Abstract
BACKGROUND: IGFBP-3 is a multifunctional protein that inhibits growth and induces apoptosis of cancer cells. Hypermethylation of the promoter represses expression of the IGFBP-3 gene. We undertook this study to assess the impact of IGFBP-3 methylation on survival of early stage gastric cancer patients. METHODS: Of the 482 tissue samples from gastric cancer patients who underwent curative surgery, IGFBP-3 methylation was tested in 138 patients with stage IB/II gastric cancer. We also analyzed IGFBP-3 methylation in 26 gastric cancer cell lines. IGFBP-3 methylation was evaluated by methylation-specific polymerase chain reaction (MethyLight). Statistical analyses, all two-sided, were performed to investigate the prognostic effects of methylation status of the IGFBP-3 promoter on various clinical parameters. RESULTS: Hypermethylation of IGFBP-3 was observed in 26 (19%) of the 138 stage IB/II gastric cancer patients. Clinicopathological factors such as age, Lauren classification, sex, tumor infiltration, lymph node metastasis, and histologic grade did not show a statistically significant association with the methylation status of the IGFBP-3 promoter. Patients with a hypermethylated IGFBP-3 promoter had similar 8-year disease-free survival compared with those without a hypermethylated IGFBP-3 promoter (73% vs 75%, P = .78). In subgroup analyses, females, but not males, seemed to have poorer prognosis for DFS and OS in the subset of patients with IGFBP-3 methylation as compared with those without IGFBP-3 methylation (8-year DFS: 55.6% vs 71.6%, P = .3694 and 8-year overall survival: 55.6% vs 68.4%, P = .491, respectively) even with no statistical significance. CONCLUSIONS: The status of IGFBP-3 methylation as measured by methylation-specific polymerase chain reaction proposed the modest role for predicting survival in specific subgroups of patients with early-stage gastric cancer who undergo curative surgery. However, this needs further investigation.
Introduction
Gastric cancer is the leading cause of cancer death worldwide, with an incidence of 18.9 of 100,000 per year [1]. The incidence of gastric cancer is estimated to be 934,000 cases, with 56% of new cases occurring in East Asia [2]. According to the Central Tumor Registry data for 2002, gastric cancer accounts for 20.8% of all cancers in Korea [3]. Although the overall survival (OS) of gastric cancer has been improved owing to the application of a national fiber optic esophagogastroduodenoscopy screening program in adults older than 40 years in Korea, a large proportion of patients are still diagnosed at the metastatic stage. The median survival time following cytotoxic chemotherapy is still less than 1 year; and thus metastatic gastric cancer remains a therapeutic challenge for medical oncologists [1]. The role of molecularly targeted therapy has not been adequately explored in gastric cancer compared with other common solid tumors such as breast, colorectal, or non–small cell lung cancer.
Insulin-like growth factors (IGF-I and -II) and their receptors play a pivotal role in regulating cell proliferation, differentiation, and apoptosis [4]. Aberrant expression and regulation of these proteins have been implicated in the development and prognosis of many human cancers [5,6]. There are at least six IGF-binding proteins (IGFBPs). IGFBP-3 is the most abundant IGFBP found in serum and binds the majority of serum IGFs [7]. IGFBP-3 regulates IGF bioactivity by sequestering IGF in the extracellular milieu, thereby inhibiting its mitogenic and antiapoptotic actions. In addition to its IGF-dependent function, IGFBP-3 also has IGF-independent antiproliferative and proapoptotic effects that seem to involve cell surface receptors for IGFBP-3 [4,8–11]. IGFBP-3 is expressed in most human tissue and in a variety of cell lines, including those of gastric origin [12,13]. Studies of gastric cancer cell lines have shown that IGFBP-3 can markedly inhibit the response of gastric cancer cells to IGF [12]. A recent study also reported that IGFBP-3 is protective against the development of gastric carcinoma by preventing the formation of intestinal metaplasia, and improves the prognosis of gastric cancer [6].
The integral role of epigenetic mechanisms such as promoter hypermethylation of tumor suppressor genes has become ever more apparent over the past decade [14]. Promoter hypermethylation is widespread in some cancers, occurs in the early stages of cancer development, and has been correlated with clinicopathologic features indicative of a poor prognosis, thus indicating the potential of gene hypermethylation as a marker of clinically significant disease [15]. Promoter hypermethylation has been proposed as a mechanism for the transcriptional silencing of IGFBP-3 in hepatocellular carcinomas [16], early-stage non–small cell lung cancer [17], prostate cancer [18,19], and in cancers of the bladder and ovary [20,21].
We undertook this study to assess the impact of IGFBP-3 methylation on survival of early-stage gastric cancer patients. Statistical analysis was performed to investigate the correlations between methylation and clinical and pathologic parameters.
Methods
Patients and Tissues
We previously reported the outcomes of 544 stage II to IV (M0) gastric cancer patients who received adjuvant chemoradiation therapy after curative surgery [22]. All patients were Korean and were staged by American Joint Committee on Cancer sixth edition. The postoperative adjuvant treatment adopted was the same as that used for the INT-0116 (SWOG-9008) trial, the results of which were previously reported [23]. Between these patients and an additional 23 stage IB patients who were included in our previous study, formalin-fixed, paraffin-embedded primary tumor tissues were available from 482 patients. All patients provided written informed consent according to institutional guidelines, and the study was approved by the Institutional Review Board. Among 482 patients, 138 stage IB/II gastric cancer patients were analyzed in this study.
Gastric Cancer Cell Lines
Human gastric carcinoma cells AGS, KATO-III, MKN-1, MKN-28, MKN-45, MKN-74, N87, SNU-1, SNU-5, SNU-16, SNU-216, SNU-484, SNU-601, SNU-620, SNU-638, SNU-668, and SNU-719 were purchased from the Korean Cell Line Bank (Seoul, Korea). YCC-1, YCC2, YCC-3, and YCC-7 were kindly provided by Dr. Hyun Cheol Chung (Yonsei Cancer Center, Seoul, Korea). OCUM-2M was kindly provided by Dr. Masakazu Yashiro (Osaka City University, Osaka, Japan). YCC-1, YCC-2, YCC-3, and YCC-7 were maintained in Dulbecco’s modified Eagle’s medium (Gibco-BRL, Carlsbad, CA) supplemented with 10% heat-inactivated FBS, 100 U/ml penicillin, 100 U/ml streptomycin, and 2 mM glutamine. All other cell lines were cultured in RPMI-1640 medium (Gibco-BRL) supplemented with 10% heat-inactivated FBS, 100 U/ml penicillin, 100 U/ml streptomycin, and 2 mM glutamine. All cells were incubated in a humidified atmosphere contained 5% CO2 at 37°C.
Bisulfite Modification
Bisulfite treatment of DNA was performed using the EpiTect Bisulfite Kit (QIAGEN) according to manufacturer’s instructions. Briefly, 1 μg of genomic DNA in 20 μl of water was combined with 85 μl of bisulfate mix and 35 μl of DNA protection buffer. Bisulfate conversion of DNA was performed using the following conditions: denaturation at 99°C for 5 minutes, incubation at 60°C for 25 minutes, denaturation at 99°C for 5 minutes, incubation at 60°C for 85 minutes, denaturation at 99°C for 5 minutes, incubation at 60°C for 175 minutes, and hold at 20°C. The bisulfite-converted DNA was mixed with 560 μl of Buffer BL, applied to a spin column, and centrifuged at 12,000 rpm for 1 minute. The flow-through was discarded and the column was washed with 500 μl of Buffer BW. Buffer BD (500 μl) was applied to the column, which was incubated at room temperature for 15 minutes. The column was centrifuged to remove Buffer BD and then washed twice with Buffer BW (500 μl). Residual Buffer BW was removed by an additional spin (12,000 rpm for 1 minute). Buffer EB (20 μl) was added to the column to elute the DNA. The DNA concentration was determined using a Nano-drop spectrophotometer.
Methylation-Specific Polymerase Chain Reaction (MS-PCR) for Quantitative DNA Methylation Analysis
The primers and probe for IGFBP-3 were as follows: IGFBP3-F, 5ʹ-GTT TCG GGC GTG AGT ACG A-3ʹ; IGFBP3-R, 5ʹ-GAA TCG ACG CAA ACA CGA CTA C-3ʹ; and IGFBP3-probe, 6FAM-5ʹ-TCG GTT GTT TAG GGC GAA GTA CGG G-3ʹ-TAMRA. The collagen 2A1 gene (COL2A1) was used to normalize for the amount of input bisulfite-converted DNA. The primers and probe for COL2A1 were as follows: COL2A1-F, 5ʹ-TCT AAC AAT TAT AAA CTC CAA CCA CCA A-3ʹ; COL2A1-R, 5ʹ-GGG AAG ATG GGA TAG AAG GGA ATA T-3ʹ; and COL2A1-probe, VIC-5ʹ-CCT TCA TTC TAA CCC AAT ACC TAT CCC ACC TCT AAA-TAMRA-3ʹ. Reactions were done in triplicate with genomic DNA at 5 ng, primers at 900 nmol/l, probes at 250 nmol/l, and TaqMan Universal PCR reagent under standard thermocycling conditions. After initial denaturating for 10 minutes at 95°C, 45 cycles at 95°C for 15 seconds, 60°C for 1 minute, and 72°C for 1 minute were carried out, followed by a final extension for 5 minutes at 72°C. AsPC-1 cell line was included as negative control sample to compare methylation results. The percentage of methylated reference at a specific locus was calculated by dividing the ratio of GENE/COL2A1 template in a sample by the ratio of GENE/COL2A1 template in SssI-treated human genomic DNA (presumably fully methylated) and multiplying this value by 100.
DNA Extraction
One hundred thirty-eight stage IB/II gastric cancer patients were evaluated for the methylation status of the IGFBP-3 promoter. Fifty-four of those patients had nonneoplastic and noninfiltrated gastric mucosa also evaluated. DNA was extracted from five 10-μm formalin-fixed, paraffin-embedded sections containing a representative portion of each tumor block using the QIAamp DNA Mini kit (Qiagen, Hilden, Germany). A pathologist reviewed each slide and verified the presence of adequate tumor tissue with greater than 50% representative malignant cells.
Statistical Analyses
Disease-free survival (DFS) was defined as the time from surgery to the first relapse of cancer or death of any cause. Overall survival was calculated from the date of surgery to the date of death. OS and DFS were calculated using the Kaplan-Meier method. Correlation analyses were performed using the two-sided χ2 test or Fisher exact test. Differences in DFS and OS were compared using log-rank tests and Cox proportional hazard analysis. A P value of less than .05 was considered statistically significant.
Results
The Status of IGFBP-3 Methylation in Gastric Cancer Cell Lines
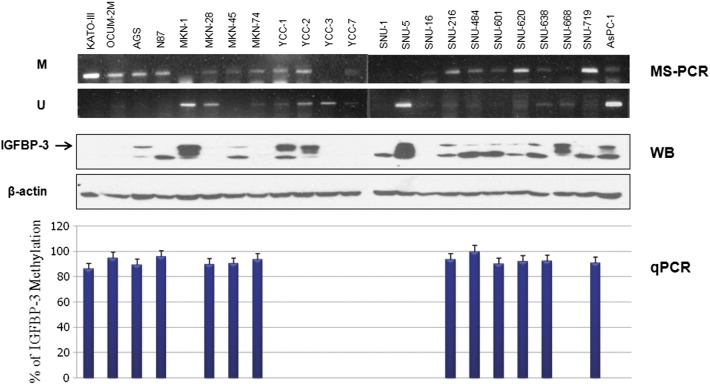
IGFBP-3 methylation pattern was measured by MS-PCR in 22 known gastric cell lines (KATO-III, OCUM-2M, AGS, N87, MKN-1, MKN-28, MKN-45, MKN-74, YCC-1, YCC-2, YCC-3, YCC-7, SNU-1, SNU-5, SNU-16, SNU-216, SNU-484, SNU-601, SNU-620, SNU-638, SNU-668, and SNU-719). The clinical information of these gastric cell lines was summarized in Table 1. In methylation analysis by MSP of IGFBP3, all gastric cell lines with methylated band revealed IGFBP3 methylation of more than 80% in quantitative PCR. The representative results are shown in Figure 1.
Table 1.
The Information of 23 Gastric Cell Lines Analyzed for IGFBP-3 Methylation
| Status | Gender | Histologic Subtype | Origin |
|---|---|---|---|
| Methylated IGFBP-3 | |||
| KATO-III | Male | Poorly | Pleural effusion, metastatic |
| OCUM-2 M | Female | Poorly | Stomach, primary |
| AGS | Female | Poorly | Stomach, primary |
| N87 | Male | Well | Liver, metastatic |
| MKN-28 | Female | Moderate | Lymph node, metastatic |
| MKN-45 | Female | Poorly | Liver, metastatic |
| MKN-74 | Male | Moderate | Liver, metastasis |
| SNU-16 | Female | Poorly | Ascites, metastatic |
| SNU-216 | Female | Moderate | Lymph node, metastatic |
| SNU-484 | Male | Poorly | Stomach, primary |
| SNU-601 | Male | Poorly | Ascites, metastatic |
| SNU-620 | Female | Poorly | Ascites, metastatic |
| SNU-638 | Male | Poorly | Ascites, metastatic |
| SNU-719 | Male | Moderate | Stomach, primary |
| Un-Methylated IGFBP-3 | |||
| MKN-1 | Male | Adenosquamous | Lymph node, metastatic |
| YCC-1 | Male | NA | Ascites, metastatic |
| YCC-2 | Male | NA | Ascites, metastatic |
| YCC-3 | Male | NA | Ascites, metastatic |
| YCC-7 | Male | NA | Ascites, metastatic |
| SNU-1 | Male | Poorly | Stomach, primary |
| SNU-5 | Female | Poorly | Ascites, metastatic |
| SNU-668 | Male | Poorly | Ascites, metastatic |
Figure 1.
IGFBP-3 methylation as measured by MS-PCR and MethyLight quantitativePCR in gastric cancer cell lines.
Correlation Between IGFBP-3 Methylation and Clinical Variables
The general clinical characteristics of 138 patients analyzed for the status of IGFBP-3 methylation are presented in Table 2. The IGFBP-3 promoter was methylated in 26 of 138 stage IB/II gastric cancer samples. In univariate analysis, there was no statistically significant association between the hypermethylation of IGFBP-3 and clinicopathological factors such as age, Lauren classification, sex, tumor infiltration, lymph node metastasis, and histologic grade. The age of 138 patients with stage IB/II gastric cancer ranged from 23 to 70 years (median, 54.5 years). Eighty-one (58.7%) of the patients were diffuse type according to Lauren classification, and the majority (70.3%) of patients were male.
Table 2.
Clinical Features and IGFBP-3 Methylation in Stage IB/II
| No. of Cases (N = 138) (%) |
IGFBP-3 Methylation |
|||
|---|---|---|---|---|
| Positive (N = 26) | Negative (N = 112) | P Value | ||
| Age | ||||
| ≤ 60 | 100(72.5) | 21 | 79 | .293 |
| > 60 | 38 (27.5) | 5 | 33 | |
| Lauren classification | ||||
| Intestinal | 56 (40.6) | 8 | 48 | .109 |
| Diffuse | 81 (58.7) | 17 | 64 | |
| Intermediate | 1 (0.7) | 1 | 0 | |
| Sex | ||||
| Male | 97 (70.3) | 17 | 80 | .543 |
| Female | 41 (29.7) | 9 | 32 | |
| Tumor infiltration | ||||
| T1/T2 | 118 (85.5) | 23 | 95 | .766 |
| T3/T4 | 20 (14.5) | 3 | 17 | |
| Lymph node metastasis | ||||
| N0/N1 | 135 (97.8) | 26 | 109 | 1.000 |
| N2/N3 | 3 (2.2) | 0 | 3 | |
| Histologic grade (adenocarcinoma only) | ||||
| Well/moderately differentiated | 52 (37.7) | 8 | 44 | .419 |
| Poorly differentiated/Other types | 86 (62.3) | 18 | 68 | |
Impact of IGFBP-3 Methylation on Recurrence and Survival
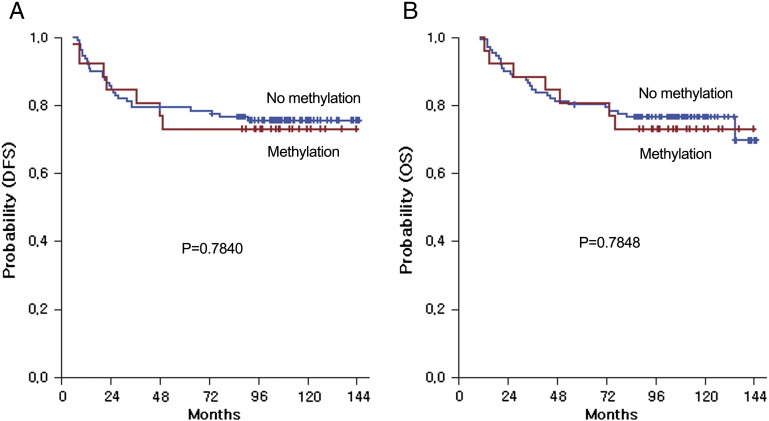
We performed survival analyses according to the methylation status of the IGFBP-3 promoter (Figure 2). Disease recurrence was observed in 34 (24.6%) of 138 patients during the median follow-up period of 110.7 months. Stage IB/II gastric cancer patients with a hypermethylated IGFBP-3 promoter had a similar DFS following curative surgery as compared with those without IGFBP-3 methylation (5-year DFS: 73.1% vs 79.5%; IGFBP-3 methylation (+) vs IGFBP-3 methylation (−); P = .7840). Moreover, gastric cancer patients with IGFBP-3 methylation also demonstrated similar OS following curative surgery as compared with those without IGFBP-3 methylation (5-year OS: 80.7% vs 80.3%; IGFBP-3 methylation (+) vs IGFBP-3 methylation (−); P = .7848). The following variables were tested using backward stepwise Cox proportional hazards regression modeling: age (≤ 60 vs > 60), Lauren classification, sex (male vs female), tumor infiltration (T1/T2 vs T3/T4), lymph node (N0/N1 vs N2/N3), histologic grade, and the methylation status of the IGFBP-3 promoter. For DFS and OS in all patients, none of the factors tested had a predictive role with statistical significance at the univariate and multivariate level.
Figure 2.
Survival analysis: Disease-free (A) and overall (B) survival curves according to the status of IGFBP-3 methylation.
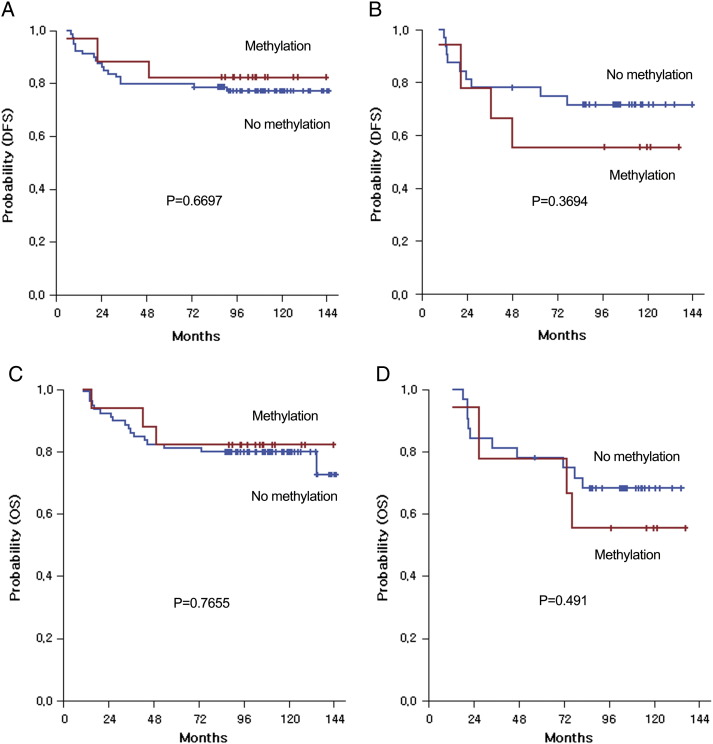
In subgroup analyses, women, but not men, showed a trend toward poorer prognosis for DFS and OS in the subset of patients with IGFBP-3 methylation as compared with those without IGFBP-3 methylation (8-year DFS: 55.6% vs 71.6%, P = .3694 and 8-year OS: 55.6% vs 68.4%, P = .491, respectively; Figure 3) even with no statistical significance.
Figure 3.
Survival analysis: Disease-free (A, B) and overall (C, D) survival curves by gender according to methylation status.
Discussion
It has been known that the IGF-IGFBP system might play an important role in the initiation, progression, and metastasis of gastric cancer [6,12]. IGFBP-3, one of six members of the IGFBP family, regulates IGF bioactivity by sequestering IGFs away from the IGF receptor in the extracellular milieu, thereby inhibiting the mitogenic and antiapoptotic action of IGFs [4,10]. A negative correlation between serum IGFBP-3 levels and human cancer risk suggests a protective role of IGFBP-3 against the effects of systemic IGFs [24,25]. Zhang et al. [6] reported a significantly higher percentage of positive IGFBP-3 staining in the tumor tissues of patients with well or moderately differentiated tumors than those with poorly differentiated tumors, indicating that IGFBP-3 may be associated with a better prognosis.
DNA methylation, which is the major form of epigenetic information in mammalian cells, has profound effects on the mammalian genome, including transcriptional repression, chromatin structure modulation, X-chromosome inactivation, genomic imprinting, and suppression of the detrimental effects of repetitive and parasitic DNA sequence on genomic integrity [26–28]. A recent study using a monoclonal antibody specific for 5-methylcytosine to evaluate for the status of global DNA methylation suggests that alteration in DNA methylation is an important epigenetic difference in susceptibility to the development of human cancer [29]. Genomic methylation patterns are frequently altered in tumor cells, with global hypomethylation accompanying region-specific hypermethylation events. CpG islands in specific regions of the promoter have been described as a common epigenetic mechanism for the silencing of tumor suppressor genes in cancer and as a regulator of growth in cancers [28]. Promoter hypermethylation has been proposed as a mechanism for transcriptional silencing of IGFBP-3 in hepatocellular carcinoma [16], early-stage non–small cell lung cancer [17], prostate cancer [18,19], and cancers of the bladder and ovary [20,21]. Moreover, it was shown that this hypermethylation of IGFBP-3 occurs as an early event in cancer carcinogenesis. However, the role of IGFBP-3 methylation in early-stage gastric cancer had not been evaluated until now.
The present study is the first to assess the impact of IGFBP-3 methylation on survival in early-stage gastric cancer patients. We found that clinicopathological factors such as age, Lauren classification, sex, tumor infiltration, lymph node metastasis, and histologic grade did not show a statistically significant association with the methylation status of the IGFBP-3 promoter. In addition, no significant difference in survival outcomes of stage IB/II gastric cancer patients according to the status of IGFBP-3 methylation was observed. However, this finding should be interpreted with caution. In our study, most patients (87%), with the exception of 18 patients with stage IB cancers, received adjuvant chemoradiation therapy after curative surgery. Treatments such as adjuvant chemoradiation may change the impact of IGFBP-3 methylation on survival of early-stage gastric cancer patients. In this study, the 5-year survival rate for patients with IB/II gastric cancer was about 80%. Therefore, the impact of IGFBP-3 methylation on survival may have been diluted by good survival outcomes in the analyzed patients.
The most interesting finding of this study was that women, but not men, showed a trend toward poorer prognosis for DFS and OS in the subset of patients with IGFBP-3 methylation as compared with those without IGFBP-3 methylation. This observation may be caused by a difference in IGFBP-3 values according to sex. Lin et al. reported that women had significantly higher IGFBP-3 values than men and that women older than 50 years showed a significant reduction in IGF-I/IGFBP-3 molar ratio. Thus, IGFBP-3 methylation may have a more powerful influence on survival outcomes in female than male patients [30].
Previous studies have shown that the transcriptional activity of the methylated IGFBP-3 promoter is restored by treatment with the demethylating agent 5’-aza-dC in some human cancers, including hepatocellular carcinoma and non–small cell lung cancer [16,17]. However, 5’-aza-dC treatment restores IGFBP-3 expression in only a portion of those cancer cell lines in which the promoter is methylated [17]. These findings suggest that the mechanisms regulating IGFBP-3 expression in human cancer cells are diverse and complex. Thus, the multifaceted nature of the processes involved in the regulation of IGFBP-3 expression might affect the results of our study. Future research will investigate other mechanisms related to IGFBP-3 expression and silencing in gastric cancer.
The mechanisms of IGFBP-3 expression and silencing in gastric cancer have not been fully elucidated. However the status of IGFBP-3 methylation as measured by MS-PCR may be good predictive factor for survival in specific subgroups of patients with early-stage gastric cancer undergoing curative surgery. Further research is needed to better understand the role of IGFBP-3 in gastric cancer.
Disclosure of Potential Conflicts of Interest
The authors have declared no conflict of interest.
Authors’ contributions
All authors made substantial contributions to the conception and design of the study, and the acquisition, analysis, and interpretation of the data. All authors were involved in drafting the manuscript (or revising it), and all read and approved the final manuscript.
Acknowledgements
This study was supported by a grant from the Korea Healthcare Technology R&D Project, Ministry for Health, Welfare, & Family Affairs, Republic of Korea (J.O.P.; A090425). Support was also provided by a grant from the 20 by 20 project of Samsung Medical Center (GF01140111).
Footnotes
This study was supported by a grant from the Korea Healthcare Technology R&D Project, Ministry for Health, Welfare, & Family Affairs, Republic of Korea (J.O.P.; A090425). Support was also provided by a grant from the 20 by 20 project of Samsung Medical Center (GF01140111).
References
- 1.Cunningham D, Jost LM, Purkalne G, Oliveira J. ESMO Minimum Clinical Recommendations for diagnosis, treatment and follow-up of gastric cancer. Ann Oncol. 2005;16(Suppl. 1):i22–i23. doi: 10.1093/annonc/mdi812. [DOI] [PubMed] [Google Scholar]
- 2.Inoue M, Tsugane S. Epidemiology of gastric cancer in Japan. Postgrad Med J. 2005;81:419–424. doi: 10.1136/pgmj.2004.029330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bae JM, Won YJ, Jung KW, Park JG. Annual report of the Korea Central Cancer Registry Program 2000: Based on registered data from 131 hospital. Cancer Res Treat. 2002;34:77–83. doi: 10.4143/crt.2002.34.2.77. [DOI] [PubMed] [Google Scholar]
- 4.Butt AJ, Firth SM, King MA, Baxter RC. Insulin-like growth factor-binding protein-3 modulates expression of Bax and Bcl-2 and potentiates p53-independent radiation-induced apoptosis in human breast cancer cells. J Biol Chem. 2000;275:39174–39181. doi: 10.1074/jbc.M908888199. [DOI] [PubMed] [Google Scholar]
- 5.Yu H, Berkel H. Insulin-like growth factors and cancer. J La State Med Soc. 1999;151:218–223. [PubMed] [Google Scholar]
- 6.Zhang ZW, Newcomb PV, Moorghen M, Gupta J, Feakins R, Savage P, Hollowood A, Alderson D, Holly JM. Insulin-like growth factor binding protein-3: relationship to the development of gastric pre-malignancy and gastric adenocarcinoma (United Kingdom) Cancer Causes Control. 2004;15:211–218. doi: 10.1023/B:CACO.0000019510.96285.e9. [DOI] [PubMed] [Google Scholar]
- 7.Bond JJ, Meka S, Baxter RC. Binding characteristics of pro-insulin-like growth factor-II from cancer patients: binary and ternary complex formation with IGF binding proteins-1 to -6. J Endocrinol. 2000;165:253–260. doi: 10.1677/joe.0.1650253. [DOI] [PubMed] [Google Scholar]
- 8.Oh Y, Muller HL, Pham H, Rosenfeld RG. Demonstration of receptors for insulin-like growth factor binding protein-3 on Hs578T human breast cancer cells. J Biol Chem. 1993;268:26045–26048. [PubMed] [Google Scholar]
- 9.Oh BH, Pandit J, Kang CH, Nikaido K, Gokcen S, Ames GF, Kim SH. Three-dimensional structures of the periplasmic lysine/arginine/ornithine-binding protein with and without a ligand. J Biol Chem. 1993;268:11348–11355. [PubMed] [Google Scholar]
- 10.Rajah R, Valentinis B, Cohen P. Insulin-like growth factor (IGF)-binding protein-3 induces apoptosis and mediates the effects of transforming growth factor-beta1 on programmed cell death through a p53- and IGF-independent mechanism. J Biol Chem. 1997;272:12181–12188. doi: 10.1074/jbc.272.18.12181. [DOI] [PubMed] [Google Scholar]
- 11.Valentinis B, Bhala A, DeAngelis T, Baserga R, Cohen P. The human insulin-like growth factor (IGF) binding protein-3 inhibits the growth of fibroblasts with a targeted disruption of the IGF-I receptor gene. Mol Endocrinol. 1995;9:361–367. doi: 10.1210/mend.9.3.7539889. [DOI] [PubMed] [Google Scholar]
- 12.Yi HK, Hwang PH, Yang DH, Kang CW, Lee DY. Expression of the insulin-like growth factors (IGFs) and the IGF-binding proteins (IGFBPs) in human gastric cancer cells. Eur J Cancer. 2001;37:2257–2263. doi: 10.1016/s0959-8049(01)00269-6. [DOI] [PubMed] [Google Scholar]
- 13.Guo YS, Beauchamp RD, Jin GF, Townsend CM, Jr., Thompson JC. Insulinlike growth factor-binding protein modulates the growth response to insulinlike growth factor 1 by human gastric cancer cells. Gastroenterology. 1993;104:1595–1604. doi: 10.1016/0016-5085(93)90634-o. [DOI] [PubMed] [Google Scholar]
- 14.Herman JG, Baylin SB. Gene silencing in cancer in association with promoter hypermethylation. N Engl J Med. 2003;349:2042–2054. doi: 10.1056/NEJMra023075. [DOI] [PubMed] [Google Scholar]
- 15.Perry AS, Foley R, Woodson K, Lawler M. The emerging roles of DNA methylation in the clinical management of prostate cancer. Endocr Relat Cancer. 2006;13:357–377. doi: 10.1677/erc.1.01184. [DOI] [PubMed] [Google Scholar]
- 16.Hanafusa T, Yumoto Y, Nouso K, Nakatsukasa H, Onishi T, Fujikawa T, Taniyama M, Nakamura S, Uemura M, Takuma Y. Reduced expression of insulin-like growth factor binding protein-3 and its promoter hypermethylation in human hepatocellular carcinoma. Cancer Lett. 2002;176:149–158. doi: 10.1016/s0304-3835(01)00736-4. [DOI] [PubMed] [Google Scholar]
- 17.Chang YS, Wang L, Suh YA, Mao L, Karpen SJ, Khuri FR, Hong WK, Lee HY. Mechanisms underlying lack of insulin-like growth factor-binding protein-3 expression in non-small-cell lung cancer. Oncogene. 2004;23:6569–6580. doi: 10.1038/sj.onc.1207882. [DOI] [PubMed] [Google Scholar]
- 18.Perry AS, Loftus B, Moroose R, Lynch TH, Hollywood D, Watson RW, Woodson K, Lawler M. In silico mining identifies IGFBP3 as a novel target of methylation in prostate cancer. Br J Cancer. 2007;96:1587–1594. doi: 10.1038/sj.bjc.6603767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Liu B, Lee KW, Anzo M, Zhang B, Zi X, Tao Y, Shiry L, Pollak M, Lin S, Cohen P. Insulin-like growth factor-binding protein-3 inhibition of prostate cancer growth involves suppression of angiogenesis. Oncogene. 2007;26:1811–1819. doi: 10.1038/sj.onc.1209977. [DOI] [PubMed] [Google Scholar]
- 20.Wiley A, Katsaros D, Chen H, Rigault de la Longrais IA, Beeghly A, Puopolo M, Singal R, Zhang Y, Amoako A, Zelterman D. Aberrant promoter methylation of multiple genes in malignant ovarian tumors and in ovarian tumors with low malignant potential. Cancer. 2006;107:299–308. doi: 10.1002/cncr.21992. [DOI] [PubMed] [Google Scholar]
- 21.Christoph F, Weikert S, Kempkensteffen C, Krause H, Schostak M, Miller K, Schrader M. Regularly methylated novel pro-apoptotic genes associated with recurrence in transitional cell carcinoma of the bladder. Int J Cancer. 2006;119:1396–1402. doi: 10.1002/ijc.21971. [DOI] [PubMed] [Google Scholar]
- 22.Kim S, Lim DH, Lee J, Kang WK, MacDonald JS, Park CH, Park SH, Lee SH, Kim K, Park JO. An observational study suggesting clinical benefit for adjuvant postoperative chemoradiation in a population of over 500 cases after gastric resection with D2 nodal dissection for adenocarcinoma of the stomach. Int J Radiat Oncol Biol Phys. 2005;63:1279–1285. doi: 10.1016/j.ijrobp.2005.05.005. [DOI] [PubMed] [Google Scholar]
- 23.Macdonald JS, Smalley SR, Benedetti J, Hundahl SA, Estes NC, Stemmermann GN, Haller DG, Ajani JA, Gunderson LL, Jessup JM. Chemoradiotherapy after surgery compared with surgery alone for adenocarcinoma of the stomach or gastroesophageal junction. N Engl J Med. 2001;345:725–730. doi: 10.1056/NEJMoa010187. [DOI] [PubMed] [Google Scholar]
- 24.Hankinson SE. Overview of breast cancer population studies. Growth Horm IGF Res. 2000;10(Suppl. A):S22–S23. doi: 10.1016/s1096-6374(00)90010-8. [DOI] [PubMed] [Google Scholar]
- 25.Ma J, Giovannucci E, Pollak M, Chan JM, Gaziano JM, Willett W, Stampfer MJ. Milk intake, circulating levels of insulin-like growth factor-I, and risk of colorectal cancer in men. J Natl Cancer Inst. 2001;93:1330–1336. doi: 10.1093/jnci/93.17.1330. [DOI] [PubMed] [Google Scholar]
- 26.Baylin SB, Herman JG. DNA hypermethylation in tumorigenesis: epigenetics joins genetics. Trends Genet. 2000;16:168–174. doi: 10.1016/s0168-9525(99)01971-x. [DOI] [PubMed] [Google Scholar]
- 27.Jones PA, Laird PW. Cancer epigenetics comes of age. Nat Genet. 1999;21:163–167. doi: 10.1038/5947. [DOI] [PubMed] [Google Scholar]
- 28.Robertson KD. DNA methylation, methyltransferases, and cancer. Oncogene. 2001;20:3139–3155. doi: 10.1038/sj.onc.1204341. [DOI] [PubMed] [Google Scholar]
- 29.Piyathilake CJ, Frost AR, Bell WC, Oelschlager D, Weiss H, Johanning GL, Niveleau A, Heimburger DC, Grizzle WE. Altered global methylation of DNA: an epigenetic difference in susceptibility for lung cancer is associated with its progression. Hum Pathol. 2001;32:856–862. doi: 10.1053/hupa.2001.26471. [DOI] [PubMed] [Google Scholar]
- 30.Lin CM, Huang YL, Lin ZY. Influence of gender on serum growth hormone, insulin-like growth factor-I and its binding protein-3 during aging. Yonsei Med J. 2009;50:407–413. doi: 10.3349/ymj.2009.50.3.407. [DOI] [PMC free article] [PubMed] [Google Scholar]