the antibody can’t be identified by its name, use the search interface where information related to the antibody can be entered as search terms.
-
16. To search for an antibody, begin on the lower left side of the Antibody Wiki home page (Fig. 37). Click the View All ZFIN Antibodies hyperlink to launch a new window with a list of antibodies found in ZFIN.
The two sections on the lower left of the Antibody Wiki home page have a brief listing of the first five antibodies in each category so, in most cases, users will click the link to browse the entire list.
-
17. Under Finding An Antibody on the middle left of the Antibody Wiki home page, enter the antibody name zn-5 in the search field.
An antibody may have several names or the name of the antibody record may not be very informative. Using the search interface to locate an antibody allows the use of alternate names, target gene symbols and other information related to the antibody as search terms.
-
18. Select the linked name of the antibody from the menu of search results that appears as you type or click the Search button after entering the term, then click on the zn-5 link in your search results to view the antibody record (Fig. 38).
The antibody wiki pages have a table, including names, IDs, host organism, isotype, target molecules and genes, and supplier. Information on whether the antibody works in zebrafish is reported, with a separate table showing the assays tested and notes if available. The zebrafish anatomical structures labeled by the antibody are listed, with links to the anatomy terms in ZFIN. At the bottom is a Notes field where additional information may be found.
-
19. Click on the zn-5 link in the Antibody Name field to view the antibody record in ZFIN.
The antibody records in the wiki are linked to their companion ZFIN records. Similarly, the ZFIN antibody records have a link in their Antibody Name field that launches the companion wiki page in a new window.
Figure 38.
The individual antibody pages in the Antibody Wiki contain a table view of information, including the name and other aliases and IDs, host organism, isotype, structures labeled, genes and molecules targeted, and suppliers. Information on whether the antibody works on zebrafish and the assays tested are also included on the record. This zn-5 antibody (Fashena and Westerfield, 1999) record shows what a registered user would see, including the Edit button and Add menu on the upper right.
Contributing to the Community Wiki
The level of participation on the wiki is integral to its success. The more each member of the zebrafish community contributes to the wiki, the greater its benefit to all. Registered users can create new protocols, add antibodies, and comment on existing content to pass on their knowledge and experience to others.
-
20. To contribute to the wiki by creating pages or commenting on the current content, click the links at the upper right side of any wiki page to Log In or Sign Up to be a registered user.
Registered users who have signed in will see an Edit link at the top right of each antibody, protocol, and recipe page (Fig. 38). Additional options will appear in the Tools menu.
21. To add a comment to an antibody, protocol or recipe page, click the Add Comment hyperlink at the top or bottom of the page.
-
22. From the Comment edit page, enter your comment in the text field.
The default view is the Wiki Markup format. Use the icons to insert links or images into your comment. Use the Help Tips on the right for tips on formatting text. Alternatively, click on the Rich Text tab to enter and format the comment using the menu icons. From the Rich Text tab, click the Make Rich Text Default hyperlink to switch the default from Wiki Markup. Click the Preview tab to see how the comment will be displayed before posting.
23. Click the Post button to add the comment or click Cancel to return to the page without posting a comment.
The creation of new protocols and addition of antibodies to the wiki is the best way to share information with the zebrafish research community and augment publications and curation of data by ZFIN staff.
24. To create a new protocol, click on the appropriate Section for inclusion from the lower left of the Protocol Wiki home page (Fig. 35). As an example, click the hyperlink for General Methods for Zebrafish Care.
25. In the New Protocol Name text field, enter the name of the protocol you want to add to this section and click the Create New Protocol button.
-
26. Enter the protocol content in the text field or cut and paste from another document, then click the Save button on the upper right.
The text can be added with the Wiki Markup tab using the tips on the right or the Rich Text tab can be selected to use the Rich Text editor. Click the Preview tab to view the protocol before submitting. To create a protocol from a Word document, name and create an empty protocol in the appropriate section. View the new protocol and select Doc Import from the Tools menu. Click Choose File, select file from computer, and click Import. The page should be rendered from your document, although some edits to formatting may be necessary.
27. To add a new antibody to the wiki, enter the antibody name in the New Antibody Name text field on the right side of the Antibody Wiki home page (Fig. 37) and click the Create New Antibody button.
-
28. Add all available data to the fields in the table.
Do not edit the Antibody Name field. To see an example page with data, click the Antibody Example hyperlink, which will launch a new window.
29. Click the green + (plus) button under Assays Tested to add information to this table.
30. Add Notes or Comments to the appropriate fields then click the Preview tab to see how the record will be displayed before submitting.
31. Click the Save button to create the new antibody record.
-
32. Click Edit to update or edit the antibody record. Use the Tools menu to Copy, Move, or Remove an antibody record.
Registered users can add comments to any existing page but can only modify pages they have created.
GUIDELINES FOR UNDERSTANDING RESULTS
ZFIN is a curated, web-based resource and some general guidelines should be followed when interpreting search results. Information in ZFIN is constantly being updated with new data from literature as well as through data exchange with other web-based resources.
Data revisions in ZFIN
Data in ZFIN is revised or reassigned as additional information becomes available. Improvements to the genome assembly and genome annotation can result in annotations being merged, deleted, or withdrawn. This is commonly observed in the case of genes, transcripts, clones, and other markers. Assignments that were made based on limited sequence information are corrected with the availability of new sequence and clone data. These changes can affect the association of genes and other markers to related information such as sequence IDs and orthologies. In instances of merged records at ZFIN, the users will be directed to the most current record because all records in ZFIN can be searched for by their previous name or ZFIN ID. In the case of a split, which involves one ZFIN record being separated into two or more markers, data associated with the original gene may now be associated with only one of the two or more resulting genes. Markers that are obsolete have WITHDRAWN displayed as part of the name and symbol, or Annotation Status in the case of transcripts. The Nomenclature History link on the gene page provides a summary of all changes made to the gene name.
Literature References in ZFIN
ZFIN strives to attribute every annotation to the source of the data. In the majority of cases, there is an explicit hyperlink to a primary literature reference that is the source of the information. In other instances, the reference points to a ZFIN-created reference, that provides details about the data exchange or download process. These references provide the user with a starting point for further analysis. The references may contain additional content or caveats that cannot be captured by ZFIN but could be valuable for the user.
Use of Evidence Codes
Gene Ontology (GO) annotations
Each Gene Ontology (GO) annotation is attributed to a specific reference as well as an evidence code to indicate how the annotation is supported. Evidence codes are extremely useful in interpreting GO annotations. A description of the complete set of evidence codes is available at http://www.geneontology.org/GO.evidence.shtml. It is important that the user understand the differences between manually assigned evidence codes and the computationally assigned Inferred from Electronic Annotation (IEA) code as well as differences among the various types of manually assigned evidence codes. The evidence codes do not provide a measure of the quality of annotation but indicate the type of analysis (experimental, computational or curatorial) that produced the annotation. Understanding the structure of ontologies and how they are used will allow users to avoid errors in interpretation (Rhee et al., 2008)
Orthology Annotations
ZFIN also uses evidence codes to support assertion of orthology. The evidence for orthology is presented in the orthology section on the gene page, as well as on the Orthology Details page. The evidence is displayed as a two-letter code, which is hyperlinked to a page with the full list of abbreviations and their definitions (http://zfin.org/zf_info/oev.html).
COMMENTARY
ZFIN Contents
ZFIN is a dynamic resource. A table summarizing our growth in data types and content since 1998 is available at http://zfin.org/zf_info/zfin_stats.html.
Nomenclature
ZFIN serves as a member of the Zebrafish Nomenclature Committee, playing a key role in the naming of zebrafish genes and mutants. Good nomenclature practices are integral to cross-species comparisons, unambiguous communication, and effective database querying. Zebrafish nomenclature conventions are based on rules applied to naming human and mouse genes and on input from the zebrafish community. These guidelines are available at http://zfin.org/zf_info/nomen.html. We encourage researchers to contact us prior to publishing new genes or mutants. Links are provided on the ZFIN home page for the submission of mutant and transgenic line names and to obtain approval for gene names. The researcher must be logged in to submit requests (contact zfinadmn@zfin.org for a ZFIN account). Researchers may contact the nomenclature committee by email (nomenclature@zfin.org).
Use of ontologies
Standardization of data annotations is key to robust searching of data and promotes comparisons between species and among databases (Washington et al., 2009). Ontologies provide a common vocabulary of terms (names) within a domain and their relationships to one another. For instance, the zebrafish anatomical ontology contains the term called eye, which has the child term retina. Retina is part of the eye. The use of ontologies provides common terms for annotations and searches ensuring retrieval of the appropriate set of results (see Basic Protocol 6 for the use of the Zebrafish Anatomical Ontology in a search). Ontologies, through the use of defined relationships, provide the ability to perform more comprehensive searches. For instance a search for eye could also include retina.
ZFIN makes use of ontologies that are created and maintained by the larger bioinformatics community. These ontologies are under continual development and need input from the community to ensure completeness and accuracy. Ontologies used include the Zebrafish Anatomical Ontology (AO; see Basic Protocol 6), Gene Ontology (GO), Entity Quality (PATO; http://obofoundry.org/wiki/index.php/PATO:Main_Page), Cell (CL; Bard et al., 2005) and Sequence (SO; (Eilbeck et al., 2005). Further discussion of GO can be found in UNIT 7.2. Many of these ontologies are available from the Open Biological Ontologies site (OBO; Smith et al., 2007).
Creating a ZFIN account
To optimize their experience, users are encouraged to contact ZFIN to request a ZFIN account and password. A ZFIN login allows users to create records that can be searched using the People, Lab and Company search forms. Records created for individuals, laboratories and companies can be updated with name, affiliation, research interests, and publications, thereby facilitating contact and collaboration within the research community. In addition, having a ZFIN login will allow access to areas of ZFIN where registration is required, including online forms for obtaining approval for official gene names and for submitting mutant and transgenic line names.
Submitting data to ZFIN
ZFIN encourages individual laboratories to share unpublished data with the community using Phenote, a software package that facilitates annotation of mutant phenotypes and gene expression. Phenote annotations are comprised of zebrafish AO, GO, and PATO terms allowing for easy integration with ZFIN data. The advantages of using Phenote for submissions include basic browsing of terms, which assists in locating the correct term and discovering new terms, and streamlining of submission, where all data submitted to ZFIN is attributed directly to their sources. Phenote is available at http://www.phenote.org/download.shtml.
Ordering from the Zebrafish International Resource Center (ZIRC)
ZFIN works in close collaboration with ZIRC (http://zebrafish.org/zirc), both sharing a similar mission to serve the zebrafish research community (Henken et al., 2004). ZFIN is a repository for data, whereas ZIRC acquires, maintains and redistributes resources for the research community, including zebrafish lines, ESTs/cDNAs and antibodies. From ZFIN’s home page, users can find a link to ZIRC’s home page, as well as more direct links to individual pages at ZIRC where various resources can be ordered. Links to ZIRC are located in the ZIRC section at the top right of the main section of the ZFIN home page and on the ZIRC navigation tab (Fig. 2c) from any ZFIN page. In addition, many of the ZFIN detail pages for mutant fish lines, ESTs, and other resources have Order This links in the Current Source section, directing users to ZIRC for ordering fish lines and reagents.
Exploring community resources
ZFIN provides a wealth of news, information, and community resources to those involved in zebrafish research. Links to many resources used by the community are provided on our home page (http://zfin.org). On the left side of the home page is a Community section that includes links to the latest job postings and meeting announcements. ZFIN also hosts a moderated newsgroup for online discussion of research, protocols and topical questions for the zebrafish community. Recent topics on the newsgroup include water quality and disease issues, experimental protocols and funding opportunities. ZFIN keeps an archive of news relevant to the research community and publishes a bi-annual newsletter that includes announcements regarding additions and updates to ZFIN, as well as other newsworthy items submitted by the community. Additional links to and descriptions of genome resources, cDNA and EST resources, stock centers, mutagenesis projects, the gene and enhancer trap database and pathways can be found at http://zfin.org/zf_info/catch/catch.html.
Research benefits greatly from the exchange of information between community members. ZFIN hosts a Community Wiki that includes a Protocol Wiki and Antibody Wiki where zebrafish researchers can share and discuss laboratory protocols and antibody information (see Basic Protocol 9). The Protocol Wiki consists of protocols from The Zebrafish Book, 5th edition (Westerfield, 2007), and protocols submitted by the community. The Antibody Wiki contains all antibodies in the ZFIN database, as well as antibody records created by the community. The Community Wiki can be found at http://wiki.zfin.org.
Critical Parameters and Troubleshooting
No data found
It is possible to submit a query, via the ZFIN search forms, that does not retrieve any results. There are several possible reasons for this. (1) The data may not be available. ZFIN adds new data types in response to research trends (see http://zfin.org/zf_info/zfin_stats.html -for a display of ZFIN data contents over time). Unfortunately, resources restrict our ability to back curate the literature as data types are added, consequently, not all data is present. Curation of current literature is our priority but there may be a short lag period between publication date and curation. (2) Data may also not be found as a result of a poorly formed query. The use of too many query form options may inadvertently place too many restrictions on the result set. It can be useful to resubmit the query with fewer specified parameters. You can then make use of the ‘modify your query’ feature to add options back in one at a time to see what may be causing the unintended consequence. (3) ZFIN search form name fields accept a single entry. Multiple entries will result in no records found. For instance, specifying hsp70 and cre simultaneously in the mutant search form name field will result in no records found. Our site search does accept multiple entries and may be used in lieu of the specific search form.
Too much data found
On occasion, a query may return more results than you can realistically review. In these cases it is advisable to make use of the various options provided on the search form. Descriptions of search form options are found in this document (retrieving information about zebrafish genes (Basic Protocol 2); finding and viewing comprehensive gene expression data (Basic Protocol 3); finding antibodies (Basic Protocol 4); finding phenotype data (Basic Protocol 5)). Alternatively, data may be downloaded from http://zfin.org/zf_info/downloads.html as tab-delimited text files and imported into another tool, such as Excel. Once imported, the data may be explored according to parameters of your choosing.
Data dependency on other databases
ZFIN maintains extensive links to other web resources such as NCBI, UniProt, and Ensembl to provide the user with the most comprehensive view of available information. These links are maintained through an intricate schedule of daily, weekly, or monthly data exchanges followed by curator analysis of potential problems. In some instances, this process can cause a synchronization lag between data at different resources. Accession IDs listed in one resource may not yet be available at ZFIN or could be associated with a different gene at ZFIN. There may also be instances where data from a publication is not in ZFIN, because the publication was not entered into ZFIN or has not yet been curated. User queries and suggestions are appreciated and allow ZFIN to maintain accurate and current information.
Asking for help, information, or new features
ZFIN is a community resource and input from the community on ways to add to and improve ZFIN is encouraged. In addition to the contact link found on the navigation bar and General Information navigation tab, each data page has a Your Input Welcome button. Clicking the button launches a page with a form for users to provide their input and comments. Questions and comments will be reviewed and, if contact information is provided, the user will receive a reply from ZFIN staff. Comments, suggestions, and feedback are always welcome and appreciated and may also be submitted directly to zfinadmn@zfin.org. In addition to these regular avenues for feedback, ZFIN conducts periodic user surveys to help establish goals.
Suggestions for Further Analysis
Downloading bulk data
Additional analysis of ZFIN data can be performed locally by downloading ZFIN data reports at http://zfin.org/zf_info/downloads.html. These reports are updated nightly. All files use tab as the field delimiter. The format for each file is provided on the ZFIN Data Reports page listed above. Questions or special requests for data should be directed to zfinadmn@zfin.org.
Searching ZFIN Gene Ontology (GO) annotations using AmiGO
ZFIN curates data from publications using GO terms (see Unit 7.2); however, searching via these annotations is not yet available at ZFIN. ZFIN regularly provides zebrafish GO annotations to the GO Consortium where the entire GO database can be searched using AmiGO (http://wiki.geneontology.org/index.php/AmiGO_Manual:_Overview). AmiGO provides many functionalities including:
Search for a gene or gene product, or a list of gene or gene products, and view the GO term associations
Perform a sequence identity BLAST search and view the GO term associations for the genes or proteins returned
Search for GO terms and view the genes or gene products to which they are annotated
Browse the GO ontology and view terms
Map the granular annotations of the query set of genes to one or more high-level using the slimmer tool
Run custom SQL queries against the GO database using GOOSE
Acknowledgments
The authors of this unit would like to thank the zebrafish community for their continued support, suggestions, and data submissions which are vital to ZFIN’s growth and success. We would also like to thank Monte Westerfield, Yvonne Bradford, Tom Conlin, Nathan Dunn, David Fashena, Ken Frazer, Doug Howe, Jonathan Knight, Prita Mani, Ryan Martin, Sierra Moxon, Holly Paddock, Leyla Ruzicka, Kevin Schaper, Holle Bauer Schaper, Xiang Shao, Amy Singer, Brock Sprunger, and Ceri Van Slyke whose expertise and dedication make ZFIN work. ZFIN, located at the University of Oregon, is supported by the National Human Genome Research Institute (P41 HG002659) of the National Institutes of Health.
Footnotes
Internet Resources
http://zfin.org/zf_info/catch/catch.html
ZFIN’s external links page includes web resources and stock centers for zebrafish.
http://zfin.org/zf_info/downloads.html
ZFIN’s site for downloading large data sets.
http://zfin.org/zf_info/nomen.html
Zebrafish Nomenclature guidelines.
http://zfin.org/cgi-bin/webdriver?MIval=aa-new_line.apg
ZFIN’s submission form for proposed mutant/transgenic line names. ZFIN login is required.
http://zfin.org/cgi-bin/webdriver?MIval=aa-new_gene.apg
ZFIN’s submission form for proposed gene names. ZFIN login required.
http://zfin.org/zf_info/anatomy/dict/sum.html
ZFIN’s links to zebrafish atlases and anatomical resources.
The ZFIN Community Wiki, where zebrafish researchers can share information with the rest of the community.
The Zebrafish International Resource Center (ZIRC) acquires, maintains and redistributes zebrafish resources.
OpenHelix provides tutorial and training materials that introduce new users to ZFIN.
http://www.sanger.ac.uk/Projects/D_rerio/
The Danio rerio Sequencing Project at the Sanger Institute describes the genome project and its status.
The GMOD wiki page for GBrowse provides a description, and links to further documentation.
The Open Biomedical Ontologies website provides science-based ontologies in the biomedical domain.
Literature Cited
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215:403–10. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- Ashurst JL, Chen CK, Gilbert JG, Jekosch K, Keenan S, Meidl P, Searle SM, Stalker J, Storey R, Trevanion S, Wilming L, Hubbard T. The Vertebrate Genome Annotation (Vega) database. Nucleic Acids Res. 2005;33:D459–65. doi: 10.1093/nar/gki135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bard J, Rhee SY, Ashburner M. An ontology for cell types. Genome Biol. 2005;6:R21. doi: 10.1186/gb-2005-6-2-r21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrett T, Troup D, Wilhite S, Ledoux P, Rudnev D, Evangelista C, Kim I, Soboleva A, Tomashevsky M, Marshall K, Phillippy K, Sherman P, Muertter R, Edgar R. NCBI GEO archive for high-throughput functional genomic data. Nucleic Acid Res. 2009;37:D885–890. doi: 10.1093/nar/gkn764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bill BR, Petzold AM, Clark KJ, Schimmenti LA, Ekker SC. A primer for morpholino use in zebrafish. Zebrafish. 2009;6:69–77. doi: 10.1089/zeb.2008.0555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eilbeck K, Lewis S, Mungall CJ, Yandell M, Stein L, Durbin R, Ashburner M. The Sequence Ontology: A tool for the unification of genome annotations. Genome Biol. 2005;6:R44. doi: 10.1186/gb-2005-6-5-r44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fashena D, Westerfield M. Secondary motoneuron axons localize DM-GRASP on their fasciculated segments. J Comp Neurol. 1999;406:415–24. doi: 10.1002/(sici)1096-9861(19990412)406:3<415::aid-cne9>3.0.co;2-2. [DOI] [PubMed] [Google Scholar]
- Henken DB, Rasooly RS, Javois L, Hewitt AT, National Institutes of Health Trans-NIH Zebrafish Coordinating Committee The National Institutes of Health and the growth of the zebrafish as an experimental model organism. Zebrafish. 2004;1:105–10. doi: 10.1089/zeb.2004.1.105. [DOI] [PubMed] [Google Scholar]
- Hubbard TJP, Aken BL, Ayling S, Ballester B, Beal K, Bragin E, Brent S, Chen Y, Clapham P, Clarke L, Coates G, Fairley S, Fitzgerald S, Fernandez-Banet J, Gordon L, Gräf S, Haider S, Hammond M, Holland R, Howe K, Jenkinson A, Johnson N, Kähäri A, Keefe D, Keenan S, Kinsella R, Kokocinski F, Kulesha E, Lawson D, Longden I, Megy K, Meidl P, Overduin B, Parker A, Pritchard B, Rios D, Schuster M, Slater G, Smedley D, Spooner W, Spudich G, Trevanion S, Vilella A, Vogel J, White S, Wilder S, Zadissa A, Birney E, Cunningham F, Curwen V, Durbin R, Fernandez-Suarez XM, Herrero J, Kasprzyk A, Proctor G, Smith J, Searle S, Flicek P. Ensembl 2009. Nucleic Acids Res. 2009;37:D690–D697. doi: 10.1093/nar/gkn828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jekosch K. The zebrafish genome project: sequence analysis and annotation. Methods Cell Biol. 2004;77:225–39. doi: 10.1016/s0091-679x(04)77012-0. [DOI] [PubMed] [Google Scholar]
- Kent WJ. BLAT–the BLAST-like alignment tool. Genome Res. 2002;12:656–64. doi: 10.1101/gr.229202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimmel CB, Ballard WW, Kimmel SR, Ullmann B, Schilling TF. Stages of embryonic development of the zebrafish. Dev Dyn. 1995;203:253–310. doi: 10.1002/aja.1002030302. [DOI] [PubMed] [Google Scholar]
- Plaster N, Sonntag C, Schilling TF, Hammerschmidt M. REREa/Atrophin-2 interacts with histone deacetylase and Fgf8 signaling to regulate multiple processes of zebrafish development. Dev Dyn. 2007;236:1891–1904. doi: 10.1002/dvdy.21196. [DOI] [PubMed] [Google Scholar]
- Ramel MC, Buckles GR, Baker KD, Lekven AC. WNT8 and BMP2B co-regulate non-axial mesoderm patterning during zebrafish gastrulation. Dev Biol. 2005;287(2):237–248. doi: 10.1016/j.ydbio.2005.08.012. [DOI] [PubMed] [Google Scholar]
- Rhee SY, Wood V, Dolinski K, Draghici S. Use and misuse of gene ontology annotations. Nat Rev Genet. 2008;9:509–515. doi: 10.1038/nrg2363. [DOI] [PubMed] [Google Scholar]
- Sayers EW, Barrett T, Benson DA, Bolton E, Bryant SH, Canese K, Chetvernin V, Church DM, Dicuccio M, Federhen S, Feolo M, Geer LY, Helmberg W, Kapustin Y, Landsman D, Lipman DJ, Lu Z, Madden TL, Madej T, Maglott DR, Marchler-Bauer A, Miller V, Mizrachi I, Ostell J, Panchenko A, Pruitt KD, Schuler GD, Sequeira E, Sherry ST, Shumway M, Sirotkin K, Slotta D, Souvorov A, Starchenko G, Tatusova TA, Wagner L, Wang Y, John WW, Yaschenko E, Ye J. Database resources of the National Center for Biotechnology Information. Nucleic Acids Res. 2010;38:D5–16. doi: 10.1093/nar/gkp967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith B, Ashburner M, Rosse C, Bard J, Bug W, Ceusters W, Goldberg LJ, Eilbeck K, Ireland A, Mungall CJ, OBI Consortium, Leontis N, Rocca-Serra P, Ruttenberg A, Sansone SAA, Scheuermann RH, Shah N, Whetzel PL, Lewis S. The OBO Foundry: coordinated evolution of ontologies to support biomedical data integration. Nat Biotechnol. 2007;25:1251–1255. doi: 10.1038/nbt1346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sprague J, Bayraktaroglu L, Bradford Y, Conlin T, Dunn N, Fashena D, Frazer K, Haendel M, Howe DG, Knight J, Mani P, Moxon SA, Pich C, Ramachandran S, Schaper K, Segerdell E, Shao X, Singer A, Song P, Sprunger B, Van Slyke CE, Westerfield M. The Zebrafish Information Network: the zebrafish model organism database provides expanded support for genotypes and phenotypes. Nucleic Acids Res. 2008;36:D768–772. doi: 10.1093/nar/gkm956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stein LD, Mungall C, Shu S, Caudy M, Mangone M, Day A, Nickerson E, Stajich JE, Harris TW, Arva A, Lewis S. The generic genome browser: A building block for a model organism system database. Genome Res. 2002;12:1599–1610. doi: 10.1101/gr.403602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- UniProt Consortium. The Universal Protein Resource (UniProt) in 2010. Nucleic Acids Res. 2010;38:D142–8. doi: 10.1093/nar/gkp846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Washington NL, Haendel MA, Mungall CJ, Ashburner M, Westerfield M, Lewis SE. Linking human diseases to animal models using ontology-based phenotype annotation. PLoS Biol. 2009;7:e1000247. doi: 10.1371/journal.pbio.1000247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilming LG, Gilbert JG, Howe K, Trevanion S, Hubbard T, Harrow JL. The vertebrate genome annotation (Vega) database. Nucleic Acids Res. 2008;36:D753–760. doi: 10.1093/nar/gkm987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westerfield M. The Zebrafish Book: A guide for the laboratory use of zebrafish (Danio rerio) 5th. University of Oregon Press; Eugene: 2007. [Google Scholar]