Abstract
Protein acetylation is a well-studied regulatory mechanism for several cellular processes, ranging from gene expression to metabolism. Recent discoveries of new post-translational modifications, including malonylation, succinylation, and glutarylation, have expanded our understanding of the types of modifications found on proteins. These three acidic lysine modifications are structurally similar but have the potential to regulate different proteins in different pathways. The deacylase sirtuin 5 (SIRT5) catalyzes the removal of these modifications from a wide range of proteins in different subcellular compartments. Here, we review these new modifications, their regulation by SIRT5, and their emerging role in cellular regulation and diseases.
Lysine acetylation (Kac)1 is the addition of acetyl group to the ε-amine of a lysine side chain. This chemical modification is a reversible, dynamic, and evolutionarily conserved protein post-translational modification (PTM) (Fig. 1). Early studies on the biology of lysine acetylation focused on nuclear proteins, such as histones and transcriptional factors, which clearly demonstrated an important function of acetylation for chromatin structure and dynamics (1, 2). These studies led to the notion that lysine acetylation was restricted to the nucleus. The discovery of lysine acetylation on tubulin and on mitochondrial proteins demonstrated non-nuclear localization and suggested an important role for acetylation in an expanded array of cellular biology (3–5). Using mass-spectrometry-based proteomics, the cellular acetyl proteomes (a.k.a. “acetylomes”) have been extensively characterized and have revealed high abundance of lysine acetylation outside the nucleus (6–10). For example, lysine acetylation plays key roles in regulating mitochondrial enzymes and other metabolic and cellular processes in addition to chromatin biology (6, 11–13). Lysine acetylation is abundant on most metabolic pathways, a finding that is conserved from bacteria to mammals (6, 7, 9, 14–21). Dysregulation of lysine acetylation plays a pathogenic role in diverse conditions such as metabolic syndrome, age-associated stem cell dysfunction, cardiac failure, and cancer (22–25).
Fig. 1.
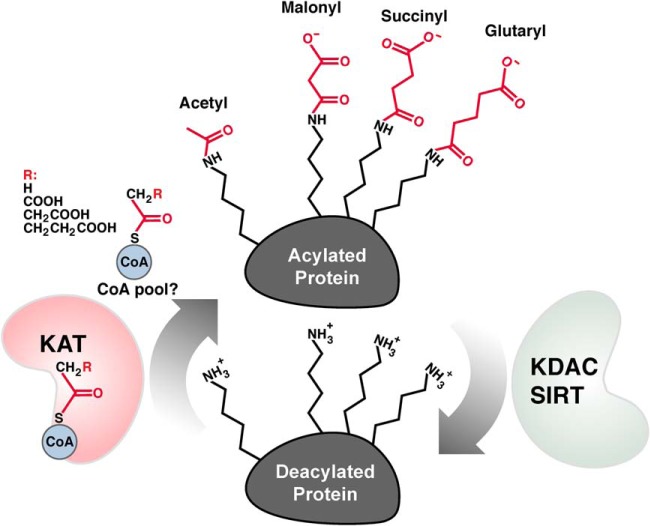
Protein acylation is balanced by KDACs and KATs. Protein acylation can be enzymatically catalyzed by lysine acyltransferases (KATs) and removed by lysine deacylases (KDACs).
Beyond protein acetylation, the landscape of protein modifications is rapidly expanding. Using mass spectrometry as a discovery tool, in combination chemical biology and biochemistry as validation tools, we recently identified and validated three new types of lysine modifications: lysine malonylation (Kmal) (26, 27), lysine succinylation (Ksucc) (27, 28), and lysine glutarylation (Kglu) (29), which are collectively referred to as lysine acylation (Fig. 2). Similar to lysine acetylation, emerging evidence suggests that these new lysine acylations are important in regulating cellular metabolism in physiological and pathophysiological states. In this review, we describe these new modifications and their regulation and highlight their emerging role in cellular regulation.
Fig. 2.
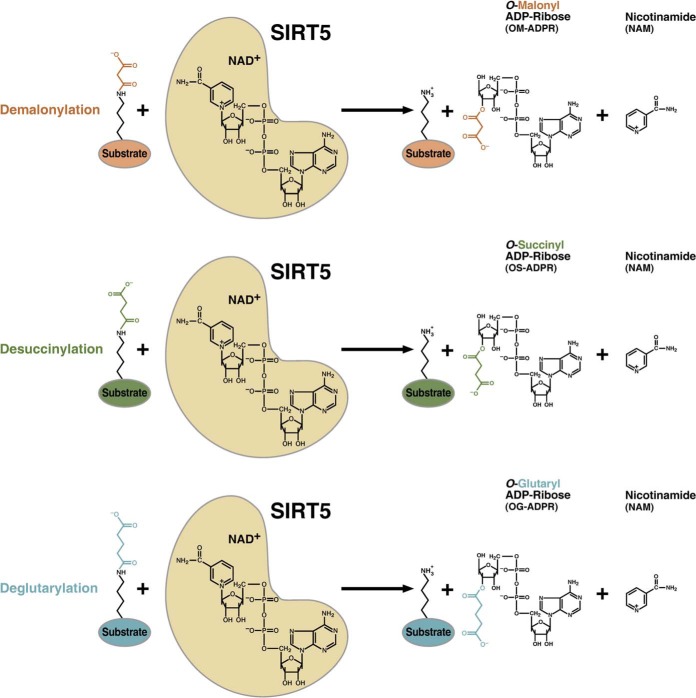
Regulation of lysine malonylation, succinylation, and glutarylation. Lysine malonylation, succinylation, and glutarylation is targeted for removal by the NAD+-dependent deacylase SIRT5.
Lysine Malonylation, Succinylation, and Glutarylation Pathways
Recently identified lysine acyl modifications, Kmal (26,27), Ksucc (27,28), and Kglu (29) (Fig. 2), are evolutionarily conserved and dynamic under diverse cellular conditions, such as stressors, metabolic substrates and availability, and genetic mutations (29–31). Since these new modifications have an acidic carboxylic group under physiological pH, they are referred to collectively as acidic lysine acyl modifications; furthermore, the sirtuins are more appropriately referred to as protein deacylases (Box1) (32).
Short-chain acyl-CoAs, malonyl-CoA, succinyl-CoA, and glutaryl-CoA, are the substrates for the corresponding lysine acylation reactions. These CoAs can possibly be synthesized by their corresponding short-chain acyl salts, malonate, succinate, and glutarate, catalyzed by acyl-CoA synthetases, such as succinyl-CoA synthetase and malonyl-CoA synthetase (33, 34). Additionally, these CoAs can be generated and consumed in the TCA cycle, as well as by metabolism of amino acids and lipids. These CoAs are highly regulated in cells and tissues. For example, dysregulation of malonyl-CoA is associated with diseases, such as ischemic heart disease and diabetes (35, 36). Additionally, these short-chain CoAs are thermodynamically favorable for their corresponding lysine acylation reactions (37). Further, these CoAs are structurally similar to acetyl-CoA, leading to apparent enzymatically catalyzed acylation by promiscuous acetyltransferases. For example, the binding pocket of Hat1 has a large enough CoA binding pocket to accommodate CoAs larger than acetyl CoA (38), such as malonyl-CoA, succinyl-CoA, or glutaryl-CoA.
Acidic modifications are chemically different from lysine acetylation in two distinct ways. First, they add a bulkier acyl group to lysines compared with acetylation: three, four, or five carbons for malonylation, succinylation, or glutarylation, respectively, compared with two carbons for acetylation. Second, acidic modifications change the charge on lysine from +1 to −1 under physiological conditions (28). Interestingly, protein phosphorylation also generates a −1 charge on proteins. In contrast, lysine acetylation changes the charge on lysine from +1 to 0. Because of this difference, it is conceivable that acidic modifications could disrupt any ionic interactions between the positively charged, unmodified lysine side chains with a negative charged chemical moiety in other molecules, such as nucleotides, proteins, or small molecules. Alternatively, changes in lysine charge could impart structural alterations on proteins, as acidic modifications would be predicted to have a more profound impact on protein structure and function compared with lysine acetylation, if located in the same lysine site. Future studies will address these possibilities and determine how acidic acyl modifications influence protein function.
Regulatory Enzymes for Lysine Malonylation, Succinylation, and Glutarylation
Two groups of enzymes with opposing activities, lysine acetyltransferases and deacetylases (Box1), are known to regulate the status of lysine acetylation (Fig. 1) (39). Eighteen known class I, class II, and class IV histone deacetylases (HDAC1–11) and seven class III HDACs (the sirtuin family SIRT1–7) make up the entire HDAC family (Box 1). The HDACs have been a subject of intense study since their discovery in 1996 (40). Indeed, they have important roles in several areas of biology and disease and are increasingly used as targets for drug design. For example, deacetylase inhibitors, such as vorinnostat (trade name Zolinza) and romidepsin (trade name Istodax), have been approved by the U.S. Food and Drug Administration for clinical use in cancer therapy (41, 42).
The sirtuin family of HDACs has seven highly conserved NAD+-dependent deacetylases, which were discovered in 1999/2000 (43, 44). Sirtuins regulate key biological processes in mammals, including many aspects of metabolism, stress response, and signaling (45). Three mammalian sirtuins (SIRT3, SIRT4, and SIRT5) localize mostly or exclusively to the mitochondrial matrix. SIRT3 has robust deacetylase activity and is the major enzyme in the mitochondria performing this reaction (46). In contrast, neither SIRT4 nor SIRT5 have been shown to regulate global mitochondrial protein acetylation levels (47). Supporting this idea, SIRT4 and SIRT5 have weak or nondetectable deacetylase activity in vitro. Low activities of these mitochondrial sirtuins against acetyl-lysine, and the absence of known substrates suggested the possibility they could be removing lysine modifications other than acetylation from proteins.
The immediate question after the discovery of acidic acyl modifications was whether they were regulated by HDACs or HDAC-like enzymes. Given the same amide bonds between lysine acetylation and the acidic acyl modifications (Fig. 2), all 18 HDACs were tested for deacylation activities against malonylation, succinylation, and glutarylation. SIRT5 was shown to have potent demalonylase and desuccinylase activities; this finding was validated using biochemical measurements, as well as quantitative proteomics in cells and tissues, with or without an expression of SIRT5, both in vitro and in vivo (26, 48). Importantly, these findings were independently discovered and verified using different, but complementary, methods, including enzymology and structural biology (49). In another recent study using quantitative proteomics, SIRT5 knockout mouse embryonic fibroblasts were shown to have an increase in lysine succinylation but not acetylation. Together, these studies demonstrate that SIRT5 is the major enzyme for lysine desuccinylation in the cells and tissues (48).
Importantly, this result, combined with emerging evidence from other studies, also suggests SIRT5 is unlikely a deacetylase (Box1) in vivo. For example, in vitro deacetylation activities of SIRT5 is low or undetectable when measured using multiple substrates such as small molecules containing a florescent acetylated lysine residue, peptide substrate, and whole cell lysate (26, 48). The current model is for SIRT5 to be a specific mammalian HDAC with potent activity to remove acidic lysine acyl groups. While mammalian SIRT5 seems to have activities restricted to acidic lysine acylation, its bacterial counterpart, CobB, the only deacetylase identified so far in prokaryotes, has both deacetylase and desuccinylase activities (50). Interestingly, CobB mutants can be generated in such a way that the two activities can be differentiated (50), and more work will be required to understand the regulation of protein succinylation in lower organisms.
In addition to lysine malonylation and lysine succinylation, a recent study demonstrated lysine glutarylation as a new lysine acylation and showed that SIRT5 is its deglutarylase (29), consistent with the idea that SIRT5 catalyzes the removal of acidic lysine modifications. Enzymatic studies and structural modeling show that SIRT5 has comparable activities among the three acyl modifications and prefers short-chain carboxylic groups, such as malonyl, succinyl, and glutaryl groups, but not acetyl (shorter) or adipoyl carboxylic groups (with a longer acidic acyl chain, e.g. six carbons or longer) (29). Thus, while SIRT5 is flexible among the three short-chain acidic lysine acylations, it does have selectivity against acidic acyl modifications.
While HDACs have been systematically tested for their activities for demalonylaiton, desuccinylation, and deglutarylation, such screens have not yet been reported in the literature for HATs (Box1). We showed that p300 has enzymatic activity for lysine glutarylation in vitro (29). More recently, Garcia and his colleagues showed that p300/CBP can catalyze lysine succinylation in vitro (51). These two lines of evidence suggest a possibility that enzyme-catalyzed lysine acylation may exist in cells. In addition, identification of lysine succinylation on N-terminal regions of histones also supports the notion that nuclear acetyltransferases could catalyze lysine succinylation.
Substrates of Lysine Malonylation, Succinylation, and Glutarylation
Identification of individual protein with acidic acyl lysines is a critical next step to understand the role of these modifications, an idea well illustrated by the history of protein acetylation. Progress in our understanding of lysine acetylation in each cellular compartment is largely synchronized with identification of modified proteins. Technological advances in mass-spectrometry-based PTM proteomics, such as improved mass spectrometry sensitivity, high-quality antibodies against PTMs of interest, and peptide fractionation procedure, make it possible to quickly identify sites of acylation in proteins, and primary substrates targeted by the HDACs/sirtuins.
Efficient affinity enrichment of the peptides containing modifications using sequence-independent anti-acyl-lysine antibodies followed by HPLC/MS/MS analysis and protein sequence alignment enables proteome-wide analysis of PTM substrates. The acylated lysine is relatively stable, and its peptide can lead to a good peptide-bond fragmentation signature in a mass spectrometer. Thus, the acylation sites can be accurately mapped in a similar fashion as those for lysine acetylation.
While succinyl-lysine was fully established few years ago, proteomic screenings have only recently been carried out to identify the proteins that contain this modification. Using a quantitative proteomic approach, a total of 2,565 succinylated lysine sites from 779 proteins in mouse cells were identified by comparing the difference of succinyl-lysine levels in MEFs with or without SIRT5 (48). Similarly, profiling of lysine succinylation has also been performed in bacteria, yeast, HeLa cells (52), and mouse liver mitochondria (48, 52, 53).
Several interesting findings emerge from these early proteomic studies. First, SIRT5 is the major enzyme for lysine desuccinylation in mammalian cells, which is supported by the findings that Sirt5 ablation (SIRT5KO) in mice potently increases global lysine succinylation levels (26), and almost all succinylated sites are increased in SIRT5KO MEFs (48). A second finding from lysine succinylation proteomic studies is that succinylation is a widespread protein post-translational modification in diverse model organisms (48, 50, 52, 53). Third, succinylation is abundant on mitochondrial proteins and is estimated to have high stoichiometry. Specifically, quantitative proteomics of both protein expression and succinyl-lysine show that 32% of the succinylated sites had stoichiometries greater than 10% in SIRT5 WT cells, whereas this figure increased to 56% in SIRT5 KO cells (48). However, these are only estimates of stoichiometry, and further work needs to be done to further test this finding. Fourth, succinyl-lysine can be more dynamic than acetyl-lysine; for example, in Escherichia coli, high glucose concentration induce a more significant change in protein succinylation than acetylation (50). This observation suggests that under some cellular conditions, succinylation may play a different role in cellular regulation than acetylation. Global lysine succinylation is dynamically changed in response growth conditions and genetic mutations that are associated succinyl-CoA homeostasis (48, 50).
In addition to succinylation, proteomic screening was carried out on lysine glutarylation in mouse liver. This study identified 683 lysine glutarylated sites in 191 proteins (29). A comparison between glutarylated and succinylated sites in mouse liver tissues show that 459 of the 683 glutarylated sites (about 67%) are overlapping with succinylated lysines, suggesting that the two modifications could be influencing the same proteins and by the same mechanism. Interestingly, carbamoyl-phosphate synthase 1 (CPS1), a protein known to be both acetylated and succinylated, was also found to be glutarylated and regulated by SIRT5 (29).
Efforts have been taken to identify substrates for lysine malonylation. Early experiments on lysine malonylation led to the identification of 17 lysine malonylated proteins in HeLa cells (26). Recent large-scale proteomic screening to characterize mammalian malonylome identified 4,042 malonylation sites on 1,426 substrate proteins in mouse liver and 4,943 malonylatioin sites in 1,822 substrate proteins in human fibrobalsts, with or without expression malonyl-CoA decarboxylase (MCD) (31). Four hundred sixty-one malonylation sites on 339 substrate proteins showed a two-fold increase or more in MCD−/− cells relative to MCD+/+ cells, and 1,452 malonylation sites on 822 proteins were only detected in MCD−/− cells. Lysine malonylation regulates mitochondrial function and fatty acid oxidation in MCD-deficient cells. Interestingly, MCD−/− cells have higher level of malonylo-CoA than MCD+/+ cells. This result indicates a possibility that MCD activity modulate cellular malonyl-CoA concentration that in turns impacts cellular lysine malonylation.
Physiological Role of Acylation
The expanding landscape of protein acylation coupled with the important role of the corresponding acyl-CoAs and the sirtuins in metabolic regulation have led to the idea that these new acyl-based lysine modifications are important regulators of metabolism (Fig. 3). Indeed, proteomic studies on lysine malonylation, succinylation, and glutarylation have shown that, like acetylation, metabolic proteins are over-represented (29, 48, 50, 52, 53). Gene ontology analyses on the proteins identified as acylated consistently find mitochondrial and metabolic pathways. While less is known about malonylation, the top pathways identified as having succinylated and glutarylated lysines are largely overlapping and include oxioreductase activity, fatty acid metabolism, and amino acid metabolism (29, 48, 53).
Fig. 3.
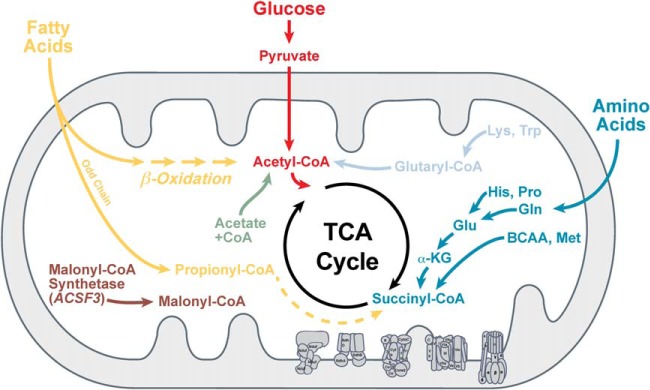
Metabolic regulation of malonyl-, succinyl-, and glutaryl-CoAs. Lipid, glucose, and amino acid metabolism all lead to the generation of acidic acyl-CoA species.
As predicted, pathway analyses showed proteins containing lysine succinylation and glutarylation are largely found within mitochondria. Succinylation is enriched in the mitochondria, but non-mitochondrial proteins are also succinylated, including ribosomes and nuclear proteins. Follow-up studies showed several sites of both succinylation and malonylation on histones (27), suggesting these modifications could be an underappreciated part of the histone code. Glutarylation was also found in both mitochondrial and extra-mitochondrial compartments (29). The small subset of proteins known to be malonylated are largely found in the cytoplasm, but whether this snapshot is representative of the overall landscape is unknown.
These observations lead to the question about the subcellular localization of SIRT5, the only known regulator of these acidic modifications. An early study on SIRT5 identified two distinct human isoforms with only minor differences in the C-terminus (54). This study proposed a model where both isoforms of SIRT5 are targeted to the mitochondria for N-terminal processing, which can translocate out of the mitochondria to the cytoplasm and have varying degrees of protein stability (54). A recent study supported this finding, showing SIRT5 was present in mitochondrial, cytoplasmic, and nuclear compartments (48). They further showed that dynamic sites of succinylation can be found outside the mitochondria (48), suggesting that SIRT5 regulates both mitochondrial and extra-mitochondrial lysine succinylation. This idea could also be extended to suggest that SIRT5 regulates sites of malonylation and glutarylation outside the mitochondria, but further studies are required to test these ideas and validate putative sites of regulation.
Indeed, a major and ongoing effort in the field is to validate sites of malonylation, succinylation, and glutarylation identified in proteomic studies and to understand how acylation affects the activity of its targets. Succinylation has been shown to regulate CPS1 (49), pyruvate dehydrogenase (PDH) (48), succinate dehydrogenase (SDH) (48), and 3-hydroxy-3-methylglutaryl-CoA synthase 2 (HMGCS2) (53). Additionally, several members of the fatty acid oxidation pathway have succinylated lysines, and this pathway is repressed in mice or cells lacking SIRT5 (53). Glutarylation has also been shown to regulate CPS1 activity and is the only target known to be regulated by this new modification (29).
No clear picture has emerged on the regulatory role of malonylation, succinylation, and glutarylation. Some proteins show reduced activity in the acylated state, either by ex vivo measurements or in vitro mimetic strategies. For example, CPS1 and HMGCS2 are suppressed, whereas PDH and SDH were shown to have increased activity in the acylated state. How these modifications regulate biological pathways, are regulated by SIRT5, and coordinate overall cellular metabolism are major questions being addressed in the field. Nonetheless, acyl-lysine modifications are emerging as important players in regulating metabolism and cellular physiology.
Pathophysiological Role of Acylation
As an extension of their role in regulating physiological processes, lysine malonylation, succinylation, and glutarylation are also associated with pathophysiological processes and disease. For example, mitochondrial protein glutarylation is significantly elevated in a mouse model of glutaric acidemia (29). Glutaric acidemia is an inborn error of metabolism (OMIM 231670) caused by a deleterious mutation in glutaryl-CoA dehydrogenase, which is a mitochondrial enzyme that degrades glutaryl-CoA. Additionally, elevated protein malonylation was recently observed in human fibroblasts of patients with MCD deficiency (OMIM 606761) (30). Supporting this idea, malonyl-CoA concentrations are elevated in MCD−/− cells compared to MCD+/+ cells, which correlates well with elevated levels of global lysine malonylation (31). Similar elevations in protein acylation were observed in inborn errors of metabolism for other modifications, including protein butyrylation and protein propionylation, which suggests that protein acylation could be a common biochemical feature of mutations in acyl-CoA-handling enzymes (30). Genetic mutations in ketoglutarate dehydrogenase and succinyl-CoA synthetase, two key enzymes for metabolism of succinyl-CoA, are associated with metabolic disesases (55, 56). It is therefore highly possible that succinyl-CoA concentration is dysregulated in the patients' cells that in turn modulates lysine succinylation. While the role of acyl-lysine modifications in inborn errors of metabolism is not known, alterations in enzymatic activity (either elevations or reductions) of key metabolic proteins could contribute to the pathogenesis of these diseases.
Future Studies
A major question remaining in the field is the mechanism by which lysine acylation occurs (for a recent review, see ref (37)). No acyltransferases have been shown to catalyze the transfer of malonyl-, succinyl-, or glutaryl-modifications to proteins in vivo, although this can be performed using a truncated p300 acetyltransferase in vitro (29). Interestingly, 35 histone lysine succinylation sites were detected in histones (27), whose succinylation status is regulated by SIRT5 in MEFs (48). Different from the lysine acetylation sites, which primarily exist at the N-terminal of histones, nearly all the lysine succinylation sites localize to the C-terminal globular domains (27). Detection of lysine succinylation in histones, but not other highly abundant nuclear proteins, such as lamin-B (48), suggests substrate specificity of lysine succinylation. This further implies the possibility of succinyltransferase-catalyzed lysine succinylation reactions in nuclei and the net histone sucinylation balanced by succinylation catalyzed by succinyltransferase and desuccinylation catalyzed by the SIRT5. Given the structural similarity among three acidic modifications with a difference of only one or two methylene groups (Fig. 1), all three could share a putative acyltransferase in a similar fashion as their deacylase.
In principle, lysine acylation could be controlled by a balance between concentrations of acyl-CoAs and activities of acyltransferases and deacylases. While structurally similar, malonyl-CoA, succinyl-CoA, glutaryl-CoA, and their corresponding short-chain acyl salts (malonate, succinate, and glutarate) are generated in different metabolic pathways (26, 28, 29) (Fig. 3). As a consequence, these three PTMs might not be redundant and have very different dynamics depending on fluctuation of metabolites in response to changes in cellular physiology and stress. It is also possible that the deacylase SIRT5 may have different activities and subcellular localization.
While enzymatic mechanisms of transfer in these conditions cannot be excluded in these studies, acyl-CoA accumulation in the setting of genetic mutations demonstrates a strong relationship between the levels of acyl-CoAs and the levels of acyl-lysines (29, 30). This relationship is further strengthened by physiological studies showing that protein succinylation is dependent upon succinyl-CoA generation (52) and that protein glutarylation fluctuates as a function of glutaryl-CoA generation (29). However, in the absence of known acyltransferases and the permissivity of these reactions in vitro (57), the idea of nonenzymatic mechanisms contributing to the global acylation landscape is gaining traction, at least in mitochondrion, where pH is higher than cytosol and nuclei. Like acetylation, both chemical and enzyme-catalyzed reactions could be occurring in cells, depending on cellular pH and chemical properties of CoAs, and likely contributes to the global acyl proteome. Additional studies on the mechanisms of acylation will lead to a deeper understanding of the role of acylation in biology.
CONCLUSION
The rapidly increasing number of protein modifications expands the known landscape of lysine acylation. Current thinking positions these modifications to be regulatory. Indeed, complementary techniques to manipulate acylation lead to reproducible changes in protein function. However, on any given protein, some acylation events will be regulated (by SIRT5, for example) and some not; furthermore, some acylation events will have a functional consequence and some not, similar to phosphorylation and acetylation. While these findings increase the complexity of protein regulation, continued research in this exciting area will clarify this important aspect of the regulatory balance between acylation and deacylation.
Boxes
Box 1: HAT, HDAC, sirtuins, acyltransferase, deacylase, lysine acylation
Histone acetyltransferase (HAT, also called lysine acetyltransferase (KAT)): an enzyme that catalyze the transferring an acetyl group form acetyl CoA to the side chain amine to form ε-N-acetyllysine.
Histone deacetylase (HDAC, also called lysine deacetylases (KDAC)): an enzyme that removes acetyl group from ε N-acetyllysine to form unmodified lysine.
Sirtuins (SIRT1–7): annonated as a subfamily of HDACs based on their protein sequence. Their enzymatic activities require NAD+ as a cofactor/cosubstrate.
Lysine acylation: introduction of an acyl group from a donor, such as acyl CoA, onto lysine side chains to from ε-N-acyl-lysine.
Lysine acyltransferase (KAT): an enzyme that catalyze the transferring an acyl group from acyl-CoA to the side chain amine to form ε-N-acyl-lysine.
Lysine deacylase (KDAC): an enzyme that removes acyl group from ε-N-acyl-lysine to form unmodified lysine.
Acknowledgments
We would like to acknowledge Gozde Colak and He Huang at the Zhao lab, and other members of the Hirschey and Zhao labs for thoughtful feedback.
Footnotes
Author contributions: M.D.H. and Y.Z. wrote the paper.
* Work in the Hirschey lab is supported by the American Heart Association Grants 12SDG8840004 and 12IRG9010008, The Ellison Medical Foundation, the National Institutes of Health (R01AA022146 and R01AG045351), the Duke O'Brien Center for Kidney Research (5P30DK096493-02), and the Duke Pepper Older Americans Independence Center (OAIC) Program in Aging Research supported by the National Institute of Aging (P30AG028716-01). Work in the Zhao lab is supported by the National Institutes of Health (U54RR020389, U24CA160036, and R01GM105933) and American Cancer Society Grant (RSG-13-198-01-DDC).
1 The abbreviations used are:
- Kac
- Lysine acetylation
- PTM
- post-translational modification
- Kmal
- lysine malonylation
- Ksucc
- lysine succinylation
- Kglu
- lysine glutarylation
- MCD
- malonyl-CoA decarboxylase
- CPS1
- carbamoyl-phosphate synthase 1
- PDH
- pyruvate dehydrogenase
- SDH
- succinate dehydrogenase
- HMGCS2
- 3-hydroxy-3-methylglutaryl-CoA synthase 2
- HAT
- Histone acetyltransferase
- HDAC
- Histone deacetylase.
REFERENCES
- 1. Roth S. Y., Denu J. M., Allis C. D. (2001) Histone acetyltransferases. Annu. Rev. Biochem. 70, 81–120 [DOI] [PubMed] [Google Scholar]
- 2. Gu W., Roeder R. G. (1997) Activation of p53 sequence-specific DNA binding by acetylation of the p53 C-terminal domain. Cell 90, 595–606 [DOI] [PubMed] [Google Scholar]
- 3. Hubbert C., Guardiola A., Shao R., Kawaguchi Y., Ito A., Nixon A., Yoshida M., Wang X. F., Yao T. P. (2002) HDAC6 is a microtubule-associated deacetylase. Nature 417, 455–458 [DOI] [PubMed] [Google Scholar]
- 4. Onyango P., Celic I., McCaffery J. M., Boeke J. D., Feinberg A. P. (2002) SIRT3, a human SIR2 homologue, is an NAD-dependent deacetylase localized to mitochondria. Proc. Natl. Acad. Sci. U.S.A. 99, 13653–13658 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Schwer B., North B. J., Frye R. A., Ott M., Verdin E. (2002) The human silent information regulator (Sir)2 homologue hSIRT3 is a mitochondrial nicotinamide adenine dinucleotide-dependent deacetylase. J. Cell Biol. 158, 647–657 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Kim S. C., Sprung R., Chen Y., Xu Y., Ball H., Pei J., Cheng T., Kho Y., Xiao H., Xiao L., Grishin N. V., White M., Yang X. J., Zhao Y. (2006) Substrate and functional diversity of lysine acetylation revealed by a proteomics survey. Mol. Cell 23, 607–618 [DOI] [PubMed] [Google Scholar]
- 7. Chen Y., Zhao W., Yang J. S., Cheng Z., Luo H., Lu Z., Tan M., Gu W., Zhao Y. (2012) Quantitative acetylome analysis reveals the roles of SIRT1 in regulating diverse substrates and cellular pathways. Mol. Cell. Proteomics 11, 1048–1062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Choudhary C., Kumar C., Gnad F., Nielsen M. L., Rehman M., Walther T. C., Olsen J. V., Mann M. (2009) Lysine acetylation targets protein complexes and co-regulates major cellular functions. Science 325, 834–840 [DOI] [PubMed] [Google Scholar]
- 9. Zhao S., Xu W., Jiang W., Yu W., Lin Y., Zhang T., Yao J., Zhou L., Zeng Y., Li H., Li Y., Shi J., An W., Hancock S. M., He F., Qin L., Chin J., Yang P., Chen X., Lei Q., Xiong Y., Guan K. L. (2010) Regulation of cellular metabolism by protein lysine acetylation. Science 327, 1000–1004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Hebert A. S., Dittenhafer-Reed K. E., Yu W., Bailey D. J., Selen E. S., Boersma M. D., Carson J. J., Tonelli M., Balloon A. J., Higbee A. J., Westphall M. S., Pagliarini D. J., Prolla T. A., Assadi-Porter F., Roy S., Denu J. M., Coon J. J. (2013) Calorie restriction and SIRT3 trigger global reprogramming of the mitochondrial protein acetylome. Mol. Cell 49, 186–199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Chalkiadaki A., Guarente L. (2012) Sirtuins mediate mammalian metabolic responses to nutrient availability. Nat. Rev. Endocrinol. 8, 287–296 [DOI] [PubMed] [Google Scholar]
- 12. Xiong Y., Guan K. L. (2012) Mechanistic insights into the regulation of metabolic enzymes by acetylation. J. Cell Biol. 198, 155–164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Huang J. Y., Hirschey M. D., Shimazu T., Ho L., Verdin E. (2010) Mitochondrial sirtuins. Biochim. Biophys. Acta 1804, 1645–1651 [DOI] [PubMed] [Google Scholar]
- 14. Wang Q., Zhang Y., Yang C., Xiong H., Lin Y., Yao J., Li H., Xie L., Zhao W., Yao Y., Ning Z. B., Zeng R., Xiong Y., Guan K. L., Zhao S., Zhao G. P. (2010) Acetylation of metabolic enzymes coordinates carbon source utilization and metabolic flux. Science 327, 1004–1007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Zhang J., Sprung R., Pei J., Tan X., Kim S., Zhu H., Liu C. F., Grishin N. V., Zhao Y. (2009) Lysine acetylation is a highly abundant and evolutionarily conserved modification in Escherichia coli. Mol. Cell. Proteomics 8, 215–225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Starai V. J., Celic I., Cole R. N., Boeke J. D., Escalante-Semerena J. C. (2002) Sir2-dependent activation of acetyl-CoA synthetase by deacetylation of active lysine. Science 298, 2390–2392 [DOI] [PubMed] [Google Scholar]
- 17. Finley L. W., Carracedo A., Lee J., Souza A., Egia A., Zhang J., Teruya-Feldstein J., Moreira P. I., Cardoso S. M., Clish C. B., Pandolfi P. P., Haigis M. C. (2011) SIRT3 opposes reprogramming of cancer cell metabolism through HIF1alpha destabilization. Cancer Cell 19, 416–428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Hallows W. C., Yu W., Smith B. C., Devries M. K., Ellinger J. J., Someya S., Shortreed M. R., Prolla T., Markley J. L., Smith L. M., Zhao S., Guan K. L., Denu J. M. (2011) SIRT3 promotes the urea cycle and fatty acid oxidation during dietary restriction. Mol. Cell 41, 139–149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Smith B. C., Settles B., Hallows W. C., Craven M. W., Denu J. M. (2011) SIRT3 substrate specificity determined by peptide arrays and machine learning. ACS Chem. Biol. 6, 146–157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Hirschey M. D., Shimazu T., Jing E., Grueter C. A., Collins A. M., Aouizerat B., Stančáková A., Goetzman E., Lam M. M., Schwer B., Stevens R. D., Muehlbauer M. J., Kakar S., Bass N. M., Kuusisto J., Laakso M., Alt F. W., Newgard C. B., Farese R. V., Jr., Kahn C. R., Verdin E. (2011) SIRT3 deficiency and mitochondrial protein hyperacetylation accelerate the development of the metabolic syndrome. Mol. Cell 44, 177–190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Hebert A. S., Dittenhafer-Reed K. E., Yu W., Bailey D. J., Selen E. S., Boersma M. D., Carson J. J., Tonelli M., Balloon A. J., Higbee A. J., Westphall M. S., Pagliarini D. J., Prolla T. A., Assadi-Porter F., Roy S., Denu J. M., Coon J. J. (2013) Calorie restriction and SIRT3 trigger global reprogramming of the mitochondrial protein acetylome. Mol. Cell 49, 186–199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Brown K., Xie S., Qiu X., Mohrin M., Shin J., Liu Y., Zhang D., Scadden D. T., Chen D. (2013) SIRT3 reverses aging-associated degeneration. Cell Reports 3, 319–327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Hirschey M. D., Shimazu T., Jing E., Grueter C. A., Collins A. M., Aouizerat B., Stančáková A., Goetzman E., Lam M. M., Schwer B., Stevens R. D., Muehlbauer M. J., Kakar S., Bass N. M., Kuusisto J., Laakso M., Alt F. W., Newgard C. B., Farese R. V., Jr., Kahn C. R., Verdin E. (2011) SIRT3 deficiency and mitochondrial protein hyperacetylation accelerate the development of the metabolic syndrome. Mol. Cell 44, 177–190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Finley L. W., Haigis M. C. (2012) Metabolic regulation by SIRT3: Implications for tumorigenesis. Trends Mol. Med. 18, 516–523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Haberland M., Montgomery R. L., Olson E. N. (2009) The many roles of histone deacetylases in development and physiology: implications for disease and therapy. Nat. Rev. Genet. 10, 32–42 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Peng C., Lu Z., Xie Z., Cheng Z., Chen Y., Tan M., Luo H., Zhang Y., He W., Yang K., Zwaans B. M., Tishkoff D., Ho L., Lombard D., He T. C., Dai J., Verdin E., Ye Y., Zhao Y. (2011) The first identification of lysine malonylation substrates and its regulatory enzyme. Mol. Cell. Proteomics 10, M111 012658 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Xie Z., Dai J., Dai L., Tan M., Cheng Z., Wu Y., Boeke J. D., Zhao Y. (2012) Lysine succinylation and lysine malonylation in histones. Mol. Cell. Proteomics 11, 100–107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Zhang Z., Tan M., Xie Z., Dai L., Chen Y., Zhao Y. (2011) Identification of lysine succinylation as a new post-translational modification. Nat. Chem. Biol. 7, 58–63 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Tan M., Peng C., Anderson K. A., Chhoy P., Xie Z., Dai L., Park J., Chen Y., Huang H., Zhang Y., Ro J., Wagner G. R., Green M. F., Madsen A. S., Schmiesing J., Peterson B. S., Xu G., Ilkayeva O. R., Muehlbauer M. J., Braulke T., Mühlhausen C., Backos D. S., Olsen C. A., McGuire P. J., Pletcher S. D., Lombard D. B., Hirschey M. D., Zhao Y. (2014) Lysine glutarylation is a protein post-translational modification regulated by SIRT5. Cell Metab. 19, 605–617 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Pougovkina O., te Brinke H., Wanders R. J., Houten S. M., de Boer V. C. (2014) Aberrant protein acylation is a common observation in inborn errors of acyl-CoA metabolism. J. Inherit. Metab. Dis. 37, 709–714 [DOI] [PubMed] [Google Scholar]
- 31. Colak G., Pougovkina O., Dai L., Tan M., te Brinke H., Huang H., Cheng Z., Park J., Wan X. L., Liu X., Yue W. W., Wanders R. J. A., Loccasale J. W., Lombard D. B., de Boer V. C., Zhao Y. (2015) Proteomics of lysine malonylation suggests its malonic aciduria-associated regulatory role in mitochondrial function and fatty acid oxidation. Submitted [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Hirschey M. D. (2011) Old enzymes, new tricks: Sirtuins are NAD(+)-dependent de-acylases. Cell Metab. 14, 718–719 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Fraser M. E., Hayakawa K., Hume M. S., Ryan D. G., Brownie E. R. (2006) Interactions of GTP with the ATP-grasp domain of GTP-specific succinyl-CoA synthetase. J. Biol. Chem. 281, 11058–11065 [DOI] [PubMed] [Google Scholar]
- 34. Witkowski A., Thweatt J., Smith S. (2011) Mammalian ACSF3 protein is a malonyl-CoA synthetase that supplies the chain extender units for mitochondrial fatty acid synthesis. J. Biol. Chem. 286, 33729–33736 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Ruderman N., Prentki M. (2004) AMP kinase and malonyl-CoA: targets for therapy of the metabolic syndrome. Nat. Rev. Drug Discov. 3, 340–351 [DOI] [PubMed] [Google Scholar]
- 36. Wolfgang M. J., Lane M. D. (2011) Hypothalamic malonyl-CoA and CPT1c in the treatment of obesity. FEBS J. 278, 552–558 [DOI] [PubMed] [Google Scholar]
- 37. Wagner G. R., Hirschey M. D. (2014) Nonenzymatic protein acylation as a carbon stress regulated by sirtuin deacylases. Mol. Cell 54, 5–16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Dutnall R. N., Tafrov S. T., Sternglanz R., Ramakrishnan V. (1998) Structure of the yeast histone acetyltransferase Hat1: Insights into substrate specificity and implications for the Gcn5-related N-acetyltransferase superfamily. Cold Spring Harb. Symp. Quant. Biol. 63, 501–507 [DOI] [PubMed] [Google Scholar]
- 39. Yang X. J., Seto E. (2007) HATs and HDACs: From structure, function and regulation to novel strategies for therapy and prevention. Oncogene 26, 5310–5318 [DOI] [PubMed] [Google Scholar]
- 40. Taunton J., Hassig C. A., Schreiber S. L. (1996) A mammalian histone deacetylase related to the yeast transcriptional regulator Rpd3p. Science 272, 408–411 [DOI] [PubMed] [Google Scholar]
- 41. Marks P. A., Breslow R. (2007) Dimethyl sulfoxide to vorinostat: development of this histone deacetylase inhibitor as an anticancer drug. Nat. Biotechnol. 25, 84–90 [DOI] [PubMed] [Google Scholar]
- 42. Guan P., Fang H. (2010) Clinical development of histone deacetylase inhibitor romidepsin. Drug Discov. Ther 4, 388–391 [PubMed] [Google Scholar]
- 43. Frye R.A. (2000) Phylogenetic classification of prokaryotic and eukaryotic Sir2-like proteins. Biochem. Biophys. Res. Commun. 273, 793–798 [DOI] [PubMed] [Google Scholar]
- 44. Frye R.A. (1999) Characterization of five human cDNAs with homology to the yeast SIR2 gene: Sir2-like proteins (sirtuins) metabolize NAD and may have protein ADP-ribosyltransferase activity. Biochem. Biophys. Res. Commun. 260, 273–279 [DOI] [PubMed] [Google Scholar]
- 45. Hall J. A., Dominy J. E., Lee Y., Puigserver P. (2013) The sirtuin family's role in aging and age-associated pathologies. J. Clin. Invest. 123, 973–979 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Lombard D. B., Zwaans B. M. (2014) SIRT3: As simple as it seems? Gerontology 60, 56–64 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Lombard D. B., Alt F. W., Cheng H. L., Bunkenborg J., Streeper R. S., Mostoslavsky R., Kim J., Yancopoulos G., Valenzuela D., Murphy A., Yang Y., Chen Y., Hirschey M. D., Bronson R. T., Haigis M., Guarente L. P., Farese R. V., Jr., Weissman S., Verdin E., Schwer B. (2007) Mammalian Sir2 homolog SIRT3 regulates global mitochondrial lysine acetylation. Mol. Cell. Biol. 27, 8807–8814 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Park J., Chen Y., Tishkoff D. X., Peng C., Tan M., Dai L., Xie Z., Zhang Y., Zwaans B. M., Skinner M. E., Lombard D. B., Zhao Y. (2013) SIRT5-mediated lysine desuccinylation impacts diverse metabolic pathways. Mol. Cell 50, 919–930 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Du J., Zhou Y., Su X., Yu J. J., Khan S., Jiang H., Kim J., Woo J., Kim J. H., Choi B. H., He B., Chen W., Zhang S., Cerione R. A., Auwerx J., Hao Q., Lin H. (2011) Sirt5 is a NAD-dependent protein lysine demalonylase and desuccinylase. Science 334, 806–809 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Colak G., Xie Z., Zhu A. Y., Dai L., Lu Z., Zhang Y., Wan X., Chen Y., Cha Y. H., Lin H., Zhao Y., Tan M. (2013) Identification of lysine succinylation substrates and the succinylation regulatory enzyme CobB in Escherichia coli. Mol. Cell. Proteomics 12, 3509–3520 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Hu A., Britton L. M., Garcia B. A. (2014) Investigating the specificity of histone acetyltransferase activity for producing rare modifications on histones using mass spectrometry. in The 62nd Annual American Society for Mass Spectrometry Conference on Mass Spectrometry and Allied Topics, Baltimore, MD [Google Scholar]
- 52. Weinert B. T., Schölz C., Wagner S. A., Iesmantavicius V., Su D., Daniel J. A., Choudhary C. (2013) Lysine succinylation is a frequently occurring modification in prokaryotes and eukaryotes and extensively overlaps with acetylation. Cell Rep. 4, 842–851 [DOI] [PubMed] [Google Scholar]
- 53. Rardin M. J., He W., Nishida Y., Newman J. C., Carrico C., Danielson S. R., Guo A., Gut P., Sahu A. K., Li B., Uppala R., Fitch M., Riiff T., Zhu L., Zhou J., Mulhern D., Stevens R. D., Ilkayeva O. R., Newgard C. B., Jacobson M. P., Hellerstein M., Goetzman E. S., Gibson B. W., Verdin E. (2013) SIRT5 regulates the mitochondrial lysine succinylome and metabolic networks. Cell Metab. 18, 920–933 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Matsushita N., Yonashiro R., Ogata Y., Sugiura A., Nagashima S., Fukuda T., Inatome R., Yanagi S. (2011) Distinct regulation of mitochondrial localization and stability of two human Sirt5 isoforms. Genes Cells 16, 190–202 [DOI] [PubMed] [Google Scholar]
- 55. Bonnefont J. P., Chretien D., Rustin P., Robinson B., Vassault A., Aupetit J., Charpentier C., Rabier D., Saudubray J. M., Munnich A. (1992) Alpha-ketoglutarate dehydrogenase deficiency presenting as congenital lactic acidosis. J. Pediatr. 121, 255–258 [DOI] [PubMed] [Google Scholar]
- 56. Ostergaard E. (2008) Disorders caused by deficiency of succinate-CoA ligase. J. Inherit. Metab. Dis. 31, 226–229 [DOI] [PubMed] [Google Scholar]
- 57. Wagner G. R., Payne R. M. (2013) Widespread and enzyme-independent Nepsilon-acetylation and Nepsilon-succinylation of proteins in the chemical conditions of the mitochondrial matrix. J. Biol. Chem. 288, 29036–29045 [DOI] [PMC free article] [PubMed] [Google Scholar]