Abstract
Inhibition of de novo palmitate synthesis via fatty acid synthase (FASN) inhibition provides an unproven approach to cancer therapy with a strong biological rationale. FASN expression increases with tumor progression and associates with chemoresistance, tumor metastasis, and diminished patient survival in numerous tumor types. TVB-3166, an orally-available, reversible, potent, and selective FASN inhibitor induces apoptosis, inhibits anchorage-independent cell growth under lipid-rich conditions, and inhibits in-vivo xenograft tumor growth. Dose-dependent effects are observed between 20–200 nM TVB-3166, which agrees with the IC50 in biochemical FASN and cellular palmitate synthesis assays. Mechanistic studies show that FASN inhibition disrupts lipid raft architecture, inhibits biological pathways such as lipid biosynthesis, PI3K–AKT–mTOR and β-catenin signal transduction, and inhibits expression of oncogenic effectors such as c-Myc; effects that are tumor-cell specific. Our results demonstrate that FASN inhibition has anti-tumor activities in biologically diverse preclinical tumor models and provide mechanistic and pharmacologic evidence that FASN inhibition presents a promising therapeutic strategy for treating a variety of cancers, including those expressing mutant K-Ras, ErbB2, c-Met, and PTEN. The reported findings inform ongoing studies to link mechanisms of action with defined tumor types and advance the discovery of biomarkers supporting development of FASN inhibitors as cancer therapeutics.
Research in context
Fatty acid synthase (FASN) is a vital enzyme in tumor cell biology; the over-expression of FASN is associated with diminished patient prognosis and resistance to many cancer therapies. Our data demonstrate that selective and potent FASN inhibition with TVB-3166 leads to selective death of tumor cells, without significant effect on normal cells, and inhibits in vivo xenograft tumor growth at well-tolerated doses. Candidate biomarkers for selecting tumors highly sensitive to FASN inhibition are identified. These preclinical data provide mechanistic and pharmacologic evidence that FASN inhibition presents a promising therapeutic strategy for treating a variety of cancers.
Abbreviations1: NADPH, nicotinamide adenine dinucleotide phosphate; HUVEC, human umbilical vein endothelial cells; NSCLC, non-small-cell lung cancer; CRC, colorectal cancer; TGI, tumor growth inhibition; MEM, minimal essential media; DMEM, Dulbecco's Modified Eagle's Medium; FBS, fetal bovine serum; LC–MS, liquid chromatography–mass spectrometry; PBS, phosphate buffered saline; FITC, fluorescein isothiocyanate
Keywords: Fatty acid synthase, Inhibitor, Beta-catenin, MYC, KRAS, Lipid raft
Graphical abstract
Highlights
-
•
TVB-3166 is an orally available, reversible, potent, and selective FASN inhibitor.
-
•
TVB-3166 inhibition of FASN induces apoptosis in tumor cells but not normal cells.
-
•
TVB-3166 inhibits in vivo xenograft tumor growth.
-
•
Candidate biomarkers to select tumors sensitive to FASN inhibition are identified.
1. Introduction
Fatty acid synthase (FASN) is a homodimeric and multi-functional enzyme that catalyzes the biosynthesis of palmitate in a NADPH-dependent reaction (Maier et al., 2006). Normal cells in adult tissue ubiquitously express low to moderate levels of FASN; however, these cells, which primarily import lipids from the extracellular milieu, do not have a strict requirement for FASN activity. This is demonstrated in a variety of mouse models with tissue-specific knockout of FASN expression that are characterized by the absence of an effect under non-stress conditions (Chirala et al., 2003, Shearn et al., 2014). In contrast, tumor cells have an increased requirement for lipids in functions such as membrane biosynthesis, protein modification, and as signaling molecules. Consequently, tumor cells are more dependent on de novo palmitate synthesis catalyzed by FASN than normal cells (Menendez and Lupu, 2007, Flavin et al., 2010). Accordingly, FASN is overexpressed in many solid and hematopoietic tumors, including breast, ovarian, prostate, colon, lung, and pancreatic (Ueda et al., 2010, Shah et al., 2006, Zaytseva et al., 2012, Witkiewicz et al., 2008, Sebastiani et al., 2006). Moreover, FASN tumor expression is increased in a stage-dependent manner that is associated with diminished patient survival (Ueda et al., 2010, Tao et al., 2013, Nguyen et al., 2010, Notarnicola et al., 2012, Witkiewicz et al., 2008, Zaytseva et al., 2012). This expression–prognosis relationship suggests that FASN plays an important role in affecting tumor cell biology and therapeutic response across a wide range of cancer types.
Alteration of energy and macromolecular biosynthetic metabolism in tumor cells compared to non-tumor cells is well established and known as the Warburg effect, in recognition of Otto Warburg's hypothesis that extended from his observation that ascites tumor cells convert the majority of their glucose carbon to lactose in oxygen-rich environments (Ward and Thompson, 2012). Tumor cell survival, growth, and proliferation demand increased energy in the form of NADPH and increased macromolecular biosynthesis of DNA, RNA, protein, and lipids. Reprogramming of tumor cell mitochondrial metabolism to support these requirements occurs directly through growth factor signaling and the PI3K–AKT–mTOR pathway. AKT activation drives both glycolytic metabolism of glucose and mitochondrial metabolism that generates acetyl-CoA, the biosynthetic precursor of fatty acids, cholesterol, and isoprenoid synthesis. As a critical aspect of tumor cell metabolic reprogramming, mTORC1 complex activation occurs via AKT signal transduction. A central component of the mTORC1 cell growth program is stimulation of de novo lipogenesis via regulation of SREBP-mediated FASN expression (Shackelford and Shaw, 2009, Lupu and Menendez, 2006). In the synthesis of fatty acids, FASN consumes NADPH, acetyl-CoA, and malonyl-CoA. The consumption of these substrates as well as the production of fatty acids contributes to the sustained altered metabolic state that tumor cells require for growth and survival.
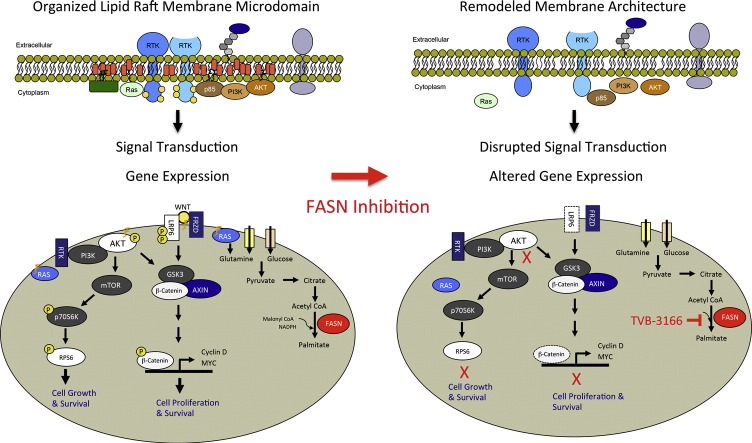
Palmitate and additional fatty acids derived from its function in diverse, vital biological processes. Fatty acids serve as precursors for synthesis of cellular lipids, as lipid bilayer constituents that affect membrane fluidity and architecture, and as substrates for post-translational protein modification that affect protein localization and activity. Palmitate affects membrane architecture at specialized plasma membrane microdomains known as lipid rafts. Lipid rafts are localized regions that contain high concentrations of lipids such as palmitate, cholesterol, and sphingosine, and also are rich in lipid-modified membrane-associated proteins that function in receiving, localizing, and transmitting cell growth signals (Simons and Sampaio, 2011, Staubach and Hanisch, 2011). Depletion of palmitate and other cellular lipids is expected to cause reorganization of membrane architecture and disruption of lipid raft domains. Growth factor and intracellular signal transduction require intricate membrane-associated protein–protein interactions that are dependent upon lipid raft architecture and protein lipidation. These lipid rafts facilitate the co-localization of proteins that must associate to form functional signaling complexes, and thereby regulate the efficiency of signal transduction as rafts increase and decrease in number and size. By disruption of membrane structure, FASN inhibition may disable signal transduction networks and biological processes required for cell growth, proliferation, and response to cellular stress. Activation of these pathways is a hallmark of cancer, and enables FASN inhibition to affect multiple points within a tumor cell that can produce anti-tumor activity.
FASN activity is intimately linked to receptor tyrosine kinase (RTK), PI3K–AKT–mTOR and MAPK signaling pathways, and activation of these pathways is a hallmark of aggressively growing tumor cells. Activation of the PI3K–AKT–mTOR pathway is among the most frequent aberrations in human cancers, and occurs through numerous different genetic lesions (Vivanco and Sawyers, 2002). The PI3K–AKT–mTOR pathway controls many biological processes that include glucose uptake and metabolism, protein synthesis, cell growth, and cell survival (Hollander et al., 2011). FASN gene expression is activated downstream of the PI3K–AKT–mTOR signal transduction pathway in response to cell metabolism and growth signals, and is driven by SREBP-1, ZBTB7A, and p53 family transcription factors (Van De Sande et al., 2005, Choi et al., 2008). Increased FASN activity promotes the tumorigenic capacity of cells via multiple mechanisms that include supporting enhanced macromolecular biosynthesis and glucose metabolism, cell growth and survival signal transduction, cellular stress response, and resistance to chemotherapeutic agents. In tumor cells, the connection between signal transduction pathways and FASN often becomes inextricably linked. Tumors with activated RTKs such as ERBB2 provide an example; whereby, the ERBB2–PI3K–AKT–FASN signaling axis results in continued stimulation of ErbB2 activity (Grunt et al., 2009). The inter-dependence enables tumor cells of this type to be killed with either ErbB2 or FASN inhibitors.
FASN inhibition using siRNAs and small molecules with varied biochemical mechanisms and selectivity profiles have been shown to inhibit Akt phosphorylation, induce tumor cell apoptosis, sensitize chemotherapy-resistant tumor cells to drug activity, and inhibit mouse xenograft tumor growth (Chuang et al., 2011, Kant et al., 2012, Kridel, 2004, Puig et al., 2008, Puig et al., 2009, Tomek et al., 2011). These activities of FASN inhibition have been reported in different tumor cell types that overexpress FASN, including, breast, ovary, prostate, and colorectal tumors. Despite the compelling support for FASN as an oncology therapeutic target, to date no compounds have progressed into clinical studies. Some compounds previously described in the literature suffered from significant pharmaceutical liabilities, including off-target activities such as stimulation of fatty acid oxidation that leads to significant and rapid weight loss in animal model studies and confounds interpretation of study results (Liu et al., 2010, Menendez and Lupu, 2007, Flavin et al., 2010). In vitro studies have shown that inhibition of Akt phosphorylation and induction of tumor cell apoptosis occur when FASN inhibition is uncoupled from CPT1 stimulation (Puig et al., 2008); thus suggesting that selective FASN inhibition can achieve the desired tumoricidal effects without inducing the rapid weight loss associated with activation of fatty acid oxidation. These and other observations have spurred the discovery and development of ‘next generation’ FASN inhibitors with optimized pharmacological properties and in vivo tolerability.
We report studies that characterize the anti-tumor activity of TVB-3166, a highly selective, potent, reversible, and oral FASN inhibitor discovered and developed by 3-V Biosciences. Using in vitro and in vivo models of human cancer we find that FASN inhibition has multiple mechanisms of action that can operate in specific types of tumors to cause tumor cell apoptosis. These mechanisms include inhibition of signal transduction through the PI3K–AKT–mTOR and β-catenin pathways that regulate tumor cell growth and survival. Our studies provide insights into how these pathways are affected by FASN inhibition and guide the discovery of biomarkers to select tumors with the greatest susceptibility to the tumoricidal effects of FASN inhibition. We also demonstrate that oral dosing of TVB-3166 modulates the target enzyme in vivo, is well tolerated, and inhibits mouse xenograft tumor growth in a dose-dependent manner.
2. Materials and Methods
2.1. Cell Lines and Antibodies
The cell lines used were obtained from ATCC and ECACC, except for OVCAR-8 cells (Biotox Sciences) The following antibodies were used: FASN (3180; Cell Signaling), pAKT-S473 (4060; Cell Signaling), pRPS6-S240 and 244 (2215; Cell Signaling), PARP-cleaved (9541, Cell Signaling), β-catenin (9582, Cell Signaling), pβ-catenin-S675 (4176, Cell Signaling), LRP6 (2560, Cell Signaling) pLRP6-S1490 (2568, Cell Signaling), c-Myc (5605, Cell Signaling), α-tubulin (2125; Cell Signaling), N-Ras (sc-519, Santa Cruz Biotechnology), and cholera-toxin subunit-B (227040; EMD Millipore).
2.2. Palmitate Synthesis Assays
Cells were seeded into 96-well culture plates at a density of 30,000 cells per well. After an overnight incubation, media were removed from all wells and replaced with media containing both 1 mM 13C-acetate and TVB-3166 across a range of concentrations in 0.5% DMSO. After 18 h incubation, the cells were washed and saponified with sodium hydroxide, neutralized, and prepared for LC–MS bioanalytical analysis using an API4000 mass spectrometer in SRM mode.
2.3. Viability Assays
Screening assays using 2 different drug concentrations of cell line panels were conducted at Biotox Sciences (San Diego, CA) and extended dose-response assays were conducted at 3-V Biosciences. For drug treatment, tumor cell lines were plated in 96-well plates in growth medium (DMEM containing 10% FBS and 1% l-glutamine). 24-h after cell plating, DMEM growth medium was removed and Advanced MEM containing 1% charcoal-stripped FBS, 1% l-glutamine, and the diluted compound was added to each well. The final drug concentrations in assay wells were 10,000, 3300, 1100, 370, 123, 41, 14, 5, and 0 nM. Cell viability was measured 7 days after compound addition using the Cell Titer-Glo assay (Promega). IC50 determination was performed using a non-linear regression curve-fitting algorithm: log (inhibitor) vs. response-variable slope (four parameter) within Graph Pad Prism 6.0a.
2.4. Lipidomics Analysis
Tumor cell lines were plated in growth media. 24-h after cell plating, the growth media were removed and a final concentration of either 0.01% DMSO or 200 nM TVB-3166 was added to each flask containing Advanced MEM supplemented with 1% Charcoal Stripped FBS and 1% l-glutamine. Cells were treated for 24-h, 48-h, or 72-h. Cells were collected and washed in DPBS. Quantitative analysis of lipids was performed at Metabolon (Sacramento, CA and Durham, NC) using LC–MS-based methods and proprietary software.
2.5. Flow Cytometry
Cells were seeded in growth media. One day after seeding, the media was changed to the Advanced formulation treatment media. Cells were treated with TVB-3166 for 24 to 96 h and harvested by Accutase (Gibco, cat.# A11105), washed with cold PBS, and stained for Annexin V staining (FITC Annexin V Apoptosis Detection Kit II; BD Pharmingen cat. # 556570). Flow cytometry analysis was performed at Stanford University's Shared FACS Facility using the LSRII.UV instrument that was obtained by Stanford University using NIH S10 Shared Instrument Grant (S10RR027431-01). Data were analyzed at 3-V Biosciences using FlowJo software (Ashland, OR).
2.6. Western Blot Analysis
For Western blot analysis cells were seeded in growth media. One day after cell seeding, growth media was replaced by Advanced formulation treatment media containing TVB-3166. Cells were incubated with TVB-3166 for 96 h. For analysis of β-catenin pathway markers following stimulation of cells with Wnt3A, cells were incubated with TVB-3166 for 78 h followed by addition of recombinant hWnt3A at 200 ng/mL (R&D; cat# 5036-WNP) for an additional 18 h. Whole cell lysates were prepared by adding RIPA buffer (Cell Signaling, 9806) supplemented with HALT phosphatase and protease inhibitor cocktail (Pierce, 78442). Proteins were separated using SDS-PAGE, transferred to nitrocellulose membranes, blocked, and incubated with primary antibodies overnight at 4 °C. Following washes in TBST, blots were probed with HRP-conjugated secondary antibodies (Cell Signaling). ECL prime chemiluminescent substrate (GE healthcare, RPN2232) was added and blots were developed using LAS-4000 imager (Fujifilm). Quantitation of protein bands was performed using Multigauge software (version 3.2, Fujifilm).
2.7. TCF Reporter Assay
Cells were transduced with the TCF-LEF Luciferase Cignal Lenti Reporter construct (Qiagen, 336851 CLS) 72 h prior the signaling experiment. Cells were seeded and one day later exposed overnight to the lenti-viral particles together with Sure ENTRY reagent (Qiagen, 336921). After stable integration of the TCF-LEF reporter 2–3 days later, cells were harvested and 5–10 × 103 cells per well were seeded in 96-well white clear bottom plates in growth media. After one day, media was changed to the Advanced formulation treatment media containing TVB-3166. After 48 h, hWnt3A (R&D, 5036-WNP) was added to A-549 cells at 200 ng/mL and cells were incubated further for 8 h. In experiments with COLO-205 cells, hWnt3A was not added. Luciferase was assayed using Dual-Glo Luciferase Assay System (Promega, E2940).
2.8. Immunofluorescence
CALU-6 cells were seeded in growth media at 200 cells per chamber in Permanox 4-chamber slides (Fisher Scientific, 1256521). The following day, media was replaced with treatment media containing TVB-3166 and cells were incubated for 95.5 h. Fluorescent staining of lipid raft domains was performed using cholera toxin patching method as described (Janes et al., 1999, Abulrob et al., 2004). Fixation was performed in 4% formaldehyde for 20 min on ice. Permeabilization was performed in PBS with 0.1% Triton X-100 for 10 min at room temperature. Blocking was performed in PBS with 1% BSA for 30 min at room temperature. N-Ras antibody (Santa Cruz Biotechnology, sc-519) diluted 1:250 in PBS with 0.1% BSA was added and incubated overnight at 4 °C. Anti-rabbit Alexa-fluor 594 antibody (Life Technologies, ab150076) diluted 1:500 in PBS with 0.1% BSA was added and incubated for 1 h at room temperature. After 4 × PBS washes, slides were mounted with coverslips using Prolong Gold Antifade mounting medium with DAPI (Life Technologies, P36935) and hardened overnight. Imaging was performed using a Zeiss LSM510 confocal microscope.
2.9. Soft Agar Colony Growth Assays
4 × 104 cells per well (in a 12-well plate) were resuspended in a 42 °C mixture of 0.35% ultrapure agarose (Life Technologies, 12200036), IMDM growth medium, and compound. The cell mixture was laid over a solidified base layer comprised of 0.6% bacteriological agar (Sigma, A5306) and IMDM growth medium and allowed to harden at room temperature for 1 h. A feeder layer comprised of IMDM growth medium and compound was poured on top of the hardened cell mixture. The feeder layer was replaced every 7 days and the number of colonies was counted after 3–4 weeks using OpenCFU colony counting software (http://opencfu.sourceforge).
2.10. Gene Expression Analysis
PANC-1 and 22Rv1 cells were seeded in growth media. The following day, treatment media with DMSO alone or with TVB-3166 (0.1 or 1.0 μM) was added and cells were incubated for an additional 48 (22Rv1) or 72 h (PANC-1). All treatments were performed in triplicate. For PANC-1 Affymetrix microarray studies (HG-U133 Plus 2.0), cells were lysed using Qiazol (Qiagen, 79306) and processed using the RNeasy microarray kit (Qiagen, 73304). For 22Rv1 RNA sequencing studies, cells were lysed using RLT buffer supplemented with β-mercaptoethanol and processed using the RNeasy mini kit (Qiagen, 74104). Microarray and RNAseq analysis was performed at Expression Analysis (Durham, NC). Data analysis was performed at 3-V Biosciences using Partek Genomics Suite software (St. Louis, MO). The PANC-1 Affymetrix gene expression data is available through the NCBI-GEO database using the accession number GSE64337.
2.11. Tumor Cell Line Xenograft Studies
Female BALB-c-nude mice were inoculated subcutaneously at the right flank with PANC-1 or OVCAR-8 tumor cells. TVB-3166 treatment via oral gavage administration was initiated when the mean tumor size reached approximately 150 mm3. Mice were assigned into treatment groups (N = 10) using a randomized block design based upon their tumor volumes. Tumor sizes were measured twice weekly in two dimensions using a caliper, and the volumes were expressed in mm3 using the formula: V = width2 × length × 0.5. The study was terminated 6 h after the final dose. Tumor growth inhibition (TGI) was calculated as the percentage of tumor growth, relative to tumor size at the start of treatment, in drug-treated groups compared to vehicle-treated groups. The Mann–Whitney U test was used to assess statistical significance of the mean tumor size between the drug and vehicle-treated groups. The in-life phase of the studies was conducted by Crown Biosciences (Santa Clara, CA, U.S.A. and Beijing, China). The experimenters were not blinded to group assignments.
2.12. Patient-derived Xenograft Studies
Female BALB-c-nude mice implanted unilaterally in the flank region with tumor fragments harvested from donor animals. When tumors reached approximately 100–300 mm3, animals were matched by tumor volume into the treatment and control groups and TVB-3166 dosing by oral gavage was initiated. Tumor dimensions were measured twice weekly by a digital caliper and data including individual and mean estimated tumor volumes (mean TV ± SEM) recorded for each group; tumor volume was calculated using the formula (1): TV = width2 × length × 0.52. Tumor growth inhibition (TGI) was calculated as the percentage of tumor growth, relative to tumor size at the start of treatment, in drug-treated groups compared to vehicle-treated groups. The in-life phase of the studies was conducted by Champions Oncology (Baltimore, MD). The experimenters were not blinded to group assignments.
2.13. Animal Work Statement
CrownBio IACUC follows the “Guide of Animal Care and Use” NRC 2011, the Chinese National Standard and Local government's regulations as well as animal welfare assurance number (A5896-01TC and A5895-01BJ). As an AAALAC accreditation facility, Crown Bioscience IACUC agree to play a role as monitoring the animal activities to ensure the laws and regulations being well implemented in our facility. SoBran Inc.'s Institutional Animal Care and Use Committee has approved all animal use protocols at Champions Oncology and ensures that all protocols meet the guidelines described in the Guide for the Care and Use of Laboratory Animals.
3. Results
3.1. TVB-3166 Reversibly Inhibits Fatty Acid Synthase
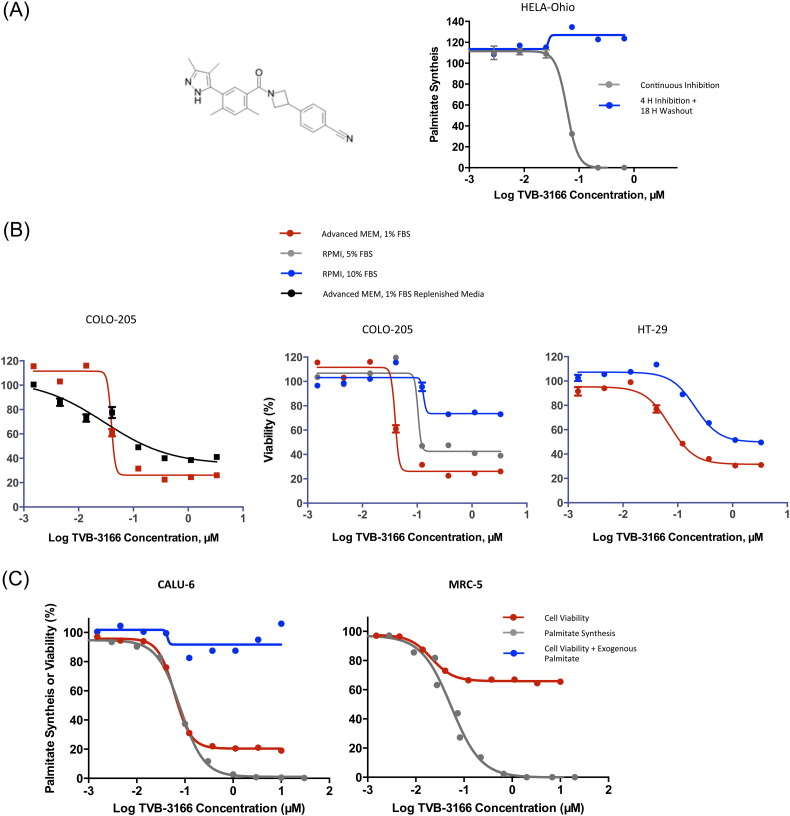
In order to evaluate the anti-tumor properties of pharmacological FASN inhibition, we characterized the properties of TVB-3166 (Fig. 1A), which has a lower molecular weight and increased solubility than previously reported orally available, reversible, potent, and selective FASN inhibitors discovered and developed by 3-V Biosciences (Oslob et al., 2013). In particular, the physicochemical properties were optimized by replacing the benzimidazole moiety with a more polar pyrrazole group (Oslob et al., 2014). These changes improved both systemic and tumor exposures. TVB-3166 inhibits the keto-reductase enzymatic function of FASN. The activity and potency of this compound were demonstrated by growing HeLa cells in the presence of 13C-acetate for 18 h and observing dose dependent inhibition of 13C-labeled-palmitate synthesis measured by lipid extraction and mass spectrometry analysis (Fig. 1A). Very similar results were obtained using 13C-glucose in the place of 13C-acetate (data not shown). The cellular palmitate synthesis IC50 of 0.060 μM aligns with the in vitro FASN biochemical IC50 of 0.042 μM that measures the catalytic activity of purified FASN. FASN inhibition by TVB-3166 is reversible; this was demonstrated in a cell-based assay. HeLa cells were exposed to TVB-3166 for 4 h, followed by removal of the drug and addition of 13C-acetate for 18 h. Palmitate synthesis was not inhibited under these conditions compared to cells treated continuously with the inhibitor. These data demonstrated potent and reversible cellular FASN inhibition with TVB-3166.
Fig. 1.
FASN inhibition with TVB-3166 inhibits palmitate synthesis and tumor cell viability. (A) TVB-3166 chemical structure (left) and reversible dose response inhibition of palmitate synthesis in HeLa-Ohio cells (right). (B) Effects on cell viability in varied medium and serum conditions as shown. (C) TVB-3166 inhibits palmitate synthesis (gray) and viability (red) with the same dose response and IC50 values in CALU-6 tumor cells. In the presence of 25 μM exogenous palmitate (blue) tumor cell viability is not affected. MRC-5 fibroblasts (right) have the same palmitate synthesis IC50 value as CALU-6 tumor cells but without the effect on cell viability.
3.2. FASN Inhibition Causes Selective Death of Tumor Cells
Palmitate, the enzymatic product of FASN, is a critical fatty acid that functions directly in many vital cellular processes and also serves as a precursor for the biosynthesis of longer and shorter chain fatty acids and lipids. In order to fully characterize the impact of TVB-3166 on a number of different tumor and normal cell lines we first established media conditions that supported robust growth across an array of diverse cell types. Many different tumor cell lines including COLO-205 and HT-29 cells were cultured 7 days in either Advanced MEM supplemented with 1% charcoal-stripped FBS and 1% l-glutamine or RPMI supplemented with 5–10% FBS and 1% l-glutamine. Cell growth and proliferation were comparable in the Advanced media (1% FBS) and RPMI (5–10% FBS) formulations (data not shown). Next, a TVB-3166 dose response evaluation demonstrated similar effects on cell viability when cells were treated in Advanced (1% FBS) or RPMI media with 5% FBS (less than 15% difference). Increasing the FBS content in RPMI to 10% resulted in a 20–50% reduction in sensitivity depending upon the cell type (Fig. 1B). The basis for this reduction could be effects on the cells or nonspecific protein binding of the drug. From these combined data, it was determined that Advanced media containing 1% FBS supported robust cell growth that enabled the measurement of TVB-3166-mediated cell killing. To demonstrate the overall suitability of Advanced media with 1% FBS for measuring the effects of TVB-3166 on cell viability, COLO-205 cells were incubated continuously for 7 days in the presence of TVB-3166 or had the media and drug replenished after 4 days of growth. The overall reduction of cell viability was similar under both conditions; thus, indicating the effect on cell viability is due to impact of the drug and not from cumulative nutrient and metabolite alterations in the media. These data supported the use of the Advanced media formulation and it was used throughout unless otherwise stated.
The on-target effect of TVB-3166 was evaluated in cells by the addition of exogenous palmitate. FASN inhibition using TVB-3166 caused cell death in CALU-6 non-small-cell lung tumor cells with a cellular IC50 value of 0.10 μM. This is in agreement with the concentrations required to inhibit cellular palmitate synthesis (IC50 0.081 μM) and FASN in an in vitro biochemical assay (0.042 μM). Supplementing the cell growth media with 25 μM palmitate ameliorated the tumor cell killing activity of TVB-3166, although potent inhibition of FASN still occurred under these conditions. Lower, more physiologically relevant concentrations of palmitate did not impact significantly the ability of FASN inhibition to induce tumor cell death in our in vitro assay. The ability of supraphysiological levels of exogenously applied palmitate to prevent the phenotypic consequences of FASN inhibition as well as the similar IC50 values in cell-based and in vitro assays demonstrated that TVB-3166-induced cell death is an on-target effect of FASN inhibition (Fig. 1C).
TVB-3166 mediated cell killing is restricted to tumor cells; not normal cells. Inhibition of FASN-catalyzed palmitate synthesis in MCF-10A breast epithelial cells, MRC-5 lung fibroblasts, and endothelial cells (HUVEC) was indistinguishable from that observed in CALU-6 tumor cells; however, the inhibition of palmitate synthesis in these cells failed to induce cell death (Fig. 1C). The observed 20–30% reduction in ATP levels was due to inhibition of cell proliferation.
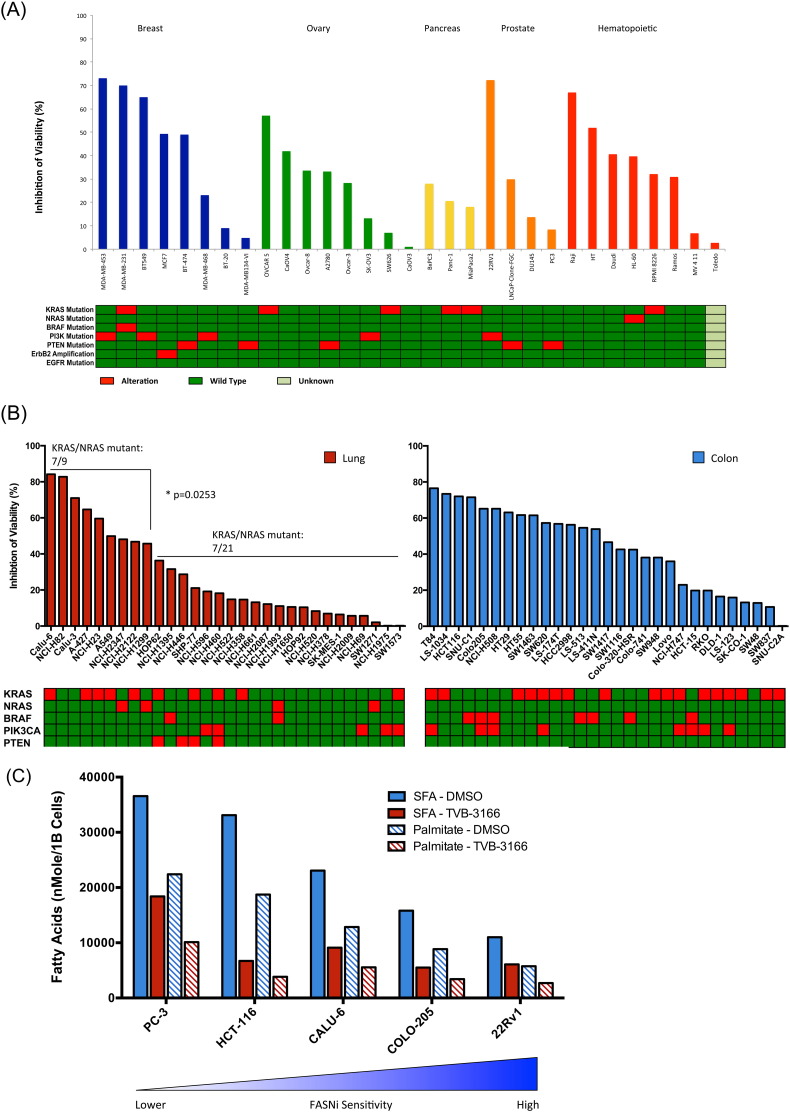
FASN inhibition induces cell death in diverse tumor cell. A panel of 90 different tumor cell lines representing six solid tumor types and hematopoietic tumors were assayed for cell viability following treatment with 0.02 or 0.20 μM TVB-3166 for 7 days (Fig. 2A–B). Dose-dependent induction of cell death was observed in all tumor cell lines; however, the percentage of cell death varied from 72% in the CALU-6 NSCLC cell line to less than 10% in the NCI-H1975 NSCLC cell line (Fig. 2 B). 14 of the 50 cell lines show greater than 50% cell death in this assay; whereas, 10 cell lines showed less than 10% cell death. Variability in sensitivity to FASN inhibition was found between cell lines from different solid tumor types and also in tumor cell lines of hematopoietic origin, indicating that features determining sensitivity occur across tumor types.
Fig. 2.
FASN inhibition inhibits tumor cell viability and sensitivity associates with saturated fatty acid levels. (A and B) 90 tumor cell lines from solid and hematopoietic tumor types were profiled for sensitivity to FASN inhibition with TVB-3166 in the Cell Titer Glo assay for cell viability. Cells were treated with 0.2 μM TVB-3166 for 7 days in Advanced MEM media with 1% charcoal-stripped FBS. (B) KRAS mutations associate with sensitivity in lung tumor cell lines. (C) TVB-3166 decreases the absolute levels of palmitate and saturated fatty acids in tumor cells. Tumor cells with lower pre-inhibition levels of palmitate and saturated fatty acids have increased sensitivity to FASN inhibition with TVB-3166 in a cell viability assay.
FASN expression levels did not correlate with sensitivity to FASN inhibition in our studies. We measured FASN mRNA and protein expression in many of the 50 cell lines and did not establish a correlation between FASN expression and TVB-3166 sensitivity. To investigate other possible markers of FASN sensitivity, the effect of TVB-3166 on the relative quantities of 36 different lipid species was measured in several tumor cell lines and compared to the extent of cell killing that results from FASN inhibition. In this analysis, TVB-3166 treatment for 72 h significantly decreased both palmitate and total saturated fatty acid levels; there was no discernable trend on the impact of FASN inhibition on polyunsaturated fatty acids. Tumor cell lines that had lower levels of palmitate and total saturated fatty acids prior to FASN inhibitor treatment had increased sensitivity to TVB-3166 in cell viability assays (Fig. 2C). KRAS mutation is associated with altered tumor cell metabolism, and analysis of 30 lung tumor cell lines, in which approximately 50% harbors a KRAS mutation, showed enrichment for mutant KRAS in cell lines with increased sensitivity to FASN inhibition (p < 0.05) (Fig. 2B). In contrast, an association of FASN inhibitor sensitivity with KRAS mutation status was not observed in analysis of 29 colon tumor cell lines.
3.3. FASN Inhibition Prevents Anchorage-Independent Tumor Cell Growth
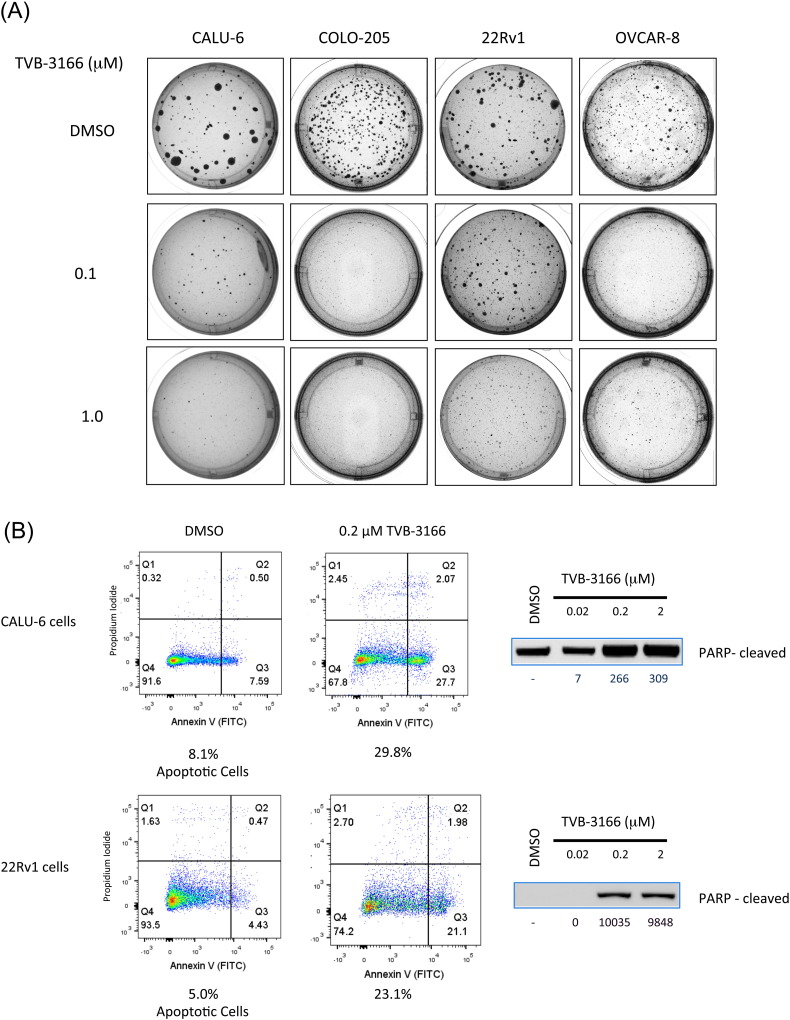
To characterize the effects of FASN inhibition on tumor cell growth and survival further, studies were performed to assess the effect of FASN inhibition on soft agar colony growth under conditions of complete media with 10% FBS. Several tumor cell lines, representing lung, ovary, prostate, and colorectal tumors, were examined at 0.1 or 1.0 μM TVB-3166. In each of the different tumor cells, dose dependent inhibition of colony growth was observed. At a lower dose of 0.1 μM TVB-3166 the most prominent effect is reduced colony size; however, at a concentration of 1.0 μM both colony size and number were inhibited. Colony growth of COLO-205, CALU-6, and OVCAR-8 cells showed heightened sensitivity to FASN inhibition compared to 22Rv1 cells (Fig. 3A). Together the ATP and soft agar assay results demonstrate that FASN inhibition interferes with cell growth, proliferation, and viability under varying medium and serum conditions that include 1–10% FBS.
Fig. 3.
FASN inhibition inhibits and anchorage-independent growth in soft agar induces apoptosis. (A) Soft agar colony growth assays. Cells were treated with TVB-3166 for 21–28 days in IMDM plus 10% FBS. Cells were plated in 0.35% ultrapure agarose over a solidified base layer of 0.6% bacteriological agar. (B) TVB-3166 induces apoptosis in CALU-6 (lung) and 22Rv1 (prostate) tumor cells. Cells were treated with TVB-3166 for 72 (22Rv1 cells) or 96 h (CALU-6 cells) in Advanced MEM media with 1% CF FBS. Annexin V and propidium iodide staining were measured by flow cytometry. Cleaved PAPR was detected by Western blot analysis.
3.4. FASN Inhibition Induces Tumor-Cell Apoptosis
To determine whether FASN inhibition causes tumor cell death by apoptosis, tumor cells were exposed to TVB-3166 and evaluated by both FACS analysis for Annexin V staining and Western blot analysis for PARP expression. Following treatment of lung (CALU-6) or prostate (22Rv1) tumor cells with TVB-3166, a 4–5 fold increase in Annexin V staining was observed for TVB-3166 treated cells compared to vehicle, respectively (Fig. 3B). In this analysis, the lower right quadrant with high Annexin V and low propidium iodide staining showed significantly more cells than the upper right quadrant with high staining for both markers. This profile indicated that cells were undergoing active apoptosis rather than dying via necrosis. Further demonstrating the apoptotic mechanism of cell death, cleaved-PARP expression was found to increase across a range of tumor cell lines by a magnitude of approximately 2-fold (MDA-MB-468 cells) to 100-fold (22Rv1 cells) (Fig. 3B).
3.5. FASN Inhibition Disrupts Lipid Raft Architecture and Localization of Membrane-associated Signaling Proteins
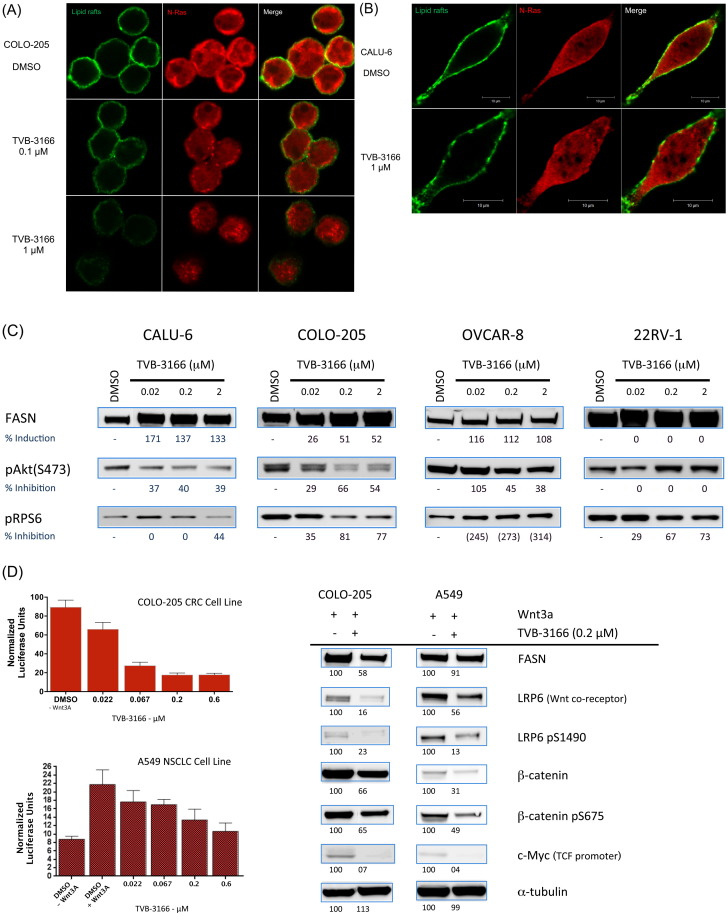
Having demonstrated that FASN inhibition inhibits tumor cell growth and survival under varied growth conditions; it was of interest to characterize the cellular mechanisms that contribute to these activities including those acting directly on lipid components. We examined the effect of FASN inhibition on lipid raft architecture and palmitoylated protein localization. CALU-6 or COLO-205 tumor cells were treated with TVB-3166 for 96 h, at which time lipid raft architecture and palmitoylated protein localization were imaged by immuno-fluorescent confocal microscopy. Lipid rafts were imaged using FITC-conjugated cholera-toxin subunit-B, which specifically binds to the cell surface raft-associated protein ganglioside GM1. As an example of palmitoylated protein localization, N-Ras localization was determined using the Alexa-fluor-conjugated antibody. FASN inhibition with TVB-3166 significantly disrupted lipid raft organization and membrane localization of N-Ras in both tumor cell types. In vehicle-treated cells lipid rafts were seen as a largely continuous ring around the cell with infrequent spots of lower intensity staining. N-Ras showed similar membrane-associated staining; however, was also found with lower intensity and partially punctate staining in the cytoplasm. Fig. 4A–B shows representative control and drug treated cells. Following FASN inhibition by 96 h of treatment with TVB-3166, lipid raft staining of the plasma membrane was discontinuous and overall had a lower intensity than that observed in control cells; sporadic staining was observed at irregular intervals. N-Ras staining also was altered following FASN inhibition. The plasma–membrane-associated pattern was discontinuous and irregular like the lipid rafts and the cytoplasmic distribution showed an increased number of regions without any staining. In addition to a greater number of cytoplasmic regions lacking N-Ras staining, these regions were larger following FASN inhibition than those in vehicle-treated cells. The effects were dose dependent; treatment with 0.1 μM TVB-3166 (near the FASN IC50) induced moderate changes that were more pronounced at 1 μM TVB-3166. Similar effects of FASN inhibition were observed in both the CALU-6 and COLO-205 tumor cells. Together the lipid raft and N-Ras immunofluorescence data show that FASN activity is required for normal lipid raft architecture and localization of membrane-associated signaling proteins such as N-Ras.
Fig. 4.
TVB-3166 inhibits lipid raft architecture and signal transduction of the AKT–mTOR and β-catenin pathways. In vitro immunofluorescent staining of lipid rafts (cholera toxin) and N-Ras in COLO-205 (A) and CALU-6 (B) tumor cell lines. (C) Western blot analysis of CALU6, COLO-205, OVCAR-8, and 22Rv1 tumor cell lines. Cells were treated with 0.02, 0.2, or 2.0 μM TVB-3166 for 96 h. (D) Inhibition of β-catenin pathway activity by FASN inhibition. COLO-205 and A549 cells were treated with 0.2 μM TVB-3166 for 48 h for TCF promoter-driven luciferase expression analysis (left) or 96 h for Western blot analysis (right). COLO-205 cells have constitutive pathway activity. Where indicated, cells were stimulated with 200 ng/ml Wnt3A for 18 h before treating with TVB-3166.
3.6. FASN Inhibition Blocks Signal Transduction Pathways that Regulate Tumor Cell Proliferation and Survival
Many signal transduction complexes are associated with lipid raft membrane microdomains. Disruption of lipid rafts and raft-associated protein localization provides a potential mechanism for FASN inhibition-mediated modulation of signal transduction. To investigate the possible consequences of tumor cell membrane remodeling following FASN inhibition, effects on signal transduction pathways involved with control of cell proliferation and survival were examined in several tumor cell lines. Dose-dependent inhibition of the PI3K–AKT–mTOR and β-catenin pathways was observed by Western blot analysis following in vitro treatment of several tumor cell lines with 0.02, 0.2, or 2 μM TVB-3166. FASN inhibition with TVB-3166 resulted in the inhibition of Akt (S743) and ribosomal protein S6 (S240 and 244) phosphorylation in cell lines from different tumor types, including lung, colon, prostate, ovary, and breast. The relative inhibition of Akt and S6 phosphorylation differed between some cell lines. Inhibition of both Akt and S6 phosphorylation was observed frequently; examples include CALU-6, COLO-205, and OVCAR-8. In some tumor cells; however, inhibition of Akt or S6 phosphorylation was observed but not both. The prostate tumor cell line 22Rv1 provides an example where TVB-3166 treatment resulted in inhibition of S6 phosphorylation without effect on Akt (Fig. 4C). Many tumor cells lines were investigated with the aim of identifying an association between known genetic features and the Akt-S6 inhibition profile. Additional cell lines that were profiled include A-549, OVCAR-5, HT-29, MDA-MB-231, MDA-MB-453, and MDA-MB-468. Inhibition of pAkt (S473) was observed in each of these cell lines; whereas, inhibition of pS6 (240 and 244) was observed in a subset. The data suggest that tumor cells with mutations in RAS, BRAF, PTEN, or ERBB2 are likely to show pAkt (S473) inhibition (Table 1). The strongest inhibition of pS6 (240 and 244) was observed in a PIK3CA-mutant cell line, 22Rv1.
Table 1.
TVB-3166 inhibits Akt and S6 phosphorylation in diverse tumor cell lines.
| Tumor cell line | Tissue of origin | RAS, BRAF, PIK3CA, PTEN, ERBB2 | % inhibition pAkt (S473) | % inhibition pS6 (S240/244) |
|---|---|---|---|---|
| CALU-6 | Lung | KRAS | 40 | 0 |
| A-549 | Lung | KRAS | 40 | 40 |
| MDA-MB-231 | Breast | KRAS, BRAF | 2 | 20 |
| MDA-MB-453 | Breast | PIK3CA, PTEN, ERBB2a | 70 | 40 |
| MDA-MB-468 | Breast | PTEN | 60 | 0 |
| COLO-205 | Colorectal | BRAF, APC | 66 | 81 |
| HT-29 | Colorectal | BRAF, APC | 95 | 0 |
| 22RV1 | Prostate | PIK3CA | 0 | 73 |
| OVCAR-5 | Ovary | KRAS | 77 | 50 |
| OVCAR-8 | Ovary | ERBB2b | 54 | 0 |
Mutations reported in the COSMIC database.
Copy number gain.
Activating mutation.
FASN inhibition with TVB-3166 inhibits β-catenin pathway signal transduction and transcriptional activity (Fig. 4D). Phosphorylation of β-catenin on serine 675 is associated with β-catenin protein stability, nuclear localization, and transcriptional activation. TVB-3166 treatment results in dose-dependent inhibition of β-catenin S675 phosphorylation and β-catenin protein expression levels in COLO-205 and A-549 tumor cell lines (Fig. 4D); similar results were observed in the MDA-MB-468 and OVCAR-8 breast and ovarian tumor cell lines, respectively (data not shown). To further investigate the mechanism and consequences of β-catenin inhibition by TVB-3166, transcriptional activity was measured using the TCF-TOPFLASH reporter system, and protein expression of TCF-regulated genes such as c-Myc was measured by Western blot analyses. Data from these readouts showed that FASN inhibition blocks β-catenin pathway activity at multiple levels: phosphorylation and expression of the Wnt ligand co-receptor LRP6 is inhibited, TCF transcriptional activity is lowered to basal levels, and c-Myc protein expression is reduced by 50%. Inhibition of AKT–mTOR and β-catenin pathway signal transduction by FASN inhibition with TVB-3166 was not observed in MRC5 lung fibroblasts, demonstrating that tumor and fibroblast cells have differential dependence on FASN for regulation of these proliferation and survival pathways.
3.7. FASN Inhibition Modulates Gene Expression of Tumor Cell Metabolism, Proliferation, and Survival Pathways
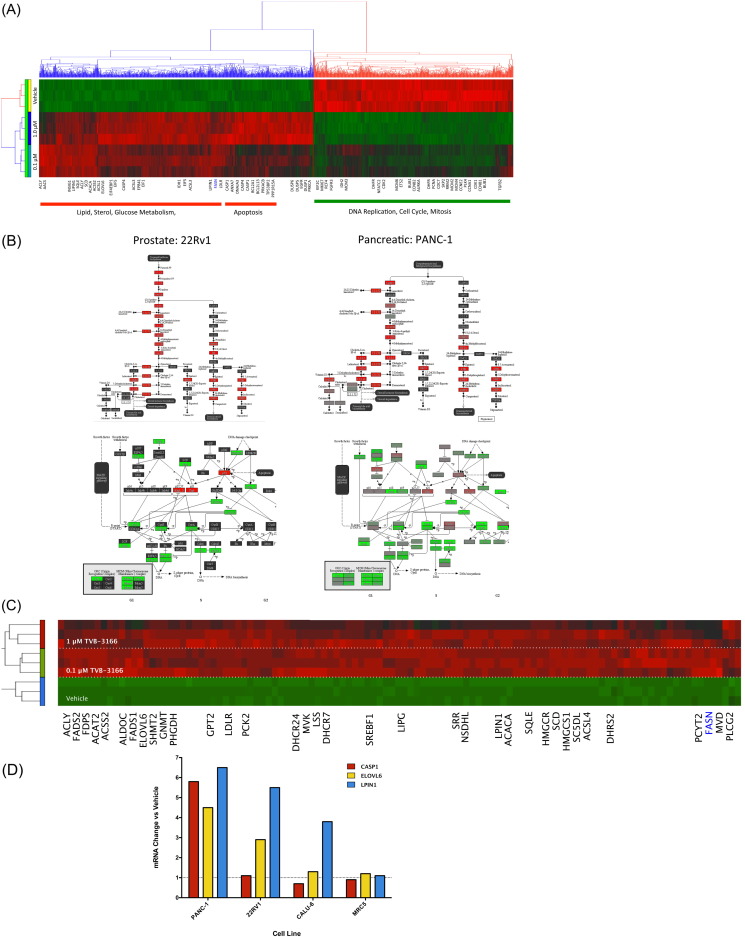
The significant changes that TVB-3166 exerts on membrane structure and signal transduction pathways, combined with the observation that tumor cells undergo apoptosis in response to drug treatment, caused us to evaluate of the impact of FASN inhibition on tumor cell gene expression. PANC-1 or 22Rv1 tumor cells growing in vitro were treated with 0.1 or 1 μM TVB-3166 and analyzed for changes in gene expression. Genome-wide analysis using microarray and RNA sequencing technology platforms showed extensive, coordinated, dose-dependent effects of FASN inhibition on the transcriptional activity of genes within pathways controlling tumor cell metabolism, proliferation, and survival. Genes with significant changes in mRNA abundance were identified using the statistical (p < 0.01) and fold change (> 1.6-fold) criteria (Fig. 5A; Supplemental Tables 1–2). Highly coordinated changes were evident in this analysis; the expression levels of multiple genes within a pathway were all coordinately increased or decreased. Pathway modulation by the induction of concerted gene expression changes is a remarkable feature of FASN inhibition. Examples of regulated pathways include up-regulation of sterol biosynthesis, pyruvate, amino acid, and glucose metabolism pathways and down-regulation of cell cycle, DNA replication, DNA-repair, and mitosis pathways (Table 2). Additional pathways with significantly increased gene expression include P53 signaling and apoptosis; whereas, the β-catenin, Notch, and the unfolded protein response pathways are down-regulated. Highly similar changes were found in both PANC-1 and 22Rv-1 cells (Fig. 5B). Many of the genes showing modulation in response to FASN inhibition are regulated downstream of the AKT–mTOR pathway via SREBP, e.g., genes within the sterol and lipid biosynthesis pathways (Fig. 5C), or the β-catenin pathway, e.g., genes involved in control of cell cycle progression and cell proliferation.
Fig. 5.
TVB-3166 modulates metabolic and growth-associated pathways in tumor cells. PANC-1 cells treated with TVB-3166 for 72 h were analyzed for gene expression changes using Affymetrix HU133 Plus 2.0 microarrays. Analysis includes unsupervised hierarchical clustering (A) and pathway enrichment analysis (B). Prostate and pancreatic tumor cell lines have similar changes: sterol biosynthesis pathway (top) and cell cycle pathway (bottom). 22Rv1 prostate tumor cells were treated with TVB-3166 for 48 h and gene expression changes were determined using RNA sequencing (RNASeq-25, Illumina, Inc). (C) Heat map showing 22Rv1 gene expression changes in lipid and sterol biosynthesis pathway genes. (D) RT-PCR validation of microarray and RNA sequencing data in PANC-1 and 22Rv1 cells and extension of results to CALU-6 and MRC5 cells.
Table 2.
Pathways with significant gene expression modulation by TVB-3166.
| Pathway namea | Enrichment score | Enrichment p-value |
|---|---|---|
| Steroid biosynthesis | 19.01 | 5.55E − 09 |
| Cell cycle | 14.42 | 5.47E − 07 |
| Metabolic pathways | 9.13 | 0.0001 |
| DNA replication | 8.65 | 0.0002 |
| p53 signaling pathway | 8.06 | 0.0003 |
| One carbon pool by folate | 7.95 | 0.0004 |
| Terpenoid backbone biosynthesis | 7.60 | 0.0005 |
| Insulin signaling pathway | 7.06 | 0.0009 |
| Viral carcinogenesis | 6.45 | 0.0016 |
| Aminoacyl-tRNA biosynthesis | 6.07 | 0.0023 |
| HIF-1 signaling pathway | 5.61 | 0.0037 |
| NF-kappa B signaling pathway | 5.24 | 0.0053 |
| Pyruvate metabolism | 4.77 | 0.0085 |
| Glycine, serine and threonine metabolism | 4.28 | 0.0138 |
| Biosynthesis of unsaturated fatty acids | 4.17 | 0.0155 |
| Folate biosynthesis | 4.16 | 0.0156 |
| Glycolysis/gluconeogenesis | 4.10 | 0.0166 |
| Glutathione metabolism | 4.08 | 0.0169 |
| 2-Oxocarboxylic acid metabolism | 3.91 | 0.0200 |
| Mismatch repair | 3.78 | 0.0228 |
| Propanoate metabolism | 3.65 | 0.0261 |
| Dorso-ventral axis formation | 3.61 | 0.0271 |
| Selenocompound metabolism | 3.47 | 0.0311 |
| Pyrimidine metabolism | 3.39 | 0.0336 |
| Gap junction | 2.98 | 0.0506 |
| Mineral absorption | 2.76 | 0.0632 |
| Vitamin B6 metabolism | 2.73 | 0.0655 |
| Fatty acid biosynthesis | 2.73 | 0.0655 |
KEGG database.
FASN inhibition-mediated effects on gene expression were measured using selected RT-PCR assays in additional tumor cell lines as well as in non-transformed cell types, including fibroblasts and endothelial cells. Tumor cell lines such as the NSCLC-derived line CALU-6 showed modulation of gene expression similar to that observed in the PANC-1 and 22Rv1 cells, with increased expression of CASP1, LPIN1, and ELOVL6 (Fig. 5D). In contrast, expression of these genes was not affected by FASN inhibition in MRC5 lung fibroblast and HUVEC endothelial cells.
3.8. Once-daily, Oral Dosing of TVB-3166 Inhibits Xenograft Tumor Growth
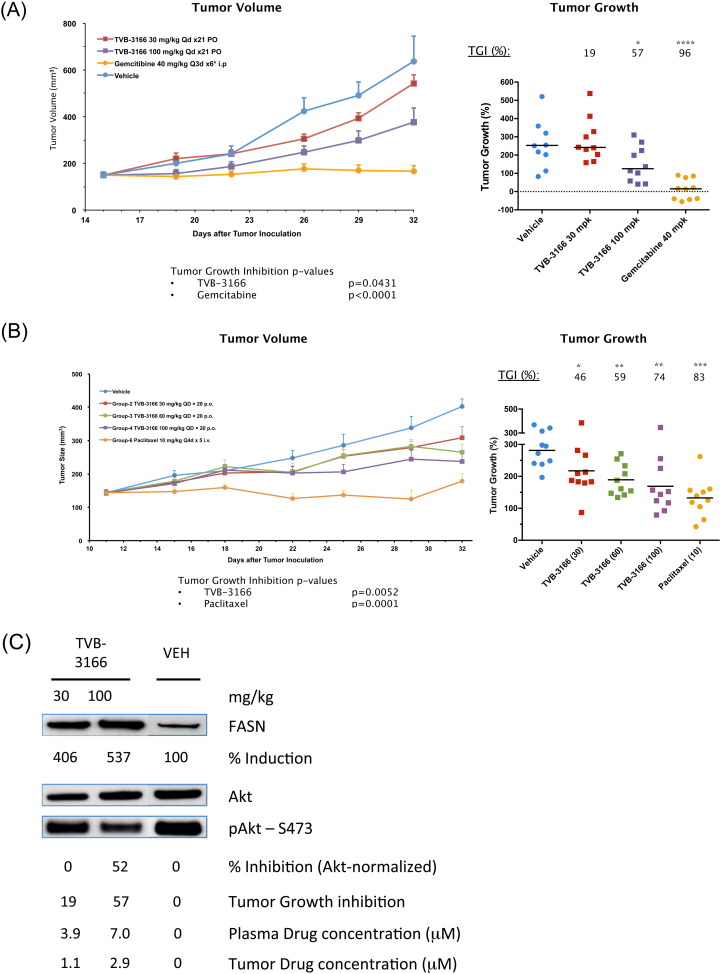
In vivo efficacy of FASN inhibition on tumor growth was investigated in tumor cell line and patient-derived xenograft tumor models. Dose responsive tumor growth inhibition was observed in multiple tumor models (Fig. 6, Fig. 7). In the PANC-1 tumor cell line xenograft (Fig. 6A), once-daily, oral dosing of TVB-3166 at 100 mg/kg for 18 days resulted in statistically significant 57% tumor growth inhibition compared to vehicle (p = 0.04). A once-daily dose of 30 mg/kg TVB-3166 showed non-significant 19% tumor growth inhibition. Tumor growth inhibition was associated with dose-dependent inhibition of Akt phosphorylation (S473) and induction of FASN expression (Fig. 6C). Inhibition of Akt phosphorylation in FASN inhibitor-treated tumors, relative to vehicle treatment, was 53% and 0% in the 100 and 30 mg/kg treatment groups, respectively. FASN protein expression was increased 5.4 fold and 4-fold in the high and low dose groups, respectively. Pharmacokinetic analysis of plasma and tumor samples collected 6 h post-dosing showed the concentration of TVB-3166 to be approximately 3-fold higher in plasma than tumor. The 100 and 30 mg/kg groups had plasma and tumor concentrations of 7 and 2.9 μM, respectively. Following normalization for free, non-protein-bound drug, the TVB-3166 plasma concentration in the 100 mg/kg group at 6 h post-dosing was equivalent to the FASN IC50. The overall PK of TVB-3166 in mice predicts that most of the drug is cleared by 12 h after dosing. Body weight loss was not significant in the TVB-3166-treated mice and was similar to changes observed in the vehicle-treated mice. Together, the results demonstrated that TVB-3166 has excellent oral bioavailability and manageable pharmacokinetic properties, which resulted in well-tolerated dose-dependent inhibition of Akt phosphorylation and tumor growth.
Fig. 6.
TVB-3166 inhibits growth of cell-line-derived xenograft tumors. TVB-3166 was dosed once daily by oral gavage at 30, 60, or 100 mg/kg. Animals were randomized according to tumor size and drug treatment was started when the mean tumor size was 150–200 mm3. (A) PANC-1 pancreatic tumor cell line. (B) OVCAR-8 ovarian tumor cell line. (C) PD analysis of AKT phosphorylation and FASN expression by Western blot. Tumors were harvested 6 h after the last dose. TVB-3166 plasma and tumor drug concentrations were determined by mass spectrometry. The in-life phase for both studies was performed at Crown Biosciences (Santa Clara, CA; Beijing, China).
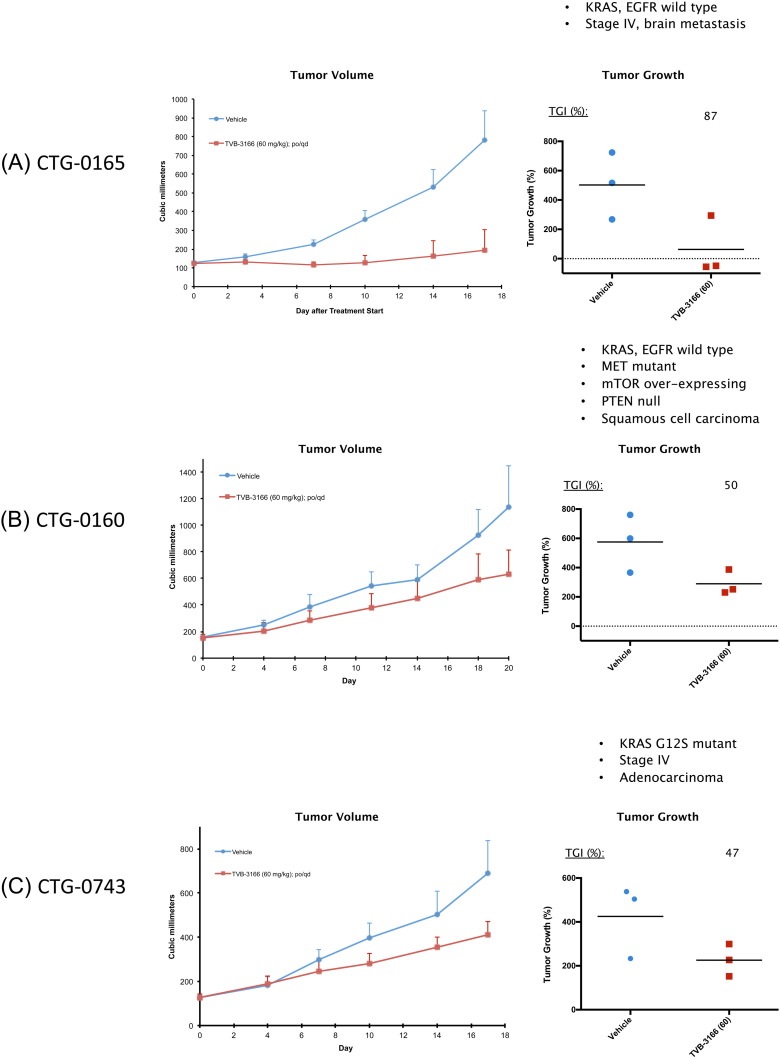
Fig. 7.
TVB-3166 inhibits growth of patient-derived non-small-cell lung cancer xenograft tumors: (A) CTG-0165, (B) CTG-0160 and (C) CTG-0743. TVB-3166 was dosed once daily by oral gavage at 60 mg/kg. Animals were randomized according to tumor size and drug treatment was started when the mean tumor size was 150–200 mm3. The in-life phase for all studies was performed at Champions, Oncology (Baltimore, MD).
Dose-dependent efficacy in the OVCAR-8 ovarian tumor cell line xenograft was observed following once daily dosing of TVB-3166 (Fig. 6B). In the OVCAR-8 tumor model, once daily, oral dosing of TVB-3166 at 30, 60, or 100 mg/kg resulted in significant tumor growth inhibition compared to vehicle treatment. Following 21 days of dosing, tumor growth inhibition values were 46, 59, and 74% (p = 0.0052) compared to vehicle, respectively for the 30, 60, and 100 mg/kg dose groups. Body weight loss was not significant in the TVB-3166-treated mice and was similar to changes observed in the vehicle-treated mice. These results demonstrated that well-tolerated doses of TVB-3166 were associated with tumor growth inhibition efficacy in K-Ras-mutant (Panc-1) and ErbB2-mutant (OVCAR-8) tumor xenografts.
FASN inhibition with TVB-3166 showed tumor growth inhibition efficacy in patient-derived xenografts (Fig. 7). Once-daily, oral dosing of TVB-3166 at 60 mg/kg was found to inhibit tumor growth compared to vehicle at 87%, 50%, and 47% in the CTG-0165, CTG-0160, and CTG-0743 tumor models, respectively. Body weight changes were not significant in TVB-3166-treated mice and were similar to changes observed in vehicle-treated mice. CTG-0743 expresses mutant K-Ras (G12S); whereas, CTG-0165 and CTG-0160 are KRAS-wild-type tumors. All three tumors have wild type alleles of EGFR; however, CTG-0160 harbors a c-MET mutation. The CTG-0165 and CTG-0743 tumors are adenocarcinomas; whereas, CTG-0160 is a squamous cell carcinoma. These data show that FASN inhibition is well tolerated and inhibits tumor growth in genetically and histologically diverse patient-derived NSCLC tumor models, including squamous cell and adenocarcinomas as well as tumors with varied genetic profiles that include KRAS and c-MET mutations.
4. Discussion
We describe the activity of a highly selective, potent, reversible, and orally available small molecule FASN inhibitor that has not been described previously and characterize the biological consequences of FASN inhibition using in vitro and in vivo preclinical tumor models. Our findings show that FASN inhibition selectively inhibits growth and viability of tumor cells both in vitro and in vivo, while having minimal effects on non-tumor cells such as fibroblasts and endothelial cells, and demonstrate that tumor cells have a unique dependence on FASN function for survival that is not present in normal cells. The potency and selectivity of TVB-3166 for FASN as well as its reversible mechanism of action differentiates it from earlier FASN inhibitors such as C75, Orlistat®, and cerulenin that often are limited by their in vivo toxicity. Our data show TVB-3166 to be well tolerated and to exhibit potent and broad antitumor activity in diverse preclinical xenograft tumor models.
FASN inhibition by TVB-3166 inhibits tumor growth and induces tumor cell apoptosis via multiple mechanisms of action. This may account in part for the finding in our data and of others (Benjamin et al., in press) that FASN expression alone does not predict sensitivity to FASN inhibition. Additional factors that likely contribute to the disconnect between FASN expression and sensitivity to FASN inhibition include: (1) the presence of post-translational mechanisms for regulating lipogenesis (e.g., ACC1 phosphorylation) in the face of FASN overexpression; (2) the association of FASN overexpression with a variety of genetic alterations (e.g., RTK overexpression and PI3K mutation) that promote tumor cell growth and survival through mechanism and pathways that can act in concert with or independent of FASN. Thus, a combination of factors likely governs cellular saturated fatty acid levels, which were found to associate with sensitivity to FASN inhibition.
Using flow cytometry to measure Annexin V staining and Western blot analyses to measure the expression of cleaved PARP, we show that FASN inhibition induces apoptosis in many different tumor cell lines. To investigate the mechanism of apoptosis induction, we characterized the effects of FASN inhibition on lipid raft organization, cellular localization of palmitoylated proteins, signal transduction activity through pathways that regulate tumor cell proliferation and viability, and whole-genome gene expression analysis. Our data reveal that FASN inhibition disrupts lipid raft distribution in the plasma membrane, alters the localization of raft-associated proteins such as N-Ras, inhibits multiple signal transduction pathways including PI3K–AKT–mTOR and β-catenin, and modulates the expression of many genes in metabolic, proliferation and apoptosis pathways. These findings are consistent with a mechanistic model where FASN inhibition leads to the depletion of palmitate and palmitate-derived lipids required for proper protein trafficking and localization and lipid raft organization and architecture. Protein palmitoylation is required for trafficking and localization of many membrane-associated proteins, including all Ras and Wnt isoforms (Ahearn et al., 2012, Song et al., 2013, Clevers and Nusse, 2012, Fiorentino et al., 2008). Mutant N-Ras that cannot be palmitoylated localizes to intracellular membranes delimiting the Golgi and shows impaired plasma membrane localization (Eisenberg et al., 2011, Eisenberg et al., 2013). The localization of N-Ras in our studies following FASN inhibition showed this same pattern. Disruption of membrane structure and lipid raft distribution results in impaired signaling by preventing protein access to these critical lipid domains that bring complex signaling machinery together at the appropriate subcellular location. Lipid rafts are plasma membrane microdomains characterized by high local concentrations of sphingolipids, glycolipids, sterols, and lipid-modified proteins (Lingwood and Simons, 2010, Simons and Sampaio, 2011). Additionally, raft domains are enriched for lipid species with saturated and longer hydrocarbon fatty acid chains. Our data show that palmitate and total saturated fatty acid levels are depleted by FASN inhibition and, moreover, that tumor cell lines with lower levels of saturated fatty acids exhibit increased sensitivity to FASN inhibition in viability assays. Raft domains function as dynamic, organizing centers within the plasma membrane and play a vital role in facilitating protein–protein interactions that promote mitotic and cell survival-associated signal transduction. Thus, inhibition of de novo fatty acid synthesis leads to the disruption of lipid raft architecture and organization that is required for cell growth and survival signal transduction, and this effect is expected to be greater in tumor cells than in resting or non-tumor cells where lipid raft domains are smaller and less abundant. In agreement with this model, the PI3K–AKT pathway, which is inhibited in our studies by TVB-3166, has been shown to require lipid rafts for signaling activity in multiple tumor cell lines (Reis-Sobreiro et al., 2013, Lasserre et al., 2008).
Multiple signaling pathways that regulate tumor cell proliferation and survival, including PI3K–AKT–mTOR and β-catenin, are inhibited by FASN inhibition. Our data show that the effects on these pathways vary between tumor cell lines and that particular cell lines are more susceptible to inhibition of Akt, mTOR, or Wnt-β-catenin signaling. We find inhibition of the PI3K–AKT–mTOR and β-catenin pathways by FASN inhibition in cell lines derived from a variety of tumor types, including breast, colon, lung, prostate and ovarian tumors. Additional work remains to identify strongly predictive markers of pathway susceptibility to FASN inhibition; however, our data suggest that Akt (S473) inhibition is associated with RAS, BRAF, PTEN, or ERBB2 alterations. Cell lines with activating PI3K mutations show mTOR inhibition (as measured by phosphorylation of RPS6) and, less reliably, inhibition of Akt. In an extended analysis of tumor cell lines for sensitivity to FASN inhibition using an in vitro assay for cell viability, we find that KRAS and NRAS mutations associate with sensitivity to FASN inhibition in non-small-cell lung cancer cell lines. Consistent with these data, in vivo tumor growth inhibition was observed in a KRAS mutant patient-derived NSCLC xenograft tumor. Highly sensitive colorectal tumor cell lines included KRAS mutant lines but were not enriched for such mutations, indicating that multiple factors influence FASN inhibitor sensitivity. Taken together, our in vitro and in vivo data indicate that FASN inhibition has potential utility in KRAS and NRAS mutant tumors, and that this activity may be mediated, at least in part, by inhibition of AKT signaling.
FASN inhibition results in Wnt–β-catenin pathway inhibition (Fiorentino et al., 2008), including decreased c-Myc expression, in cell lines from a variety of tumor types, including colon, lung, breast and ovarian tumors. Inhibition of the β-catenin pathway and c-Myc expression is a feature of FASN inhibition in cell lines with high constitutive TCF promoter activity due to APC or β-catenin mutations, and also in cell lines without these mutations that increase promoter activity in response to stimulation with exogenous Wnt3A. Our data show that FASN inhibition blocks activation of the Wnt receptor and co-receptor complex as we observe inhibition of co-receptor (LRP6) phosphorylation in response to FASN inhibitor treatment, and further demonstrate that FASN inhibition decreases β-catenin phosphorylation at serine 675 and expression levels of the β-catenin protein. Phosphorylation at S675 is associated with β-catenin stability and nuclear localization (Zhu et al., 2012). Therefore, inhibition of phosphorylation at this site is expected to result in the loss of β-catenin protein expression, and consequently TCF promoter activity, as observed in our data. The underlying mechanism of pathway inhibition remains to be fully elucidated; however, it may depend on the palmitoylation of Wnt, a modification requisite for the activity of this protein (Clevers and Nusse, 2012, Fiorentino et al., 2008). Lipid raft disruption may also contribute to inhibition of frizzled-LRP6 activation. Together our data show that FASN activity is required for Wnt–β-catenin pathway activity and expression of vital oncogenic proteins such as c-Myc, and suggests that tumors driven by β-catenin and c-Myc activity may be susceptible to treatment by FASN inhibition.
In agreement with the effects on the PI3K–AKT–mTOR and Wnt–β-catenin signal transduction pathways, global gene expression analyses revealed that FASN inhibition modulates many biological processes important for tumor cell proliferation and survival. In response to FASN inhibitor treatment, highly similar gene expression changes are observed in PANC-1 and 22Rv1 tumor cells, indicating FASN inhibition can affect tumor cells from different tissues and with different genetic backgrounds similarly. Modulated pathways include lipid, sterol and glucose metabolism, DNA replication and repair, cell cycle progression, cell proliferation and survival, and apoptosis. Remarkably, highly concerted changes in expression of numerous genes within several pathways are observed, showing that the related biological processes are strongly affected and, further, that master regulators of pathway activity are likely impacted by FASN inhibition. SREBP regulation of the lipid and sterol biosynthesis pathways provides a clear example of this in our data. These gene expression data are guiding further elucidation of FASN inhibition mechanisms of action and the discovery of biomarkers for selection of tumors types most likely to respond to FASN targeted therapy.
TVB-3166 is a potent and selective FASN inhibitor that shows in vitro and in vivo activity in diverse preclinical tumor models. Our data show that once-daily, oral dosing of TVB-3166 is well tolerated and inhibits growth of patient-derived and cell-line-derived xenograft tumors. Previous studies by others using FASN inhibitors such as polyphenolic compounds including G28UCM, orlistat and analogs of C75 have demonstrated in vivo xenograft tumor growth inhibition with limited toxicity (Kridel, 2004, Puig et al., 2009, Puig et al., 2011). Our studies advance these findings by characterizing the activity of reversible FASN inhibition on many diverse tumor types using a highly selective FASN inhibitor that does not cause activation of fatty-acid oxidation or other undesirable effects through off-target activities. In vivo TVB-3166 exposure with once-daily dosing inhibits FASN approximately 10–12 h each day. This is in contrast to continuous inhibition in our in vitro studies, and shows that continuous target engagement is not required for in vivo tumor growth inhibition. Xenograft tumor growth inhibition was observed in diverse tumor models representing lung, ovarian, and pancreatic tumors with varied molecular genetic features, including KRAS, ERBB2, c-MET, and PTEN mutations. The OVCAR-8 tumor cell line harbors an activating mutation in ERBB2, and in these xenograft tumors TVB-3166 displayed dose-dependent inhibition of tumor growth. Activation of receptor tyrosine kinases such as EFGFR, ERBB2, and c-MET can promote over-expression of FASN and other genes involved in lipid and sterol biosynthesis, and this may sensitize these types of tumors to FASN inhibition.
TVB-3166 inhibits tumor growth in patient-derived non-small cell lung xenograft tumors with both mutant and wild type KRAS mutation status, as well as with adenocarcinoma and squamous cell carcinoma histology. Both in vitro and in vivo data suggest that FASN inhibition may be effective in K-Ras-mutant non-small-cell lung tumors. KRAS mutations are prevalent in other tumor types such as colorectal and pancreatic adenocarcinomas. Our data shows that TVB-3166 also inhibits growth of the KRAS-mutant PANC-1 xenograft tumor. Mutations in KRAS have been linked to altered metabolic features of tumor cells, including pancreatic adenocarcinomas where KRAS mutations are found in more than 95% of all tumors (Donahue et al., 2012, Jones et al., 2008, Yachida et al., 2010, Ying et al., 2012). Mutant K-Ras has been shown to reprogram tumor cell metabolism and cause increased dependency on glutamine and glucose metabolism (Son et al., 2013). A consequence of this altered metabolic state is the increased production of pyruvate. Pyruvate can be metabolized to lactate to generate ATP and acetyl-CoA or, alternatively, can be shunted to citrate and then acetyl-CoA, which is a substrate for palmitate biosynthesis by FASN (Fig. 8). Thus, mutant K-Ras may increase the dependency of tumor cells on de novo lipogenesis and provide a biological basis for targeting these tumors, or a subset of them, with a FASN inhibitor.
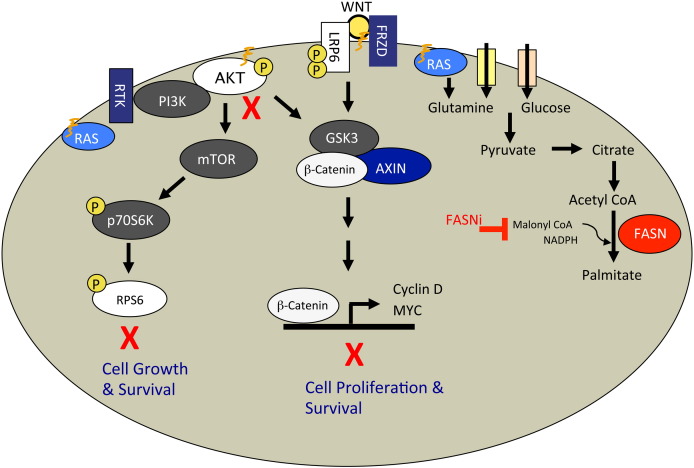
Fig. 8.
FASN inhibition blocks metabolic and signal transduction pathways vital to cancer cell growth, proliferation, and survival. FASN inhibition results in inhibition of Akt and S6 phosphorylation in the AKT–mTOR signal transduction pathway. In the Wnt–β-catenin pathway, FASN inhibition results in the inhibition of Lrp6 and β-catenin phosphorylation as well as the expression of TCF promoter-driven genes such as c-Myc. FASN inhibition impairs the plasma membrane localization of palmitoylated and other lipid-raft-associated proteins such as N-Ras. Our data fit a model whereby the disruption of lipid rafts and mislocalization of membrane-associated proteins can drive inhibition of signaling through cellular growth and survival pathways such as AKT–mTOR and Wnt–β-catenin. Signal transduction through molecules such as K-Ras or pathways such as AKT–mTOR is tightly linked with tumor cell metabolism of glucose and glutamine as well as lipid biosynthesis.
In conclusion, our data show FASN inhibition inhibits xenograft tumor growth in K-Ras-mutant and wild-type tumor models as well as tumors from multiple different diseases with mutations in other commonly mutated or over-expressed oncogenes. Mechanism of action studies demonstrated that TVB-3166 rapidly shuts down de novo palmitate synthesis, which later results in a range of different outcomes in tumor cells. Following the depletion of palmitate, gene expression in tumor cells is altered on a global scale, lipid raft architecture is disrupted, signaling pathways are inhibited and ultimately the tumor cell dies through an apoptotic mechanism. Oncogenic signal transduction pathways including PI3K–AKT–mTOR and β-catenin are inhibited and importantly, FASN inhibition significantly decreases c-Myc expression and induces apoptosis of c-Myc expressing tumor models. In vivo activity in Wnt–β-catenin-activated and c-Myc over-expressing tumors models is under investigation. Taken together, the wide range of cellular mechanistic consequences of FASN inhibition provides potential for effective treatment of a large number of cancers of unmet need.
Funding
All studies were funded privately by 3-V Biosciences.
Conflict of Interest
All authors are full-time employees of 3-V Bioscience and shareholders of 3-V Biosciences stock.
Acknowledgments
We thank Jia Li, Zhun Wang, and Guanping Mao at Crown Biosciences, Stanley Parish and Elizabeth Bruckheimer at Champions Oncology, and Francesca Incardona at Biotox Sciences. We thank Marie O'Farrell, Allan Wagman, Greg Duke, Steve Smith, and Merdad Parsey at 3-V Biosciences for the scientific discussions.
Footnotes
Gene symbols and common abbreviations such as DNA and RNA are not defined.
Supplementary data to this article can be found online at http://dx.doi.org/10.1016/j.ebiom.2015.06.020.
Appendix A. Supplementary data
Supplemental materials and methods.
Table 1: PANC-1 cell differentially expressed gene list.
Table 2: 22Rv1 cell differentially expressed gene list.
PANC-1 Affymetrix gene expression data: NCBI-GEO accession number: GSE64337.
References
- Abulrob A., Giuseppin S., Andrade M.F., McDermid A., Moreno M., Stanimirovic D. Interactions of EGFR and caveolin-1 in human glioblastoma cells: evidence that tyrosine phosphorylation regulates EGFR association with caveolae. Oncogene. 2004;23:6967–6979. doi: 10.1038/sj.onc.1207911. [DOI] [PubMed] [Google Scholar]
- Ahearn I.M., Haigis K., Bar-Sagi D., Philips M.R. Regulating the regulator: post-translational modification of RAS. Nat. Rev. Mol. Cell Biol. 2012;13:39–51. doi: 10.1038/nrm3255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benjamin D.I., Li D.S., Lowe W., Heuer T., Kemble G., Nomura D.K. Diacylglycerol metabolism and signaling is a driving force underlying FASN inhibitor sensitivity in cancer cells. ACS Chem. Biol. 2015 doi: 10.1021/acschembio.5b00240. (in press) (Apr 17, Epub ahead of print) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chirala S.S., Chang H., Matzuk M., Abu-Elheiga L., Mao J., Mahon K., Finegold M., Wakil S.J. Fatty acid synthesis is essential in embryonic development: fatty acid synthase null mutants and most of the heterozygotes die in utero. Proc. Natl. Acad. Sci. U. S. A. 2003;100:6358–6363. doi: 10.1073/pnas.0931394100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi W.I., Jeon B.N., Park H., Yoo J.Y., Kim Y.S., Koh D.I., Kim M.H., Kim Y.R., Lee C.E., Kim K.S., Osborne T.F., Hur M.W. Proto-oncogene FBI-1 (Pokemon) and SREBP-1 synergistically activate transcription of fatty-acid synthase gene (FASN) J. Biol. Chem. 2008;283:29341–29354. doi: 10.1074/jbc.M802477200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chuang H.Y., Chang Y.F., Hwang J.J. Antitumor effect of orlistat, a fatty acid synthase inhibitor, is via activation of caspase-3 on human colorectal carcinoma-bearing animal. Biomed. Pharmacother. 2011;65:286–292. doi: 10.1016/j.biopha.2011.02.016. [DOI] [PubMed] [Google Scholar]
- Clevers H., Nusse R. Wnt/beta-catenin signaling and disease. Cell. 2012;149:1192–1205. doi: 10.1016/j.cell.2012.05.012. [DOI] [PubMed] [Google Scholar]
- Donahue T.R., Tran L.M., Hill R., Li Y., Kovochich A., Calvopina J.H., Patel S.G., Wu N., Hindoyan A., Farrell J.J., Li X., Dawson D.W., Wu H. Integrative survival-based molecular profiling of human pancreatic cancer. Clin. Cancer Res. 2012;18:1352–1363. doi: 10.1158/1078-0432.CCR-11-1539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eisenberg S., Beckett A.J., Prior I.A., Dekker F.J., Hedberg C., Waldmann H., Ehrlich M., Henis Y.I. Raft protein clustering alters N-Ras membrane interactions and activation pattern. Mol. Cell. Biol. 2011;31:3938–3952. doi: 10.1128/MCB.05570-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eisenberg S., Laude A.J., Beckett A.J., Mageean C.J., Aran V., Hernandez-Valladares M., Henis Y.I., Prior I.A. The role of palmitoylation in regulating Ras localization and function. Biochem. Soc. Trans. 2013;41:79–83. doi: 10.1042/BST20120268. [DOI] [PubMed] [Google Scholar]
- Fiorentino M., Zadra G., Palescandolo E., Fedele G., Bailey D., Fiore C., Nguyen P.L., Migita T., Zamponi R., Di Vizio D., Priolo C., Sharma C., Xie W., Hemler M.E., Mucci L., Giovannucci E., Finn S., Loda M. Overexpression of fatty acid synthase is associated with palmitoylation of Wnt1 and cytoplasmic stabilization of beta-catenin in prostate cancer. Lab. Investig. 2008;88:1340–1348. doi: 10.1038/labinvest.2008.97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flavin R., Peluso S., Nguyen P.L., Loda M. Fatty acid synthase as a potential therapeutic target in cancer. Future Oncol. 2010;6:551–562. doi: 10.2217/fon.10.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grunt T.W., Wagner R., Grusch M., Berger W., Singer C.F., Marian B., Zielinski C.C., Lupu R. Interaction between fatty acid synthase- and ErbB-systems in ovarian cancer cells. Biochem. Biophys. Res. Commun. 2009;385:454–459. doi: 10.1016/j.bbrc.2009.05.085. [DOI] [PubMed] [Google Scholar]
- Hollander M.C., Blumenthal G.M., Dennis P.A. PTEN loss in the continuum of common cancers, rare syndromes and mouse models. Nat. Rev. Cancer. 2011;11:289–301. doi: 10.1038/nrc3037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janes P.W., Ley S.C., Magee A.I. Aggregation of lipid rafts accompanies signaling via the T cell antigen receptor. J Cell Biol. 1999;147:447–461. doi: 10.1083/jcb.147.2.447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones S., Zhang X., Parsons D.W., Lin J.C., Leary R.J., Angenendt P., Mankoo P., Carter H., Kamiyama H., Jimeno A., Hong S.M., Fu B., Lin M.T., Calhoun E.S., Kamiyama M., Walter K., Nikolskaya T., Nikolsky Y., Hartigan J., Smith D.R., Hidalgo M., Leach S.D., Klein A.P., Jaffee E.M., Goggins M., Maitra A., Iacobuzio-Donahue C., Eshleman J.R., Kern S.E., Hruban R.H., Karchin R., Papadopoulos N., Parmigiani G., Vogelstein B., Velculescu V.E., Kinzler K.W. Core signaling pathways in human pancreatic cancers revealed by global genomic analyses. Science. 2008;321:1801–1806. doi: 10.1126/science.1164368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kant S., Kumar A., Singh S.M. Fatty acid synthase inhibitor orlistat induces apoptosis in T cell lymphoma: role of cell survival regulatory molecules. Biochim. Biophys. Acta. 2012;1820:1764–1773. doi: 10.1016/j.bbagen.2012.07.010. [DOI] [PubMed] [Google Scholar]
- Kridel S.J. Orlistat is a novel inhibitor of fatty acid synthase with antitumor activity. Cancer Res. 2004;64:2070–2075. doi: 10.1158/0008-5472.can-03-3645. [DOI] [PubMed] [Google Scholar]
- Lasserre R., Guo X.J., Conchonaud F., Hamon Y., Hawchar O., Bernard A.M., Soudja S.M., Lenne P.F., Rigneault H., Olive D., Bismuth G., Nunes J.A., Payrastre B., Marguet D., He H.T. Raft nanodomains contribute to Akt/PKB plasma membrane recruitment and activation. Nat. Chem. Biol. 2008;4:538–547. doi: 10.1038/nchembio.103. [DOI] [PubMed] [Google Scholar]
- Lingwood D., Simons K. Lipid rafts as a membrane-organizing principle. Science. 2010;327:46–50. doi: 10.1126/science.1174621. [DOI] [PubMed] [Google Scholar]
- Liu H., Liu J.Y., Wu X., Zhang J.T. Biochemistry, molecular biology, and pharmacology of fatty acid synthase, an emerging therapeutic target and diagnosis/prognosis marker. Int. J. Biochem. Mol. Biol. 2010;1:69–89. [PMC free article] [PubMed] [Google Scholar]
- Lupu R., Menendez J.A. Targeting fatty acid synthase in breast and endometrial cancer: an alternative to selective estrogen receptor modulators? Endocrinology. 2006;147:4056–4066. doi: 10.1210/en.2006-0486. [DOI] [PubMed] [Google Scholar]
- Maier T., Jenni S., Ban N. Architecture of mammalian fatty acid synthase at 4.5 A resolution. Science. 2006;311:1258–1262. doi: 10.1126/science.1123248. [DOI] [PubMed] [Google Scholar]
- Menendez J.A., Lupu R. Fatty acid synthase and the lipogenic phenotype in cancer pathogenesis. Nat. Rev. Cancer. 2007;7:763–777. doi: 10.1038/nrc2222. [DOI] [PubMed] [Google Scholar]
- Nguyen P.L., Ma J., Chavarro J.E., Freedman M.L., Lis R., Fedele G., Fiore C., Qiu W., Fiorentino M., Finn S., Penney K.L., Eisenstein A., Schumacher F.R., Mucci L.A., Stampfer M.J., Giovannucci E., Loda M. Fatty acid synthase polymorphisms, tumor expression, body mass index, prostate cancer risk, and survival. J. Clin. Oncol. 2010;28:3958–3964. doi: 10.1200/JCO.2009.27.0793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Notarnicola M., Tutino V., Calvani M., Lorusso D., Guerra V., Caruso M.G. Serum levels of fatty acid synthase in colorectal cancer patients are associated with tumor stage. J. Gastrointest. Cancer. 2012;43:508–511. doi: 10.1007/s12029-011-9300-2. [DOI] [PubMed] [Google Scholar]
- Oslob J.D., Johnson R.J., Cai H., Feng S.Q., Hu L., Kosaka Y., Lai J., Sivaraja M., Tep S., Yang H., Zaharia C.A., Evanchik M.J., McDowell R.S. Imidazopyridine-based fatty acid synthase inhibitors that show anti-HCV activity and in vivo target modulation. ACS Med. Chem. Lett. 2013;4:113–117. doi: 10.1021/ml300335r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oslob J.D.M., Robert S., Johnson Russell, Yang Hanbiao, Evanchik Marc, Zaharia Cristiana A., Cai Haiying, Hu Lily W. 2014. (Acylaryl)imidazoles as modulators of lipid synthesis and their preparation WO 2014008197 A1 20140109. [Google Scholar]
- Puig T., Vazquez-Martin A., Relat J., Petriz J., Menendez J.A., Porta R., Casals G., Marrero P.F., Haro D., Brunet J., Colomer R. Fatty acid metabolism in breast cancer cells: differential inhibitory effects of epigallocatechin gallate (EGCG) and C75. Breast Cancer Res. Treat. 2008;109:471–479. doi: 10.1007/s10549-007-9678-5. [DOI] [PubMed] [Google Scholar]
- Puig T., Turrado C., Benhamu B., Aguilar H., Relat J., Ortega-Gutierrez S., Casals G., Marrero P.F., Urruticoechea A., Haro D., Lopez-Rodriguez M.L., Colomer R. Novel inhibitors of fatty acid synthase with anticancer activity. Clin. Cancer Res. 2009;15:7608–7615. doi: 10.1158/1078-0432.CCR-09-0856. [DOI] [PubMed] [Google Scholar]
- Puig T., Aguilar H., Cufi S., Oliveras G., Turrado C., Ortega-Gutierrez S., Benhamu B., Lopez-Rodriguez M.L., Urruticoechea A., Colomer R. A novel inhibitor of fatty acid synthase shows activity against HER2 + breast cancer xenografts and is active in anti-HER2 drug-resistant cell lines. Breast Cancer Res. 2011;13:R131. doi: 10.1186/bcr3077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reis-Sobreiro M., Roue G., Moros A., Gajate C., De La Iglesia-Vicente J., Colomer D., Mollinedo F. Lipid raft-mediated Akt signaling as a therapeutic target in mantle cell lymphoma. Blood Cancer J. 2013;3:e118. doi: 10.1038/bcj.2013.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sebastiani V., Botti C., Di Tondo U., Visca P., Pizzuti L., Santeusanio G., Alo P.L. Tissue microarray analysis of FAS, Bcl-2, Bcl-x, ER, PgR, Hsp60, p53 and Her2-neu in breast carcinoma. Anticancer Res. 2006;26:2983–2987. [PubMed] [Google Scholar]
- Shackelford D.B., Shaw R.J. The LKB1–AMPK pathway: metabolism and growth control in tumour suppression. Nat. Rev. Cancer. 2009;9:563–575. doi: 10.1038/nrc2676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shah U.S., Dhir R., Gollin S.M., Chandran U.R., Lewis D., Acquafondata M., Pflug B.R. Fatty acid synthase gene overexpression and copy number gain in prostate adenocarcinoma. Hum. Pathol. 2006;37:401–409. doi: 10.1016/j.humpath.2005.11.022. [DOI] [PubMed] [Google Scholar]
- Shearn C.T., Mercer K.E., Orlicky D.J., Hennings L., Smathers-McCullough R.L., Stiles B.L., Ronis M.J., Petersen D.R. Short term feeding of a high fat diet exerts an additive effect on hepatocellular damage and steatosis in liver-specific PTEN knockout mice. PLoS One. 2014;9:e96553. doi: 10.1371/journal.pone.0096553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simons K., Sampaio J.L. Membrane organization and lipid rafts. Cold Spring Harb. Perspect. Biol. 2011;3:a004697. doi: 10.1101/cshperspect.a004697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Son J., Lyssiotis C.A., Ying H., Wang X., Hua S., Ligorio M., Perera R.M., Ferrone C.R., Mullarky E., Shyh-Chang N., Kang Y., Fleming J.B., Bardeesy N., Asara J.M., Haigis M.C., Depinho R.A., Cantley L.C., Kimmelman A.C. Glutamine supports pancreatic cancer growth through a KRAS-regulated metabolic pathway. Nature. 2013;496:101–105. doi: 10.1038/nature12040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song S.P., Hennig A., Schubert K., Markwart R., Schmidt P., Prior I.A., Bohmer F.D., Rubio I. Ras palmitoylation is necessary for N-Ras activation and signal propagation in growth factor signalling. Biochem. J. 2013;454:323–332. doi: 10.1042/BJ20121799. [DOI] [PubMed] [Google Scholar]
- Staubach S., Hanisch F.G. Lipid rafts: signaling and sorting platforms of cells and their roles in cancer. Expert Rev. Proteomics. 2011;8:263–277. doi: 10.1586/epr.11.2. [DOI] [PubMed] [Google Scholar]
- Tao B.B., He H., Shi X.H., Wang C.L., Li W.Q., Li B., Dong Y., Hu G.H., Hou L.J., Luo C., Chen J.X., Chen H.R., Yu Y.H., Sun Q.F., Lu Y.C. Up-regulation of USP2a and FASN in gliomas correlates strongly with glioma grade. J. Clin. Neurosci. 2013;20:717–720. doi: 10.1016/j.jocn.2012.03.050. [DOI] [PubMed] [Google Scholar]
- Tomek K., Wagner R., Varga F., Singer C.F., Karlic H., Grunt T.W. Blockade of fatty acid synthase induces ubiquitination and degradation of phosphoinositide-3-kinase signaling proteins in ovarian cancer. Mol. Cancer Res. 2011;9:1767–1779. doi: 10.1158/1541-7786.MCR-10-0467. [DOI] [PubMed] [Google Scholar]
- Ueda S.M., Yap K.L., Davidson B., Tian Y., Murthy V., Wang T.L., Visvanathan K., Kuhajda F.P., Bristow R.E., Zhang H., Shih Ie M. Expression of fatty acid synthase depends on NAC1 and is associated with recurrent ovarian serous carcinomas. J. Oncol. 2010;2010:285191. doi: 10.1155/2010/285191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van De Sande T., Roskams T., Lerut E., Joniau S., Van Poppel H., Verhoeven G., Swinnen J.V. High-level expression of fatty acid synthase in human prostate cancer tissues is linked to activation and nuclear localization of Akt/PKB. J. Pathol. 2005;206:214–219. doi: 10.1002/path.1760. [DOI] [PubMed] [Google Scholar]
- Vivanco I., Sawyers C.L. The phosphatidylinositol 3–kinase AKT pathway in human cancer. Nat. Rev. Cancer. 2002;2:489–501. doi: 10.1038/nrc839. [DOI] [PubMed] [Google Scholar]
- Ward P.S., Thompson C.B. Metabolic reprogramming: a cancer hallmark even Warburg did not anticipate. Cancer Cell. 2012;21:297–308. doi: 10.1016/j.ccr.2012.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Witkiewicz A.K., Nguyen K.H., Dasgupta A., Kennedy E.P., Yeo C.J., Lisanti M.P., Brody J.R. Co-expression of fatty acid synthase and caveolin-1 in pancreatic ductal adenocarcinoma: implications for tumor progression and clinical outcome. Cell Cycle. 2008;7:3021–3025. doi: 10.4161/cc.7.19.6719. [DOI] [PubMed] [Google Scholar]
- Yachida S., Jones S., Bozic I., Antal T., Leary R., Fu B., Kamiyama M., Hruban R.H., Eshleman J.R., Nowak M.A., Velculescu V.E., Kinzler K.W., Vogelstein B., Iacobuzio-Donahue C.A. Distant metastasis occurs late during the genetic evolution of pancreatic cancer. Nature. 2010;467:1114–1117. doi: 10.1038/nature09515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ying H., Kimmelman A.C., Lyssiotis C.A., Hua S., Chu G.C., Fletcher-Sananikone E., Locasale J.W., Son J., Zhang H., Coloff J.L., Yan H., Wang W., Chen S., Viale A., Zheng H., Paik J.H., Lim C., Guimaraes A.R., Martin E.S., Chang J., Hezel A.F., Perry S.R., Hu J., Gan B., Xiao Y., Asara J.M., Weissleder R., Wang Y.A., Chin L., Cantley L.C., Depinho R.A. Oncogenic Kras maintains pancreatic tumors through regulation of anabolic glucose metabolism. Cell. 2012;149:656–670. doi: 10.1016/j.cell.2012.01.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaytseva Y.Y., Rychahou P.G., Gulhati P., Elliott V.A., Mustain W.C., O'Connor K., Morris A.J., Sunkara M., Weiss H.L., Lee E.Y., Evers B.M. Inhibition of fatty acid synthase attenuates CD44-associated signaling and reduces metastasis in colorectal cancer. Cancer Res. 2012;72:1504–1517. doi: 10.1158/0008-5472.CAN-11-4057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu G., Wang Y., Huang B., Liang J., Ding Y., Xu A., Wu W. A Rac1/PAK1 cascade controls beta-catenin activation in colon cancer cells. Oncogene. 2012;31:1001–1012. doi: 10.1038/onc.2011.294. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental materials and methods.
Table 1: PANC-1 cell differentially expressed gene list.
Table 2: 22Rv1 cell differentially expressed gene list.
PANC-1 Affymetrix gene expression data: NCBI-GEO accession number: GSE64337.