Abstract
Background
Diabetes and metabolic syndromes are chronic, devastating diseases with increasing prevalence. Human pluripotent stem cells are gaining popularity in their usage for human in vitro disease modeling. With recent rapid advances in genome editing tools, these cells can now be genetically manipulated with relative ease to study how genes and gene variants contribute to diabetes and metabolic syndromes.
Scope of review
We highlight the diabetes and metabolic genes and gene variants, which could potentially be studied, using two powerful technologies – human pluripotent stem cells (hPSCs) and genome editing tools – to aid the elucidation of yet elusive mechanisms underlying these complex diseases.
Major conclusions
hPSCs and the advancing genome editing tools appear to be a timely and potent combination for probing molecular mechanism(s) underlying diseases such as diabetes and metabolic syndromes. The knowledge gained from these hiPSC-based disease modeling studies can potentially be translated into the clinics by guiding clinicians on the appropriate type of medication to use for each condition based on the mechanism of action of the disease.
Keywords: Diabetes, Metabolic disease, Pluripotent stem cells, Genome editing, CRISPR/Cas, Disease modeling
Abbreviations
- APC
antigen presenting cell
- CGL
congenital generalized lipodystrophy
- CRISPR
clustered regularly interspaced short palindromic repeat
- DSB
double-strand break
- DSBR
double-strand break repair
- ER
endoplasmic reticulum
- FPL
familial partial lipodystrophy
- GWAS
genome-wide association study
- hESC
human embryonic stem cell
- hiPSC
human induced pluripotent stem cell
- HLA
human leukocyte antigen
- hPSC
human pluripotent stem cell
- HR
homologous recombination
- MODY
maturity onset diabetes of the young
- NHEJ
non-homologous end joining
- RVD
repeat-variable diresidue
- SDSA
synthesis-dependent strand annealing
- SsODN
single-stranded oligodeoxyribonucleotide
- TALEN
transcription activator-like effector nuclease
- T1D
type 1 diabetes
- T2D
type 2 diabetes
- UPR
unfolded protein response
- ZFN
zinc finger nuclease
1. Introduction
Diabetes and metabolic syndromes are chronic devastating diseases with increasing prevalence. Although a subset of patients develops diabetes and metabolic syndromes due to monogenic mutations, a majority are due to complex gene-environment interactions. Human pluripotent stem cells are gaining popularity in their usage for human in vitro disease modeling. With recent rapid advances in genome editing tools, these cells can now be genetically manipulated with relative ease. In this review, we highlight the diabetes and, metabolic genes and gene variants which could potentially be studied using these two powerful technologies (human pluripotent stem cells and genome editing tools) to aid the elucidation of currently elusive mechanisms underlying these complex diseases.
2. Increased usage of human pluripotent stem cells (hPSCs) for human in vitro disease modeling
Since their discovery in 2007 [1], human induced pluripotent stem cells (hiPSCs) have become a major focus of research [2]. In terms of using hiPSCs for understanding human disease mechanisms, numerous hiPSCs have been generated from patients with a wide variety of diseases [3]. Many of these hiPSCs have been derived from patients with single gene mutations as these monogenic diseases are those that can be modeled with the greatest ease. However, complex diseases such as diabetic cardiomyopathy can also be potentially modeled in vitro [4].
Diabetes, one of the most common metabolic disorders, is a major cause of increased morbidity and mortality for which a cure remains elusive. Classical methodologies for studying diabetes disease mechanisms, such as the use of rodent models, provide significant insights into disease biology. However, it is also known that rodent models do not fully recapitulate the diabetes phenotype in humans, for instance in several types of maturity onset diabetes of the young (MODY) (autosomal dominant) conditions (mice with heterozygous mutations do not develop diabetes) [5] and Wolfram Syndrome [6]. Therefore, hiPSCs could be derived from patients with these forms of diabetes and differentiated into pancreatic beta cells to serve as suitable alternative human models for diabetes research [7,8,105]. To further understand the etiology of more common forms of diabetes, hiPSCs have been derived from both type 1 diabetes (T1D) and type 2 diabetes (T2D) patients [9–11]. However, due to its complexity, T1D and T2D have yet to be convincingly modeled in vitro with these hiPSCs [12]. In this review, we propose and discuss the use of genome editing tools to generate hiPSCs carrying specific gene variants associated with diabetes along with isogenic controls. Once these hiPSCs have been derived, they can then be differentiated into various cell types relevant to diabetes, such as pancreatic cells, skeletal myocytes, adipocytes and hepatocytes, in order to study their biology and for drug screening purposes.
3. Advances in genome editing tools that can be applied to human pluripotent stem cell biology
Genome editing, the genetic engineering (insertion, deletion or replacement) of DNA, is an approach to understand the function of a gene of interest. Genome editing tools such as the conventional homologous recombination (HR), or the more advanced (and advancing) tools such as zinc finger nucleases (ZFNs), transcription activator-like effector nucleases (TALENs) or clustered regularly interspaced short palindromic repeat (CRISPR)/Cas system [13] can be applied to hPSCs in the presence of a donor template to either 1) create naturally occurring mutations or 2) repair a mutation to generate isogenic controls in hPSCs. Compared to ZFNs, TALENs are simpler to engineer [14] and may exhibit greater specificity with less cytotoxicity [15]. The CRISPR/Cas9 system has a significant advantage over ZFN and TALEN as it relies on RNA to home onto DNA whereas ZFN and TALEN depend on custom-making proteins for each specific DNA target. Thus, CRISPR/Cas9 is technically easier to use and is more efficient at cutting target DNA [16]. Given these advantages, coupled with the fact that the CRISPR/Cas9 system is cost-effective and easily accessible in repositories such as Addgene (https://www.addgene.org/), this system has led to an exponential increase in genome engineering of cells and model organisms to study disease biology. The subsequent differentiation of these genetically engineered hPSCs into cell types of interest would thus facilitate the study of disease biology. We will briefly introduce each of these genome editing methodologies.
3.1. Homologous recombination (HR)/Gene targeting
Homologous recombination (HR), or gene targeting, is a method for introducing a particular DNA sequence into a host genome (Figure 1) [17]. When DNA double-strand breaks (DSBs) occur in cells (e.g. due to DNA damage), the DNA can be repaired either via HR or via (error-prone) non-homologous end joining (NHEJ) [18]. DNA repair by HR occurs either via the double-strand break repair (DSBR) pathway or the synthesis-dependent strand annealing (SDSA) pathway. The former commonly results in an exchange of nucleotide sequence(s) between the two strands of DNA (crossover), leading to genetic recombination in contrast to the SDSA pathway, which does not result in a crossover. The natural process of DNA damage and DSBR pathway is the conventional gene targeting method to insert transgenes into the genome without the use of nucleases (ZFNs, TALENs or CRISPR/Cas).
Figure 1.
Genome editing methodologies which can be applied to human pluripotent stem cells. Homologous recombination (HR), or the more advanced tools such as zinc finger nucleases (ZFNs), transcription activator-like effector nucleases (TALENs) or clustered regularly interspaced short palindromic repeat (CRISPR)/Cas system can be applied to human pluripotent stem cells (hPSCs) either to 1) create naturally occurring mutations or 2) repair a mutation to generate isogenic controls in hPSCs, to understand the function of a gene of interest.
3.2. Zinc finger nuclease (ZFN)
The first advancement in genome editing arose from the discovery of zinc finger nucleases (ZFNs), synthetic restriction enzymes containing DNA-binding domain(s) fused to a DNA-cleavage domain (typically a non-specific cleavage domain from bacterial type IIS FokI restriction enzyme; located at the C-terminus) [19]. These DNA-binding domains, which contain 3–6 zinc finger repeats and are able to recognize 9–18 base pairs, allow homing to target DNA followed by DNA cleavage (Figure 1) [20]. For DNA cleavage to occur, however, the cleavage domain needs to dimerize, necessitating a pair of ZFNs to bind opposite strands of DNA and flank a region of interest. ZFNs induce DNA DSBs and stimulate the endogenous DNA repair machinery [21], which, in the presence of a homologous donor template, can result in a replacement of the target allele. In contrast, NHEJ operates in the absence of a homologous donor template. This strategy may be suitable for disabling autosomal dominant mutations present in hiPSCs derived from diseased patients.
3.3. Transcription activator-like effector nuclease (TALEN)
Following on the heels of the use of ZFNs, transcription activator-like effector nucleases (TALENs) were discovered. TALE is a Xanthomonas bacterial protein containing a domain of tandem repeats (33–34 amino acids), which can target DNA. These tandem repeats contain repeat-variable diresidues (RVDs; 12th and 13th amino acid), which dictate DNA-binding specificity. By combining the appropriate RVDs, TALEs can be custom-made to target DNA. When fused to a DNA-cleavage domain (e.g. FokI endonuclease), TALE nucleases (TALENs) are generated [22]. Like ZFNs, a pair of TALENs induces DSBs and relies upon HR or NHEJ, to either introduce foreign DNA for gene repair or disrupt endogenous gene function, respectively (Figure 1).
3.4. Clustered regularly interspaced short palindromic repeat (CRISPR)/Cas
Very recently, another technological breakthrough led to the discovery of clustered regularly interspaced short palindromic repeats (CRISPRs), a repetitive pattern (between 24 and 48 base pairs) in which DNA sequences would be followed by nearly the same sequence in reverse orientation followed by spacer DNA. This unique pattern, first identified in E. coli bacteria, which integrated the sequence of phages (spacer DNA) to fend off future attacks, was often found to be associated with CRISPR-associated (Cas) genes. Upon phage attack, the bacterium first transcribes the spacer and palindromic DNA into a long RNA molecule. This RNA molecule is then cleaved by RNase III, involving Cas9 (a nuclease which has two active sites for cutting each strand of the DNA double helix) and trans-activating CRISPR RNA (tracrRNA), into short spacer-derived RNA (CRISPR RNA; crRNA) in the cell [23].
tracrRNA and crRNA can now be intelligently combined into a “guide RNA” molecule, and when combined with Cas9 could target specific DNA and cut accurately (Figure 1) [24]. CRISPR/Cas9 has also been modified into CRISPR interference (CRISPRi), which can reversibly turn off genes by targeting a site without cutting it (Cas9 nuclease rendered inactive), or the inclusion of activators or repressors (synthetic transcription factors) to regulate gene activity at promoter regions [25,26]. Besides the constitutive expression of CRISPR/Cas9, there are alternative systems, which allow for conditional induction via the doxycycline- or Tet-regulated Cas9 system. Current CRISPR/Cas systems are from Streptococcus pyogenes, Streptococcus thermophilus, Neisseria meningitides and Treponema denticola.
3.5. Caveats of advanced genome editing tools
Off-target effects. The DNA-binding domains of ZFNs and TALENs need to be very specific for the target site to avoid off-target cleavage, which results in unwanted mutations and potentially cytotoxic effects [27]. CRISPR/Cas9 is also known to generate off-target alterations, albeit apparently at low incidence [28,29], since mispairing is allowed between the guide RNA and the genomic DNA. Nonetheless, caution is required in their design and use. Some strategies involving the optimization of the guide RNA/Cas9 include using of software tools to predict potential off-target sites (http://omictools.com/crispr-cas9-c1268-p1.html), truncating the guide RNA (<20 nucleotides) to decrease off-target mutagenesis [30], lowering the dosage of guide RNA and Cas9 plasmids, and decreasing the number of mismatches between the guide RNA and the genomic DNA. A “double nick” system with Cas9 nickase, which contains a single inactive catalytic domain, may also be used [31–33].
Repeated cleavage of a repaired site. Due to the high cleavage efficiency of these genome editing tools, there is a possibility of re-cutting a repaired site [34]. One way to mitigate this is to introduce silent mutations in the donor template to minimize subsequent nuclease binding [35].
3.6. Single-stranded oligodeoxyribonucleotide (ssODN)
Since the generation of conventional donor template gene targeting constructs for conventional HR is time consuming, single-stranded oligodeoxyribonucleotides (ssODNs) are currently being employed in concert with these advanced genome editing tools for high frequency “footprint-free” genome editing [36]. These genome editing tools act by introducing a sequence-specific DSB within the ssODN homology region (thus increasing recombination frequency) and facilitating HR via the DSBR pathway using ssODN as a template [37].
SsODNs are short synthetic nucleotides designed to be either exactly complementary to the target DNA sequence (for gene correction) or contain nucleotide mismatch(es), insertions or deletions. SsODNs are very simple to use, as opposed to conventional donor template constructs (plasmids or viral vectors), predicted to have high safety and fidelity with minimal immunotoxicity, and do not appear to have off-target effects [36]. Studies combining ZFNs with ssODN were among the first to suggest the possibility of high frequency genome editing [38] and have been successfully applied to Parkinson's disease-hiPSCs [39].
4. Coupling human pluripotent stem cells and genome editing tools to investigate diabetes and metabolic disease mechanisms
4.1. Examples of studying disease mechanisms with hPSCs and genome editing tools
Traditional HR can be used for gene correction to demonstrate genotype-phenotype causality albeit with very low rates of efficiency. ZFNs were first demonstrated in 2007 to be capable of gene editing in human embryonic stem cells (hESCs) [40]. Their ability to enhance HR-mediated gene targeting [41,42] has led to their use in multiple studies including targeted gene disruption at the CCR5 locus (HIV-1 resistance) [43] and correction in the α-synuclein gene (susceptibility variant for Parkinson's disease) [39], α1-antitrypsin [44], sickle cell anemia mutation [35,45], gp91(phox) (X-linked chronic granulomatous disease) [46], α-thalassemia genes [47] and glucokinase (GCK) [7]. Shortly after, TALENs were employed for genetic engineering in hPSCs [48,49] for correcting the COL7A1 [50] and α1-antitrypsin genes [51]. Given the superior cutting efficiency, CRISPR/Cas9 is increasingly becoming the favored choice for genome editing in hPSCs [16,52].
4.2. Employing hPSCs and genome editing tools to study diabetes and metabolic syndromes
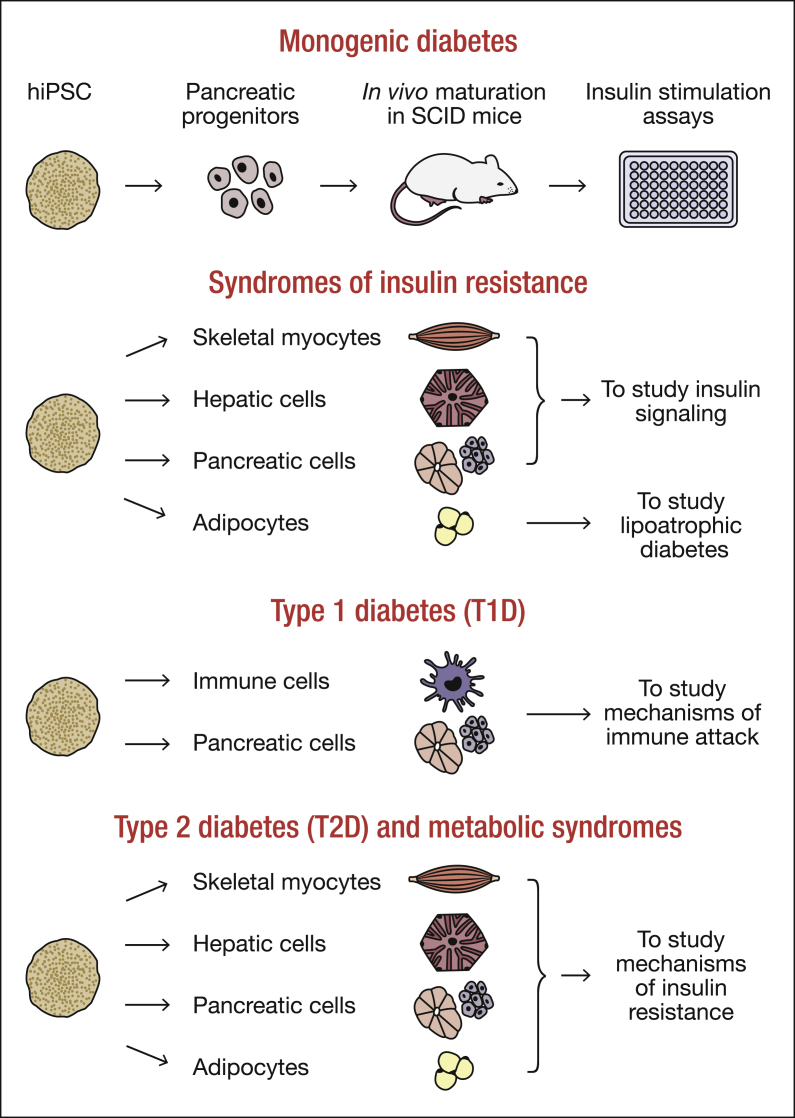
In general, the strategy to carry out in vitro disease modeling of diabetes and related metabolic syndromes with hPSCs and genome editing tools would be to 1) derive hiPSCs from patients with these conditions, 2) generate “repaired/corrected” isogenic controls [53] and then 3) differentiate them into pancreatic cells or target cells of relevance, such as immune cells in the case of T1D or myocytes, adipocytes and hepatocytes in the case of T2D (Figure 2). If patient material is inaccessible, one could introduce (naturally occurring) mutations or gene variants into hESCs and differentiate them accordingly to study disease mechanisms. Since excellent reviews have been published recently, we will provide a brief overview to familiarize the reader with the classification of diabetes and metabolic disorders.
Figure 2.
Schematic showing the use of human pluripotent stem cells from patients with diabetes and metabolic diseases to study disease mechanisms in vitro. Human induced pluripotent stem cells (hiPSCs) derived from patients with monogenic diabetes, syndromes of insulin resistance, type 1 diabetes (T1D), type 2 diabetes (T2D) and metabolic syndromes can be differentiated into pancreatic cells, skeletal myocyotes, hepatic cells, adipocytes and immune cells, where relevant, to study disease mechanisms in vitro.
4.3. Genetic determinants of diabetes and metabolic syndromes
Diabetes is a metabolic disease characterized by uncontrolled hyperglycemia resulting from the variable combination of dysfunctional insulin secretion by pancreatic beta cells and insulin resistance. It is generally classified into monogenic diabetes (maturity onset diabetes of the young [MODY], neonatal diabetes, mitochondrial diabetes [54,55], syndromes of insulin resistance) [56], type 1 diabetes (T1D) and type 2 diabetes (T2D). The metabolic syndrome is a combination of conditions including central obesity (as measured by waist circumference), dyslipidemia (high triglycerides, reduced high density lipoprotein cholesterol), hypertension and elevated fasting plasma glucose. These conditions increase the risk of diabetes and atherosclerotic vascular disease. The genetic variants responsible for monogenic diabetes are typically rare and have high penetrance whereas the variants involved in the etiology of the complex forms of T1D and T2D are relatively common and have low penetrance. Some of the known genetic determinants of each of these types of diabetes and metabolic syndromes are summarized in Table 1.
Table 1.
Genetic determinants of diabetes and metabolic syndromes.
| Monogenic forms of diabetes | ||
| Type of monogenic diabetes | Gene(s) | |
| MODY | MODY1 (HNF4A)(>30 mutations) MODY2 (GCK)(>600 mutations) MODY3 (HNF1A)(>200 mutations) MODY4 (PDX1) MODY5 (HNF1B)(>65 mutations) MODY6 (NEUROD1), MODY7 (KLF11), MODY8 (CEL), MODY9 (PAX4), MODY10 (INS), MODY11 (BLK), MODYX (GATA6), MODYX (ABCC8) |
|
| Neonatal diabetes | KCNJ11, ABCC8 and many others | |
| Mitochondrial diabetes | tRNA(Leu)(UUR)(A3243G) | |
| Syndromes of insulin resistance | Defects in insulin signaling pathway | Donohue syndrome, Rabson-Mendenhall syndrome, Type A insulin resistance (INSR) AKT2, TBC1D4 |
| Lipoatrophic diabetes | Congenital generalized lipodystrophy (AGPAT2, BSCL2, PTRF, CAV1) Familial partial lipodystrophy (LMNA, PPARG, AKT2, CIDEC, PLIN1) |
|
| Monogenic forms of obesity | LEP, LEPR, POMC, PCSK1, MC4R, SIM1, BDNF, NTRK2 | |
| Type 1 diabetes (T1D) | ||
| Candidate gene(s) | Specific associated variant(s)/single nucleotide polymorphisms (SNPs) | |
| HLA DR-DQ, HLA-B, HLA-A, INS | ||
| PTPN22 | R620W, 1858C/T | |
| IL2RA/CD25 | rs706778, rs3118470, rs41295061, rs35285258 | |
| CTLA4 | A17T | |
| IFIH1/MDA5 | rs1990760 | |
| CLEC16A/KIAA0350 | rs2903692, rs725613, rs17673553 | |
| ERBB3 | rs2271189, rs11171747, rs2292399 | |
| PTPN2 | rs2542151, rs1893217 | |
| UBASH3A/STS2 | T946A | |
| IL18 | rs1946519, rs1946518, rs187238 | |
| RANTES | rs4251719, rs2306630, rs2107538 | |
| VDR | rs1544410, rs2228570 | |
| CIITA | rs11074932, rs3087456 | |
| CYP27B1 | rs4646536 | |
| EFR3B | rs478222 | |
| CD226 | rs763361 | |
| LMO7 | rs539514 | |
| DLK1 | rs941576 | |
| SLC11A1 | rs3731685 | |
| SH2B3, IL27, BACH2, PRKCQ, IL10, CD69, CYP27B1, CTSH, TNFAIP3, STAT4, IL18RAP, TAGAP, RNLS, IL2, C1QTNF6, CRYAB, CTBR2, ERAP1/ARTS1, FHOD3, GLIS3, HERC2, HTR1A, IL19, IL20, IL26, ITPR3, PIP4K2C, RASGRP1, RFN180, RGS1, SIRPG, SKAP2, STAT3, TNFA, TNFB, Xp22 | ||
| Type 2 diabetes (T2D) | ||
| Candidate gene(s) | Specific associated variant(s)/single nucleotide polymorphisms (SNPs) | |
| TCF7L2 (strongest) | rs7903146, rs7901695, rs12255372, rs10885409, rs12573128 | |
| PPARG | Pro12Ala (P12A)/rs1801282 | |
| KCNJ11 | Glu23Lys (E23K)/rs5219, C42R | |
| HHEX/IDE | rs1111875, rs7923837, rs5015480, rs7923837 | |
| SLC30A8 | Arg325Trp/rs13266634, rs3802177, rs2466293 | |
| CDKAL1 | rs7756992, rs7754840, rs10946398, rs9465871, rs4712523, rs4712524, rs6931514 | |
| IGF2BP2 | rs4402960, rs1470579, rs6769511 | |
| CDKN2A/B | rs10811661, rs1412829 | |
| ABCC8 | Ala1369Ser (A1369S)/rs757110, Y356C | |
| MTNR1B/ADCY5 | rs10830963, rs1387153, rs2877716, rs1374645, rs2166706, rs10930963 | |
| KCNQ1 | rs2237892, rs2237895, rs2237897, rs231362, rs2283228, rs163182, rs2299620 | |
| GCKR | P446L, rs780094, rs1260326 | |
| GCK | rs1799884, -30G/A polymorphism in promoter, rs4607517 | |
| HNF1A | G319S, rs7957197 | |
| ADAMTS9 | rs4607103 | |
| G6PC2 | rs560887, rs552976 | |
| JAZF1 | rs864745 | |
| CDC123/CAMK1D | rs12779790, rs10906115 | |
| IRS1 | rs2943641 | |
| WFS1 | rs4689388, rs1801214 | |
| DUSP9 | rs5945326 | |
| KLF14 | rs972283, rs4731702 | |
| C2CD4A/C2CD4B | rs1436955, rs1370176, rs1436953, rs7172432 | |
| PTPRD | rs17584499 | |
| PROX1 | rs340874 | |
| BCL11A | rs10490072 | |
| RBMS1 | rs7593730 | |
| THADA | Thr1187Ala/rs7578597 | |
| ARAP1 (formerly CENTD2) | rs1552224 | |
| IGFBP2 | rs1470579 | |
| PAX4 | rs10229583 | |
| PAX6 | rs685428 | |
| VEGFA | rs9472138 | |
| VEGF | rs6921438 | |
| HNF1B, HNF4A, DGKB/TMEM195, HMGA2, MADD, AP3S2, CHCHD9, GRB14, HMG20A, LARP6, SGSM2, ST6GAL1, TP53INP1, VPS26A, ZBED3, UBE2E2, ZFAN6, TSPAN8/LGR5, NOTCH2/ADAMS30, ZFAND3 | ||
| Obesity/Metabolic syndrome | ||
| Candidate gene(s) | Specific associated variant(s)/single nucleotide polymorphisms (SNPs) | |
| FTO | rs8050136, rs1121980, rs17817449, rs9941349, rs3751812, rs1421085, rs17820875, rs860713, rs1558902, rs9935401, rs9930506, rs6499640, rs11642015, rs62048402, rs57103849, rs16945088, rs9930333, rs8057044, rs12447427, rs7202116 | |
| MC4R | rs17782313, rs12970134, rs17700633, rs571312, rs17773430, rs11872992, rs17782312, rs1942872 | |
| TMEM18 | rs6548238, rs7561317, rs4854344, rs2867125 | |
| NEGR1 | rs2815752, rs3101336, rs9424977, rs2568958 | |
| GNPDA2 | rs10938397 | |
| SH2B1 | rs7498665, rs4788102, rs7201929, rs147094247, rs60604881, rs62037368, rs62037369 | |
| KCTD15 | rs29941 | |
| MTCH2 | rs4752856, rs10838738, rs3817334, rs7120548 | |
| BDNF | rs925946, rs6265, rs4923461, rs10968576, rs988712 | |
| SEC16B | rs10913469, rs543874 | |
| APOA5 | rs662799 | |
| FAIM2/BCDIN3 | rs7138803, rs7132908 | |
| TFAP2B | rs987237 | |
| QPCTL/GIPR | rs2287019, rs10423928 | |
| LYPLAL1/SLC30A10 | rs4846567, rs2605100, rs11118316 | |
| HMGCR | rs7703051, rs12654264, rs3846663, rs3846662 | |
| PTP-1B/PTPN1 | rs718049, rs2282146, rs1885177, 1484insG, P303P, P387L | |
| CD36 | rs1049673, rs3211931, rs3211938, rs1194197, rs1761667 | |
| SFRS10-ETV5-DGKG | rs7647305 | |
| MSRA/TNKS | rs545854 | |
| NPC1 | rs1805081 | |
| MAP2K5/SKOR1 | rs2241423 | |
| NRXN3 | rs10146997, rs11624704 | |
| PCSK1 | rs6235, rs1799904 | |
| GRB14 | rs13389219 | |
| VEGFA | rs6905288, rs9472138 | |
| INPPL1/SHIP2 | rs2276047, rs9886, rs2276048 | |
| AHSG | rs2077119, rs4917 | |
| GHRL | rs696217, rs26802 | |
| NISCH/STAB1 | rs6784615 | |
| LEPR | rs1137101 | |
| ADAMTS9 | rs13060013 | |
| NUDT3 | rs206936 | |
| CDKAL1 | rs2206734 | |
| CPEB4 | rs6861681 | |
| MAF | rs7192960 | |
| TNNI3K | rs1514175 | |
| LPIN1 | rs11693809 | |
| IL6 | −597 G/A, −572 G/C, −174 G/C | |
| LEP | rs10954174, rs6966536 | |
| ENPP1 | rs7754561 | |
| GNAT3 | rs11760281 | |
| GPR120 | R270H | |
| KLF9 | rs11142387 | |
| LRP1B | rs2890652 | |
| MTIF3 | rs4771122 | |
| NPY | L7P | |
| PFKP | rs6602024 | |
| PRKD1 | rs10144903 | |
| THNSL2 | rs1659258 | |
| RSPO3, ITPR2/SSPN, INSIG2, TBX15/WARS2, LY86, NFE2L3, SDCCAG8, PTER, BEACON/UBL5, ADCY3/DNAJC27, ADIPOQ, AIF1, AKT1, BAT2, CADM2, DNM3/PIGC, GPRC5B, GP2, HNF4A, HOXC13, KLF14, LRRN6C, PACS1, PAX6, POMC, PRKCH, PRL, PTBP2, RBJ, RMST, SLC39A8, TMEM160, TRK, ZNF608, ZNRF3/KREMEN1, 1q25 | ||
4.3.1. MODY, neonatal diabetes and mitochondrial diabetes
Patients with MODY, neonatal diabetes and mitochondrial diabetes (Table 1) commonly exhibit impairments in one or more of the three main beta cell functions: 1) glucose sensing, 2) making and storing insulin, and 3) secreting insulin on demand. For instance, mutations in genes such as GCK [57] and glucose transporter SLC2A2 (also known as GLUT2) [58] result in altered glucose sensing, which, in turn, results in an increased blood glucose threshold for insulin secretion, decreased hepatic glycogen storage, and increased hepatic gluconeogenesis [59–61]. Mutations in several transcription factors result in defects in pancreatic beta cell development, which directly impact insulin synthesis and function while mutations in the insulin gene (INS) obviously affect the key hormone made by pancreatic beta cells [62]. ATP synthesis defect (mitochondrial diabetes) and mutations in ATP-sensitive potassium channel subunits (channel-building Kir6.2 [potassium inwardly-rectifying channel, subfamily J, member 11; KCNJ11] and regulatory SUR1 [ATP-binding cassette transporter subfamily C member 8], ABCC8) all affect insulin secretion [63].
One strategy to study these monogenic syndromes would be to derive hiPSCs from these patients [105], differentiate them into pancreatic progenitors and then transplant these progenitors into immunocompromised (SCID-Beige or NSG) mice for in vivo maturation (Figure 2). This methodology has been recently used to successfully model MODY2, demonstrating that beta cells derived from hiPSCs with GCK mutation are indeed less sensitive to glucose levels [7]. Endoplasmic reticulum (ER) stress-related diabetes in patients with Wolfram syndrome has also been modeled using hiPSC-derived beta cells, demonstrating that WFS1 protein maintains ER function in beta cells by acting upstream of the unfolded protein response (UPR) pathways [8]. These two studies indicate that hiPSC-derived beta cells can indeed recapitulate diabetic phenotypes occurring in humans. Likewise, the stepwise analysis of human pancreatic development with this strategy would likely provide mechanistic insights into the ability of a single gene mutation (PDX1, PTF1A, HNF1B, GATA6 and GATA4) to promote pancreatic agenesis/atrophy. Further, studying mutations in KCNJ11 and ABCC8 using hiPSC-derived beta cells may elucidate the mechanistic differences between permanent and transient neonatal diabetes [64]. Overall, insulin production and secretion could be compared between diseased and gene-corrected pancreatic cells to understand the underlying cause of each type of monogenic diabetes (Figure 2).
4.3.2. Syndromes of insulin resistance
Syndromes due to defects in the insulin signaling pathway as well as lipoatrophic diabetes could be modeled using hiPSC and genome editing technologies. hiPSCs from patients with defective insulin signaling molecules such as INSR [65], v-akt murine thymoma viral oncogene homolog 2 (AKT2) and TBC1 domain family member 4 (TBC1D4) could be differentiated into skeletal myocytes, adipocytes, hepatic and pancreatic cell types to study abnormalities in development [66] (Figure 2) and to distinguish cell autonomous versus non-cell autonomous effects. Outstanding questions that could be addressed include: 1) studying mechanisms that underlie hyperglycemia and hyperinsulinemia in individuals with INSR mutations, 2) dissecting the mechanistic differences between autosomal dominant and recessive gene mutations that cause insulin resistance of varying degrees, 3) interrogating the role of AKT2 in beta cell function, and 4) exploring why INSR mutations do not result in hepatosteatosis and dyslipidemia in contrast to other forms of severe insulin resistance. hiPSCs from patients with lipoatrophic diabetes could be differentiated into adipocytes to study developmental defects (Figure 2). Monogenic forms of insulin resistance that result into lipoatrophic diabetes can be subclassified into congenital generalized lipodystrophy (CGL) and familial partial lipodystrophy (FPL) [67] (Table 1). In the former, gene mutations typically affect various aspects of adipocyte biology such as the biosynthesis of triglycerides [68] or adipocyte differentiation [69], and hiPSCs from these patients could be a source for investigating the role of the mutated genes in adipocyte biology, lipoatrophic diabetes and insulin resistance. Considering the significant genotypic and phenotypic heterogeneity in FPL, hiPSCs with LMNA and PPARG mutations can be used to study dysfunction in fat metabolism whereas cells with CIDEC and PLIN1 mutations can be used to study triglyceride mobilization and lipid droplet dynamics.
4.3.3. Monogenic forms of obesity
Several gene mutations involved in these syndromes affect satiety and body weight [70], making the brain-fat inter-organ cross-talk challenging to model. However, one could consider examining the effects on adipocyte development using hiPSCs.
4.4. Employing hPSCs and genome editing tools to study type 1 diabetes (T1D)
Patients with T1D are unable to secrete insulin due to near complete destruction of their pancreatic beta cells. More than 50 risk variants/susceptibility alleles have been found to be associated with susceptibility to this disease [71] (https://www.niddkrepository.org/studies/t1dgc/) (Table 1). The strongest association is with the human leukocyte antigens (HLAs), which accounts for a large proportion of the genetic risk for T1D [71]. Most of the T1D genes affect adaptive and innate autoimmunity leading to incomplete self-tolerance to beta cell antigens and immune-mediated destruction of beta cells [71]. T1D-hiPSCs can be differentiated into T lymphocytes [72–74] and pancreatic beta cells [75–77] to allow co-culture experiments aimed at progressively evaluating their interactions in vitro (Figure 2) [78]. A similar strategy can be applied to hiPSCs derived from T1D-susceptible patients to examine the impact of susceptible gene variants (Table 1) on the vulnerability of pancreatic beta cells to immune attack. For instance, hiPSCs derived from patients with a gene variant in PTPN22 can be differentiated into lymphocytes to study lymphocyte function [79–81]. hiPSCs from subjects with gene variants in ERBB3, which is expressed in monocytes and dendritic cells, and may affect antigen presenting cell (APC) function [82], can be differentiated into selective immune cells to study how they affect APC function. hiPSCs from patients with gene variants in UBASH3A (also known as STS2), which is specifically expressed in lymphocytes [83], are well suited for differentiation into lymphocytes to study the function of this gene.
4.5. Employing hPSCs and genome editing tools to study type 2 diabetes (T2D) and metabolic syndromes
T2D results from a combination of genetic and environmental effects. T2D patients, characterized by early insulin resistance followed by a failure of pancreatic beta cells to compensate, develop uncontrolled hyperglycemia. The association of obesity with insulin resistance suggests that excess adipose tissue could contribute to the insulin resistance [84]. The poor ability of skeletal myocytes and adipocytes to utilize glucose and fatty acids, and to store them as glycogen and triglycerides (respectively) in states of insulin resistance lead to hyperglycemia and dyslipidemia. Insulin resistant hepatocytes respond poorly to insulin, fail to synthesize and store glycogen, and, with rising glucagon levels, continue to produce glucose leading to hyperglycemia. Patients with T2D exhibit defects in glucose-stimulated insulin secretion and a gradual decrease in pancreatic beta cell mass that, combined, fail to sustain the increased demands of insulin resistance. Thus, T2D-hiPSCs can be differentiated into skeletal myocytes, adipocytes and hepatocytes to study insulin resistance (independent of organ cross-talk) in each of these tissues, and into pancreatic beta cells to study mechanisms underlying cell death, de-differentiation and/or altered secretory function in T2D [85,86] (Figure 2).
To date, at least 100 risk variants/susceptibility alleles have been associated with T2D and/or quantitative glycemic traits (fasting glucose, fasting insulin, HOMA-B and HOMA-IR) [87] (Table 1). Although the cumulative effects of these variants appear to explain only 5–10 % of the familial aggregation of T2D [88], understanding the mechanisms by which they increase T2D risk has important implications for the development of new therapies. The use of hPSCs and advanced genome editing tools can potentially facilitate this goal. For example, the transcription factor-7 like 2 (TCF7L2) gene, a transcription factor reported to be important for beta cell survival and function [89], contains variants that exhibit the strongest association with T2D [90]. Gene variants in TCF7L2 influence the therapeutic response to sulfonylureas [91] and have been demonstrated to modulate other aspects of pancreatic islet function [92]. The poorly understood mechanisms underlying these effects potentially could be investigated by using hPSCs and genome editing. For instance, one can leverage upon hiPSCs derived from T2D-susceptible patients based on their TCF7L2 genotype, and differentiate them into pancreatic beta cells to answer whether TCF7L2 gene variants affect development and provide clues to the initial beta cell mass at adulthood. Related to the two cases of hiPSC-based modeling of MODY2 [7] and Wolfram Syndrome [8], gene variants found in GCK, GCKR and WFS1 have been linked to T2D risk. Thus, it may be possible to extrapolate these findings and begin investigating the functional effects of associated gene variants. A similar approach could be adapted to investigate the role of other variants (Table 1) with appropriate readouts. For instance, hiPSCs with a defined gene variant and their gene-corrected isogenic control hiPSCs could be differentiated simultaneously and then subjected to high throughput analyses to determine developmental or expression profile differences. This is expected to reveal gene targets affected by the presence of the specific gene variant and could thereby shed light on mechanistic insights into the predisposition towards T2D. One caveat, though, would be the specific cell type of action of the identified genome-wide association study (GWAS) gene. For example, gene variants in the HHEX gene could possibly affect pancreatic beta cell, delta cell or even hepatocyte biology, thereby predisposing individuals toward developing T2D.
More than 30 risk variants/susceptibility alleles have been associated with waist circumference, waist-hip ratio, BMI and obesity (Table 1), and are estimated to account for 2% of the occurrence of obesity [87]. The fat mass and obesity-associated (FTO) locus exhibits a strong association with BMI and obesity [93]. Other variants including those found near melanocortin 4 receptor (MC4R) which can explain 2–3 % of cases of severe obesity [94] and proopiomelanocortin (POMC) [95], are strong biological candidates for BMI and obesity.
Compared to gene variants that affect satiety and body weight [96], those affecting adipocyte development and function are relatively easier to model because of a definitive readout in the latter. For instance, FTO is a potential candidate for studies in hiPSC-derived adipocytes since its expression has been reported to decrease during adipocyte differentiation [97]. In common obesity, circulating leptin (LEP) levels are high, indicating leptin resistance [98,99]. Differentiating hiPSCs derived from those with risk variants in the LEP gene into adipocytes can aid in increased understanding of leptin regulation of adipocyte function. Genes such as TMEM18, KCTD15, MTCH2, BCDIN3, SULT1A1, PTER, TUFM, FAIM2, BDNF, NEGR1 and NPC1 with potential regulatory roles in the adipose tissue [100,101] could be studied by independently differentiating hiPSCs into adipocytes to study the specific gene function(s). In addition, TMEM18, PTER, SULT1A1 and TUFM may be studied in hepatocytes whereas SH2B1 and TUFM could be studied in skeletal myocytes [102]. However, it should be cautioned that genes currently associated with the risk-conferring polymorphisms may not actually be the only genes involved in the polymorphism, as in the case of FTO and IRX3 [103].
5. Conclusions
hPSCs and the advancing genome editing tools appear to be a timely and potent combination for probing molecular mechanism(s) underlying diseases such as diabetes and metabolic syndromes. Studying monogenic forms of diabetes and syndromes of insulin resistance using these tools would be extremely useful given the lack of an autoimmune attack and confounding effects of insulin resistance and obesity. One caveat of this methodology at the moment is the “low” efficiency of deriving human beta cells in vitro [75,76], possibly due to our incomplete knowledge on human pancreatic development. Another explanation would be the lack of in vivo environmental cues emanating from proximal tissues such as the vasculature. Nonetheless, successful disease modeling of MODY2 [7] and Wolfram Syndrome [8] already suggests a high possibility of success. These technologies have the potential to elucidate the underlying pathophysiology that stem from defects in 1) beta cell development, metabolism or survival or 2) development of adipocyte. For instance in the case of MODY2, it is now clear that GCK mutation affects glucose-stimulated insulin secretion but not insulin synthesis or beta cell proliferation [7]. With the latest advances in the derivation of mature and functional human pancreatic beta-like cells from hPSCs in vitro [75–77], eventually circumventing the requirement for in vivo maturation, disease modeling of diabetes is expected to progress exponentially. The knowledge gained from these hiPSC-based disease modeling studies can potentially be translated into the clinics by guiding clinicians on the appropriate type of medication to use for each condition based on the mechanism of action of the disease.
Findings from these proposed studies could also offer clues to the pathophysiology of the “garden variety” of type 2 diabetes which is known to manifest defects in each of these tissues. hPSCs and genome editing tools may also provide an opportunity to better understand the relevance of gene variants identified from GWAS studies, in causing T1D, T2D, obesity and metabolic syndromes, given that they exhibit only modest effects and ∼85% of the variants map onto non-coding regions such as enhancers or regulatory elements [104]. Investment into hPSCs and genome editing would allow a better mechanistic understanding of the pathophysiology of monogenic and complex diseases relevant for organismal homeostasis and therefore an improved approach to stratified personalized medicine. By identifying the impact of gene variants on disease predisposition, prophylactic measures in the form of lifestyle alterations or medication could be adopted early on in life to delay or even prevent the onset of diabetes and/or metabolic diseases. It is also likely that these hiPSC-based disease modeling studies would provide insights into approaches to predict the susceptibility of disease. Henceforth, the translational potential of studying human diabetes and metabolic syndrome disease mechanisms is huge, with opportunities for early prophylactic intervention that could have long-term implications for global health care and reduction of economic burden. While the derivation of hiPSCs from human tissues is relatively easier and gaining popularity compared to just a few years ago [2], it is likely that the modern technology of generating site-specific nucleases will also rapidly mature to make in vitro disease modeling a routine approach.
Contribution statements
A.K.K.T. conceived the idea and wrote the manuscript. M.K.G. contributed to the manuscript. A.D. edited and commented on the manuscript. R.N.K. edited and approved the manuscript.
Acknowledgments
A.K.K.T. was supported by a Juvenile Diabetes Research Foundation Postdoctoral Fellowship (3-2013-236) and is currently supported by the Institute of Molecular and Cell Biology (IMCB), A*STAR, NHG-KTPH SIG/14033 and NUHSRO/2014/033. A.D. is supported by the National Institutes of Health grant RO1 DK 55523. R.N.K. is supported by the Harvard Stem Cell Institute, the National Institutes of Health grants RO1 DK 67536, DK 55523 and DK103215, and a grant from AstraZeneca. The funding sources had no involvement in the study design, writing of report or submission of article for publication.
Contributor Information
Adrian Kee Keong Teo, Email: drainteo@gmail.com, ateo@imcb.a-star.edu.sg.
Manoj K. Gupta, Email: Manoj.Gupta@joslin.harvard.edu.
Alessandro Doria, Email: Alessandro.Doria@joslin.harvard.edu.
Rohit N. Kulkarni, Email: rohit.kulkarni@joslin.harvard.edu.
Conflict of interest
The authors declare that they have no conflict of interest.
References
- 1.Takahashi K., Tanabe K., Ohnuki M., Narita M., Ichisaka T., Tomoda K. Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell. 2007;131:861–872. doi: 10.1016/j.cell.2007.11.019. [DOI] [PubMed] [Google Scholar]
- 2.Trounson A., Shepard K.A., DeWitt N.D. Human disease modeling with induced pluripotent stem cells. Current Opinion in Genetics and Development. 2012;22:509–516. doi: 10.1016/j.gde.2012.07.004. [DOI] [PubMed] [Google Scholar]
- 3.Robinton D.A., Daley G.Q. The promise of induced pluripotent stem cells in research and therapy. Nature. 2012;481:295–305. doi: 10.1038/nature10761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Drawnel F.M., Boccardo S., Prummer M., Delobel F., Graff A., Weber M. Disease modeling and phenotypic drug screening for diabetic cardiomyopathy using human induced pluripotent stem cells. Cell Reports. 2014;9:810–820. doi: 10.1016/j.celrep.2014.09.055. [DOI] [PubMed] [Google Scholar]
- 5.Servitja J.M., Ferrer J. Transcriptional networks controlling pancreatic development and beta cell function. Diabetologia. 2004;47:597–613. doi: 10.1007/s00125-004-1368-9. [DOI] [PubMed] [Google Scholar]
- 6.Riggs A.C., Bernal-Mizrachi E., Ohsugi M., Wasson J., Fatrai S., Welling C. Mice conditionally lacking the Wolfram gene in pancreatic islet beta cells exhibit diabetes as a result of enhanced endoplasmic reticulum stress and apoptosis. Diabetologia. 2005;48:2313–2321. doi: 10.1007/s00125-005-1947-4. [DOI] [PubMed] [Google Scholar]
- 7.Hua H., Shang L., Martinez H., Freeby M., Gallagher M.P., Ludwig T. iPSC-derived beta cells model diabetes due to glucokinase deficiency. Journal of Clinical Investigation. 2013;123(7):3146–3153. doi: 10.1172/JCI67638. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 8.Shang L., Hua H., Foo K., Martinez H., Watanabe K., Zimmer M. beta-cell dysfunction due to increased ER stress in a stem cell model of Wolfram syndrome. Diabetes. 2014;63:923–933. doi: 10.2337/db13-0717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Maehr R., Chen S., Snitow M., Ludwig T., Yagasaki L., Goland R. Generation of pluripotent stem cells from patients with type 1 diabetes. Proceedings of the National Academy of Sciences of the United States of America. 2009;106:15768–15773. doi: 10.1073/pnas.0906894106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kudva Y.C., Ohmine S., Greder L.V., Dutton J.R., Armstrong A., De Lamo J.G. Transgene-free disease-specific induced pluripotent stem cells from patients with type 1 and type 2 diabetes. Stem Cells Translational Medicine. 2012;1:451–461. doi: 10.5966/sctm.2011-0044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Park I.H., Arora N., Huo H., Maherali N., Ahfeldt T., Shimamura A. Disease-specific induced pluripotent stem cells. Cell. 2008;134:877–886. doi: 10.1016/j.cell.2008.07.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Thatava T., Kudva Y.C., Edukulla R., Squillace K., De Lamo J.G., Khan Y.K. Intrapatient variations in type 1 diabetes-specific iPS cell differentiation into insulin-producing cells. Molecular Therapy. 2012;21(1):228–239. doi: 10.1038/mt.2012.245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gaj T., Gersbach C.A., Barbas C.F., 3rd ZFN, TALEN, and CRISPR/Cas-based methods for genome engineering. Trends in Biotechnology. 2013;31:397–405. doi: 10.1016/j.tibtech.2013.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Reyon D., Khayter C., Regan M.R., Joung J.K., Sander J.D. Engineering designer transcription activator-like effector nucleases (TALENs) by REAL or REAL-Fast assembly. Current Protocols in Molecular Biology. 2012;12 doi: 10.1002/0471142727.mb1215s100. Unit 12 15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mussolino C., Morbitzer R., Lutge F., Dannemann N., Lahaye T., Cathomen T. A novel TALE nuclease scaffold enables high genome editing activity in combination with low toxicity. Nucleic Acids Research. 2011;39:9283–9293. doi: 10.1093/nar/gkr597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ding Q., Regan S.N., Xia Y., Oostrom L.A., Cowan C.A., Musunuru K. Enhanced efficiency of human pluripotent stem cell genome editing through replacing TALENs with CRISPRs. Cell Stem Cell. 2013;12:393–394. doi: 10.1016/j.stem.2013.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.San Filippo J., Sung P., Klein H. Mechanism of eukaryotic homologous recombination. Annual Review of Biochemistry. 2008;77:229–257. doi: 10.1146/annurev.biochem.77.061306.125255. [DOI] [PubMed] [Google Scholar]
- 18.Liang F., Han M., Romanienko P.J., Jasin M. Homology-directed repair is a major double-strand break repair pathway in mammalian cells. Proceedings of the National Academy of Sciences of the United States of America. 1998;95:5172–5177. doi: 10.1073/pnas.95.9.5172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kim Y.G., Cha J., Chandrasegaran S. Hybrid restriction enzymes: zinc finger fusions to Fok I cleavage domain. Proceedings of the National Academy of Sciences of the United States of America. 1996;93:1156–1160. doi: 10.1073/pnas.93.3.1156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Porteus M.H., Carroll D. Gene targeting using zinc finger nucleases. Nature Biotechnology. 2005;23:967–973. doi: 10.1038/nbt1125. [DOI] [PubMed] [Google Scholar]
- 21.Urnov F.D., Rebar E.J., Holmes M.C., Zhang H.S., Gregory P.D. Genome editing with engineered zinc finger nucleases. Nature Reviews Genetics. 2010;11:636–646. doi: 10.1038/nrg2842. [DOI] [PubMed] [Google Scholar]
- 22.Joung J.K., Sander J.D. TALENs: a widely applicable technology for targeted genome editing. Nature Reviews Molecular Cell Biology. 2013;14:49–55. doi: 10.1038/nrm3486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Deltcheva E., Chylinski K., Sharma C.M., Gonzales K., Chao Y., Pirzada Z.A. CRISPR RNA maturation by trans-encoded small RNA and host factor RNase III. Nature. 2011;471:602–607. doi: 10.1038/nature09886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jinek M., Chylinski K., Fonfara I., Hauer M., Doudna J.A., Charpentier E. A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science. 2012;337:816–821. doi: 10.1126/science.1225829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Maeder M.L., Linder S.J., Cascio V.M., Fu Y., Ho Q.H., Joung J.K. CRISPR RNA-guided activation of endogenous human genes. Nat Methods. 2013;10:977–979. doi: 10.1038/nmeth.2598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pennisi E. The CRISPR craze. Science. 2013;341:833–836. doi: 10.1126/science.341.6148.833. [DOI] [PubMed] [Google Scholar]
- 27.Pattanayak V., Ramirez C.L., Joung J.K., Liu D.R. Revealing off-target cleavage specificities of zinc-finger nucleases by in vitro selection. Nat Methods. 2011;8:765–770. doi: 10.1038/nmeth.1670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Veres A., Gosis B.S., Ding Q., Collins R., Ragavendran A., Brand H. Low incidence of off-target mutations in individual CRISPR-Cas9 and TALEN targeted human stem cell clones detected by whole-genome sequencing. Cell Stem Cell. 2014;15:27–30. doi: 10.1016/j.stem.2014.04.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Smith C., Gore A., Yan W., Abalde-Atristain L., Li Z., He C. Whole-genome sequencing analysis reveals high specificity of CRISPR/Cas9 and TALEN-based genome editing in human iPSCs. Cell Stem Cell. 2014;15:12–13. doi: 10.1016/j.stem.2014.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Fu Y., Sander J.D., Reyon D., Cascio V.M., Joung J.K. Improving CRISPR-Cas nuclease specificity using truncated guide RNAs. Nature Biotechnology. 2014;32:279–284. doi: 10.1038/nbt.2808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cho S.W., Kim S., Kim Y., Kweon J., Kim H.S., Bae S. Analysis of off-target effects of CRISPR/Cas-derived RNA-guided endonucleases and nickases. Genome Research. 2014;24:132–141. doi: 10.1101/gr.162339.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Fu Y., Foden J.A., Khayter C., Maeder M.L., Reyon D., Joung J.K. High-frequency off-target mutagenesis induced by CRISPR-Cas nucleases in human cells. Nature Biotechnology. 2013;31:822–826. doi: 10.1038/nbt.2623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ran F.A., Hsu P.D., Lin C.Y., Gootenberg J.S., Konermann S., Trevino A.E. Double nicking by RNA-guided CRISPR Cas9 for enhanced genome editing specificity. Cell. 2013;154:1380–1389. doi: 10.1016/j.cell.2013.08.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Isalan M. Zinc-finger nucleases: how to play two good hands. Nature Methods. 2012;9:32–34. doi: 10.1038/nmeth.1805. [DOI] [PubMed] [Google Scholar]
- 35.Sebastiano V., Maeder M.L., Angstman J.F., Haddad B., Khayter C., Yeo D.T. In situ genetic correction of the sickle cell anemia mutation in human induced pluripotent stem cells using engineered zinc finger nucleases. Stem Cells. 2011;29:1717–1726. doi: 10.1002/stem.718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Papaioannou I., Simons J.P., Owen J.S. Oligonucleotide-directed gene-editing technology: mechanisms and future prospects. Expert Opinion on Biological Therapy. 2012;12:329–342. doi: 10.1517/14712598.2012.660522. [DOI] [PubMed] [Google Scholar]
- 37.Strouse B., Bialk P., Niamat R.A., Rivera-Torres N., Kmiec E.B. Combinatorial gene editing in mammalian cells using ssODNs and TALENs. Scientific Reports. 2014;4:3791. doi: 10.1038/srep03791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chen F., Pruett-Miller S.M., Huang Y., Gjoka M., Duda K., Taunton J. High-frequency genome editing using ssDNA oligonucleotides with zinc-finger nucleases. Nature Methods. 2011;8:753–755. doi: 10.1038/nmeth.1653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Soldner F., Laganière J., Cheng A.W., Hockemeyer D., Gao Q., Alagappan R. Generation of isogenic pluripotent stem cells differing exclusively at two early onset Parkinson point mutations. Cell. 2011;146:318–331. doi: 10.1016/j.cell.2011.06.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lombardo A., Genovese P., Beausejour C.M., Colleoni S., Lee Y.L., Kim K.A. Gene editing in human stem cells using zinc finger nucleases and integrase-defective lentiviral vector delivery. Nature Biotechnology. 2007;25:1298–1306. doi: 10.1038/nbt1353. [DOI] [PubMed] [Google Scholar]
- 41.Zou J., Maeder M.L., Mali P., Pruett-Miller S.M., Thibodeau-Beganny S., Chou B.K. Gene targeting of a disease-related gene in human induced pluripotent stem and embryonic stem cells. Cell Stem Cell. 2009;5:97–110. doi: 10.1016/j.stem.2009.05.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hockemeyer D., Soldner F., Beard C., Gao Q., Mitalipova M., DeKelver R.C. Efficient targeting of expressed and silent genes in human ESCs and iPSCs using zinc-finger nucleases. Nature Biotechnology. 2009;27:851–857. doi: 10.1038/nbt.1562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lei Y., Lee C.L., Joo K.I., Zarzar J., Liu Y., Dai B. Gene editing of human embryonic stem cells via an engineered baculoviral vector carrying zinc-finger nucleases. Molecular Therapy. 2011;19:942–950. doi: 10.1038/mt.2011.12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yusa K., Rashid S.T., Strick-Marchand H., Varela I., Liu P.Q., Paschon D.E. Targeted gene correction of alpha1-antitrypsin deficiency in induced pluripotent stem cells. Nature. 2011;478:391–394. doi: 10.1038/nature10424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zou J., Mali P., Huang X., Dowey S.N., Cheng L. Site-specific gene correction of a point mutation in human iPS cells derived from an adult patient with sickle cell disease. Blood. 2011;118:4599–4608. doi: 10.1182/blood-2011-02-335554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zou J., Sweeney C.L., Chou B.K., Choi U., Pan J., Wang H. Oxidase-deficient neutrophils from X-linked chronic granulomatous disease iPS cells: functional correction by zinc finger nuclease-mediated safe harbor targeting. Blood. 2011;117:5561–5572. doi: 10.1182/blood-2010-12-328161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Chang C.J., Bouhassira E.E. Zinc-finger nuclease-mediated correction of alpha-thalassemia in iPS cells. Blood. 2012;120:3906–3914. doi: 10.1182/blood-2012-03-420703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hockemeyer D., Wang H., Kiani S., Lai C.S., Gao Q., Cassady J.P. Genetic engineering of human pluripotent cells using TALE nucleases. Nature Biotechnology. 2011;29:731–734. doi: 10.1038/nbt.1927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ding Q., Lee Y.K., Schaefer E.A., Peters D.T., Veres A., Kim K. A TALEN genome-editing system for generating human stem cell-based disease models. Cell Stem Cell. 2013;12:238–251. doi: 10.1016/j.stem.2012.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Osborn M.J., Starker C.G., McElroy A.N., Webber B.R., Riddle M.J., Xia L. TALEN-based gene correction for epidermolysis bullosa. Molecular Therapy. 2013;21:1151–1159. doi: 10.1038/mt.2013.56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Choi S.M., Kim Y., Shim J.S., Park J.T., Wang R.H., Leach S.D. Efficient drug screening and gene correction for treating liver disease using patient-specific stem cells. Hepatology. 2013;57:2458–2468. doi: 10.1002/hep.26237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.González F., Zhu Z., Shi Z.D., Lelli K., Verma N., Li Q.V. An iCRISPR platform for rapid, multiplexable, and inducible genome editing in human pluripotent stem cells. Cell Stem Cell. 2014;15:215–226. doi: 10.1016/j.stem.2014.05.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wong G.K., Chiu A.T. Gene therapy, gene targeting and induced pluripotent stem cells: applications in monogenic disease treatment. Biotechnology Advances. 2011;29:1–10. doi: 10.1016/j.biotechadv.2010.07.005. [DOI] [PubMed] [Google Scholar]
- 54.McCarthy M.I., Hattersley A.T. Learning from molecular genetics: novel insights arising from the definition of genes for monogenic and type 2 diabetes. Diabetes. 2008;57:2889–2898. doi: 10.2337/db08-0343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Teo A.K., Wagers A.J., Kulkarni R.N. New opportunities: harnessing induced Pluripotency for discovery in diabetes and metabolism. Cell Metabolism. 2013;18(6):775–791. doi: 10.1016/j.cmet.2013.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Semple R.K., Savage D.B., Cochran E.K., Gorden P., O'Rahilly S. Genetic syndromes of severe insulin resistance. Endocrine Reviews. 2011;32:498–514. doi: 10.1210/er.2010-0020. [DOI] [PubMed] [Google Scholar]
- 57.Osbak K.K., Colclough K., Saint-Martin C., Beer N.L., Bellanné-Chantelot C., Ellard S. Update on mutations in glucokinase (GCK), which cause maturity-onset diabetes of the young, permanent neonatal diabetes, and hyperinsulinemic hypoglycemia. Human Mutation. 2009;30:1512–1526. doi: 10.1002/humu.21110. [DOI] [PubMed] [Google Scholar]
- 58.Sansbury F.H., Flanagan S.E., Houghton J.A., Shuixian Shen F.L., Al-Senani A.M., Habeb A.M. SLC2A2 mutations can cause neonatal diabetes, suggesting GLUT2 may have a role in human insulin secretion. Diabetologia. 2012;55:2381–2385. doi: 10.1007/s00125-012-2595-0. [DOI] [PubMed] [Google Scholar]
- 59.Velho G., Froguel P., Clement K., Pueyo M.E., Rakotoambinina B., Zouali H. Primary pancreatic beta-cell secretory defect caused by mutations in glucokinase gene in kindreds of maturity onset diabetes of the young. Lancet. 1992;340:444–448. doi: 10.1016/0140-6736(92)91768-4. [DOI] [PubMed] [Google Scholar]
- 60.Velho G., Petersen K.F., Perseghin G., Hwang J.H., Rothman D.L., Pueyo M.E. Impaired hepatic glycogen synthesis in glucokinase-deficient (MODY-2) subjects. Journal of Clinical Investigation. 1996;98:1755–1761. doi: 10.1172/JCI118974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Byrne M.M., Sturis J., Clément K., Vionnet N., Pueyo M.E., Stoffel M. Insulin secretory abnormalities in subjects with hyperglycemia due to glucokinase mutations. Journal of Clinical Investigation. 1994;93:1120–1130. doi: 10.1172/JCI117064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Støy J., Edghill E.L., Flanagan S.E., Ye H., Paz V.P., Pluzhnikov A. Insulin gene mutations as a cause of permanent neonatal diabetes. Proceedings of the National Academy of Sciences of the United States of America. 2007;104:15040–15044. doi: 10.1073/pnas.0707291104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.McTaggart J.S., Clark R.H., Ashcroft F.M. The role of the KATP channel in glucose homeostasis in health and disease: more than meets the islet. Journal of Physiology. 2010;588:3201–3209. doi: 10.1113/jphysiol.2010.191767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Hattersley A.T., Ashcroft F.M. Activating mutations in Kir6.2 and neonatal diabetes: new clinical syndromes, new scientific insights, and new therapy. Diabetes. 2005;54:2503–2513. doi: 10.2337/diabetes.54.9.2503. [DOI] [PubMed] [Google Scholar]
- 65.Iovino S., Burkart A.M., Kriauciunas K., Warren L., Hughes K.J., Molla M. Genetic insulin resistance is a potent regulator of gene expression and proliferation in human iPS cells. Diabetes. 2014;63:4130–4142. doi: 10.2337/db14-0109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Magner N.L., Jung Y., Wu J., Nolta J.A., Zern M.A., Zhou P. Insulin and IGFs enhance hepatocyte differentiation from human embryonic stem cells via the PI3K/AKT pathway. Stem Cells. 2013;31:2095–2103. doi: 10.1002/stem.1478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Garg A. Lipodystrophies. American Journal of Medicine. 2000;108:143–152. doi: 10.1016/s0002-9343(99)00414-3. [DOI] [PubMed] [Google Scholar]
- 68.Agarwal A.K., Garg A. Congenital generalized lipodystrophy: significance of triglyceride biosynthetic pathways. Trends in Endocrinology and Metabolism. 2003;14:214–221. doi: 10.1016/s1043-2760(03)00078-x. [DOI] [PubMed] [Google Scholar]
- 69.Payne V.A., Grimsey N., Tuthill A., Virtue S., Gray S.L., Dalla Nora E. The human lipodystrophy gene BSCL2/seipin may be essential for normal adipocyte differentiation. Diabetes. 2008;57:2055–2060. doi: 10.2337/db08-0184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.O'Rahilly S. Human genetics illuminates the paths to metabolic disease. Nature. 2009;462:307–314. doi: 10.1038/nature08532. [DOI] [PubMed] [Google Scholar]
- 71.Polychronakos C., Li Q. Understanding type 1 diabetes through genetics: advances and prospects. Nature Reviews Genetics. 2011;12:781–792. doi: 10.1038/nrg3069. [DOI] [PubMed] [Google Scholar]
- 72.Timmermans F., Velghe I., Vanwalleghem L., De Smedt M., Van Coppernolle S., Taghon T. Generation of T cells from human embryonic stem cell-derived hematopoietic zones. Journal of Immunology. 2009;182:6879–6888. doi: 10.4049/jimmunol.0803670. [DOI] [PubMed] [Google Scholar]
- 73.Kennedy M., Awong G., Sturgeon C.M., Ditadi A., LaMotte-Mohs R., Zúñiga-Pflücker J.C. T lymphocyte potential marks the emergence of definitive hematopoietic progenitors in human pluripotent stem cell differentiation cultures. Cell Reports. 2012;2:1722–1735. doi: 10.1016/j.celrep.2012.11.003. [DOI] [PubMed] [Google Scholar]
- 74.Lei F., Haque R., Xiong X., Song J. Directed differentiation of induced pluripotent stem cells towards T lymphocytes. Journal of Visualized Experiments. 2012:e3986. doi: 10.3791/3986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Pagliuca F.W., Millman J.R., Gürtler M., Segel M., Van Dervort A., Ryu J.H. Generation of functional human pancreatic beta cells in vitro. Cell. 2014;159:428–439. doi: 10.1016/j.cell.2014.09.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Rezania A., Bruin J.E., Arora P., Rubin A., Batushansky I., Asadi A. Reversal of diabetes with insulin-producing cells derived in vitro from human pluripotent stem cells. Nature Biotechnology. 2014;32:1121–1133. doi: 10.1038/nbt.3033. [DOI] [PubMed] [Google Scholar]
- 77.Takeuchi H., Nakatsuji N., Suemori H. Endodermal differentiation of human pluripotent stem cells to insulin-producing cells in 3D culture. Science Reports. 2014;4:4488. doi: 10.1038/srep04488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Eizirik D.L., Sammeth M., Bouckenooghe T., Bottu G., Sisino G., Igoillo-Esteve M. The human pancreatic islet transcriptome: expression of candidate genes for type 1 diabetes and the impact of pro-inflammatory cytokines. PLoS Genetics. 2012;8:e1002552. doi: 10.1371/journal.pgen.1002552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Santin I., Eizirik D.L. Candidate genes for type 1 diabetes modulate pancreatic islet inflammation and beta-cell apoptosis. Diabetes, Obesity and Metabolism. 2013;15(Suppl 3):71–81. doi: 10.1111/dom.12162. [DOI] [PubMed] [Google Scholar]
- 80.Cerosaletti K., Buckner J.H. Protein tyrosine phosphatases and type 1 diabetes: genetic and functional implications of PTPN2 and PTPN22. Review of Diabetic Studies. 2012;9:188–200. doi: 10.1900/RDS.2012.9.188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Rieck M., Arechiga A., Onengut-Gumuscu S., Greenbaum C., Concannon P., Buckner J.H. Genetic variation in PTPN22 corresponds to altered function of T and B lymphocytes. Journal of Immunology. 2007;179:4704–4710. doi: 10.4049/jimmunol.179.7.4704. [DOI] [PubMed] [Google Scholar]
- 82.Wang H., Jin Y., Reddy M.V., Podolsky R., Liu S., Yang P. Genetically dependent ERBB3 expression modulates antigen presenting cell function and type 1 diabetes risk. PLoS One. 2010;5:e11789. doi: 10.1371/journal.pone.0011789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Tsygankov A.Y. TULA-family proteins: an odd couple. Cellular and Molecular Life Sciences. 2009;66:2949–2952. doi: 10.1007/s00018-009-0071-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Kasuga M. Insulin resistance and pancreatic beta cell failure. Journal of Clinical Investigation. 2006;116:1756–1760. doi: 10.1172/JCI29189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Donath M.Y., Ehses J.A., Maedler K., Schumann D.M., Ellingsgaard H., Eppler E. Mechanisms of beta-cell death in type 2 diabetes. Diabetes. 2005;54(Suppl 2):S108–S113. doi: 10.2337/diabetes.54.suppl_2.s108. [DOI] [PubMed] [Google Scholar]
- 86.Talchai C., Xuan S., Lin H.V., Sussel L., Accili D. Pancreatic beta cell dedifferentiation as a mechanism of diabetic beta cell failure. Cell. 2012;150:1223–1234. doi: 10.1016/j.cell.2012.07.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Drong A.W., Lindgren C.M., McCarthy M.I. The genetic and epigenetic basis of type 2 diabetes and obesity. Clinical Pharmacology & Therapeutics. 2012;92:707–715. doi: 10.1038/clpt.2012.149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Travers M.E., McCarthy M.I. Type 2 diabetes and obesity: genomics and the clinic. Human Genetics. 2011;130:41–58. doi: 10.1007/s00439-011-1023-8. [DOI] [PubMed] [Google Scholar]
- 89.Shu L., Sauter N.S., Schulthess F.T., Matveyenko A.V., Oberholzer J., Maedler K. Transcription factor 7-like 2 regulates beta-cell survival and function in human pancreatic islets. Diabetes. 2008;57:645–653. doi: 10.2337/db07-0847. [DOI] [PubMed] [Google Scholar]
- 90.Grant S.F., Thorleifsson G., Reynisdottir I., Benediktsson R., Manolescu A., Sainz J. Variant of transcription factor 7-like 2 (TCF7L2) gene confers risk of type 2 diabetes. Nature Genetics. 2006;38:320–323. doi: 10.1038/ng1732. [DOI] [PubMed] [Google Scholar]
- 91.Pearson E.R., Donnelly L.A., Kimber C., Whitley A., Doney A.S., McCarthy M.I. Variation in TCF7L2 influences therapeutic response to sulfonylureas: a GoDARTs study. Diabetes. 2007;56:2178–2182. doi: 10.2337/db07-0440. [DOI] [PubMed] [Google Scholar]
- 92.Lyssenko V., Lupi R., Marchetti P., Del Guerra S., Orho-Melander M., Almgren P. Mechanisms by which common variants in the TCF7L2 gene increase risk of type 2 diabetes. Journal of Clinical Investigation. 2007;117:2155–2163. doi: 10.1172/JCI30706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Frayling T.M., Timpson N.J., Weedon M.N., Zeggini E., Freathy R.M., Lindgren C.M. A common variant in the FTO gene is associated with body mass index and predisposes to childhood and adult obesity. Science. 2007;316:889–894. doi: 10.1126/science.1141634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Larsen L.H., Echwald S.M., Sorensen T.I., Andersen T., Wulff B.S., Pedersen O. Prevalence of mutations and functional analyses of melanocortin 4 receptor variants identified among 750 men with juvenile-onset obesity. Journal of Clinical Endocrinology and Metabolism. 2005;90:219–224. doi: 10.1210/jc.2004-0497. [DOI] [PubMed] [Google Scholar]
- 95.Speliotes E.K., Willer C.J., Berndt S.I., Monda K.L., Thorleifsson G., Jackson A.U. Association analyses of 249,796 individuals reveal 18 new loci associated with body mass index. Nature Genetics. 2010;42:937–948. doi: 10.1038/ng.686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.O'Rahilly S., Farooqi I.S. Human obesity: a heritable neurobehavioral disorder that is highly sensitive to environmental conditions. Diabetes. 2008;57:2905–2910. doi: 10.2337/db08-0210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Tews D., Fischer-Posovszky P., Wabitsch M. Regulation of FTO and FTM expression during human preadipocyte differentiation. Hormone and Metabolic Research. 2011;43:17–21. doi: 10.1055/s-0030-1265130. [DOI] [PubMed] [Google Scholar]
- 98.Spiegelman B.M., Flier J.S. Adipogenesis and obesity: rounding out the big picture. Cell. 1996;87:377–389. doi: 10.1016/s0092-8674(00)81359-8. [DOI] [PubMed] [Google Scholar]
- 99.Heymsfield S.B., Greenberg A.S., Fujioka K., Dixon R.M., Kushner R., Hunt T. Recombinant leptin for weight loss in obese and lean adults: a randomized, controlled, dose-escalation trial. JAMA. 1999;282:1568–1575. doi: 10.1001/jama.282.16.1568. [DOI] [PubMed] [Google Scholar]
- 100.Bernhard F., Landgraf K., Klöting N., Berthold A., Büttner P., Friebe D. Functional relevance of genes implicated by obesity genome-wide association study signals for human adipocyte biology. Diabetologia. 2013;56:311–322. doi: 10.1007/s00125-012-2773-0. [DOI] [PubMed] [Google Scholar]
- 101.Yoganathan P., Karunakaran S., Ho M.M., Clee S.M. Nutritional regulation of genome-wide association obesity genes in a tissue-dependent manner. Nutrition & Metabolism (London) 2012;9:65. doi: 10.1186/1743-7075-9-65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Gutierrez-Aguilar R., Kim D.H., Woods S.C., Seeley R.J. Expression of new loci associated with obesity in diet-induced obese rats: from genetics to physiology. Obesity (Silver Spring) 2012;20:306–312. doi: 10.1038/oby.2011.236. [DOI] [PubMed] [Google Scholar]
- 103.Smemo S., Tena J.J., Kim K.H., Gamazon E.R., Sakabe N.J., Gómez-Marín C. Obesity-associated variants within FTO form long-range functional connections with IRX3. Nature. 2014;507:371–375. doi: 10.1038/nature13138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Hindorff L.A., Sethupathy P., Junkins H.A., Ramos E.M., Mehta J.P., Collins F.S. Potential etiologic and functional implications of genome-wide association loci for human diseases and traits. Proceedings of the National Academy of Sciences of the United States of America. 2009;106:9362–9367. doi: 10.1073/pnas.0903103106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Teo A.K., Windmueller R., Johansson B.B., Dirice E., Njolstad P.R., Tjora E. Derivation of human induced pluripotent stem cells from patients with maturity onset diabetes of the young. The Journal of Biological Chemistry. 2013;288:5353–5356. doi: 10.1074/jbc.C112.428979. [DOI] [PMC free article] [PubMed] [Google Scholar]