Abstract
Appropriate regulation of regional uterine stromal cell decidualization in implantation, at the mesometrial triangle and secondary decidual zone (SDZ) locations, is critical for successful pregnancy, although the regulatory mechanisms remain poorly understood. In this regard, the available animal models that would specifically allow mechanistic analysis of site-specific decidualization are strikingly limited. Our study found that heightened expression of FoxM1, a Forkhead box transcription factor, is regulated during decidualization, and its conditional deletion in mice reveals failure of implantation with regional decidualization defects such as a much smaller mesometrial decidua with enlarged SDZ. Analysis of cell cycle progression during decidualization both in vivo and in vitro demonstrates that the loss of FoxM1 elicits diploid cell deficiency with enhanced arrests prior to mitosis and concomitant upregulation of polyploidy. We further showed that Hoxa10 and cyclin D3, two decidual markers, control transcriptional regulation and intra-nuclear protein translocation of FoxM1 in polyploid cells, respectively. Overall, we suggest that proper regional decidualization and polyploidy development requires FoxM1 signaling downstream of Hoxa10 and cyclin D3.
Uterine stromal cells undergo transformation into morphologically and functionally distinct cells called decidual cells (decidualization), which occurs in women during the secretory phase of the menstrual cycle as well as in pregnancy; in rodents, this process only occurs during pregnancy. The onset of decidualization following embryo implantation is essential for successful pregnancy1,2. In the receptive uterus on day 4 (D4) of pregnancy (D1 = vaginal plug) in mice, uterine stromal cells experience proliferation under the coordinated control of both ovarian estrogen and progesterone. However, following embryonic attachment to the uterine luminal epithelium, which occurs at 24:00 h on D4, stromal cells proximally surrounding the implantation chamber exhibit rapid proliferation and spreading. By D5 morning, these cells can be found throughout the stromal bed. The first sign of stromal differentiation, forming of the primary decidual zone (PDZ), occurs in the first few layers of cells at the antimesometrial location of the implantation site (IS) in the afternoon on D53,4. PDZ is avascular and epithelioid in nature5. From D6 through D8, stromal cells next to the PDZ continue to proliferate and differentiate to form polyploidy in the secondary decidual zone (SDZ), which develops both at the lateral and antimesometrial locations of the IS. In contrast to SDZ development, mesometrial stromal cells continue to proliferate and differentiate to form the non-polyploid decidual zone, a presumptive site for placentation.
Decidual polyploidization is a hallmark of terminally differentiated cells and has been well characterized in rodents3,4,6,7,8,9 and recently recognized in humans [Hirota Y and Dey SK (unpublished observations)]. These cells undergo endoreduplication cycle to develop as giant mono- or bi-nuclear cells with multiple copies of chromosomes3,4,6,7,8,9 and possess increased mitochondrial activity6. The loss of decidual polyploidy in association with pregnancy failure by mid-gestation has been reported in Dedd null mice10.
Uterine decidualization in implantation is believed to be regulated through complex signaling mechanisms that involve homeobox transcription factors, cell-cycle genes, cytokines, growth factors, lipid mediators, and other regulatory molecules1,2,11,12. However, there remains a major gap in understanding the mechanisms that control regional (mesometrial vs. antimesometrial) decidual development in implantation. The homeobox transcription factor Hoxa10 has been shown to play an important role in directing proper regional decidual development11,13. It has been shown the Hoxa10 null mutation in mice produces a lack of uterine stromal cell proliferation in response to progesterone and consequentially results in the failure of proper decidua growth14,15,16. Consistently, cyclin D3—a G1 phase cell cycle regulator for stromal cell proliferation, differentiation, and polyploidy development3,4,17—exhibits severe downregulation of expression during decidual progression in Hoxa10 null mice13,17. Moreover, studies have shown that adenovirus-driven overexpression of cyclin D3 at the site of implantation improves decidualization defects in Hoxa10−/− mice18, indicating cyclin D3 plays an important role downstream of Hoxa10 during decidualization.
FoxM1, a member of the large family of Forkhead box transcription factors, is highly expressed in proliferating cells and plays pivotal roles in DNA replication and mitosis through modulation of diverse regulatory genes involved in transitions between G1-S and G2-M phases of the cell cycle19. It has been well recognized that FoxM1 is robustly expressed by oncogenic signals in almost all types of malignant tumor tissues and cancer cell lines20, and is highly expressed in a broad range of tissues during embryo development19,21. However, its expression is found in few normal adult tissues19. Our findings as reported here have provided new evidence that FoxM1 is expressed and regulated in the early post-implantation uteri during decidualization. By utilizing genetic knockout mouse models, we have provided novel evidence that FoxM1 is regulated during stromal cell decidualization and uterine conditional deletion of FoxM1 reveals regional decidualization defects via impaired stromal cell mitosis and aberrantly upregulated polyploidy at the site of implantation. Further, we showed that FoxM1 is regulated at the transcriptional level by Hoxa10 and in its intra-nuclear protein localization by cyclin D3.
Results
FoxM1 is regulated during uterine stromal cell proliferation and differentiation for decidualization
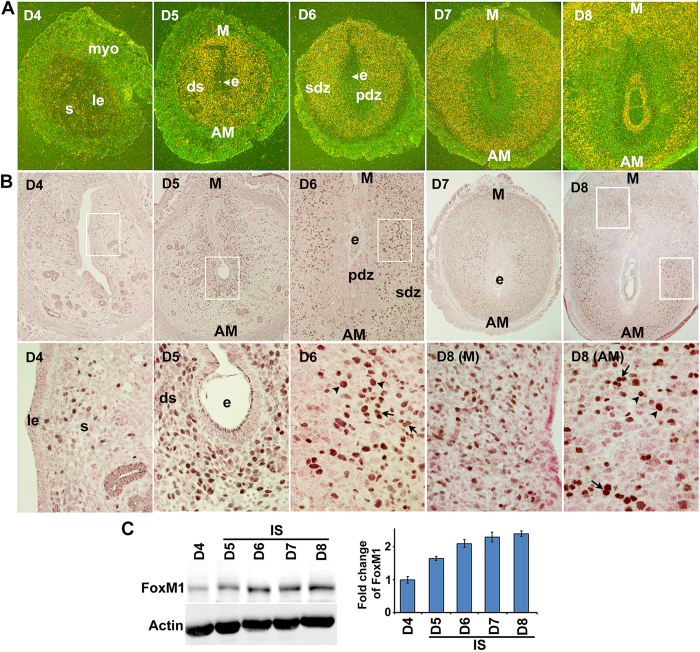
To better understand the role of uterine FoxM1 during the periimplantation period, we examined the spatiotemporal expression of FoxM1 mRNA and protein on the receptive day (D4) and postimplantation uteri on D5-8. Our in situ hybridization results show a moderate expression with scattered distribution within the endometrial stroma on D4. In contrast, a heightened expression was noted in decidualizing stromal cells throughout the endometrium at the IS on D5 (Fig. 1A). However, at the IS on D6-8, FoxM1 expression was predominantly localized in the SDZ, as well as in the M-polar decidual bed (Fig. 1A), and this pattern of expression was enhanced with the progression of pregnancy. Cell-specific localization of FoxM1 immunoreactive protein was consistent with that of mRNAs (Fig. 1B). More specifically, nuclear FoxM1 was noted with a scattered distribution in the stromal bed on D4 (Fig. 1B). On D5, nuclear signals were distinctly upregulated in the sub-luminal stromal cells at the IS (Fig. 1B). However, on D6–8, both decidual polyploid (in SDZ) and non-polyploid (in M-polar bed) cells were positive for nuclear FoxM1 localization (Fig. 1B). Consistent with the above results, further quantitative analyses of FoxM1 by western blotting also revealed an increased gradual accumulation of protein levels at the IS on D5-8, while D4 also had low levels (Fig. 1C).
Figure 1. FoxM1 expression in peri-implantation uteri.
(A) In situ hybridization of FoxM1 expression on D4-8 of pregnancy. le, luminal epithelium; s, stroma; myo, myometrium; e, embryo; ds, decidualizing stroma; pdz, primary decidual zone; sdz, secondary decidual zone; M, mesometrial; AM, antimesometrial. The magnifications are at 40X. (B) Immunostaining of FoxM1 on D4-8 of pregnancy. Dark brown staining indicates positive signals; nuclei are counterstained with fast red. le, luminal epithelium; s, stroma; e, embryo; ds, decidualizing stroma; pdz, primary decidual zone; sdz, secondary decidual zone; M, mesometrial; AM, antimesometrial. Arrows and arrowheads indicate mono and bi-nucleated polyploid cells, respectively. Lower panels show magnified images for selected areas shown in corresponding upper panels. The magnifications in the upper panels are at 100X (D4-6) and 40X (D7-8), while in the lower panels are at 400X. (C) Western blotting for FoxM1 analysis. Bar diagram shows relative levels of expression after normalization with actin.
Since FoxM1 is critical for cell cycle progression, we next analyzed FoxM1 expression by combined immunofluorescence with cell cycle phase-specific markers, incorporating BrdU (S-phase for DNA synthesis) and phosphorylated histone H3 (pHH3) (M-phase for mitosis) in periimplantation uterine sections on D4 as well as at D5-D8 IS. In general, FoxM1 positive stromal/decidual cells were co-localized either with BrdU or pHH3 on D4-D8 of pregnancy, indicating that FoxM1 indeed plays a role in cell cycle progression during the S- and M-phases (Supplementary Fig. S1A, representative data on D6-IS is shown for mesometrial and antimesometrial locations). However, quantitative analyses of localization showed that although dual positive FoxM1/BrdU cells were significantly upregulated at the mesometrial location from D5 through D8, there was a slight decline in levels on D7 and D8 (Supplementary Fig. S1B). Similarly, FoxM1/pHH3 cells were also upregulated between D6 through D8, but with a decline on D8 (Supplementary Fig. S1B). In contrast, at the SDZ for both lateral (L) and antimesometrial (AM) locations, FoxM1/BrdU double labelled cells were significantly increased from D6 through D8 with a peak on D6, while FoxM1/pHH3 cells did not reveal any change (Supplementary Fig. S1B) This suggests that FoxM1 positive SDZ cells may lack mitotic progression despite an increase in S-phase activity, a condition that is conducive for polyploidy development3,4. Overall, FoxM1 expression is closely associated with stromal cell proliferation and differentiation, including terminal differentiation with polyploidy development.
Uterine deletion of FoxM1 leads to decidualization defects in early pregnancy
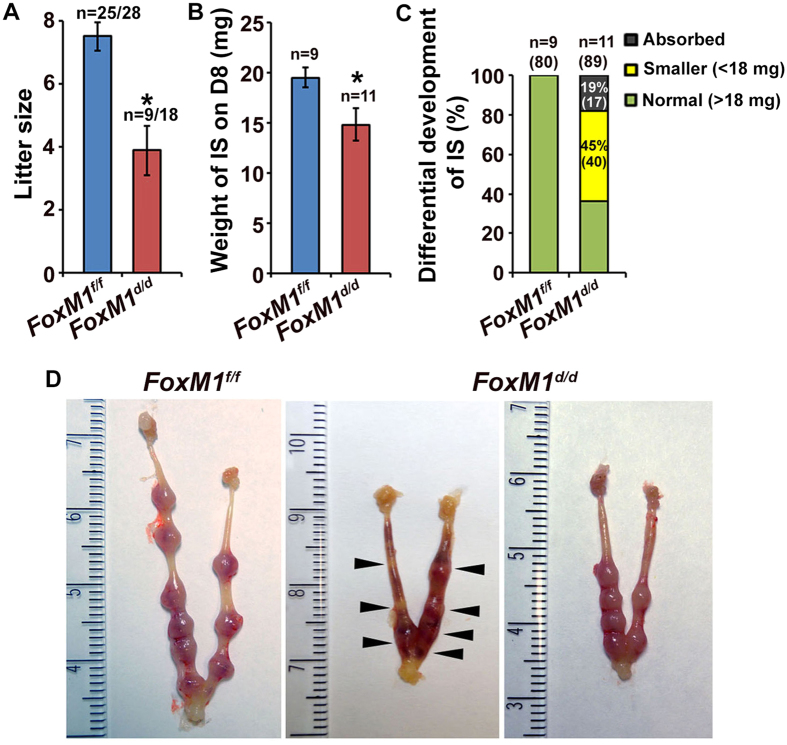
To explore the role of uterine FoxM1 in early pregnancy, we used PR-Cre mice to specifically delete FoxM1 in the uterus. Our analyses clearly revealed deletion of FoxM1 in null (FoxM1d/d) as compared to control (FoxM1f/f) at the IS on D8, as examined by qRT-PCR (Supplementary Fig. S2A). We next analyzed pregnancy outcome between null and control mice. We observed that the homozygous deletion of FoxM1 causes severe subfertility, as compared to control (Fig. 2A). These results were consistent with a significant decrease in average IS weight on D8 for null mice, as compared to control (Fig. 2B). However, based on uterine gross morphology and distribution of D8 IS weight, we arbitrarily divided into three parts for null: ∼19% (17 out of 89) of IS were primarily absorbed and darkly bloody while another ∼45% (40 out of 89) of IS were much smaller (<18 mg) without showing any sign of resorption compared to normal (>18 mg) (Fig. 2C,D), indicating that loss of FoxM1 differentially affects IS’s developmental progression during decidualization. Consistent with these observations, after the induction of artificial decidualization, we also noted that only 37.5% of null mice were responsive to the onset of decidualization, but with a reduction in response by ∼3-fold, as compared to control (Supplementary Fig. S3A–C). In addition, the above defects were not associated with any disturbances in ovarian hormone levels in serum (Supplementary Fig. S2B) or ovarian corpora luteal structures (Supplementary Fig. S2C). Whereas only ovarian granulosa cells in the preantral/antral follicles for control mice had detectable FoxM1 expression and this was not affected in null mice (Supplementary Fig. S2C); this was consistent with the findings of a prior study that found only corpus luteal Cre-expression using a Pgr-driven promoter22. Overall, our results suggest that insufficient decidual progression in implantation is a primary cause for early pregnancy defects in FoxM1d/d mice.
Figure 2. Uterine deletion of FoxM1 results in female subfertility with loss of embryo implantation during decidualization.
(A) Pregnancy outcome in null (FoxM1d/d) and control (FoxM1f/f) littermates after mating with fertile males. n, indicates number of females with pups out of total plug-positive females for each genotype. Results shown as Mean ± SEM. *Significantly different (p < 0.05). (B) Weight of IS on D8 for null and control littermates. n, indicates number of females with IS for each genotype. Results shown as Mean ± SEM. (C) Developmental competence of IS between null and control on D8. n, indicates number of pregnant females, with a total number of IS (in parentheses) for each group. The percent with total number of IS for each category (absorbed, smaller, and normal) also indicated by color codes. (D) Uterine IS morphology for FoxM1d/d and FoxM1f/f mice is shown on D8. Arrowheads point to resorption sites.
FoxM1 deficiency leads to mesometrial decidua shrinkage with increased SDZ and polyploidy development in early pregnancy
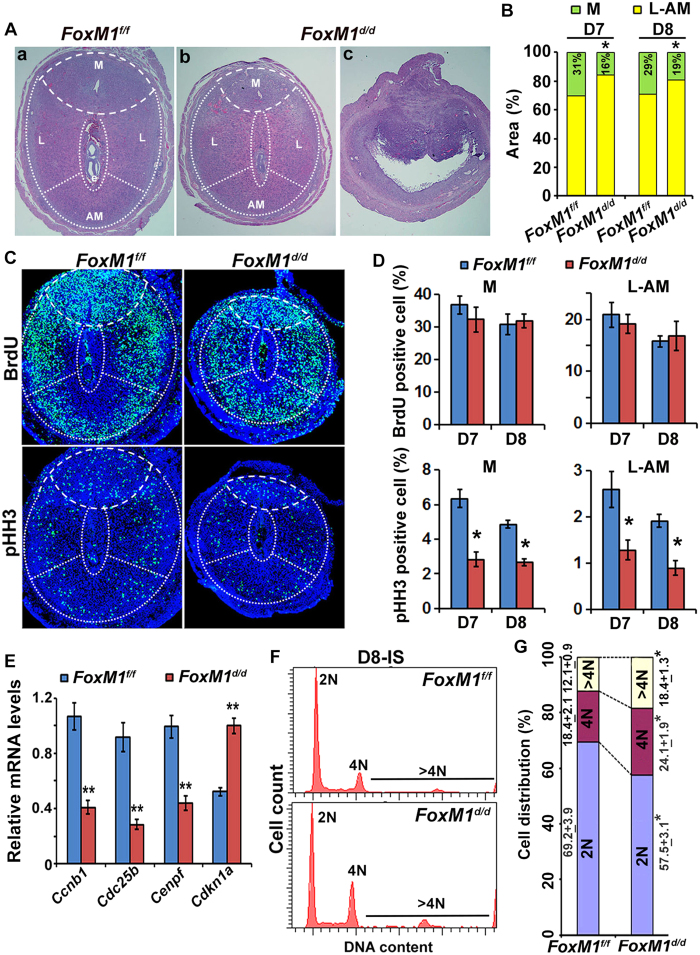
To determine the cause for early implantation defects, we compared implantation progression histologically on serial sections between FoxM1 null and control mice. As expected, null ISs were found to be in the process of resorption, as clearly revealed by embryo loss and collapse of decidua structure (Fig. 3Ac). However, the analysis of partially compromised null sites revealed that the mesometrial decidual bed was significantly smaller with a concomitant increase in the SDZ (lateral and antimesometrial regions) than control on D8 (Fig. 3Aa,b,B), as well as on D7 (Fig. 3B). Embryonic growth was also significantly smaller for null as compared to control (Fig. 3Aa,b). Despite compromised decidualization, gene expression for stromal cell decidualization (Bmp2 and Hoxa-10)12,16, local vascularization (Ang2 and Ptgs2)23,24, and polyploidy development (Ccnd3 and Trp53)3,25,26 was not affected at the IS on D7 or D8 between control and null mice, indicating FoxM1 is not involved with the regulation of these molecular events. Overall, FoxM1 essentially controls proper regional decidualization without affecting many implantation specific genes.
Figure 3. Defective regional decidualization with abnormal cell cycle activity results from the loss of uterine FoxM1.
(A) (a,b), Histological analyses by hematoxylin and eosin staining of D8-IS between null (FoxM1d/d) and control (FoxM1f/f) mice. Mesometrial (M), lateral (L) and antimesometrial (AM) locations are demarcated by broken lines. (c), A representative IS shows signs of resorption for null mice. (B) Comparison of developmental area (%) for M and SDZ (L+AM) locations on D7-8 IS between null and control mice. *Significantly different (p < 0.001) for corresponding M or SDZ locations between null and control on particular day of pregnancy. (C) Immunofluorescence analyses of BrdU and pHH3 at D7-IS between null and control. (D) Quantitative analyses of BrdU or pHH3 positively stained cells (%) in M and SDZ locations of D7-8 IS between null and control mice. *Significantly different (p < 0.001) between null vs. control. (E) Quantitative RT-PCR analyses of cell cycle regulatory genes [Ccnb1, Cdc25b, Cenpf, and Cdkn1a (p21)] at D7-IS for control and null mice. **Significantly different (p < 0.001) between null vs. control. (F) A representative flow cytometric analysis of cell cycle distribution based on DNA content for decidual cells isolated on D8-IS between null vs. control. (G) Cellular distribution (%) for 2N, 4N, and >4N populations at the D8-IS between null vs. control. *Significantly different (p < 0.001) for corresponding groups between control vs null mice.
We next evaluated whether cell cycle activity was affected by the loss of FoxM1 during decidualization. Analyses of BrdU (for S-phase) and pHH3 (for M-phase) at the IS on D7 (Fig. 3C,D) and D8 (Fig. 3D) revealed that although accumulation of BrdU positive cells was not affected, pHH3 labelled cells at the mesometrial and SDZ (L+M) locations were significantly reduced in null as compared to control, indicating loss of FoxM1 crucially affects cell cycle progression in M-phase, but not S-phase, during decidual progression. Consistently, quantitative RT-PCR analyses demonstrated that expression of G2/M phase regulators Ccnb1, Cdc25b, and Cenpf were significantly downregulated, whereas the cyclin-dependent kinase inhibitor/differentiation regulator Cdkn1a (p21) was upregulated in null as compared to control (Fig. 3E). Interestingly, all aforementioned mitotic genes have been known to act as transcriptional targets upregulated by FoxM127,28,29. P21 has been found to be downregulated by FoxM1 in other cell types29,30. However, the expression of several G1/S phase regulators (Ccnd3, Ccna1, Ccna2, Ccne1, Ccne2, Cdc25a, cdk4, cdk6, cdk2, E2f1, Cks1b, Skp2 and Gas1), other G2/M phase regulators (Ccnb2, cdk1, Plk, Aurkb, Birc5 and Nek2), and cdk inhibitors (cdkn1b and cdkn1c) were not affected between the mice, although many of these genes have been shown as FoxM1 transcriptional targets in other cell types30.
The arrest of decidual cell development prior to entering M-phase is crucially linked with the onset of polyploidy3,4; thus, we next analyzed ploidy levels by flow cytometric analyses of DNA content between null and control mice during decidualization. Our analyses revealed that 2N cells were significantly decreased with a concomitant enhancement of 4N and >4N cells within the decidual bed for null mice as compared to control on D8-IS (Fig. 3F,G), suggesting FoxM1 is critical for the balanced progression of diploid cell and polyploidy levels during decidualization. This phenomenon was also consistent with our observation of increased bi-nuclear polyploid cells in the SDZ for null mice as compared to control on D8 (Supplementary Fig. S4). Overall, the proper development of mesometrial decidua and SDZ at the site of implantation necessitates uterine FoxM1, primarily to appropriately control diploid vs. polyploid decidual cell status during decidualization.
FoxM1 is critical to appropriately balance mitosis and polyploidy development during stromal cell decidualization in vitro
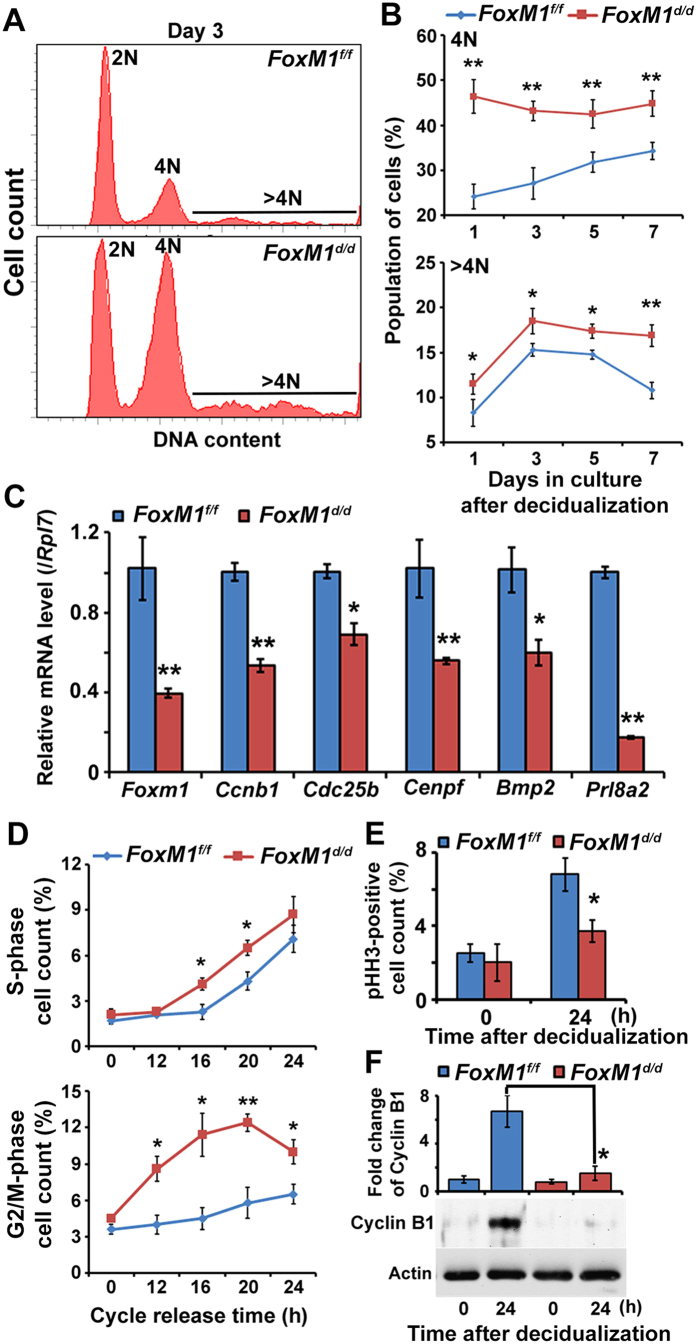
To analyze the mechanism for aberrant polyploidy development in FoxM1d/d mice, we examined cell cycle progression during stromal cell decidualization in vitro. Following the induction of decidualization31, our analysis revealed that FoxM1 null stromal cells, as opposed to control, were able to show increased accumulation of 4N and >4N (polyploid) cells during days 1–7 in culture (Fig. 4A,B), indicating FoxM1-deficiency results in aberrant upregulation of cells with polyploidy. Meanwhile, 2N cells decreased in null compared to control. FoxM1 null cells exhibited aberrant downregulation of expression for G2/M-phase cell cycle regulators (Ccnb1, Cdc25b, and Cenpf) and markers of decidualization (Bmp2 and Prl8a2), as compared to control (Fig. 4C), indicating FoxM1-deficient cells probably suffer from cell cycle arrest during G2/M with defective decidual progression. Because decidualization process depends on simultaneous progression of proliferation and differentiation, to specifically define FoxM1 regulation in proliferation, synchronized FoxM1 null or control stromal cells were reactivated in order to re-enter the cell cycle by serum for 0–24 h in culture. Our analyses revealed that despite significantly enhanced accumulation of FoxM1 null cells compared to control, either in S (at 16–20 h) or G2/M (at 12–24 h) (Fig. 4D), the level of pHH3 (M-phase marker)-positive cells (Fig. 4E) and expression of cyclin B1 (M-phase regulator) (Fig. 4F) were strikingly lower at 24 h post reactivation for null cells compared to control, indicating FoxM1 deficiency results in cell cycle arrest prior to the M phase. Our analysis of FoxM1 in control cells revealed that the expression was induced both at mRNA and protein levels, during cell cycle phase progression, particularly at 12–24 h following the cycle release (Supplementary Fig. S5). In addition, FoxM1 expression by immunofluorescence localization did not reveal any obvious intracellular trafficking, since the expression remained in nuclear location during this period. Overall, the loss of FoxM1 in stromal cells leads to enhanced cell cycle arrest prior to mitosis with aberrantly upregulated polyploidy during decidualization in vitro.
Figure 4. FoxM1 deficiency in stromal cells leads to enhanced cell cycle arrest prior to mitosis with aberrantly upregulated polyploidy during decidualization in vitro.
(A) A representative flow cytometric analysis of cell cycle distribution based on DNA content for cells after in vitro decidualization for 3-days between null (FoxM1d/d) vs. control (FoxM1f/f) stromal cells. (B) Cellular distribution (%) for 4N and >4N populations after in vitro decidualization at indicated days (1–7 days) between null vs. control cells. **p < 0.001 and *p < 0.05. (C) Quantitative RT-PCR analyses of FoxM1, Ccnb1, Cdc25b, Cenpf, Bmp2, and Prl8a2 after in vitro decidualization for 3 days between null and control cells. **p < 0.001 and *p < 0.05. (D) Cell cycle phase-specific count (%) in S and G2/M after the release in cell cycle for proliferation in vitro at indicated times (0–24 h) between null and control cells. **p < 0.001 and *p < 0.05. E. Quantitative analysis of pHH3-positive cell count (%) after in vitro decidualization at indicated times (0 and 24 h) between null and control cells. *p < 0.05. F. Analysis of Cyclin B1 protein after in vitro decidualization at indicated times (0 and 24 h) between null and control cells. *p < 0.001.
FoxM1 expression is regulated at the transcription level by Hoxa10 during decidualization
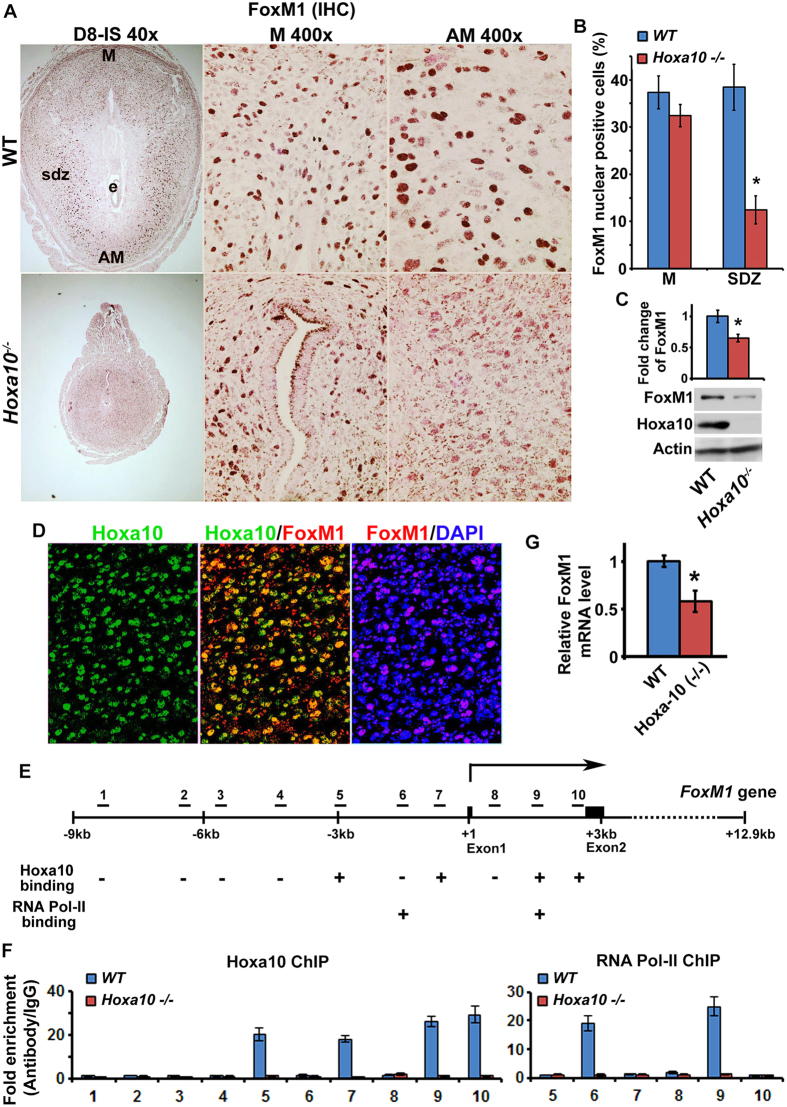
Hoxa10 is a major regulator of decidualization in early pregnancy11,13,14. To determine whether FoxM1 is regulated downstream of Hoxa10 during decidualization, we examined FoxM1 expression at IS between Hoxa10−/− mice as compared to wild-type (WT) on D8. Immunohistochemical analyses show that nuclear FoxM1 expression at the SDZ was significantly reduced in Hoxa10−/− mice as compared to WT (Fig. 5A,B). In contrast, mesometrial FoxM1 expression was comparable between WT and Hoxa10 null mice (Fig. 5A,B). Consistently, quantitation of protein levels by western blotting also revealed downregulation of FoxM1 expression in Hoxa10 null, as compared to WT (Fig. 5C). Since Hoxa10 is a transcriptional factor, we next examined whether the above regulation of FoxM1 expression is directly controlled by Hoxa10. To examine this notion, we first evaluated dual expression analysis of FoxM1 and Hoxa10 at IS on D8 for WT. Based on dual immunofluorescence studies, we observed that FoxM1 expression was co-localized with Hoxa10 at the SDZ (Fig. 5D). Next, to analyze a possibility for direct regulation of FoxM1 by Hoxa10 during decidualization, we examined the relationship by chromatin immunoprecipitation (ChIP) studies. We identified several Hoxa10 binding consensus sequences by computational analysis of the 10 kb region for the 5′-flanking, first exon, and first intron of the FoxM1 gene (Fig. 5E). Our analyses of Hoxa10 ChIP followed by qPCR showed that several distinct regions within the promoter and gene body are responsible for Hoxa10 binding, since they are undetected in Hoxa10−/− mice (Fig. 5F). These results are consistent with analyses for RNA polymerase II recruitment by ChIP (Fig. 5F) and the status of FoxM1 mRNAs by RT and qPCR (Fig. 5G) between WT and Hoxa10 null mice. Overall, these results suggest that Hoxa10 is necessary to control FoxM1 expression in SDZ cells via transcriptional mechanism.
Figure 5. Hoxa10 regulates FoxM1 in SDZ cells via a transcriptional mechanism.
(A) Immunohistochemical analysis of FoxM1 at IS on D8 between wild-type (WT) and Hoxa10−/− mice. High magnification pictures (at 400X) are shown for mesometrial (M) and antimesometrial (AM) locations. Note: Data show reduced expression of FoxM1 at the SDZ location (lateral or antimesometrial) in Hoxa10−/− compared to that of WT. e, embryo; sdz, secondary decidual zone; M, mesometrial; AM, antimesometrial. (B) Quantitation of nuclear FoxM1 positive cells in M and SDZ locations at IS on D8 between WT and Hoxa10 null mice. (C) Western blot analyses of FoxM1, Hoxa10, and actin at IS on D8 between WT and Hoxa10 null mice. Quantitative analysis of FoxM1 expression shows in the Bar plot. (D) Dual immunofluorescence analysis of Hoxa10 (green) and FoxM1 (red) at the SDZ on D8-IS. DAPI was used for nuclear staining. (E) Diagrammatic illustration of putative Hoxa10 binding regions (1–10) in 5’-flanking, exon 1, and intron 1 of FoxM1 gene. “+” or “–” indicates actual detection of positive or negative binding of Hoxa10 or RNA Pol-II. (F) Quantitative ChIP-PCR analysis of Hoxa10 or RNA Pol II binding for WT and Hoxa10 null in deciduoma tissues on D7, as described in Materials and Methods. (G) Analysis of FoxM1 expression in deciduoma tissues on D7 by quantitative RT-PCR between WT and Hoxa10 null mice, after normalization with Rpl7, a housekeeping gene.
Intra-nuclear translocation of FoxM1 protein is regulated by cyclin D3 via the cyclin-dependent kinase pathway
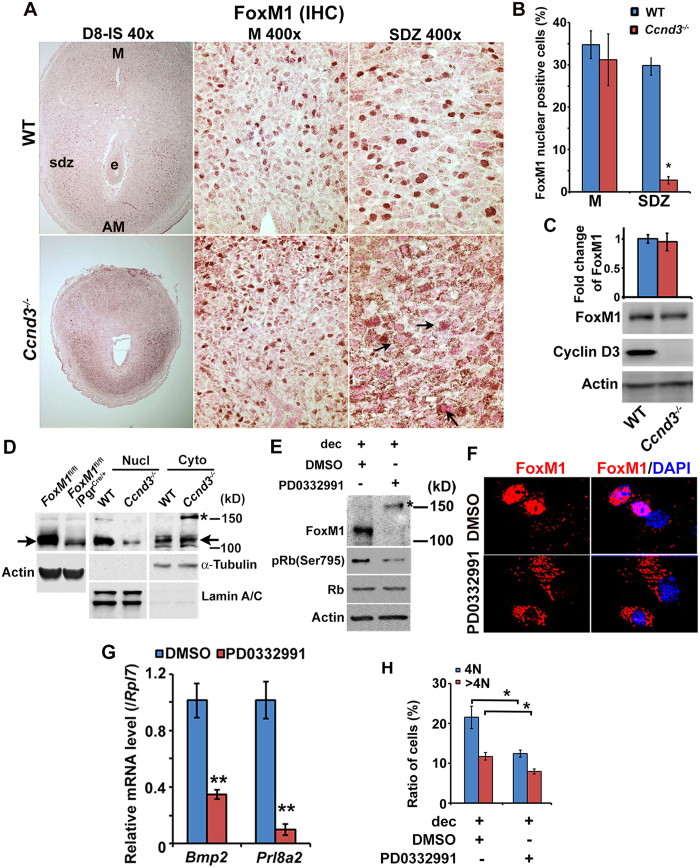
Cyclin D3 plays a major role in cell cycle regulation during stromal cell decidualization3,4,17, and since FoxM1 also plays roles in cell cycle events19, we next wanted whether cyclin D3 controls FoxM1 during decidualization. FoxM1 localization at IS on D8 between Ccnd3 null and WT mice was examined by immunohistochemical analyses. Results showed that although nuclear localization of FoxM1 protein at the mesometrial decidual cells was similar between mice, nuclear positivity of FoxM1 in polyploid cells at the SDZ was significantly lower along with cytoplasmic accumulation (shown by arrows) in Ccnd3−/− as compared to WT (Fig. 6A,B). However, quantitative analysis of FoxM1 protein levels by western blotting did not reveal any change between mice (Fig. 6C). Furthermore, fractionation of SDZ cells into nuclear and cytoplasmic protein extracts followed by western blotting analyses revealed that the level of FoxM1 was downregulated in nuclear fraction, but concomitantly upregulated in cytoplasmic fraction with detection of an additional band ∼150 kDa [shown by asterisk (*)], an indication of protein modification, in Ccnd3−/− as compared to WT (Fig. 6D). In addition, the inclusion of western blot results for FoxM1 null vs. control cells show the specificity of antibody detected bands for comparison (Fig. 6D). FoxM1 full-length protein size ∼109 kDa is shown by arrows (Fig. 6D). The purity of the above fractions was apparently normal, as judged by the analyses of lamin A/C and α-tubulin for corresponding nuclear and cytoplasmic proteins (Fig. 6D). In order to evaluate the mechanism for this regulation, we next used in vitro uterine stromal cell decidualization31 following the inhibition of cdk4/cdk6 kinase activity. The application of an inhibitor (PD0332991) as compared to control (DMSO as a vehicle) caused inhibition of FoxM1 nuclear accumulation, yet a high molecular protein band was still clearly visible [shown by asterisk (*)] (Fig. 6E). These results were also consistent with immunofluorescence studies that found a loss of nuclear FoxM1 localization (Fig. 6F), downregulation of decidualization marker expression (Fig. 6G), and lesser accumulation of 4N and >4N (polyploid) cell populations (Fig. 6H), with the application of an inhibitor versus the control. Overall, these results suggest that cyclin D3 crucially controls nuclear localization of FoxM1 with polyploidy in a cdk4/6 kinase dependent manner.
Figure 6. Cyclin D3 regulates nuclear FoxM1 localization in SDZ cells via cyclin-dependent kinase mechanism.
(A) Immunohistochemical analysis of FoxM1 at D8-IS between WT and Ccnd3−/− mice. High magnification pictures (at 400X) are shown for mesometrial (M) and antimesometrial (AM) locations. Note: Cyclin D3 deficiency causes loss of nuclear FoxM1 at SDZ location (lateral or antimesometrial) as compared to WT. e, embryo; sdz, secondary decidual zone; M, mesometrial; AM, antimesometrial. Arrows indicate cytoplasmic accumulation of immunostained signals. (B) Quantitation of nuclear FoxM1 positive cells in M and SDZ locations at D8-IS between WT and Ccnd3 null mice. (C) Western blot analyses of FoxM1, cyclin D3, and actin at IS on D8 between WT and Ccnd3 null mice. Quantitative analysis of FoxM1 expression shows in the Bar plot. (D) Western blot analyses of FoxM1, α-Tubulin and Lamin A/C in nuclear (Nucl) and cytoplasmic (Cyto) fractions of SDZ between WT and Ccnd3−/− mice. The expression of α-Tubulin and Lamin A/C was used to judge purity of cytoplasmic and nuclear fractions, respectively. Western blot analyses for FoxM1 null vs. control cells show the specificity of antibody detected bands for comparison. Actin was used as control. Note: The level of FoxM1 was downregulated in nuclear fraction, but concomitantly upregulated in cytoplasmic fraction with detection of an additional band ∼150 kDa [shown by asterisk (*)], an indication of protein modification. FoxM1 full-length protein size ∼109 kDa is shown by arrows. E. Western blot analyses of FoxM1, pRb(Ser795), Rb, and Actin during in vitro decidualization after inhibition of Cdk4/6 activity by PD0332991 (10 μM), as compared to control (DMSO). (F) Immunofluorescence analyses of FoxM1 localization after addition of PD0332991 or DMSO during in vitro decidualization. (G) Quantitative RT-PCR analyses of decidual marker genes (Bmp2 and Prl8a2). **Significantly different (p < 0.001) between the treated groups. (H) Flow cytometric analysis cell cycle distribution for 4N and >4N cells during in vitro decidualization after addition of PD0332991 or DMSO. *Significantly different (p < 0.05) between the compared groups.
Discussion
The successful regional decidualization in implantation has been recognized by development of the diploid mesometrial decidual triangle (a site for placentation), together with polyploid SDZ in antimesometrial location. Although polyploidization occurs in terminally differentiated decidual cells and its development during implantation has shown to be critical for pregnancy10,26, the underlying regulatory mechanisms for establishing these two regions remain poorly studied. The highlight of the present investigation is that FoxM1 is critical for controlling regional (mesometrial vs. antimesometrial) development in implantation through a finely tuned, cell cycle-based regulatory balance between diploid and polyploid decidual cell status (Fig. 7). Furthermore, we showed that two decidual markers - Hoxa10 and cyclin D3 - control respective FoxM1 gene transcription and protein translocation to polyploid cell nuclei. Uterine conditional deletion of FoxM1 causes a reduction in the mesometrial decidual area, thereby lowering diploid cell content primarily due to increased arrests prior to mitosis and enhancing the commitment for polyploidy and thus producing aberrant expansion of the SDZ.
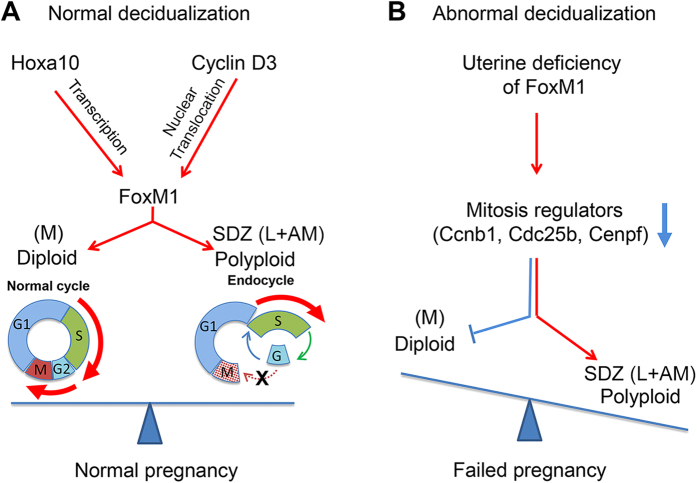
Figure 7. A proposed model depicting the role of FoxM1, downstream of Hoxa10 or cyclin D3, in mediating appropriate control for stromal cell diploid vs polyploid status during decidualization.
Hoxa10 plays a major role in decidualization primarily through upregulation of cyclin D3 in decidualizing stromal cells and also exerts critical regulation in regional decidualization in implantation; mesometrial (M) vs. SDZ development. Here, we provide evidence that the expression of FoxM1 is regulated during decidualization and is controlled by Hoxa10 for transcription and by cyclin D3, a target of Hoxa10, for nuclear FoxM1 protein localization (A). Based on FoxM1 deletion studies, we noted that FoxM1 is critical for normal female fertility, primarily to control appropriate regional progression of decidualization [M vs SDZ development]. The loss of FoxM1 leads to the failure of implantation with decidualization defects; smaller decidual in M-bed, due to 2N cell deficiency with increased arrests prior to mitosis and a bigger SDZ with aberrantly upregulated polyploidy, indicating FoxM1 is critical to appropriately balance diploid vs. polyploid cell status during decidualization (B).
Regulation of cell proliferation, both at the S- and M-phases of the cell cycle, is consistent with FoxM1 expression; however, FoxM1−/− cardiomyocytes, hepatocytes, and smooth muscle cells show normal proliferation19. In this study, we observed that FoxM1 expression is linked with the S-phase for both diploid (mesometrial) and polyploid (SDZ) decidual cells at the IS (Fig. 1 and Supplementary Fig. S1). However, FoxM1 deficiency had no impact on decidual cell progression during S-phase (Figs 3C,D and 4D), indicating it is not crucial during this period. Despite this, FoxM1 may still play an important role during S-phase. As many M-phase genes are induced during S-phase and then silenced in anaphase27,32, it is possible that FoxM1 mediates an interdependent function between S- and M-phases. Interestingly, we have observed that several M-phase regulatory genes, as well as M-phase cells, were strikingly downregulated by FoxM1 deficiency in both regions (Fig. 3). However, thus far FoxM1 expression has primarily been reported in connection with malignant tumor cells, while deletion of FoxM1 is mostly detrimental to cancer progression20, suggesting that FoxM1 plays an essential role under malignancy as opposed to normal physiological conditions. In this regard, decidual growth has been shown to share many tumor-like features such as rapid expansion, vascularization, inflammatory reaction, immune-suppressive microenvironment, and an aberrant number of chromosome copies4. Consistently, it is not surprising in the present study that we found the loss of FoxM1 significantly affects cell cycle progression with enhanced polyploidy development (Figs 3 and 4), suggesting that FoxM1 may potentially control a regulatory balance between diploid vs. polyploid cell development (Fig. 7).
It has been well recognized that decidual polyploidy develops as mono- or bi-nucleated cells due to blockage in G2/M phase of the cell cycle3,7. Here, we specifically noted that FoxM1 deficiency perturbs normal cell cycle progression resulting from increased arrest prior to cytokinesis, and leads to enhanced endoreduplication for polyploidy development, as judged by the analyses of DNA content from cells in vivo (Fig. 3G) and in vitro (Fig. 4A,B), as well as by the analyses of bi-nucleation in vivo (Supplementary Fig. S4). These results clearly document that the loss of FoxM1 promotes transition from diploid into polyploid cells. Polyploid decidual cells predominantly constitute the development of SDZ, so the overall increase in polyploid (both mono- and bi-nucleated) cells could explain the expansion of SDZ at the site of implantation. The decrease in IS weight (Fig. 2B,C) and smaller cross section (Fig. 3A) of FoxM1d/d mice suggest that the net effect of relative expansion of SDZ is inhibition of overall decidual growth. The increase in both mono- and bi-nucleated polyploid cells has been reported in breast cancer or pancreatic cells after suppression of FoxM127,33,34,35, and this phenomenon appears to be common with deficiency of other mitotic regulators, such as Cdh1 and Plk136,37. It is known that the cell cycle regulatory factors Ccnb1 (also known as cyclin B) and Cenpf possess diverse roles in M phase, such as chromosome segregation and cytokinesis38,39. The loss of FoxM1 has been shown to cause pleiotropic cell-cycle defects, including delay in G2, aberration in chromosome segregation, and frequent failure of cytokinesis27. Since, both Ccnb1 and Cenpf were affected by FoxM1 deficiency at the site of implantation and in vitro decidualization (Figs 3E and 4C), we suggest their downregulation plays a major contribution to cease cytokinesis, but enhance polyploidy and bi-nucleation in FoxM1d/d mice as compared to control. In this regard, it is worth mentioning that the accelerated entry into S-phase with increased polyploidization and failure of mitosis has also been shown in FoxM1 null fetal cardiomyocytes40 and hepatocytes41. In addition, it is worth mentioning that the size of polyploid cells in FoxM1d/d mice did not reveal any significant difference as compared to FoxM1f/f mice, although the size of uterine polyploid cells are much larger than that of diploid cell6,31,42. Therefore, we believe that the expansion of SDZ as revealed in our study was not caused by the change in cell size, but rather by increase in polyploid cell population.
The development of decidual polyploidy has been shown to be beneficial for the support of implantation progression and successful pregnancy. For example, decidua with insufficient polyploidy is inhibitory for implantation progression, as shown by deletion of DEDD, Hoxa10, or Ccnd3 in mice4,10,18. However, over-amplification of decidual polyploid cells is also detrimental to the aspect of implantation, as shown by Trp53 null mice26. In the present study, the increased accumulation of polyploidy (both mono- and bi-nucleation) in FoxM1 null mice also resulted in defects, as evidenced by insufficient growth of embryo implantation and lower decidual tissue weight. Therefore, we suggest that enhancement of polyploidy, but lack of complete mitosis in FoxM1 null cells, could reduce the pool of diploid cells, which will severely affect the repertoire of diploid cell proliferation and further decidual growth, which in turn leads to failed implantation sites in severe cases. Thus, balanced progression of proliferation and polyploidy development appears to be critical to support formation of functional decidua in implantation. Together, either deficiency or overabundance of polyploidy is detrimental to successful pregnancy.
Previously, it has been shown that the increase in decidual polyploidy at the site of implantation is associated with enhanced decidual senescence, as in the case of Trp53 null mice26, although we did not see any alteration in the expression of Trp53 or decidual senescence at the IS for FoxM1 null vs control mice. Previously, studies have shown that enhanced polyploidy due to suppression of FoxM1 could lead to cell death by mitotic catastrophe33, suggesting polyploidy mechanism limits the lifespan of cells. However, based on our analyses by immunofluorescence studies using cleaved caspase 3, a well-established proapoptotic cell marker43, on sections of implantation sites between FoxM1d/d and FoxM1f/f mice, did not reveal any increase in apoptosis in the decidual bed by FoxM1 deficiency, as compared to control (Supplementary Fig. S6). Apoptosis is barely detected by TUNEL or other assays in normal decidual bed as previously reported by us25 and others44.
Previously, in an induced pneumonia model, expression of FoxM1 in the knockout mice restored the proliferation and trans-differentiation defects of alveolar epithelium progenitor cells45. The regulatory function of FoxM1 at G2-M phase was also been rescued by its steady expression in neural plate cells46. Similarly, it will be interesting to study the rescue experiments whether FoxM1 overexpression reverses the defects with enhanced cell cycle arrest prior to mitosis with aberrantly upregulated polyploidy during decidualization, which will be the subject of research in future.
FoxM1 is highly expressed in polyploid decidual cells, although its expression also revealed in mesometrial decidual cells (Fig. 1). The periimplantation uterine expression of FoxM1 appeared to be similar with Hoxa10 or cyclin D3 during decidual progression3,14,17. For example, all three genes exhibit weak to moderate expression on D4 in uterine stroma, but are markedly induced in decidualizing stromal cells from D5 through D83,15,17,47, suggesting these genes may have a regulatory connection. Consistently, we observed regulation of FoxM1 gene transcription or its nuclear protein translocation in polyploid cells by Hoxa10 or Ccnd3 deficiency, respectively.
Hoxa10 mutant mice are sterile due to failure of decidualization and implantation15, and these defects have been primarily implicated with impaired progesterone signaling for stromal cell proliferation14 and differentiation13, loss of regional decidualization13 and polyploidy development18, as well as aberrant upregulation of cell cycle inhibitory genes (Cdkn1c, Cdkn2b, Ccng1, and Ccng2)25,48 and downregulation of growth promoting cyclin (Ccnd3/cyclin D3)18. In this study, we observed that Hoxa10 directly binds to the promoter of FoxM1 (Fig. 5), identifying as Hoxa10 downstream transcriptional target mediating cell cycle regulatory functions during decidualization. In this regard, it should be noted that Hoxa10−/− mice show severe decidualization defects, as compared to Ccnd3−/− or FoxM1d/d mice, indicating Hoxa10 plays a pivotal upstream role for the expression of genes which effect cell proliferation and differentiation in the decidual bed. Although we noted that the loss of Hoxa10 results in reduction to the half of FoxM1 protein levels than control (WT) (Fig. 5G), our analysis revealed that the phenotype of FoxM1f/d mice, as compared to FoxM1f/f (control), was normal in terms of litter size or uterine morphology, histology, and weight of implantation sites.
Cyclin D3 specifically exhibits upregulated expression during decidualization3,17 and the loss of Ccnd3 in mice showed decidualization defects and failure of polyploidy development4,31, which could be mediated through compromised DNA synthesis49. We showed that the Ccnd3 mutant mice exhibits loss of nuclear FoxM1 localization in polyploid cells (Fig. 6), while the loss of FoxM1 showed a greater induction of polyploidy (Figs 3 and 4) and no effect on Ccnd3 expression, indicating that cyclin D3 acting upstream for nuclear FoxM1 during decidualization, and these two regulators may participate in a feed-back loop to control appropriate polyploidy levels. Studies have shown that cyclinD3/cdk4/6 activity can target FoxM1 phosphorylation in cancer cell lines50. Consistent to the above notion, pharmacological inhibition of cdk4/6 kinase activity in vitro causes inhibition of nuclear FoxM1 translocation and loss of polyploidy, indicating nuclear FoxM1, acting downstream of cyclin D3-cdk4/6 activity. We tested the possibility of interaction between cyclin D3 and FoxM1 using immunoprecipitation with either antibody followed by immunoblotting with both antibodies. However, we were unable to see any interaction between cyclin D3 and FoxM1. Collectively, we suggest that cyclin D3 contributes to the nuclear localization of FoxM1 through cyclin D3/cdk4/6 kinase dependent pathway, without any direct protein-protein interaction between cyclin D3 and FoxM1.
Cyclin D3 is known to act only during G1 phase, while FoxM1 functions in later phases of the cell cycle, thus lacking cyclin D3 elicits defects at earlier stage than FoxM1 deficiency. Moreover, studies have shown that delay in DNA synthesis affects both cell proliferation and polyploidization49. Indeed, our previous studies have shown that suppression of cyclin D3 lowers the level of decidual polyploidy31 and overexpression of cyclin D3 rescue the defects of polyploidization in Hoxa10 null stromal cells18. In contrast, the defects of FoxM1 deficiency mainly occurred during M phase, without affecting normal DNA synthesis (Figs 3C,D and 4D). Thus, this may explain further why cyclin D3-deficient mice reveal shrinkage of SDZ that is opposite to that observed in FoxM1-deficient uterus.
Subcellular distribution of FoxM1 is known to be regulated by diverse post-translational mechanisms. For example, ERK1/2 initiated phosphorylation promotes FoxM1 translocation to the nucleus51. SUMO1-mediated SUMOylation promotes FoxM1 translocation to the cytoplasm with inhibition of its activity and enhances ubiquitination and degradation52. SUMO2-mediated SUMOylation helps increase FoxM1 activity and nuclear localization53. The mechanism by which cyclin D3-cdk4/6 controls FoxM1 nuclear translocation in decidual cells is still not known. In this regard, it is worth mentioning that we were unable to detect modification of FoxM1 protein by SUMOylation or ubiquitination in Ccnd3 null or cyclin D-cdk4/6 pathway inhibited cells. In our study, the immunoblotting was performed in reducing condition to prevent formation of non-specific protein aggregates, but without affecting true protein modifications. Additionally, the inclusion of 1 mM DTT to lysis buffer or β-mercaptoethanol to sample buffer, should not affect SUMO or ubiquitin mediated modifications under reducing condition, as previously reported54,55. Moreover, we were unable to ascertain phosphorylation status of FoxM1 in vivo, because availability of phosphorylated site-specific FoxM1 antibodies is currently limited. Overall, FoxM1 is an important effector acts downstream of cyclin D3 in controlling nuclear activity for gene regulation (Figs 3E and 4C,F).
Although loss of FoxM1 can lead to defects in decidual development, certain embryo implantation sites are able to overcome these defects and survive through term labor. We do not know the precise reason why certain sites in the mutant uterus can accommodate embryos while others cannot, but this is not an uncommon phenomenon in implantation4. To our knowledge there is no report to suggest that FoxM1 can have a redundancy with other Forkhead box proteins. In this regard, it is interesting to note that FoxO proteins (FoxO1/FoxO1A and FoxO3a) have been shown to play roles in human decidualization56,57, although their roles in mouse decidualization remain unknown.
In conclusion, we have presented novel comprehensive evidence supporting the hypothesis that the mesometrial decidual bed (a presumptive site for placentation) is developmentally interlinked with the appropriate progression of the SDZ at the site of implantation. In this regard, we have provided evidence that FoxM1 is regulated during decidualization and necessary for appropriate control of stromal cell cytokinesis and polyploidy development during decidual progression. Also, Hoxa10 and cyclin D3 utilize FoxM1 as a downstream effector in this process.
Methods
Animals
Hoxa10−/− 15, Ccnd3−/− 58, PR-Cre (PgrCre/+)22, and FoxM1f/f lines28 were generated as previously described. FoxM1 deleted (FoxM1d/d) mice were generated by mating FoxM1f/f mice with PR-Cre mice22. Induction of pregnancy, artificial decidualization, tissue collection, and examination of implantation sites in early pregnancy were performed as previously described13,18. All mice were housed in the animal care facility at Cincinnati Children’s Hospital Medical Center according to National Institutes of Health and institutional guidelines for the use of laboratory animals. All protocols were approved by the Institutional Animal Care and Use Committee (IACUC) (Approval number: IACUC2013-0059).
In situ hybridization
The procedures for 35S-labeled antisense or sense cRNA probes and in situ hybridization were previously described59.
Reverse transcription (RT) and quantitative PCR (qPCR)
RNA (0.5–2 μg) was primed with random-hexamers in a volume of 20 μl and reverse transcribed into cDNA with MMLV Reverse Transcriptase (Promega, cat# M1701). Comparative cDNAs were quantitatively analyzed by real-time PCR using Fast SYBR® Green Master Mix reagent (Life Tech, Grand Island, NY, cat# 4385610) and the ABI StepOnePlus System (Applied Biosystems, Foster City, CA) according to the manufacturer’s instructions. All samples were run for 40 cycles of 3 sec at 95 °C and 30 sec at 60 °C, followed by melting curve stage consisting of temperature increase at 0.3 °C per min to 95 °C. Melting curves for all products showed single peaks. The relative target gene expression was quantified by the ΔΔCt method60, using ribosomal protein l7 (Rpl7, housekeeping gene) for normalization. Primer sequence information for analyzed genes is listed in Supplementary Table S1.
Immunohistochemistry and immunofluorescence
Sections of paraffin-embedded neutralized buffered formalin fixed tissue were used for Ni-DAB (FoxM1) or DAB (other antibodies) colorimetric immunohistochemistry (IHC), while paraformaldehyde fixed frozen tissue sections or cells grown on coverslips were subjected to immunofluorescence (IF). Antibodies include rabbit anti-FoxM1 (1:1000 for IHC or 1:500 for IF; Santa Cruz, cat# sc-502), rabbit anti-cyclin D3 (1:500; Santa Cruz), rabbit anti-Ki67 (1:500; Thermo), rat anti-BrdU (1:200; Abcam), rat anti-phosphorylated histone H3 (Ser 10) (1: 1000; Millipore), rabbit anti-PR (1:300; Santa Cruz), rabbit anti-ERα (1:300; Santa Cruz), and rabbit anti-cleaved caspase 3 (1:300 for IF; Santa Cruz). Counting of immunostaining-positive cells were determined using the Image J program available at http://imagej.nih.gov/ij (NIH, USA), and the analyses were based on examination of serial sections for at least 5–6 IS samples collected from 3–5 different mice for each group.
Chromatin immunoprecipitation
This procedure was followed with some modifications of previously described methods61. Deciduoma tissues collected after 3 days of induction were minced into small pieces and incubated in 1% paraformaldehyde/ PBS for 15 min to crosslink DNA and proteins. After termination of reaction by 0.125 M glycine, tissues were grinded in 1% SDS Tris buffer, followed by sonication (at 50% output) for 8 pulses at 10 sec each using Sonic Dismembrator (Fisher Scientific). Sonicated chromatin was incubated with goat anti-Hoxa10 (1:50; Santa Cruz), mouse anti-RNA polymerase II (1:200; Millipore) or corresponding IgGs on a rotator overnight at 4 °C. The immunoprecipitated chromatins were retrieved by binding to protein G-coupled Dynabeads (Invitrogen). After low salt, high salt, LiCl and TE buffer washing, DNA was recovered after elution, RNase and proteinase K digestion and column based purification as described commercially (Qiagen). DNA was analyzed by qPCR and binding levels were determined as percentage against the input samples and expressed as fold enrichment against IgG. Primer sequence information for analyzed genes is listed in Supplementary Table S2.
Preparation of cellular extracts and western blotting
Whole uterine tissues on D4 or ISs on D5-D8 were used for protein extraction. For subcellular fraction analysis, deciduomal tissues separated from myometrium were divided into mesometrial and SDZ (lateral + antimesometrial) and minced into small pieces, incubated with hypotension buffer (10 mM HEPES pH 8.0, 1.5 mM MgCl2, 10 mM KCl, 0.1 mM EDTA, 1 mM DTT), then passed successively through 18, 22, and 25 gauge needles followed by 40 μm cell strainers and centrifuge to separate the nuclear and cytoplasmic fractions. Protein was extracted with RIPA buffer and subjected to western blotting as described previously3. All antibodies were purchased from Santa Cruz unless specifically stated and they include rabbit anti-FoxM1 (1:2000), goat anti-Actin (1:1000), rabbit anti-Lamin A/C (1: 1000), mouse anti-tubulin α (1: 5000), rabbit anti-cyclin D3 (1: 1000), goat anti-Hoxa10 (1: 1000), and rabbit anti-cyclin B1 (1: 500).
Flow cytometry analysis
This procedure was followed as described previously6. Decidual cells were isolated from implantation sites after removal of embryos and passed through various gauges of needles to prepare single-cell suspensions using Cycle Test Plus DNA Reagent Kit (BD). Cultured uterine stromal/decidual cells were fixed in 70% ethanol, treated with RNase A (500 μg/ml) for 30 min at 37 °C, and then stained with propidium iodide (PI, 50 μg/ml). Analyses of DNA content was done by flow cytometry (BD FACSCanto II). At least 5,000 to 10,000 cells were subjected for each analysis.
Cell culture and treatment
The procedures for uterine stromal cell isolation, culture and induction of decidualization were followed as we previously described31. In brief, stroma cells at around 50% confluence were cultured in DMEM/F-12 (1:1) medium containing 1% charcoal-stripped-fetal bovine serum (CS-FBS), and decidualization was induced by addition of 1 μM P4, 10 nM E2 and 10 ng/ml HB-EGF. Cdk4/6 inhibitor PD0332991 (10 μM; Sigma) and dimethyl sulfoxide (DMSO, 0.1%), a vehicle control, were added to cells at the time of decidual stimulation and cells were analyzed after 48 h. For cell cycle proliferation analysis by flow cytometry, seeded stromal cells were starved for synchronization in absence of FBS (overnight), then cultured in 1% charcoal stripped (CS)-FBS containing DMEM/F-12 (1:1), and cells were collected at 12, 16, 20, and 24 h.
Measurement of serum estradiol-17β and progesterone levels
Sera were collected on day 8 of pregnancy, and estradiol-17β and progesterone levels were measured by EIA kits (Cayman Chemical).
Additional Information
How to cite this article: Gao, F. et al. Control of regional decidualization in implantation: Role of FoxM1 downstream of Hoxa10 and cyclin D3. Sci. Rep. 5, 13863; doi: 10.1038/srep13863 (2015).
Supplementary Material
Acknowledgments
We are grateful to Drs. John B. Lydon and Francesco J. DeMayo (Baylor College of Medicine, Houston, Texas, USA) for providing us with the original breeding pair of PR-Cre mice. We thank Serenity Curtis (Cincinnati Children’s Hospital Medical Center, Cincinnati, Ohio, USA) for editing this manuscript. This work was supported in part by grants from National Institute of Health (NIH) (HD56044 and ES07814 to SKD; HL84151 and HL123490 to VVK) and March of Dimes (#22-FY13-543).
Footnotes
Author Contributions Conceived and designed the experiments: F.G. and S.K.D.; Performed the experiments: F.G., F.B. and X.M.; Analyzed the data: F.G., V.V.K. and S.K.D.; Contributed reagents/materials/analysis tools: F.G., V.V.K. and S.K.D.; Contributed to the writing of the manuscript: F.G., V.V.K. and S.K.D. All authors read and approved the final manuscript.
References
- Cha J., Sun X. & Dey S. K. Mechanisms of implantation: strategies for successful pregnancy. Nat Med 18, 1754–1767 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dey S. K. et al. Molecular cues to implantation. Endocr Rev 25, 341–373 (2004). [DOI] [PubMed] [Google Scholar]
- Tan J. et al. Evidence for coordinated interaction of cyclin D3 with p21 and cdk6 in directing the development of uterine stromal cell decidualization and polyploidy during implantation. Mech Dev 111, 99–113 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das S. K. Cell cycle regulatory control for uterine stromal cell decidualization in implantation. Reproduction 137, 889–899 (2009). [DOI] [PubMed] [Google Scholar]
- Paria B. C., Zhao X., Das S. K., Dey S. K. & Yoshinaga K. Zonula occludens-1 and E-cadherin are coordinately expressed in the mouse uterus with the initiation of implantation and decidualization. Developmental biology 208, 488–501 (1999). [DOI] [PubMed] [Google Scholar]
- Ma X. et al. Decidual cell polyploidization necessitates mitochondrial activity. PloS one 6, e26774 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sroga J. M., Ma X. & Das S. K. Developmental regulation of decidual cell polyploidy at the site of implantation. Front Biosci (Schol Ed) 4, 1475–1486 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ansell J. D., Barlow P. W. & McLaren A. Binucleate and polyploid cells in the decidua of the mouse. J Embryol Exp Morphol 31, 223–227 (1974). [PubMed] [Google Scholar]
- Sachs L. & Shelesnyak M. C. The development and suppression of polyploidy in the developing and suppressed deciduoma in the rat. The Journal of endocrinology 12, 146–151 (1955). [DOI] [PubMed] [Google Scholar]
- Mori M. et al. Death effector domain-containing protein (DEDD) is required for uterine decidualization during early pregnancy in mice. Journal of Clinical Investigation 121, 318–327 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das S. K. Regional Development of Uterine Decidualization: Molecular Signaling by Hoxa-10. Molecular Reproduction and Development 77, 387–396 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee K. Y. et al. Bmp2 is critical for the murine uterine decidual response. Molecular and cellular biology 27, 5468–5478 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rahman M. A. et al. Hoxa-10 deficiency alters region-specific gene expression and perturbs differentiation of natural killer cells during decidualization. Developmental biology 290, 105–117 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim H., Ma L., Ma W. G., Maas R. L. & Dey S. K. Hoxa-10 regulates uterine stromal cell responsiveness to progesterone during implantation and decidualization in the mouse. Mol Endocrinol 13, 1005–1017 (1999). [DOI] [PubMed] [Google Scholar]
- Satokata I., Benson G. & Maas R. Sexually dimorphic sterility phenotypes in Hoxa10-deficient mice. Nature 374, 460–463 (1995). [DOI] [PubMed] [Google Scholar]
- Benson G. V. et al. Mechanisms of reduced fertility in Hoxa-10 mutant mice: uterine homeosis and loss of maternal Hoxa-10 expression. Development 122, 2687–2696 (1996). [DOI] [PubMed] [Google Scholar]
- Das S. K., Lim H., Paria B. C. & Dey S. K. Cyclin D3 in the mouse uterus is associated with the decidualization process during early pregnancy. Journal of molecular endocrinology 22, 91–101 (1999). [DOI] [PubMed] [Google Scholar]
- Sroga J. M., Gao F., Ma X. H. & Das S. K. Overexpression of Cyclin D3 Improves Decidualization Defects in Hoxa-10(−/−) Mice. Endocrinology 153, 5575–5586 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalin T. V., Ustiyan V. & Kalinichenko V. V. Multiple faces of FoxM1 transcription factor: lessons from transgenic mouse models. Cell Cycle 10, 396–405 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wierstra I. FOXM1 (Forkhead box M1) in tumorigenesis: overexpression in human cancer, implication in tumorigenesis, oncogenic functions, tumor-suppressive properties, and target of anticancer therapy. Adv Cancer Res 119, 191–419 (2013). [DOI] [PubMed] [Google Scholar]
- Ye H. G. et al. Hepatocyte nuclear factor 3/fork head homolog 11 is expressed in proliferating epithelial and mesenchymal cells of embryonic and adult tissues. Molecular and cellular biology 17, 1626–1641 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soyal S. M. et al. Cre-mediated recombination in cell lineages that express the progesterone receptor. Genesis 41, 58–66 (2005). [DOI] [PubMed] [Google Scholar]
- Matsumoto H. et al. Cyclooxygenase-2 differentially directs uterine angiogenesis during implantation in mice. The Journal of biological chemistry 277, 29260–29267 (2002). [DOI] [PubMed] [Google Scholar]
- Lim H. et al. Multiple female reproductive failures in cyclooxygenase 2-deficient mice. Cell 91, 197–208 (1997). [DOI] [PubMed] [Google Scholar]
- Yue L. et al. Cyclin G1 and cyclin G2 are expressed in the periimplantation mouse uterus in a cell-specific and progesterone-dependent manner: evidence for aberrant regulation with Hoxa-10 deficiency. Endocrinology 146, 2424–2433 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirota Y. et al. Uterine-specific p53 deficiency confers premature uterine senescence and promotes preterm birth in mice. The Journal of clinical investigation 120, 803–815 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laoukili J. et al. FoxM1 is required for execution of the mitotic programme and chromosome stability. Nature cell biology 7, 126–136 (2005). [DOI] [PubMed] [Google Scholar]
- Wang X. H., Kiyokawa H., Dennewitz M. B. & Costa R. H. The Forkhead Box m1b transcription factor is essential for hepatocyte DNA replication and mitosis during mouse liver regeneration. Proceedings of the National Academy of Sciences of the United States of America 99, 16881–16886 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan Y., Raychaudhuri P. & Costa R. H. Chk2 mediates stabilization of the FoxM1 transcription factor to stimulate expression of DNA repair genes. Molecular and cellular biology 27, 1007–1016 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wierstra I. The transcription factor FOXM1 (Forkhead box M1): proliferation-specific expression, transcription factor function, target genes, mouse models, and normal biological roles. Adv Cancer Res 118, 97–398 (2013). [DOI] [PubMed] [Google Scholar]
- Tan Y. et al. HB-EGF directs stromal cell polyploidy and decidualization via cyclin D3 during implantation. Developmental biology 265, 181–195 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackman M., Firth M. & Pines J. Human cyclins B1 and B2 are localized to strikingly different structures: B1 to microtubules, B2 primarily to the Golgi apparatus. The EMBO journal 14, 1646–1654 (1995). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wonsey D. R. & Follettie M. T. Loss of the forkhead transcription factor FoxM1 causes centrosome amplification and mitotic catastrophe. Cancer research 65, 5181–5189 (2005). [DOI] [PubMed] [Google Scholar]
- Fu Z. et al. Plk1-dependent phosphorylation of FoxM1 regulates a transcriptional programme required for mitotic progression. Nature cell biology 10, 1076–1082 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H. et al. The FoxM1 transcription factor is required to maintain pancreatic beta-cell mass. Mol Endocrinol 20, 1853–1866 (2006). [DOI] [PubMed] [Google Scholar]
- Engelbert D., Schnerch D., Baumgarten A. & Wasch R. The ubiquitin ligase APC(Cdh1) is required to maintain genome integrity in primary human cells. Oncogene 27, 907–917 (2008). [DOI] [PubMed] [Google Scholar]
- Seong Y. S. et al. A spindle checkpoint arrest and a cytokinesis failure by the dominant-negative polo-box domain of Plk1 in U-2 OS cells. The Journal of biological chemistry 277, 32282–32293 (2002). [DOI] [PubMed] [Google Scholar]
- Wheatley S. P. et al. CDK1 inactivation regulates anaphase spindle dynamics and cytokinesis in vivo. The Journal of cell biology 138, 385–393 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holt S. V. et al. Silencing Cenp-F weakens centromeric cohesion, prevents chromosome alignment and activates the spindle checkpoint. Journal of cell science 118, 4889–4900 (2005). [DOI] [PubMed] [Google Scholar]
- Korver W. et al. Uncoupling of S phase and mitosis in cardiomyocytes and hepatocytes lacking the winged-helix transcription factor Trident. Current biology : CB 8, 1327–1330 (1998). [DOI] [PubMed] [Google Scholar]
- Krupczak-Hollis K. et al. The mouse Forkhead Box m1 transcription factor is essential for hepatoblast mitosis and development of intrahepatic bile ducts and vessels during liver morphogenesis. Developmental biology 276, 74–88 (2004). [DOI] [PubMed] [Google Scholar]
- Qi Q. R. et al. Involvement of atypical transcription factor E2F8 in the polyploidization during mouse and human decidualization. Cell Cycle 14, 1842–1858 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salvesen G. S. Caspases: opening the boxes and interpreting the arrows. Cell death and differentiation 9, 3–5 (2002). [DOI] [PubMed] [Google Scholar]
- Nagashima T. et al. BMPR2 is required for postimplantation uterine function and pregnancy maintenance. The Journal of clinical investigation 123, 2539–2550 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y. et al. FoxM1 mediates the progenitor function of type II epithelial cells in repairing alveolar injury induced by Pseudomonas aeruginosa. The Journal of experimental medicine 208, 1473–1484 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ueno H., Nakajo N., Watanabe M., Isoda M. & Sagata N. FoxM1-driven cell division is required for neuronal differentiation in early Xenopus embryos. Development 135, 2023–2030 (2008). [DOI] [PubMed] [Google Scholar]
- Daikoku T. et al. Proteomic analysis identifies immunophilin FK506 binding protein 4 (FKBP52) as a downstream target of Hoxa10 in the periimplantation mouse uterus. Mol Endocrinol 19, 683–697 (2005). [DOI] [PubMed] [Google Scholar]
- Yao M. W. et al. Gene expression profiling reveals progesterone-mediated cell cycle and immunoregulatory roles of Hoxa-10 in the preimplantation uterus. Mol Endocrinol 17, 610–627 (2003). [DOI] [PubMed] [Google Scholar]
- Kozar K. et al. Mouse development and cell proliferation in the absence of D-cyclins. Cell 118, 477–491 (2004). [DOI] [PubMed] [Google Scholar]
- Anders L. et al. A Systematic Screen for CDK4/6 Substrates Links FOXM1 Phosphorylation to Senescence Suppression in Cancer Cells. Cancer Cell 20, 620–634 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma R. Y. et al. Raf/MEK/MAPK signaling stimulates the nuclear translocation and transactivating activity of FOXM1c. Journal of cell science 118, 795–806 (2005). [DOI] [PubMed] [Google Scholar]
- Myatt S. S. et al. SUMOylation inhibits FOXM1 activity and delays mitotic transition. Oncogene 33, 4316–4329 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schimmel J. et al. Uncovering SUMOylation dynamics during cell-cycle progression reveals FoxM1 as a key mitotic SUMO target protein. Mol Cell 53, 1053–1066 (2014). [DOI] [PubMed] [Google Scholar]
- Alontaga A. Y., Bobkova E. & Chen Y. Biochemical analysis of protein SUMOylation. Current protocols in molecular biology Unit10. 29, 1–37 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laney J. D. & Hochstrasser M. Analysis of protein ubiquitination. Current protocols in protein science 66, 14.15.11–14.15.13 (2011). [DOI] [PubMed] [Google Scholar]
- Grinius L., Kessler C., Schroeder J. & Handwerger S. Forkhead transcription factor FOXO1A is critical for induction of human decidualization. The Journal of endocrinology 189, 179–187 (2006). [DOI] [PubMed] [Google Scholar]
- Kajihara T. et al. Differential expression of FOXO1 and FOXO3a confers resistance to oxidative cell death upon endometrial decidualization. Mol Endocrinol 20, 2444–2455 (2006). [DOI] [PubMed] [Google Scholar]
- Sicinska E. et al. Requirement for cyclin D3 in lymphocyte development and T cell leukemias. Cancer Cell 4, 451–461 (2003). [DOI] [PubMed] [Google Scholar]
- Das S. K. et al. Heparin-Binding Egf-Like Growth-Factor Gene Is Induced in the Mouse Uterus Temporally by the Blastocyst Solely at the Site of Its Apposition - a Possible Ligand for Interaction with Blastocyst Egf-Receptor in Implantation. Development 120, 1071–1083 (1994). [DOI] [PubMed] [Google Scholar]
- Livak K. J. & Schmittgen T. D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 25, 402–408 (2001). [DOI] [PubMed] [Google Scholar]
- Ray S., Xu F., Wang H. & Das S. K. Cooperative control via lymphoid enhancer factor 1/T cell factor 3 and estrogen receptor-alpha for uterine gene regulation by estrogen. Mol Endocrinol 22, 1125–1140 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.