Abstract
We investigated the hypothesis that the varying treatment efficacy of adjuvant 5-fluorouracil (5FU) in stage III colon cancer is linked to the TP53 mutational status.
ABCSG-90 was a prospective randomized trial in which effect of adjuvant 5FU was studied in stage III colon cancer patients. Tumor material of 70% of these patients (389/572) was available for analysis of the biomarker TP53 using a TP53-gene-specific Sanger sequencing protocol.
Median follow-up was 88 months. TP53 mutation frequency was 33%. A significant interaction between TP53 status, outcomes and nodal category was found (P = 0.0095). In the N1 category, TP53 wildtype patients had significantly better overall survival than TP53 mutated (81.0% vs. 62.0% overall survival at 5 years; HR = 2.131; 95% CI: 1.344–3.378; P = 0.0010). In the N2 category, the TP53 status did not affect survival (P = 0.4992). In TP53 wildtype patients, the prognostic significance of N category was significantly enhanced (P = 0.0002). In TP53 mutated patients, survival curves of N1 and N2 patients overlapped and nodal category was no longer prognostic.
The biomarker TP53 independently predicted effect of adjuvant 5FU in N1 colon cancer patients. TP53 was not predictive in N2 patients, in whom 5FU is known to have no effect.
Abbreviations: 5FU, 5-fluorouracil; ABCSG, Austrian Breast and Colorectal Cancer Study Group; LEV, levamisole; INF, interferon; FFPE, formalin-fixed paraffin-embedded; HR, hazard ratios; CI, confidence intervals
Keywords: TP53, Predictive biomarker, Stage III colon cancer, Adjuvant 5-fluorouracil, Varying treatment efficacy
Highlights
-
•
The TP53 status was found to be an independent predictive marker for the effect of adjuvant 5FU in stage III colon cancer.
-
•
In the N1 category, patients with wildtype TP53 experienced a significant survival benefit from adjuvant 5FU.
-
•
In TP53 mutant patients survival curves of N1 and N2 patients overlapped and nodal category was no longer prognostic.
Postoperative chemotherapy is recommended for all patients with lymph node positive colon cancer. In colon cancer, mutations in the TP53 gene are present in more than 30% of tumors. We found that the most commonly used postoperative chemotherapy resulted in a marked survival benefit for patients with normal TP53 status while it was associated with significant survival disadvantage in TP53 mutant patients. Overall the survival disadvantage of the mutated patients almost balanced the survival benefit of the TP53 normal patients. This might be an explanation why chemotherapy resulted in less progress in survival of colon cancer than we would have expected.
1. Introduction
Adjuvant chemotherapy is recommended for all patients with lymph-node positive colon cancer (Gill et al., 2004, Greene et al., 2002a). For decades, 5-fluorouracil (5FU) was the only drug demonstrating meaningful activity in colorectal cancer and is still used in any modern adjuvant chemotherapy regimens for colon cancer (Chau and Cunningham, 2006). At the beginning of this century, oxaliplatin, irinotecan, capecitabine, cetuximab, and bevacizumab became available. But the major, dramatic progress promised by new biological and targeted agents failed to emerge (Brenner et al., 2014, Cassidy et al., 2008, Saltz et al., 2008, Tol et al., 2009, Hecht et al., 2009). Still no drug shows greater single agent activity than 5FU. But it is general knowledge that treatment efficacy varies substantially among 5FU-treated patients of similar stage and baseline risk factors. The question why remained unanswered although it is burning since the early days of 5FU.
5FU induces DNA-damage in rapidly growing cells. It is generally accepted that wild type TP53 is responsible for apoptosis induction in response to DNA-damage. In more than 30% of colon cancers TP53 is mutated and apoptosis cannot be induced. However, clinical reports about the role of the TP53 status for response to adjuvant chemotherapy in colon cancer are not consistent (Elsaleh et al., 2001, Russo et al., 2005, Petersen et al., 2001, Ahmed et al., 2008, Bleeker et al., 2001).
The aim of this study was to evaluate whether and how the TP53 mutational status is linked to the varying efficacy of 5FU treatment in stage III colon cancer. For this purpose, we performed a retrospective biomarker analysis in a prospective randomized cohort of stage III colon cancer patients treated with adjuvant 5FU. The prospective randomized trial (ABCSG-90) was initially reported by the Austrian Breast and Colorectal Cancer Study Group (ABCSG) in 2005 (Schippinger et al., 2005).
2. Materials and Methods
2.1. Patient Cohort
Between 1991 and 1999, 572 stage III colon cancer patients were recruited for ABCSG-90 in order to investigate the influence of adding levamisole (LEV) and/or interferon (INF) alpha to adjuvant fluorouracil. In 2005 the results of ABCSG-90 were published. It was reported that neither LEV nor INF improved the efficacy of adjuvant 5FU in stage III colon cancer (Schippinger et al., 2005). Patient inclusion criteria were histologically proven lymph node positive carcinoma of the colon (stage III), age between 18 and 80 and curative resection. Rectal cancers were excluded.
2.2. Tumor Material
Formalin-fixed paraffin-embedded (FFPE) tissue samples of the surgical specimens from 389 patients were available for the retrospective TP53 biomarker analysis, corresponding to 70% of the original 572 study patients. The 389 analyzed patients were equally distributed between the randomized treatment arms of ABCSG-90.
2.3. Ethics
The retrospective biomarker analysis was approved by the local ethics committee of the Medical University of Vienna (EK 761/2008).
2.4. TP53 Sequencing
The mutational status of TP53 was assessed using DNA extracts from archived FFPE material from the surgical specimens. TP53-gene-specific Sanger sequencing was used for gene analysis. (MARK53gssx6, p53-gene-specific sequencing kit; Mark53 Ltd.; www.mark53.com; Vienna). Genetic variations in the TP53 gene were reported and interpreted according to recommendations of the Human Genome Variation Society (www.hgvs.org) (den Dunnen and Antonarakis, 2000).
The reference genomic sequence used for TP53 was NM_000546.5 and U94788 (NCBI GenBank).
The functional activity of TP53 mutations was classified based on the overall transcriptional activity on eight different promoters as measured in yeast assays by Kato et al. (Kato et al., 2003, Petitjean et al., 2007) : TP53 mutants considered to have a median transcriptional activity > 75% were classified as wildtype for the calculation. A median transcriptional activity of < 75% was considered partially functional and was classified as mutant for calculation.
2.5. Statistical Methods
Continuous data are described by median, minimum, and maximum. Categorical data are described with absolute and relative frequencies. A chi-square test was used to assess for group differences for binary and nominal variables. For ordinal variables, a trend chi-square test was used. In the case of sparse data, Fisher's Exact test or exact chi-square tests were used.
The primary outcome measure was overall survival. Overall survival is defined as the time between the date of randomization until death from any cause or censored at last time known to be alive. Median follow-up time was calculated using the Kaplan–Meier method, in which deaths are censored for further follow-up. Survival probabilities were estimated by Kaplan–Meier graphs, and group differences were assessed by log-rank test. A Cox regression model was used to assess group differences by hazard ratios (HR) and corresponding 95% confidence intervals (CI). A Cox regression model was used to assess interactions between prognostic factors and TP53 mutation status. In the case of a significant interaction, separate subgroup analyses were performed. Multiple Cox regression models were fitted to assess the impact of TP53 on survival above that of standard prognostic factors.
Statistical calculations were performed with SAS® (SAS Institute Inc., Cary, NC, USA). All p-values are two-sided, and P ≤ 0.05 was considered statistically significant.
Colon cancers were classified according to the UICC/AJCC staging system (N1 = metastasis in one to three regional lymph nodes; N2 = metastasis to four or more regional lymph nodes) (Greene et al., 2002b). No rectal cancers were included.
3. Results
TP53 sequencing analysis was successful in all 389 specimens. TP53 mutations were detected in 129 patients (33%). Mutations are characterized in Table 1. Three mutants had a median transcriptional activity > 75% and were classified as wildtype for the calculation. Six mutants were only partially functional and were classified as mutant for calculation. Four mutations, though located in the introns, had a predicted effect on splicing and were classified as mutant (NNsplice 0.9, HSF_V2.3) (Petitjean et al., 2007). Four silent mutations had a predicted effect on splicing and were classified as mutant. Seven patients had two mutations in the TP53 gene.
Table 1.
TP53 mutations.
| Type of TP53 mutationa | Nr. of patients with that mutation | Exon |
|---|---|---|
| c.102dupC p.(Pro36fs) | 1 | 4 |
| c.216dupC p.(Val73fs) | 1 | 4 |
| c.326T>C p.(Phe109Ser) | 1 | 4 |
| c.332T>G p.(Leu111Arg) | 1 | 4 |
| c.338T>G p.(Phe113Cys) | 1 | 4 |
| c.341T>A p.(Leu114Ter) | 1 | 4 |
| c.375G>A p.? | 1 | 4 |
| c.380C>T p.(Ser127Phe) | 1 | 5 |
| c.402T>A p.(Phe134Leu) | 1 | 5 |
| c.404G>T p.(Cys135Phe) | 1 | 5 |
| c.405C>G p.(Cys135Trp) | 1 | 5 |
| c.406C>T p.(Gln136Ter) | 1 | 5 |
| c.413C>T p.(Ala138Val) | 1 | 5 |
| c.452C>A p.(Pro151His) | 1 | 5 |
| c.454_466del p.(Pro152fs) | 1 | 5 |
| c.455C>T p.(Pro152Leu) | 1 | 5 |
| c.470T>G p.(Val157Gly) | 1 | 5 |
| c.475G>C p.(Ala159Pro) | 1 | 5 |
| c.485T>G p.(Ile162Ser) | 1 | 5 |
| c.493C>T p.(Gln165Ter) | 1 | 5 |
| c.514_524del p.(Val172fs) | 1 | 5 |
| c.517G>A p.(Val173Met) | 1 | 5 |
| c.524G>A p.(Arg175His) | 10 | 5 |
| c.527G>T p.(Cys176Phe) | 1 | 5 |
| c.536A>C p.(His179Pro) | 1 | 5 |
| c.553delA p.(Ser185fs) | 1 | 5 |
| c.559 + 1G>T p.? | 1 | Intron 5 |
| c.571_573delCCT p.(Pro191del) | 1 | 6 |
| c.576_577delinsCA p.[(Gln192His(;)His193Asn)] | 1 | 6 |
| c.578A>G p.(His193Arg) | 1 | 6 |
| c.578A>T p.(His193Leu) | 1 | 6 |
| c.580C>T p.(Leu194Phe) | 1 | 6 |
| c.583A>T p.(Ile195Phe) | 1 | 6 |
| c.586C>T p.(Arg196Ter) | 1 | 6 |
| c.612_613insA p.(Tyr205fs) | 1 | 6 |
| c.626_627delGA p.(Arg209fs) | 1 | 6 |
| c.637C>T p.(Arg213Ter) | 9 | 6 |
| c.638G>A p.(Arg213Gln) | 1 | 6 |
| c.641A>G p.(His214Arg) | 1 | 6 |
| c.646G>A p.(Val216Met) | 1 | 6 |
| c.650_658del p.(Val217_Tyr220delinsAsp) | 1 | 6 |
| c.672 + 1G>C p.? | 1 | Intron 6 |
| c.672 + 1G>T p.? | 1 | Intron 6 |
| c.700T>G p.(Tyr234Asp) | 1 | 7 |
| c.701A>G p.(Tyr234Cys) | 1 | 7 |
| c.706T>G p.(Tyr236Asp) | 1 | 7 |
| c.707A>C p.(Tyr236Ser) | 1 | 7 |
| c.711G>T p.(Met237Ile) | 1 | 7 |
| c.712T>C p.(Cys238Arg) | 1 | 7 |
| c.713G>A p.(Cys238Tyr) | 1 | 7 |
| c.716A>G p.(Asn239Ser) | 1 | 7 |
| c.723delC p.(Cys242fs) | 1 | 7 |
| c.726C>T p.? | 2 | 7 |
| c.733G>A p.(Gly245Ser) | 2 | 7 |
| c.737T>C p.(Met246Thr) | 1 | 7 |
| c.742C>T p.(Arg248Trp) | 3 | 7 |
| c.743G>A p.(Arg248Gln) | 5 | 7 |
| c.746G>T p.(Arg249Met) | 2 | 7 |
| c.747G>T p.(Arg249Ser) | 1 | 7 |
| c.750delC p.(251Ilefs) | 1 | 7 |
| c.770T>C p.(Leu257Pro) | 1 | 7 |
| c.772G>A p.(Glu258Lys) | 1 | 7 |
| c.773A>G p.(Glu258Gly) | 1 | 7 |
| c.782_782 + 1delGG p.? | 1 | 7 |
| c.794T>C p.(Leu265Pro) | 1 | 8 |
| c.799C>A p.? | 1 | 8 |
| c.800G>A p.(Arg267Gln) | 2 | 8 |
| c.809T>C p.(Phe270Ser) | 1 | 8 |
| c.811G>A p.(Glu271Lys) | 1 | 8 |
| c.814G>A p.(Val272Met) | 3 | 8 |
| c.814G>C p.(Val272Leu) | 1 | 8 |
| c.817C>T p.(Arg273Cys) | 3 | 8 |
| c.818G>A p.(Arg273His) | 7 | 8 |
| c.824G>A p.(Cys275Tyr) | 1 | 8 |
| c.832C>T p.(Pro278Ser) | 1 | 8 |
| c.833C>T p.(Pro278Leu) | 1 | 8 |
| c.844C>T p.(Arg282Trp) | 3 | 8 |
| c.851_852delCA p.(Thr284fs) | 1 | 8 |
| c.856G>A p.(Glu286Lys) | 1 | 8 |
| c.880delG p.(Glu294fs) | 1 | 8 |
| c.892G>T p.(Glu298Ter) | 1 | 8 |
| c.916C>T p.(Arg306Ter) | 6 | 8 |
| c.920-2A>G p.? | 1 | Intron 8 |
| c.920-7_923delTTCCTAGCACT p.? | 1 | 9/Intron 8 |
| c.1009C>T p.(Arg337Cys) | 1 | 10 |
| c.1024C>T p.(Arg342Ter) | 1 | 10 |
| c.1025_1041dup17 p.(Leu348fs) | 1 | 10 |
| c.1027G>A p.(Glu343Lys) | 1 | 10 |
| c.1045G>T p.(Glu349Ter) | 1 | 10 |
| c.274C>T p.(Pro92Ser)b | 1 | 4 |
| c.548C>T p.(Ser183Leu)b | 1 | 5 |
| c.629A>G p.(Asn210Ser)b | 1 | 6 |
| c.277C>T p.(=)c | 1 | 4 |
| c.291C>T p.(=)c | 1 | 4 |
| c.525C>A p.(=)c | 1 | 5 |
| c.624C>T p.(=)c | 1 | 6 |
| c.699C>T p.(=)c | 1 | 7 |
| c.717C>T p.(=)c | 1 | 7 |
| c.1014C>T p.(=)c | 1 | 10 |
| c.1023C>T p.(=)c | 1 | 10 |
| c.1098C>T p.(=)c | 1 | 10 |
Mutations are reported according to Recommendations of the Human Genome Variation Society (www.hgvs.org).
Mutations were considered and calculated as wildtype due to transactivation activity > 75%.
Mutations were considered and calculated as wildtype as they were characterized as silent mutations without affecting transcription, splicing or translation.
The prognostic covariates are shown in Table 2. The frequency of TP53 mutations did not differ between nodal status (N1, N2; P = 0.187), tumor stages (IIIA, IIIB, IIIC; P = 0.445), or between the treatment arms of the ABCSG-90 (fluorouracil, fluorouracil + LEV, fluorouracil + INF, fluorouracil + LEV + INF; P = 0.148).
Table 2.
Prognostic covariates.
| Cohorts: | Study cohort n = 389 |
TP53 wildtype n = 260 |
TP53 mutant n = 129 |
P-value |
|---|---|---|---|---|
| Stage IIIA | 32 | 23 (71.9%) | 9 (28.1%) | 0.445 |
| Stage IIIB | 191 | 120 (62.8%) | 71 (37.2%) | |
| Stage IIIC | 166 | 117 (70.5%) | 49 (29.5%) | |
| N1 (1-3) | 223 | 143 (64.1%) | 80 (35.9%) | 0.1878 |
| N2 (≥ 4) | 166 | 117 (70.5%) | 49 (29.5%) | |
| Female | 190 | 132 (69.5%) | 58 (30.5%) | 0.2806 |
| Male | 199 | 128 (64.3%) | 71 (35.7%) | |
| age | 63.3 | 62.1 (20.2–79.1) | 65.1 (32.8–79.2) | 0.0062 |
| Tumor grade | ||||
| G1-G2 | 262 | 169 (64.5%) | 93 (35.5%) | 0.1751 |
| G3 | 126 | 90 (71.4%) | 36 (28.6%) | |
| Location | ||||
| Proximal | 190 | 135 (71.1%) | 55 (28.9%) | 0.0845 |
| Distal | 199 | 125 (62.8%) | 74 (37.2%) | |
| Treatment arms | ||||
| 5FU | 94 | 57 (60.6%) | 37 (39.4%) | 0.1482 |
| 5FU/interferon | 102 | 76 (74.5%) | 26 (25.5%) | |
| 5FU/levamisol | 91 | 63 (69.2%) | 28 (30.8%) | |
| 5FU/interferon/levamisol | 102 | 64 (62.7%) | 38 (37.3%) | |
Age is given as median (minimum–maximum), other categorical variables are described with absolute frequencies and percentages.
Median age was 63.3 years (min 20.2 and max 79.2 years). Median follow-up time was 88 months. The five-year overall survival of the analyzed 389 patients was 71.2%, which is comparable to five-year overall survival reported for the original cohort of the ABCSG-90, which included 572 patients.
3.1. Nodal Category Specific Interaction Between TP53 Status and Survival
Overall, TP53 wildtype patients performed better than TP53 mutant patients, but this difference was not significant (P = 0.0640). A significant interaction between TP53 status and lymph node categories was seen in Cox regression models (P = 0.0095) and justified the examination of the effect of TP53 status in the nodal categories N1 versus N2 separately.
In N1 patients, the five-year overall survival was 81.0% for TP53 wildtype patients compared to 62.0% in TP53 mutant patients (HR = 2.131 (95 % CI: 1.344–3.378); P = 0.0010) (Fig. 1A). In N2 patients, the TP53 status did not affect the overall survival (P = 0.4992). The five-year overall survival was 66.3% for TP53 wildtype patients and 69.3% for mutant patients (HR = 0.834 (95% CI: 0.493–1.412)) (Fig. 1B).
Fig. 1.
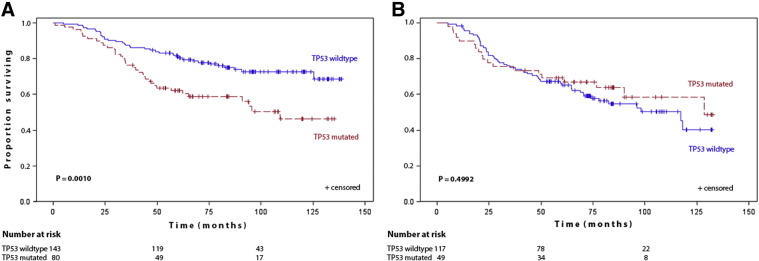
(A,B): Kaplan–Maier estimates of cumultative survival by TP53 status (A) in N1 patients (B) in N2 patients.
After adjustment for other prognostic factors, including sex, age as continuous variable, grading, therapy, and tumor location, the TP53 mutational status remained a significant factor for overall survival in N1 patients (HR = 1.886 (95% CI: 1.172–3.036) P = 0.0090). In N2 patients, TP53 remained a non-significant factor for overall survival.
As expected, N1 patients had a significant better survival than N2 patients (P = 0.0117) (Fig. 2). In TP53 wildtype patients, discrimination between favorable and poor survival prognosis by nodal category was improved (P = 0.0002) (Fig. 3A). In TP53 mutant patients, survival curves of N1 and N2 patients overlapped (P = 0.6393) and nodal category was no longer prognostic in TP53 mutant patients (Fig. 3B).
Fig. 2.
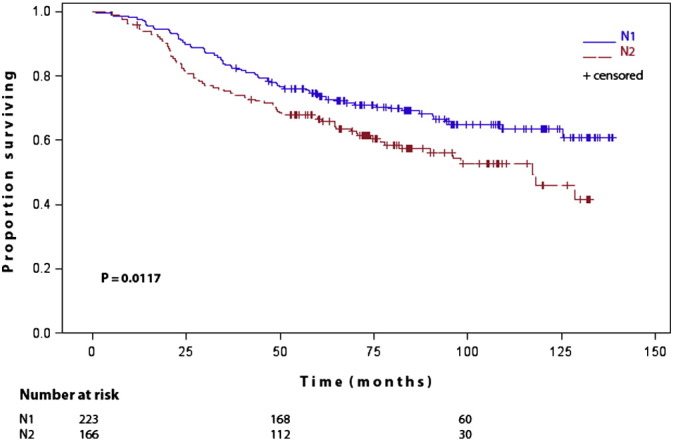
Kaplan–Maier estimates of cumultative survival of N1 patients versus N2 patients.
Fig. 3.
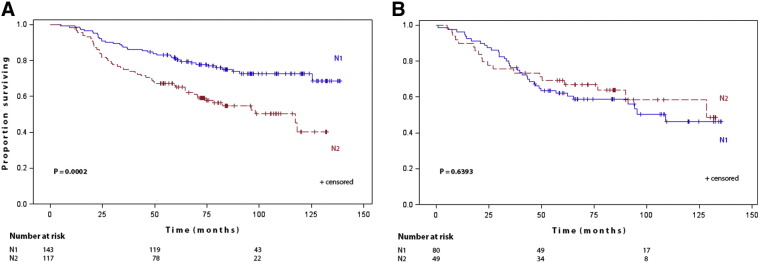
(A,B): Kaplan–Maier estimates of cumultative survival by lymphnode category (A) in TP53 wildtype patients (B) in TP53 mutant patients.
3.2. Tumor-stage Specific Interaction of TP53 Status and Survival
The Cox regression models consistently showed a significant interaction between TP53 status and traditional tumor stage (P = 0.0374). Due to this significant interaction, the effect of TP53 status was also investigated separately in the different tumor stages (IIIA, IIIB, and IIIC).
In stage IIIA, the five-year survival for patients with wildtype TP53 was 91.3% versus 77.8% for TP53 mutant stage IIIA patients. The HR was 3.008 (95% CI: 0.598–15.128; P = 0.1609). Statistical significance was not reached due to the small number of stage IIIA patients (n = 32) and the small number of events (n = 6) in this subgroup.
In stage IIIB, the five-year survival for TP53 wildtype patients was 79.0% versus 60.1% for TP53 mutant patients (HR = 2.014 (95% CI: 1.246–3.258); P = 0.0036).
Stage IIIC was identical to N2, with no differential benefit in relation to TP53 status.
Factors evaluated in a multiple Cox regression model included sex, age, grading, therapy, and tumor location. After adjustment for the prognostic factors, the effect of TP53 on survival increased for stage IIIA patients (HR = 8.775 (95% CI: 0.608–126.619); P = 0.1108), although this was still non-significant due to the small numbers of patients and events as mentioned above. For stage IIIB patients, the effect of TP53 on survival remained significant (HR = 1.731 (95% CI: 1.048–2.859); P = 0.0321). For stage IIIC patients, TP53 still had no effect on survival (HR = 1.023 (95% CI: 0.584–1.791); P = 0.9379).
4. Discussion
Here we report the presence of a significant interaction between TP53 status and survival in stage III colon cancer patients treated with adjuvant 5FU. We found that adjuvant 5FU resulted in a marked survival benefit in TP53 wildtype N1 patients, while N1 patients with mutant TP53 performed as bad as N2 patients. The TP53 status independently predicted the effect of adjuvant 5FU on survival in N1 patients while there was no prediction of 5FU effect in N2 patients.
Colon cancer patients analyzed in this study were treated in the 1990s, when adjuvant 5FU was the standard of care in stage III colon cancer. In the meantime it was recognized that adjuvant 5FU alone is too week as chemotherapy at least for patients with advanced stage of disease (N2 disease). Our results reflect this clinical experience on a molecular level: TP53 predicted the effect of 5FU in N1 patients but not in N2 patients, in whom we do not await a therapeutic effect from 5FU alone. It makes sense that in the absence of a therapeutic effect, a predictive marker does not predict an effect.
Moreover, the here reported results support the hypothesis that TP53 is not a prognostic but purely predictive type of biomarker (Pilat et al., 2015). A purely predictive marker is considered to affect survival only in the presence of a treatment. In the absence of a treatment or a treatment effect, a pure predictive marker will not interfere with survival (Pilat et al., 2015, Akiyama et al., 2014). In N2 patients TP53 was not predictive for an effect of 5FU most likely because there is no effect of 5FU in these advanced patients.
TP53 has been intensively studied in colorectal cancer. Some currently unexplained findings of meta-analyses appear in a new light based upon our findings. For instance, it has been reported that alterations in the TP53 gene have little or no influence on survival in patients treated with surgery alone, but were associated with worse survival for patients treated with chemotherapy (Iacopetta, 2003). This again supports our observation that TP53 is not a prognostic but a predictive biomarker, influencing survival only in the presence of effective chemotherapy, as mentioned above. Another meta-analysis including more than 18,000 patients was published 2005, when 5FU was standard adjuvant chemotherapy (Munro et al., 2005). The analysis found “the adverse impact of abnormal TP53 to be greater in colon cancer patients with a lower baseline risk of dying”. This is consistent with our finding that TP53 was predictive for the effect of 5FU in N1 patients but not in N2 patients.
The traditional T and N classification, originally developed in the 1950s, still provides the most important prognostic information in colon cancer and is guiding treatment decisions (Brenner et al., 2014, Denoix and Schwartz, 1959). But in stage III colon cancer, prognostic inconsistency is a well-known phenomenon. By contrast to stage II, stage III patients usually are treated with adjuvant chemotherapy. Our IIIA/B patients with wildtype TP53 status experienced a marked benefit from adjuvant 5FU, resulting in survival rates, which are comparable to those of stage II colon cancer patients (Greene et al., 2002b, Gunderson et al., 2010). A positive treatment effect occurred in TP53 wild type patients with stage IIIA/B but not IIIC as well as in wild type N1 but not in N2 patients. Therefore the prognostic significance of the N category shown in Fig. 2 appeared to be enhanced in TP53 wild type patients as demonstrated in Fig. 3A. Our TP53 mutated stage IIIA/B or N1 patients however, did as bad as our stage IIIC or N2 patients. Thus a negative treatment effect occurred in TP53 mutated N1 patients but not in N2 patients. Therefore N category was no longer prognostic in TP53 mutated patients which represent 33% of stage III patients. We suggest that the TP53 marker status is linked to varying treatment effects, which are likely to cause prognostic inconsistency in low risk stage III colon cancer.
Biomarker research is complex in that proof of the clinical utility of a biomarker is crucially linked to the analytical validity of the respective biomarker-test. TP53 has not yet been recommended for clinical use as a marker. Thus demonstration of the validity of TP53 tests was not top of the list (Petersen et al., 2001, Munro et al., 2005, Roth et al., 2012). Even though immunohistochemistry has been recognized as an insufficient method for indicating the presence of TP53 mutations, and even though sequencing has been generally accepted as the ‘gold standard’ for investigating the TP53 status, methodology is still far from being standardized. Various sequencing technologies and protocols are in use worldwide, and there is evidence for dramatic variations in the sensitivity and specificity of these methods (Petitjean et al., 2007, Edlund et al., 2012, Soussi et al., 2006).
In recent years, we have identified numerous pitfalls while sequencing the TP53 gene. We believe that they arise from the special characteristics of the TP53 gene itself and its mutations, as well as from technical conditions such as FFPE material, low copy numbers of tumor DNA in biopsy material, or from the presence of normal cells in tumor material, to list only a few. Over the years, we developed a p53-gene-specific sequencing protocol to addressing these and other conditions in a step-wise fashion. We believe that this was a crucial step that allowed us to uncover the here reported interaction between TP53 and chemotherapy effect. Meanwhile, we proceeded to validating this standardized, TP53 gene-specific sequencing protocol in prospective biomarker trials (www.ClinicalTrials.gov; PANCHO NCT00525200, PART NCT02140723).
ABCSG-90 was a prospective randomized adjuvant colon cancer trial investigating the influence of adding levamisole (LEV) and/or interferon (INF) alpha to adjuvant fluorouracil. Tumor material from only 70% of patients from the ABCSG-90 was available for TP53 analysis in the here reported study. Thus study limitations may arise from incomplete analysis of the ABCSG-90 cohort as well as from the treatment combinations, which were investigated in ABCSG-90. However, we believe that random patient selection as well as different treatment effects is unlikely to bias our results: The ABCSG-90 reported no difference in survival among the investigated treatment arms. The patients included in the retrospective biomarker analysis were distributed equally among treatment arms as demonstrated in Table 2. The TP53 mutations were also distributed equally among treatment arms as well as among tumor stages and lymph node categories. And finally the five-year overall survival of ABCSG-90 was comparable to the smaller cohort analyzed for the biomarker.
In conclusion, TP53 was found to be a strong, independent marker for predicting the effect of adjuvant 5FU chemotherapy in lymph node positive colon cancer patients.
We found that adjuvant 5FU resulted in a marked survival benefit in TP53 wildtype N1 patients, while TP53 mutated N1 patients performed as bad as N2 patients. The TP53 status independently predicted the effect of adjuvant 5FU on survival in N1 patients while there was no prediction of 5FU effect in N2 patients. The significant interaction between TP53 status, nodal category and treatment effect offers an explanation for the known prognostic inconsistency in stage III colon cancer patients. Further studies are needed to validate these findings.
Funding
This work was supported by the Medical Scientific Fund of the Mayor of the Capital City of Vienna (#2093, #2297 Kandioler) and by the Anniversary Fund of the Oesterreichische Nationalbank (#8916 Kandioler, #12557 Teleky).
The funders had no role in the design of the study, in the collection, analysis, and interpretation of data, in the writing of the report, or in the decision to submit the paper for publication.
Conflict of Interest Statement
Leadership position, associate: Daniela Kandioler (Mark53 Ltd).
Employment: Sonja Kappel (half time) (Mark53 Ltd).
Consultant or Advisory Role: Daniela Kandioler (uncompensated) (academic sponsored clinical TP53 trials).
Honoraria: None.
Research Funding (governmental): Daniela Kandioler, Bela Teleky.
Expert Testimony: None.
Other Remuneration: None.
Author Contributions
Conception and design: D. Kandioler.
Development of methodology: S. Kappel, B. Wolf.
Acquisition of data: H. Puhalla, F. Herbst, C. Langner, J. Tschmelitsch, W. Schippinger, G. Steger, F. Hofbauer, H. Samonigg, B. Teleky, I. Kührer.
Analysis and interpretation of data: D. Kandioler, M. Mittlböck, I. Kührer.
Writing, review and/or revision of the manuscript: D. Kandioler, M. Mittlböck, S. Kappel, H. Puhalla, C. Langner, M. Gnant.
Administrative, technical, or material support: S. Kappel, H. Puhalla, B. Wolf.
Study supervision: D. Kandioler, M. Gnant (ABCSG-90).
Acknowledgments
This article is based on tumor material and clinical data of the ABCSG-90, published in 2005 by Schippinger et al. (Schippinger et al., 2005).
We would like to thank the authors of the above article and the members of the ABCSG participating in the initial trial as listed in Appendix A of the above article.
References
- Ahmed I.A., Kelly S.B., Anderson J.J., Angus B., Challen C., Lunec J. The predictive value of p53 and p33(ING1b) in patients with Dukes'C colorectal cancer. Colorectal Dis. 2008;10(4):344–351. doi: 10.1111/j.1463-1318.2007.01317.x. [DOI] [PubMed] [Google Scholar]
- Akiyama J., Alexandre L., Baruah A. Strategy for prevention of cancers of the esophagus. Ann. N. Y. Acad. Sci. 2014;1325:108–126. doi: 10.1111/nyas.12529. [DOI] [PubMed] [Google Scholar]
- Bleeker W.A., Hayes V.M., Karrenbeld A. Prognostic significance of K-ras and TP53 mutations in the role of adjuvant chemotherapy on survival in patients with Dukes C colon cancer. Dis. Colon Rectum. 2001;44(3):358–363. doi: 10.1007/BF02234733. [DOI] [PubMed] [Google Scholar]
- Brenner H., Kloor M., Pox C.P. Colorectal cancer. Lancet. 2014;383(9927):1490–1502. doi: 10.1016/S0140-6736(13)61649-9. [DOI] [PubMed] [Google Scholar]
- Cassidy J., Clarke S., Diaz-Rubio E. Randomized phase III study of capecitabine plus oxaliplatin compared with fluorouracil/folinic acid plus oxaliplatin as first-line therapy for metastatic colorectal cancer. J. Clin. Oncol. 2008;26(12):2006–2012. doi: 10.1200/JCO.2007.14.9898. [DOI] [PubMed] [Google Scholar]
- Chau I., Cunningham D. Adjuvant therapy in colon cancer—what, when and how? Ann. Oncol. 2006;17(9):1347–1359. doi: 10.1093/annonc/mdl029. [DOI] [PubMed] [Google Scholar]
- den Dunnen J.T., Antonarakis S.E. Mutation nomenclature extensions and suggestions to describe complex mutations: a discussion. Hum. Mutat. 2000;15(1):7–12. doi: 10.1002/(SICI)1098-1004(200001)15:1<7::AID-HUMU4>3.0.CO;2-N. [DOI] [PubMed] [Google Scholar]
- Denoix P.F., Schwartz D. General rules for classification of cancers and presentation of the therapeutic results. Mem. Acad. Chir. 1959;85(15–16):415–424. [PubMed] [Google Scholar]
- Edlund K., Larsson O., Ameur A. Data-driven unbiased curation of the TP53 tumor suppressor gene mutation database and validation by ultradeep sequencing of human tumors. Proc. Natl. Acad. Sci. U. S. A. 2012;109(24):9551–9556. doi: 10.1073/pnas.1200019109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elsaleh H., Powell B., McCaul K. P53 alteration and microsatellite instability have predictive value for survival benefit from chemotherapy in stage III colorectal carcinoma. Clin. Cancer Res. 2001;7(5):1343–1349. [PubMed] [Google Scholar]
- Gill S., Loprinzi C.L., Sargent D.J. Pooled analysis of fluorouracil-based adjuvant therapy for stage II and III colon cancer: who benefits and by how much? J. Clin. Oncol. 2004;22(10):1797–1806. doi: 10.1200/JCO.2004.09.059. [DOI] [PubMed] [Google Scholar]
- Greene F.L., Stewart A.K., Norton H.J. A new TNM staging strategy for node-positive (stage III) colon cancer: an analysis of 50,042 patients. Ann. Surg. 2002;236(4):416–421. doi: 10.1097/00000658-200210000-00003. (discussion 21) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greene F.L., Page D.L., Fleming I.D. 6th ed. Springer; New York, NY: 2002. AJCC Cancer Staging Manual. [Google Scholar]
- Gunderson L.L., Jessup J.M., Sargent D.J., Greene F.L., Stewart A.K. Revised TN categorization for colon cancer based on national survival outcomes data. J. Clin. Oncol. 2010;28(2):264–271. doi: 10.1200/JCO.2009.24.0952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hecht J.R., Mitchell E., Chidiac T. A randomized phase IIIB trial of chemotherapy, bevacizumab, and panitumumab compared with chemotherapy and bevacizumab alone for metastatic colorectal cancer. J. Clin. Oncol. 2009;27(5):672–680. doi: 10.1200/JCO.2008.19.8135. [DOI] [PubMed] [Google Scholar]
- Iacopetta B. TP53 mutation in colorectal cancer. Hum. Mutat. 2003;21(3):271–276. doi: 10.1002/humu.10175. [DOI] [PubMed] [Google Scholar]
- Kato S., Han S.Y., Liu W. Understanding the function–structure and function–mutation relationships of p53 tumor suppressor protein by high-resolution missense mutation analysis. Proc. Natl. Acad. Sci. U. S. A. 2003;100(14):8424–8429. doi: 10.1073/pnas.1431692100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munro A.J., Lain S., Lane D.P. P53 abnormalities and outcomes in colorectal cancer: a systematic review. Br. J. Cancer. 2005;92(3):434–444. doi: 10.1038/sj.bjc.6602358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petersen S., Thames H.D., Nieder C., Petersen C., Baumann M. The results of colorectal cancer treatment by p53 status: treatment-specific overview. Dis. Colon Rectum. 2001;44(3):322–333. doi: 10.1007/BF02234727. (discussion 33-4) [DOI] [PubMed] [Google Scholar]
- Petitjean A., Mathe E., Kato S. Impact of mutant p53 functional properties on TP53 mutation patterns and tumor phenotype: lessons from recent developments in the IARC TP53 database. Hum. Mutat. 2007;28(6):622–629. doi: 10.1002/humu.20495. [DOI] [PubMed] [Google Scholar]
- Pilat N., Grünberger T., Längle F. Assessing the TP53 marker type in patients treated with or without neoadjuvant chemotherapy for resectable colorectal liver metastases: a p53 Research Group study. EJSO. 2015;41:683–689. doi: 10.1016/j.ejso.2015.02.003. [DOI] [PubMed] [Google Scholar]
- Roth A.D., Delorenzi M., Tejpar S. Integrated analysis of molecular and clinical prognostic factors in stage II/III colon cancer. J. Natl. Cancer Inst. 2012;104(21):1635–1646. doi: 10.1093/jnci/djs427. [DOI] [PubMed] [Google Scholar]
- Russo A., Bazan V., Iacopetta B. The TP53 colorectal cancer international collaborative study on the prognostic and predictive significance of p53 mutation: influence of tumor site, type of mutation, and adjuvant treatment. J. Clin. Oncol. 2005;23(30):7518–7528. doi: 10.1200/JCO.2005.00.471. [DOI] [PubMed] [Google Scholar]
- Saltz L.B., Clarke S., Diaz-Rubio E. Bevacizumab in combination with oxaliplatin-based chemotherapy as first-line therapy in metastatic colorectal cancer: a randomized phase III study. J. Clin. Oncol. 2008;26(12):2013–2019. doi: 10.1200/JCO.2007.14.9930. [DOI] [PubMed] [Google Scholar]
- Schippinger W., Jagoditsch M., Sorre C. A prospective randomised trial to study the role of levamisole and interferon alfa in an adjuvant therapy with 5-FU for stage III colon cancer. Br. J. Cancer. 2005;92(9):1655–1662. doi: 10.1038/sj.bjc.6602555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soussi T., Asselain B., Hamroun D. Meta-analysis of the p53 mutation database for mutant p53 biological activity reveals a methodologic bias in mutation detection. Clin. Cancer Res. 2006;12(1):62–69. doi: 10.1158/1078-0432.CCR-05-0413. [DOI] [PubMed] [Google Scholar]
- Tol J., Koopman M., Cats A. Chemotherapy, bevacizumab, and cetuximab in metastatic colorectal cancer. N. Engl. J. Med. 2009;360(6):563–572. doi: 10.1056/NEJMoa0808268. [DOI] [PubMed] [Google Scholar]


