Abstract
Macrophages clear millions of apoptotic cells daily and, during this process, take up large quantities of cholesterol. The membrane transporter ABCA1 is a key player in cholesterol efflux from macrophages and has been shown via human genetic studies to provide protection against cardiovascular disease. How the apoptotic cell clearance process is linked to macrophage ABCA1 expression is not known. Here, we identified a plasma membrane–initiated signaling pathway that drives a rapid upregulation of ABCA1 mRNA and protein. This pathway involves the phagocytic receptor brain-specific angiogenesis inhibitor 1 (BAI1), which recognizes phosphatidylserine on apoptotic cells, and the intracellular signaling intermediates engulfment cell motility 1 (ELMO1) and Rac1, as ABCA1 induction was attenuated in primary macrophages from mice lacking these molecules. Moreover, this apoptotic cell–initiated pathway functioned independently of the liver X receptor (LXR) sterol–sensing machinery that is known to regulate ABCA1 expression and cholesterol efflux. When placed on a high-fat diet, mice lacking BAI1 had increased numbers of apoptotic cells in their aortic roots, which correlated with altered lipid profiles. In contrast, macrophages from engineered mice with transgenic BAI1 overexpression showed greater ABCA1 induction in response to apoptotic cells compared with those from control animals. Collectively, these data identify a membrane-initiated pathway that is triggered by apoptotic cells to enhance ABCA1 within engulfing phagocytes and with functional consequences in vivo.
Introduction
A majority of the approximately 200 billion cells turned over daily as part of normal homeostasis in various tissues of our body die via apoptosis (1–3). These dying cells are subsequently cleared by professional phagocytes (such as macrophages) and by nonprofessional neighboring cells (such as epithelial cells). When a phagocyte ingests an apoptotic cell, it increases its cellular contents and metabolic load. Since macrophages can often engulf multiple apoptotic cells, processing of the ingested contents has important implications for many metabolic disorders (4, 5). Most cells, including macrophages, lack the capacity to break down cholesterol, one of the major apoptotic cell–derived components, thus making the efflux of cellular-free cholesterol critical for lipid homeostasis (6–9).
Macrophages actively export their cellular cholesterol via ABC transporters, with ABCA1 and ABCG1 being the best studied (10–13). ABCA1 exports cellular phospholipids and cholesterol to lipid-poor apolipoprotein A1 (ApoA1), which is critical for the biogenesis of HDL (14); on the other hand, ABCG1 transfers cholesterol primarily to lipid-rich HDL (15). These HDL moieties are then taken up by the liver and excreted through the bile, a process termed “reverse cholesterol transport,” which is a major mechanism for lowering the cholesterol load in cells throughout the body (16).
Atherosclerosis, which can progress to cardiovascular disease, has been a leading cause of death in the United States for almost a century (17). While the etiology of atherosclerosis is complex, macrophages play a key role in the development of atherosclerotic plaques in the vessel wall and the perpetuation of inflammation within the lesions (18–22). In humans and mice, multiple studies have shown that higher levels of ABCA1 and higher HDL correlate with reduced risk for cardiovascular disease (23–25). Patients with genetic ABCA1 deficiencies show severe dyslipidemia (26). It has also been reported that HDL generated by ABCA1 can have beneficial antiinflammatory effects in different tissues (27). Therefore, defining the modalities by which ABCA1 levels are regulated in physiological settings becomes important.
Previously, using macrophage cell lines in vitro, we observed that apoptotic cells induce cholesterol efflux, which was linked to higher levels of ABCA1 protein expression induced in the phagocytes (28). This induction of cholesterol efflux by the macrophages was found to be dependent on the recognition of phosphatidylserine (PtdSer), a key eat-me signal on the apoptotic cells, by the phagocytes (28). However, how the PtdSer recognition triggers ABCA1 in phagocytes and the in vivo relevance of this apoptotic cell–induced ABCA1 induction is not known. Here, using primary macrophages, we identify a membrane-initiated pathway by which recognition of apoptotic cells triggers ABCA1 upregulation in phagocytes. Using gain-of-function and loss-of-function mouse models, we show that the membrane receptor brain-specific angiogenesis inhibitor 1 (BAI1), along with its cytoplasmic intermediaries engulfment cell motility 1 (ELMO1) and Rac1, represents a new signaling pathway to induce ABCA1 under physiological conditions.
Results
Apoptotic cells induce a transcriptional upregulation of ABCA1.
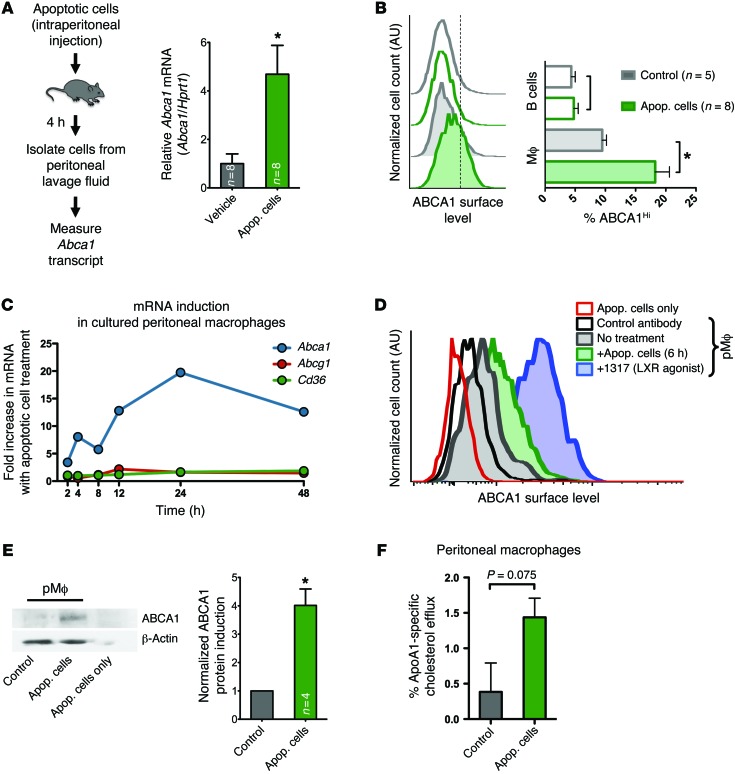
Due to the reported differences in cholesterol homeostasis between macrophage cell lines and primary macrophages (29), we first asked whether the upregulation of ABCA1 during apoptotic cell recognition also occurs in primary resident peritoneal macrophages and in an in vivo context. We injected apoptotic cells into the peritoneum of WT mice and assessed ABCA1 in the cells recovered from the peritoneal lavage. As murine macrophages can recognize and engulf apoptotic cells of human or murine origin comparably (30), we used apoptotic human T cells as targets to assess the upregulation of murine ABCA1 in the phagocytes. Four hours after intraperitoneal apoptotic cell injection, the cells isolated from the peritoneal lavage showed nearly 5-fold upregulation of murine Abca1 mRNA (Figure 1A). When we analyzed ABCA1 surface levels by flow cytometry 6 hours after intraperitoneal injection of apoptotic thymocytes or vehicle control, we found that peritoneal macrophages (IgM–, CD11b+) increased their ABCA1 surface levels, whereas B cells (IgM+) had no change in ABCA1 levels (Figure 1B). To further characterize the effects of apoptotic cell treatment on ABCA1 levels in resident peritoneal macrophages, we cultured peritoneal lavage cells for 3 days, at which time 90% of the remaining cells were macrophages (Supplemental Figure 1; supplemental material available online with this article; doi:10.1172/JCI80300DS1). Cultured primary peritoneal macrophages incubated with apoptotic cells demonstrated a rapid upregulation of ABCA1 in as little as 2 hours in response to apoptotic cells. The higher Abca1 levels induced by apoptotic cells remained for many hours; under the same conditions, Abcg1 and Cd36, two other genes regulated by sterol sensing, were largely unaffected (Figure 1C). The increase in Abca1 transcript levels corresponded with greater surface expression of ABCA1 protein in macrophages (Figure 1, D and E). Furthermore, there was an increase in cholesterol efflux to ApoA1, a measure of ABCA1 function (Figure 1F). These data suggested that apoptotic cells induce Abca1 mRNA and protein in primary macrophages.
Figure 1. Primary resident peritoneal macrophages rapidly upregulate ABCA1 in response to apoptotic cell recognition.
(A) Apoptotic Jurkat cells or control medium was injected intraperitoneally into C57BL/6 mice (n = 8/group). Abca1 mRNA in peritoneal cells was measured after 4 hours (mean ± SEM). Representative of 2 experiments. *P < 0.05 (t test). Apop, apoptotic. (B) Apoptotic murine thymocytes (tinted) or control medium (open) was injected intraperitoneally into C57BL/6 mice. After 6 hours, peritoneal lavage cells were analyzed by flow cytometry for IgM, CD11b, and ABCA1. Representative histograms of ABCA1 staining of B cells (IgM+) and macrophages (IgM–, CD11b+) are shown with quantification from 2 independent experiments on the right (mean ± SEM). *P < 0.05 (t test). (C) Cultured WT peritoneal macrophages incubated with apoptotic human Jurkat cells were analyzed for mRNA levels of murine Abca1, Abcg1, and Cd36 over a time course. Representative of 2 independent experiments. (D) Surface expression of ABCA1 on primary peritoneal macrophages by flow cytometry before (gray trace) and 6 hours after (green) treatment with apoptotic thymocytes or the LXR agonist 1317 (positive control [see Figure 2A], purple). ABCA1 staining on thymocytes (red) and control antibody staining on peritoneal macrophages (black) are also shown. Representative of 3 independent experiments. (E) Representative immunoblot of ABCA1 expression in primary peritoneal macrophages after 6-hour incubation with apoptotic Jurkat cells (left) and densitometry quantification from 4 independent experiments (right, mean ± SEM). *P < 0.05 (paired t test). (F) Cholesterol efflux to the acceptor ApoA1 by primary peritoneal macrophages after apoptotic cell treatment was measured over 4 hours (mean ± SD).
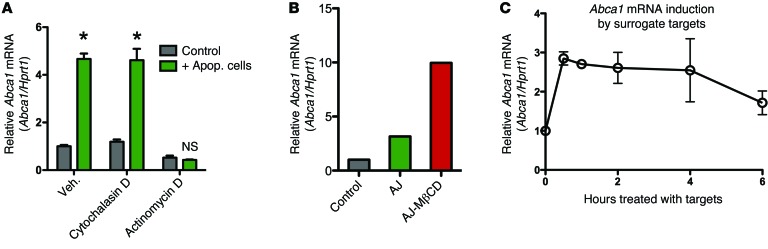
We initially considered that sensing of apoptotic cell–derived cholesterol within the engulfing phagocytes was the most likely mode by which ABCA1 upregulation occurred. Surprisingly, when we treated phagocytes with cytochalasin D, a blocker of actin polymerization that allows corpse binding but not internalization, the enhanced Abca1 upregulation was still observed (Figure 2A and Supplemental Figure 2A). In contrast, blocking transcription in phagocytes with actinomycin D strongly inhibited Abca1 induction by apoptotic cells, suggesting that apoptotic cells induce new transcription rather than a change in the rate of Abca1 message turnover (Figure 2A). This suggested that perhaps an event initiated at the membrane, independent of intracellular cholesterol sensing, might trigger Abca1 gene transcription. To test whether the cholesterol content of the apoptotic targets was important for the upregulation of Abca1, as has been previously shown, we treated apoptotic cells with methyl-β-cyclodextran (MβCD) to extract their membrane cholesterol before adding to phagocytes. Surprisingly, MβCD-treated apoptotic cells were still able to induce Abca1 upregulation, and any enhancement by MβCD was likely due to greater cell death among the treated cells (Figure 2B and Supplemental Figure 2B). As a third means of testing this possibility, we incubated the peritoneal macrophages with surrogate targets lacking sterols. These targets were carboxylate-modified beads that are thought to mimic certain negative charges characteristic of apoptotic cells and can compete with apoptotic cells for binding to phagocytes (31). These surrogate targets also induced Abca1 transcriptional upregulation, with this effect detectable in as little as 30 minutes (Figure 2C). The more rapid kinetics of Abca1 induction by the beads, compared with apoptotic cell induction, was likely due to the beads being more uniform in their charge expression, facilitating better binding to macrophages.
Figure 2. Primary resident peritoneal macrophages rapidly upregulate Abca1 in response to apoptotic cell recognition.
(A) Abca1 mRNA levels from primary peritoneal macrophages treated with vehicle control, 1 μM cytochalasin d, or 5 μg/ml actinomycin d, and incubated with apoptotic Jurkat cells (2 hours) (mean ± SD). Representative of 3 independent experiments. *P < 0.05 (2-way ANOVA, Bonferroni’s post tests comparing control treated to apoptotic cell treated). (B)Primary peritoneal macrophages were left untreated, treated with 2 × 106 apoptotic Jurkat (AJ) cells, or treated with 2 × 106 apoptotic Jurkat cells that had been treated with MβCD (AJ-MβCD). Macrophages were treated for 2 hours before RNA was isolated. Representative of 2 independent experiments. (C) Peritoneal macrophages were incubated with surrogate targets (carboxylate-modified latex beads), and the upregulation of Abca1 upregulation was assessed in a time course (2 independent experiments, mean ± SD).
The Abca1 upregulation induced by apoptotic cells is independent of LXR.
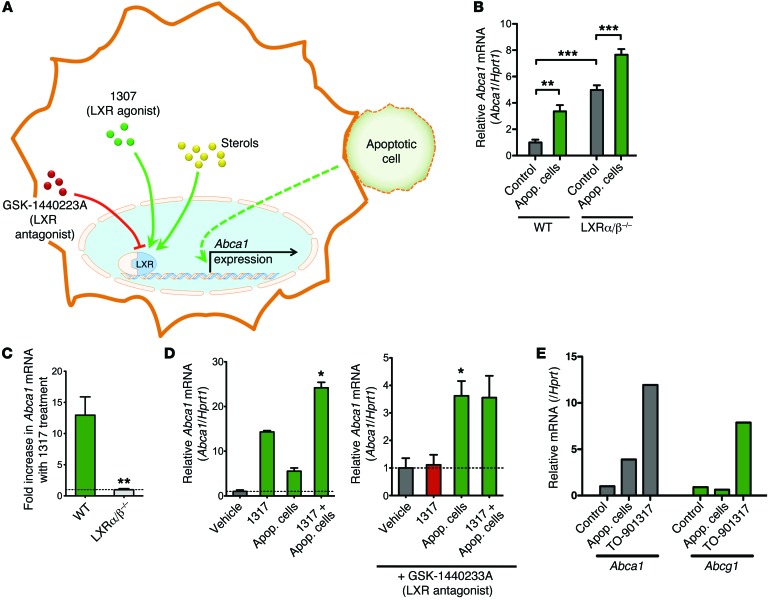
Liver X receptors (LXRs) are crucial regulators of sterol-dependent upregulation of ABCA1, and the binding of LXR to specific sites on the ABCA1 promoter has been well described (32, 33). It has also been reported that in thioglycollate-elicited macrophages, LXR-regulated messages (including Abca1) are upregulated at 24 hours after apoptotic cell engulfment (34, 35). In contrast, the rapid time frame of Abca1 upregulation in our system, the lack of a need for corpse internalization, and the dispensability of cholesterol in the targets suggested that apoptotic cell–initiated Abca1 upregulation might be independent of LXR (Figure 3A). To test this directly, we assessed the response of primary peritoneal macrophages from mice lacking LXR α and β (Lxra/b–/–) (33, 35). Although the LXR-deficient macrophages had higher basal levels of ABCA1, as previously reported (36), incubation with apoptotic cells significantly increased ABCA1 transcript levels in the Lxra/b–/– macrophages (Figure 3B). In contrast, the Lxra/b–/– macrophages were completely deficient in increasing the Abca1 message in response to the LXR agonist TO-901317 (denoted 1317) (Figure 3C) (37).
Figure 3. Upregulation of Abca1 in peritoneal macrophages in response to apoptotic cells is independent of LXR sterol-sensing.
(A) Schematic of Abca1 upregulation by intracellular sterol-sensing by LXR versus a potential independent pathway by apoptotic cells. LXR agonist TO-901317 (1317) and antagonist GSK-1440223A are shown. (B) Abca1 mRNA upregulation in response to apoptotic Jurkat cells in peritoneal macrophages from Lxra/b–/– mice or control C57BL/6 mice (n = 5 mice/group, for each genotype). As previously reported, ABCA1 message is basally higher in the Lxra/b–/– mice, but is enhanced further by apoptotic cells (mean ± SEM). **P < 0.01; ***P < 0.001 (t test). (C) Abca1 upregulation in response to the LXR agonist 1317 is abolished in peritoneal macrophages from Lxra/b–/– mice. The horizontal line is set at 1 (no change) (n = 5/group, mean ± SEM). **P < 0.01 (t test). (D) Treatment with 1317 and apoptotic cells shows an additive effect (left panel). While Abca1 upregulation in response to 1317 is blocked by the LXR antagonist, the antagonist does not affect apoptotic cell–induced Abca1 upregulation (right panel). Graph shown is representative (mean ± SD). *P < 0.05 (paired t test from 4 independent experiments). (E) Peritoneal macrophages were treated with apoptotic cells, the LXR agonist 1317, or vehicle control for 2 hours and analyzed for mRNA levels of Abca1 and Abcg1. Apoptotic cells and 1317 upregulated Abca1, whereas only 1317 but not apoptotic cells upregulated Abca1. Data shown are representative of 2 experiments.
To further explore the role of LXR in apoptotic cell–induced Abca1 upregulation, we assessed Abca1 levels in WT macrophages after treatment with agonists or antagonists of LXR (Figure 3A). Adding the LXR activator 1317 along with apoptotic cells had an additive effect on Abca1 transcript levels in macrophages (Figure 3D). Importantly, the LXR antagonist GSK-1440233A was unable to prevent apoptotic cell–induced upregulation of Abca1, but it was able to potently block Abca1 induction by the LXR agonist 1317 (Figure 3D). Furthermore, addition of the LXR antagonist did not alter the kinetics of Abca1 upregulation after apoptotic cell treatment (Supplemental Figure 3). Also, while Abca1 was upregulated by both apoptotic cells and LXR agonists, Abcg1 was only upregulated by TO-901317 (Figure 3E). These data suggest that apoptotic cell recognition utilizes an LXR-independent mechanism for upregulating Abca1. Collectively, these data suggest that the rapid ABCA1 induction in response to apoptotic cell recognition involves a membrane-initiated signaling pathway that functions independently of the LXR sterol–sensing mechanism.
The BAI1/ELMO1/Rac pathway is necessary for efficient upregulation of ABCA1 in response to apoptotic cells.
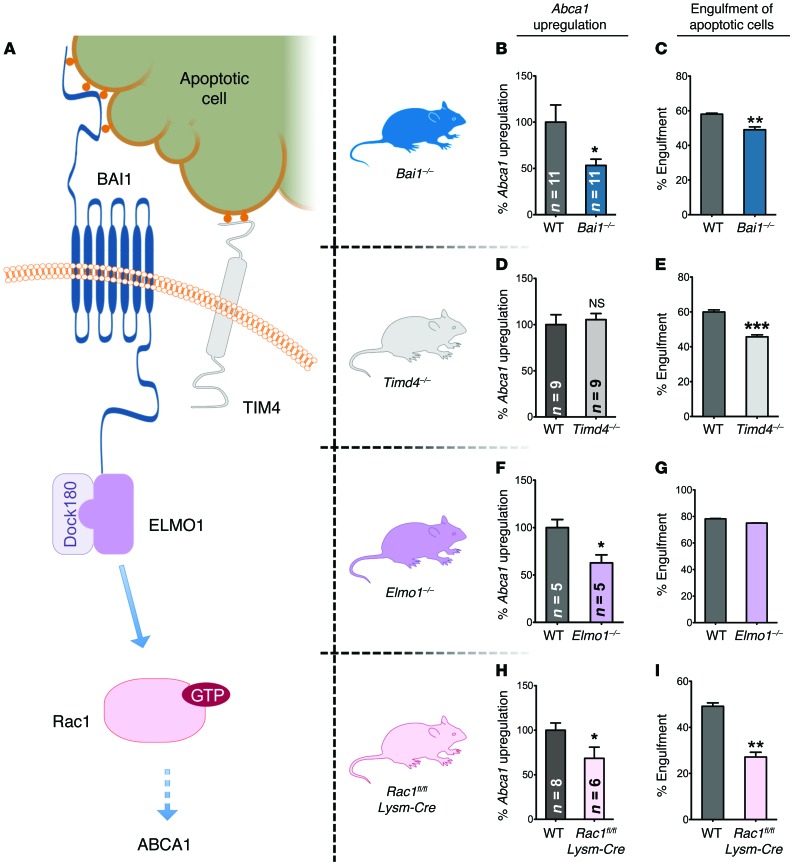
Phagocytes engage ligands on apoptotic cells either directly or indirectly via multiple engulfment receptors. Two pieces of evidence led us to hypothesize that a direct PtdSer receptor might initiate the signaling event. First, carboxylate-modified beads, which lack most characteristics of apoptotic cells other than the negative charge due to PtdSer exposure, were sufficient to induce Abca1 upregulation. Second, these short-term assays for Abca1 upregulation were carried out in X-VIVO 10 medium (serum free and devoid of exogenous serum proteins known to bridge PtdSer on apoptotic cells to the certain engulfment receptors on phagocytes). BAI1 and TIM-4 are 2 engulfment receptors expressed on macrophages that directly engage PtdSer on apoptotic cells, but are thought to signal by independent modalities. To test their relevance in Abca1 upregulation, we isolated resident peritoneal macrophages from mice lacking BAI1 (38) or TIM-4 (39) or from their littermate controls and incubated them with apoptotic cells (Figure 4A and Supplemental Figure 4, A and B). There was no noticeable difference in the number of recovered macrophages in either of the mouse genotypes, and basally, the morphology and growth properties of these macrophages were indistinguishable from control macrophages isolated in parallel from WT mice. While the loss of either BAI1 or TIM4 led to a partial defect in apoptotic cell engulfment, only the BAI1-deficient macrophages showed an attenuated upregulation of Abca1 (Figure 4, B–E), despite the known role of TIM-4 in corpse binding (40). These data suggest that Abca1 upregulation induced by apoptotic cells utilizes a pathway that, at least in part, depends on the PtdSer recognition receptor BAI1.
Figure 4. The BAI1/ELMO/Rac pathway signals to upregulate ABCA1 in response to apoptotic cells.
(A) Schematic of the BAI1/ELMO1/Rac1 signaling pathway that is further tested using the mice that lack expression of BAI1, ELMO1, or Rac1. A second PtdSer recognition receptor TIM-4, which also recognizes PtdSer, is shown. (B, D, F, and H) Apoptotic cell–induced Abca1 upregulation, as measured by mRNA levels, in the peritoneal macrophages from the indicated KO mice. The Abca1 upregulation in the littermate controls (WT) analyzed in parallel was set to 100% for comparison. Number of mice analyzed for each genotype is shown in the bar graph (mean ± SEM). *P < 0.05 (t test). (C, E, G, and I) Representative assays (of at least 2 independent experiments) for the engulfment of apoptotic thymocytes by peritoneal macrophages for each genotype used (mean ± SD). Combined Bai1 KO (n = 6–8 mice), Tim4 KO (n = 3 mice), Elmo1 KO (n = 2 mice), and Rac1 KO (n = 4–5 mice) statistics are shown. **P < 0.01; ***P < 0.001 (t test).
The signaling downstream of BAI1 occurs via an evolutionarily conserved module involving the cytoplasmic proteins ELMO and DOCK that act as a bipartite guanine nucleotide exchange factor (GEF) for the small GTPase Rac1, which then promotes corpse uptake (41, 42) (see schematic in Figure 4A). To test the relevance of these cytoplasmic proteins, we used mice that we previously generated, either globally lacking Elmo1 (43) or conditionally lacking Rac1 in macrophages (as Rac1 global deletion leads to embryonic lethality) (44) (Figure 4A and Supplemental Figure 4, C and D). Peritoneal macrophages from Elmo1–/– mice showed a significant reduction in ABCA1 upregulation in response to apoptotic cells compared with control littermates (Figure 4F). Similarly, loss of Rac1 expression in macrophages also significantly attenuated apoptotic cell–induced Abca1 upregulation compared with that of littermates (Figure 4H). As has been shown previously, the corpse internalization per se by Elmo1–/– macrophages was largely unaffected, due to functional compensation by ELMO2 in engulfment (43) (Figure 4G). Loss of Rac1 led to a strong decrease in apoptotic cell engulfment (Figure 4I). There was still residual Abca1 upregulation in these Rac1- and ELMO1-deficient macrophages, suggesting that other signals initiated by apoptotic cell recognition likely also contribute to Abca1 upregulation. This is not dissimilar to what is normally seen with engulfment, where elimination of single or multiple engulfment-related proteins does not completely eliminate the capacity of phagocytes to clear apoptotic cells due to partial redundancy among pathways. Collectively, these data using primary macrophages from mice lacking specific engulfment proteins identify the BAI1/ELMO/Rac1 signaling module as part of a membrane-initiated signaling pathway that drives Abca1 upregulation upon apoptotic cell recognition.
Loss of BAI1 affects corpse clearance and serum lipid levels in vivo.
Since one of the primary functions of macrophages under homeostatic conditions involves removal of apoptotic cells and BAI1 seems to affect both cell clearance and ABCA1 upregulation, we next tested the loss of BAI1 in vivo. Mutations in ABCA1 were previously found to be causative for Tangier disease, and these patients have very low serum HDL levels and accumulate lipid-laden macrophages in tissues (45). Furthermore, in Tg mice, small changes in ABCA1 have been shown to systemically affect serum lipid levels (46). To test whether this BAI1-initiated Abca1 upregulation might be relevant in vivo, we placed the BAI1 KO mice on a high-fat Western diet to assess the development of dyslipidemia and atherosclerosis. Interestingly, the Bai1 KO mice fed Western diet (assessed at 30 weeks) displayed a mild increase in the levels of TUNEL-positive apoptotic cells in aortic root sections compared with WT littermates (Supplemental Figure 5A). However, when the serum lipids were assessed over 30 weeks, we did not see a significant change in serum lipids in the BAI1 KO mice (Supplemental Figure 5, B–G).
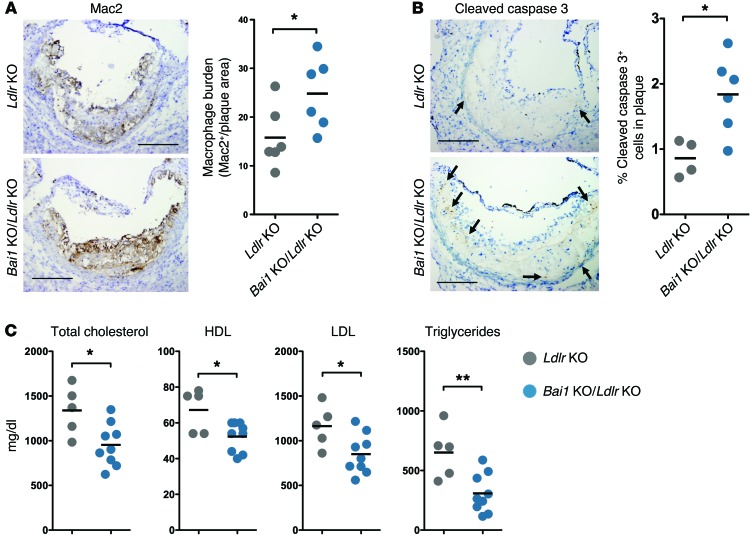
Since phenotypes due to genes linked to dyslipidemia are often better detected when the mice also lack LDL receptor (LDLR), with features that resemble certain aspects of the human disease (47), we crossed the Bai1 KO mice with Ldlr–/– (denoted Ldlr KO) mice. Abca1 deletion in phagocytes on this background has yielded mixed results in atherosclerosis; bone marrow chimeras using Abca1–/– donors and Ldlr KO recipients resulted in increased atherosclerotic plaque area and decreased serum lipids (48–50), whereas more recent studies using myeloid cell–specific ABCA1 deletion bred onto the Ldlr KO background resulted in little change in atherosclerosis, but significant decreases in serum lipids (51). We placed the Bai1–/–Ldlr–/– (denoted Bai1 KO/Ldlr KO) mice on a Western diet for 15 weeks and assessed their phenotypes. We found an increased macrophage burden (as measured by Mac2 staining) within the plaques of the Bai1 KO/Ldlr KO mice. However, despite the presence of more macrophages, the Bai1 KO/Ldlr KO mice showed a greater number of uncleared apoptotic cells in atherosclerotic plaques formed in their aortic roots compared with their control littermates (as measured by cleaved caspase 3 staining), suggesting a defect in corpse clearance within the plaques of mice lacking BAI1 (Figure 5, A and B). The Bai1 KO/Ldlr KO mice also had significantly lower serum levels of total cholesterol, HDL, LDL, and triglycerides compared with the littermate Ldlr KO mice (Figure 5C), but did not show a statistically significant difference in atherosclerotic lesions (Supplemental Figure 6, A–D). This phenotype of the Bai1 KO/Ldlr KO null mice is remarkably similar to that observed with specific Abca1 deletion in macrophages (in the LDLR-deficient background) (51). The latter mice also did not show a difference in atherosclerotic lesions compared with their littermates, but did show a decrease in serum lipids (51), essentially similar to that observed here in the Bai1-null mice. These data suggest that the decrease in apoptotic cell–induced Abca1 upregulation seen with the loss of BAI1 could translate to dyslipidemia in vivo that is analogous to that seen in mice lacking ABCA1 specifically in macrophages.
Figure 5. BAI1 signaling affects serum lipid levels in dyslipidemic Ldlr-deficient mice.
(A) Immunohistochemistry for the macrophage marker Mac2 within the aortic roots of Bai1 KO/Ldlr KO mice and control Ldlr–/– littermates after 15 weeks on a Western diet. Quantification of Mac2+ area normalized to oil red O+ area (Supplemental Figure 6D) (both measured using ImageJ software) as an indication of macrophage burden is shown on right. *P < 0.05 (t test). (B) Immunohistochemistry for cleaved caspase 3 (indicative of cells undergoing apoptosis) within the aortic roots of Bai1 KO/Ldlr KO mice and control Ldlr–/– littermates after 15 weeks on Western diet. Scale bars: 200 μm. Graph represents the number of cleaved caspase 3–positive cells (arrows) counted in the atherosclerotic plaques of each mouse in a representative cohort (of 2 independent experiments producing similar results). *P < 0.05 (t test). (C) Analysis of serum levels of total cholesterol, HDL, LDL, and triglycerides in Ldlr–/– mice compared with Bai1 KO/Ldlr KO mice after 15 weeks on a Western diet. *P < 0.05; **P < 0.01 (t test).
Overexpression of BAI1 enhances ABCA1 expression in response to apoptotic cells.
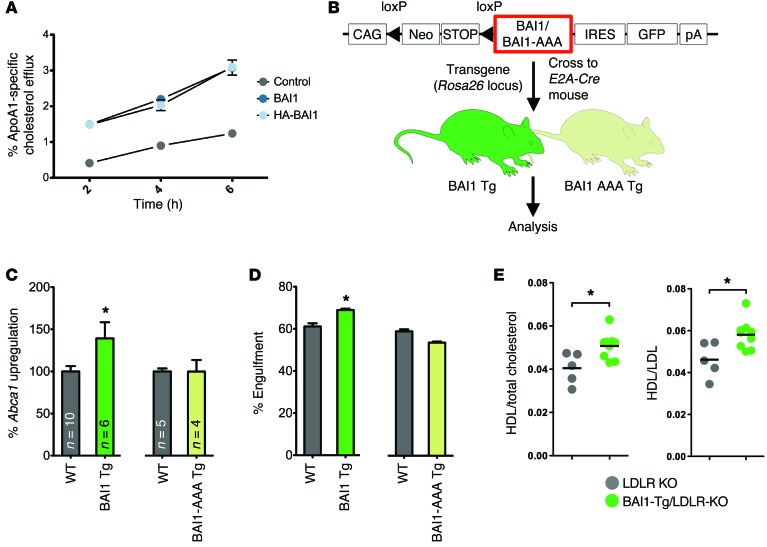
We next asked whether boosting the signaling via the BAI1-initiated signaling pathway can lead to higher ABCA1 upregulation in response to apoptotic cells and whether this can translate to a “gain of function” in vivo. We first tested this in vitro using LR73 cell lines overexpressing WT BAI1 or HA-tagged BAI1. Cells overexpressing BAI1 showed enhanced cholesterol efflux to ApoA1 when exposed to apoptotic cells compared with control LR73 cells (Figure 6A). To test this in primary macrophages, we used Tg mice that we recently engineered to globally overexpress WT BAI1 (R26CTV-Bai1, herein referred to as Tg BAI1) (C.S. Lee, unpublished observations) (Figure 6B and Supplemental Figure 4E). When we isolated peritoneal macrophages from the Tg BAI1 mice and incubated them with apoptotic cells, they showed enhanced engulfment compared with macrophages from non-Tg littermate mice (Figure 6D). When Abca1 upregulation was assessed, the peritoneal macrophages from the Tg BAI1 mice had enhanced Abca1 expression in response to apoptotic cells compared with control mice (Figure 6C).
Figure 6. BAI1 signaling affects serum lipid levels in dyslipidemic Ldlr-deficient mice.
(A) Control LR73 (gray), BAI1-overexpressing LR73 (dark blue), or HA-tagged BAI1-overexpressing (HA-BAI1) LR73 (light blue) cells were treated with apoptotic Jurkat cells and their efflux 3H-cholesterol to ApoA1 was assessed (mean ± SD). Representative of 3 independent experiments. (B) Schematic for the generation of Tg mice overexpressing BAI1. The Tg constructs engineered to express WT BAI1 and BAI1-AAA mutant (that fail to engage ELMO1-mediated downstream signaling) contain a loxP-flanked STOP cassette preceding the coding sequence for BAI1. These constructs were knocked into the Rosa26 locus of C57BL/6 ES cells, from which Tg mice were generated. Upon crossing with E2A-Cre–expressing mice and Cre-mediated removal of the STOP cassette, BAI1 and BAI1-AAA expression were attained. (C) Peritoneal macrophages from Tg BAI1 or Tg BAI1-AAA mice were incubated with apoptotic Jurkat cells. Abca1 upregulation (compared with untreated controls) was measured, and data are shown as normalized to littermate controls (WT) (mean ± SEM). *P < 0.05 (t test). (D) Representative assay of at least 2 independent experiments for the engulfment of apoptotic thymocytes by peritoneal macrophages isolated from Tg BAI1 or Tg BAI1-AAA mice (mean ± SD). Combined Tg BAI1 (n = 9–10 mice) and Tg BAI1-AAA (n = 6–7 mice) statistics are shown. *P < 0.05, P = 0.887 for BAI1-AAA vs. WT (t test). (E) Tg BAI1 Ldlr–/– mice were fed a Western diet for 15 weeks, and the ratios of HDL/total cholesterol and HDL/LDL in the serum were analyzed. *P < 0.05 (t test).
To test whether this effect of BAI1 required downstream signaling via ELMO1, we used another line of Tg mice that we generated in parallel, where the mice overexpressed a mutant form of BAI1 (R26CTV-Bai1-AAA, herein referred to as Tg BAI1-AAA) (C.S. Lee, unpublished observations). Although the BAI1-AAA protein gets expressed on the surface at levels comparable to that of the WT BAI1, due to the cytoplasmic point mutations, this BAI1-AAA fails to bind ELMO and link to ELMO/Rac-mediated downstream signaling. Macrophages overexpressing the BAI1-AAA did not show a detectable increase in Abca1 upregulation when incubated with apoptotic cells and had no increase in their engulfment capacity compared with littermate controls (Figure 6, C and D). These data further support the concept that BAI1-initiated signaling events can upregulate Abca1 and also highlight the importance of ELMO in mediating the signal downstream of BAI1.
We then crossed the WT Tg BAI1 mice onto the LDLR-deficient background and placed them on a Western diet to test whether gain of function in BAI1 signaling and Abca1 upregulation had an effect in vivo. Under these settings, while the overexpression of WT BAI1 per se did not reduce the number of apoptotic cells within the plaques or the size of the lesions, the Tg BAI1 Ldlr KO mice displayed higher ratios of HDL to total cholesterol and HDL to LDL (Figure 6E), suggesting an effect of Tg BAI1 expression on lipid homeostasis.
Discussion
Collectively, the data presented here suggest several important insights. First, this work identifies a plasma membrane–initiated pathway in phagocytes, driven by apoptotic cell recognition, that can lead to upregulation of ABCA1 and in turn regulate cholesterol homeostasis in vivo. Although ABCA1 is recognized as a critical cholesterol transporter that is essential for HDL biogenesis, the major known mechanism for ABCA1 upregulation has been intracellular sterol sensing and subsequent LXR-dependent ABCA1 transcription. Our studies using macrophages lacking BAI1, ELMO1, or Rac1 and macrophages from Tg mice overexpressing BAI1 demonstrate that the BAI1/ELMO1/Rac1 module is part of what we believe to be a novel membrane-initiated pathway leading to Abca1 gene transcription that is LXR independent. Apoptotic cell recognition occurs via more than one membrane receptor (1, 52). While our data do not rule out other engulfment receptors that might also contribute to ABCA1 upregulation, the data presented in this report clearly suggest that the BAI1/ELMO1/Rac1 signaling pathway contributes to ABCA1 upregulation in phagocytes as they engage apoptotic cells.
Second, there is accumulating evidence that clearance of apoptotic cells, which occurs in essentially all tissues throughout life, is an important homeostatic process, but how this might influence lipid homeostasis has not been addressed. The attenuation of ABCA1 upregulation due to loss of BAI1 translates to a greater number of uncleared apoptotic cells in the aortic roots; it also correlated with altered lipid levels in vivo. Conversely, enhancing the signaling via BAI1 (in Tg mice) improves the HDL/LDL ratio, suggesting a beneficial effect of this signaling. HDL, in addition to its role in getting rid of excess cholesterol through reverse cholesterol transport, has also been linked to antiinflammatory effects in different tissues and contexts. Therefore, in addition to the known antiinflammatory mediators elicited in macrophages by apoptotic cell recognition (such as TGF-β, IL-10, and PGE2) (9), the ABCA1-dependent generation of HDL may provide additional antiinflammatory benefits.
Third, there is extensive correlation in humans of higher HDL levels with lower cardiovascular disease, which forms the basis for attempts to increase HDL levels in patients to provide a benefit against cardiovascular disease; however, recent efforts to increase HDL levels have not yielded a benefit (53). This may be due in part to the initial emphasis placed on raising HDL levels in isolation and/or HDL being raised via pharmacological or supraphysiological mechanisms (53). Similarly, LXR agonist drugs have also faced problems with toxicity issues, likely due to the “sledgehammer” nature of this approach, with many LXR target genes being simultaneously activated or inhibited (54). The cell clearance–initiated pathway identified here leads to ABCA1 expression (and in turn HDL levels), which occurs routinely in many tissues as part of normal homeostasis and may provide a safer alternative. It is possible that targeting the LXR-independent BAI1/ELMO1/Rac1 pathway at the membrane (either by directly activating BAI1 or perhaps with liposomes mimicking features of apoptotic cells) might be of benefit by triggering a natural cellular pathway with less adverse effects.
Methods
Mice.
C57BL/6J mice were obtained from Jackson Laboratories. Lxra/b–/– mice were provided by David Mangelsdorf (University of Texas Southwestern Medical Center, Dallas, Texas, USA) (55). We have previously reported the generation of Bai1–/– mice (38) and Elmo1–/– mice (43). Tim4–/– mice were provided by Vijay Kuchroo (Harvard Medical School, Boston, Massachusetts, USA) (39). To conditionally delete Rac1 in the myeloid lineage, we crossed Rac1fl/fl mice (56) with Lysm-Cre mice to generate Rac1fl/fl Lysm-Cre mice (44). Tg BAI1 mice were generated by engineering either the WT BAI1 or the BAI1-AAA mutant coding sequences within the CTV vector and knocking them into the Rosa26 locus of C57BL/6 embryonic stem (ES) cells. The ES cells were then used to generate the Tg mice. The fidelity of the STOP cassette and the expression of BAI1 or BAI1-AAA mutants after crossing to E2A mice were confirmed by PCR and protein expression (38). Bai1–/– mice were bred to Ldlr KO mice (on a C57BL/6 background) to obtain Bai1+/–Ldlr–/– mice, which were then crossed to produce cohorts of Bai1–/–Ldlr–/– mice (denoted Bai1 KO/Ldlr KO) and the control Bai1+/+Ldlr–/– mice (denoted Ldlr KO). Female mice were used in these experiments.
Quantitative RT-PCR.
Total RNA was isolated from cells using the Quick RNA MiniPrep Kit (Zymo Research), and cDNA was generated using the QuantiTect Reverse Transcriptase Kit (QIAGEN) according to the manufacturer’s instructions. Gene expression for mouse Abca1 normalized to Hprt was measured using TaqMan probes (Applied Biosystems) by Real-Time PCR with a StepOnePlus qPCR system (Applied Biosystems).
Analysis of Abca1 upregulation in vivo.
For RNA measurements, 80 million human Jurkat T cells (from ATCC, clone E6-1) in 4 ml of X-VIVO 10 (Lonza) were UV irradiated and incubated for 4 hours to induce apoptosis. 300 μl of apoptotic cells (6 million cells) or X-VIVO 10 alone were injected intraperitoneally per mouse, with 8 mice per treatment condition. After 4 hours, the mice were euthanized, the peritoneum was lavaged with 10 ml of PBS plus 5% FBS, and cells collected were lysed for RNA isolation. For cell-surface ABCA1 expression, 250 μl of X-VIVO 10 or X-VIVO 10 containing 10 million unstained apoptotic thymocytes (prepared as described below) were intraperitoneally injected into WT female mice. After 6 hours, mice were euthanized and peritoneal cells were collected as described above and stained for CD11b (eBioscience, 17-0112-82), IgM (Life Technologies, A21042), Ly6G (eBioscience, 35-5931-81), and ABCA1 (as described below). Flow cytometry was performed using a FACSCanto (BD Biosciences) and the data analyzed using FlowJo software (TreeStar Inc.).
Analysis of Abca1 upregulation in primary peritoneal macrophages.
To isolate the macrophages, the peritoneum was lavaged with 10 ml PBS with 5% FBS; the collected cells were resuspended in X-VIVO 10, and the required number of cells were plated. The next day, the cells were washed 3 times with PBS, and the media was changed every day. The peritoneal macrophages were assayed on the third day after isolation. Peritoneal macrophages were plated at 250,000 cells per well in a 24-well plate. On the day of the assay, Jurkat T cells at 4 × 106 cells/ml in X-VIVO 10 medium were UV irradiated to induce apoptosis and incubated for 4 hours. Two million apoptotic Jurkat cells or X-VIVO 10, along with any pharmacological treatments, were added to duplicate wells for 2 hours. At the end of the treatment period, the wells were washed 3 times with PBS and lysed for RNA isolation. All drugs used were diluted from stocks of 1 mM cytochalasin d (Sigma-Aldrich, C-8273) in DMSO, 5 mg/ml actinomycin d (Sigma-Aldrich, A9415) in ethanol, 1 mM TO-901317 (Sigma-Aldrich, T-2320) in ethanol, and 1 mM GSK-1440233A in DMSO. For the time-course analysis (Figure 1B and Supplemental Figure 2C), peritoneal macrophages were given a single treatment of 2 million apoptotic cells for the indicated time duration before all samples were lysed simultaneously for RNA isolation. Apoptotic cells were generated as described above independently for each time point.
ABCA1 surface expression.
To evaluate ABCA1 surface expression on peritoneal macrophages, isolated peritoneal macrophages were treated with apoptotic murine thymocytes, 1 μM TO-901317, or left untreated for 6 hours. The cells were then washed, trypsinized, and stained for flow cytometry. Antibodies used were anti-ABCA1 (Abcam, ab48264), anti-rat IgG2a-PE (eBioscience, 12-4817-82), and anti-CD5 as an isotype control (Santa Cruz Biotechnology Inc., 18912). For detection of peritoneal lavage cells, cells were either stained immediately after isolation or after 3 days of culture. The antibodies used were CD11b, Ly6G (both described above), CD19 (eBioscience, 45-0193-80), and CD3 (eBioscience, 17-0031-82). Similar results were obtained using antibodies against F4/80 (eBioscience, 11-4801-81) and B220 (eBioscience, 17-0452-82).
Cholesterol efflux assay.
Cholesterol efflux was measured as previously described (28). Briefly, peritoneal macrophages or LR73 CHO cells (57) were loaded with 3H-cholesterol overnight, allowed to equilibrate, and then treated with either regular media or apoptotic Jurkat-containing media for 4 hours. At the end of the treatment period, the macrophages were washed and treated with either BSA-containing media or ApoA1-containing media for 4 hours. Radioactivity was measured in the media and in the cells to get a percentage of efflux. Efflux to ApoA1 was calculated relative to efflux to BSA (% ApoA1-specific cholesterol efflux).
Immunoblotting.
Peritoneal macrophages were plated in 6-cm plates and treated with 20 million apoptotic Jurkat cells or the media alone. After 6 hours, the wells were washed extensively to remove unengulfed Jurkat cells, and then the macrophages were lysed. The antibodies used were anti-ABCA1 (Novus, NB400-105) and anti–β-actin HRP (Sigma-Aldrich, A3854).
Engulfment assay.
Engulfment assays were carried out as previously described (58) using apoptotic thymocytes stained with either TAMRA (Invitrogen, C-1171) or CypHer5E (GE Healthcare, PA15401). Thymocytes isolated from 4- to 6-week-old C57BL/6 mice were induced to undergo apoptosis via incubation with 10 μM dexamethasone for 4 hours, followed by staining. Peritoneal macrophages were incubated at a 1:10 ratio with apoptotic thymocytes for 1 hour and then washed and analyzed by flow cytometry.
Aortic root analysis.
Bai1–/– mice or WT littermate controls were given a Western diet (Harlan, TD.88137) for 30 weeks before the mice were euthanized. The aortic root was dissected and frozen in OCT media. Aortic root blocks were cut into 5-μm–thick sections. TUNEL analysis was done using the In Situ Cell Death Detection Kit from Roche according to the manufacturer’s protocol. TUNEL images were taken with an AxioImager 2 equipped with an ApoTome attachment (Zeiss). The resulting images were tiled to reassemble the entire aortic root, and analyzed for the percentage of TUNEL+ events per total DAPI+ events using CellProfiler software (59).
Ldlr KO and Bai1 KO/Ldlr KO littermate mice or Ldlr KO and Tg BAI1/LDLR KO littermate mice were given a Western diet for 15 weeks before aortic root analysis. Immunohistochemistry for cleaved caspase 3 was performed by the UVA Biorepository and Tissue Research Facility (BTRF). The number of cleaved caspase 3–positive cells was counted with ImageJ software (http://imagej.nih.gov/ij/) using low magnification images taken of the entire aortic root. Each aortic root was counted 3 times and then averaged, with a single value plotted for each mouse. Images of staining were taken with an Olympus SZX12 microscope.
Serum lipid analysis.
After euthanasia, blood was collected from mice by cardiac puncture. The blood was centrifuged in a tabletop microcentrifuge to obtain serum. Total cholesterol, HDL, LDL, and triglyceride concentrations were measured by the UVA Health System Clinical Core Laboratories.
Statistical analysis.
All statistical analysis was done using Graphpad Prism 5 using Student’s 2-tailed t test, 2-way ANOVA, or paired 2-tailed t test, as indicated in legends. Outliers were determined using the Grubbs’ outlier test with an α of 0.05. Statistical significance was defined as P < 0.05.
Study approval.
All animal procedures were approved by the Institutional Animal Care and Use Committee (IACUC) of UVA.
Supplementary Material
Acknowledgments
We thank the members of the Ravichandran, Schulman, and Kiss labs for their valuable input, especially S. Arandjelovic, L. Haney, and J. Angdisen. We also thank the UVA BTRF and the UVA Cardiovascular Research Center Histology Core, especially P. Pramoonjago and M. Brevard, for their assistance in histological services and tissue processing. This work was supported by grants to K.S. Ravichandran (GM064709 and HL120840, funds from the Center for Cell Clearance, and the UVa-AZ Alliance) and support to A.M. Fond via the UVA Medical Scientist Training Program and a T32 Immunology Training Grant.
Footnotes
Conflict of interest: The authors have declared that no conflict of interest exists.
Reference information:J Clin Invest. 2015;125(7):2748–2758. doi:10.1172/JCI80300.
References
- 1.Nagata S, Hanayama R, Kawane K. Autoimmunity and the clearance of dead cells. Cell. 2010;140(5):619–630. doi: 10.1016/j.cell.2010.02.014. [DOI] [PubMed] [Google Scholar]
- 2.Janssen WJ, Henson PM. Cellular regulation of the inflammatory response. Toxicol Pathol. 2012;40(2):166–173. doi: 10.1177/0192623311428477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gregory CD, Pound JD. Microenvironmental influences of apoptosis in vivo and in vitro. Apoptosis. 2010;15(9):1029–1049. doi: 10.1007/s10495-010-0485-9. [DOI] [PubMed] [Google Scholar]
- 4.Han CZ, Ravichandran KS. Metabolic connections during apoptotic cell engulfment. Cell. 2011;147(7):1442–1445. doi: 10.1016/j.cell.2011.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Green DR, Galluzzi L, Kroemer G. Metabolic control of cell death. Science. 2014;345(6203):1250256. doi: 10.1126/science.1250256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Phillips MC. Molecular mechanisms of cellular cholesterol efflux. J Biol Chem. 2014;289(35):24020–24029. doi: 10.1074/jbc.R114.583658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Curtiss LK, Kubo N, Schiller NK, Boisvert WA. Participation of innate and acquired immunity in atherosclerosis. Immunol Res. 2000;21(2):167–176. doi: 10.1385/IR:21:2-3:167. [DOI] [PubMed] [Google Scholar]
- 8.Oram JF. Molecular basis of cholesterol homeostasis: lessons from Tangier disease and ABCA1. Trends Mol Med. 2002;8(4):168–173. doi: 10.1016/S1471-4914(02)02289-X. [DOI] [PubMed] [Google Scholar]
- 9.Ravichandran KS, Lorenz U. Engulfment of apoptotic cells: signals for a good meal. Nat Rev Immunol. 2007;7(12):964–974. doi: 10.1038/nri2214. [DOI] [PubMed] [Google Scholar]
- 10.Wang M-D, Franklin V, Marcel YL. In vivo reverse cholesterol transport from macrophages lacking ABCA1 expression is impaired. Arterioscler Thromb Vasc Biol. 2007;27(8):1837–1842. doi: 10.1161/ATVBAHA.107.146068. [DOI] [PubMed] [Google Scholar]
- 11.Tarling EJ, de Aguiar Vallim TQ, Edwards PA. Role of ABC transporters in lipid transport and human disease. Trends Endocrinol Metab. 2013;24(7):342–350. doi: 10.1016/j.tem.2013.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Westerterp M, Bochem AE, Yvan-Charvet L, Murphy AJ, Wang N, Tall AR. ATP-binding cassette transporters, atherosclerosis, and inflammation. Circ Res. 2014;114(1):157–170. doi: 10.1161/CIRCRESAHA.114.300738. [DOI] [PubMed] [Google Scholar]
- 13.Tall AR. Cholesterol efflux pathways and other potential mechanisms involved in the athero-protective effect of high density lipoproteins. J Intern Med. 2008;263(3):256–273. doi: 10.1111/j.1365-2796.2007.01898.x. [DOI] [PubMed] [Google Scholar]
- 14.Van Eck M. ATP-binding cassette transporter A1: key player in cardiovascular and metabolic disease at local and systemic level. Curr Opin Lipidol. 2014;25(4):297–303. doi: 10.1097/MOL.0000000000000088. [DOI] [PubMed] [Google Scholar]
- 15.Marcel YL, Ouimet M, Wang M-D. Regulation of cholesterol efflux from macrophages. Curr Opin Lipidol. 2008;19(5):455–461. doi: 10.1097/MOL.0b013e32830f4a1d. [DOI] [PubMed] [Google Scholar]
- 16.Maxfield FR, Tabas I. Role of cholesterol and lipid organization in disease. Nature. 2005;438(7068):612–621. doi: 10.1038/nature04399. [DOI] [PubMed] [Google Scholar]
- 17.Go AS, et al. Heart disease and stroke statistics--2014 update: a report from the American Heart Association. Circulation. 2014;129(3):e28–e292. doi: 10.1161/01.cir.0000441139.02102.80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gerrity RG, Naito HK, Richardson M, Schwartz CJ. Dietary induced atherogenesis in swine. Morphology of the intima in prelesion stages. Am J Pathol. 1979;95(3):775–792. [PMC free article] [PubMed] [Google Scholar]
- 19.Fenyo IM, Gafencu AV. The involvement of the monocytes/macrophages in chronic inflammation associated with atherosclerosis. Immunobiology. 2013;218(11):1376–1384. doi: 10.1016/j.imbio.2013.06.005. [DOI] [PubMed] [Google Scholar]
- 20.Shibata N, Glass CK. Regulation of macrophage function in inflammation and atherosclerosis. J Lipid Res. 2009;50(suppl):S277–S281. doi: 10.1194/jlr.R800063-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Koltsova EK, Hedrick CC, Ley K. Myeloid cells in atherosclerosis: a delicate balance of anti-inflammatory and proinflammatory mechanisms. Current Opinion in Lipidology. 2013;24(5):371–380. doi: 10.1097/MOL.0b013e328363d298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Thorp E, Subramanian M, Tabas I. The role of macrophages and dendritic cells in the clearance of apoptotic cells in advanced atherosclerosis. Eur J Immunol. 2011;41(9):2515–2518. doi: 10.1002/eji.201141719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tarling EJ, Edwards PA. Dancing with the sterols: critical roles for ABCG1, ABCA1, miRNAs, and nuclear and cell surface receptors in controlling cellular sterol homeostasis. Biochim Biophys Acta. 2012;1821(3):386–395. doi: 10.1016/j.bbalip.2011.07.011. [DOI] [PubMed] [Google Scholar]
- 24.Brunham LR, Singaraja RR, Hayden MR. Variations on a gene: rare and common variants in ABCA1 and their impact on HDL cholesterol levels and atherosclerosis. Annu Rev Nutr. 2006;26:105–129. doi: 10.1146/annurev.nutr.26.061505.111214. [DOI] [PubMed] [Google Scholar]
- 25.Oram JF, Heinecke JW. ATP-binding cassette transporter A1: a cell cholesterol exporter that protects against cardiovascular disease. Physiol Rev. 2005;85(4):1343–1372. doi: 10.1152/physrev.00005.2005. [DOI] [PubMed] [Google Scholar]
- 26. Assmann G, von Eckardstein A, Brewer HB. Familial high density lipoprotein deficiency: Tangier disease. In Scriver CR, ed. The Metabolic and Molecular Basis of Inherited Disease. New York, New York, USA: McGraw-Hill;1995:2053–2072. [Google Scholar]
- 27.Moore KJ, Fisher EA. High-density lipoproteins put out the fire. Cell Metab. 2014;19(2):175–176. doi: 10.1016/j.cmet.2014.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kiss RS, Elliott MR, Ma Z, Marcel YL, Ravichandran KS. Apoptotic cells induce a phosphatidylserine-dependent homeostatic response from phagocytes. Curr Biol. 2006;16(22):2252–2258. doi: 10.1016/j.cub.2006.09.043. [DOI] [PubMed] [Google Scholar]
- 29.Annema W, Tietge UJ. Regulation of reverse cholesterol transport - a comprehensive appraisal of available animal studies. Nutr Metab (Lond). 2012;9(1):25. doi: 10.1186/1743-7075-9-25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pattabiraman G, Lidstone EA, Palasiewicz K, Cunningham BT, Ucker DS. Recognition of apoptotic cells by viable cells is specific, ubiquitous, and species independent: analysis using photonic crystal biosensors. Mol Biol Cell. 2014;25(11):1704–1714. doi: 10.1091/mbc.E13-11-0700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tóth B, et al. Transglutaminase 2 is needed for the formation of an efficient phagocyte portal in macrophages engulfing apoptotic cells. J Immunol. 2009;182(4):2084–2092. doi: 10.4049/jimmunol.0803444. [DOI] [PubMed] [Google Scholar]
- 32.Hong C, Tontonoz P. Liver X receptors in lipid metabolism: opportunities for drug discovery. Nat Rev Drug Discov. 2014;13(6):433–444. doi: 10.1038/nrd4280. [DOI] [PubMed] [Google Scholar]
- 33.Venkateswaran A, et al. Control of cellular cholesterol efflux by the nuclear oxysterol receptor LXR alpha. Proc Natl Acad Sci U S A. 2000;97(22):12097–12102. doi: 10.1073/pnas.200367697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.A-Gonzalez N, Hidalgo A. Nuclear receptors and clearance of apoptotic cells: stimulating the macrophage’s appetite. Front Immunol. 2014;5:211. doi: 10.3389/fimmu.2014.00211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.A-Gonzalez N, et al. Apoptotic cells promote their own clearance and immune tolerance through activation of the nuclear receptor LXR. Immunity. 2009;31(2):245–258. doi: 10.1016/j.immuni.2009.06.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wagner BL, et al. Promoter-specific roles for liver X receptor/corepressor complexes in the regulation of ABCA1 and SREBP1 gene expression. Mol Cell Biol. 2003;23(16):5780–5789. doi: 10.1128/MCB.23.16.5780-5789.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zuercher WJ, et al. Discovery of tertiary sulfonamides as potent liver X receptor antagonists. J Med Chem. 2010;53(8):3412–3416. doi: 10.1021/jm901797p. [DOI] [PubMed] [Google Scholar]
- 38.Hochreiter-Hufford AE, et al. Phosphatidylserine receptor BAI1 and apoptotic cells as new promoters of myoblast fusion. Nature. 2013;97(7448):263–267. doi: 10.1038/nature12135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rodriguez-Manzanet R, et al. T and B cell hyperactivity and autoimmunity associated with niche-specific defects in apoptotic body clearance in TIM-4-deficient mice. Proc Natl Acad Sci U S A. 2010;107(19):8706–8711. doi: 10.1073/pnas.0910359107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Park D, Hochreiter-Hufford A, Ravichandran KS. The phosphatidylserine receptor TIM-4 does not mediate direct signaling. Curr Biol. 2009;19(4):346–351. doi: 10.1016/j.cub.2009.01.042. [DOI] [PubMed] [Google Scholar]
- 41.Brugnera E, et al. Unconventional Rac-GEF activity is mediated through the Dock180-ELMO complex. Nat Cell Biol. 2002;4(8):574–582. doi: 10.1038/ncb824. [DOI] [PubMed] [Google Scholar]
- 42.Kinchen JM, et al. Two pathways converge at CED-10 to mediate actin rearrangement and corpse removal in C. elegans. Nature. 2005;434(7029):93–99. doi: 10.1038/nature03263. [DOI] [PubMed] [Google Scholar]
- 43.Elliott MR, et al. Unexpected requirement for ELMO1 in clearance of apoptotic germ cells in vivo. Nature. 2010;467(7313):333–337. doi: 10.1038/nature09356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Juncadella IJ, et al. Apoptotic cell clearance by bronchial epithelial cells critically influences airway inflammation. Nature. 2013;493(7433):547–551. doi: 10.1038/nature11714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Brooks-Wilson A, et al. Mutations in ABC1 in Tangier disease and familial high-density lipoprotein deficiency. Nat Genet. 1999;22(4):336–345. doi: 10.1038/11905. [DOI] [PubMed] [Google Scholar]
- 46.Vaisman BL, et al. Endothelial expression of human ABCA1 in mice increases plasma HDL cholesterol and reduces diet-induced atherosclerosis. J Lipid Research. 2012;53(1):158–167. doi: 10.1194/jlr.M018713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ishibashi S, Brown MS, Goldstein JL, Gerard RD, Hammer RE, Herz J. Hypercholesterolemia in low density lipoprotein receptor knockout mice and its reversal by adenovirus-mediated gene delivery. J Clin Invest. 1993;92(2):883–893. doi: 10.1172/JCI116663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.van Eck M, et al. Leukocyte ABCA1 controls susceptibility to atherosclerosis and macrophage recruitment into tissues. Proc Natl Acad Sci U S A. 2002;99(9):6298–6303. doi: 10.1073/pnas.092327399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Aiello RJ, et al. Increased atherosclerosis in hyperlipidemic mice with inactivation of ABCA1 in macrophages. Arterioscler Thromb Vasc Biol. 2002;22(4):630–637. doi: 10.1161/01.ATV.0000014804.35824.DA. [DOI] [PubMed] [Google Scholar]
- 50.Meurs I, et al. Effects of deletion of macrophage ABCA7 on lipid metabolism and the development of atherosclerosis in the presence and absence of ABCA1. PLoS ONE. 2012;7(3):e30984. doi: 10.1371/journal.pone.0030984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Bi X, et al. Myeloid cell-specific ATP-binding cassette transporter A1 deletion has minimal impact on atherogenesis in atherogenic diet-fed low-density lipoprotein receptor knockout mice. Arterioscler Thromb Vasc Biol. 2014;34(9):1888–1899. doi: 10.1161/ATVBAHA.114.303791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Korns D, Frasch SC, Fernandez-Boyanapalli R, Henson PM, Bratton DL. Modulation of macrophage efferocytosis in inflammation. Front Immun. 2011;2:57. doi: 10.3389/fimmu.2011.00057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Rader DJ. Illuminating HDL--is it still a viable therapeutic target? N Engl J Med. 2007;357(21):2180–2183. doi: 10.1056/NEJMe0707210. [DOI] [PubMed] [Google Scholar]
- 54.Ono K. Current concept of reverse cholesterol transport and novel strategy for atheroprotection. J Cardiol. 2012;60(5):339–343. doi: 10.1016/j.jjcc.2012.07.014. [DOI] [PubMed] [Google Scholar]
- 55.Uppal H, et al. Activation of LXRs prevents bile acid toxicity and cholestasis in female mice. Hepatology. 2007;45(2):422–432. doi: 10.1002/hep.21494. [DOI] [PubMed] [Google Scholar]
- 56.Glogauer M, et al. Rac1 deletion in mouse neutrophils has selective effects on neutrophil functions. J Immunol. 2003;170(11):5652–5657. doi: 10.4049/jimmunol.170.11.5652. [DOI] [PubMed] [Google Scholar]
- 57.Su HP, et al. Identification and characterization of a dimerization domain in CED-6, an adapter protein involved in engulfment of apoptotic cells. J Biol Chem. 2000;275(13):9542–9549. doi: 10.1074/jbc.275.13.9542. [DOI] [PubMed] [Google Scholar]
- 58.Park D, et al. BAI1 is an engulfment receptor for apoptotic cells upstream of the ELMO/Dock180/Rac module. Nature. 2007;450(7168):430–434. doi: 10.1038/nature06329. [DOI] [PubMed] [Google Scholar]
- 59.Carpenter AE, et al. CellProfiler: image analysis software for identifying and quantifying cell phenotypes. Genome Biol. 2006;7(10):R100. doi: 10.1186/gb-2006-7-10-r100. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.