Abstract
Background
Quantifying latently infected cells is critical to evaluate the efficacy of therapeutic strategies aimed at reducing the size of the long-lived viral reservoir, but the low frequency of these cells makes this very challenging.
Methods
We developed TILDA (Tat/rev Induced Limiting Dilution Assay) to measure the frequency of cells with inducible multiply-spliced HIV RNA, as these transcripts are usually absent in latently infected cells but induced upon viral reactivation. TILDA requires less than a million cells, does not require RNA extraction and can be completed in two days.
Findings
In suppressed individuals on ART, we found the median frequency of latently infected CD4 + T cells as estimated by TILDA to be 24 cells/million, which was 48 times more than the frequency measured by the quantitative viral outgrowth assay, and 6–27 times less than the frequencies of cells harbouring viral DNA measured by PCR-based assays. TILDA measurements strongly correlated with most HIV DNA assays. The size of the latent reservoir measured by TILDA was lower in subjects who initiated ART during the early compared to late stage of infection (p = 0.011). In untreated HIV disease, the frequency of CD4 + cells carrying latent but inducible HIV largely exceeded the frequency of actively producing cells, demonstrating that the majority of infected cells are transcriptionally silent even in the absence of ART.
Interpretations
Our results suggest that TILDA is a reproducible and sensitive approach to measure the frequency of productively and latently infected cells in clinical settings. We demonstrate that the latent reservoir represents a substantial fraction of all infected cells prior to ART initiation.
Research in context
In this manuscript, we describe the development of a novel assay that measures the magnitude of the latent HIV reservoir, the main barrier to HIV eradication. This novel assay, termed TILDA for Tat/rev Induced Limiting Dilution Assay, requires only 10 ml of blood, does not necessitate extraction of viral nucleic acids, is highly reproducible, covers a wide dynamic range of reservoir sizes and can be completed in two days. As such, TILDA may represent an alternative to existing assays used to evaluate the efficacy of therapeutic strategies aimed at reducing the size of the latent HIV reservoir.
Keywords: HIV, Latency, Inducible virus, Reservoir, Multiply spliced RNA, Eradication, TILDA
Highlights
-
•
We developed TILDA (Tat/rev Induced Limiting Dilution Assay) to measure the frequency of cells with inducible multiply-spliced HIV RNA in HIV-infected individuals on suppressive ART.
-
•
Our results suggest that TILDA is a reproducible and sensitive approach to measure the frequency of productively and latently infected cells in clinical settings.
-
•
Using TILDA, We demonstrate that the latent reservoir represents a substantial fraction of all infected cells prior to ART initiation.
1. Introduction
Advances in the treatment of HIV infection have dramatically reduced AIDS mortality and improved the quality of life of many people living with HIV (Palella et al., 1998). The initiation of ART results in a rapid drop in plasma viral load and in a substantial reduction in the number of cells carrying proviral DNA in both blood and tissues (Murray et al., 2014, Garrigue et al., 2000). However, ART alone does not eradicate HIV. The virus persists in long-lived latently infected CD4 + T cells that can produce infectious particles following T cell stimulation (Finzi et al., 1997, Wong et al., 1997, Chun et al., 1997a). The magnitude of the latent reservoir is highly variable among virally suppressed individuals and is influenced by a variety of factors including the nadir CD4 + T cell count (Boulassel et al., 2012), the CD4/CD8 ratio (Chun et al., 2002), the time between infection and initiation of ART (Josefsson et al., 2013, Strain et al., 2005, Archin et al., 2012, Buzon et al., 2014, Ananworanich et al., 2012, Chomont et al., 2009) and, to a lesser extent, the duration of therapy (Siliciano et al., 2003). Because strategies targeting this latent reservoir are now being tested in clinical trials, well-validated high-throughput assays that quantify this reservoir are urgently needed to evaluate the effectiveness of these interventions.
Several assays have been developed to measure HIV persistence in virally suppressed subjects who have received ART for years (Lewin et al., 1999, Li et al., 2010, Siliciano and Siliciano, 2005, Yu et al., 2008, Palmer et al., 2003, Vandergeeten et al., 2014, Pasternak et al., 2008, Eriksson et al., 2013, Brussel and Sonigo, 2003, Rouzioux et al., 2005, Strain et al., 2013, O'Doherty et al., 2002). The quantitative viral outgrowth assay (Q-VOA) performed on purified resting CD4 + T cells isolated from patients on suppressive ART is considered the gold standard method to measure the minimum frequency of latently infected cells with replication competent virus (Finzi et al., 1997, Siliciano and Siliciano, 2005, Chun et al., 1997b). However, this assay is time-consuming, labour intensive, expensive and requires large volumes of blood (120–180 ml). Recently, a more rapid and simplified version of Q-VOA has been described (Laird et al., 2013), but this novel assay still requires large number of cells and has not been adapted to a large scale clinical trial. In addition, Ho et al. identified intact proviruses from p24 negative wells of the virus outgrowth assay, which raises the possibility that the Q-VOA underestimates the size of the in vivo latent reservoir (Ho et al., 2013). This could be attributed to the fact that HIV reactivation in this system is inherently stochastic, as recently proposed (Weinberger and Weinberger, 2013).
An alternative to the Q-VOA is the use of PCR-based methods that measure the frequency of cells harbouring HIV genomes (either total or integrated HIV DNA) (Yu et al., 2008, Vandergeeten et al., 2014, Brussel and Sonigo, 2003, Strain et al., 2013, O'Doherty et al., 2002). Although these methods can be used in large cohort studies, they are often criticized, as a large number of the viral genomes quantified by these assays are not replication-competent. Indeed, total HIV DNA in PBMCs and resting CD4 + T cells generally yields infected cell frequencies that are 2–3 logs higher than the Q-VOA, reflecting the high occurrence of defective and non-inducible viral genomes (Eriksson et al., 2013). Notwithstanding this limitation, measuring viral DNA, and particularly integrated HIV DNA, has provided crucial information that have contributed to the understanding of the mechanisms of HIV persistence during ART (Chomont et al., 2009, Agosto et al., 2011, Graf et al., 2011, Mexas et al., 2012, Vandergeeten et al., 2013). Levels of HIV DNA predict viral rebound after structured treatment interruption (Williams et al., 2014, Yerly et al., 2004) and integrated HIV DNA is the only assay that appears to correlate with Q-VOA (Eriksson et al., 2013) although it is likely that frequencies of cells bearing HIV DNA greatly overestimates the size of the inducible latent HIV reservoir (Eriksson et al., 2013). These studies thus emphasize the need to develop novel assays that would measure the size of the latent and inducible HIV reservoir in a simple, reproducible and cost-effective manner.
An ideal assay would measure the frequency of latently infected CD4 + T cells without relying on the amplification of viral replication, which is difficult to control and requires at least a week of cell culture. Measuring the production of viral particles in culture supernatants of stimulated cells is attractive (Cillo et al., 2014), but would require ultracentrifugation and RNA extraction steps that are not desirable for a clinical trial scalable assay. Cell associated RNA is an alternative virological marker that can be easily measured in a limiting dilution assay.
Low amounts of cell-associated unspliced HIV RNA are frequently detected in PBMCs from virally suppressed subjects on ART (Lewin et al., 1999, Furtado et al., 1999, Fischer et al., 2002, Schmid et al., 2010, Pasternak et al., 2009), as well as in latently infected CD4 + T cells that do not produce HIV particles (Fischer et al., 2004, Peng et al., 1995, Hermankova et al., 2003), and therefore, cannot be used as a surrogate for viral release (Cillo et al., 2014, Kearney, 2015). In contrast, tat/rev multiply spliced RNA (msRNA) reflects the ability of a cell to produce virus (Lewin et al., 1999, Pasternak et al., 2008, Pasternak et al., 2009, Fischer et al., 2002, Fischer et al., 2004, Schmid et al., 2010, Peng et al., 1995, Hermankova et al., 2003, Vesanen et al., 1997, Sonza et al., 2002). Of note, recent data indicate that tat positive feedback alone may be sufficient in reversing latency, independent of cellular activation (Razooky et al., 2015), suggesting that tat transcripts may be used as a surrogate for productive infection. This is consistent with the fact that direct addition of tat activates viral expression in resting CD4 + T cells without requiring cellular activation (Lassen et al., 2006, Lin et al., 2003). In addition, many defective HIV genomes have deletions that encompass the tat and rev genes (Ho et al., 2013), indicating that transcripts from these genes are very unlikely to be generated in cells harbouring defective proviruses. This provides a rationale for the development of a sensitive assay that can detect tat/rev msRNA upon maximal stimulation.
Here, we report the development of TILDA, a novel assay that measures the frequency of cells harbouring viral genomes that produce tat/rev multiply-spliced HIV RNA upon maximal stimulation. By combining ultrasensitive detection of msRNA and maximal activation of CD4 + T cells in a limiting dilution format, this method allowed us to measure the frequency of cells capable of being induced to produce HIV RNA transcripts. Significantly, this assay has potential for application in larger clinical trials and cohort studies as it requires only 10 ml of blood, is robust, sensitive, covers a wide dynamic range of reservoir size and can be completed in two days.
2. Methods
2.1. Participants and Samples
Samples from 20 subjects on stably suppressive ART and from 13 viremic subjects not receiving ART at the time of sampling were used in this study. None of the participants under ART experienced any detectable plasma viremia at the time of study, as assessed by viral load measurement using the Ampliprep/Cobas Taqman HIV-1 Test v 2.0 (Roche), with a detection limit of 20 copies/ml of plasma. These subjects had an undetectable viral load for an average of 6 years (range 3–17 years). All participants underwent leukapheresis to collect large numbers of PBMCs. Subjects characteristics are summarized in Table 1.
Table 1.
Participant characteristics.
| Sexa | CD4 count (cells/μl) | CD8 count (cells/μl) | HIV viral load (copies/ml) | Duration of HIV infection (years)b | Duration of ART (years)c | ART regimend | |
|---|---|---|---|---|---|---|---|
| Virally suppressed subjects | |||||||
| ART1 | NA | 1177 | 935 | < 20 | NA | 6.5 | NA |
| ART2 | M | 1183 | 912 | < 20 | 25.6 | 3.4 | ABC/3TC/ATV |
| ART3 | M | 1112 | 594 | < 20 | 12.9 | 12.9 | ABC/3TC/NVP |
| ART4 | M | 326 | 1085 | < 20 | 6.2 | 6.1 | TDF/FTC/EFV |
| ART5 | M | 612 | 834 | < 20 | 25.7 | 16.9 | NA |
| ART6 | M | 1065 | 1408 | < 20 | 15.0 | 8.2 | TDF/FTC/EFV |
| ART7 | M | 1145 | 1410 | < 20 | 6.1 | 5.8 | TDF/FTC/LPV/r |
| ART8 | M | 1095 | 736 | < 20 | 22.1 | 6.2 | TDF/FTC/EFV |
| ART9 | M | 876 | 1389 | < 20 | 5.7 | 5.0 | ABC/3TC/EFV |
| ART10 | F | 522 | 238 | < 20 | 22.1 | 7.5 | TDF/FTC/ATV/r |
| ART11 | F | 460 | 1879 | < 20 | 5.4 | 4.4 | TDF/FTC/RAL/LPV/r |
| ART12 | F | 355 | 815 | < 20 | 21.1 | 4.7 | TDF/FTC/DRV/r |
| ART13 | M | 289 | 512 | < 20 | 16.2 | 3.0 | TDF/FTC/LPV/r |
| ART14 | F | 351 | 344 | < 20 | 15.1 | 3.6 | TDF/FTC/ATV/r |
| ART15 | M | 734 | 703 | < 20 | 7.2 | 2.5 | TDF/FTC/RAL |
| ART16 | M | 267 | 518 | < 20 | NA | 3.8 | TDF/FTC/EFV |
| ART17 | M | 1102 | 875 | < 20 | NA | NA | TDF/FTC/ATV |
| ART18 | M | 291 | 491 | < 20 | 10.4 | 2.9 | ABC/3TC/ATV/r |
| ART19 | M | 1897 | 830 | < 20 | NA | 6.1 | TDF/FTC/EFV |
| ART20 | M | 387 | 625 | < 20 | NA | 6.3 | ABC/3TC/ATV |
| Median [IQR] |
734 [353–1107] | 815 [556–1010] | < 20 [< 20– < 20] | 15 [6.7–21.6] | 5.8 [3.7–6.4] | ||
| Viremic subjects | |||||||
| VIR1 | F | 319 | 470 | 51,000 | 11.8 | – | – |
| VIR2 | M | 434 | 2497 | 51,000 | 2.5 | – | – |
| VIR3 | F | 548 | 751 | 23,000 | 18.0 | – | – |
| VIR4 | M | 480 | 1552 | 3300 | 5.7 | – | – |
| VIR5 | M | 258 | 1704 | 400,000 | 5.1 | – | – |
| VIR6 | M | 185 | 647 | 50,000 | 9.0 | – | – |
| VIR7 | F | 284 | 605 | 21,000 | 5.1 | – | – |
| VIR8 | F | 681 | 543 | 15,000 | 8.0 | – | – |
| VIR9 | M | 499 | 1590 | 52,915 | 9.9 | – | – |
| VIR10 | NA | 543 | 896 | 6077 | 4.0 | – | – |
| VIR11 | M | 732 | 1506 | 51,647 | 0.4 | – | – |
| VIR12 | M | 506 | 948 | 2959 | 1.1 | – | – |
| VIR13 | M | 1400 | 2040 | 8082 | 0.4 | – | – |
| Median [IQR] |
499 [319–548] | 948 [647–1590] | 23,000 [8082–51,000] | 5.1 [2.5–9.0] | |||
NA: not available.
Duration of HIV infection was calculated from the date of diagnosis of HIV infection.
Time of documented suppression of viremia to < 50 copies/ml on ART.
Drug abbreviation: 3TC, lamivudine; ABC, abacavir; ATV, atazanavir; ATV/r, atazanavir boosted with ritonavir; DRV/r, darunavir boosted with ritonavir; EFV, efavirenz; FTC, emtricitabine; LPV/r, lopinavir boosted with ritonavir; NVP, nevirapine; RAL, raltegravir; TDF, tenofovir disoproxil fumarate.
Data presented in Fig. 2, Fig. 3 were generated by analysing samples obtained from individuals who participated in a previous study aimed at comparing different assays to measure HIV persistence (Eriksson et al., 2013). Briefly, this study enrolled 30 subjects from two well-established cohorts at the University of California San Francisco (UCSF). Samples from 27 of these participants were used for the present study (17 from the SCOPE cohort and 10 from the OPTIONS cohort). All of the 27 subjects had at least 36 months of continuous ART at study entry with no regimen changes in the preceding 24 weeks and maintenance of plasma HIV RNA levels below the limit of detection of conventional assays for at least 36 months (intermittent isolated episodes of detectable low-level viremia were allowed). Primary or recent HIV infection was defined if one or several of these 3 criteria were met: (Palella et al., 1998) repeated plasma HIV RNA > 5000 copies/ml combined with a negative or indeterminate HIV antibody test; (Murray et al., 2014) seroconversion within 6 months of a documented negative HIV antibody test; (Garrigue et al., 2000) a history compatible with primary HIV infection (including no prior positive HIV antibody tests) and laboratory testing consistent with recent infection on the “detuned” antibody EIA. Characteristics of these participants can be found in Eriksson et al. (2013).
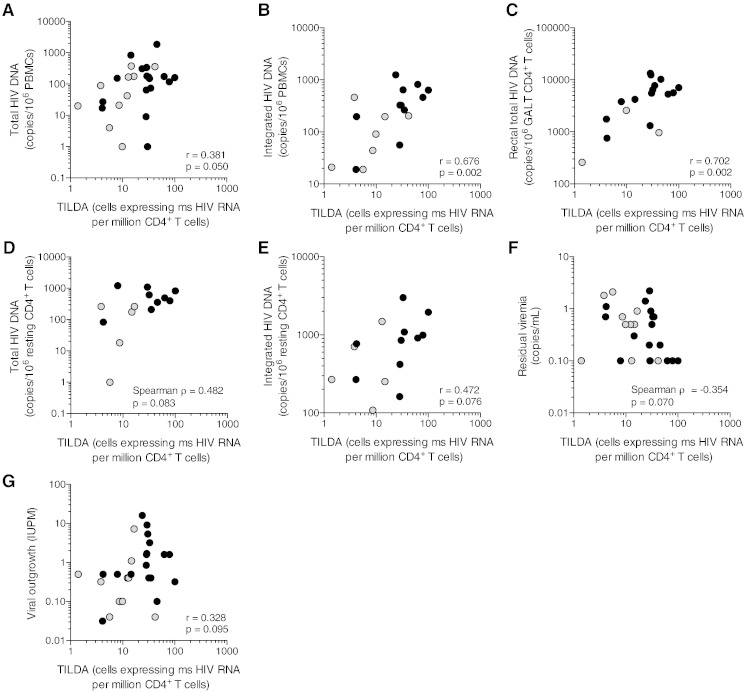
Fig. 2.
TILDA correlates with several measures of HIV persistence. (A) Correlation between TILDA and the droplet digital PCR assay for HIV DNA in PBMCs. (B) Correlation between TILDA and the Alu-PCR assay for HIV DNA in PBMCs. (C) Correlation between TILDA and the droplet digital PCR assay for HIV DNA in rectal CD4 + T cells. (D) Correlation between TILDA and the droplet digital PCR assay for HIV DNA in resting CD4 + T cells. (E) Correlation between TILDA and the Alu-PCR assay for HIV DNA in resting CD4 + T cells. (F) Correlation between TILDA and single copy assay for residual viremia. (G) Correlation between TILDA and Q-VOA in resting CD4 + T cells. Black circles and grey circles denote subjects who started ART during chronic and early infection, respectively. P values were obtained from the Pearson (A, B, C, E, G) or Spearman test (D, F), according to the results of the normality test (see Methods).
Fig. 3.
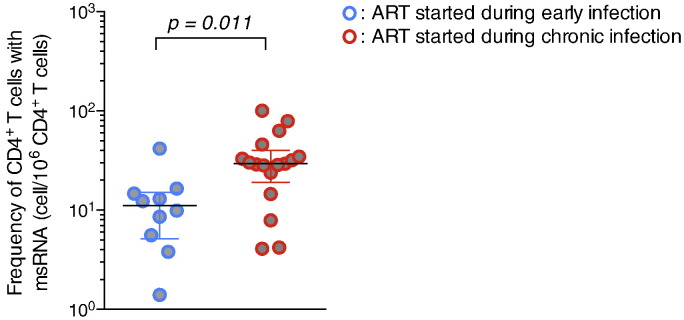
Initiation of ART during early HIV infection leads to a restricted size of the reservoir measured by TILDA. The frequency of cells harbouring inducible msRNA was measured by TILDA on CD4 + T cells obtained from subjects who started ART during chronic (n = 17, red circles) or recent (n = 10, blue circles) infection. Horizontal bars indicate median values. P value was obtained from the Mann–Whitney test.
This study was approved by the Martin Health System Institutional review board and by the UCSF Committee on Human Research. All study subjects provided written informed consent prior to participation in the study.
2.2. Isolation of CD4 T cells
PBMCs were isolated from the leukapheresis product by Ficoll Hypaque density gradient centrifugation and cryopreserved in liquid nitrogen. Upon thawing of the PBMCs, total CD4 + T cells were isolated by negative magnetic selection (Stemcell technologies). The purity of enriched CD4 + T cells was generally greater than 95%, as assessed by flow cytometry.
2.3. TILDA
Enriched CD4 + T cells were resuspended at 2 × 106 cells/ml in RPMI + 10% foetal bovine serum and rested for 3–5 h at 37 °C, 5% CO2. In some experiments, antiretroviral drugs were added to the culture medium (200 nM raltegravir, 100 nM efavirenz and 180 nM AZT). CD4 + T cells were stimulated for 12 h with 100 ng/ml PMA and 1 μg/ml ionomycin (both from Sigma). The duration of 12 h was based on kinetic experiments demonstrating maximal production of tat/rev RNA and preserved cell viability at this time point (not shown). After stimulation, cells were washed in RPMI, counted and serially diluted to 18 × 106 cells/ml, 9 × 106 cells/ml, 3 × 106 cells/ml and 1 × 106 cells/ml in culture medium. 1 μl of the cell suspension from each serial dilution was distributed in 22 to 24 wells of a 96 well plate containing 5 μl of 2 × reaction buffer from the SuperScript III Platinum One-Step qRT-PCR Kit (Life Technologies) corresponding to 18,000, 9000, 3000 and 1000 cells per well. Different cell numbers were used for samples obtained from viremic, untreated HIV-infected individuals (9000; 3000; 1000 and 333 cells per well). In some experiments, a fraction of the isolated CD4 + T cells were immediately distributed in limiting dilutions after thawing to quantify the frequency of cells that spontaneously produce HIV msRNA (no stimulation). Similar frequencies of positive cells were measured by TILDA whether the cells were distributed immediately after thawing or after 12 h of resting (not shown). Pre-amplification was carried out by adding 5 μl of a PCR mix containing 0.2 μl Superscript III Platinum Taq (Life Technologies), 0.1 μl RNase inhibitor (Life Technologies), 0.125 μl of each primer (tat1.4 and rev both at 20 μM), 2.2 μl Tris–EDTA (TE) buffer and 2.25 μl H2O to each well (final reaction volume = 11 μl). The sequences of the oligonucleotides were slightly adapted from Pasternak et al. (2008): tat1.4: 5′-TGG CAG GAA GAA GCG GAG A-3′; rev: 5′-GGA TCT GTC TCT GTC TCT CTC TCC ACC-3′. Pre-amplification was carried out using the following steps: reverse transcription at 50 °C for 15 min, denaturation at 95 °C for 2 min, 24 cycles of amplification (95 °C 15 s, 60 °C 4 min) on a C1000 PCR instrument (Biorad). At the end of the pre-amplification, 40 μl of TE buffer was added to each well and 1 μl of the diluted PCR products was used as template for the tat/rev real-time PCR reaction. This reaction was performed by adding 5 μl of the Lightcycler Probe Master buffer (Roche Applied Sciences), 0.2 μl of each primer (tat2 and rev, both at 20 μM), 0.2 μl of the probe HIV FamZen at 5 μM and 3.4 μl H2O to each well (final reaction volume = 10 μl). Sequence of tat2 and the HIV probe were also adapted from (Pasternak et al., 2008): tat2: 5′- ACA GTC AGA CTC ATC AAG TTT CTC TAT CAA AGC A -3′. Probe HIV: 5′-/56-FAM/TTC CTT CGG /ZEN/GCC TGT CGG GTC CC/3IABkFQ/-3′. All primers and probes were synthesized by IDT. The real-time PCR reaction was carried out in a Light Cycler 480 II (Roche Life Science) using the following programme: Preincubation 95 °C for 10 min, 45 cycles of 95 °C 10 s, 60 °C 30 s, 72 °C 1 s and a cooling step at 40 °C fro 30 s. Positive wells at each dilution were counted and the maximum likelihood method was used to calculate the frequency of cells with inducible HIV msRNA (http://bioinf.wehi.edu.au/software/elda).
2.4. Statistical Analysis
Baseline and induced TILDA values were compared using the Wilcoxon matched-pairs signed rank test. For correlations between values obtained with different assays, normality of the log transformed virologic data was tested with the D'Agostino–Pearson test. With the exception of total HIV DNA in resting CD4 + T cells and residual viremia by single copy assay, all virologic data met the normality test and correlations were performed on log-transformed values using the Pearson test. When the data did not meet the normality test, the Spearman test was used. When the result of an assay was negative (below the limit of detection), an imputed value representing the lower of the limit of detection was used in the calculation of the Pearson correlation coefficient. In the case of the single-copy assay for HIV-1 RNA in plasma, for which one-third of the measurements was below the limit of detection (0.2 copies/ml), a low imputed value of 0.1 was used to calculate the Spearman rank correlation coefficient. Virologic data obtained from subjects who started ART during the early and chronic phases of infection were compared using a two-tailed Mann–Whitney test. Data analysis was done using Microsoft Excel and Prism 6.0.
3. Results
3.1. Principle of TILDA
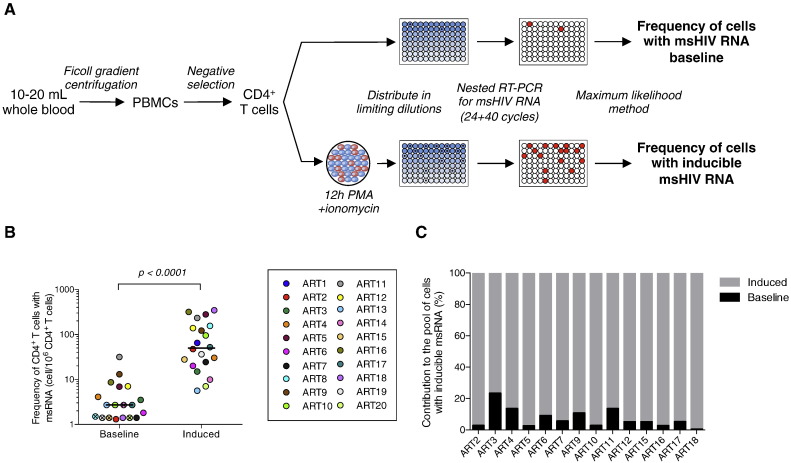
We sought to develop an assay that detects tat/rev msRNA (adapted from Pasternak et al., 2008) in maximally activated CD4 + T cells, using a limiting dilution format, postulating that this method will allow us to measure the frequency of cells harbouring inducible viruses. We chose to activate the cells with phorbol 12-myristate 13-acetate (PMA), which has been shown to induce a rapid onset of increased transcription from the HIV LTR in latently infected cell lines (Li et al., 1991), induces higher levels of viral production than PHA (Tong-Starkesen et al., 1989) and activates rare T cell subsets (Wang et al., 2013). Ionomycin, a calcium ionophore, is added to trigger calcium release, a requirement for N-FAT signalling. PMA ionomycin has been shown to induce high levels of viral production in several primary cell models of HIV latency (Spina et al., 2013) as well as in CD4 + T cells from virally suppressed individuals (Bullen et al., 2014, Laird et al., 2014). After stimulation, cells are distributed in 24 replicate wells in a 96 well plate and a nested PCR amplifying tat/rev transcripts is directly performed without RNA extraction (Fig. 1A). Using the maximum likelihood method, the frequency of cells producing msRNA is calculated. TILDA can also be used to measure the frequency of CD4 + T cells that spontaneously produce msRNA ex vivo by omitting the stimulation step.
Fig. 1.
TILDA can be used to measure the frequency of persistently infected cells in blood samples from virally suppressed subjects. (A) Principle of TILDA: PBMCs were isolated from 10–20 ml of blood and CD4 + T cells were enriched by negative magnetic selection. CD4 + T cells were precisely counted and directly distributed in a 96 well plate or stimulated for 12 h with PMA/ionomycin. Tat/rev msRNA were quantified by an ultrasensitive nested RT-PCR and the frequency of cells with inducible msRNA was determined using the maximum likelihood method. (B) Frequency of CD4 + T cells producing msRNA spontaneously (baseline, n = 19) or after 12 h of stimulation (induced, n = 20) in samples obtained from virally suppressed subjects. Horizontal bars indicate median values. For samples in which no positive cells were detected at baseline, the limit of detection (based on cell input) is plotted and represented as a crossed circle. P value was obtained from the Wilcoxon matched-pairs signed rank test. (C) Proportion of CD4 + T cells that produce msRNA spontaneously or after stimulation in virally suppressed subjects on ART. Only samples with detectable values at baseline (panel B) are represented.
3.2. TILDA is Specific, Sensitive and Reproducible
We first evaluated the sensitivity and specificity of TILDA. We infected activated CD4 + T cells with an infectious HIV clone containing a GFP reporter (NL4-3 nef IRES GFP). Two days postinfection, we sorted GFP + and GFP- cells and used our assay to detect tat/rev msRNA in wells containing single cells (Fig. S1A). Among 48 well containing GFP + cells, 42 were positive for HIV DNA and GAPDH (88%). 79% (33/42) of these were positive by TILDA. In contrast, none of the 48 wells containing GFP negative cells were positive for msRNA, in spite of a significant fraction of these well containing HIV DNA (42/48, 88%). These results suggested that the presence of msRNA is a specific surrogate for the production of viral proteins, at least in this in vitro system of HIV infection.
Using samples obtained from 4 virally suppressed subjects, we determined the robustness of TILDA i.e., intra- and inter-assay coefficients of variation; we measured the frequency of CD4 + T cells producing tat/rev msRNA upon stimulation with PMA/ionomycin in duplicate plates or in up to 4 independent experiments performed by different operators. The intra and inter-assay coefficients of variation of TILDA were 0.15 and 0.21, respectively (Fig. S1B) confirming the robustness of this assay. The theoretical dynamic range of TILDA is > 3logs10 with minimal and maximal detectable values of 1.4 and 3185 cells/106 CD4 + T cells, respectively. Of note, all the experimental values determined in this study ranged over 3 logs, a dynamic range of TILDA that is large enough to efficiently measure low frequency of infected cells as is the case in the vast majority of virally suppressed individuals (Fig. S1C). Importantly, the limit of detection of TILDA indicated above (1.4 cells/106 CD4 + T cells) is based on the number of cells plated in a typical experiment (744,000 cells corresponding to 24 × 18,000 + 24 × 9000 + 24 × 3000 + 24 × 1000) and can further be lowered by increasing the number of cells assayed in multiple plates.
3.3. TILDA Measures the Frequency of CD4 + T cells With Inducible msRNA in Virally Suppressed Individuals on ART
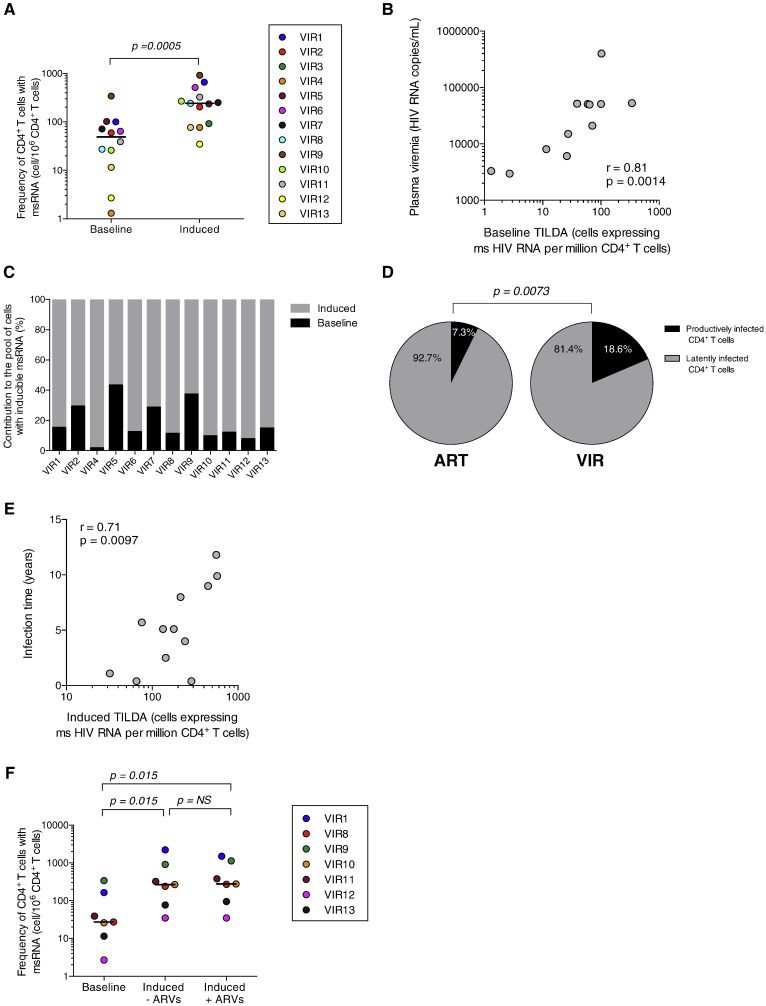
We used TILDA to measure the frequency of CD4 + T cells producing tat/rev msRNA in samples from 20 virally suppressed subjects on suppressive ART for a median time of 6 years (Table 1). As expected, the frequency of cells spontaneously producing msRNA was low (median = 2.7 cells/106 CD4 + T cells, Fig. 1B), and even below the limit of detection in 5 out of 19 samples (26%). CD4 + T cells producing tat/rev RNA were detected in all samples upon stimulation, with a median frequency of cells producing msRNA of 50 cells/106 CD4 + T cells (p < 0.0001). By subtracting the baseline from the induced values, we calculated the frequency of cells carrying inducible HIV. We estimated that only 7.3% of CD4 + T cells with the ability to generate msRNA produced these transcripts spontaneously, whereas the majority (92.7%) of cells were devoid of msRNA ex vivo but produced them upon stimulation with PMA/Ionmycin (Fig. 1C). These results are in line with previous reports demonstrating that the majority of infected cells in individuals on suppressive ART, harbour latent HIV and do not spontaneously produce viral particles (Chun et al., 1997b, Finzi et al., 1999, Chun et al., 2003). These results demonstrated that TILDA can be used to measure the frequency of CD4 + T cells that have the ability to produce HIV msRNA upon stimulation in virally suppressed subjects.
3.4. TILDA Values Correlate With Several Assays Measuring HIV Persistence During ART
We next compared the frequency of latently infected CD4 + T cells measured by TILDA with values obtained from a variety of well-established assays (Siliciano and Siliciano, 2005, Yu et al., 2008, Palmer et al., 2003, Strain et al., 2013, Yukl et al., 2010). For this purpose, we used samples that were originally used to compare Q-VOA with several PCR-based assays (Table 2) (Eriksson et al., 2013). In these samples obtained from 27 participants of the SCOPE and Options studies, we found that the median frequency of latently infected CD4 + T cells measured by TILDA was 24 cells/106 CD4 + T cells (range 1.4–101 cells/million), which was 48 times higher than frequencies measured by Q-VOA (median = 0.5, range 0.03–16 cells/106 CD4 + T cells), and 6–27 times lower than frequencies of infected cells measured by PCR-based assays (Table 2).
Table 2.
Results from the virological assays used in this study.
| Q-VOA | TILDA | Tot. HIV DNA in PBMCs | Tot. HIV DNA in CD4 | Int. HIV DNA in PBMCs | Int. HIV DNA in CD4 | Tot. HIV DNA in GALT cells | Residual viremia plasma | |
|---|---|---|---|---|---|---|---|---|
| Methoda | Cell culture in limiting dilution | Stimulation and PCR in limiting dilution | ddPCR | ddPCR | alu-PCR | alu-PCR | ddPCR | SCA |
| Sample | Resting CD4 + T cells | Total CD4 + T cells | PBMCs | Resting CD4 + T cells | PBMCs | Resting CD4 + T cells | GALT CD4 + T cells | Plasma |
| Unit | Infectious unit/106 cells | Cells expressing msRNA/106 cells | Copies/106 cells | Copies/106 cells | Copies/106 cells | Copies/106 cells | Copies/106 cells | Copies/ml |
| n samples | 27 | 27 | 27 | 14 | 18 | 15 | 17 | 27 |
| Median | 0.5 | 23.8 | 150 | 307.5 | 234.5 | 767 | 5301 | 0.5 |
| Ratio with TILDAb | 0.021 | – | 6.3 | 13.4 | 9.0 | 27.1 | 226c | NA |
| Max. value | 16 | 100.6 | 1858 | 1200 | 1248 | 2997 | 13,388 | 2.2 |
| Min. value | 0.0315 | 1.4 | 1 | 1 | 19 | 108 | 259 | 0.1 |
ddPCR: droplet digital PCR; SCA: single copy assay. NA: not applicable.
Median values were used to calculate the ratio. Samples for which one of the 2 measurements used in each comparison was not performed were excluded.
Ratio between GALT DNA and TILDA measured in blood.
We then determined correlations between the values obtained by TILDA and the other assays used to measure the size of the HIV reservoir using samples from the same group of subjects. TILDA values were positively correlated with the frequency of PBMCs harbouring total HIV DNA (r = 0.38, p = 0.05, Fig. 2A) and integrated HIV DNA (r = 0.68, p = 0.002, Fig. 2B) as well as with the frequency of rectal CD4 + T cells harbouring total HIV DNA (r = 0.70, p = 0.002, Fig. 2C). Although TILDA values tended to correlate with the frequencies of resting CD4 + T cells harbouring total and integrated HIV DNA, these correlations did not reach statistical significance (ρ = 0.48, p = 0.083 and r = 0.47, p = 0.076, respectively, Fig. 2D and E). Of note, the number of samples in which total and integrated HIV DNA were measured in resting CD4 + T cells was limited (n = 14 and 15, respectively, Table 2), which curtailed our capacity to detect significant associations between TILDA and these two assays. TILDA values did not correlate significantly with residual viremia, although a trend for a negative association was noted (ρ = − 0.35, p = 0.070, Fig. 2F). The frequency of resting CD4 + T cells harbouring replication competent HIV measured by Q-VOA tended to correlate with TILDA, but this was not statistically significant in this relatively small population of virally suppressed individuals (r = 0.33, p = 0.095, Fig. 2G); this may be attributed to the fact that Q-VOA uses resting CD4 + T cells, whereas TILDA is performed on total CD4 + T cells that include homeostatically proliferating cells. Altogether, these results demonstrate that TILDA can be efficiently used to measure the frequency of CD4 + T cells producing msRNA upon stimulation, a frequency that correlates with several assays that measure HIV persistence.
3.5. Initiation of ART During Early HIV Infection Leads to a Restricted Size of the Reservoir During Therapy Measured by TILDA
We measured the frequency of infected cells with inducible msRNA in virally suppressed subjects who initiated ART during recent (n = 10) or chronic (n = 17) infection (Fig. 3). Using TILDA, we observed that the size of the reservoir was significantly smaller in subjects who started ART during recent infection (< 6 months) when compared to those who initiated therapy at a later stage of the disease (median frequencies of 11 and 29 cells/106 CD4 + T cells in early and chronic infection respectively, p = 0.011). This was in line with the results form previous studies (Strain et al., 2005, Archin et al., 2012, Buzon et al., 2014, Ananworanich et al., 2012, Ananworanich et al., 2015, Eriksson et al., 2013), in which a lower frequency of reservoir cells was measured in subjects who started ART early when using Q-VOA, integrated HIV DNA in PBMCs and total HIV DNA in resting CD4 + T cells.
3.6. TILDA Reveals the Existence of a Large Pool of Latently Infected CD4 + T cells in HIV-infected Individuals who do not Receive ART
Our results confirm that the latent HIV reservoir is established early in infection (Strain et al., 2005, Archin et al., 2012, Buzon et al., 2014, Ananworanich et al., 2012, Chomont et al., 2009). However, the relative proportion of productively and latently infected CD4 + T cells during untreated HIV infection is not known. As TILDA can measure the frequency of cells spontaneously producing msRNA as well as the frequency of cells that has the ability to do so upon maximal stimulation (Fig. 1), we used our assay to measure both frequencies in CD4 + T cells obtained from HIV-infected individuals who did not receive ART (Table 1). CD4 + T cells containing tat/rev transcripts at baseline were readily detected in all samples from viremic individuals (Fig. 4A). Overnight stimulation with PMA/ionomycin led to a significant increase in the frequency of CD4 + T cells producing msRNA (median fold increase of 7.4 in the frequency of cells with HIV msRNA, p = 0.0005). The frequency of cells spontaneously producing these transcripts was strongly correlated with plasma viral load (r = 0.81, p = 0.0014, Fig. 4B). Our calculations indicate that only a minor fraction (18.6%) of CD4 + T cells with inducible HIV genomes were spontaneously producing tat/rev transcripts in vivo (Fig. 4C), whereas the majority of infected cells (81.4%) were latently infected and produced msRNA only upon stimulation. Therefore, similar to virally suppressed subjects, ART-naïve individuals harbour a large pool of CD4 + T cells in which msRNA can be induced upon stimulation (Fig. 4D). The frequency of latently infected CD4 + T cells in untreated HIV-infected individuals was positively correlated with the duration of HIV infection (r = 0.71, p = 0.0097, Fig. 4E), suggesting that the size of the latent and inducible reservoir gradually increases in untreated viremic subjects, a consequence of continuous seeding of the HIV reservoir.
Fig. 4.
TILDA reveals that untreated HIV-infected individuals harbour a large pool of latently infected CD4 + T cells. (A) Frequency of CD4 + T cells producing tat/rev msRNA spontaneously (baseline) or after 12 h of stimulation (induced) in 13 samples obtained from viremic, untreated HIV-infected individuals. Horizontal bars indicate median values. P value was obtained from the Wilcoxon matched-pairs signed rank test. (B) Correlation between the frequency of CD4 + T cells spontaneously producing HIV msRNA and HIV plasma viremia. P value was obtained from the Pearson test. (C) Proportion of CD4 + T cells that produce tat/rev msRNA spontaneously or after stimulation in untreated HIV-infected subjects. (D) Pie charts show the average relative proportions of productively and latently infected CD4 + T cells in virally suppressed (ART) and untreated viremic (VIR) HIV-infected subjects. P value was obtained from the Mann–Whitney test. (E) Correlation between the frequency of CD4 + T cells producing HIV msRNA after stimulation and duration of HIV infection. P value was obtained from the Pearson test. (F) Frequency of CD4 + T cells producing HIV msRNA before and after stimulation in the presence (+ ARVs) or absence (− ARVs) of antiretrovirals (raltegravir, efavirenz and AZT). P values were obtained from the Wilcoxon matched-pairs signed rank test.
Non-integrated HIV DNA molecules are present at high frequencies in CD4 + T cells isolated from viremic HIV-infected individuals (Chomont et al., 2009, Koelsch et al., 2008). Integration and transcription of these linear genomes may account for the increase in the frequency of CD4 + T cells producing msRNA we observed upon stimulation with PMA/ionomycin. To test for this possibility, we repeated these experiments in the presence of antiretroviral drugs, including the integrase inhibitor raltegravir. We observed that the addition of raltegravir to the culture medium had no impact on the increase in the frequency of cells producing msRNA upon stimulation. This indicated that the increase in the frequency of CD4 + T cells producing msRNA is attributable to viral genomes that were already integrated before stimulation, demonstrating that post-integration latency can be detected and measured in viremic HIV-infected subjects who do not receive ART.
4. Discussion
The definitive test of an HIV cure will require the interruption of ART. It is necessary, however, to measure the impact of eradication strategies on the size of the viral reservoir even as therapy is continued if clinical trials are to proceed in an efficient and ethical manner (International ASSWGoHIVC et al., 2012). A large number of assays have been developed to measure the frequency of infected cells that persist in virally suppressed subjects who have received ART for years (Lewin et al., 1999, Li et al., 2010, Siliciano and Siliciano, 2005, Yu et al., 2008, Palmer et al., 2003, Vandergeeten et al., 2014, Pasternak et al., 2008, Eriksson et al., 2013, Brussel and Sonigo, 2003, Rouzioux et al., 2005, Strain et al., 2013, O'Doherty et al., 2002). Each assay measures different parameters (replication competent HIV, total and integrated HIV DNA, cell associated HIV RNA, viral particles) in different cells types (PBMCs, CD4 + T cells and resting CD4 + T cells). These assays provide a frequency of persistently infected cells that can range over 2 logs in the same individual (Eriksson et al., 2013). For example, the size of the reservoir is likely to be overestimated by PCR assays, which can include a large proportion of defective viruses. On the other hand, the reservoir is likely to be underestimated by the Q-VOA assay as it does not capture all cells carrying replication competent HIV (Ho et al., 2013). This prompted us to develop an assay that would not measure the background attributed to defective genomes and that would not rely on the amplification of viral replication, a step that requires at least seven days of cell culture. Unlike cells that produce unspliced and often short HIV gag RNA (Lassen et al., 2004), infected cells that are actively producing msRNA (particularly tat/rev transcripts) can produce virions (Lewin et al., 1999, Pasternak et al., 2008, Pasternak et al., 2009, Schmid et al., 2010, Fischer et al., 2004, Peng et al., 1995, Hermankova et al., 2003, Vesanen et al., 1997). This well-characterized process suggests that quantification of tat/rev msRNA may prove to be a sensitive and reproducible manner to estimate the frequency of cells harbouring transcriptionally silent – nonetheless inducible – viruses after maximal stimulation.
TILDA does not rely on the amplification of viral replication and offers similar advantages of the DNA PCR assays with regard to simplicity and small blood volume requirement. It does not require RNA extraction and 96-well plates with serially diluted cells can be stored at − 80 °C for future quantification in batch analyses, making TILDA well-suited for longitudinal clinical studies. Therefore, TILDA has a number of characteristics that make it attractive for clinical trials, particularly in paediatric studies in which the number of cells that can be obtained are often limited. In addition, TILDA could be used to measure the frequency of infection in rare population of cells, which may facilitate the measurement of the HIV reservoir in tissues.
Using TILDA, we found the median frequency of latently infected CD4 + T cells during ART to be 24 cells/million, which is 48 times more than the frequency measured by the quantitative viral outgrowth assay. Ho et al. recently reported that the median frequency of intact noninduced proviruses is around 60-fold higher than the frequency of induced proviruses detected in Q-VOA (Ho et al., 2013). Therefore, TILDA may provide an accurate estimate of the size of the latent HIV reservoir, although the precise fraction of the replication competent latent and inducible HIV reservoir captured by TILDA will require further studies. Similarly, evaluating the ability of TILDA to predict viral rebound upon ART interruption will necessitate the development of well-powered clinical trials and cannot be inferred from our current observations.
TILDA correlated with several PCR-based assays that measure the size of the latent reservoir, including the quantification of total and integrated HIV DNA in PBMCs. TILDA tended to correlate with the size of the reservoir measured by Q-VOA, but this was not statistically significant. Our data are consistent with a lack of strong correlation between these two measures although the lack of statistical significance may reflect, in part, the sample size. The lack of correlation between the two assays may also result from the fact that Q-VOA is performed on purified resting CD4 + T cells, whereas total CD4 + T cells are used for TILDA determinations. Total CD4 + T cells include cells undergoing homeostatic proliferation which express some activation markers and are not captured in the Q-VOA assay. Ho et al. reported that Q-VOA does not correlate strongly with the frequency of cells harbouring intact non-induced proviruses (Ho et al., 2013), suggesting that it may not provide an accurate estimate of the frequency of latently infected CD4 + T cells. Whether TILDA could provide a better estimate of the size of the replication competent reservoir remains to be determined by using full-length sequencing approaches, similar to the ones used by Ho et al. and others (Ho et al., 2013, Cohn et al., 2015).
Using TILDA, we observed that early initiation of ART leads to a reduced size of the viral reservoir after years of viral suppression. This is in line with several reports that have clearly demonstrated the benefit of early therapy on the size of the latent HIV reservoir (Strain et al., 2005, Archin et al., 2012, Buzon et al., 2014, Ananworanich et al., 2012, Ananworanich et al., 2015, Chomont et al., 2009). In addition, we demonstrated that before the initiation of ART, the latent HIV reservoir already represents a substantial fraction of all infected CD4 + T cells and that the size of the reservoir increases with time during untreated HIV infection. This observation further argues for early ART intervention to prevent the establishment and continuous seeding of an increasingly large pool of latently infected CD4 + T cells.
Although TILDA constitutes an attractive alternative to other methods to measure the size of the latent HIV reservoir, there are potential limitations associated with the use of our assay. First, although all cells that are releasing viral particles are producing tat/rev msRNA, the opposite may not be necessarily true. We cannot exclude the possibility that some cells that generate a positive signal in TILDA do not produce virions. Indeed, very low levels of nuclear msRNA can be detected in resting CD4 + T cells using ultrasensitive approaches (Lassen et al., 2004, Lassen et al., 2006). Therefore, TILDA is likely to overestimate the size of the latent and replication competent reservoir. Notwithstanding this limitation, TILDA is more likely to capture the functional reservoir than other existing PCR-based assays: TILDA positive events result from the transcription of integrated HIV genomes that have an intact LTR (to ensure transcription) and that are not deleted in the two most commonly deleted regions identified in defective genomes namely tat and rev (Ho et al., 1995). Of note, TILDA measures frequencies of latently infected CD4 + T cells close to those predicted to harbour replication competent virus in the study by Ho et al. (2013). Another potential caveat of TILDA is the fact that it relies on the amplification of a highly variable region of the HIV genome, suggesting that the primers and probes used in our assay may not recognize all viral quasispecies. We anticipate that the use of TILDA on HIV clades other than B will require further validation and optimization.
In conclusion, we report the development and validation of TILDA, a novel assay to measure the size of the latent reservoir that is quantitative, sensitive, robust and requires only 10 ml of blood. Moreover, results can be generated in two days. As such, TILDA may represent an alternative to existing assays used to evaluate the efficacy of therapeutic strategies aimed at reducing the size of the latent HIV reservoir.
The following is the supplementary data related to this article.
Performance of TILDA. (A) Activated CD4 + T cells were infected with HIV pNL4.3-GFP. GFP + and GFP- cells were sorted and tat/rev RNA were detected in single cells. The amplifications curves obtained from 24 GFP + and 24 GFP- cells are shown. (B) TILDA measurements were performed in duplicate in the same experiments or 3 to 4 times in independent experiments to calculate the intra and inter-assay coefficients of variation of the assay. Horizontal bars show means and standard deviations. (C) Dynamic range of TILDA: All TILDA values determined in samples obtained from virally suppressed subjects participating in the present study are represented. Unstimulated (spontaneous) and stimulated TILDA values are represented by grey and black circles, respectively. Horizontal dotted lines show the lower and upper limits of detection of the assay.
Author Contributions
Conceived and designed the experiments: FAP RF RPS NC. Performed the experiments: FAP RF DK JB AGB MCS. Analysed the data: FAP RF JB RPS NC. Contributed reagents/materials/analysis tools: MCS DDR UOD SP FH RJOB MDM DJH SGD. Wrote the paper: FAP RF RPS NC.
Financial Disclosure
This work was supported by NIH grant 1R21AI113096 (NC), by the Delaney AIDS Research Enterprise (DARE) to Find a Cure1U19AI096109 and by the Foundation for AIDS Research (amfAR Research Consortium on HIV Eradication (108928-56-RGRL)). R.F. is supported by an amfAR fellowship (108264-51-RFRL). The funders had no role in study design, data collection, data analysis, interpretation and writing of the report.
Declaration of Interests
RJOB, MDM and DJH are employees and shareholders of Merck & Co., Inc., manufacturer of raltegravir and efavirenz.
Acknowledgements
The authors thank the study participants for their important contribution to this study. The authors also thank Patrick Yeramian, Moti Ramgopal, Rebeka Bordi, Brenda Jacobs, and Kathyrin Penniman for their recruitment and clinical assistance with study participants, Amanda McNulty and Stephanie Santos for technical assistance and Janet D. Siliciano, Robert F. Siliciano and Jun Lai for Q-VOA measurements. The authors thank the members of the Cleveland Immunopathogenesis Consortium for their advice and helpful discussions.
Contributor Information
Rafick-Pierre Sékaly, Email: rafick.sekaly@case.edu.
Nicolas Chomont, Email: nicolas.chomont@umontreal.ca.
References
- Agosto L.M., Liszewski M.K., Mexas A. Patients on HAART often have an excess of unintegrated HIV DNA: implications for monitoring reservoirs. Virology. 2011;409(1):46–53. doi: 10.1016/j.virol.2010.08.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ananworanich J., Schuetz A., Vandergeeten C. Impact of multi-targeted antiretroviral treatment on gut T cell depletion and HIV reservoir seeding during acute HIV infection. PLoS One. 2012;7(3):e33948. doi: 10.1371/journal.pone.0033948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ananworanich J., Dube K., Chomont N. How does the timing of antiretroviral therapy initiation in acute infection affect HIV reservoirs? Curr. Opin. HIV AIDS. 2015;10(1):18–28. doi: 10.1097/COH.0000000000000122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Archin N.M., Vaidya N.K., Kuruc J.D. Immediate antiviral therapy appears to restrict resting CD4 + cell HIV-1 infection without accelerating the decay of latent infection. Proc. Natl. Acad. Sci. U. S. A. 2012;109(24):9523–9528. doi: 10.1073/pnas.1120248109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boulassel M.R., Chomont N., Pai N.P., Gilmore N., Sekaly R.P., Routy J.P. CD4 T cell nadir independently predicts the magnitude of the HIV reservoir after prolonged suppressive antiretroviral therapy. J. Clin. Virol. 2012;53(1):29–32. doi: 10.1016/j.jcv.2011.09.018. [DOI] [PubMed] [Google Scholar]
- Brussel A., Sonigo P. Analysis of early human immunodeficiency virus type 1 DNA synthesis by use of a new sensitive assay for quantifying integrated provirus. J. Virol. 2003;77(18):10119–10124. doi: 10.1128/JVI.77.18.10119-10124.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bullen C.K., Laird G.M., Durand C.M., Siliciano J.D., Siliciano R.F. New ex vivo approaches distinguish effective and ineffective single agents for reversing HIV-1 latency in vivo. Nat. Med. 2014;20(4):425–429. doi: 10.1038/nm.3489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buzon M.J., Martin-Gayo E., Pereyra F. Long-term antiretroviral treatment initiated in primary HIV-1 infection affects the size, composition and decay kinetics of the reservoir of HIV-1 infected CD4 T cells. J. Virol. 2014;88:10056–10065. doi: 10.1128/JVI.01046-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chomont N., El-Far M., Ancuta P. HIV reservoir size and persistence are driven by T cell survival and homeostatic proliferation. Nat. Med. 2009;15(8):893–900. doi: 10.1038/nm.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chun T.W., Stuyver L., Mizell S.B. Presence of an inducible HIV-1 latent reservoir during highly active antiretroviral therapy. Proc. Natl. Acad. Sci. U. S. A. 1997;94(24):13193–13197. doi: 10.1073/pnas.94.24.13193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chun T.W., Carruth L., Finzi D. Quantification of latent tissue reservoirs and total body viral load in HIV-1 infection. Nature. 1997;387(6629):183–188. doi: 10.1038/387183a0. [DOI] [PubMed] [Google Scholar]
- Chun T.W., Justement J.S., Pandya P. Relationship between the size of the human immunodeficiency virus type 1 (HIV-1) reservoir in peripheral blood CD4 + T cells and CD4 +:CD8 + T cell ratios in aviremic HIV-1-infected individuals receiving long-term highly active antiretroviral therapy. J. Infect. Dis. 2002;185(11):1672–1676. doi: 10.1086/340521. [DOI] [PubMed] [Google Scholar]
- Chun T.W., Justement J.S., Lempicki R.A. Gene expression and viral production in latently infected, resting CD4 + T cells in viremic versus aviremic HIV-infected individuals. Proc. Natl. Acad. Sci. U. S. A. 2003;100(4):1908–1913. doi: 10.1073/pnas.0437640100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cillo A.R., Sobolewski M.D., Bosch R.J. Quantification of HIV-1 latency reversal in resting CD4 + T cells from patients on suppressive antiretroviral therapy. Proc. Natl. Acad. Sci. U. S. A. 2014;111:7078–7083. doi: 10.1073/pnas.1402873111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohn L.B., Silva I.T., Oliveira T.Y. HIV-1 integration landscape during latent and active infection. Cell. 2015;160(3):420–432. doi: 10.1016/j.cell.2015.01.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eriksson S., Graf E.H., Dahl V. Comparative analysis of measures of viral reservoirs in HIV-1 eradication studies. PLoS Pathog. 2013;9(2):e1003174. doi: 10.1371/journal.ppat.1003174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finzi D., Hermankova M., Pierson T. Identification of a reservoir for HIV-1 in patients on highly active antiretroviral therapy. Science. 1997;278(5341):1295–1300. doi: 10.1126/science.278.5341.1295. [DOI] [PubMed] [Google Scholar]
- Finzi D., Blankson J., Siliciano J.D. Latent infection of CD4 + T cells provides a mechanism for lifelong persistence of HIV-1, even in patients on effective combination therapy. Nat. Med. 1999;5(5):512–517. doi: 10.1038/8394. [DOI] [PubMed] [Google Scholar]
- Fischer M., Wong J.K., Russenberger D. Residual cell-associated unspliced HIV-1 RNA in peripheral blood of patients on potent antiretroviral therapy represents intracellular transcripts. Antivir. Ther. 2002;7(2):91–103. [PubMed] [Google Scholar]
- Fischer M., Joos B., Wong J.K. Attenuated and nonproductive viral transcription in the lymphatic tissue of HIV-1-infected patients receiving potent antiretroviral therapy. J. Infect. Dis. 2004;189(2):273–285. doi: 10.1086/380797. [DOI] [PubMed] [Google Scholar]
- Furtado M.R., Callaway D.S., Phair J.P. Persistence of HIV-1 transcription in peripheral-blood mononuclear cells in patients receiving potent antiretroviral therapy. N. Engl. J. Med. 1999;340(21):1614–1622. doi: 10.1056/NEJM199905273402102. [DOI] [PubMed] [Google Scholar]
- Garrigue I., Pellegrin I., Hoen B. Cell-associated HIV-1-DNA quantitation after highly active antiretroviral therapy-treated primary infection in patients with persistently undetectable plasma HIV-1 RNA. AIDS. 2000;14(18):2851–2855. doi: 10.1097/00002030-200012220-00006. [DOI] [PubMed] [Google Scholar]
- Graf E.H., Mexas A.M., Yu J.J. Elite suppressors harbor low levels of integrated HIV DNA and high levels of 2-LTR circular HIV DNA compared to HIV + patients on and off HAART. PLoS Pathog. 2011;7(2):e1001300. doi: 10.1371/journal.ppat.1001300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hermankova M., Siliciano J.D., Zhou Y. Analysis of human immunodeficiency virus type 1 gene expression in latently infected resting CD4 + T lymphocytes in vivo. J. Virol. 2003;77(13):7383–7392. doi: 10.1128/JVI.77.13.7383-7392.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho D.D., Neumann A.U., Perelson A.S., Chen W., Leonard J.M., Markowitz M. Rapid turnover of plasma virions and CD4 lymphocytes in HIV-1 infection. Nature. 1995;373(6510):123–126. doi: 10.1038/373123a0. [DOI] [PubMed] [Google Scholar]
- Ho Y.-C., Shan L., Hosmane N.N. Replication-competent noninduced proviruses in the latent reservoir increase barrier to HIV-1 cure. Cell. 2013;155(3):540–551. doi: 10.1016/j.cell.2013.09.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- International ASSWGoHIVC, Deeks S.G., Autran B. Towards an HIV cure: a global scientific strategy. Nat. Rev. Immunol. 2012;12(8):607–614. doi: 10.1038/nri3262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Josefsson L., von Stockenstrom S., Faria N.R. The HIV-1 reservoir in eight patients on long-term suppressive antiretroviral therapy is stable with few genetic changes over time. Proc. Natl. Acad. Sci. U. S. A. 2013;110(51):E4987–E4996. doi: 10.1073/pnas.1308313110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kearney M. Analysis of HIV RNA in single cells reveals clonal expansions and defective genomes. CROI. 2015;2015 [Google Scholar]
- Koelsch K.K., Liu L., Haubrich R. Dynamics of total, linear nonintegrated, and integrated HIV-1 DNA in vivo and in vitro. J. Infect. Dis. 2008;197(3):411–419. doi: 10.1086/525283. [DOI] [PubMed] [Google Scholar]
- Laird G.M., Eisele E.E., Rabi S.A. Rapid quantification of the latent reservoir for HIV-1 using a viral outgrowth assay. PLoS Pathog. 2013;9(5):e1003398. doi: 10.1371/journal.ppat.1003398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laird G.M., Eisele E.E., Rabi S.A., Nikolaeva D., Siliciano R.F. A novel cell-based high-throughput screen for inhibitors of HIV-1 gene expression and budding identifies the cardiac glycosides. J. Antimicrob. Chemother. 2014;69(4):988–994. doi: 10.1093/jac/dkt471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lassen K.G., Bailey J.R., Siliciano R.F. Analysis of human immunodeficiency virus type 1 transcriptional elongation in resting CD4 + T cells in vivo. J. Virol. 2004;78(17):9105–9114. doi: 10.1128/JVI.78.17.9105-9114.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lassen K.G., Ramyar K.X., Bailey J.R., Zhou Y., Siliciano R.F. Nuclear retention of multiply spliced HIV-1 RNA in resting CD4 + T cells. PLoS Pathog. 2006;2(7):e68. doi: 10.1371/journal.ppat.0020068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewin S.R., Vesanen M., Kostrikis L. Use of real-time PCR and molecular beacons to detect virus replication in human immunodeficiency virus type 1-infected individuals on prolonged effective antiretroviral therapy. J. Virol. 1999;73(7):6099–6103. doi: 10.1128/jvi.73.7.6099-6103.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y.C., Ross J., Scheppler J.A., Franza B.R., Jr. An in vitro transcription analysis of early responses of the human immunodeficiency virus type 1 long terminal repeat to different transcriptional activators. Mol. Cell. Biol. 1991;11(4):1883–1893. doi: 10.1128/mcb.11.4.1883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li P., Ruel T., Fujimoto K. Novel application of locked nucleic acid chemistry for a Taqman assay for measuring diverse human immunodeficiency virus type 1 subtypes. J. Virol. Methods. 2010;170(1–2):115–120. doi: 10.1016/j.jviromet.2010.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin X., Irwin D., Kanazawa S. Transcriptional profiles of latent human immunodeficiency virus in infected individuals: effects of Tat on the host and reservoir. J. Virol. 2003;77(15):8227–8236. doi: 10.1128/JVI.77.15.8227-8236.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mexas A.M., Graf E.H., Pace M.J. Concurrent measures of total and integrated HIV DNA monitor reservoirs and ongoing replication in eradication trials. AIDS. 2012;26(18):2295–2306. doi: 10.1097/QAD.0b013e32835a5c2f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murray J.M., Zaunders J.J., McBride K.L. HIV DNA subspecies persist in both activated and resting memory CD4 + T cells during antiretroviral therapy. J. Virol. 2014;88(6):3516–3526. doi: 10.1128/JVI.03331-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Doherty U., Swiggard W.J., Jeyakumar D., McGain D., Malim M.H. A sensitive, quantitative assay for human immunodeficiency virus type 1 integration. J. Virol. 2002;76(21):10942–10950. doi: 10.1128/JVI.76.21.10942-10950.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palella F.J., Jr., Delaney K.M., Moorman A.C. Declining morbidity and mortality among patients with advanced human immunodeficiency virus infection. HIV outpatient study investigators. N. Engl. J. Med. 1998;338(13):853–860. doi: 10.1056/NEJM199803263381301. [DOI] [PubMed] [Google Scholar]
- Palmer S., Wiegand A.P., Maldarelli F. New real-time reverse transcriptase-initiated PCR assay with single-copy sensitivity for human immunodeficiency virus type 1 RNA in plasma. J. Clin. Microbiol. 2003;41(10):4531–4536. doi: 10.1128/JCM.41.10.4531-4536.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pasternak A.O., Adema K.W., Bakker M. Highly sensitive methods based on seminested real-time reverse transcription-PCR for quantitation of human immunodeficiency virus type 1 unspliced and multiply spliced RNA and proviral DNA. J. Clin. Microbiol. 2008;46(7):2206–2211. doi: 10.1128/JCM.00055-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pasternak A.O., Jurriaans S., Bakker M., Prins J.M., Berkhout B., Lukashov V.V. Cellular levels of HIV unspliced RNA from patients on combination antiretroviral therapy with undetectable plasma viremia predict the therapy outcome. PLoS One. 2009;4(12):e8490. doi: 10.1371/journal.pone.0008490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng H., Reinhart T.A., Retzel E.F., Staskus K.A., Zupancic M., Haase A.T. Single cell transcript analysis of human immunodeficiency virus gene expression in the transition from latent to productive infection. Virology. 1995;206(1):16–27. doi: 10.1016/s0042-6822(95)80015-8. [DOI] [PubMed] [Google Scholar]
- Razooky B.S., Pai A., Aull K., Rouzine I.M., Weinberger L.S. A hardwired HIV latency program. Cell. 2015;160(5):990–1001. doi: 10.1016/j.cell.2015.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rouzioux C., Hubert J.B., Burgard M. Early levels of HIV-1 DNA in peripheral blood mononuclear cells are predictive of disease progression independently of HIV-1 RNA levels and CD4 + T cell counts. J. Infect. Dis. 2005;192(1):46–55. doi: 10.1086/430610. [DOI] [PubMed] [Google Scholar]
- Schmid A., Gianella S., von Wyl V. Profound depletion of HIV-1 transcription in patients initiating antiretroviral therapy during acute infection. PLoS One. 2010;5(10):e13310. doi: 10.1371/journal.pone.0013310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siliciano J.D., Siliciano R.F. Enhanced culture assay for detection and quantitation of latently infected, resting CD4 + T-cells carrying replication-competent virus in HIV-1-infected individuals. Methods Mol. Biol. 2005;304:3–15. doi: 10.1385/1-59259-907-9:003. [DOI] [PubMed] [Google Scholar]
- Siliciano J.D., Kajdas J., Finzi D. Long-term follow-up studies confirm the stability of the latent reservoir for HIV-1 in resting CD4 + T cells. Nat. Med. 2003;9(6):727–728. doi: 10.1038/nm880. [DOI] [PubMed] [Google Scholar]
- Sonza S., Mutimer H.P., O'Brien K. Selectively reduced tat mRNA heralds the decline in productive human immunodeficiency virus type 1 infection in monocyte-derived macrophages. J. Virol. 2002;76(24):12611–12621. doi: 10.1128/JVI.76.24.12611-12621.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spina C.A., Anderson J., Archin N.M. An in-depth comparison of latent HIV-1 reactivation in multiple cell model systems and resting CD4 + T cells from aviremic patients. PLoS Pathog. 2013;9(12):e1003834. doi: 10.1371/journal.ppat.1003834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strain M.C., Little S.J., Daar E.S. Effect of treatment, during primary infection, on establishment and clearance of cellular reservoirs of HIV-1. J. Infect. Dis. 2005;191(9):1410–1418. doi: 10.1086/428777. [DOI] [PubMed] [Google Scholar]
- Strain M.C., Lada S.M., Luong T. Highly precise measurement of HIV DNA by droplet digital PCR. PLoS One. 2013;8(4):e55943. doi: 10.1371/journal.pone.0055943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tong-Starkesen S.E., Luciw P.A., Peterlin B.M. Signaling through T lymphocyte surface proteins, TCR/CD3 and CD28, activates the HIV-1 long terminal repeat. J. Immunol. 1989;142(2):702–707. [PubMed] [Google Scholar]
- Vandergeeten C., Fromentin R., DaFonseca S. Interleukin-7 promotes HIV persistence during antiretroviral therapy. Blood. 2013;121(21):4321–4329. doi: 10.1182/blood-2012-11-465625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vandergeeten C., Fromentin R., Merlini E. Cross-clade ultrasensitive PCR-based assays to measure HIV persistence in large-cohort studies. J. Virol. 2014;88(21):12385–12396. doi: 10.1128/JVI.00609-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vesanen M., Markowitz M., Cao Y., Ho D.D., Saksela K. Human immunodeficiency virus type-1 mRNA splicing pattern in infected persons is determined by the proportion of newly infected cells. Virology. 1997;236(1):104–109. doi: 10.1006/viro.1997.8718. [DOI] [PubMed] [Google Scholar]
- Wang H., Daniel V., Sadeghi M., Opelz G. Differences in the induction of induced human CD4(+) CD25(+) FoxP3(+) T-regulatory cells and CD3(+) CD8(+) CD28(−) T-suppressor cells subset phenotypes in vitro: comparison of Phorbol 12-myristate 13-acetate/ionomycin and phytohemagglutinin stimulation. Transplant. Proc. 2013;45(5):1822–1831. doi: 10.1016/j.transproceed.2012.10.061. [DOI] [PubMed] [Google Scholar]
- Weinberger A.D., Weinberger L.S. Stochastic fate selection in HIV-infected patients. Cell. 2013;155(3):497–499. doi: 10.1016/j.cell.2013.09.039. [DOI] [PubMed] [Google Scholar]
- Williams J.P., Hurst J., Stohr W. HIV-1 DNA predicts disease progression and post-treatment virological control. eLife. 2014;3:e03821. doi: 10.7554/eLife.03821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong J.K., Hezareh M., Gunthard H.F. Recovery of replication-competent HIV despite prolonged suppression of plasma viremia. Science. 1997;278(5341):1291–1295. doi: 10.1126/science.278.5341.1291. [DOI] [PubMed] [Google Scholar]
- Yerly S., Gunthard H.F., Fagard C. Proviral HIV-DNA predicts viral rebound and viral setpoint after structured treatment interruptions. AIDS. 2004;18(14):1951–1953. doi: 10.1097/00002030-200409240-00011. [DOI] [PubMed] [Google Scholar]
- Yu J.J., Wu T.L., Liszewski M.K. A more precise HIV integration assay designed to detect small differences finds lower levels of integrated DNA in HAART treated patients. Virology. 2008;379(1):78–86. doi: 10.1016/j.virol.2008.05.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yukl S.A., Gianella S., Sinclair E. Differences in HIV burden and immune activation within the gut of HIV-positive patients receiving suppressive antiretroviral therapy. J. Infect. Dis. 2010;202(10):1553–1561. doi: 10.1086/656722. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Performance of TILDA. (A) Activated CD4 + T cells were infected with HIV pNL4.3-GFP. GFP + and GFP- cells were sorted and tat/rev RNA were detected in single cells. The amplifications curves obtained from 24 GFP + and 24 GFP- cells are shown. (B) TILDA measurements were performed in duplicate in the same experiments or 3 to 4 times in independent experiments to calculate the intra and inter-assay coefficients of variation of the assay. Horizontal bars show means and standard deviations. (C) Dynamic range of TILDA: All TILDA values determined in samples obtained from virally suppressed subjects participating in the present study are represented. Unstimulated (spontaneous) and stimulated TILDA values are represented by grey and black circles, respectively. Horizontal dotted lines show the lower and upper limits of detection of the assay.