Abstract
Central nervous system (CNS) myelination by oligodendrocytes (OLs) is a highly orchestrated process involving well-defined steps from specification of neural stem cells into proliferative OL precursors followed by terminal differentiation and subsequent maturation of these precursors into myelinating OLs. These specification and differentiation processes are mediated by profound global changes in gene expression, which are in turn subject to control by both extracellular signals and regulatory networks intrinsic to the OL lineage. Recently, basic transcriptional mechanisms that control OL differentiation and myelination have begun to be elucidated at the molecular level and on a genome scale. The interplay between transcription factors activated by differentiation-promoting signals and master regulators likely exerts a crucial role in controlling stage-specific progression of the OL lineage. In this review, we describe the current state of knowledge regarding the transcription factors and the epigenetic programs including histone methylation, acetylation, chromatin remodeling, micro-RNAs, and noncoding RNAs that regulate development of OLs and myelination.
The transcription factors and epigenetic mechanisms that control oligodendrocyte differentiation and myelination continue to be elucidated. In particular, genome-scale analyses have elucidated the relationships between key transcription factors and their direct targets.
Along with neurons and astrocytes, oligodendrocyte (OL) precursor cells (OPCs) arise from multipotent neuroepithelial progenitor cells in the neurogenic niches of the developing and adult central nervous system (CNS). Once specified, OPCs remain highly proliferative and motile, dividing as they migrate out throughout the CNS. Once in their final position, they can undergo a terminal differentiation event before myelinating adjacent axons (see Simons and Nave 2015). These highly dynamic cellular processes, which occur on an ongoing basis during both development and in adulthood, are mediated by equally dynamic changes in the expression and activity of transcription factors and epigenetic programs. Perhaps more so than for any other CNS cell type, these transcriptional and epigenetic programs have been mapped out for the OL lineage, using combinations of expression profiling, electroporation of the developing neural tube, and knockout/transgenic experiments. More recently, genome-wide analysis of transcription factor binding, histone methylation, and acetylation patterns has been effectively used to elucidate the direct targets and relationships between key factors. This review summarizes some of the major transcriptional and epigenetic programs and key molecules that mediate development of the OL lineage and myelination in the CNS.
TRANSCRIPTION FACTORS MEDIATING SPECIFICATION TO THE OL LINEAGE
During both embryonic development and also within neurogenic niches in the adult, OPCs are specified from neuroepithelial precursor cells, which also give rise to neurons and astrocytes. The initial transcriptional control of specification of neural progenitor cells to the OL lineage is tightly related to the transcriptional control of dorsoventral patterning of the neural tube, largely established by gradients of Sonic hedgehog (Shh) and bone morphogenic proteins (BMPs). During early development of the spinal cord, OPCs arise from the ventral pMN domain, which gives rise first to motor neurons and then to OPCs. This pMN domain is established and defined by the transcription factor Olig2, which is therefore essential for the generation of these early ventrally derived OPCs (Novitch et al. 2001; Zhou et al. 2001; Fu et al. 2002; Lu et al. 2002; Zhou and Anderson 2002). Less directly, other transcription factors that are involved in defining the borders of the pMN domain, such as Nkx6-1 (Liu et al. 2003) and Gli2 (Qi et al. 2003), also influence the production of OPCs, as both the extent of the pMN domain and OPC specification are reduced in their absence.
Although Olig2 is vital for this early ventral production of OPCs and is a consistent marker of the OL lineage, at later stages of embryogenesis, OPCs also arise from more dorsal regions of the neural tube that do not initially express Olig2 or the Nkx6 genes (Cai et al. 2005; Vallstedt et al. 2005; Kessaris et al. 2006; Richardson et al. 2006). To some extent, this probably reflects compensation by the closely related Olig1, especially within the hindbrain (Zhou and Anderson 2002), but it does suggest that Olig2 is not absolutely essential for specification to the OL lineage. Nevertheless, Olig2 most likely has a relatively broad role in promoting neural precursors toward an OL fate, given forced expression of Olig2 either in the developing neural tube (Zhou et al. 2001; Liu et al. 2007) or cultured embryonic stem (ES) cells (Du et al. 2006) promotes oligodendrogliogenesis.
Like the pMN domain in general, Olig2 has a dual role in promoting both motor neuron and OL fate. Phosphorylation of Olig2 at a serine residue (Ser147) promotes motor neuron specification, whereas, at later points in development, dephosphorylation at this site shifts the balance toward production of OLs, largely through sequestration of the proneural transcription factor Ngn2 (Li et al. 2011). More broadly, during development, several other transcription factors are involved in this neural–glial switch throughout the nervous system. Ascl1 (also known as Mash1) appears to have a broad role in promoting specification to the OL lineage, as Ascl1 knockout mice display a severe reduction in the number of OPCs (Parras et al. 2007; Sugimori et al. 2008). At early ages, Ascl1 promotes OL specification at the expense of neurons (Petryniak et al. 2007); at later ages, it promotes OL specification at the expense of generation of astrocytes (Nakatani et al. 2013). Sox9 also promotes glial specification, including to the OL lineage, as generation of OPCs is substantially delayed in Sox9 conditional knockout embryos (Stolt et al. 2003). At least some of Sox9’s gliogenic role is likely to be mediated through its direct induction of the proglial/antineuronal transcription factor nuclear factor I/A (NFIA) (Kang et al. 2012), as Nfia null mice likewise display substantial reduction specification of neural progenitors to the OL lineage (Deneen et al. 2006).
TRANSCRIPTION FACTORS IN OPCs
Transcriptional Control of Maintenance of OPCs
Following their specification to the OL lineage, OPCs express a number of transcription factors as they migrate from their birthplace to colonize the CNS. These included continued expression of Olig1 and Olig2 as well as induced expression of Nkx2-2 and Sox10. Electroporation of Olig2 expression vectors into the developing chick spinal cord strongly induces the expression of both Nkx2-2 (Liu et al. 2007) and Sox10 (Zhou et al. 2001; Liu et al. 2007), suggesting that Olig2 acts to induce these two factors following specification. This was confirmed with the later discovery that Olig2 directly targets upstream enhancers of the Sox10 gene (Kuspert et al. 2011; Yu et al. 2013). Despite its early expression within the OL lineage, Nkx2-2 does not appear to be vital for the maintenance of OPCs. Instead, Nkx2-2 null mice show defects in the terminal differentiation of OPCs into OLs (Qi et al. 2001) (see below), a finding consistent with the transient up-regulation of Nkx2-2 protein observed during differentiation (Fu et al. 2002). Similarly, despite the early expression of Sox10 in the lineage, mice lacking Sox10 display a relatively normal complement of OPCs (Stolt et al. 2002). However, deletion of both Sox10 and the closely related Sox9 within the OL lineage results in a marked reduction in the density of Olig2-positive OPCs within the developing spinal cord, indicating that the two SoxE proteins have partially redundant but important roles within OPCs (Finzsch et al. 2008). The loss of OPCs seen in the Sox9/10 double mutants is associated with a substantial decrease in the expression of PDGFR-α within the remaining Olig2+ OPCs, suggesting that a major role of SoxE proteins Sox9 and Sox10 within the OPCs is to promote the expression of PDGFR-α (Finzsch et al. 2008), which in turn promotes survival and proliferation of the OPCs via binding of its secreted ligand PDGF (Barres et al. 1993; Calver et al. 1998).
Transcriptional Regulation of Terminal Differentiation
One of the most important points of regulation of the OL lineage and of CNS myelination is the stage at which an OPC exits the cell cycle and differentiates into an OL. Although mature OLs are capable of dedifferentiating in vitro in the presence of basic fibroblast growth factor (bFGF) (Grinspan et al. 1996), there appears to be little evidence of this occurring in vivo, where the OL differentiation step seems to be a terminal event. As such, the differentiation step needs to be tightly controlled both to regulate the timing of myelination during development and to maintain a pool of OPCs capable of subsequent division and differentiation throughout life.
Negative Regulators of Terminal Differentiation
OPCs express high levels of the transcription factors Sox5, Sox6, Hes5, Id2, and Id4 (Fig. 1). Together, these factors mediate the influences of inhibitory extracellular signals, which act on the OPCs to prevent their differentiation and myelination. For example, Id2 and Id4 are downstream from the BMPs (Samanta and Kessler 2004) and the antidifferentiation G protein–coupled receptor Gpr17 (Chen et al. 2009). Modulation of Id2 levels strongly influences the propensity of OPCs to differentiate; overexpression inhibits OPC differentiation in vitro, whereas Id2 knockout OPCs show accelerated differentiation in vitro even in conditions that would normally support their continued proliferation (Wang et al. 2001). Some mechanistic insight into how Id2 and Id4 may inhibit OL differentiation comes from the finding that they can physically interact with the bHLH proteins Olig1 and Olig2 (Samanta and Kessler 2004), presumably blocking their activity in the nucleus. In addition to the mRNA for Id2 and Id4 being strongly down-regulated during differentiation (Fig. 1), the expression of Id2 protein also shifts from nuclear to cytoplasmic (Wang et al. 2001), indicating that differentiation involves regulation of the Id proteins at both the transcriptional and posttranslational level.
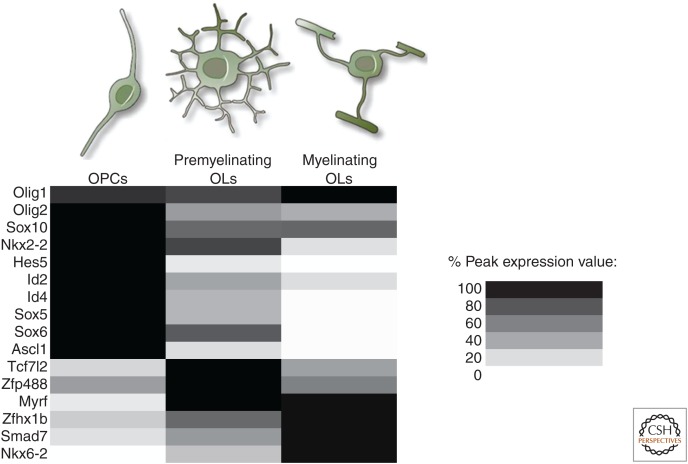
Figure 1.
Expression of transcription factors during oligodendrocyte (OL) differentiation. Expression levels of relevant transcription factors in OPCs, premyelinating OLs, and myelinating OLs. The intensity of the shading reflects the expression level relative to the gene’s peak level within the lineage. Note the high expression of inhibitors of differentiation and myelination (Hes5, Id2, Id4, Sox5, and Sox6) in OPCs, all of which are rapidly down-regulated at the premyelinating stage. Tcf7l2 and Zfp488 show a more transient expression during differentiation, suggesting a relatively specific role during differentiation. Factors including myelin regulatory factor (Myrf), Zfhx1b, Smad7, and Nkx6-2 show later expression, consistent with their roles in myelination and beyond. (Expression data from Cahoy et al. 2008.)
The Notch signaling pathway, in which extracellular ligands cause the proteolytic cleavage and nuclear translocation of the membrane-associated transcription factor Notch, mediates numerous aspects of neural development (Louvi and Artavanis-Tsakonas 2006). Among these, the pathway is a major limiting factor in OPC differentiation during development; axons initially express high levels of the Notch ligand Jagged, with this being down-regulated coinciding with myelination (Wang et al. 1998). Treatment of OPCs in culture with Notch ligands Delta or Notch strongly inhibits OPC differentiation (Wang et al. 1998). Conversely, Notch1 haploinsufficiency in vivo results in precocious myelination within the CNS (Givogri et al., 2002). Notch largely acts to limit OPC differentiation through its transcriptional target Hes5. Just as Id2 and Id4 have been found to interact with and block the transcriptional activity of the Olig proteins (Samanta and Kessler 2004), Hes5 binds to and competes with the promyelination factor Sox10, preventing its activity at the myelin basic protein (MBP) promoter (Liu et al. 2006). Consistent with this, the brains of Hes5 null mice show accelerated expression of markers of myelinating cells including MBP and proteolipid protein (PLP), and forced expression of Hes5 in cultured OPCs blocks their differentiation into GalC-positive OLs (Liu et al. 2006).
The Wnt pathway is a key extracellular inhibitor of OL differentiation, both in the contexts of normal development and injury/remyelination (Fancy et al. 2009; Ye et al. 2009). Consistent with a key role during the differentiation step, one of the main partners of β-catenin, the Tcf7l2 transcription factor, is relatively transiently expressed during differentiation at the late-OPC/premyelinating OL step (Fig. 1) (Fancy et al. 2009; Fu et al. 2009; Ye et al. 2009). Tcf7l2 and β-catenin form a transcriptional complex that inhibits differentiation, although the direct targets of this complex in the OL lineage likely include the inhibitory Id2 (Ye et al. 2009), as is the case in carcinoma cells (Rockman et al. 2001). Nevertheless, OL differentiation is reduced rather than enhanced in Tcf7l2 null mice (Fu et al. 2009; Ye et al. 2009) and delayed in mice in which β-catenin is inactivated in the OL lineage (Dai et al. 2014), indicating that the roles of the β-catenin and Tcf7l2 are more complex than simply mediating the inhibitory effects on Wnt signaling. This could be partially explained by Tcf7l2 interacting with histone deacetylases HDAC1 and HDAC2 following down-regulation of Wnt signaling and β-catenin degradation (see below), thus repressing rather than activating the expression of inhibitors of differentiation (Ye et al. 2009). In addition, the role of Wnt/β-catenin signaling may be highly stage specific, inhibiting specification, promoting some aspects of differentiation, but ultimately needing to be down-regulated for full differentiation (Dai et al. 2014).
Although Sox10 (along with Sox9) has a role in the up-regulation of PDGFR-α, the predominant role of Sox10 within the lineage appears to be prodifferentiation and myelination. Within OPCs, the SoxD proteins Sox5 and Sox6 act to antagonize Sox10’s prodifferentiation role. Both Sox5 and Sox6 are expressed in neural progenitor cells and OPCs, being down-regulated during differentiation (Stolt et al. 2006). This down-regulation is at least partially mediated by induction of several micro-RNAs during OL differentiation that target Sox6 and other OPC genes (Dugas et al. 2010; Zhao et al. 2010). Ablation of both the Sox5 and Sox6 genes in the OL lineage results in marked precocious expression of mature markers, such as PLP and MBP in the developing spinal cord (Stolt et al. 2006), highlighting the importance of their role in limiting differentiation in vivo. At the molecular level, Sox5 and Sox6 are capable of binding the promoter region of the MBP gene, antagonizing the activity of Sox10 (Stolt et al. 2006)—presumably this antagonism extends to other Sox10 targets as well.
Prodifferentiation Factors
In contrast to the above factors, which serve to inhibit OL differentiation, several transcription factors induced early in the specification in the lineage appear to have a role in promoting differentiation. One of these is Olig2. Because of its importance in specification of the pMN domain and the paucity of OPCs in Olig2 null mice (Lu et al. 2002; Zhou and Anderson 2002), its subsequent role in OL differentiation was initially difficult to show, although its continued expression in the lineage suggested a likely role. Using a conditional knockout strategy to bypass the initial OPC specification defect (Cai et al. 2007; Mei et al. 2013), it was found that deletion of Olig2 in 3′-cyclic nucleotide 3′-phosphodiesterase (CNP+) postmitotic OPCs and differentiating OLs substantially reduced the density of mature (CC1+) OLs and myelin postnatally. Surprisingly, Olig2 deletion in PLP+ OLs at the neonatal stage enhanced OL myelination (Mei et al. 2013), although the fate of these Olig2-ablated cells requires more detailed evaluation. Given Olig2 has generally been considered to be a transcriptional repressor (Novitch et al. 2001), the molecular mechanisms by which it might act to promote differentiation were initially unclear, but are now thought to be highly dependent on its relationship with the chromatin-remodeling enzyme Brg1, with which it acts on OL enhancers in a stage-specific manner (Yu et al. 2013; and see below).
The zinc finger homeobox transcription factor Zfhx1b (also known as Sip1) was initially identified as a direct transcriptional target of Olig2, also being downstream from Olig1 (Weng et al. 2012). Despite having normal numbers of OPCs, Zfhx1b conditional knockout mice fail to express myelin genes or myelinate—conversely, forced expression of Zfhx1b in cultured OPCs promotes their differentiation (Weng et al. 2012). At least part of Zfhx1b’s activity is likely to be through antagonism of the p300/SMAD complex at the promoter regions of Id2, Id4, and Hes5, as well as induction of the Smad7, a negative-feedback regulator of the BMP signaling complexes (Weng et al. 2012). In this way, induction of Zfhx1b would be well placed to antagonize the inhibitory effects of a number of the inhibitory signals that converge on Id2, Id4, and Hes5, including the BMPs, Notch, and Wnt. The Wnt pathway is also inhibited by Sox17, which is transiently expressed in the lineage and acts to promote exit of the cell cycle and differentiation (Sohn et al. 2006). Sox17 promotes the degradation of β-catenin, thus relieving the Wnt/β-catenin /Tcf7l2 brake on differentiation (Chew et al. 2011).
REGULATORS OF OL MATURATION AND MYELINATION
Following terminal differentiation, postmitotic OLs undergo profound changes in their morphology, ceasing migration and extending elaborate networks of processes to contact adjacent axons. They subsequently withdraw many of these processes, but dramatically increase their total surface area as they ensheath some of the adjacent axons with myelin and assemble the axoglial structure of the node of Ranvier. This change in cellular function and morphology is underpinned by an equally profound change in gene expression, with genes involved in cell cycle being down-regulated and genes required for myelination and paranodal formation being induced in several distinct “waves” (Dugas et al. 2006; Cahoy et al. 2008). Perhaps most conspicuous among the genes induced during myelination are the genes encoding myelin-associated proteins, such as PLP, myelin-associated glycoprotein (MAG), and MBP, which together are strongly induced following OL differentiation (Dubois-Dalcq et al. 1986). Accordingly, many studies on transcription factors in myelination have focused on their effects on the promoter regions of these genes (e.g., Berndt et al. 2001; Qi et al. 2001; Stolt et al. 2002; Gokhan et al. 2005; Xin et al. 2005; Li et al. 2007a). Nevertheless, a wide variety of genes are induced as part of the molecular machinery for myelination including genes encoding cytoskeletal proteins and regulators, lipid metabolism enzymes, and components of the axoglial junction; the full extent of these genes and their regulation is the current focus of genome-wide expression and binding studies. Strikingly, OL differentiation essentially stalls at either the differentiation or early myelination stage in knockouts for a number of transcription factors, including Nkx2-2, Olig1, Ascl1, YY1, Zfhx1b, Sox10, and Myrf (Qi et al. 2001; Stolt et al. 2002; Xin et al. 2005; He et al. 2007; Sugimori et al. 2008; Emery et al. 2009; Weng et al. 2012). Given the severity of the phenotype of all these knockouts, it would seem that all of these factors must act nonredundantly, either in parallel or downstream from one another, to drive the maturation and myelination program (see Fig. 2 for a proposed model of how some of these factors interact).
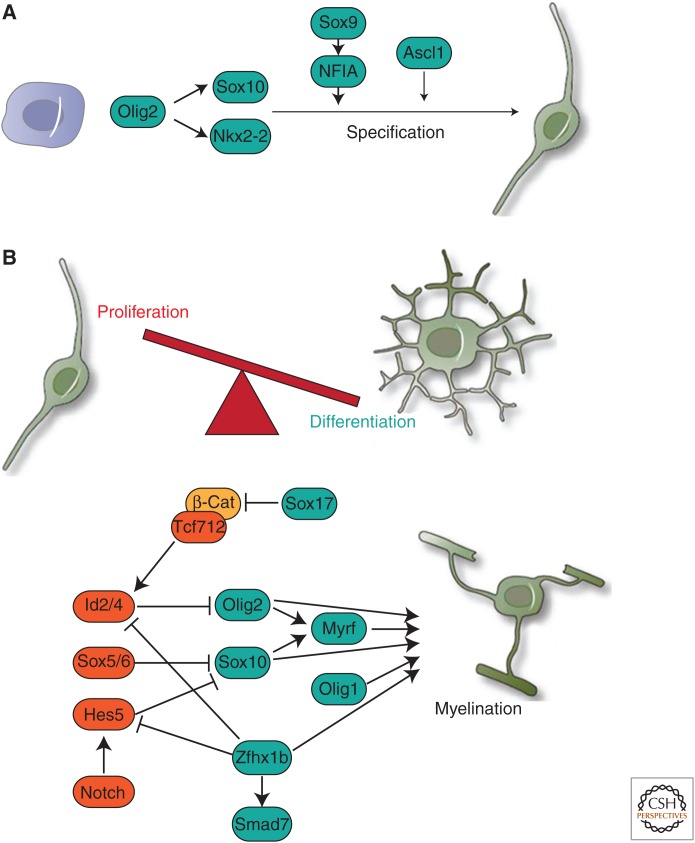
Figure 2.
Transcription factors in OL specification and differentiation. (A) Within neural precursor cells, Olig2 acts to induce the expression of Sox10 and Nkx2-2, promoting specification to the OL lineage, also inhibiting the proneuronal Ngn2 in its dephosphorylated form. Sox9 induction of the NFIA promotes gliogenesis at the expense of neurogenesis, Ascl1 also broadly promotes specification of progenitors to the OL lineage. (B) Within OPCs, the balance between proliferation/maintenance and terminal differentiation is maintained by anti- and prodifferentiation factors. Antidifferentiation factors including Id2, Sox5, Sox6, and Hes5 (orange) mediate the effects of a number of extracellular inhibitors of differentiation, antagonizing the activity of factors that have predominantly prodifferentiation roles (blue). On the balance being tipped toward terminal differentiation, a cascade is initiated in which factors including Olig2, Sox10, Olig1, and Myrf mediate the myelination program. β-Cat, β-Catenin.
Although Olig1 is expressed from early in the lineage, in Olig1 knockout mice, defects are largely limited to the postmitotic stage. The original Olig1-null mouse strain Olig1-Cre-PGK-neo−/− shows a developmental delay in OL maturation (Lu et al. 2002), while a subsequent strain of Olig1 null mice that removed the PGK-Neo cassette (Olig1-Cre−/−) developed severe myelination deficits and died postnatally (Xin et al. 2005). Two more recently generated Olig1 null strains, Olig1-PGK-neo-NICD−/− and an Olig−/−:Olig2Tg strain carrying Olig1-PGK-hygromycin, show a similar phenotype to the original Olig1-Cre-PGK-neo−/− line, displaying delayed OL differentiation embryonically (de Faria et al. 2014). At present, the exact mechanisms for the weak versus strong knockout phenotypes in these different lines are not fully understood, but may be because of differences in the cassettes used in the targeting constructs or unintended disruption of nearby genes. Irrespective of these differences, consistent with the observed delay or loss of mature markers, such as MBP in Olig1 null mice, Olig1 promotes transcription from the MBP promoter in luciferase assays (Xin et al. 2005; Li et al. 2007a), implicating MBP as a direct target gene. Perhaps paradoxically, as the OLs mature during myelination the subcellular distribution of the Olig1 protein shifts from nuclear to cytoplasmic (Arnett et al. 2004). This shift is dependent on phosphorylation of the Olig1 Serine138 residue (Niu et al. 2012). Although forced expression of nuclear-targeted Olig1 is sufficient to drive MBP expression, cytoplasmic shift of Olig1 is required to promote membrane expansion, a vital requirement of myelin sheath formation (Niu et al. 2012).
As with many of the OL lineage transcription factors, Nkx2-2’s importance is underscored by the severity of the null phenotype. In Nkx2-2 null mice, there is a substantial decrease in expression of MBP and PLP in the developing nervous system (Qi et al. 2001). Consistent with the differentiation defect seen in Nkx2-2 knockouts, it can promote GFP expression from the PLP promoter in heterologous cells (Qi et al. 2001). Somewhat paradoxically, however, it has also been shown to repress activity at the MBP promoter (Gokhan et al. 2005; Wei et al. 2005) or the Sirt2 promoter in the CG-4 oligodendroglial cell line (Ji et al. 2011).
Unlike Olig1, Sox10, and Nkx2-2, all of which are strongly expressed by OPCs, myelin regulatory factor (Myrf, previously known as GM98, C11Orf9, and MRF) is strongly induced during the early stages of OL differentiation (Fig. 1). This induction of Myrf requires the activity of SoxE proteins Sox8 and Sox10 at an enhancer found in the first intron of the Myrf gene during differentiation (Hornig et al. 2013). This regulatory region of the Myrf gene is also targeted by the Olig2/Brg1 chromatin-remodeling complex (Yu et al. 2013), suggesting that Myrf is a common target of both Olig2 and Sox10. Myrf then acts to promote OL maturation and myelination; in Myrf conditional knockout mice, OL differentiation stalls at the early premyelinating stage resulting in a complete failure of CNS myelination and postnatal death (Emery et al. 2009). This phenotype is remarkably similar to the stalling of differentiation before myelination seen in Olig1 null mice (Xin et al. 2005) and in Sox10 mutant zebrafish (Takada et al. 2010), suggesting that all three factors, either alone or in combination, are required for the initiation of myelination. In addition to be required for the initiation of myelination, Myrf is also required on an ongoing basis for the maintenance of the myelin; conditional ablation of the gene in mature OLs of the adult CNS results in a loss of myelin gene expression that ultimately results in a degeneration of the myelin sheaths (Koenning et al. 2012). This contrasts with the effects of conditional ablation of Olig2 in PLP-positive OLs, which instead causes an increase in Olig1 and myelin gene expression (Mei et al. 2013).
Genome-wide binding studies (ChIP-Seq) found that Myrf directly targets a wide range of genes underpinning the morphological development and myelination by OLs, including cytoskeletal genes, lipid metabolism genes, other transcription factors, such as Smad7 (see above) and Nkx6-2 (see below), and the more well-studied myelin proteins, such as MBP and PLP (Bujalka et al. 2013). Notably, independently performed ChIP-Seq studies have indicated that there is substantial overlap between the binding sites for Olig2, Myrf, and Sox10, suggesting a close functional relationship between the three factors in regulation of myelin gene expression (Bujalka et al. 2013; Yu et al. 2013). Consistent with this, Myrf and Sox10 show synergy at a subset of myelin gene regulatory regions in luciferase assays (Hornig et al. 2013), although many regulatory regions appeared to be selectively targeted by either Sox10 or Myrf (Bujalka et al. 2013; Hornig et al. 2013). This gives rise to a model in which Olig2 and Brg1 would transiently act on the regulatory elements of key genes during differentiation to make them accessible, with factors including Sox10 and Myrf subsequently acting on these genes during the myelination phase.
The use of genome-wide occupancy (ChIP-Seq) studies to elucidate the direct targets of Myrf and Olig2 highlights the usefulness of this technology, which has similarly been successfully used to define the direct targets of Sox10 and Egr2 in Schwann cells of the peripheral nervous system (PNS) (Srinivasan et al. 2012). It seems likely that the field will continue to benefit from this approach for some of the other transcription factors for which the immediate and most relevant targets are yet to be established. For example, in addition to its role in OL specification (see above), Ascl1 knockout mice display a substantial reduction in mature OLs (Sugimori et al. 2008), suggesting an important role in differentiation. Consistent with this, conditional ablation of the Ascl1 gene in OPCs reduces OL differentiation, instead favoring symmetric division events resulting in two OPCs (Nakatani et al. 2013). Nevertheless, the genes that Ascl1 targets that mediate this effect are yet to be determined, as are the case for the equally important Nkx2-2 (see above). Similarly, the transcription factor Nkx6-2 (also known as Gtx) is up-regulated at the late stages of OL differentiation (Awatramani et al. 1997) where it is required for proper formation of the paranodes (Southwood et al. 2004). Again, the majority of Nkx6-2 targets are yet to be identified. Presumably, future genome-wide occupancy studies for these factors and others will be important in systematically determining the broad range of targets for each transcription factor and fully characterizing the transcriptional network underpinning myelination.
EPIGENETIC REGULATORS IN OL DEVELOPMENT
The above transcriptional program of OL differentiation and myelination is highly influenced and reliant on a parallel process of epigenetic regulation. Epigenetic regulation, which by definition is mediated through mechanisms other than changes in the targeted DNA sequence, is crucial for cell growth, differentiation, and organogenesis. It comprises highly interconnected processes including at least four main classes of epigenetic marks: histone modifications, ATP-dependent chromatin remodeling, a network of noncoding RNAs (e.g., microRNAs and lncRNAs), and DNA methylation (Fig. 3) (Hsieh and Gage 2004). The transition from progenitor to mature OL shows rapid and substantial chromatin remodeling (Nielsen et al. 2002), suggesting that chromatin reorganization is an event crucial for OL differentiation. Activators and repressors of gene transcription during differentiation must negotiate through the underlying organization and structure of chromatin. There is emerging evidence that epigenetic regulation plays an important role in OL development, myelination, and remyelination.
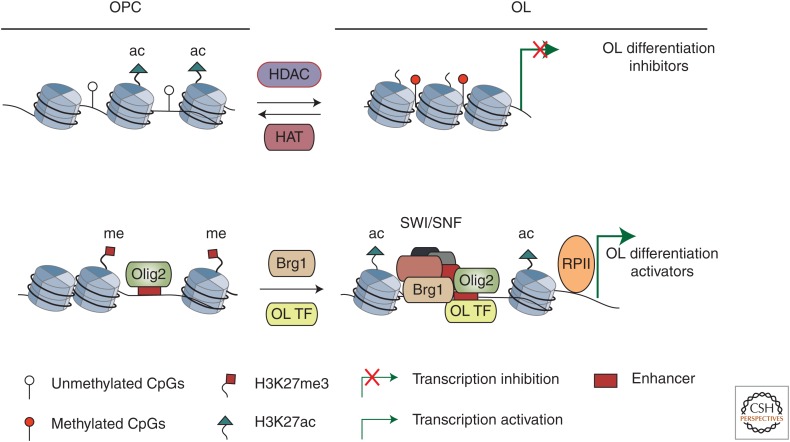
Figure 3.
Chromatin remodeling coupled with transcriptional control of oligodendrocyte (OL) differentiation. (Upper panel) In OL precursor cells (OPCs), histone-modifying enzymes histone deacetylases (HDACs), which oppose the function of histone acetyltransferases (HATs), remove the acetyl group on lysine 27 of histone 3, leading to chromatin compaction. Histone deacetylation, coupling with promoter methylation, inhibits expression of pro-OPC/progenitor genes or OL differentiation inhibitors. (Lower panel) In progenitors, Olig2 may act as a pioneer factor to target and keep the OL-lineage-specific enhancers in a poised state marked by H3K27me3. At the onset of OL differentiation, Olig2 recruits chromatin-remodeling enzymes, such as the Brg1-associated SWI/SNF complex, to acquire an open chromatin conformation on enhancers (e.g., marked by H3K27ac). This process, in cooperation with OL-specific transcription factors, such as Sox10 and Myrf, ensures proper transcription of differentiation-associated genes. ac, Acetyl group; me, methyl group; TF, transcription factor.
HISTONE-MODIFYING ENZYMES HDAC AND HAT
Histone modifications by methylation, acetylation, phosphorylation, and ubiquitylation modulate gene transcription at specific genomic loci by controlling accessibility of RNAPII and transcription regulators (Kouzarides 2007). One of the best-characterized histone modifications is histone acetylation on lysine residues regulated by histone acetyltransferases (HATs) and histone deacetylases (HDACs). Histone deacetylation by HDACs is capable of causing chromatin compaction, leading to gene silencing (Fig. 3). HDACs can be classified into four broad classes based on sequence homology with the yeast homologs and domain organization: class I (HDAC1–3, and 8), class II (HDAC4–7 and 9–10), NAD-dependent class III (SIRT1–7), and class IV (HDAC11) (McKinsey et al. 2000; Gao et al. 2002; Grozinger and Schreiber 2002; Gregoretti et al. 2004; Haberland et al. 2009). At present, the role of HATs in OL development remains elusive; however, a series of studies indicate that HDACs or their activities are required for OPC differentiation and myelination processes.
Treatment of pharmacological HDAC inhibitors (e.g., trichostatin A [TSA] or valproic acid) blocks OL differentiation in culture and during animal development (Marin-Husstege et al. 2002; Shen et al. 2005). Similarly, HDAC inhibition by TSA and sodium butyrate shows an inhibitory effect on differentiation of human primary OPCs (Conway et al. 2012).
Effects of HDAC inhibition on OL development appear to be temporally specific. HDAC inhibitors impair the early phase of OPC differentiation, but do not affect myelin gene expression after the onset of myelination (Shen et al. 2005). These inhibitors also block the remyelination process that follows cuprizone-induced demyelination. Impeded HDAC1 and HDAC2 recruitment to myelin gene promoters in aging animals is likely associated with a decrease in myelination potential, suggesting an age-dependent role for HDAC (Shen et al. 2008).
RNAi-mediated gene silencing suggests that class I HDACs (HDAC1 and 2) are critical for OPC differentiation in vitro (Shen et al. 2008). In vivo loss-of-function studies with HDAC1/2 knocked-out in the OL lineage directed by an Olig1-promoter driven Cre show an essential role for both HDAC1 and HDAC2 in OL differentiation and myelination (Ye et al. 2009), although HDAC1 or HDAC2 single mutations do not yield detectable myelination defects. This suggests functional redundancy of HDAC1 and HDAC2 in myelination (Ye et al. 2009). Similarly, both HDAC1 and HDAC2 have been shown to regulate the transcriptional program of myelination in Schwann cells and control their survival (Chen et al. 2011; Jacob et al. 2011), indicating a conserved function of HDAC1 and HDAC2 in central and peripheral myelination.
HDACs show multifaceted actions to regulate target gene expression. Despite the lack of DNA-binding activity, HDACs interact with other transcriptional regulators to form repressive complexes to inhibit expression of OPC differentiation inhibitors or block the activation of neuronal differentiation genes (He et al. 2007; Ye et al. 2009). For instance, HDAC1 can recruit YY1 to the promoter of OL differentiation inhibitors, including Id2, Id4, and Hes5, to repress their transcription (Fig. 3) (Shen et al. 2005; He et al. 2007). HDAC1 and HDAC2 can hijack TCF effectors, such as Tcf7l2/Tcf4, and disrupt the β-catenin/TCF transcription complex (Ye et al. 2009), thereby preventing expression of Wnt signaling target genes Id2 and Id4, the potent inhibitors of OL differentiation.
HDACs also reportedly compete with Notch intracellular domain (NCID) for binding to canonical C-promoter binding factor (CBF), thereby inhibiting transcription of Notch effectors, such as Hes5, an inhibitor of OL differentiation (Kadam and Emerson 2003; Sekiya and Zaret 2007). Moreover, HDACs can be recruited by the Groucho-related corepressors (GRO/TLE/GRG) to inhibit downstream events of Notch and Wnt signaling pathways to control timely OL myelination and remyelination after demyelinating injury (Fancy et al. 2009). HDAC1 and HDAC2 are also able to modulate the acetylation state of nonhistone protein substrates, such as NF-κB, to orchestrate myelin gene expression, suggesting that HDAC1 and HDAC2 serve a developmental switch through regulating NF-κB activity to control myelination in the PNS (Chen et al. 2011).
A class III HDAC (NAD+)-dependent deacetylase sirtuin 2 (SIRT2) is enriched in OLs and localized to the outer and juxtanodal loops in the myelin sheath (Li et al. 2007b; Werner et al. 2007). Loss of SIRT2 in PLP1 null mice suggests that the tetraspan protein PLP is required for its transport into myelin compartments, which could be necessary for stabilization/protection from degradation (Werner et al. 2007). The main substrate for the SIRT2 deacetylase has been identified as acetylated α-tubulins rather than histones proteins (Li et al. 2007b). An RNAi-mediated knockdown study reveals that SIRT2 has a role in OL cytoskeleton remodeling and process arborization (Li et al. 2007b). In Schwann cells, SIRT2 may regulate myelin formation by deacetylating Par-3, a regulator of cell polarity (Beirowski et al. 2011). Class IV HDAC11 reportedly regulates OL-specific gene expression and cell development in an oligodendroglial cell line (Liu et al. 2009), although its function in OL development in vivo is currently unknown.
At present, the role of class II HDACs in OL development remains elusive. Class IIa HDACs have no enzymatic activity owing to the lack of the key catalytic Tyr residue (Jones et al. 2008). Interestingly, deacetylase-dead HDAC3 mutants retain the same degree of function in repressing lipogenic gene expression and rescuing the hepatosteatosis phenotype in HDAC3-depleted mouse liver (Sun et al. 2013; You et al. 2013). These studies point to an important nonenzymatic role of HDACs. Essentially all existing HDAC inhibitors exert their inhibition by chelating the zinc metal ion in their active site (Gryder et al. 2012). Because there are more than 300 zinc-dependent enzymes in each cell, HDAC inhibitors may interfere their activities besides HDACs. Whether and, if so, to what extent the deacetylase enzyme activity of HDACs contributes to biological functions in OL differentiation and CNS myelination in vivo may call for further investigation.
How the specificity of HDACs in regulating OL-specific gene expression is achieved remains elusive. In neural progenitor cells, transcriptional corepressors, such as REST/NRSF, NcoR, or noncoding RNAs, have been shown to recruit HDACs to repress neuronal gene expression while facilitating glial gene expression (Kuwabara et al. 2004; Abrajano et al. 2009). In addition, distinct HDACs may control the timing of OL linage progression by repressing differentiation inhibitors in a stage-specific manner. How HDAC activity, expression or targeting specificity is regulated during myelination remains to be defined in this field. Nonetheless, through modulating multiple pathways such as Wnt and Notch signaling, HDACs could function as a convergent point that controls the myelination program in the CNS.
ATP-DEPENDENT CHROMATIN REMODELING COMPLEX
In addition to histone modification, chromatin remodeling is also regulated by ATP-dependent chromatin remodeling enzyme complexes, which use energy derived from ATP hydrolysis to alter chromatin structures. Chromatin remodelers impart specific chromatin states and regulate the accessibility of transcription factors to DNA as a prerequisite for transcriptional activation (Ho and Crabtree 2010). Depending on the core ATPase in the complexes, they can be divided into three broad subfamilies, which include SWI2/SNF2 (or Brg1/Brm1-associated factors [BAF]), imitation switch (ISWI), chromodomain helicase DNA-binding protein (CHD), and INO80 in vertebrates (Ho and Crabtree 2010). The BAF complexes use either Brahma-related gene-1 (Brg1/Smarca4) or Brahma (Brm), two Drosophila Brm homologs, to provide ATPase activity (Reyes et al. 1998; Bultman et al. 2000). The core ATPase can form a complex with other protein subunits, which can be assembled combinatorially. The subunit compositions have been proposed to define the functional specificity of chromatin remodeler complexes (Yoo and Crabtree 2009).
SWI/SNF complexes achieve their functional specificity by dynamic exchange of subunits (Lemon et al. 2001; Wu et al. 2009). A neural progenitor-specific BAF complex contains the PHD-domain BAF45a and the actin-related protein BAF53a and is required for self-renewal and proliferation of neural progenitors (Olave et al. 2002; Lessard et al. 2007), whereas a neuron-specific BAF complex (nBAF) contains BAF45b/c and BAF53b subunits, which plays a role in neurite outgrowth and synapse formation (Wu et al. 2007). The subunit exchange allows BAF complexes to interact with distinct sets of transcription factors at different stages of neural cell lineage development. The deletion of Brg1 in mouse neural stem cells results in the failure of neural progenitor maintenance and the reduction of gliogenesis (Seo et al. 2005; Matsumoto et al. 2006; Lessard et al. 2007). Currently, the glial subtype-specific subunit(s) for the BAF complex have not been identified.
Whole-genome RNA polymerase II (RNAPII) targeting analysis has been used to map gene transcripts that are activated during the onset of OPC differentiation (Yu et al. 2013). This study identifies Smarca4/Brg1 as the gene most significantly targeted by RNAPII at the onset of differentiation. The Brg1 chromatin remodeler is necessary for expression of myelination-associated genes and promotes the transition from progenitors to differentiating OLs. Remarkably, Brg1 targeting specificity on the enhancers is transcriptionally prepatterned by Olig2, which function as a potential pioneer factor, to facilitate initiation and establishment of the transcriptional program that promotes OL differentiation (Yu et al. 2013).
Further targeting studies of Brg1 and Olig2 along with active and repressive histone marks at different developmental stages reveal a temporally regulated mechanism in controlling the stage-specific transcriptional program during OL lineage progression. During the early phase of differentiation, Brg1/Olig2 are recruited to the enhancers of differentiation-promoting genes, such as Sox10 and Myrf, whereas at the late stage of differentiation, they are recruited to the enhancers of cellular morphogenesis-associated regulators, such as Cdc42 and Rac1, critical for cytoskeleton reorganization and curvature-dependent actin polymerization (Thurnherr et al. 2006; Benninger et al. 2007). Thus, OL lineage progression requires chromatin remodeling on myelination-promoting genes in a stage-specific manner (Yu et al. 2013). Moreover, Brg1/BAF chromatin remodelers may couple with transcriptionally linked chromatin activation (e.g., H3k27ac) and repression (e.g., H3k9me3) marks to activate the transcriptional program for myelination (Yu et al. 2013).
In the PNS, Brg1 can be recruited by Sox10 to the promoters of Oct6 and Krox20, the key transcription factors in Schwann cell differentiation, and stimulates their expression to promote myelination (Weider et al. 2012). In addition, Brg1 forms a complex with NF-κB to regulate Schwann cell differentiation, suggesting a critical conserved role of Brg1 in regulating central and peripheral myelination (Limpert et al. 2013).
At present, other ATP-dependent chromatin remodeling complexes in regulation of CNS myelination remain to be defined. The CHD family member, CHD4, reportedly regulates myelination in the PNS (Hung et al. 2012). CHD4 conditional mutant mice show a delay in myelination and an increase of immature Schwann cell proliferation. CHD4 acts together with another NuRD component, Mta2, to target the gene loci that are also regulated by a myelin-promoting factor, Egr2/Krox20, during myelination. This study suggests that CHD4 is required for proper timing of PNS myelination. Interestingly, CHD4 can cooperate with the PRC2 polycomb silencer Ezh2 to inhibit astroglial differentiation (Sparmann et al. 2013), indicating a role of CHD4 in glial cell fate specification.
MicroRNAs AND NONCODING RNAs
MicroRNAs (miRNAs) are a class of ∼21–23 nucleotide small noncoding RNAs. The primary precursors of miRNAs (pri-miRNAs) are transcribed by RNA polymerase II. In animals, most pri-miRNAs are processed by the endonuclease Drosha/DGCR8 in the nucleus and then by the RNaseIII-like endonuclease, Dicer1, in the cytoplasm (Bartel 2009). They form a RISC complex (RNA-induced silencing complex) through either perfect or imperfect base-pairing of their 5′ “seed” sequence with the 3′ untranslational regions (UTR) of target genes, and inhibit gene expression mainly by either translational inhibition or mRNA destabilization.
Deletion of Dicer1 in neural progenitor cells blocks neurogenesis while facilitating gliogenesis (Kawase-Koga et al. 2009; Zheng et al. 2010). Importantly, inactivation of Dicer1 by Cre recombinases in the OL lineage directed by Olig1, Olig2, or CNP promoters causes severe dysmyelination and motor deficits including tremors and seizures (Dugas et al. 2010; Zhao et al. 2010). Dicer1 deletion leads to an increase of OPC proliferation, and a drastic reduction in myelination. Although mice with Dicer1 deletion mediated by Olig1-Cre (Zhao et al. 2010) or Olig2-Cre-tva (avian tumor virus receptor A) (Dugas et al. 2010) show similar dysmyelination, the former mutants lose the majority of myelin in the CNS and die around postnatal week three, whereas the latter shows a developmental delay (He et al. 2012). Nonetheless, despite the different extent of dysmyelination in two Dicer1 mutants, these studies suggest that miRNAs are required for OPC differentiation while inhibiting OPC proliferation. Deletion of Dicer1 in mature OLs by inducible OL-expressing PLPCreERT resulted in demyelination and progressive axonal degeneration, leading to shorter animal lifespan (Shin et al. 2009). Thus, miRNAs are required for not only for OPC differentiation, but also myelin maintenance and homeostasis in OLs.
A series of miRNA microarray profiling studies using isolated OLs (Lau et al. 2008; Dugas et al. 2010) or the spinal cord from myelin-deficient Dicer1 knockout mice (Zhao et al. 2010) have identified a cohort of miRNAs that are highly enriched in mature OLs. Most notably, miR-219 and miR-338 are substantially increased at the onset of OL differentiation. Overexpression of miR-219 or miR-338 is sufficient to accelerate OPC differentiation in culture and promote precocious expression of OL lineage markers in the developing chick neural tube and mouse CNS (Zhao et al. 2010). Consistently, knockdown of miR-219 in OPCs (Dugas et al. 2010; Zhao et al. 2010) or in zebrafish embryos (Zhao et al. 2010) inhibits OPC differentiation, suggesting that miR-219 is both sufficient and essential for OL differentiation. It is worth noting that miR-338 is not detected in the CNS of zebrafish embryos, and miR-338 knockdown does not appear to affect myelination in zebrafish (Zhao et al. 2010). A cohort of miRNAs with a transient expression pattern has been identified including miR-138, which is expressed in the early differentiation phase but down-regulated in mature OLs. Overexpression of miR-138 appears to enhance differentiation initiation but inhibits OL terminal differentiation (Dugas et al. 2010). The targets and functions of miR-138 in vivo remained elusive at present.
Target predictions based on computational algorithms reveal a cohort of potential miR-219 targets, such as differentiation inhibitory factors, including PDGFR-α, Hes5, and Sox6 (Dugas et al. 2010; Zhao et al. 2010). Despite the lack of sequence homology between miR-219 and miR-338, both miRNAs are predicted to target the negative regulators of OL differentiation, such as Hes5 and Sox6 (Wang et al. 1998; Stolt et al. 2006). miR-219 and miR-338 are sufficient to repress expression of Hes5 and Sox6. Thus, miR-219 and miR-338 may act synergistically to alleviate differentiation brakes to facilitate OPC differentiation (Nave 2010). miR-219 expression likely permits or promotes OL differentiation by suppressing PDGFR-α expression in OPCs or causing OPCs less responsive to mitogen signals like PDGF (Fig. 4) (Dugas et al. 2010; Zhao et al. 2010). Interestingly, miR-219 may also target other factors involved in neurogenesis, such as Zfp238, FoxJ3, NeuroD1, Isl1, and Otx2 (Dugas et al. 2010; Zhao et al. 2010), which are the opposing cues to gliogenesis. Similarly, miR-7a can induce OPC specification by directly targeting proneural genes (e.g., Pax6, NeuroD4), and it also prevents further OPC differentiation through repressing differentiation regulators (Zhao et al. 2012).
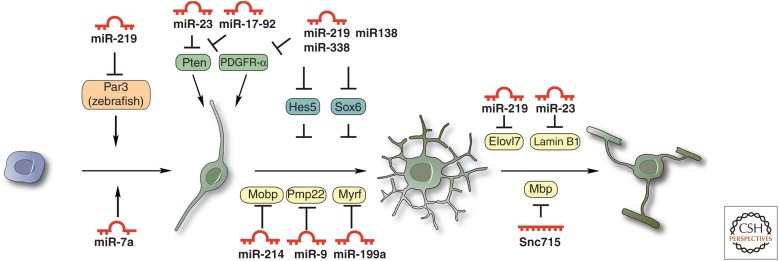
Figure 4.
Stage-specific miRNA control of OL lineage progression. The expression pattern of miRNAs is correlated with their functions at different stages of OL development. OPC-enriched miRNAs (e.g., miR-9, miR-214, miR-199a-5p, and miR-7a) target myelin-associated components including Mobp and PMP22 and differentiation-promoting factors including Myrf to inhibit OL differentiation. OL-enriched miRNAs, such as miR-138, miR-219, and miR-338, promote OL differentiation by inhibiting differentiation inhibitors (e.g., Hes5, Sox6, and Sox5). miR-219 likely permits or promotes OL differentiation by suppressing PDGFR-α expression in OPCs or causing OPCs less responsive to PDGF mitogen. Besides controlling OPC differentiation, miR-219 may also have multifaceted functions in OPC specification (e.g., by targeting cell polarity protein Par3 in zebrafish) and myelin maintenance (e.g., by targeting fatty acid elongase Elvol7).
OPC-enriched miRNAs, miR-214 and miR-199a-5p, can target and inhibit expression of Mobp and C11Orf/Myrf in OPCs derived from human ES cells, respectively, to prevent precocious differentiation (Letzen et al. 2010). Oncomir-1 encoding polycistronic miR-17–92 cluster shows an important role in controlling OPC number and proliferation. miR-19b and miR-17 are highly enriched in the OL lineage including A2B5+ OPCs and GalC+ OL (Lau et al. 2008; Budde et al. 2010). Targeted inactivation of the miR-17–92 cluster resulted in a reduction of Olig2-positive OLs in the brain (Budde et al. 2010). miR-19b appears to target the tumor suppressor gene Pten, leading to the activation of downstream Akt signaling, and promotes OPC proliferation. Recently, another miRNA, miR-223, has been shown to suppress glial precursor proliferation by repressing NFIA. Overexpression of miR-223 inhibits tumorigenesis in a human glioma cell line, suggesting a role for miR-223 in gliogenesis and glioma formation (Glasgow et al. 2013). Thus, OL-expressing miRNAs may prevent the fate of neural progenitors from neurogenesis while promoting OL lineage commitment and maintaining cell identity at proper stages. A recent study in zebrafish indicates that miR-219 promotes precursor differentiation by inhibiting Par polarity proteins (Hudish et al. 2013). miR-219 targets the 3′ UTRs of Pard3/Par-3 and Prkci mRNAs and reduces their expression to promote cell-cycle exit and differentiation of neural precursors (Hudish et al. 2013). It remains to be determined whether miR-219-targeted Par proteins have a conserved role in regulating OL development in mice. Besides regulating the differentiation program, miR-219 also regulates myelin homeostasis, such as by targeting fatty acid elongase Elovl7, which synthesizes very long chain of fatty acid, to modulate lipid metabolism (Shin et al. 2009).
MiR-23 is identified as a negative regulator of lamin B1, of which the overexpression causes severe myelin loss (Lin and Fu 2009). miR-23a overexpression in transgenic mice enhances both OL differentiation and myelin thickness by modulating the PTEN/Akt/mTOR signaling axis. miR-23a may activate a long noncoding RNA, 2700046G09Rik, to inhibit PTEN (Lin et al. 2013). Interestingly, an increased level of the neuronal miR-124 appears to be associated with hippocampal demyelination and memory dysfunction (Dutta et al. 2013), suggesting that maintenance of proper miRNA levels is required for myelin integrity. Currently, the in vivo role of distinct miRNAs in OL development remains to be further determined by gene targeting in animals. Nonetheless, a network of miRNAs would likely individually or cooperatively regulate distinct stages of OL lineage development, as well as safeguards against the expression of neuronal and other cell lineage genes (Fig. 4).
A small nucleolar noncoding RNA (snc715) is identified as an inhibitor of MBP translation by associating with MBP mRNA transport granule components (Bauer et al. 2012). Recently, long noncoding RNAs (lncRNAs) are increasingly being documented as important epigenetic regulators of gene expression. Although a cohort of lncRNAs that are expressed in distinct cell types of the CNS have been identified recently (Cahoy et al. 2008; Mercer et al. 2010), their functions in OL development and myelination remain to be defined. Different regulatory noncoding RNAs may act cooperatively and combinatorially to modulate the activities of transcription factors, chromatin remodelers, and signaling networks at the posttranscriptional level and thereby regulate OL fate specification and differentiation.
DNA METHYLATION
DNA methylation is an important epigenetic regulatory mechanism during the nervous system development. The majority of DNA methylation occurs on cytosines that precede a guanine nucleotide or CpG sites in the 5′ promoter of genes. The majority of gene promoters, roughly 70%, reside within CpG islands, which are unmethylated (Saxonov et al. 2006). The establishment, recognition, and removal of DNA methylation are regulated by three classes of enzymes, namely, the writers, DNA methyltransferases (e.g., Dnmt1 and Dnmt3), the erasers, ten-eleven translocation (TET) family enzymes (TET1, TET2 and TET3) that can oxidize 5-methylcytosine (Kohli and Zhang 2013), and the readers, methyl-CpG-binding domain-containing proteins (MBDs).
DNA methylation shows a cell-type-specific distribution and pattern in neurons (Guo et al. 2013) and glia-enriched white matter (Mastronardi et al. 2007). The differentiation switch in which neural progenitors transition from neurogenesis to astrocyte differentiation coincides with DNA methylation (Teter et al. 1996; Takizawa et al. 2001). Dnmt1 and Dnmt3a have been shown to control the timing of astrogliogenesis (Fan et al. 2005; Nguyen et al. 2007). A recent study reports that 5-hydroxymethylcytosine (5hmC) is enriched in active gene regions where 5-methylcytosine (5mC) is strongly depleted in neurons (Mellen et al. 2012). Interestingly, Rett-syndrome-causing mutation R133C preferentially disrupts MeCP2 binding to 5hmC but not 5mC (Mellen et al. 2012). At present, the function of DNA methylation in OL development is not fully understood.
Interestingly, brains from multiple sclerosis (MS) patients show specific alterations in DNA methylation (Mastronardi et al. 2007; Huynh et al. 2014). Studies on DNA isolated from white matter of MS brains reveal hypomethylation (Mastronardi et al. 2007), which is likely caused by a higher DNA demethylase activity in MS brains compared with normal controls rather than a decrease of DNA methyltransferase activity. One of hypomethylated targets identified in MS brains is on the promoter of PAD2, which encodes is a peptidylarginine deiminase that catalyzes the conversion of the guanidine group of arginine to citrulline in MBP (Mastronardi et al. 2007). Hypomethylation leads to elevation of PAD2 levels in MS brains (Mastronardi et al. 2007) and in a transgenic animal model of MS (Moscarello et al. 2013). Abnormal citrullination of MBP could potentially sensitize T cells and lead to subsequent enhancement of the autoimmune response during MS pathogenesis (D’Souza et al. 2005; Harauz and Musse 2007). Thus, there is a possibility that specific sets of genes or proteins are altered in MS brains because epigenetic modifications may induce or potentiate MS pathogenesis.
Recently, a gene-chip array has been used to examine genome-wide DNA methylation in MS brains (Huynh et al. 2014). The study identifies subtle but reproducible differences between pathology-free white-matter regions derived from human MS-affected and control brains. Integrated DNA methylation and RNA-sequencing analysis shows hypermethylation of the genes that regulate cell survival, such as BCL2L2 and NDRG1, that are expressed at lower levels in MS brains, whereas genes involved in proteolytic processing (e.g., LGMN, CTSZ) were hypomethylated and elevated. These studies suggest an epigenomic change in genes affecting the vulnerability of OLs to pathological insults during MS pathogenesis.
The studies of epigenomic alterations on DNA methylation, histone modifications, and microRNA deregulation will contribute to our understanding of demyelinating disease pathogenesis. Although much of the studies have been conducted during normal development, there are several challenges that we will be facing in understanding the role of epigenetic regulation of gene expression in demyelinating diseases. First, epigenetic marks are cell-type specific, yet much of the research in this area could mainly be performed at a tissue level, such as the white matter, because isolation of enough material to scrutinize specific cell types is often infeasible in human subjects. One approach to address this concern may be to identify cell-specific epigenetic marks to decompose complex tissues into cell-specific components; this approach may prove useful in identifying pathogenesis in a cell-type-specific manner, for instance, by using a combination of single-cell real-time polymerase chain reaction (RT-PCR) and bisulfite sequencing. Second, another challenge is the dynamic nature of epigenetic marks that are context dependent. Thus, the epigenetic alteration needs to be considered in the context of environment, such as diet, exposures, disease states, and age. Finally, a major challenge with epigenomics is to experimentally and analytically integrate the epigenetic mechanisms that affect gene transcription and translation. Evidence for cross talk between DNA methylation and histone modifications in promoter and enhancer regions and miRNAs/noncoding RNAs that target to the 3′ untranslated regions has been rapidly accumulating. The complexity of these epigenetic interactions presents unique experimental and analytic challenges. Using transcriptional regulation and epigenetic mechanisms to understand the dynamic biology in demyelinating diseases and applying this knowledge to the development of novel diagnostic and therapeutic approaches presents promising opportunities for our patients with demyelinating diseases, such as MS.
Footnotes
Editors: Ben A. Barres, Marc R. Freeman, and Beth Stevens
Additional Perspectives on Glia available at www.cshperspectives.org
REFERENCES
*Reference is also in this collection.
- Abrajano JJ, Qureshi IA, Gokhan S, Zheng D, Bergman A, Mehler MF. 2009. Differential deployment of REST and CoREST promotes glial subtype specification and oligodendrocyte lineage maturation. PLoS ONE 4: e7665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arnett HA, Fancy SP, Alberta JA, Zhao C, Plant SR, Kaing S, Raine CS, Rowitch DH, Franklin RJ, Stiles CD. 2004. bHLH transcription factor Olig1 is required to repair demyelinated lesions in the CNS. Science 306: 2111–2115. [DOI] [PubMed] [Google Scholar]
- Awatramani R, Scherer S, Grinspan J, Collarini E, Skoff R, O’Hagan D, Garbern J, Kamholz J. 1997. Evidence that the homeodomain protein Gtx is involved in the regulation of oligodendrocyte myelination. J Neurosci 17: 6657–6668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barres BA, Schmid R, Sendnter M, Raff MC. 1993. Multiple extracellular signals are required for long-term oligodendrocyte survival. Development 118: 283–295. [DOI] [PubMed] [Google Scholar]
- Bartel DP. 2009. MicroRNAs: Target recognition and regulatory functions. Cell 136: 215–233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bauer NM, Moos C, van Horssen J, Witte M, van der Valk P, Altenhein B, Luhmann HJ, White R. 2012. Myelin basic protein synthesis is regulated by small non-coding RNA 715. EMBO Rep 13: 827–834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beirowski B, Gustin J, Armour SM, Yamamoto H, Viader A, North BJ, Michan S, Baloh RH, Golden JP, Schmidt RE, et al. 2011. Sir-two-homolog 2 (Sirt2) modulates peripheral myelination through polarity protein Par-3/atypical protein kinase C (aPKC) signaling. Proc Natl Acad Sci 108: E952–E961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benninger Y, Thurnherr T, Pereira JA, Krause S, Wu X, Chrostek-Grashoff A, Herzog D, Nave K-A, Franklin RJ, Meijer D, et al. 2007. Essential and distinct roles for cdc42 and rac1 in the regulation of Schwann cell biology during peripheral nervous system development. J Cell Biol 177: 1051–1061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berndt JA, Kim JG, Tosic M, Kim C, Hudson LD. 2001. The transcriptional regulator Yin Yang 1 activates the myelin PLP gene. J Neurochem 77: 935–942. [DOI] [PubMed] [Google Scholar]
- Budde H, Schmitt S, Fitzner D, Opitz L, Salinas-Riester G, Simons M. 2010. Control of oligodendroglial cell number by the miR-17–92 cluster. Development 137: 2127–2132. [DOI] [PubMed] [Google Scholar]
- Bujalka H, Koenning M, Jackson S, Perreau VM, Pope B, Hay CM, Mitew S, Hill AF, Lu QR, Wegner M, et al. 2013. MYRF is a membrane-associated transcription factor that autoproteolytically cleaves to directly activate myelin genes. PLoS Biol 11: e1001625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bultman S, Gebuhr T, Yee D, La Mantia C, Nicholson J, Gilliam A, Randazzo F, Metzger D, Chambon P, Crabtree G, et al. 2000. A Brg1 null mutation in the mouse reveals functional differences among mammalian SWI/SNF complexes. Mol Cell 6: 1287–1295. [DOI] [PubMed] [Google Scholar]
- Cahoy JD, Emery B, Kaushal A, Foo LC, Zamanian JL, Christopherson KS, Xing Y, Lubischer JL, Krieg PA, Krupenko SA, et al. 2008. A transcriptome database for astrocytes, neurons, and oligodendrocytes: A new resource for understanding brain development and function. J Neurosci 28: 264–278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai J, Qi Y, Hu X, Tan M, Liu Z, Zhang J, Li Q, Sander M, Qiu M. 2005. Generation of oligodendrocyte precursor cells from mouse dorsal spinal cord independent of Nkx6 regulation and Shh signaling. Neuron 45: 41–53. [DOI] [PubMed] [Google Scholar]
- Cai J, Chen Y, Cai WH, Hurlock EC, Wu H, Kernie SG, Parada LF, Lu QR. 2007. A crucial role for Olig2 in white matter astrocyte development. Development 134: 1887–1899. [DOI] [PubMed] [Google Scholar]
- Calver AR, Hall AC, Yu WP, Walsh FS, Heath JK, Betsholtz C, Richardson WD. 1998. Oligodendrocyte population dynamics and the role of PDGF in vivo. Neuron 20: 869–882. [DOI] [PubMed] [Google Scholar]
- Chen Y, Wu H, Wang S, Koito H, Li J, Ye F, Hoang J, Escobar SS, Gow A, Arnett HA, et al. 2009. The oligodendrocyte-specific G protein-coupled receptor GPR17 is a cell-intrinsic timer of myelination. Nat Neurosci 12: 1398–1406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y, Wang H, Yoon SO, Xu X, Hottiger MO, Svaren J, Nave K-A, Kim HA, Olson EN, Lu QR. 2011. HDAC-mediated deacetylation of NF-κB is critical for Schwann cell myelination. Nat Neurosci 14: 437–441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chew LJ, Shen W, Ming X, Senatorov VV Jr, Chen HL, Cheng Y, Hong E, Knoblach S, Gallo V. 2011. SRY-box containing gene 17 regulates the Wnt/β-catenin signaling pathway in oligodendrocyte progenitor cells. J Neurosci 31: 13921–13935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conway GD, O’Bara MA, Vedia BH, Pol SU, Sim FJ. 2012. Histone deacetylase activity is required for human oligodendrocyte progenitor differentiation. Glia 60: 1944–1953. [DOI] [PubMed] [Google Scholar]
- Dai ZM, Sun S, Wang C, Huang H, Hu X, Zhang Z, Lu QR, Qiu M. 2014. Stage-specific regulation of oligodendrocyte development by Wnt/β-catenin signaling. J Neurosci 34: 8467–8473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Faria JP, Kessaris N, Andrew P, Richardson WD, Li H. 2014. New Olig1 null mice confirm a non-essential role for Olig1 in oligodendrocyte development. BMC Neurosci 15: 12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deneen B, Ho R, Lukaszewicz A, Hochstim CJ, Gronostajski RM, Anderson DJ. 2006. The transcription factor NFIA controls the onset of gliogenesis in the developing spinal cord. Neuron 52: 953–968. [DOI] [PubMed] [Google Scholar]
- D’Souza CA, Wood DD, She YM, Moscarello MA. 2005. Autocatalytic cleavage of myelin basic protein: An alternative to molecular mimicry. Biochemistry 44: 12905–12913. [DOI] [PubMed] [Google Scholar]
- Du ZW, Li XJ, Nguyen GD, Zhang SC. 2006. Induced expression of Olig2 is sufficient for oligodendrocyte specification but not for motoneuron specification and astrocyte repression. Mol Cell Neurosci 33: 371–380. [DOI] [PubMed] [Google Scholar]
- Dubois-Dalcq M, Behar T, Hudson L, Lazzarini RA. 1986. Emergence of three myelin proteins in oligodendrocytes cultured without neurons. J Cell Biol 102: 384–392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dugas JC, Tai YC, Speed TP, Ngai J, Barres BA. 2006. Functional genomic analysis of oligodendrocyte differentiation. J Neurosci 26: 10967–10983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dugas JC, Cuellar TL, Scholze A, Ason B, Ibrahim A, Emery B, Zamanian JL, Foo LC, McManus MT, Barres BA. 2010. Dicer1 and miR-219 are required for normal oligodendrocyte differentiation and myelination. Neuron 65: 597–611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dutta R, Chomyk AM, Chang A, Ribaudo MV, Deckard SA, Doud MK, Edberg DD, Bai B, Li M, Baranzini SE, et al. 2013. Hippocampal demyelination and memory dysfunction are associated with increased levels of the neuronal microRNA miR-124 and reduced AMPA receptors. Ann Neurol 73: 637–645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emery B, Agalliu D, Cahoy JD, Watkins TA, Dugas JC, Mulinyawe SB, Ibrahim A, Ligon KL, Rowitch DH, Barres BA. 2009. Myelin gene regulatory factor is a critical transcriptional regulator required for CNS myelination. Cell 138: 172–185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan G, Martinowich K, Chin MH, He F, Fouse SD, Hutnick L, Hattori D, Ge W, Shen Y, Wu H, et al. 2005. DNA methylation controls the timing of astrogliogenesis through regulation of JAK-STAT signaling. Development 132: 3345–3356. [DOI] [PubMed] [Google Scholar]
- Fancy SP, Baranzini SE, Zhao C, Yuk DI, Irvine KA, Kaing S, Sanai N, Franklin RJ, Rowitch DH. 2009. Dysregulation of the Wnt pathway inhibits timely myelination and remyelination in the mammalian CNS. Genes Dev 23: 1571–1585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finzsch M, Stolt CC, Lommes P, Wegner M. 2008. Sox9 and Sox10 influence survival and migration of oligodendrocyte precursors in the spinal cord by regulating PDGF receptor α expression. Development 135: 637–646. [DOI] [PubMed] [Google Scholar]
- Fu H, Qi Y, Tan M, Cai J, Takebayashi H, Nakafuku M, Richardson W, Qiu M. 2002. Dual origin of spinal oligodendrocyte progenitors and evidence for the cooperative role of Olig2 and Nkx2.2 in the control of oligodendrocyte differentiation. Development 129: 681–693. [DOI] [PubMed] [Google Scholar]
- Fu H CJ, Clevers H, Fast E, Gray S, Greenberg R, Jain MK, Ma Q, Qiu M, Rowitch DH, Taylor CM, et al. 2009. A genome-wide screen for spatially restricted expression patterns identifies transcription factors that regulate glial development. J Neurosci 29: 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao L, Cueto MA, Asselbergs F, Atadja P. 2002. Cloning and functional characterization of HDAC11, a novel member of the human histone deacetylase family. J Biol Chem 277: 25748–25755. [DOI] [PubMed] [Google Scholar]
- Givogri MI, Costa RM, Schonmann V, Silva AJ, Campagnoni AT, Bongarzone ER. 2002. Central nervous system myelination in mice with deficient expression of Notch1 receptor. J Neurosci Res 67: 309–320. [DOI] [PubMed] [Google Scholar]
- Glasgow SM, Laug D, Brawley VS, Zhang Z, Corder A, Yin Z, Wong ST, Li XN, Foster AE, Ahmed N, et al. 2013. The miR-223/nuclear factor I-A axis regulates glial precursor proliferation and tumorigenesis in the CNS. J Neurosci 33: 13560–13568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gokhan S, Marin-Husstege M, Yung SY, Fontanez D, Casaccia-Bonnefil P, Mehler MF. 2005. Combinatorial profiles of oligodendrocyte-selective classes of transcriptional regulators differentially modulate myelin basic protein gene expression. J Neurosci 25: 8311–8321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gregoretti IV, Lee YM, Goodson HV. 2004. Molecular evolution of the histone deacetylase family: Functional implications of phylogenetic analysis. J Mol Biol 338: 17–31. [DOI] [PubMed] [Google Scholar]
- Grinspan JB, Reeves MF, Coulaloglou MJ, Nathanson D, Pleasure D. 1996. Re-entry into the cell cycle is required for bFGF-induced oligodendroglial dedifferentiation and survival. J Neurosci Res 46: 456–464. [DOI] [PubMed] [Google Scholar]
- Grozinger CM, Schreiber SL. 2002. Deacetylase enzymes: Biological functions and the use of small-molecule inhibitors. Chem Biol 9: 3–16. [DOI] [PubMed] [Google Scholar]
- Gryder BE, Sodji QH, Oyelere AK. 2012. Targeted cancer therapy: Giving histone deacetylase inhibitors all they need to succeed. Future Med Chem 4: 505–524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo JU, Su Y, Shin JH, Shin J, Li H, Xie B, Zhong C, Hu S, Le T, Fan G, et al. 2013. Distribution, recognition and regulation of non-CpG methylation in the adult mammalian brain. Nat Neurosci 17: 215–222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haberland M, Montgomery RL, Olson EN. 2009. The many roles of histone deacetylases in development and physiology: Implications for disease and therapy. Nat Rev Genet 10: 32–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harauz G, Musse AA. 2007. A tale of two citrullines—Structural and functional aspects of myelin basic protein deimination in health and disease. Neurochem Res 32: 137–158. [DOI] [PubMed] [Google Scholar]
- He Y, Dupree J, Wang J, Sandoval J, Li J, Liu H, Shi YN, Nave K-A, Casaccia-Bonnefil P. 2007. The transcription factor Yin Yang 1 is essential for oligodendrocyte progenitor differentiation. Neuron 55: 217–230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He X, Yu Y, Awatramani R, Lu QR. 2012. Unwrapping myelination by microRNAs. Neuroscientist 18: 45–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho L, Crabtree GR. 2010. Chromatin remodelling during development. Nature 463: 474–484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hornig J, Frob F, Vogl MR, Hermans-Borgmeyer I, Tamm ER, Wegner M. 2013. The transcription factors Sox10 and Myrf define an essential regulatory network module in differentiating oligodendrocytes. PLoS Genet 9: e1003907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsieh J, Gage FH. 2004. Epigenetic control of neural stem cell fate. Curr Opin Genet Dev 14: 461–469. [DOI] [PubMed] [Google Scholar]
- Hudish LI, Blasky AJ, Appel B. 2013. miR-219 regulates neural precursor differentiation by direct inhibition of apical par polarity proteins. Dev Cell 27: 387–398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hung H, Kohnken R, Svaren J. 2012. The nucleosome remodeling and deacetylase chromatin remodeling (NuRD) complex is required for peripheral nerve myelination. J Neurosci 32: 1517–1527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huynh JL, Garg P, Thin TH, Yoo S, Dutta R, Trapp BD, Haroutunian V, Zhu J, Donovan MJ, Sharp AJ, et al. 2014. Epigenome-wide differences in pathology-free regions of multiple sclerosis-affected brains. Nat Neurosci 17: 121–130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacob C, Christen CN, Pereira JA, Somandin C, Baggiolini A, Lotscher P, Ozcelik M, Tricaud N, Meijer D, Yamaguchi T, et al. 2011. HDAC1 and HDAC2 control the transcriptional program of myelination and the survival of Schwann cells. Nat Neurosci 14: 429–436. [DOI] [PubMed] [Google Scholar]
- Ji S, Doucette JR, Nazarali AJ. 2011. Sirt2 is a novel in vivo downstream target of Nkx2.2 and enhances oligodendroglial cell differentiation. J Mol Cell Biol 3: 351–359. [DOI] [PubMed] [Google Scholar]
- Jones P, Altamura S, De Francesco R, Gallinari P, Lahm A, Neddermann P, Rowley M, Serafini S, Steinkuhler C. 2008. Probing the elusive catalytic activity of vertebrate class IIa histone deacetylases. Bioorg Med Chem Lett 18: 1814–1819. [DOI] [PubMed] [Google Scholar]
- Kadam S, Emerson BM. 2003. Transcriptional specificity of human SWI/SNF BRG1 and BRM chromatin remodeling complexes. Mol Cell 11: 377–389. [DOI] [PubMed] [Google Scholar]
- Kang P, Lee HK, Glasgow SM, Finley M, Donti T, Gaber ZB, Graham BH, Foster AE, Novitch BG, Gronostajski RM, et al. 2012. Sox9 and NFIA coordinate a transcriptional regulatory cascade during the initiation of gliogenesis. Neuron 74: 79–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawase-Koga Y, Otaegi G, Sun T. 2009. Different timings of dicer deletion affect neurogenesis and gliogenesis in the developing mouse central nervous system. Dev Dyn 238: 2800–2812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kessaris N, Fogarty M, Iannarelli P, Grist M, Wegner M, Richardson WD. 2006. Competing waves of oligodendrocytes in the forebrain and postnatal elimination of an embryonic lineage. Nat Neurosci 9: 173–179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koenning M, Jackson S, Hay CM, Faux C, Kilpatrick TJ, Willingham M, Emery B. 2012. Myelin gene regulatory factor is required for maintenance of myelin and mature oligodendrocyte identity in the adult CNS. J Neurosci 32: 12528–12542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohli RM, Zhang Y. 2013. TET enzymes, TDG and the dynamics of DNA demethylation. Nature 502: 472–479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kouzarides T. 2007. Chromatin modifications and their function. Cell 128: 693–705. [DOI] [PubMed] [Google Scholar]
- Kuspert M, Hammer A, Bosl MR, Wegner M. 2011. Olig2 regulates Sox10 expression in oligodendrocyte precursors through an evolutionary conserved distal enhancer. Nucleic Acids Res 39: 1280–1293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuwabara T, Hsieh J, Nakashima K, Taira K, Gage FH. 2004. A small modulatory dsRNA specifies the fate of adult neural stem cells. Cell 116: 779–793. [DOI] [PubMed] [Google Scholar]
- Lau P, Verrier JD, Nielsen JA, Johnson KR, Notterpek L, Hudson LD. 2008. Identification of dynamically regulated microRNA and mRNA networks in developing oligodendrocytes. J Neurosci 28: 11720–11730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemon B, Inouye C, King DS, Tjian R. 2001. Selectivity of chromatin-remodelling cofactors for ligand-activated transcription. Nature 414: 924–928. [DOI] [PubMed] [Google Scholar]
- Lessard J, Wu JI, Ranish JA, Wan M, Winslow MM, Staahl BT, Wu H, Aebersold R, Graef IA, Crabtree GR. 2007. An essential switch in subunit composition of a chromatin remodeling complex during neural development. Neuron 55: 201–215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Letzen BS, Liu C, Thakor NV, Gearhart JD, All AH, Kerr CL. 2010. MicroRNA expression profiling of oligodendrocyte differentiation from human embryonic stem cells. PLoS ONE 5: e10480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Lu Y, Smith HK, Richardson WD. 2007a. Olig1 and Sox10 interact synergistically to drive myelin basic protein transcription in oligodendrocytes. J Neurosci 27: 14375–14382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W, Zhang B, Tang J, Cao Q, Wu Y, Wu C, Guo J, Ling EA, Liang F. 2007b. Sirtuin 2, a mammalian homolog of yeast silent information regulator-2 longevity regulator, is an oligodendroglial protein that decelerates cell differentiation through deacetylating α-tubulin. J Neurosci 27: 2606–2616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, de Faria JP, Andrew P, Nitarska J, Richardson WD. 2011. Phosphorylation regulates OLIG2 cofactor choice and the motor neuron-oligodendrocyte fate switch. Neuron 69: 918–929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Limpert AS, Bai S, Narayan M, Wu J, Yoon SO, Carter BD, Lu QR. 2013. NF-κB forms a complex with the chromatin remodeler BRG1 to regulate Schwann cell differentiation. J Neurosci 33: 2388–2397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin ST, Fu YH. 2009. miR-23 regulation of lamin B1 is crucial for oligodendrocyte development and myelination. Dis Model Mech 2: 178–188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin ST, Huang Y, Zhang L, Heng MY, Ptacek LJ, Fu YH. 2013. MicroRNA-23a promotes myelination in the central nervous system. Proc Natl Acad Sci 110: 17468–17473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu R, Cai J, Hu X, Tan M, Qi Y, German M, Rubenstein J, Sander M, Qiu M. 2003. Region-specific and stage-dependent regulation of Olig gene expression and oligodendrogenesis by Nkx6.1 homeodomain transcription factor. Development 130: 6221–6231. [DOI] [PubMed] [Google Scholar]
- Liu A, Li J, Marin-Husstege M, Kageyama R, Fan Y, Gelinas C, Casaccia-Bonnefil P. 2006. A molecular insight of Hes5-dependent inhibition of myelin gene expression: Old partners and new players. EMBO J 25: 4833–4842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Z, Hu X, Cai J, Liu B, Peng X, Wegner M, Qiu M. 2007. Induction of oligodendrocyte differentiation by Olig2 and Sox10: Evidence for reciprocal interactions and dosage-dependent mechanisms. Dev Biol 302: 683–693. [DOI] [PubMed] [Google Scholar]
- Liu H, Hu Q, D’Ercole AJ, Ye P. 2009. Histone deacetylase 11 regulates oligodendrocyte-specific gene expression and cell development in OL-1 oligodendroglia cells. Glia 57: 1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Louvi A, Artavanis-Tsakonas S. 2006. Notch signalling in vertebrate neural development. Nat Rev Neurosci 7: 93–102. [DOI] [PubMed] [Google Scholar]
- Lu QR, Sun T, Zhu Z, Ma N, Garcia M, Stiles CD, Rowitch DH. 2002. Common developmental requirement for Olig function indicates a motor neuron/oligodendrocyte connection. Cell 109: 75–86. [DOI] [PubMed] [Google Scholar]
- Marin-Husstege M, Muggironi M, Liu A, Casaccia-Bonnefil P. 2002. Histone deacetylase activity is necessary for oligodendrocyte lineage progression. J Neurosci 22: 10333–10345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mastronardi FG, Noor A, Wood DD, Paton T, Moscarello MA. 2007. Peptidyl argininedeiminase 2 CpG island in multiple sclerosis white matter is hypomethylated. J Neurosci Res 85: 2006–2016. [DOI] [PubMed] [Google Scholar]
- Matsumoto S, Banine F, Struve J, Xing R, Adams C, Liu Y, Metzger D, Chambon P, Rao MS, Sherman LS. 2006. Brg1 is required for murine neural stem cell maintenance and gliogenesis. Dev Biol 289: 372–383. [DOI] [PubMed] [Google Scholar]
- McKinsey TA, Zhang CL, Lu J, Olson EN. 2000. Signal-dependent nuclear export of a histone deacetylase regulates muscle differentiation. Nature 408: 106–111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mei F, Wang H, Liu S, Niu J, Wang L, He Y, Etxeberria A, Chan JR, Xiao L. 2013. Stage-specific deletion of Olig2 conveys opposing functions on differentiation and maturation of oligodendrocytes. J Neurosci 33: 8454–8462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mellen M, Ayata P, Dewell S, Kriaucionis S, Heintz N. 2012. MeCP2 binds to 5hmC enriched within active genes and accessible chromatin in the nervous system. Cell 151: 1417–1430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mercer TR, Qureshi IA, Gokhan S, Dinger ME, Li G, Mattick JS, Mehler MF. 2010. Long noncoding RNAs in neuronal-glial fate specification and oligodendrocyte lineage maturation. BMC Neurosci 11: 14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moscarello MA, Lei H, Mastronardi FG, Winer S, Tsui H, Li Z, Ackerley C, Zhang L, Raijmakers R, Wood DD. 2013. Inhibition of peptidyl-arginine deiminases reverses protein-hypercitrullination and disease in mouse models of multiple sclerosis. Dis Model Mech 6: 467–478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakatani H, Martin E, Hassani H, Clavairoly A, Maire CL, Viadieu A, Kerninon C, Delmasure A, Frah M, Weber M, et al. 2013. Ascl1/Mash1 promotes brain oligodendrogenesis during myelination and remyelination. J Neurosci 33: 9752–9768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nave K-A. 2010. Oligodendrocytes and the “micro brake” of progenitor cell proliferation. Neuron 65: 577–579. [DOI] [PubMed] [Google Scholar]
- Nguyen S, Meletis K, Fu D, Jhaveri S, Jaenisch R. 2007. Ablation of de novo DNA methyltransferase Dnmt3a in the nervous system leads to neuromuscular defects and shortened lifespan. Dev Dyn 236: 1663–1676. [DOI] [PubMed] [Google Scholar]
- Nielsen JA, Hudson LD, Armstrong RC. 2002. Nuclear organization in differentiating oligodendrocytes. J Cell Sci 115: 4071–4079. [DOI] [PubMed] [Google Scholar]
- Niu J, Mei F, Wang L, Liu S, Tian Y, Mo W, Li H, Lu QR, Xiao L. 2012. Phosphorylated olig1 localizes to the cytosol of oligodendrocytes and promotes membrane expansion and maturation. Glia 60: 1427–1436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novitch BG, Chen AI, Jessell TM. 2001. Coordinate regulation of motor neuron subtype identity and pan-neuronal properties by the bHLH repressor Olig2. Neuron 31: 773–789. [DOI] [PubMed] [Google Scholar]
- Olave I, Wang W, Xue Y, Kuo A, Crabtree GR. 2002. Identification of a polymorphic, neuron-specific chromatin remodeling complex. Genes Dev 16: 2509–2517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parras CM, Hunt C, Sugimori M, Nakafuku M, Rowitch D, Guillemot F. 2007. The proneural gene Mash1 specifies an early population of telencephalic oligodendrocytes. J Neurosci 27: 4233–4242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petryniak MA, Potter GB, Rowitch DH, Rubenstein JL. 2007. Dlx1 and Dlx2 control neuronal versus oligodendroglial cell fate acquisition in the developing forebrain. Neuron 55: 417–433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi Y, Cai J, Wu Y, Wu R, Lee J, Fu H, Rao M, Sussel L, Rubenstein J, Qiu M. 2001. Control of oligodendrocyte differentiation by the Nkx2.2 homeodomain transcription factor. Development 128: 2723–2733. [DOI] [PubMed] [Google Scholar]
- Qi Y, Tan M, Hui CC, Qiu M. 2003. Gli2 is required for normal Shh signaling and oligodendrocyte development in the spinal cord. Mol Cell Neurosci 23: 440–450. [DOI] [PubMed] [Google Scholar]
- Reyes JC, Barra J, Muchardt C, Camus A, Babinet C, Yaniv M. 1998. Altered control of cellular proliferation in the absence of mammalian brahma (SNF2α). EMBO J 17: 6979–6991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richardson WD, Kessaris N, Pringle N. 2006. Oligodendrocyte wars. Nat Rev Neurosci 7: 11–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rockman SP, Currie SA, Ciavarella M, Vincan E, Dow C, Thomas RJ, Phillips WA. 2001. Id2 is a target of the β-catenin/T cell factor pathway in colon carcinoma. J Biol Chem 276: 45113–45119. [DOI] [PubMed] [Google Scholar]
- Samanta J, Kessler JA. 2004. Interactions between ID and OLIG proteins mediate the inhibitory effects of BMP4 on oligodendroglial differentiation. Development 131: 4131–4142. [DOI] [PubMed] [Google Scholar]
- Saxonov S, Berg P, Brutlag DL. 2006. A genome-wide analysis of CpG dinucleotides in the human genome distinguishes two distinct classes of promoters. Proc Natl Acad Sci 103: 1412–1417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sekiya T, Zaret KS. 2007. Repression by Groucho/TLE/Grg proteins: Genomic site recruitment generates compacted chromatin in vitro and impairs activator binding in vivo. Mol Cell 28: 291–303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seo S, Richardson GA, Kroll KL. 2005. The SWI/SNF chromatin remodeling protein Brg1 is required for vertebrate neurogenesis and mediates transactivation of Ngn and NeuroD. Development 132: 105–115. [DOI] [PubMed] [Google Scholar]
- Shen S, Li J, Casaccia-Bonnefil P. 2005. Histone modifications affect timing of oligodendrocyte progenitor differentiation in the developing rat brain. J Cell Biol 169: 577–589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen S, Sandoval J, Swiss VA, Li J, Dupree J, Franklin RJ, Casaccia-Bonnefil P. 2008. Age-dependent epigenetic control of differentiation inhibitors is critical for remyelination efficiency. Nat Neurosci 11: 1024–1034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin D, Shin JY, McManus MT, Ptacek LJ, Fu YH. 2009. Dicer ablation in oligodendrocytes provokes neuronal impairment in mice. Ann Neurol 66: 843–857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *.Simons M, Nave K-A. 2015. Oligodendrocytes: Myelination and axonal support. Cold Spring Harb Perspect Biol 10.1101/cshperspect.a020479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sohn J, Natale J, Chew LJ, Belachew S, Cheng Y, Aguirre A, Lytle J, Nait-Oumesmar B, Kerninon C, Kanai-Azuma M, et al. 2006. Identification of Sox17 as a transcription factor that regulates oligodendrocyte development. J Neurosci 26: 9722–9735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Southwood C, He C, Garbern J, Kamholz J, Arroyo E, Gow A. 2004. CNS myelin paranodes require Nkx6-2 homeoprotein transcriptional activity for normal structure. J Neurosci 24: 11215–11225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sparmann A, Xie Y, Verhoeven E, Vermeulen M, Lancini C, Gargiulo G, Hulsman D, Mann M, Knoblich JA, van Lohuizen M. 2013. The chromodomain helicase Chd4 is required for polycomb-mediated inhibition of astroglial differentiation. EMBO J 32: 1598–1612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srinivasan R, Sun G, Keles S, Jones EA, Jang SW, Krueger C, Moran JJ, Svaren J. 2012. Genome-wide analysis of EGR2/SOX10 binding in myelinating peripheral nerve. Nucleic Acids Res 40: 6449–6460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stolt CC, Rehberg S, Ader M, Lommes P, Riethmacher D, Schachner M, Bartsch U, Wegner M. 2002. Terminal differentiation of myelin-forming oligodendrocytes depends on the transcription factor Sox10. Genes Dev 16: 165–170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stolt CC, Lommes P, Sock E, Chaboissier MC, Schedl A, Wegner M. 2003. The Sox9 transcription factor determines glial fate choice in the developing spinal cord. Genes Dev 17: 1677–1689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stolt CC, Schlierf A, Lommes P, Hillgartner S, Werner T, Kosian T, Sock E, Kessaris N, Richardson WD, Lefebvre V, et al. 2006. SoxD proteins influence multiple stages of oligodendrocyte development and modulate SoxE protein function. Dev Cell 11: 697–709. [DOI] [PubMed] [Google Scholar]
- Sugimori M, Nagao M, Parras CM, Nakatani H, Lebel M, Guillemot F, Nakafuku M. 2008. Ascl1 is required for oligodendrocyte development in the spinal cord. Development 135: 1271–1281. [DOI] [PubMed] [Google Scholar]
- Sun Z, Feng D, Fang B, Mullican SE, You SH, Lim HW, Everett LJ, Nabel CS, Li Y, Selvakumaran V, et al. 2013. Deacetylase-independent function of HDAC3 in transcription and metabolism requires nuclear receptor corepressor. Mol Cell 52: 769–782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takada N, Kucenas S, Appel B. 2010. Sox10 is necessary for oligodendrocyte survival following axon wrapping. Glia 58: 996–1006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takizawa T, Nakashima K, Namihira M, Ochiai W, Uemura A, Yanagisawa M, Fujita N, Nakao M, Taga T. 2001. DNA methylation is a critical cell-intrinsic determinant of astrocyte differentiation in the fetal brain. Dev Cell 1: 749–758. [DOI] [PubMed] [Google Scholar]
- Teter B, Rozovsky I, Krohn K, Anderson C, Osterburg H, Finch C. 1996. Methylation of the glial fibrillary acidic protein gene shows novel biphasic changes during brain development. Glia 17: 195–205. [DOI] [PubMed] [Google Scholar]
- Thurnherr T, Benninger Y, Wu X, Chrostek A, Krause SM, Nave K-A, Franklin RJ, Brakebusch C, Suter U, Relvas JB. 2006. Cdc42 and Rac1 signaling are both required for and act synergistically in the correct formation of myelin sheaths in the CNS. J Neurosci 26: 10110–10119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vallstedt A, Klos JM, Ericson J. 2005. Multiple dorsoventral origins of oligodendrocyte generation in the spinal cord and hindbrain. Neuron 45: 55–67. [DOI] [PubMed] [Google Scholar]
- Wang S, Sdrulla AD, diSibio G, Bush G, Nofziger D, Hicks C, Weinmaster G, Barres BA. 1998. Notch receptor activation inhibits oligodendrocyte differentiation. Neuron 21: 63–75. [DOI] [PubMed] [Google Scholar]
- Wang S, Sdrulla A, Johnson JE, Yokota Y, Barres BA. 2001. A role for the helix-loop-helix protein Id2 in the control of oligodendrocyte development. Neuron 29: 603–614. [DOI] [PubMed] [Google Scholar]
- Wei Q, Miskimins WK, Miskimins R. 2005. Stage-specific expression of myelin basic protein in oligodendrocytes involves Nkx2.2-mediated repression that is relieved by the Sp1 transcription factor. J Biol Chem 280: 16284–16294. [DOI] [PubMed] [Google Scholar]
- Weider M, Kuspert M, Bischof M, Vogl MR, Hornig J, Loy K, Kosian T, Muller J, Hillgartner S, Tamm ER, et al. 2012. Chromatin-remodeling factor brg1 is required for Schwann cell differentiation and myelination. Dev Cell 23: 193–201. [DOI] [PubMed] [Google Scholar]
- Weng Q, Chen Y, Wang H, Xu X, Yang B, He Q, Shou W, Higashi Y, van den Berghe V, Seuntjens E, et al. 2012. Dual-mode modulation of Smad signaling by Smad-interacting protein Sip1 is required for myelination in the central nervous system. Neuron 73: 713–728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Werner HB, Kuhlmann K, Shen S, Uecker M, Schardt A, Dimova K, Orfaniotou F, Dhaunchak A, Brinkmann BG, Mobius W, et al. 2007. Proteolipid protein is required for transport of sirtuin 2 into CNS myelin. J Neurosci 27: 7717–7730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu JI, Lessard J, Olave IA, Qiu Z, Ghosh A, Graef IA, Crabtree GR. 2007. Regulation of dendritic development by neuron-specific chromatin remodeling complexes. Neuron 56: 94–108. [DOI] [PubMed] [Google Scholar]
- Wu JI, Lessard J, Crabtree GR. 2009. Understanding the words of chromatin regulation. Cell 136: 200–206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xin M, Yue T, Ma Z, Wu FF, Gow A, Lu QR. 2005. Myelinogenesis and axonal recognition by oligodendrocytes in brain are uncoupled in Olig1-null mice. J Neurosci 25: 1354–1365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye F, Chen Y, Hoang T, Montgomery RL, Zhao XH, Bu H, Hu T, Taketo MM, van Es JH, Clevers H, et al. 2009. HDAC1 and HDAC2 regulate oligodendrocyte differentiation by disrupting the β-catenin-TCF interaction. Nat Neurosci 12: 829–838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoo AS, Crabtree GR. 2009. ATP-dependent chromatin remodeling in neural development. Curr Opin Neurobiol 19: 120–126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- You SH, Lim HW, Sun Z, Broache M, Won KJ, Lazar MA. 2013. Nuclear receptor co-repressors are required for the histone-deacetylase activity of HDAC3 in vivo. Nat Struct Mol Biol 20: 182–187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu Y, Chen Y, Kim B, Wang H, Zhao C, He X, Liu L, Liu W, Wu LM, Mao M, et al. 2013. Olig2 targets chromatin remodelers to enhancers to initiate oligodendrocyte differentiation. Cell 152: 248–261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao X, He X, Han X, Yu Y, Ye F, Chen Y, Hoang T, Xu X, Mi QS, Xin M, et al. 2010. MicroRNA-mediated control of oligodendrocyte differentiation. Neuron 65: 612–626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao X, Wu J, Zheng M, Gao F, Ju G. 2012. Specification and maintenance of oligodendrocyte precursor cells from neural progenitor cells: Involvement of microRNA-7a. Mol Biol Cell 23: 2867–2878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng K, Li H, Zhu Y, Zhu Q, Qiu M. 2010. MicroRNAs are essential for the developmental switch from neurogenesis to gliogenesis in the developing spinal cord. J Neurosci 30: 8245–8250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Q, Anderson DJ. 2002. The bHLH transcription factors OLIG2 and OLIG1 couple neuronal and glial subtype specification. Cell 109: 61–73. [DOI] [PubMed] [Google Scholar]
- Zhou Q, Choi G, Anderson DJ. 2001. The bHLH transcription factor Olig2 promotes oligodendrocyte differentiation in collaboration with Nkx2.2. Neuron 31: 791–807. [DOI] [PubMed] [Google Scholar]