Abstract
Pancreatic β cells secrete insulin in response to postprandial increases in glucose levels to prevent hyperglycemia and inhibit insulin secretion under fasting conditions to protect against hypoglycemia. β cells lack this functional capability at birth and acquire glucose-stimulated insulin secretion (GSIS) during neonatal life. Here, we have shown that during postnatal life, the de novo DNA methyltransferase DNMT3A initiates a metabolic program by repressing key genes, thereby enabling the coupling of insulin secretion to glucose levels. In a murine model, β cell–specific deletion of Dnmt3a prevented the metabolic switch, resulting in loss of GSIS. DNMT3A bound to the promoters of the genes encoding hexokinase 1 (HK1) and lactate dehydrogenase A (LDHA) — both of which regulate the metabolic switch — and knockdown of these two key DNMT3A targets restored the GSIS response in islets from animals with β cell–specific Dnmt3a deletion. Furthermore, DNA methylation–mediated repression of glucose-secretion decoupling genes to modulate GSIS was conserved in human β cells. Together, our results reveal a role for DNA methylation to direct the acquisition of pancreatic β cell function.
Introduction
Adult pancreatic β cells are uniquely designed to couple the rate of intracellular glucose metabolism with insulin secretion in proportion to postprandial rise in blood glucose (1–8). Neonatal β cells, by contrast, secrete insulin irrespective of changes in extracellular glucose levels (9, 10). The molecular mechanisms that drive the transition of β cells to functional maturity during the postnatal period are not known. Temporal expression of specific transcription factors has been associated with this transition. In mice, transcription factor MAFB is expressed in β cells only during the neonatal period, while MAFA, a related transcription factor, is upregulated in mature β cells (11, 12). Stage-specific activity of several transcription factors, such as MAFA and NeuroD, has been implicated in directing the establishment of glucose-stimulated insulin secretion during β cell maturation (11–14). Recent work also suggests the involvement of cellular signals, such as thyroid hormone and TGF-β, in the regulation of β cell functional maturation (15, 16). Molecular profiling studies show that neonatal and adult β cells are marked by distinct molecular signatures, but how these signatures correlate with the acquisition of glucose-stimulated insulin secretion (GSIS) is unclear.
Adult β cells are also designed to repress insulin secretion under fasting conditions to protect against hypoglycemia. This coupling of metabolic flux with extracellular availability of glucose in adult β cells is facilitated by the absence of low-Km hexokinases, so the first step of glycolysis is mediated by a high-Km variant, glucokinase. This, together with low expression of lactate dehydrogenase, and the lactate pyruvate transporter Slc16a1 (also known as Mct1) in β cells results in the virtual absence of anaerobic glycolysis to ensure that the extent of pyruvate production and its subsequent entry into the tricarboxylic acid (TCA) cycle is directly proportional to the postprandial rise in blood glucose. These genes were first identified as a group of housekeeping genes specifically disallowed in β cells, and it was proposed that their aberrant expression would interfere with glucose sensing (17). Work using RNA expression analysis of cultured islets, compared with RNA expression data from other tissues, validated the selective repression of these genes in β cells (18). Subsequent studies comparing the mRNA profiles from cultured islets with many more tissue types using rigorous microarray screens further highlighted the β cell–specific inhibition of these genes (19). The repression of these genes is critical in the context of maintaining mature behavior of β cells, and aberration in the process could potentially result in loss of glucose homeostasis (20, 21). This is illustrated by ectopic expression of lactate dehydrogenase and the lactate pyruvate transporter Slc16a1, which results in impaired insulin secretion (17, 20). Mutations with β cell misexpression of the disallowed Slc16a1 in β cells are associated with a disease called exercise-induced hyperinsulinism (EIHI), characterized by inappropriate insulin secretion and associated with hypoglycemia during anerobic exercise (22). Together, these studies suggest that critical mechanisms must be in place to prevent the expression of these genes in β cells toward appropriate function.
Examination of epigenetic marks histone H3K27 tri-methylation (H3K27me3), and H3K9 acetylation (H3K9Ac) in islets compared with other tissues suggested that the disallowed genes are regulated epigenetically by a repressive chromatin state (19). Comprehensive genome-wide analysis of activating and repressive histone marks during β cell development further underscored the importance of epigenetic regulation in islet maturation, showing that the establishment of a differentiated expression profile also includes specific repression of genes in β cells (23, 24). It has been shown that polycomb-mediated gene repression is key for the establishment of repression of these genes during pancreas organogenesis but is not responsible for maintenance of their repression in the differentiated β cells (19, 23, 24). This suggests the existence of polycomb-independent mechanisms in the differentiated β cell to mediate repression of these genes.
In this study, we describe an essential role for DNA methylation in the establishment of pancreatic β cell maturation. We show that the de novo DNA methyltransferase DNMT3A directs a metabolic program by repressing key genes to enable the coupling of insulin secretion to glucose levels during β cell maturation. Loss of Dnmt3a in pancreatic β cells prevents this developmental metabolic reprogramming, resulting in loss of GSIS. Furthermore, we show that the DNA methylation-mediated pathway regulating GSIS is conserved in human β cells. Our results highlight a epigenetic mechanism that governs β cell maturation during development and could provide new insights into impaired β cell function in diabetes.
Results
To determine whether the levels of metabolic enzymes involved in regulation of GSIS were altered during the postnatal maturation period of β cells, we used mice in which GFP is driven by the mouse insulin promoter (MIP) to sort β cells by FACS. Comparison of the gene expression patterns of metabolic enzymes in sorted β cells from postnatal day 4 (P4) and P25 animals revealed that many genes that regulate glycolysis, the pentose phosphatase pathway, and lipid biosynthesis were dramatically downregulated in adult β cells (Supplemental Figure 1A; supplemental material available online with this article; doi:10.1172/JCI79956DS1). The changes in the levels of many of these metabolic genes could be attributed to shifts in energy metabolism, as β cells during the early postnatal period transition from high rates of replication to relative quiescence (25). To distinguish metabolic genes related to shifts in energy metabolism from genes specifically related to β cell maturation, we compared β cells to proliferating and quiescent mouse embryonic fibroblasts (MEFs) (Supplemental Figure 1B; ref. 26). We identified a set of genes that regulate intracellular glucose metabolism — namely, hexokinases 1 and 2 (Hk1, Hk2), lactate dehydrogenase a (Ldha), and aldolase B (AldoB) — that were relatively unaltered in proliferating and quiescent MEFs but were selectively repressed during the transition of immature β cell to a functionally mature β cell. Ectopic expression of Hk1 and Ldha in β cells has been shown to affect coupling of glucose metabolism to secretion of insulin, resulting in high basal insulin secretion at low glucose levels (27–31). Prior studies also identified the absence of a few of these genes in adult β cells as disallowed genes (18). This group of genes share a common attribute of permitting activation of glycolytic flux at lower extracellular glucose thresholds, and we refer to these genes as glucose-secretion decoupling genes. Together, these results are consistent with the idea that repression of these glucose-secretion decoupling genes can modulate functional maturity of β cells and prevent inappropriate insulin release (14).
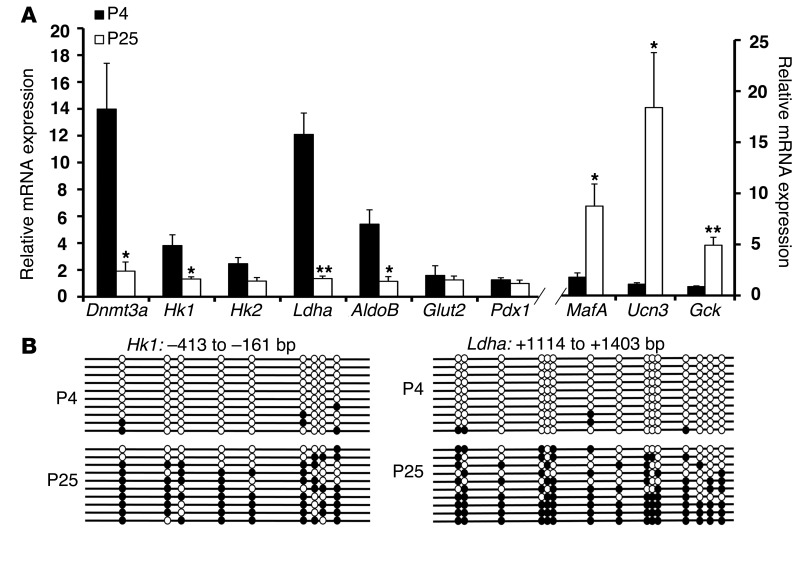
To confirm the downregulation of these glucose-secretion decoupling genes in β cells during postnatal life, we analyzed sorted β cells from multiple neonatal and adult mice and assessed expression by quantitative PCR (qPCR). The glucose-secretion decoupling genes were expressed in neonatal β cells but downregulated during the early postnatal period, which is characterized by the upregulation of markers associated with mature β cells such as MafA (32, 33) and urocortin3 (Ucn3) (9). The levels of β cell–specific transcription factors such as PDX1 were unaltered during this period (Figure 1A; ref. 34). IHC of GLUT2 (35) did not display any changes in the postnatal period, indicating that acquisition of GSIS did not involve the cell-surface glucose transporter (Supplemental Figure 1C). To address the mechanism by which the glucose-secretion decoupling genes were downregulated, we assessed whether DNA methylation was involved in the repression of glucose metabolism genes. We carried out bisulfite sequencing analysis of the CpG-rich regions of the glucose-secretion decoupling genes in sorted β cells from neonatal and adult mice. We found that the CpG-rich regions in the Hk1 (–413 to –161 bp) and Ldha (–417 to –179 bp; +1119 to +1419 bp) loci were predominantly unmethylated in β cells sorted from neonatal mice but mostly methylated in β cells sorted from adult mice (Figure 1B and Supplemental Figure 1D). Another key glycolytic gene that is involved in regulating the rate of glucose metabolism AldoB also showed increased methylation in adult β cells (Supplemental Figure 1D).
Figure 1. DNA methylation directs metabolic programming during β cell maturation.
(A) Relative mRNA expression of indicated genes in sorted β cells from P4 and P25 MIP-GFP mice. Cyclophilin A was used as a housekeeping gene. n = 3 independent experiments. Error bars represent SEM. *P < 0.05, **P < 0.01, Student’s t test. (B) Bisulfite sequencing analysis for the Hk1 and Ldha loci at indicated regions comparing sorted β cells from P4 and P25 MIP-GFP mice (representative clones from n = 3 mice). Each horizontal line with dots is an independent clone, and 10 clones are shown here. These regions are almost fully DNA methylated (filled circles) in β cells from P25 mice, but largely hypomethylated (open circles) in β cells from P4 mice.
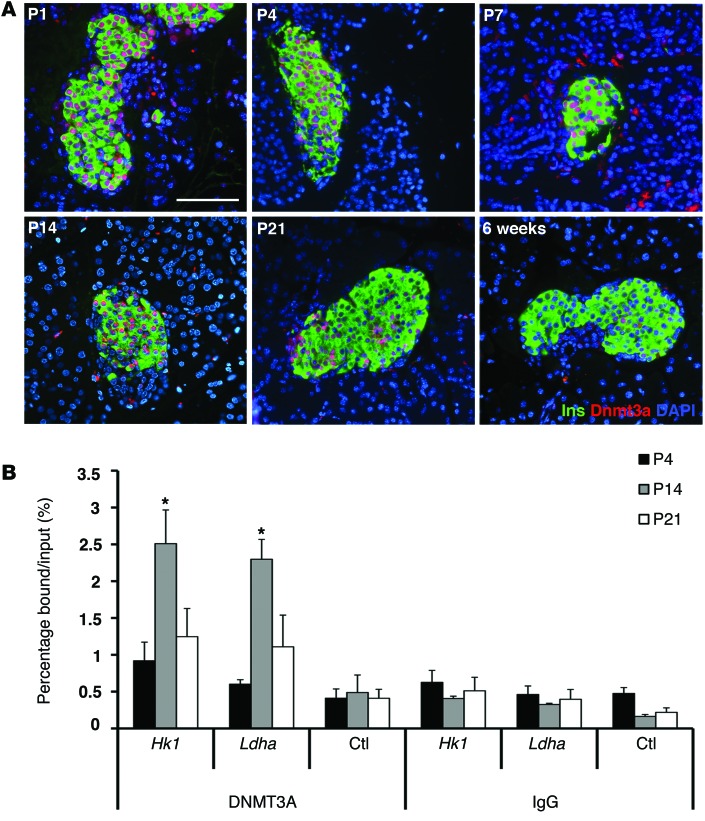
To investigate which enzymes mediate de novo methylation of the regulatory regions of glucose-secretion decoupling genes in β cells, we examined the expression of de novo DNA methytranferases DNMT3A and DNMT3B, in the postnatal period, using IHC and qPCR analyses. DNMT3b was not detectable in the endocrine cells in these experiments (data not shown). Analysis of RNA expression showed a decline in the levels of de novo DNA methyltransferase DNMT3A during the transition from early neonatal life to after-weaning period (Figure 1A). IHC analysis on pancreatic sections from early postnatal period revealed strong nuclear staining for DNMT3A in the islets (Figure 2A). Costaining with insulin showed that DNMT3A was expressed exclusively in β cells, and the number of β cells expressing DNMT3A declined with increased postnatal age. In 6-week-old pancreas, very few β cells showed weak staining for DNMT3A (Figure 2A). Expression of another de novo DNA methyltransferase, DNMT3B was not detected in the β cells during the postnatal period (data not shown). To address whether DNMT3A directly regulated glucose-secretion decoupling genes, we examined the binding of DNMT3A to the CpG-rich regulatory regions Hk1 and Ldha loci by ChIP assays. ChIP assay on sorted β cells from P4, P14, and P25 animals revealed that Dnmt3a was enriched at the CpG-rich differentially methylated regions Hk1 and Ldha loci in β cells from P14 mice. The binding of DNMT3A to Hk1 and Ldha loci decreased in β cells sorted from P25 mice (Figure 2B). Together, the expression pattern and binding of DNMT3A to Hk1 and Ldha loci is consistent with DNMT3A mediating de novo DNA methylation of glucose metabolic genes in β cells during the neonatal period.
Figure 2. De novo DNA methyltransferase DNMT3A targets specific metabolic gene loci during β cell maturation.
(A) Expression profile of DNMT3A in representative pancreatic sections from WT mice at indicated ages (P1 to 6 weeks) using immunostaining for DNMT3A (red) and insulin (Ins; green). DAPI (blue) counter-stains the nuclei. (Representative from 3 independent experiments). Scale bar: 100 μm. (B) ChIP analysis showing the binding of DNMT3A to the Ldha (+1114 to +1220 bp) and Hk1 (–324 to –215 bp) loci, along with a negative-control primer pair (ctl) in the Arx locus (+1348 to +1470 bp) in sorted β cells from MIP-GFP mice at indicated ages. ChIP with IgG is shown as a negative control. n = 3 independent experiments. The error bars represent SEM. *P < 0.05, Student’s t test.
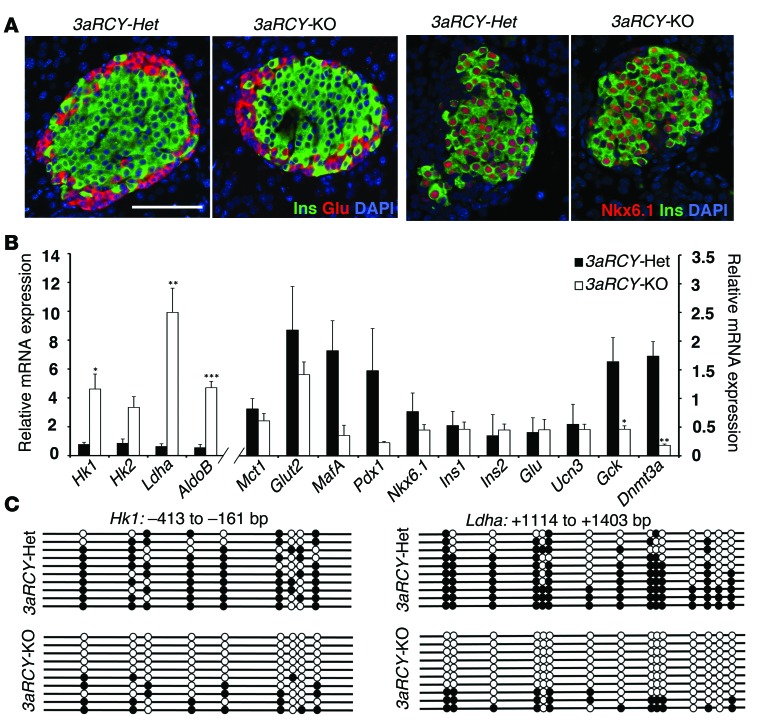
To address the requirement for DNMT3A in de novo DNA methylation of glucose-secretion decoupling genes during β cell functional maturation, we crossed mice transgenic for Cre recombinase under the control of rat insulin promoter (RIP-Cre) with Dnmt3afl/fl mice to selectively inactivate Dnmt3a in β cells. In addition, the Dnmt3afl/fl mice were maintained in the R26R-eYFP reporter background to allow isolation of heritably labeled cells that transcribed the insulin gene. Immunohistology confirmed Cre recombinase activity specifically in β cells in pancreatic sections from Dnmt3afl/fl R26R-eYFP RIP-Cre mice (Supplemental Figure 2A). The recombined β cells were marked by the absence of detectable DNMT3A protein, while abundant DNMT3A protein was observed within the islets of pancreatic tissue from control littermates (Supplemental Figure 2B ). Pancreatic tissue isolated from 6-week-old Dnmt3afl/fl R26R-eYFP RIP-Cre (herein referred to as 3aRCY-KO) and Dnmt3afl/+ R26R-eYFP RIP-Cre (herein referred to as 3aRCY-Het) littermate mice were immunostained for insulin and glucagon, endocrine hormones that mark β and α cells, respectively. No overt differences were apparent, and islets from both 3aRCY-KO and 3aRCY-Het littermate mice displayed a characteristic distribution of endocrine cells, with β cells forming the core and α cells at the periphery forming the mantle (Figure 3A). Staining for transcription factors NKX6.1 (Figure 3A) and PDX1 (not shown), as well as GLUT2 on the cell surface (Supplemental Figure 2C), showed no difference between 3aRCY-KO and 3aRCY-Het littermate pancreatic sections. No differences were found in the proliferation of β cells between 3aRCY-KO and 3aRCY-Het littermates, as examined by immunostaining for Ki-67 (Supplemental Figure 2D). Transcriptional profile of sorted β cells from 6-week-old 3aRCY-KO and 3aRCY-Het littermate mice revealed that the glucose-secretion decoupling genes were de-repressed in the absence of Dnmt3a (Supplemental Figure 2E). We confirmed using qPCR that these glucose-secretion decoupling genes were upregulated in sorted β cells from 3aRCY-KO mice, while transcription factors and hormones characteristic of β cells, by contrast, were not significantly altered (Figure 3B), compared with β cells from 3aRCY-Het. No significant changes were observed in the RNA levels of β cell transcription factors Pdx1, MafA, and Nkx6.1.
Figure 3. De novo DNA methyltransferase DNMT3A is required for metabolic programming during β cell maturation.
(A) Representative pancreatic sections from 2-week-old 3aRCY-KO and littermate-control 3aRCY-Het animals, immunostained for insulin (Ins; green) and glucagon (Glu; red), with DAPI (blue) (left 2 panels), or NKX6.1 (red), insulin (Ins; green) and DAPI (blue) (right 2 panels). Scale bar: 100 μm. (B) Relative mRNA expression of indicated genes in sorted β cells from 6-week-old 3aRCY-KO and control 3aRCY-Het littermates. Cyclophilin A was used as a housekeeping gene. (C) Bisulfite sequencing analysis for the Hk1 and Ldha loci at indicated regions comparing sorted β cells from 6-week-old 3aRCY-KO and control 3aRCY-Het littermates (representative clones from n = 3 mice). Each horizontal line with dots is an independent clone, and 10 clones are shown here. These regions are almost fully DNA-methylated (filled circles) in β cells from 3aRCY-Het mice, but largely hypomethylated (open circles) in β cells from 3aRCY-KO mice. For all experiments, n = 3 independent experiments. The error bars represent SEM. *P < 0.05, **P < 0.01, ***P < 0.005, Student’s t test.
To investigate whether β cells deficient in Dnmt3a displayed changes in DNA methylation at CpG-rich regions within the loci of the glucose-secretion decoupling genes, we used the R26R-eYFP reporter to isolate heritably labeled cells that transcribed the insulin gene in 3aRCY-KO and 3aRCY-Het littermates. Bisulfite sequencing analysis on YFP-positive cells isolated from 3aRCY-Het 6-week-old mice showed hypermethylation in the differentially methylated regions of Hk1, Ldha, and AldoB loci similar to the pattern observed in adult WT β cells. In contrast, YFP-positive cells isolated from conditional Dnmt3a mutant littermates revealed that differentially methylated regions of these loci were predominantly hypomethylated and characteristic of WT neonatal β cells (Figure 3C, Supplemental Figure 2F, and Figure 1B). To rule out any potential artifacts arising from the expression of Cre recombinase alone, the changes in RNA expression and DNA methylation profiles in the β cells from 3aRCY-KO were further validated and confirmed by comparing them with β cells from RCY (R26R-eYFP RIP-Cre) animals (Supplemental Figure 2, G and H).
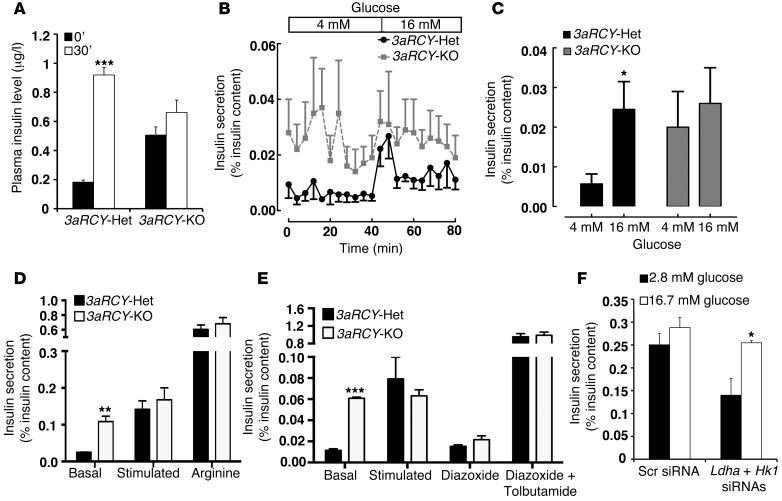
To assess β cell function, we measured plasma insulin levels at fasting and after an intra-peritoneal glucose challenge. 3aRCY-Het mice showed a 5-fold increase in plasma insulin levels 30 minutes after glucose stimulation. In contrast, 3aRCY-KO mice displayed high plasma insulin levels at fasting and no significant increased after glucose stimulation (Figure 4A). We next measured dynamic insulin secretion in isolated islets from 3aRCY-KO and 3aRCY-Het littermate mice. 3aRCY-Het islets treated with single-step increase in glucose concentration from 4–16 mM produced a robust insulin secretory response to glucose. By contrast, 3aRCY-KO islets displayed increased basal insulin secretion at low glucose concentration and did not respond to stepwise increases in glucose concentration (Figure 4, B and C). Islets from both 3aRCY-KO and 3aRCY-Het mice did not display any difference in insulin secretion in response to depolarizing secretogogue arginine, suggesting that the defect was likely to precede membrane depolarization (Figure 4D). Furthermore, using drugs that act on KATP channels, we showed that insulin secretion in islets isolated from both 3aRCY-KO and 3aRCY-Het mice was abolished by diazoxide and reversibly restored by tolbutamide (29), suggesting that the defects due to loss of Dnmt3a are upstream of the KATP channel (Figure 4E). This, together with our observations on changes in the metabolic program, support the idea that these defects in β cell function are likely due to changes in glucose metabolism.
Figure 4. β Cells lacking Dnmt3a are functionally immature.
(A) Plasma insulin levels in 6-week-old 3aRCY-KO mice and littermate-control 3aRCY-Het mice (fasted 6 hours) at 0 and 30 minutes after i.p. injection of 2 g/kg glucose (n = 3 for each genotype). (B) Insulin secretion (percentage of insulin content) in a dynamic islet perifusion assay comparing islets from 6-week-old 3aRCY-KO or control 3aRCY-Het littermates at 4 mM and 16 mM glucose. (C) Average insulin secretion (percentage of content) in the dynamic islet perifusion GSIS at 4 mM and 16 mM glucose, comparing 3aRCY-KO islets with control 3aRCY-Het islets from n = 3 mice aged 6 weeks. (D) Static incubation GSIS assay, comparing insulin secretion (percentage of insulin content) in islets from 3aRCY-KO mice with control 3aRCY-Het islets (n = 3 mice), at 2.8 mM glucose, 16.7 mM glucose, and 20 mM arginine. (E) Static incubation GSIS assay, comparing insulin secretion (percentage of insulin content) in islets from 6-week-old 3aRCY-KO mice with control 3aRCY-Het islets, at 2.8 mM glucose, 16.7 mM glucose, 100 μM diazoxide, and a 100 mM each of diazoxide and tolbutamide (n = 3 mice). (F) Static incubation GSIS assay, comparing insulin secretion (percentage of insulin content) in islets from 6-week-old 3aRCY-KO mice, treated either with a combination of siRNAs targeting Ldha and Hk1, or scrambled (Scr) siRNA, at 2.8 mM glucose and 16.7 mM glucose (n = 3 mice, islets from each mice divided into the 2 experimental groups). Error bars indicate ± SEM; *P < 0.05, **P < 0.01, ***P < 0.005, Student’s t test.
The increased levels of Ldha in the 3aRCY-KO islets translated into increased lactate release from these islets, in comparison to islets from 3aRCY-Het control mice (Supplemental Figure 3A). As Hk1 and Ldha, 2 key glucose metabolism genes, were upregulated due to the loss of DNMT3A, we asked whether inhibition of Hk1 and Ldha in islets that lacked Dnmt3a could restore GSIS. We used specific siRNAs to knock down both Hk1 and Ldha in islets from 3aRCY-KO mice before assessing GSIS. Treatment of islets from 3aRCY-KO mice with a combination of Hk1 and Ldha siRNA specifically and efficiently reduced the levels of these transcripts, without affecting the levels of other glycolytic genes tested (Supplemental Figure 3B). Knockdown using scrambled siRNA showed no change in insulin secretion in response to stepwise increases in glucose concentration. However, knockdown of Hk1 and Ldha resulted in reduction in basal insulin secretion at low glucose concentration and increased insulin secretory response at high glucose concentration (Figure 4F).
To address whether DNMT3A was required during embryogenesis, we crossed mice transgenic for a tamoxifen-responsive Cre recombinase–estrogen receptor fusion under the control of a Pdx1 promoter (Pdx1-CreERT) with Dnmt3afl/fl mice to inactivate Dnmt3a in β cells during neonatal life and drive a R26R-eYFP reporter (Pdx1-CreERT Dnmt3afl/fl R26R-eYFP, herein referred to as 3aPCERY-KO). Real-time PCR for Dnmt3a on islets isolated from 3aPCERY-KO mice showed efficient deletion of Dnmt3a levels, compared with islets from Dnmt3afl/+ R26R-eYFP Pdx1-CreERT (herein referred to as 3aPCERY-Het) (Supplemental Figure 3C). A comparison of YFP-positive cells isolated from 3aPCERY-KO animals with those from 3aPCERY-Het showed increased expression of Hk1, Ldha, and AldoB in a manner similar to the 3aRCY model, while no significant changes were observed in the transcript levels of Nkx6.1 and Pdx1 (Supplemental Figure 3, D and E). These observations confirmed a stage-specific requirement for DNMT3A in directing methylation and repression of these genes during β cell maturation. In vivo insulin secretion experiments revealed no difference from the 3aRCY model (Supplemental Figure 3F), suggesting that the acute requirement of Dnmt3a during the maturation period and the changes in β cell function upon loss of Dnmt3a are not a consequence of leakiness from the RIP-Cre driver (36).
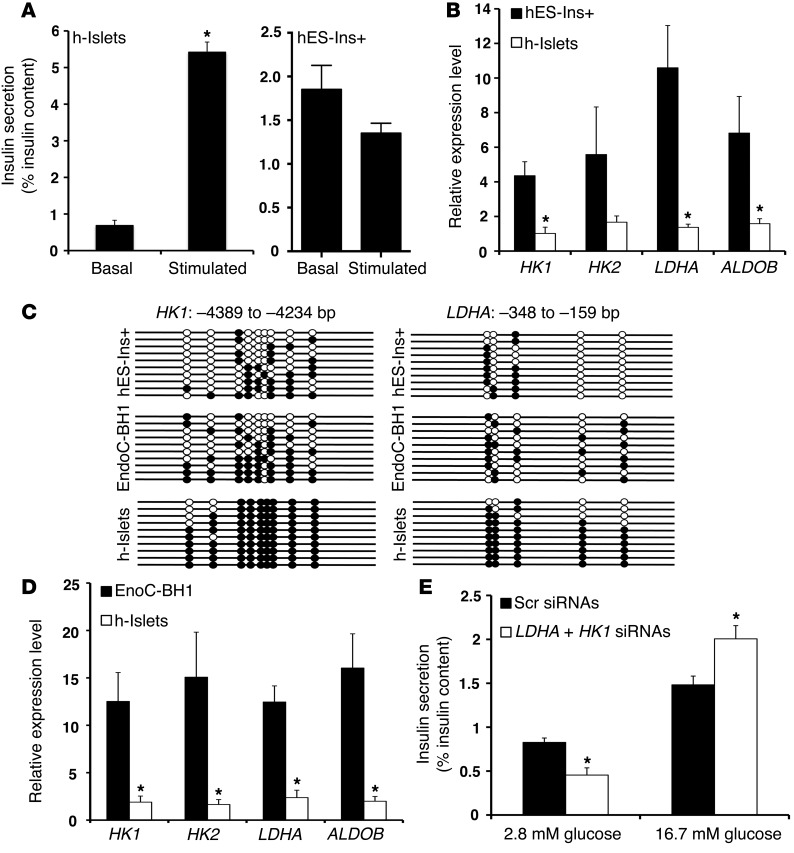
We next investigated the regulatory regions of glucose-secretion decoupling genes in human islets and insulin-expressing cells derived from pluripotent cells (37). As most pluripotent cell differentiation protocols generate insulin-positive cells that resemble immature β cells (9, 37), we used these cells as a proxy for fetal human β cells. In contrast to human islets, which showed GSIS response to stepwise increase in glucose concentration, the response of insulin-expressing cells from pluripotent cells to increases in glucose concentration was reminiscent of neonatal and 3aRCY-KO islets (Figure 5A). Analysis of the DNMT3A transcript levels in pluripotent-derived insulin-positive cells showed higher DNMT3A levels in these cells, compared with mature human islets, reminiscent of the DNMT3A expression profile in neonatal β cells (Supplemental Figure 4A). Transcriptional profile of pluripotent-derived insulin-expressing cells showed increased levels of HK1, HK2, LDHA, and ALDOB, compared with levels seen in human islets (Figure 5B and Supplemental Figure 4B). Bisulfite sequencing analysis revealed that CpG-rich regions with the HK1, HK2, LDHA, and ALDOB loci were hypomethylated in pluripotent-derived insulin-expressing cells. By contrast, these CpG-rich regions were hypermethylated in human islets (Figure 5C and Supplemental Figure 4C).
Figure 5. The metabolic programming directed by DNA methylation to regulate GSIS is conserved in human β cells.
(A) Static incubation GSIS assay, showing insulin secretion (percentage of insulin content) in differentiated insulin-positive (Ins+) human ES cells (hES-Ins+ cells; left panel), and human islets (h-islets; right panel) at 2.8 mM glucose and 16.7 mM glucose. (B) Relative mRNA expression of indicated genes in sorted GFP+ hES-Ins+ cells, compared with human islets from cadaveric donors (average donor age 43 years; n = 3). CYCLOPHILIN A was used as a housekeeping gene. The error bars represent SEM. *P < 0.05. (C) Bisulfite sequencing analysis for the HK1 and LDHA loci at indicated regions comparing sorted GFP+ hES-Ins+ cells, and EndoC-BH1 human β cell line, compared with human islets from cadaveric donors (representative clones from n = 3 samples per group). Each horizontal line with dots is an independent clone These regions are almost fully DNA methylated (filled circles) in human islets but largely hypomethylated (open circles) in hES-Ins+ cells and the EndoC-BH1 human β cell line. (D) Relative mRNA expression of indicated genes in EndoC-BH1 human β cell line, compared with human islets from cadaveric donors (average donor age 43 years). CYCLOPHILIN A was used as a housekeeping gene. n = 3 independent experiments. The error bars represent SEM. *P < 0.05. (E) Static incubation GSIS assay, showing insulin secretion (percentage of insulin content) in comparing EndoCBH1 cells treated with either a combination of siRNAs targeting HK1 and LDHA, or a scrambled (Scr) siRNA, at 2.8 mM glucose and 16.7 mM glucose. (n = 3). Error bars indicate ± SEM, *P < 0.05, Student’s t test.
The transcriptional profile of EndoC-βH1 human pancreatic β cell line derived from fetal pancreas (38) also showed high levels of DNMT3A, HK1, HK2, and LDHA, compared with human islets levels (Figure 5D and Supplemental Figure 5A). Bisulfite sequencing analysis in EndoC-βH1 cells showed that the CpG-rich regions with the HK1 and LDHA loci showed reduced methylation, compared with human islets (Figure 5C). We next used specific siRNAs to knock down the expression of HK1 and LDHA in the EndoC-βH1 cells (Supplemental Figure 5B). GSIS measurements showed that the loss of HK1 and LDHA resulted in improved insulin secretion in response to stepwise increases in glucose concentration, compared with cells transfected with a scrambled siRNA (Figure 5E). These results indicate that the repression of glucose-secretion decoupling genes to modulate functional maturity is conserved in human β cells. In addition, the expression and DNA methylation patterns of glucose-secretion decoupling genes could serve as molecular markers for deriving functional human β cells for diabetes therapy. Overall, our results indicate that DNA methylation-mediated repression of glucose-secretion coupling genes occurs during postnatal maturation and is critical for regulating GSIS in rodent and human β cells.
Discussion
Neonatal and adult β cells differ in the ability to properly regulate insulin in response to glucose (39–41). Our study here shows that the functional maturation of β cells requires remodeling of the metabolic infrastructure to support the acquisition of GSIS within a narrow range of glucose concentration. This transition from neonatal to adult β cells is accompanied by a shift in energy metabolism from a predominantly glycolytic to oxidative phosphorylation. While many other cell types undergo a similar metabolic shift during cellular differentiation and phenotypic maturation (42–45), β cells are unique in the absence of a specific set of metabolic genes that we refer to as glucose-secretion decoupling genes, such as low Km hexokinases, Ldha, and the lactate pyruvate transporter Slc16a1 (17, 19, 21). β cell–specific repression of these key metabolic genes is essential toward maintenance of the mature, functional phenotype. Ectopic expression of Hk1 and Ldha in β cells has been shown to disrupt the coupling of glucose metabolism to secretion of insulin and impaired GSIS (27, 28, 30, 31). The physiological importance of such mechanisms is further highlighted by disease pathologies resulting from the aberrant expression of these genes in β cells. Gain-of-function mutations in the promoter region of Slc16a1 result in EIHI, which is characterized by inappropriate insulin secretion and consequent hypoglycemia during anerobic exercise (22), with similar observations made in a mouse model with ectopic expression of Slc16a1 (46). Similarly, aberrant expression of low Km hexokinase variant HK1 in β cells in humans has been associated with a form of congenital hyperinsulinism (CHI) (29). Thus, understanding the mechanisms that result in the repression of glucose-secretion decoupling genes to regulate β cell function is crucial.
There is intense interest in the generation of functional β cells from pluripotent cells for use in replacement strategies in diabetic patients. Recent work has highlighted the development of protocols to generate insulin-expressing cells derived from human pluripotent cells that resemble human primary β cells in several ways, and while these cells display glucose sensing, they are still not fully equivalent to mature primary human β cells (47, 48). The expression levels and DNA methylation patterns of glucose-secretion decoupling genes could be useful as a molecular benchmark in facilitating the generation of pluripotent-derived cells that closely resemble human primary β cells for replacement strategies in diabetic patients.
Our previous work has shown that DNA methylation is an important regulatory mechanism in pancreatic organogenesis during embryonic development, as well as in the specification and maintenance of β cell fate (49–51). In this manuscript, we show that de novo DNA methylation directs metabolic reprogramming of pancreatic β cells toward their functional maturation, and pancreatic β cells lacking DNA methyltransferase Dnmt3a metabolically and physiologically resemble functionally immature neonatal β cells. Compared with WT adult β cells, the Dnmt3a KO β cells display higher lactate production and basal insulin secretion, similar to neonatal immature β cells. This suggests that de novo DNA methylation plays a key role in pancreatic β cell maturation. Several previous studies have indicated the involvement of H3K27me3 in the repression of metabolic genes, such as Ldha and Slc16a1 in mature β cells, in a polycomb protein–directed mechanism (19, 23, 24). There is evidence in literature indicating that H3K27me3 may be required for proper placement of DNA methylation marks (52). It is likely that epigenetic repression of these genes is initiated via histone methylation in early development, during β cell formation, and this histone methylation coordinates with DNA methylation during β cell maturation to establish the mature β cell metabolic program. Interestingly, there is no statistically significant difference in the levels of Slc16a1, MafA, and Pdx1 in the Dnmt3a KO β cells compared with control adult β cells, further suggesting the involvement of other regulatory mechanisms. The levels of glucokinase are unaffected in Dnmt3a KO β cells, also suggesting that transcriptional activation, in addition to DNA methylation–mediated repression of glucose-secretion decoupling genes, is needed for the establishment of a functional β cell phenotype. Together, these observations further underscore the importance of multiple regulatory layers to maintain a mature β cell phenotype.
Type 2 diabetes is characterized by impaired insulin secretion and loss of β cell identity (reviewed in ref. 53). Multiple studies have highlighted changes in DNA methylation at loci associated with β cell identity and insulin secretion in islets from subjects with type 2 diabetes (54–56). Altered metabolic program due to defects in DNA methylation have also been implicated in impaired insulin secretion in type 2 diabetes (57). This work, coupled with our previous work on the role of DNA methylation in the maintenance of β cell fate (49), could provide crucial insights into the loss of β cell fate and function in type 2 diabetes (34, 58).
Methods
Animals, genotyping, and cell lines.
We used Cre/loxP system to conditionally delete Dnmt3a in pancreatic β cells. The following mice have been described previously: Dnmt3afl/fl mice with loxP sites at exon 19 of the Dnmt3a gene, RIP-Cre mice expressing Cre recombinase from rat insulin promoter, and Pdx-CreERT mice expressing Cre recombinase from Pdx1 promoter upon tamoxifen treatment (59, 60). The RIP-Cre and Pdx1-CreERT mice were bred into the Rosa26R-YFP or Rosa26R-Tomato-EGFP (denoted Tom, marked by membrane localized EGFP upon recombination) background to indelibly mark all cells that were derived from insulin-expressing cells (The Jackson Laboratory) (61, 62). The animals were maintained by mating WT males and females on a C57BL/6J background. Mice were fed ad libitum and kept under a 12-hour light/12-hour dark cycle. DNA extracted from the tails was used for PCR-based genotyping, as previously described, for individual lines using standard methods. Mouse MEFs were generated as described previously (63) and maintained on dishes coated with 0.2% porcine gelatin, in DMEM-based medium containing 10% FBS at 37°C in 5% CO2 environment. EndoC-BH1 cell line was provided by Philippe Ravassard (ICM, Biotechnology & biotherapy group, Hôpital Pitié Salpêtrière, Paris, France) and cultured as described (38).
Tamoxifen-dependent induction of recombination in Pdx-CreERT line.
For induction of recombination during neonatal life, nursing mothers were injected i.p. with 5 mg tamoxifen (Sigma-Aldrich) a total of 3 times, once every 48 hours, starting 1 day after giving birth. Pups were weaned normally at 21 days, and the pancreatic tissue from these weanlings was harvested at 6 weeks of age.
Human embryonic stem cell culture and differentiation.
Mel1ins/GFP human embryonic stem cells were maintained on nonproliferative MEFs in standard DMEM/F12 medium supplemented with 10 ng/ml FGF2. Cells were then seeded at 150,000–175,000 cell/cm2 on matrigel and started differentiation 48 hours after seeding. Day 1–5 differentiation was carried out with RPMI 1640 (Corning), 0.2% FBS (HyClone), 1× GlutaMAX-1 (Invitrogen) plus Activin A (100 ng/ml, R&D Systems), Wnt3a (50 ng/ml, R&D Systems), and insulin-transferrin-selenium (ITS, 1:5,000, Invitrogen) for day 1; Activin A (100 ng/ml) and ITS (1:5000) for day 2; KGF (25 ng/ml, R&D Systems), TGF-βiIV (2.5 μM, EMD Millipore), and ITS (1:1,000) for day3; KGF (25 ng/ml) and ITS (1:1,000) for days 4–5. Day 6–20 differentiation was carried out with DMEM/HIGH Glucose (HyClone) supplemented with 0.5× B-27 supplement (Invitrogen) plus KAAD-cyclopamine (0.25 μM, Toronto Research Chemicals Inc.), Noggin (50 ng/ml, R&D Systems), and TTNPB (3 nM, Sigma-Aldrich) for days 6–8; KGF (50 ng/ml), EGF (50 ng/ml, R&D Systems), and Noggin (50 ng/ml) for days 9–11; and ALK5 inhibitor II (1 μM, Axxora), dorsomorphin (1 μM, EMD Millipore), TPB ([2S,5S]-[E,E]-8-[5-(4-[trifluoromethyl]phenyl)-2,4-pentadienoylamino]benzolactam) (50 nM, EMD Millipore), and Noggin (50 ng/ml) for days 12–20.
Mouse islet isolation, culture and transfection, cell sorting, and human islet culture.
Islets were isolated using the Liberase/DNase enzyme mix (Roche Diagnostics) as described (49). In brief, the pancreas was inflated with the enzyme mix through the common bile duct and digested at 37°C. Islets were enriched by gradient and were hand picked. One hundred to 200 islets were cultured in easy-grip tissue culture dishes (35 mm × 10 mm, BD Falcon) in DMEM (Corning) supplemented with 10% FBS. The knockdown of Hk1 and Ldha in isolated mouse islets was performed by transfecting isolated mouse islets with a specific targeting siRNA. The siRNA targeting Hk1 and Ldha (siGenome SMARTpool) and a control scrambled siRNA (targeting sequence: 5′-CTGACGCGGAATACTTCGA-3′) were purchased from Dharmacon. Briefly, isolated islets were cultured for 24 hours prior to transfection, and then transfected using Lipofectamine-2000 (Invitrogen), according to the manufacturer’s instructions. The islets were harvested 48 hours after transfection and processed for insulin secretion studies, as described below. For purification of β cells, islets from neonatal (P4) or post-weaning (P25) MIP-GFP transgenic mice were digested into single cells and sorted for GFP by FACS to an average percentage purity of 85%–95%. Cells from WT islets were used as negative control for FACS gating. β cell–derived endocrine cells lineage traced with YFP driven by RIP-Cre were purified similarly by FACS sorting to a purity of 85%–95%. Sorted cells were then processed for RNA extraction, bisulfite sequencing, or ChIP studies. Human islet samples were obtained from healthy, adult, nondiabetic organ donors deceased due to acute traumatic or anoxic death and obtained from Integrated Islet Distribution Program (IIDP) as previously described (64). Equilibrated human islets were processed for qPCR and bisulfite sequencing analysis, similar to the mouse islets.
Immunofluorescence staining and morphometric analysis, β galactosidase staining, and antibodies.
Standard immunofluorscence protocol was used for immunodetection of various proteins in pancreatic sections (64). Briefly, pancreatic tissue was dissected in PBS, fixed in 4% formaldehyde for 4 hours to overnight, dehydrated in grades of ethanol, and stored in –20°C until processed for paraffin embedding. Primary antibodies were diluted in the blocking solution at the following dilutions: mouse anti-glucagon (diluted 1:1,000, 0.2 ml, Sigma-Aldrich, catalog G2654); rabbit anti-glucagon (diluted 1:500, ImmunoStar Inc, catalog 20076); guinea pig anti-insulin (diluted 1:400, Dako, catalog A0564); mouse anti-DNMT3a (diluted 1:1,000, Novus Biologicals, catalog IMG-268A); chicken anti-GFP (1:500, Aves Labs Inc., catalog 1020); mouse anti-NKX6.1 (diluted 1:50, Developmental Studies Hybridoma Bank, catalog F55A10-S); rabbit anti-GLUT2 (diluted 1:1000, Millipore, catalog 07-1402), and mouse anti–Ki-67 (1:40; BD Biosciences, catalog 550609). Donkey- and goat-derived secondary antibodies conjugated to FITC or Cy3 were diluted 1:500 (Jackson ImmunoResearch Laboratories Inc.). Slides were viewed using a Leica DM6000 microscope, and images were acquired using Openlab software (Improvision). For analysis of the β cell mass and proliferation index, 3–6 mice were analyzed in each group. Proliferation index was calculated using Ki-67 staining as a marker for proliferation and insulin staining for β cells, defined as a percentage of Ki-67–positive β cells.
RNA isolation, RT-PCR, and real-time PCR.
RNA was isolated from different cells and tissues (pancreatic islets, sorted β cells, human insulin-positive differentiated cells, human islets or MEFs) using RNeasyPlus Mini Kit (QIAGEN) and digested with RNase-free DNase (Fermentas) to eliminate DNA. RNA (0.5 mg) was used for preparation of single-stranded cDNA using Superscript III Reverse transcriptase (Invitrogen) by the oligo(dT) priming method. qPCR was performed using the 1× Fast SYBR Green Mix (Applied Biosystems) in an ABI7900HT system (Applied Biosystems). The expression levels of each transcript were normalized to the housekeeping gene cyclophilin. The primer sequences used are listed in Supplemental Tables 1 and 2. Each real-time PCR experiment shown is a representative from at least 3 independent experiments; for each experiment, islets were pooled from 3–4 mice per specified group. For a pilot, comprehensive analysis of mRNA expression to study the changes in different groups of metabolic genes, we also used a custom designed real-time PCR array (CAPM12177) of selected genes (QIAGEN), according to the manufacturer’s instructions, using the standard DDCt method with cyclophilin A (PpiA) as a housekeeping gene, and the data were presented as fold changes on a logarithmic scale. The gene list for this array is detailed in Supplemental Table 3.
Metabolic and physiological analyses.
Glucose tolerance test (GTT) and plasma insulin measurements were performed following overnight fasting of the mice, according to established protocols (64). To assess GSIS, we used islet perifusion and static incubation protocols. Islet perifusion experiments were performed (ACUSYST-S, Cellex Biosciences Inc.) as previously described (65). Batches of 20 islets were first exposed to 40 minutes of 4 mM glucose in Krebs Ringer bicarbonate buffer supplemented with 0.2% serum albumin, preheated to 37°C, and oxygenized with 95% O2 and 5% CO2, followed by 40 minutes of hyperglycemic perifusate (16 mM glucose). Effluent was collected in 4-minute intervals and assayed for insulin by ELISA (Mouse Insulin ELISA, Mercodia) for subsequent determination of basal and glucose-stimulated insulin secretion. For static-incubation GSIS assays, batches of 15 islets were first incubated in 2.8 mM glucose, followed by transfer to 16.7 mM glucose buffer in Krebs Ringer bicarbonate buffer, supplemented with 0.2% serum albumin at 37°C for 30 minutes each, and supernatant was collected after each incubation. Insulin was then extracted from islets by overnight incubation with acid ethanol. Insulin in supernatants and islet extracts was measured by ELISA (Mouse Insulin ELISA, Mercodia). To measure lactate production, isolated islets were incubated at 37°C, 5% CO2 in RPMI 1640 with 11.1 mM glucose overnight, followed by incubation in Krebs buffer with either 2.8 mM or 16.7 mM glucose for 1 hour (100 islets in 200 μl), and the supernatant was collected, snap frozen on dry ice, and analyzed with a fluorometric L-Lactate assay kit (Abcam, catalog ab65330) according to the manufacturer’s instructions. DNA was extracted from these islets by sonication, and DNA content was quantified by using Qubit fluorometric quantification (Invitrogen).
DNA methylation analyses and ChIP analyses.
Bisulfite conversion of DNA from sorted β cells was performed as described previously (66). DNA samples were incubated with sodium bisulfite for 4–5 hours. Bisulfite-treated DNA was then desalted and precipitated. To generate PCR products, 1/10 of the precipitated DNA was used for each PCR using primers described in supplemental tables 4 and 5. Primers were designed using the MethPrimer software (Urogene). PCR products were gel purified and used for TOPO-TA cloning (Invitrogen), followed by sequencing. Bisulfite sequencing data were quantified and represented as percentage of methylated residues in each clone analyzed. Averages were derived from independent triplicates of each, and 10 clones were analyzed per group. ChIP assays on the purified β cells were carried out according to the micro-ChIP protocol (67), using minor modifications. Sorted cells were treated with 1% paraformaldehyde at room temperature for 20 minutes to crosslink the DNA with bound proteins. Formaldehyde was quenched by adding glycine to a final concentration of 125 mM, followed by cell lysis in 100 μl lysis buffer consisting of 50 mM Tris HCl pH 8.0, 10 mM EDTA, and 1% SDS, supplemented with 1 × complete proteinase inhibitors (EMD Biosciences) and sonicated to yield DNA fragments with an average size of 500 bp using Bioruptor (Diagenode). A total of 2.5 μg of antibody was bound to 20 μl Protein-A/G Dynabeads (Invitrogen), depending on the antibody isotype, for 2 hours at 4°C with agitation. The following antibodies were used: Dnmt3a (Imgenex, catalog IMG-268A) and control mouse IgG (Diagenode, catalog kch-819-015). The primers used to amplify the different loci are listed in Supplemental Table 6.
Statistics.
All data were expressed as mean ± SEM. Mean and SEM values were calculated from at least triplicates of a representative experiment. The statistical significance of differences was measured by 2-tailed Student’s t test. A P value < 0.05 indicated statistical significance. *P < 0.05, **P < 0.01, ***P < 0.005.
Study approval.
All animal experiments were performed in accordance with NIH policies on the use of laboratory animals and approved by the Animal Research Committee of the Office for the Protection of Research Subjects at UCLA.
Supplementary Material
Acknowledgments
We are grateful to Peter Butler (UCLA) for discussions and input on these studies, and to Emily Snyder and James Maksymetz (UCLA) for technical support. This work was supported by an Advanced Postdoctoral Fellowship from the Juvenile Diabetes Research Foundation and a start-up grant from Larry L. Hillblom Foundation to S. Dhawan, as well as grants from National Institute of Diabetes and Digestive and Kidney Diseases (DK068763 and DK080996), Juvenile Diabetes Research Foundation, and the Helmsley Trust to M. Hebrok and A. Bhushan.
Footnotes
Conflict of interest: The authors have declared that no conflict of interest exists.
Reference information:J Clin Invest. 2015;125(7):2851–2860. doi:10.1172/JCI79956.
See the related Commentary beginning on page 2565.
References
- 1.Ashcroft SJ. Glucoreceptor mechanisms and the control of insulin release and biosynthesis. Diabetologia. 1980;18(1):5–15. doi: 10.1007/BF01228295. [DOI] [PubMed] [Google Scholar]
- 2.German MS. Glucose sensing in pancreatic islet β cells: the key role of glucokinase and the glycolytic intermediates. Proc Natl Acad Sci U S A. 1993;90(5):1781–1785. doi: 10.1073/pnas.90.5.1781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hedeskov CJ. Mechanism of glucose-induced insulin secretion. Physiol Rev. 1980;60(2):442–509. doi: 10.1152/physrev.1980.60.2.442. [DOI] [PubMed] [Google Scholar]
- 4.Malaisse WJ, Sener A, Herchuelz A, Hutton JC. Insulin release: the fuel hypothesis. Metabolism. 1979;28(4):373–386. doi: 10.1016/0026-0495(79)90111-2. [DOI] [PubMed] [Google Scholar]
- 5.Malmgren S, et al. Tight coupling between glucose and mitochondrial metabolism in clonal β-cells is required for robust insulin secretion. J Biol Chem. 2009;284(47):32395–32404. doi: 10.1074/jbc.M109.026708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Matschinsky F, et al. Glucokinase as pancreatic β cell glucose sensor and diabetes gene. J Clin Invest. 1993;92(5):2092–2098. doi: 10.1172/JCI116809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Meglasson MD, Matschinsky FM. Pancreatic islet glucose metabolism and regulation of insulin secretion. Diabetes Metab Rev. 1986;2(3):163–214. doi: 10.1002/dmr.5610020301. [DOI] [PubMed] [Google Scholar]
- 8.Schuit FC, Huypens P, Heimberg H, Pipeleers DG. Glucose sensing in pancreatic β-cells: a model for the study of other glucose-regulated cells in gut, pancreas, and hypothalamus. Diabetes. 2001;50(1):1–11. doi: 10.2337/diabetes.50.1.1. [DOI] [PubMed] [Google Scholar]
- 9.Blum B, Hrvatin SS, Schuetz C, Bonal C, Rezania A, Melton DA. Functional β-cell maturation is marked by an increased glucose threshold and by expression of urocortin 3. Nat Biotechnol. 2012;30(3):261–264. doi: 10.1038/nbt.2141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jermendy A, et al. Rat neonatal β cells lack the specialised metabolic phenotype of mature β cells. Diabetologia. 2011;54(3):594–604. doi: 10.1007/s00125-010-2036-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Artner I, et al. MafB is required for islet β cell maturation. Proc Natl Acad Sci U S A. 2007;104(10):3853–3858. doi: 10.1073/pnas.0700013104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nishimura W, et al. A switch from MafB to MafA expression accompanies differentiation to pancreatic β-cells. Dev Biol. 2006;293(2):526–539. doi: 10.1016/j.ydbio.2006.02.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Aye T, Toschi E, Sharma A, Sgroi D, Bonner-Weir S. Identification of markers for newly formed β-cells in the perinatal period: a time of recognized β-cell immaturity. J Histochem Cytochem. 2010;58(4):369–376. doi: 10.1369/jhc.2009.954909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gu C, et al. Pancreatic β cells require NeuroD to achieve and maintain functional maturity. Cell Metab. 2010;11(4):298–310. doi: 10.1016/j.cmet.2010.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Aguayo-Mazzucato C, et al. Thyroid hormone promotes postnatal rat pancreatic beta-cell development and glucose-responsive insulin secretion through MAFA. Diabetes. 2013;62(5):1569–1580. doi: 10.2337/db12-0849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Blum B, et al. Reversal of β cell de-differentiation by a small molecule inhibitor of the TGFbeta pathway. Elife. 2014;3:e02809. doi: 10.7554/eLife.02809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Quintens R, Hendrickx N, Lemaire K, Schuit F. Why expression of some genes is disallowed in β-cells. Biochem Soc Trans. 2008;36(pt 3):300–305. doi: 10.1042/BST0360300. [DOI] [PubMed] [Google Scholar]
- 18.Pullen TJ, Khan AM, Barton G, Butcher SA, Sun G, Rutter GA. Identification of genes selectively disallowed in the pancreatic islet. Islets. 2010;2(2):89–95. doi: 10.4161/isl.2.2.11025. [DOI] [PubMed] [Google Scholar]
- 19.Thorrez L, et al. Tissue-specific disallowance of housekeeping genes: the other face of cell differentiation. Genome Res. 2011;21(1):95–105. doi: 10.1101/gr.109173.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pullen TJ, Rutter GA. When less is more: the forbidden fruits of gene repression in the adult beta-cell. Diabetes Obes Metab. 2013;15(6):503–512. doi: 10.1111/dom.12029. [DOI] [PubMed] [Google Scholar]
- 21.Schuit F, et al. β-Cell-specific gene repression: a mechanism to protect against inappropriate or maladjusted insulin secretion? Diabetes. 2012;61(5):969–975. doi: 10.2337/db11-1564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Otonkoski T, et al. Physical exercise-induced hypoglycemia caused by failed silencing of monocarboxylate transporter 1 in pancreatic β cells. Am J Hum Genet. 2007;81(3):467–474. doi: 10.1086/520960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.van Arensbergen J, et al. Ring1b bookmarks genes in pancreatic embryonic progenitors for repression in adult β cells. Genes Dev. 2013;27(1):52–63. doi: 10.1101/gad.206094.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.van Arensbergen J, et al. Derepression of Polycomb targets during pancreatic organogenesis allows insulin-producing β-cells to adopt a neural gene activity program. Genome Res. 2010;20(6):722–732. doi: 10.1101/gr.101709.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Georgia S, Bhushan A. β Cell replication is the primary mechanism for maintaining postnatal β cell mass. J Clin Invest. 2004;114(7):963–968. doi: 10.1172/JCI22098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lemons JM, et al. Quiescent fibroblasts exhibit high metabolic activity. PLoS Biol. 2010;8(10):e1000514. doi: 10.1371/journal.pbio.1000514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Alcazar O, Tiedge M, Lenzen S. Importance of lactate dehydrogenase for the regulation of glycolytic flux and insulin secretion in insulin-producing cells. Biochem J. 2000;352(pt 2):373–380. [PMC free article] [PubMed] [Google Scholar]
- 28.Becker TC, BeltrandelRio H, Noel RJ, Johnson JH, Newgard CB. Overexpression of hexokinase I in isolated islets of Langerhans via recombinant adenovirus. Enhancement of glucose metabolism and insulin secretion at basal but not stimulatory glucose levels. J Biol Chem. 1994;269(33):21234–21238. [PubMed] [Google Scholar]
- 29.Henquin JC, et al. Congenital hyperinsulinism caused by hexokinase I expression or glucokinase-activating mutation in a subset of β-cells. Diabetes. 2013;62(5):1689–1696. doi: 10.2337/db12-1414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ishihara H, et al. Overexpression of hexokinase I but not GLUT1 glucose transporter alters concentration dependence of glucose-stimulated insulin secretion in pancreatic β-cell line MIN6. J Biol Chem. 1994;269(4):3081–3087. [PubMed] [Google Scholar]
- 31.Zhao C, Rutter GA. Overexpression of lactate dehydrogenase A attenuates glucose-induced insulin secretion in stable MIN-6 β-cell lines. FEBS Lett. 1998;430(3):213–216. doi: 10.1016/S0014-5793(98)00600-0. [DOI] [PubMed] [Google Scholar]
- 32.Artner I, et al. MafA and MafB regulate genes critical to β-cells in a unique temporal manner. Diabetes. 2010;59(10):2530–2539. doi: 10.2337/db10-0190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Matsuoka TA, et al. MafA regulates expression of genes important to islet β-cell function. Mol Endocrinol. 2007;21(11):2764–2774. doi: 10.1210/me.2007-0028. [DOI] [PubMed] [Google Scholar]
- 34.Talchai C, Xuan S, Lin HV, Sussel L, Accili D. Pancreatic β cell dedifferentiation as a mechanism of diabetic β cell failure. Cell. 2012;150(6):1223–1234. doi: 10.1016/j.cell.2012.07.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Guillam MT, et al. Early diabetes and abnormal postnatal pancreatic islet development in mice lacking Glut-2. Nat Genet. 1997;17(3):327–330. doi: 10.1038/ng1197-327. [DOI] [PubMed] [Google Scholar]
- 36.Magnuson MA, Osipovich AB. Pancreas-specific Cre driver lines and considerations for their prudent use. Cell Metab. 2013;18(1):9–20. doi: 10.1016/j.cmet.2013.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Guo T, Landsman L, Li N, Hebrok M. Factors expressed by murine embryonic pancreatic mesenchyme enhance generation of insulin-producing cells from hESCs. Diabetes. 2013;62(5):1581–1592. doi: 10.2337/db12-0167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ravassard P, et al. A genetically engineered human pancreatic β cell line exhibiting glucose-inducible insulin secretion. J Clin Invest. 2011;121(9):3589–3597. doi: 10.1172/JCI58447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Boschero AC, Bordin S, Sener A, Malaisse WJ. D-glucose and L-leucine metabolism in neonatal and adult cultured rat pancreatic islets. Mol Cell Endocrinol. 1990;73(1):63–71. doi: 10.1016/0303-7207(90)90045-a. [DOI] [PubMed] [Google Scholar]
- 40.Rozzo A, Meneghel-Rozzo T, Delakorda SL, Yang SB, Rupnik M. Exocytosis of insulin: in vivo maturation of mouse endocrine pancreas. Ann N Y Acad Sci. 2009;1152:53–62. doi: 10.1111/j.1749-6632.2008.04003.x. [DOI] [PubMed] [Google Scholar]
- 41.Tu J, Tuch BE. Glucose regulates the maximal velocities of glucokinase and glucose utilization in the immature fetal rat pancreatic islet. Diabetes. 1996;45(8):1068–1075. doi: 10.2337/diab.45.8.1068. [DOI] [PubMed] [Google Scholar]
- 42.Agathocleous M, et al. Metabolic differentiation in the embryonic retina. Nat Cell Biol. 2012;14(8):859–864. doi: 10.1038/ncb2531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Alaynick WA, et al. ERRgamma directs and maintains the transition to oxidative metabolism in the postnatal heart. Cell Metab. 2007;6(1):13–24. doi: 10.1016/j.cmet.2007.06.007. [DOI] [PubMed] [Google Scholar]
- 44.Stringari C, Edwards RA, Pate KT, Waterman ML, Donovan PJ, Gratton E. Metabolic trajectory of cellular differentiation in small intestine by Phasor Fluorescence Lifetime Microscopy of NADH. Sci Rep. 2012;2:568. doi: 10.1038/srep00568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zhang J, Nuebel E, Daley GQ, Koehler CM, Teitell MA. Metabolic regulation in pluripotent stem cells during reprogramming and self-renewal. Cell Stem Cell. 2012;11(5):589–595. doi: 10.1016/j.stem.2012.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Pullen TJ, Sylow L, Sun G, Halestrap AP, Richter EA, Rutter GA. Overexpression of monocarboxylate transporter-1 (SLC16A1) in mouse pancreatic β-cells leads to relative hyperinsulinism during exercise. Diabetes. 2012;61(7):1719–1725. doi: 10.2337/db11-1531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Pagliuca FW, et al. Generation of functional human pancreatic β cells in vitro. Cell. 2014;159(2):428–439. doi: 10.1016/j.cell.2014.09.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Rezania A, et al. Reversal of diabetes with insulin-producing cells derived in vitro from human pluripotent stem cells. Nat Biotechnol. 2014;32(11):1121–1133. doi: 10.1038/nbt.3033. [DOI] [PubMed] [Google Scholar]
- 49.Dhawan S, Georgia S, Tschen SI, Fan G, Bhushan A. Pancreatic β cell identity is maintained by DNA methylation-mediated repression of Arx. Dev Cell. 2011;20(4):419–429. doi: 10.1016/j.devcel.2011.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Georgia S, Kanji M, Bhushan A. DNMT1 represses p53 to maintain progenitor cell survival during pancreatic organogenesis. Genes Dev. 2013;27(4):372–377. doi: 10.1101/gad.207001.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Papizan JB, et al. Nkx2.2 repressor complex regulates islet β-cell specification and prevents β-to-α-cell reprogramming. Genes Dev. 2011;25(21):2291–2305. doi: 10.1101/gad.173039.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Rose NR, Klose RJ. Understanding the relationship between DNA methylation and histone lysine methylation. Biochim Biophys Acta. 2014;1839(12):1362–1372. doi: 10.1016/j.bbagrm.2014.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Puri S, Hebrok M. Diabetic β cells: to be or not to be? Cell. 2012;150(6):1103–1104. doi: 10.1016/j.cell.2012.08.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Dayeh T, et al. Genome-wide DNA methylation analysis of human pancreatic islets from type 2 diabetic and non-diabetic donors identifies candidate genes that influence insulin secretion. PLoS Genet. 2014;10(3):e1004160. doi: 10.1371/journal.pgen.1004160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Yang BT, et al. Insulin promoter DNA methylation correlates negatively with insulin gene expression and positively with HbA(1c) levels in human pancreatic islets. Diabetologia. 2011;54(2):360–367. doi: 10.1007/s00125-010-1967-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Yang BT, et al. Increased DNA methylation and decreased expression of PDX-1 in pancreatic islets from patients with type 2 diabetes. Mol Endocrinol. 2012;26(7):1203–1212. doi: 10.1210/me.2012-1004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Prentki M, Nolan CJ. Islet β cell failure in type 2 diabetes. J Clin Invest. 2006;116(7):1802–1812. doi: 10.1172/JCI29103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Guo S, et al. Inactivation of specific β cell transcription factors in type 2 diabetes. J Clin Invest. 2013;123(8):3305–3316. doi: 10.1172/JCI65390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Herrera PL. Adult insulin- and glucagon-producing cells differentiate from two independent cell lineages. Development. 2000;127(11):2317–2322. doi: 10.1242/dev.127.11.2317. [DOI] [PubMed] [Google Scholar]
- 60.Jackson-Grusby L, et al. Loss of genomic methylation causes p53-dependent apoptosis and epigenetic deregulation. Nat Genet. 2001;27(1):31–39. doi: 10.1038/83730. [DOI] [PubMed] [Google Scholar]
- 61.Soriano P. Generalized lacZ expression with the ROSA26 Cre reporter strain. Nat Genet. 1999;21(1):70–71. doi: 10.1038/5007. [DOI] [PubMed] [Google Scholar]
- 62.Muzumdar MD, Tasic B, Miyamichi K, Li L, Luo L. A global double-fluorescent Cre reporter mouse. Genesis. 2007;45(9):593–605. doi: 10.1002/dvg.20335. [DOI] [PubMed] [Google Scholar]
- 63.Zhou JX, et al. Combined modulation of polycomb and trithorax genes rejuvenates β cell replication. J Clin Invest. 2013;123(11):4849–4858. doi: 10.1172/JCI69468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Zhong L, Georgia S, Tschen SI, Nakayama K, Bhushan A. Essential role of Skp2-mediated p27 degradation in growth and adaptive expansion of pancreatic β cells. J Clin Invest. 2007;117(10):2869–2876. doi: 10.1172/JCI32198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Song SH, et al. Pulsatile insulin secretion by human pancreatic islets. J Clin Endocrinol Metab. 2002;87(1):213–21. doi: 10.1210/jcem.87.1.8181. [DOI] [PubMed] [Google Scholar]
- 66.Millar DS, Warnecke PM, Melki JR, Clark SJ. Methylation sequencing from limiting DNA: embryonic, fixed, and microdissected cells. Methods. 2002;27(2):108–113. doi: 10.1016/S1046-2023(02)00061-0. [DOI] [PubMed] [Google Scholar]
- 67.Dahl JA, Collas P. MicroChIP — a rapid micro chromatin immunoprecipitation assay for small cell samples and biopsies. Nucleic Acids Res. 2008;36(3):e15. doi: 10.1093/nar/gkm1158. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.