Abstract
Gut microbiota has been implicated as a pivotal contributing factor in diet-related obesity; however, its role in development of disease phenotypes in human genetic obesity such as Prader–Willi syndrome (PWS) remains elusive. In this hospitalized intervention trial with PWS (n = 17) and simple obesity (n = 21) children, a diet rich in non-digestible carbohydrates induced significant weight loss and concomitant structural changes of the gut microbiota together with reduction of serum antigen load and alleviation of inflammation. Co-abundance network analysis of 161 prevalent bacterial draft genomes assembled directly from metagenomic datasets showed relative increase of functional genome groups for acetate production from carbohydrates fermentation. NMR-based metabolomic profiling of urine showed diet-induced overall changes of host metabotypes and identified significantly reduced trimethylamine N-oxide and indoxyl sulfate, host-bacteria co-metabolites known to induce metabolic deteriorations. Specific bacterial genomes that were correlated with urine levels of these detrimental co-metabolites were found to encode enzyme genes for production of their precursors by fermentation of choline or tryptophan in the gut. When transplanted into germ-free mice, the pre-intervention gut microbiota induced higher inflammation and larger adipocytes compared with the post-intervention microbiota from the same volunteer. Our multi-omics-based systems analysis indicates a significant etiological contribution of dysbiotic gut microbiota to both genetic and simple obesity in children, implicating a potentially effective target for alleviation.
Research in context
Poorly managed diet and genetic mutations are the two primary driving forces behind the devastating epidemic of obesity-related diseases. Lack of understanding of the molecular chain of causation between the driving forces and the disease endpoints retards progress in prevention and treatment of the diseases. We found that children genetically obese with Prader–Willi syndrome shared a similar dysbiosis in their gut microbiota with those having diet-related obesity. A diet rich in non-digestible but fermentable carbohydrates significantly promoted beneficial groups of bacteria and reduced toxin-producers, which contributes to the alleviation of metabolic deteriorations in obesity regardless of the primary driving forces.
Keywords: Prader–Willi syndrome, Obesity, Gut microbiota, Metagenomics, Metabolomics, Genome interaction network
Highlights
-
•
Genetic and simple obesity in children shared a similar dysbiotic gut microbiota.
-
•
A diet rich in non-digestible carbohydrates significantly improved gut microbiota and alleviated genetic and simple obesity.
-
•
Specific bacterial genomes for producing obesity-related metabolites were identified.
1. Introduction
Prader–Willi syndrome (PWS) is the most common form of human genetic obesity caused by mutations in the paternal genes on chromosome 15q11.2–q13 (Butler, 2011). Bodyweight management of children with PWS has proven to be extraordinarily difficult as they have a low muscle tone that tends to result in less physical activity than normal, a chronic and uncontrollable feeling of hunger, and a deficit of satiety that drives their constant food-craving behavior. The molecular chain of causation between these drivers and obesity development in PWS children remains largely uncharacterized (Lacroix et al., 2014).
Accumulating evidence suggests that a dysbiotic gut microbiota may work as a contributing factor in diet-related obesity (Backhed et al., 2004, Zhao, 2013). Germfree mice were shown to be resistant to high-sugar, high-fat, “Western” diet-induced obesity (Backhed et al., 2007). Transplantation of gut microbiota from obese mice or humans increased fat deposits in germfree mouse recipients (Turnbaugh et al., 2008, Ridaura et al., 2013). Development of adiposity and insulin resistance in mice could be significantly reduced by diminishing the gut microbiota with broad-spectrum antibiotics (Cani et al., 2008). Transplantation of gut microbiota from healthy donors improved insulin resistance in the first six weeks in obese human volunteers (Vrieze et al., 2012). A diet based on whole-grains, traditional Chinese medicinal foods and prebiotics (WTP diet), has been shown to reduce endotoxin producers and enrich beneficial bifidobacteria in the gut of obese adult human volunteers, leading to decreased endotoxin in the bloodstream and significant alleviation of inflammation, adiposity and insulin resistance (Xiao et al., 2014, Fei and Zhao, 2013). Thus, compelling evidence suggests that the gut microbiota serves as a pivotal contributing factor in the development of diet-related obesity in both mice and humans.
Intriguingly, gut microbiota may even be involved in genetically predisposed obesity in mice. Removal of most of the gut microbiota with broad-spectrum antibiotics effectively reduced the development of adiposity and insulin resistance in genetically obese mice including leptin-deficient ob/ob mice and toll-like receptor 5 knockout mice (Cani et al., 2008, Vijay-Kumar et al., 2010). Transplantation of gut microbiota from these two strains of genetically obese animals into germ free wild type mice also conferred parts of the obesity phenotype on the recipients (Vijay-Kumar et al., 2010, Turnbaugh et al., 2006). However, the role of gut microbiota in genetically predisposed obesity in humans has not been characterized so far. Whether modulation of the gut microbiota can contribute to alleviation of metabolic deteriorations in human genetic obesity becomes an interesting question.
To tackle this question, we recruited children morbidly obese with PWS or diet-related simple obesity (SO) for a hospitalized intervention with the WTP diet (Xiao et al., 2014). By using a top-down systems strategy to combine metagenomic characterization of the gut microbiota, metabolomic profiling of co-metabolites between host and gut bacteria, and transplantation of human gut microbiota to germfree mice, we showed a potentially significant contribution of dysbiotic gut microbiota to the metabolic deteriorations associated with genetically predisposed obesity, as with simple obesity. These results imply a gut microbiota-mediated path towards obesity-related metabolic diseases, common to diet-related and genetically predisposed obesity.
2. Materials and Methods
2.1. Clinical Investigation
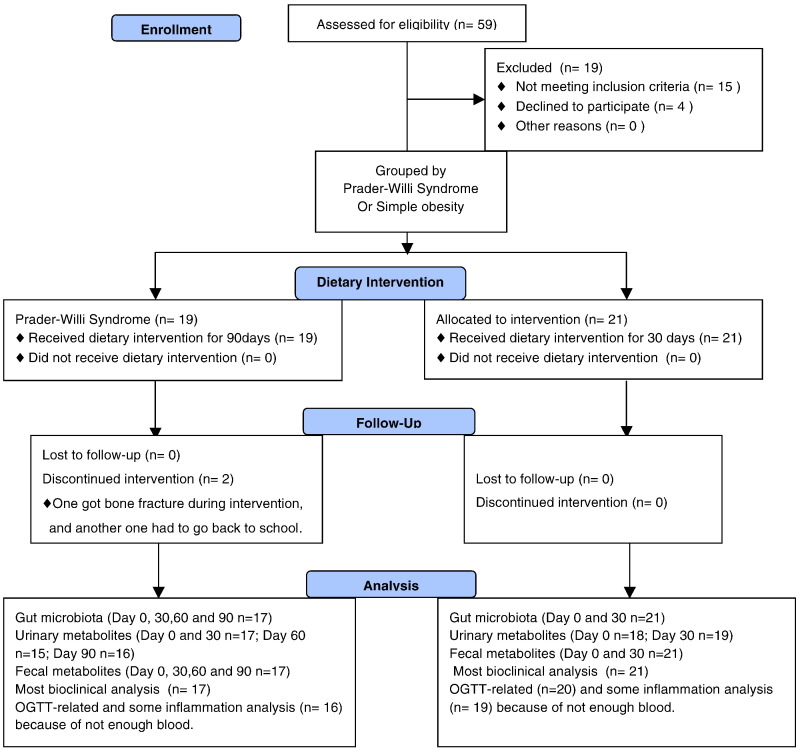
The open-labeled and self-controlled study was performed under the approval of the Ethics Committee of the School of Life Sciences and Biotechnology, Shanghai Jiao Tong University (No. 2012-016). The clinical trial was registered at Chinese Clinical Trial Registry (No. ChiCTR-ONC-12002646). Written informed consents were obtained from the guardians of all the participant children. The design and process of clinical trial was shown in Fig. 1.
Fig. 1.
The flow diagram of participants.
Obese children (3–16 years old), who were diagnosed with Prader–Willi syndrome or simple obesity, were recruited into a hospitalized dietary intervention for 30 days in Guangdong Women and Children Hospital, Guangzhou, China (Table S1). Upon the request of parents, all PWS patients continued the intervention for 60 more days. One volunteer (GD02) stayed in the hospital for 285 days on this intervention. The volunteers did not take any exercise program. Subjects were excluded if they had gastrointestinal pathologies, gastrointestinal surgery, chronic pathologies except overweight, antibiotics administration lasting more than 3 days in the previous 3 months or participation in other weight-loss programs in the previous 3 months. General questionnaires were implemented to collect information on demographic characteristics, health status, disease history, gastrointestinal conditions, dietary habit, and physical activity. A meal-based food frequency questionnaire and a 24-hour dietary record were used to calculate baseline nutrient intake based on China Food Composition 2002 (Yang et al., 2002). A hyperphagia questionnaire designed for Prader–Willi syndrome was administered to the parents of the children with PWS to assess hyperphagia before and after the intervention (Dykens et al., 2007).
A diet based on whole grains, traditional Chinese medicinal foods, and prebiotics (WTP diet) (Xiao et al., 2014) (The three ready-to-consume pre-prepared foods, Formula No. 1, Formula No. 2 and Formula No. 3 in the diet were manufactured by PERFECT (CHINA) CO., LTD.) was administered in combination with appropriate amounts of vegetables, fruits and nuts according to dietician's advice. Subjects were allowed to consume enough of Formula No. 1 to satisfy hunger pangs. The intake of macronutrients was balanced according to their age standard nutritional requirements provided by Chinese Dietary Reference Intakes (DRIs) recommended by the Chinese Nutrition Society (CNS, 2012). More specifically, Formula No. 1 was a pre-cooked mixture of 12 component materials from whole grains and TCM food plants that are rich in dietary fiber, including adlay (Coix lachrymal-jobi L.), oat, buckwheat, white bean, yellow corn, red bean, soybean, yam, peanut, lotus seed, and wolfberry, which was prepared in the form of canned gruel (370 g wet weight per can) by a contract food manufacturer. Each can contained 100 g of ingredients (59 g carbohydrate, 15 g protein, 5 g fat, and 6 g fiber) and 336 kcal (70% carbohydrate, 17% protein, 13% fat). Formula No. 2 was a powder preparation for infusion (20 g per bag) containing bitter melon (Momordica charantia) and oligosaccharides, including fructo-oligosaccharides and oligoisomaltoses. Each volunteer had on pack of Formula No. 2 a day. Formula No. 3 contained soluble prebiotics, including Fibersol-2, fructo-oligosaccharides and oligoisomaltoses, and was administered in the form of powder for infusion (30 g per bag). Each volunteer had at least one pack of Formula No. 3 a day. The intake of No. 3 may be increased to enhance bowl movement by producing more gas. The detailed composition of formulas 1, 2 and 3 was shown in Table 1. The dietary record of each subject was used to calculate nutrient intakes based on China Food Composition 2002 (Yang et al., 2002) (Tables S2 and S3).
Table 1.
The components of the three ready-to-consume formula of WTP diet in the study.
| Formula 1a | Formula 2b | Formula 3b | |
|---|---|---|---|
| Ash content (g/100 g) | 0.93 | 4.8 | 0.067 |
| Water (g/100 g) | – | 3.62 | 4.2 |
| Protein (g/100 g) | 5.7 | 9.76 | < 0.1 |
| Fat (g/100 g) | 1.7 | 1.2 | < 0.1 |
| Fiber (g/100 g) | 3.0 | 18.9 | 24.5 |
| Carbohydrate (g/100 g) | 20.2 | 61.7 | 71.2 |
| Soluble fiber (g/100 g) | 0.14 | 5.6 | 24.5 |
| Unsoluble fiber (g/100 g) | 2.8 | 13.3 | < 0.1 |
| Vitamin A (mg/kg) | – | – | – |
| Vitamin D (mg/kg) | – | – | – |
| Vitamin E (mg/kg) | 0.54 | 15.2 | – |
| Vitamin K1 (μg/100 g) | 0.0036 | 26.8 | – |
| Vitamin B1 (mg/100 g) | 0.054 | – | – |
| Vitamin B2 (mg/100 g) | 0.025 | 0.324 | 0.052 |
| Vitamin B6 (μg/100 g) | 0.051 | – | – |
| Vitamin B12 (μg/100 g) | – | 0.48 | – |
| Vitamin C (mg/100 g) | – | < 0.3 | < 0.3 |
| Biotin (μg/100 g) | – | 2.67 | – |
| Niacin (μg/100 g) | 0.18 | – | – |
| Vitamin B5 (μg/100 g) | – | – | – |
| Folate (μg/100 g) | – | 446 | 2.65 |
| Sodium (mg/kg) | 7.36 | 1.95 × 103 | 77 |
| Potassium (mg/kg) | 2.78 × 102 | 1.92 × 104 | 21 |
| Copper (mg/kg) | 0.21 | 4 | – |
| Magnesium (mg/kg) | 58.9 | 1.77 × 103 | 4 |
| Ferrum (mg/kg) | 1.6 | 74 | 2 |
| Zinc (mg/kg) | 0.89 | 14 | 2 |
| Manganese (mg/kg) | 0.89 | 12 | – |
| Calcium (mg/kg) | 16.3 | 264 | 4 |
| Phosphorus (mg/100 g) | 0.12 | 305 | < 2 |
| Iodine (mg/kg) | 0.021 | 0.06 | 0.05 |
| Chlorine (mg/100 g) | 31.4 | 226 | < 10 |
| Selenium (mg/kg) | 0.008 | 0.007 | – |
| Chromium (mg/kg) | – | 1.4 | – |
| Fluorine (mg/kg) | – | 4.92 | < 0.50 |
| Inositol (mg/kg) | 13.5 | 1.66 × 103 | – |
| linoleic acid (g/100 g) | – | 0.03 | – |
| α-Linolenic acid (g/100 g) | – | 0.03 | – |
| Docosahexenoic acid (g/100 g) | – | – | – |
| Eicosatetraenoic acid (g/100 g) | – | – | – |
| Cytidine (mg/100 g) | 0.014 | 6.37 | – |
| Uridine (mg/100 g) | 0.014 | 3.78 | – |
| Carnine (mg/100 g) | 0.014 | 2.2 | – |
| Guanosine (mg/100 g) | 0.014 | 4.1 | – |
| Adenosine (mg/100 g) | 0.014 | 1.54 | – |
| Choline (mg/100 g) | 30.3 | 92 | 9 |
| l-Carnitine (mg/kg) | – | – | – |
| Taurine (mg/100 g) | – | 336 | – |
| Molybdenum (mg/kg) | 0.33 | – | – |
| Cobalt (mg/kg) | – | – | – |
| Aspartic acid (g/100 g) | 0.38 | 0.59 | – |
| Threonine (g/100 g) | 0.1 | 0.29 | – |
| Serine (g/100 g) | 0.22 | 0.33 | – |
| Glutamic acid (g/100 g) | 0.68 | 0.95 | – |
| Proline (g/100 g) | 0.27 | 0.27 | – |
| Glycine (g/100 g) | 0.18 | 0.33 | 0.01 |
| Alanine (g/100 g) | 0.24 | 0.17 | – |
| Valine (g/100 g) | 0.09 | 0.39 | – |
| Cystine (g/100 g) | – | 0.17 | – |
| Methionine (g/100 g) | 0.03 | 0.09 | – |
| Isoleucine (g/100 g) | 0.06 | 0.3 | – |
| Leucine (g/100 g) | 0.26 | 0.18 | – |
| Tyrosine (g/100 g) | 0.09 | 0.26 | – |
| Phenylalanine (g/100 g) | 0.16 | 0.34 | – |
| Histidine (g/100 g) | 0.07 | 0.19 | – |
| Trytophan (g/100 g) | 0.03 | 0.07 | – |
| Lysine (g/100 g) | 0.14 | 0.37 | – |
| Arginine (g/100 g) | 0.3 | 0.8 | – |
| Total amino acid (g/100 g) | 3.26 | 6.09 | 0.01 |
Formula 1 is a pre-cooked, ready to eat food. The concentration of each nutrient is the amount in each 100 g wet weight.
Formulas 2 and 3 are ready-to consume dry powder. The concentration of each nutrient is the amount in each 100 g dry weight.
Biological samples, anthropometric data and clinical laboratory analysis were obtained at baseline and every 30 days during the intervention. Blood samples were collected after overnight fasting and centrifuged at 3000 rpm for 20 min after standing at room temperature for 30 min to obtain serum. The feces and morning urine were collected on the same day. Feces, morning urine and serum samples were immediately frozen on dry ice upon collection and stored at − 80 °C until further analysis.
Serum glucose, CRP, total cholesterol, triglycerides, free fatty acids, ALT and AST were analyzed with the automatic biochemical analyzer (ADVIA® 1800 Clinical Chemistry System, Siemens A.G., Germany). Insulin was measured by immunochemiluminometric assays (ADVIA Centaur, Siemens A.G., German). Plasma HbAlc was measured by HPLC (Bio-rad D-10, Bio-Rad Laboratories Co., Ltd., Germany). Blood routine tests were performed on Swelab® Alfa Cell analyzer (Boule Diagnostics AB, Sweden).
Enzyme-linked immunosorbent assays (ELISAs) were conducted according to the manufacturer's instructions. The ELISA kit for Human LBP was from Hycult Biotech Inc. (HK315-01, Plymouth Meeting, PA, USA.). The kits for leptin, adiponectin, and high sensitivity IL-6 were all purchased from R&D Systems, Inc. (DLP00, DRP300, HS600B, Minneapolis, MN, USA). The kits for AGP and SAA were from Assaypro Inc. (EG5001-1, St. Charles, MO, USA) and Invitrogen Corporation (HS600B, Camarillo, CA, USA.), respectively. The intra-assay and inter-assay coefficients of variation were < 5% and < 10%, respectively.
2.1.1. Statistics
Statistical analysis was carried out using the SPSS Statistics 17.0 Software Package (SPSS Inc.). Wilcoxon matched-pairs signed rank test (two-tailed) was performed to analyze changes between various time points in the same cohort, while Independent Mann–Whitney U test (two-tailed) was performed to analyze the differences between groups
3. Molecular Diagnosis of Prader–Willi Syndrome
Genomic DNA was isolated from the peripheral blood specimens of the volunteers using the DNA isolation mini kit (ZEESAN, Xiamen, China) and Lab-Aid 820 nucleic acid extraction instrument (ZEESAN, Xiamen, China) according to the manufacturer's instructions. DNA samples were inspected on a Nanodrop 2000 (Thermo, US).
Genomic DNA was treated with sulfite using the CpGenome Turbo Bisulfite Modification Kit (Millipore, CA, USA) according to the manufacturer's instructions. Using methylation-specific primers in the CpG island of the SNRPN gene (Kubota et al., 1997), different amplicons were obtained from paternal and maternal chromosomal DNA in the core region associated with PWS. Two amplicons of different sizes were amplified from paternal and maternal chromosome in control samples. No amplification occurred in untreated samples. Only the amplicon from the mother was detectable in the PWS patient samples.
The methylation-specific and non-methylation primers are as follows (Askree et al., 2011): MF 5′-TAAATAAGTACGTTTGCGCGGTC-3′ and MR 5′-AACCTTACCCGCTCCATCGCG-3′ were used to generate the 174 bp methylation product. Primers, PF 5′-GTAGGTTGGTGTGTATGTTTAGGT-3′ and PR 5′-ACATCAAACATCTCCAACAACCA-3′ were used to amplify 100 bp of the non-methylated allele.
The PCR reaction mixture was prepared as follows: Taq-HS (TaKaRa, Tokyo, Japan) 12.5 μL, primer forward 0.8 nmol, primer reverse 0.8 nmol, template DNA 60 ng, ddH2O to a total volume of 25 μL. The PCR program was 95 °C for 5 min, 95 °C for 30 s, 61 °C for 30 s, 72 °C for 30 s, cycled 40 times, 72 °C for 7 min. Electrophoresis was performed using a 3% agarose gel (Fig. S1).
4. Metabolomic Analysis
The fecal metabolite extraction (Wu et al., 2010), and the urine sample preparation (Jiang et al., 2012, Xiao et al., 2009) were performed as described previously.
All one-dimensional (1D) 1H NMR spectra of fecal water and urine samples were acquired on a Bruker AVIII 600 MHz NMR spectrometer equipped with a cryogenic probe (Bruker Biospin, Germany). The first increment of NOESY pulse sequence was employed with continuous-wave irradiation on water peak during recycle delay and mixing time for water suppression. The 90° pulse was adjusted to about 10 μs, and 64 scans were collected into 32 k data points for each spectrum with the spectral width of 20 ppm. To assist metabolite assignments, two-dimensional (2D) NMR spectra were acquired including 1H–1H COSY, 1H–1H TOCSY, 1H J-resolved, 1H–13C HSQC and 1H–13C HMBC for typical samples.
Fourier transformation of the free induction decays was performed after multiplying by an exponential function with a line-width factor of 1 Hz. The phase- and baseline-corrections were achieved manually, and chemical shift was calibrated to the TSP signal at δ0.00 with software TOPSPIN (v3.0, Bruker Biospin).
For the spectra of fecal water, the spectral region δ0.5–9.5 was integrated into bins with the width of 0.004 ppm using the AMIX package (v3.8, Bruker Biospin), and the region δ 4.75–4.93 for water peaks was removed. Each bin area was normalized to the dry weight of feces used for the extraction.
For the urine spectra, the spectral region δ0.5–9.35 was integrated into bins with the width of 0.004 ppm. The regions for water (δ4.71–5.10) and urea (δ5.43–6.20) peaks were removed, and each bin area was normalized to the total area of the respective spectrum. Some regions, including δ7.00–7.25, δ7.86–8.00 and δ8.05–8.20, were discarded after the normalization because of imperfect peak alignment.
4.1. Statistics
Orthogonal projection to latent structure-discriminant analysis (OPLS-DA) was performed with the software SIMCA-P + (v12.0, Umetrics, Sweden) with unit variance (UV) scaling and a 7-fold cross validation method. The qualities of all OPLS-DA models were assessed with R2X, the total variation being explained by the model, and Q2, denoting the predictability of the model. The significance of the models were further validated by variance analysis of the cross-validated residuals (CV-ANOVA) (P < 0.05) (Eriksson et al., 2008). The loadings generated from the models were plotted with color-coded OPLS-DA coefficients in MATLAB 7.1 using an in-house script after back-transformation (Cloarec et al., 2005). The color code means the absolute value of Pearson correlation coefficients (|r|), which indicate the weight of each variable contributing to the inter-group differentiation. The changed metabolites discovered by OPLS-DA were also confirmed by univariate analysis. Signal areas for specific metabolites were integrated with deconvolution using MestReNova (v8.0.0, Mestrelab Research S. L., Spain) and analyzed by t-test or Mann–Whitney U test using SPSS 13.0 as appropriate.
5. Gut Microbiota Profiling
5.1. 454 Pyrosequencing
DNA extraction from fecal samples was conducted as previously described (Godon et al., 1997) and purified by QIAamp DNA mini kit (51304, QIAGEN, Germany). The primers 5′-CGTATCGCCTCCCTCGCGCCATCAGACGAGTGCGTAGAGTTTGATYMTGGCTCAG-3′ and 5′-CTATGCGCCTTGCCAGCCCGCTCAGNNNNNNNNNNATTACCGCGGCTGCTGG-3′, with a sample-unique DNA barcode of ten-mer sequences represented as N, were used to amplify the V1–V3 region of each fecal sample by PCR as previously described (Zhang et al., 2012). PCR products were mixed at equal ratio for pyrosequencing using the GS FLX platform (Roche, Branford, CT, USA). The details of sequencing data analysis were shown in Supplementary Materials.
5.2. Metagenomic Sequencing
110 samples (For PWS, n = 17 at Day 0, 30, 60, and 90; For SO, n = 21 at Day 0 and 30) were sequenced using Illumina Hiseq 2000 platform at Shanghai Biotechnology Co., Ltd. DNA library preparation followed Illumina's instructions. Cluster generation, template hybridization, isothermal amplification, linearization, blocking, and denaturing and hybridization of the sequencing primers were performed according to the workflow indicated by the provider.
Libraries were constructed with an insert size of approximately 300 bp, followed by high-throughput sequencing to obtain paired-end reads with 100 bp in the forward and reverse directions. The library construction for the Day 30 sample of GD10 failed after repeated attempts and yielded no sequencing results. For the 109 successfully sequenced samples, an average of 80.8 ± 20.0 million (mean ± s.d.) reads were obtained for each sample.
5.3. Data Quality Control
Flexbar (Dodt et al., 2012) was used to trim the adapter from the reads; Prinseq (Le Chatelier et al., 2013) was employed a) to trim the reads from the 3′ end until reaching the first nucleotide with a quality threshold of 20; b) to remove read pairs if either read was shorter than 60 bp or contained ‘N’ bases, and c) to de-duplicate the reads. Reads that could be aligned to the human genome (H. sapiens, UCSC hg19) were removed (aligned with Bowtie2 (Langmead and Salzberg, 2012), using — reorder–no-hd–no-contain–dovetail). On average, 76.0 million ± 18.0 million (mean ± s.d.) reads for each sample were retained and used for further analysis.
5.4. De Novo Assembly and Non-Redundant Metagenomic Gene Catalogue Construction
High-quality reads were used for de novo assembly with IDBA-UD (Peng et al., 2012) (an iterative De Bruijn Graph De Novo Assembler) into contigs of at least 500 bp. Reads from all samples from the same volunteer were assembled. Genes were predicted using MetaGeneMark (Zhu et al., 2010). A non-redundant gene catalogue of 2,077,766 was constructed with CD-HIT (Li and Godzik, 2006) using the parameters “− c 0.95 − aS 0.9”.
5.5. Abundance Profile of Gene Catalogue
High quality reads were mapped to the gene catalogue with SOAPaligner (R. Li et al., 2009) (− M 4 − l 50 − r 1 − v 5). To adjust for sequencing depth, we sampled the alignment results and downsized the number of mapped reads to 28 million for each sample and then used soap.covearge.script (http://soap.genomics.org.cn/down/soap.coverage.tar.gz) to calculate gene-length normalized base counts. The sampling procedure was repeated 30 times and the mean value of abundance was used for further analysis.
5.6. Co-Abundance Gene Groups (CAGs)
All the genes were clustered into CAGs based on their abundance data using the Canopy-based algorithm with default parameters. CAGs with more than 700 genes were regarded as bacterial CAGs for further analysis. CAG abundance profiles were calculated as the sample-wise median gene abundance (Nielsen et al., 2014). The PCoA of bacteria CAGs based on different distance and MANOVA was performed using QIIME. Procrustes superimposition was then performed from the PCoA of CAGs and principal component analysis (PCA) of bioclinical data.
5.7. Genome Interaction Groups (GIGs)
Bacterial CAGs shared by more 20% of the samples were regarded as prevalent species. Associations between these prevalent bacterial CAGs were determined by bootstrapped Spearman correlation coefficient based on their abundance profiles. The correlations were converted to a correlation distance (1-correlation coefficient) and then clustered using the Ward clustering algorithm. From the top of the clustering tree, we used Permutational MANOVA (9999 permutations, P < 0.001) to sequentially determine whether the two clades were significantly different to cluster the prevalent bacterial CAGs into GIGs. The bootstrapped Spearman correlation coefficient between GIGs and bioclinical parameters was calculated to identify the relationship between gut microbiota and host phenotype. Species within the same GIG decreased or increased their abundance together during the dietary intervention. Therefore we added their individual abundance together to calculate group-level abundance changes during the trial. Spearman correlation coefficient and ward clustering were performed using MATLAB®. Permutational MANOVA was done in the vegan package in R. The networks were then visualized in Cytoscape v3.1.1. The abundance of each GIG was calculated as the sum of the abundance of all the bacterial CAGs in the GIG. Wilcoxon matched-pairs signed rank test (two-tailed) was performed to analyze changes of each GIG between various time points in the same cohort. The PCoA of GIGs based on Bray–Curtis distance was performed using the vegan package in R. Procrustes superimposition was then performed from the PCoA of GIGs and principal component analysis (PCA) of bioclinical data.
5.8. Assembly of Bacterial CAGs
For each of the 161 prevalent bacterial CAGs, we performed a de novo bacterial CAG-augmented and sample-augmented assembly (Nielsen et al., 2014). For a given sample, the reads were aligned to the bacterial CAG-specific contigs with Bowtie2, the mapped reads, including unmapped mates, were extracted. These reads were then de novo assembled with Velvet (Zerbino and Birney, 2008) using kmer from 45 to 75 with parameters ‘− cov_cutoff auto’ and ‘− exp_cov auto’. As each bacterial CAG had several assemblies derived from several samples independently, the assembly with highest N50 was selected. The minimum lengths of contigs and scaffolds were filtered to 100- and 500-bp respectively. And the gaps in the scaffolds were filled by using SOAPdenovo GapCloser (1.12).
5.9. Assembly Statistics
To assess the quality of the assemblies, the six high-quality draft genome assembly criteria from the Human Microbiome Project (HMP) were adopted. The criteria are (i) 90% of the genome assembly must be included in contigs > 500 bp, (ii) 90% of the assembled bases must be at > 5 × read coverage, (iii) The contig N50 must be > 5 kb, (iv) scaffold N50 must be > 20 kb, (v) average contig length must be > 5 kb and (vi) > 90% of the core genes must be present in the assembly. The general assembly statistics were calculated with an in-house Perl script based on BioPerl. The coverage was obtained by aligning reads to the contigs with Bowtie2 and calculating with BEDtools (Quinlan and Hall, 2010). The core gene ratios were determined with HMP standard operating procedures.
5.10. Comparison of Assemblies and Reference Genomes
118 high quality assemblies passing 5 or 6 HMP criteria for reference genomes were compared to 352 draft reference genomes from the human gastrointestinal tract HMP DACC database and 2645 complete reference genomes from the NCBI database (downloaded on May 6, 2014). The comparison was performed with MUMmer3.0 (Kurtz et al., 2004). The contigs were aligned to the reference genomes with numcer (default parameters). Delta-filter (parameters: − i 95 − o 80) was used to filter the low identity matches. Finally, the comparison of the assemblies and the reference genomes and the estimation of mean identity and coverage were performed with dnadiff.
5.11. Phylogeny of the Bacterial CAG Assemblies
We used CVtree3.0 web server (Xu and Hao, 2009), which applies a composition vector to perform phylogenetic analysis, to construct a phylogenetic tree of the 118 bacterial CAGs with high quality assemblies, 352 reference gastrointestinal tract genomes from the HMP DACC database, and the server's inbuilt database, to identify the phylogenetic position of the bacterial CAGs. We also applied SpecI (Mende et al., 2013), which is a method to group organisms into species clusters based on 40 universal and single-copy phylogenetic marker genes, to delineate the bacterial CAGs. 64 bacterial CAGs could be delineated by SpecI, and only 5 of the 64 showed disagreement with CVtree (CAG00178 Streptococcus gallolyticus (SpecI), Streptococcus pasteurianus (CVtree); CAG00047 Ruminococcus gnavus (SpecI), Lachnospiraceae bacterium 2 1 58FAA (CVtree); CAG00120 Ruminococcus gnavus (SpecI), Lachnospiraceae bacterium (CVtree); CAG00226 Parasutterella excrementihominis (SpecI), Burkholderiales bacterium 1_1 _47 (CVtree); CAG00020 Bacteroides uniformis (SpecI), Bacteroides sp. D20 (CVtree)).
5.12. Comparison of Strains of the Same Species
Within the same species, strains were compared pair-wise using MUMer3.0 based on their high-quality draft genomes. For strains of the same species, Mann–Whitney test was used to compare the identity of strains in the same GIG with that in different GIGs.
5.13. Metagenomic CAZy Profiling
A local-version database was downloaded from the dbCAN website (http://csbl.bmb.uga.edu/dbCAN/annotate.php, version: 11 May, 2013). Genes in the non-redundant gene catalogue were aligned to the database with HMMscan (Finn et al., 2011). The alignment result was parsed by the hmmscan-parser.sh, which was provided by dbCAN, and the best-hit alignment was retained. The CAZy family profile was calculated by summing the relative abundance of genes aligned to it.
6. Global Structural Analysis of Gut Microbiota
6.1. Gene Richness Analysis
Gene richness was compared between subjects at the baseline using the same number of mapped reads based on the downsizing results mentioned above. The distribution of gene count was calculated in MATLAB® using histfit (kernel).
6.2. Enterotypes Analysis
Enterotypes were determined using the abundance of genus, OTUs and CAGs separately using the methods suggested in http://enterotype.embl.de/. The package ade4 in R was used in this analysis.
7. Functional Annotation
7.1. Global Level Based on All High-Quality Reads and Non-Redundant Gene Catalogue
Metagenomic data were metabolically profiled by using HUMAnN. All high quality reads were aligned with Bowtie2 to the KEGG database (Kanehisa et al., 2012) (Release: February, 24, 2014), from which sequences of eukaryotes had been excluded. The alignments were transformed into bam format with SAMtools (H. Li et al., 2009) and input into HUMAnN to obtain the abundance of KOs and pathways. The linear discriminant analysis (LDA) effect size (LEfSe) (Segata et al., 2011) was used to identify enzymatic pathways that were significantly different between pre- and post-intervention.
The protein sequences of formate-tetrahydrofolate ligase (GI|293416147|), Propionyl-CoA:succinate-CoA transferase (GI|218353245), propionate CoA-transferase (GI|7242549|), and butyryl-CoA transferase (GI|71081820|) (Claesson et al., 2012) were downloaded from NCBI database. The non-redundant gene catalogue was aligned to these sequences by using BLASTP (best-hit with E-value < 1E − 5, identity > 30% and coverage > 70%).
7.2. GIGs Level
To analyze the function of each GIG, the reads from the sample group that contained the highest abundance of the GIG and aligned to the GIG-specific genes were gathered. As described before, these reads were aligned to KEGG database and the alignments were parsed with HUMAnN, thus obtaining the relative abundance of pathways in each GIG.
7.3. Bacterial CAGs Level
For each of the 118 bacterial CAGs, the open reading frames (ORFs) were identified using myRAST (Overbeek et al., 2014). The reference protein sequences of tryptophanase (K01667) were collected from the KEGG database. Sequences of choline TMA-lyase (GI|219868924|) and choline TMA-lyase-activating enzyme (GI|219868925|) (Craciun and Balskus, 2012) were downloaded from the NCBI database. The potential tryptophanase, choline TMA-lyase and choline TMA-lyase-activating enzyme in bacterial CAGs were identified by aligning the ORF protein sequences against the references with BLASTP (best-hit with E-value < 1E-5, identity > 30% and coverage > 70%). Additionally, for identification of choline TMA-lyase and choline TMA-lyase-activating enzyme, we required that they co-exist as components in a cluster.
7.4. Association Between Microbiota and Urine Metabolome
To aid interpretation of the relationship between the bacterial CAG-level microbiota and metabolomics datasets, we integrated them using a multivariate method known as co-inertia analysis (CIA) (Fagan et al., 2007). PCA was applied to each of the individual datasets as inputs for the CIA. Ordinations of the two datasets that are maximally co-variant were identified to find the shared biological trends within the two datasets. CIA was performed by ade4 package in R. The bootstrapped Spearman correlation coefficients between bacterial CAGs and indoxyl sulfate or TMAO based on their abundance in all the samples were calculated.
7.5. Accession Numbers
The raw pyrosequencing and Illumina read data for all samples has been deposited in the Sequence Read Archive (SRA) under the accession number SRP045211.
8. Gut Microbiota Transplantation
8.1. Animal Trial
Fecal samples were collected from a Prader–Willi syndrome patient (GD58) before (Day 0) and after (Day 90) the intervention as described above. The samples were preserved nearly one year at − 80 °C. 500 μg of fecal sample was diluted in 25 mL of sterile Ringer working buffer (containing 9 g of sodium chloride, 0.4 g of potassium chloride, 0.25 g of calcium chloride dihydrate and 0.05% l-cysteine-HCl in 1 L buffer) in an anaerobic chamber (whitely DG500; Don Whitley Scientific; atmosphere composed of 80% N2/10% CO2/10% H2). The fecal material was suspended by thorough vortexing (5 min) and settled by gravity for 5 min. The clarified supernatant was transferred to new tubes and an equal volume of 20% skim milk (LP0031, Oxoid, UK) was added. The fresh inoculum was used on the day of preparation, and the rest was preserved at − 80 °C until the second inoculation.
All experimental procedures and protocols were approved by the Institutional Animal Care and Use Committee of SLAC Inc. Nineteen weaned, germ-free (GF) male C57BL/6J mice were maintained in flexible-film plastic isolators under a regular 12-h light cycle (lights on at 06:00 h) in SLAC Inc. (Shanghai, China). Mice were fed with a sterile normal chow diet (D12450B, Research Diets, Inc., New Brunswick, NJ) ad libitum. Surveillance for bacterial contamination was performed by periodic bacteriologic examination of feces, food and padding.
When they reached 8 weeks of age, the GF animals were randomly divided into 2 groups. For the Day 0 group, mice were inoculated by oral gavage with 100 μL of fecal suspension collected from patient GD58 before the dietary intervention (n = 10), and for the Day 90 group, mice were inoculated with 100 μL of fecal suspension collected from patient GD58 after 90 days of intervention (n = 9). A repeat inoculation was conducted the next day to reinforce the microbiota transplantation. The body weight of each mouse was measured twice a week.
8.2. Tissue Sample Collection
At the end of the 2nd and 4th weeks, half of the mice were sacrificed, and blood and tissues samples were collected. After exsanguination, white adipose tissues (epididymal, mesenteric, subcutaneous inguinal and retroperitoneal), liver, total gut and muscle were collected, sectioned, weighed and stored in liquid nitrogen.
8.3. RT-qPCR
Total RNA from liver, ileum and colon tissue was isolated using an RNeasy lipid tissue mini kit (75842, QIAGEN, Germany) according to the manufacturer's protocol. 2 μg of each total RNA sample was treated with RNase-free Dnase (18068-015, Invitrogen, USA). First-strand cDNA was synthesized using random hexamers and Superscript II reverse transcriptase (18080-051, Invitrogen, USA). PCRs were performed using the Eppendorf Realplex thermocycler. Primer sequences for the targeted mouse genes were as follows:
- Tnfα,
- F: 5′ACGGCATGGATCTCAAAGAC3′, R: 5′AGATAGCAAATCGGCTGACG3′;
- Il6,
- F: 5′-GTTCTCTGGGAAATCGTGGA-3′, R: 5′-TGTACTCCAGGTAGCTA-3′;
- Tlr4,
- F: 5′ATGGCATGGCTTACACCACC3′, R: 5′GAGGCCAATTTTGTCTCCACA3′;
- Gapdh
- F: 5′GTGTTCCTACCCCCAATGTGT3′,
- R: 5′ATTGTCATACCAGGAAATGAGCTT3′.
Cycle threshold (Ct) values were converted to quantification values based on the standard curves.
8.4. H&E Staining of Fat Tissue and Histopathologic Analysis
Fresh epididymal fat pads were fixed with 4% formaldehyde for 48 h and dehydrated through a series of graded ethanol baths to displace the water before being embedded in paraffin. Samples were sectioned at 5 μm and stained by hematoxylin and eosin. Digital images of sections were acquired with a Leica DMRBE microscope. Adipocyte size (cross-sectional area) was counted by Image Pro Plus 6.0. For each mouse, adipocytes areas were determined in at least three histologic sections and 300 total adipocytes.
8.5. Statistics
Statistical analysis was carried out using the SPSS Statistics 17.0 Software Package (SPSS Inc.). Student t-test or Mann–Whitney test was used for detecting variation between gnotobiotic mice receiving the fecal microbiota from Day 0 compared to Day 90 samples, depending on data distribution.
9. Results
9.1. Dietary Alleviation of Genetic and Simple Obesity
The WTP diet was used for this hospitalized intervention study performed on morbidly obese children with PWS or SO. The two cohorts (PWS, n = 17, average age 9.26 yrs, range 5–16 yrs; SO, n = 21, average age 10.52 yrs, range 3–16 yrs) showed no significant difference in age range (Table S1). Both cohorts received the hospitalized intervention for 30 days. At their parents' request, the PWS cohort continued for another 60 days. One PWS volunteer (GD02) stayed in the hospital for 285 days. During the dietary intervention, both cohorts of children had their total calorie intake cut by about 30% compared to their pre-intervention diets. Protein intake remained at 13–14% of total kcal consumed. Carbohydrate intake increased from 52% to 62% of total calories in PWS and from 57% to 62% in SO. The form of carbohydrates changed from primarily white rice and wheat flour to whole grains. Fat intake decreased from 34% to 20% of total calories in PWS and from 30% to 20% in SO. The most substantial change was the total dietary fiber intake, which increased from 6 g to 49 g per day in PWS and from 9 g to 51 g per day in SO (Tables S2 and S3). Anthropometric measurements and metabolic panel blood testing were used to track changes.
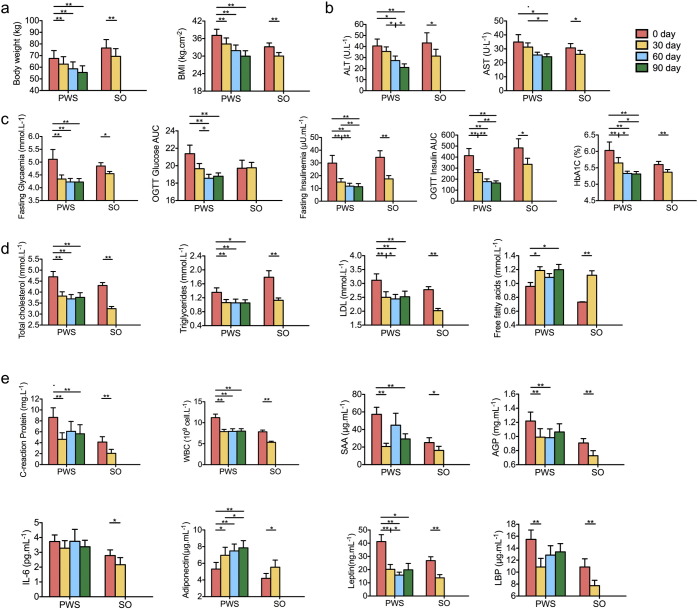
Almost all relevant bioclinical parameters indicate a significant alleviation of the metabolic deteriorations in children with both genetic and simple obesity after 30 days of the dietary intervention (Fig. 2). With one-month intervention, the SO cohort lost 9.5 ± 0.4% (mean ± s.e.m.) of their initial bodyweight, and the PWS cohort 7.6 ± 0.6% (Fig. 2a). Both PWS and SO children showed significant improvement in markers of metabolic health (Tables S4 and S5). Aspartate aminotransferase (AST) and alanine aminotransferase (ALT) levels in the blood were reduced, indicating improved liver condition (Fig. 2b). Glucose homeostasis was significantly improved (Fig. 2c). Blood levels of total cholesterol, triglycerides, and low-density lipoprotein (LDL) were decreased (Fig. 2d). The PWS cohort was followed for two more months on the WTP diet. They lost a total of 18.3 ± 1.0% of their initial bodyweight and showed continued improvement in several metabolic parameters (Fig. 2a–d). In addition, the PWS cohort showed a modest improvement in their overall hyperphagia behavior (Table S6). GD02 reduced his bodyweight from 140.1 kg to 83.6 kg after 285 days in the hospital. He then continued this intervention at home and reduced to 73.2 kg after 430 days on this diet. All his metabolic parameters came to normal range (Table S7). This extended dietary intervention can thus significantly alleviate the metabolic deteriorations in human genetic obesity, in which the diet-induced weight loss can be comparable to that achievable by gastric bypass surgery (Papavramidis et al., 2006).
Fig. 2.
Improved bioclinical parameters and inflammatory conditions after the intervention. (a) Anthropometric markers. (b) Hepatic function markers. (c) Plasma glucose homeostasis. (d) Plasma lipid homeostasis. (e) Inflammation related markers. Data are shown as mean ± s.e.m. Wilcoxon matched-pairs signed rank test (two-tailed) was used to analyze variation between each two-time points in PWS or SO children. *P < 0.05, **P < 0.01. For most of the bioclinical variables, PWS n = 17 and SO n = 21; For OGTT Glycaemia AUC and OGTT Insulinemia AUC, PWS n = 16 and SO n = 20; For CRP, W.B.C., SAA, AGP, Adiponectin and IL-6, PWS n = 16 and SO n = 19. BMI: body mass index; ALT: Alanine aminotransferase; AST: Aspartate aminotransferase; OGTT: Oral glucose tolerance test; LDL: low-density lipoprotein; CRP: C reactive protein; W.B.C.: White blood cell count; SAA: serum amyloid A protein; AGP: α-acid glycoprotein; LBP: Lipopolysaccharide binding protein.
Several markers of systemic inflammation were also improved in PWS and SO cohorts after 30 days of dietary intervention, including C-reactive protein (CRP), serum amyloid A protein (SAA), α-acid glycoprotein (AGP) and white blood cell count (WBC) (Fig. 2e). The level of adiponectin, an anti-inflammatory adipokine, was increased, and leptin was decreased, indicating an alleviation of the “at-risk” phenotype (Labruna et al., 2011). Lipopolysaccharide binding protein (LBP), a surrogate marker for bacterial antigen load in the blood (Zweigner et al., 2006), was decreased (Fig. 2e). This suggests that the gut microbiota of both cohorts may have changed after the intervention (Xiao et al., 2014, Fei and Zhao, 2013).
9.2. Structural Modulation of the Gut Microbiota
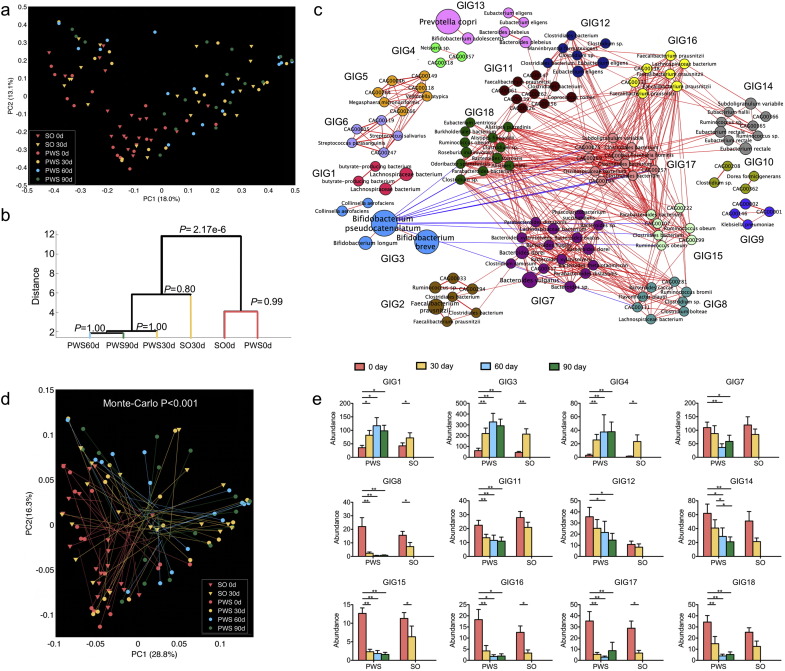
To determine how the overall structure of the gut microbiota was modulated during the dietary intervention, we performed Illumina-based shotgun metagenomic sequencing on 110 fecal samples collected from 21 SO (Day 0 and 30) and 17 PWS (Day 0, 30, 60, and 90) subjects. On average, 76.0 ± 18.0 million (mean ± s.d.) high-quality paired-end reads from each sample were used for de novo assembly and for predicting genes (Table S8), from which a non-redundant gene catalogue of 2,077,766 microbial genes was constructed. These two million genes were binned into 28,072 co-abundance gene (CAG) groups using the canopy-based algorithm with a strict cutoff for correlation coefficients (> 0.9) to maximize chances that genes of a CAG were from the same genome (Nielsen et al., 2014). 376 CAGs each with > 700 genes were considered to represent distinct bacterial genomes, which accounted for 36.4% (775,515) of the recognized genes. Of the 376 CAGs, we focused our subsequent analyses on 161 that were shared by at least 20% of the samples. The 161 prevalent CAGs were assembled into draft genomes, and 118 of the genome assemblies met at least five of the six quality criteria of the Human Microbiome Project (HMP) for standard reference genomes (Table S9). 50 of the assemblies were closely related to known reference genomes with coverage over 80% and identity over 95% (Table S10). 10 species were represented by more than one draft genome assembled, e.g. Faecalibacterium prausnitzii having 9 assembled genomes, and Eubacterium eligens having 5, suggesting that multiple strains within these species were present in the samples that were sequenced.
The composition of the gut microbiota showed a significant shift after 30 days of the intervention in both cohorts as indicated by principal coordinates analysis (PCoA, multivariate analysis of variance (MANOVA) test, P = 2.17e − 6) based on Bray–Curtis dissimilarity of the 376 bacterial CAGs (Fig. 3a and b). There was no significant difference in gut microbiota between PWS and SO either before (P = 0.99) or after the intervention (P = 0.8), suggesting that the PWS and SO gut microbiota were similarly dysbiotic prior to the intervention and that the intervention had the same effect on both (Fig. 3b). Analyses based on other β-diversity metrics and on pyrosequencing of the V1–V3 region of 16S rRNA genes confirmed these findings (Figs. S2 and S3). For both cohorts the gene richness and diversity of the gut microbiota significantly decreased after the intervention (Figs. S4 and S5). Delineation of low gene count/high gene count groups among our volunteers before the intervention was not evident (Fig. S6). Genus-level data from 16S rRNA gene sequencing gave two enterotypes represented by Bacteroides and Prevotella (Fig. S7). OTU-level data yielded three enterotypes, represented by: Bacteroides spp. uclustout#1111, Prevotella spp. uclustotu#3124 and Streptococcus spp. uclustotu#2404 (Fig. S8). CAG-based metagenomic data also differentiate the samples into three enterotypes represented by Bacteroides, Prevotella and Bifidobacterium respectively (Fig. S9). Some individuals changed their enterotypes after one month, indicating that changing enterotypes via long-term dietary intervention is possible (Tables S11–S13). Enterotypes and their changes responding to dietary intervention showed no correlation with any of the bio-clinical parameters.
Fig. 3.
Concordance of structural shifts of gut microbiota and the improvement of the host metabolic health. (a) PCoA based on Bray–Curtis distance of all the 376 bacterial CAGs during the dietary intervention. (b) Clustering of gut microbiota based on distances between different groups calculated with MANOVA test of first 23 PCs (accounting for 80% of total variations) of PCoA based on Bray–Curtis distance of all bacterial CAGs. (c) Genome interaction groups interaction network. Network plot highlights correlation relationships between 18 GIGs of 161 prevalent bacterial CAGs at all time points from the two cohorts. Node size indicates the average abundance of the species/strains. Lines between nodes represent correlations between the nodes they connect, with line width indicating the correlation magnitude, and red and blue colors indicating positive and negative correlations, respectively. For clarity, only lines corresponding to correlations whose magnitude is greater than 0.5 are drawn, and unconnected nodes are omitted. (d) Procrustes analysis combining PCoA of GIGs (end of lines with solid symbols) with PCA of bioclinical variables presented in Fig. 1 (end of lines without solid symbols). For PWS, n = 17 at Day 0, 30, 60, and 90; For SO, n = 21 at Day 0 and n = 20 at Day 30. (e) Group level abundance shifts of GIGs that changed significantly during dietary intervention. Data are mean ± s.e.m. Wilcoxon matched-pairs signed rank test (two-tailed) was used to analyze variation between each two-time points in PWS or SO children. *P < 0.05, **P < 0.01.
More importantly, procrustes analysis combining PCoA of the 376 bacterial CAGs (Fig. 3a) with PCA of the bioclinical variables (Fig. S10) showed that the structural shifting of the gut microbiota based on the abundance of the bacterial CAGs was significantly associated with the changes of the bioclinical parameters of both PWS and SO cohorts, suggesting that the overall structural changes deep at individual bacterial genome level were significantly associated with the improvements in host metabolic health (Fig. S11, P < 0.0001, 999 times Monte-Carlo simulations).
9.3. Genome Interaction Network Analysis
To identify bacterial genomes in the gut ecosystem that responded as functional groups (guilds) to the dietary intervention (David et al., 2014), we constructed a co-abundance network across all individuals and time points based on the 161 prevalent bacterial CAGs. Ward clustering algorithm and Permutational MANOVA (9999 permutations, P < 0.001) based on bootstrapped Spearman correlation coefficients clustered these bacterial CAGs into 18 genome interaction groups (GIGs) (Figs. 3c, S12). Note that different CAGs of the same species such as the 9 Faecalibacterium prausnitzii genomes were clustered into different GIGs, suggesting that different strains of the same species might occupy different metabolic niches in the gut ecosystem. Strains of the same species in the same GIG were more similar in their genomic sequences to each other than strains of the same species clustered into different GIGs, indicating that strains of the same species in different GIGs may be functionally different (Fig. S13). Procrustes analysis showed that separations based on either group-level abundance of GIGs or host bioclinical variables before and after the intervention co-segregated along the first axis in both PWS and SO data sets, suggesting that the changes in the abundance of the various GIGs were significantly associated with the improvements in host metabolic health (Fig. 3d, P < 0.0001, 999 times Monte-Carlo simulations). The agreement between genome-level and GIG-level procrustes analysis with host bioclinical variables suggests that this strategy of organizing prevalent genomes of human gut microbiota into genome interaction groups provides a potentially useful framework for understanding their functional interactions with each other and with the hosts.
Group level abundance analysis showed that 6 GIGs, including GIG13 containing the most predominant species Prevotella copri, did not change their abundance after the intervention (Fig. S14). GIG1, 3 and 4 significantly increased their abundance after the intervention, while GIG7, 8, 11, 12, 14, 15, 16, 17 and 18 decreased (Fig. 3e). GIG3 had a negative correlation with GIG8, 15, 16, and 18 (r > 0.45, FDR < 0.01) (Fig. 3c). GIG3 became the most promoted group after the dietary intervention. Notably, the major genomes in GIG3 were in the genus Bifidobacterium. The assembly for CAG00184, the most promoted genome after the intervention, covered 81.2% of the reference genome for Bifidobacterium pseudocatenulatum DSM 20438 with 98.6% identity (Fig. S15 and Table S10). The CAG00184 genome contained pathways for fermentation of monosaccharides, disaccharides, oligosaccharides and polysaccharides to produce acetate and lactate (Fig. S16).
Spearman correlation analysis between GIGs and host bioclinical parameters showed that a few GIGs were positively associated with disease phenotypes including GIG8, 11, 14, 15, 16,17 and 18, while a few others negatively associated including GIG1, 3 and 4 (Fig. S17). Such associations between group-level abundance of GIGs and host phenotypes suggest that some of the GIGs may impact host health as a functional group.
9.4. Functional Modulation of the Gut Microbiota
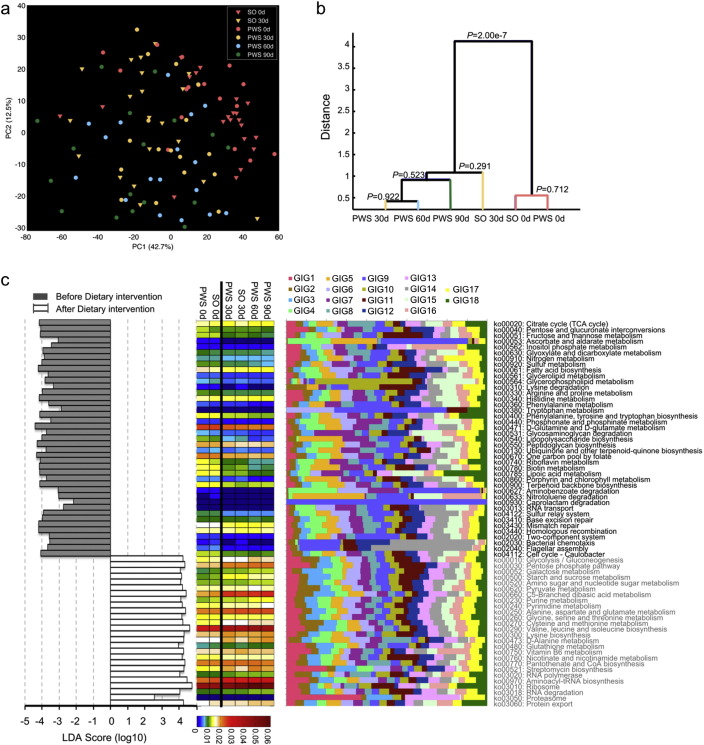
To see how the altered community structure of the gut microbiota affected its metabolic potential, we profiled the metagenomic data using HUMAnN to identify and quantify genes within biochemical pathways (Abubucker et al., 2012). In total, 5234 KEGG orthology groups (KOs) were recognized and quantified. The PCA score plot of all the KOs showed a significant shift after the intervention (MANOVA test, P = 2.00e − 7, Fig. 4a and b), indicating a modulation of the metabolic capacity of the gut microbiota concomitant with its diet-induced structural changes. There was no significant difference between the PWS and SO cohorts either before or after the intervention (MANOVA test P = 0.712 and P = 0.291, Fig. 4b). Thus, gut microbiota between PWS and SO children shared similar structural and functional features both before and after the intervention.
Fig. 4.
Functional shifts of the gut microbiome during the dietary intervention. (a) The PCA score plot of the KO Groups recognized with HUMAnN showing a significant shift of the KO profiles after the intervention (log-transformed). (b) Clustering of KO profiles based on distances between different groups calculated with MANOVA test of the first five PCs of PCA of KO. (c) Key pathways of gut microbiota responding to dietary intervention. The left histogram shows the LDA scores computed for features (on the pathway level) differentially abundant between all samples before and after the intervention. The heat map shows the abundance of the key pathways. The stacked bar chart shows relative contribution of GIGs to each pathway. For PWS, n = 17 at Day 0, 30, 60, and 90; For SO, n = 21 at Day 0 and n = 20 at Day 30. In (c) day 0 and day 30 data of PWS and SO combined together for this analysis.
Using the linear discriminant analysis (LDA) effect size (LEfSe) method (Segata et al., 2011), 67 KEGG database biochemical pathways (P < 0.05) were identified as significantly responding to the dietary intervention (Fig. 4c). 41 of these pathways were significantly decreased and 26 were enriched after the intervention. Notable among the enriched pathways were those for carbohydrate catabolism, including starch and sucrose metabolism (ko00500), and amino sugar and nucleotide sugar metabolism (ko00520). Notable among the decreased pathways were those for fat and protein metabolism, including fatty acid biosynthesis (ko00061), phenylalanine metabolism (ko00360), and tryptophan metabolism (ko00380). In addition, lipopolysaccharide biosynthesis (ko00540), peptidoglycan biosynthesis (ko00550) and flagellar assembly (ko02040) pathways were decreased, suggesting reduced bacterial antigen synthesis after the intervention. Pathways for xenobiotics biodegradation (ko00627, ko00633 and ko00930), and DNA repair-related pathways (ko03410, ko03430 and ko03440) were also decreased, perhaps reflecting reduced toxin load and mutagenic stress in the gut microbiota environment after the intervention.
9.5. Carbohydrate-Induced Metabolic Shifts in the Gut
The interventional diet contained dramatically increased levels of non-digestible carbohydrates, which may enter the colon to potentially shift the fermentation metabolism of the gut microbiota (Tables S2 and S3). Score plots of PCA and orthogonal projection to latent structure-discriminant analysis (OPLS-DA) of NMR-based metabolite profiling data of fecal water samples from the SO (Day 0 and 30) and the PWS cohorts (Day 0, 30, 60 and 90) showed a significant shift of metabolite composition after the intervention (Figs. S18 and S19). OPLS-DA coefficient plots showed dramatic increase of non-digestible carbohydrates after the intervention (Fig. S20 and Table S14). The relative abundance of 19 fecal metabolites in the SO cohort and 18 in the PWS cohort were found to be significantly reduced by the intervention (Figs. S21 and S22). Among these significantly reduced metabolites were many bacterial products. The significant decrease of these bacterial metabolites in the gut was concomitant with a significant reduction in the fecal bacterial content as determined by qPCR (Fig. S23), but fecal matter production of these children significantly increased (Table S15). Despite the decrease of bacterial metabolites, the relative concentration of acetate was increased among short chain fatty acids (SCFAs) while those of isobutyrate and isovalerate were decreased, indicating a shift of protein fermentation to carbohydrates fermentation after intervention (Fig. S24). Trimethylamine (TMA), a toxic metabolite produced by bacterial fermentation of dietary fat-derived choline, was decreased in fecal water after the intervention (Figs. S21 and S22). The cytotoxicity of fecal water samples to cultured human Caco-2 cells was significantly reduced in SO and slightly in PWS cohorts after the intervention, indicating that the post-intervention microbiota may have produced less toxic metabolites in the gut (Fig. S25).
To more closely examine how the dietary intervention changed the carbohydrate metabolism of the gut microbiota, we searched all the 2,077,766 non-redundant gut genes from our study against the dbCAN database to identify genes coding for carbohydrate-active enzymes (CAZy) (Cantarel et al., 2012, Scott et al., 2013). 84,549 genes were assigned to 299 CAZy families, which significantly differ in the pre- and post-intervention samples, indicating a shift in the genes for carbohydrate metabolism in the gut microbiome (Fig. S26). Genes for degradation of starch, inulin and cellulose were significantly enriched, while genes for degradation of glycosylated compounds of animal origin such as mucin were significantly depleted in the microbiome after the intervention (Fig. S27) (Cantarel et al., 2012). Genes for formate-tetrahydrofolate ligase participating in acetate production (Turnbaugh et al., 2006, Claesson et al., 2012) were significantly increased after the intervention, consistent with the increased relative concentration of acetate among the fecal SCFAs (Fig. S28). These shifts reflect the increased availability of plant carbohydrates in the colon, favoring proliferation of bacteria such as bifidobacteria that contain carbohydrate-fermenting genes and produce beneficial metabolites such as acetate (Flint et al., 2012).
9.6. Metabolic Interactions Between the Gut Microbiota and Human Host
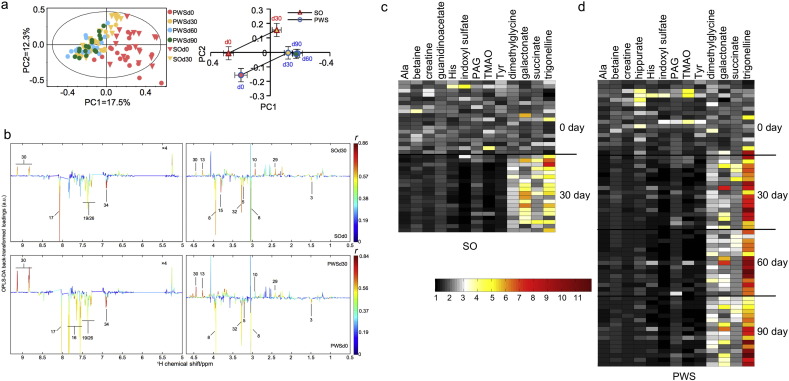
We used an NMR-based metabonomics approach in the current study to obtain unbiased profiles of urine metabolites before and after the intervention to identify those that were significantly changed by the intervention. Score plots of PCA and OPLS-DA of NMR-based metabolite profiling data of urine samples collected from the SO (Days 0 and 30) and the PWS cohorts (Day 0, 30, 60 and 90) showed significant metabolic shifts after the intervention in both cohorts (Fig. 5a). Among all the metabolites detected by NMR, only thirteen varied significantly between pre- and post-intervention samples in SO and in PWS cohorts, using OPLS-DA (Figs. 5b–d, S29–S31 and Table S14). Interestingly, 4 of the 13 significantly changed metabolites are co-metabolites between gut bacteria and host, which were all decreased after the intervention. They are trimethylamine N-oxide (TMAO), indoxyl sulfate, phenylacetylglutamine (PAG) and hippurate. TMAO is generated when TMA enters the bloodstream and is metabolized by the human liver (Wang et al., 2011). The precursors of the other three metabolites are produced by bacterial fermentation of aromatic amino acids in the gut, e.g. indole is produced by bacterial fermentation of tryptophan (Russell et al., 2011). TMAO, an independent marker for predicting clinical vascular events, has been mechanistically linked with the development of atherosclerosis in humans and mice (Wang et al., 2011, Wang et al., 2014). Likewise, indoxyl sulfate has been linked with hypertension and cardio-vascular disease in chronic kidney disease patients (Barreto et al., 2009). Importantly, the decrease in urine TMAO after the intervention was accompanied by a corresponding increase in urinary dimethylglycine (DMG) (Fig. 5c and d), an intermediate in human metabolism of dietary choline. This suggests that proportionally more of the fat-derived choline in the interventional diet was absorbed and metabolized by the human host instead of undergoing fermentation to TMA by bacteria in the gut (Dumas et al., 2006).
Fig. 5.
Altered profiles of urinary metabolites during the dietary intervention. (a) PCA score plot of urinary metabolite profiles obtained from SO and PWS groups during the dietary intervention (left) and the metabolic trajectories generated from the PCA score plot. (b) Validated OPLS-DA coefficient plots showing the alterations of metabolic profiles in the urine caused by the 30-day intervention. The plot related to the discrimination between 1H NMR spectra of urine from Day 0 and Day 30 of SO (top) groups (n = 17, r > 0.468, P < 0.05). Plot related to the discrimination between 1H NMR spectra of urine from Day 0 and Day 30 of the PWS groups (n = 17, r > 0.468, P < 0.05). See Table S14 for the metabolite identification key. (c) Heat map showing significantly changed metabolites after the 30-day intervention in the SO cohort. (d) Heat map showing the significantly changed metabolites after 30-, 60- and 90-day interventions in the PWS cohort. The significance of each statistical comparison is shown in Fig. S31.
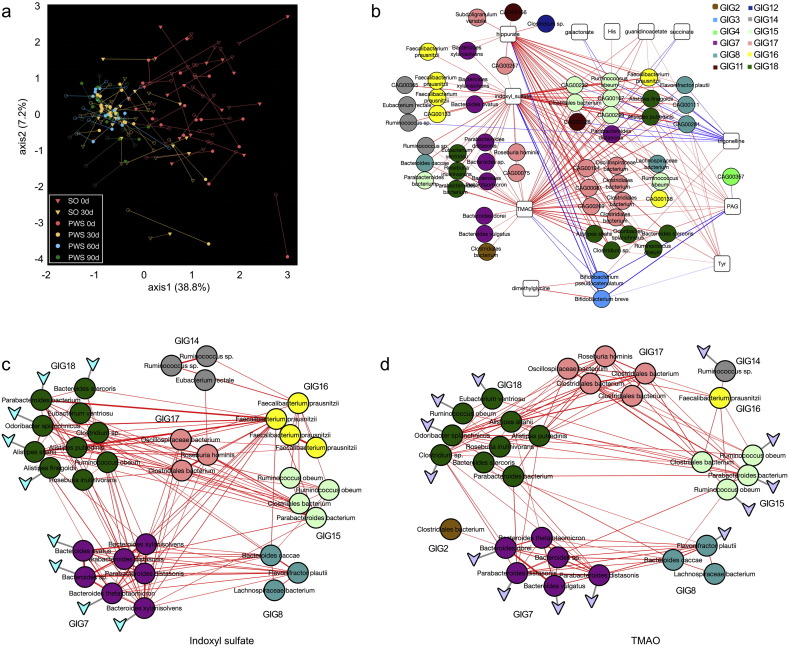
Co-inertia analysis between the 376 bacterial CAGs and the urine metabolome revealed a significant co-variation between them (R = 0.52, P < 0.01, Figs. 6a and S32). Bootstrapped Spearman correlation analysis identified all bacterial CAGs (|r| > 0.4 and FDR < 0.01) that were either positively or negatively associated with the key metabolites significantly modulated by the dietary intervention (Fig. 6b). Among the bacterial CAGs positively correlated with the potentially toxic metabolite indoxyl sulfate, nine from GIG7 and 18 (most of them are Bacteroides spp. and Alistipes spp.) contained the gene for tryptophanase (Fig. 6c), which transforms tryptophan to indole (Deeley and Yanofsky, 1982). Among those positively correlated with metabolically toxic TMAO, 13 bacterial CAGs from GIG7, 8, 14, 15, 16, and 18 (mostly in Ruminococcus spp., Parabacteroides spp. and Bacteroides spp.) had the gene cluster encoding choline TMA-lyase and choline TMA-lyase-activating enzyme (Fig. 6d), which are key for anaerobic choline degradation (Craciun and Balskus, 2012). The respective strains are the likely candidates for producing indole and TMA in the gut. They all belong to GIGs, which were reduced after the intervention (Fig. 3e). Their reduction may have contributed to the alleviation of the metabolic deteriorations in both genetic and diet-induced obesity.
Fig. 6.
Changes of co-metabolism between host and gut microbiota. (a) Co-inertia analysis (CIA) of relationships between the metabolomics PCA (end of lines with empty symbol) and microbiota CAGs PCA (end of lines with solid symbol). (b) Bacterial CAGs significantly associated with the key metabolites modulated by the intervention. The bootstrapped Spearman correlation coefficient between the 161 prevalent bacterial CAGs and the key metabolites was more than 0.4 and FDR < 0.001. Lines between nodes represent correlations between the nodes they connect, with line width indicating the correlation magnitude, and red and blue colors indicating positive and negative correlations, respectively. (c) Species interaction network of the bacterial CAGs significantly associated with indoxyl sulfate. Only the bacterial CAGs with high quality draft genomes are shown here. Lines between nodes represent correlations between the nodes they connect, with line width indicating the correlation magnitude, and red colors indicating positive correlations. The triangle represents the presence of the tryptophanase gene. (d) Species interaction network of the bacterial CAGs significantly associated with TMAO. Only the bacterial CAGs with high quality draft genomes are shown here. Lines between nodes represent correlations between the nodes they connect, with line width indicating the correlation magnitude, and red colors indicating positive correlations. The triangle represents the presence of the gene cluster encoding choline TMA-lyase and choline TMA-lyase-activating enzyme.
9.7. Gut Microbiota Transplantation
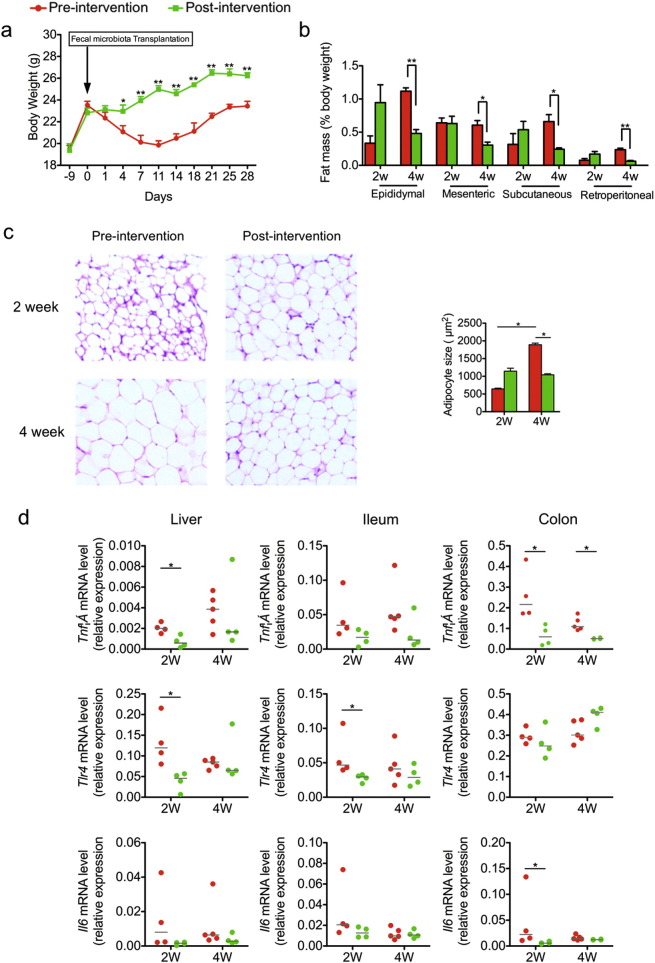
To compare the capacity of gut microbiota to induce metabolic deteriorations before and after the intervention, we transplanted the gut microbiota from the same PWS volunteer (GD58) before and after the intervention, into germ-free wild-type C57BL/6J mice. Mice that received the pre-intervention human fecal microbiota showed significantly decreased bodyweight during the first two weeks after transplantation, and then regained the lost weight in the following two weeks. Mice that received the post-intervention human fecal microbiota lost no bodyweight. Rather, they maintained weight for 4 days after transplantation and then returned to normal growth (Fig. 7a). By the end of the trial, pre-intervention microbiota recipients showed significantly greater fat mass as a percentage of body weight (Fig. 7b). Histological examination of epididymal fat pads revealed that the size of adipocytes in pre-intervention microbiota recipients was smaller than in post-intervention recipients at 2 weeks after the transplantation, consistent with toxicity of the microbiota, but then significantly bigger at 4 weeks. Adipocytes from the post-intervention microbiota recipients did not change over time (Fig. 7c). The initial weight loss was associated with appreciably higher inflammatory responses in pre-intervention transplant recipients, as measured by RT-qPCR of TNFα, IL6 and TLR4 gene expression in liver, ileum and colon at 2 weeks after transplantation (Fig. 7d), suggesting that when transplanted into germfree mice, the pre-intervention gut microbiota induced higher inflammation and larger adipocytes compared with the post-intervention gut microbiota from the same volunteer.
Fig. 7.
Impaired metabolism of gnotobiotic mice transplanted with pre-intervention gut microbiota from a PWS patient. (a) Body weight curves of gnotobiotic mice receiving the fecal microbiota from GD58 before (Day 0, red) and after (Day 90, green) the intervention. For mice receiving pre-intervention microbiota, 1 to 14 days after transplantation, n = 10, and 15 to 28 days, n = 5; For mice receiving post-intervention microbiota, 1 to 14 days after transplantation, n = 9, and 15 to 28 days, n = 4. (b) Adiposity index (%Fat mass/body weight) of gnotobiotic mice at 2 and 4 weeks after fecal transplantation. (c) Hematoxylin- and eosin-stained sections of epididymal fat pads (100 × magnification). Cell area of adipocyte in epididymal fat pad is shown as mean ± s.e.m. (d) RT-qPCR analysis of expression of Tnfα, Tlr4 and Il6 in the liver, ileum and colon. All mRNA quantification data were normalized to the housekeeping gene Glyceraldehyde-3-phosphate dehydrogenase (Gapdh). Gene expression levels are normalized to that of mice 2 weeks after inoculation with pre-intervention microbiota. The median of the data in each group is shown. Student t-test (two-tailed, in (a), (b) and (c)) or Mann–Whitney U test (two-tailed, in (d),) was used to analyze variation between gnotobiotic mice receiving pre- and post-intervention microbiota. *P < 0.05, **P < 0.01. In (b) to (d), for mice receiving pre-intervention microbiota, n = 5; for mice receiving post-intervention microbiota, n = 4.
10. Discussion
The obesity associated with PWS seems to be genetically determined, yet the primary drivers are still food craving and low calorie expenditure, similar to simple obesity. Energy restricted diets have been recommended for weight control in PWS children (Bonfig et al., 2009). Most of these diets do not improve satiety of PWS children, making it difficult for long-term adherence. Energy-restricted diet with low carbohydrates can lead to production of toxic metabolites by the gut bacteria, thus may be detrimental to health for long term use (Russell et al., 2011). In PWS children, a reduced energy diet with well balanced macronutrients and increased fiber intake yielded a better weight control than diets with just reduced energy intake (Miller et al., 2013). However, the impact of this type of diet on gut microbiota as a potential mechanism for its contribution to obesity improvement remains elusive in PWS children.
Our study shows that the gut microbiota in both genetic and simple obesity share similar structural and functional features of dysbiosis, such as 1) higher production of toxins with known potential to induce metabolic deteriorations such as TMAO and indoxyl sulfate; 2) higher abundance of genomes that encode genes for producing these toxic co-metabolites; 3) higher abundance of pathways for biosynthesis of bacterial antigens such as endotoxin. Our previous study in mice showed that diet is the major force shaping the gut microbiota. High fat diet can override the impact of host genetic mutation to gut microbiota (Zhang et al., 2010). The genetic mutation of PWS patients may have changed their dietary pattern via the hyperphagia. This over-eating behavior similar to SO volunteers may be the reason why these two cohorts share similar gut microbiota.
The WTP diet also has balanced macronutrients but much higher amount of complex carbohydrates as shown by NMR analysis of the fecal water samples. This excessive amount of carbohydrates until the end of the gut ensures that the gut bacteria have enough carbohydrates to ferment for energy extraction so that there is no need to ferment proteins or fats. Thus, the dietary intervention in this study shifted the metabolism in the gut microbiota from fermenting proteins and fats to carbohydrates, corrected the dysbiosis of the gut microbiota, which in return may have contributed to alleviation of metabolic deteriorations in genetic as well as simple obesity.
As a “proof of principal study”, gut microbiota transplantation in this study showed that the pre- and post-intervention gut microbiota from the same volunteer induced different responses in the hosts, indicating that the pre-intervention gut microbiota of a PWS volunteer indeed had higher capacity to induce gut inflammation and fat accumulation than his post-intervention gut microbiota. More trials are needed to confirm this phenomenon with samples from more donors. It would be also interesting to see if we can use the PWS microbiota-associated mice as a model to understand interactions between host and microbiota during progression of obesity.
Several structural patterns of the gut microbial community have been suggested to be associated with obesity, such as a high Firmicutes/Bacteroidetes ratio and low gene richness, but the specific relevant members of the gut microbiota and their functional interactions that contribute to obesity development and associated metabolic deteriorations remain elusive (Ley et al., 2005, Le Chatelier et al., 2013). Two or three enterotypes can be delineated from our 16S rRNA gene sequencing or metagenomic data. Short-term 10 days intervention did not change host enterotypes (Wu et al., 2011). Our dietary intervention changed the enterotypes of some volunteers but the enterotype delineation and shifts showed no correlation with any of the host phenotype changes. Our volunteers were not clustered into low gene count and high gene count groups. Different from previous report (Cotillard et al., 2013), the diversity and gene richness of their gut microbiota were significantly reduced after the intervention. The pre-intervention gut microbiota had higher diversity of toxin-producing and potentially pathogenic bacteria. The post-intervention gut microbiota were dominated by Bifidobacterium spp. These changes may have contributed to the overall reduction of diversity and gene richness after the intervention. It is thus important to move from general diversity/gene richness to functionally relevant genomes/genes for understanding the contribution of gut microbiota to host health phenotypes.
Accumulating evidence shows that important functions of the gut microbiota may be species or even strain-specific, yet many studies in metagenomics are conducted at genus or higher taxonomic levels due to the methodological limitation of assembling individual bacterial genomes directly from metagenomic data (Zhao, 2013). The recently developed “canopy-based” algorithm segregates individual genes into co-abundance gene groups based on the fact that the abundances of two genes contained on the same genomic DNA molecule will highly correlate with each other across complex metagenomic samples (Nielsen et al., 2014). With sufficient sequencing depth, reads in a CAG can be assembled into a high quality draft genome, which allowed us to perform genome-specific analysis of microbiota changes induced by the dietary intervention.
Like species in macro-ecosystems such as the rain forest (Ellison et al., 2005), bacterial species in the human gut may also survive, adapt, and decline as interdependent functional groups (guilds) responding to environmental perturbations (Claesson et al., 2012, Ellison et al., 2005, Zhou et al., 2013). Co-abundance analysis can help identify such groups but so far most studies did this analysis at the genus level (Claesson et al., 2012). In our study, all the 161 prevalent bacterial genomes were clustered into 18 Gene Interaction Groups (GIGs). The Commensurate changes of the GIGs with the overall structural changes of the gut microbiota based on 16S rRNA or CAGs data suggest that these GIGs may work as functional groups within the ecosystem. Members of the same GIG can thrive or decline together as a guild but they can come from very different taxonomic background, suggesting that we should study functional interactions of gut bacteria at the individual strain/genome level. Studies on the mechanisms of how bacteria form a GIG would lead to new insights on ecological interactions among prevalent members of the gut microbiota. More importantly, group-level abundance of some GIGs showed positive or negative correlations with host disease/health phenotypes. For example, the Bifidobacterium-dominating GIG3 was negatively associated with 12 disease phenotypes. Their interactions with the host for phenotype development in health and disease warrant further exploration.
In ecosystems such as rainforest, all species are not created equal. The tall trees are the “foundation species” as they cover the forest to create a unique environment for all other species to thrive (Prevey et al., 2010). It is an interesting question whether a microbial ecosystem in a healthy human gut would also have its “foundation species”. In this study, we found not only overall structural changes of the gut microbiota but also significant changes of specific predominant species such as the enrichment of the genome of a carbohydrate-fermenting species B. pseudocatenulatum in response to the intervention. It is a major member of GIG3, which significantly increased after intervention. This bifidobacteria-dominated GIG showed negative correlation with several other GIGs containing potentially detrimental species. The “bifid shunt” pathway gives bifidobacteria growth advantages over other carbohydrates fermentation bacteria by producing higher amount of acetate and producing more ATP from the same amount of available sugars (Pokusaeva et al., 2011). Species in GIG3 may work as “foundation species” to define much of the structure of a healthy gut ecosystem by rendering the gut environment unfavorable to pathogenic and detrimental bacteria, possibly via increased production of acetate and lactate (Ellison et al., 2005, Gibson and Wang, 1994). As such foundation species may become a powerful probiotic candidate for recovering and maintaining a healthy gut microbiota, this question warrants further exploration.
The gut microbiota may impact host health by producing toxic bacterial metabolites such as TMA and indole and delivering them into the bloodstream. However, the specific members of the gut microbiota that produce these detrimental compounds remain largely unknown, particularly in humans. In a previous “proof of principle” study of a healthy human cohort, we showed that changes in the population levels of individual bacterial species in the gut can be correlated with changes of specific metabolites in the urine to reveal “who does what in the microbiome” (Li et al., 2008). In this study, we correlated changes in the population levels of individual bacterial genomes in the gut with changes of specific metabolites in the urine to reveal “which genome does what function in the microbiome” (Li et al., 2008). Several bacterial genomes that correlated with TMAO and indoxyl sulfate levels in the urine were found to encode genes for production of their precursors by fermentation of choline or tryptophan in the gut. This opens the possibility of getting pure isolates of these key bacterial genomes for further mechanistic studies.
11. Conclusion
In summary, we have demonstrated that morbidly obese children with either genetically predisposed or simple obesity share similar dysbiosis of the gut microbiota. A dietary intervention rich in non-digestible carbohydrates shifted the dysbiotic gut microbiota to a healthier structure with relatively lower level of bacteria that can produce potentially toxic metabolites from the fermentation of dietary fats and proteins, and higher level of bacteria that can produce potentially beneficial products from the fermentation of carbohydrates. Our study shows that dietary modulation of the gut microbiota contributes to the alleviation of metabolic deteriorations in both PWS and SO cohorts, suggesting a common etiological role of dysbiotic microbiota for disease progression, regardless whether the obesity is genetically predisposed or simply diet-induced. Thus, dietary modulation of gut microbiota may become a promising strategy for being integrated into the management of metabolic diseases.
The following are the supplementary data related to this article.
Supplementary material.
De novo assembly information of 161 bacterial CAGs.
Comparison of assemblies and reference genomes.
Author Contributions
L. Z., X. Z. Z. and H. T. designed and managed the study; A. Y. and L. W. managed the clinical research; H. O., F. Y., R. W. J. S. and X. L. conducted the clinical research. Y. H., Y. Z., J. W., F. Z., R. W., Y. N. Z. and C. Z. conducted clinical and genetic tests and data analysis; H. F., F. Y. and H. O. gave dietary counseling to the patients and carried out analysis of dietary data; C. Z. and R. W. prepared the DNA for sequencing; G. W., C. Z., M. Z. H. X. and X. J. Z. established the sequence analysis pipeline; G. W. and C. Z. carried out microbial data analysis; H. L., Y. W. and H. T. conducted metabonomic analysis of urine and fecal water samples; Y. F. Z., F. Z. and X. P. conducted fecal water toxicity and SCFAs analysis; R. W., C. Z., J. S., B. Z., W. L. and H. W. conducted animal trial; L. Z., C. Z., B. H., H. T., G. W., P. B., N. Z., X. Z. Z., L. B., D. W., F. C., A. Y., J. N., J. D., K. C., G. Z., S. Y. and E. H. wrote the manuscript.
Acknowledgment
This work was supported by the grants from the National Natural Science Foundation of China (31330005, 81100632, 31121064, 20825520, 81401141 and 21221064); the Ministry of Science and Technology of China (2010CB912501); the Science and Technology Commission of Shanghai Municipality (14YF1402200) and the National Science and Technology Major Project of China (2012ZX10005001-009). A computing facility award on the PI cluster at Shanghai Jiao Tong University is acknowledged. We thank Prof. Zhu Chen from Shanghai Center for Systems Biomedicine for his insightful discussion and guidance in designing the experiment.
Footnotes
The authors have declared that no conflict of interest exists.
Contributor Information
Huiru Tang, Email: Huiru.tang@wipm.ac.cn.
Xiaozhuang Zhang, Email: zhangxiaozhuang55@126.com.
Liping Zhao, Email: lpzhao@sjtu.edu.cn.
References
- Abubucker S., Segata N., Goll J. Metabolic reconstruction for metagenomic data and its application to the human microbiome. PLoS Comput. Biol. 2012;8(6):e1002358. doi: 10.1371/journal.pcbi.1002358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Askree S.H., Hjelm L.N., Pervaiz M.A. Allelic dropout can cause false-positive results for Prader–Willi and Angelman syndrome testing. J. Mol. Diagn. 2011;13(1):108–112. doi: 10.1016/j.jmoldx.2010.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Backhed F., Ding H., Wang T. The gut microbiota as an environmental factor that regulates fat storage. Proc. Natl. Acad. Sci. U. S. A. 2004;101(44):15718–15723. doi: 10.1073/pnas.0407076101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Backhed F., Manchester J.K., Semenkovich C.F., Gordon J.I. Mechanisms underlying the resistance to diet-induced obesity in germ-free mice. Proc. Natl. Acad. Sci. U. S. A. 2007;104(3):979–984. doi: 10.1073/pnas.0605374104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barreto F.C., Barreto D.V., Liabeuf S. Serum indoxyl sulfate is associated with vascular disease and mortality in chronic kidney disease patients. Clin. J. Am. Soc. Nephrol. 2009;4(10):1551–1558. doi: 10.2215/CJN.03980609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonfig W., Dokoupil K., Schmidt H. A special, strict, fat-reduced, and carbohydrate-modified diet leads to marked weight reduction even in overweight adolescents with Prader–Willi syndrome (PWS) TheScientificWorldJOURNAL. 2009;9:934–939. doi: 10.1100/tsw.2009.105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butler M.G. Prader–Willi syndrome: obesity due to genomic imprinting. Curr. Genomics. 2011;12(3):204–215. doi: 10.2174/138920211795677877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cani P.D., Bibiloni R., Knauf C. Changes in gut microbiota control metabolic endotoxemia-induced inflammation in high-fat diet-induced obesity and diabetes in mice. Diabetes. 2008;57(6):1470–1481. doi: 10.2337/db07-1403. [DOI] [PubMed] [Google Scholar]
- Cantarel B.L., Lombard V., Henrissat B. Complex carbohydrate utilization by the healthy human microbiome. PLoS One. 2012;7(6):e28742. doi: 10.1371/journal.pone.0028742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Claesson M.J., Jeffery I.B., Conde S. Gut microbiota composition correlates with diet and health in the elderly. Nature. 2012;488(7410):178–184. doi: 10.1038/nature11319. [DOI] [PubMed] [Google Scholar]
- Cloarec O., Dumas M.E., Craig A. Statistical total correlation spectroscopy: an exploratory approach for latent biomarker identification from metabolic 1H NMR data sets. Anal. Chem. 2005;77(5):1282–1289. doi: 10.1021/ac048630x. [DOI] [PubMed] [Google Scholar]
- Cotillard A., Kennedy S.P., Kong L.C. Dietary intervention impact on gut microbial gene richness. Nature. 2013;500(7464):585–588. doi: 10.1038/nature12480. [DOI] [PubMed] [Google Scholar]
- Craciun S., Balskus E.P. Microbial conversion of choline to trimethylamine requires a glycyl radical enzyme. Proc. Natl. Acad. Sci. U. S. A. 2012;109(52):21307–21312. doi: 10.1073/pnas.1215689109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- David L.A., Maurice C.F., Carmody R.N. Diet rapidly and reproducibly alters the human gut microbiome. Nature. 2014;505(7484):559–563. doi: 10.1038/nature12820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deeley M.C., Yanofsky C. Transcription initiation at the tryptophanase promoter of Escherichia coli K-12. J. Bacteriol. 1982;151(2):942–951. doi: 10.1128/jb.151.2.942-951.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dodt M., Roehr J.T., Ahmed R., Dieterich C. FLEXBAR-flexible barcode and adapter processing for next-generation sequencing platforms. Biology. 2012;1(3):895–905. doi: 10.3390/biology1030895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dumas M.E., Barton R.H., Toye A. Metabolic profiling reveals a contribution of gut microbiota to fatty liver phenotype in insulin-resistant mice. Proc. Natl. Acad. Sci. U. S. A. 2006;103(33):12511–12516. doi: 10.1073/pnas.0601056103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dykens E.M., Maxwell M.A., Pantino E., Kossler R., Roof E. Assessment of hyperphagia in Prader–Willi syndrome. Obesity (Silver Spring) 2007;15(7):1816–1826. doi: 10.1038/oby.2007.216. [DOI] [PubMed] [Google Scholar]
- Ellison A.M., Bank M.S., Clinton B.D. Loss of foundation species: consequences for the structure and dynamics of forested ecosystems. Front. Ecol. Environ. 2005;3(9):479–486. [Google Scholar]
- Eriksson L., Trygg J., Wold S. CV-ANOVA for significance testing of PLS and OPLS (R) models. J. Chemom. 2008;22(11–12):594–600. [Google Scholar]
- Fagan A., Culhane A.C., Higgins D.G. A multivariate analysis approach to the integration of proteomic and gene expression data. Proteomics. 2007;7(13):2162–2171. doi: 10.1002/pmic.200600898. [DOI] [PubMed] [Google Scholar]
- Fei N., Zhao L. An opportunistic pathogen isolated from the gut of an obese human causes obesity in germfree mice. ISME J. 2013;7(4):880–884. doi: 10.1038/ismej.2012.153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finn R.D., Clements J., Eddy S.R. HMMER web server: interactive sequence similarity searching. Nucleic Acids Res. 2011;39(Web Server issue):W29–W37. doi: 10.1093/nar/gkr367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flint H.J., Scott K.P., Duncan S.H., Louis P., Forano E. Microbial degradation of complex carbohydrates in the gut. Gut Microbes. 2012;3(4):289–306. doi: 10.4161/gmic.19897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibson G.R., Wang X. Regulatory effects of bifidobacteria on the growth of other colonic bacteria. J. Appl. Bacteriol. 1994;77(4):412–420. doi: 10.1111/j.1365-2672.1994.tb03443.x. [DOI] [PubMed] [Google Scholar]
- Godon J.J., Zumstein E., Dabert P., Habouzit F., Moletta R. Molecular microbial diversity of an anaerobic digestor as determined by small-subunit rDNA sequence analysis. Appl. Environ. Microbiol. 1997;63(7):2802–2813. doi: 10.1128/aem.63.7.2802-2813.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang L., Huang J., Wang Y., Tang H. Eliminating the dication-induced intersample chemical-shift variations for NMR-based biofluid metabonomic analysis. Analyst. 2012;137(18):4209–4219. doi: 10.1039/c2an35392j. [DOI] [PubMed] [Google Scholar]
- Kanehisa M., Goto S., Sato Y., Furumichi M., Tanabe M. KEGG for integration and interpretation of large-scale molecular data sets. Nucleic Acids Res. 2012;40(Database issue):D109–D114. doi: 10.1093/nar/gkr988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kubota T., Das S., Christian S.L., Baylin S.B., Herman J.G., Ledbetter D.H. Methylation-specific PCR simplifies imprinting analysis. Nat. Genet. 1997;16(1):16–17. doi: 10.1038/ng0597-15. [DOI] [PubMed] [Google Scholar]
- Kurtz S., Phillippy A., Delcher A.L. Versatile and open software for comparing large genomes. Genome Biol. 2004;5(2):R12. doi: 10.1186/gb-2004-5-2-r12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Labruna G., Pasanisi F., Nardelli C. High leptin/adiponectin ratio and serum triglycerides are associated with an “at-risk” phenotype in young severely obese patients. Obesity. 2011;19(7):1492–1496. doi: 10.1038/oby.2010.309. [DOI] [PubMed] [Google Scholar]
- Lacroix D., Moutel S., Coupaye M. Metabolic and adipose-tissue signatures in adults with Prader–Willi syndrome, a model of extreme adiposity. J. Clin. Endocrinol. Metab. 2014:jc20143127. doi: 10.1210/jc.2014-3127. [DOI] [PubMed] [Google Scholar]
- Langmead B., Salzberg S.L. Fast gapped-read alignment with Bowtie 2. Nat. Methods. 2012;9(4):357–359. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Chatelier E., Nielsen T., Qin J. Richness of human gut microbiome correlates with metabolic markers. Nature. 2013;500(7464):541–546. doi: 10.1038/nature12506. [DOI] [PubMed] [Google Scholar]
- Ley R.E., Backhed F., Turnbaugh P., Lozupone C.A., Knight R.D., Gordon J.I. Obesity alters gut microbial ecology. Proc. Natl. Acad. Sci. U. S. A. 2005;102(31):11070–11075. doi: 10.1073/pnas.0504978102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W., Godzik A. Cd-hit: a fast program for clustering and comparing large sets of protein or nucleotide sequences. Bioinformatics. 2006;22(13):1658–1659. doi: 10.1093/bioinformatics/btl158. [DOI] [PubMed] [Google Scholar]
- Li M., Wang B., Zhang M. Symbiotic gut microbes modulate human metabolic phenotypes. Proc. Natl. Acad. Sci. U. S. A. 2008;105(6):2117–2122. doi: 10.1073/pnas.0712038105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li R., Yu C., Li Y. SOAP2: an improved ultrafast tool for short read alignment. Bioinformatics. 2009;25(15):1966–1967. doi: 10.1093/bioinformatics/btp336. [DOI] [PubMed] [Google Scholar]
- Li H., Handsaker B., Wysoker A. The sequence alignment/map format and SAMtools. Bioinformatics. 2009;25(16):2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mende D.R., Sunagawa S., Zeller G., Bork P. Accurate and universal delineation of prokaryotic species. Nat. Methods. 2013;10(9):881–884. doi: 10.1038/nmeth.2575. [DOI] [PubMed] [Google Scholar]
- Miller J.L., Lynn C.H., Shuster J., Driscoll D.J. A reduced-energy intake, well-balanced diet improves weight control in children with Prader–Willi syndrome. J. Hum. Nutr. Diet. 2013;26(1):2–9. doi: 10.1111/j.1365-277X.2012.01275.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nielsen H.B., Almeida M., Juncker A.S. Identification and assembly of genomes and genetic elements in complex metagenomic samples without using reference genomes. Nat. Biotechnol. 2014;32(8):822–828. doi: 10.1038/nbt.2939. [DOI] [PubMed] [Google Scholar]
- Overbeek R., Olson R., Pusch G.D. The SEED and the rapid annotation of microbial genomes using subsystems technology (RAST) Nucleic Acids Res. 2014;42(D1):D206–D214. doi: 10.1093/nar/gkt1226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papavramidis S.T., Kotidis E.V., Gamvros O. Prader–Willi syndrome-associated obesity treated by biliopancreatic diversion with duodenal switch. Case report and literature review. J. Pediatr. Surg. 2006;41(6):1153–1158. doi: 10.1016/j.jpedsurg.2006.01.065. [DOI] [PubMed] [Google Scholar]
- Peng Y., Leung H.C., Yiu S.M., Chin F.Y. IDBA-UD: a de novo assembler for single-cell and metagenomic sequencing data with highly uneven depth. Bioinformatics. 2012;28(11):1420–1428. doi: 10.1093/bioinformatics/bts174. [DOI] [PubMed] [Google Scholar]
- Pokusaeva K., Fitzgerald G.F., van Sinderen D. Carbohydrate metabolism in Bifidobacteria. Genes Nutr. 2011;6(3):285–306. doi: 10.1007/s12263-010-0206-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prevey J.S., Germino M.J., Huntly N.J. Loss of foundation species increases population growth of exotic forbs in sagebrush steppe. Ecol. Appl. 2010;20(7):1890–1902. doi: 10.1890/09-0750.1. [DOI] [PubMed] [Google Scholar]
- Quinlan A.R., Hall I.M. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics. 2010;26(6):841–842. doi: 10.1093/bioinformatics/btq033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ridaura V.K., Faith J.J., Rey F.E. Gut microbiota from twins discordant for obesity modulate metabolism in mice. Science. 2013;341(6150):1241214. doi: 10.1126/science.1241214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russell W.R., Gratz S.W., Duncan S.H. High-protein, reduced-carbohydrate weight-loss diets promote metabolite profiles likely to be detrimental to colonic health. Am. J. Clin. Nutr. 2011;93(5):1062–1072. doi: 10.3945/ajcn.110.002188. [DOI] [PubMed] [Google Scholar]
- Scott K.P., Gratz S.W., Sheridan P.O., Flint H.J., Duncan S.H. The influence of diet on the gut microbiota. Pharmacol. Res. 2013;69(1):52–60. doi: 10.1016/j.phrs.2012.10.020. [DOI] [PubMed] [Google Scholar]
- Segata N., Izard J., Waldron L. Metagenomic biomarker discovery and explanation. Genome Biol. 2011;12(6):R60. doi: 10.1186/gb-2011-12-6-r60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turnbaugh P.J., Ley R.E., Mahowald M.A., Magrini V., Mardis E.R., Gordon J.I. An obesity-associated gut microbiome with increased capacity for energy harvest. Nature. 2006;444(7122):1027–1031. doi: 10.1038/nature05414. [DOI] [PubMed] [Google Scholar]
- Turnbaugh P.J., Backhed F., Fulton L., Gordon J.I. Diet-induced obesity is linked to marked but reversible alterations in the mouse distal gut microbiome. Cell Host Microbe. 2008;3(4):213–223. doi: 10.1016/j.chom.2008.02.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vijay-Kumar M., Aitken J.D., Carvalho F.A. Metabolic syndrome and altered gut microbiota in mice lacking Toll-like receptor 5. Science. 2010;328(5975):228–231. doi: 10.1126/science.1179721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vrieze A., Van Nood E., Holleman F. Transfer of intestinal microbiota from lean donors increases insulin sensitivity in individuals with metabolic syndrome. Gastroenterology. 2012;143(4):913–916. doi: 10.1053/j.gastro.2012.06.031. e7. [DOI] [PubMed] [Google Scholar]
- Wang Z., Klipfell E., Bennett B.J. Gut flora metabolism of phosphatidylcholine promotes cardiovascular disease. Nature. 2011;472(7341):57–63. doi: 10.1038/nature09922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z., Tang W.H., Buffa J.A. Prognostic value of choline and betaine depends on intestinal microbiota-generated metabolite trimethylamine-N-oxide. Eur. Heart J. 2014;35(14):904–910. doi: 10.1093/eurheartj/ehu002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu J., An Y., Yao J., Wang Y., Tang H. An optimised sample preparation method for NMR-based faecal metabonomic analysis. Analyst. 2010;135(5):1023–1030. doi: 10.1039/b927543f. [DOI] [PubMed] [Google Scholar]
- Wu G.D., Chen J., Hoffmann C. Linking long-term dietary patterns with gut microbial enterotypes. Science. 2011;334(6052):105–108. doi: 10.1126/science.1208344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao C., Hao F., Qin X., Wang Y., Tang H. An optimized buffer system for NMR-based urinary metabonomics with effective pH control, chemical shift consistency and dilution minimization. Analyst. 2009;134(5):916–925. doi: 10.1039/b818802e. [DOI] [PubMed] [Google Scholar]
- Xiao S., Fei N., Pang X. A gut microbiota-targeted dietary intervention for amelioration of chronic inflammation underlying metabolic syndrome. FEMS Microbiol. Ecol. 2014;87(2):357–367. doi: 10.1111/1574-6941.12228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Z., Hao B.L. CVTree update: a newly designed phylogenetic study platform using composition vectors and whole genomes. Nucleic Acids Res. 2009;37:W174–W178. doi: 10.1093/nar/gkp278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y.X., Wang G.Y., C PX. Peking University Medical Press; Beijing: 2002. China Food Composition 2002. [Google Scholar]
- Zerbino D.R., Birney E. Velvet: algorithms for de novo short read assembly using de Bruijn graphs. Genome Res. 2008;18(5):821–829. doi: 10.1101/gr.074492.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang C., Zhang M., Wang S. Interactions between gut microbiota, host genetics and diet relevant to development of metabolic syndromes in mice. ISME J. 2010;4(2):232–241. doi: 10.1038/ismej.2009.112. [DOI] [PubMed] [Google Scholar]
- Zhang C., Zhang M., Pang X., Zhao Y., Wang L., Zhao L. Structural resilience of the gut microbiota in adult mice under high-fat dietary perturbations. ISME J. 2012;6(10):1848–1857. doi: 10.1038/ismej.2012.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao L. The gut microbiota and obesity: from correlation to causality. Nat. Rev. Microbiol. 2013;11(9):639–647. doi: 10.1038/nrmicro3089. [DOI] [PubMed] [Google Scholar]
- Zhou Y., Gao H., Mihindukulasuriya K.A. Biogeography of the ecosystems of the healthy human body. Genome Biol. 2013;14(1):R1. doi: 10.1186/gb-2013-14-1-r1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu W., Lomsadze A., Borodovsky M. Ab initio gene identification in metagenomic sequences. Nucleic Acids Res. 2010;38(12):e132. doi: 10.1093/nar/gkq275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zweigner J., Schumann R.R., Weber J.R. The role of lipopolysaccharide-binding protein in modulating the innate immune response. Microb. Inf./Instit. Pasteur. 2006;8(3):946–952. doi: 10.1016/j.micinf.2005.10.006. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material.
De novo assembly information of 161 bacterial CAGs.
Comparison of assemblies and reference genomes.