Abstract
Ischemic heart disease is the leading cause of heart failure. Both clinical trials and experimental animal studies demonstrate that chronic hypoxia can induce contractile dysfunction even before substantial ventricular damage, implicating a direct role of oxygen in the regulation of cardiac contractile function. Prolyl hydroxylase domain (PHD) proteins are well recognized as oxygen sensors and mediate a wide variety of cellular events by hydroxylating a growing list of protein substrates. Both PHD2 and PHD3 are highly expressed in the heart, yet their functional roles in modulating contractile function remain incompletely understood. Here, we report that combined deletion of Phd2 and Phd3 dramatically decreased expression of phospholamban (PLN), resulted in sustained activation of calcium/calmodulin-activated kinase II (CaMKII), and sensitized mice to chronic β-adrenergic stress–induced myocardial injury. We have provided evidence that thyroid hormone receptor-α (TR-α), a transcriptional regulator of PLN, interacts with PHD2 and PHD3 and is hydroxylated at 2 proline residues. Inhibition of PHDs increased the interaction between TR-α and nuclear receptor corepressor 2 (NCOR2) and suppressed Pln transcription. Together, these observations provide mechanistic insight into how oxygen directly modulates cardiac contractility and suggest that cardiac function could be modulated therapeutically by tuning PHD enzymatic activity.
Introduction
Hypoxia is associated with many disease conditions, including coronary artery disease (CAD) and myocardial infarction (MI). Despite improvements in the diagnosis and treatment of these cardiac pathologies, MI remains the leading cause of death and disability in the United States (1). During the acute phase of MI, blockage of a coronary artery results in a shortage of the oxygen and nutrition required for cellular metabolism, which eventually leads to irreversible myocardial cell death in the infarct area and impairment of cardiac contractile function. The reduction in contractile function is initially compensated by increased local secretion of catecholamines (2). However, the resulting sustained activation of the sympathetic nervous system, which is commonly observed in patients with MI and heart failure, promotes cardiac arrhythmia and left ventricular remodeling. This adverse remodeling includes cardiomyocyte apoptosis, cardiac hypertrophy, and contractile dysfunction (2, 3). In response to catecholamines, cardiac β-adrenergic receptors (β-ARs) activate both protein kinase A (PKA) and calcium/calmodulin-dependent kinase II (CaMKII) pathways. Blockage of the β-AR pathway with beta blockers is one of the most common and standard therapeutic approaches for patients with ST-segment elevation MI and heart failure (4–6). PKA and CaMKII pathways share many common substrates involved in excitation-contraction (E-C) coupling and Ca2+ cycling such as phospholamban (PLN) and sarcoplasmic/endoplasmic reticulum Ca2+ ATPase 2a (SERCA2a) (7). It is well documented that the canonical cAMP/PKA pathway plays a central role in cardiac E-C coupling and is responsible for the “fight-or-flight” response (4). In contrast, the CaMKII pathway is particularly prominent under chronic adrenergic stress conditions and promotes cardiomyocyte apoptosis, cardiac hypertrophy, and arrhythmia (8). Inhibition of CaMKII, through either pharmacological or genetic means, attenuates cardiomyocyte apoptosis, prevents arrhythmia, and protects the heart from adverse ventricular remodeling after MI (8, 9). Although it is well recognized that chronic hypoxia and β-adrenergic stress are 2 major risk factors for heart failure induced by MI, it remains largely unknown whether and how hypoxia cooperates with chronic β-AR stress to contribute to the adverse progression observed in ischemic cardiomyopathy.
Hypoxia can also induce a set of physiologically beneficial adaptive responses such as increased erythropoiesis, angiogenesis, and glycolysis to maintain oxygen homeostasis (10). A conserved pathway regulated by oxygen-dependent prolyl hydroxylation of hypoxia-inducible factor-α subunit (HIF-α) plays a crucial role in these processes (10). In mammals, there are 3 isoforms of HIF-α prolyl-4 hydroxylase, termed PHD1–3 (10). They belong to a superfamily of Fe+2 and 2-oxoglutarate–dependent dioxygenases (11, 12). Under normoxic conditions, 2 conserved proline residues of HIF-α are hydroxylated by prolyl hydroxylase domain (PHD) proteins, which promote HIF-α polyubiquitination and degradation via the proteasomal pathway (13, 14). Under hypoxic conditions, hydroxylation is inhibited, resulting in the accumulation of HIF-α protein and activation of the HIF pathway. Both PHD2 and PHD3 are highly expressed in the heart (15). Marked induction of HIF-α protein was observed in the region close to the infarct area in a rat model of MI (16). Interestingly, pretreatment with prolyl hydroxylase inhibitor or depletion of PHD2 or PHD3 attenuates myocardial injury induced by myocardial ischemia in several rodent models (16–20). It was suggested that the HIF pathway plays a central role in this cardioprotective effect (18, 21). However, concerns have also been raised over the safety of long-term PHD inhibition. It was recently reported that long-term depletion of both PHD2 and PHD3 induced many of the hallmarks of ischemic cardiomyopathy such as the loss of myocardial contractility and cardiac hypertrophy (22, 23). Although involvement of the HIF pathway was also implicated, the underlying mechanism remained largely unclear (22, 23). Nevertheless, these findings suggest that PHD2 and PHD3 may play a dual role in myocardial ischemia. Recently, we and other groups have identified a growing list of novel substrates of PHDs and demonstrated that prolyl hydroxylation mediated by PHDs is involved in a broad range of signaling pathways (24–26). Thus, we were intrigued and decided to investigate whether other potential substrates of PHD2 and PHD3 exist in the heart and whether PHD2 and PHD3 are directly involved in the regulation of cardiac contractile function.
Ca2+ cycling, which refers to the release and reuptake of intracellular Ca2+, drives cardiac sarcomere contraction and relaxation. With each beat of the heart, Ca2+ is released from the sarcoplasmic reticulum (SR) via cardiac ryanodine receptor 2 (RyR2), raising the cytoplasmic Ca2+ concentration and causing cardiac muscle contraction. To relax the muscles, Ca2+ is pumped back into the SR by SERCA2a, which lowers the cytoplasmic Ca2+ concentration. In addition, the association of PLN with SERCA2a decreases the apparent affinity of SERCA2a for Ca2+ and inhibits Ca2+ reuptake into the SR (27). Dysfunction of calcium regulators due to either genetic mutation or pathological stimuli, such as myocardial ischemia, is one of the major causes of heart failure (27, 28). In this report, we demonstrate that combined deletion of Phd2 and Phd3 dramatically decreases the expression of PLN, results in sustained activation of CaMKII, and sensitizes mice to myocardial injury induced by chronic β-AR stress. We provide evidence suggesting that thyroid hormone receptor-α (TR-α), a transcriptional regulator of Pln (29), interacts with PHD2 and PHD3 and can be hydroxylated at 2 proline residues. Hypoxia, which inhibits TR-α hydroxylation, increases the interaction between TR-α and nuclear receptor corepressor 2 (NCOR2) and suppresses Pln transcription. Thus, we reveal what we believe to be a novel role of PHD2 and PHD3 in cardiac function via regulation of PLN/Ca2+ cycling through the TR-α pathway.
Results
Depletion of PHD2/3 decreases PLN protein levels.
To understand the functional role of PHD2 and PHD3 in the heart, we obtained Phd2flox/flox (Phd2fl/fl) and Phd3flox/flox (Phd3fl/fl) mice (provided by Guohua Fong, University of Connecticut, Storrs, Connecticut, USA) (30) and CAG-Cre (Cre+/–) transgenic mice (on a C57BL/6 background; The Jackson Laboratory), which have a tamoxifen-inducible and Cre-mediated recombination system. After appropriate breeding, we generated Phd2/3fl/fl Cre+/– and Phd2/3fl/fl Cre–/– mice. To initiate the study, we deleted Phd2 and Phd3 by infusing mice i.p. with tamoxifen (40 mg/kg/d) for 5 consecutive days (Figure 1A). On the seventh day after the start of tamoxifen infusion, mouse hearts were harvested, and the protein levels of crucial cardiac Ca2+ cycling regulators were examined by immunoblot analysis. Surprisingly, although we observed no significant changes in heart rate or blood pressure (Supplemental Figure 1, A and B; supplemental material available online with this article; doi:10.1172/JCI80369DS1), we found that PLN protein levels in the hearts from Phd2/3fl/fl Cre+/– mice were significantly lower than those in Phd2/3fl/fl Cre–/– littermate control hearts (Figure 1, B and C). In addition, there was no significant change in SERCA2a or RyR2 protein levels in Phd2/3fl/fl Cre+/– hearts as compared with those in Phd2/3fl/fl Cre–/– mice (Figure 1B). The decrease in PLN protein levels was not a side effect of tamoxifen administration, since we observed no significant change in PLN protein levels in Phd2/3fl/fl mice infused with tamoxifen (Supplemental Figure 2A). Furthermore, we examined whether depletion of either PHD2 or PHD3 alone could alter PLN protein levels. As shown in Supplemental Figure 2B, depletion of PHD3 alone had no effect on PLN levels. In contrast, depletion of PHD2 moderately reduced PLN protein levels compared with combined depletion of PHD2 and PHD3 (Figure 1C and Supplemental Figure 2C), suggesting that PHD2 is the major regulator of PLN in the heart and that PHD3 may play a compensatory role in the absence of PHD2.
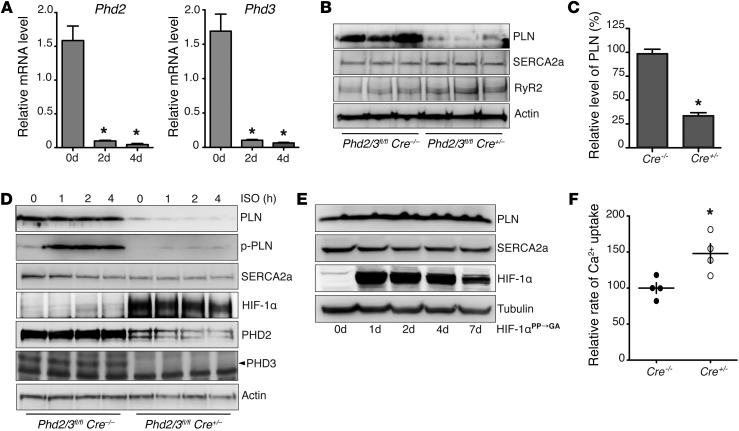
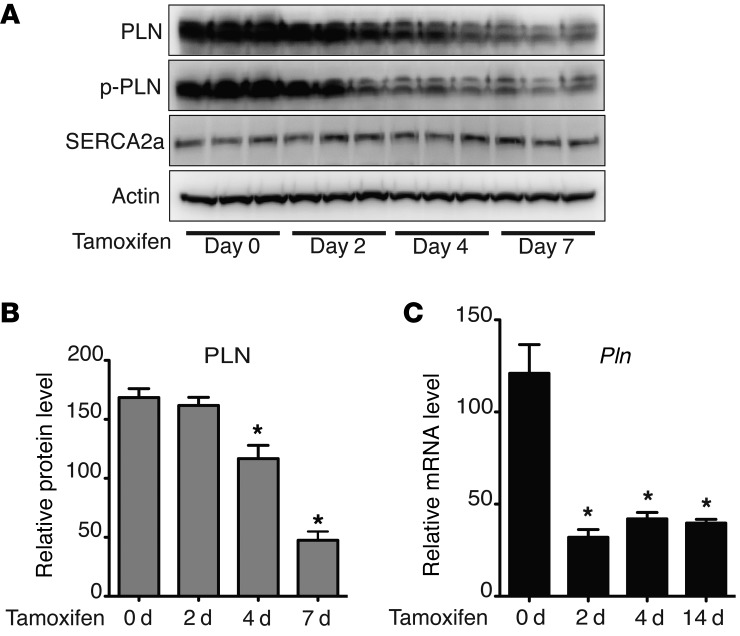
Figure 1. Depletion of PHD2/3 results in decreased PLN protein levels and increased SR Ca2+ uptake.
Phd2/3fl/flCre+/– mice were i.p. injected with tamoxifen once daily for 5 consecutive days. (A) Hearts were harvested on the indicated day after the first injection of tamoxifen, and real-time PCR was performed to determine the relative mRNA levels of Phd2 and Phd3. Phd2 and Phd3 mRNA levels were significantly decreased after 2 days of tamoxifen injection. n = 4, *P < 0.01, compared with day 0, 1-way ANOVA. (B and C) Seven days after the first injection of tamoxifen, mouse hearts were harvested, and Western blot analysis was performed. Densitometric analysis of PLN protein levels is shown in C. PLN, but not SERCA2a or RyR2, protein levels were significantly decreased. n = 6, *P < 0.01, 2-tailed Student’s t test. (D) Neonatal ventricular myocytes were isolated from Phd2/3fl/fl Cre+/– and Phd2/3fl/fl Cre–/– mice. After treatment with 4-OHT for 5 days, myocytes were treated with ISO for 0, 1, 2, or 4 hours. Western blot analysis was then performed. (E) After infection with adenovirus expressing normoxia-stable HIF-1α for 0, 1, 2, 4, and 7 days, neonatal rat ventricular myocytes were harvested, and Western blot analysis was performed. p-PLN, phosphorylated PLN. (F) SR Ca2+ uptake rates were measured using heart homogenates from Phd2/3fl/fl Cre+/– and Phd2/3fl/fl Cre–/– mice after treatment with tamoxifen for 7 days. The Ca2+ uptake rate was increased in mice lacking PHD2 and PHD3. n = 4, *P < 0.05, 2-tailed Student’s t test.
To avoid potential complications raised by using the global KO system, we isolated neonatal ventricular cardiomyocytes from Phd2/3fl/fl Cre+/– and Phd2/3fl/fl Cre–/– mice. Isolated cardiomyocytes were then treated with 4-hydroxyl-tamoxifen (4-OHT) for 5 days to deplete PHD2 and PHD3. Consistently, depletion of both PHD2 and PHD3 in primary cardiomyocytes also dramatically decreased the protein levels of PLN and had no effect on SERCA2a protein levels (Figure 1D). Not surprisingly, the phosphorylation of PLN at Thr17 induced by the β-AR agonist isoproterenol (ISO) was also decreased accordingly (Figure 1D). In addition, overexpression of either PHD2 or PHD3 was sufficient to restore PLN protein levels in Phd2/3-null cardiomyocytes, further suggesting the redundant role of PHD2 and PHD3 in regulating PLN protein levels in the heart (Supplemental Figure 2D).
It is well documented that depletion of PHD2 and PHD3 stabilizes HIF-α protein levels (23). Consistently, we observed a dramatic increase in HIF-1α protein levels in Phd2/3fl/fl Cre+/– cardiomyocytes as compared with those in Phd2/3fl/fl Cre–/– controls (Figure 1D), suggesting the successful depletion of PHD2 and PHD3. Since hundreds of genes are transcriptionally regulated by the HIF pathway, it is interesting to examine whether the HIF pathway is involved in PLN regulation. To test this hypothesis, we activated the HIF pathway in primary cardiomyocytes by infecting them with adenovirus expressing normoxia-stable HIF-1α (HIF-1αPP→GA) (24). As shown in Figure 1E, overexpression of HIF-1αPP→GA had no effect on protein levels of PLN or SERCA2a in neonatal rat ventricular cardiomyocytes, suggesting that an HIF-independent mechanism was involved in the regulation of PLN protein levels.
It has been documented that an increase in SERCA2a-to-PLN stoichiometry in cardiomyocytes leads to an increase in the SR Ca2+ uptake rate (31). As expected, we demonstrated that the Ca2+ uptake rate of crude heart homogenates from Phd2/3fl/fl Cre+/– mice was significantly higher than that of their littermate controls (Figure 1F). Taken together, our data show that depletion of PHD2 and PHD3 dramatically decreases PLN protein levels through an HIF-independent pathway, resulting in an increase in SR Ca2+ uptake rates.
Depletion of PHD2/3 results in sustained activation of CaMKII and promotes cardiomyocyte apoptosis induced by chronic treatment with ISO.
Previous reports detail that depletion of PLN leads to increased SR Ca2+ load and promotes cardiomyocyte apoptosis induced by ISO treatment through the CaMKII pathway (31, 32). Inhibition of CaMKII significantly decreased cardiomyocyte apoptosis induced by ISO in Pln-KO mice in vivo (31). Consistently, we observed a significant increase in SR Ca2+ load in cardiomyocytes isolated from Phd2/3-KO mice (Supplemental Figure 3A). In addition, we observed increased activation of CaMKII in hearts from Phd2/3fl/fl Cre+/– mice upon treatment with ISO (Figure 2A). It is well documented that CaMKII-dependent RyR phosphorylation increases RyR open probability and spontaneous SR Ca2+ release (or Ca2+ spark) (33, 34). Indeed, we demonstrated that the Ca2+ spark frequency was significantly increased in Phd2/3-null cardiomyocytes under basal conditions and after ISO treatment (Supplemental Figure 3, B and C). To determine whether depletion of PHD2 and PHD3 promoted cardiomyocyte apoptosis induced by chronic treatment with ISO, male Phd2/3fl/fl Cre+/– and Phd2/3fl/fl Cre–/– mice were injected i.p. with tamoxifen once daily for 5 days, followed by ISO infusion with a miniosmotic pump for 2 days. We then performed TUNEL staining to assess apoptosis in myocardium. As expected, we found that extensive apoptosis was evident in the myocardium of Phd2/3fl/fl Cre+/– mice, but not in the myocardium of Phd2/3fl/fl Cre–/– mice after ISO treatment (Figure 2, B and C). To further confirm that depletion of PHD2 and PHD3 sensitized cardiomyocytes to ISO-induced apoptosis, we isolated neonatal mouse ventricular myocytes (NMVMs) from Phd2/3fl/fl mice and infected them with Ad-Cre to deplete PHD2 and PHD3. Consistent with our results described above, deletion of Phd2 and Phd3 substantially increased NMVM apoptosis induced by ISO (Figure 2, D and E). It is worth noting that depletion of PHD2 and PHD3 had no effect on NMVM apoptosis under basal conditions (Figure 2, D and E). In summary, these results indicated that depletion of PHD2 and PHD3 leads to increased activation of CaMKII and promotes cardiomyocyte apoptosis induced by chronic β-AR stimulation.
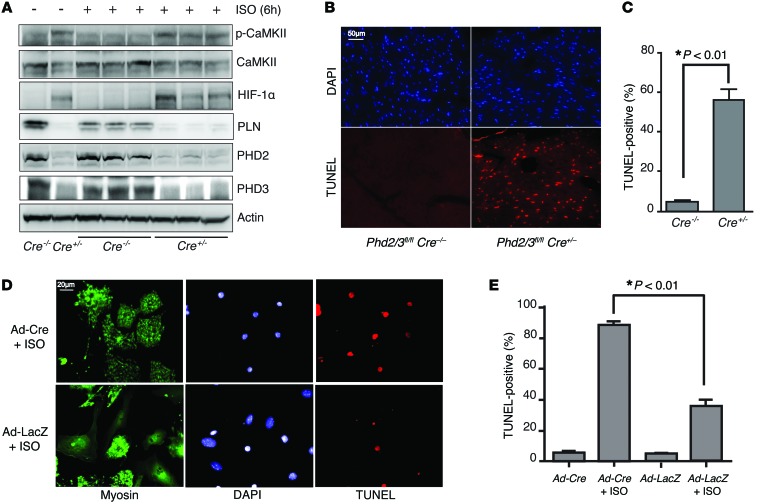
Figure 2. Depletion of PHD2/3 leads to abnormal activation of CaMKII and increases cardiomyocyte apoptosis induced by ISO.
(A) Mice lacking PHD2 and PHD3 have elevated CaMKII activation compared with WT mice. Phd2/3fl/fl Cre+/– and Phd2/3fl/fl Cre–/– mice were i.p. injected with tamoxifen once daily for 5 days. Mice were then treated with ISO or PBS for 6 hours on day 7 after the first tamoxifen injection. Heart lysates were immunoblotted with the indicated antibodies. Each lane represents heart lysate from 1 mouse. (B and C) Deletion of PHD2 and PHD3 potentiates ISO-induced apoptosis in cardiomyocytes. Male Phd2/3fl/fl Cre+/– and Phd2/3fl/fl Cre–/– mice were i.p. injected with tamoxifen once daily for 5 days, followed by ISO infusion with miniosmotic pumps (20 mg/kg/d) for 2 days. Hearts were harvested and fixed for H&E or TUNEL staining (B). Graph in C shows quantitative analyses. n = 5, P < 0.01, 2-tailed Student’s t test. (D and E) Primary isolated cardiomyocytes deficient in PHD2 and PHD3 were also sensitized to ISO-induced apoptosis. Phd2/3fl/fl cardiomyocytes were isolated and infected with Ad-Cre or LacZ for 2 days and then treated with PBS or ISO for another 24 hours. Myosin and TUNEL staining was then performed. Representative images are shown in D. Quantitative analysis of TUNEL staining from 3 experiments is shown in E. *P < 0.01, 2-way ANOVA. Scale bars: 50 μm (B), 20 μm (D).
Depletion of PHD2 and PHD3 exaggerates myocardial injury and potentiates cardiac hypertrophy induced by chronic β-adrenergic stress.
It was reported that increased contractility induced by PLN ablation in mouse heart significantly increased ischemic energy demand and exaggerated myocardial injury induced by ischemia (35). Since we have demonstrated that depletion of PHD2 and PHD3 results in a significant decrease in PLN protein levels and that chronic β-adrenergic stress plays an essential role in adverse ventricular remodeling including cardiac hypertrophy after MI (4), we decided to test whether depletion of PHD2 and PHD3 exacerbates myocardial injury induced by chronic β-AR stimulation. To examine the in vivo response of Phd2/3-null mice to chronic β-AR stimulation, we deleted Phd2 and Phd3, as described above, then administered ISO via miniosmotic pumps for 7 days. Cardiac function was then evaluated by ECG. We observed no significant change in cardiac function 2 weeks after Phd2 and Phd3 deletion (Figure 3A), which is consistent with a previous report suggesting that short-term depletion of PHD2 and PHD3 has no effect on cardiac function (23). ECG analyses indicated that infusion of ISO for 7 days significantly impaired cardiac function in female Phd2/3fl/fl Cre+/– mice compared with that observed in their littermate controls (Figure 3, B and C). Notably, most of the male Phd2/3fl/fl Cre+/– mice died within 4 days after ISO infusion (Figure 3D), and severe cardiac arrhythmia might have been responsible for their death (Figure 3E). Furthermore, we found that cardiac hypertrophy induced by ISO was also significantly increased in Phd2/3-null mice compared with their littermate controls (Figure 4, A and B), and the cardiomyocyte cross-sectional area was also significantly increased by PHD2/3 depletion (Figure 4, C and D).
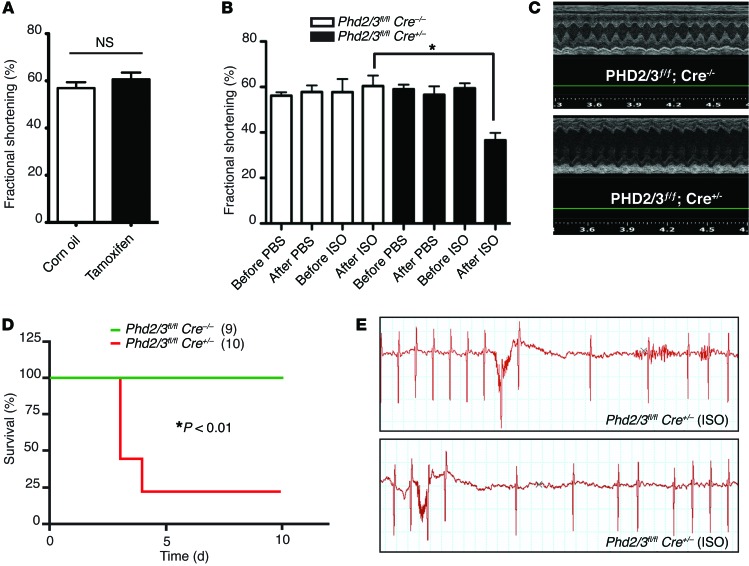
Figure 3. Depletion of PHD2/3 exaggerates myocardial injury induced by chronic treatment with ISO.
(A) Short-term deletion of Phd2 and Phd3 had no significant effect on cardiac function. ECG analyses of Phd2/3fl/fl Cre+/– mice were performed 2 weeks after day 1 of i.p. injection of tamoxifen or corn oil control (n = 5/group). NS, 2-tailed Student’s t test. (B and C) Deletion of PHD2 and PHD3 exacerbated cardiac dysfunction induced by ISO in female mice. Female Phd2/3fl/fl Cre+/– and Phd2/3fl/fl Cre–/– mice were i.p. injected with tamoxifen once daily for 5 days, followed by PBS or ISO infusion with miniosmotic pumps (20 mg/kg/d) for 7 days, and cardiac function was measured by ECG. Quantitative analysis of fractional shortening is shown in B (n = 5/group). *P < 0.05, 2-way ANOVA. Representative M-mode ECGs of Phd2/3fl/fl Cre+/– and Phd2/3fl/fl Cre–/– mice after 7 days’ treatment with ISO are shown in C. (D and E) Survival of male Phd2- and Phd3-null mice was significantly lower than that of WT mice. Male Phd2/3fl/fl Cre+/– and Phd2/3fl/fl Cre–/– mice were i.p. injected with tamoxifen once daily for 5 days, followed by infusion of PBS or ISO for 7 days. Kaplan-Meier survival curves for mice with the indicated genotypes are shown in D. *P < 0.01, log-rank test. Representative ECGs with severe cardiac arrhythmias observed in male Phd2/3fl/fl Cre+/– mice are shown in E.
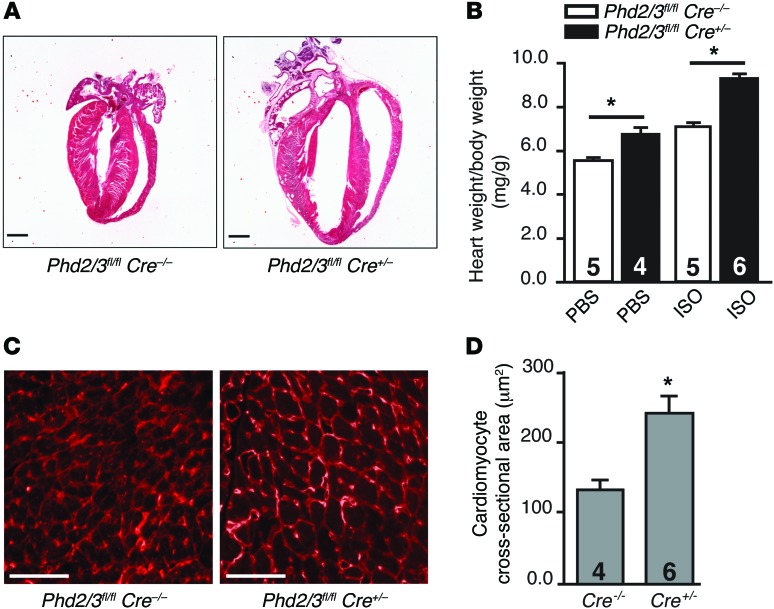
Figure 4. Depletion of PHD2/3 potentiates cardiac hypertrophy.
Phd2/3fl/flCre+/– and Phd2/3fl/fl Cre–/– mice were infused with tamoxifen for 5 consecutive days. Miniosmotic pumps were then implanted to chronically deliver PBS or ISO (20 mg/kg/d) for another 7 days. (A) Hearts were fixed and stained with H&E. Representative heart images of Phd2/3fl/fl Cre+/– and Phd2/3fl/fl Cre–/– mice treated with ISO are shown. The chamber size in Phd2- and Phd3-null hearts was dramatically increased compared with that in WT hearts. (B) The heart weight/body weight ratio is shown for the indicated group. n = 4–6 mice/group, *P < 0.05, 2-way ANOVA. (C and D) Representative cross sections of left ventricles from Phd2/3fl/fl Cre–/– and Phd2/3fl/fl Cre+/– mice treated with ISO and stained with WGA showed increased cardiomyocyte size in Phd2- and Phd3-null mice compared with that in WT mice (C). Cross-sectional areas of cardiomyocytes are shown in D. n = 4 or 6 mice/group, *P < 0.02, 2-tailed Student’s t test. Scale bars: 50 μm.
To better understand the processes and pathways affected by the loss of PHD2 and PHD3 in the heart, we surveyed genome-wide transcriptional changes in Phd2/3 double-KO hearts by microarray analysis (Supplemental Figure 4). We identified 2,500 differentially expressed genes including numerous cardiac contractile genes (Supplemental Table 1). Likewise, pathway enrichment analysis identified glycolytic and gluconeogenic pathways as well as fatty acid β oxidation and apoptosis (Supplemental Table 2).
Taken together, we conclude that depletion of PHD2/3 increases the myocardial damage and cardiac arrhythmia induced by chronic β-AR stimulation. Since chronic β-AR stress is commonly observed in patients with MI, these data suggest that hypoxia, a condition under which PHD2 and PHD3 enzymatic activity is inhibited, may cooperate with β-AR stress, leading to exaggerated myocardial dysfunction and cardiac arrhythmia after MI.
Pln transcription is decreased by both hypoxia and deletion of Phd2 and Phd3.
We observed a dramatic decrease in PLN protein levels in Phd2/3-null hearts (Figure 1, B and C). However, the underlying mechanism remained to be determined. To investigate the mechanisms by which depletion of PHD2 and PHD3 decreases PLN protein levels, we depleted PHD2 and PHD3 in mice as described above and harvested the hearts at 0, 2, 4, 7, or 14 days after the initial tamoxifen infusion. PLN protein and mRNA levels were then analyzed by Western blotting and real-time PCR, respectively. As shown in Figure 5, A and B, PLN protein levels decreased gradually with time, reaching the lowest level by day 7. In contrast, Pln mRNA levels reached their lowest levels by day 2 and maintained this low level even at day 14 (Figure 5C). We observed no significant change in SERCA2a protein or mRNA levels (Figure 5A and Supplemental Figure 5A). The increased expression of PDK1 (an HIF target gene) on day 2 suggests an activation of the HIF pathway and also confirms the depletion of PHD2 and PHD3 (Supplemental Figure 5B). Taken together, these data suggest that the decrease in PLN protein levels observed in Phd2/3-null hearts is caused by the suppression of Pln transcription. The slower rate of reduction of PLN protein levels compared with that of Pln mRNA suggests that PLN protein may have a long half-life in the heart in vivo.
Figure 5. Dynamic regulation of PLN expression by depletion of PHD2/3.
Phd2 and Phd3 deletion induced a reduction of both PLN protein and Pln mRNA transcript levels. On days 0, 2, 4, 7, or 14 after tamoxifen infusion, Phd2/3fl/fl Cre+/– mouse hearts were harvested for Western blot (A) (each lane represents 1 heart) and real-time PCR (C) analyses. PLN protein levels were reduced in the absence of PHD2 and PHD3 compared with those in untreated mice. Densitometric analysis of PLN protein levels is shown in B. n = 4, *P < 0.05, compared with day 0, 1-way ANOVA. (C) Real-time PCR was performed to determine Pln mRNA levels in the hearts. n = 4, *P < 0.05, compared with day 0, 1-way ANOVA.
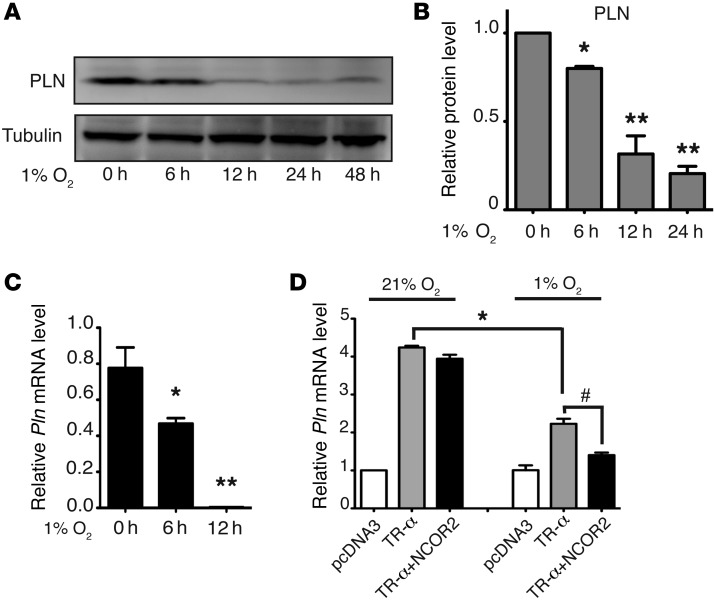
The enzymatic activity of PHD requires oxygen and is thereby inhibited under hypoxic conditions (36). To determine whether PHD2 and PHD3 enzymatic activity is required for decreased expression of PLN, we cultured neonatal rat ventricular cardiomyocytes under hypoxic conditions (1% O2) to inhibit PHD enzymatic activity. Cells were then harvested at the indicated time points, and PLN protein levels were examined via Western blot analysis. As shown in Figure 6, A and B, hypoxia significantly decreased PLN protein levels. In addition, the prolyl hydroxylase inhibitor dimethyloxaloylglycine (DMOG) also dramatically decreased PLN protein levels in primary cardiomyocytes (Supplemental Figure 6). Furthermore, we demonstrated that Pln mRNA levels were also significantly decreased under hypoxic conditions (Figure 6C), suggesting that hypoxia may suppress the transcription of Pln in primary cardiomyocytes in culture. To further confirm the suppressive effect of hypoxia on Pln transcription, we performed a dual-luciferase assay using a reporter construct containing the core promoter region of the Pln gene in HL-1 cardiomyocytes. It has been reported that there are several thyroid hormone response elements (TREs) on the core promoter region of the Pln gene and that TR-α, the major isoform expressed in cardiomyocytes, directly binds to the Pln core promoter (29). Consistent with these reports, we found that overexpression of TR-α dramatically increased Pln reporter activity and that reporter activity induced by TR-α could be significantly inhibited by hypoxia (Figure 6D). Taken together, these results suggest that loss of prolyl hydroxylase activity, due to either Phd2/3 depletion or to hypoxia, is crucial for the transcriptional repression of Pln and that PHD2 and PHD3 may regulate PLN expression through the TR-α pathway.
Figure 6. Hypoxia decreases PLN protein levels and suppresses Pln transcription.
Neonatal rat ventricular myocytes were treated with hypoxia (1% O2) as indicated. Cells were then harvested for protein or mRNA analysis. (A and B) Western blot analyses were performed with the indicated antibodies (A). Densitometric analysis from 3 experiments is shown in B. *P < 0.05; **P < 0.01, compared with 0 hours, 1-way ANOVA. (C) Real-time PCR was performed to analyze relative Pln mRNA levels. n = 3, *P < 0.05; **P < 0.01, compared with 0 hours, 1-way ANOVA. (D) HL-1 cardiomyocytes were transfected with a pGL3-Pln promoter (–156 to +64) luciferase construct, together with pcDNA3, or with a construct expressing TR-α or NCOR2, as indicated. Cells were also cotransfected with a construct expressing Renilla luciferase as the internal control. Twenty-four hours after transfection, cells were cultured under 21% or 1% O2 conditions for 24 hours. Cells were then harvested and luciferase activity determined with a luminometer. Relative luciferase activity was calculated from 3 separate experiments. *P < 0.05; #P < 0.01, 2-way ANOVA.
Hydroxylation of TR-α at P160 regulates its transcriptional activity by blocking the recruitment of NCOR2.
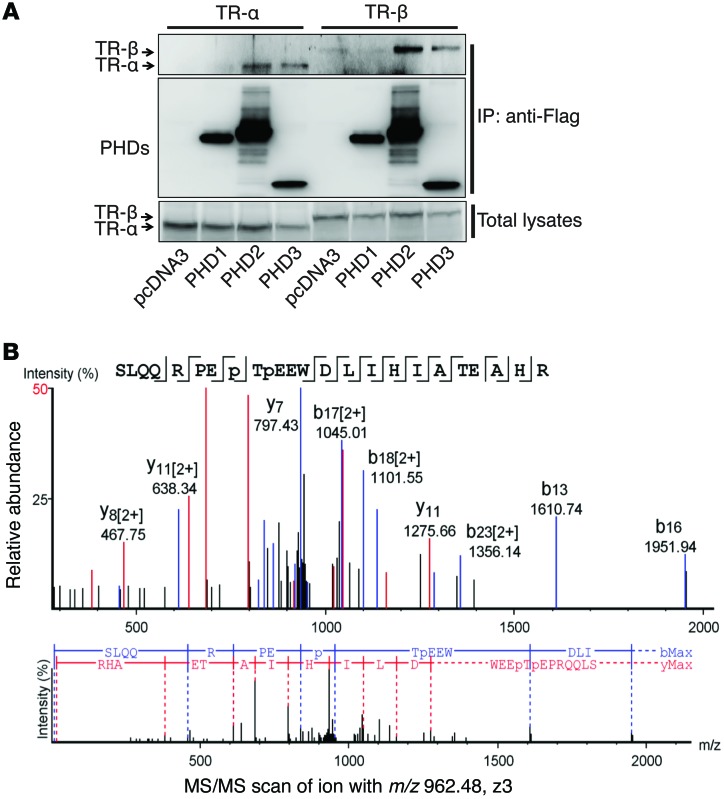
A growing body of evidence suggests that prolyl hydroxylation mediated by PHD is involved in a variety of signaling pathways (24–26). We demonstrate here that both PHD2 and PHD3, but not PHD1, interacted with TR-α (Figure 7A). Therefore, we hypothesized that TR-α might be a novel substrate of PHD2 and PHD3 and that the hydroxylation of TR-α might play an important role in regulating TR-α transcriptional activity. To confirm this hypothesis, we first examined TR-α prolyl hydroxylation by LTQ Orbitrap Elite mass spectrometry. Flag-TR-α was exogenously overexpressed in HEK293 cells and purified with Flag resin. An in vitro hydroxylation assay was then performed with recombinant PHD2 and PHD3. Mass spectrometric analysis revealed that 2 proline residues of TR-α, P160 and P162, were hydroxylated in the presence of PHD2 and PHD3 (Figure 7B and Supplemental Figures 7 and 8).
Figure 7. TR-α is hydroxylated at proline residues P160 and P162.
(A) HL-1 cells were transfected with constructs expressing TR-α or TR-β, together with constructs expressing Flag-PHD1, -2, or -3, as indicated. Co-IP was performed with anti-Flag resin, and Western blot experiments were performed with the indicated antibodies. Both PHD2 and PHD3, but not PHD1, were able to pull down TR-α or TR-β. Representative blots from 3 experiments are shown. (B) An in vitro hydroxylation assay was performed with Flag–TR-α and PHD2/3. The protein band corresponding to TR-α was cut out for trypsin digestion. LC-MS/MS analysis was then performed. Tandem mass spectra of the precursor ion at m/z = 962.48 (Z = 3) for the human TR-α 153-176 sequence SLQQRPEP(+15.99)TP(+15.99)EEWDLIHIATEAHR are shown. The peak heights are the relative abundances of the corresponding fragment ions, with annotation of the identified matched N terminus–containing ions (b ions) shown in blue and C terminus–containing ions (y ions) shown in red. For clarity, only the major identified peaks are labeled (a complete table of fragment ions is presented in Supplemental Figure 3). Fragment ions at m/z = 952.66 (b8) and m/z = 1024.01 (y17)2+ represent characteristic ions that unambiguously identified P160–P162 double hydroxylation.
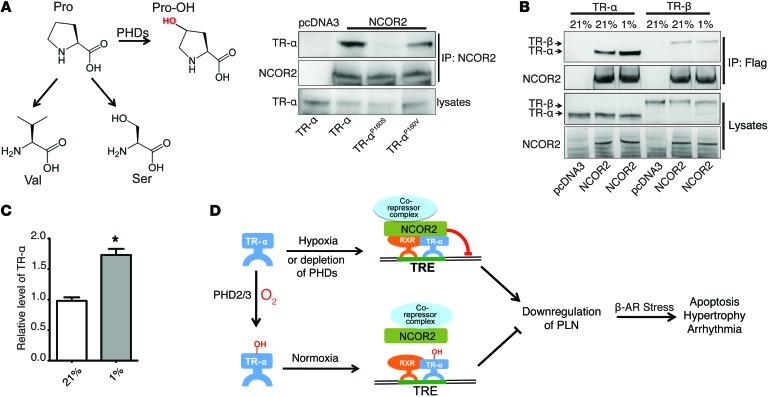
Posttranslational modifications are able to alter the biophysical properties of a protein by the addition of a chemical group to one or more amino acid residues, resulting in changes such as enzymatic activity or protein-protein interaction (37). Prolyl hydroxylation, which is expected to increase the polarity of proline residues, has been shown to regulate protein degradation and signaling transduction by altering various protein-protein interactions (13, 24, 25). It has been reported that TR-α is able to recruit NCOR2 to the promoter region of its responsive genes and suppress their transcription, and P160 of TR-α is crucial for its interaction with NCOR2 (38). Consistent with this finding, we observed that replacing P160 of TR-α with different classes of amino acids altered its binding affinity for NCOR2. Replacing P160 of TR-α with serine, a polar amino acid, dramatically decreased the interaction between TR-α and NCOR2 (Figure 8A). In contrast, replacing P160 of TR-α with valine, a hydrophobic amino acid, only moderately changed its interaction with NCOR2 (Figure 8A). This result suggests that a polar side chain of the residue at position 160 of TR-α decreases the binding affinity between TR-α and NCOR2. Since hydroxyl proline contains a polar hydroxyl group, we hypothesized that prolyl hydroxylation of TR-α P160 might inhibit its interaction with NCOR2. To test this, we cultured HEK293 cells ectopically expressing both TR-α and NCOR under normoxic (21% O2) and hypoxic (1% O2) conditions, in which we found that hypoxia significantly increased the amount of TR-α that co-IP with NCOR2, suggesting that inhibition of prolyl hydroxylation by hypoxia potentiates the interaction between TR-α and NCOR2 (Figure 8, B and C). Furthermore, we demonstrated that the interaction between endogenous TR-α and NCOR2 was also increased in Phd2/3-null heart (Supplemental Figure 9A). Taken together with our results demonstrating that hypoxia significantly potentiated the inhibitory effect of NCOR2 on Pln reporter activity induced by TR-α (Figure 6D), this suggests that hypoxia can increase the recruitment of NCOR2 to the core promoter region of the Pln gene. To further confirm this hypothesis, we infected neonatal ventricular myocytes with lentivirus expressing TR-αP160V, which cannot be hydroxylated but still binds to NCOR2 (Figure 8A). As shown in Supplemental Figure 9B, TR-αP160V dramatically decreased PLN protein levels, even under normoxic conditions, and hypoxia had no further suppressive effect on PLN. Although PHD2 and PHD3 also associated with TR-β (Figure 7A), hypoxia had no effect on the interaction between TR-β and NCOR2 (Figure 8B). Protein sequence alignment revealed a conserved PEP motif in TR-α, however, the upstream sequence is almost completely different from that of TR-α (Supplemental Figure 10). Therefore, it is possible that either the 2 conserved proline residues of TR-β cannot be hydroxylated by PHD2 or PHD3, or these residues may not be crucial for TR-β and NCOR2 interaction. In summary, we found that P160 and P162 of TR-α can be hydroxylated by PHD2 and PHD3 and that the inhibition of PHD-mediated TR-α hydroxylation at the P160 residue, either by hypoxia or depletion of PHD2 and PHD3, increases recruitment of NCOR2 to the promoter region of Pln, resulting in transcriptional suppression of Pln (Figure 8D).
Figure 8. Hypoxia increases the interaction between TR-α and NCOR2.
(A) Replacement of the P160 residue of TR-α alters NCOR2-binding affinity. Molecular structures of proline, 4-hydroxy-proline, valine, and serine are shown. HEK293 cells were transfected with TR-α, TR-αP160S, or TR-αP160V, together with pcDNA3 or NCOR2, as indicated, followed by IP with NCOR2. Western blot analyses were then performed with the indicated antibodies. (B and C) Hypoxia potentiates the interaction between NCOR2 and TR-α. HEK293 cells cultured under 21% O2 conditions were transfected with constructs expressing TR-α or TR-β, together with pcDNA3 or NCOR2, as indicated, and were cultured under 21% or 1% O2 conditions for 8 hours. Flag-NCOR2 was IP, and Western blotting was performed with the indicated antibodies. Densitometric analysis of the IP TR-α is shown in C. n = 3, *P < 0.01, 2-tailed Student’s t test. (D) Schematic illustrating that PLN downregulation in the heart by hypoxia or PHD2 and PHD3 depletion contributes to myocardial injury induced by chronic β-AR stress. PHD2- and PHD3-mediated hydroxylation of TR-α blocks recruitment of the transcriptional repressor NCOR2 to the promoter region of Pln, resulting in transcription of Pln. Inhibition of TR-α hydroxylation, either through PHD2 and PHD3 depletion or through hypoxia, leads to increased recruitment of NCOR2 and suppression of Pln transcription. This decrease in PLN expression exacerbates cardiomyocyte apoptosis, cardiac hypertrophy, and arrhythmia induced by chronic β-AR stress.
Discussion
In this study, we demonstrate that TR-α is a hydroxylation substrate of PHD2 and PHD3. Inhibition of PHD-mediated hydroxylation on P160 of TR-α increases the interaction of TR-α with NCOR2, resulting in transcriptional suppression of Pln in cardiomyocytes. Decreased expression of PLN in the heart eventually leads to abnormal CaMKII activation, which potentiates the cardiomyocyte apoptosis, hypertrophy, and arrhythmia observed in our Phd2- and Phd3-null mice under chronic β-AR stress. Thus, our findings provide insight into the pathogenesis of ischemic heart disease.
Ischemic heart disease accounts for approximately 70% of all cases of heart failure and remains the leading cause of mortality in the United States (39). Stenotic plaques formed in the wall of coronary arteries reduce the perfusion of myocardium downstream of the stenosis, resulting in myocardial ischemia. Unstable plaques may rupture and induce thrombosis, which may lead to complete occlusion of a coronary artery and, ultimately, to MI (40). Notably, clinical trials and registries demonstrate that over 20% of patients with acute MI had already been showing symptoms of heart failure at the time of their presentation to the hospital (41), suggesting that myocardial ischemia may be sufficient to induce contractile dysfunction even before substantial ventricular damage has occurred. This concept is further supported by a study using genetically modified mice, in which reversible myocardial ischemia without overt infarction and cardiomyocyte death can be induced with a tunable transgene that encodes a vascular endothelial growth factor trap (42). It was demonstrated that chronic myocardial ischemia leads to metabolic reprogramming, mitophagy, and systolic dysfunction. Importantly, the degree of myocardial dysfunction is linearly related to the degree of myocardial ischemia (42), suggesting a more direct role of tissue oxygen tension in ventricular maladaptive remodeling.
Molecular oxygen plays a key role in energy metabolism and is essential for survival in multicellular organisms. An inadequate supply of oxygen will trigger a series of HIF-mediated adaptive responses (43). Activation of the HIF pathway can activate a large number of genes involved in the regulation of neovascularization, vascular remodeling, and metabolic reprogramming to combat hypoxia-induced damage. Partial depletion of HIF-1α significantly reduces the recovery of blood flow and exacerbates tissue damage after femoral artery ligation in Hif1a+/– mice (44). HIF-1α is also required to maintain oxygen homeostasis during compensatory myocardial hypertrophy in response to pressure overload (45). Rapidly increasing cardiac muscle mass leads to hypoxia in the myocardium. This in turn activates the HIF-1 pathway and promotes cardiac angiogenesis to ensure adequate oxygen delivery. Depletion of HIF-1α impairs this adaptive angiogenesis and results in the accelerated onset of heart failure induced by chronic transverse aortic constriction (45), further suggesting the cardioprotective effect of the HIF-mediated adaptive response. It is well documented that PHD family proteins play a central role in regulating the HIF pathway and are well recognized as oxygen sensors in metazoans (10). These dioxygenases require oxygen as a cosubstrate to catalyze the prolyl hydroxylation of protein substrates. HIF-1α was the first identified substrate of PHDs (15). Prolyl hydroxylation of HIF-1α facilitates its interaction with von Hippel-Lindau (VHL) E3 ligase complex and subsequent polyubiquitination, leading to proteasomal degradation. When oxygen availability is limited, PHD enzymatic activity will be inhibited, resulting in stabilization of HIF-1α protein and activation of the HIF-1 pathway (10). Therefore, it is not surprising that pretreatment with prolyl hydroxylase inhibitor or depletion of PHD2, the major prolyl hydroxylator of HIF-α, activates the HIF pathway and elicits HIF-mediated adaptive responses before the onset of hypoxia, attenuating cardiac injury induced by subsequent myocardial ischemia (16–19). However, it was recently reported that long-term depletion of both PHD2 and PHD3 in the heart resulted in myocardial dysfunction and produced many hallmarks of ischemic cardiomyopathy (23). Consistent with this, we have demonstrated here that the combined deletion of Phd2 and Phd3 potentiates cardiomyocyte apoptosis, hypertrophy, and cardiac arrhythmia induced by chronic β-AR stress, which is commonly observed in patients with MI. This suggests that the inhibition of PHD enzymatic activity under hypoxic conditions may be the driving force of the maladaptive ventricular remodeling induced by myocardial ischemia. The next question is to determine the underlying molecular mechanisms by which loss of PHD activity induces myocardial dysfunction. Notably, it was shown that long-term transgenic overexpression of normoxia-stable mutants of HIF-1α or HIF-2α leads to cardiomyopathy (23, 46). It was suggested that HIF-mediated metabolic reprogramming and angiogenesis might contribute to cardiac dysfunction induced by forced activation of the HIF pathway. However, it remains largely unclear how these HIF-mediated adaptive responses eventually lead to maladaptive ventricular remodeling (23). In addition, transgenic overexpression of normoxia-stable HIF-1α or HIF-2α may not be a faithful model of ischemic disease. Although it may be challenging, further genetic studies involving deletion of HIF-1α or HIF-2α are clearly necessary to determine the detrimental role of the HIF pathway in the context of PHD depletion.
Abnormal Ca2+ cycling in cardiomyocytes is well recognized as a major contributor to impaired contractile function and cardiac arrhythmia in both humans and experimental animal models of heart failure (27). During each cycle of cardiac contraction and relaxation, Ca2+ is released from the SR through cardiac RyR2, increasing cytosolic Ca2+ concentrations and resulting in cardiac muscle contraction. Calcium is then pumped back into the SR through cardiac SERCA2a, lowering cytosolic Ca2+ concentrations and causing muscle relaxation (47). The activity of SERCA2a is negatively regulated by PLN, which binds to SERCA2a and inhibits its Ca2+ pump activity (48). Although the physiological role of PLN is well established, the pathogenic role of PLN in cardiac contractile dysfunction is conflicting, possibly due to the use of different human disease and experimental animal models. Depletion of PLN in mice improves SR Ca2+ uptake and enhances cardiac contractile function without development of heart failure (49). In addition, depletion of PLN rescues the defects in contractility seen in muscle-specific LIM protein–KO (MLP-KO) mice and calsequestrin transgenic mice (50, 51). In contrast, humans lacking PLN develop lethal dilated cardiomyopathy at an early age (52), suggesting a significant difference in cardiac physiology and Ca2+ cycling regulation between human and mouse hearts. Mouse hearts beat approximately 600 times per minute, operating close to their theoretical maximum. The enhanced inotropic and lusitropic effects induced by the loss of PLN in mouse heart may be less deleterious than in human heart (52). Furthermore, other pathophysiological conditions may also contribute to the potential pathogenic role of PLN depletion. It was shown that persistently increased contractility induced by PLN ablation in mouse heart significantly increased the ischemic energy demand and exaggerated myocardial injury induced by cardiac ischemia (35). Depletion of PLN also results in Ca2+ overload in the SR, leading to abnormal activation of CaMKII, and promotes myocardial injury induced by the β-agonist ISO (31). Our finding that Phd2 and Phd3 deletion decreases PLN expression provides, to our knowledge, the first genetic evidence suggesting that PLN may be downregulated in ischemic heart tissue. If our hypothesis is indeed correct, then during myocardial infarction, when local levels of catecholamine are high and PLN expression is decreased due to hypoxia, the result would be a “perfect storm” for a negative feedback loop in the remaining viable myocardium, synergistically contributing to the further deterioration of myocardial function.
Since long-term depletion of PLN in the mouse heart has no adverse effects on cardiac function, it remains unclear whether decreased expression of PLN in Phd2/3-null mice contributes to the long-term defects in cardiac function observed in the Phd2/3-null mice under normal, unchallenged conditions. It is possible that persistently increased contractility and energy expenditure induced by the loss of PLN may not be favorable for overall cardiac function because of the altered cardiac energy metabolism induced by the HIF pathway in Phd2/3-null mice (23). Other potential substrates or pathways regulated by PHD2 and PHD3 are also likely to exist in the heart and contribute to myocardial dysfunction. Nevertheless, considering the deleterious effect of loss of PLN in human dilated cardiomyopathy, further research is required to examine whether this oxygen-dependent PHD2/3 negative regulation of PLN expression is conserved in humans. These potential findings could provide novel insights and therapeutic strategies to combat ischemic heart disease and heart failure.
Methods
Animals and tamoxifen injection.
Phd2fl/fl and Phd3fl/fl mice were obtained from Guohua Fong (University of Connecticut, Storrs, Connecticut, USA) and described previously (30). The CAG-Cre/Esr1 (Cre+/–) transgenic mice, which have a tamoxifen-inducible and Cre-mediated recombination system driven by the chicken β-actin promoter, were obtained from The Jackson Laboratory and mated with Phd2fl/fl and Phd3fl/fl mice to generate Phd2fl/fl CAG-Cre+/–, Phd3fl/fl CAG-Cre+/–, or Phd2/3fl/fl CAG-Cre+/– mice. The genotypes of these mice were determined by PCR, as described previously, and with a protocol provided by The Jackson Laboratory (30).
Tamoxifen (Sigma-Aldrich) in corn oil at 20 mg/ml was aliquoted and maintained at –80°C prior to use. Six- to 8-week-old Phd2fl/fl CAG-Cre+/–, Phd3fl/fl CAG-Cre+/–, or Phd2/3fl/fl CAG-Cre+/– mice were i.p. injected with tamoxifen (40 mg/kg/day) once daily for 5 consecutive days to generate Phd2–/–, Phd3–/–, and Phd2/3–/– mice (53). Deletion of the Phd2 and Phd3 genes was confirmed by real-time PCR, using primers described previously (30).
Cell culture.
HL-1 cardiomyocytes (derived from murine atrial cardiomyocytes) were maintained as previously described (54). Primary neonatal rat or mouse ventricular cardiomyocytes were isolated from 1- to 2-day-old rats or mice using a neonatal cardiomyocyte isolation kit (Worthington Biochemical) and maintained in MEM supplemented with 10% horse serum, 5% FBS, antibiotics (100 U/ml penicillin, 68.6 mol/l streptomycin), and BrdU (100 μM). With this protocol, over 95% of the remaining cells were cardiomyocytes as determined by staining with MF-20 antibody. The percentage of oxygen (pO2) was controlled by incubating cells at 37°C in a humidified, O2/CO2-regulated incubator (Coy Laboratory Products) adjusted to 5% CO2 and the indicated pO2 (Figures 6 and 8).
Western blotting, immunofluorescence, wheat germ agglutinin, and TUNEL staining.
Cells were washed once with cold PBS and harvested in cold lysis buffer (1% Triton X-100, 50 mmol/l Tris, pH 7.4, 150 mmol/l NaCl, protease and phosphatase inhibitors) on ice. Cell lysates were then clarified by centrifugation at 14,000 g for 10 minutes. Equal amounts of protein were used for Western blotting with antibodies against phospholamban (8495S; Cell Signaling Technology); phospholamban phospho-threonine-17 (catalog A010-13AP; Badrilla); SERCA2a (catalog A010-20; Badrilla); PHD2 (catalog NB100-2219; Novus Biologicals); RyR2 (catalog A010-35AP; Badrilla); HIF-1α (catalog NB100-479; Novus Biologicals); tubulin (catalog T6074; Sigma-Aldrich); CaMKII (3362S; Cell Signaling Technology); CaMKII phospho-threonine-286 (catalog A010-50AP; Badrilla); TR-α/β (catalog sc-772; Santa Cruz Biotechnology Inc.); TR-α (catalog PA1-211A; Thermo Scientific); NCOR2 (catalog PA1-843; Thermo Scientific); and MF20 (University of Iowa Developmental Studies Hybridoma Bank).
Freshly isolated hearts were fixed with 10% formalin and embedded in paraffin. Heart sections were mounted on glass slides and then deparaffinized and hydrated for immunostaining, wheat germ agglutinin (WGA) staining (Alexa Fluor 594 conjugate; Life Technologies), or TUNEL staining. Cultured cells fixed in 4% paraformaldehyde and tissue slides were permeabilized with 0.2% Triton X-100 for 5 minutes at room temperature, blocked with 5% goat serum in PBS for 1 hour and then with the primary antibodies overnight at 4°C in blocking solution. Cells and slides were then washed in PBS and incubated in the dark with a secondary antibody conjugated with Alexa Fluor 488 (Life Technologies) in blocking solution for 60 minutes at room temperature, followed by counterstaining with DAPI. TUNEL staining was performed following the manufacturer’s protocol (catalog S7100; EMD Millipore). Images were taken with a fluorescence microscope.
Luciferase reporter assays.
HL-1 cells seeded in 12-well plates in Claycomb Medium were transfected with either the rat pGL3-Pln (–156 to +64) promoter luciferase reporter or an empty pGL3 vector along with plasmids expressing TR-α or NCOR2. pGL2-Renilla luciferase was used as an internal control. Cells were cultured under 21% or 1% O2 conditions for an additional 48 hours after transfection. Cells were then harvested, and lysates were assayed for luciferase activity using a dual-luciferase assay kit (Promega) via a luminometer. Luciferase activity was normalized to Renilla luciferase activity.
Real-time PCR.
cDNA was synthesized from 1 μg RNA purified from heart tissues using an iScript cDNA synthesis kit (Bio-Rad). Gene-specific mRNA levels were measured using the LightCycler 480 Real-Time PCR system (Roche Diagnostics, Roche Applied Science) with LightCycler 480 Probe Master (Roche Diagnostics) and their specific primers and probes (software designed by the Roche Universal ProbeLibrary Assay Design Center). Samples were analyzed in triplicate. Results are expressed as the ratio of the gene of interest corrected to the housekeeping gene Gapdh mRNA level and then normalized to WT. The following primer sequences were used: pyruvate dehydrogenase kinase 1 (Pdk1), forward: GTTGAAACGTCCCGTGCT, reverse: GCGTGATATGGGCAATCC; Serca2a, forward: TGTGTAATGCCCTCAACAGC, reverse: CCCACGAGCCAGATATTCTC; and Pln, forward: CTGTGACGATCACCGAAGC, reverse: TGGTCAAGAGAAAGATAAAAAGTTGA.
In vitro hydroxylation assay.
Approximately 1 μg Flag–TR-α captured on Flag resin (Sigma-Aldrich) was incubated with 1 μg recombinant PHD2 and PHD3 in a reaction buffer containing 10 μM FeSO4, 100 μM 2-oxo-glutarate, 1 mM ascorbate, 60 μg catalase, 100 μM DTT, 2 mg BSA, and 50 μM Tris-HCl buffer, adjusted to pH 7.8. The enzyme reaction was carried out at 37°C for 30 minutes.
Co-IP.
Cell or heart lysates in lysis buffer (50 mM Tris, pH 7.4, 150 mM NaCl, 5 mM EDTA, 1% NP-40, 10% glycerol and protease inhibitor cocktail) were clarified by centrifugation at 14,000 g for 10 minutes. Anti-Flag resin (25 μl) or 10 μl anti–TR-α antibody was added to the lysates, and the reactions were incubated with rotation overnight at 4°C. The anti–TR-α antibody complex was precipitated with protein A beads and collected by centrifugation (2,500 g, 5 minutes). Beads were washed 5 times with lysis buffer. Eluted proteins were analyzed by Western blotting.
45Ca2+ uptake.
ATP-dependent 45Ca2+ uptake by crude mouse heart membrane fractions was determined using a filtration method we described previously (55). 45Ca2+ uptake was initiated by placing membranes in 0.15 M KCl; 20 mM imidazole, pH 7.0; solution containing 5 mM ATP, 8 mM Mg2+, and 5 mM potassium oxalate (a Ca2+ precipitating agent to increase Ca2+ uptake capacity); 10 μM ruthenium red (to inhibit RyRs); 5 mM NaN3 (to inhibit mitochondrial Ca2+ uptake); 1 mM EGTA; and Ca2+ and 45Ca2+ to yield a free Ca2+ concentration of 0.5 μM. To obtain 45Ca2+ uptake rates, aliquots were placed at 2.5, 5, and 10 minutes on 0.45-μM Millipore filters (EMD Millipore) under vacuum and rinsed with three 3-ml volumes of ice-cold solution containing 0.175 M KCl and 5 mM imidazole, pH 7.0. Radioactivity remaining with the vesicles on the filters was determined by liquid scintillation counting.
Adult cardiomyocyte isolation and confocal Ca2+ imaging.
Following isoflurane anesthesia, hearts were removed from mice approximately 6 to 8 weeks of age, rinsed in KB solution (90 mM KCl, 30 mM pyruvic acid, 5 mM β-hydroxybutyric acid, 5 mM creatine, 20 mM taurine, 10 mM glucose, 0.5 mM EGTA, 5 mM HEPES, pH 7.2), cannulated through the aorta, and perfused on a Langendorff apparatus with 0 Ca2+ Tyrode (3–5 minutes, 37°C), then 0 Ca2+ Tyrode containing Liberase TH Research Grade (Roche Applied Science) for 10 to 15 minutes at 37°C. After digestion, the heart was perfused with 3 ml KB solution to wash out the collagenase, then minced in KB solution, agitated gently, and filtered through a 210-μm polyethylene mesh. After settling, the isolated myocytes were washed once with KB solution and stored in KB solution at room temperature until use a few hours later.
Isolated adult ventricular myocytes were incubated at room temperature for 30 minutes with 2 μmol/l fluo-4-acetoxymethyl ester (Fluo-4 AM; Life Technologies) in KB solution before being incubated for 20 minutes with dye-free normal Tyrode solution (1.8 mM Ca2+) for de-esterification. Cells were then transferred to a pacing chamber equipped with a pair of parallel platinum electrodes on a laser-scanning confocal microscope (LSM 510; Carl Zeiss). The cells were visualized through a ×40 oil immersion objective lens, and only cells that showed clear striation and normal contractility after being paced at 1 Hz for at least 2 minutes were selected for experiments. Fluo-4 was excited at 488 nm, with emission collected through a 515-nm-long pass filter, while fluorescence images were recorded in line-scan mode with 1,024 pixels per line at 500 Hz. Caffeine (10 mM) was rapidly applied toward the end of the recording for estimation of steady-state SR Ca2+ content.
Blood pressure measurements.
Tail blood pressure measurements were obtained by photoplethysmography with the Visitech Systems BP-2000 Blood Pressure Analysis System. The study took place in a quiet room and according to the manufacturer’s protocol. Mice were placed on a 37.5°C (99.5°F) heated surface, and individual isolation chambers were placed on top of each mouse (4 subjects recorded at a time) for immobilization. Systolic blood pressure (SBP) readings were obtained by placing the mouse’s tail in its corresponding tail cuff. Measurements were made at least 25 times at baseline and 5 minutes after ISO treatment (3 mg/kg).
Transthoracic ECG.
Anesthetized mice were restrained on a temperature-controlled mouse board (Indus Instruments), and ECG was performed using a Vevo 660 ultrasound system (VisualSonics) equipped with a 30 MHz transducer. An echocardiographer blinded to animal genotype captured 2D parasternal long-axis views of the left ventricle. From this view, an M-mode cursor was positioned perpendicularly to the interventricular septum and the posterior wall of the left ventricle at the level of the papillary muscles. Fractional shortening measurements were obtained for systole and diastole using the average of 4 cardiac cycles.
Liquid chromatography/tandem mass spectrometric analysis.
Flag–TR-α IP from HeLa cells was excised from a Coomassie blue–stained SDS gel, followed by DTT reduction, iodoacetamide alkylation, and in-gel digestion (12.5 ng/μl trypsin overnight). The resulting tryptic peptides were concentrated and analyzed by C18 capillary reverse-phase liquid chromatography coupled with tandem mass spectrometry (LC-MS/MS) using a LTQ Orbitrap Elite mass spectrometer (ThermoFisher Scientific) operating in the optimized high-resolution mode (56). The acquired MS/MS data were searched against a database to identify hydroxylated proline sites in TR-α, through a dynamic mass shift for the modified proline (+15.9949 Da), using PEAKS software (PEAKS7, build 20140321; Bioinformatics Solutions Inc.) (57). The identified modified peptides were filtered by mass accuracy and matching scores to reduce protein FDRs to less than 1% and accepted after manually examining the MS/MS spectra. Moreover, additional LC-MS/MS runs were performed, in which high-resolution MS/MS data were collected in the Orbitrap, and modified sites were further validated by independent de novo sequencing of raw spectra and confirmed on the basis of the unambiguous assignment of characteristic site-specific fragment ions.
Microarray analysis.
Total RNA was purified via tissue homogenization (TissueLyser LT; QIAGEN) and extracted using the RNeasy Microarray Tissue Mini Kit (QIAGEN). cRNA was amino-allyl labeled with Cy3, hybridized to Agilent G3 Mouse GE 8 × 60K arrays, and scanned using a GeneTAC UC-4 Microarray Analyzer (Genomic Solutions/PerkinElmer). Principal component analysis, hierarchical clustering, significance analysis of microarrays, and pathway enrichment were performed as described in the figure legends. The gene expression data discussed herein are deposited in the NCBI’s Gene Expression Omnibus database (GEO GSE67726).
Statistics.
Data are shown as the mean ± SEM for 3 or 4 separate experiments. Differences were analyzed with a 2-tailed Student’s t test or ANOVA and post-hoc analyses as appropriate. P values of 0.05 or less were considered statistically significant.
Study approval.
The mice were handled according to NIH guidelines for the care and use of experimental animals. All experiments were approved by the IACUCs of Baylor College of Medicine and the University of North Carolina at Chapel Hill.
Supplementary Material
Acknowledgments
This work was supported by American Heart Association (AHA) National Center Research Program (NCRP) Scientist Development Grant 10SDG3860014 (to L. Xie); NIH grants R01 HL061656 and HL112890 (to X. Pi); and NIH grant R37 65619 (to C. Patterson). J. Peng is supported by the American Lebanese Syrian Associated Charities (ALSAC).
Footnotes
Note regarding evaluation of this manuscript: Manuscripts authored by scientists associated with Duke University, The University of North Carolina at Chapel Hill, Duke-NUS, and the Sanford-Burnham Medical Research Institute are handled not by members of the editorial board but rather by the science editors, who consult with selected external editors and reviewers.
Conflict of interest: The authors have declared that no conflict of interest exists.
Reference information:J Clin Invest. 2015;125(7):2759–2771. doi:10.1172/JCI80369.
References
- 1.Roger VL, et al. Heart disease and stroke statistics — 2012 update: a report from the American Heart Association. Circulation. 2012;125(1):e2–e220. doi: 10.1161/CIR.0b013e31823ac046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Schomig A. Adrenergic mechanisms in myocardial infarction: cardiac and systemic catecholamine release. J Cardiovasc Pharmacol. 1988;12(suppl 1):S1–S7. [PubMed] [Google Scholar]
- 3.Desantiago J, et al. Arrhythmogenic effects of β2-adrenergic stimulation in the failing heart are attributable to enhanced sarcoplasmic reticulum Ca load. Circ Res. 2008;102(11):1389–1397. doi: 10.1161/CIRCRESAHA.107.169011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bristow MR. β-Adrenergic receptor blockade in chronic heart failure. Circulation. 2000;101(5):558–569. doi: 10.1161/01.CIR.101.5.558. [DOI] [PubMed] [Google Scholar]
- 5.Kushner FG, et al. 2009 Focused Updates: ACC/AHA Guidelines for the Management of Patients With ST-Elevation Myocardial Infarction (updating the 2004 Guideline and 2007 Focused Update) and ACC/AHA/SCAI Guidelines on Percutaneous Coronary Intervention (updating the 2005 Guideline and 2007 Focused Update): a report of the American College of Cardiology Foundation/American Heart Association Task Force on Practice Guidelines. Circulation. 2009;120(22):2271–2306. doi: 10.1161/CIRCULATIONAHA.109.192663. [DOI] [PubMed] [Google Scholar]
- 6.Van de Werf F, et al. Management of acute myocardial infarction in patients presenting with persistent ST-segment elevation: the Task Force on the Management of ST-Segment Elevation Acute Myocardial Infarction of the European Society of Cardiology. Eur Heart J. 2008;29(23):2909–2945. doi: 10.1093/eurheartj/ehn416. [DOI] [PubMed] [Google Scholar]
- 7.Grimm M, Brown JH. β-Adrenergic receptor signaling in the heart: role of CaMKII. J Mol Cell Cardiol. 2010;48(2):322–330. doi: 10.1016/j.yjmcc.2009.10.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Swaminathan PD, Purohit A, Hund TJ, Anderson ME. Calmodulin-dependent protein kinase II: linking heart failure and arrhythmias. Circ Res. 2012;110(12):1661–1677. doi: 10.1161/CIRCRESAHA.111.243956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rokita AG, Anderson ME. New therapeutic targets in cardiology: arrhythmias and Ca2+/calmodulin-dependent kinase II (CaMKII). Circulation. 2012;126(17):2125–2139. doi: 10.1161/CIRCULATIONAHA.112.124990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kaelin WG, Jr, Ratcliffe PJ. Oxygen sensing by metazoans: the central role of the HIF hydroxylase pathway. Mol Cell. 2008;30(4):393–402. doi: 10.1016/j.molcel.2008.04.009. [DOI] [PubMed] [Google Scholar]
- 11.Aravind L, Koonin EV. The DNA-repair protein AlkB, EGL-9, and leprecan define new families of 2-oxoglutarate- and iron-dependent dioxygenases. Genome Biol. 2001;2(3):RESEARCH0007. doi: 10.1186/gb-2001-2-3-research0007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bruick RK, McKnight SL. A conserved family of prolyl-4-hydroxylases that modify HIF. Science. 2001;294(5545):1337–1340. doi: 10.1126/science.1066373. [DOI] [PubMed] [Google Scholar]
- 13.Jaakkola P, et al. Targeting of HIF-α to the von Hippel-Lindau ubiquitylation complex by O2-regulated prolyl hydroxylation. Science. 2001;292(5516):468–472. doi: 10.1126/science.1059796. [DOI] [PubMed] [Google Scholar]
- 14.Ivan M, et al. HIFalpha targeted for VHL-mediated destruction by proline hydroxylation: implications for O2 sensing. Science. 2001;292(5516):464–468. doi: 10.1126/science.1059817. [DOI] [PubMed] [Google Scholar]
- 15.Freeman RS, Hasbani DM, Lipscomb EA, Straub JA, Xie L. SM-20, EGL-9, and the EGLN family of hypoxia-inducible factor prolyl hydroxylases. Mol Cells. 2003;16(1):1–12. [PubMed] [Google Scholar]
- 16.Willam C, et al. HIF prolyl hydroxylases in the rat; organ distribution and changes in expression following hypoxia and coronary artery ligation. J Mol Cell Cardiol. 2006;41(1):68–77. doi: 10.1016/j.yjmcc.2006.04.009. [DOI] [PubMed] [Google Scholar]
- 17.Nwogu JI, Geenen D, Bean M, Brenner MC, Huang X, Buttrick PM. Inhibition of collagen synthesis with prolyl 4-hydroxylase inhibitor improves left ventricular function and alters the pattern of left ventricular dilatation after myocardial infarction. Circulation. 2001;104(18):2216–2221. doi: 10.1161/hc4301.097193. [DOI] [PubMed] [Google Scholar]
- 18.Holscher M, et al. Cardiomyocyte-specific prolyl-4-hydroxylase domain 2 knock out protects from acute myocardial ischemic injury. J Biol Chem. 2011;286(13):11185–11194. doi: 10.1074/jbc.M110.186809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Eckle T, Kohler D, Lehmann R, El Kasmi K, Eltzschig HK. Hypoxia-inducible factor-1 is central to cardioprotection: a new paradigm for ischemic preconditioning. Circulation. 2008;118(2):166–175. doi: 10.1161/CIRCULATIONAHA.107.758516. [DOI] [PubMed] [Google Scholar]
- 20.Xie L, Pi X, Wang Z, He J, Willis MS, Patterson C. Depletion of PHD3 protects heart from ischemia/reperfusion injury by inhibiting cardiomyocyte apoptosis. J Mol Cell Cardiol. 2015;80:156–165. doi: 10.1016/j.yjmcc.2015.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kido M, et al. Hypoxia-inducible factor 1-α reduces infarction and attenuates progression of cardiac dysfunction after myocardial infarction in the mouse. J Am Coll Cardiol. 2005;46(11):2116–2124. doi: 10.1016/j.jacc.2005.08.045. [DOI] [PubMed] [Google Scholar]
- 22.Minamishima YA, et al. A feedback loop involving the Phd3 prolyl hydroxylase tunes the mammalian hypoxic response in vivo. Mol Cell Biol. 2009;29(21):5729–5741. doi: 10.1128/MCB.00331-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Moslehi J, et al. Loss of hypoxia-inducible factor prolyl hydroxylase activity in cardiomyocytes phenocopies ischemic cardiomyopathy. Circulation. 2010;122(10):1004–1016. doi: 10.1161/CIRCULATIONAHA.109.922427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Xie L, et al. Oxygen-regulated beta(2)-adrenergic receptor hydroxylation by EGLN3 and ubiquitylation by pVHL. Sci Signal. 2009;2(78):ra33. doi: 10.1126/scisignal.2000444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Xie L, Pi X, Mishra A, Fong G, Peng J, Patterson C. PHD3-dependent hydroxylation of HCLK2 promotes the DNA damage response. J Clin Invest. 2012;122(8):2827–2836. doi: 10.1172/JCI62374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Luo W, et al. Pyruvate kinase M2 is a PHD3-stimulated coactivator for hypoxia-inducible factor 1. Cell. 2011;145(5):732–744. doi: 10.1016/j.cell.2011.03.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Marks AR. Calcium cycling proteins and heart failure: mechanisms and therapeutics. J Clin Invest. 2013;123(1):46–52. doi: 10.1172/JCI62834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yano M, Ikeda Y, Matsuzaki M. Altered intracellular Ca2+ handling in heart failure. J Clin Invest. 2005;115(3):556–564. doi: 10.1172/JCI24159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Belakavadi M, Saunders J, Weisleder N, Raghava PS, Fondell JD. Repression of cardiac phospholamban gene expression is mediated by thyroid hormone receptor-{alpha}1 and involves targeted covalent histone modifications. Endocrinology. 2010;151(6):2946–2956. doi: 10.1210/en.2009-1241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Takeda K, et al. Regulation of adult erythropoiesis by prolyl hydroxylase domain proteins. Blood. 2008;111(6):3229–3235. doi: 10.1182/blood-2007-09-114561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yang Y, et al. Calmodulin kinase II inhibition protects against myocardial cell apoptosis in vivo. Am J Physiol Heart Circ Physiol. 2006;291(6):H3065–H3075. doi: 10.1152/ajpheart.00353.2006. [DOI] [PubMed] [Google Scholar]
- 32.Wu Y, et al. Suppression of dynamic Ca(2+) transient responses to pacing in ventricular myocytes from mice with genetic calmodulin kinase II inhibition. J Mol Cell Cardiol. 2006;40(2):213–223. doi: 10.1016/j.yjmcc.2005.11.005. [DOI] [PubMed] [Google Scholar]
- 33.Wehrens XH, Lehnart SE, Reiken SR, Marks AR. Ca2+/calmodulin-dependent protein kinase II phosphorylation regulates the cardiac ryanodine receptor. Circ Res. 2004;94(6):e61–70. doi: 10.1161/01.RES.0000125626.33738.E2. [DOI] [PubMed] [Google Scholar]
- 34.Maier LS, Bers DM. Role of Ca2+/calmodulin-dependent protein kinase (CaMK) in excitation-contraction coupling in the heart. Cardiovasc Res. 2007;73(4):631–640. doi: 10.1016/j.cardiores.2006.11.005. [DOI] [PubMed] [Google Scholar]
- 35.Cross HR, Kranias EG, Murphy E, Steenbergen C. Ablation of PLB exacerbates ischemic injury to a lesser extent in female than male mice: protective role of NO. Am J Physiol Heart Circ Physiol. 2003;284(2):H683–H690. doi: 10.1152/ajpheart.00567.2002. [DOI] [PubMed] [Google Scholar]
- 36.Epstein AC, et al. C. elegans EGL-9 and mammalian homologs define a family of dioxygenases that regulate HIF by prolyl hydroxylation. Cell. 2001;107(1):43–54. doi: 10.1016/S0092-8674(01)00507-4. [DOI] [PubMed] [Google Scholar]
- 37.Deribe YL, Pawson T, Dikic I. Post-translational modifications in signal integration. Nat Struct Mol Biol. 2010;17(6):666–672. doi: 10.1038/nsmb.1842. [DOI] [PubMed] [Google Scholar]
- 38.Chen JD, Evans RM. A transcriptional co-repressor that interacts with nuclear hormone receptors. Nature. 1995;377(6548):454–457. doi: 10.1038/377454a0. [DOI] [PubMed] [Google Scholar]
- 39.Gheorghiade M, Bonow RO. Chronic heart failure in the United States: a manifestation of coronary artery disease. Circulation. 1998;97(3):282–289. doi: 10.1161/01.CIR.97.3.282. [DOI] [PubMed] [Google Scholar]
- 40.Libby P, Theroux P. Pathophysiology of coronary artery disease. Circulation. 2005;111(25):3481–3488. doi: 10.1161/CIRCULATIONAHA.105.537878. [DOI] [PubMed] [Google Scholar]
- 41.Spencer FA, Meyer TE, Gore JM, Goldberg RJ. Heterogeneity in the management and outcomes of patients with acute myocardial infarction complicated by heart failure: the National Registry of Myocardial Infarction. Circulation. 2002;105(22):2605–2610. doi: 10.1161/01.CIR.0000017861.00991.2F. [DOI] [PubMed] [Google Scholar]
- 42.May D, et al. Transgenic system for conditional induction and rescue of chronic myocardial hibernation provides insights into genomic programs of hibernation. Proc Natl Acad Sci U S A. 2008;105(1):282–287. doi: 10.1073/pnas.0707778105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Semenza GL. Hypoxia-inducible factor 1 and cardiovascular disease. Annu Rev Physiol. 2014;76:39–56. doi: 10.1146/annurev-physiol-021113-170322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bosch-Marce M, et al. Effects of aging and hypoxia-inducible factor-1 activity on angiogenic cell mobilization and recovery of perfusion after limb ischemia. Circ Res. 2007;101(12):1310–1318. doi: 10.1161/CIRCRESAHA.107.153346. [DOI] [PubMed] [Google Scholar]
- 45.Sano M, et al. p53-induced inhibition of Hif-1 causes cardiac dysfunction during pressure overload. Nature. 2007;446(7134):444–448. doi: 10.1038/nature05602. [DOI] [PubMed] [Google Scholar]
- 46.Bekeredjian R, et al. Conditional HIF-1α expression produces a reversible cardiomyopathy. PLoS One. 2010;5(7):e11693. doi: 10.1371/journal.pone.0011693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Bers DM. Calcium cycling and signaling in cardiac myocytes. Annu Rev Physiol. 2008;70:23–49. doi: 10.1146/annurev.physiol.70.113006.100455. [DOI] [PubMed] [Google Scholar]
- 48.Frank K, Kranias EG. Phospholamban and cardiac contractility. Ann Med. 2000;32(8):572–578. doi: 10.3109/07853890008998837. [DOI] [PubMed] [Google Scholar]
- 49.Luo W, et al. Targeted ablation of the phospholamban gene is associated with markedly enhanced myocardial contractility and loss of β-agonist stimulation. Circ Res. 1994;75(3):401–409. doi: 10.1161/01.RES.75.3.401. [DOI] [PubMed] [Google Scholar]
- 50.Minamisawa S, et al. Kranias EG, et al. Chronic phospholamban-sarcoplasmic reticulum calcium ATPase interaction is the critical calcium cycling defect in dilated cardiomyopathy. Cell. 1999;99(3):313–322. doi: 10.1016/S0092-8674(00)81662-1. [DOI] [PubMed] [Google Scholar]
- 51.Sato Y, et al. Rescue of contractile parameters and myocyte hypertrophy in calsequestrin overexpressing myocardium by phospholamban ablation. J Biol Chem. 2001;276(12):9392–9399. doi: 10.1074/jbc.M006889200. [DOI] [PubMed] [Google Scholar]
- 52.Haghighi K, et al. Human phospholamban null results in lethal dilated cardiomyopathy revealing a critical difference between mouse and human. J Clin Invest. 2003;111(6):869–876. doi: 10.1172/JCI17892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Moresi V, et al. Myogenin and class II HDACs control neurogenic muscle atrophy by inducing E3 ubiquitin ligases. Cell. 2010;143(1):35–45. doi: 10.1016/j.cell.2010.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Claycomb WC, et al. HL-1 cells: a cardiac muscle cell line that contracts and retains phenotypic characteristics of the adult cardiomyocyte. Proc Natl Acad Sci U S A. 1998;95(6):2979–2984. doi: 10.1073/pnas.95.6.2979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Yamaguchi N, Takahashi N, Xu L, Smithies O, Meissner G. Early cardiac hypertrophy in mice with impaired calmodulin regulation of cardiac muscle Ca release channel. J Clin Invest. 2007;117(5):1344–1353. doi: 10.1172/JCI29515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Xu P, Duong DM, Peng J. Systematical optimization of reverse-phase chromatography for shotgun proteomics. J Proteome Res. 2009;8(8):3944–3950. doi: 10.1021/pr900251d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ma B, et al. PEAKS: powerful software for peptide de novo sequencing by tandem mass spectrometry. Rapid Commun Mass Spectrom. 2003;17(20):2337–2342. doi: 10.1002/rcm.1196. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.