Abstract
With the development of genomics and bioinformatics, especially the extensive applications of high-throughput sequencing technology, more transcriptional units with little or no protein-coding potential have been discovered. Such RNA molecules are called non-protein-coding RNAs (npcRNAs or ncRNAs). Among them, long npcRNAs or ncRNAs (lnpcRNAs or lncRNAs) represent diverse classes of transcripts longer than 200 nucleotides. In recent years, the lncRNAs have been considered as important regulators in many essential biological processes. In plants, although a large number of lncRNA transcripts have been predicted and identified in few species, our current knowledge of their biological functions is still limited. Here, we have summarized recent studies on their identification, characteristics, classification, bioinformatics, resources, and current exploration of their biological functions in plants.
Keywords: Long non-coding RNA (lncRNA), Long non-coding natural antisense transcripts (lncNATs), Epigenetic, Small RNA, MicroRNA, Target mimicry
Introduction
As a class of RNAs that have no or little protein-coding potential, the mechanism underlying the functions of non-protein coding RNAs (ncRNAs or npcRNAs) is a fascinating area of research [1]. The recent wide applications of the high-throughput RNA-sequencing (RNA-seq) approaches have facilitated the identification of thousands of novel ncRNAs (or npcRNAs) in many organisms, such as humans, animals, and plants [2–6]. The ncRNAs are a heterogeneous group of RNA molecules, which can be classified in different ways according to their location, length, and biological functions [1,7–10].
The canonical ncRNAs such as ribosomal RNAs (rRNAs) and transfer RNAs (tRNAs) were discovered earlier owing to their important functions in protein synthesis in all living organisms. Small RNAs (sRNAs), for instance, small nucleolar and small nuclear RNAs (snoRNAs and snRNAs) are found in specific cellular locations, which can function through modification of other RNAs (e.g., rRNAs and tRNAs) and processing of pre-mRNA [5]. Besides RNAs with specific functions, other ncRNAs are mainly classified based on the length of their mature products. Small ncRNAs of 20−30 nucleotides (nt) in length are mainly microRNAs (miRNAs) and small interfering RNAs (siRNAs), commonly found as transcriptional and translational regulators [11]. Medium ncRNAs of 50−200 nt in length and long ncRNAs (lncRNAs) with size beyond 200 nt are usually involved in other processes, such as splicing, gene inactivation, and translation [1,10,12,13]. To date, the best-characterized ncRNAs are sRNAs [14,15].
As mentioned above, lncRNAs are arbitrarily defined as RNA transcripts that contain > 200 nt but lack protein-coding potential [16]. lncRNAs are transcribed by RNA polymerase II or III, and additionally, by polymerase IV/V in plants [17–19]. They are processed by splicing or non-splicing, polyadenylation or non-polyadenylation, and can be located in the nucleus or cytoplasm. Functional analyses of lncRNAs have shown that they are potent cis- and trans-regulators of gene transcription, and act as scaffolds for chromatin-modifying complexes. As potent regulatory components involved in gene regulation from various aspects, lncRNAs can exert their effects during tissue development and in response to external stimuli [20]. lncRNAs are classified primarily based on four major features, namely, genomic location, functions exerted on DNA or RNA, functioning mechanisms, and targeting mechanisms [12].
Although lncRNAs have received more attention in recent years, the research in this field is still in its infancy. Thus far, only a few lncRNAs have been sufficiently described [21–23]. In particular, research in this area in plants is far behind that in humans and animals [9,24,25]. Nonetheless, studies available suggest that plant lncRNAs exert regulatory functions similar to those in animals [9,24]. In order to gain a better understanding of recent progress in the research of plant lncRNAs, we provide a brief overview on their discovery and functional analyses in the following context.
Discovery of lncRNAs in plants
Novel ncRNAs can be detected and discovered by both experimental and computational screenings [26]. Genome-wide approaches used for transcriptomic analyses such as microarrays and RNA sequencing in model organisms have revealed that non-protein coding transcripts occupy most of the eukaryote transcriptome, much higher than that previously believed [7,27–33]. Especially, next-generation sequencing (NGS)-based technology provides us with a more complex perspective and a much closer and complete view of the RNA world. lncRNAs have been discovered in yeast and other higher eukaryotes [34–36]. For instance, genome-wide analyses have discovered more than 50,000 lncRNAs in the human genome [35,37–39].
About 6480 lncRNAs were identified from 200 Arabidopsis thaliana transcriptomic data sets, with either organ-specific or stress-induced expression profiles [7]. Wang et al. discovered 37,238 long non-coding natural antisense transcripts (lncNATs) in A. thaliana, with antisense transcripts associated with 70% of annotated mRNAs [28]. Using a strand-specific RNA sequencing approach, Zhu et al. [40] identified lncRNAs in A. thaliana induced by Fusarium oxysporum infection. Results showed that antisense transcripts existed in about 20% of the annotated genes, and most newly-identified transcriptionally-active regions (TARs) were adjacent to or located as an extension of the annotated genes. Besides poly(A)+ lncRNAs, lncRNAs without poly(A) tails (poly(A)− lncRNAs) were also identified in humans [41]. In plants, the presence of poly(A)− lncRNAs was revealed in seedlings of A. thaliana under different stress conditions using RNA-seq [42]. Compared to poly(A)+ lncRNAs, poly(A)− lncRNAs are shorter, have lower expression, and are more specific in response to stresses.
Combining both computational and experimental analyses, Xin et al. [43] identified 125 putative stress responsive lncRNAs in wheat. These lncRNAs were tissue-specific and can be induced by powdery mildew infection and heat stress. lncRNAs were also reported in maize. Li et al. [27] identified 20,163 putative lncRNAs in maize by integrating the available EST data, annotated information of maize genome, and RNA-seq datasets obtained from 30 different experiments. By comparing these putative lncRNAs to a comprehensive set of maize sRNAs, they found that more than 90% of these lncRNAs are potential precursors of sRNAs, while only 1704 are high-confidence lncRNAs. It is of note that half of the high-confidence lncRNAs were tissue specific, as supported by the tissue-specific H3K27me3 heterochromatin epigenetic mark. In addition, Zhang et al. [44] performed strand-specific RNA sequencing of rice anthers, pistils, seeds, and shoots. In combination with the analysis of other available rice RNA-seq datasets, they systematically identified 2224 lncRNAs from rice and showed that rice lncRNAs were highly tissue-specific or stage-specific. Studies integrating strand-specific RNA sequencing and sRNA sequencing data were also reported in detecting NATs in rice under normal and different stress conditions. In total 2292 putative cis-NATs were shown to be expressed, among which 503 cis-NATs were expressed under specific conditions [45]. In addition, sRNAs were also detected from their corresponding overlapping regions.
Besides lncRNAs identified in the model plant Arabidopsis, rice, and maize, Qi et al. identified 584 lncRNAs that were responsive to simulated drought stress in foxtail millet by using a deep transcriptomic sequencing approach [46]. Ye et al. identified many endogenous target mimics (eTM, a class of lncRNAs which are complementary to miRNAs as decoy RNAs to prevent miRNAs from binding to their authentic targets) and phased siRNA (phasiRNA, phased secondary siRNAs which function in trans to suppress the expression of target transcripts)-producing loci (PHAS) genes in soybean [47]. They found that miRNAs potentially regulate lipid metabolism-related genes and trigger the production of phasiRNAs from PHAS genes, although some of these miRNAs can be further regulated by eTMs [47]. Additional efforts to identify more novel lncRNAs have also been exerted in other plants such as peach [48], populus [49,50], and Brassica rapa [51] by employing RNA sequencing strategy.
Other than direct transcriptomics analysis, chromatin signature-based approach was also used to define TARs. K4–K36 domain is usually used to define TARs, since active promoter that is marked by H3K4me3 usually combines with TARs that are marked by H3K36me3. In humans and mice, many lncRNAs were identified by the presence of K4–K36 domains in the intergenic regions [4,35]. However, this approach has only been adopted in Arabidopsis among model plants. Information on chromatin states should be obtained in other model plants and crops to assist the identification of TARs in the future [52,53].
Plant lncRNAs as precursors of miRNAs and other sRNAs
As an emerging class of riboregulators, lncRNAs either act directly or are processed to shorter ncRNAs for functioning [54]. Some lncRNAs are primary transcripts of small regulatory RNAs such as miRNAs and siRNAs. Similar to protein-coding genes and some lncRNAs, primary transcripts of miRNA (pri-miRNA) genes are transcribed by RNA polymerase II (Pol II) [55]. In contrast to vertebrates and flies, miRNAs in plants are minor constituents because plants have more complex small regulatory RNA pools. Such complexity of sRNA pools in plants can be exemplified by the presence of plant-specific RNA polymerase IV/V (Pol IV/Pol V)-dependent siRNAs and secondary endogenous siRNAs [56]. Biogenesis pathway of the Pol IV/Pol V-dependent siRNAs also produces a plant-specific class of lncRNAs called the Pol IV/V-dependent lncRNAs, which are required for RNA-directed DNA methylation (RdDM) [20].
Analysis of the full-length cDNA databases led to the identification of numerous 24-nt siRNAs that were matched with five lncRNAs including npc34, npc351, npc375, npc520, and npc523 in Arabidopsis. Most siRNAs derived from these five lncRNAs are mapped to both strands of the lncRNA region, suggesting that these lncRNAs are siRNA precursors [1]. Mapping sRNA present in databases [57] to the complete collection of 76 lncRNAs [54] revealed that 34 lncRNAs are potential precursors of sRNAs. For example, miRNAs miR869a and miR160c mature from npc83 and npc521, respectively. Based on sRNA sequencing and degradome sequencing data in Arabidopsis, Ma et al. [58] identified 43 regions that have the potential to form highly-complementary long-stem structures, which can be potentially recognized by Dicer-like 1 (DCL1) for further cropping, suggesting that these regions may function as sRNA precursors [58]. It is noteworthy that Lauressergues et al. recently discovered that peptides can be encoded by the non-coding regions of miRNA precursors, indicating that some lncRNAs may still possess coding potential [59,60].
Plant lncRNAs as miRNA target mimics
Target mimicry was first found in plants, rising as a novel mechanism for regulating miRNA functions [61]. During target mimicry, interactions between miRNAs and their authentic targets are blocked by the binding of decoy RNAs to miRNAs via partially-complementary sequences [61,62]. Recently, competing endogenous RNAs (ceRNAs) with similar mechanisms were also identified in human and animal cells, indicating that inhibition of miRNA activity by target mimicry may be a widespread phenomenon [38,63,64].
As an endogenous lncRNA, Induced by Phosphate Starvation 1 (IPS1) was first identified in A. thaliana, which functioned as an eTM of miR399 [61,62]. Pairing with a three-nucleotide bulge, IPS1 binds to miR399 and destroys the miR399-mediated cleavage of its target genes. Thus, IPS1 interferes with the binding of ath-miR399 to its authentic targets as a decoy [61]. Genome-wide analyses have identified some candidate eTMs in several plant species with completely-sequenced genomes [65–67]. However, most predictions of eTMs were mainly performed on annotated genes. eTMs for 20 miRNAs conserved in Arabidopsis and rice were systematically identified in intergenic or non-coding gene regions by Wu and colleagues [62]. They show that different eTMs can bind to the same miRNA and the binding sites were well conserved among eTMs, while sequences flanking the miRNA binding sites varied a lot. Using agroinfiltration-based transient expression assay, they identified the important regulatory roles of functional target mimics for miR160 and miR166 in plant development and validated the effectiveness of eTMs for three other miRNAs including ath-miR156, ath-miR159, and ath-miR172 [62].
Target mimicry effects can be induced by both endogenous and engineered artificial miRNA TMs [62,68,69]. Therefore, in addition to their important biological significance, discovery of miRNA target mimics has provided an alternative method for functional studies on miRNAs. For instance, artificial TMs imported into transgenic plants were capable of attenuating the functions of corresponding miRNAs [65,68,70].
Plant lncRNAs and vernalization
Flowering time is one of the most important adaptive traits to ensure the transition of reproductive growth and development that occurs under favorable conditions during a plant’s life cycle [71]. Vernalization is an important mechanism controlling flowering in some plant species that grow in a vegetative state during the cold winter seasons and begin to flower in the warmer spring [72,73]. Vernalization is the best-studied regulatory process in plants that is known to involve lncRNAs, primarily in the regulation of FLOWERING LOCUS C (FLC) gene [74,75].
FLC is a key regulator of flowering time in A. thaliana [76], which acts as a repressor to inhibit flowering under cold temperature [76]. FLC gene is located at a complex locus. Recent studies have shown that at least two types of lncRNAs are present in this locus. A group of long antisense RNAs, called COLD INDUCED LONG ANTISENSE INTRAGENIC RNAs (COOLAIR) are transcribed in antisense orientation in relation to FLC [23,77,78], whereas another lncRNA COLD ASSISTED INTRONIC NONCODING RNA (COLDAIR), is transcribed from the intron of FLC gene in the sense orientation [22]. Both lncRNAs can help recruit PHD-PRC2 complex to enable histone modifications of FLC via epigenetic regulation. Considering that there have been several excellent review articles on this topic [79–81], we do not go into too much detail in this paper.
Plant lncRNAs and fertility
Rice is an important crop as well as an important model organism. Breeding of hybrid rice is one of the evolutionary applications of heterosis in agriculture and male sterility lines are essential for this process. However, little is known about the regulatory genes and molecular mechanisms underlying plant male sterility. Ding et al. and Zhou et al. [82,83] cloned the gene controlling photoperiod-sensitive genetic male sterility (PSMF) independently, and found that the cloned gene was a lncRNA. However, the action modes of this gene are not consistent in the two studies. Ding et al. [82] suggested that sufficient amount of the lncRNA, which they termed as long day (LD)-specific male-fertility-associated RNA (LDMAR), is necessary for rice fertility under LD conditions. The transcription level of LDMAR is reduced specifically under LD conditions, which results in programmed cell death (PCD) during rice anther development and causes male sterility [82]. They also identified a spontaneous point mutation, which led to alteration in RNA secondary structure and increased DNA methylation in its promoter region. Further investigation [84] by the same group showed that an siRNA, Psi–LDMAR, is produced in the promoter region of LDMAR. Overexpression of Psi–LDMAR induced RdDM in the promoter region of LDMAR and resulted in reduced expression of LDMAR. Zhou et al. [83] found that P/TMS12-1 (another name for LDMAR) encodes a unique ncRNA, which produces a 21-nt sRNA, osa-smR5864w. A C-to-G point mutation present in osa-smR5864w may lead to loss-of-function of the sRNA, eventually resulting in the production of light- and temperature-sensitive male sterile rice [83]. The different explanations of the action mechanisms between the two groups illustrate the complex functions of lncRNAs. Therefore, further detailed mechanistic study is required.
Plant lncRNAs and photomorphogenesis
Light is regarded as an important ecological factor to regulate almost all processes of growth and development in plants [85]. The mechanistic study of photomorphogenesis is one of the hotspots in plant molecular biology. The sophisticated regulatory processes of photomorphogenesis have been thoroughly elucidated, and many important mechanisms are well understood at the molecular level. However, the regulatory factors identified so far were mainly proteins. The involvement of lncRNAs in this process is yet to be explored and is an interesting area of research.
As aforementioned, Wang et al. had identified genome-wide lncNATs in model plant A. thaliana [28]. They focused on the roles of lncRNAs in response to light and identified 626 concordant and 766 discordant NAT pairs in A. thaliana, with many light-responsive lncNATs related to histone modifications. It would be very interesting to explore the functions of lncRNAs in phototropic responses. Deng et al. [86] identified and functionally characterized a novel 236-nt lncRNA, HIDDEN TREASURE 1 (HID1), which is involved in the sophisticated photomorphogenic process. By screening their T-DNA insertion mutant collection, they identified a mutant named hid1 later on, which exhibits a hypo-photomorphogenic phenotype under continuous red light (cR). The mutant results from the loss-of-function of the lncRNA gene HID1. Through detailed analyses, the authors discovered that HID1 may function by regulating the expression of the transcription factor, PHYTOCHROME-INTERACTING FACTOR 3 (PIF3), one of the key repressors in photomorphogenesis that modulates light response [86]. Genetic analyses showed that HID1 could negatively regulate the expression of PIF3 gene. HID1 is located in the nucleus and can associate with chromatin and may bind directly to the promoter region of PIF3 to repress its expression [86]. It is of note that HID1 homologs are found in many plant species [86] and may possess conserved functions in different species. For instance, OsHID1, the rice homolog, can rescue the phenotype of hid1 mutant in A. thaliana [86]. HID1 is the first known lncRNA involved in photomorphogenesis, shedding light on the association of ncRNAs and light response in plants [86].
Plant lncRNAs and phosphate homeostasis
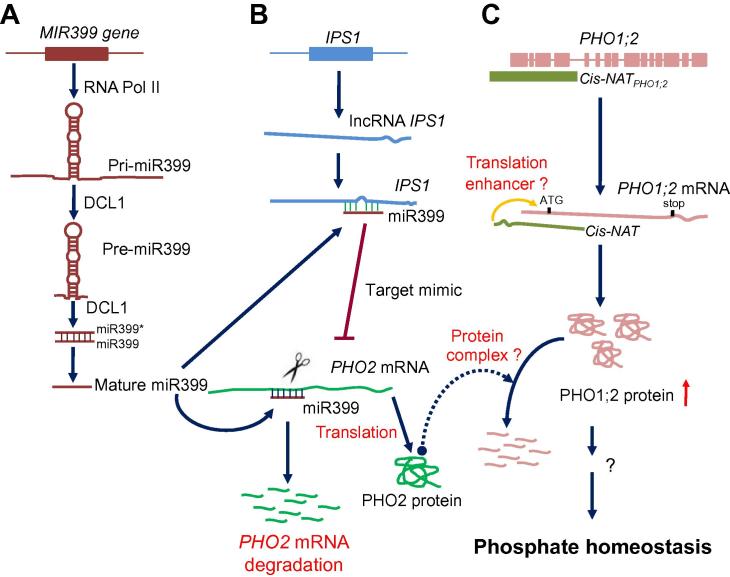
Phosphate is an essential mineral nutrient for plant growth and development [87,88]. Several lines of evidence have suggested that lncRNAs are involved in the phosphate homeostasis [61,89]. First, some miRNAs have been reported to exert effects in regulating phosphate homeostasis [88,90,91]. The well-studied miR399 [91,92] can suppress the expression of its target gene, PHOSPHATE2 (PHO2), which encodes a ubiquitin-conjugating E2 enzyme (Figure 1A). PHO2 can interact with PHOSPHATE1 (PHO1), a membrane protein involved in phosphate loading to the xylem and a key regulator for phosphate homeostasis conserved in plants [93], to control phosphate homeostasis [92]. Since plant miRNAs are mainly encoded by lncRNAs [10,15], involvement of miRNAs in phosphate homeostasis is indicative of the involvement of lncRNAs in phosphate homeostasis. In addition, the aforementioned eTM-type lncRNAs IPS1 exemplifies the direct involvement of lncRNAs in phosphate homeostasis [61]. IPS1 is induced under phosphate deficiency and acts as a target mimic for miR399 (Figure 1B) [61]. Jabnoune et al. reported another layer of regulation in plants. They found that in rice, the cis-natural antisense RNA, cis-NATPHO1;2, can act as a translational enhancer for the expression of its sense gene, PHOSPHATE1;2 (PHO1;2) (Figure 1C) [89], the functional ortholog of PHO1 in Arabidopsis [94]. These findings reveal that there exists complex RNA regulatory network to control phosphate homeostasis in plants. Other lncRNAs related to phosphate homeostasis in tomato and rice are listed in Table 1, together with other important lncRNAs reported in plants [95–99].
Figure 1.
The lncRNA-related regulatory networks for phosphate homeostasis in plants
A. miR399 can suppress the expression of its target gene, PHOSPHATE2 (PHO2), to control phosphate homeostasis [91,92]. miR399 is encoded by MIR399 genes, for which primary transcripts are lncRNAs. B. lncRNA IPS1 is induced under phosphate deficiency and acts as a target mimic for miR399 to regulate the phosphate homeostasis [61]. C.Cis-natural antisense RNA, cis-NATPHO1;2, can act as a translational enhancer for the expression of its sense gene, PHOSPHATE1;2 (PHO1;2), to control phosphate homeostasis in rice [89].
Table 1.
Summary of the lncRNAs reported in plants
| Name | Species | Biological function | Regulation mechanism | Refs. |
|---|---|---|---|---|
| COLDAIR | Arabidopsis (Arabidopsis thaliana) | Flowering time | Histone modification | [22] |
| COOLAIR | Arabidopsis (Arabidopsis thaliana) | Flowering time | Promoter interference | [23,77,78] |
| LDMAR (P/TMS12-1) | Rice (Oryza sativa) | Fertility | Promoter methylation | [82–84] |
| HID1 | Arabidopsis (Arabidopsis thaliana) | Photomorphogenesis | Chromatin association | [86] |
| IPS1 | Arabidopsis (Arabidopsis thaliana) | Phosphate homeostasis | Target mimicry | [61] |
| Cis-NATPHO1;2 | Rice (Oryza sativa) | Phosphate homeostasis | Translational enhancer | [89] |
| OsPI1 | Rice (Oryza sativa) | Phosphate homeostasis | Unknown | [95] |
| TPS11 | Tomato (Solanum lycopersicum) | Phosphate homeostasis | Unknown | [96] |
| asHSFB2a | Arabidopsis (Arabidopsis thaliana) | Vegetative and gametophytic development | Antisense transcription | [97] |
| HvCesA6 lnc-NAT | Barley (Hordeum vulgare) | Cell-wall synthesis | siRNA precursor | [98] |
| SHO lnc-NAT | Petunia (Petunia hybrida) | Local cytokinin synthesis | dsRNA degradation | [99] |
| GmENOD40 | Soybean (Glycine max) | Nodule formation | Protein re-localization | [102] |
| OsENOD40 | Rice (Oryza sativa) | Nodule formation | Unknown | [104] |
| MtENOD40 | Barrel medic (Medicago truncatula) | Nodule formation | Protein re-localization | [107] |
| ASCO-lncRNA | Arabidopsis (Arabidopsis thaliana) | Lateral root development | Alternative splicing regulators | [111] |
| APOLO | Arabidopsis (Arabidopsis thaliana) | Auxin-controlled development | Chromatin loop dynamics | [114] |
Note: COLDAIR, cold assisted intronic noncoding RNA; COOLAIR, cold induced long antisense intragenic RNAs; LDMAR, long day-specific male-fertility-associated RNA; HID1, hidden treasure 1; IPS1, induced by phosphate starvation 1; PHO1;2, PHOSPHATE1;2; PI1, phosphate-limitation inducible gene 1; OsPI1, Oryza sativa phosphate-limitation inducible gene 1; TPS11, tomato phosphate starvation-induced gene; asHSFB2a, natural long non-coding antisense RNA of heat stress transcription factor B; CesA6 lncNAT, natural antisense of CesA6 cellulose synthase gene; SHO, an enzyme responsible for the synthesis of plant cytokinins; ENOD40, early nodulin 40; ASCO, alternative splicing competitor; APOLO, auxin-regulated promoter loop.
Plant lncRNAs and protein re-localization
Early nodulin 40 (ENOD40) [100] is conserved in legumes [101,102] and is found in several non-legume species [103,104] as well. ENOD40 participates in the regulation of symbiosis between bacteria or fungi and leguminous plants [104,105]. During symbiotic interaction, ENOD40 expression is rapidly induced by rhizobia in the nodule primordium [106]. Although the underlying molecular mechanisms are not clear, ENOD40 can play roles in transporting metabolites necessary for cell growth in non-symbiotic plants [105].
ENOD40 encodes two short peptides but there lacks long open reading frame [107,108]. ENOD40 exerts its biological activity directly by translation of these two short peptides in barrel medic (Medicago truncatula) [107], while in soybean, the two peptides of ENOD40 bind specifically to sucrose synthase, suggesting its role in sucrose utilization [108]. Five conserved domains in ENOD40 mRNA are found in various leguminous and non-leguminous species. Notably, one structural domain conserved in Enod40 is similar to the expansion segments in some structural RNAs [109]. Structural elements of ENOD40 mRNA are much more conserved than the encoded short peptides, which suggest that the RNA structure determines the principal functions of ENOD40, whereas more diverse functions, revealed in a minority of plant families, are exerted by short peptides. This hypothesis has been proved in M. truncatula [110].
M. truncatula RNA-binding protein 1 (MtRBP1) directly interacts with ENOD40 in mature nodules, where ENOD40 is expressed at high levels [110]. MtRBP1 re-localizes from nuclear speckles to cytoplasmic granules with the aid of ENOD40 during nodulation in M. truncatula. MtRBP1 protein could be found localized in the cytoplasm only when ENOD40 was co-expressed in these cells. Induction of MtRBP1 relocalization was similarly achieved by ENOD40 transcripts with the initial ATG mutated, indicating that ENOD40-encoded peptides are not involved in this activity. Hence, for the re-localization activity of MtRBP1, RNA structures, and not the encoded short peptides of ENOD40, are required.
Plant lncRNAs and alternative splicing
Alternative splicing is an important regulatory layer in gene expression. Multiple variants of proteins or transcripts can be generated from a single gene via alternative splicing, thus increasing the complexity of proteome and transcriptome. Bardou et al. reported the involvement of lncRNA in alternative splicing in Arabidopsis [111]. They found that an lncRNA acts as an alternative splicing competitor (ASCO). The ASCO-lncRNA and the nuclear speckle RNA-binding protein (NSR) could form an alternative splicing regulatory module. AtNSR is mainly expressed in primary and lateral root meristems and regulates development of lateral roots. Transgenic plants overexpressing the ASCO-lncRNA exhibit an altered ability to form lateral roots, which is similar to the phenotypes of the double Atnsr mutants. AtNSRs interact with the ASCO-lncRNA in vivo and affect the splicing patterns of NSR-regulating mRNA targets. It seems that lncRNA can recruit the alternative splicing regulators to modulate the related processes [111,112].
Plant lncRNAs and modulation of chromatin loop dynamics
Dynamic chromatin topology can affect the pattern of gene expression [113]. Ben Amor et al. identified 76 lncRNAs from the Arabidopsis full-length cDNA databases by using bioinformatics approach [1]. Among them, npc34, was renamed as AUXIN REGULATED PROMOTER LOOP (APOLO) in a recent study [114]. This intergenic lncRNA is encoded by a gene located about 5 kb upstream of PINOID (PID) and is transcribed by two RNA polymerases, RNA Pol II and Pol V [114]. It was reported that the dual APOLO transcription could control the chromatin loop dynamics to regulate the promoter activity of the neighbor PID gene [114], which is an important regulator of polar auxin transport [114,115]. The phytohormone auxin regulates expression of APOLO and PID. Exogenous auxin treatment can activate the demethylation of the APOLO−PID genomic region and the chromatin loop encompassing the promoter region of PID. When the loop is opened, RNA Pol II transcribes the two genes and the accumulation of both PID and APOLO RNAs is increased. Then, the APOLO transcripts produced by RNA Pol II gradually recruit the polycomb repressive complex 1 (PRC1) to close the loop. Then the APOLO transcripts produced by RNA Pol V are recruited by ARGONAUTE4 (AGO4) and trigger DNA methylation. Finally, the APOLO lncRNAs-mediated chromatin loop is reformed and PID expression is down-regulated [114]. It seems that the dual lncRNA transcription influencing the local chromatin topology is a new layer of the regulation of gene expression [114,116].
Databases of plant lncRNAs
Mammalian lncRNAs, especially human and mouse lncRNAs, were recorded elaborately in public databases [117,118]. In addition to basic annotation information, the expression level and imprinting information of mammalian lncRNAs were also deposited in specific databases [17,119,120]. Unlike mammalian lncRNAs, lncRNAs identified in plants were not comprehensively and timely recorded in public databases. Currently, only six databases are available for depositing plant lncRNAs. These include TAIR—Arabidopsis gene structure and function annotation [121], PlantNATsDB—a comprehensive database of plant NATs [122], lncRNAdb—a reference database for lncRNAs [123,124], NONCODE—integrative annotation of lncRNAs [125,126], PLncDB—plant lncRNA database [8], and PNRD—a plant ncRNA database [127]. The functions, features, and links to these databases are listed in Table 2. Among these databases, NONCODE and lncRNAdb are comprehensive databases but not specifically designed for recording plant lncRNAs. PlantNATsDB deposits about 2 million NAT pairs from 70 plant species, but providing no genomic view. Although initially designed for plants, PLncDB currently deposits lncRNA information only of Arabidopsis, and aims to contain comprehensive information including genomic, transcriptomic, and epigenomic information related to plant lncRNAs. PNRD aims to provide lncRNAs of 150 plant species and now contains 5571 lncRNAs of A. thaliana, Oryza sativa, Zea mays, and Populus trichocarpa only.
Table 2.
Summary of databases depositing plant lncRNAs
| Name | Main features | Link | Refs. |
|---|---|---|---|
| TAIR | The Arabidopsis Information Resource; serves as a comprehensive data repository; multiple analysis tools available | https://www.arabidopsis.org/ | [121] |
| PlantNATsDB | Plant NATs database; contains NATs of 70 plant species; provides prediction of NATs; deposits networks formed by NATs; GO annotation and gene set analysis available | http://bis.zju.edu.cn/pnatdb/ | [122] |
| lncRNAdb | A reference database for lncRNAs; deposits all known functional lncRNAs and manual annotation information of lncRNAs; sequence analysis tools available | http://www.lncrnadb.org/ | [123,124] |
| NONCODE | An integrated knowledge database of ncRNAs; deposits all kinds of ncRNAs except tRNAs and rRNAs; all sequences information were confirmed manually; provides expression profile of lncRNA genes by graphs; provides an ID conversion tool from RefSeq or Ensembl ID to NONCODE ID and a service of lncRNA identification | http://www.noncode.org/ | [125,126] |
| PLncDB | A plant lncRNA database; currently just contains Arabidopsis lncRNAs; provides genome browser of lncRNAs | http://chualab.rockefeller.edu/gbrowse2/homepage.html | [8] |
| PNRD | A plant ncRNA database; aims to provide information of both sRNAs and lncRNAs for 150 species; multiple analysis tools available | http://structuralbiology.cau.edu.cn/PNRD/ | [127] |
Note: TAIR, The Arabidopsis Information Resource; PlantNATsDB, Plant Natural Antisense Transcripts DataBase; lncRNAdb, A reference database for lncRNAs; NONCODE, An integrated knowledge database of ncRNAs; PLncDB, A plant lncRNA database; PNRD, A plant ncRNA database.
Concluding remarks and future perspectives
The rapid development of high-throughput RNA-seq and related bioinformatics methods provides revolutionary ways for discovering novel lncRNAs [39,128]. In recent years, many more lncRNA transcripts have been identified. Their number and types are far beyond previous expectations. lncRNA studies have become one of the new hotspots in current molecular biology. However, compared to the studies on humans and animals, the research in plants is still premature [25,129].
lncRNAs reported in plant species are limited to only a few model angiosperm plants such as Arabidopsis, rice, maize, wheat, foxtail millet, and soybean [7,13,27,28,40,42–44,46,130]. The task of discovering new plant lncRNAs is still very arduous. In recent years, DNA sequencing of the plant genome has developed rapidly, and genome sequencing data of dozens of plant species have been reported [131]. However, the annotations of most of the plant species lack information of lncRNAs. With the improved quality of plant genomic sequences, discovery of new lncRNAs will be more thorough and convenient. Many novel lncRNAs will be identified in plants with the increasingly-sophisticated high-throughput sequencing technology, especially strand-specific RNA-seq technology [132].
Functional studies on plant lncRNAs are a challenge. As described in this review, the current functional studies in plants are confined to a few cases [22,86,89]. Compared to protein-coding genes, mutants corresponding to lncRNAs are rare and not easy to be identified, which poses difficulties for functional studies. Systematic discovery and identification of mutant plants will help resolve the biological functions of lncRNAs. A recent endeavor in functional identification and prediction of novel lncRNAs in rice on a large scale by screening of a mutant library has been a promising example [44]. Besides, the traditional reverse genetics, such as over-expression and RNAi, as well as the lately popular CRISPR/cas9 genome editing technology [133], may also play their roles in promoting functional analysis.
Competing interests
The authors have declared that no competing interests exist.
Acknowledgments
We thank other members of the Zhu’s and Hu’s labs for their valuable discussion. We apologize to all colleagues whose work contributed to our understanding of plant lncRNAs but could not be cited here only due to the space constraints. We thank Dr. Jun Fang (Institute of Genetics and Developmental Biology, Chinese Academy of Sciences) for his critical reading and helpful discussion. We are also thankful to the reviewers for their important advice. This work was supported by the China Postdoctoral Science Foundation (Grant No. 2013M530694 to XL) and the National Natural Science Foundation of China (Grant Nos. 31271385 to SH, 31100915 to LH, and 31123007 to LZ). This work was also supported by the State Key Laboratory of Plant Genomics of China (Grant No. 2015B0129-03).
Handled by Ming Chen
Footnotes
Peer review under responsibility of Beijing Institute of Genomics, Chinese Academy of Sciences and Genetics Society of China.
Contributor Information
Lihuang Zhu, Email: lhzhu@genetics.ac.cn.
Songnian Hu, Email: husn@big.ac.cn.
References
- 1.Ben Amor B., Wirth S., Merchan F., Laporte P., d’Aubenton-Carafa Y., Hirsch J. Novel long non-protein coding RNAs involved in Arabidopsis differentiation and stress responses. Genome Res. 2009;19:57–69. doi: 10.1101/gr.080275.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ravasi T., Suzuki H., Pang K.C., Katayama S., Furuno M., Okunishi R. Experimental validation of the regulated expression of large numbers of non-coding RNAs from the mouse genome. Genome Res. 2006;16:11–19. doi: 10.1101/gr.4200206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Consortium E.P., Birney E., Stamatoyannopoulos J.A., Dutta A., Guigo R., Gingeras T.R. Identification and analysis of functional elements in 1% of the human genome by the ENCODE pilot project. Nature. 2007;447:799–816. doi: 10.1038/nature05874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Guttman M., Amit I., Garber M., French C., Lin M.F., Feldser D. Chromatin signature reveals over a thousand highly conserved large non-coding RNAs in mammals. Nature. 2009;458:223–227. doi: 10.1038/nature07672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Matera A.G., Terns R.M., Terns M.P. Non-coding RNAs: lessons from the small nuclear and small nucleolar RNAs. Nat Rev Mol Cell Biol. 2007;8:209–220. doi: 10.1038/nrm2124. [DOI] [PubMed] [Google Scholar]
- 6.Ponting C.P., Oliver P.L., Reik W. Evolution and functions of long noncoding RNAs. Cell. 2009;136:629–641. doi: 10.1016/j.cell.2009.02.006. [DOI] [PubMed] [Google Scholar]
- 7.Liu J., Jung C., Xu J., Wang H., Deng S., Bernad L. Genome-wide analysis uncovers regulation of long intergenic noncoding RNAs in Arabidopsis. Plant Cell. 2012;24:4333–4345. doi: 10.1105/tpc.112.102855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jin J., Liu J., Wang H., Wong L., Chua N.H. PLncDB: plant long non-coding RNA database. Bioinformatics. 2013;29:1068–1071. doi: 10.1093/bioinformatics/btt107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhu Q.H., Wang M.B. Molecular functions of long non-coding RNAs in plants. Genes (Basel) 2012;3:176–190. doi: 10.3390/genes3010176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Costa F.F. Non-coding RNAs: new players in eukaryotic biology. Gene. 2005;357:83–94. doi: 10.1016/j.gene.2005.06.019. [DOI] [PubMed] [Google Scholar]
- 11.Gomes A.Q., Nolasco S., Soares H. Non-coding RNAs: multi-tasking molecules in the cell. Int J Mol Sci. 2013;14:16010–16039. doi: 10.3390/ijms140816010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ma L., Bajic V.B., Zhang Z. On the classification of long non-coding RNAs. RNA Biol. 2013;10:925–933. doi: 10.4161/rna.24604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Liu T.T., Zhu D., Chen W., Deng W., He H., He G. A global identification and analysis of small nucleolar RNAs and possible intermediate-sized non-coding RNAs in Oryza sativa. Mol Plant. 2013;6:830–846. doi: 10.1093/mp/sss087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zamore P.D., Haley B. Ribo-gnome: the big world of small RNAs. Science. 2005;309:1519–1524. doi: 10.1126/science.1111444. [DOI] [PubMed] [Google Scholar]
- 15.Bonnet E., Van de Peer Y., Rouze P. The small RNA world of plants. New Phytol. 2006;171:451–468. doi: 10.1111/j.1469-8137.2006.01806.x. [DOI] [PubMed] [Google Scholar]
- 16.Ng S.Y., Lin L., Soh B.S., Stanton L.W. Long noncoding RNAs in development and disease of the central nervous system. Trends Genet. 2013;29:461–468. doi: 10.1016/j.tig.2013.03.002. [DOI] [PubMed] [Google Scholar]
- 17.Dinger M.E., Pang K.C., Mercer T.R., Crowe M.L., Grimmond S.M., Mattick J.S. NRED: a database of long noncoding RNA expression. Nucleic Acids Res. 2009;37:D122–D126. doi: 10.1093/nar/gkn617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dinger M.E., Pang K.C., Mercer T.R., Mattick J.S. Differentiating protein-coding and noncoding RNA: challenges and ambiguities. PLoS Comput Biol. 2008;4:e1000176. doi: 10.1371/journal.pcbi.1000176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wierzbicki A.T., Haag J.R., Pikaard C.S. Noncoding transcription by RNA polymerase Pol IVb/Pol V mediates transcriptional silencing of overlapping and adjacent genes. Cell. 2008;135:635–648. doi: 10.1016/j.cell.2008.09.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kim E.D., Sung S. Long noncoding RNA: unveiling hidden layer of gene regulatory networks. Trends Plant Sci. 2012;17:16–21. doi: 10.1016/j.tplants.2011.10.008. [DOI] [PubMed] [Google Scholar]
- 21.Rinn J.L., Kertesz M., Wang J.K., Squazzo S.L., Xu X., Brugmann S.A. Functional demarcation of active and silent chromatin domains in human HOX loci by noncoding RNAs. Cell. 2007;129:1311–1323. doi: 10.1016/j.cell.2007.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Heo J.B., Sung S. Vernalization-mediated epigenetic silencing by a long intronic noncoding RNA. Science. 2011;331:76–79. doi: 10.1126/science.1197349. [DOI] [PubMed] [Google Scholar]
- 23.Swiezewski S., Liu F., Magusin A., Dean C. Cold-induced silencing by long antisense transcripts of an Arabidopsis polycomb target. Nature. 2009;462:799–802. doi: 10.1038/nature08618. [DOI] [PubMed] [Google Scholar]
- 24.Zhang Y.C., Chen Y.Q. Long noncoding RNAs: new regulators in plant development. Biochem Biophys Res Commun. 2013;436:111–114. doi: 10.1016/j.bbrc.2013.05.086. [DOI] [PubMed] [Google Scholar]
- 25.Bai Y., Dai X., Harrison A.P., Chen M. RNA regulatory networks in animals and plants: a long noncoding RNA perspective. Brief Funct Genomics. 2015;14:91–101. doi: 10.1093/bfgp/elu017. [DOI] [PubMed] [Google Scholar]
- 26.Huttenhofer A., Brosius J., Bachellerie J.P. RNomics: identification and function of small, non-messenger RNAs. Curr Opin Chem Biol. 2002;6:835–843. doi: 10.1016/s1367-5931(02)00397-6. [DOI] [PubMed] [Google Scholar]
- 27.Li L., Eichten S.R., Shimizu R., Petsch K., Yeh C.T., Wu W. Genome-wide discovery and characterization of maize long non-coding RNAs. Genome Biol. 2014;15:R40. doi: 10.1186/gb-2014-15-2-r40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wang H., Chung P.J., Liu J., Jang I.C., Kean M.J., Xu J. Genome-wide identification of long noncoding natural antisense transcripts and their responses to light in Arabidopsis. Genome Res. 2014;24:444–453. doi: 10.1101/gr.165555.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Okazaki Y., Furuno M., Kasukawa T., Adachi J., Bono H., Kondo S. Analysis of the mouse transcriptome based on functional annotation of 60,770 full-length cDNAs. Nature. 2002;420:563–573. doi: 10.1038/nature01266. [DOI] [PubMed] [Google Scholar]
- 30.Numata K., Kanai A., Saito R., Kondo S., Adachi J., Wilming L.G. Identification of putative noncoding RNAs among the RIKEN mouse full-length cDNA collection. Genome Res. 2003;13:1301–1306. doi: 10.1101/gr.1011603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ota T., Suzuki Y., Nishikawa T., Otsuki T., Sugiyama T., Irie R. Complete sequencing and characterization of 21,243 full-length human cDNAs. Nat Genet. 2004;36:40–45. doi: 10.1038/ng1285. [DOI] [PubMed] [Google Scholar]
- 32.Rinn J.L., Euskirchen G., Bertone P., Martone R., Luscombe N.M., Hartman S. The transcriptional activity of human Chromosome 22. Genes Dev. 2003;17:529–540. doi: 10.1101/gad.1055203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chekanova J.A., Gregory B.D., Reverdatto S.V., Chen H., Kumar R., Hooker T. Genome-wide high-resolution mapping of exosome substrates reveals hidden features in the Arabidopsis transcriptome. Cell. 2007;131:1340–1353. doi: 10.1016/j.cell.2007.10.056. [DOI] [PubMed] [Google Scholar]
- 34.Bumgarner S.L., Dowell R.D., Grisafi P., Gifford D.K., Fink G.R. Toggle involving cis-interfering noncoding RNAs controls variegated gene expression in yeast. Proc Natl Acad Sci U S A. 2009;106:18321–18326. doi: 10.1073/pnas.0909641106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Khalil A.M., Guttman M., Huarte M., Garber M., Raj A., Rivea Morales D. Many human large intergenic noncoding RNAs associate with chromatin-modifying complexes and affect gene expression. Proc Natl Acad Sci U S A. 2009;106:11667–11672. doi: 10.1073/pnas.0904715106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ulitsky I., Shkumatava A., Jan C.H., Sive H., Bartel D.P. Conserved function of lincRNAs in vertebrate embryonic development despite rapid sequence evolution. Cell. 2011;147:1537–1550. doi: 10.1016/j.cell.2011.11.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chen L.L., Carmichael G.G. Long noncoding RNAs in mammalian cells: what, where, and why? Wiley Interdiscip Rev RNA. 2010;1:2–21. doi: 10.1002/wrna.5. [DOI] [PubMed] [Google Scholar]
- 38.Cabili M.N., Trapnell C., Goff L., Koziol M., Tazon-Vega B., Regev A. Integrative annotation of human large intergenic noncoding RNAs reveals global properties and specific subclasses. Genes Dev. 2011;25:1915–1927. doi: 10.1101/gad.17446611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Iyer M.K., Niknafs Y.S., Malik R., Singhal U., Sahu A., Hosono Y. The landscape of long noncoding RNAs in the human transcriptome. Nat Genet. 2015;47:199–208. doi: 10.1038/ng.3192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhu Q.H., Stephen S., Taylor J., Helliwell C.A., Wang M.B. Long noncoding RNAs responsive to Fusarium oxysporum infection in Arabidopsis thaliana. New Phytol. 2014;201:574–584. doi: 10.1111/nph.12537. [DOI] [PubMed] [Google Scholar]
- 41.Yang L., Duff M.O., Graveley B.R., Carmichael G.G., Chen L.L. Genomewide characterization of non-polyadenylated RNAs. Genome Biol. 2011;12:R16. doi: 10.1186/gb-2011-12-2-r16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Di C., Yuan J., Wu Y., Li J., Lin H., Hu L. Characterization of stress-responsive lncRNAs in Arabidopsis thaliana by integrating expression, epigenetic and structural features. Plant J. 2014;80:848–861. doi: 10.1111/tpj.12679. [DOI] [PubMed] [Google Scholar]
- 43.Xin M., Wang Y., Yao Y., Song N., Hu Z., Qin D. Identification and characterization of wheat long non-protein coding RNAs responsive to powdery mildew infection and heat stress by using microarray analysis and SBS sequencing. BMC Plant Biol. 2011;11:61. doi: 10.1186/1471-2229-11-61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhang Y.C., Liao J.Y., Li Z.Y., Yu Y., Zhang J.P., Li Q.F. Genome-wide screening and functional analysis identify a large number of long noncoding RNAs involved in the sexual reproduction of rice. Genome Biol. 2014;15:512. doi: 10.1186/s13059-014-0512-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lu T., Zhu C., Lu G., Guo Y., Zhou Y., Zhang Z. Strand-specific RNA-seq reveals widespread occurrence of novel cis-natural antisense transcripts in rice. BMC Genomics. 2012;13:721. doi: 10.1186/1471-2164-13-721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Qi X., Xie S., Liu Y., Yi F., Yu J. Genome-wide annotation of genes and noncoding RNAs of foxtail millet in response to simulated drought stress by deep sequencing. Plant Mol Biol. 2013;83:459–473. doi: 10.1007/s11103-013-0104-6. [DOI] [PubMed] [Google Scholar]
- 47.Ye C.Y., Xu H., Shen E., Liu Y., Wang Y., Shen Y. Genome-wide identification of non-coding RNAs interacted with microRNAs in soybean. Front Plant Sci. 2014;5:743. doi: 10.3389/fpls.2014.00743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wang L., Zhao S., Gu C., Zhou Y., Zhou H., Ma J. Deep RNA-Seq uncovers the peach transcriptome landscape. Plant Mol Biol. 2013;83:365–377. doi: 10.1007/s11103-013-0093-5. [DOI] [PubMed] [Google Scholar]
- 49.Chen J., Quan M., Zhang D. Genome-wide identification of novel long non-coding RNAs in Populus tomentosa tension wood, opposite wood and normal wood xylem by RNA-seq. Planta. 2015;241:125–143. doi: 10.1007/s00425-014-2168-1. [DOI] [PubMed] [Google Scholar]
- 50.Shuai P., Liang D., Tang S., Zhang Z., Ye C.Y., Su Y. Genome-wide identification and functional prediction of novel and drought-responsive lincRNAs in Populus trichocarpa. J Exp Bot. 2014;65:4975–4983. doi: 10.1093/jxb/eru256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Yu X., Yang J., Li X.R., Liu X.X., Sun C.B., Wu F.J. Global analysis of cis-natural antisense transcripts and their heat-responsive nat-siRNAs in Brassica rapa. BMC Plant Biol. 2013;13:208. doi: 10.1186/1471-2229-13-208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zhang X., Bernatavichute Y.V., Cokus S., Pellegrini M., Jacobsen S.E. Genome-wide analysis of mono-, di- and trimethylation of histone H3 lysine 4 in Arabidopsis thaliana. Genome Biol. 2009;10:R62. doi: 10.1186/gb-2009-10-6-r62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Roudier F., Ahmed I., Berard C., Sarazin A., Mary-Huard T., Cortijo S. Integrative epigenomic mapping defines four main chromatin states in Arabidopsis. EMBO J. 2011;30:1928–1938. doi: 10.1038/emboj.2011.103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Hirsch J., Lefort V., Vankersschaver M., Boualem A., Lucas A., Thermes C. Characterization of 43 non-protein-coding mRNA genes in Arabidopsis, including the MIR162a-derived transcripts. Plant Physiol. 2006;140:1192–1204. doi: 10.1104/pp.105.073817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Lee Y., Kim M., Han J., Yeom K.H., Lee S., Baek S.H. MicroRNA genes are transcribed by RNA polymerase II. EMBO J. 2004;23:4051–4060. doi: 10.1038/sj.emboj.7600385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Chen X.M. Small RNAs and their roles in plant development. Annu Rev Cell Dev Biol. 2009;25:21–44. doi: 10.1146/annurev.cellbio.042308.113417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Rajagopalan R., Vaucheret H., Trejo J., Bartel D.P. A diverse and evolutionarily fluid set of microRNAs in Arabidopsis thaliana. Genes Dev. 2006;20:3407–3425. doi: 10.1101/gad.1476406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ma X., Shao C., Jin Y., Wang H., Meng Y. Long non-coding RNAs: a novel endogenous source for the generation of Dicer-like 1-dependent small RNAs in Arabidopsis thaliana. RNA Biol. 2014;11:373–390. doi: 10.4161/rna.28725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Waterhouse P.M., Hellens R.P. Plant biology: Coding in non-coding RNAs. Nature. 2015;520:41–42. doi: 10.1038/nature14378. [DOI] [PubMed] [Google Scholar]
- 60.Lauressergues D., Couzigou J.M., Clemente H.S., Martinez Y., Dunand C., Becard G. Primary transcripts of microRNAs encode regulatory peptides. Nature. 2015;520:90–93. doi: 10.1038/nature14346. [DOI] [PubMed] [Google Scholar]
- 61.Franco-Zorrilla J.M., Valli A., Todesco M., Mateos I., Puga M.I., Rubio-Somoza I. Target mimicry provides a new mechanism for regulation of microRNA activity. Nat Genet. 2007;39:1033–1037. doi: 10.1038/ng2079. [DOI] [PubMed] [Google Scholar]
- 62.Wu H.J., Wang Z.M., Wang M., Wang X.J. Widespread long noncoding RNAs as endogenous target mimics for microRNAs in plants. Plant Physiol. 2013;161:1875–1884. doi: 10.1104/pp.113.215962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Rubio-Somoza I., Weigel D. MicroRNA networks and developmental plasticity in plants. Trends Plant Sci. 2011;16:258–264. doi: 10.1016/j.tplants.2011.03.001. [DOI] [PubMed] [Google Scholar]
- 64.Salmena L., Poliseno L., Tay Y., Kats L., Pandolfi P.P. A ceRNA hypothesis: the Rosetta stone of a hidden RNA language? Cell. 2011;146:353–358. doi: 10.1016/j.cell.2011.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Ivashuta S., Banks I.R., Wiggins B.E., Zhang Y., Ziegler T.E., Roberts J.K. Regulation of gene expression in plants through miRNA inactivation. PLoS One. 2011;6:e21330. doi: 10.1371/journal.pone.0021330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Banks I.R., Zhang Y., Wiggins B.E., Heck G.R., Ivashuta S. RNA decoys: an emerging component of plant regulatory networks? Plant Signal Behav. 2012;7:1188–1193. doi: 10.4161/psb.21299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Meng Y., Shao C., Wang H., Jin Y. Target mimics: an embedded layer of microRNA-involved gene regulatory networks in plants. BMC Genomics. 2012;13:197. doi: 10.1186/1471-2164-13-197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Todesco M., Rubio-Somoza I., Paz-Ares J., Weigel D. A collection of target mimics for comprehensive analysis of microRNA function in Arabidopsis thaliana. PLoS Genet. 2010;6:e1001031. doi: 10.1371/journal.pgen.1001031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Eamens A.L., Wang M.B. Alternate approaches to repress endogenous microRNA activity in Arabidopsis thaliana. Plant Signal Behav. 2011;6:349–359. doi: 10.4161/psb.6.3.14340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Yan J., Gu Y., Jia X., Kang W., Pan S., Tang X. Effective small RNA destruction by the expression of a short tandem target mimic in Arabidopsis. Plant Cell. 2012;24:415–427. doi: 10.1105/tpc.111.094144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Song Y.H., Ito S., Imaizumi T. Flowering time regulation: photoperiod- and temperature-sensing in leaves. Trends Plant Sci. 2013;18:575–583. doi: 10.1016/j.tplants.2013.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Song J., Angel A., Howard M., Dean C. Vernalization—a cold-induced epigenetic switch. J Cell Sci. 2012;125:3723–3731. doi: 10.1242/jcs.084764. [DOI] [PubMed] [Google Scholar]
- 73.Amasino R.M. Vernalization and flowering time. Curr Opin Biotechnol. 2005;16:154–158. doi: 10.1016/j.copbio.2005.02.004. [DOI] [PubMed] [Google Scholar]
- 74.He Y. Chromatin regulation of flowering. Trends Plant Sci. 2012;17:556–562. doi: 10.1016/j.tplants.2012.05.001. [DOI] [PubMed] [Google Scholar]
- 75.Heo J.B., Sung S. Encoding memory of winter by noncoding RNAs. Epigenetics. 2011;6:544–547. doi: 10.4161/epi.6.5.15235. [DOI] [PubMed] [Google Scholar]
- 76.Michaels S.D., Amasino R.M. FLOWERING LOCUS C encodes a novel MADS domain protein that acts as a repressor of flowering. Plant Cell. 1999;11:949–956. doi: 10.1105/tpc.11.5.949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Sun Q.W., Csorba T., Skourti-Stathaki K., Proudfoot N.J., Dean C. R-loop stabilization represses antisense transcription at the Arabidopsis FLC locus. Science. 2013;340:619–621. doi: 10.1126/science.1234848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Liu F., Marquardt S., Lister C., Swiezewski S., Dean C. Targeted 3’ processing of antisense transcripts triggers Arabidopsis FLC chromatin silencing. Science. 2010;327:94–97. doi: 10.1126/science.1180278. [DOI] [PubMed] [Google Scholar]
- 79.Gitschier J. How cool is that: an interview with Caroline Dean. PLoS Genet. 2013;9:e1003593. doi: 10.1371/journal.pgen.1003593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.De Lucia F., Dean C. Long non-coding RNAs and chromatin regulation. Curr Opin Plant Biol. 2011;14:168–173. doi: 10.1016/j.pbi.2010.11.006. [DOI] [PubMed] [Google Scholar]
- 81.Crevillen P., Dean C. Regulation of the floral repressor gene FLC: the complexity of transcription in a chromatin context. Curr Opin Plant Biol. 2011;14:38–44. doi: 10.1016/j.pbi.2010.08.015. [DOI] [PubMed] [Google Scholar]
- 82.Ding J., Lu Q., Ouyang Y., Mao H., Zhang P., Yao J. A long noncoding RNA regulates photoperiod-sensitive male sterility, an essential component of hybrid rice. Proc Natl Acad Sci U S A. 2012;109:2654–2659. doi: 10.1073/pnas.1121374109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Zhou H., Liu Q., Li J., Jiang D., Zhou L., Wu P. Photoperiod- and thermo-sensitive genic male sterility in rice are caused by a point mutation in a novel noncoding RNA that produces a small RNA. Cell Res. 2012;22:649–660. doi: 10.1038/cr.2012.28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Ding J., Shen J., Mao H., Xie W., Li X., Zhang Q. RNA-directed DNA methylation is involved in regulating photoperiod-sensitive male sterility in rice. Mol Plant. 2012;5:1210–1216. doi: 10.1093/mp/sss095. [DOI] [PubMed] [Google Scholar]
- 85.Jiao Y., Lau O.S., Deng X.W. Light-regulated transcriptional networks in higher plants. Nat Rev Genet. 2007;8:217–230. doi: 10.1038/nrg2049. [DOI] [PubMed] [Google Scholar]
- 86.Wang Y., Fan X., Lin F., He G., Terzaghi W., Zhu D. Arabidopsis noncoding RNA mediates control of photomorphogenesis by red light. Proc Natl Acad Sci U S A. 2014;111:10359–10364. doi: 10.1073/pnas.1409457111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Lv Q.D., Zhong Y.J., Wang Y.G., Zhang L., Shi J., Wu Z.C. SPX4 negatively regulates phosphate signaling and homeostasis through its interaction with PHR2 in rice. Plant Cell. 2014;26:1586–1597. doi: 10.1105/tpc.114.123208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Secco D., Jabnoune M., Walker H., Shou H., Wu P., Poirier Y. Spatio-temporal transcript profiling of rice roots and shoots in response to phosphate starvation and recovery. Plant Cell. 2013;25:4285–4304. doi: 10.1105/tpc.113.117325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Jabnoune M., Secco D., Lecampion C., Robaglia C., Shu Q.Y., Poirier Y. A rice cis-natural antisense RNA acts as a translational enhancer for its cognate mRNA and contributes to phosphate homeostasis and plant fitness. Plant Cell. 2013;25:4166–4182. doi: 10.1105/tpc.113.116251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Lin W.Y., Huang T.K., Chiou T.J. Nitrogen limitation adaptation, a target of microRNA827, mediates degradation of plasma membrane-localized phosphate transporters to maintain phosphate homeostasis in Arabidopsis. Plant Cell. 2013;25:4061–4074. doi: 10.1105/tpc.113.116012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Chiou T.J., Aung K., Lin S.I., Wu C.C., Chiang S.F., Su C.L. Regulation of phosphate homeostasis by microRNA in Arabidopsis. Plant Cell. 2006;18:412–421. doi: 10.1105/tpc.105.038943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Bari R. Datt Pant B, Stitt M, Scheible WR. PHO2, microRNA399, and PHR1 define a phosphate-signaling pathway in plants. Plant Physiol. 2006;141:988–999. doi: 10.1104/pp.106.079707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Hamburger D., Rezzonico E. MacDonald-Comber Petetot J, Somerville C, Poirier Y. Identification and characterization of the Arabidopsis PHO1 gene involved in phosphate loading to the xylem. Plant Cell. 2002;14:889–902. doi: 10.1105/tpc.000745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Secco D., Baumann A., Poirier Y. Characterization of the rice PHO1 gene family reveals a key role for OsPHO1;2 in phosphate homeostasis and the evolution of a distinct clade in dicotyledons. Plant Physiol. 2010;152:1693–1704. doi: 10.1104/pp.109.149872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Wasaki J., Yonetani R., Shinano T., Kai M., Osaki M. Expression of the OsPI1 gene, cloned from rice roots using cDNA microarray, rapidly responds to phosphorus status. New Phytol. 2003;158:239–248. [Google Scholar]
- 96.Liu C., Muchhal U.S., Raghothama K.G. Differential expression of TPS11, a phosphate starvation-induced gene in tomato. Plant Mol Biol. 1997;33:867–874. doi: 10.1023/a:1005729309569. [DOI] [PubMed] [Google Scholar]
- 97.Wunderlich M., Gross-Hardt R., Schoffl F. Heat shock factor HSFB2a involved in gametophyte development of Arabidopsis thaliana and its expression is controlled by a heat-inducible long non-coding antisense RNA. Plant Mol Biol. 2014;85:541–550. doi: 10.1007/s11103-014-0202-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Held M.A., Penning B., Brandt A.S., Kessans S.A., Yong W., Scofield S.R. Small-interfering RNAs from natural antisense transcripts derived from a cellulose synthase gene modulate cell wall biosynthesis in barley. Proc Natl Acad Sci U S A. 2008;105:20534–20539. doi: 10.1073/pnas.0809408105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Zubko E., Meyer P. A natural antisense transcript of the Petunia hybrida Sho gene suggests a role for an antisense mechanism in cytokinin regulation. Plant J. 2007;52:1131–1139. doi: 10.1111/j.1365-313X.2007.03309.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Crespi M.D., Jurkevitch E., Poiret M., d’Aubenton-Carafa Y., Petrovics G., Kondorosi E. Enod40, a gene expressed during nodule organogenesis, codes for a non-translatable RNA involved in plant growth. EMBO J. 1994;13:5099–5112. doi: 10.1002/j.1460-2075.1994.tb06839.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Matvienko M., Van de Sande K., Yang W.C., van Kammen A., Bisseling T., Franssen H. Comparison of soybean and pea ENOD40 cDNA clones representing genes expressed during both early and late stages of nodule development. Plant Mol Biol. 1994;26:487–493. doi: 10.1007/BF00039559. [DOI] [PubMed] [Google Scholar]
- 102.Yang W.C., Katinakis P., Hendriks P., Smolders A., de Vries F., Spee J. Characterization of GmENOD40, a gene showing novel patterns of cell-specific expression during soybean nodule development. Plant J. 1993;3:573–585. doi: 10.1046/j.1365-313x.1993.03040573.x. [DOI] [PubMed] [Google Scholar]
- 103.Imaizumi-Anraku H., Kouchi H., Syono K., Akao S., Kawaguchi M. Analysis of ENOD40 expression in alb1, a symbiotic mutant of Lotus japonicus that forms empty nodules with incompletely developed nodule vascular bundles. Mol Gen Genet. 2000;264:402–410. doi: 10.1007/s004380000330. [DOI] [PubMed] [Google Scholar]
- 104.Kouchi H., Takane K., So R.B., Ladha J.K., Reddy P.M. Rice ENOD40: isolation and expression analysis in rice and transgenic soybean root nodules. Plant J. 1999;18:121–129. doi: 10.1046/j.1365-313x.1999.00432.x. [DOI] [PubMed] [Google Scholar]
- 105.Flemetakis E., Kavroulakis N., Quaedvlieg N.E., Spaink H.P., Dimou M., Roussis A. Lotus japonicus contains two distinct ENOD40 genes that are expressed in symbiotic, nonsymbiotic, and embryonic tissues. Mol Plant Microbe Interact. 2000;13:987–994. doi: 10.1094/MPMI.2000.13.9.987. [DOI] [PubMed] [Google Scholar]
- 106.Compaan B., Yang W.C., Bisseling T., Franssen H. ENOD40 expression in the pericycle precedes cortical cell division in Rhizobium-legume interaction and the highly conserved internal region of the gene does not encode a peptide. Plant Soil. 2001;230:1–8. [Google Scholar]
- 107.Sousa C., Johansson C., Charon C., Manyani H., Sautter C., Kondorosi A. Translational and structural requirements of the early nodulin gene enod40, a short-open reading frame-containing RNA, for elicitation of a cell-specific growth response in the alfalfa root cortex. Mol Cell Biol. 2001;21:354–366. doi: 10.1128/MCB.21.1.354-366.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Rohrig H., Schmidt J., Miklashevichs E., Schell J., John M. Soybean ENOD40 encodes two peptides that bind to sucrose synthase. Proc Natl Acad Sci U S A. 2002;99:1915–1920. doi: 10.1073/pnas.022664799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Gultyaev A.P., Roussis A. Identification of conserved secondary structures and expansion segments in enod40 RNAs reveals new enod40 homologues in plants. Nucleic Acids Res. 2007;35:3144–3152. doi: 10.1093/nar/gkm173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Campalans A., Kondorosi A., Crespi M. Enod40, a short open reading frame-containing mRNA, induces cytoplasmic localization of a nuclear RNA binding protein in Medicago truncatula. Plant Cell. 2004;16:1047–1059. doi: 10.1105/tpc.019406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Bardou F., Ariel F., Simpson C.G., Romero-Barrios N., Laporte P., Balzergue S. Long noncoding RNA modulates alternative splicing regulators in Arabidopsis. Dev Cell. 2014;30:166–176. doi: 10.1016/j.devcel.2014.06.017. [DOI] [PubMed] [Google Scholar]
- 112.Kornblihtt A.R. A long noncoding way to alternative splicing in plant development. Dev Cell. 2014;30:117–119. doi: 10.1016/j.devcel.2014.07.010. [DOI] [PubMed] [Google Scholar]
- 113.Cavalli G., Misteli T. Functional implications of genome topology. Nat Struct Mol Biol. 2013;20:290–299. doi: 10.1038/nsmb.2474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Ariel F., Jegu T., Latrasse D., Romero-Barrios N., Christ A., Benhamed M. Noncoding transcription by alternative RNA polymerases dynamically regulates an auxin-driven chromatin loop. Mol Cell. 2014;55:383–396. doi: 10.1016/j.molcel.2014.06.011. [DOI] [PubMed] [Google Scholar]
- 115.Kakar K., Zhang H., Scheres B., Dhonukshe P. CLASP-mediated cortical microtubule organization guides PIN polarization axis. Nature. 2013;495:529–533. doi: 10.1038/nature11980. [DOI] [PubMed] [Google Scholar]
- 116.Ariel F., Romero-Barrios N., Jegu T., Benhamed M., Crespi M. Battles and hijacks: noncoding transcription in plants. Trends Plant Sci. 2015;20:362–371. doi: 10.1016/j.tplants.2015.03.003. [DOI] [PubMed] [Google Scholar]
- 117.Volders P.J., Verheggen K., Menschaert G., Vandepoele K., Martens L., Vandesompele J. An update on LNCipedia: a database for annotated human lncRNA sequences. Nucleic Acids Res. 2015;43:D174–D180. doi: 10.1093/nar/gku1060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Ma L., Li A., Zou D., Xu X., Xia L., Yu J. LncRNAWiki: harnessing community knowledge in collaborative curation of human long non-coding RNAs. Nucleic Acids Res. 2015;43:D187–D192. doi: 10.1093/nar/gku1167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Liao Q., Xiao H., Bu D., Xie C., Miao R., Luo H. NcFANs: a web server for functional annotation of long non-coding RNAs. Nucleic Acids Res. 2011;39:W118–W124. doi: 10.1093/nar/gkr432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Zhang Y., Guan D.G., Yang J.H., Shao P., Zhou H., Qu L.H. NcRNAimprint: a comprehensive database of mammalian imprinted noncoding RNAs. RNA. 2010;16:1889–1901. doi: 10.1261/rna.2226910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Swarbreck D., Wilks C., Lamesch P., Berardini T.Z., Garcia-Hernandez M., Foerster H. The Arabidopsis Information Resource (TAIR): gene structure and function annotation. Nucleic Acids Res. 2008;36:D1009–D1014. doi: 10.1093/nar/gkm965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Chen D., Yuan C., Zhang J., Zhang Z., Bai L., Meng Y. PlantNATsDB: a comprehensive database of plant natural antisense transcripts. Nucleic Acids Res. 2012;40:D1187–D1193. doi: 10.1093/nar/gkr823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Amaral P.P., Clark M.B., Gascoigne D.K., Dinger M.E., Mattick J.S. LncRNAdb: a reference database for long noncoding RNAs. Nucleic Acids Res. 2011;39:D146–D151. doi: 10.1093/nar/gkq1138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Quek X.C., Thomson D.W., Maag J.L., Bartonicek N., Signal B., Clark M.B. lncRNAdb v2.0: expanding the reference database for functional long noncoding RNAs. Nucleic Acids Res. 2015;43:D168–73. doi: 10.1093/nar/gku988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Bu D., Yu K., Sun S., Xie C., Skogerbo G., Miao R. NONCODE v3.0: integrative annotation of long noncoding RNAs. Nucleic Acids Res. 2012;40:D210–D215. doi: 10.1093/nar/gkr1175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Xie C., Yuan J., Li H., Li M., Zhao G., Bu D. NONCODEv4: exploring the world of long non-coding RNA genes. Nucleic Acids Res. 2014;42:D98–D103. doi: 10.1093/nar/gkt1222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Yi X., Zhang Z., Ling Y., Xu W., Su Z. PNRD: a plant non-coding RNA database. Nucleic Acids Res. 2015;43:D982–D989. doi: 10.1093/nar/gku1162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Wang Z., Gerstein M., Snyder M. RNA-Seq: a revolutionary tool for transcriptomics. Nat Rev Genet. 2009;10:57–63. doi: 10.1038/nrg2484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Liu J., Wang H., Chua N.H. Long noncoding RNA transcriptome of plants. Plant Biotechnol J. 2015;13:319–328. doi: 10.1111/pbi.12336. [DOI] [PubMed] [Google Scholar]
- 130.Lertampaiporn S., Thammarongtham C., Nukoolkit C., Kaewkamnerdpong B., Ruengjitchatchawalya M. Identification of non-coding RNAs with a new composite feature in the Hybrid Random Forest Ensemble algorithm. Nucleic Acids Res. 2014;42:e93. doi: 10.1093/nar/gku325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Michael TP, Jackson S. The First 50 Plant Genomes. Plant Genome 2013;6:10.3835/plantgenome2013.03.0001in.
- 132.Levin J.Z., Yassour M., Adiconis X., Nusbaum C., Thompson D.A., Friedman N. Comprehensive comparative analysis of strand-specific RNA sequencing methods. Nat Methods. 2010;7:709–715. doi: 10.1038/nmeth.1491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Shan Q.W., Wang Y.P., Li J., Gao C.X. Genome editing in rice and wheat using the CRISPR/Cas system. Nat Protoc. 2014;9:2395–2410. doi: 10.1038/nprot.2014.157. [DOI] [PubMed] [Google Scholar]