Abstract
Gut microbiota of higher vertebrates is host-specific. The number and diversity of the organisms residing within the gut ecosystem are defined by physiological and environmental factors, such as host genotype, habitat, and diet. Recently, culture-independent sequencing techniques have added a new dimension to the study of gut microbiota and the challenge to analyze the large volume of sequencing data is increasingly addressed by the development of novel computational tools and methods. Interestingly, gut microbiota maintains a constant relative abundance at operational taxonomic unit (OTU) levels and altered bacterial abundance has been associated with complex diseases such as symptomatic atherosclerosis, type 2 diabetes, obesity, and colorectal cancer. Therefore, the study of gut microbial population has emerged as an important field of research in order to ultimately achieve better health. In addition, there is a spontaneous, non-linear, and dynamic interaction among different bacterial species residing in the gut. Thus, predicting the influence of perturbed microbe–microbe interaction network on health can aid in developing novel therapeutics. Here, we summarize the population abundance of gut microbiota and its variation in different clinical states, computational tools available to analyze the pyrosequencing data, and gut microbe–microbe interaction networks.
Keywords: Disease, Sequencing, 16S rRNA, Operational taxonomic unit, Microbial interaction network
Introduction
Metagenomics is the study of genetic material retrieved directly from environmental samples including the gut, soil, and water. Typically, human gut microbiota behaves like a multicellular organ, which consists of nearly 200 prevalent bacterial species and approximately 1000 uncommon species [1]. Several factors, such as diet and genetic background of the host and immune status, affect the composition of the microbiota [2,3]. It is also shown that early environmental exposure and the maternal inoculums have a large impact on gut microbiota in adulthood [4]. Gut microbiota complements the biology of an organism in ways that are mutually beneficial [5].
Gut microbiota can be studied using different approaches. For instance, descriptive metagenomics can reveal community structure and variation of the microbiome and microbial relative abundance is estimated based on different physiological and environmental conditions [6,7]. On the other hand, functional metagenomics is the study of host–microbe and microbe–microbe interactions toward a predictive, dynamic ecosystem model. Such studies reflect connections between the identity of a microbe or a community and their respective functions in the environment (terms are defined in Box 1) [8,9]. However, a major challenge in the study of gut microbiota is the inability to culture most of the gut microbial species [10]. Several efforts have been previously made in this regard. Gordon et al. identified 86 culturable species in human colonic microbiota from three healthy adults (http://www.genome.gov/Pages/Research/Sequencing/SeqProposals/HGMISeq.pdf). Gut ecosystems are currently being studied in the native state using 16S rRNA gene amplicon sequencing or whole genome sequencing (WGS) techniques [11]. 16S rRNA gene sequencing is widely used for phylogenetic reconstruction, nucleic acid-based detection, and quantification of microbial diversity. In contrast, WGS additionally explores the functions of the metagenome. The gut microbial community structure and function have been studied in different host species, including mouse [12], human [13], canine, [14], feline [14], cow [15], and yak [15]. Despite inter-species differences in community structure and function, gut microbiota frequently play a beneficial role in host metabolism and immunity across different species [16].
Large numbers of metagenomic sequence datasets have been generated, thanks to the advances in WGS and 16S rRNA pyrosequencing techniques [17]. These datasets are available in different repositories including the National Center for Biotechnology Information (NCBI) Sequence Read Archive (SRA) (http://www.ncbi.nlm.nih.gov/sra), the Data Analysis and Coordination Center (DACC) under the Human Microbiome Project (HMP) (http://hmpdacc.org) supported by the National Institutes of Health (NIH), metagenomic data resource from the European Bioinformatics Institute (EBI) (https://www.ebi.ac.uk/metagenomics/) and the UniProt Metagenomic and Environmental Sequences (UniMES) database (http://www.uniprot.org/help/unimes). All these sequence archives also provide different tools for the analysis of metagenomic sequences. Starting with the first-generation Sanger (e.g., Applied Biosystems) platforms to the second-generation 454 Life Sciences Roche (e.g., GS FLX Titanium) and Illumina (e.g., GA II, MiSeq, and HiSeq) platforms and finally, the recently developed Ion Torrent Personal Genome Machines (PGM) and Single-Molecule Real-Time (SMRT) third generation sequencing techniques introduced by Pacific Bioscience have evolved according to the need for generating cost-effective and faster metagenomic sequencing techniques. The Roche-454 Titanium platform generates consistently longer reads compared to the latest PGM platform. Whereas the MiSeq platform from Illumina produces consistently higher sequence coverage in both depth and breadth, the Ion Torrent is unique for its speed of sequencing. However, the short read length, higher complexity, and inherent incompleteness make metagenomic sequences difficult to assemble and annotate [18]. The sequences obtained from metagenomic studies are fragmented (lies between 20 and 700 base pairs) and incomplete, because of the limitations in the available sequencing techniques. Each genomic fragment is sequenced from a single species, but within a sample there are many different species, and for most of them, a full genome is absent. It becomes impossible to determine the species of origin of a particular sequence. Moreover, the volume of sequence data acquired by environmental sequencing is several orders of magnitude higher than that acquired by sequencing of a single genome [19].
Box 1. Glossary.
| Microbiome: the ecological community of commensal, symbiotic, and pathogenic microorganisms that literally share our body space. |
| Metagenome: all the genetic material present in an environmental sample, consisting of the genomes of many individual organisms. |
| Metagenomic sequencing: the high-throughput sequencing of metagenome using next-generation sequencing technology. |
| Metagenomics: the study of genetic material or the variation of species recovered directly from environmental samples. |
| Descriptive metagenomics: estimation of microbial relative abundance based on different physiological and environmental conditions to reveal community structure and variation of the microbiome. |
| Functional metagenomics: the study of host–microbe and microbe–microbe interactions toward a predictive dynamic ecosystem model to reflect a connection between the identity of a microbe or a community. |
It is well established that gut microbes constantly interact among themselves and with the host tissues. Different types of interactions are present, but most are of commensal nature. The composition of the microbial community varies significantly between and within the host species. For example, there is similarity of the microbiota between humans and mice at the super kingdom level, but significant difference exists at the phylum level [20]. In this review, we focus on different gut microbial communities residing within various host species, different software used for metagenomic data analysis, clinical importance of metagenomic studies, and importance of the microbial network toward predicting ecosystem structure and relationship among different species.
Gut microbiota studied in mammals
The gut microbial composition of only a few host species has been investigated with respect to diet, genetic potential, and disease conditions (Table 1). It was reported that human gut microbial communities were transplanted into gnotobiotic animal models, such as germ-free C57BL/6J mice, to examine the effects of diet on the human gut microbiome [3,21]. Diet plays a vital role in determining the composition of the resident gut microbes [3]. Turnbaugh et al. found that the human gut microbiome is shared among family members, who have similar microbiota even if they live at different locations [4]. In a study, Tap et al. identified 66 dominant and prevalent operational taxonomic units (OTUs) from human fecal samples, which included members of the genera Faecalibacterium, Ruminococcus, Eubacterium, Dorea, Bacteroides, Alistipes, and Bifidobacterium [22]. Another study in mice showed that host genetics along with diet is important in shaping the gut microbiota [23]. Using 16S rRNA sequencing, common microbes that belong to the Cytophaga-Flavobacterium-Bacteroides (CFB) phylum had been identified in the intestines of mice, rats, and humans [24]. Diversity in the fecal bacterial and fungal communities was also reflected in studies on canine and feline gut samples [25]. The most abundant phyla in canine gut microbiota were found to be Firmicutes, followed by Actinobacteria and Bacteroidetes, whereas the most common orders were Clostridiales, Erysipelotrichales, Lactobacillales (Firmicutes), and Coriobacteriales (Actinobacteria). In ruminants, the common rumen microbes are Fibrobacter succinogenes, Ruminococcus albus, Ruminococcus flavefaciens, Butyrivibrio fibrisolvens, and Prevotella [26].
Table 1.
Gut microbiota studies in different species using pyrosequencing technology
| Host | Sample source | Sequencing method | Amount of data retrieved | GenBank ID | Ref. |
|---|---|---|---|---|---|
| Mouse | Cecum | 16S rRNA-based sequencing | 5088 16S rRNA sequences | DQ014552−DQ015671; AY989911−AY993908 | [20] |
| Mouse | Cecum and feces | 16S rRNA-based sequencing | 2878 16S rRNA sequences | GQ491120−GQ493997 | [3] |
| Mouse | Feces | 16S rRNA-based sequencing | 4172 16S rRNA sequences | FJ032696−FJ036849 ; EU584214−EU584231 | [23] |
| Mouse and zebrafish | Zebrafish intestine and mouse cecum | 16S rRNA-based sequencing | 5545 16S rRNA sequences | DQ813844−DQ819377 | [35] |
| Human | Colonic mucosa and feces | 16S rRNA-based sequencing | 11,831 16S rRNA sequences | AY916135−AY916390; AY974810−AY986384 | [13] |
| Human | Feces | 16S rRNA-based sequencing | 9773 16S rRNA sequences | FJ362604−FJ372382 | [4] |
| Human | Feces | 16S rRNA-based sequencing | 2064 16S rRNA sequences | DQ325545−DQ327606 | [36] |
| Cat | Feces | 454 pyrosequencing | 187,396 reads | SRA012231.1 | [37] |
| Dog | Feces | 454 pyrosequencing | 201,642 reads | SRA012231.1 | [37] |
| Cow | Rumen | Whole genome sequencing | 268 G of metagenomic DNA | HQ706005−HQ706094; SRA023560 | [38] |
| Yak | Rumen | 454 pyrosequencing | 88 Mb genomic DNA | NA | [15] |
Gut metagenomics and disease: implications, scopes and limitations
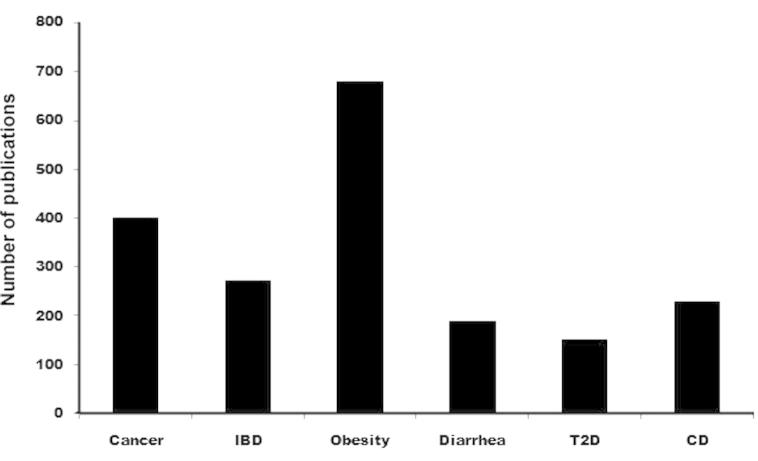
Commensal microbiota of the intestine play a key role in normal anatomical development and physiological function of the human intestine as well as other organs or systems, such as the brain [27] and the metabolic [28] and immune systems [29]. Gut microbiota exerts a major impact on an organism’s health by providing essential nutrients like vitamins and short chain fatty acids, digesting complex polysaccharides, harvesting energy and metabolizing drugs and environmental toxins [30–34]. Although microbiota composition is relatively stable in the adult, permanent changes in terms of diversity of the community and/or abundance of individual phylotypes (dysbiosis) may occur due to dietary and environmental alterations and genetic mutation of the host [30,31]. This has been associated with the development of various diseases related to the digestive system, such as inflammatory bowel disease (IBD) [39,40], irritable bowel syndrome (IBS) [41], and non-alcoholic hepatitis; obesity and obesity-related metabolic diseases like atherosclerosis [42] and type 2 diabetes (T2D); neurological disorders like Alzheimer’s disease [43–45]; atopy and asthma [46]; and cancer [47,48]. The number of publications in PubMed could reflect the importance of gut microbiota in different diseases to some extent. As shown in Figure 1, association of gut microbiota is highest with obesity followed by cancer. Bacterial species that were reported with increased abundance under certain disease conditions are mentioned in Table 2. It is interesting to note that in different disease conditions, distinct types of bacterial species become abundant.
Figure 1.
Association of gut microbiota with disease in PubMed publications
PubMed publications on different diseases involving gut microbiota were searched on February 09, 2015. IBD, inflammatory bowel disease; T2D, type 2 diabetes; CD, Crohn’s disease.
Table 2.
Highly-abundant bacterial species under different disease conditions
| Disease | Name of prevalent bacteria | Ref. |
|---|---|---|
| Symptomatic atherosclerosis | Escherichia coli | [42] |
| Eubacterium rectale | ||
| Eubacterium siraeum | ||
| Faecalibacterium prausnitzii | ||
| Ruminococcus bromii | ||
| Ruminococcus sp. 5_1_39BFAA | ||
| Type 2 diabetes | Akkermansia muciniphila | [49] |
| Bacteroides intestinalis | ||
| Bacteroides sp. 20_3 | ||
| Clostridium bolteae | ||
| Clostridium ramosum | ||
| Clostridium sp. HGF2 | ||
| Clostridium symbiosum | ||
| Colstridium hathewayi | ||
| Desulfovibrio sp. 3_1_syn3 | ||
| Eggerthella lenta | ||
| Escherichia coli | ||
| Obesity/IBD/CD | Acidimicrobidae ellin 7143 | [50] |
| Actinobacterium GWS-BW-H99 | ||
| Actinomyces oxydans | ||
| Bacillus licheniformis | ||
| Drinking water bacterium Y7 | ||
| Gamma proteobacterium DD103 | ||
| Nocardioides sp. NS/27 | ||
| Novosphingobium sp. K39 | ||
| Pseudomonas straminea | ||
| Sphingomonas sp. AO1 | ||
| Colorectal cancer | Acinetobacter johnsonii | [47,51–53] |
| Anaerococcus murdochii | ||
| Bacteroides fragilis | ||
| Bacteroides vulgatus | ||
| Butyrate-producing bacterium A2-166 | ||
| Dialister pneumosintes | ||
| Enterococcus faecalis | ||
| Fusobacterium nucleatum E9_12 | ||
| Fusobacterium periodonticum | ||
| Gemella morbillorum | ||
| Lachnospira pectinoschiza | ||
| Parvimonas micra ATCC 33270 | ||
| Peptostreptococcus stomatis | ||
| Shigella sonnei | ||
Note: IBD, inflammatory bowel disease; CD, Crohn’s disease.
It is critical to define healthy microbiota and the deviations related to etiopathogenesis of diseases. This would allow us to predict the development and/or progression of diseases and foster the idea of microbiota-targeted therapy. Metagenomic sequencing has revealed that bacteria constitute the overwhelming majority of gut microbiota in health and there is remarkable inter-individual conservation at the phylum level. For example, in more than 90% of healthy individuals, gut bacteria belong to two major phyla, Bacteroides and Firmicutes [54]. However, efforts to define a core microbiome resulted in mixed outcomes. Qin et al. analyzed 3.3 million non-redundant microbial genes from intestinal samples of 124 Europeans [55]. They found that 18 species were present in all individuals, while 57 and 75 species were detected in >75% and >50% of the population, respectively [55]. In contrast, Turnbaugh et al. reported that a functional core microbiome exists in human gut [4], since gut microbiota serves critical metabolic and immunological functions to maintain homeostasis. In fact, studies with discrete population groups have indicated that the super-kingdom level conservation rapidly disappears lower in the phylogenetic hierarchy, giving rise to a “microbiota fingerprint” of an individual at the levels of genus, species, and strain. This is underscored by the sharing of only approximately 40% species by monozygotic twins [12]. Interestingly, the individual gut microbiota is more unique under healthy conditions than during disease, when the diversity generally decreases. It is believed that the ratio of potentially pathogenic to beneficial commensal microbes, rather than the presence of a specific organism or a group, is more crucial for disease development [56]. However, a single pathobiont (commensal turned into a pathogen) has also been reported to cause disease under specific genetic and environmental conditions. Bloom et al. demonstrated that commensal Bacteroides isolates induce disease in genetically-modified (il10r2−/− with dominant-negative TGF-betaR2 expression in T cells) IBD-susceptible mice, but not in IBD-nonsusceptible mice [57].
Importantly, metagenomic sequencing has unearthed a separate kingdom of resident viral species, many of which were unknown so far, constituting the “gut virome” [58]. Reyes et al. sequenced the viromes isolated from fecal samples of monozygotic twins and their mothers, and compared them with the total fecal DNA. This experiment revealed that the bacterial community present in the mother and the twins was highly similar, whereas individual viromes were unique despite their genetic similarity. They also performed a longitudinal study for one year on the fecal samples collected from the same individuals at different time points and found that >95% of virotypes were constant, but the abundance of bacterial population changed over time [58]. Although the role of viral species in human diseases is far from fully appreciated, inter-kingdom interactions between bacteria, viruses, and eukaryotes in the intestine have been shown to influence virulence of the organisms and pathogenesis [59].
Altered diversity and abundance of the so-called ‘normal flora’ during disease development and progression were unknown before the introduction of metagenomic sequencing, since most of these organisms are non-culturable. 16S rRNA sequencing has indicated a decrease in Bacteroides and Firmicutes numbers in the colon and an increase in Enterobacteriaceae, such as adherent-invasive E. coli and other Proteobacteria in Crohn’s disease [60]. In contrast, obesity is associated with fermenting bacterial species, such as Bacteroides and Firmicutes, which can harvest energy from complex polysaccharides [54]. Although the association of bacterial flora with etiopathogenesis of disease is not fully established, development of colitis and obesity following transfer of disease-associated microbiota to gnotobiotic mice strongly suggests disease association [61,62]. Animal models indeed have emerged as invaluable tools to establish the underlying mechanisms related to altered microflora in disease development. Altered flora may be the consequence of inflammation, which may be demonstrated by reconstitution of germ-free mice or piglets with the human disease flora. Furthermore, study of temporal changes in the microbiota by metagenomic sequencing of genetically-predisposed individuals or their first-degree relatives may be helpful. Such information may be therapeutically important, since an early intervention appears to be critical to restore normal flora [63].
Although various sequencing techniques have been used to map the diversity of microbial communities that exist during health and disease, microbiota-associated genes and gene products that may protect from or predispose to disease remain largely unknown. Metagenomic sequencing data provide genetic composition of the whole microbiome, but give little information about functioning of gene expression. Functional metagenomics may be useful, but currently the objective of sequencing is to identify functionally-important non-abundant genes. Insights into the cellular and molecular interactions between the host and the microbiota necessitate integration of metagenomics with metatranscriptomics (gene expression profile), metaproteomics (protein mapping profile), and metabolomics (metabolic profile) data. For example, combination of metagenomics and metabolomics identified the role of microbiota in dietary phospholipid metabolism, contributing to atherosclerosis [64]. Multiple omics platforms integrating metabolic changes in the host, including the metabolism of drugs and environmental toxins, with microbiota diversity have highlighted the necessity of personalized medicine. Gut microbial enzymes for the metabolism of commonly-prescribed drugs, such as acetaminophen and cholesterol-lowering agent simvastatin, were identified [65,66]. In addition, microbiota plays a critical role in the generation of more- (e.g., sulfasalazine) or less-active (e.g., digoxin) drug metabolites [58]. Therapeutically active metabolite 5-aminosalicylate is released from the prodrug sulfasalazine, while digoxin may be converted to less active reduced derivatives by the action of colonic microflora [34,67]. This implies that there may be significant inter-individual variability in the drug response and/or adverse events. Similarly, toxin exposure may have very different outcomes due to the variability in the microbiota composition of the exposed individuals. Several neurotoxins and carcinogenic metabolites may be generated by resident microbes such as E. coli [68]. Identification of individual microbial species or the specific enzymes they produce with the metabolites generated would make it possible to target the microbiota for therapeutic purposes. This is best exemplified by the successful treatment of chemotherapy-associated diarrhea following administration of CPT-11, a drug used in colon cancers, by the use of bacterial β-glucuronidase enzyme inhibitor [69].
Intestinal microbiota is emerging as the target for next-generation therapeutics. On the one hand, it may be considered as a repository of potential drugs or drug-like molecules, such as antimicrobial peptide bacteriocin or thuricin CD, and anti-inflammatory molecules like the cell wall polysaccharide (Bacteroides fragilis) and peptidoglycan (Lactobacillus) [34]. Metagenomics coupled with bioinformatics may spearhead the ‘bugs to drugs’ research. On the other hand, ‘disease microbiota’ may be targeted for treatment. Current therapies are limited to non-specifically targeting the microbiota with probiotics, prebiotics, and synbiotics to restore the ‘healthy flora’ [70–73]. Probiotics therapy has shown promise in the treatment of acute diarrhea and prophylaxis against necrotizing enterocolitis [74]. Although the exact mechanism of action remains unknown, these organisms may render the host resistant to colonization by pathogens through competing with them for the intestinal niche, in addition to their bactericidal function, thus creating an environment for the lost flora to re-establish. Fecal transplantation of the healthy flora has been successfully employed for the treatment of drug-resistant or recurrent Clostridium difficile-associated diarrhea [24]. However, the results are less-encouraging in obesity and chronic diseases like diabetes mellitus, IBD, and IBS [53]. In these conditions, early institution of therapy before an altered flora is established in the affected individuals or treatment of the high-risk groups, such as first-degree relatives of the patients, may be more helpful. It is unlikely that a single probiotic or a specific combination would be effective in all conditions and subjects. Therefore, a more personalized treatment may be required based on the microbiota composition to ensure a predicted outcome.
A major bottleneck to the specificity of microbiota-targeted therapies is our limited knowledge about the resident organisms and their interactions with the host. Moreover, microbe–microbe cross-talk may influence the disease outcome. Naturally, members of the microbiota with known genome sequences or biochemical functions will be the initial targets for drug or vaccine development. However, non-specificity of the effects, which potentially results in removal of beneficial flora and development of resistance, may be issues that will require further attention. A systems biology approach may be required with a therapeutic goal to restore the biochemical, proteomic, and metagenomic profiles of an individual.
Importance of microbial interaction network
Gut microbiota is an example of a complex ecological community involving interactions with the host cells as well as among hundreds of bacterial species. These interactions may be of five different types including (i) mutualism, where both the participants are benefited; (ii) amensalism, where one organism is inhibited or destroyed and the other is unaffected; (iii) commensalism, where one partner gets the advantage without any help or harm to the other; (iv) competition, where both the participants harm each other; and (v) parasitism, where one gets benefited out of the other [8].
Establishing a model of the gut microbial interaction network is a major challenge for the scientific community and little progress has been made in this area. Predictions of microbial associations may include a simple binary mode or complex relationship, where more than two species are involved in an absence–presence relationship (1 or 0 mode) or abundance data (quantitative values obtained from OTU). It is possible to predict the simple binary or pair-wise microbial relationship using a similarity-based network inference, while the complex microbial relationship can be predicted using regression and a rule-based modeling approach. The similarity-based network inferences are based on co-occurrence and/or mutual exclusion pattern of two species over different sampling conditions. Pair-wise relationship scores are computed and further compared with the random co-occurrence scores using a similar sampling approach. Faust et al. recently built a gut microbiota network with co-occurrence relationship using Spearman rank correlation method. Here, 16S rRNA marker genes were used for compromised gut in children with anti-islet cell autoimmunity [75]. This network established a strong association between microbiota and their body niches. The dominant species at a specific body site emerged as a “hub” in the network and was found to act as the signature taxa, which was responsible for the composition of each microcommunity. Examples for hubs include Bacteroides in the gut and Streptococcus in the oral cavity. This microbial association is also reflected in their phylogenetic and functional relatedness. Especially, phylogenetically related microbes have been found to co-occur at environmentally similar body sites [75]. However, this type of approach cannot be applied to complex, nonlinear, and evolving systems, where more than one dominant species are present at any point of time and the abundance changes over time. In such cases, the regression model and rule-based model are used, where the abundance of one species is predicted from combined abundances of the organisms in the system [76]. Generalized Lotka–Volterra (gLV) equations are used to study these complex types of dynamic microbial community interactions [77]. Few examples are present where gut microbiota is used to develop diet-induced predictive models [63]. In this model, a linear equation connects microbiota changes to given concentrations of each of the four dietary ingredients (Casein, Starch, Sucrose, and oil). There is still limited knowledge about the gut microbial interactions and interactions between the microbes and the host. In-depth investigation is required to model these interactions in a better way and predict the outcome of community-level microbial interactions after external disturbance of the gut system due to diseases or the use of drugs.
Whole genome sequencing of gut microbiota
16S rRNA-based sequencing of metagenomes is an established approach for the identification of known bacteria, based on the reference sequences. However, most bacterial species of the gut microbiota are novel, for which no reference sequence is available. Moreover, 16S sequencing does not provide any functional input about the community, since the sequence is not strain-specific. Gene contents may differ between bacterial strains with identical 16S rRNA gene sequence and underlie their functional difference related to genes responsible for toxicity and pathogenesis [78]. WGS of the microbiota (e.g., Human Microbiome Project Consortium, 2012) is preferred over 16S rRNA-based analysis to elucidate taxonomic classification and bacterial diversity within members of the microbial community. WGS is also useful for a detailed understanding of the functional potential of the microbiome. For example, fecal metagenomic data obtained from WGS of 124 unrelated individuals along with six monozygotic twin pairs and their mothers were analyzed by the construction of community level metabolic networks of the microbiome. It was observed that gene-level and network-level topological differences are strongly associated with obesity and IBD [79]. WGS of 252 fecal metagenomic samples in another study showed huge variations at the metagenomic level, in which authors identified 107,991 short insertions/deletions, 10.3 million single nucleotide polymorphisms (SNPs) and 1051 structural variants. In addition, they found that despite considerable changes in the composition of the gut microbiota, the individual specific SNP variation pattern showed a temporal stability. This further suggests that every individual carries a unique metagenome, which can be exploited further for personalized medicine or dietary modifications [80]. Many 16S rRNA-based studies have reported a connection between the gut microbiota and health [24,39,59]. A detailed WGS based analysis of the gut metagenome may help to better understand the disease pathogenesis and identify new targets for therapy, because it may reveal minor genomic variations within species that cause altered phenotypes, leading to pathogenesis. For instance, WGS studies with Citrobacter spp. showed that genomic variations within species altered their phenotype and environmental adaptation [81].
Currently, Illumina shotgun sequencing of stool samples is widely used for WGS studies of the gut microbiome. Since the gut contains diverse microbial species, a deep sequencing (20 × coverage) is required to study individual communities with low abundance [81]. However, analyzing the large volume of WGS data (short reads) is very challenging, as there may be from hundreds to thousands of bacterial species present with different abundances, especially as there is no taxonomic identification available for most of the species.
Tools/web-servers related to gut microbiota studies
To overcome the challenges in metagenomic data analysis, several standalone software, web servers, and R packages have been developed and are available in the public domain (Table 3). Here, we focus on the popular software, which can be used in studying gut microbiota. There are many standalone tools, which may be used for the analysis of 16S rRNA marker gene sequencing data and the WGS data. Quantitative Insights Into Microbial Ecology (QIIME), investigates microbial diversity using 16S rRNAs data. It provides the users with taxonomy assignments to phylogenetic analysis along with demultiplexing and quality filtering of the raw reads generated from Illumina or other platforms. But the installation of QIIME needs some expertise in Linux and Windows systems, and it lacks parallel processing at the OTU picking step [82]. mothur is a software package with several functions, including identification of OTUs and description of alpha (within a specific sample) and beta (between different samples) diversity between different samples [83]. RAMMCAP is a GUI-based tool, which performs metagenomic sequence clustering and analysis and can process a huge number of sequences in a very short time compared to other tools and software. RAMMCAP also includes protein family annotation tool and a novel GUI-based metagenome comparison method based on statistical analysis [84]. For WGS-based sequencing data analysis (mainly for taxonomy binning), several approaches are available, which integrates Basic Local Alignment Search Tool (BLAST) for species identification. The tool MEtaGenome ANalyzer (MEGAN) uses BLAST search against a reference sequence database like non-redundant sequence database from NCBI NR database and provides results in a graphical user interface (GUI). It allows large datasets to be dissected without further assembly or the targeting of specific 16S rRNA marker gene. It can also compare different datasets based on statistical analysis and provides graphical output [85]. Metagenomic Phylogenetic Analysis (MetaPhlAn) is another tool that provides faster taxonomic assignments by removing redundant sequences [86]. Short reads need to be assembled into contigs, which are similar in length to a gene, so that they may be annotated for function inference. Such assembly can be performed using tools such as MetaVelvet [87] and Short Oligonucleotide Analysis Package (SOAPdenovo2) [88]. Moreover, simultaneous assembly and annotation are also possible with some software packages, such as MOCAT, which assembles metagenomic short reads into contigs along with quality control and performs gene prediction from contigs [89]. For functional analysis of the metagenomic reads, predicted genes from the assembled contigs or raw sequence reads with long read length may be used. To annotate functions to the sequences or genes, Kyoto Encyclopedia of Genes and Genomes (KEGG) organizes genes into KEGG enzymes, pathways, and orthologs appropriate for the elucidation of metabolic potential of the community. Certain pipelines, such as SmashCommunity [90], Microbiome Project Unified Metabolic Analysis Network (HUMAnN) [91], and Functional Annotation and Taxonomic Analysis of Metagenomes (FANTOM) [92], which are easy-to-use GUIs for metagenomic data analysis, are also available to automate the process of assembly and annotation.
Table 3.
Tools/webservers related to gut microbiota studies
| Name | Platform | Website | Main features | Ref. |
|---|---|---|---|---|
| QIIME | Stand alone | http://qiime.sourceforge.net/ | Network analysis, histograms of within- or between-sample diversity | [82] |
| mothur | Stand alone | http://www.mothur.org/ | Fast processing of large sequence data | [83] |
| RAMMCAP | Stand alone | http://weizhonglab.ucsd.edu/rammcap/cgibin/rammcap.cgi | Ultra fast sequence clustering and protein family annotation | [84] |
| MEGAN | Stand alone | http://www-ab.informatik.unituebingen.de/software/megan/ | Laptop analysis of large metagenomic shotgun sequencing data sets | [85] |
| MetaPhlAn | Stand alone | http://huttenhower.sph.harvard.edu/metaphlan | Faster profiling of the composition of microbial communities using unique clade-specific marker genes | [86] |
| MetaVelvet | Stand alone | http://metavelvet.dna.bio.keio.ac.jp/ | High quality metagenomic assembler | [87] |
| SOAPdenovo2 | Stand alone | http://soap.genomics.org.cn/soapdenovo.html | Metagenomic assembler, specifically for Illumina GA short reads | [88] |
| MOCAT | Stand alone | http://vmlux.embl.de/~kultima/MOCAT/ | Generate taxonomic profiles and assemble metagenomes | [89] |
| SmashCommunity | Stand alone | http://www.bork.embl.de/software/smash/ | Performs assembly and gene prediction mainly for data from Sanger and 454 sequencing technologies | [90] |
| HUMAnN | Stand alone | http://huttenhower.sph.harvard.edu/humann | Analysis of metagenomic shotgun data from the Human Microbiome Project | [91] |
| FANTOM | Stand alone | http://www.sysbio.se/Fantom/ | Comparative analysis of metagenomics abundance data integrated with databases like KEGG Orthology, COG, PFAM and TIGRFAM, etc. | [92] |
| MetaCV | Stand alone | http://metacv.sourceforge.net/ | Classification short metagenomic reads (75–100 bp) into specific taxonomic | [94] |
| Phymm | Stand alone | http://www.cbcb.umd.edu/software/phymm/ | Phylogenetic classification of metagenomic short reads using interpolated Markov models | [97] |
| PhyloPythiaS | Web server | http://binning.bioinf.mpiinf.mpg.de/ | Fast and accurate sequence composition-based classifier that utilizes the hierarchical relationships between clades | [96] |
| TETRA | Web server | http://www.megx.net/tetra | Correlation of tetranucleotide usage patterns in DNA | [93] |
| METAREP | Web server | http://www.jcvi.org/metarep/ | Flexible comparative metagenomics framework | [98] |
| CD-HIT | Web server | http://weizhonglab.ucsd.edu/cd-hit/ | Identity-based clustering of sequences | [99] |
| METAGENassist | Web server | http://www.metagenassist.ca/ | Performs comprehensive multivariate statistical analyses on the data from different host and environment sites | [100] |
| CoMet | Web server | http://comet.gobics.de/ | ORF finding and subsequent Pfam domain assignment to protein sequences | [101] |
| WebCARMA | Web server | http://webcarma.cebitec.unibielefeld.de/ | Unassembled reads as short as 35 bp can be used for the taxonomic classification with less false positive prediction | [102] |
| MG-RAST | Web server | https://metagenomics.anl.gov/ | High-throughput pipeline for functional metagenomic analysis | [103] |
| CAMERA | Web server | https://portal.camera.calit2.net/gridsphere/gridsphere | Provides list of workflows for WGS data analysis | [104] |
| WebMGA | Web server | http://weizhonglilab.org/metagenomic-analysis/ | Implemented to run in parallel on local computer cluster | [105] |
Most of the aforementioned tools use known 16S rRNA reference sequence databases like RDP (http://rdp.cme.msu.edu/) and Greengenes (http://greengenes.lbl.gov) to assign taxonomy information to the unknown sequence. Nonetheless, some WGS-based unsupervised tools, such as TETRA [93], MetaCV [94], and PhyloPythia [95], are also available. They use different sequence features for taxonomy binning. TETRA is a DNA-based fingerprinting technique for genomic fragment correlation based on tetranucleotide usage pattern, while MetaCV is an algorithm based on composition and phylogeny to classify short metagenomic reads (75–100 bp) into specific taxonomic and functional groups. Similarly, PhyloPythiaS web server [96] is also is a fast and accurate classifier based on sequence composition utilizing the hierarchical relationships between clades. Among these composition-based classification methods, Phymm [97] is another classifier for metagenomic data that has been trained on 539 complete, curated bacterial and archaeal genomes, and can accurately classify reads as short as 100 bp. Along with TETRA and PhyloPythiaS web servers, several other online web-servers are also available for metagenomic analysis. METAREP is a web 2.0 application, which provides graphical summaries for top taxonomic and functional classifications. It also provides Gene Ontology (GO), NCBI Taxonomy and KEGG Pathway Browser-based comparison of multiple datasets at various functional and taxonomic levels [98]. Another online tool, CD-HIT, can be used in identification of non-redundant sequences and gene-families by clustering raw reads [99]. METAGENassist, a web server for comparative metagenomics, can be used for comprehensive multivariate statistical analyses on the bacterial census data from different environment sites or different biological hosts selected by the users [100]; CoMet, another web-based comparative metagenomics platform is used for the analysis of metagenomic short read data resulting from WGS-based studies. It integrates ORF finder, Pfam domain detection software and statistical analysis tools to a user-friendly web interface for functional comparison of metagenomic data from multiple samples [101]. WebCARMA is a web application for taxonomic classification of ultra-short reads as 35 bp [102]. MG-RAST (the Metagenomics RAST) server is an automated platform for the analysis of microbial metagenomes to get the quantitative insights of the microbial populations . Modularity of MG-RAST allows new analysis steps or comparative data to be added during the analysis according to the user’s need. It enables the user to annotate multiple metagenomes at a time and also to compare the metabolic data [103].
CAMERA [104] and WebMGA [105] are also frequently used web servers for metagenomic data analysis. CAMERA offers a list of workflows, but many useful tools are missing, such as Filter-HUMAN, RDP-binning, FR-HIT-binning, and CD-HIT-OTU, which are otherwise available with WebMGA. Filter-HUMAN is a tool for filtering human sequences from human microbiome samples. RDP-binning uses the binning tool from Ribonsomal Database Project (RDP) to classify rRNA sequences. FR-HIT-binning first aligns the query metagenomic reads to NCBI’s Refseq database and then classifies reads to the specific taxon, which is the lowest common ancestor (LCA) of the hits. CD-HIT-OTU is a clustering program able to process millions of rRNAs in a few minutes. Moreover, both MG-RAST and CAMERA require user registration and login, so it is difficult to access their web servers using scripts. However, WebMGA has resolved these issues and allows a fast, easy and flexible solution for metagenomic data analysis. The user can perform data analysis through customized annotation pipeline and it does not require any login information. In addition, metaphor package is also available for users having expertise in R statistical language (http://CRAN.R-project.org/package=metafor). Although these programs are widely used for metagenomic data analysis, there is still a bottleneck to identify novel bacteria, as a majority of them are unknown.
Conclusion and future prospects
We have reached a level of saturation regarding 16S rRNA sequence catalogs of gut microbiota from the Western population. This is exemplified by the fact that we are fairly close to identifying all gene families encoded by the human gut microbiota of the Western population. It has been observed that the bacterial phylogeny obtained from the gut microbial DNA sequencing of 124 individuals is not much different from that of the first 70 individuals [55]. While the above findings need to be extended to diverse phenotypes (populations, diseases, age, etc.), more efforts should be directed to compile reference genomes, which will require WGS, and perhaps, culturing individual organisms. In addition, there are multiple ecosystems along the length of the gut, which remain unexplored in terms of metagenomic diversity. An increasing number of studies in the future will be directed toward understanding the functions of the microbiome and RNA-seq may play a critical role. However, preparing high quality representative RNAs for sequencing to generate metatranscriptome is a challenge.
As opposed to the sequencing data, functional annotations of the genes are grossly incomplete due to the unavailability of suitable computational tools and we have only limited knowledge about the metabolic functions of the microbiota. Germ-free animals are valuable tools for functional assessment of the microbiota and their association with diseases, but high variability between facilities is a major problem for data interpretation. Microbiota has great potential for the identification of genetic biomarkers of disease, but proper statistical analysis is extremely difficult.
Finally, the association of gut microbiota with human diseases has obliterated the boundary between infectious and non-infectious diseases. While the manipulation of microbiota has immense therapeutic potential, techniques need to be developed to manipulate individual bacteria within a community and for targeted therapy, such as designer probiotics. There is an urgent need for novel approaches toward the construction of gut ecosystem-wide association networks to develop global models of gut ecosystem dynamics. Such models may then, predict the outcome of perturbation effects in the gut and eventually aid in therapeutic intervention.
Competing interests
The authors have declared no competing interests.
Acknowledgments
This study was supported by Indian Council of Medical Research (Grant No. 2013-1551G). SS thanks Department of Biotechnology, India for providing Ramalingaswami Fellowship (BT/RLF/Re-entry/11/2011).
Handled by Fangqing Zhao
Footnotes
Peer review under responsibility of Beijing Institute of Genomics, Chinese Academy of Sciences and Genetics Society of China.
Contributor Information
Sudipto Saha, Email: ssaha4@jcbose.ac.in.
Santasabuj Das, Email: dasss@icmr.org.in.
References
- 1.Ley R.E., Hamady M., Lozupone C., Turnbaugh P.J., Ramey R.R., Bircher J.S. Evolution of mammals and their gut microbes. Science. 2008;320:1647–1651. doi: 10.1126/science.1155725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Benson A.K., Kelly S.A., Legge R., Ma F., Low S.J., Kim J. Individuality in gut microbiota composition is a complex polygenic trait shaped by multiple environmental and host genetic factors. Proc Natl Acad Sci U S A. 2010;107:18933–18938. doi: 10.1073/pnas.1007028107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Turnbaugh P.J., Ridaura V.K., Faith J.J., Rey F.E., Knight R., Gordon J.I. The effect of diet on the human gut microbiome: a metagenomic analysis in humanized gnotobiotic mice. Sci Transl Med. 2009;1:6ra14. doi: 10.1126/scitranslmed.3000322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Turnbaugh P.J., Hamady M., Yatsunenko T., Cantarel B.L., Duncan A., Ley R.E. A core gut microbiome in obese and lean twins. Nature. 2009;457:480–484. doi: 10.1038/nature07540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Backhed F., Ley R.E., Sonnenburg J.L., Peterson D.A., Gordon J.I. Host-bacterial mutualism in the human intestine. Science. 2005;307:1915–1920. doi: 10.1126/science.1104816. [DOI] [PubMed] [Google Scholar]
- 6.Xia L.C., Cram J.A., Chen T., Fuhrman J.A., Sun F. Accurate genome relative abundance estimation based on shotgun metagenomic reads. PLoS ONE. 2011;6:e27992. doi: 10.1371/journal.pone.0027992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Garmendia L., Hernandez A., Sanchez M.B., Martinez J.L. Metagenomics and antibiotics. Clin Microbiol Infect. 2012;18:27–31. doi: 10.1111/j.1469-0691.2012.03868.x. [DOI] [PubMed] [Google Scholar]
- 8.Faust K., Raes J. Microbial interactions: from networks to models. Nat Rev Microbiol. 2012;10:538–550. doi: 10.1038/nrmicro2832. [DOI] [PubMed] [Google Scholar]
- 9.Chistoserdovai L. Functional metagenomics: recent advances and future challenges. Biotechnol Genet Eng Rev. 2010;26:335–352. [PubMed] [Google Scholar]
- 10.Siezen R.J., Kleerebezem M. The human gut microbiome: are we our enterotypes? Microb Biotechnol. 2011;4:55053. doi: 10.1111/j.1751-7915.2011.00290.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Eisen J.A. Environmental shotgun sequencing: its potential and challenges for studying the hidden world of microbes. PLoS Biol. 2007;5:e82. doi: 10.1371/journal.pbio.0050082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Turnbaugh P.J., Quince C., Faith J.J., McHardy A.C., Yatsunenko T., Niazi F. Organismal genetic and transcriptional variation in the deeply sequenced gut microbiomes of identical twins. Proc Natl Acad Sci U S A. 2010;107:7503–7508. doi: 10.1073/pnas.1002355107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Eckburg P.B., Bik E.M., Bernstein C.N., Purdom E., Dethlefsen L., Sargent M. Diversity of the human intestinal microbial flora. Science. 2005;308:1635–1638. doi: 10.1126/science.1110591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Suchodolski J.S. Companion animals symposium: microbes and gastrointestinal health of dogs and cats. J Anim Sci. 2011;89:1520–1530. doi: 10.2527/jas.2010-3377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dai X., Zhu Y., Luo Y., Song L., Liu D., Liu L. Metagenomic insights into the fibrolytic microbiome in yak rumen. PLoS ONE. 2012;7:e40430. doi: 10.1371/journal.pone.0040430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tilg H., Kaser A. Gut microbiome obesity and metabolic dysfunction. J Clin Invest. 2011;121:2126–2132. doi: 10.1172/JCI58109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jumpstart Consortium Human Microbiome Project Data Generation Working G Evaluation of 16S rDNA-based community profiling for human microbiome research. PLoS ONE. 2012;7:e39315. doi: 10.1371/journal.pone.0039315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Markowitz V.M., Chen I.M., Chu K., Szeto E., Palaniappan K., Grechkin Y. IMG/M: the integrated metagenome data management and comparative analysis system. Nucleic Acids Res. 2012;40:D123–D129. doi: 10.1093/nar/gkr975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wooley J.C., Godzik A., Friedberg I. A primer on metagenomics. PLoS Comput Biol. 2010;6:e1000667. doi: 10.1371/journal.pcbi.1000667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ley R.E., Backhed F., Turnbaugh P., Lozupone C.A., Knight R.D., Gordon J.I. Obesity alters gut microbial ecology. Proc Natl Acad Sci U S A. 2005;102:11070–11075. doi: 10.1073/pnas.0504978102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Goodman A.L., Kallstrom G., Faith J.J., Reyes A., Moore A., Dantas G. Extensive personal human gut microbiota culture collections characterized and manipulated in gnotobiotic mice. Proc Natl Acad Sci U S A. 2011;108:6252–6257. doi: 10.1073/pnas.1102938108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tap J., Mondot S., Levenez F., Pelletier E., Caron C., Furet J.P. Towards the human intestinal microbiota phylogenetic core. Environ Microbiol. 2009;11:2574–2584. doi: 10.1111/j.1462-2920.2009.01982.x. [DOI] [PubMed] [Google Scholar]
- 23.Zhang C., Zhang M., Wang S., Han R., Cao Y., Hua W. Interactions between gut microbiota host genetics and diet relevant to development of metabolic syndromes in mice. ISME J. 2010;4:232–241. doi: 10.1038/ismej.2009.112. [DOI] [PubMed] [Google Scholar]
- 24.Kinross J.M., Darzi A.W., Nicholson J.K. Gut microbiome-host interactions in health and disease. Genome Med. 2011;3:14. doi: 10.1186/gm228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Handl S., Dowd S.E., Garcia-Mazcorro J.F., Steiner J.M., Suchodolski J.S. Massive parallel 16S rRNA gene pyrosequencing reveals highly diverse fecal bacterial and fungal communities in healthy dogs and cats. FEMS Microbiol Ecol. 2011;76:301–310. doi: 10.1111/j.1574-6941.2011.01058.x. [DOI] [PubMed] [Google Scholar]
- 26.Flint H.J., Bayer E.A., Rincon M.T., Lamed R., White B.A. Polysaccharide utilization by gut bacteria: potential for new insights from genomic analysis. Nat Rev Microbiol. 2008;6:121–131. doi: 10.1038/nrmicro1817. [DOI] [PubMed] [Google Scholar]
- 27.Mayer E.A., Tillisch K., Gupta A. Gut/brain axis and the microbiota. J Clin Invest. 2015;125:926–938. doi: 10.1172/JCI76304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Li M., Wang B., Zhang M., Rantalainen M., Wang S., Zhou H. Symbiotic gut microbes modulate human metabolic phenotypes. Proc Natl Acad Sci U S A. 2008;105:2117–2122. doi: 10.1073/pnas.0712038105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Round J.L., Mazmanian S.K. The gut microbiota shapes intestinal immune responses during health and disease. Nat Rev Immunol. 2009;9:313–323. doi: 10.1038/nri2515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Clemente J.C., Ursell L.K., Parfrey L.W., Knight R. The impact of the gut microbiota on human health: an integrative view. Cell. 2012;148:1258–1270. doi: 10.1016/j.cell.2012.01.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Blumberg R., Powrie F. Microbiota, disease, and back to health: a metastable journey. Sci Transl Med. 2012;4:137rv7. doi: 10.1126/scitranslmed.3004184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jia W., Li H., Zhao L., Nicholson J.K. Gut microbiota: a potential new territory for drug targeting. Nat Rev Drug Discov. 2008;7:123–129. doi: 10.1038/nrd2505. [DOI] [PubMed] [Google Scholar]
- 33.Shanahan F. Therapeutic implications of manipulating and mining the microbiota. J Physiol. 2009;587:4175–4179. doi: 10.1113/jphysiol.2009.174649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Shanahan F. The gut microbiota-a clinical perspective on lessons learned. Nat Rev Gastroenterol Hepatol. 2012;9:609–614. doi: 10.1038/nrgastro.2012.145. [DOI] [PubMed] [Google Scholar]
- 35.Rawls J.F., Mahowald M.A., Ley R.E., Gordon J.I. Reciprocal gut microbiota transplants from zebrafish and mice to germ-free recipients reveal host habitat selection. Cell. 2006;127:423–433. doi: 10.1016/j.cell.2006.08.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gill S.R., Pop M., Deboy R.T., Eckburg P.B., Turnbaugh P.J., Samuel B.S. Metagenomic analysis of the human distal gut microbiome. Science. 2006;312:1355–1359. doi: 10.1126/science.1124234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Garcia-Mazcorro J.F., Lanerie D.J., Dowd S.E., Paddock C.G., Grützner N., Steiner J.M. Effect of a multi-species synbiotic formulation on fecal bacterial microbiota of healthy cats and dogs as evaluated by pyrosequencing. FEMS Microbiol Ecol. 2011;78:542–554. doi: 10.1111/j.1574-6941.2011.01185.x. [DOI] [PubMed] [Google Scholar]
- 38.Hess M., Sczyrba A., Egan R., Kim T.W., Chokhawala H., Schroth G. Metagenomic discovery of biomass-degrading genes and genomes from cow rumen. Science. 2011;331:463–467. doi: 10.1126/science.1200387. [DOI] [PubMed] [Google Scholar]
- 39.Schippa S., Conte M.P. Dysbiotic events in gut microbiota: impact on human health. Nutrients. 2014;6:5786–5805. doi: 10.3390/nu6125786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Asquith M., Elewaut D., Lin P., Rosenbaum J.T. The role of the gut and microbes in the pathogenesis of spondyloarthritis. Best Pract Res Clin Rheumatol. 2014;28:687–702. doi: 10.1016/j.berh.2014.10.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kennedy P.J., Cryan J.F., Dinan T.G., Clarke G. Irritable bowel syndrome: a microbiome-gut-brain axis disorder? World J Gastroenterol. 2014;20:14105–14125. doi: 10.3748/wjg.v20.i39.14105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Karlsson F.H., Fak F., Nookaew I., Tremaroli V., Fagerberg B., Petranovic D. Symptomatic atherosclerosis is associated with an altered gut metagenome. Nat Commun. 2012;3:1245. doi: 10.1038/ncomms2266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Moreno-Indias I., Cardona F., Tinahones F.J., Queipo-Ortuño M.I. Impact of the gut microbiota on the development of obesity and type 2 diabetes mellitus. Front Microbiol. 2014;5:190. doi: 10.3389/fmicb.2014.00190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Alam M.Z., Alam Q., Kamal M.A., Abuzenadah A.M., Haque A. A possible link of gut microbiota alteration in type 2 diabetes and Alzheimer’s disease pathogenicity: an update. CNS Neurol Disord Drug Targets. 2014;13:383–390. doi: 10.2174/18715273113126660151. [DOI] [PubMed] [Google Scholar]
- 45.Chen X., D’Souza R., Hong S.T. The role of gut microbiota in the gut-brain axis: current challenges and perspectives. Protein Cell. 2013;4:403–414. doi: 10.1007/s13238-013-3017-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Azad M.B., Konya T., Maughan H., Guttman D.S., Field C.J., Sears M.R. Infant gut microbiota and the hygiene hypothesis of allergic disease: impact of household pets and siblings on microbiota composition and diversity. Allergy Asthma Clin Immunol. 2013;9:15. doi: 10.1186/1710-1492-9-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wang T., Cai G., Qiu Y., Fei N., Zhang M., Pang X. Structural segregation of gut microbiota between colorectal cancer patients and healthy volunteers. ISME J. 2012;6:320–329. doi: 10.1038/ismej.2011.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Greenhill C. Gut microbiota: anti-cancer therapies affected by gut microbiota. Nat Rev Gastroenterol Hepatol. 2014;11:1. doi: 10.1038/nrgastro.2013.238. [DOI] [PubMed] [Google Scholar]
- 49.Qin J., Li Y., Cai Z., Li S., Zhu J., Zhang F. A metagenome-wide association study of gut microbiota in type 2 diabetes. Nature. 2012;490:55–60. doi: 10.1038/nature11450. [DOI] [PubMed] [Google Scholar]
- 50.Frank D.N., St Amand A.L., Feldman R.A., Boedeker E.C., Harpaz N. Pace NR. Molecular-phylogenetic characterization of microbial community imbalances in human inflammatory bowel diseases. Proc Natl Acad Sci U S A. 2007;104:13780–13785. doi: 10.1073/pnas.0706625104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wu S., Rhee K.J., Albesiano E., Rabizadeh S., Wu X., Yen H.R. A human colonic commensal promotes colon tumorigenesis via activation of T helper type 17 T cell responses. Nat Med. 2009;15:1016–1022. doi: 10.1038/nm.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Balamurugan R., Rajendiran E., George S., Samuel G.V., Ramakrishna B.S. Real-time polymerase chain reaction quantification of specific butyrateproducing bacteria, Desulfovibrio and Enterococcus faecalis in the feces of patients with colorectal cancer. J Gastroenterol Hepatol. 2008;23:1298–1303. doi: 10.1111/j.1440-1746.2008.05490.x. [DOI] [PubMed] [Google Scholar]
- 53.Hemarajata P., Versalovic J. Effects of probiotics on gut microbiota: mechanisms of intestinal immunomodulation and neuromodulation. Therap Adv Gastroenterol. 2013;6:39–51. doi: 10.1177/1756283X12459294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Fernandes J., Su W., Rahat-Rozenbloom S., Wolever T.M., Comelli E.M. Adiposity, gut microbiota and faecal short chain fatty acids are linked in adult humans. Nutr Diabetes. 2014;4:e121. doi: 10.1038/nutd.2014.23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Qin J., Li R., Raes J., Arumugam M., Burgdorf K.S., Manichanh C. A human gut microbial gene catalogue established by metagenomic sequencing. Nature. 2010;464:59–65. doi: 10.1038/nature08821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Lupp C., Robertson M.L., Wickham M.E., Sekirov I., Champion O.L., Gaynor E.C. Host-mediated inflammation disrupts the intestinal microbiota and promotes the overgrowth of Enterobacteriaceae. Cell Host Microbe. 2007;2:204. doi: 10.1016/j.chom.2007.08.002. [DOI] [PubMed] [Google Scholar]
- 57.Bloom S.M., Bijanki V.N., Nava G.M., Sun L., Malvin N.P., Donermeyer D.L. Commensal Bacteroides species induce colitis in host-genotype-specific fashion in a mouse model of inflammatory bowel disease. Cell Host Microbe. 2011;9:390–403. doi: 10.1016/j.chom.2011.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Reyes A., Haynes M., Hanson N., Angly F.E., Heath A.C., Rohwer F. Viruses in the faecal microbiota of monozygotic twins and their mothers. Nature. 2010;466:334–338. doi: 10.1038/nature09199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kelder T., Stroeve J.H., Bijlsma S., Radonjic M., Roeselers G. Correlation network analysis reveals relationships between diet-induced changes in human gut microbiota and metabolic health. Nutr Diabetes. 2014;4:e122. doi: 10.1038/nutd.2014.18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Peterson D.A., Frank D.N., Pace N.R., Gordon J.I. Metagenomic approaches for defining the pathogenesis of inflammatory bowel diseases. Cell Host Microbe. 2008;3:417–427. doi: 10.1016/j.chom.2008.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Elinav E., Strowig T., Kau A.L., Henao-Mejia J., Thaiss C.A., Booth C.J. NLRP6 inflammasome regulates colonic microbial ecology and risk for colitis. Cell. 2011;145:745–757. doi: 10.1016/j.cell.2011.04.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Vijay-Kumar M., Aitken J.D., Carvalho F.A., Cullender T.C., Mwangi S., Srinivasan S. Metabolic syndrome and altered gut microbiota in mice lacking Toll-like receptor 5. Science. 2010;328:228–231. doi: 10.1126/science.1179721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Faith J.J., McNulty N.P., Rey F.E., Gordon J.I. Predicting a human gut microbiota’s response to diet in gnotobiotic mice. Science. 2011;333:101–104. doi: 10.1126/science.1206025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Wang Z., Klipfell E., Bennett B.J., Koeth R., Levison B.S., Dugar B. Gut flora metabolism of phosphatidylcholine promotes cardiovascular disease. Nature. 2011;472:57–63. doi: 10.1038/nature09922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Clayton T.A., Baker D., Lindon J.C., Everett J.R., Nicholson J.K. Pharmacometabonomic identification of a significant host-microbiome metabolic interaction affecting human drug metabolism. Proc Natl Acad Sci U S A. 2009;106:14728–14733. doi: 10.1073/pnas.0904489106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Aura A.M., Mattila I., Hyotylainen T., Gopalacharyulu P., Bounsaythip C., Oresic M. Drug metabolome of the simvastatin formed by human intestinal microbiota in vitro. Mol Biosyst. 2011;7:437–446. doi: 10.1039/c0mb00023j. [DOI] [PubMed] [Google Scholar]
- 67.Saha J.R., Butler V.P., Jr, Neu H.C., Lindenbaum J. Digoxin-inactivating bacteria: identification in human gut flora. Science. 1983;220:325–327. doi: 10.1126/science.6836275. [DOI] [PubMed] [Google Scholar]
- 68.Jia W., Li H., Zhao L., Nicholson J.K. Gut microbiota: a potential new territory for drug targeting. Nat Rev Drug Discov. 2008;7:123–129. doi: 10.1038/nrd2505. [DOI] [PubMed] [Google Scholar]
- 69.Wallace B.D., Wang H., Lane K.T., Scott J.E., Orans J., Koo J.S. Alleviating cancer drug toxicity by inhibiting a bacterial enzyme. Science. 2010;330:831–835. doi: 10.1126/science.1191175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Vitali B., Ndagijimana M., Cruciani F., Carnevali P., Candela M., Guerzoni M.E. Impact of a synbiotic food on the gut microbial ecology and metabolic profiles. BMC Microbiol. 2010;10:4. doi: 10.1186/1471-2180-10-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Jones S.E., Versalovic J. Probiotic Lactobacillus reuteri biofilms produce antimicrobial and anti-inflammatory factors. BMC Microbiol. 2009;9:35. doi: 10.1186/1471-2180-9-35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Pagnini C., Saeed R., Bamias G., Arseneau K.O., Pizarro T.T., Cominelli F. Probiotics promote gut health through stimulation of epithelial innate immunity. Proc Natl Acad Sci U S A. 2010;107:454–459. doi: 10.1073/pnas.0910307107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Wolvers D., Antoine J.M., Myllyluoma E., Schrezenmeir J., Szajewska H., Rijkers G.T. Guidance for substantiating the evidence for beneficial effects of probiotics: prevention and management of infections by probiotics. J Nutr. 2010;140:698S–712S. doi: 10.3945/jn.109.113753. [DOI] [PubMed] [Google Scholar]
- 74.Floch M.H., Walker W.A., Madsen K., Sanders M.E., Macfarlane G.T., Flint H.J. Recommendations for probiotic use-2011 update. J Clin Gastroenterol. 2011;45:S168–S171. doi: 10.1097/MCG.0b013e318230928b. [DOI] [PubMed] [Google Scholar]
- 75.Faust K., Sathirapongsasuti J.F., Izard J., Segata N., Gevers D., Raes J. Microbial co-occurrence relationships in the human microbiome. PLoS Comput Biol. 2012;8:e1002606. doi: 10.1371/journal.pcbi.1002606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Chaffron S., Rehrauer H., Pernthaler J., von Mering C. A global network of coexisting microbes from environmental and whole-genome sequence data. Genome Res. 2010;20:947–959. doi: 10.1101/gr.104521.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Mounier J., Monnet C., Vallaeys T., Arditi R., Sarthou A.S., Helias A. Microbial interactions within a cheese microbial community. Appl Environ Microbiol. 2008;74:172–181. doi: 10.1128/AEM.01338-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Morowitz M.J., Denef V.J., Costello E.K., Thomas B.C., Poroyko V., Relman D.A. Strain-resolved community genomic analysis of gut microbial colonization in a premature infant. Proc Natl Acad Sci U S A. 2011;108:1128–1133. doi: 10.1073/pnas.1010992108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Greenblum S., Turnbaugh P.J., Borenstein E. Metagenomic systems biology of the human gut microbiome reveals topological shifts associated with obesity and inflammatory bowel disease. Proc Natl Acad Sci U S A. 2012;109:594–599. doi: 10.1073/pnas.1116053109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Schloissnig S., Arumugam M., Sunagawa S., Mitreva M., Tap J., Zhu A. Genomic variation landscape of the human gut microbiome. Nature. 2013;493:45–50. doi: 10.1038/nature11711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Karlsson F., Tremaroli V., Nielsen J., Bäckhed F. Assessing the human gut microbiota in metabolic diseases. Diabetes. 2013;62:3341–3349. doi: 10.2337/db13-0844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Caporaso J.G., Kuczynski J., Stombaugh J., Bittinger K., Bushman F.D., Costello E.K. QIIME allows analysis of high-throughput community sequencing data. Nat Methods. 2010;7:335–336. doi: 10.1038/nmeth.f.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Schloss P.D., Westcott S.L., Ryabin T., Hall J.R., Hartmann M., Hollister E.B. Introducing mothur: open-source platform-independent community-supported software for describing and comparing microbial communities. Appl Environ Microbiol. 2009;75:7537–7541. doi: 10.1128/AEM.01541-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Li W. Analysis and comparison of very large metagenomes with fast clustering and functional annotation. BMC Bioinformatics. 2009;10:359. doi: 10.1186/1471-2105-10-359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Huson D.H., Mitra S., Ruscheweyh H.J., Weber N., Schuster S.C. Integrative analysis of environmental sequences using MEGAN4. Genome Res. 2011;21:1552–1560. doi: 10.1101/gr.120618.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Segata N., Waldron L., Ballarini A., Narasimhan V., Jousson O., Huttenhower C. Metagenomic microbial community profiling using unique clade-specific marker genes. Nat Methods. 2012;9:811–814. doi: 10.1038/nmeth.2066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Namiki T., Hachiya T., Tanaka H., Sakakibara Y. MetaVelvet: an extension of Velvet assembler to de novo metagenome assembly from short sequence reads. Nucleic Acids Res. 2012;40:e155. doi: 10.1093/nar/gks678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Luo R., Liu B., Xie Y., Li Z., Huang W., Yuan J. SOAPdenovo2: an empirically improved memory-efficient short-read de novo assembler. Gigascience. 2012;1:18. doi: 10.1186/2047-217X-1-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Kultima J.R., Sunagawa S., Li J., Chen W., Chen H., Mende D.R. MOCAT: a metagenomics assembly and gene prediction toolkit. PLoS ONE. 2012;7:e47656. doi: 10.1371/journal.pone.0047656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Arumugam M., Harrington E.D., Foerstner K.U., Raes J., Bork P. SmashCommunity: a metagenomic annotation and analysis tool. Bioinformatics. 2010;26:2977–2978. doi: 10.1093/bioinformatics/btq536. [DOI] [PubMed] [Google Scholar]
- 91.Abubucker S., Segata N., Goll J., Schubert A.M., Izard J., Cantarel B.L. Metabolic reconstruction for metagenomic data and its application to the human microbiome. PLoS Comput Biol. 2012;8:e1002358. doi: 10.1371/journal.pcbi.1002358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Sanli K., Karlsson F.H., Nookaew I., Nielsen J. FANTOM: functional and taxonomic analysis of metagenomes. BMC Bioinformatics. 2013;14:38. doi: 10.1186/1471-2105-14-38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Teeling H., Waldmann J., Lombardot T., Bauer M., Glockner F.O. TETRA:a web-service and a stand-alone program for the analysis and comparison of tetranucleotide usage patterns in DNA sequences. BMC Bioinformatics. 2004;5:163. doi: 10.1186/1471-2105-5-163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Liu J., Wang H., Yang H., Zhang Y., Wang J., Zhao F. Composition-based classification of short metagenomic sequences elucidates the landscapes of taxonomic and functional enrichment of microorganisms. Nucleic Acids Res. 2013;41:e3. doi: 10.1093/nar/gks828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.McHardy A.C., Martin H.G., Tsirigos A., Hugenholtz P., Rigoutsos I. Accurate phylogenetic classification of variable-length DNA fragments. Nat Methods. 2007;4:63–72. doi: 10.1038/nmeth976. [DOI] [PubMed] [Google Scholar]
- 96.Patil K.R., Roune L., McHardy A.C. The PhyloPythiaS web server for taxonomic assignment of metagenome sequences. PLoS ONE. 2012;7:e38581. doi: 10.1371/journal.pone.0038581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Brady A., Salzberg S.L. Phymm and PhymmBL: metagenomic phylogenetic classification with interpolated Markov models. Nat Methods. 2009;6:673–676. doi: 10.1038/nmeth.1358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Goll J., Rusch D.B., Tanenbaum D.M., Thiagarajan M., Li K., Methe B.A. METAREP: JCVI metagenomics reports–an open source tool for high-performance comparative metagenomics. Bioinformatics. 2010;26:2631–2632. doi: 10.1093/bioinformatics/btq455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Li W., Godzik A. Cd-hit:a fast program for clustering and comparing large sets of protein or nucleotide sequences. Bioinformatics. 2006;22:1658–1659. doi: 10.1093/bioinformatics/btl158. [DOI] [PubMed] [Google Scholar]
- 100.Arndt D., Xia J., Liu Y., Zhou Y., Guo A.C., Cruz J.A. METAGENassist: a comprehensive web server for comparative metagenomics. Nucleic Acids Res. 2012;40:W88–W95. doi: 10.1093/nar/gks497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Lingner T., Asshauer K.P., Schreiber F., Meinicke P. CoMet–a web server for comparative functional profiling of metagenomes. Nucleic Acids Res. 2011;39:W518–W523. doi: 10.1093/nar/gkr388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Gerlach W., Junemann S., Tille F., Goesmann A., Stoye J. WebCARMA:a web application for the functional and taxonomic classification of unassembled metagenomic reads. BMC Bioinformatics. 2009;10:430. doi: 10.1186/1471-2105-10-430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Meyer F., Paarmann D., D’Souza M., Olson R., Glass E.M., Kubal M. The metagenomics RAST server - a public resource for the automatic phylogenetic and functional analysis of metagenomes. BMC Bioinformatics. 2008;9:386. doi: 10.1186/1471-2105-9-386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Seshadri R., Kravitz S.A., Smarr L., Gilna P., Frazier M. CAMERA: a community resource for metagenomics. PLoS Biol. 2007;5:e75. doi: 10.1371/journal.pbio.0050075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Wu S., Zhu Z., Fu L., Niu B., Li W. WebMGA: a customizable web server for fast metagenomic sequence analysis. BMC Genomics. 2011;12:444. doi: 10.1186/1471-2164-12-444. [DOI] [PMC free article] [PubMed] [Google Scholar]