Abstract
Analysis of microarray data is associated with the methodological problems of high dimension and small sample size. Various methods have been used for variable selection in high-dimension and small sample size cases with a single survival endpoint. However, little effort has been directed toward addressing competing risks where there is more than one failure risks. This study compared three typical variable selection techniques including Lasso, elastic net, and likelihood-based boosting for high-dimensional time-to-event data with competing risks. The performance of these methods was evaluated via a simulation study by analyzing a real dataset related to bladder cancer patients using time-dependent receiver operator characteristic (ROC) curve and bootstrap .632+ prediction error curves. The elastic net penalization method was shown to outperform Lasso and boosting. Based on the elastic net, 33 genes out of 1381 genes related to bladder cancer were selected. By fitting to the Fine and Gray model, eight genes were highly significant (P < 0.001). Among them, expression of RTN4, SON, IGF1R, SNRPE, PTGR1, PLEK, and ETFDH was associated with a decrease in survival time, whereas SMARCAD1 expression was associated with an increase in survival time. This study indicates that the elastic net has a higher capacity than the Lasso and boosting for the prediction of survival time in bladder cancer patients. Moreover, genes selected by all methods improved the predictive power of the model based on only clinical variables, indicating the value of information contained in the microarray features.
Keywords: Microarray, Elastic net, Lasso, Competing risks, Subdistribution hazard, Cause-specific hazard
Introduction
Bladder cancer is the fourth most common cancer with more than 350,000 new cases and causing 145,000 deaths worldwide annually [1,2]. Despite improved surgical procedures and aggressive treatments, muscle invasive bladder carcinoma continues to have a high mortality rate [2]. The prevalence of bladder cancer is 3–8 folds higher than its incidence, which makes it one of the most prevalent neoplasms and a major burden for health care systems [1]. Approximately 20%–30% of patients exhibit muscle-invasive (stages T2–T4) or metastatic disease at the time of diagnosis, while about one third of patients with initially non-muscle-invasive disease (stages Ta/T1/Tis) will progress to muscle-invasive or metastatic disease [3,4]. Clinical variables such as stage and grade (high or low) are highly associated with the outcomes and play a substantial role in determining treatment [4]. Despite the important role of these variables in predicting outcome, significant variability remains in the prognosis of patients with analogous characteristics. Due to this discrepancy, it is necessary to gain additional information about tumor characteristics that predict clinical behaviors [4].
The advent of genome-wide transcriptome profiling and advances in experimental technologies in molecular biology have greatly impacted the discovery of new molecular markers or gene expression signatures for classifying and predicting disease outcome in various cancers, including bladder cancer [1]. Analysis of such data is of particular interest in dealing with some types of phenotypic data such as patient survival time or time to cancer relapse, which contain censored observations. In such instances, the main purpose is typically to identify a subset of genes that have significant correlation with time-to-event response [5]. To this end, a major problem arises from the high dimensionality of these data (i.e., the number of genomic variables is usually much larger than the number of subjects), due to the inability to apply standard statistical methods. The microarray time-to-event data become more complicated when there are competing events, such as “progression” versus “death from non-cancer cause”, i.e., the failure of a patient can occur due to one of multiple distinct causes. Furthermore, the influence of one selected variable on different causes might vary [6].
Different variable selection methods have been employed for the analysis of high dimension and small sample size time-to-event data [7–10]. However, given the large number of variables compared to the small sample size of microarray data, there are only a small number of effective genes that can be identified reliably [6]. Therefore, techniques with sparse results are more desirable, where a small amount of non-zero variables were selected as the important ones [6].
To achieve this goal, penalized methods such as least absolute shrinkage and selection operator (Lasso) [11,12] and likelihood-based boosting methods [13] can be applied. However, due to the sparseness of these methods, if there is a group of highly-correlated variables related to the response, typically only one or two of them will receive non-zero estimates and others will be ignored [5,6]. One possible solution of this drawback is the elastic net penalization approach, which may be applied in analyzing microarray time-to-event data [5], since it simultaneously performs automatic variable selection and continuous shrinkage similar to the Lasso method. In addition, the elastic net penalization approach is capable of performing “grouped selection”. Thus it can identify an entire set of correlated genes [5,14], while remaining computationally efficient [14].
These techniques have been used in several studies for a single survival endpoint [13,15,16]. To our knowledge, the only effort in the context of high-dimensional time-to-event data with competing risks was made by Binder et al. [6], who employed a likelihood-based boosting technique for variable selection [6]. However, the performance of other methods like elastic net and Lasso for gene selection remains uninvestigated for competing risks.
In the present study, we aimed to compare the performance of three variable selection methods including Lasso, elastic net, and likelihood-based boosting for analysis of high-dimensional time-to-event data with competing risks based on the commonly-used cause-specific hazard model to predict survival time in patients with bladder cancer. Moreover, we also identified significant genes among those that were selected by the best variable selection method and to determine their effect on the survival time in patients with bladder cancer, according to the subdistribution hazard model.
Results
Bladder cancer data analysis
The cause-specific Cox proportional hazards model was fitted to bladder cancer microarray data by the elastic net and Lasso penalization techniques for the event of interest “progression or death from bladder cancer”. In this regard, the hazard of the patients who progressed or died from bladder cancer was modeled using a Cox model by treating individuals failing from other or unknown causes as censored observations. The results of the component-wise likelihood-based boosting utilized by Binder et al. [6] are also provided for comparison. The genes selected by elastic net and Lasso approaches, along with those identified by Binder et al. [6] are shown in Table 1. The number of selected genes that were significantly (P ⩽ 0.05) associated with progression or death from bladder cancer varied remarkably among the three methods. In general, elastic net identified a greater number of significant genes than Lasso and boosting methods. Elastic net identified 33 genes in total, whereas the Lasso and boosting methods identified 6 and 8 genes, respectively.
Table 1.
Genes selected by three methods for bladder cancer event included in Dyrskjøt dataset
| Gene ID |
Method |
||
|---|---|---|---|
| Elastic net | Lasso | Boosting | |
| SEQ1014 | + | − | − |
| SEQ1038 | − | + | − |
| SEQ1082 | + | − | − |
| SEQ1111 | + | − | − |
| SEQ1164 | + | − | − |
| SEQ1197 | + | − | − |
| SEQ1225 | + | − | − |
| SEQ1226 | + | − | − |
| SEQ1262 | + | − | − |
| SEQ1330 | + | − | − |
| SEQ1381 | + | − | + |
| SEQ1384 | + | − | + |
| SEQ162 | + | − | + |
| SEQ164 | + | + | + |
| SEQ183 | + | − | − |
| SEQ213 | + | − | − |
| SEQ240 | + | − | − |
| SEQ251 | − | + | − |
| SEQ265 | + | − | + |
| SEQ279 | + | − | − |
| SEQ287 | + | − | − |
| SEQ34 | + | − | + |
| SEQ347 | + | + | + |
| SEQ370 | + | + | − |
| SEQ377 | + | − | − |
| SEQ410 | + | − | − |
| SEQ424 | − | + | − |
| SEQ634 | + | − | − |
| SEQ681 | + | − | − |
| SEQ785 | + | − | − |
| SEQ813 | + | − | − |
| SEQ820 | + | − | + |
| SEQ833 | + | − | − |
| SEQ940 | + | − | − |
| SEQ972 | + | − | − |
| SEQ973 | + | − | − |
| No. of genes | 33 | 6 | 8 |
Note: Genes for bladder cancer event listed in Dyrskjøt dataset [1] were selected using three methods. “+” indicates that the gene was selected by the respective method and genes not selected by the respective methods are indicated with “−”.
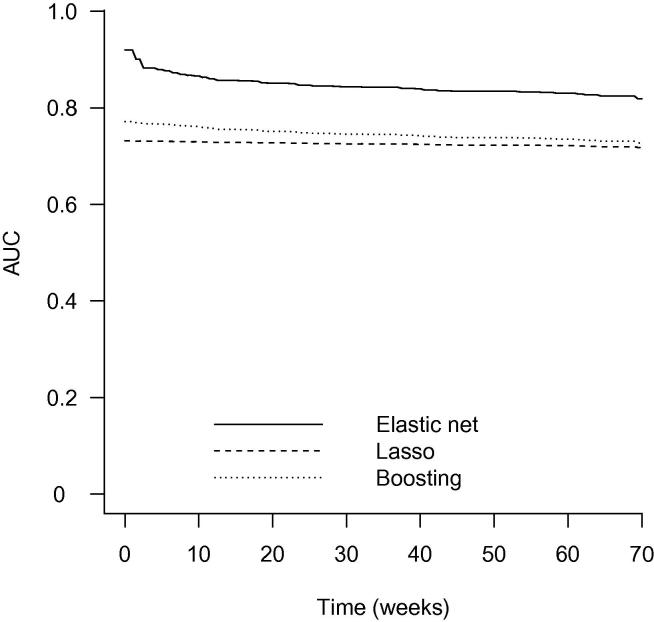
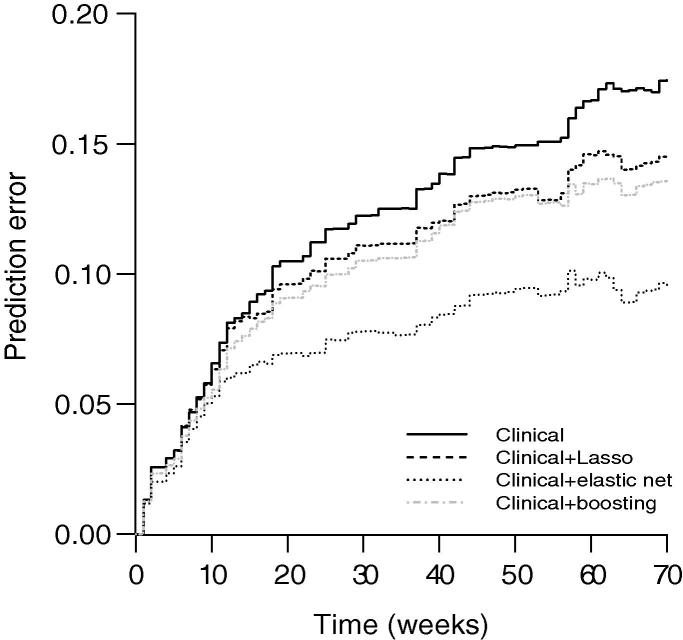
To assess predictive performance, the median area under the curve (AUC) was calculated and plotted for each method. The results are presented in Figure 1. The average median AUC (across all time points) were 0.808, 0.695, and 0.729 for the elastic net, Lasso, and boosting methods, respectively. As shown in Figure 1, in terms of prediction, the predictive performance of elastic net was superior to the Lasso and boosting in the analysis of this dataset, whereas the predictive performance of boosting was slightly better than the Lasso method. Moreover, bootstrap .632+ prediction error curves were plotted for the three methods (Figure 2). The data clearly indicated that the elastic net outperformed the Lasso and boosting methods, which agreed well with the AUC analysis.
Figure 1.
The area under the ROC curve for bladder cancer data
AUC value over time was presented in y-axis, survival time on x-axis was time to progression or death from bladder cancer (in week).
Figure 2.
The prediction error curves for bladder cancer data
Clinical model used age, sex, stage, grade and treatment as predictors. The elastic net, Lasso, and boosting used microarray features in addition to the clinical parameters as predictors.
As a result, 8 out of 33 genes selected by elastic net, including SEQ1082, SEQ1197, SEQ1262, SEQ1330, SEQ162, SEQ377, SEQ634, and SEQ940, were significant based on the Fine and Gray model (P < 0.05). The coefficients, standard errors, hazard ratios, and P values for these genes are listed in Table 2. The survival time increased with the gene expression of SEQ940 (P = 0.004) and decreased with the expression of the remaining seven significant genes.
Table 2.
Genes affecting bladder cancer patients’ survival as selected by elastic net
| Gene ID | GenBank accession No. | Gene symbol | Gene description | Coefficient | Hazard ratio | P value |
|---|---|---|---|---|---|---|
| SEQ1082 | NM_207521.1 | RTN4 | Homo sapiens reticulon 4 | 0.745 ± 0.250 | 2.11 | 0.00290 |
| SEQ1197 | NM_003103.5 | SON | Homo sapiens (human) SON, DNA binding protein | 1.335 ± 0.364 | 3.80 | 0.00024 |
| SEQ1262 | NM_000875.2 | IGF1R | Homo sapiens insulin-like growth factor 1 receptor, mRNA | 1.364 ± 0.510 | 3.85 | 0.00750 |
| SEQ1330 | NM_003094.1 | SNRPE | Homo sapiens small nuclear ribonucleoprotein polypeptide E | 0.789 ± 0.193 | 2.2 | 0.00005 |
| SEQ162 | NM_001146108 | PTGR1 | Homo sapiens prostaglandin reductase 1 | 1.386 ± 0.395 | 3.99 | 0.00045 |
| SEQ377 | NM_002664 | PLEK | Homo sapiens pleckstrin, mRNA | 1.058 ± 0.315 | 2.88 | 0.00078 |
| SEQ634 | NM_004453 | ETFDH | Homo sapiens electron-transferring-flavoprotein dehydrogenase, transcript variant 1, mRNA | 1.400 ± 0.399 | 4.06 | 0.00045 |
| SEQ940 | NM_020159.1 | SMARCAD1 | Homo sapiens SWI/SNF-related, matrix-associated actin-dependent regulator of chromatin, subfamily a, containing DEAD/H box 1, transcript variant 3, mRNA | −1.000 ± 0.348 | 0.37 | 0.00400 |
Note: Genes affecting bladder cancer patients’ survival were selected by elastic net based on Fine and Gray model. Coefficient is indicated as average ± standard error.
Simulation study
In order to evaluate the performance of the three methods, a simulation study was performed. The results of the simulation study with over 100 runs including true positive (TP), false negative (FN), false positive (FP), and true negative (TN) for the three methods of Lasso and elastic net are provided in Table 3. In each run, we simulated a competing risks dataset including two possible failure types (type 1 and type 2), censoring rate of 35%, and p = 5000 covariate with a fixed number of 400 observations. Sixteen informative covariates corresponding to a sparse true model were considered with coefficients equal to 0.5 and −0.5 for increasing and decreasing effects, respectively. In addition, the coefficient of covariates with no direct effect on the hazards were considered zero. The values of performance criteria were computed for the three methods. The results showed that the average number of selected genes by elastic net (53.89 ± 51.85 and 69.28 ± 76.29 for events type 1 and 2, respectively) were greater than those selected by the Lasso and boosting methods (Table 3). In addition, as shown in Table 3, considering either failure type, the proportion of covariates that had no effect but received non-zero parameter estimates (FP) was slightly greater for the elastic net (1.03% for event type 1 and 1.30% for event type 2) due to the greater number of selected genes. On the other hand, with respect to the selection of those covariates with effects on the hazards (TP), the performance of the elastic net was better (31.58% and 31.80% for event 1 and event 2, respectively) than that of Lasso (12.5% and 16.41% for event 1 and event 2, respectively) and boosting (13.58% and 16.67% for event 1 and event 2, respectively). In addition, the results showed that the elastic net tended to select the informative covariates. In the simulation study, sixteen informative variables were selected. The informative covariates were selected from three different blocks of covariates with correlation coefficient of 0.5, 0.35, and 0.05. The first four informative covariates were selected from the first block that the correlation between its variables was 0.5 and the related coefficients were β1, β2, β3, and β4. These four variables had an increasing effect on both event types. The second four informative variables were selected from the second block with the correlation of 0.35 (they had an increasing effect on the first event hazard and a decreasing effect on the second event hazard). Finally, four informative variables were selected from third block that had a decreasing effect on the first event hazard and another four informative variables with an increasing effect on the second event hazard. For example, every time that β1 received a non-zero coefficient, the other three covariates (β2, β3, and β4) were selected. This was also the case for the coefficients from the second block.
Table 3.
Results of simulation study using the three methods
| Event type | No. of selected variables | TP (%) | FN (%) | FP (%) | TN (%) | |
|---|---|---|---|---|---|---|
| Elastic net | 1 | 53.89 ± 0.52 | 31.58 | 68.42 | 1.03 | 98.97 |
| 2 | 69.28 ± 0.76 | 31.80 | 68.20 | 1.30 | 98.67 | |
| Lasso | 1 | 15.16 ± 0.93 | 12.50 | 87.50 | 0.30 | 99.70 |
| 2 | 15.90 ± 0.85 | 16.41 | 83.59 | 0.32 | 99.68 | |
| Boosting | 1 | 23.86 ± 0.12 | 13.58 | 86.42 | 0.58 | 99.42 |
| 2 | 23.90 ± 0.12 | 16.67 | 83.33 | 0.58 | 99.42 |
Note: Type 1 is the first simulated event and type 2 is the competing event. Number of selected variables is indicated as average ± standard error. TP, true positive, the proportion of correctly-included variables; FN, false negative, the proportion of incorrectly-excluded variables; FP, false positive, the proportion of incorrectly-included variables; TN, true negative, the proportion of correctly-excluded variables.
Discussion
This study compared the performance of three variable selection approaches including elastic net, Lasso, and boosting in high dimension and with a small sample size setting with two competing events using real and simulation datasets.
Based on the criteria of AUC and .632+ prediction error curves, the elastic net penalty resulted in higher capability of prediction than the Lasso. The same dataset was also analyzed by Binder et al. [6] using a likelihood-based boosting technique. All their selected genes were also selected by the present study including SEQ34, SEQ162, SEQ164, SEQ265, SEQ347, SEQ820, SEQ1381, and SEQ1384. These data indicate that the performance of the elastic net method was superior to the boosting method. Furthermore, the results of the simulation study indicated that the ability of recovering informative variables was lower in Lasso and boosting methods than in the elastic net method, while the covariates wrongly selected as informative ones by the three methods were fairly similar. In summary, the elastic net exhibited better performance in the simulation study and real dataset implementation, followed by boosting and then Lasso.
As expected, the elastic net penalization method exhibited the grouping effect and identified correlated gene expression while Lasso did not. In this regard, both elastic net and Lasso methods selected SEQ164, while the former method also selected SEQ162, which was highly correlated with gene SEQ164 (ρ = 0.65, P < 0.001). In addition, SEQ972 and SEQ973 were selected by the elastic net method with a high correlation (ρ = 0.85). For the vast majority of the genes selected by the elastic net, the observed correlation was >0.35, whereas the observed correlation between most of genes selected by the Lasso method was lower than 0.30. This was also the case for the boosting method. Due to the sparseness of the boosting and Lasso methods, only one or two of the genes were selected from a group of highly-correlated genes [6,14]. In addition, the results of the simulation study confirmed that the elastic net exhibited the grouping effect in the competing risks setting, while the other two methods did not. This is a substantial property in the analysis of microarray data, because the understanding of the biological pathway may be improved by the identification of an entire set of correlated genes [5].
There was also an overlap with the 88-gene progression classifiers proposed by Dyrskjøt et al. [1] using a univariate Cox regression model. Four genes including SEQ183, SEQ213, SEQ833 and SEQ820 were identified in the present study as well as Dyrskjøt’s study [1].
Based on the Fine and Gray model, 8 out of 33 genes that were selected by the elastic net method were diagnosed as influential genes on bladder cancer survival. Accordingly, the expression of these genes was related to the survival time of patients with bladder cancer. The expression of RTN4, SON, IGF1R, SNRPE, PTGR1, PLEK, and ETFDH appeared related to a decrease in survival time, whereas the expression of SMARCAD1 may be related to an increase in survival time.
Previous studies have shown that RTN4, a myelin-associated endoplasmic reticulum protein, may play a role in apoptosis particularly in cancerous cells [17]. Alternative splicing of genes involved in apoptosis and epigenetic modification can be regulated by SON and its absence will disrupt expression of these genes [18]. Another gene, IGF1R, which encodes insulin-like growth factor 1, plays an important role in regulating cellular proliferation and apoptosis through signaling pathway [19]. Several studies have confirmed that IGF1R is over-expressed in invasive bladder cancer tissues and promotes motility and invasion of urothelial carcinoma cells [20–24]. In addition, over-expression of SNRPE and PLEK could play some important roles in different types of cancers such as prostate, lung, and breast cancers [25–27]. The spliceosome is a dynamic macromolecular ribonucleoprotein (RNP) complex that catalyzes the splicing of nuclear pre-mRNAs into mRNAs. The splicing process plays an important role in the control of expression of a number of genes including those involved in cell cycle, signal transduction, angiogenesis, apoptosis, and invasion [26]. PLEK protein may play a dual role in tumorigenesis and chemoresistance, depending on the tissue specificity [28]. PTGR1 encodes prostaglandin reductase 1, which is a highly-inducible enzyme involved in the inactivation of the chemotactic factor leukotriene B4 [29–31]. The electron-transferring-flavoprotein dehydrogenase (ETFDH) in the inner mitochondrial membrane accepts electrons from ETF. Being an energy pathway gene, ETFDH is overexpressed in different cancers [32]. SMARCAD1 encodes a member of the SNF subfamily of helicase proteins and is involved in restoring heterochromatin organization and propagating epigenetic patterns following DNA replication by mediating histone H3/H4 deacetylation [33].
Our findings were consistent with several previous studies. Engler and Li [5] compared the Cox elastic net and Cox Lasso variable selection methods for a single point survival data in a high-dimensional setting with the AUC criteria over time and relative frequency of variable selection. They found that the elastic net method performed better than the Lasso method with both the correlated and uncorrelated covariates, as shown in the current study. Similarly, Zou and Hastie [14] also showed that the performance of the elastic net in linear regression was superior to the Lasso based on mean square error (MSE) criteria, and Ogutu et al. reported similar accuracies for Lasso and elastic net methods for handling linear regression [34]. In addition, Lin and Lv [35] showed similar performance of Lasso and elastic net penalties based on the additive hazards model with the simulation data. The present study introduced a new set of influential microarray features for predicting bladder cancer survival. According to the results of the present study, the genes selected by the three methods of boosting, Lasso and elastic net the selected genes improved prediction performance over a pure clinical model, which reflects valuable information contained in the microarray features. These results suggest the potential to characterize bladder cancer based on influential gene expression features, and that the expression levels of these genes could be correlated with the patient survival time. Hence, such information can be considered as a prognostic factor in secondary prevention.
This study indicated that the elastic net outperformed the Lasso and boosting methods for the prediction of survival time in patients with bladder cancer in the presence of competing risks. This superiority was also confirmed by a simulation study. Moreover, including microarray features selected by the three methods in the models resulted in improvement over the pure clinical model, indicating that valuable information is contained in the microarray features.
Although the elastic net was shown to perform more efficiently when compared to the Lasso and boosting methods, this method evaluates the effects of genes individually. It is suggested that other methods such as path analysis and random survival forests be utilized to examine the simultaneous effects of genes on each other as well as on the response variable.
Materials and methods
In this study, a public available time-to-event dataset (GSE5479) related to 1381 preprocessed custom platform microarray features extracted from patients with bladder cancer was utilized. This dataset contains information about tumor samples from 404 patients with pTa or pT1 tumors but with no previous or synchronous muscle-invasive tumors, which was used by Dyrskjøt et al. [1] to validate a signature for predicting tumor progression. In addition to gene expression data, this dataset contained information about clinical covariates including age, sex, stage (pTa vs. pT1), grade (PUNLMP/low vs. high), and treatment. Since complete information was available for 301 patients, only this subset of patients was analyzed in the current study. There were two competing events: (a) the event of interest (event 1 or cause 1), which was the time to progression or death from bladder cancer and (b) the competing event (event 2 or cause 2), which was death from other or unknown causes. Progression or death from bladder cancer occurred in 74 patients, death from other or unknown causes was observed in 33 patients, and censoring occurred in 194 patients.
In the case of competing risks, the observations are shown by (ti, Δiεi, Xi), i = 1,…n, where ti is the observed time, is the type of event, xi = (xi1, …, xip) is a vector of covariates, and Δi is the censoring indicator, which takes the value 1 for occurrence of an event and value 0 for occurrence of censoring, i.e., Δ = I (T∗ ⩽ C), where T∗ and C are the event time and the censoring time, respectively [6].
There are different approaches to handle competing risks. The most commonly used method is the cause-specific hazard method, which is utilized in many microarray literatures [36]. We used the cause-specific approach to make the results of the present study comparable with those of the previous studies conducted [1,6] on the same dataset.
Under the Cox proportional hazards (PH) model that considers predictors the cause-specific hazard model is specified as:
| (1) |
where is the coefficient of the i-th predictor and is the unspecified baseline hazard of the event type ε [37].
Also, the Cox PH model for the subdistribution hazard is defined as:
| (2) |
where h1,0(t) is an unspecified baseline, is the instantaneous risk of an event occurring in the absence of competing events [37], and is the cumulative incidence function for event of interest ε = 1 (the expected proportion of patients experiencing event 1 over time). In the present study, the cause-specific approach was used in the gene selection stage. The Fine and Gray model was then utilized to analyze the dataset based on the genes selected by the better variable selection technique of Lasso, elastic net, and boosting.
The Lasso variable selection method [38] is a regularized estimation method for regression models including the Cox PH model. In this method, an L1 norm constraint of is added to the regression coefficients (s is a positive user-specified value and is the coefficient corresponding to the jth covariate) [39]. This constraint shrinks the coefficients toward zero and results in coefficients with values of exactly zero. In general, the Lasso estimate of the vector of regression coefficients in terms of Lagrange multiplier is defined as:
| (3) |
where Rp is a p-dimensional space of covariates, is the Cox log partial likelihood for cause and is defined as , where D denotes the set of indices for observed events of type ε and is the risk set of cause .
The elastic net estimate of is also defined as:
| (4) |
where is the elastic net estimate of the vector of regression coefficients, is the Cox log partial likelihood, and λ1 and λ2 are fixed non-negative values [5]. Due to the strict convexity of the penalty function for 0 < λ2 ⩽ 1, the elastic net method can identify entire sets of highly-correlated variables [5]. In addition, the optimum values of the tuning parameters related to the methods were determined by 10-fold cross validation.
The likelihood-based boosting approach is based on two main parameters: penalty term and number of boosting steps. At each boosting step, only one element of the parameter vector is updated, and the previous boosting steps are included as an offset [6]. In the cause-specific Cox model, the objective function is a penalized partial log likelihood function as follows:
| (5) |
where λ is the penalty parameter, which was selected to avoid boosting steps to be too large, is the parameter vector, and is the corresponding linear predictors [13]. Once is calculated for the best candidate model j∗, the following update is performed:
| (6) |
Evaluation of the predictive performance of the three models using the bladder cancer dataset was performed via time-dependent receiver operator characteristic (ROC) curves and bootstrap .632+ prediction error curves (to assess prediction performance improvement by including selected genes over a pure clinical model) [6]. To obtain AUC over time, 10-fold cross validation was utilized and the average AUC over time was calculated.
Moreover, to evaluate and compare the performance of the boosting, Lasso, and elastic net methods for identifying true important variables, a simulation study was conducted. Competing risks data, in which there is more than one cause of failure with two possible failure types, were simulated. To ensure the comparability of the results with the boosting method [6], a similar strategy was considered to implement the simulation study. Since it was of interest to assess the performance of the methods in high-dimensional settings, p = 5000 covariates were considered following the design employed by Binder et al. [6] to produce correlations with a fixed number of 400 observations. Sixteen informative covariates corresponding to a sparse true model were considered with an effect on the cause-specific hazards for events of type 1 (the first cause of failure from disease) and/or type 2 (the second cause of failure from disease). Each informative covariate was selected from one of three blocks of correlated covariates, where the correlations in blocks were 0.5, 0.35, and 0.05, respectively. The informative covariates were selected so that four covariates had an increasing effect on type 1 and type 2 hazards that were selected from the first block, four other covariates had an increasing effect on the cause-specific hazard for type 1 hazard and a decreasing effect on the type 2 hazard and were selected from the second block, four covariates had a decreasing effect on the event type-1 hazard only, and four other covariates had an increasing effect on the event type-2 hazard that all were selected from the third block. The true coefficient , with , took values 0.5 (for increasing effects) and −0.5 (for decreasing effects). Therefore, the hazard ratios of positive and negative coefficients were 1.65 and 0.61, respectively. The remaining covariates had no direct effect on the hazards with . Survival time was generated based on the cause-specific hazard Cox-exponential models for each cause with baseline hazards equal to 0.1 [6]. Censoring time was generated from a uniform distribution in interval 0 to 9 (U(0,9)), which led to an overall censoring rate of 35%. The ratio between the number of observations of the event I and event II was fixed at 6:4. One hundred datasets were generated to determine the prediction performance of the methods.
To investigate the performance of the methods, the non-zero estimates of informative and non-informative covariates were used as described by Binder et al. [6]. In this regard, the proportion of correctly-included variables or TP, the proportion of incorrectly-excluded variables or FN, the proportion of incorrectly-included variables or FP, and the proportion of correctly-excluded variables or TN were calculated.
In this study, all analyses were implemented using the R software packages including “fastcox”, “CoxBoost”, “cmprsk”, “survAUC”, and “pec” (http://www.r-project.org).
Authors’ contributions
LT and HM were involved in the study design, simulation study, and data analysis. MSa participated in the simulation study and data analysis. MSj and JP were involved in consultation about genes and bladder cancer, respectively. LT, HM, and JP were involved in manuscript writing. All authors read and approved the final manuscript.
Competing interests
The authors declare that there is no conflict of interest.
Acknowledgments
This paper was a part of PhD thesis of LT in Biostatistics and funded by the Vice Chancellor for Research and Technology of Hamadan University of Medical Sciences (grant No. 9210173382). We gratefully acknowledge the editors and referees for their constructive suggestions to improve our manuscript.
Handled by Shaoqi Rao
Footnotes
Peer review under responsibility of Beijing Institute of Genomics, Chinese Academy of Sciences and Genetics Society of China.
References
- 1.Dyrskjøt L., Zieger K., Real F.X., Malats N., Carrato A., Hurst C. Gene expression signatures predict outcome in non-muscle-invasive bladder carcinoma: a multicenter validation study. Clin Cancer Res. 2007;13:3545–3551. doi: 10.1158/1078-0432.CCR-06-2940. [DOI] [PubMed] [Google Scholar]
- 2.Hecker N., Stephan C., Mollenkopf H.-J., Jung K., Preissner R., Meyer H.-A. A new algorithm for integrated analysis of miRNA-mRNA interactions based on individual classification reveals insights into bladder cancer. PLoS One. 2013;8:e64543. doi: 10.1371/journal.pone.0064543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kaufman D.S., Shipley W.U., Feldman A.S. Bladder cancer. Lancet. 2009;374:239–249. doi: 10.1016/S0140-6736(09)60491-8. [DOI] [PubMed] [Google Scholar]
- 4.Riester M., Taylor J.M., Feifer A., Koppie T., Rosenberg J.E., Downey R.J. Combination of a novel gene expression signature with a clinical nomogram improves the prediction of survival in high-risk bladder cancer. Clin Cancer Res. 2012;18:1323–1333. doi: 10.1158/1078-0432.CCR-11-2271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Engler D., Li Y. Survival analysis with high-dimensional covariates: an application in microarray studies. Stat Appl Genet Mol Biol. 2009;8:1–22. doi: 10.2202/1544-6115.1423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Binder H., Allignol A., Schumacher M., Beyersmann J. Boosting for high-dimensional time-to-event data with competing risks. Bioinformatics. 2009;25:890–896. doi: 10.1093/bioinformatics/btp088. [DOI] [PubMed] [Google Scholar]
- 7.Antoniadis A., Fryzlewicz P., Letué F. The Dantzig selector in Cox’s proportional hazards model. Scand J Stat. 2010;37:531–552. [Google Scholar]
- 8.Fan J., Li R. Variable selection via nonconcave penalized likelihood and its oracle properties. J Am Stat Assoc. 2001;96:1348–1360. [Google Scholar]
- 9.Gui J., Li H. Threshold gradient descent method for censored data regression, with applications in pharmacogenomics. Pac Symp Biocomput. 2005;10:272–283. doi: 10.1142/9789812702456_0026. [DOI] [PubMed] [Google Scholar]
- 10.Li H., Luan Y. Kernel Cox regression models for linking gene expression profiles to censored survival data. Pac Symp Biocomput. 2003;8:65–76. [PubMed] [Google Scholar]
- 11.Gui J., Li H. Penalized Cox regression analysis in the high-dimensional and low-sample size settings, with applications to microarray gene expression data. Bioinformatics. 2005;21:3001–3008. doi: 10.1093/bioinformatics/bti422. [DOI] [PubMed] [Google Scholar]
- 12.Park M.Y., Hastie T. L1 regularization path algorithm for generalized linear models. J R Stat Soc Series B Stat Methodol. 2007;69:659–677. [Google Scholar]
- 13.Binder H., Schumacher M. Allowing for mandatory covariates in boosting estimation of sparse high-dimensional survival models. BMC Bioinformatics. 2008;9:14. doi: 10.1186/1471-2105-9-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zou H., Hastie T. Regularization and variable selection via the elastic net. J R Stat Soc Series B Stat Methodol. 2005;67:301–320. [Google Scholar]
- 15.Yang J.-Y., Yoshihara K., Tanaka K., Hatae M., Masuzaki H., Itamochi H. Predicting time to ovarian carcinoma recurrence using protein markers. J Clin Invest. 2013;123:3740. doi: 10.1172/JCI68509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tibshirani R.J. Univariate shrinkage in the Cox model for high dimensional data. Stat Appl Genet Mol Biol. 2009;8:1–18. doi: 10.2202/1544-6115.1438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chen C.-L., Lai Y.-F., Tang P., Chien K.-Y., Yu J.-S., Tsai C.-H. Comparative and targeted proteomic analyses of urinary microparticles from bladder cancer and hernia patients. J Proteome Res. 2012;11:5611–5629. doi: 10.1021/pr3008732. [DOI] [PubMed] [Google Scholar]
- 18.Hickey C.J., Kim J.H., Ahn E.Y.E. New discoveries of old son: a link between RNA splicing and cancer. J Cell Biochem. 2014;115:224–231. doi: 10.1002/jcb.24672. [DOI] [PubMed] [Google Scholar]
- 19.Quan H., Tang H., Fang L., Bi J., Liu Y., Li H. IGF1 (CA) 19 and IGFBP-3-202A/C gene polymorphism and cancer risk: a meta-analysis. Cell Biochem Biophys. 2014;69:169–178. doi: 10.1007/s12013-013-9784-4. [DOI] [PubMed] [Google Scholar]
- 20.Moreira A., Meira-Machado L. SurvivalBIV: estimation of the bivariate distribution function for sequentially ordered events under univariate censoring. J Stat Softw. 2012;46:1–16. [Google Scholar]
- 21.Pineda S., Milne R.L., Calle M.L., Rothman N., de Maturana E.L., Herranz J. Genetic variation in the TP53 pathway and bladder cancer risk. A comprehensive analysis. PLoS One. 2014;9:e89952. doi: 10.1371/journal.pone.0089952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Morrione A., Neill T., Iozzo R.V. Dichotomy of decorin activity on the insulin-like growth factor-I system. FEBS J. 2013;280:2138–2149. doi: 10.1111/febs.12149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Metalli D., Lovat F., Tripodi F., Genua M., Xu S.-Q., Spinelli M. The insulin-like growth factor receptor I promotes motility and invasion of bladder cancer cells through Akt-and mitogen-activated protein kinase-dependent activation of paxillin. Am J Pathol. 2010;176:2997–3006. doi: 10.2353/ajpath.2010.090904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rochester M.A., Patel N., Turney B.W., Davies D.R., Roberts I.S., Crew J. The type 1 insulin-like growth factor receptor is over-expressed in bladder cancer. BJU Int. 2007;100:1396–1401. doi: 10.1111/j.1464-410X.2007.06931.x. [DOI] [PubMed] [Google Scholar]
- 25.Tamura K., Furihata M., Tsunoda T., Ashida S., Takata R., Obara W. Molecular features of hormone-refractory prostate cancer cells by genome-wide gene expression profiles. Cancer Res. 2007;67:5117–5125. doi: 10.1158/0008-5472.CAN-06-4040. [DOI] [PubMed] [Google Scholar]
- 26.Quidville V., Alsafadi S., Goubar A., Commo F., Scott V., Pioche-Durieu C. Targeting the deregulated spliceosome core machinery in cancer cells triggers mTOR blockade and autophagy. Cancer Res. 2013;73:2247–2258. doi: 10.1158/0008-5472.CAN-12-2501. [DOI] [PubMed] [Google Scholar]
- 27.Cardous-Ubbink M., Heinen R., Bakker P., Van Den Berg H., Oldenburger F., Caron H. Risk of second malignancies in long-term survivors of childhood cancer. Eur J Cancer. 2007;43:351–362. doi: 10.1016/j.ejca.2006.10.004. [DOI] [PubMed] [Google Scholar]
- 28.Liu Y., Zeng L., Zhang S., Zeng S., Huang J., Tang Y. Identification of differentially expressed proteins in chemotherapy-sensitive and chemotherapy-resistant diffuse large B cell lymphoma by proteomic methods. Med Oncol. 2013;30:1–10. doi: 10.1007/s12032-013-0528-5. [DOI] [PubMed] [Google Scholar]
- 29.Sharron Lin X, Hu L, Sandy K, Correll M, Quackenbush J, Wu C-L, et al. Differentiating progressive from nonprogressive T1 bladder cancer by gene expression profiling: applying RNA-sequencing analysis on archived specimens. Urol Oncol: Seminars and original investigations: Elsevier; 2013. [DOI] [PMC free article] [PubMed]
- 30.Lai K.-C., Lu C.-C., Tang Y.-J., Chiang J.-H., Kuo D.-H., Chen F.-A. Allyl isothiocyanate inhibits cell metastasis through suppression of the MAPK pathways in epidermal growth factor-stimulated HT29 human colorectal adenocarcinoma cells. Oncol Rep. 2014;31:189–196. doi: 10.3892/or.2013.2865. [DOI] [PubMed] [Google Scholar]
- 31.Yu X., Erzinger M.M., Pietsch K.E., Cervoni-Curet F.N., Whang J., Niederhuber J. Up-regulation of human prostaglandin reductase 1 improves the efficacy of hydroxymethylacylfulvene, an antitumor chemotherapeutic agent. J Pharmacol Exp Ther. 2012;343:426–433. doi: 10.1124/jpet.112.195768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Schuetz A.N., Yin-Goen Q., Amin M.B., Moreno C.S., Cohen C., Hornsby C.D. Molecular classification of renal tumors by gene expression profiling. J Mol Diagn. 2005;7:206–218. doi: 10.1016/S1525-1578(10)60547-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Adra C.N., Donato J.-L., Badovinac R., Syed F., Kheraj R., Cai H. SMARCAD1, a novel human helicase family-defining member associated with genetic instability: cloning, expression, and mapping to 4q22–q23, a band rich in breakpoints and deletion mutants involved in several human diseases. Genomics. 2000;69:162–173. doi: 10.1006/geno.2000.6281. [DOI] [PubMed] [Google Scholar]
- 34.Ogutu J.O., Schulz-Streeck T., Piepho H.-P. Genomic selection using regularized linear regression models: ridge regression, lasso, elastic net and their extensions. BMC Proc. 2012;6(Suppl. 2):S10. doi: 10.1186/1753-6561-6-S2-S10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lin W., Lv J. High-dimensional sparse additive hazards regression. J Am Stat Assoc. 2013;108:247–264. [Google Scholar]
- 36.Wu P., Walker B.A., Brewer D., Gregory W.M., Ashcroft J., Ross F.M. A gene expression–based predictor for myeloma patients at high risk of developing bone disease on bisphosphonate treatment. Clin Cancer Res. 2011;17:6347–6355. doi: 10.1158/1078-0432.CCR-11-0994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Fine J.P., Gray R.J. A proportional hazards model for the subdistribution of a competing risk. J Am Stat Assoc. 1999;94:496–509. [Google Scholar]
- 38.Tibshirani R. Regression shrinkage and selection via the lasso. J R Stat Soc Series B Methodol. 1996:267–288. [Google Scholar]
- 39.Goeman J.J. L1 penalized estimation in the cox proportional hazards model. Biom J. 2010;52:70–84. doi: 10.1002/bimj.200900028. [DOI] [PubMed] [Google Scholar]