Abstract
In mass spectrometry-based bottom-up proteomics, data-independent acquisition is an emerging technique because of its comprehensive and unbiased sampling of precursor ions. However, current data-independent acquisition methods use wide precursor isolation windows, resulting in cofragmentation and complex mixture spectra. Thus, conventional database searching tools that identify peptides by interpreting individual tandem MS spectra are inherently limited in analyzing data-independent acquisition data. Here we discuss an alternative approach, peptide-centric analysis, which tests directly for the presence and absence of query peptides. We discuss how peptide-centric analysis resolves some limitations of traditional spectrum-centric analysis, and we outline the unique characteristics of peptide-centric analysis in general.
Tandem mass spectrometry has become the technology of choice for proteome characterization. In a typical bottom-up proteomic experiment, a mixture of proteins is proteolytically digested into peptides, separated by liquid chromatography, and analyzed using tandem mass spectrometry. The ultimate goal is to identify and quantify proteins by detecting and quantifying individual peptides, thereby shedding light on the underlying cellular mechanisms or phenotype. Several modes of data acquisition have been developed for bottom-up proteomics. The most commonly applied mode uses data-dependent acquisition (DDA)1, in which tandem MS (MS/MS) spectra are acquired from the dissociation of precursor ions selected from an MS survey spectrum. Constrained by the speed of instrumentation, DDA can sample only a subset of precursor ions for MS/MS characterization, generally targeting the top-N most abundant ions detected in the most recent survey spectrum. In addition, DDA is typically coupled with a method referred to as “dynamic exclusion” (1) that attempts to prevent reselection of the same m/z for some specified period of time. These acquisition strategies greatly increase proteome coverage and extend the dynamic range of detection for shotgun proteomics. The resulting MS/MS spectra are typically analyzed using sequence database searching software such as SEQUEST, Mascot, X!Tandem, MaxQuant, Comet, MS-GF+, or OMSSA (2–8). Because these algorithms identify peptides by first associating each individual spectrum with a matching peptide sequence and then aggregating the thus matched spectra into a list of identified peptides, we refer to them as “spectrum-centric analyses.” In spectrum-centric analysis, spectra are most commonly interpreted using database searching, but can also be interpreted using de novo sequencing (9–11), or by searching against a spectrum library (12–14). For the past two decades, spectrum-centric analysis has been an essential driving force for the development of large-scale shotgun proteomics using DDA.
DDA is a powerful and well-established technique for LC-MS/MS data acquisition. By targeting precursor ions observed in MS survey scans with highly selective MS/MS scans, DDA generates a large set of high quality MS/MS spectra, which can be automatically interpreted by database searching to identify thousands of proteins in a complex sample. When DDA was introduced, instrumentation was not fast enough to sample every observed precursor in the survey scan; thus, high-intensity precursors were preferentially targeted because they tend to generate higher quality MS/MS spectra that lead to peptide identification. Although this stochastic sampling approach results in a large amount of peptide identification in a single sample run, it comes at the cost of reproducibility of MS/MS acquisition between sample analyses and an inherent bias against low abundance analytes that are less likely to be sampled (15). On modern instrumentation, the speed of MS/MS acquisition has dramatically improved to the point where the majority of MS precursors that are not already in the dynamic exclusion list can be sampled for MS/MS analysis. However, even if every precursor observed in each survey MS scan is sampled, DDA is still biased against low abundance analytes that fall below the limit of detection in the MS analysis and will never be sampled. This bias is a practical limitation in the analysis of a complex mixture with high dynamic range in which many analytes will be below the limit of detection in MS analysis, but remain detectable by the more selective and more sensitive MS/MS analysis (16).
DDA remains a powerful method for identifying a large number of proteins in a sample. However, because of the incomplete sampling, when a peptide is not identified in a conventional shotgun experiment using DDA, it is incorrect to conclude that the peptide is missing from the sample, or even below the limit of detection of MS/MS, because the peptide ions may have never been sampled for MS/MS analysis. To overcome such limitations, targeted acquisition approaches such as selected reaction monitoring (SRM, also commonly referred to as multiple reaction monitoring) are often the methods of choice. In targeted acquisition approaches, a set of predetermined precursor ions are systematically subjected to MS/MS characterization throughout the LC time domain. The collision energy for each targeted ion can be optimized for fragmentation efficiency. The resulting data are typically analyzed using “targeted analysis” (17–19), in which the software looks for the co-eluting patterns from a group of predetermined pairs of precursor–product ions (called transitions). With systematic MS/MS sampling and the combined specificity of chromatographic retention time, precursor ion mass, and the distribution of product ions, targeted acquisition allows highly sensitive and reproducible detection of the targeted analytes within a complex mixture.
Modern targeted acquisition approaches are the gold standard for sensitively and reproducibly measuring hundreds of peptides in a single LC-MS/MS run (20–22). However, data acquired in this manner is only informative for the set of peptides targeted for analysis. Because of this narrow focus, iterative testing of different hypotheses (i.e. a different set of target peptides) also requires iterative acquisition of additional data. Moreover, assay development often requires retention time scheduling and/or refinement steps to find the optimal peptides and transitions for testing a particular hypothesis.
With the existence of two complementary but distinct approaches—DDA for broad sample characterization and targeted acquisition for interrogation of a specific hypothesis—the natural question is if the benefits of both techniques may be combined in a single technique. A potential solution is an alternative mode of bottom-up proteomics referred to as data-independent acquisition (DIA) that has been described and realized with various implementations (16, 23–32). In DIA, the instrument acquires MS/MS spectra systematically and independently from the content of MS survey spectra. These DIA approaches differ from DDA methods, targeted acquisition methods, and from each other in MS/MS isolation window width, total range of precursor m/z covered, duration of completing one cycle of isolation scheme (called the cycle time), single or multiple isolation windows per MS/MS analysis, and instrument platform. Because of the benefits of systematic sampling of the precursor m/z range by MS/MS, data from a single DIA experiment can be useful for both peptide detection and quantification in a complex mixture. Similar to DDA approaches, DIA data is broadly informative because the MS/MS characterization is not specific to a predefined set of peptide targets. Similar to targeted approaches, MS/MS information of peptides across the entire LC time domain can be extracted from DIA data to test a particular hypothesis. As the acquisition speed of modern instrumentation continues to increase, DIA has become more popular because of its comprehensive and unbiased sampling.
Although DIA resolves the problem of biased or incomplete MS/MS sampling, current DIA methods come with compromises (33), where the most common compromise is precursor selectivity. Constrained by the speed and accuracy of instrumentation, DIA methods typically use five- to 10-fold wider isolation windows compared with DDA to achieve the same breadth and depth of a single LC-MS/MS run. Because of this reduction in precursor selectivity, MS/MS spectra from DIA are noisier than DDA spectra. In particular, DIA by design generates mixture spectra, each containing product ions from multiple analytes with various abundance and different charge states. Fragmenting multiple analytes together also precludes DIA from tailoring collision energy for every analyte, a standard optimization in DDA and targeted acquisition.
The low precursor selectivity and resulting complexity of DIA spectra severely challenges the performance of traditional spectrum-centric analysis, which generally assumes that the detected product ions were derived from a single, isolated precursor. The major challenges in interpreting mixture spectra lie in allowing for multiple contributing precursor ions, assessing the dynamic range of mixture peptides, distributing intensities of product ions shared by contributing peptides, and adjusting statistical confidence estimates. Because almost every spectrum is mixed in DIA data, it is poorly suited for analysis by classic spectrum-centric approaches initially designed for DDA data. Some sophisticated spectrum-centric approaches (34–39) address these challenges by deconvolving mixture spectra into pseudo spectra or by matching mixture spectra to combinations of product ions from multiple candidate peptides. However, identification of low abundance analytes by interpreting mixture spectra is inherently difficult because the MS/MS signals from low abundance analytes are naturally overwhelmed by the signals from high abundance ones.
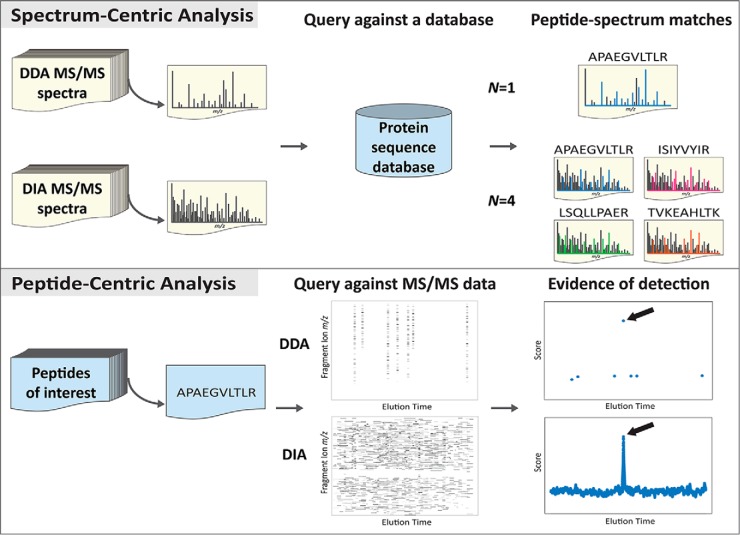
Recently, Gillet et al. demonstrated an alternative approach that analyzes DIA data in a targeted fashion (24), opening a new door for the investigation of tandem mass spectrometry data. Much like targeted analysis of transitions used in targeted acquisition methods, Gillet et al. use extracted ion chromatograms to detect and quantify query peptides. Similarly, Weisbrod et al. identify peptides by searching peptide fragmentation patterns against DIA data (25). Instead of interpreting individual spectra in a spectrum-centric fashion, these alternative approaches take each peptide of interest and ask: “Is this peptide detected in the data?” We refer to this approach as “peptide-centric analysis” in contrast with “spectrum-centric analysis.” In peptide-centric analysis, each peptide is detected by searching the MS and MS/MS data for signals selective for the query peptide. Peptide-centric analysis covers all methods that use peptides as an independent query unit, including but not limited to the targeted analysis. Peptide-centric analysis is intrinsically very different from spectrum-centric analysis (Table I, Fig. 1) and better suited for addressing many biological problems. This perspective discusses the analytical advantages of peptide-centric analysis and how they could translate to improvements in protein inference, and the analysis of DIA data.
Table I. Analytical comparison of spectrum-centric analysis versus peptide-centric analysis.
| Spectrum-centric analysis | Peptide-centric analysis | |
|---|---|---|
| Query unit | MS/MS spectrum | Peptide |
| Assumption | Each spectrum is generated from at least one peptide | Each peptide elutes once (for a short period of time) during liquid chromatography |
| Goal | Identify peptide(s) from each spectrum | Find evidence of detection for each peptide |
| Scoring | Candidate peptides from the sequence database compete with each other for the best scoring PSM | Candidate spectra from the acquired data compete with each other for the best scoring evidence of detection |
| Example tools | SEQUEST, Comet, MASCOT, X!Tandem, OMSSA, ProbID, MS-GF+, MaxQuant | FT-ARM, OpenSWATH, Skyline, SALSA |
Fig. 1.
Spectrum-centric analysis and peptide-centric analysis. In spectrum-centric analysis, each MS/MS spectrum from either a DDA or DIA experiment is queried against a protein sequence database. The peptides that yield the best scoring N statistically significant PSMs are assigned to the corresponding MS/MS spectrum. Typically N is one for a DDA spectrum and multiple for a DIA spectrum (showing N = 4 here). In peptide-centric analysis, every peptide of interest is queried against the acquired MS/MS data. The bottom-middle panel shows the extracted MS/MS signal of the query peptide over time in which the signal is extracted from any MS/MS spectrum generated from isolating the query precursor m/z. The extraction window width corresponds to the acquisition method, showing here 2 m/z for DDA and 10 m/z for DIA. The precursor m/z of the query peptide is sampled stochastically and sparsely in DDA but systematically in DIA. The MS/MS signal that provides the best scoring evidence of detection is assigned to the query peptide (indicated by the arrows).
Unique Characteristics of Peptide-Centric Analysis
I. Direct Statistical Measurements of Query Peptides
A drawback of spectrum-centric analysis is that the confidence estimates for peptides are indirect. In spectrum-centric analysis, each MS/MS spectrum is first assigned at least one peptide identity, yielding a large set of peptide-spectrum matches (PSMs). These PSMs are classified into accepted or not accepted by methods (40–43) that assign to each PSM statistical confidence estimates, indicating the confidence of either a set of PSMs being correct (e.g. FDR) or an individual PSM being correct (e.g. p values and E-values). Subsequently, peptide-level confidence estimates can be assigned by aggregating the best PSM per peptide in a postprocessing step (43, 44). Because the query unit for spectrum-centric analysis is an MS/MS spectrum, only the peptides that are matched to at least one spectrum are subject to the peptide level statistical tests. As a result, only this subset of peptides is assigned statistical confidence estimates, and the remaining peptides are implicitly considered missing.
Peptide-centric analysis, on the other hand, tests every peptide queried, providing direct and complete statistical measurements. The goal of peptide-centric analysis is to ascertain whether a query peptide was detected in an experiment. Thus, in a given data set, all of the query peptides can be separated into those with or without evidence of detection (i.e. detected or not detected). An empirical null can be estimated by generating decoy query peptides with shuffled sequences, measuring the null score distribution, and calculating p values and q-values for every query peptide using common statistical methods (40, 43, 45). With peptide-centric analysis, direct peptide-level testing makes answering biological questions more straightforward, and the completeness of statistical measurements makes subsequent comparison and quantification much easier.
Peptide-centric analysis could be very useful when considering the protein inference problem, which involves estimating the set of detected proteins from the set of detected peptides (46). Protein inference is heavily affected by the observed peptides. The value of peptide-centric analysis is that each peptide in a database can be directly assigned a confidence estimate of being detected/not detected because each peptide is directly investigated. In contrast, spectrum-centric analysis implicitly assigns all “missing” peptides equal, very low confidence estimates. These imputed confidence estimates could lead to biases in the inferred set of detected proteins. This includes peptides that distinguish splice isoforms or paralogs. Therefore, when comparing the result from a peptide-centric analysis to the detectability of such a peptide (47, 48), it is possible to begin to probabilistically evaluate the presence/absence of a protein isoform. With directly tested peptide probabilities, peptide-centric analysis makes the input of protein inference more straightforward and transparent.
II. Considerations for Mixture Spectra
When investigating a complex proteome with shotgun proteomics, mixture spectra are a common occurrence. Although conventional DDA uses narrow isolation windows (typically ∼2 m/z-wide) targeting single precursor ion species for fragmentation, as many as 50% of the MS/MS spectra are mixed (35, 39, 49). The frequency and impact of mixture spectra in a DDA experiment vary with the sample complexity, LC separation, acquisition parameters, and instrumentation. Some studies used isolation windows as narrow as 0.7 m/z-wide to minimize unwanted precursor ions from being co-isolated and cofragmented (15, 50). In the context of DIA, all spectra are essentially mixture spectra because DIA isolates and fragments all precursor ions within a wide m/z range. As discussed previously, identification of multiple components in a mixture spectrum is challenging: Most spectrum-centric software is designed to identify a single component from each spectrum.
Peptide-centric analysis excels in handling mixture spectra because it does not interpret individual spectra. Rather than deconvolving each individual spectrum, peptide-centric analysis searches for evidence of detection for individual peptides, explicitly tolerating cofragmentation. Although spectrum-centric analysis struggles to identify multiple components with wide dynamic range from each mixture spectrum, peptide-centric analysis queries each peptide independently from other peptides. This subtle but significant change of query unit (Table I) shifts the problem from “peptides competing with each other to explain the mixture spectrum” to “spectra competing with each other to represent the query peptide.” With peptide-centric analysis, a single spectrum can be the top-scoring evidence of detection for multiple distinct peptides, as expected in the case of mixture spectra. In addition, peptide-centric analysis readily benefits from the systematic sampling of DIA when each analyte is sampled multiple time across its chromatographic peak. Conversely, even if the product ions of the query peptide comprise the minority of the mixture spectra, peptide detection can still be achieved using peptide-centric analysis.
III. Roles of Precursor Ion Signals
Precursor information is a powerful component of MS/MS data analysis. Inherently designed to identify DDA spectra, spectrum-centric approaches typically use precursor information as a “filter” to constrain peptide candidates for PSMs (2–8). These approaches assign precursor ion(s) to each spectrum in various ways spanning from using the un-processed precursor ion target, considering multiple monoisotopic ions in the isolation window, to detecting peptide features in the MS space. With high mass measurement accuracy and high resolution instruments, spectrum-centric searches could allow for only ±10 ppm of monoisotopic mass tolerance, thus greatly reducing the number of peptide candidates for PSMs and reducing the false discovery rate.
In the context of analyzing DIA data there is no clear consensus on how to use precursor information. Recent DIA methods emphasize the systematic measurement of both precursor and product ions, allowing for the detection of precursor and product ions that covary over elution time and likely are derived from the same analyte (26). This concept of detecting covarying precursor-product ion groups has been used to generate deconvolved spectra from DIA spectra. Each deconvolved spectrum contains precursor and product ions ostensibly derived from a single analyte and are thus more compatible with spectrum-centric analysis (51, 52).
Peptide-centric approaches could also use precursor information as evidence of detection. Rardin et al. recently demonstrated improved quantification from DIA data using Skyline with precursor ion filtering and transition filtering by correlation analysis (18, 53). Although filtering with precursor ions and precursor-product groups improves selectivity and specificity, the detection process could reduce sensitivity because analytes may have no MS signal, or an MS signal with substantial chemical noise despite having an MS/MS signal amenable for quantification. One way to incorporate precursor information without reducing sensitivity is to use it as a scoring feature rather than a filter, which is employed in some peptide-centric approaches such as the algorithms used in Skyline (18). When analyzing complex mixtures, incorporating precursor information without filtering may provide greater confidence in peptide detection for analytes with a signal in MS spectra without compromising sensitivity by eliminating analytes that may have an MS/MS signal but no detectable MS signal.
Applications of Peptide-Centric Analysis
Peptide-centric analysis is particularly suited for DIA experiments given its advantages in handling mixture spectra. In addition, peptide-centric analysis can easily incorporate valuable properties from DIA data, such as retention time and elution profile, that are commonly ignored by spectrum-centric analysis. For example, Gillet et al. demonstrated peptide detection and quantification by extracting peptide-specific product ion chromatograms, or extracted ion chromatograms, from DIA data using 26-m/z SWATH acquisition (24). Weisbrod et al. demonstrated peptide detection and quantification by searching theoretical or empirical peptide fragmentation patterns against the DIA data acquired using high mass accuracy Fourier transform-all reaction monitoring (FT-ARM) of 100- m/z wide isolation windows (25). With low precursor selectivity and high intrascan dynamic range in both cases, correctly interpreting the spectra using spectrum-centric analysis is extremely challenging.
Peptide-centric analysis can also be applied to DDA data. For example, Liebler et al. used a pattern recognition algorithm (SALSA) to search for peptide-specific ion series against the DDA MS/MS spectra (54). Because of the stochastic nature of the DDA data, the evidence for peptide detection appears sparse and scattered compared with analyzing DIA data (Fig. 1). Nonetheless, peptide-centric analysis provides statistical measure for every query peptide regardless of whether the data is sparse or dense. In addition, given that many DDA spectra are mixed, peptide-centric analysis retains the benefits of handling of mixture spectra when analyzing DDA data.
Extensible Framework for Mass Spectrometry
This concept of defining peptides as analytes and directly searching for their evidence of detection generalizes into a broader paradigm, which we call “analyte-centric analysis.” Analyte-centric analysis comprises any method that uses the analyte as the query unit to ask whether the analyte is detected or not. It includes the traditional targeted data analysis, but is not limited to the methods that scores based on transitions or extracted ion chromatograms. The analyte of interest can be naturally extended from peptides to include small molecules, peptides with modifications, intact proteins, lipids and metabolites. In this analyte-centric paradigm, any properties of an analyte can be naturally incorporated into the score that summarizes the evidence supporting an analyte being detected. For example, “Does the discovered fragmentation evidence coincide with chromatographic expectations?” Also, as mass spectrometer resolution continues to improve toward fine-scale isotope resolution (55), the analyte-centric approach can discriminate an isotopic profile based on the elemental composition of the analyte.
One of the subtle but significant benefits of analyte-centric analysis is the change in the query unit and null hypothesis. In the spectrum-centric approach, validation programs that modeled a false distribution of decoy hits were in reality posing the null hypothesis as, “This spectrum is made up of a random analyte.” For analyte-centric analysis, the null hypothesis is, “The analyte is not detected in the data.” This more direct hypothesis is better suited for answering most biological problems.
CONCLUSIONS
In this perspective, we discuss the analytically unique characteristics of peptide-centric analysis compared with traditional spectrum-centric analysis in analyzing shotgun proteomics data. Specifically, peptide-centric analysis provides direct statistical measurements for every peptide, and could improve the analysis of mixture spectra common in DIA data. We also discussed how peptide-centric approaches could use precursor signals as essential or supporting evidence of detection. As mass spectrometry instruments continue to improve in acquisition speed, DDA will be able to sample deeper for lower abundance analytes and DIA will be able to systematically acquire MS/MS spectra with improved precursor selectivity or a shorter cycle time. Analysis of the resulting large collections of data could benefit from the alternative peptide-centric approaches. Specifically, changing the perspective from identifying as many spectra as possible to confidently detecting peptides from an experiment greatly benefits protein inference and quantitative comparison. The fact that the same peptide is fragmented in DIA data sets multiple times generating a chromatographic elution profile for each product ion further increases the achievable quantitative accuracy. Furthermore, a peptide-centric perspective can be naturally extended to other analytes such as intact proteins, lipids, and metabolites. We hope the analytical advantages of analyte-centric analysis over spectrum-centric analysis will incite the field to further advance bioinformatics and statistical solutions for analyzing mass spectrometry data.
Acknowledgments
We thank J. D. Storey, S. J. McIlwain, C. R. Weisbrod, H. Y. Yang, and J. K. Eng for insightful discussions.
Footnotes
Author contributions: Y.S.T., W.S.N., and M.J.M. designed research; Y.S.T. performed research; Y.S.T. and B.M. contributed new reagents or analytic tools; Y.S.T. and S.K. analyzed data; Y.S.T. wrote the paper; Y.S.T., J.D.E., S.H.P., L.K., R.H.A., R.D.S., W.S.N., and M.J.M. contribute perspectives in details.
* This work was supported by the National Institutes of Health Grants R01 GM103551, R01 GM096306, P41 GM103533, R21 CA192983, and F31 AG037265, an Early Career Award from the US Department of Energy (to S.H.P.) and the European Research Council (Grant# ERC-2008-AdG 233226).
1 The abbreviations used are:
- DDA
- data-dependent acquisition
- DIA
- data-independent acquisition
- FDR
- false discovery rate
- PSM
- peptide-spectrum match.
REFERENCES
- 1. Gatlin C. L., Eng J. K., Cross S. T., Detter J. C., Yates J. R., 3rd (2000) Automated identification of amino acid sequence variations in proteins by HPLC/microspray tandem mass spectrometry. Anal. Chem. 72, 757–763 [DOI] [PubMed] [Google Scholar]
- 2. Eng J. K., Fischer B., Grossmann J., MacCoss M. J. (2008) A fast SEQUEST cross correlation algorithm. J. Proteome Res. 7, 4598–4602 [DOI] [PubMed] [Google Scholar]
- 3. Koenig T., Menze B. H., Kirchner M., Monigatti F., Parker K. C., Patterson T., Steen J. J., Hamprecht F. A., Steen H. (2008) Robust prediction of the MASCOT score for an improved quality assessment in mass spectrometric proteomics. J. Proteome Res. 7, 3708–3717 [DOI] [PubMed] [Google Scholar]
- 4. Craig R., Beavis R. C. (2004) TANDEM: matching proteins with tandem mass spectra. Bioinformatics 20, 1466–1467 [DOI] [PubMed] [Google Scholar]
- 5. Cox J., Mann M. (2008) MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat. Biotechnol. 26, 1367–1372 [DOI] [PubMed] [Google Scholar]
- 6. Eng J. K., Jahan T. A., Hoopmann M. R. (2013) Comet: An open-source MS/MS sequence database search tool. Proteomics 13, 22–24 [DOI] [PubMed] [Google Scholar]
- 7. Kim S., Pevzner P. A. (2014) MS-GF+ makes progress towards a universal database search tool for proteomics. Nat. Commun. 5, 5277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Geer L. Y., Markey S. P., Kowalak J. A., Wagner L., Xu M., Maynard D. M., Yang X., Shi W., Bryant S. H. (2004) Open mass spectrometry search algorithm. J. Proteome Res. 3, 958–964 [DOI] [PubMed] [Google Scholar]
- 9. Taylor J. A., Johnson R. S. (1997) Sequence database searches via de novo peptide sequencing by tandem mass spectrometry. Rapid Commun. Mass Spectrom. RCM 11, 1067–1075 [DOI] [PubMed] [Google Scholar]
- 10. Ma B., Zhang K., Hendrie C., Liang C., Li M., Doherty-Kirby A., Lajoie G. (2003) PEAKS: powerful software for peptide de novo sequencing by tandem mass spectrometry. Rapid Commun. Mass Spectrom. 17, 2337–2342 [DOI] [PubMed] [Google Scholar]
- 11. Frank A. M., Savitski M. M., Nielsen M. L., Zubarev R. A., Pevzner P. A. (2007) De novo peptide sequencing and identification with precision mass spectrometry. J. Proteome Res. 6, 114–123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Frewen B. E., Merrihew G. E., Wu C. C., Noble W. S., MacCoss M. J. (2006) Analysis of peptide MS/MS spectra from large-scale proteomics experiments using spectrum libraries. Anal. Chem. 78, 5678–5684 [DOI] [PubMed] [Google Scholar]
- 13. Lam H., Deutsch E. W., Eddes J. S., Eng J. K., Stein S. E., Aebersold R. (2008) Building consensus spectral libraries for peptide identification in proteomics. Nat. Methods 5, 873–875 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Yen C. Y., Houel S., Ahn N. G., Old W. M. (2011) Spectrum-to-spectrum searching using a proteome-wide spectral library. Mol. Cell. Proteomics 10, M111.007666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Michalski A., Cox J., Mann M. (2011) More than 100,000 Detectable peptide species elute in single shotgun proteomics runs but the majority is inaccessible to data-dependent LC-MS/MS. J. Proteome Res. 10, 1785–1793 [DOI] [PubMed] [Google Scholar]
- 16. Panchaud A., Scherl A., Shaffer S. A., von, Haller P. D., Kulasekara H. D., Miller S. I., Goodlett D. R. (2009) Precursor acquisition independent from ion count: How to dive deeper into the proteomics ocean. Anal. Chem. 81, 6481–6488 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Prakash A., Tomazela D. M., Frewen B., MacLean B., Merrihew G., Peterman S., Maccoss M. J. (2009) Expediting the development of targeted SRM assays: Using data from shotgun proteomics to automate method development. J. Proteome Res. 8, 2733–2739 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. MacLean B., Tomazela D. M., Shulman N., Chambers M., Finney G. L., Frewen B., Kern R., Tabb D. L., Liebler D. C., MacCoss M. J. (2010) Skyline: an open source document editor for creating and analyzing targeted proteomics experiments. Bioinformatics 26, 966–968 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Picotti P., Aebersold R. (2012) Selected reaction monitoring-based proteomics: workflows, potential, pitfalls and future directions. Nat. Methods 9, 555–566 [DOI] [PubMed] [Google Scholar]
- 20. Escher C., Reiter L., MacLean B., Ossola R., Herzog F., Chilton J., MacCoss M. J., Rinner O. (2012) Using iRT, a normalized retention time for more targeted measurement of peptides. Proteomics 12, 1111–1121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Marx V. (2013) Targeted proteomics. Nat. Methods 10, 19–22 [DOI] [PubMed] [Google Scholar]
- 22. Burgess M. W., Keshishian H., Mani D. R., Gillette M. A., Carr S. A. (2014) Simplified and efficient quantification of low-abundance proteins at very high multiplex via targeted mass spectrometry. Mol. Cell. Proteomics 13, 1137–1149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Chapman J. D., Goodlett D. R., Masselon C. D. (2014) Multiplexed and data-independent tandem mass spectrometry for global proteome profiling. Mass Spectrom. Rev. 33, 452–470 [DOI] [PubMed] [Google Scholar]
- 24. Gillet L. C., Navarro P., Tate S., Röst H., Selevsek N., Reiter L., Bonner R., Aebersold R. (2012) Targeted data extraction of the MS/MS spectra generated by data-independent acquisition: A new concept for consistent and accurate proteome analysis. Mol. Cell. Proteomics 11, O111.016717 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Weisbrod C. R., Eng J. K., Hoopmann M. R., Baker T., Bruce J. E. (2012) Accurate peptide fragment mass analysis: Multiplexed peptide identification and quantification. J. Proteome Res. 11, 1621–1632 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Purvine S., Eppel J. T., Yi E. C., Goodlett D. R. (2003) Shotgun collision-induced dissociation of peptides using a time of flight mass analyzer. Proteomics 3, 847–850 [DOI] [PubMed] [Google Scholar]
- 27. Venable J. D., Dong M. Q., Wohlschlegel J., Dillin A., Yates J. R. (2004) Automated approach for quantitative analysis of complex peptide mixtures from tandem mass spectra. Nat. Methods 1, 39–45 [DOI] [PubMed] [Google Scholar]
- 28. Silva J. C., Denny R., Dorschel C., Gorenstein M. V., Li G. Z., Richardson K., Wall D., Geromanos S. J. (2006) Simultaneous qualitative and quantitative analysis of the Escherichia coli proteome a sweet tale. Mol. Cell. Proteomics 5, 589–607 [DOI] [PubMed] [Google Scholar]
- 29. Plumb R. S., Johnson K. A., Rainville P., Smith B. W., Wilson I. D., Castro-Perez J. M., Nicholson J. K. (2006) UPLC/MSE; a new approach for generating molecular fragment information for biomarker structure elucidation. Rapid Commun. Mass Spectrom. 20, 1989–1994 [DOI] [PubMed] [Google Scholar]
- 30. Bern M., Finney G., Hoopmann M. R., Merrihew G., Toth M. J., MacCoss M. J. (2010) Deconvolution of mixture spectra from ion-trap data-independent-acquisition tandem mass spectrometry. Anal. Chem. 82, 833–841 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Carvalho P. C., Han X., Xu T., Cociorva D., Carvalho, Mda G., Barbosa V. C., Yates J. R., 3rd (2010) XDIA: Improving on the label-free data-independent analysis. Bioinformatics 26, 847–848 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Egertson J. D., Kuehn A., Merrihew G. E., Bateman N. W., MacLean B. X., Ting Y. S., Canterbury J. D., Marsh D. M., Kellmann M., Zabrouskov V., Wu C. C., MacCoss M. J. (2013) Multiplexed MS/MS for improved data-independent acquisition. Nat. Methods 10, 744–746 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Egertson J. D., MacLean B., Johnson R., Xuan Y., MacCoss M. J. (2015) Multiplexed peptide analysis using data-independent acquisition and Skyline. Nat. Protoc. 10, 887–903 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Zhang N., Li X. J., Ye M., Pan S., Schwikowski B., Aebersold R. (2005) ProbIDtree: An automated software program capable of identifying multiple peptides from a single collision-induced dissociation spectrum collected by a tandem mass spectrometer. Proteomics 5, 4096–4106 [DOI] [PubMed] [Google Scholar]
- 35. Houel S., Abernathy R., Renganathan K., Meyer-Arendt K., Ahn N. G., Old W. M. (2010) Quantifying the impact of chimera MS/MS spectra on peptide identification in large-scale proteomics studies. J. Proteome Res. 9, 4152–4160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Hsieh E. J., Hoopmann M. R., MacLean B., MacCoss M. J. (2010) Comparison of database search strategies for high precursor mass accuracy MS/MS data. J. Proteome Res. 9, 1138–1143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Wang J., Bourne P. E., Bandeira N. (2011) Peptide identification by database search of mixture tandem mass spectra. Mol. Cell. Proteomics, mcp.M111.010017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Wang J., Bourne P. E., Bandeira N. (2014) MixGF: Spectral probabilities for mixture spectra from more than one peptide. Mol. Cell. Proteomics 13, 3688–3697 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Zhang B., Pirmoradian M., Chernobrovkin A., Zubarev R. A. (2014) DeMix workflow for efficient identification of cofragmented peptides in high resolution data-dependent tandem mass spectrometry. Mol. Cell. Proteomics 13, 3211–3223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Keller A., Nesvizhskii A. I., Kolker E., Aebersold R. (2002) Empirical statistical model to estimate the accuracy of peptide identifications made by MS/MS and database search. Anal. Chem. 74, 5383–5392 [DOI] [PubMed] [Google Scholar]
- 41. Tabb D. L., McDonald W. H., Yates J. R., 3rd (2002) DTASelect and contrast: Tools for assembling and comparing protein identifications from shotgun proteomics. J. Proteome Res. 1, 21–26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Elias J. E., Gygi S. P. (2007) Target-decoy search strategy for increased confidence in large-scale protein identifications by mass spectrometry. Nat. Methods 4, 207–214 [DOI] [PubMed] [Google Scholar]
- 43. Käll L., Canterbury J. D., Weston J., Noble W. S., MacCoss M. J. (2007) Semi-supervised learning for peptide identification from shotgun proteomics datasets. Nat. Methods 4, 923–925 [DOI] [PubMed] [Google Scholar]
- 44. Shteynberg D., Deutsch E. W., Lam H., Eng J. K., Sun Z., Tasman N., Mendoza L., Moritz R. L., Aebersold R., Nesvizhskii A. I. (2011) iProphet: Multi-level integrative analysis of shotgun proteomic data improves peptide and protein identification rates and error estimates. Mol. Cell. Proteomics 10, M111.007690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Reiter L., Rinner O., Picotti P., Hüttenhain R., Beck M., Brusniak M. Y., Hengartner M. O., Aebersold R. (2011) mProphet: Automated data processing and statistical validation for large-scale SRM experiments. Nat. Methods 8, 430–435 [DOI] [PubMed] [Google Scholar]
- 46. Nesvizhskii A. I., Aebersold R. (2005) Interpretation of shotgun proteomic data the protein inference problem. Mol. Cell. Proteomics 4, 1419–1440 [DOI] [PubMed] [Google Scholar]
- 47. Mallick P., Schirle M., Chen S. S., Flory M. R., Lee H., Martin D., Ranish J., Raught B., Schmitt R., Werner T., Kuster B., Aebersold R. (2007) Computational prediction of proteotypic peptides for quantitative proteomics. Nat. Biotechnol. 25, 125–131 [DOI] [PubMed] [Google Scholar]
- 48. Li Y. F., Arnold R. J., Tang H., Radivojac P. (2010) The importance of peptide detectability for protein identification, quantification, and experiment design in MS/MS proteomics. J. Proteome Res. 9, 6288–6297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Luethy R., Kessner D. E., Katz J. E., Maclean B., Grothe R., Kani K., Faça V., Pitteri S., Hanash S., Agus D. B., Mallick P. (2008) Precursor-ion mass re-estimation improves peptide identification on hybrid instruments. J. Proteome Res. 7, 4031–4039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Hebert A. S., Richards A. L., Bailey D. J., Ulbrich A., Coughlin E. E., Westphall M. S., Coon J. J. (2014) The one hour yeast proteome. Mol. Cell. Proteomics 13, 339–347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Li G. Z., Vissers J. P., Silva J. C., Golick D., Gorenstein M. V., Geromanos S. J. (2009) Database searching and accounting of multiplexed precursor and product ion spectra from the data independent analysis of simple and complex peptide mixtures. Proteomics 9, 1696–1719 [DOI] [PubMed] [Google Scholar]
- 52. Tsou C. C., Avtonomov D., Larsen B., Tucholska M., Choi H., Gingras A. C., Nesvizhskii A. I. (2015) DIA-Umpire: Comprehensive computational framework for data-independent acquisition proteomics. Nat. Methods 12, 258–264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Rardin M. J., Schilling B., Cheng L.-Y., Maclean B. X., Sorenson D. J., Sahu A. K., MacCoss M. J., Vitek O., Gibson B. W. (2015) MS1 peptide ion intensity chromatograms in MS2 (SWATH) data independent acquisitions. Improving post acquisition analysis of proteomic experiments. Mol. Cell. Proteomics, mcpO115.048181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Liebler D. C., Hansen B. T., Davey S. W., Tiscareno L., Mason D. E. (2002) Peptide sequence motif analysis of tandem MS data with the SALSA algorithm. Anal. Chem. 74, 203–210 [DOI] [PubMed] [Google Scholar]
- 55. Rose C. M., Merrill A. E., Bailey D. J., Hebert A. S., Westphall M. S., Coon J. J. (2013) Neutron encoded labeling for peptide identification. Anal. Chem. 85, 5129–5137 [DOI] [PMC free article] [PubMed] [Google Scholar]