Abstract
The discovery of neuropeptides as signaling molecules with paracrine or hormonal regulatory functions has led to trailblazing advances in physiology and fostered the characterization of numerous neuropeptide-binding G-protein coupled receptors (GPCRs) as potential drug targets. The impact on human health has been tremendous: approximately 30% of commercial drugs act via the GPCR pathway. However, about 25% of the GPCRs encoded by the mammalian genome still lack their pharmacological identity. Searching for the orphan GPCR endogenous ligands that likely are neuropeptides has proved to be a formidable task. Here we describe the mass spectrometry-based technologies and experimental strategies that have been successful in achieving high throughput characterization of endogenous peptides in nervous and endocrine systems.
Keywords: bioinformatics, endogenous peptides, sequencing, mass spectrometry, nervous system, quantitation
What is peptidomics?
Genome and transcriptome sequencing is upon us, so why we are still looking for ways to identify bioactive peptides in living systems? Bold genomic research efforts have provided extraordinary insights into the inventory of GPCR-receptor genes, the most coveted targets in drug development [1]. Gene association and knockout studies have illuminated the roles of various peptide genes in pathological conditions and diseases in animal models [2, 3]. Yet genetic investigations do not determine the actual peptide gene products, neuropeptides, and hormones that mediate vital body functions and complex behaviors. The immense challenge is due to the complexity of molecular readout; a single gene can produce many products as a result of single nucleotide polymorphisms, alternative gene splicing, post-translational processing of precursor proteins, and the addition of chemical post-translational modifications (PTMs) of cleaved peptides that oftentimes cannot be inferred from genomic data. Even annotating peptide coding genes and in silico neuropeptidome prediction requires specialized expertise and bioinformatics tools [4–6].
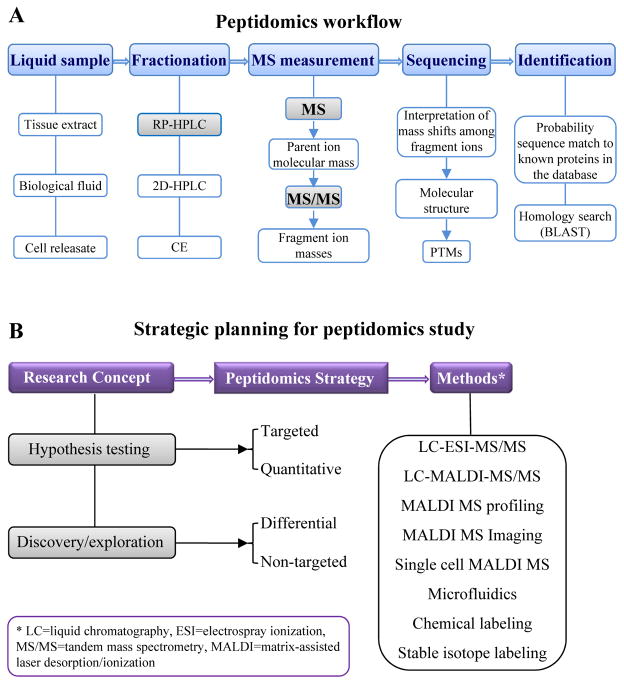
At the dawn of the new millennium, the term ‘peptidomics’ (see the workflow shown in Figure 1A) was formally adopted to describe a method for high throughput, direct measurement and structural characterization of the endogenous peptides present in a given biological sample (see detailed historical review by Schrader and co-authors [7]). In the intervening 15 years since the ground-breaking publications in the field, peptidomics has blossomed into a multitude of distinct approaches (Figure 1B), designed to accommodate a range of sample-related issues (chemical and anatomical complexity, difficulty of sampling, size and/or volume) and a lack of prior knowledge on the peptides expressed in the sample [8–10]. The quantitative capability of peptidomics has become more refined and reliable [11–13]. The use of bioanalytical methods, powered by mass spectrometry (MS) aided by liquid chromatography (LC) and bioinformatics, has steadily increased in the medical and life sciences. At the same time, technological refinement continues to push the boundaries of the limits of detection, resolution, mass accuracy, throughput, and efficiency of data processing.
Figure 1.
A simplified flow chart showing (A) a general peptidomics workflow and (B) a strategy that helps to explain the wide variety of mass spectrometry platforms and for which experiments to use them.
Here we review the current state of MS-based peptidomic technologies and provide guidelines on their application, while also highlighting examples of how to make optimum methodological choices defined by the specific study goals and available resources. Our analysis of the literature covers peer-reviewed publications from the past two years, with a few important exceptions, and focuses exclusively on the analysis of endogenous bioactive peptides in nervous and endocrine systems. Other applications of peptidomic technologies are thoroughly reviewed elsewhere [11, 14–18].
Choosing your peptidomics modus operandi: a guide on methodological approaches
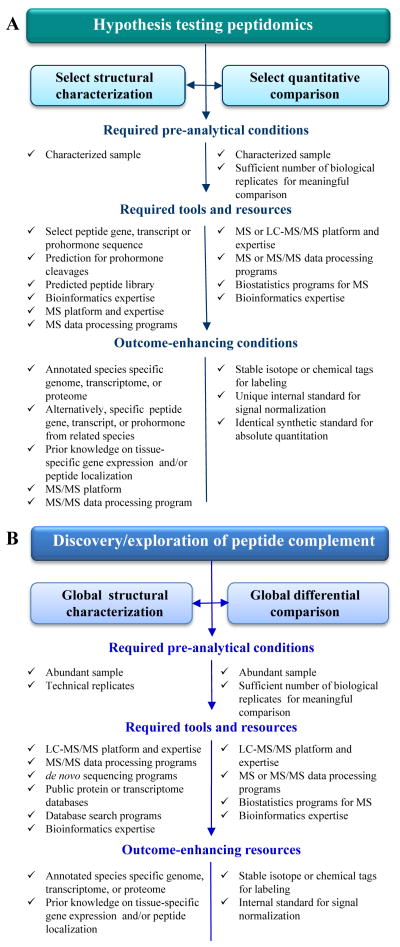
There are two main approaches to bioactive peptide discovery and functional characterization. The first, more traditional experimental pipeline targets a specific (or a few) compound(s) of interest within a biochemical pathway or neural circuit that have already been investigated using molecular probes or expression techniques. The success of this tactic greatly depends on in silico data mining and prior information (Figure 2A). The second, contrasting approach casts a broader net and aims to structurally characterize “all” soluble peptides present in detectable amounts in tissue or organs. One issue is that no current technology can actually measure “all” of the peptides present; yet even with this caveat, this untargeted strategy may lead to the discovery of unexpected molecules and is especially useful as an initial hypothesis-generating study (Figure 2B). Importantly, having a working knowledge of the available methods and selecting the appropriate approaches determines the success of the measurement and furthers investigative outcomes. This is a great way to chemically test a new, unexplored sample type that is abundant and accessible for peptide extraction procedures. In the following sections, we highlight applications and specific methods for both peptidomics strategies.
Figure 2.
Synergy of tools and resources in peptidomics research showing both (A) non-targeted approaches and (B) targeted approaches.
Untargeted high throughput peptide exploration
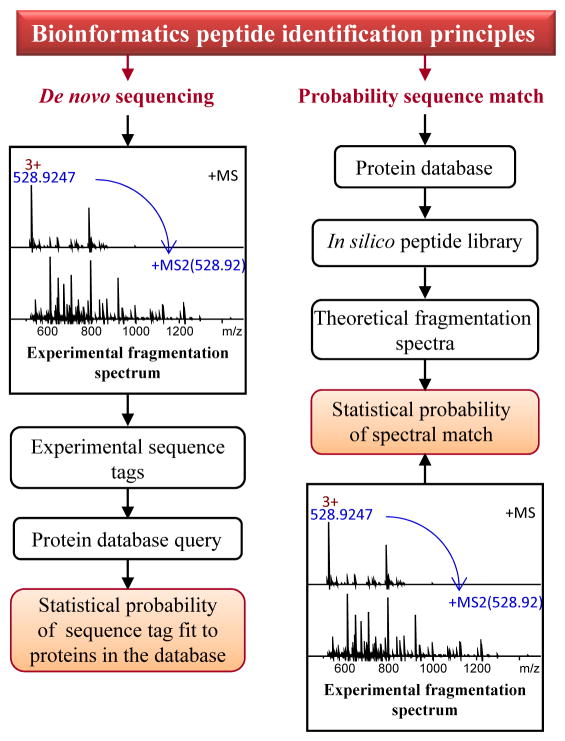
Peptidomics is powered by a hyphenated technique (Figure 1A), liquid chromatography (LC)-tandem mass spectrometry (MS/MS), which can handle chemically complex mixtures and the wide dynamic range of concentrations typically found in biological samples. Chromatographic methods most often include reversed-phase high-performance liquid chromatography (RP-HPLC), or orthogonal, two-dimensional LC. Downstream sequencing MS platforms measure and fragment a set number of ions, usually from the most abundant peptides, eluting from the chromatographic column in a small time frame, which are then temporarily excluded from analysis to allow detection of less abundant peptides within the same time frame. The method is known as data-dependent acquisition (DDA); its effectiveness is influenced by the LC separation, and the sensitivity, spectral rate, and resolution of the mass spectrometer. The resulting data are a combination of intact peptide profiles in the sample and their respective fragmentation spectra (e.g., MS/MS), both needed for follow-up bioinformatics-guided peptide identification (Figure 3). Another separation technique, capillary electrophoresis (CE), is less frequently used as a front-end approach in peptidomics applications because of fewer available commercial systems; CE-MS does allow the down-scaling of the measurement to smaller-volume or low-abundant samples [19, 20]. When the required pre-analytical conditions are met and essential resources are available (Figure 2B), robust characterization of the peptide complement in most peptidergic tissues, peptide extracts, and biological fluids generates a wealth of information for further investigation. Numerous model and socio-economically important organisms have benefited from high-throughput queries of their peptidomes [4, 5, 21–29], providing molecular detail to link peptides to environmental factors, nutrition, physiological states and behaviors. Discovery peptidomics usually involves larger samples, often comprised of many individual samples, which ensures detectable levels of low-abundant peptides and a broader prohormone coverage of detected peptides. Pooling hundreds of individual samples, such as insect brains [4, 28], neural ganglia [23], or defined mammalian brain regions [30–32], is common in these types of untargeted peptidomics studies.
Figure 3.
Bioinformatics approaches for MS-based peptide identification. Two categories of approaches are distinct: the database search approach depends on generating theoretical spectra in silico from protein sequences in a database and querying experimental spectra against those to find the closest matches; the de novo tag approach infers peptide sequence directly from experimental MS/MS data by calculating mass shifts between series of peptide fragment ions, and then aligns the tag to protein in the database. Unassigned de novo tags can be used to interrogate EST depositories or databases of homologous species.
Neuroendocrine tissue and select types of neuronal populations often contain high local concentrations of neuropeptides and thus are suitable for direct analysis either by matrix-assisted laser desorption/ionization (MALDI) MS profiling [33] or MALDI MS imaging (MSI) with minimal sample manipulation [34–38]. Mass spectrometers with standard or interchangeable MALDI ion sources usually have MS/MS capabilities, which allow automatic or manual sequencing of peptides in solid tissue samples [39, 40]. MALDI MSI has an added benefit of being capable of mapping peptides to specific loci within the tissue section [34, 38]. Combined with in situ or immunohistological analyses of selected neuropeptides, direct MS measurements reveal neuropeptide distribution patterns in organs or tissues of interest [27]. Although direct analysis of peptides in tissue is convenient and effective, it has been applied to only a limited range of animal models. The approach has been used more frequently to investigate invertebrates due to the relative simplicity of their nervous and endocrine system organization.
Targeted peptide characterization
This approach relies on the same high sensitivity, high throughput MS-based technology described above, but focuses on specific peptides [41, 42] or peptide families [43–45] that are usually implicated in biological pathways of interest to the researchers (Figure 2A). Identification of PTM sites on known peptides is another example of a targeted peptidomics approach [46].
An unusual PTM is the enzymatic single amino acid d-isomerization in a peptide, as has been observed in frog skin antimicrobial peptides, spider, mollusk and mammalian venom toxins, mollusk neuroexcitatory peptides, and crustacean neurohormones (comprehensively reviewed here [47]). Compared with the all-l-amino acid epimer, a d-amino acid in the peptide can confer distinct and dramatically enhanced bioactivity, as is the case with the newly characterized GdFFD peptide from the marine mollusk Aplysia californica [48]. Targeting d-amino acid-containing peptides for investigation has been notoriously difficult via molecular techniques, or even MS, due to the lack of a sequence change or mass defect associated with this PTM. However, new multidisciplinary peptidomics methods promise to facilitate the discovery of other putative d-amino acid-containing peptides in many animal models by measuring the distinct molecular fragmentation patterns among peptide diastereomers with MS/MS [48–50].
Differential and quantitative peptidomics
It has become evident that prohormone levels do not always correlate with their coding mRNA levels. Although microarray techniques can be useful analytical tools, they do not provide definitive information on neuropeptide dynamics in perturbed biological systems. The demand for peptide-relevant, in-depth quantitative assays has stimulated the development of quantitative MS approaches (reviewed by Romanova and co-authors [11]). Differential peptidomics compares qualitatively or quantitatively detectable peptides (Figure 2) between experimental sample groups, either to test or generate hypotheses on the functional connections of detected peptides, or to correlate peptide levels to the biological paradigms under investigation [10]; these investigations can be performed on a global scale [30–32, 51–53] or selectively [54]. The intensity of the observed intact peptide ions, or frequency of fragmentation events during chromatographic separation, serves as a basis for peptide level comparisons. In either case, the approach requires careful experimental design and thoughtful interpretation of results.
Although it is tempting to associate differential peptide profiles or levels in the samples to peptide expression in the phenotype of interest, they may not reflect true in vivo expression status. First, only peptides that remain stable under the tissue collection protocols used, and are soluble in the extraction media, retained under the selected separation condition, and detectable with the chosen MS approach, can be assessed by peptidomics. Second, perturbation of the biological system leading to in vivo peptide changes may also affect in vitro detection of peptides, irrespective of the perturbing event, simply by altering the balance of molecular homeostasis within the sample. This can lead to changes in the solubility, charge balance, and ultimately, the ionization efficiency of different ion species during the MS measurement. In other words, at the measurement stage, we deal with a subset of biomolecules that are only as representative of their physiological state as our sampling protocol allows. Recent developments in sampling approaches, such as focused microwave irradiation and heat stabilization [55], minimize post-mortem protein degradation products and thus, may provide information that more closely reflects the in vivo peptidome. Additionally, the measurement strategy has a profound effect on experimental outcome. The versatile method of MS-based structural characterization, DDA, may restrict global quantitative analysis because it relies on real-time decisions about which precursor ions from a survey scan should be directed for fragmentation, and so its performance declines as sample complexity increases. Experimenting with alternative methods, the Li group [53] conducted a proof-of-principle study using data-independent acquisition (DIA) to quantify feeding-related peptides from crab. A brute force method, DIA [56], which acquires fragmentation spectra independently of precursor ion information, has gained acceptance for both identification of tryptic peptides and targeted quantification of proteins. However, quantitation of endogenous neuropeptides by DIA remains challenging. Finally, processing of MS data for quantitation is critical and should take multiple variables into account, ranging from simple instrument performance drift over time to the presence of multiple charge state ions for the same peptide, peptide co-elution, partial labeling with chemical or isotopic tags (if used), high background noise, small numbers, and natural variability between biological replicates. In our opinion, integrated studies where differential peptidomics data are validated by independent approaches deliver the most biologically relevant discoveries [57–59].
Recently, differential comparisons have been used to probe unknown mechanisms in the physiological response to environmental [51], developmental [60] or pharmacological perturbation [31, 61, 62], disease states [36, 63–65], phenotypes [39, 66], or even various sample preparation methods crucial for successful neuropeptide detection [67, 68]. It is safe to say that relative or differential MS quantitation has already become routine due to the availability of commercial reagents, sensitive analytical instrumentation, and software. With this said, absolute quantitation of endogenous peptides continues to challenge the bioanalytical community. Just as with radioimmunoassay and ELISA, absolute MS quantitation is most effectively performed for selected and known neuropeptides for which synthetic or isotopically labeled standards are available [54, 69–71].
Probing cellular diversity by single-cell peptidomics
Locating a specific cell from within a relatively uniform cell population to determine its chemical content presents a demanding bioanalytical task. MS measurement at the single-cell level can identify neuropeptides co-localized within the cell soma, even if they are encoded by different co-expressed genes. Single-cell peptidomics is most effective when working with well-characterized neuronal circuits that underlie defined behaviors or physiological functions in suitable neurobiological models having accessible neurons or other cells of interest, which rarely includes mammals. Historically, the most common application of single-cell peptidomics has been geared towards determination of the actual peptide products of a cloned gene or transcript in functionally characterized cells. Detecting multiple predicted peptides by their molecular mass often is sufficient to assign peptides to a certain prohormone, in the same way as the peptide mass fingerprint method is widely used in proteomics for protein identification. This approach allows matching of the individual cell peptide profiles to electrophysiological activity [72], localization of molecular probes [73], or both [74, 75]. A unique two-step strategy hyphenates MS to targeted chemical analysis of immunocytochemical-selected peptidergic neurons containing selected biomarkers of interest [76]. While robust, single-cell peptidomics is difficult to mainstream for comprehensive chemical analysis, primarily due to technical challenges in adapting chemically complex nanoliter-volume samples to compatible nanoseparation platforms and hyphenating those with MS [77]. Therefore, direct targeted MALDI MS continues to be the most successful approach for single-cell measurement. We have reviewed numerous inspiring examples during the past decade elsewhere [77].
Microfluidic platforms for peptidomics of cellular releasates
Given the tremendous chemical diversity of neurons and neuroendocrine cells, it is logical to focus on peptides released in response to physiological stimulation to gain insights into the mechanisms of intercellular communication. In vitro neuronal networks present the opportunity to collect and characterize intercellular signaling peptides released by the neurons upon physiologically relevant stimulation, but small dimensions as well as a high degree of dilution of released compounds in media has long limited the investigative process. The challenges can be partially circumvented by using microfluidic systems suited to the study of released neuropeptides [78, 79]. Microfluidic designs allow the user to selectively apply chemical stimulations to neurons maintained in these devices. The resulting neuropeptide releasates can be collected off-line and detected with MALDI-time-of-flight (TOF) MS [57, 78, 80]. An additional benefit of the off-line coupling of microfluidic devices with MS for the characterization of small-volume extracellular releasates is the capability for label-free, absolute quantitation of peptides [81].
Synergy of tools and resources in peptidomics research
While MS technology is often marketed as a complete solution for comprehensive protein/peptide characterization because of its unprecedented structural characterization capabilities, speed, sensitivity, and throughput, it often works best as a part of multifaceted or integrative characterization effort. A multifaceted approach can be defined as a combination of complementary separation and MS techniques having specific advantages for peptides of a certain mass range [82], or other characteristics or modifications. The synergetic effect may be achieved by combining, in one study, different ionization techniques for generating different subsets of peptide ions from the same sample [37], multiple complementary fragmentation methods (ETD, HCD, and CID) performed on liquid chromatographic time scales, which ultimately enhance peptide structure determination or PTM localization [82–84], or different mass analyzers [82, 85]. Liquid separation methods on the front end of a peptidomics pipeline offer additional leverage for improved peptide coverage. Respectively, various separation optimization strategies, such as multidimensional LC [29, 62, 86, 87] and alternative separation methods, have been actively explored in the peptidomics area [40, 88–90].
Assignment/identification of neuropeptides in peptidomics showcases this interdisciplinary synergy. Existing identification methods (Figure 3) are facilitated by the ever-growing transcriptomic resources and protein databases obtained by translation from the coding sequences in public nucleic acid databases. However, a robust and widely popular identification approach via database search that relies on in silico data is effective for assigning peptides from known prohormones that are already in the utilized database, and when the deposited prohormone sequences are free from mistakes. Moreover, only peptides resulting from conventional enzymatic processing for which theoretical MS/MS spectra can be predicted may be found via database search. In contrast, de novo sequence tagging is independent of databases and thus offers more discovery power, even with partially correct tags. It is currently the only means for detecting mistakes in deposited protein sequences, and revealing single amino acid substitutions, PTMs, transcript variants, and homologous sequences. Overall, there are numerous interactions between peptidomics, genomics, and transcriptomics that ultimately drive neuropeptide discovery [5, 21, 25, 26, 28, 29].
Of utmost importance to accurate and effective peptide detection and identification in a peptidomics experiment are the bioinformatics tools used for interpretation of the MS and MS/MS spectra, employing a search engine that queries data against a protein database to find statistically validated peptide-spectrum matches (Figure 3). Due to the multitude of commercially available and open source programs, often built upon different mathematical algorithms for interpretation of the MS/MS data [91, 92], multilayered or combinatorial analysis of MS/MS data sets have proved to be beneficial [93, 94]. Likewise, quantitative assessment of peptide levels may not be executed without advanced bioinformatics algorithms for signal normalization, noise reduction, correct feature extraction (m/z, charge state, peak intensity/area, retention time), and data conversion from vendor proprietary formats to open formats that are compatible with stand-alone statistical tools.
Quo vadimus: Future perspectives and concluding remarks
Owing to the rapid and dramatic advancements within the past two decades, highly sophisticated mass spectrometric instrumentation and auxiliary resources have become useable and affordable to many laboratories. Because of this availability, neuropeptide discovery has skyrocketed in terms of the diversity of the biological systems investigated and the number of identified peptides reported by individual studies. However, enthusiasm about the long lists of peptides sometimes prevails over scientific wisdom and hinders thoughtful interpretation of chemically exciting results relevant to their biological significance. Peptidomics methods do not differentiate between naturally occurring peptides, post-mortem degradation products, and sampling artifacts: any peptide has a chance to be detected and identified. Reporting a few hundred chemically unique peptides from a few dozen prohormones is often a simple mathematical exercise that overlooks the fact that many of the detected peptides are sequentially truncated forms of mature peptides positioned between conventional cleavage sites on the prohormone. This is especially common to mammalian neuropeptidomes because of their astonishing complexity. Should these shorter forms be accounted for as independent peptides, or be considered as redundancies? While in a few cases, such as with angiotensin, the truncated forms have distinct and important biological roles, we feel that often, many of the truncated peptides reported in peptidomics studies are low-level peptides arising from sampling artifacts. We hope that in the coming years the awareness of such factors becomes more widespread in the bioanalytical community.
Even with the recent technical advances, the functional characterization of novel peptidomes is still a slow and often multi-laboratory collaborative process, with newly discovered putative neuropeptides needing to be individually evaluated. There is a pressing need for reliable approaches that can focus the peptide functional annotation effort on a subset of the most probable candidate compounds. Existing differential and quantitative MS methods help with the selection process, but to become broadly accessible, they have to be streamlined. Nonetheless, we expect in the coming years that breakthroughs in MS technologies will allow absolute quantitation and easier implementation of complex peptidomic studies.
Thinking about peptidomics from the functional perspective emphasizes an idea that only secreted peptides are cell-to-cell signaling molecules. Although cellular releasates have been successfully probed by microdialysis and microfluidics in combination with MS, these approaches are still being optimized to provide more universal and dependable technical solutions to sampling and sample pre-concentration. Additional methods involve brain slice chambers to sample from specific release sites [95]. We envision wider implementation of lab-on-chip platforms, which may dramatically reduce the amount of releasate sample required, minimize sample manipulation, and shorten analysis times.
In conclusion, MS-based methods provide an effective toolset for the discovery and characterization of neuropeptides in biological samples. Due to its high sensitivity, multiplexed detection, small sample size, and compatibility with a wide array of sampling approaches and complimentary measurement techniques, MS allows broad scale quantitative and qualitative investigations, from the organismal to single-cell level. Ultimately, it is the flexibility of MS that enables the elucidation of inherent chemical and functional complexities within the nervous and endocrine systems.
Highlights.
Alternative splicing, posttranslational processing challenge neuropeptide prediction
Mass spectrometry sequences and quantifies neuropeptides directly in tissues and cells
Synergy of mass spectrometry, genomics and bioinformatics drives neuropeptide discovery
Acknowledgments
The project described was supported by Award Nos. P30 DA018310 from the National Institute on Drug Abuse, RO1 NS031609 from the National Institute of Neurological Disorders and Stroke, and R21 MH100704 from the National Institute of Mental Health. The content is solely the responsibility of the authors and does not necessarily represent the official views of the funding agencies.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Zalewska M, et al. G protein-coupled receptors: abnormalities in signal transmission, disease states and pharmacotherapy. Acta Pol Pharm. 2014;71:229–243. [PubMed] [Google Scholar]
- 2.Bali A, et al. Neuropeptides as therapeutic targets to combat stress-associated behavioral and neuroendocrinological effects. CNS Neurol Disord Drug Targets. 2014;13:347–368. doi: 10.2174/1871527313666140314163920. [DOI] [PubMed] [Google Scholar]
- 3.Giguere PM, et al. Tuning up the right signal: chemical and genetic approaches to study GPCR functions. Curr Opin Cell Biol. 2014;27:51–55. doi: 10.1016/j.ceb.2013.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Audsley N, et al. Genomic and peptidomic analyses of the neuropeptides from the emerging pest, Drosophila suzukii. Peptides. 2015;68:33–42. doi: 10.1016/j.peptides.2014.08.006. [DOI] [PubMed] [Google Scholar]
- 5.Stewart MJ, et al. Neuropeptides encoded by the genomes of the Akoya pearl oyster Pinctata fucata and Pacific oyster Crassostrea gigas: a bioinformatic and peptidomic survey. BMC Genomics. 2014;15:840. doi: 10.1186/1471-2164-15-840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Christie AE. In silico characterization of the neuropeptidome of the Western black widow spider Latrodectus hesperus. Gen Comp Endocrinol. 2015;210:63–80. doi: 10.1016/j.ygcen.2014.10.005. [DOI] [PubMed] [Google Scholar]
- 7.Schrader M, et al. Historical perspective of peptidomics. EuPA Open Proteomics. 2014;3:171–182. [Google Scholar]
- 8.Li L, Sweedler JV. Peptides in the brain: mass spectrometry-based measurement approaches and challenges. Annu Rev Anal Chem. 2008;1:451–483. doi: 10.1146/annurev.anchem.1.031207.113053. [DOI] [PubMed] [Google Scholar]
- 9.Finoulst I, et al. Sample preparation techniques for the untargeted LC-MS-based discovery of peptides in complex biological matrices. J Biomed Biotechnol. 2011;2011:245291. doi: 10.1155/2011/245291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.De Haes W, et al. Functional neuropeptidomics in invertebrates. Biochim Biophys Acta. 2015;1854:812–826. doi: 10.1016/j.bbapap.2014.12.011. [DOI] [PubMed] [Google Scholar]
- 11.Romanova EV, et al. Quantitation of endogenous peptides using mass spectrometry based methods. Curr Opin Chem Biol. 2013;17:801–808. doi: 10.1016/j.cbpa.2013.05.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Craft GE, et al. Recent advances in quantitative neuroproteomics. Methods. 2013;61:186–218. doi: 10.1016/j.ymeth.2013.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.OuYang C, et al. Mass spectrometric analysis of spatio-temporal dynamics of crustacean neuropeptides. Biochim Biophys Acta. 2015;1854:798–811. doi: 10.1016/j.bbapap.2014.10.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lai ZW, et al. The emerging role of the peptidome in biomarker discovery and degradome profiling. Biol Chem. 2014;396:185–192. doi: 10.1515/hsz-2014-0207. [DOI] [PubMed] [Google Scholar]
- 15.Bauca JM, et al. Peptidomics of urine and other biofluids for cancer diagnostics. Clin Chem. 2014;60:1052–1061. doi: 10.1373/clinchem.2013.211714. [DOI] [PubMed] [Google Scholar]
- 16.Lone AM, et al. Peptidomics methods for the identification of peptidase-substrate interactions. Curr Opin Chem Biol. 2013;17:83–89. doi: 10.1016/j.cbpa.2012.10.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dallas DC, et al. Current peptidomics: Applications, purification, identification, quantification and functional analysis. Proteomics. 2015;15:1026–1038. doi: 10.1002/pmic.201400310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sapio MR, Fricker LD. Carboxypeptidases in disease: insights from peptidomic studies. Proteomics Clin Appl. 2014;8:327–337. doi: 10.1002/prca.201300090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yin P, et al. Peptidomic analyses of mouse astrocytic cell lines and rat primary cultured astrocytes. J Proteome Res. 2012;11:3965–3973. doi: 10.1021/pr201066t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhong X, et al. Recent advances in coupling capillary electrophoresis-based separation techniques to ESI and MALDI-MS. Electrophoresis. 2014;35:1214–1225. doi: 10.1002/elps.201300451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Toullec JY, et al. Transcriptome and peptidome characterisation of the main neuropeptides and peptidic hormones of a euphausiid: the Ice Krill, Euphausia crystallorophias. PLoS One. 2013;8:e71609. doi: 10.1371/journal.pone.0071609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rong M, et al. Peptidomics combined with cDNA library unravel the diversity of centipede venom. J Proteomics. 2015;114:28–37. doi: 10.1016/j.jprot.2014.10.014. [DOI] [PubMed] [Google Scholar]
- 23.Hui L, et al. Mass spectrometric characterization of the neuropeptidome of the ghost crab Ocypode ceratophthalma (Brachyura, Ocypodidae) Gen Comp Endocrinol. 2013;184:22–34. doi: 10.1016/j.ygcen.2012.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhang X, et al. High identification rates of endogenous neuropeptides from mouse brain. J Proteome Res. 2012;11:2819–2827. doi: 10.1021/pr3001699. [DOI] [PubMed] [Google Scholar]
- 25.Collins JJ, 3rd, et al. Genome-wide analyses reveal a role for peptide hormones in planarian germline development. PLoS Biol. 2010;8:e1000509. doi: 10.1371/journal.pbio.1000509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Xie F, et al. The zebra finch neuropeptidome: prediction, detection and expression. BMC Biol. 2010;8:28. doi: 10.1186/1741-7007-8-28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Binzer M, et al. Neuropeptidome of Tribolium castaneum antennal lobes and mushroom bodies. J Comp Neurol. 2014;522:337–357. doi: 10.1002/cne.23399. [DOI] [PubMed] [Google Scholar]
- 28.Rahman MM, et al. Neuropeptidomics of the Australian sheep blowfly Lucilia cuprina (Wiedemann) and related Diptera. Peptides. 2013;41:31–37. doi: 10.1016/j.peptides.2012.12.021. [DOI] [PubMed] [Google Scholar]
- 29.Ranc V, et al. Broad characterization of endogenous peptides in the tree shrew visual system. J Proteomics. 2012;75:2526–2535. doi: 10.1016/j.jprot.2012.01.028. [DOI] [PubMed] [Google Scholar]
- 30.Lee JE, et al. Quantitative peptidomics for discovery of circadian-related peptides from the rat suprachiasmatic nucleus. J Proteome Res. 2013;12:585–593. doi: 10.1021/pr300605p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Romanova EV, et al. Comparative peptidomics analysis of neural adaptations in rats repeatedly exposed to amphetamine. J Neurochem. 2012;123:276–287. doi: 10.1111/j.1471-4159.2012.07912.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Southey BR, et al. Comparing label-free quantitative peptidomics approaches to characterize diurnal variation of peptides in the rat suprachiasmatic nucleus. Anal Chem. 2014;86:443–452. doi: 10.1021/ac4023378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Siju KP, et al. Neuropeptides in the antennal lobe of the yellow fever mosquito, Aedes aegypti. J Comp Neurol. 2014;522:592–608. doi: 10.1002/cne.23434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chen R, et al. In situ identification and mapping of neuropeptides from the stomatogastric nervous system of Cancer borealis. Rapid Commun Mass Spectrom. 2014;28:2437–2444. doi: 10.1002/rcm.7037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ye H, et al. Mapping of neuropeptides in the crustacean stomatogastric nervous system by imaging mass spectrometry. J Am Soc Mass Spectrom. 2013;24:134–147. doi: 10.1007/s13361-012-0502-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hanrieder J, et al. MALDI imaging mass spectrometry of neuropeptides in Parkinson’s disease. J Vis Exp. 2012:3445. doi: 10.3791/3445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sturm RM, et al. Comparison of NIMS and MALDI platforms for neuropeptide and lipid mass spectrometric imaging in brain tissue. Anal Methods. 2013;5:1623–1628. doi: 10.1039/C3AY26067D. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chatterji B, Pich A. MALDI imaging mass spectrometry and analysis of endogenous peptides. Expert Rev Proteomics. 2013;10:381–388. doi: 10.1586/14789450.2013.814939. [DOI] [PubMed] [Google Scholar]
- 39.Salisbury JP, et al. A rapid MALDI-TOF mass spectrometry workflow for Drosophila melanogaster differential neuropeptidomics. Mol Brain. 2013;6:60. doi: 10.1186/1756-6606-6-60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Holtta M, et al. Peptidome analysis of cerebrospinal fluid by LC-MALDI MS. PLoS One. 2012;7:e42555. doi: 10.1371/journal.pone.0042555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Marciniak P, et al. Identification and localisation of selected myotropic neuropeptides in the ventral nerve cord of tenebrionid beetles. Comp Biochem Physiol A Mol Integr Physiol. 2013;166:44–51. doi: 10.1016/j.cbpa.2013.05.008. [DOI] [PubMed] [Google Scholar]
- 42.Pratavieira M, et al. MALDI imaging analysis of neuropeptides in the Africanized honeybee (Apis mellifera) brain: effect of ontogeny. J Proteome Res. 2014;13:3054–3064. doi: 10.1021/pr500224b. [DOI] [PubMed] [Google Scholar]
- 43.Gellerer A, et al. Identification and distribution of SIFamide in the nervous system of the desert locust Schistocerca gregaria. J Comp Neurol. 2015;523:108–125. doi: 10.1002/cne.23671. [DOI] [PubMed] [Google Scholar]
- 44.Jia C, et al. High-definition de novo sequencing of crustacean hyperglycemic hormone (CHH)-family neuropeptides. Mol Cell Proteomics. 2012;11:1951–1964. doi: 10.1074/mcp.M112.020537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sturm S, Predel R. Mass spectrometric identification, sequence evolution, and intraspecific variability of dimeric peptides encoded by cockroach akh genes. Anal Bioanal Chem. 2015:1685–1693. doi: 10.1007/s00216-014-8382-7. [DOI] [PubMed] [Google Scholar]
- 46.Sturm S, Predel R. Serine phosphorylation of CAPA pyrokinin in cockroaches-a taxon-specific posttranslational modification. Peptides. 2014;57:52–58. doi: 10.1016/j.peptides.2014.04.012. [DOI] [PubMed] [Google Scholar]
- 47.Ollivaux C, et al. Biogenesis of D-amino acid containing peptides/proteins: where, when and how? J Pept Sci. 2014;20:595–612. doi: 10.1002/psc.2637. [DOI] [PubMed] [Google Scholar]
- 48.Bai L, et al. Characterization of GdFFD, a D-amino acid-containing neuropeptide that functions as an extrinsic modulator of the Aplysia feeding circuit. J Biol Chem. 2013;288:32837–32851. doi: 10.1074/jbc.M113.486670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Jia C, et al. Site-specific characterization of (D)-amino acid containing peptide epimers by ion mobility spectrometry. Anal Chem. 2014;86:2972–2981. doi: 10.1021/ac4033824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Bai L, et al. Distinguishing endogenous D-amino acid-containing neuropeptides in individual neurons using tandem mass spectrometry. Anal Chem. 2011;83:2794–2800. doi: 10.1021/ac200142m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Chen R, et al. Quantitative neuropeptidomics study of the effects of temperature change in the crab Cancer borealis. J Proteome Res. 2014;13:5767–5776. doi: 10.1021/pr500742q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Fouillen L, et al. Neuropeptide alterations in the tree shrew hypothalamus during volatile anesthesia. J Proteomics. 2013;80:311–319. doi: 10.1016/j.jprot.2012.11.002. [DOI] [PubMed] [Google Scholar]
- 53.Schmerberg CM, et al. Data-independent MS/MS quantification of neuropeptides for determination of putative feeding-related neurohormones in microdialysate. ACS Chem Neurosci. 2015;6:174–180. doi: 10.1021/cn500253u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Gozdowska M, et al. Neuropeptides isotocin and arginine vasotocin in urophysis of three fish species. Fish Physiol Biochem. 2013;39:863–869. doi: 10.1007/s10695-012-9746-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Svensson M, et al. Heat stabilization of the tissue proteome: a new technology for improved proteomics. J Proteome Res. 2009;8:974–981. doi: 10.1021/pr8006446. [DOI] [PubMed] [Google Scholar]
- 56.Egertson JD, et al. Multiplexed MS/MS for improved data-independent acquisition. Nat Methods. 2013;10:744–746. doi: 10.1038/nmeth.2528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Murphy D, et al. The hypothalamic-neurohypophyseal system: from genome to physiology. J Neuroendocrinol. 2012;24:539–553. doi: 10.1111/j.1365-2826.2011.02241.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Holtta M, et al. An integrated workflow for multiplex CSF proteomics and peptidomics-identification of candidate cerebrospinal fluid biomarkers of Alzheimer’s disease. J Proteome Res. 2014;14:654–663. doi: 10.1021/pr501076j. [DOI] [PubMed] [Google Scholar]
- 59.Castro LM, et al. Peptidomic analysis of the neurolysin-knockout mouse brain. J Proteomics. 2014;111:238–248. doi: 10.1016/j.jprot.2014.03.043. [DOI] [PubMed] [Google Scholar]
- 60.Jiang X, et al. Mass spectral charting of neuropeptidomic expression in the stomatogastric ganglion at multiple developmental stages of the lobster Homarus americanus. ACS Chem Neurosci. 2012;3:439–450. doi: 10.1021/cn200107v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Petruzziello F, et al. Chronic nicotine treatment impacts the regulation of opioid and non-opioid peptides in the rat dorsal striatum. Mol Cell Proteomics. 2013;12:1553–1562. doi: 10.1074/mcp.M112.024828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Nilsson A, et al. Neuropeptidomics of mouse hypothalamus after imipramine treatment reveal somatostatin as a potential mediator of antidepressant effects. Neuropharmacology. 2012;62:347–357. doi: 10.1016/j.neuropharm.2011.08.004. [DOI] [PubMed] [Google Scholar]
- 63.Cafe-Mendes CC, et al. Using mass spectrometry-based peptidomics to understand the brain and disorders such as Parkinson’s disease and schizophrenia. Curr Top Med Chem. 2014;14:369–381. doi: 10.2174/1568026613666131204120747. [DOI] [PubMed] [Google Scholar]
- 64.Ishigami N, et al. Cerebrospinal fluid proteomic patterns discriminate Parkinson’s disease and multiple system atrophy. Mov Disord. 2012;27:851–857. doi: 10.1002/mds.24994. [DOI] [PubMed] [Google Scholar]
- 65.Gelman JS, et al. Quantitative peptidomics to measure neuropeptide levels in animal models relevant to psychiatric disorders. Methods Mol Biol. 2012;829:487–503. doi: 10.1007/978-1-61779-458-2_31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Berezniuk I, et al. Quantitative peptidomics of Purkinje cell degeneration mice. PLoS One. 2013;8:e60981. doi: 10.1371/journal.pone.0060981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Sturm RM, et al. Mass spectrometric evaluation of neuropeptidomic profiles upon heat stabilization treatment of neuroendocrine tissues in crustaceans. J Proteome Res. 2013;12:743–752. doi: 10.1021/pr300805f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Gelman JS, et al. Analysis of peptides secreted from cultured mouse brain tissue. Biochim Biophys Acta. 2013;1834:2408–2417. doi: 10.1016/j.bbapap.2013.01.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Zhang Z, et al. Liquid chromatography-matrix-assisted laser desorption/ionization mass spectrometric imaging with sprayed matrix for improved sensitivity, reproducibility and quantitation. Analyst. 2013;138:6600–6606. doi: 10.1039/c3an01225e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Pailleux F, Beaudry F. Evaluation of multiple reaction monitoring cubed for the analysis of tachykinin related peptides in rat spinal cord using a hybrid triple quadrupole-linear ion trap mass spectrometer. J Chromatogr B Analyt Technol Biomed Life Sci. 2014;947–948:164–167. doi: 10.1016/j.jchromb.2013.12.025. [DOI] [PubMed] [Google Scholar]
- 71.Li Y, et al. Quantitative multiple reaction monitoring of peptide abundance introduced via a capillary zone electrophoresis-electrospray interface. Anal Chem. 2012;84:6116–6121. doi: 10.1021/ac300926h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Neupert S, et al. Toward a single-cell-based analysis of neuropeptide expression in Periplaneta americana antennal lobe neurons. J Comp Neurol. 2012;520:694–716. doi: 10.1002/cne.22745. [DOI] [PubMed] [Google Scholar]
- 73.Jarecki JL, et al. Three independent techniques localize expression of transcript afp-11 and its bioactive peptide products to the paired AVK neurons in Ascaris suum: in situ hybridization, immunocytochemistry, and single cell mass spectrometry. ACS Chem Neurosci. 2013;4:418–434. doi: 10.1021/cn3001334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Jing J, et al. Feedforward compensation mediated by the central and peripheral actions of a single neuropeptide discovered using representational difference analysis. J Neurosci. 2010;30:16545–16558. doi: 10.1523/JNEUROSCI.4264-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Romanova EV, et al. Urotensin II in invertebrates: from structure to function in Aplysia californica. PLoS One. 2012;7:e48764. doi: 10.1371/journal.pone.0048764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Neupert S, et al. Targeted single-cell microchemical analysis: MS-based peptidomics of individual paraformaldehyde-fixed and immunolabeled neurons. Chem Biol. 2012;19:1010–1019. doi: 10.1016/j.chembiol.2012.05.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Romanova EV, et al. Small-volume analysis of cell-cell signaling molecules in the brain. Neuropsychopharmacology. 2014;39:50–64. doi: 10.1038/npp.2013.145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Croushore CA, et al. Microfluidic device for the selective chemical stimulation of neurons and characterization of peptide release with mass spectrometry. Anal Chem. 2012;84:9446–9452. doi: 10.1021/ac302283u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Croushore CA, Sweedler JV. Microfluidic systems for studying neurotransmitters and neurotransmission. Lab Chip. 2013;13:1666–1676. doi: 10.1039/c3lc41334a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Fan Y, et al. Stimulation and release from neurons via a dual capillary collection device interfaced to mass spectrometry. Analyst. 2013;138:6337–6346. doi: 10.1039/c3an01010d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Zhong M, et al. Label-free quantitation of peptide release from neurons in a microfluidic device with mass spectrometry imaging. Lab Chip. 2012;12:2037–2045. doi: 10.1039/c2lc21085a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Jia C, et al. A multi-scale strategy for discovery of novel endogenous neuropeptides in the crustacean nervous system. J Proteomics. 2013;91:1–12. doi: 10.1016/j.jprot.2013.06.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Sasaki K, et al. Large-scale identification of endogenous secretory peptides using electron transfer dissociation mass spectrometry. Mol Cell Proteomics. 2013;12:700–709. doi: 10.1074/mcp.M112.017400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Hayakawa E, et al. Improving the identification rate of endogenous peptides using electron transfer dissociation and collision-induced dissociation. J Proteome Res. 2013;12:5410–5421. doi: 10.1021/pr400446z. [DOI] [PubMed] [Google Scholar]
- 85.Hui L, et al. Mass spectrometric elucidation of the neuropeptidome of a crustacean neuroendocrine organ. Peptides. 2012;36:230–239. doi: 10.1016/j.peptides.2012.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Hu L, et al. Analysis of the endogenous human serum peptides by on-line extraction with restricted-access material and HPLC-MS/MS identification. Talanta. 2014;127:191–195. doi: 10.1016/j.talanta.2014.04.011. [DOI] [PubMed] [Google Scholar]
- 87.Tharakan R, et al. Integrated microfluidic chip and online SCX separation allows untargeted nanoscale metabolomic and peptidomic profiling. J Proteome Res. 2015;14:1621–1626. doi: 10.1021/pr5011422. [DOI] [PubMed] [Google Scholar]
- 88.Zhang Z, et al. Pressure-assisted capillary electrophoresis coupling with matrix-assisted laser desorption/ionization-mass spectrometric imaging for quantitative analysis of complex peptide mixtures. Anal Chem. 2012;84:7684–7691. doi: 10.1021/ac300628s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Zhang Z, et al. Neuropeptide analysis with liquid chromatography-capillary electrophoresis-mass spectrometric imaging. J Sep Sci. 2012;35:1779–1784. doi: 10.1002/jssc.201200051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Zhang Z, et al. Semi-automated liquid chromatography-mass spectrometric imaging platform for enhanced detection and improved data analysis of complex peptides. J Chromatogr. 2013;1293:44–50. doi: 10.1016/j.chroma.2013.03.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Akhtar MN, et al. Evaluation of database search programs for accurate detection of neuropeptides in tandem mass spectrometry experiments. J Proteome Res. 2012;11:6044–6055. doi: 10.1021/pr3007123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Akhtar MN, et al. Identification of best indicators of peptide-spectrum match using a permutation resampling approach. J Bioinf Comput Biol. 2014;12:1440001. doi: 10.1142/S0219720014400010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Shteynberg D, et al. Combining results of multiple search engines in proteomics. Mol Cell Proteomics. 2013;12:2383–2393. doi: 10.1074/mcp.R113.027797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Akhtar MN, et al. Accurate assignment of significance to neuropeptide identifications using Monte Carlo k-permuted decoy databases. PLoS One. 2014;9:e111112. doi: 10.1371/journal.pone.0111112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Mitchell JW, et al. Direct cellular peptidomics of hypothalamic neurons. Front Neuroendocrinol. 2011;32:377–386. doi: 10.1016/j.yfrne.2011.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]