Abstract
We analyzed the genome of a rhabdoid glioblastoma (R-GBM) tumor, a very rare variant of GBM. A surgical specimen of R-GBM from a 20-year-old woman was analyzed using whole exome sequencing (WES), whole transcriptome sequencing (WTS), single nucleotide polymorphism array, and array comparative genomic hybridization. The status of gene expression in R-GBM tissue was compared with that of normal brain tissue and conventional GBM tumor tissue. We identified 23 somatic non-synonymous small nucleotide variants with WES. We identified the BRAF V600E mutation and possible functional changes in the mutated genes, ISL1 and NDRG2. Copy number alteration analysis revealed gains of chromosomes 3, 7, and 9. We found loss of heterozygosity and focal homozygous deletion on 9q21, which includes CDKN2A and CDKN2B. In addition, WTS revealed that CDK6, MET, EZH2, EGFR, and NOTCH1, which are located on chromosomes 7 and 9, were over-expressed, whereas CDKN2A/2B were minimally expressed. Fusion gene analysis showed 14 candidate genes that may be functionally involved in R-GBM, including TWIST2, and UPK3BL. The BRAF V600E mutation, CDKN2A/2B deletion, and EGFR/MET copy number gain were observed. These simultaneous alterations are very rarely found in GBM. Moreover, the NDRG2 mutation was first identified in this study as it has never been reported in GBM. We observed a unique genomic signature in R-GBM compared to conventional GBM, which may provide insight regarding R-GBM as a distinct disease entity among the larger group of GBMs.
Introduction
Rhabdoid glioblastoma (R-GBM) is a very rare disease with few cases reported [1–8]. R-GBM is characterized by tumor cells that resemble rhabdomyoblasts [2], which robustly express vimentin, epithelial membrane antigen (EMA), and SMARCB1 (INI-1), but only faintly express glial fibrillary acidic protein (GFAP) [5,7,9,10]. Clinically, R-GBMs can occur at any age but most commonly occur in teenagers younger than 20 years old [1,2,5].
Chromosome 22, which is frequently lost in atypical teratoid rhabdoid tumors (ATRT), is often deleted in these tumors [3,8], although this finding is inconsistent [1]. In one case, copy number gains were noted for chromosomes 3, 7, 9, 12, 17q, and 21q [1] in R-GBM. Also, in a case series, copy number gain or amplification of EGFR on chromosome 7 was noted [9]. Regarding genetic changes prevalent in brain tumors, CDKN2A hemizygous deletion was reported in one case [2]. Otherwise, BRAF mutations were absent in two cases that were examined [11], and SMARCB1 (INI-1) [12,13], which is important in ATRT, was not mutated in R-GBM [2].
Presently, R-GBM is not recognized as a distinct disease entity by the World Health Organization [14] classification system because accumulated information on this rare variety is still rudimentary. To our knowledge, no study has evaluated the genome-wide profile of this disease except for one case that was evaluated using array comparative genomic hybridization (CGH) [1]. To determine whether R-GBM should be recognized as a disease that is distinct from conventional glioblastoma (GBM) or other tumors with similar characteristics such as ATRT, comprehensive genomic data will be fundamental for diagnostic, prognostic, and therapeutic decisions.
A 20-year-old female presented with a rim-enhanced tumor that was pathologically proven to be an R-GBM. She underwent two extensive surgeries and concurrent chemoradiotherapy combined with oral temozolomide treatment. She was free of disease for 25 months after the treatment. Using next generation sequencing techniques, we studied this tumor to obtain novel insight into identifying distinctive genetic changes in an R-GBM compared to conventional GBM as well as normal brain tissue. We performed whole exome sequencing (WES), whole transcriptome sequencing (WTS), single nucleotide polymorphism (SNP) array, and array-CGH. The aims of this study were to investigate the genomic profile of R-GBM and to explore whether R-GBM had a distinct genomic signature that could be used as a therapeutic target.
Materials and Methods
Study Patient
A 20-year-old female patient was seen in an outpatient clinic at Seoul National University Hospital because of headache, nausea, and vomiting in April 2011. Brain magnetic resonance imaging showed a 5-cm sized, well-enhanced mass in the right temporal lobe. The mass also showed diffusion restriction with increased perfusion at the peripheral enhanced portion. She underwent a craniotomy for tumor removal in May 2011. The molecular genetic characteristics of the surgical specimen were evaluated as follows. Immunohistochemical staining revealed focal expression of GFAP and strong expression of EMA and INI-1 (Figure 1). Fluorescence in situ hybridization (FISH) showed no EGFR amplification and no deletion of chromosomes 1p, 9p21, or 19q. In addition, methylation-specific PCR showed hypermethylation of the MGMT promoter, and the MIB-1 labeling index was measured as 36.5% with an Aperio Spectrum plus image analyzer. The study patient received adjuvant concurrent chemoradiotherapy with oral temozolomide treatment after the surgery. However, the tumor recurred on the ipsilateral side of the frontal lobe, and she underwent a second operation to remove the recurrent tumor. The final pathology confirmed that, as with the initial mass, the recurrent tumor was an R-GBM. The recurrent tumor had a MIB-1 labeling index of 37.5%. She has been free from disease for 25 months as of December 2013. The study protocol was reviewed and approved by the institutional review board of the Seoul National University Hospital, and informed consent was obtained from the study patient. The recommendations of the Declaration of Helsinki for biomedical research involving human subjects were followed.
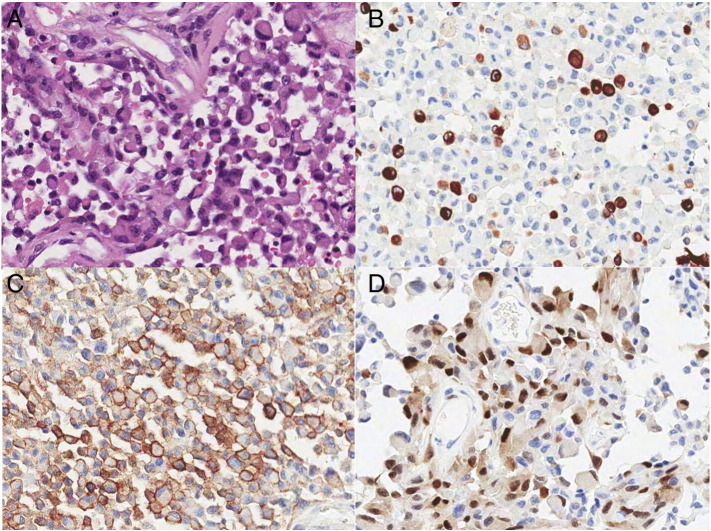
Figure 1.
Pathology of rhabdoid glioblastoma. (A) A representative H&E picture shows non-cohesive rhabdoid cells with eccentrically located pleomorphic nuclei and eosinophilic globular cytoplasm (H&E, original magnification × 200). (B) GFAP is robustly positive in some, but not all, tumor cells (GFAP immunostaining, original magnification × 200). (C) EMA is strongly positive in a cytoplasmic membrane pattern in almost all tumor cells (EMA immunohistochemistry, original magnification × 200). (D) Cyclin D1 (CCND1) staining is strongly positive in the nuclei of the tumor cells (cyclin D1 immunohistochemistry, original magnification × 200).
DNA and RNA Preparation
Fresh frozen tumor tissue and 5 ml peripheral blood were obtained at the time of the first surgery. The DNeasy® Blood & Tissue Kit (Qiagen, Hilden, Germany) was used to extract genomic DNA and tumor DNA, according to the manufacturer’s recommendations. Extracted DNA was quantified using a NanoDrop ND-1000 spectrophotometer (NanoDrop Technologies, Wilmington, DE). RNA was extracted from the tumor tissue using TRIzol (Invitrogen, Grand Island, NY) and eluted in RNAse-free water. RNA quantity and quality were assessed using an Agilent 2100 Bioanalyzer (Agilent Technologies Inc., Santa Clara, CA).
Whole Exome Sequencing
We used the Agilent SureSelect50-Mb ExomeCapture Kit for exon target enrichment (Agilent Technologies Inc.). Sequencing was performed using the Illumina HiSeq2000 (Illumina Inc., San Diego, CA) with 100-bp paired-end reads. Using UCSC hg19 as a reference genome, mapping and pairing were performed with the Burrows-Wheeler Aligner (BWA) algorithm [15]. Local realignment was performed using Genome Analysis ToolKit (GATK) [16], and duplication removal was conducted using Picard.
Somatic calling of somatic single nucleotide variants (SNVs) and indels is described in Supplement 1. Using SnpEff [17], we selected variations that were non-synonymous and rare in the general population (defined as < 1% in the 1000 genome project (http://www.1000genomes.org/)). For copy number alteration (CNA) analysis of WES data, we used the Copy Number Analysis for Targeted Resequencing (CONTRA) tool [18] and summarized the exon-level log2 fold changes of read depth between the normal and tumor samples into gene-level log2 fold changes. Loss of heterozygosity (LOH: heterozygous in normal tissue but homozygous in the tumor) analysis also was performed using WES. We used variant allele fraction values of normal and tumor samples to determine the LOH region.
Whole Transcriptome Sequencing
The 200- to 500-bp double-stranded cDNA fragments were purified by agarose gel electrophoresis and amplified using PCR to produce the library. Raw sequencing reads were produced by Illumina HiSeq 2000 with 100-bp paired-end reads. After removing noisy raw reads, which contained the adaptor sequence and more than 10% unknown bases or low quality bases, the remaining reads were aligned with the human reference genome (UCSC hg19). To find fusion transcripts, we utilized three types of fusion discovery software: deFuse [19], BreakFusion [20], and ChimeraScan [21]. UniGene clusters were downloaded from the National Center for Biotechnology Information (http://www.ncbi.nlm.nih.gov/) to assist in locating potential gene fusions. To quantify the gene expression level, the number of reads that mapped to the exons of each RefSeq gene was calculated, and the corresponding reads per kilobase per million reads (RPKM) [22] value was derived. Possible functional fusions were annotated using Oncofuse [23] and went through further analysis.
SNP Array
We applied a genome-wide SNP array (Illumina HumanOmni5-Quad BeadChip, Illumina) using the genomic DNA sample. With B allele frequency data from GenomeStudio (Illumina) analysis results for SNP array data, we used the paired parent-specific circular binary segmentation method for LOH and CNA analysis.
Array Comparative Genomic Hybridization and Identification of CNAs
We used the Agilent aCGH G3 Human 1×1M array with tumor and matched normal genomic DNA samples. Raw data were acquired and normalized using the locally weighted scatterplot smoothing (LOWESS) algorithm using Feature Extraction software ver10.7 (Agilent software). The significance test for each CNV region used the Z-statistic calculated by DNA Analytics ver4.0.81 (Agilent software), which sets the window size to 1M and Z-score threshold to 4.0.
Use of the Public Database as a Reference
We used gene expression data estimated from WTS to select possible functional genetic changes in our study. Because R-GBM is a rare disease and obtaining control samples is not easy, we used a public database as a reference. First, we compared the RPKM value of specific genetic changes found in our analysis with normal brain expression values. Then, we compared the RPKM value of specific genetic changes found in our analysis with GBM data to determine whether R-GBM is simply a subtype of GBM. For the normal brain data, we used the normalized expression dataset from BrainSpan (http://www.brainspan.org/). For the GBM data, we used datasets from TCGA (https://tcga-data.nci.nih.gov) and cBioPortal for Cancer Genomics (http://www.cbioportal.org).
Results
Tumor Purity, Alignment, and Coverage Statistics
The purity of the tumor samples was estimated using SNP array data with the Allele-specific copy number analysis of tumors (ASCAT) algorithm [24]. The proportion and the ploidy of tumor cells in the sample were about 89% and 2.17, respectively (Online Resource Section 1: Supplementary Figure 1). In WES, the total numbers of uniquely mapped reads were 181,350,341 and 186,695,100 for normal and tumor samples, respectively. These data yielded mean target coverages of 210 and 197 for the samples, respectively (Online Resource Section 2: Supplementary Table 1).
Somatic SNVs and Small Indels Found With WES
We found 46,468 (45,045 in dbSNP138) and 46,191 (44,748 in dbSNP138) SNVs from the paired normal DNA and tumor DNA, respectively. A total of 45,542 (44,264 in dbSNP138) SNVs were commonly observed in both samples. We identified 3753 (3362 in dbSNP137) and 3678 (3314) small indels from the paired normal and tumor DNA, respectively. A total of 3594 (3273 in dbSNP138) small indels were common in both samples (Online Resource Section 3 and 4: Supplementary Figures 2–4). The somatic calling method is described in the Supplement Text (Online Resource Section 4). As a result, 38 somatically mutated SNVs and one small indel were detected with WES. Twenty-three non-synonymous SNVs (Table 1) were found, 13 of which were also found with WTS.
Table 1.
List of 23 Candidate Non-Synonymous Somatic SNVs
| Chr | Position | dbSNP | Ref | Alt | Transcript | Gene | Effect | AA Change | DepthN | DepthT | VAFN | VAFT | RNA-Seq (confirmed) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Chr1 | 27876290 | . | G | C | NM_001029882.2 | AHDC1 | MISSENSE | H779Q | 81 | 66 | 0 | 0.409 | O |
| Chr1 | 104093621 | . | C | G | NM_017619.3 | RNPC3 | MISSENSE | P474A | 169 | 192 | 0 | 0.432 | O |
| Chr2 | 30748467 | . | G | A | NM_182551.3 | LCLAT1 | NONSENSE | W42* | 266 | 259 | 0 | 0.409 | X |
| Chr2 | 209201623 | . | G | A | NM_015040.3 | PIKFYVE | MISSENSE | G1528R | 182 | 174 | 0 | 0.379 | O |
| Chr3 | 52413954 | . | G | A | NM_015512.4 | DNAH1 | MISSENSE | E2471K | 15 | 34 | 0 | 0.294 | O |
| Chr3 | 122247474 | . | T | C | NM_031458.2 | PARP9 | MISSENSE | T768A | 234 | 300 | 0 | 0.563 | O |
| Chr5 | 50685703 | . | C | G | NM_002202.2 | ISL1 | MISSENSE | C234W | 78 | 77 | 0 | 0.455 | X |
| Chr6 | 124604235 | . | G | A | NM_001040214.1 | NKAIN2 | MISSENSE | V47I | 278 | 264 | 0 | 0.371 | X |
| Chr7 | 42962956 | . | C | T | NM_002787.4 | PSMA2 | MISSENSE | G142R | 169 | 176 | 0 | 0.477 | O |
| Chr7 | 99170311 | . | A | T | NM_001083956.1 | ZNF655 | MISSENSE | M229L | 183 | 204 | 0 | 0.309 | O |
| Chr7 | 140453136 | rs113488022 | A | T | NM_004333.4 | BRAF | MISSENSE | V600E | 208 | 294 | 0 | 0.551 | O |
| Chr9 | 14720261 | . | G | A | NM_005454.2 | CER1 | MISSENSE | P211S | 163 | 183 | 0.006 | 0.874 | X |
| Chr10 | 100189389 | . | C | G | NM_000195.3 | HPS1 | MISSENSE | S293T | 64 | 63 | 0 | 0.397 | O |
| Chr11 | 799344 | . | G | A | NM_145886.3 | PIDD | MISSENSE | S899F | 53 | 67 | 0 | 0.373 | O |
| Chr11 | 124766873 | . | G | A | NM_019055.5 | ROBO4 | MISSENSE | R119W | 25 | 23 | 0 | 0.478 | X |
| Chr12 | 49237760 | . | C | G | NM_004818.2 | DDX23 | MISSENSE | D95H | 345 | 269 | 0 | 0.353 | O |
| Chr14 | 21490291 | . | T | A | NM_201537.1 | NDRG2 | MISSENSE | I92F | 148 | 148 | 0 | 0.338 | X |
| Chr18 | 54424349 | . | G | A | NM_015285.2 | WDR7 | MISSENSE | G842D | 317 | 318 | 0.003 | 0.374 | O |
| Chr19 | 6772990 | . | C | T | NM_005428.3 | VAV1 | MISSENSE | R58C | 154 | 139 | 0 | 0.36 | X |
| Chr19 | 13211542 | rs149285767 | C | T | NM_005583.4 | LYL1 | MISSENSE | G119E | 283 | 238 | 0 | 0.176 | O |
| Chr22 | 40417337 | . | C | A | NM_138435.2 | FAM83F | MISSENSE | L275I | 283 | 257 | 0 | 0.416 | X |
| ChrX | 49840621 | . | T | A | NM_001127899.2 | CLCN5 | MISSENSE | I196N | 165 | 141 | 0 | 0.142 | X |
| ChrX | 111698369 | . | A | G | NM_001004308.2 | ZCCHC16 | MISSENSE | K138R | 298 | 290 | 0 | 0.41 | X |
Chr, Chromosome; Ref, Reference; Alt, Alternative; AA, amino acid; VAF, Variant allele frequency.
Loss of Function SNVs and Analysis of Small Indels (Online Resource Section 5)
The candidates for loss of function were selected from the nonsense, splice junction, and frameshift variants (Supplementary Table 3 of Online Resource Section 5).
Whole Chromosome Copy Gains and Losses
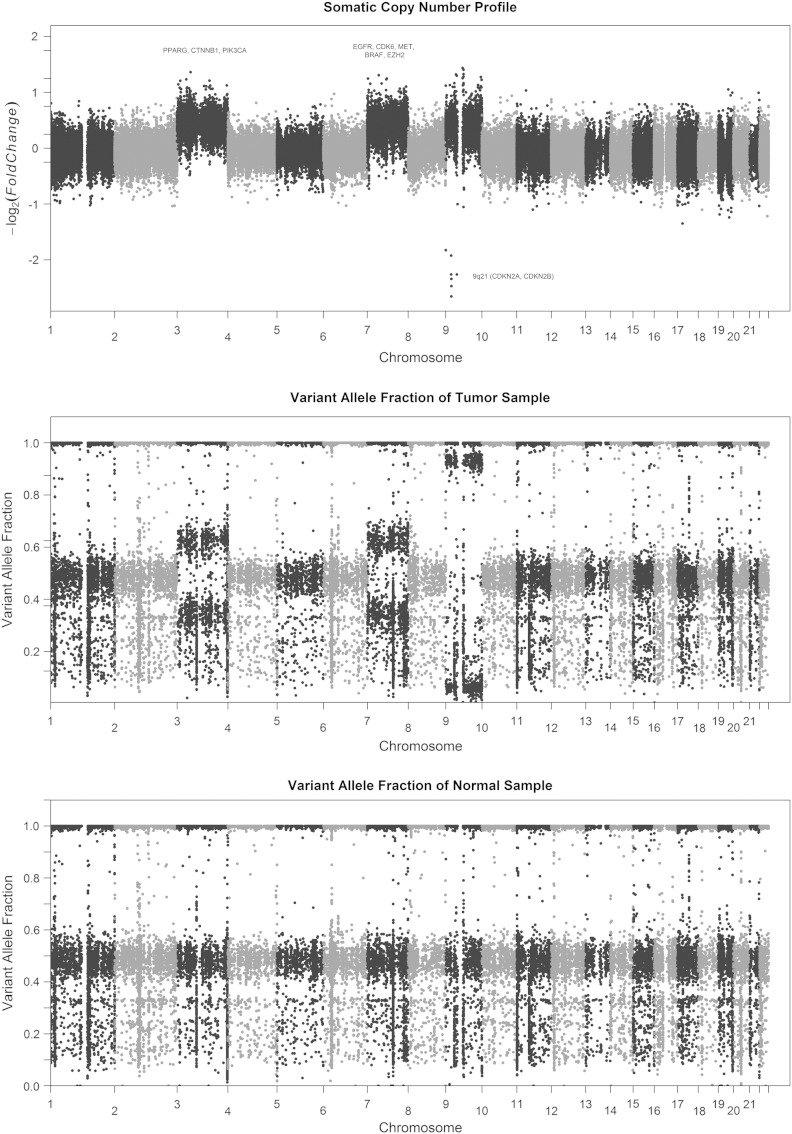
Gains were identified in chromosomes 3, 7, and 9 from the SNP array and WES data (Figure 2). Interestingly, chromosome 9 showed a homozygous deletion of the 9p21 locus that contains the tumor suppressor genes CDKN2A and CDKN2B (Figure 2B).
Figure 2.
Copy number status of R-GBM. (A) Gross copy number changes, (B) variant allele frequency in the tumor sample, and (C) variant allele frequency in the normal sample.
CNA Analysis Using WES and Array-CGH
CNA data were generated from array-CGH and WES. First, CNAs were analyzed with array-CGH with probe-based CNA and interval-based CNA. The probe-based method revealed that more than 80,000 CNA regions were present in tumor tissue compared to paired normal tissue. The interval-based method revealed 370 tumor-specific CNA regions. WES analysis identified 323 regions with CNAs, which included 11 genes that are well-known tumor suppressors and oncogenes [25,26]: VHL, CTNNB1, PIK3CA, EGFR, CDK6, MET, EZH2, MLL3, CDKN2A, CDKN2B, and NOTCH1 (Table 2). In addition, WES, SNP array, and array-CGH analyses showed that CDKN2A/2B were homozygously deleted. Copy number gain was observed for CTNNB1, CDK6, VHL, MLL3, EZH2, PIK3CA, EGFR, NOTCH1, and MET with WES and SNP array analyses.
Table 2.
Genes with Copy Number Alterations that Are Well Known to be Associated with Cancer Development and/or Progression
| Chr | Start | End | Gene | Genetic alteration | aCGH | WES | SNP array | Classification | Expression Ratio1⁎ | Expression Ratio2# |
|---|---|---|---|---|---|---|---|---|---|---|
| Chr3 | 9022276 | 9291369 | VHL | Gain | Yes | Yes | Yes | TSG | − 0.28 | 3.07 |
| Chr3 | 41240942 | 41281939 | CTNNB1 | Gain | Yes | Yes | Yes | Oncogene | − 0.04 | 5.83 |
| Chr3 | 178916609 | 178922393 | PIK3CA | Gain | Yes | Yes | Yes | Oncogene | 1.93 | 2.92 |
| Chr7 | 55086951 | 55214485 | EGFR | Gain | Yes | Yes | Yes | Oncogene | 3.02 | 1.54 |
| Chr7 | 92234235 | 92465941 | CDK6 | Gain | Yes | Yes | Yes | Oncogene | 3.05 | 1.86 |
| Chr7 | 116312459 | 116438440 | MET | Gain | Yes | Yes | Yes | Oncogene | 4.32 | 3.83 |
| Chr7 | 148504464 | 148581441 | EZH2 | Gain | Yes | Yes | Yes | Oncogene | 5.29 | 0.12 |
| Chr7 | 151832010 | 152133090 | MLL3 | Gain | Yes | Yes | Yes | Oncogene | 0.84 | 1.94 |
| Chr9 | 21967751 | 21994490 | CDKN2A | Loss | Yes | Yes | Yes | TSG | − 2.38 | − 1.03 |
| Chr9 | 22005935 | 22009013 | CDKN2B | Loss | Yes | Yes | Yes | TSG | − 3.02 | − 8.89 |
| Chr9 | 139388896 | 139440238 | NOTCH1 | Gain | Yes | Yes | Yes | Oncogene | 2.37 | 2.77 |
Chr, chromosome; aCGH, array comparative genomic hybridization; WES, whole exome sequencing; Del, deletion; TSG, tumor suppressor gene.
Expression ratio 1 is the log2 ratio of the expression level (value in RPKM) in our patient over the mean expression level (value in RPKM) in normal brain (http://www.brainspan.org).
Expression ratio 2 is the log2 ratio of the expression level (value in RPKM) in our patient over the mean expression level (value in RPKM) in glioblastoma multiforme (https://tcga-data.nci.nih.gov/tcga/tcgaHome2.jsp).
Fusions Found With WTS
A total of 376 fusions were observed with WTS with deFuse [19], BreakFusion [20], and ChimeraScan [21]. Interchromosomal fusions and intrachromosomal fusions > 50 kb were selected, and thus, 24 fusions (Online Resource Section 6: Supplementary Table 4) were analyzed further. Among these 24 fusions, in-frame fusions were selected for candidate genetic hallmarks in R-GBM.
Selection of Genetic Hallmarks in R-GBM
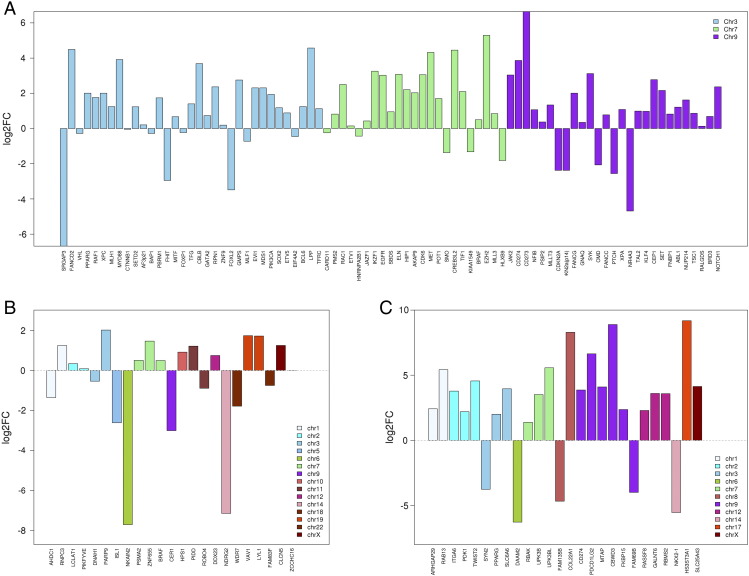
We used WTS data to investigate functional genetic changes in R-GBM, and the public database was used as a reference. First, we compared the RPKM values of specific genetic changes found between tumor and normal brain tissue (Figure 3). We focused on affected genes with more than a 4-fold change in expression and integrated the results among WES, WTS, and array-CGH. Several genes had significant SNVs, CNAs, or fusions. Among genes with SNVs, NDRG2, NKAIN2, CER1, and ISL1 were downregulated, whereas PARP9 was upregulated in the tumor sample of the study patient compared to normal brain tissue. Among genes with CNAs, NOTCH1, EGFR, CDK6, EZH2, and MET were upregulated, whereas CDKN2A and 2B were downregulated in the tumor sample of the study patient compared to normal brain tissue. These results are summarized in Table 3.
Figure 3.
Expression status of selected genetic changes found in the R-GBM sample in comparison with normal brain. (A) genetic changes with copy number alteration, (B) genetic changes with single nucleotide variation, and (C) genetic changes with gene fusion (y axis shows the log2 ratio of the expression level (value in RPKM) in our patient over the mean expression level (value in RPKM) in normal brain (http://www.brainspan.org)).
Table 3.
Genetic Hallmarks of Rhabdoid Glioblastoma.
| Chr | Position | Change | Gene | Amino Acid Change | Ratio1⁎ | Ratio2# | Frequency in GBM | Other Changes Involving This Gene in GBM |
|---|---|---|---|---|---|---|---|---|
| Chr5 | 50678958 | SNV | ISL1 | C234W | − 2.62 | − 6.78 | 0% | Deletion (0.4%) |
| Chr7 | 140453136 | SNV | BRAF | V600E | 0.50 | 1.85 | 1.7% | Amplification (4.4%), deletion (0.2%) |
| Chr14 | 21490291 | SNV | NDRG2 | I92F | − 7.16 | 4.80 | 0% | Amplification (0.4%), deletion (0.6%) |
| Chr3 | 178916609 | CN gain | CDK6 | NA | 3.05 | 1.86 | 7% | Mutation (26%) |
| Chr7 | 55086725 | CN gain | EGFR | NA | 3.02 | 1.54 | 49% | None |
| Chr7 | 116335706 | CN gain | MET | NA | 4.32 | 3.83 | 8.9% | Mutation (0.7%) |
| Chr7 | 148504464 | CN gain | EZH2 | NA | 5.29 | 0.12 | 4.4% | Mutation (1.1%) |
| Chr9 | 139388896 | CN gain | NOTCH1 | NA | 2.37 | 2.77 | 0.6% | None |
| Chr9 | 21968144 | CN loss | CDKN2A | NA | − 2.38 | − 1.03 | 62% | Mutation (0.7%) |
| Chr9 | 22005935 | CN loss | CDKN2B | NA | − 3.02 | − 8.89 | 61% | Mutation (0.4%) |
Chr, chromosome; CN, copy number; GBM, glioblastoma multiforme; SNV, single nucleotide variant; NA, not applicable.
Ratio 1 is the log2 ratio of the expression level (value in RPKM) in our patient over the mean expression level (value in RPKM) in normal brain (http://www.brainspan.org).
Ratio 2 is the log2 ratio of the expression level (value in RPKM) in our patient over the mean expression level (value in RPKM) in glioblastoma multiforme (https://tcga-data.nci.nih.gov/tcga/tcgaHome2.jsp).
The aforementioned analysis was assumed to have identified functional genetic changes in the selected genes. In addition, RPKM values of these selected genes in GBM and R-GBM were compared (genetic changes in GBM were obtained from the TCGA database). We listed genes with more than a 2-fold difference in genetic expression between conventional GBM and R-GBM. The following significant alterations between study samples (R-GBM) and conventional GBMs were found: 1) CER1 and ISL1 had SNVs that were significantly downregulated. 2) CDKN2A and 2B were genes with CNAs and were significantly downregulated. 3) NOTCH1, EGFR, CDK6, PIK3CA, and MET were genes with CNAs and were significantly upregulated. 4) PDK1, RASSF8, FKBP15, GALNT6, ITGA6, SLC6A6, TWIST2,and UPK3BL were significantly up-regulated fusion genes correlated to R-GBM.
Search for GBMs With Similar Genetic Hallmarks in the TCGA Database
In our case, genetic hallmarks excluding fusions are summarized below (Table 3): 1) BRAF V600E; 2) NDRG2 I92F and ISL1 C234W mutation; 3) CDKN2A/2B homozygous loss; and 4) EGFR, CDK6, EZH2, NOTCH1, and MET copy number gain. Subsequently, we searched the cBioportal (TCGA provisional data) for GBM cases that also harbor the above mutations. For mutations, we used WES data (284 samples), and for gene copy number gain/loss, we used array-CGH data (497 samples).
Among 284 GBM cases with sequencing data, the BRAF V600E mutation was found in 1.7% of GBM cases. The NDRG2 mutation was not found in GBM. However, when we searched for NDRG2 genetic changes in the array-CGH database, NDRG2 was amplified in two cases (2/497 = 0.4%) and homozygously deleted in three cases (3/497 = 0.6%). For ISL1, no mutation was found in GBM cases. However, the ISL1 deletion was observed in two GBM cases (2/497 = 0.4%).
Based on array-CGH data from 497 GBM cases, homozygous deletions of CDKN2A and CDKN2B were found in 62% and 61% of the cases, respectively. Copy number gains of EGFR, CDK6, EZH2, NOTCH1, and MET were found in 49%, 7%, 4.4%, 0.6%, and 8.9% of the cases, respectively. Interestingly, NOTCH1 copy number gain, and that of EGFR, CDK6, EZH2, and MET, were mutually exclusive. Two out of three NOTCH1-amplified cases accompanied the CDKN2A/B homozygous deletion. On the other hand, copy number gains in EGFR, CDK6, EZH2, and MET were not mutually exclusive, and co-amplification of these genes was frequently seen.
In three GBM cases with the NDRG2 homozygous deletion, two had simultaneous homozygous CDKN2A and CDKN2B deletions. In addition, two of the three GBM cases with deleted NDRG2 had a NKX2-1 homozygous deletion. Regarding gene amplification, one of the three patients had MET and EZH2 amplifications, and the other patient harbored EGFR amplification.
Among 198 GBM samples with both array-CGH and WES data, only one sample harbored the BRAF mutation, MET, EGFR, and CDK6 amplifications, and CDKN2A/B homozygous deletion at the same time. However, no patient harbored the BRAF mutation and NOTCH1 amplification at the same time. The BRAF mutation and EZH2 amplification were also mutually exclusive. To summarize, although our case of R-GBM is not representative of all R-GBMs, coexistence of the genetic hallmarks found in our patient is a very rare event in GBM.
Discussion
In this study, we addressed the genomic profile of R-GBM, a very rare disease entity. At the chromosomal level, we found copy number gains in chromosomes 3, 7, and 9, and the deletion of 9p21. When we correlate this karyotypic abnormality with genetic changes, we made the following observations.
On chromosome 3, PIK3CA was amplified, and its corresponding expression was elevated compared to normal brain. PIK3CA is frequently altered in GBM, and indeed, amplification of this gene is found in 13% of primary GBMs [27]. Hence, the PIK3CA copy number gain found in our sample was not surprising and implies that a common genetic denominator exists between GBM and R-GBM.
On chromosome 7, EGFR, EZH2, CDK6, and MET had copy number gain, and their expression was elevated compared to that in normal brain tissue. In fact, the gain of chromosome 7 along with EGFR and MET gene amplification is relatively common in adult brain tumors including GBM [28]. In addition, EGFR copy number gain and amplification were observed in a series of R-GBM cases [9,29]. On the other hand, amplification of EZH2 and CDK6 is not commonly observed; only 4% and 7% of GBM cases had amplification of these genes, respectively. Moreover, 1.2% of GBM cases had co-amplification of EZH2 and CDK6, and 0.4% (2 out of 497) of GBM cases also had co-amplification of MET, EZH2, and CDK6 according to the TCGA database. We reviewed the pathology slides of these two cases, which are detailed on the websites (cBioportal case_id=TCGA-06-0187 and cBioportal case ID=TCGA-19-1390). A pathology review of these two cases did not provide a definite diagnostic clue regarding R-GBM. Therefore, we could not draw a definite conclusion regarding whether amplification of one or more of EZH2, CDK6, or MET may be an irrelevant event or an oncogenic driver in the pathogenesis of R-GBM.
On chromosome 9, NOTCH1 copy number gain and associated over-expression were observed. Although the role of the Notch pathway in brain tumors is an area of active investigation, Notch1 signaling is known to promote survival of GBM cells via EGFR-mediated signaling [30]. In addition, Notch signaling has oncogenic potential in a model of medulloblastoma [31]. Hence, we believe that the NOTCH1 copy number gain found in our case may have substantially contributed to oncogenesis and tumor progression. However, only three among 497 cases had NOTCH1 amplification in the TCGA database, which implies that this alteration in NOTCH1 is not a common event in GBM. Regarding the genomic profile of these three cases, two harbored the CDKN2A/2B homozygous deletion as in our case, and one case harbored TP53 and IDH1 missense mutations. As mentioned in the Results section, NOTCH1 amplification was mutually exclusive with EGFR, MET, EZH2, and CDK6 amplification in the TCGA database. Hence, the simultaneous copy number gain in, and over-expression of, NOTCH1, EGFR, MET, EZH2, and CDK6 in our sample is a very interesting phenomenon. What is most interesting regarding NOTCH1 amplification is that one case with NOTCH1 amplification in the TCGA database (cBioportal case id=TCGA-02-2483) had rhabdoid features upon pathology review. Therefore, we believe that further testing for NOTCH1 copy number gain in other R-GBM samples is necessary to confirm whether NOTCH1 is a key factor for rhabdoid morphogenesis.
For chromosome 9, the 9p21 deletion (rather than chromosome 9 copy number gain) was found using WES and SNP microarray. This alteration was not detected with conventional FISH, which confirms the high sensitivity of WES and SNP microarray compared to conventional FISH. Chromosome 9p21 contains CDKN2A and CDKN2B, which are well-known tumor suppressor genes that play an important role in GBM. CDKN2A and CDKN2B were homozygously deleted in this patient, and their expression was correspondingly low. Thus, CDKN2A and CDKN2B may play an important role in our patient.
As for non-synonymous SNVs excluding BRAF V600E, we designated the ISL1 and NDRG2 mutations as genetic hallmarks of R-GBM. We selected these genes for the following reasons. First, gene expression of ISL1 and NDRG2 was significantly reduced compared to expression in normal brain, which implies that these genetic changes are functional. Second, both ISL1 and NDRG2 are biologically relevant to brain tumor development. ISL1 is required for neural development, and expression of this gene is associated with neuroendocrine carcinoma [32,33]. NDRG2 is a well-known tumor suppressor in brain tumors [34]. In contrast to our sample, ISL1 and NDRG2 mutations were not found in the GBM TCGA database. Instead, homozygous deletion of ISL1 (n = 2) and NDRG2 (n = 3) was identified in a small subset (0.4% and 0.6%, respectively) of GBM cases in the TCGA database. As for the NDRG2 deleted cases (n = 3) in the GBM (TCGA database), KIT, PDGFRA, and CHIC2 amplifications were found in two cases (67%). Amplification of other oncogenes including MET, EZH2, CDK4, and EGFR was also identified. Interestingly, for tumor suppressor genes, CDKN2A/2B homozygous deletion (n = 2) and NKX2-1 homozygous deletion (n = 2) were found in NDRG2-deleted GBMs. NKX2-1 was also downregulated in our sample and was fused with ARL6IP4 (Supplementary Table 4). Hence, these phenomena observed in the TCGA database coincide with the genetic changes found in our sample. More importantly, one case with the NDRG2 deletion in the TCGA database (cBioportal case_id=TCGA-02-0281) showed possible GBM with rhabdoid features when we reviewed the histological images. Hence, we believe that loss of NDRG2 function may play an important role in R-GBM pathogenesis.
Finally, comparison of genetic changes in our case with those of ATRT is valuable because ATRT and R-GBM share common morphologic features. First, genes in the SWItch/Sucrose NonFermentable (SWI/SNF) complex, which is a genetic hallmark of ATRT [12,35,36], were not altered in R-GBM as had previously been shown. This finding suggests that although ATRT and R-GBM share common morphologic features, the SWI/SNF complex abnormality is not a key factor for rhabdoid morphogenesis. However, EZH2 over-expression, which was recently shown to be important in ATRT [37], was observed in our sample. EZH2 was both amplified and over-expressed in our sample. Therefore, EZH2 copy number gain and over-expression may play an important role in rhabdoid tumor generation.
Here, we addressed genetic hallmarks found in our R-GBM case including BRAF V600E, ISL1 C234W, NDRG2 I92F, CDKN2A/2B deletion, NOTCH1 copy number gain, and gain of chromosome 7 (including CDK6, MET, EZH2, and EGFR copy number gain). The patterns of mutation and gene expression in R-GBM are rather unique compared to conventional GBM, suggesting that R-GBM is a distinct disease entity. Among these genetic changes, NOTCH1 copy number gain and NDRG2 mutation, which are rare events in the TCGA GBM database, appear to be important genetic markers in R-GBM formation. Furthermore, EZH2 copy number gain and over-expression may play an important role in rhabdoid tumorigenesis.
Footnotes
Funding: This study was supported by a grant from the Ministry for Health, Welfare & Family Affairs, Republic of Korea (HI13C0015) and by grants from the Innovative Research Institute for Cell Therapy, Republic of Korea (A062260).
Conflicts of Interest: The authors have nothing to declare regarding this study.
Location of raw data: The raw data from whole exome and whole transcriptome sequencing are located at ftp gbm.snu.ac.kr.
Supplementary data to this article can be found online at http://dx.doi.org/10.1016/j.tranon.2015.05.003.
Contributor Information
Hongseok Yun, Email: hongseok.yun@samsung.com.
Se-Hoon Lee, Email: shlee119@snu.ac.kr.
Appendix A. Supplementary data
Supplementary materials.
References
- 1.Mutou J, Hirose Y, Ikeda E, Yoshida K, Nakazato Y, Kawase T. Malignant brain tumor with rhabdoid features in an adult. Neurol Med Chir. 2011;51:449–454. doi: 10.2176/nmc.51.449. [DOI] [PubMed] [Google Scholar]
- 2.Momota H, Iwami K, Fujii M, Motomura K, Natsume A. Rhabdoid glioblastoma in a child: case report and literature review. Brain Tumor Pathol. 2011;28:65–70. doi: 10.1007/s10014-010-0010-4. [DOI] [PubMed] [Google Scholar]
- 3.Wyatt-Ashmead J, Kleinschmidt-DeMasters BK, Hill DA, Mierau GW, McGavran L. Rhabdoid glioblastoma. Clin Neuropathol. 2001;20:248–255. [PubMed] [Google Scholar]
- 4.Lath R, Unosson D, Blumbergs P, Stahl J, Brophy BP. Rhabdoid glioblastoma: a case report. J Clin Neurosci. 2003;10:325–328. doi: 10.1016/s0967-5868(03)00030-4. [DOI] [PubMed] [Google Scholar]
- 5.He MX, Wang JJ. Rhabdoid glioblastoma: case report and literature review. Neuropathology. 2011;31:421–426. doi: 10.1111/j.1440-1789.2010.01166.x. [DOI] [PubMed] [Google Scholar]
- 6.Fung KM, Perry A, Payner TD, Shan Y. Rhabdoid glioblastoma in an adult. Pathology. 2004;36:585–587. doi: 10.1080/00313020400010930. [DOI] [PubMed] [Google Scholar]
- 7.Endo S, Terasaka S, Yamaguchi S, Ikeda H, Kato T. Primary rhabdoid tumor with low grade glioma component of the central nervous system in a young adult. Neuropathology. 2013;33:185–191. doi: 10.1111/j.1440-1789.2012.01336.x. [DOI] [PubMed] [Google Scholar]
- 8.Kleinschmidt-DeMasters BK, Alassiri AH, Birks DK, Newell KL, Moore W, Lillehei KO. Epithelioid versus rhabdoid glioblastomas are distinguished by monosomy 22 and immunohistochemical expression of INI-1 but not claudin 6. Am J Surg Pathol. 2010;34:341–354. doi: 10.1097/PAS.0b013e3181ce107b. [DOI] [PubMed] [Google Scholar]
- 9.Babu R, Hatef J, McLendon RE, Cummings TJ, Sampson JH. Clinicopathological characteristics and treatment of rhabdoid glioblastoma. J Neurosurg. 2013;119:412–419. doi: 10.3171/2013.3.JNS121773. [DOI] [PubMed] [Google Scholar]
- 10.Hiroyuki M, Ogino J, Takahashi A, Hasegawa T, Wakabayashi T. Rhabdoid glioblastoma: an aggressive variaty of astrocytic tumor. Nagoya J Med Sci. 2015;77:321–328. [PMC free article] [PubMed] [Google Scholar]
- 11.Kleinschmidt-DeMasters BK, Aisner DL, Birks DK, Foreman NK. Epithelioid GBMs show a high percentage of BRAF V600E mutation. Am J Surg Pathol. 2013;37:685–698. doi: 10.1097/PAS.0b013e31827f9c5e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lee RS, Stewart C, Carter SL, Ambrogio L, Cibulskis K. A remarkably simple genome underlies highly malignant pediatric rhabdoid cancers. J Clin Invest. 2012;122:2983–2988. doi: 10.1172/JCI64400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hasselblatt M, Isken S, Linge A, Eikmeier K, Jeibmann A. High-resolution genomic analysis suggests the absence of recurrent genomic alterations other than SMARCB1 aberrations in atypical teratoid/rhabdoid tumors. Genes Chromosomes Cancer. 2013;52:185–190. doi: 10.1002/gcc.22018. [DOI] [PubMed] [Google Scholar]
- 14.Louis DN. WHO classification of tumours of the central nervous system. International Agency for Research on Cancer; Lyon: 2007. International Agency for Research on Cancer. [309 p. pp.] [Google Scholar]
- 15.Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25:1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.McKenna A, Hanna M, Banks E, Sivachenko A, Cibulskis K. The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010;20:1297–1303. doi: 10.1101/gr.107524.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cingolani P, Platts A, Wang le L, Coon M, Nguyen T. A program for annotating and predicting the effects of single nucleotide polymorphisms, SnpEff: SNPs in the genome of Drosophila melanogaster strain w1118; iso-2; iso-3. Fly. 2012;6:80–92. doi: 10.4161/fly.19695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Huillard E, Hashizume R, Phillips JJ, Griveau A, Ihrie RA. Cooperative interactions of BRAFV600E kinase and CDKN2A locus deficiency in pediatric malignant astrocytoma as a basis for rational therapy. Proc Natl Acad Sci U S A. 2012;109:8710–8715. doi: 10.1073/pnas.1117255109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.McPherson A, Hormozdiari F, Zayed A, Giuliany R, Ha G. deFuse: an algorithm for gene fusion discovery in tumor RNA-Seq data. PLoS Comput Biol. 2011;7:e1001138. doi: 10.1371/journal.pcbi.1001138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chen K, Wallis JW, Kandoth C, Kalicki-Veizer JM, Mungall KL. BreakFusion: targeted assembly-based identification of gene fusions in whole transcriptome paired-end sequencing data. Bioinformatics. 2012;28:1923–1924. doi: 10.1093/bioinformatics/bts272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Iyer MK, Chinnaiyan AM, Maher CA. ChimeraScan: a tool for identifying chimeric transcription in sequencing data. Bioinformatics. 2011;27:2903–2904. doi: 10.1093/bioinformatics/btr467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mortazavi A, Williams BA, McCue K, Schaeffer L, Wold B. Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nat Methods. 2008;5:621–628. doi: 10.1038/nmeth.1226. [DOI] [PubMed] [Google Scholar]
- 23.Shugay M, Ortiz de Mendibil I, Vizmanos JL, Novo FJ. Oncofuse: a computational framework for the prediction of the oncogenic potential of gene fusions. Bioinformatics. 2013;29:2539–2546. doi: 10.1093/bioinformatics/btt445. [DOI] [PubMed] [Google Scholar]
- 24.Van Loo P, Nordgard SH, Lingjaerde OC, Russnes HG, Rye IH. Allele-specific copy number analysis of tumors. Proc Natl Acad Sci U S A. 2010;107:16910–16915. doi: 10.1073/pnas.1009843107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Vogelstein B, Papadopoulos N, Velculescu VE, Zhou S, Diaz LA, Jr., Kinzler KW. Cancer genome landscapes. Science. 2013;339:1546–1558. doi: 10.1126/science.1235122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sottoriva A, Spiteri I, Piccirillo SG, Touloumis A, Collins VP. Intratumor heterogeneity in human glioblastoma reflects cancer evolutionary dynamics. Proc Natl Acad Sci U S A. 2013;110:4009–4014. doi: 10.1073/pnas.1219747110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kita D, Yonekawa Y, Weller M, Ohgaki H. PIK3CA alterations in primary (de novo) and secondary glioblastomas. Acta Neuropathol. 2007;113:295–302. doi: 10.1007/s00401-006-0186-1. [DOI] [PubMed] [Google Scholar]
- 28.Fuller GN, Scheithauer BW. The 2007 Revised World Health Organization (WHO) Classification of Tumours of the Central Nervous System: newly codified entities. Brain Pathol. 2007;17:304–307. doi: 10.1111/j.1750-3639.2007.00084.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Byeon SJ, Cho HJ, Baek HW, Park CK, Choi SH. Rhabdoid glioblastoma is distinguishable from classical glioblastoma by cytogenetics and molecular genetics. Hum Pathol. 2014;45:611–620. doi: 10.1016/j.humpath.2013.08.024. [DOI] [PubMed] [Google Scholar]
- 30.Fassl A, Tagscherer KE, Richter J, Berriel Diaz M, Alcantara Llaguno SR. Notch1 signaling promotes survival of glioblastoma cells via EGFR-mediated induction of anti-apoptotic Mcl-1. Oncogene. 2012;31:4698–4708. doi: 10.1038/onc.2011.615. [DOI] [PubMed] [Google Scholar]
- 31.Natarajan S, Li Y, Miller EE, Shih DJ, Taylor MD. Notch1-induced brain tumor models the sonic hedgehog subgroup of human medulloblastoma. Cancer Res. 2013;73:5381–5390. doi: 10.1158/0008-5472.CAN-13-0033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ehrman LA, Mu X, Waclaw RR, Yoshida Y, Vorhees CV. The LIM homeobox gene Isl1 is required for the correct development of the striatonigral pathway in the mouse. Proc Natl Acad Sci U S A. 2013;110:E4026–E4035. doi: 10.1073/pnas.1308275110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Agaimy A, Erlenbach-Wunsch K, Konukiewitz B, Schmitt AM, Rieker RJ. ISL1 expression is not restricted to pancreatic well-differentiated neuroendocrine neoplasms, but is also commonly found in well and poorly differentiated neuroendocrine neoplasms of extrapancreatic origin. Mod Pathol. 2013;26:995–1003. doi: 10.1038/modpathol.2013.40. [DOI] [PubMed] [Google Scholar]
- 34.Deng Y, Yao L, Chau L, Ng SS, Peng Y. N-Myc downstream-regulated gene 2 (NDRG2) inhibits glioblastoma cell proliferation. Int J Cancer. 2003;106:342–347. doi: 10.1002/ijc.11228. [DOI] [PubMed] [Google Scholar]
- 35.Versteege I, Sevenet N, Lange J, Rousseau-Merck MF, Ambros P. Truncating mutations of hSNF5/INI1 in aggressive paediatric cancer. Nature. 1998;394:203–206. doi: 10.1038/28212. [DOI] [PubMed] [Google Scholar]
- 36.Roberts CW, Leroux MM, Fleming MD, Orkin SH. Highly penetrant, rapid tumorigenesis through conditional inversion of the tumor suppressor gene Snf5. Cancer Cell. 2002;2:415–425. doi: 10.1016/s1535-6108(02)00185-x. [DOI] [PubMed] [Google Scholar]
- 37.Knutson SK, Warholic NM, Wigle TJ, Klaus CR, Allain CJ. Durable tumor regression in genetically altered malignant rhabdoid tumors by inhibition of methyltransferase EZH2. Proc Natl Acad Sci U S A. 2013;110:7922–7927. doi: 10.1073/pnas.1303800110. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary materials.