Abstract
Streptococcus dysgalactiae, the long recognized mammalian pathogen, has currently received a major concern regarding fish bacterial infection. Adhesion to host epithelial cells and the presence of wall-associated plasminogen binding proteins are prerequisites to Streptococcus infection. This is the first study of the occurrence of nephritis-associated plasminogen-binding receptor (NAPlr) and α-enolase genes in piscine S. dysgalactiae subspecies dysgalactiae (SDSD) isolates. Further characterization of surface localized NAPlr of fish SDSD revealed a similar immune-reactive band of 43 KDa as that from porcine S. dysgalactiae subsp. equisimilis (SDSE). The phylogenetic analysis revealed that NAPlr of fish SDSD is more associated with those of mammalian SDSE and Streptococcus pyogenes rather than of other streptococci. Our findings warrant public attention to the possible implication of these virulence genes in dissemination of SDSD to different tissues of infected hosts and to get advantage to new niches. The SDSD adherence patterns were also studied to better understand their pathogenicity. The patterns of adherence of SDSD on two different cell lines showed a different pattern of adherence. Such difference gives an insight about the variance in host susceptibility to infection.
Keywords: NAPlr gene, α-enolase gene, Piscine S. dysgalactiae subsp. dysgalactiae, Virulence traits, Adherence pattern
Introduction
Streptococcus dysgalactiae was discovered by Diernhofer in 1932 [1], and officially recognized as a new species in 1983 [2]. S. dysgalactiae was subdivided into two genetically similar subspecies: the animal subspecies dysgalactiae (belongs to Lancefield group C (GCS)) and human subspecies equisimilis (belongs to GCS or GGS or GLS) [3]. The α-hemolytic S. dysgalactiae subsp. dysgalactiae (SDSD) is a strict animal pathogen of pyrogenic streptococcus [4]. SDSD is responsible for diverse problems such as mastitis, toxic shock like syndrome, subcutaneous cellulitis in cows [5], extensive fibrinous pleurisy in ewes [6], suppurative polyarthritis in lambs [7], neonatal mortalities in dogs [8], severe septicemia in fish [9], and bacteremia and meningitis in immunocompromised individuals [10,11]. SDSD is potentially considered as an emerging zoonotic agent since it is implicated in cutaneous cellulitis in humans engaged either in cleaning fish [12] or handling livestock [13].
SDSD has been associated with high mortalities in Kingfish (Seriola lalandi), amberjack (S. dumerili) and yellowtail (S. quinqueradiata) in Japan [9,14–17], Nile tilapia (Oreochromis niloticus) in Brazil [18], Amur sturgeon (Acipenser schrenckii), the Siberian sturgeon (A. baerii), golden pomfret (Trachinotus ovatus), Soiny mullet (Liza haematocheila) grass carp (Ctenopharyngodon idella), crucian carp (Carassius carassius) and pompano (Trachinotus blochii) in China [19–22]. It has been recovered from cobia (Rachycentron canadum), basket mullet (Liza alata) and grey mullet (Mugil cephalus) in Taiwan, hybrid red tilapia (Oreochromis sp.) in Indonesia, white spotted snapper (Lutjanus stellatus) and pompano (T. blochii) in Malaysia [9,16,17,23], and rainbow trout (Oncorhynchus mykiss) in Iran [24]. The infected fish revealed systemic pyrogranulomatous inflammation with a severe necrotic lesion in their caudal peduncles [25]. Despite its clinical significance, the complete sequence revelation and virulence characterization are generally unknown for SDSD. Fish SDSD was found to possess some virulence factors such as streptolysin S structural gene (sagA), streptococcal pyrogenic exotoxin G gene (spegg) and serum opacity factor (SOF-FD) [17,26]. Fish SDSD strongly adheres to and invades fish epithelial cell line as Epithelial Papiloma of Carp (EPC) in vitro [14]. However, the adherence patterns and the surface structures implicated in adhesion are still uncovered. The M/M-like proteins (emm), surface dehydrogenase (SDH) and α-enolase are the most important wall-associated plasminogen-binding proteins of pathogenic streptococci [27]. The ability of pathogenic streptococci to bind host plasminogen system empowers their invasiveness through utilizing the fibrinolytic activity of plasmin and promoting the adherence of streptococci to host cells [27]. Plasminogen-binding glycoproteins, such as α-enolase and SDH, are generally found in the cytosolic compartment and are transported to the bacterial cell wall by a yet unknown mechanism that comprised moonlighting functions [28–30]. The surface protein SDH displays ADP-ribosylating activities and glyceraldehyde 3-phosphate dehydrogenase (GAPDH) [31], and has been recognized as a potential nephritogenic protein under the name nephritis-associated plasminogen-binding receptor (NAPlr) [32]. Streptococcal cell wall α-enolase is associated with streptococcal infection and post-streptococcal autoimmune disease in human [28,30].
Hence, NAPlr and α-enolase genes are important virulence factors in Streptococcus pyogenes [33,34], S. agalactiae [35], S. iniae [30], and S. pneumoniae [28,29] due to its contribution to the establishment of infections and colonization by bacterial pathogens [27,36]. This is the first study to investigate the occurrence of gapdh/naplr/sdh and α-enolase genes in piscine isolates of SDSD. We also investigated the adherence patterns of selected SDSD strains to EPC and CHSE-214 (Chinook salmon embryo) cell lines in vitro.
Material and methods
Bacterial isolates
Twenty-three bacterial isolates were used in this study. The α-hemolytic SDSD isolates (n = 18) were recovered from moribund fishes obtained from various fish farms in Japan (n = 9; three from king fish, three from amberjack and three from yellowtail), Taiwan (n = 5; three from grey mullet, one from cobia and one from basket mullet), Malaysia (n = 2; one from pompano and one from snapper), China (n = 1; one from pompano) and Indonesia (n = 1; one from tilapia). For comparative purpose, β-hemolytic S. dysgalactiae subsp. equisimilis (SDSE) isolates (n = 5) were collected from pigs with endocarditis (Kumamoto meat inspection office in Japan).
DNA extraction
The pure stock isolates were stored in Todd-Hewitt broth (THB; Difco, Sparks, MD, USA) at −80 °C. All isolates were cultured aerobically on Todd Hewitt agar (THA; Difco, Sparks, MD, USA), and on 5% sheep blood agar (Columbia agar base; Becton Dickinson, Cockeysville, MD, USA), and then incubated at 37 °C for 24 h. Genomic DNA was extracted from cultivated strains using a DNAzol® reagent (Invitrogen, Carlsbad, USA) [37]. The fish SDSD isolates were discriminated from pig SDSE isolates by using sodA gene primers specific for fish SDSD detection. PCR was performed as described previously [37].
PCR detection of virulence genes
PCR amplification of emm was performed using specific primer pairs; A: (5′-TATTAGCTTAGAAAATTAA-3′) and B: (5′-GCAAGTTCTTCAGCTTGTTT-3′) as described previously by Zhao et al., [38]. To amplify a 963-bp fragment of NAPlr; the specific primer pairs of Plr 1: 5′-GTTAAAGTTGGTATTAACGGT-3′, and Plr 2: 5′-TTGAGCAGTGTAAGACATTTC-3′ were designed based on nephritis associated plasminogen receptor gene of SDSE (GenBank accession number AB217852). PCR was performed with the following parameters: an initial denaturation cycle at 94 °C for 5 min, followed by 35 cycles of denaturation at 94 °C for 30 s, primer annealing at 52 °C for 30 s, elongation at 72 °C for 50 s, and a final cycle at 72 °C for 10 min. To amplify a 1308-bp fragment of α-enolase; the primer pairs of Eno1: 5′-ATGTCAATTATTACTGATGT-3′, and Eno2: 5′-CTATTTTTTTAAGTTATAGA-3′ were designed based on α-enolase gene of SDSE (AP012976). The thermal scheme of PCR was performed as described for the NAPlr gene, except that the primer annealing was adjusted at 50 °C and the primer extension was set for 1 min.
Cloning and sequencing of NAPlr and α-enolase
The NAPlr and α-enolase genes were sequenced according to Abdelsalam et al. [17]. The amplified products were cloned into pGEM-T easy vector (Promega, Madison, WI, USA), and the recombinant plasmid was introduced into Escherichia coli DH5α. The QIAprep Spin Miniprep kit (Qiagen, Germantown, MD, USA) was used to purify the plasmid DNA. Sequencing reactions were performed by using the oligonucleotide primers SP6 (5-ATTTAGGTGACACTATAGAA-3) and T7 (5-TAATACGACTCACTATAGGG-3) with the GenomeLab DTCS Quick Start Kit (Beckman Coulter, Fullerton, CA, USA). The samples were then loaded into the CEQ 8000 Genetic Analysis System (Beckman Coulter) and the nucleotide sequence was determined. The nucleotide sequences were analyzed by using BioEdit version 7.0 [39]. The phylogenetic analysis was performed by the neighbor joining method using MEGA version 5 [40]. The nucleotide sequences of the NAPlr and α-enolase genes were submitted to the DNA Data Bank of Japan (DDBJ) nucleotide sequence database.
Surface protein extraction
Bacterial surface proteins were extracted according to the protocol described by Fujino et al. [32] with some modifications. Briefly, bacteria were inoculated onto Todd Hewitt agar and the culture was incubated for 16 h at 37 °C. Then, bacterial colonies were harvested from the surface of the grow medium/agar plates by loops and were suspended in phosphate-buffered saline (PBS, pH 7.5) in a tube. The bacterial cells were then centrifuged at 10,000g for 20 min. The bacterial cell pellet was then resuspended in PBS. Bacterial cell pellets were washed three times with sterile PBS, and surface proteins were extracted using sodium dodecyl sulfate (SDS; Bio-Rad, Hercules, CA, USA, 30 mg wet weight of bacteria per 100 μl of 0.2% SDS) for 1 h at 4 °C. Extraction mixture was centrifuged and supernatant protein samples were recovered. The SDS extract of bacterial surface proteins was filtered consecutively through 0.45-μm (Millex-HV, Millipore) and 0.22-μm (Millex-GX, Millipore) sterile Millipore filters to remove bacteria. Protein concentration was determined using Bradford assay kit (Bio-Rad, Hercules, CA, USA).
Production of anti-NAPlr monoclonal antibody
Anti-NAPlr monoclonal antibody (mAb) was produced as previously described [32]. Briefly, the specific pathogen-free BALB/c mice were injected intraperitoneal (IP) with 100 mg recombinant NAPlr emulsified in Freund’s complete adjuvant. Three weeks later, the mice were given a booster immunization with 100 mg of recombinant Plr emulsified in Freund’s incomplete adjuvant. Thirty days later, the mice were injected intraperitoneally with 100 mg of recombinant PH in PBS. After 3 days, the mice were sacrificed and their spleens removed. The splenocytes were fused with P3U1 myeloma cells. Hybridoma cultures that secreted anti-Plr antibody were cloned by limiting dilution and the resulting monoclonal antibodies (mAbs) were rescreened to determine the specificity and reactivity with Plr.). The gained Anti-NAPlr mAb from hybridoma cultured cells was evaluated by ELISA using rNAPlr. All Institutional and National Guidelines for the care and use of animals were followed.
Western blots for NAPlr
Protein extract (10 μg protein/lane) of three SDSD isolates (12–06, KNH07808, T11358) and another (5 μg protein/lane) of three SDSE isolates (PAGU656, PAGU706, PAGU707) were separated by sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS–PAGE) on 12.5% polyacrylamide gels (SuperSep 12.5%. Wako Pure Chemical, Osaka, Japan), and then transferred to PVDF (Millipore, Bedford, MA, USA) using a semi-dry blotter (ATTO Bioscience, Tokyo, Japan). SDS–PAGE ‘‘wide range” (200–6.5 kDa) molecular weight standard was purchased from Sigma. NAPlr was identified by the use of the previously prepared anti-NAPlr mAb combined with peroxidase-labeled anti-mouse IgG (American Qualex, San Clemente, CA, USA) and ECL Advance Western Blotting Detection Kit (GE Healthcare, Buckinghamshire, UK). Blot of E. coli was included as a negative control. NAPlr expression in each strain was quantified based on the strength of the luminescence of the mAb – specific band with the densitometry system (Atto).
Adherence pattern of SDSD
This assay was performed according to the method described by Duary et al. [41] with some modifications. Briefly, a sterile 12 mm diameter glass cover-slip coated with poly-l-lysine (NeuVitro, El Monte, CA, USA) was placed in each well of the 24-well tissue culture plate (Costar, Corning, Inc., NY, USA) and the wells were seeded with EPC or CHSE-214 cells. The seeded cells of the EPC or CHSE-214 were grown in Leibovitz-15 (L-15) medium (Gibco Invitrogen, USA) containing 10% (v/v) fetal bovine serum and penicillin (5 μg/ml; Sigma–Aldrich Inc., USA), and incubated at 25 °C and 18 °C respectively, in 5% CO2, and inspected daily until they attained semi-Confluency (2 × 105 cells/well). The SDSD isolates (12–06, KNH07808, T11358) were incubated in THB overnight at 37 °C to midlogarithmic phase (108 CFU/ml), and then centrifuged at 2190g for 30 min. Pellets were washed twice with phosphate-buffered saline (PBS; pH 7.2), and the cell concentration/counts were adjusted to approximately 108 CFU/ml. 100 μl of the bacterial suspension was inoculated to the wells containing EPC and CHSE-214 cells (final bacterial cell concentration in the wells was approximately 107 CFU/ml) and the culture plates were incubated for 30 min at 25 °C and 18 °C for EPC and CHSE-214, respectively. The monolayers were then carefully washed several times with L-15 medium to remove non-adherent bacteria by gentle pippeting. The cells were then fixed with 70% methanol for 10 min and fixed cells were stained with 10% Giemsa stock solution for 2 h. Finally, the glass cover-slips were thoroughly washed with PBS and mounted onto glass slides before being examined by light microscope and photographed. The bacterial adherence patterns were categorized according to the following criteria: localized-like-adherence (LAL), when the bacteria adhered to the cell surface, forming loose clusters; localized adherence (LA), when the bacteria adhered to the cell surface as tight clusters; diffuse adherence (DA), when the bacteria adhered diffuse to the cell surface; and aggregative adherence (AA), when the bacteria adhered to the cell surface and to the cover slip in a stacked-brick pattern [42]. The adherence rate was expressed as the number of adhering bacteria per 50 cells of EPC or CHSE-214. The results were expressed as a weak adherence (⩽100 adherent bacteria), moderate adherence (100–200 adherent bacteria) and strong adherence (⩾200 adherent bacteria) [43].
Nucleotide sequence accession numbers
The nucleotide sequences determined in this study were submitted to the DDBJ nucleotide sequence database. The accession numbers of sequenced Gapdh/sdh/naplr and α-enolase genes are AB470099 and AB758245, respectively.
Results
Occurrence of emm, NAPlr and α-enolase genes
All fish SDSD isolates were PCR negative for emm. However, three SDSE isolates (PAGU656, PAGU706, PAGU707) were PCR positive for emm. All SDSD and SDSE isolates contained homologous segments of NAPlr and α-enolase (Table 1). The PCR products of distinct strains were of the expected size, 963 bp and 1308 bp, respectively.
Table 1.
The α-hemolytic fish SDSD and β-hemolytic pig SDSE strains used in this study.
| No. | Isolates | Source | Country | Hemolysis | NAPlra | enob | emmc | |
|---|---|---|---|---|---|---|---|---|
| Fish SDSD strains | 1 | 12-06 | Amberjack | Japan | α | + | + | − |
| 2 | Kdys0412 | Amberjack | Japan | α | + | + | − | |
| 3 | Kdys0429 | Amberjack | Japan | α | + | + | − | |
| 4 | Kdys0728 | Yellowtail | Japan | α | + | + | − | |
| 5 | Kdys0719 | Yellowtail | Japan | α | + | + | − | |
| 6 | Kdys0707 | Yellowtail | Japan | α | + | + | − | |
| 7 | KNH07808 | King fish | Japan | α | + | + | − | |
| 8 | KNH07901 | King fish | Japan | α | + | + | − | |
| 9 | KNH07902 | King fish | Japan | α | + | + | − | |
| 10 | 95980 | Mullet | Taiwan | α | + | + | − | |
| 11 | 95921 | Mullet | Taiwan | α | + | + | − | |
| 12 | 95900 | Mullet | Taiwan | α | + | + | − | |
| 13 | 951003 | Basket mullet | Taiwan | α | + | + | − | |
| 14 | AOD-96086-K | Cobia | Taiwan | α | + | + | − | |
| 15 | PF880 | Pompano | Malaysia | α | + | + | − | |
| 16 | PP1564 | Pompano | China | α | + | + | − | |
| 17 | WSSN1609 | Snapper | Malaysia | α | + | + | − | |
| 18 | T11358 | Tilapia | Indonesia | α | + | + | − | |
| Pig SDSE strains | 19 | PAGU656 | Pig | Japan | β | + | + | +d |
| 20 | PAGU657 | Pig | Japan | β | + | + | − | |
| 21 | PAGU699 | Pig | Japan | β | + | + | − | |
| 22 | PAGU706 | Pig | Japan | β | + | + | +d | |
| 23 | PAGU707 | Pig | Japan | β | + | + | +d | |
NAPlr: Nephritis associated plasminogen receptor.
eno: α-enolase gene.
emm: M protein gene.
The sequences of emm locus of positive SDSE isolates not determined.
Nucleotide sequence analyses of NAPlr
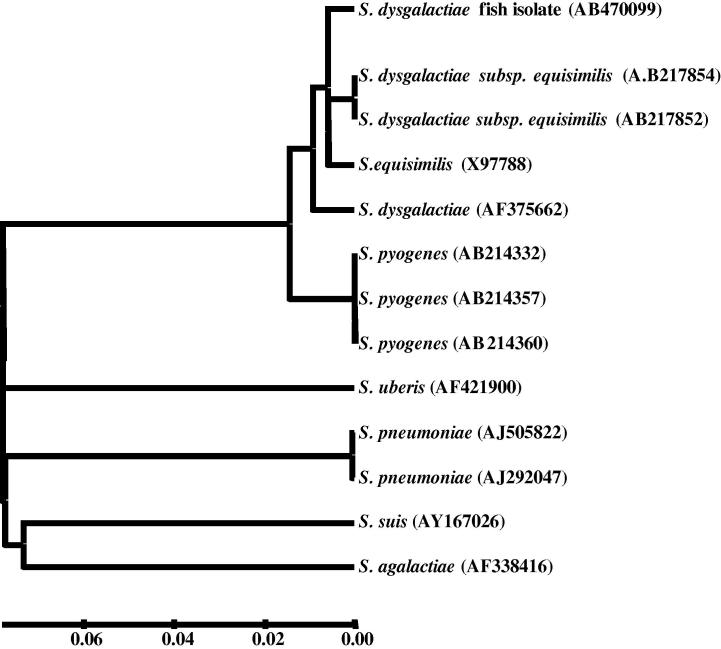
The NAPlr gene of SDSD collected from diseased fish was sequenced under the GenBank accession number AB470099. The NAPlr gene obtained from SDSD strain (T11358) was 963 bp long. The NAPlr was found to have 99% similarity to NAPlr (AB217852) of SDSE and 97% similarity to NAPlr (AB214357) of S. pyogenes, and has one ORF encoding 336 amino acids. Therefore, phylogenetic analysis revealed that NAPlr of piscine SDSD isolate was related to that of SDSE and S. pyogenes and separated from other gapdh/sdh/naplr clusters of other streptococci (Fig. 1). The deduced amino acid sequence of fish SDSD NAPlr was identical to the previous investigated nephritogenic strains of SDSE and S. pyogenes (Fig. 2).
Fig. 1.
Phylogenetic tree of NAPlr of fish SDSD and related species of the genus Streptococcus.
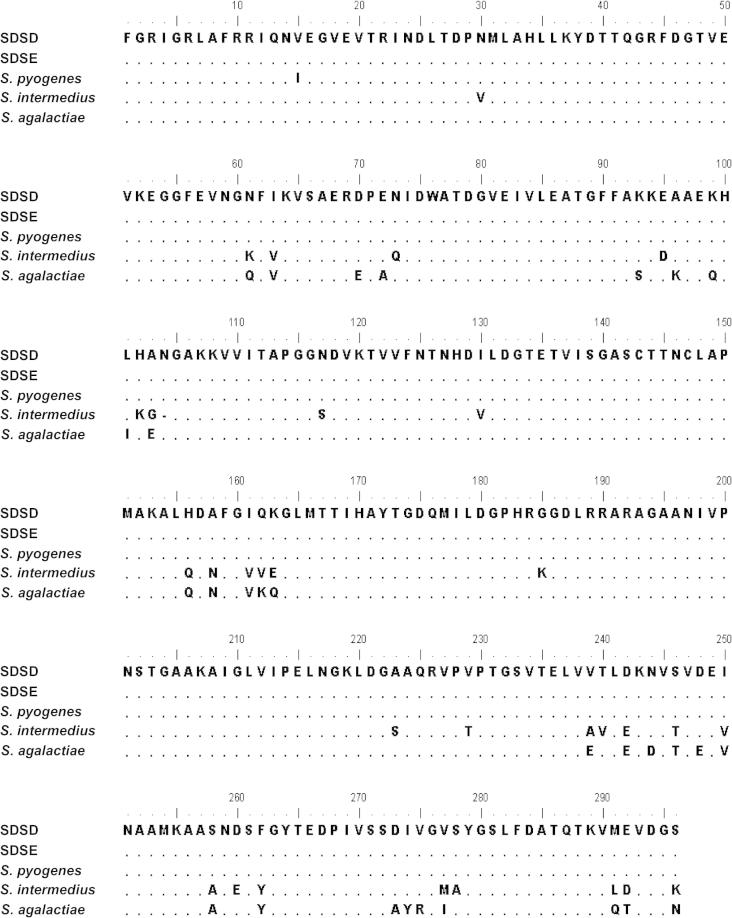
Fig. 2.
Alignment of the deduced amino acid sequences of the NAPlr from fish SDSD (accession No. AB470099), S. pyogenes (accession No. AB088214), and SDSE (accession No. AB217852), S. intermedius (accession No. NC022244) and S. agalactiae (accession No. AB221040). The dots represent identical residues. NAPlr from SDSD shares 100%, 99%, 91% and 90% identity with its homologous from SDSE, S. pyogenes, S. agalactiae and S. intermedius respectively. The numbering of the residues is indicated above the amino acids.
Nucleotide sequence analyses of α-enolase
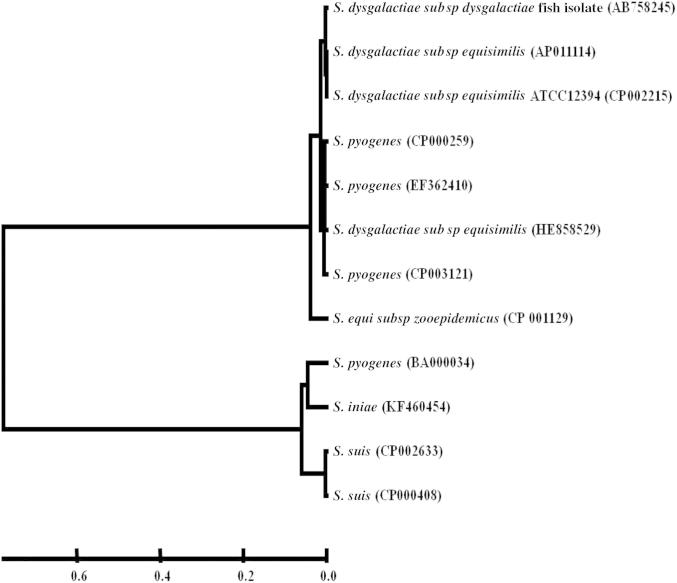
The α-enolase gene of SDSD from diseased fish was sequenced under the GenBank accession number AB758245. The α-enolase locus obtained from fish SDSD strain (KNH07808) was 1308 bp long. The α-enolase was found to have 99% similar to that of SDSE (AP011114), 97% similarity to S. pyogenes (EF362410), and 91% similarity to S. iniae (KF460454), and has one ORF encoding 435 amino acids. Therefore, phylogenetic analysis revealed that α-enolase of fish SDSD isolate was related to that of SDSE and S. pyogenes and separated from other α-enolase clusters of other streptococci (Fig. 3).
Fig. 3.
Phylogenetic tree of enolase of fish SDSD and related species of the genus Streptococcus.
Western blots
The presence of NAPlr in the cell wall was analyzed by Western blotting using anti-NAPlr mAb. As expected, a 43-kDa band corresponds to the molecular weight of NAPlr of SDSD was clearly detected (Fig. 4). All fish isolates of SDSD and pig isolates of SDSE expressed a similar 43-kDa NAPlr band (Fig. 4).
Fig. 4.
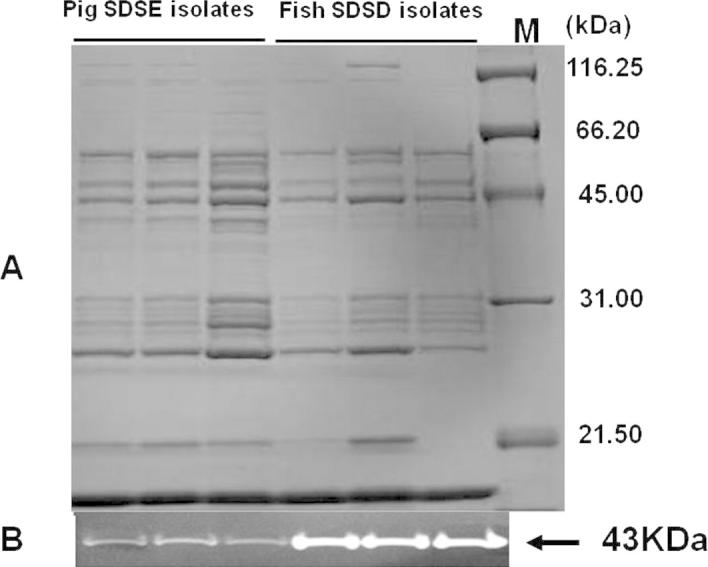
Expression of NAPlr. (A) Protein extracts of fish SDSD (10 μg protein/lane) and pig SDSE (5 μg protein/lane) were separated by 10% polyacrylamide SDS–PAGE and subjected to Western blotting. The similar densities of the 43-kDa NAPlr protein bands and other protein bands on the same blot indicate that the samples contain equal amounts of proteins. (B) Western blot analysis of NAPlr of three isolates of fish SDSD (12-06, KNH07808, T11358) and three isolates of pig SDSE (PAGU656, PAGU706, PAGU707) probed with anti-NAPlr mAb.
Cell adherence pattern
Fish SDSD has a localized adherence pattern (Fig. 5) characterized by the presence of one chain of bacterial cells attached to the surface of CHSE-214 at a focal point. On the other hand, fish SDSD has an aggregated adherence pattern (Fig. 5) characterized by clumps or clusters of bacterial cells on the EPC cells. SDSD were also attached to the surfaces of the cultured EPC and to exposed areas of the glass slide around the EPC cells. EPC and CHSE-214 cells that were infected with SDSD showed cytoplasmic vacuoles.
Fig. 5.
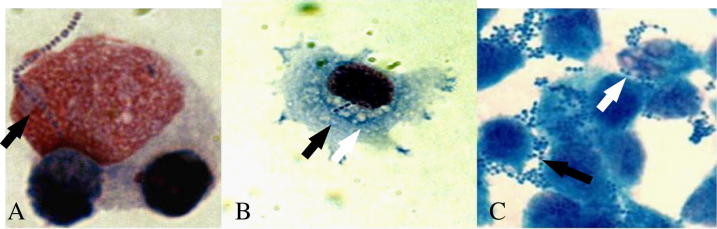
Microscopic appearances of SDSD adhered to: (A) CHSE-214; (B) CHSE-214; and (C) EPC. Both cell lines were exposed to 107 CFU/well and stained with Giemsa (X1000). Black arrows showed cells of SDSD adhered to CHSE-214 and EPC. The white arrow showed cytoblasmic vacuoles.
Tested 12-06, KNH07808 and T11358 isolates were categorized as strongly adhesive (⩾200 adherent bacteria) on EPC culture, but weak adhesive (⩽100 adherent bacteria) on CHSE-214 culture.
Discussion
S. dysgalactiae was found in human and porcine β-hemolytic SDSE isolates and in piscine, bovine, and porcine α-hemolytic SDSD isolates [17,37]. Recently, SDSD infection has been observed in farmed fish resulting in severely necrotizing caudal peduncles [23,25]. SDSD caused either an opportunistic infection in immunocompromised patients [12,13,44], or invasive infection in individuals handling livestock and seafood [10,11]. Pathogenesis and clinical signs of different Streptococcus species appear highly similar among a variety of infected hosts. This means that similar virulence traits may exist [45]. However, little is known about the virulence factors of fish SDSD when compared with other streptococci.
Pathogenic streptococci can use host plasminogen for adherence to cell surfaces, dissemination in the body, and protects against immune defense [27,34]. This complex pathogenic scenario reveals the complicated adaptation of streptococci in invading their host environments. Streptococci harbor a broad variety of different plasminogen binding and activation mechanisms. The M/M like protein, gapdh/sdh/naplr and α-enolase have been described as proteins associated with virulence in several pathogenic bacteria [27–31,34]. In this study, fish SDSD strains were found to be PCR negative for emm gene. This indicates either the absence of this gene within the investigated isolates or the isolates possess gene variants that could not be detected by S. pyogenes-based primers used in this study. On the other hand, three SDSE isolates were positive for the presence of the emm gene. These findings concur with previous investigations that proved the presence of emm gene in clinical isolates of β-hemolytic SDSE, but not in bovine SDSD [45].
The present study also confirms the presence of NAPlr and α-enolase genes in all examined fish SDSD and pig SDSE isolates using their specific primers. These findings go parallel with previous reports that detected the presence of GAPDH and α-enolase in bovine SDSD [45]. Interestingly, the sequenced fragments of NAPlr and α-enolase genes revealed 99% similarity with those of SDSE. Moreover, the partial predicted amino acid sequence of NAPlr of fish SDSD shows no difference from that of SDSE. Most of amino acid variants observed in fish SDSD are structurally relevant and functionally compatible with their corresponding substitution residues in other isolates (e.g. the replacement of non-polar Valine (V) amino acid residue with non-polar Isoleucine (I) at 16, and substitution of I with V at positions 63, 130, 161 and 250). These results agree with Madureira et al. [35]. Recently, gapdh/sdh/naplr and α-enolase genes play multiple roles in virulence of pathogenic streptococci such as adhesions, helping the bacteria escape detection by neutrophils, and allowing the evasion of the complement system [33,34,36]. It has been also reported that gapdh/sdh/naplr induces clot formation, disrupts intracellular signaling in the host, promotes bacterial adherence to host cells, and binding to various host proteins, including plasmin, actin, fibronectin and myosin [27,31–35]. The recent studies have provided definitive proof that NAPlr is a potent nephritogenic antigen [31]. NAPlr gene was thought to be a factor leading to the pathogenesis of acute post-streptococcal glomerulonephritis (APSGN). Kim et al. [30] proposed that α-enolase might facilitate the invasion and dissemination of S. iniae in infected fish. Our findings signify that α-hemolytic fish SDSD isolates carried homologous genes that may be responsible for pathogenesis and virulence of SDSE and S. pyogenes. Consequently, α-hemolytic fish SDSD isolates should not be neglected as putative infectious disease agents in mammals and humans. Further studies are needed to clarify the role of NAPlr and α-enolase in the pathogenesis of SDSD among cultured fish.
It has been postulated that the portal of entry of SDSD into fish is mainly through the skin rather than the oral route. Therefore, adherence of streptococci to epithelial cells was tested since adherence capacity is correlated with the pathogenesis of streptococci. Here the adherence of tested isolates to cell lines, CHSE-214 and EPC cells, was generally varied. Fish SDSD was previously found to adhere strongly to EPC due to the high hydrophobic character of SDSD [14]. In this study, the isolates that adhered strongly to EPC cells were the same ones that adhered weakly to CHSE-214 cells. This variation may occur due to either different cell lines were used or it was associated with the host susceptibility to bacterial infection. The SDSD with the same surface hydrophobic property might employ different mechanisms in adherence upon growth on different cell lines.
Conclusions
In conclusion, this is the first study on molecular characterization of NAPlr and α-enolase – as two virulence-related genes – in fish SDSD isolates. Our finding clearly demonstrates the immune-reactive similarity of NAPlr protein as that from SDSE. With more conserved nature, the phylogenetic analysis proved that NAPlr of fish SDSD is more related to SDSE and S. pyogenes and separated from other gapdh/sdh/naplr clusters of other streptococci.
Conflict of interest
The authors have declared no conflict of interest.
Acknowledgements
The first author would like to thank the Egyptian Ministry of High Education for the financial support of his studies abroad. The authors are appreciative to Dr. Lauke Labrie for gently providing Streptococcus dysgalactiae isolates.
Footnotes
Peer review under responsibility of Cairo University.
Contributor Information
M. Abdelsalam, Email: m.abdelsalam@staff.cu.edu.eg.
M. Warda, Email: maawarda@eun.eg.
References
- 1.Diernhofer K. Aesculinbouillon als Hilfsmittel für die Differenzierung von Euter- und Milchstreptokokken bei Massenuntersuchungen. Milchwirts Forsch. 1932;13:368–374. [Google Scholar]
- 2.Garvie E.I., Farrow J.A.E., Bramley A.J. Streptococcus dysgalactiae (Diernhofer) nom. Rev Int J Syst Bacteriol. 1983;33:404–405. [Google Scholar]
- 3.Vandamme P., Pot B., Falsen E., Kersters K., Devriese L.A. Taxonomic study of Lancefield streptococcal groups C, G, and L (Streptococcus dysgalactiae) and proposal of S. dysgalactiae subsp. equisimilis subsp. nov. Int J Syst Bacteriol. 1996;46:774–781. doi: 10.1099/00207713-46-3-774. [DOI] [PubMed] [Google Scholar]
- 4.Facklam R. What happened to the streptococci: overview of taxonomic and nomenclature changes. Clin Microbiol Rev. 2002;15:613–630. doi: 10.1128/CMR.15.4.613-630.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chénier S., Lecle‘re M., Messier S., Fecteau G. Streptococcus dysgalactiaecellulitis and toxic shock like syndrome in a Brown Swiss cow. J Vet Diagn Invest. 2008;20:99–103. doi: 10.1177/104063870802000122. [DOI] [PubMed] [Google Scholar]
- 6.Scott P.R. Extensive fibrinous pleurisy associated with Streptococcus dysgalactiae mastitis in two ewes. Vet Rec. 2000;146:347–349. doi: 10.1136/vr.146.12.347. [DOI] [PubMed] [Google Scholar]
- 7.Lacasta D., Ferrer L.M., Ramos J.J., Loste A., Bueso J.P. Digestive pathway of infection in Streptococcus dysgalactiae polyarthritis in lambs. Small Ruminant Res. 2008;78:202–205. [Google Scholar]
- 8.Vela A.I., Falsen E., Simarro I., Rollan E., Collins M.D., Domınguez L. Neonatal mortality in puppies due to bacteremia by Streptococcus dysgalactiae subsp. dysgalactiae. J Clin Microbiol. 2006;44:666–668. doi: 10.1128/JCM.44.2.666-668.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Abdelsalam M., Asheg A., Eissa A.E. Streptococcus dysgalactiae: an emerging pathogen of fishes and mammals. Int J Vet Sci Med. 2013;1:1–6. [Google Scholar]
- 10.Bert F., Lambert-Zechovsky N. A case of bacteremia caused by Streptococcus dysgalactiae. Eur J Clin Microbiol Infect Dis. 1997;16:324–325. doi: 10.1007/BF01695642. [DOI] [PubMed] [Google Scholar]
- 11.Quinn R.J.M., Hallett A.F., Appelbaum P.C., Cooper R.C. Meningitis caused by Streptococcus dysgalactiae in a preterm infant. Am J Clinical Pathol. 1978;70:948–950. doi: 10.1093/ajcp/70.6.948. [DOI] [PubMed] [Google Scholar]
- 12.Koh T.H., Sng L.H., Yuen S.M., Thomas C.K., Tan P.L., Tan S.H. Streptococcal cellulitis following preparation of fresh raw seafood. Zoonoses Public Health. 2009;56:206–208. doi: 10.1111/j.1863-2378.2008.01213.x. [DOI] [PubMed] [Google Scholar]
- 13.Fernández-Aceñero J., Fernández-López P. Cutaneous lesions associated with bacteremia by Streptococcus dysgalactiae. J Am Acad Dermatol. 2006;55:91–92. doi: 10.1016/j.jaad.2005.01.097. [DOI] [PubMed] [Google Scholar]
- 14.Abdelsalam M., Chen S.C., Yoshida T. Surface properties of Streptococcus dysgalactiae strains isolated from marine fish. Bull Eur Assoc Fish Pathol. 2009;29:15–23. [Google Scholar]
- 15.Abdelsalam M., Nakanishi K., Yonemura K., Itami T., Chen S.C., Yoshida T. Application of Congo red agar for detection of Streptococcus dysgalactiae isolated from diseased fish. J Appl Ichthyol. 2009;25:442–446. [Google Scholar]
- 16.Abdelsalam M., Chen S.C., Yoshida T. Phenotypic and genetic characterization of Streptococcus dysgalactiae strains isolated from fish collected in Japan and other Asian countries. FEMS Microbiol Lett. 2010;302:32–38. doi: 10.1111/j.1574-6968.2009.01828.x. [DOI] [PubMed] [Google Scholar]
- 17.Abdelsalam M., Chen S.C., Yoshida T. Dissemination of streptococcal pyrogenic exotoxin G (spegg) with an IS-like element in fish isolates of Streptococcus dysgalactiae. FEMS Microbiol Lett. 2010;309:105–113. doi: 10.1111/j.1574-6968.2010.02024.x. [DOI] [PubMed] [Google Scholar]
- 18.Netto L.N., Leal C.A.G., Figueiredo H.C.P. Streptococcus dysgalactiae as an agent of septicaemia in Nile tilapia, Oreochromis niloticus (L.) J Fish Dis. 2011;34:254. doi: 10.1111/j.1365-2761.2010.01220.x. [DOI] [PubMed] [Google Scholar]
- 19.Zhou S.M., Li A.X., Ma Y., Liu R.M. Isolation, identification and characteristics of 16S rRNA gene sequences of the pathogen responsible for the streptococcosis in cultured fish. Acta Sci Nat Univ Sunyatseni. 2007;46:71. [in Chinese with English abstract] [Google Scholar]
- 20.Pan H.J., Liu X.Y., Chang O.Q., Wang Q., Sun H.W., Liu R.M. Isolation, identification and pathogenicity of Streptococcus dysgalactiae from Siberian Sturgeon, Acipenser baerii. J Fish Sci China. 2009;16:904. [In Chinese with English abstract] [Google Scholar]
- 21.Yang W., Li A. Isolation and characterization of Streptococcus dysgalactiae from diseased Acipenser schrenckii. Aquaculture. 2009;294:14–17. [Google Scholar]
- 22.Qi Z.T., Tian J.Y., Zhang Q.H., Shao R., Qiu M., Wang Z.S. Susceptibility of Soiny Mullet (Liza haematocheila) to Streptococcus dysgalactiae and physiological response to formalin inactivated S. dysgalactiae. Pak Vet J. 2013;33:237. [Google Scholar]
- 23.Abdelsalam M., Eissa A.E., Chen S.C. Genetic diversity of geographically distinct Streptococcus dysgalactiae isolates from fish. J Adv Res. 2015;6(2):233–238. doi: 10.1016/j.jare.2013.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pourgholam R., Laluei F., Saeedi A.A., Zahedi A., Safari R., Taghavi M.J. Distribution and molecular identification of some causative agents of streptococcosis isolated farmed rainbow trout (Oncorhynchus mykiss, Walbaum) in Iran. Iran J Fish Sci. 2011;10:109–122. [Google Scholar]
- 25.Hagiwara H., Takano R., Noguchi M., Narita M. A study of the lesions induced in Seriola dumerili by intradermal or intraperitoneal injection of Streptococcus dysgalactiae. J Comp Pathol. 2009;140:35–40. doi: 10.1016/j.jcpa.2008.09.005. [DOI] [PubMed] [Google Scholar]
- 26.Nishiki I., Minami T., Chen S.C., Itami T., Yoshida T. Expression of the serum opacity factor gene and the variation in its upstream region in Streptococcus dysgalactiae isolates from fish. J Gen Appl Microbiol. 2012;58:457–463. doi: 10.2323/jgam.58.457. [DOI] [PubMed] [Google Scholar]
- 27.Fulde M., Steinert M., Bergmann S. Interaction of streptococcal plasminogen binding proteins with the host fibrinolytic system. Front Cell Infect Microbiol. 2013;3:85. doi: 10.3389/fcimb.2013.00085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bergmann S., Rohde M., Chhatwal G.S., Hammerschmidt S. Alpha-Enolase of Streptococcus pneumoniae is a plasmin(ogen)-binding protein displayed on the bacterial cell surface. Mol Microbiol. 2001;40:1273–1287. doi: 10.1046/j.1365-2958.2001.02448.x. [DOI] [PubMed] [Google Scholar]
- 29.Bergmann S., Rohde M., Hammerschmidt S. Glyceraldehyde-3-phosphate dehydrogenase of Streptococcus pneumoniae is a surface-displayed plasminogen-binding protein. Infect Immun. 2004;72:2416–2419. doi: 10.1128/IAI.72.4.2416-2419.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kim M.S., Choi S.H., Lee E.H., Nam Y.K., Kim S.K., Kim K.H. α-enolase, a plasmin(ogen) binding protein and cell wall associating protein from a fish pathogenic Streptococcus iniae strain. Aquaculture. 2007;265:55–60. [Google Scholar]
- 31.Pancholi V., Fischetti V.A. Glyceraldehyde-3-phosphate dehydrogenase on the surface of group A streptococci is also an ADP-ribosylating enzyme. Proc Natl Acad Sci USA. 1993;90:8154–8158. doi: 10.1073/pnas.90.17.8154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Fujino M., Yamakami K., Oda T., Omasu F., Murai T., Yoshizawa N. Sequence and expression of NAPlr is conserved among group A streptococci isolated from patients with acute poststreptococcal glomerulonephritis (APSGN) and non-APSGN. J Nephrol. 2007;20:364–369. [PubMed] [Google Scholar]
- 33.Terao Y., Yamaguchi M., Hamada S., Kawabata S. Multifunctional glyceraldehyde-3-phosphate dehydrogenase of Streptococcus pyogenes is essential for evasion from neutrophils. J Biol Chem. 2006;281:14215–14223. doi: 10.1074/jbc.M513408200. [DOI] [PubMed] [Google Scholar]
- 34.Jin H., Agarwal S., Pancholi V. Surface export of GAPDH/SDH, a glycolytic enzyme, is essential for Streptococcus pyogenes virulence. MBio. 2011;2:e00011–e00068. doi: 10.1128/mBio.00068-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Madureira P., Baptista M., Vieira M., Magalhaes V., Camelo A., Oliveira L. Streptococcus agalactiae GAPDH is a virulence-associated immunomodulatory protein. J Immunol. 2007;178:1379–1387. doi: 10.4049/jimmunol.178.3.1379. [DOI] [PubMed] [Google Scholar]
- 36.Pancholi V. Multifunctional α-enolase: its role in diseases. Cell Mol Life Sci. 2001;58:902–920. doi: 10.1007/PL00000910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Nomoto R., Kagawa H., Yoshida T. Partial sequencing of sodA Gene and its application to identification of Streptococcus dysgalactiae subsp. dysgalactiae isolated from farmed fish. Lett Appl Microbiol. 2008;46:95–100. doi: 10.1111/j.1472-765X.2007.02272.x. [DOI] [PubMed] [Google Scholar]
- 38.Zhao J., Hayashi T., Saarinen S., Papageorgiou A.C., Kato H., Imanishi K. Cloning, expression, and characterization of the superantigen streptococcal pyrogenic exotoxin G from Streptococcus dysgalactiae. Infect Immun. 2007;75:1721–1729. doi: 10.1128/IAI.01183-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hall T.A. Bioedit: a user friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp Ser. 1999;41:95–98. [Google Scholar]
- 40.Tamura K., Peterson D., Peterson N., Stecher G., Nei M., Kumar S. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol. 2011;28:2731–2739. doi: 10.1093/molbev/msr121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Duary R.K., Rajput Y.S., Batish V.K., Grover S. Assessing the adhesion of putative indigenous probiotic lactobacilli to human colonic epithelial cells. Indian J Med Res. 2011;134:664–671. doi: 10.4103/0971-5916.90992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mora A., Blanco M., Yamamoto D., Dahbi G., Blanco J.E., López C. HeLa-cell adherence patterns and actin aggregation of enteropathogenic Escherichia coli (EPEC) and Shiga-toxin-producing E. coli (STEC) strains carrying different eae and tir alleles. Int Microbiol. 2009;12:243–251. [PubMed] [Google Scholar]
- 43.Allgaier A., Wisselink H.J., Smith H.E., Valentin-Weigand P. Relatedness of Streptococcus suis isolates of various serotypes and clinical backgrounds as evaluated by macrorestriction analysis and expression of potential virulence traits. J Clin Microbiol. 2001;39:445–453. doi: 10.1128/JCM.39.2.445-453.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Park M.-J., Eun I.-S., Jung C.-Y., Ko Y.-C., Kim Y.-J., Kim C.-k. Streptococcus dysgalactiae subspecies dysgalactiae infection after total knee arthroplasty: a case report. Knee Surg Relat Res. 2012;24:120–123. doi: 10.5792/ksrr.2012.24.2.120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Rato M.G., Nerlich A., Bergmann R., Bexiga R., Nunes S.F., Vilela C.L. Virulence gene pool detected in bovine group C Streptococcus dysgalactiae subsp. dysgalactiae solates by use of a group A S. pyogenes virulence microarray. J Clin Microbiol. 2011;49:2470–2479. doi: 10.1128/JCM.00008-11. [DOI] [PMC free article] [PubMed] [Google Scholar]