Abstract
The sinoatrial node (SAN) maintains a rhythmic heartbeat; therefore, a better understanding of factors that drive SAN development and function is crucial to generation of potential therapies, such as biological pacemakers, for sinus arrhythmias. Here, we determined that the LIM homeodomain transcription factor ISL1 plays a key role in survival, proliferation, and function of pacemaker cells throughout development. Analysis of several Isl1 mutant mouse lines, including animals harboring an SAN-specific Isl1 deletion, revealed that ISL1 within SAN is a requirement for early embryonic viability. RNA-sequencing (RNA-seq) analyses of FACS-purified cells from ISL1-deficient SANs revealed that a number of genes critical for SAN function, including those encoding transcription factors and ion channels, were downstream of ISL1. Chromatin immunoprecipitation assays performed with anti-ISL1 antibodies and chromatin extracts from FACS-purified SAN cells demonstrated that ISL1 directly binds genomic regions within several genes required for normal pacemaker function, including subunits of the L-type calcium channel, Ank2, and Tbx3. Other genes implicated in abnormal heart rhythm in humans were also direct ISL1 targets. Together, our results demonstrate that ISL1 regulates approximately one-third of SAN-specific genes, indicate that a combination of ISL1 and other SAN transcription factors could be utilized to generate pacemaker cells, and suggest ISL1 mutations may underlie sick sinus syndrome.
Introduction
Rhythmic contraction of the heart is controlled by electrical impulses generated by pacemaker cells of the sinoatrial node (SAN). Abnormalities in SAN development lead to cardiac arrhythmia and sudden death (1). Sick sinus syndrome is a common age-related SAN dysfunction that ultimately requires implantation of a permanent pacemaker (2, 3). Despite its marked clinical relevance, genes and signaling pathways required for progressive development of the SAN remain unexplored, preventing development of effective therapies for sinus arrhythmias, including the possibility of biological pacemakers (4).
Pacemaker activity of SAN cells is due to slow diastolic depolarization generated by several ion channels, including calcium channels and hyperpolarization-activated cyclic nucleotide-gated potassium/sodium channels (HCN channels), which are modulated by autonomic neurons (5, 6). Molecular mechanisms underlying automaticity of the SAN are a subject of intensive study, and a complex coupled-clock system has been proposed: the voltage clock generated by HCN channels (the current referred to as If, the “funny” current); and the calcium clock generated by rhythmic Ca2+ release from sarcoplasmic reticulum. Both mechanisms act interdependently and synergistically to initiate the heartbeat (5, 7, 8).
Of 4 HCN channels, HCN4 is the most highly expressed in the SAN. During development, HCN4 expression is initiated in the cardiac crescent and is progressively confined to and later maintained in the SAN during later development and in the adult (9–11). Mutations in the human HCN4 gene lead to sinus bradycardia and have been associated with inherited sick sinus syndrome, long QT syndrome with bradycardia, and ventricular tachycardia (12–17). Mouse embryos that are null for Hcn4 exhibit long pauses in heartbeat and die around E10.5, demonstrating a critical requirement for Hcn4 in early pacemaker function of the heart (11). However, mice with Hcn4 deleted during later developmental stages and postnatal life survive, exhibiting normal basal heart rate with periodic long pauses. This observation, together with other in vitro physiological studies, suggest a role for If in stabilizing the pacemaker rhythm in later-stage hearts (18, 19).
Calcium release and cycling via the ryanodine receptor (RyR2), the sarcoplasmic reticulum calcium ATPase (SERCA2), the sodium calcium exchanger (NCX), and associated regulatory proteins play an essential role in pacemaker automaticity (20–23). Deletion of Ryr2 or Ncx1 leads to early embryonic lethality and substantial impairment in pacemaker function (22, 24). Phosphorylation of RyR, SERCA2, and its binding protein phospholamban modulate calcium cycling characteristics. SERCA2 inhibition leads to a reduced rate of beating in SAN cells in culture (25). However, deletion or overexpression of SERCA2 leads to abnormalities in loading sarcoplasmic reticulum calcium, as well as impaired cardiac contractility, but it has minimal effects on cardiac rhythm (26–28).
SAN formation is a complex and highly regulated process that involves multiple cell types with distinct developmental origins. During mouse development, the first heartbeat is recorded in the inflow tract as early as E8 (29, 30), and later, the sinus venosus (SV; the inflow tract) of the forming heart tube functions as a primitive pacemaker region. The first morphologically discernable SAN is formed at E11.5, and it becomes further mature and fully functional at E13.5 (31, 32). Lineages of the second heart field marked by Isl1-Cre contribute to SAN formation, with a posterior-most subset marked by TBX18 also contributing to SAN formation (33–35). SAN cell proliferation continues until shortly before birth (36). A number of signaling pathways, including neuregulin/ErbB, endothelin, and Notch signaling pathways, have been implicated in various aspects of atrioventricular and ventricular conduction system development (37–42); however, our understanding of signaling pathways regulating proliferation and differentiation of SAN cells and their progenitors is limited.
Genetic studies have revealed requirements for a number of transcription factors in various aspects of SAN development (43, 44). Several T-box genes are expressed in the SAN during development, including Tbx3 and Tbx18 (44). Deletion of Tbx3 in mouse results in bradycardia, reduced size of SAN, and upregulation of Cx43, Cx40, and other atrial myocyte–specific genes in the prospective SAN region, indicating a role for TBX3 in SAN development and suppression of working myocardial gene expression within the SAN (34, 36, 43, 45, 46). Overexpression of TBX3 is sufficient to induce mouse cardiomyocytes to acquire a pacemaker-like phenotype (36, 43, 47). TBX18 is expressed in sinus horn myocardium, part of which is the substratum for the forming SAN, and TBX18 is required for formation of the SAN (33, 34). Mice deficient in Tbx18 exhibit markedly smaller SANs. However, segregation of the SAN and atrial gene programs is complete, and no transdifferentiation of atrial myocytes or SAN pacemaker cells is observed in Tbx18 mutant hearts (34). Reduced size of the SAN in Tbx18 mutants has been attributed to delayed recruitment of SAN progenitors into the SAN. Ectopic overexpression of TBX18 in the ventricle of adult guinea pigs, or pigs, reprograms ventricular cardiomyocytes to pacemaker cells that can function as a de novo pacemaker (35, 48). Despite perturbations in SAN differentiation and morphogenesis, deletion of Tbx18 in mice appears to have minimal effect on pacemaker function (34). In addition, no changes in cell death or proliferation of SAN cells were observed in Tbx3 or Tbx18 mutants (34).
SHOX2 is a homeodomain transcription factor that is expressed in the SV and SAN (49, 50). Ablation of Shox2 in mice leads to embryonic lethality between E11.5–E13.5. Shox2 mutant embryos exhibit reduced size of the SAN with ectopic expression of Cx40, Cx43, and Nkx2.5 within the SAN, suggesting a critical role of SHOX2 in establishment of SAN identity (49–51). Shox2 hypomorphic mice die a few days after birth and exhibit bradycardia and arrhythmia (51).
ISL1 is a LIM homeodomain transcriptional factor that marks undifferentiated cardiac progenitors of the second heart field and is required for these progenitors to contribute to the heart (52). ISL1-expressing progenitors have the potential to develop into multiple cell types within the heart, including cardiomyocytes, smooth muscle cells, pacemaker cells, and endothelial cells (52–54). Recent studies have shown that ISL1 expression is maintained in the SAN (54–56). In zebrafish, Isl1 mutation results in bradycardia and irregular heartbeat with frequent pauses (57, 58). In Shox2-deficient mice, expression of ISL1 is diminished in SAN. ChIP-PCR revealed that SHOX2 binds to and regulates expression of an Isl1 enhancer. Overexpression of ISL1 can rescue the bradycardia phenotype in Shox2-deficient zebrafish (59). Due to early lethality and loss of cardiac cells derived from ISL1 progenitors in Isl1-null mice, a specific role for ISL1 in the SAN remains unknown. Recent transcriptome studies demonstrated changes in gene expression in response to Isl1 ablation in the SAN (60). However, no studies have been performed to address phenotypic or physiological consequences of Isl1 ablation. Furthermore, direct targets of ISL1 action in SAN cells remain unexplored.
In this study, we generated several Isl1 mutant mouse lines with reduced ISL1 expression, with Isl1 ablation specifically in differentiated cardiomyocytes, or with Isl1 ablation specifically in SAN cells. Our studies uncover a cell-autonomous requirement for ISL1 within the SAN in regulating proliferation, survival, and pacemaker function. Furthermore, we have performed RNA-sequencing (RNA-seq) studies of Isl1 mutant and control purified SAN cells, intersecting these data with genome-wide chromatin immunoprecipitation studies utilizing antibodies to ISL1 on chromatin from purified SAN cells to gain insight into mechanisms by which ISL1 regulates SAN formation and function.
Results
ISL1 is expressed in cardiac pacemaker cells of the SAN.
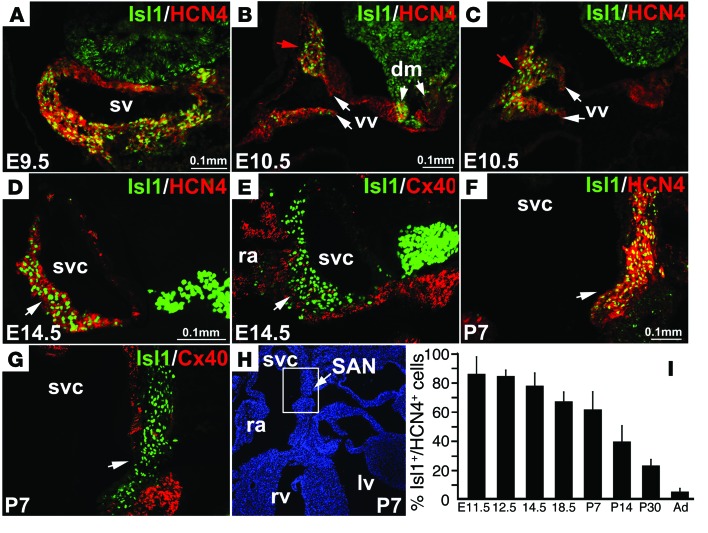
To investigate the role of ISL1 in SAN formation and function, we first analyzed ISL1 expression during SAN development and performed coimmunostaining utilizing antibodies for markers of SAN and surrounding atrial myocardium. HCN4 is a pacemaker channel expressed within the heart during early developmental stages, with its expression progressively being confined to and specifically marking the SAN (9, 10, 61, 62). At E9.5, ISL1 was expressed in myocardium of the SV, the primitive pacemaker region of early-stage embryos, where it was strongly coexpressed with HCN4 (Figure 1A). At E10.5, ISL1 and HCN4 were coexpressed in the head (Figure 1B) and tail (Figure 1C) of the SAN, venous valves (vv), and atrial myocardium surrounding the dorsal mesocardium (DM) (Figure 1, A and B). During later developmental stages and early postnatal life, ISL1 was expressed in a majority of SAN cells marked by HCN4 (Figure 1, D, F, and H). ISL1 expression in the SAN gradually decreased with age. The percentage of ISL1-expressing cells relative to the total number of HCN4+ pacemaker cells decreased markedly 2 weeks after birth and continued to decrease with age (Figure 1, I and H). Cx40 was expressed in atrial myocardium complementary to ISL1 expression in the SAN (Figure 1, E and G). These observations suggested a role for ISL1 in SAN development and function.
Figure 1. Expression of ISL1 in pacemaker cells of the SAN during development and after birth.
ISL1 was coexpressed with HCN4 in myocardium of the SV at E9.5 (A), and in the majority of SAN cells from E10.5–P7 (B–G). ISL1 expression did not overlap with Cx40, which is expressed in atrial myocardium (E and G). The boxed area in H delineates regions depicted in F and G. The fraction of HCN4 cells that expressed Isl1 remained constant at early stages from E11.5–E14.5, but decreased at E18 (I). After birth, the fraction of HCN4 cells that expressed Isl1 decreased significantly (I). n = 4, P < 0.05, 2-tailed t test. svc, superior vena cava; ra, right atria; lv, left ventricle; rv, right ventricle. Scale bars as shown.
Reduced Isl1 expression in Isl1 compound mutant embryo leads to sinus arrhythmia and loss of SAN cells.
To better visualize ISL1 expression during development and investigate the role of ISL1 in SAN formation and function, we generated Isl1 compound mutants with a less severe cardiac phenotype than that of Isl1 global null mutants (52), allowing for later survival. Isl1 compound mutant mice were generated by crossing an Isl1 nuclear LacZ knockin/knockout mouse line (Isl1nLacZ) (54, 63) and an Isl1 hypomorphic mouse line (floxed Isl1 allele with neomycin, Isl1fl:Neo/+), in which the presence of the neomycin cassette interferes with Isl1 expression (64). The Isl1 compound mutant (Isl1nLacZ/fl:Neo) exhibits further reduction in Isl1 expression and is embryonic lethal around E11.5 (63).
X-gal staining of tissue sections from control Isl1nLacZ mice revealed that Isl1-nLacZ expression recapitulated endogenous Isl1 expression, as previously published (52). Consistent with data shown in Figure 1, Isl1-nLacZ was expressed in DM, the SAN, and surrounding atrial myocardium at E9.5 (Figure 2, A and C). At E11.5, Isl1-nLacZ expression was observed in the SAN (Figure 2, G and I). However, in Isl1 compound mutants at E9.5 and E11.5, the number of cells expressing Isl1-nLacZ in SV myocardium, the SAN, and DM was substantially reduced (Figure 2, B, D, H, and J). Immunostaining with ISL1 and HCN4 antibodies revealed markedly reduced expression of ISL1 and HCN4 in the SV of Isl1 compound mutant embryos at E9.5 compared with littermate controls (Figure 2, E and F).
Figure 2. Bradycardia and loss of SAN cells in Isl1 compound mutants.
ISL1-nLacZ was expressed in SV myocardium, including the SAN region (red arrow), and mesocardium at E9.5 (A and C) and E11.5 (G and I). Expression of ISL1 and HCN4 in the SV region of Isl1 compound mutant embryos was significantly reduced (E and F). Expression of ISL1 and the number of ISL1-expressing cells in the SV, SAN (red arrow), and DM was markedly reduced in Isl1 compound mutant embryos at E9.5 (B and D) and E11.5 (H and J). BrdU staining revealed significantly reduced proliferation of SV myocardium in Isl1 compound mutants at E9.5 (K–M). TUNEL labeling showed significantly increased cell death in the SV of Isl1 compound mutant embryos at E10.5 (N–P) (n = 4 per group. Scale bars as shown). Echocardiography revealed a significant reduction in the heart rate of Isl1 compound mutant embryos at E9.5 and E11.5 (Q). n = 15 per group; *P < 0.05, 2-tailed t test.
These observations suggested that ISL1 may be required for proliferation and/or survival of SAN cells, SV cardiomyocytes, and/or cardiac progenitors of the posterior heart field. Therefore, we analyzed proliferation by BrdU labeling and cell death by TUNEL staining. The SV at E9.5 functions as a pacemaker region. We observed a significant reduction in the number of BrdU-labeled cells in SV myocardium (Figure 2, K–M) and a significant increase in cell death in the right horn of the SV at E9.5 (Figure 2, N–P).
To test whether pacemaker function was compromised in Isl1 compound mutants, we performed echocardiography to examine heart rate from E9.5–E11.5. Individual embryos were mapped in utero and analyzed by echocardiography to evaluate heart rate. The heart rate of Isl1 compound mutant embryos was significantly slower at E9.5 and further decreased at E11.5 (Figure 2Q). We also observed periodic heart-rate variability with long pauses, the frequency and duration of which were increased with embryonic age. Some embryos with long pauses in heartbeat died during the course of the examination, strongly suggesting that Isl1 mutant embryos died of cardiac arrhythmia.
Reduced Isl1 expression in Isl1 compound mutant embryo results in reduced expression of genes critical to SAN function.
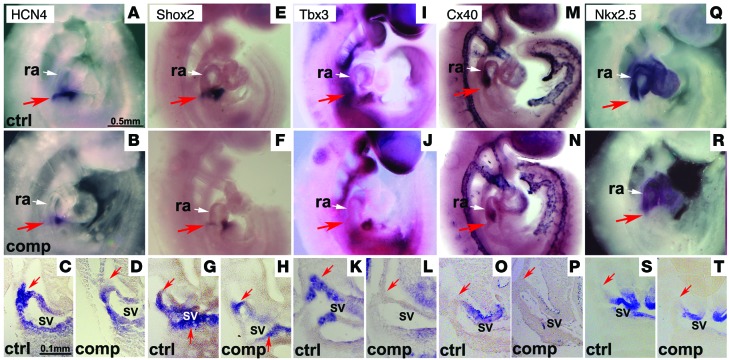
HCN4, SHOX2, and TBX3 are critical factors for SAN development and function. In situ hybridization revealed that Hcn4, Shox2, and Tbx3 were expressed in SV of control littermates at E9.5 (Figure 3, A, C, E, G, I, and K), but their expression was markedly reduced in SV of Isl1 compound mutant embryos (Figure 3, B, D, F, H, J, and L). Cx40 and Nkx2.5 were expressed in myocardium of control embryos but not in the SAN (Figure 3, M, O, Q, and S). In contrast to ectopic expression previously observed in Shox2 and Tbx3 mutants, we did not observe ectopic expression of Cx40 or Nkx2.5 in Isl1 compound mutants (Figure 3, N, P, R, and T). Together, these observations suggested a general loss of cells in this region in the compound mutant.
Figure 3. Reduced expression of Hcn4, Tbx3, and Shox2 in the SAN region of Isl1 compound mutant embryos.
At E9.5, Hcn4 and Shox2 were expressed in the SV, and SAN region (red arrow; A, C, E, and G). Tbx3 was expressed in the SV and surrounding mesenchyme (red arrow; I and K ). In Isl1 compound mutant embryos, expression of Hcn4, Shox2, and Tbx3 in the SV and SAN region was markedly reduced (B, D, F, H, J, and L). Cx40 and Nkx2-5 were expressed in working myocardium but not in the SAN region (M, O, Q, and S In Isl1 compound mutant embryos, expression of Cx40 and Nkx2-5 was markedly reduced in atrial myocardium, but no expansion or ectopic expression of Cx40 or Nkx2-5 was observed in the SAN region (N, P, R, and T). n = 4 per group, Scale bars as shown.
Specific ablation of Isl1 in differentiated cardiomyocytes.
Reduced ISL1 expression in Isl1 compound mutants might affect both differentiated SAN pacemaker cells and progenitors of the second heart field that contribute to SV myocardium and the SAN. To investigate a potential direct role for ISL1 in differentiated cardiomyocytes, we ablated Isl1 using cardiac Troponin T-Cre (cTnT-Cre) mice (65). Similar to phenotypes observed in Isl1 compound mutants, cTnT-Cre Isl1fl/fl mutants died from E9.5–E11.5 and displayed severe bradycardia, increased heart-rate variability, and long pauses (Supplemental Figure 1; supplemental material available online with this article; doi:10.1172/JCI68257DS1), demonstrating that ISL1 is required within differentiated myocardial cells for pacemaker function. Pulse-wave Doppler revealed well-coupled outflow and inflow wave, indicating there was no block in atrioventricular conduction (Supplemental Figure 1).
Ablation of Isl1 in SAN cells at E9.5 using Hcn4-CreERT2 leads to sinus arrhythmia, fewer SAN cells by E11.5, and embryonic lethality.
As Isl1 expression persists in the SAN well beyond stages that Isl1 compound and cTnT-Cre Isl1fl/fl mutant embryos can survive, a requirement for ISL1 in the SAN during later developmental stages remained to be addressed. The pacemaker channel HCN4 begins to be expressed in the cardiac crescent and specifically marks pacemaker cells in the SAN during later development and in adult heart (9–11). Therefore, we utilized an HCN4-inducible Cre mouse line (Hcn4-CreERT2) generated in our lab to specifically ablate Isl1 in pacemaker cells of the SAN. Upon tamoxifen induction, Hcn4-CreERT2 lineage–labeled cells are found in the cardiac conduction system, including the SAN (61). Our data are consistent with those of a similar Hcn4-CreERT2 mouse published by others (66). We crossed Hcn4-CreERT2 with Isl1fl/fl mice into Rosa-LacZ or Rosa-tdTomato indicator backgrounds to allow for Cre lineage tracing.
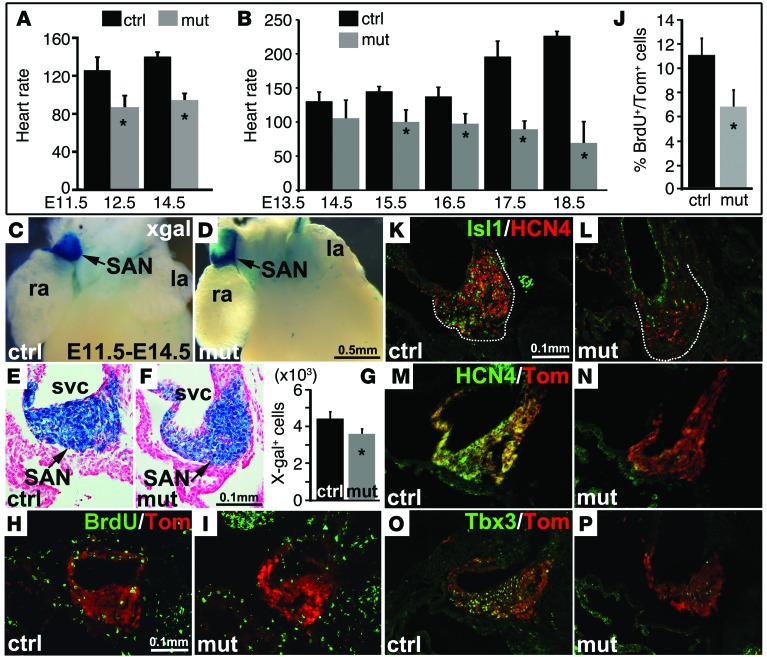
To determine temporal requirements for ISL1 in formation and function of the SAN, we ablated Isl1 specifically in pacemaker cells of the SAN using Hcn4-CreERT2 by giving tamoxifen at distinct times during development. At E9.5, the SV at the posterior pole of the heart functions as a primitive pacemaker. The first structurally discernable SAN is formed at E11.5, which becomes further matured and fully functional by E13.5 (31, 32). Therefore, we focused our study on these critical windows of SAN development. Tamoxifen was given at E9.5, and embryonic heart rate was examined daily by echocardiography. We found that, compared with control littermates, the heart rate of Hcn4-CreERT2 Isl1fl/fl mutants was significantly slower at E10.5. The heart rate of Hcn4-CreERT2 Isl1fl/fl mutants was further reduced at E11.5 with periodic long pauses, and a majority of these Hcn4-CreERT2 Isl1fl/fl mutants died around E11.5 (Figure 4A).
Figure 4. Bradycardia and loss of SAN cells following ablation of Isl1 in SAN during early developmental stages using Hcn4-CreERT2.
Isl1 mutant (Hcn4-CreERT2 Isl1fl/fl) and control (Hcn4-CreERT2 Isl1fl/+ or +/+) embryos were given tamoxifen at E9.5. Embryos were analyzed 36 and 48 hours after induction. (A) Echocardiography revealed that the heart rate of Isl1 mutants was significantly reduced at E11 and was further reduced at E11.5 (n = 20 per group). (B–D) Whole-mount X-gal staining and quantitative analysis revealed a significantly reduced number of X-gal+ and Tomato+ cells in the SAN (red arrow) of Isl1 mutants relative to control littermates at E11.5 (n = 4. Scale bars as shown). (D–H) Immunostaining demonstrated significantly reduced expression of HCN4 and TBX3 in the SAN of Isl1 mutants compared with controls marked by Tomato+ at E11.5. However, a slight but not significant reduction in the number of Hcn4 lineage–labeled cells in Isl1 mutant SAN region was observed when analyzed at E11 (D, I, and J). (K–M) TUNEL revealed increased cell death in Isl1 mutant SAN marked by Tomato+. (N–P) BrdU revealed decreased proliferation in Isl1 mutant SAN marked by Tomato. n = 4; *P < 0.05, 2-tailed t test. Scale bars as shown.
To examine the number and distribution of SAN cells in Hcn4-CreERT2 Isl1fl/fl mutants and somite-matched littermates, we performed lineage-tracing experiments in a Rosa-LacZ or Rosa-tdTomato reporter background. Tamoxifen was given at E9.5, and embryos were analyzed at E11.5 (48 hours after induction) by X-gal staining (Figure 4, B and C). In control littermates at 11.5, Hcn4-CreERT2 lineage–labeled cells (X-gal+) were found in the SV and SAN (Figure 4B), in a pattern that recapitulated endogenous expression of Hcn4 mRNA (Figure 3A). At E11.5, the location of X-gal+ cells was similar to that of control littermates, but the number of Hcn4-CreERT2 Isl1fl/fl lineage–labeled cells in the SV of mutants was somewhat reduced (Figure 4, C and D). Furthermore, in Hcn4-CreERT2 Isl1fl/fl mutants, expression of HCN4 and TBX3 was markedly reduced in remaining SAN cells, as revealed by coimmunofluorescence studies utilizing antibodies to HCN4 and TBX3 in an Hcn4-CreERT2 Rosa-tdTomato lineage–labeled background (Figure 4, E–H).
When Hcn4-CreERT2 Rosa-LacZ embryos were induced at E9.5 and examined at E11 (36 hours after induction), no significant changes in the number of X-gal+ cells were observed in mutant embryos compared with those of controls (Figure 4, D, I, and J). Expression of ISL1 was examined by immunostaining at E11, when major cell loss was not yet evident. Results demonstrated that tamoxifen induction performed at E9.5 effectively eliminated ISL1 expression, specifically in the SV and SAN, while ISL1 expression in the pharyngeal region and DM were not changed (Supplemental Figure 2). We observed a significantly increased number of TUNEL-labeled cells (Figure 4, K–M, and Supplemental Figure 2) and a reduced number of BrdU-labeled cells (Figure 4, N–P) in the SAN marked by Hcn4-CreERT2 Rosa-tdTomato labeling, suggesting a requirement for ISL1 in both survival and proliferation of SAN cells at these stages.
Ablation of Isl1 in SAN cells at E11.5 or E13.5 using Hcn4-CreERT2 leads to sinus arrhythmias and reduced proliferation of SAN cells.
To examine a potential later role for ISL1 in SAN development and pacemaker function, we ablated Isl1 in SAN at later development stages utilizing Hcn4-CreERT2 Rosa-LacZ. When tamoxifen was given at E11.5, Isl1 mutant embryos displayed a significantly slower heart rate when examined by echocardiography at 24 and 72 hours after induction (Figure 5A). In contrast to ablation of Isl1 at earlier stages, however, a majority of mutant embryos survived. When tamoxifen was given at E13.5, a significant reduction in heart rate was observed 48 hours after induction (Figure 5B). During these stages of development, the heart rate of control embryos increased gradually; however, the heart rate of Hcn4-CreERT2 Isl1fl/fl mutants did not increase and exhibited more variability (Figure 5B). Whole mount and section X-gal staining of E14.5 hearts (tamoxifen induced at E11.5) showed a slightly reduced intensity of X-gal+ cells in SAN of Isl1 mutants (Figure 5, D and F) when compared with SAN of littermate controls (Figure 5, C and E). Quantitative analysis revealed a slight but significant decrease in the number of X-gal+ cells in the SAN of Isl1 mutants (3,635 ± 254 per SAN) compared with controls (4,441 ± 368 per SAN) (Figure 5G). Consistent with this, we observed a significant decrease in the number of proliferating cells (BrdU+) in Isl1 mutant SAN marked by Hcn4-CreERT2 Rosa-tdTomato (Figure 5, H–J). However, in contrast to earlier stages, no significantly increased cell death was observed in the SAN of Hcn4-CreERT2 Isl1fl/fl mutants (not shown). At E14.5, ISL1 and HCN4 coimmunostaining revealed effective loss of ISL1 expression when tamoxifen was given at E11.5 (Figure 5, K and L). Furthermore, expression of HCN4 and TBX3 was markedly reduced in Isl1 mutant SAN marked by the Rosa-tdTomato reporter (Figure 5, M–P). Despite significant reduction in the expression of TBX3, no ectopic expression of NKX2.5 was observed in Isl1 mutant SAN (not shown).
Figure 5. Bradycardia and reduced TBX3 and HCN4 expression following ablation of Isl1 during later SAN morphogenesis.
(A) Ablation of Isl1 at E11.5 led to significantly slower heart rate at E12.5 and E14.5 (n = 15 per group, P < 0.05, 2-tailed t test). (B) During development, the heart rate of control embryos is gradually increased. When Isl1 was ablated at E13.5 and the heartbeat was examined daily after tamoxifen induction by echocardiography, no significant reduction in heart rate was observed 24 hours after induction. However, thereafter, the heart rate of Isl1 mutant embryos was gradually reduced (n = 15 per group). (C–G) Whole-mount and section X-gal staining showed a slight but significant reduction in the number of X-gal+ cells in Isl1 mutant SAN relative to littermate control SAN. (G) Quantitative analysis revealed a significant decrease in the number of X-gal+ and Tomato+ cells in Isl1 mutant SAN (3,635 ± 254 per SAN) compared with controls (4,441 ± 368 per SAN) (n = 4 per group. Scale bar as shown). (H–J) BrdU staining revealed a decreased number of BrdU+ cells in Isl1 mutant SAN marked by Tomato (n = 4 per group. Scale bar as shown). (K–P) Immunostaining demonstrated a marked reduction in the expression of ISL1, HCN4, and TBX3 in the SANs of Isl1 mutant embryos compared with control embryos. n = 4; *P < 0.05, 2-tailed t test. Scale bars as shown.
RNA-seq analyses reveal dysregulation of a number of genes important for SAN function in Hcn4-CreERT2 Isl1fl/fl mutants.
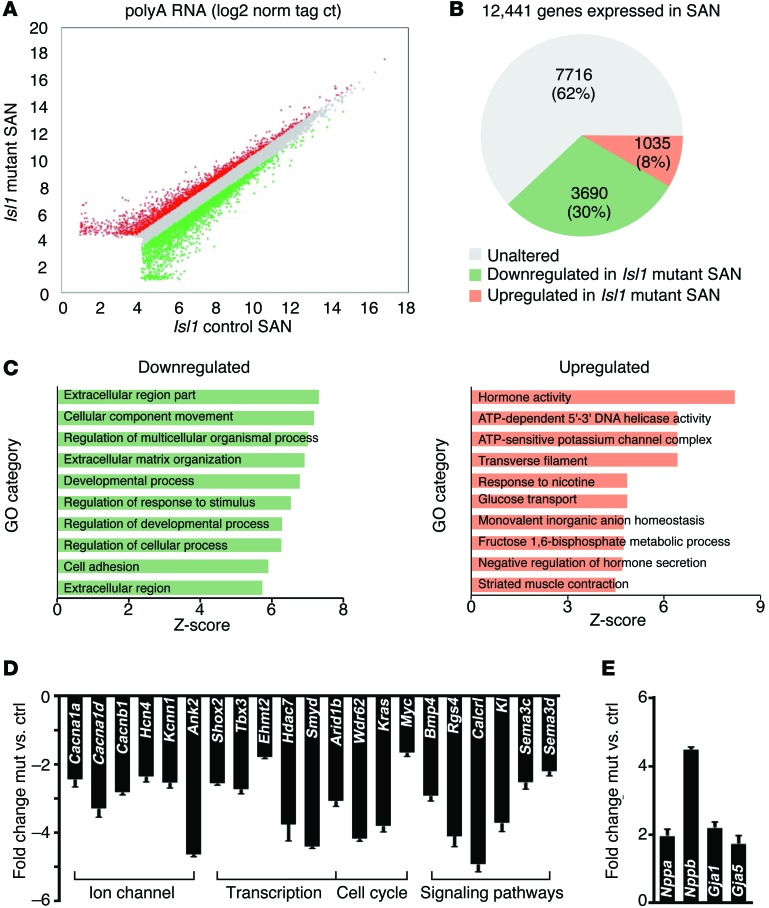
To investigate potential direct or indirect downstream targets of ISL1 that could account for observed phenotypes, RNA-seq analyses were performed on samples from Hcn4-CreERT2 Isl1fl/fl mutant and control SANs induced with tamoxifen at E10 and harvested at E12.5 (Figure 6). Results demonstrated that, of 12,441 genes expressed at significant levels in the SAN, transcripts for 3,690 (30%) of them were downregulated, and transcripts for 1,035 (8%) of them were upregulated in Isl1 mutants ( |fold-change Hcn4-CreERT2 Isl1fl/fl mutant vs. ctrl| ≥ 1.5) (Figure 6, A and B, and Supplemental Table 1). Gene ontology (GO) and reactome pathway enrichment analysis demonstrated significantly affected categories in genes downregulated or upregulated in Isl1 mutants. For downregulated genes, extracellular matrix components, cell adhesion, and cell guidance categories were prominent. For upregulated genes, categories included potassium channel complex components and striated muscle contraction (Figure 6C, Supplemental Figure 3, and Supplemental Table 2). We examined significantly altered genes for those that might contribute to the observed Isl1 mutant SAN phenotypes, confirming alterations in their expression by quantitative PCR (qPCR) analyses (Figure 6, D and E, and Supplemental Table 3). These included ion channels and associated genes (Cacna1a, Cacna1d, Cacnb1, Hcn4, Kcnn1, and Ank2), genes involved in transcriptional regulation (Shox2, Tbx3, Ehmt2, Hdac7, Smyd, and Arid1b), cell cycle (Arid1b, Wdr62, Kras, and Myc), and signaling pathways (Bmp4, Rgs4, Calcitonin receptor-like [Calcrl], Klotho [Kl], Sema3c, and Sema3d), many of which play a role in heart development or are critical for SAN function (Figure 6D). Consistent with previous RNA-seq data (60), we observed significant upregulation of a number of atrial myocardial specific genes, including Nppa, Nppb, Gja1/Cx43, and Gja5/Cx40 (Figure 6E).
Figure 6. RNA-seq analyses reveal dysregulation of a number of genes important for SAN function in Hcn4-CreERT2 Isl1fl/fl mutants.
(A) Scatterplot illustrating relative gene expression of polyA-selected RNA transcripts from RNA-seq comparison of control and Hcn4-CreERT2 Isl1fl/fl mutant SAN cells. Genes upregulated or downregulated 1.5-fold in Isl1 mutant SAN cells are shown in red and green, respectively. Values are presented as log2 of tag counts normalized to 107 uniquely mapped tags. (B) RNA-seq comparison of control and Hcn4-CreERT2 Isl1fl/fl mutant SAN transcriptomes revealed a total of 12,441 genes expressed (RPKM ≥ 1) in SAN cells, of which 1,035 upregulated and 3,690 downregulated in Isl1 mutant SAN cells ( |fold-change mutant vs. ctrl| ≥ 1.5). (C) GO functional clustering of genes down- and upregulated in Isl1 mutant, highlighting cellular processes most significantly affected in mutant SAN (top 10 not redundant categories are shown). (D) qPCR validation analysis. mRNA expression of ion channels and associated genes, and genes involved in transcription regulation, cell cycle, and signaling pathways are shown. (E) qRT-PCR validation analysis. mRNA expression of atrial myocardial specific genes. Results are shown as fold-change Isl1 mutant vs. ctrl. n = 4 per group, P < 0.05, 2-tailed t test. See also Supplemental Figure 3 and Supplemental Tables 1 and 2.
ISL1 directly regulates a number of genes required for normal pacemaker function in mice and human.
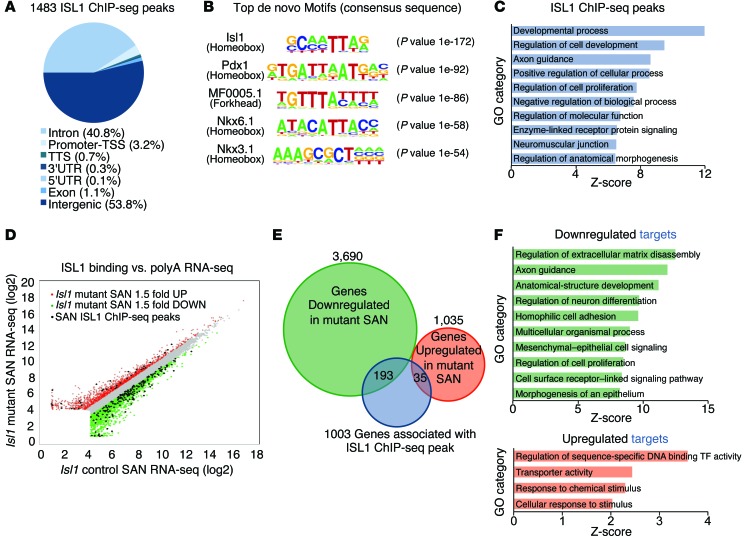
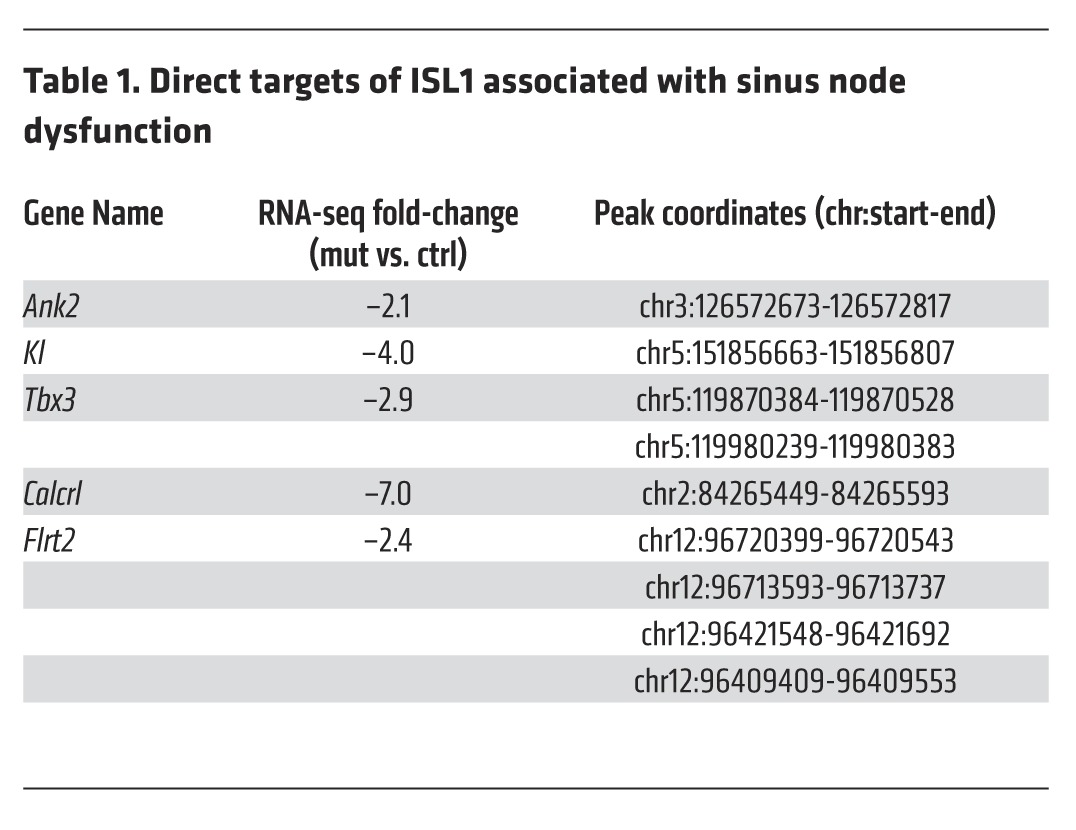
To gain insight into direct downstream targets of ISL1 that could account for the observed SAN phenotype, we performed genome-wide ChIP-seq analyses on FACS-purified Hcn4-H2BGFP–expressing neonatal SAN cells (61). Analyses of these data revealed 1,483 ChIP-seq peaks for ISL1, with the majority of binding, 94.6%, occurring at intergenic or intronic sites (Figure 7A). The spatial distribution for ISL1 occupancy is consistent with other reports of cell type–specific binding patterns of transcription factors, localizing to transcription start site distal enhancer regions (67, 68). This demonstrates that ISL1 is preferentially localizing to SAN-specific regulatory regions, conferring its role in driving gene expression profiles of these pacemaker cells. De novo motif analysis revealed that the most highly enriched DNA binding motif among ISL1-bound regions was a consensus element for ISL1 (P value 1 × 10–172), followed closely by forkhead and other homeobox binding motifs (Figure 7B and Supplemental Tables 4 and 5), suggesting that transcription factors from these families may act as collaborating factors with ISL1 to regulate SAN gene expression profiles. Spatial annotation of ISL1 binding sites, via nearest transcription start site, reveals 1,003 potential ISL1 targets including those involved in important developmental processes, axon guidance, and cell proliferation (Supplemental Table 1, Figure 7C, and Supplemental Figure 4). Intersection of ChIP-seq data with RNA-seq data from Isl1 mutants and controls revealed genes that were directly regulated positively (193 targets) or negatively (35 targets) by ISL1, suggesting that ISL1 acts mainly as an activator in SAN cells but that ISL1 can also act as a repressor in this context (Figure 7, D and E, and Supplemental Table 1). GO and Reactome pathway enrichment analysis of direct targets of ISL1 revealed that, for direct targets downregulated in Hcn4-CreERT2 Isl1fl/fl mutant SANs, overrepresented categories included extracellular matrix disassembly, axon guidance, and cell proliferation — categories overall similar to those overrepresented in genes found to be downregulated by RNA-seq analyses. For direct targets of ISL1 upregulated in Hcn4-CreERT2 Isl1fl/fl mutants, overrepresented categories included regulation of sequence-specific DNA binding activity and transporter activity (Figure 7F, Supplemental Figure 4, and Supplemental Table 2). Examination of direct targets downregulated in Isl1 mutants revealed several genes known to be critical for SAN function, including Ank2 and Kl (refs. 69, 70, and Table 1). Although Tbx3 was one of the most highly downregulated genes, and Tbx3 mutation has been shown to result in bradycardia and heart-rate variability (34, 46) — phenotypes similar to those observed in Isl1 mutant SANs — analyses of our ChIP-seq data did not assign any ISL1 ChIP-seq peaks to the Tbx3 promoter, based on proximity to the nearest transcription start site. Because enhancers can regulate genes at long distances, we postulated that ISL1 might bind to a long-range enhancer to regulate expression of Tbx3. Recently, 4C-seq chromatin conformation capture experiments have been performed for Tbx3 in E10.5 mouse hearts and have indicated multiple long-range potential enhancers looping to the Tbx3 promoter region (71). Using these and a recently published genome-wide, high-resolution chromatin conformation capture dataset (72) in conjunction with our ISL1 ChIP-seq data, we identified 2 ISL1 peaks within domains that loop to the Tbx3 promoter (Chr5:119870384-119870528 and Chr5:119980239-119980383), suggesting that ISL1 may bind to these sites within SAN cells to regulate Tbx3 expression. In addition to directly regulating Kl, Ank2, and potentially Tbx3, we also found that ISL1 directly regulated genes, including Calcrl and Flrt2, for which human variants have been found associated with irregular heart-rate phenotypes (Table 1 and ref. 73).
Figure 7. ISL1 directly regulates a number of genes required for normal pacemaker function in mice and humans.
(A) ChIP-seq ISL1-binding regions were mapped relative to their nearest TSS. Annotation includes whether a peak is in the TSS (defined as from –1 kb to +100 bp), transcription termination site (TTS; defined as from –100 bp to +1 kb), exon (coding), 5′ UTR, 3′ UTR, intronic, or intergenic. (B) Top motifs enriched in the vicinity of ISL1-binding sites. (C) GO functional clustering of genes associated with ISL1 ChIP-seq peaks (top 10 not redundant categories are shown). (D) Overlay of RNA-seq and ChIP-seq results correlates ISL1 binding with gene regulation in SAN cells. Scatterplot of RNA-Seq from Figure 6A with genes upregulated or downregulated 1.5-fold in Isl1 mutant SAN cells, colored in red and green, respectively. Up- and downregulated genes demonstrating vicinal ISL1 binding in SAN cells are black. (E) Overlay of RNA-seq and ChIP-seq results revealed 228 genes as potential direct targets of ISL1 in SAN cells. (F) GO functional clustering of these genes allowed for identification of cellular functions directly regulated by ISL1 (top 10 not redundant categories are shown). See also Supplemental Figure 4, Table 1, and Supplemental Tables 1, 2, 4, and 5.
Table 1. Direct targets of ISL1 associated with sinus node dysfunction.
Discussion
ISL1 is required for proliferation, survival, and function of SAN cells at distinct stages throughout SAN development.
Reduced ISL1 expression in Isl1 compound mutants, or ablation of Isl1 by tamoxifen injection at E9.5, resulted in severe bradycardia, increased heart-rate variability, and prolonged sinus pauses, with Isl1 mutant embryos dying in utero with severely slowed heart rates around E11.5. The foregoing demonstrated an indispensable role for ISL1 in cardiac pacemaking and embryonic viability at these early stages. Lineage tracing of Hcn4-CreERT2 cells revealed a reduced number of pacemaker cells at E11.5 when Isl1 ablation was induced at E9.5, which could be accounted for by increased cell death and reduced proliferation in mutants relative to control littermates. Marked reduction of TBX3 and HCN4 was also observed in remaining lineage-traced pacemaker cells in Hcn4-CreERT2 Isl1fl/fl mutants, suggesting that regulation of these genes is directly or indirectly downstream of ISL1 in SAN cells.
Ablation of Isl1 at later stages with Hcn4-CreERT2 (by administration of tamoxifen at E11.5 or E13.5) resulted in similar bradycardia, increased heart-rate variability, and reduced Hcn4, Tbx3, and Shox2 expression with only a slight loss of pacemaker cells. Cell loss was correlated with reduced rates of proliferation but no increased apoptosis. Hcn4-CreERT2 Isl1fl/fl mutants induced at these later stages were able to survive.
ISL1 is upstream of ion channels and transcription factors required for SAN function.
To gain insight into genes downstream of ISL1 that might account for observed SAN phenotypes, we performed transcriptome analyses on RNA from FACS-purified SAN cells from Hcn4-CreERT2 Isl1fl/fl mutants and littermate controls that had been tamoxifen induced at E10 and harvested at E12. Bioinformatics analyses of these data revealed decreased expression of a number of key ion channel and cardiac transcription factor genes in Hcn4-CreERT2 Isl1fl/fl mutant SAN.
SAN function is thought to be dependent on a 2-clock mechanism: the calcium clock and a voltage clock (5, 6). Decreased expression of mRNA encoding critical components of each of these clocks was observed in Isl1 mutant SAN cells, including mRNA for genes encoding subunits of the L-type calcium channel, Cacna1a, Cacna1b, Cacnb1, and Hcn4, a key component of the voltage clock. Germline ablation of Hcn4 results in severe bradycardia and early embryonic lethality, with embryos dying around E10–E11 (11). However, later ablation of Hcn4 utilizing an Hcn4-inducible Cre does not affect viability but results in sinus pauses and increased heart-rate variability (19). Mutations in the human Hcn4 gene also lead to sinus bradycardia and have been associated with inherited sick sinus syndrome (12–17).
Transcription factors that are critical for SAN function were significantly downregulated in Hcn4-CreERT2 Isl1fl/fl mutants, including Shox2 and Tbx3. Germline ablation of Shox2 results in SAN hypoplasia and bradycardia (49–51). TBX3 is selectively expressed in the cardiac conduction system, and aberrations in Tbx3 are associated with human arrhythmias (74). Hypomorphic and conditional mutants of Tbx3 in mice exhibit sinus bradycardia and sinus pauses (46).
We also observed significant upregulation of transcripts from several genes characteristic of atrial phenotype, including Nppa, Nppb, Gja1/Cx43, and Gja5/Cx40 (44) in Hcn4-CreERT2 Isl1fl/fl mutant SANs. Interestingly, Tbx3 and Shox2 mutants also display upregulation of Gja5/Cx40 in the SAN (34, 36, 46, 49–51).
Decreased expression of a number of other genes associated with aberrant pacemaker function was also observed in Hcn4-CreERT2 Isl1fl/fl mutant SANs. These included Ank2, Kl, Wdr62, and Calcrl. Two families with highly penetrant and severe sinus node dysfunction have been mapped to the Ank2 locus, and mice heterozygous for Ank2 display severe sinus node dysfunction, including severe bradycardia and heart-rate variability (69), as observed in our Isl1 SAN mutants. ANK2 is required for normal membrane trafficking and organization of ion channels and transporters essential for SAN function. Kl is selectively expressed in SAN within the heart, and ablation of Kl results in an inability of the SAN to respond to stress (70). Human mutations in WDR62, required for mitotic spindle formation, result in a recessive syndrome of microcephaly, cerebellar hypoplasia, and congenital bradycardia with irregular heart rate (75). Human variants in Calcrl, encoding the adrenomedullin receptor, have been associated with irregular heart rate (73). Mice with global knockout of Calcrl die between E13.5–E14.5 and exhibit cardiac defects (76).
Both early- and later-stage ablations of Isl1 adversely affected proliferation of SAN cells. Expression of 2 widely utilized cell cycle genes, Kras and Myc, was significantly downregulated in SANs from Hcn4-CreERT2 Isl1fl/fl mutants. The potential role of these genes in SAN remains to be explored. The foregoing results suggest that aberrant regulation of a number of genes important for SAN phenotype and function contributes to the SAN phenotype of Hcn4-CreERT2 Isl1fl/fl mutants.
Genome-wide ChIP-seq studies of purified SAN cells reveal critical direct targets of ISL1 and suggest potential cofactor families for ISL1 action in SAN cells.
To gain insight into mechanisms by which ISL1 regulates SAN phenotype and function, we performed ChIP-seq studies utilizing antibodies to ISL1 on purified SAN cells. Intersection of ChIP-seq and RNA-seq data demonstrated that several genes critical to SAN function in mouse and/or human were directly activated by ISL1 in SAN cells, including Ank2, Kl, Tbx3, Calcrl, and Flrt2 (46, 69, 70, 73, 74). Human variants in FLRT2 have been associated with irregular heart rate, although the potential role of FLRT2 in SAN function has not yet been validated in experimental models (73).
Bioinformatics analyses of DNA binding motifs enriched within ISL1 ChIP-seq peaks in SAN cells revealed enrichment for other homeodomain and FOX transcription factor binding sites, suggesting that members of these transcription factor families may cooperate with ISL1 to regulate expression of SAN genes. Notably, SHOX2 is another homeodomain transcription factor known to be important for SAN function (49–51, 59). FOX factors that may be important for SAN function remain to be identified.
Sick sinus syndrome accounts for the necessity of approximately half of the pacemaker implants within the United States (2, 3). Gene transfer and reprogramming of working cardiomycytes or stem cells into pacemaker cells to generate a biological pacemaker represents a promising alternative therapy for sick sinus syndrome. Overexpression of HCN4 is sufficient to induce pacemaker function and has been successful in generation of a biological pacemaker (77). Overexpression of TBX3 in embryonic mouse atrial myocytes is sufficient to induce an SAN gene program (36). However, in mature cardiomyocytes, overexpression of TBX3 induces only a subset of pacemaker-specific genes, and neither ectopic pacemaker activity nor pacemaker current (If) was observed (47). Recent studies have demonstrated that overexpression of ISL1 in either embryonic stem cells or Xenopus embryos results in upregulation of nodal-specific genes and downregulation of transcripts of working myocardium (78), consistent with our observations of ISL1 action with loss-of-function studies in SAN cells. However, overexpression of ISL1 resulted in partial activation of the SAN program. Together, these observations suggest that generation of a biological pacemaker might benefit from combinatorial expression of transcription factors important to SAN identity and function, including TBX3, ISL1, and TBX18 (35, 48).
Altogether, results of these studies have given mechanistic insight into distinct cell autonomous requirements for ISL1 throughout SAN development, identifying a number of key downstream targets that can account for observed defects in SAN phenotype and function. Our studies represent the first in vivo ChIP-seq studies for SAN cells, which provide a basis for further exploration of factors critical to SAN formation and function.
Methods
Transgenic mice.
Isl1nLacZ knockin, floxed Isl1 (Isl1fl/fl), and Isl1 hypomorphic (Isl1fl:Neo/+) mouse lines were generated as described (54, 64, 79). Strategy to generate the tamoxifen-inducible Hcn4-Cre mice (Hcn4-CreERT2) will be published separately. Briefly, a targeting cassette with CreERT2 cDNA was inserted immediately before translation initiation site (ATG) of the Hcn4 gene. Isl1 compound mutant mice (Isl1nLacZ/fl:Neo) were generated by crossing Isl1fl:Neo/+ mice with Isl1nLacZ/+ mice (63). To specifically delete Isl1 in cardiomyocytes or in the SAN, we crossed cTnT-Cre mice (65) or Hcn4-CreERT2 mice to homozygous floxed Isl1 mice on Rosa-LacZ or tdTomato backgrounds. To induce Cre activity of Hcn4-CreERT2, depending on embryonic age, pregnant mice were fed 150–300 μl of tamoxifen (10 mg/ml) by oral gavage at desired time points. For the early stage, tamoxifen was given at E9.5 and samples were harvested 32–36 hours after induction at around E11. For the later stage, tamoxifen was given at E11.5, and samples were harvested at E14.5. The presence of a Rosa-tdTomato allele allowed the embryos expressing Hcn4-CreERT2 to be visually genotyped upon tamoxifen induction.
IHC and in situ hybridization.
X-gal staining, immunostaining, and in situ hybridization were performed as described (80, 81). The following primary antibodies were used: mouse monoclonal anti-ISL1/2 (39.4D5, Developmental Studies Hybridoma Bank [DSHB]), rabbit anti-ISL1 (ab20670, Abcam), rat anti-HCN4 (ab32675, Abcam), rabbit anti-Cx40 (sc-28658, Santa Cruz), goat anti-TBX3 (sc-17871, Santa Cruz Biotechnology Inc.), and rat anti-BrdU (ab6326, Abcam). The secondary antibodies were Alexa 488 or 594 labeled (Invitrogen). TUNEL staining was performed as recommended (Roche Diagnostics). For BrdU staining, pregnant mice were injected with 500 μl of BrdU (proprietary mixture, Ambion) every 3 hours for 3 times, and BrdU staining was performed as described (79). For cell counting, sections of the SAN (region) of the appropriated developmental stages were cut at 10 μm, every fourth section was stained, and positive cells were counted. To compare cell number of control and Isl1 mutant SAN at E14.5 (tamoxifen induced at 11.5), X-gal+ or Tomato+ cells were counted every fourth section, and total number of SAN cells per SAN are expressed. For proliferation and cell death assay, total BrdU+ or TUNEL+ cells in the region of right SV or SAN (marked by Tomato+) of the control and mutant samples were counted and expressed as percentage of total Tomato+ SAN cells. To assess ISL1 expression during development, ISL1+ and HCN4+ cells were counted and data were expressed as percentage of total HCN4+ cells. At least 4–6 matched sections were analyzed, and 3 samples per genotype per time point were analyzed.
RNA-seq and qPCR.
RNA-seq was performed using the pacemaker cells FACS sorted from embryos of genotype Hcn4-CreERT2 Isl1fl/fl (mutant) and Hcn4-CreERT2 Isl1+/+ (control). Tamoxifen was given at E10.5, and samples were harvested 36 hours after induction at E12–E12.5. The presence of a Rosa-tdTomato allele allowed the embryos expressing Hcn4-CreERT2 to be visually genotyped upon tamoxifen induction. The Tomato+ SANs of Isl1 mutant and control embryos were dissected under fluorescence microscopy. The samples for the same genotype were pooled and digested with a mixture of collagenase II (1 mg/ml)/trypsin (0.1%) for 10 minutes with periodical pipetting, and clear supernatants were collected in a 15-ml tube. The digestion was repeated until SAN tissue was totally digested. The cell suspension was filtered through a 40-um filter unit (Fisher Scientific), and endothelial cells were removed by incubation with CD31 microbeads (Miltenyi Biotec). SAN cells were resuspended in 0.5 ml DMEM medium and kept on ice until sorting. Hcn4-CreERT2–expressing pacemaker cells (Tomato+) were FACS sorted (BD FACSAria, BD Biosciences) into RNAlater (QIAGEN) and stored overnight at 4°C. The following day, the samples were centrifuged. RNAlater was removed, and the cells were frozen at –80°C. After multiple rounds of cell sorting, cells from individual collections were pooled at the time of RNA extraction. RNA was prepared using the RNeasy Mini kit (QIAGEN) following the manufacturer’s instructions and were quantified using a NanoDrop ND-1000 spectrophotometer.
RNA-seq was performed as described (82). The complete RNA-seq datasets are available from the Gene Expressing Omnibus database (http://www.ncbi.nlm.nih.gov/geo/) under the accession number GSE69097. Briefly, total RNA (10 ng) was incubated with Oligo(dT) magnetic beads to isolated mRNA. RNA-seq libraries were prepared with the SMARTer cDNA library construction kit (Clontech) according to manufacturer’s instructions and sequenced using Illumina HiSeqTM 2000 with paired-end sequencing at 90-bp read length (BGI). Adapter and poor-quality read sequences were trimmed with Trim Galore (Babraham Bioinformatics) using default settings for paired-end reads. Trimmed reads were then quality controlled with FastQC (http://www.bioinformatics.bbsrc.ac.uk/projects/fastqc). On average, each read pair resulted in approximately 22 million uniquely mapped reads after mapping to the mm9 reference genome with TopHat2 (83) and Bowtie2 (84). Transcript expression values were determined after transcript normalization (reads per kilobase per million; RPKM) with AltAnalyze (85). Transcripts were considered significantly expressed if RPKM ≥ 1 in either one of the 2 conditions. Differential expression analysis was performed considering |fold-change Isl1 mutant vs. ctrl| ≥ 1.5 fold as a cut-off. For scatterplot representation, values are log2 transformed tag counts normalized to 10 million uniquely mapped tags. GO and Reactome pathway enrichment analysis was conducted with GO-Elite (http://www.genmapp.org/go_elite) (86).
qPCR validation of RNA-seq targets were performed using SAN pacemaker cells as described above. qPCR was performed using SYBR green detection. Primer pairs are listed in Supplemental Table 3.
ChIP-sequencing and data analysis.
For ChIP-seq assay, the SANs from Hcn4-nGFP mice at postnatal days 1–3 (P1–P3) were digested and FACS-sorted as describe above.
ChIP of ISL1 was performed with modified protocol described previously (87). The complete ChIP-seq datasets are available from the GEO database under the accession number GSE68974. Briefly, for ISL1 ChIP, 2 × 106 cells were first crosslinked in 2 mM disuccinimidyl glutarate (Pierce Biotechnology) in PBS for 30 minutes, then subsequently in 1% formaldehyde (Sigma-Aldrich) in PBS for 10 minutes, all at room temperature. The reactions were quenched by adding glycine (Sigma-Aldrich) to a final concentration of 125 mM. The cells were immediately centrifuged (5 minutes, 700 × g, 4°C) and washed twice with ice-cold PBS. Cells were resuspended in swelling buffer (10 mM HEPES/KOH pH7.9, 85 mM KCl, 1 mM EDTA, 0.5% IGEPAL CA-630 [Sigma-Aldrich], 1× protease inhibitor cocktail [Roche Applied Science], 1 mM PMSF) for 5 minutes. Cell pellets were spun down and resuspended in 1 ml RIPA buffer (10 mM Tris/HCl pH7.6, 1 mM EDTA, 1 mM EGTA, 0.1% SDS, 0.1% Na-Deoxycholate, 1% Triton X-100, 1× protease inhibitor cocktail [Roche Applied Science], 1 mM PMSF). Chromatin was sheared to an average DNA size of 100–400 bp by administering 10 pulses of 30-second duration at 12 W power output with 60-second pause on wet ice using a Misonix 3000 sonicator. The lysate was cleared by centrifugation (5 minutes, 16,000 × g, 4°C). Supernatant (1%) was kept as ChIP input. Meanwhile, Dynabeads Protein G were prepared with the ISL1 antibody (39.4D5, DSHB) by incubating Dynabeads Protein G and 5 μg specific antibody in 0.5% BSA/PBS for 1 hour at 4°C on rotator, then washed twice with 0.5% BSA/PBS and brought up to the original volume with 0.1% BSA/PBS. The protein-DNA complex of interest was immunoprecipitated by rotating the supernatant with 30 μl Dynabeads Protein G coated with specific antibody overnight at 4°C (Invitrogen). Beads were washed with each buffer by rotating in 1 ml buffer at 4°C for 5 minutes: RIPA buffer (10 mM Tris/HCl pH7.6, 1 mM EDTA, 1 mM EGTA, 0.1% SDS, 0.1% Na-Deoxycholate, 1% Triton X-100, 1× protease inhibitor cocktail [Roche Applied Science], 1 mM PMSF), LiCl buffer (0.25 M LiCl, 1% NP40, 1% NaDOC), Tris-EDTA (TE) plus 0.2% Triton X-100, and TE plus 50 mM NaCl. Immunoprecipitated chromatin was eluted twice with 100 μl elution buffer each (TE, 2% SDS) into fresh tubes for 30 minutes and 10 minutes, respectively. Eluates were pooled, the Na+ concentration was adjusted to 300 mM with 5 M NaCl, and crosslinks were reversed overnight at 65°C in a hybridization oven. The samples were sequentially incubated at 37°C for 1 hour each with 0.33 mg/ml RNase A and 0.5 mg/ml proteinase K (Sigma-Aldrich). The DNA was isolated using the ChIP DNA Clean & Concentrator (Zymo Research Corp.) according to the manufacturer’s instructions.
Sequencing libraries were prepared from collected ISL1 ChIP and corresponding input DNA by blunting, A-tailing, adaptor ligation as previously described (88) using NEXTFlex barcoded adapters from Bioo Scientific. Libraries were PCR-amplified for 12–15 cycles, size selected for 225–375 bp fragments by gel extraction, and single-end sequenced on a Hi-Seq 2500 (Illumina) for 50 cycles. Reads were aligned to the mouse mm9 genome assembly (NCBI Build 37) using Bowtie allowing up to 2 mismatches. Only tags that mapped uniquely to the genome were considered for further analysis. Genomic binding peaks for transcription factor ISL1 were identified using the findPeaks command from HOMER (http://homer.salk.edu/homer/) with 8-fold enrichment over the input sample, 4-fold enrichment over local background, a minimal tag number of 16, and normalization to 10 million mapped reads per experiment (88). Peaks were annotated using the annotatePeaks command, assigning to gene targets based on the closest RefSeq-defined transcription start site (TSS). Identification of Isl1-associated transcription factor binding motifs was carried out with command findMotifsGenome.pl using a standard background as random genomic sequence sampled according to GC content of input sequences. GO and Reactome pathway enrichment analysis was conducted with GO-Elite (86).
Echocardiography.
Pregnant mice were anesthetized with isoflurane. Echocardiography was performed utilizing a VisualSonics Vevo770 high-resolution ultrasound system with a RMV704 probe (40 MHz). The position of individual embryos was scanned and identified, and a map with individual embryos was drawn. B-mode and pulse-wave Doppler images were obtained to allow for assessment of heart-rate and stroke volume. After echocardiography, embryos were dissected, correlated to the map, and genotyped for phenotype-genotype correlations.
Statistics.
Data are presented as mean ± SEM, and a 2-tailed t test was used for 2-group comparisons. Differences were considered statistically significant at a value of P < 0.05.
Study approval.
All the experiments involving mice were carried out in accordance with protocols approved by the Institutional Animal Care and Use Committee of USCD (A3033-01) and by the Animal Committee of Tongji University School of Medicine (TJmed-010-10).
Supplementary Material
Acknowledgments
Y. Sun was supported by grants from the Ministry of Science and Technology China (2013CB967400) and the National Natural Science Foundation of China (NSFC) (31071280, 81171069), and a GBIA grant from American Heart Association; X. Liang was supported by grants from the NSFC (31171393, 81370196); H-S.V. Chen was supported by a NIH grant (RO1 HL105194); and S.M. Evans was supported by grants from the NIH (HL123747, HL117649, and HL074066). Thanks to Karl Laugwitz, Alessandra Moretti, and Cornelis J. Boogerd for helpful discussions and sharing of data prior to publication.
Footnotes
Conflict of interest: The authors have declared that no conflict of interest exists.
Reference information:J Clin Invest. 2015;125(8):3256–3268. doi:10.1172/JCI68257.
References
- 1.Chugh SS, et al. Epidemiology of sudden cardiac death: clinical and research implications. Prog Cardiovasc Dis. 2008;51(3):213–228. doi: 10.1016/j.pcad.2008.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lamas GA, et al. Ventricular pacing or dual-chamber pacing for sinus-node dysfunction. N Engl J Med. 2002;346(24):1854–1862. doi: 10.1056/NEJMoa013040. [DOI] [PubMed] [Google Scholar]
- 3.Dobrzynski H, Boyett MR, Anderson RH. New insights into pacemaker activity: promoting understanding of sick sinus syndrome. Circulation. 2007;115(14):1921–1932. doi: 10.1161/CIRCULATIONAHA.106.616011. [DOI] [PubMed] [Google Scholar]
- 4.Gepstein L. Stem cells as biological heart pacemakers. Expert Opin Biol Ther. 2005;5(12):1531–1537. doi: 10.1517/14712598.5.12.1531. [DOI] [PubMed] [Google Scholar]
- 5.Lakatta EG, Maltsev VA, Vinogradova TM. A coupled SYSTEM of intracellular Ca2+ clocks and surface membrane voltage clocks controls the timekeeping mechanism of the heart’s pacemaker. Circ Res. 2010;106(4):659–673. doi: 10.1161/CIRCRESAHA.109.206078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mangoni ME, Nargeot J. Genesis and regulation of the heart automaticity. Physiol Rev. 2008;88(3):919–982. doi: 10.1152/physrev.00018.2007. [DOI] [PubMed] [Google Scholar]
- 7.Lakatta EG, et al. The integration of spontaneous intracellular Ca2+ cycling and surface membrane ion channel activation entrains normal automaticity in cells of the heart’s pacemaker. Ann N Y Acad Sci. 2006;1080:178–206. doi: 10.1196/annals.1380.016. [DOI] [PubMed] [Google Scholar]
- 8.Maltsev VA, Lakatta EG. Dynamic interactions of an intracellular Ca2+ clock and membrane ion channel clock underlie robust initiation and regulation of cardiac pacemaker function. Cardiovasc Res. 2008;77(2):274–284. doi: 10.1093/cvr/cvm058. [DOI] [PubMed] [Google Scholar]
- 9.Garcia-Frigola C, Shi Y, Evans SM. Expression of the hyperpolarization-activated cyclic nucleotide-gated cation channel HCN4 during mouse heart development. Gene Expr Patterns. 2003;3(6):777–783. doi: 10.1016/S1567-133X(03)00125-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ludwig A, Zong X, Jeglitsch M, Hofmann F, Biel M. A family of hyperpolarization-activated mammalian cation channels. Nature. 1998;393(6685):587–591. doi: 10.1038/31255. [DOI] [PubMed] [Google Scholar]
- 11.Stieber J, et al. The hyperpolarization-activated channel HCN4 is required for the generation of pacemaker action potentials in the embryonic heart. Proc Natl Acad Sci U S A. 2003;100(25):15235–15240. doi: 10.1073/pnas.2434235100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Laish-Farkash A, et al. A novel mutation in the HCN4 gene causes symptomatic sinus bradycardia in moroccan jews. J Cardiovasc Electrophysiol. 2010;21(12):1365–1372. doi: 10.1111/j.1540-8167.2010.01844.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ueda K, et al. Role of HCN4 channel in preventing ventricular arrhythmia. J Hum Genet. 2009;54(2):115–121. doi: 10.1038/jhg.2008.16. [DOI] [PubMed] [Google Scholar]
- 14.Nof E, et al. Point mutation in the HCN4 cardiac ion channel pore affecting synthesis, trafficking, and functional expression is associated with familial asymptomatic sinus bradycardia. Circulation. 2007;116(5):463–470. doi: 10.1161/CIRCULATIONAHA.107.706887. [DOI] [PubMed] [Google Scholar]
- 15.Milanesi R, Baruscotti M, Gnecchi-Ruscone T, DiFrancesco D. Familial sinus bradycardia associated with a mutation in the cardiac pacemaker channel. N Engl J Med. 2006;354(2):151–157. doi: 10.1056/NEJMoa052475. [DOI] [PubMed] [Google Scholar]
- 16.Ueda K, et al. Functional characterization of a trafficking-defective HCN4 mutation, D553N, associated with cardiac arrhythmia. J Biol Chem. 2004;279(26):27194–27198. doi: 10.1074/jbc.M311953200. [DOI] [PubMed] [Google Scholar]
- 17.Schulze-Bahr E, et al. Pacemaker channel dysfunction in a patient with sinus node disease. J Clin Invest. 2003;111(10):1537–1545. doi: 10.1172/JCI16387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Schweizer PA, et al. cAMP sensitivity of HCN pacemaker channels determines basal heart rate but is not critical for autonomic rate control. Circ Arrhythm Electrophysiol. 2010;3(5):542–552. doi: 10.1161/CIRCEP.110.949768. [DOI] [PubMed] [Google Scholar]
- 19.Herrmann S, Stieber J, Stockl G, Hofmann F, Ludwig A. HCN4 provides a ‘depolarization reserve’ and is not required for heart rate acceleration in mice. EMBO J. 2007;26(21):4423–4432. doi: 10.1038/sj.emboj.7601868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sasse P, Zhang J, Cleemann L, Morad M, Hescheler J, Fleischmann BK. Intracellular Ca2+ oscillations, a potential pacemaking mechanism in early embryonic heart cells. J Gen Physiol. 2007;130(2):133–144. doi: 10.1085/jgp.200609575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yang HT, et al. The ryanodine receptor modulates the spontaneous beating rate of cardiomyocytes during development. Proc Natl Acad Sc U S A. 2002;99(14):9225–9230. doi: 10.1073/pnas.142651999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Koushik SV, et al. Targeted inactivation of the sodium-calcium exchanger (Ncx1) results in the lack of a heartbeat and abnormal myofibrillar organization. FASEB J. 2001;15(7):1209–1211. doi: 10.1096/fj.00-0696fje. [DOI] [PubMed] [Google Scholar]
- 23.Mangoni ME, Couette B, Marger L, Bourinet E, Striessnig J, Nargeot J. Voltage-dependent calcium channels and cardiac pacemaker activity: from ionic currents to genes. Prog Biophys Mol Biol. 2006;90(1–3):38–63. doi: 10.1016/j.pbiomolbio.2005.05.003. [DOI] [PubMed] [Google Scholar]
- 24.Takeshima H, Komazaki S, Hirose K, Nishi M, Noda T, Iino M. Embryonic lethality and abnormal cardiac myocytes in mice lacking ryanodine receptor type 2. EMBO J. 1998;17(12):3309–3316. doi: 10.1093/emboj/17.12.3309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Narayanan N, Xu A. Phosphorylation and regulation of the Ca(2+)-pumping ATPase in cardiac sarcoplasmic reticulum by calcium/calmodulin-dependent protein kinase. Basic Res Cardiol. 1997;92(suppl 1):25–35. doi: 10.1007/BF00794065. [DOI] [PubMed] [Google Scholar]
- 26.Meyer M, Dillmann WH. Sarcoplasmic reticulum Ca(2+)-ATPase overexpression by adenovirus mediated gene transfer and in transgenic mice. Cardiovasc Res. 1998;37(2):360–366. doi: 10.1016/S0008-6363(97)00270-8. [DOI] [PubMed] [Google Scholar]
- 27.Periasamy M, et al. Impaired cardiac performance in heterozygous mice with a null mutation in the sarco(endo)plasmic reticulum Ca2+-ATPase isoform 2 (SERCA2) gene. J Biol Chem. 1999;274(4):2556–2562. doi: 10.1074/jbc.274.4.2556. [DOI] [PubMed] [Google Scholar]
- 28.Ver Heyen M, et al. Replacement of the muscle-specific sarcoplasmic reticulum Ca(2+)-ATPase isoform SERCA2a by the nonmuscle SERCA2b homologue causes mild concentric hypertrophy and impairs contraction-relaxation of the heart. Circ Res. 2001;89(9):838–846. doi: 10.1161/hh2101.098466. [DOI] [PubMed] [Google Scholar]
- 29.Hirota A, Fujii S, Kamino K. Optical monitoring of spontaneous electrical activity of 8-somite embryonic chick heart. Jpn J Physiol. 1979;29(5):635–639. doi: 10.2170/jjphysiol.29.635. [DOI] [PubMed] [Google Scholar]
- 30.Kamino K, Hirota A, Fujii S. Localization of pacemaking activity in early embryonic heart monitored using voltage-sensitive dye. Nature. 1981;290(5807):595–597. doi: 10.1038/290595a0. [DOI] [PubMed] [Google Scholar]
- 31.Van Mierop LH. Location of pacemaker in chick embryo heart at the time of initiation of heartbeat. Am J Physiol. 1967;212(2):407–415. doi: 10.1152/ajplegacy.1967.212.2.407. [DOI] [PubMed] [Google Scholar]
- 32.Viragh S, Challice CE. The development of the conduction system in the mouse embryo heart. Dev Biol. 1980;80(1):28–45. doi: 10.1016/0012-1606(80)90496-0. [DOI] [PubMed] [Google Scholar]
- 33.Christoffels VM, et al. Formation of the venous pole of the heart from an Nkx2-5-negative precursor population requires Tbx18. Circ Res. 2006;98(12):1555–1563. doi: 10.1161/01.RES.0000227571.84189.65. [DOI] [PubMed] [Google Scholar]
- 34.Wiese C, et al. Formation of the sinus node head and differentiation of sinus node myocardium are independently regulated by Tbx18 and Tbx3. Circ Res. 2009;104(3):388–397. doi: 10.1161/CIRCRESAHA.108.187062. [DOI] [PubMed] [Google Scholar]
- 35.Kapoor N, Liang W, Marban E, Cho HC. Direct conversion of quiescent cardiomyocytes to pacemaker cells by expression of Tbx18. Nat Biotechnol. 2013;31(1):54–62. doi: 10.1038/nbt.2465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hoogaars WM, et al. Tbx3 controls the sinoatrial node gene program and imposes pacemaker function on the atria. Genes Dev. 2007;21(9):1098–1112. doi: 10.1101/gad.416007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Patel R, Kos L. Endothelin-1 and Neuregulin-1 convert embryonic cardiomyocytes into cells of the conduction system in the mouse. Dev Dyn. 2005;233(1):20–28. doi: 10.1002/dvdy.20284. [DOI] [PubMed] [Google Scholar]
- 38.Gourdie RG, Wei Y, Kim D, Klatt SC, Mikawa T. Endothelin-induced conversion of embryonic heart muscle cells into impulse-conducting Purkinje fibers. Proc Natl Acad Sci U S A. 1998;95(12):6815–6818. doi: 10.1073/pnas.95.12.6815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hyer J, et al. Induction of Purkinje fiber differentiation by coronary arterialization. Proc Natl Acad Sci U S A. 1999;96(23):13214–13218. doi: 10.1073/pnas.96.23.13214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rentschler S, et al. Neuregulin-1 promotes formation of the murine cardiac conduction system. Proc Natl Acad Sci U S A. 2002;99(16):10464–10469. doi: 10.1073/pnas.162301699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Milan DJ, Giokas AC, Serluca FC, Peterson RT, MacRae CA. Notch1b and neuregulin are required for specification of central cardiac conduction tissue. Development. 2006;133(6):1125–1132. doi: 10.1242/dev.02279. [DOI] [PubMed] [Google Scholar]
- 42.Rentschler S, et al. Notch signaling regulates murine atrioventricular conduction and the formation of accessory pathways. J Clin Invest. 2011;121(2):525–533. doi: 10.1172/JCI44470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mommersteeg MT, et al. Molecular pathway for the localized formation of the sinoatrial node. Circ Res. 2007;100(3):354–362. doi: 10.1161/01.RES.0000258019.74591.b3. [DOI] [PubMed] [Google Scholar]
- 44.Christoffels VM, Smits GJ, Kispert A, Moorman AF. Development of the pacemaker tissues of the heart. Circ Res. 2010;106(2):240–254. doi: 10.1161/CIRCRESAHA.109.205419. [DOI] [PubMed] [Google Scholar]
- 45.Hoogaars WM, et al. The transcriptional repressor Tbx3 delineates the developing central conduction system of the heart. Cardiovasc Res. 2004;62(3):489–499. doi: 10.1016/j.cardiores.2004.01.030. [DOI] [PubMed] [Google Scholar]
- 46.Frank DU, et al. Lethal arrhythmias in Tbx3-deficient mice reveal extreme dosage sensitivity of cardiac conduction system function and homeostasis. Proc Natl Acad Sci U S A. 2012;109(3):E154–E163. doi: 10.1073/pnas.1115165109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Bakker ML, et al. T-box transcription factor TBX3 reprogrammes mature cardiac myocytes into pacemaker-like cells. Cardiovasc Res. 2012;94(3):439–449. doi: 10.1093/cvr/cvs120. [DOI] [PubMed] [Google Scholar]
- 48.Hu YF, Dawkins JF, Cho HC, Marban E, Cingolani E. Biological pacemaker created by minimally invasive somatic reprogramming in pigs with complete heart block. Sci Transl Med. 2014;6(245):245ra94. doi: 10.1126/scitranslmed.3008681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Blaschke RJ, et al. Targeted mutation reveals essential functions of the homeodomain transcription factor Shox2 in sinoatrial and pacemaking development. Circulation. 2007;115(14):1830–1838. doi: 10.1161/CIRCULATIONAHA.106.637819. [DOI] [PubMed] [Google Scholar]
- 50.Espinoza-Lewis RA, et al. Shox2 is essential for the differentiation of cardiac pacemaker cells by repressing Nkx2-5. Dev Biol. 2009;327(2):376–385. doi: 10.1016/j.ydbio.2008.12.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Liu H, et al. Functional redundancy between human SHOX and mouse Shox2 genes in the regulation of sinoatrial node formation and pacemaking function. J Biol Chem. 2011;286(19):17029–17038. doi: 10.1074/jbc.M111.234252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Cai CL, et al. Isl1 identifies a cardiac progenitor population that proliferates prior to differentiation and contributes a majority of cells to the heart. Dev Cell. 2003;5(6):877–889. doi: 10.1016/S1534-5807(03)00363-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Moretti A, et al. Multipotent embryonic isl1+ progenitor cells lead to cardiac, smooth muscle, and endothelial cell diversification. Cell. 2006;127(6):1151–1165. doi: 10.1016/j.cell.2006.10.029. [DOI] [PubMed] [Google Scholar]
- 54.Sun Y, et al. Islet 1 is expressed in distinct cardiovascular lineages, including pacemaker and coronary vascular cells. Dev Biol. 2007;304(1):286–296. doi: 10.1016/j.ydbio.2006.12.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Sizarov A, Devalla HD, Anderson RH, Passier R, Christoffels VM, Moorman AF. Molecular analysis of patterning of conduction tissues in the developing human heart. Circ Arrhythm Electrophysiol. 2011;4(4):532–542. doi: 10.1161/CIRCEP.111.963421. [DOI] [PubMed] [Google Scholar]
- 56.Mommersteeg MT, et al. The sinus venosus progenitors separate and diversify from the first and second heart fields early in development. Cardiovasc Res. 2010;87(1):92–101. doi: 10.1093/cvr/cvq033. [DOI] [PubMed] [Google Scholar]
- 57.de Pater E, et al. Distinct phases of cardiomyocyte differentiation regulate growth of the zebrafish heart. Development. 2009;136(10):1633–1641. doi: 10.1242/dev.030924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Tessadori F, et al. Identification and functional characterization of cardiac pacemaker cells in zebrafish. PLoS One. 2012;7(10):e47644. doi: 10.1371/journal.pone.0047644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Hoffmann S, et al. Islet1 is a direct transcriptional target of the homeodomain transcription factor Shox2 and rescues the Shox2-mediated bradycardia. Basic Res Cardiol. 2013;108(2):339. doi: 10.1007/s00395-013-0339-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Vedantham V, Galang G, Evangelista M, Deo RC, Srivastava D. RNA sequencing of mouse sinoatrial node reveals an upstream regulatory role for islet-1 in cardiac pacemaker cells. Circ Res. 2015;116(5):797–803. doi: 10.1161/CIRCRESAHA.116.305913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Liang X, et al. HCN4 dynamically marks the first heart field and conduction system precursors. Circ Res. 2013;113(4):399–407. doi: 10.1161/CIRCRESAHA.113.301588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Spater D, et al. A HCN4+ cardiomyogenic progenitor derived from the first heart field and human pluripotent stem cells. Nat Cell Biol. 2013;15(9):1098–1106. doi: 10.1038/ncb2824. [DOI] [PubMed] [Google Scholar]
- 63.Liang X, et al. Isl1 is required for multiple aspects of motor neuron development. Mol Cell Neurosci. 2011;47(3):215–222. doi: 10.1016/j.mcn.2011.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Song MR, Sun Y, Bryson A, Gill GN, Evans SM, Pfaff SL. Islet-to-LMO stoichiometries control the function of transcription complexes that specify motor neuron and V2a interneuron identity. Development. 2009;136(17):2923–2932. doi: 10.1242/dev.037986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Jiao K, et al. An essential role of Bmp4 in the atrioventricular septation of the mouse heart. Genes Dev. 2003;17(19):2362–2367. doi: 10.1101/gad.1124803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Hoesl E, et al. Tamoxifen-inducible gene deletion in the cardiac conduction system. J Mol Cell Cardiol. 2008;45(1):62–69. doi: 10.1016/j.yjmcc.2008.04.008. [DOI] [PubMed] [Google Scholar]
- 67.Heinz S, et al. Effect of natural genetic variation on enhancer selection and function. Nature. 2013;503(7477):487–492. doi: 10.1038/nature12615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Gosselin D, et al. Environment drives selection and function of enhancers controlling tissue-specific macrophage identities. Cell. 2014;159(6):1327–1340. doi: 10.1016/j.cell.2014.11.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Le Scouarnec S, et al. Dysfunction in ankyrin-B-dependent ion channel and transporter targeting causes human sinus node disease. Proc Natl Acad Sci U S A. 2008;105(40):15617–15622. doi: 10.1073/pnas.0805500105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Takeshita K, et al. Sinoatrial node dysfunction and early unexpected death of mice with a defect of klotho gene expression. Circulation. 2004;109(14):1776–1782. doi: 10.1161/01.CIR.0000124224.48962.32. [DOI] [PubMed] [Google Scholar]
- 71.van Weerd JH, et al. A large permissive regulatory domain exclusively controls Tbx3 expression in the cardiac conduction system. Circ Res. 2014;115(4):432–441. doi: 10.1161/CIRCRESAHA.115.303591. [DOI] [PubMed] [Google Scholar]
- 72.Jin F, et al. A high-resolution map of the three-dimensional chromatin interactome in human cells. Nature. 2013;503(7475):290–294. doi: 10.1038/nature12644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.den Hoed M, et al. Identification of heart rate-associated loci and their effects on cardiac conduction and rhythm disorders. Nat Genet. 2013;45(6):621–631. doi: 10.1038/ng.2610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Bamshad M, et al. Mutations in human TBX3 alter limb, apocrine and genital development in ulnar-mammary syndrome. Nat Genet. 1997;16(3):311–315. doi: 10.1038/ng0797-311. [DOI] [PubMed] [Google Scholar]
- 75.Zaki MS, et al. New recessive syndrome of microcephaly, cerebellar hypoplasia, and congenital heart conduction defect. Am J Med Genet Part A. 2011;155A(12):3035–3041. doi: 10.1002/ajmg.a.34078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Dackor RT, Fritz-Six K, Dunworth WP, Gibbons CL, Smithies O, Caron KM. Hydrops fetalis, cardiovascular defects, and embryonic lethality in mice lacking the calcitonin receptor-like receptor gene. Mol Cell Biol. 2006;26(7):2511–2518. doi: 10.1128/MCB.26.7.2511-2518.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Yang XJ, Zhou YF, Li HX, Han LH, Jiang WP. Mesenchymal stem cells as a gene delivery system to create biological pacemaker cells in vitro. J Int Med Res. 2008;36(5):1049–1055. doi: 10.1177/147323000803600523. [DOI] [PubMed] [Google Scholar]
- 78.Dorn T, et al. Direct nkx2-5 transcriptional repression of isl1 controls cardiomyocyte subtype identity. Stem Cells. 2015;33(4):1113–1129. doi: 10.1002/stem.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Sun Y, Dykes IM, Liang X, Eng SR, Evans SM, Turner EE. A central role for Islet1 in sensory neuron development linking sensory and spinal gene regulatory programs. Nat Neurosci. 2008;11(11):1283–1293. doi: 10.1038/nn.2209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Liang X, et al. PINCH1 plays an essential role in early murine embryonic development but is dispensable in ventricular cardiomyocytes. Mol Cell Biol. 2005;25(8):3056–3062. doi: 10.1128/MCB.25.8.3056-3062.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Liang X, et al. Pinch1 is required for normal development of cranial and cardiac neural crest-derived structures. Circ Res. 2007;100(4):527–535. doi: 10.1161/01.RES.0000259041.37059.8c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Guo G, et al. Whole-genome and whole-exome sequencing of bladder cancer identifies frequent alterations in genes involved in sister chromatid cohesion and segregation. Nat Genet. 2013;45(12):1459–1463. doi: 10.1038/ng.2798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Kim D, Pertea G, Trapnell C, Pimentel H, Kelley R, Salzberg SL. TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol. 2013;14(4):R36. doi: 10.1186/gb-2013-14-4-r36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Langmead B, Salzberg SL. Fast gapped-read alignment with Bowtie 2. Nat Methods. 2012;9(4):357–359. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Salomonis N, et al. Alternative splicing regulates mouse embryonic stem cell pluripotency and differentiation. Proc Natl Acad Sci U S A. 2010;107(23):10514–10519. doi: 10.1073/pnas.0912260107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Zambon AC, et al. GO-Elite: a flexible solution for pathway and ontology over-representation. Bioinformatics. 2012;28(16):2209–2210. doi: 10.1093/bioinformatics/bts366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Li P, et al. NCoR repression of LXRs restricts macrophage biosynthesis of insulin-sensitizing omega 3 fatty acids. Cell. 2013;155(1):200–214. doi: 10.1016/j.cell.2013.08.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Heinz S, et al. Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol Cell. 2010;38(4):576–589. doi: 10.1016/j.molcel.2010.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.