Abstract
Pancreatic β cells support glucose homeostasis with great precision by matching insulin release to the metabolic needs of the moment. Previous gene-expression analysis indicates that adult β cells not only produce cell-specific proteins, but also repress a small set of housekeeping genes — such as those encoding lactate dehydrogenase A (LDHA), solute transporter MCT1, and hexokinase 1 (HK1) — that would otherwise interfere with normal β cell function. In this issue of the JCI, Dhawan et al. elucidate a molecular mechanism involved in β cell–specific repression of Ldha and Hk1 that is mediated by induction of the de novo DNA methyltransferase DNMT3A during the first weeks after birth. Failure to induce DNMT3A-dependent methylation disrupts normal glucose-induced insulin release in adult life. The results of this study reinforce the idea that the phenotype of adult β cells has two faces and that failure to achieve selective gene repression undermines β cell support of normal glucose homeostasis.
Two faces of differentiated β cells
Insulin release is regulated as a function of nutritional supplies and metabolic demands, both of which fluctuate over time and depend on physical activity, pregnancy status, age, and a variety of other factors. As too little or too much circulating insulin can be harmful, mature β cells are equipped with an efficient delivery apparatus that rapidly mobilizes a fraction of stored insulin within secretory granules via the secretory pathway (1). Moreover, β cells possess a precise metabolic detection system (Figure 1) that couples the extracellular glucose concentration to the rate of secretory granule exocytosis (2). Several of these critical phenotypic features are acquired shortly after birth, when β cells mature (3). The transition from the neonatal to the adult β cell involves not only upregulation of genes — such as the transcription factor MAFA and insulin genes, required for the specialized tasks of β cells — but also repression of a set of genes that are detrimental to pancreatic islet formation and are widely used in other mammalian cell types (4). The scale of this tissue-specific repression (a phenomenon referred to as disallowed genes) was examined by genome-wide analyses that compared gene expression in adult islets of Langerhans with a panel of other tissues (4, 5). By looking at the overlap between the above-mentioned microarray studies, Pullen and Rutter identified a core set of about 10 disallowed genes in pancreatic islets (6). Among the genes in this core set are those encoding the lactate pyruvate transporter MCT1 (also known as SLC16A1) and lactate dehydrogenase A (LDHA). MCT1 and LDHA closely collaborate (Figure 1) to allow anaerobic glycolysis in most tissues and cells.
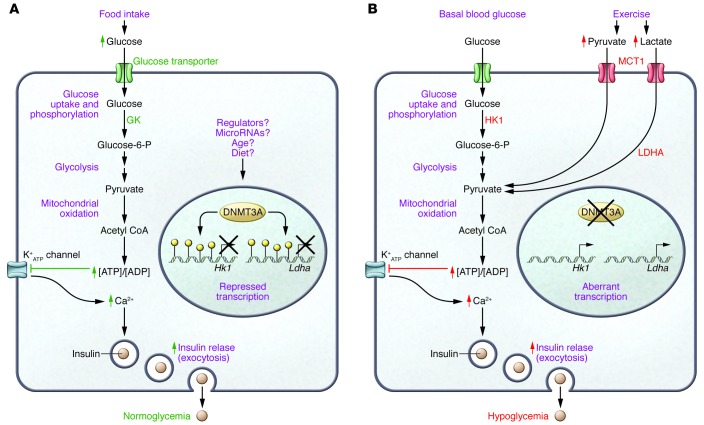
Figure 1. DNMT3A mediates repression of genes that disrupt normal glucose-regulated insulin release in β cells.
(A) Physiological, nutrient-induced insulin release during a carbohydrate-containing meal depends on the expression of key proteins in pancreatic β cells that couple a rise in circulating glucose to an accelerated metabolic flux, leading to an increased [ATP]/[ADP] ratio, closure of ATP-sensitive potassium channels and a rise in cytosolic calcium (1), and appropriate levels of insulin secretion that support normoglycemia. In this issue, Dhawan et al. (18) show that the DNA methyltransferase DNMT3A is critical for repression of disallowed genes Hk1 and Ldha that would otherwise interfere with β cell function. The factors that induce and maintain DNMT3A in β cells have not been fully elucidated. (B) Inappropriate insulin secretion, as in too much insulin under basal conditions or activation of induction of secretion by nonphysiological stimuli, can cause hypoglycemia (15). In the absence of DNMT3A, expression of HK1 elevates basal insulin release because the enzyme has a high affinity for its substrate glucose, and elevated LDHA promotes conversion of lactate into pyruvate. Moreover, increased levels of the pyruvate transporter MCT1 promote inappropriate insulin secretion (10). Dhawan and colleagues show that murine β cells switch off Hk1 and Ldha expression when cells mature the first weeks after birth, as the result of DNMT3A-mediated cytosine methylation (yellow circles) in the Hk1 and Ldha gene promoters.
It is known that β cells avoid anaerobic glycolysis during metabolic glucose sensing (7). Instead, normal β cells funnel glucose to mitochondrial pyruvate uptake and metabolism (8). The selective absence of MCT1 in β cells explains why insulin secretion is unresponsive to a rise in extracellular pyruvate, a situation that is reversed in individuals with a Mendelian disorder called exercise-induced hyperinsulinism (9), which results from loss of regulation at the level of the human MCT1 promoter (10). The presence of MCT1 in β cells would confuse the physiology of glucose homeostasis in that β cells would no longer discriminate elevated glucose (after meals) from elevated pyruvate (after exercise) and would therefore fail to match circulating insulin levels to metabolic demands (Figure 1).
Although the number of genes specifically repressed in islets is low, the scale of the phenomenon increases when genes that are repressed not only in islets but also in one or two additional tissues are considered. One example is the group of low-Km hexokinases (HK1–HK3), which catalyze the first step of glycolysis in most cells. For pancreatic β cells and hepatocytes, however, these isoforms are ill adapted to sustain glucose homeostasis because they are saturated by micromolar amounts of their substrate glucose. In contrast, glucokinase (GK, Figure 1) is well adapted for precise control of glucose homeostasis, as its enzymatic affinity for glucose is in the millimolar range (11). β Cells efficiently repress HK1, and what little enzymatic activity that has been measured in islets was attributed to contaminating exocrine pancreatic cells (12). Forced expression of low-Km HK1 in β cells (13) elevates basal insulin secretion, rendering β cells dangerous in terms of hypoglycemic risk (Figure 1). Hk1, Ldha, and Mct1 therefore represent examples of a class of disallowed genes (14) that would jeopardize normal β function when expressed (15).
The start of an epigenetic roadmap
How do β cells selectively repress genes that are active in most other cell types? Epigenetic-based mechanisms have been suggested in earlier work. Thorrez et al. detected repressive chromatin markers in a few of the genes repressed in β cells (4). A more systematic, genome-wide analysis of histone H3 modifications (16) showed that β cell maturation is associated with both activation and inactivation of specific genes. In a follow-up study, the polycomb repressor complex subunit RING1B was identified as one of the molecular components that mediate these epigenetic changes (17).
In this issue (18), Dhawan and colleagues propose that the de novo DNA methyltransferase DNMT3A plays an important role in mediating epigenetic gene repression during β cell maturation. Analysis of mRNA expression in FACS-purified β cells from MIP-GFP mice revealed that between postnatal days 4 and 25 (P4 and P25), there is an upregulation of genes encoding key β cell factors, including MAFA and GK (Figure 1), with a concurrent downregulation of Hk1, Hk2, Aldob, and Ldha. Evaluation of methylated versus nonmethylated cytosines in the Hk1 and Ldha gene loci demonstrated that there is a net increase in methylation of these 2 loci between P4 and P25. ChIP analysis showed that the DNA methyltransferase DNMT3A is maximally enriched at the Ldha and Hk1 loci in β cells two weeks after birth. Dhawan and colleagues developed a mouse strain with a β cell–specific deletion of Dnmt3a (Dnmt3afl/fl R26R-eYFP RIP-Cre mice, referred to as 3aRCY-KO) that allowed for direct evaluation for the role of DNMT3A in the methylation and repression of Hk1 and Ldha. Compared with controls, 3aRCY-KO mice had elevated basal plasma insulin and a poor insulin response after i.p. glucose injection, and isolated islets from these animals were deficient for glucose-induced insulin release. The Hk1 and Ldha loci were weakly methylated in β cells of 3aRCY-KO mice, resulting in upregulation of Hk1 and Ldha transcripts. Moreover, siRNA-mediated knockdown of Hk1 and Ldha transcripts partially restored insulin secretory competence in 3aRCY-KO β cells. Comparison of adult human pancreatic islets with immature, insulin-expressing cells derived from pluripotent cells revealed higher levels of DNA methylation and stronger repression of HK1 and LDHA in the mature islets. Together, the study by Dhawan et al. (18) supports the importance of disallowed gene repression for normal β cell function. Additionally, these results provide an explanation for how maturing β cells switch off a set of genes that would destroy normal function when expressed and show that the de novo methyltransferase DNMT3A plays an active role in this process.
New questions
The study by Dhawan and colleagues on DNMT3A (18) and previous work on RING1B (17) can be viewed as starting points that provide important fragments of information for compiling a more complete epigenetic roadmap (19) of neonatal immature β cells and adult mature β cells. It will be interesting to elucidate how DNMT3A activity in β cells is regulated and how the methyltransferase is guided toward its epigenetic target regions. One possibility is that histone H3K27 tri-methylation (H3K27me3), which is associated with polycomb repression, is a context-dependent chromatin signal that guides DNMT3A activity toward its relevant target genes (20). This raises the question of how H3K27me3 marks are selectively introduced in the disallowed target genes of maturing β cells. Interestingly, not only were Hk1 and Ldha upregulated in 3aRCY-KO β cells, but expression of key β cell genes, including Mafa, Pdx1, and Gk, was reduced, indicating that these two phenomena are interconnected. It seems possible that some of the β cell–specific transcription factors that enhance expression of a set of β cell genes could also operate as repressors in a context-dependent manner. Future experiments should address the precise link between repression of disallowed genes and upregulation of β cell genes.
It should also be noted that the neonatal period of DNMT3A activity is a continuation of intrauterine events that initiate β cell identity via DNA methylation (21). This raises the question as to whether or not environmental conditions during pregnancy influence the starting position of neonatal epigenetic events. Moreover, what happens after β cells have matured? How is the continued repression of disallowed genes at the end of the neonatal period maintained? Adult β cells are essentially a nonreplicating population, yet they remain active during the entire lifetime of rodents and humans. Therefore, it seems relevant to assess the impact of environmental factors, such as a high-fat diet, that elevate basal insulin release. Because DNMT3A acts as a de novo methylase that introduces an epigenetic landscape in newly replicated cells (22), adult, nonreplicating, β cells may utilize other mechanisms to maintain appropriate gene-expression patterns. For example, a cytosine demethylation pathway that is mediated by a family of Tet dioxygenases (23) can reactivate expression of methylated loci, but the activity of these enzymes has not been studied in adult β cells. Future studies should also address a potential role for β cell microRNA that may interfere with DNMT3A activity, in particular members of the miR-29 family (24).
It should be noted that Dhawan et al. (18) used the MIP-GFP and RIP-Cre mouse models, which harbor an artificial human growth hormone (hGH) minigene that is expressed in transgenic islets (25). Expression of the hGH minigene likely does not alter repression of disallowed genes, as hGH does not influence these genes in Pdx1-Cre mice (25). It is not clear why methylation at the Mct1 promoter was not affected by Dnmt3a KO, as Mct1 has clear H3K27me3 marks in differentiated β cells (4). Is it possible that complementary methyltransferases act on a different set of disallowed target genes?
Together, the study by Dhawan et al. (18) supports the earlier idea that β cell differentiation has two faces (4): a visible face that shows accumulation of specific β cell proteins and a hidden face that protects β cells against ubiquitous proteins that need to be repressed to prevent disturbance of normal β cell function. The facets of both faces need to be addressed in efforts to generate insulin-producing cells from pluripotent stem cells. Dhawan and colleagues (18) propose that DNMT3A is one of the waypoints in the epigenetic roadmap of β cell maturation that is essential for the transition from immature to mature β cells. Evaluation of the roles of DNMT3A in preventing β cell abnormalities during chronic metabolic disease would appear to be interesting topics for future studies.
Acknowledgments
F. Schuit is currently supported by the Flemish Foundation for Scientific Research (FWO grant G067212N) for investigation of the phenomenon of disallowed genes in β cells.
Footnotes
Conflict of interest: The author has declared that no conflict of interest exists.
Reference information:J Clin Invest. 2015;125(7):2565–2568. doi:10.1172/JCI82575.
See the related article beginning on page 2851.
References
- 1.Rorsman P, Braun M. Regulation of insulin secretion in human pancreatic islets. Annu Rev Physiol. 2013;75:155–179. doi: 10.1146/annurev-physiol-030212-183754. [DOI] [PubMed] [Google Scholar]
- 2.Rutter GA, Pullen TJ, Hodson DJ, Martinez-Sanchez A. Pancreatic β-cell identity, glucose sensing and the control of insulin secretion. Biochem J. 2015;466(2):203–218. doi: 10.1042/BJ20141384. [DOI] [PubMed] [Google Scholar]
- 3.Nishimura W, et al. A switch from MafB to MafA expression accompanies differentiation to pancreatic β-cells. Dev Biol. 2006;293(2):526–539. doi: 10.1016/j.ydbio.2006.02.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Thorrez L, et al. Tissue-specific disallowance of housekeeping genes: the other face of cell differentiation. Genome Res. 2011;21(1):95–105. doi: 10.1101/gr.109173.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Pullen TJ, Khan AM, Barton G, Butcher SA, Sun G, Rutter GA. Identification of genes selectively disallowed in the pancreatic islet. Islets. 2010;2(2):89–95. doi: 10.4161/isl.2.2.11025. [DOI] [PubMed] [Google Scholar]
- 6.Pullen TJ, Rutter GA. When less is more: the forbidden fruits of gene repression in the adult β-cell. Diabetes Obes Metab. 2013;15(6):503–512. doi: 10.1111/dom.12029. [DOI] [PubMed] [Google Scholar]
- 7.Sekine N, et al. Low lactate dehydrogenase and high mitochondrial glycerol phosphate dehydrogenase in pancreatic β-cells. Potential role in nutrient sensing. J Biol Chem. 1994;269(7):4895–4902. [PubMed] [Google Scholar]
- 8.Schuit F, et al. Metabolic fate of glucose in purified islet cells. Glucose-regulated anaplerosis in β cells. J Biol Chem. 1997;272(30):18572–18579. doi: 10.1074/jbc.272.30.18572. [DOI] [PubMed] [Google Scholar]
- 9.Otonkoski T, et al. Physical exercise-induced hyperinsulinemic hypoglycemia is an autosomal-dominant trait characterized by abnormal pyruvate-induced insulin release. Diabetes. 2003;52(1):199–204. doi: 10.2337/diabetes.52.1.199. [DOI] [PubMed] [Google Scholar]
- 10.Otonkoski T, et al. Physical exercise-induced hypoglycemia caused by failed silencing of monocarboxylate transporter 1 in pancreatic β cells. Am J Hum Genet. 2007;81(3):467–474. doi: 10.1086/520960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Matschinsky FM. Regulation of pancreatic β-cell glucokinase: from basics to therapeutics. Diabetes. 2002;51(suppl 3):S394–S404. doi: 10.2337/diabetes.51.2007.s394. [DOI] [PubMed] [Google Scholar]
- 12.Schuit F, Moens K, Heimberg H, Pipeleers D. Cellular origin of hexokinase in pancreatic islets. J Biol Chem. 1999;274(46):32803–32809. doi: 10.1074/jbc.274.46.32803. [DOI] [PubMed] [Google Scholar]
- 13.Becker TC, BeltrandelRio H, Noel RJ, Johnson JH, Newgard CB. Overexpression of hexokinase I in isolated islets of Langerhans via recombinant adenovirus. Enhancement of glucose metabolism and insulin secretion at basal but not stimulatory glucose levels. J Biol Chem. 1994;269(33):21234–21238. [PubMed] [Google Scholar]
- 14.Quintens R, Hendrickx N, Lemaire K, Schuit F. Why expression of some genes is disallowed in β-cells. Biochem Soc Trans. 2008;36(pt 3):300–305. doi: 10.1042/BST0360300. [DOI] [PubMed] [Google Scholar]
- 15.Schuit F, et al. β-Cell–specific gene repression: a mechanism to protect against inappropriate or maladjusted insulin secretion? Diabetes. 2012;61(5):969–975. doi: 10.2337/db11-1564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.van Arensbergen J, et al. Derepression of Polycomb targets during pancreatic organogenesis allows insulin-producing β-cells to adopt a neural gene activity program. Genome Res. 2010;20(6):722–732. doi: 10.1101/gr.101709.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.van Arensbergen J, et al. Ring1b bookmarks genes in pancreatic embryonic progenitors for repression in adult β cells. Genes Dev. 2013;27(1):52–63. doi: 10.1101/gad.206094.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dhawan S, et al. DNA methylation directs functional maturation of pancreatic β cells. J Clin Invest. 2015;215(7):2851–2860. doi: 10.1172/JCI79956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Roadmap Epigenomics Consortium et al. Integrative analysis of 111 reference human epigenomes. Nature. 2015;518(7539):317–330. doi: 10.1038/nature14248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rose NR, Klose RJ. Understanding the relationship between DNA methylation and histone lysine methylation. Biochim Biophys Acta. 2014;1839(12):1362–1372. doi: 10.1016/j.bbagrm.2014.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dhawan S, Georgia S, Tschen SI, Fan G Bhushan A. Pancreatic β cell identity is maintained by DNA methylation-mediated repression of Arx. Dev Cell. 2011;20(4):419–429. doi: 10.1016/j.devcel.2011.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Okano M, Bell DW, Haber DA, Li E. DNA methyltransferases Dnmt3a and Dnmt3b are essential for de novo methylation and mammalian development. Cell. 1999;99(3):247–257. doi: 10.1016/S0092-8674(00)81656-6. [DOI] [PubMed] [Google Scholar]
- 23.He YF, et al. Tet-mediated formation of 5-carboxylcytosine and its excision by TDG in mammalian DNA. Science. 2011;333(6047):1303–1307. doi: 10.1126/science.1210944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fabbri M, et al. MicroRNA-29 family reverts aberrant methylation in lung cancer by targeting DNA methyltransferases 3A and 3B. Proc Natl Acad Sci U S A. 2007;104(40):15805–15810. doi: 10.1073/pnas.0707628104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Brouwers B, et al. Impaired islet function in commonly used transgenic mouse lines due to human growth hormone minigene expression. Cell Metab. 2014;20(6):979–990. doi: 10.1016/j.cmet.2014.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]