Abstract
Increasing evidence indicates that the gut microbiota can be altered to ameliorate or prevent disease states, and engineering the gut microbiota to therapeutically modulate host metabolism is an emerging goal of microbiome research. In the intestine, bacterial urease converts host-derived urea to ammonia and carbon dioxide, contributing to hyperammonemia-associated neurotoxicity and encephalopathy in patients with liver disease. Here, we engineered murine gut microbiota to reduce urease activity. Animals were depleted of their preexisting gut microbiota and then inoculated with altered Schaedler flora (ASF), a defined consortium of 8 bacteria with minimal urease gene content. This protocol resulted in establishment of a persistent new community that promoted a long-term reduction in fecal urease activity and ammonia production. Moreover, in a murine model of hepatic injury, ASF transplantation was associated with decreased morbidity and mortality. These results provide proof of concept that inoculation of a prepared host with a defined gut microbiota can lead to durable metabolic changes with therapeutic utility.
Introduction
Dysbiosis, an abnormal and pathogenic state of the human microbiome, has been implicated in inflammatory bowel diseases, atherosclerosis, obesity, diabetes, colon cancer, and other diseases (1, 2). Fecal microbiota transplantation (FMT) is highly effective in the treatment of refractory Clostridium difficile infection (CDI), providing proof of principle that a human disease can be treated by engineering the gut microbiota (3), and further studies indicate that a healthy microbiota can prevent disease acquisition (4).
Bacteria residing in the human gut produce urease, the activity of which is beneficial in healthy hosts but pathogenic in hosts with liver disease. Urea produced by the liver as a waste product is both excreted in urine and transported into the colon, where it is hydrolyzed by bacterial urease into carbon dioxide and ammonia. Ammonia is then (a) utilized by the microbiota for protein synthesis, (b) reabsorbed by the host, where it is incorporated into the nitrogen pool by hepatic metabolism, or (c) excreted in the feces. Mammalian genomes do not encode urease genes, so ammonia production results from bacterial urease activity acting on host-produced urea (5, 6). Ammonia is also largely responsible for the alkaline pH of the colonic luminal environment, acting to buffer the short-chain fatty acids also produced by the microbiota. Systemic ammonia levels are elevated in patients with liver injury, chronic liver disease, or urea cycle defects, whose hepatic abnormalities prevent the normal processing of ammonia delivered to the liver from the intestinal tract. Circulating ammonia is correlated with damage to the CNS in patients with chronic liver disease or inborn errors of metabolism, resulting in hepatic encephalopathy (HE) (7, 8).
Current treatments for HE and hyperammonemia are inadequate (8). Antibiotics (ABX) traditionally used to treat HE, including aminoglycosides and metronidazole, are limited by side effects and concerns for safety including ototoxicity, nephrotoxicity, and peripheral neuropathy (9, 10). Although rifaximin, a minimally absorbed ABX, has shown efficacy in the treatment and prevention of HE (11, 12), potential development of antimicrobial resistance with long-term use remains a concern. Lactulose is used to acidify feces and sequester ammonia as ammonium, but lactulose is poorly tolerated, resulting in poor adherence (13). In a mouse model of thioacetamide-induced liver injury, a lactobacillus probiotic has been reported to reduce ammonia levels and mortality (14), but these benefits have not been extended to human studies (15, 16). While promising, studies of probiotic therapies in humans described to date have suffered from methodological limitations, did not document long-term effects, and showed consistently minimal effects on outcome, motivating efforts to engineer more resilient and effective bacterial communities.
Here, we show that a synthetic microbial community lacking urease activity can be installed in the gut to reduce the production of ammonia long term and thereby mitigate HE in a mouse model. For proof-of-concept studies, we used altered Schaedler flora (ASF), which consists of 8 murine gut commensal bacterial strains that were assembled in the 1970s and standardized by the National Cancer Institute in 1978 (17). The strains were originally selected on the basis of their persistence from generation to generation in germ-free mice and their ability to restore a cecal morphology that is comparable to that of conventional mice. The ASF community is innocuous, known to have a beneficial effect of inducing immune tolerance (18), and is used by commercial mouse vendors to enhance the health of immunodeficient mouse strains. We found that ASF is low in urease activity, making it useful for the studies of metabolic engineering described here. Mixed results have been reported regarding the transfer of conventional microbiota between rodents, with some studies reporting successful transfer with repetitive inoculation (19), but others demonstrating that the use of ABX prevented transfer (20). We developed methods for purging the gut microbiota from normally colonized mice that were then transplanted with ASF by gavage. Transplantation of ASF was monitored longitudinally using deep sequencing of DNA from fecal pellets, revealing highly efficient colonization in properly prepared hosts. Over a 4-week monitoring period, ASF was partially displaced by selective colonization with environmental Firmicutes, reaching a new steady state, but fecal ammonia levels remained low. The engineered gut community was tested in thioacetamide (TAA) models of acute and chronic liver injury (Figure 1A), in which we demonstrate that it reduced fecal ammonia levels, mortality, and neurobehavioral deficits. These results show that transplantation of a defined minimal microbial community can alter metabolism in a predetermined fashion by establishing a new gut microbiome, resulting in therapeutic benefits in disease models.
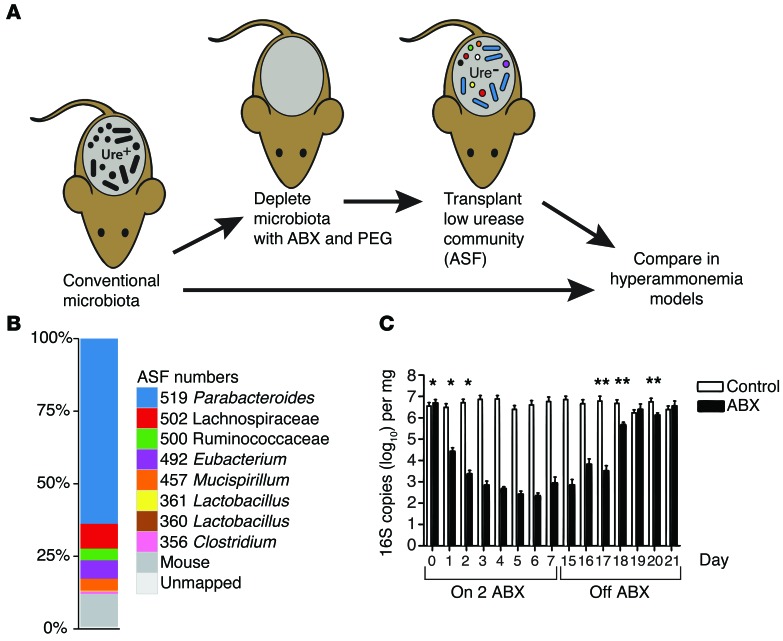
Figure 1. Transfer of ASF into a previously colonized murine host.
(A) Diagram of the experimental method. Ure, urease. (B) Shotgun metagenomic analysis of stool from ASF-colonized animals used for gavage in this study. Proportions of the different ASF lineages and other organisms are indicated by the color key. (C) Time course of 16S rRNA gene copy numbers during oral ABX treatment (14 days, vancomycin and neomycin) and upon discontinuation of ABX on day 15 (n = 3 per group). *P < 0.0001, for days 0–2 compared with the average of days 5–15 in the ABX group; **P < 0.05, between the ABX and control groups. Paired-sample t test and 2-tailed Student’s t test.
Results
Transplantation of ASF into previously colonized mice.
The original ASF was composed of 8 bacterial strains. Over time, ASF has been maintained in laboratory mice by fecal-oral transmission associated with cohousing in gnotobiotic isolators. Thus, the composition of the ASF donor material used here was first characterized by DNA isolation and shotgun metagenomic sequencing (16.9 Gb). Alignment to draft genome sequences of ASF strains (21) documented the presence of 7 of the 8 original strains in our samples (Figure 1B). Parabacteroides (ASF519) was the predominant lineage present in pellets. Additional ASF strains consisted of roughly even proportions of Clostridia (ASF356, ASF492, ASF500, ASF502) and Mucispirillum schaedleri (ASF457), accompanied by low levels of Lactobacilli (ASF361). No high-quality read pairs mapped concordantly to strain ASF360. In addition to ASF strains, 11% of reads mapped to the mouse genome, and a small fraction of alignments mapped to probable artifacts including cloning vectors, metazoans, and additional bacteria. We conclude that the bacteria in our donor material were mostly or entirely ASF strains.
To assess the persistence of ASF in mice housed under nonsterile conditions, sequential fecal pellets were collected from 10 ASF-colonized mice that were transferred to conventional SPF housing, DNA was purified from pellets, and the abundance and types of bacteria present were assessed by quantitative PCR (qPCR) and deep sequencing of 16S rRNA V1V2 gene segments (Supplemental Figure 1; supplemental material available online with this article; doi:10.1172/JCI79214DS1). Copy numbers of 16S sequences per gram of stool showed roughly similar abundance for ASF and conventionally colonized mice. Six of the eight ASF strains were resolved at the depth of sequencing performed. ASF519 (Parabacteroides) accounted for approximately 75% of sequence reads at the time of transfer of the mice. After approximately 2 months under nonsterile conditions, ASF519 remained the dominant taxon, and non-ASF taxa accounted for almost half of the sequence reads. Evidently, ASF lineages persisted but did not entirely exclude other bacteria.
We reasoned that reduction of the endogenous microbiota would promote the transfer of ASF, so we investigated the response of conventional microbiota to the oral delivery of 2 nonabsorbable ABX, neomycin and vancomycin. The 16S rRNA gene copy number was reduced by approximately 4 logs within 72 hours of oral ABX initiation (P < 0.0001 for days 0, 1, and 2 compared with the mean of days 5–15), then returned to baseline 5 days after discontinuing ABX (Figure 1C; P = 0.36 for comparison of ABX versus control at day 21), paralleling previous studies (22, 23).
Fecal slurries obtained from ASF-colonized mice (Taconic) were then gavaged into conventionally housed recipients for 7 days following a 72-hour pretreatment with oral ABX and a 12-hour intestinal purge using polyethylene glycol (PEG) (pretreatment with ABX and PEG is henceforth termed “prepared”). PEG was used in our gut-cleansing protocol, because the use of a purgative will likely be necessary to reduce bacterial load in the human intestinal tract due to high biomass. For comparison, ASF transplants were carried out on conventional mice without pretreatment and on germ-free mice. Mice that were treated with ABX and PEG and subsequently transplanted showed normal numbers of bacteria by 16S qPCR copy numbers within 10 days (data not shown).
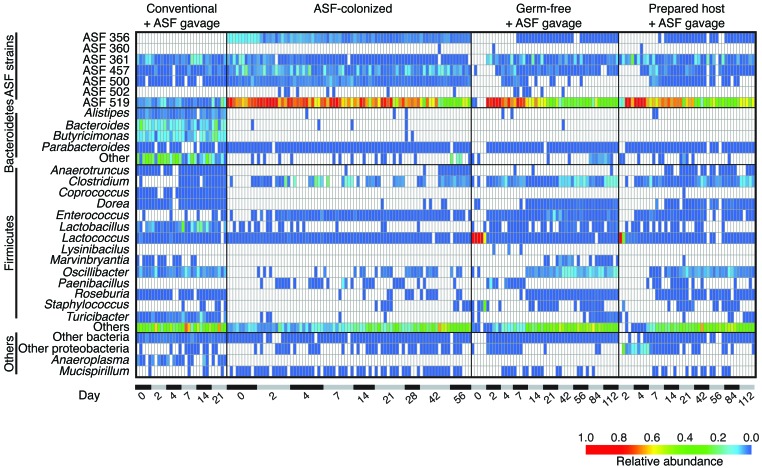
Longitudinal fecal samples were analyzed by deep sequencing of 16S rRNA gene tags, then the proportions of bacterial lineages detected were plotted as heatmaps, in which each row shows a bacterial lineage and each column a fecal specimen (Figure 2). Conventional mice that were not pretreated showed no increase in ASF lineages, despite ASF gavage (Figure 2, Conventional + ASF gavage). A few preexisting lineages were present that were indistinguishable from ASF using the V1V2 16S sequence window, but these did not increase in abundance after ASF gavage. A second group of mice (discussed above) were colonized with ASF from birth and then moved to the nonsterile facility. These mice had high levels of ASF519 and detectable levels of 4 other ASF strains (Figure 2, ASF-colonized).
Figure 2. Heatmap showing the relative abundance of bacterial lineages over time in ASF-colonized mice and controls.
Rows indicate bacterial lineages as annotated on the left. Relative abundance is indicated by the color key at the bottom of the figure. Columns summarize the sequencing results from individual fecal specimens. Elapsed time in days is shown along the bottom. The groups studied are indicated at the top of the heatmap and include (from the left) conventional mice that were gavaged with ASF stool without preparation (Conventional + ASF gavage); mice that were ASF colonized from birth, then transferred to a nonsterile SPF facility (ASF-colonized); mice that were germ-free, then gavaged with ASF (Germ-free + ASF gavage); and conventional mice that were prepared with ABX and PEG treatment, then gavaged with ASF (Prepared host + ASF gavage).
These groups were then compared with (a) germ-free mice gavaged with ASF (referred to herein as germ-free/ASF mice); and (b) conventional mice pretreated with ABX and PEG and gavaged with ASF (referred to herein as prepared/ASF mice) (Figure 2, Germ-free + ASF gavage and Prepared host + ASF gavage). Prior to ASF gavage, both the germ-free animals and ABX-treated animals showed high levels of Lactococcus, a lineage found at high levels in mouse chow (22), indicating that endogenous gut bacteria were mostly or entirely absent. Gavage of these animals with ASF resulted in establishment of ASF lineages that persisted for the duration of the sampling period. The communities were again dominated by ASF519.
Longitudinal evolution of the transplanted ASF community.
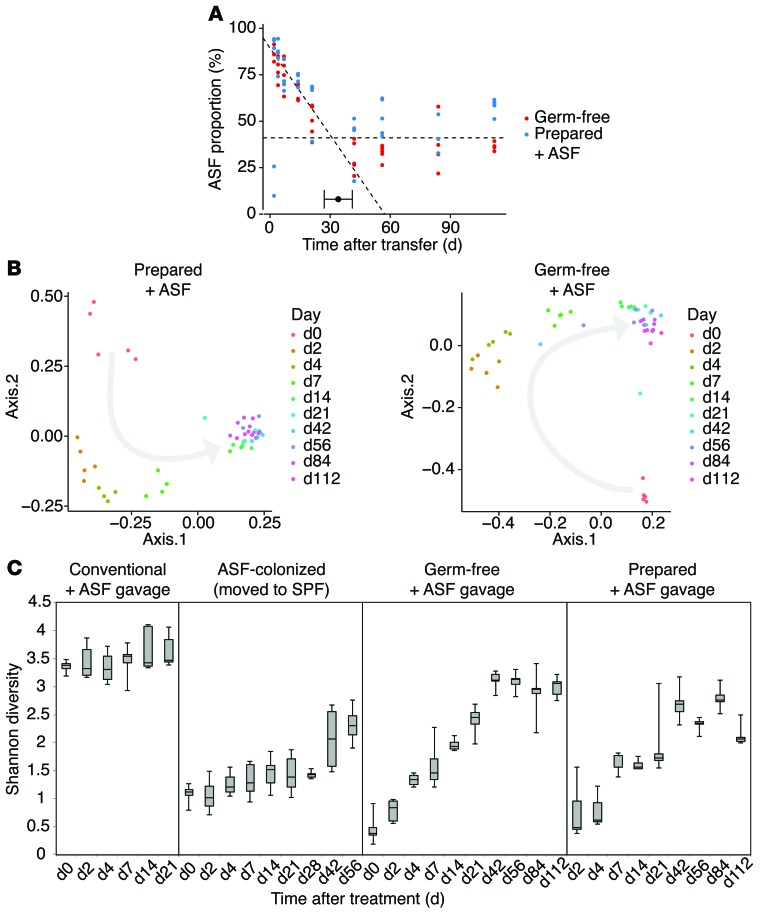
Persistence of the transplanted ASF community was quantified using segmental regression, plotting time against the proportion of ASF in each sample (Figure 3A). Prepared/ASF mice and germ-free/ASF mice transferred to nonsterile conditions were compared and found to behave similarly. Initially, ASF comprised the majority of the community. After transfer, the proportion of ASF slowly declined, constituting approximately 45% of the gut community after 34 days (Davies’ test for a change in the slope P < 0.001; 95% CI for the breakpoint was 27–41 days by segmented linear regression). After 34 days, the proportion of ASF strains did not decrease for the duration of the experiment (120 days; P = 0.86). Comparisons of community membership using unweighted UniFrac analysis of the 16S rRNA gene-sequencing data showed that non-ASF lineages had colonized by day 14, and members persisted for the duration of the study (Figure 3B), although the final abundance of non-ASF lineages was not achieved until about day 30 (Figure 3A). Diversity of this new stable state measured by the Shannon index approached that of the starting community (Figure 3C). ASF519 (Parabacteroides) was the main taxon persisting in gavage from both germ-free/ASF and prepared/ASF hosts after 6 weeks, constituting approximately 40% and 50%, respectively, in each group (Figure 2). As in humans, bacteria belonging to the Bacteroides genus were dominant taxa in conventionally housed mice (24, 25).
Figure 3. Development of a stable gut microbial community nucleated by inoculation with ASF.
(A) Segmented regression analysis of communities in mice that were either germ-free or prepared conventional mice subjected to ASF gavage. The y axis shows the proportion of ASF lineages inferred from 16S rRNA gene tag pyrosequencing data. The x axis shows the number of days after transfer. Segmented regression analysis showed 2 phases, indicating a slow decline in the ASF proportion up to about day 30, followed by establishment of a new steady state consisting of approximately 40% ASF lineages. (B) Principal coordinates analysis (PCoA) ordination over time. Changes in community membership over time were analyzed using unweighted Unifrac (61). Progression of time is indicated by a gray arrow. (C) Shannon diversity of gut microbiota over time in the 4 hosts described in Figure 2. n = 5 per group for Conventional + ASF gavage, Germ-free + ASF gavage, and Prepared + ASF gavage. n = 10 for ASF-colonized.
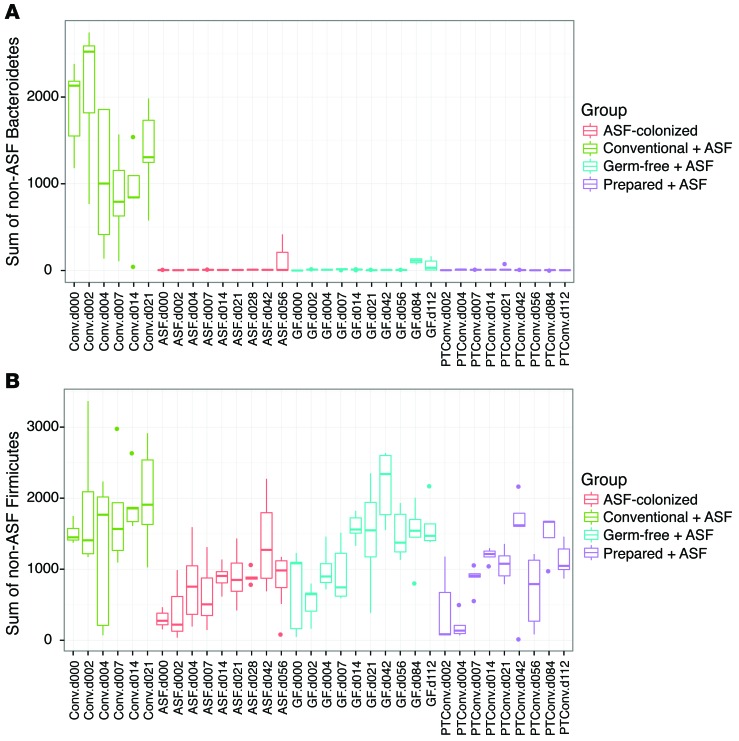
The new lineages appearing after transplantation were specific (Figure 4). Members of the Bacteroidetes phylum did not recolonize after ASF transplantation, suggesting that ASF519, a member of the closely related Parabacteroides genus, may have occupied niches available to this group. Several Firmicutes lineages did colonize over time, including lineages annotated as Oscillibacter, Clostridium, and “other” Firmicutes (mostly of the order Clostridiales). Thus, the community achieved a new steady state containing both ASF and environmentally acquired lineages.
Figure 4. Comparison of non-ASF sequence reads from either Bacteroidetes or Firmicutes, illustrating selective repopulation with environmental Firmicutes.
(A and B) Groups are color coded and represent change over time. n = 5 per group for Conventional + ASF gavage, Germ-free + ASF gavage, and Prepared + ASF gavage. n = 10 for ASF-colonized.
ASF has minimal urease gene content and activity.
An analysis of the complete ASF genomes showed a minimal presence of urease genes. No urease genes were identified in the predominant Parabacteroides ASF519. Two ASF members contained urease genes, ASF492 and ASF361, but both were minor members of the community after transplantation (Figure 2).
We then characterized urease activity, which we found readily detectable in pellets from conventionally housed mice, but undetectable in pellets from mice colonized with ASF (Figure 5A). Similar results were obtained with pellets from mice treated with oral ABX that reduced the bacterial load by 10,000-fold (Figure 5A). Intravenous delivery of 13C-urea to quantify urease activity in vivo through the production of 13CO2 in a breath test revealed minimal hydrolysis in ASF-colonized mice (Figure 5B).
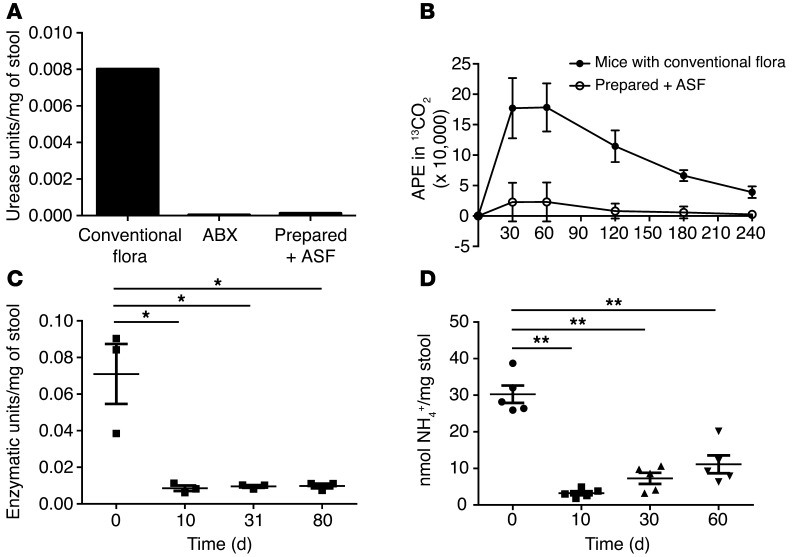
Figure 5. Transfer of ASF leads to a reduction in urease activity and fecal ammonia levels.
(A) Urease activity in the feces of a conventionally housed mouse versus a mouse treated with ABX and a mouse colonized with ASF. (B) In vivo urease activity in conventionally housed (n = 5) and ASF-colonized mice (n = 5) quantified by the release of 13CO2 after i.v. injection of 13C-urea. (C) Fecal urease activity at the indicated time points after transplantation of ASF into prepared mice fed an irradiated diet (n = 3). (D) Fecal ammonia levels before and after transplantation of ASF into prepared mice fed a nonirradiated diet (n = 5). *P < 0.01; **P < 0.001. Tukey’s test for multiple comparisons.
Transplantation of ASF into prepared hosts led to a reduction in fecal urease activity lasting for at least 80 days (Figure 5C). The novel community assembled after 30 days had low urease activity, even though new lineages became established in addition to ASF (Figure 2). Analysis of representative genomic sequences from these newly established genera showed that Oscillibacter, Dorea, Enterococcus, and Roseburia do not encode urease genes. We found that Clostridium and the group annotated as “other” Firmicutes (mostly Clostridiales) were mixed, with some representatives encoding urease genes, while others did not. It is unknown whether the newly proliferating organisms lacked urease genes, or whether they encoded urease genes but expressed them at low levels. Thus, the net effect of ASF colonization, together with the establishment of additional lineages, was the achievement of a new steady state with low urease activity. The new community persisted over the long term, as ASF-transplanted mice showed low fecal ammonia levels for over 1 year in a specific pathogen–free (SPF) housing facility (data not shown).
The resilience of this new community state following dietary stress was evaluated by placing mice transplanted with ASF on a low-protein diet similar to that used for patients with hyperammonemic inborn errors of metabolism (26). Fecal ammonia levels remained much lower than those achieved by a low-protein diet alone, an effect that was durable for several months (T.-C.D. Shen and G.D. Wu, unpublished observations). Another dietary stress, consumption of a nonirradiated diet, led to the displacement of ASF by other bacteria within weeks after FMT, with the predominant taxon Parabacteroides ASF519 being partially replaced by other taxa within the Bacteroidetes phylum (Supplemental Figure 2). Nevertheless, the reduction in fecal ammonia levels compared with those at baseline remained significant for several months, even in mice on a nonirradiated diet (Figure 5D). Reductions in fecal ammonia levels have been correlated with reductions in blood ammonia (14, 27, 28), indicating that changes in colonic ammonia production and/or absorption can be associated with blood levels. The reduction in fecal ammonia may alter levels of false neurotransmitter precursors produced by the gut microbiota and/or by the host, since ammonia is a substrate for both, and this would lead to reduced formation of the biogenic amines that are hypothesized to play a role in HE (29–31). Thus, we propose that fecal ammonia is a useful biomarker for response of the host to the treatment of hyperammonemia.
ASF transplantation reduces mortality and cognitive impairment in murine models of acute and chronic liver injury.
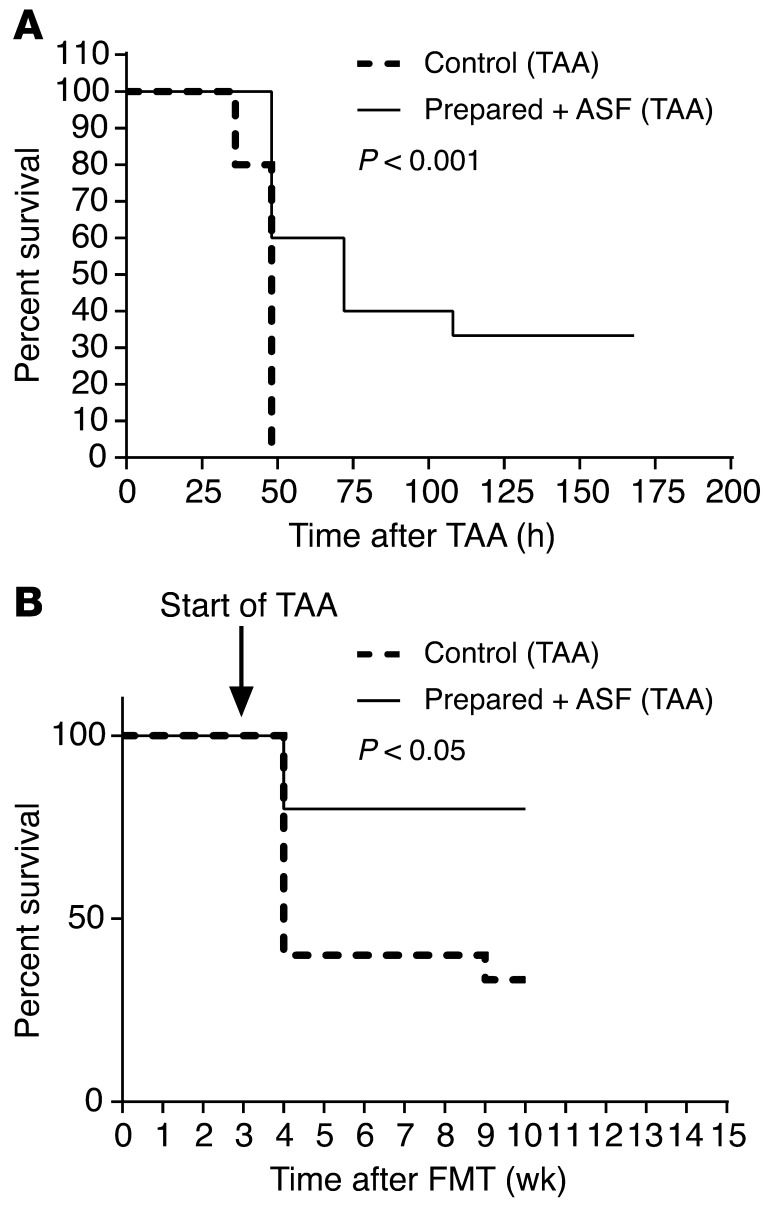
A major cause of morbidity and mortality associated with acute liver injury is the development of HE. Since hyperammonemia is associated with the development of HE in patients with impaired hepatic function (8), we asked whether the transplantation of ASF might mitigate the effects of acute hepatic injury induced by TAA (32) treatment. Prepared/ASF mice showed both a reduction in fecal ammonia levels (Supplemental Figure 3A, Prepared + ASF) and reduced mortality in response to high-dose TAA compared with mice with conventional microbiota (Figure 6A). This finding was also seen in the setting of chronic liver injury in mice transplanted 3 weeks prior to the chronic delivery of TAA at low, escalating doses for 7 weeks (Figure 6B). Compared with control mice, prepared/ASF mice demonstrated markedly reduced mortality rates that were maintained over the 7-week period during which hepatic fibrosis developed in both groups (refs. 33, 34; data not shown), consistent with a sustained reduction in fecal ammonia for several months after ASF transplantation.
Figure 6. ASF transplantation into prepared mice reduces mortality after thioacetamide-induced hepatic injury and fibrosis.
(A) Kaplan-Meier survival curves of high-dose TAA-induced acute hepatic injury in conventional versus prepared/ASF mice (n = 15 per group). (B) Kaplan-Meier survival curves of chronic, thrice weekly TAA administration at low, escalating doses, initiated 3 weeks after ASF transplantation (n = 15 per group). Survival curves were analyzed by the Kaplan-Meier method using the log-rank test.
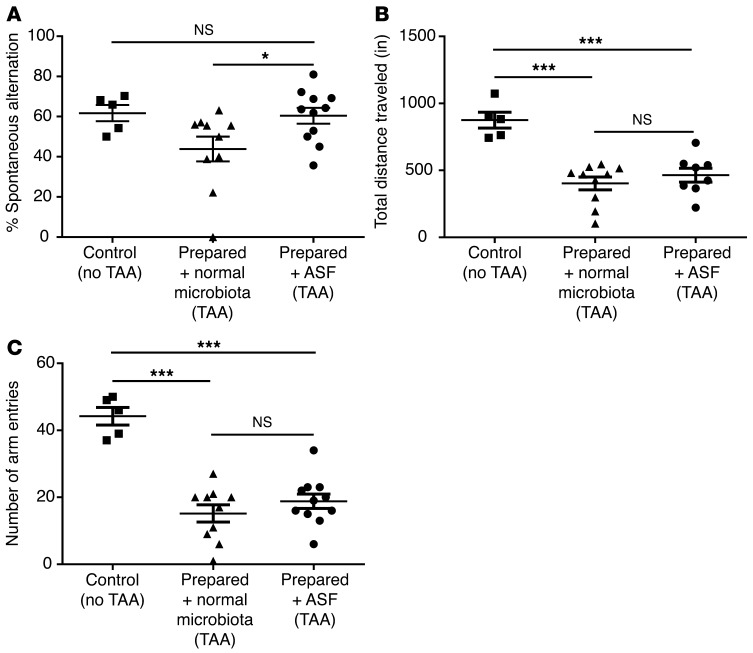
In mice, the TAA model has also been associated with neurobehavioral abnormalities resembling HE in humans (35). Using a lower dose of TAA to reduce mortality, we analyzed memory and spatial learning in a Y maze test (36) comparing prepared/ASF mice with mice transplanted with normal microbiota (referred to herein as prepared/normal microbiota mice; Figure 7 and Supplemental Figure 3, B and C) as a control. The 80%–90% survival rates were similar between the 2 groups at the lower TAA dose (Supplemental Figure 3B). Fecal ammonia levels were reduced in mice transplanted with ASF compared with those with normal microbiota (Supplemental Figure 3C). Prepared/normal microbiota mice showed a decrease in cognitive function after TAA treatment, quantified as spontaneous alternations in a Y maze test, whereas prepared/ASF mice treated with TAA were not different from untreated controls (Figure 7A). Mice transplanted with either ASF or normal microbiota did not sufficiently differ in locomotor activity after TAA treatment (as quantified by the total distance traveled and number of arm entries in the Y maze) to account for the difference in spontaneous alternations, although both groups exhibited significantly less locomotor activity compared with untreated controls (Figure 7, B and C). To exclude the possibility that ASF transplantation directly reduced liver injury, we measured plasma alanine aminotransferase (ALT) and quantified histologic evidence of hepatocyte necrosis (Supplementary Figure 4, A and B). Both revealed that liver damage induced by TAA was not reduced by either ASF transplantation or ABX treatment. Thus, improved survival and behavioral performance were associated with reduced ammonia levels and not improved locomotor activity or liver injury.
Figure 7. ASF transplantation into prepared mice restores cognitive, but not locomotor, deficits after thioacetamide-induced hepatic injury.
(A) Spontaneous alternations after TAA treatment quantified by Y maze testing in prepared mice that were transplanted with either normal microbiota (n = 10) or ASF (n = 11) compared with untreated control mice (n = 5). *P < 0.05, by 2-tailed unpaired Student’s t test; P = 0.04, by ANOVA. (B) Total distance traveled and (C) number of arm entries in the Y maze in prepared/ASF and prepared + normal microbiota mice after TAA treatment compared with untreated control mice. ***P < 0.001, by 2-tailed unpaired Student’s t test; P < 0.001, by ANOVA.
Discussion
The success of FMT in the treatment of CDI establishes that transplanting a resilient microbial community can alter a dysbiotic microbiota and thereby treat disease. Feces, however, contains not only bacteria but also a multitude of archaea, fungi, and viruses, so there is concern for safety (37), motivating the development of defined microbial consortia for human inoculation that have well-characterized biological properties and respond to the gut environment in predictable ways. Here, we show that a defined minimal consortium of bacteria, ASF (17, 18), can durably reprogram the composition and metabolic function of the gut microbiota when inoculated into a properly prepared host. By taking advantage of the minimal urease activity in ASF, we provide evidence that reprogramming the gut microbiota can lead to lower fecal ammonia levels and mitigate the morbidity and mortality associated with liver damage.
The endogenous gut microbiota needed to be depleted by treatment with oral ABX and PEG for efficient transfer of the ASF community into conventionally housed mice. After the gut purge, transfer was as effective as that into germ-free recipients. Tracking by sequencing suggested an orderly succession of lineages. After ASF transplantation, the predominant ASF519 strain appeared at the earliest times after gavage. Mucispirillum schaedleri (ASF457) and Ruminococcaceae (ASF500) also appeared early on. The last to appear was the Clostridium species (ASF356) on day 14 in some mice, 1 week after gavaging was complete, perhaps indicating the need for development of a specific niche that permitted the establishment of more fastidious taxa. Most mice contained a Lactobacillus strain indistinguishable from ASF361 prior to transplantation, so this lineage could not be tracked with the methods used. The observed succession may be similar to the succession of bacterial taxa in human infants, as consumption of oxygen by initial gut colonizers allows the expansion of bacterial clades that are obligate anaerobes (38, 39).
By monitoring the composition of the transplanted ASF community over a 4-month period using 16S rRNA gene sequencing, we were able to assess the persistence of the community in the nonsterile SPF environment. The ASF community did not fully exclude other taxa. The community appeared to achieve a new steady state, whereby both types of transplanted hosts came to resemble the mice colonized with ASF from birth and housed long term in an SPF environment. In each of these 3 groups, Parabacteroides (ASF519) remained the dominant taxon. Bacteria of the Bacteroides genus commonly dominate the human and murine gut microbiota (24, 40–43), but new Bacteroides did not accumulate over time in ASF-colonized mice, suggesting that Parabacteroides may have excluded Bacteroides, reminiscent of the trade-off between Bacteroides and Prevotella in human gut (24, 44–46). The mechanism by which Parabacteroides excludes Bacteroides is unknown but, due to their taxonomic and functional similarities, may involve competition for limiting resources (47) such as has been shown for glycans and the competition between Bacteroides species in the colonic crypt (48). Over time, the Parabacteroides and environmental Firmicutes established a new steady state approximating the composition of conventional microbiota in humans and mice (40, 41, 49), possibly involving a syntrophic relationship between these lineages (50).
There was a sustained reduction in fecal urease enzymatic activity and ammonia production upon transfer of ASF into prepared mice. There was no return of urease activity several months after transfer, despite a substantial increase of non-ASF taxa. A possible lack of urease genes in many of the Clostridium taxa that accumulated after transplantation may explain the persistently reduced urease activity, although we cannot exclude other mechanisms, since the regulation of urease enzymatic activity is known to be complex (6).
Production of ammonia by the gut microbiota has been implicated in host nitrogen balance (51–53), so there is a theoretical risk that a urease-free community might have an adverse effect on protein balance and growth of the host. However, we have tracked mice for over 1 year after ASF transplantation and observed no adverse effects on their body weight or mortality. ASF is acknowledged to be an innocuous bacterial consortium in mice with beneficial effects on immune tolerance (18). Nevertheless, additional safety studies will need to be performed using a humanized version of ASF in rodent models before human studies can be contemplated.
In summary, given that HE is a major contributor to morbidity and mortality in liver disease (54), transplantation of next-generation engineered communities based on ASF, coupled with improved preparation of the host as described here, represents a promising approach to more effective therapy.
Methods
Animals.
Feces from CB17 SCID mice colonized with ASF (Taconic) were the source of ASF in the transplantation experiments. Mice in this study were maintained in a standard SPF barrier facility and were fed irradiated AIN-76 chow (Research Diets Inc.) containing protein as 21% of the kilocalories. For the nonirradiated diet experiment, we used Laboratory Rodent Diet 5001 (LabDiet) containing protein as 29% of the kilocalories. For microbiota transfer experiments, 0.1 g feces was diluted 10-fold in PBS. For Figures 2–4, germ-free recipient mice were purchased from Taconic, as were the conventionally housed Swiss Webster mice used in the first experiment. For all other experiments, we used either male or female 8-week-old C57BL6 mice (The Jackson Laboratory). Pretreated conventional mice were prepared for inoculation by oral delivery of ABX in the drinking water (1.125 g aspartame, 0.15 g vancomycin, and 0.3 g neomycin in 300 ml sterile water) for 72 hours. During the final 12 hours, the water supply was exchanged with a 10% PEG solution (Merck), and the mice were fasted. The mice were then inoculated daily with feces by oral gavage for 5 to 7 days. Fecal pellets were collected for bacterial taxonomic and biochemical analyses at the time points indicated in the figures (Figure 1C, Figures 2–4, Figure 5, A, C, and D, Supplemental Figure 1, Supplemental Figure 2, and Supplemental Figure 3, A and C).
DNA isolation, qPCR, sequencing, and analysis.
DNA was isolated from stool as previously described (46, 55). Bacterial 16S rRNA gene sequences were PCR amplified with primers binding to the V1V2 region (46, 55) using bar-coded primers (56, 57). Shotgun metagenomic data were collected using the TruSeq library preparation method and a HiSeq instrument (both from Illumina). Sequence reads were quality controlled and analyzed using the QIIME pipeline with default parameters (58). Sequence data sets were deposited in the NCBI’s Sequence Read Archive (SRA) database (accession number SRP058968).
Urease activity and ammonia assays.
Fecal ammonia levels were determined using an Ammonia Assay Kit (ab83360; Abcam). Fecal pellets were suspended in the assay buffer provided at a concentration of 1 mg/10 μl and centrifuged at 13,000 g for 10 minutes at room temperature to remove insoluble material. Ammonia concentration was then determined according to the kit protocol.
Fecal urease activity was measured by suspending fecal pellets in 0.5 mM HEPES buffer. After sonication and centrifugation, the supernatant was incubated for 30 minutes at 37°C with 1 μCi 14C-labeled urea (19.9 mCi/mmol; catalog ARC-0150; American Radiolabeled Chemicals) in a sealed container. The air was purged into a trap containing 2.5 ml of 0.2 M benzethonium hydroxide in methanol (catalog 82156; Sigma-Aldrich), and 14CO2 activity was quantified by liquid scintillation counting. A standard curve was generated using purified E. coli urease (150 IU/mg; catalog 22060744-1; BioWORLD).
The urease breath test was performed on 4-hour fasted mice by placing them in a sealed glass chamber. A total of 4 cc air was withdrawn from the chamber using a gas-tight syringe after 10 minutes and injected into 12 ml (gas-helium) Exetainer glass vials (438B; Labco) to establish a baseline CO2 level. The mouse was then injected via the tail vein with [13C]urea (150 μg/g body weight) (CLM-311-0; Cambridge Isotope Labs). Blood was collected 15 minutes after injection to assess total urea and isotopic enrichment in [13C]urea. Enrichment in expired air was measured at 30, 60, 120, 180, and 240 minutes after injection of the labeled urea. The 13CO2/12CO2 ratio was measured in gas samples with a Finnigan DELTAplus isotope ratio mass spectrometer (Thermo Fisher Scientific). A commercial CO2 source (Airgas) was used as the standard. The measurement of isotopic abundance in [13C]urea in plasma involves the elimination of CO2 from the sample and the subsequent conversion of urea to CO2 with commercially available urease (59).
Induction of acute liver injury and hepatic fibrosis.
Mice in the high-dose TAA acute liver injury model were given a single i.p. injection of TAA at 600 mg/kg. In the low-dose TAA acute liver injury model, mice were given a single dose of TAA at 300 mg/kg by i.p. injection, with subsequent performance of the Y maze neurobehavior test 48 hours after TAA injection (see Neurobehavior test below). In the chronic liver fibrosis model, all mice were given TAA by i.p. injection 3 times weekly, with the initial 100 mg/kg dose being decreased to 50 mg/kg during the first week, given the rate of morality, followed by dose escalation to 100 mg/kg during the second week, 200 mg/kg during the third week, 300 mg/kg during the fourth week, and 400 mg/kg during the fifth to seventh weeks of TAA administration.
Neurobehavior test.
The Y maze test was conducted to assess the rodents’ memory and spatial learning (60). The maze consists of 3 identical and equally spaced arms, and the natural tendency of the mouse is to investigate a new arm of the maze rather than return to the one previously visited. A mouse with a cognitive deficit would exhibit less spontaneous alternation, defined as entering all 3 arms in 3 sequential arm entries. Control mice typically exhibit greater than 60% spontaneous alternation. In this experiment, mice were allowed to habituate to the testing room, where there were no overt visual cues, for 30 minutes prior to testing. The maze was cleaned with 70% ethanol before use and between trials to eliminate odor cues. A trial started when a mouse was released into 1 arm of the maze (same arm for each mouse). As the mouse navigated the maze, each arm entry was noted. At the end of the timed trial (8 minutes), total arm entries were summed and spontaneous alternations determined by the following formula: percentage of spontaneous alternations = [(number of alternations)/(total arm entries − 2)] × 100. Trials were video recorded as well as graded during the procedure. Image analysis software was used to measure the total distance traveled by each mouse during the trials.
Statistics.
Results are expressed as the mean ± SEM. Statistical significance among 3 or more groups was assessed by ANOVA. A 2-tailed Student’s t test and paired-sample t test were used for direct comparisons between 2 groups and within groups, respectively. Tukey’s test was used to adjust for multiple comparisons. Kaplan-Meier survival curves were compared using the log-rank test. A P value of less than 0.05 was considered statistically significant.
Study approval.
All animal studies were performed with the approval of the IACUC of the University of Pennsylvania.
Supplementary Material
Acknowledgments
This work was supported by grants from the NIH (RO1-DK089472, to G.D. Wu; UH2/3-DK083981, to G.D. Wu, F.D. Bushman, and J.D. Lewis; and HD26979, to M. Yudkoff); the Molecular Biology Core and the Molecular Pathology Imaging Core of the Penn Center for Molecular Studies in Digestive and Liver Diseases (P30 DK050306); and the Joint Penn-CHOP Center for Digestive, Liver, and Pancreatic Medicine and the Metabolomic Core at the Children’s Hospital of Philadelphia. Neurobehavior testing was conducted in collaboration with the Neurobehavior Testing Core at the University of Pennsylvania.
Footnotes
Conflict of interest: F.D. Bushman, J.D. Lewis, and G.D. Wu are cofounders of Microbiota Therapeutics LLC. No funds from this company were used to generate data for this manuscript.
Reference information:J Clin Invest. 2015;125(7):2841–2850. doi:10.1172/JCI79214.
References
- 1.Backhed F, et al. Defining a healthy human gut microbiome: current concepts, future directions, and clinical applications. Cell Host Microbe. 2012;12(5):611–622. doi: 10.1016/j.chom.2012.10.012. [DOI] [PubMed] [Google Scholar]
- 2.Lemon KP, Armitage GC, Relman DA, Fischbach MA. Microbiota-targeted therapies: an ecological perspective. Sci Transl Med. 2012;4(137):137rv135. doi: 10.1126/scitranslmed.3004183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.van Nood E, et al. Duodenal infusion of donor feces for recurrent Clostridium difficile. N Engl J Med. 2013;368(5):407–415. doi: 10.1056/NEJMoa1205037. [DOI] [PubMed] [Google Scholar]
- 4.Lawley TD, et al. Targeted restoration of the intestinal microbiota with a simple, defined bacteriotherapy resolves relapsing Clostridium difficile disease in mice. PLoS Pathog. 2012;8(10):e1002995. doi: 10.1371/journal.ppat.1002995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Walser M, Bodenlos LJ. Urea metabolism in man. J Clin Invest. 1959;38:1617–1626. doi: 10.1172/JCI103940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mobley HL, Hausinger RP. Microbial ureases: significance, regulation, and molecular characterization. Microbiol Rev. 1989;53(1):85–108. doi: 10.1128/mr.53.1.85-108.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Saudubray JM, Nassogne MC, de Lonlay P, Touati G. Clinical approach to inherited metabolic disorders in neonates: an overview. Semin Neonatol. 2002;7(1):3–15. doi: 10.1053/siny.2001.0083. [DOI] [PubMed] [Google Scholar]
- 8.Riordan SM, Williams R. Treatment of hepatic encephalopathy. N Engl J Med. 1997;337(7):473–479. doi: 10.1056/NEJM199708143370707. [DOI] [PubMed] [Google Scholar]
- 9.Leitman P. Liver disease, aminoglycoside antibiotics and renal dysfunction. Hepatology. 1988;8(4):966–968. doi: 10.1002/hep.1840080442. [DOI] [PubMed] [Google Scholar]
- 10.Hampel H, Bynum G, Zamora E, El-Serag H. Risk factors for the development of renal dysfunction in hospitalized patients with cirrhosis. Am J Gastroenterol. 2001;96(7):2206–2210. doi: 10.1111/j.1572-0241.2001.03958.x. [DOI] [PubMed] [Google Scholar]
- 11.Bass N, et al. Rifaximin treatment in hepatic encephalopathy. N Engl J Med. 2010;362(12):1071–1081. doi: 10.1056/NEJMoa0907893. [DOI] [PubMed] [Google Scholar]
- 12.Mullen K, et al. Rifaximin is safe and well tolerated for long-term maintenance of remission from overt hepatic encephalopathy. Clin Gastroenterol Hepatol. 2014;12(8):1390–1397. doi: 10.1016/j.cgh.2013.12.021. [DOI] [PubMed] [Google Scholar]
- 13.Bajaj JS, Sanyal AJ, Bell D, Gilles H, Heuman DM. Predictors of the recurrence of hepatic encephalopathy in lactulose-treated patients. Aliment Pharmacol Ther. 2010;31(9):1012–1017. doi: 10.1111/j.1365-2036.2010.04257.x. [DOI] [PubMed] [Google Scholar]
- 14.Nicaise C, et al. Control of acute, chronic, and constitutive hyperammonemia by wild-type and genetically engineered Lactobacillus plantarum in rodents. Hepatology. 2008;48(4):1184–1192. doi: 10.1002/hep.22445. [DOI] [PubMed] [Google Scholar]
- 15.McGee RG, Bakens A, Wiley K, Riordan SM, Webster AC. Probiotics for patients with hepatic encephalopathy. Cochrane Database Syst Rev. 2011;(11):CD008716. doi: 10.1002/14651858.CD008716.pub2. [DOI] [PubMed] [Google Scholar]
- 16.Lunia MK, Sharma BC, Sharma P, Sachdeva S, Srivastava S. Probiotics prevent hepatic encephalopathy in patients with cirrhosis: a randomized controlled trial. Clin Gastroenterol Hepatol. 2014;12(6):1003–1008.e1001. doi: 10.1016/j.cgh.2013.11.006. [DOI] [PubMed] [Google Scholar]
- 17.Dewhirst FE, et al. Phylogeny of the defined murine microbiota: altered Schaedler flora. Appl Environ Microbiol. 1999;65(8):3287–3292. doi: 10.1128/aem.65.8.3287-3292.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Geuking MB, et al. Intestinal bacterial colonization induces mutualistic regulatory T cell responses. Immunity. 2011;34(5):794–806. doi: 10.1016/j.immuni.2011.03.021. [DOI] [PubMed] [Google Scholar]
- 19.Willing BP, Vacharaksa A, Croxen M, Thanachayanont T, Finlay BB. Altering host resistance to infections through microbial transplantation. PLoS One. 2011;6(10):e26988. doi: 10.1371/journal.pone.0026988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Manichanh C, et al. Reshaping the gut microbiome with bacterial transplantation and antibiotic intake. Genome Res. 2010;20(10):1411–1419. doi: 10.1101/gr.107987.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wannemuehler MJ, Overstreet AM, Ward DV, Phillips GJ. Draft genome sequences of the altered schaedler flora, a defined bacterial community from gnotobiotic mice. Genome Announc. 2014;2(2):e00287-14. doi: 10.1128/genomeA.00287-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dollive S, et al. Fungi of the murine gut: episodic variation and proliferation during antibiotic treatment. PLoS One. 2013;8(8):e71806. doi: 10.1371/journal.pone.0071806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hill DA, et al. Metagenomic analyses reveal antibiotic-induced temporal and spatial changes in intestinal microbiota with associated alterations in immune cell homeostasis. Mucosal Immunol. 2010;3(2):148–158. doi: 10.1038/mi.2009.132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Arumugam M, et al. Enterotypes of the human gut microbiome. Nature. 2011;473(7346):174–180. doi: 10.1038/nature09944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hildebrandt MA, et al. High-fat diet determines the composition of the murine gut microbiome independently of obesity. Gastroenterology. 2009;137(5):1716–1724 e1711. doi: 10.1053/j.gastro.2009.08.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Singh R. Nutritional management of patients with urea cycle disorders. J Inherit Metab Dis. 2007;30(6):880–887. doi: 10.1007/s10545-007-0718-4. [DOI] [PubMed] [Google Scholar]
- 27.Alexander T, Thomas K, Cherian A. Effect of three antibacterial drugs in lowering blood & stool ammonia production in hepatic encephalopathy. Indian J Med Res. 1992;96:292–296. [PubMed] [Google Scholar]
- 28.Zhao H, Wang H, Lu Z, Xu S. Intestinal microflora in patients with liver cirrhosis. Chin J Dig Dis. 2004;5(2):64–67. doi: 10.1111/j.1443-9573.2004.00157.x. [DOI] [PubMed] [Google Scholar]
- 29.James J, Ziparo V, Jeppsson B, Fischer J. Hyperammonaemia, plasma aminoacid imbalance, and blood-brain aminoacid transport: a unified theory of portal-systemic encephalopathy. Lancet. 1979;2(8146):772. doi: 10.1016/s0140-6736(79)92119-6. [DOI] [PubMed] [Google Scholar]
- 30.Fischer J, Baldessarini R. False neurotransmitters and hepatic failure. Lancet. 1971;2(7715):75–80. doi: 10.1016/s0140-6736(71)92048-4. [DOI] [PubMed] [Google Scholar]
- 31.Holecek M. Ammonia and amino acid profiles in liver cirrhosis: effects of variables leading to hepatic encephalopathy. Nutrition. 2015;31(1):14–20. doi: 10.1016/j.nut.2014.03.016. [DOI] [PubMed] [Google Scholar]
- 32.Miranda AS, et al. A thioacetamide-induced hepatic encephalopathy model in C57BL/6 mice: a behavioral and neurochemical study. Arq Neuropsiquiatr. 2010;68(4):597–602. doi: 10.1590/S0004-282X2010000400022. [DOI] [PubMed] [Google Scholar]
- 33.Ishikawa S, et al. CD1d-restricted natural killer T cells contribute to hepatic inflammation and fibrogenesis in mice. J Hepatol. 2011;54(6):1195–1204. doi: 10.1016/j.jhep.2010.08.022. [DOI] [PubMed] [Google Scholar]
- 34.Elinav E, et al. Competitive inhibition of leptin signaling results in amelioration of liver fibrosis through modulation of stellate cell function. Hepatology. 2009;49(1):278–286. doi: 10.1002/hep.22584. [DOI] [PubMed] [Google Scholar]
- 35.Avraham Y, et al. Cannabidiol improves brain and liver function in a fulminant hepatic failure-induced model of hepatic encephalopathy in mice. Br J Pharmacol. 2011;162(7):1650–1658. doi: 10.1111/j.1476-5381.2010.01179.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hughes RN. The value of spontaneous alternation behavior (SAB) as a test of retention in pharmacological investigations of memory. Neurosci Biobehav Rev. 2004;28(5):497–505. doi: 10.1016/j.neubiorev.2004.06.006. [DOI] [PubMed] [Google Scholar]
- 37.Hecht GA, et al. What is the value of a food and drug administration investigational new drug for fecal microbiota transplantation in Clostridium difficile infection? Clin Gastroenterol Hepatol. 2014;12(2):289–291. doi: 10.1016/j.cgh.2013.10.009. [DOI] [PubMed] [Google Scholar]
- 38.Dominguez-Bello MG, Blaser MJ, Ley RE, Knight R. Development of the human gastrointestinal microbiota and insights from high-throughput sequencing. Gastroenterology. 2011;140(6):1713–1719. doi: 10.1053/j.gastro.2011.02.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Albenberg L, et al. Correlation between intraluminal oxygen gradient and radial partitioning of intestinal microbiota in humans and mice. Gastroenterology. 201414751055-63.e8 10.1053/j.gastro.2014.07.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Human Microbiome Project Consortium Structure, function diversity of the healthy human microbiome. Nature. 2012;486(7402):207–214. doi: 10.1038/nature11234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Qin J, et al. A human gut microbial gene catalogue established by metagenomic sequencing. Nature. 2010;464(7285):59–65. doi: 10.1038/nature08821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Momose Y, Park SH, Miyamoto Y, Itoh K. Design of species-specific oligonucleotide probes for the detection of Bacteroides and Parabacteroides by fluorescence in situ hybridization and their application to the analysis of mouse caecal Bacteroides-Parabacteroides microbiota. J Appl Microbiol. 2011;111(1):176–184. doi: 10.1111/j.1365-2672.2011.05039.x. [DOI] [PubMed] [Google Scholar]
- 43.Sakamoto M, Benno Y. Reclassification of Bacteroides distasonis, Bacteroides goldsteinii and Bacteroides merdae as Parabacteroides distasonis gen. nov., comb. nov., Parabacteroides goldsteinii comb. nov. and Parabacteroides merdae comb. nov. Int J Syst Evol Microbiol. 2006;56(Pt 7):1599–1605. doi: 10.1099/ijs.0.64192-0. [DOI] [PubMed] [Google Scholar]
- 44.Smith MI, et al. Gut microbiomes of Malawian twin pairs discordant for kwashiorkor. Science. 2013;339(6119):548–554. doi: 10.1126/science.1229000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Faust K, et al. Microbial co-occurrence relationships in the human microbiome. PLoS Comput Biol. 2012;8(7):e1002606. doi: 10.1371/journal.pcbi.1002606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wu GD, et al. Linking long-term dietary patterns with gut microbial enterotypes. Science. 2011;334(6052):105–108. doi: 10.1126/science.1208344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Freter R, Stauffer E, Cleven D, Holdeman LV, Moore WE. Continuous-flow cultures as in vitro models of the ecology of large intestinal flora. Infect Immun. 1983;39(2):666–675. doi: 10.1128/iai.39.2.666-675.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lee SM, Donaldson GP, Mikulski Z, Boyajian S, Ley K, Mazmanian SK. Bacterial colonization factors control specificity and stability of the gut microbiota. Nature. 2013;501(7467):426–429. doi: 10.1038/nature12447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Eckburg PB, et al. Diversity of the human intestinal microbial flora. Science. 2005;308(5728):1635–1638. doi: 10.1126/science.1110591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Fischbach MA, Sonnenburg JL. Eating for two: how metabolism establishes interspecies interactions in the gut. Cell Host Microbe. 2011;10(4):336–347. doi: 10.1016/j.chom.2011.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Torrallardona D, Harris C, Coates M, Fuller M. Microbial amino acid synthesis and utilization in rats: incorporation of 15N from 15NH4Cl into lysine in the tissues of germ-free conventional rats. Br J Nutr. 1996;75(5):689–700. doi: 10.1079/bjn19960076. [DOI] [PubMed] [Google Scholar]
- 52.Belenguer A, Balcells J, Guada J, Decoux M, Milne E. Protein recycling in growing rabbits: contribution of microbial lysine to amino acid metabolism. Br J Nutr. 2005;94(5):763–770. doi: 10.1079/BJN20051508. [DOI] [PubMed] [Google Scholar]
- 53.Metges C, et al. Incorporation of urea and ammonia nitrogen into ileal and fecal microbial proteins and plasma free amino acids in normal men and ileostomates. Am J Clin Nutr. 1999;70(6):1046–1058. doi: 10.1093/ajcn/70.6.1046. [DOI] [PubMed] [Google Scholar]
- 54.Bustamante J, et al. Prognostic significance of hepatic encephalopathy in patients with cirrhosis. J Hepatol. 1999;30(5):890–895. doi: 10.1016/S0168-8278(99)80144-5. [DOI] [PubMed] [Google Scholar]
- 55.Wu GD, et al. Sampling and pyrosequencing methods for characterizing bacterial communities in the human gut using 16S sequence tags. BMC Microbiol. 2010;10:206. doi: 10.1186/1471-2180-10-206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Hoffmann C, et al. DNA bar coding and pyrosequencing to identify rare HIV drug resistance mutations. Nucleic acids research. 2007;35(13):e91. doi: 10.1093/nar/gkm435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Hamady M, Walker JJ, Harris JK, Gold NJ, Knight R. Error-correcting barcoded primers for pyrosequencing hundreds of samples in multiplex. Nature methods. 2008;5(3):235–237. doi: 10.1038/nmeth.1184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Caporaso JG, et al. QIIME allows analysis of high-throughput community sequencing data. Nat Methods. 2010;7(5):335–336. doi: 10.1038/nmeth.f.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Tuchman M, et al. N-carbamylglutamate markedly enhances ureagenesis in N-acetylglutamate deficiency and propionic acidemia as measured by isotopic incorporation and blood biomarkers. Pediatr Res. 2008;64(2):213–217. doi: 10.1203/PDR.0b013e318179454b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Lalonde R. The neurobiological basis of spontaneous alternation. Neurosci Biobehav Rev. 2002;26(1):91–104. doi: 10.1016/S0149-7634(01)00041-0. [DOI] [PubMed] [Google Scholar]
- 61.Lozupone C, Knight R. UniFrac: a new phylogenetic method for comparing microbial communities. Appl Environ Microbiol. 2005;71(12):8228–8235. doi: 10.1128/AEM.71.12.8228-8235.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.