Abstract
Severe congenital neutropenia (SCN) is often associated with inherited heterozygous point mutations in ELANE, which encodes neutrophil elastase (NE). However, a lack of appropriate models to recapitulate SCN has substantially hampered the understanding of the genetic etiology and pathobiology of this disease. To this end, we generated both normal and SCN patient–derived induced pluripotent stem cells (iPSCs), and performed genome editing and differentiation protocols that recapitulate the major features of granulopoiesis. Pathogenesis of ELANE point mutations was the result of promyelocyte death and differentiation arrest, and was associated with NE mislocalization and activation of the unfolded protein response/ER stress (UPR/ER stress). Similarly, high-dose G-CSF (or downstream signaling through AKT/BCL2) rescues the dysgranulopoietic defect in SCN patient–derived iPSCs through C/EBPβ-dependent emergency granulopoiesis. In contrast, sivelestat, an NE-specific small-molecule inhibitor, corrected dysgranulopoiesis by restoring normal intracellular NE localization in primary granules; ameliorating UPR/ER stress; increasing expression of CEBPA, but not CEBPB; and promoting promyelocyte survival and differentiation. Together, these data suggest that SCN disease pathogenesis includes NE mislocalization, which in turn triggers dysfunctional survival signaling and UPR/ER stress. This paradigm has the potential to be clinically exploited to achieve therapeutic responses using lower doses of G-CSF combined with targeting to correct NE mislocalization.
Introduction
Severe congenital neutropenia (SCN) is a rare myelopoietic disorder resulting in recurrent life-threatening infections due to a lack of mature neutrophils (1). Individuals with SCN have myeloid hypoplasia with arrest of myelopoiesis at the promyelocyte/myelocyte stage (2). Current treatment by administration of high-dose granulocyte CSF (G-CSF) induces an increase in the neutrophil counts in the peripheral blood (PB) of most SCN patients. The lack of beneficial effect of G-CSF administration on neutrophil function (3) and the possible role of G-CSF administration in leukemogenesis (4) underscores the need to search for alternative therapies. Currently, allogeneic hematopoietic stem cell transplantation (HSCT) is the only curative treatment for SCN (5).
SCN is actually a multigene syndrome that can be caused by inherited mutations in several genes. Around 60% of SCN patients are known to carry autosomal-dominant mutations in the ELANE gene, which encodes neutrophil elastase (NE) (6, 7). NE belongs to the class of serine proteases and is expressed exclusively in mature myelomonocytic cells and their committed immature precursors (promyelocytes and promonocytes). Stored as an active protease in azurophilic granules, NE is released upon exposure of the neutrophil to inflammatory stimuli. In the extracellular environment, NE cleaves extracellular matrix proteins, while serine protease inhibitors antagonize the proteinase activity (8).
Most ELANE mutations act as dominant heterozygous mutations and are single point mutations. It is unclear how these single point mutations affect the function of the protein and the viability of phagocytes. Some data suggest that ELANE gene mutations are not sufficient to cause the SCN phenotype and other genes may act as modifiers on promyelocyte survival and their response to G-CSF (9–11). There is also an absence of correlation between the genotype and phenotype. The same ELANE mutation can induce 2 types of disease: SCN and a more benign cyclic neutropenia, with cycles of neutropenia every 21 days (6). It is possible that disturbances of a feedback circuit, in which mature neutrophils homeostatically regulate myeloid progenitor populations, are responsible for this mechanism. This hypothesis was supported by the discovery that the protein PFAAP5 interacts with NE to interfere with GFI1-controlled transcriptional regulation (12). Finally, the coexistence of various phenotypes in the same kindred may point to the existence of modifying genes that determine the severity of the clinical phenotype (13).
Early explanations of the role of mutant ELANE portrayed a potential pathophysiological role of dysregulated vesicular sorting and membrane trafficking (14) because canine cyclic neutropenia resulted from mutations in the gene that encodes a subunit of the AP3 adapter complex, which is involved in trafficking of proteins out of the Golgi complex (14). A number of indirect observations have also implicated aberrant stress response in the ER. The unfolded protein response (UPR) has evolved to protect cells from the damaging effects of improperly folded proteins. Nascent proteins destined for secretory vesicles are directed to the ER, where the protein folding takes place (15). Myeloid cell lines and primary human cells engineered to express mutant NE, as well as primary human cells from SCN patients with ELANE mutations, show increased biochemical evidence of UPR/ER stress (16, 17).
However, controversy about the pathogenetic mechanisms of this disease has extended over 20 years because neither in vitro myelopoeisis/granulopoiesis models, nor mouse models recapitulate the disease. Two different models of mutant ELANE knockin mice showed no neutropenia basally or after chemotherapy-induced stress (18, 19). One of these mice only developed neutropenia after administration of a potent proteasome inhibitor but not after silencing the most relevant UPR sensor, Perk (19). Lack of adequate modeling is compounded by the limited availability of hematopoietic progenitor materials from pediatric patients with a rare marrow-failure disorder.
The recent discovery that somatic cells can be reprogrammed to generate induced pluripotent stem cell lines (iPSC lines), and so provide a renewable source of patient-derived cells to study the cellular mechanisms of disease, has rejuvenated the application of the Koch postulate to genetic diseases that cannot be recapitulated in animal models (20). In SCN, the use of iPSCs has provided evidence of canonical Wnt signaling in disease pathogenesis (21). However, a lack of targeted therapies against Wnt signaling has limited the clinical application of this knowledge in the therapy of ELANE-mutant SCN. Our group has recently demonstrated the utility of iPSC-derived myelopoiesis models to identify the pathogenetic mechanisms of unusual ELANE mutations that affect NE translation (22). Here, by characterizing iPSC-derived granulopoiesis from SCN patients with ELANE mutations and isogenic lines generated by gene repair, we resolve the necessity of the ELANE mutation in SCN dysgranulopoiesis. Moreover, our modeling reveals the molecular details underlying SCN disease pathogenesis, linking the concepts of NE mislocalization with the induction of UPR/ER stress. We show that while high-dose G-CSF therapy rescues SCN iPSC–derived promyelocytes through C/EBPβ-dependent emergency granulopoiesis, the application of low-dose G-CSF with a small-molecule NE-protease inhibitor restores normal intracellular NE localization in primary granules, ameliorates UPR/ER stress, facilitates promyelocyte survival, and restores CEBPA expression and granulocyte differentiation. Our results underscore a central role for NE mislocalization in SCN pathogenesis and provide proof-of-principle for therapeutic intervention exploiting NE protein relocalization.
Results
SCN patient iPSC–derived myeloid progenitors display impaired granulocytic differentiation.
In the present study, we developed iPSCs from the PB of 2 healthy subjects (control 12 and control 13), and 2 children with ELANE mutations (ELANEQ97P named SCN14 and ELANEI118N named SCN15). These 2 patients were chosen because of the severity of their congenital neutropenia at diagnosis, including maintained polymorphonuclear cell counts (PMN cell counts) < 500/mm3, and both showed BM maturation arrest at the myelocytic/metamyelocytic stage with no segmented neutrophils and relative marrow eosinophilia and monocytosis. In addition, both patients responded to only high doses of G-CSF (SCN14 derives from a patient requiring 8 μg/kg/day, and SCN15 derives from a patient requiring 5 μg/kg/day to maintain PMN counts > 1,000/mm3), which represents a frequent event in mutant-ELANE SCN.
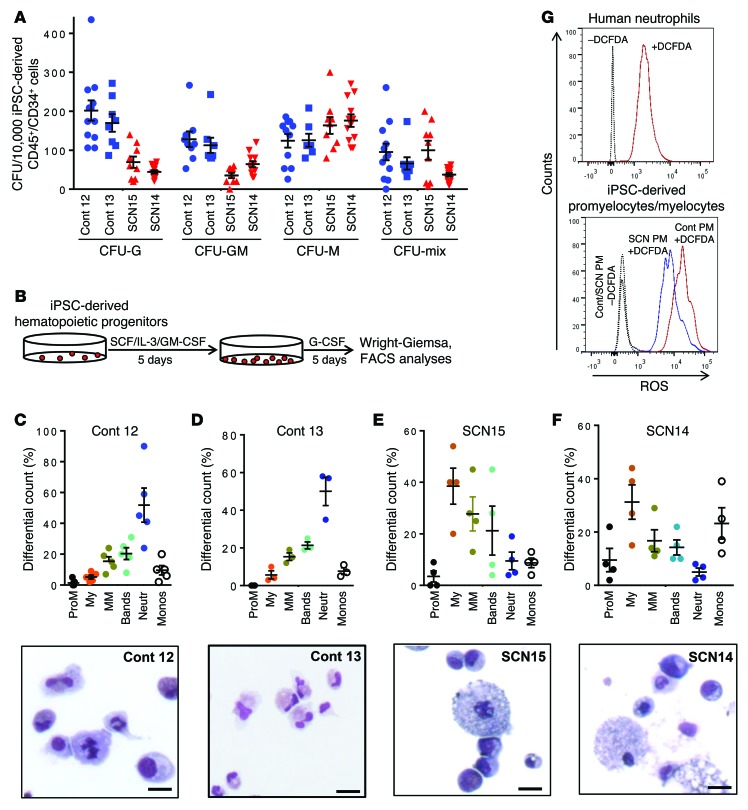
Upon transduction of PB mononuclear cells (MNCs) with Yamanaka factors in a lentiviral construct, pluripotent stem cell–like colonies appeared on the culture plate 10–15 days following transduction. The SCN iPSC lines retained their SCN point mutations after reprogramming (Supplemental Figure 1; supplemental material available online with this article; doi:10.1172/JCI80924DS1) and remained karyotypically normal throughout culture (data not shown). All iPSC lines expressed the pluripotent markers SSEA-4, Tra-1-60, Tra-1-81, CD9, and OCT-4 as analyzed by flow cytometry (Supplemental Figure 2). To investigate the ability of iPSC lines to differentiate into hematopoietic cells, we used a 10-day monolayer differentiation protocol to produce CD45+CD34+ hematopoietic progenitors (23). At the end of the monolayer differentiation protocol, the generated cells were analyzed for CD45 and CD34 expression by FACS analysis. All iPSC lines were able to differentiate into hematopoietic progenitor cells as assessed by the frequency of CD45+CD34+ cells at the end of 10 days of differentiation (control 12, 34.9% ± 11.21%; control 13, 23.4% ± 7.8%; SCN15, 17.5% ± 5.2%; SCN14, 38.8% ± 8.9%) (Supplemental Figure 3A). CD45+CD34+ cells from these cultures also expressed CD33 and CD43, indicating that these are myeloid progenitor cells derived from primitive hematopoiesis (Supplemental Figure 3, B). For the functional analyses of these myeloid progenitor cells derived from control iPSCs and ELANE-mutant SCN iPSCs, we carried out CFU assays using semisolid methyl cellulose medium containing cytokine mixtures for myeloid cells. As shown in Figure 1A, hematopoietic progenitor cells derived from the 2 ELANE-mutant SCN iPSC lines generated approximately 75% and 50% reduction in CFU-G (granulocyte) and CFU-GM (granulocyte-macrophage) colonies, respectively, in comparison to that of control iPSC–derived hematopoietic progenitor cells. However, both the SCN iPSC–derived hematopoietic progenitor cells increased modestly the generation of CFU-M (macrophagic) in SCN15. The frequency of immature CFU-Mix generated from these cultures was comparable among all the lines. The colonies were plucked, stained with Wright-Giemsa and analyzed for the presence of myeloid differentiated cells. Histologic analysis of CFU-G derived from control iPSC hematopoietic progenitors confirmed the colonies contained mature neutrophils. CFU-G colonies derived from ELANE-mutant SCN lines were smaller (controls: 835.7 μm ± 172.5 μm vs. ELANE mutant: 328.5 μm ± 121.9 μm; Supplemental Figure 4, A and B) and were mostly composed of immature myeloid cells, mainly promyelocytes and myelocytes, and a few bands and neutrophils. The CFU-GM colonies derived from the control iPSC hematopoietic progenitor cells contained mature neutrophils and macrophages. However, ELANE-mutant SCN iPSC–derived CFU-GM colonies lacked mature neutrophils and mainly contained macrophage and immature myeloid cells. The CFU-M colonies from both control and SCN iPSCs contained macrophages (Supplemental Figure 4, A and B).
Figure 1. Impaired granulopoietic differentiation of hematopoietic progenitors derived from SCN patient iPSCs.
(A) Colony forming cell assay of the hematopoietic progenitors derived from healthy donor (control 12 and control 13) and ELANE-mutant SCN iPSCs (SCN15 and SCN14). Cells were cultured in methyl cellulose semisolid medium with cytokines, and the myeloid (CFU-G, CFU-GM, CFU-M) and mix colonies were scored at day 14. (B) Schematic diagram of granulopoietic differentiation of iPSC-derived hematopoietic progenitors. (C) Top: Quantitative analyses of the granulopoietic differentiation of control iPSC–derived (control 12–derived) hematopoietic progenitors in liquid culture with myeloid differentiation cytokines. Bottom: Wright-Giemsa–stained cytospin at the end of the culture. (D) Top: Quantitative analyses of the granulopoietic differentiation of control iPSC–derived (control 13–derived) hematopoietic progenitors in liquid culture with myeloid differentiation cytokines. Bottom: Wright-Giemsa–stained cytospin at the end of the culture. (E) Top: Quantitative analyses of the granulopoietic differentiation of hematopoietic progenitors derived from SCN15 iPSCs in liquid culture condition with myeloid differentiation cytokines. Bottom: Wright-Giemsa–stained cytospin at the end of the culture. (F) Top: Quantitative analyses of the granulopoietic differentiation of hematopoietic progenitors derived from SCN14 iPSCs in liquid culture with myeloid differentiation cytokines. Bottom: Wright-Giemsa–stained cytospin at the end of the culture. (G) FACS-based ROS-generating-activity analyses of promyelocytes derived from control and SCN iPSCs. Scale bars: 10 μm. ProM, promyelocytes; My, myelocytes; MM, metamyelocytes; Neutr, neutrophils; Monos, monocytes. Data are presented as mean ± SD of individual values out of 4 or 5 independent experiments. Cont, control.
For the precise quantitation of granulocytic lineage populations, granulopoietic differentiation of myeloid progenitor cells derived from control iPSCs and ELANE-mutant SCN iPSCs was performed in a 2-step liquid culture (Figure 1B). At the end of culture, stages of differentiation were identified by morphological analysis. As shown in Figure 1, C and D, the control iPSC–derived myeloid progenitors differentiated into monocytes and increasing frequencies of differentiated neutrophils and bands. Comparative transcriptome analysis of iPSCs; iPSC-differentiated mesoderm (24); myeloid progenitors (day 10 CD45+CD34+CD33+ cells); and precursors (day 15 CD45+CD34–CD33+CD11b–CD15+/low cells) demonstrated that the myeloid progenitors and precursors were distinctly different from undifferentiated iPSCs and mesoderm-committed iPSCs. The expression of a total of 1,028 genes representing the iPSC signature is downregulated at a similar level in iPSC-derived myeloid progenitors and precursors as their primary BM myeloid progenitor and precursor counterparts. The expression of 828 hematopoietic genes was found to be upregulated in BM- or iPSC-derived myeloid progenitors and precursors. Pathway analysis using gene ontology analysis demonstrated that most of the upregulated genes were implicated in innate immunity and granulocyte physiology functions (clusters 6 and 7 of Supplemental Figure 5), and there were no major differences in the transcriptomes of primary BM myeloid progenitors or precursors and their iPSC-derived counterparts (Supplemental Figure 5). These data suggest that the expression of iPSC-derived and endogenous BM–derived myelopoiesis follow similar patterns.
Interestingly, the granulocytic population generated from ELANE-mutant SCN iPSC–derived myeloid progenitors is shifted to the left with myelocytes as the most frequently represented cell population and a largely reduced level of bands and neutrophils. These data suggest that ELANE-mutant SCN iPSC–derived myeloid progenitor cells are arrested at the myelocyte stage of granulopoietic development (Figure 1, E–F). These results mimic the precursor arrest at the myelocyte/metamyelocyte level of the BM at diagnosis from the patients from which iPSC lines were derived (data not shown). The FACS analyses of granulopoietic differentiation of control and SCN iPSC–derived cells show increased accumulation of promyelocytes/myelocytes (CD45+CD34–CD33+CD11b–CD15+/low) in SCN iPSC–derived cells (Supplemental Figure 6). There was an increased percentage of monocytes observed in the case of the SCN14-derived myelopoietic cultures (Figure 1F). Monocytopoiesis in these cultures correlates with absolute monocytosis (25) in primary cultures of BM progenitors (26) in SCN patients. The generation of reactive oxygen species (ROS) is a major functional hallmark of granulocytic lineage. Therefore, we analyzed the ROS production ability of granulocytic differentiated cells derived from control and SCN iPSC myeloid progenitors. Since the SCN iPSC–derived myeloid progenitor cells are arrested at the myelocyte stage, we analyzed the ability of promyelocytes/myelocytes derived from control or SCN iPSC myeloid progenitors to produce ROS following staining with dichlorofluorescin diacetate (DCFDA) and analysis by FACS. As shown in Figure 1G, the ROS-generating ability of SCN iPSC–derived promyelocytes/myelocytes is reduced compared with that of control iPSC–derived cells. All these results suggest that the ELANE SCN iPSC–derived hematopoietic progenitor cells can be used as a bona fide resource to study the pathophysiology of severe congenic neutropenia disease at molecular and cellular levels.
ELANE-mutant SCN iPSC–derived myeloid precursor cells undergo apoptosis that can be rescued by forced expression of active Akt or Bcl2.
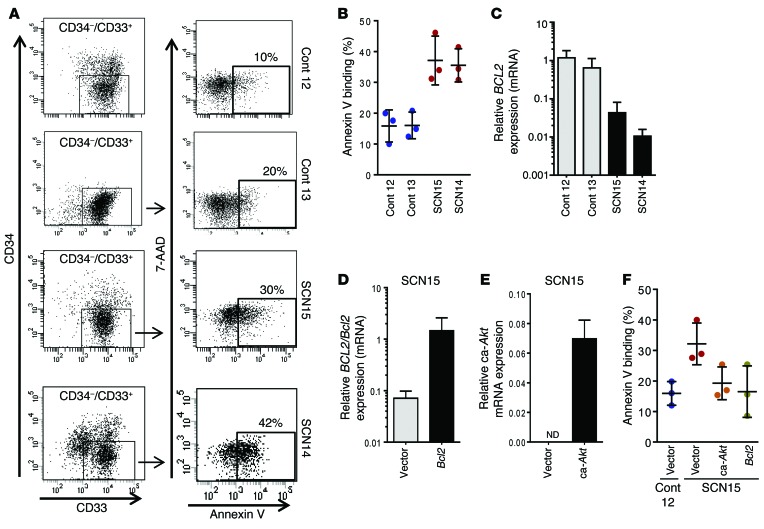
We investigated the underlying mechanisms of the impaired neutrophil development of ELANE-mutant SCN iPSC–derived myeloid progenitor cells. Survival of CD45+CD34+CD33+ myeloid progenitors and CD45+CD34–CD33+ precursors was assessed by annexin V binding assays and analyzed by FACS. Although the overall basal level of apoptosis is high in iPSC-derived myeloid progenitors for both control and SCN cells, the cells derived from ELANE-mutant SCN iPSCs show approximately 2-fold increased apoptosis in comparison to the control iPSC–derived myeloid progenitor cells (Supplemental Figure 7, A and B), suggesting enhanced apoptosis of ELANE-mutant SCN iPSC–derived myeloid progenitors. In agreement, the SCN iPSC–derived myeloid progenitors show approximately 2-fold reduced growth in myeloid expansion culture conditions (Supplemental Figure 7C).
We next analyzed the levels of apoptosis of myeloid precursor cells (CD45+CD34–CD33+) differentiated from the iPSC-derived myeloid progenitors. After 5 days of culture in stem cell factor (SCF), IL-3, and GM-CSF (as outlined in Figure 1B), the myeloid progenitor cells lost CD34 expression and differentiated to myeloid precursors, mainly promyelocytes/myelocytes or monocyte precursors. We analyzed the apoptosis in these immature myeloid cells, as SCN iPSC–derived myeloid progenitor cells are arrested at this stage in their granulopoietic development. Similar to our findings in myeloid progenitors, the level of cell death is doubled in the ELANE-mutant SCN iPSC–derived myeloid precursor cells, indicating enhanced apoptosis compared with the control iPSC–derived myeloid precursor cells (Figure 2, A and B). We investigated the expression level of the BCL2 family of prosurvival and proapoptotic proteins in the promyelocytes derived from the control and ELANE-mutant SCN iPSCs. As shown in Figure 2C, the levels of BCL2 mRNA are over 95% reduced in both the ELANE-mutant SCN iPSC–derived promyelocytes in comparison to control iPSC–derived cells. We also analyzed the expression level of BH3 domain only–containing BCL2 family proapoptotic proteins in these cells. The increased expression of BAX, BAD, and BIM, and the presence of cleaved forms of BIM, support the role of BH3-only proapoptotic molecules in the increased apoptosis in ELANE-mutant SCN iPSC–derived promyelocytes/myelocytes (Supplemental Figure 8A). To further examine the role of BCL2 in the survival of ELANE-mutant SCN iPSC–derived promyelocytes, we transduced the myeloid progenitor cells with a bicistronic retroviral construct expressing both murine Bcl2 and Thy1.1 through internal ribosomal entry site. Murine Bcl2 is highly homologous to human BCL2 and has been shown to retain the structural characteristics necessary to interface with pathways involved in the regulation of programmed cell death (27) while allowing mRNA quantitation of both endogenous and exogenous mRNA. The forced expression of Bcl2 increased approximately 2.5-fold the survival of granulocyte precursors, in comparison with vector-transduced cells, to levels similar to those found in control, mock-transduced precursor cells (Figure 2, D and F, and Supplemental Figure 8B). These data indicate that BCL2 plays a critical role in the apoptosis of ELANE-mutant SCN iPSC–derived myeloid precursor cells. Exogenous upregulation of the upstream BCL2 effector AKT was also explored through expression of constitutively active murine Akt (ca-Akt) in ELANE-mutant SCN iPSC–derived myeloid progenitors (Figure 2E). ELANE-mutant SCN iPSC–derived granulocyte precursors expressing ca-Akt had an approximately 2-fold increased survival (Figure 2F and Supplemental Figure 8B) and phenocopied the effect of Bcl2 forced expression. These data indicate that, in addition to the differentiation arrest, the deficient survival of precursors is at the basis of the poor cell output of ELANE-mutant granulopoiesis.
Figure 2. Defective differentiation of promyelocytes derived from SCN iPSCs is associated with apoptosis, which can be reversed by forced activation of Akt/Bcl2.
(A) FACS dot plot showing annexin V binding on myeloid precursors derived from control (control 12 and control 13), ELANE-mutant SCN iPSCs (SCN15 and SCN14). (B) Quantitation of the % annexin V binding. (C) Relative expression of BCL2 in promyelocytes derived from control and SCN iPSCs. (D) Ectopic expression of murine Bcl2 through retroviral transduction of myeloid precursors derived from SCN iPSCs (SCN15). (E) Ectopic expression of ca-Akt through retroviral transduction of myeloid precursors derived from SCN iPSCs (SCN15). (F) Quantitation of annexin V binding in control-, ca-Akt–, or Bcl2-expressing granulocytic precursors derived from SCN iPSCs. For B and F, data are presented as mean ± SD of 3 independent experiments. For C–E, data are presented as mean ± SD of 2 or more independent experiments in duplicate. Cont, control.
Correction of ELANE mutations by CRISPR/Cas9 gene editing corrects differentiation arrest and survival of granulocytic precursors.
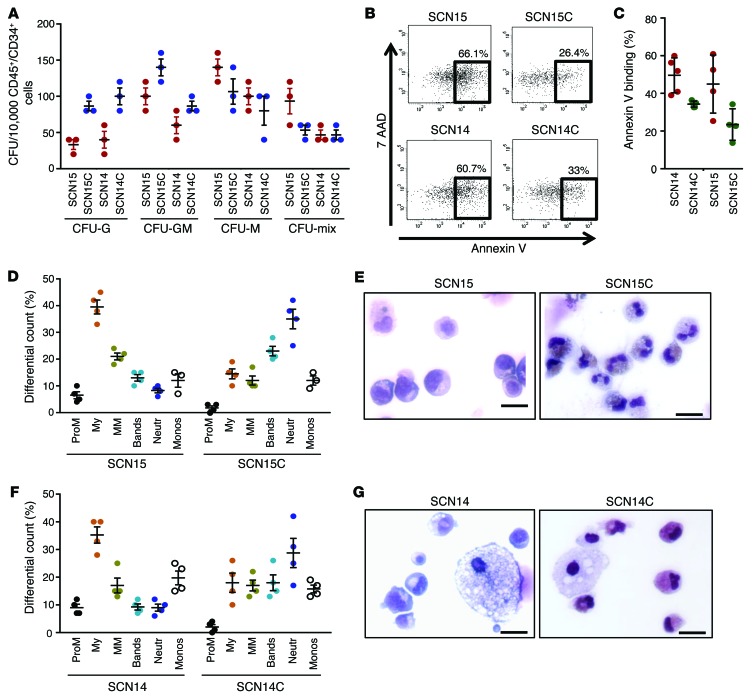
To identify whether ELANE mutations were necessary to mediate SCN, we proceeded to repair SCN14 and SCN15 by genome editing. Two guide RNA sequences (RNA-seq) were selected to target the genome around ELANE exon 3 using clustered regularly interspaced short palindromic repeats (CRISPR) design tools (28). The 20-bp guide RNA sequences were cloned into the px330 chimeric SpCas9 and guide RNA vector. A targeting donor plasmid was constructed from a WT ELANE sequence that contained 800 bp 5′ and 1,500 bp 3′ homology arms from the CRISPR cut sites. For positive selection of donor integrants, a floxed cassette containing an EFS promoter driving EGFP was inserted between the homology arms (Supplemental Figure 9, A–C). The SCN iPSCs were transfected with the CRISPRs and donor plasmid; they were expanded, and GFP+ cells were flow-sorted. GFP+ colonies were then picked and expanded, and their mRNA was converted into cDNA and sequenced (Supplemental Figure 9, D and E) for verification of sequence of corrected ELANE expression. Quantification of overall mRNA expression of ELANE in promyelocytes derived from corrected isogenic lines showed similar levels of expression (Supplemental Figure 9F). The isogenic lines expressing corrected ELANE were identified as SCN14C and SCN15C — counterparts of SCN14 and SCN15, respectively.
To determine if gene correction of ELANE mutations restores graulopoietic differentiation, CRISPR-corrected SCN iPSCs were differentiated into CD45+CD34+ hematopoietic progenitors and then to terminally differentiated neutrophils using the methods described above. Both SCN14C and SCN15C generated similar levels of CD45+/CD34+ cells as their uncorrected parental counterparts, respectively (data not shown). When plated into semisolid methyl cellulose medium, CD45+CD34+ progenitors derived from CRISPR-corrected SCN iPSCs showed an increase of CFU-G over their corresponding noncorrected iPSC line (Figure 3A). Similarly, corrected expression of NE results in improved survival (Figure 3, B and C) and granulocytic differentiation (Figure 3, D–G). These data resolve the critical role for these ELANE point mutations in the development of SCN-like disease.
Figure 3. Cellular consequences of ELANE mutation: correction of ELANE mutation rescues granulocytic differentiation.
(A) Colony forming cell assay of the hematopoietic progenitors derived from ELANE-mutant SCN iPSCs (SCN15, SCN14) and the derivative ELANE mutation–corrected iPSCs (SCN15C, SCN14C). (B) Representative example of annexin V binding analysis in ELANE mutant– and CRISPR-corrected iPSC–derived granulocytic precursors. (C) Quantitation of the % annexin V binding. (D) Quantitation of granulopoietic differentiation of myeloid progenitors derived from SCN iPSCs (SCN15) and the corrected iPSCs (SCN15C). (E) Wright-Giemsa–stained cytospin at the end of the culture (SCN15, SCN15C). (F) Quantitation of granulopoietic differentiation of myeloid progenitors derived from SCN iPSCs (SCN14) and the corrected iPSCs (SCN14C). (G) Wright-Giemsa–stained cytospin at the end of the culture (SCN14, SCN14C). Data are presented as mean ± SD of 3 or 4 independent experiments.
ELANE-mutant SCN iPSC–derived myeloid precursor cells contain mislocalized NE.
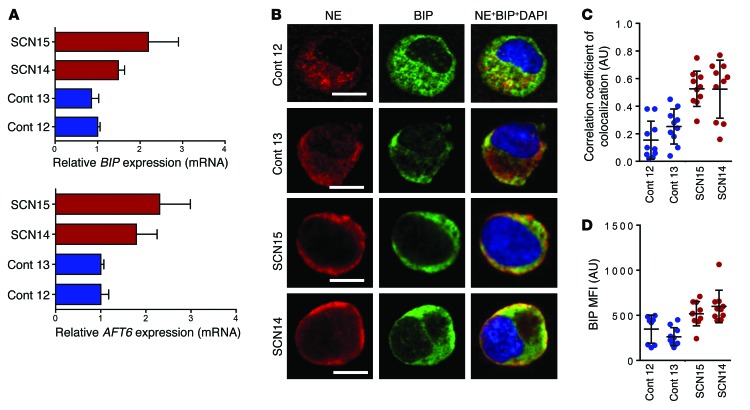
We interrogated our iPSC models to understand the possible pathobiological mechanisms of disease and to identify targets for intervention. Two hypotheses on how ELANE mutations affect NE have been proposed. In one, mutant NE is mistrafficked, while, in the other, mutant NE misfolds, resulting in activation of the UPR in the ER (16, 17, 29). We have previously demonstrated that alternative start codons in ELANE result in the translation of multiple peptides, which are susceptible to overwhelm the UPR system and induce ER stress, and in reduced clonogenic capacity of myeloid lineage cells (22). We analyzed the expression level of UPR pathway genes, and similar to the reports in primary cells, we found an approximately 2-fold increase of mRNA levels of the UPR pathway genes BIP and ATF6 in the ELANE-mutant SCN iPSC–derived promyelocytes, compared with control iPSC–derived cells (Figure 4A), which were not corrected by downstream prosurvival signals from forced expression of Bcl2 and ca-Akt (Supplemental Figure 10, A and B). This effect happened despite similar levels of expression of NE at mRNA and protein levels, and similar NE proteolytic activity in control and ELANE-mutant SCN iPSC–derived promyelocytes (Supplemental Figure 10, C–E). Analysis of the cellular distribution in the subcellular compartments of promyelocytes/myelocytes unveiled a punctate and granular distribution of NE in control iPSC–derived promyelocytes/myelocytes. In contrast, the NE was predominantly localized beneath the plasma membrane in ELANE-mutant SCN iPSC–derived promyelocytes/myelocytes (Figure 4B). NE was colocalized with BIP in both control and ELANE-mutant SCN iPSC–derived promyelocytes, but there was an approximately 2.5-fold increase in the colocalization coefficient between NE and BIP in ELANE-mutant SCN iPSC–derived promyelocytes (Figure 4C), suggestive of improper trafficking of the protein from the ER. The elevated level of BIP mean fluorescence intensity (MFI) (Figure 4D) and the colocalization with NE may support a role of ER stress–induced UPR. These data suggest that the 2 aforementioned mechanisms of neutropenia may represent a common phenomenon contributing to SCN pathobiology: the mislocalization of mutant NE, which induces UPR/ER stress.
Figure 4. ER stress and NE mislocalization are hallmarks of ELANE-mutant iPSC–derived granulocytic precursors.
(A) qPCR analyses of the mRNA for UPR pathway genes BIP and ATF6 in RNA from control and SCN iPSC–derived promyelocytes. (B) Confocal microscopic analyses of the localization of NE and ER resident chaperone BIP in control and SCN iPSC–derived promyelocytes. Scale bars: 10 μm. (C) Quantitation of the correlation of colocalization coefficient between NE and BIP. (D) MFI of BIP measured from confocal microscopic images. In A, individual values of 2 or more independent experiments in duplicate are plotted as mean ± SD. In C and D, data from more than 10 cells from 2 independent experiments are presented as mean ± SD. Cont, control.
High dose of G-CSF or lower G-CSF combined with NE inhibition rescues neutrophil maturation.
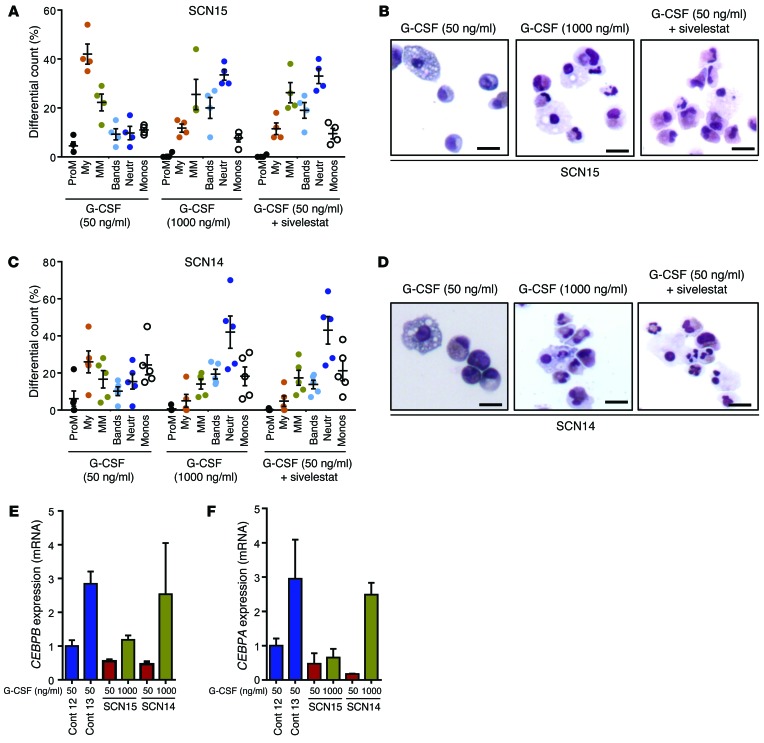
In most cases, mutant-ELANE SCN patients show a partial response to extremely high doses of G-CSF with an increase in neutrophil count in the PB and reduction of the frequency and severity of bacterial and fungal infections (4). However, frequent administration of high doses of G-CSF does not correct abnormal neutrophil function (3) and has been argued to lead to the development of myelodysplastic syndrome/acute myelogenous leukemia in more than 25% of the treated patients (4, 30). To examine the effect of high doses of G-CSF in the granulocytic maturation of ELANE-mutant SCN iPSC–derived myeloid progenitors, we added G-CSF to the culture at 1,000 ng/ml during the last 5 days of granulocytic differentiation (as depicted in Figure 1B). As shown in Figure 5, A and C, the granulocytic maturation of myeloid progenitors derived from both ELANE-mutant SCN iPSC lines cultured in the presence of a high concentration of G-CSF was rescued with an increase in the neutrophil output and a concomitant decrease in the promyelocytic and myelocytic populations, compared with the granulopoiesis in lower concentrations (50 ng/ml) of G-CSF. Cytospin analysis (shown in Figure 5, B and D) shows mature granulocytes from cultures containing 1,000 ng/ml G-CSF and only shows immature granulocytic cells when myeloid progenitors are cultured in 50 ng/ml G-CSF. The ELANE-mutant SCN iPSC–derived promyelocytes expressed similar levels of NE, retained protease activity, and accumulated in the ER lumen and other cytosolic regions outside the primary granules. We hypothesize that the inhibition of NE activity may ameliorate cell damage associated with mislocalized NE in ELANE-mutant SCN iPSC–derived myeloid precursor cells. Sivelestat (also called ONO-5046) is an NE-specific cell-permeant serine protease inhibitor used in the treatment of acute lung injury. We added sivelestat with G-CSF during the final 5 days of granulopoietic differentiation culture. Differential counts revealed that neutrophil differentiation of ELANE-mutant SCN iPSC–derived myeloid precursor cells was markedly increased and the frequency of immature myeloid cells (promyelocytes and myelocytes) was drastically decreased in the presence of 50 ng/ml G-CSF in combination with sivelestat (Figure 5, A–D). Further, the SCN iPSC–derived cultures with sivelestat also contained mature neutrophils and monocytes, indicating that the block in neutrophil differentiation was eliminated in the presence of sivelestat.
Figure 5. Cytokine rescue of granulopoietic differentiation in myeloid precursors derived from SCN iPSCs.
(A) Quantitation of granulopoietic differentiation of myeloid progenitors derived from SCN iPSCs (SCN15) in liquid culture containing G-CSF at 50 ng/ml, 1,000 ng/ml, and 50 ng/ml, along with 230 nM sivelestat (a small-molecule inhibitor of NE). (B) Wright-Giemsa–stained cytospin at the end of the culture in A. (C) Quantitation of granulopoietic differentiation of myeloid progenitors derived from SCN iPSCs (SCN14) in liquid culture containing G-CSF at 50 ng/ml, 1,000 ng/ml, and 50 ng/ml, along with 230 nM sivelestat. (D) Wright-Giemsa–stained cytospin at the end of the culture in C. (E and F) qPCR analyses of the mRNA for CEBPB (E) and CEBPA (F) in promyelocytes derived from control or ELANE-mutant SCN iPSCs. In A and C, data from 4 independent experiments are plotted as mean ± SD. In E and F, individual data from 2 or more independent experiments with duplicates are plotted as mean ± SD. Scale bars: 10 μm. Cont, control.
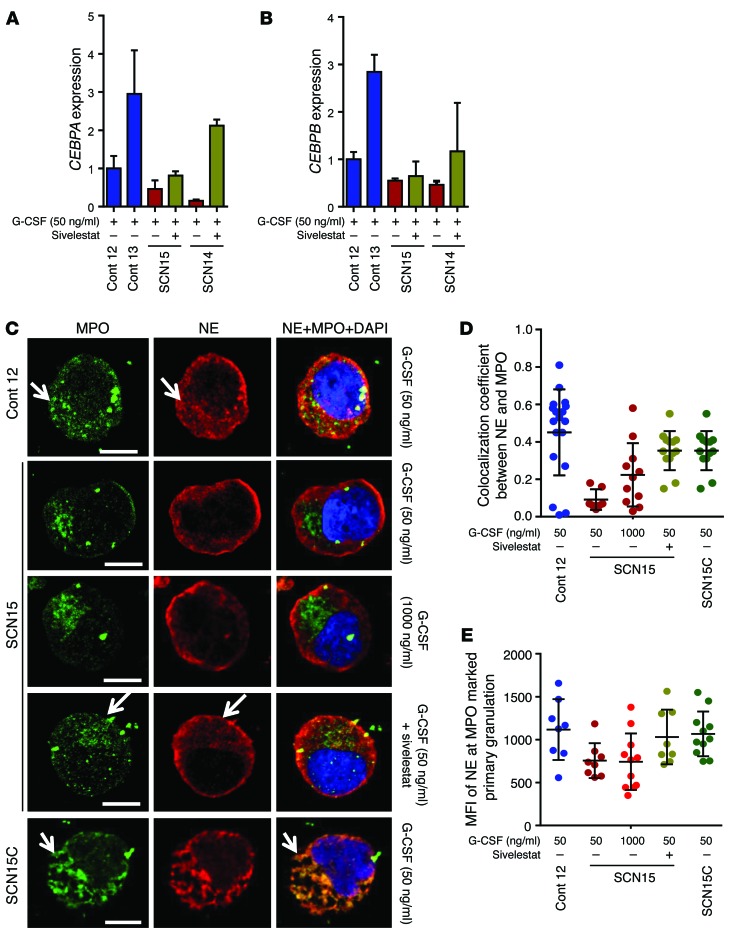
CCAAT/enhancer-binding protein alpha (C/EBPα) and its isoform C/EBPβ are crucial regulators of basal and emergent granulopoiesis, respectively (31, 32). We analyzed the expression levels of these 2 transcription factors to unravel the cellular mechanism behind sivelestat-mediated rescue and to determine whether sivelestat and high-dose G-CSF act through common or different mechanisms. Previous reports suggested that mutations in ELANE result in significant reduction in the expression levels of both CEBPA and CEBPB (33), as well as LEF1, a putative transcriptional regulator of CEBPA downstream of Wnt/β-catenin signaling (11, 34). Our data from SCN iPSC–derived granulopoiesis confirmed that the expression of these factors is low (Figure 5, E and F, and Supplemental Figure 11), but we note that the expression of the CEBPA and CEBPB was corrected in genome-edited iPSC–derived granulocytic precursors (Supplemental Figure 12, A and B). The addition of a high dose of G-CSF to ELANE-mutant iPSC–derived myelopoiesis results in the consistent upregulation of C/EBPβ, along with upregulation of C/EBPα expression, to control levels in granulocytic precursors generated from the SCN14 line but not from the SCN15 line (Figure 5, E and F). Interestingly, the addition of sivelestat to granulopoietic cultures restored expression of CEBPA, which regulates basal granulopoiesis, but did not induce CEBPB expression, which regulates emergency granulopoiesis (Figure 6, A and B). Neither treatment modified the levels of CEBPA or CEBPB in normal iPSC–derived promyelocytes (Supplemental Figure 12, C and D). These results suggest that this pharmacological approach may distinctly rescue basal granulopoiesis in the context of mutant ELANE.
Figure 6. Sivelestat rescue of SCN iPSC neutrophil maturation is associated with correction of CEBPA, but not CEBPB, expression and NE mislocalization.
(A and B) qPCR assay in sorted promyelocytes/myelocytes. (A) Relative expression of CEBPA. (B) Relative expression of CEBPB. (C) Confocal microscopic localization of primary neutrophil granule proteins NE and MPO in control and SCN iPSCs, and exon 3 mutation–corrected iPSC–derived (SCN15C-derived) promyelocytes. Arrow indicates the colocalization of NE and MPO in primary granules. Scale bars, 10 μm. (D) Quantitation of the correlation of colocalization coefficient between NE and MPO in the primary granules in C. (E) MFI of NE at MPO marked primary granules. In A and B, individual data from 2 or more independent experiments in duplicate are plotted as mean ± SD. In D and E, values of 8 or more cells from 2 independent experiments are presented as mean ± SD. Cont, control.
To further delineate the mechanism behind the amelioration of ER stress and the upregulation of prosurvival gene expression, we investigated the cellular localization of NE in the presence of sivelestat. At the promyelocyte stage, NE traffics from the ER lumen and is primarily distributed in the primary granules of promyelocytes and more differentiated granulocytic precursors and neutrophils (35, 36). Normally, NE is synthesized as a transient proform that becomes catalytically active by removal of an N-terminal propeptide after granule targeting (37). Mislocalization of activated mutant NE has been argued to be pathogenetically implicated in SCN in vitro and in vivo (14, 17, 38). To determine whether sivelestat may also modify the mislocalization of NE, we analyzed the colocalization of NE with myeloperoxidase (MPO), a bona fide primary granule marker, in the promyelocytes/myelocytes derived from control and ELANE-mutant SCN iPSCs. NE and MPO colocalized in punctate and granular patterns (Figure 6, C–E, and Supplemental Figure 13, A–C) — coinciding in form and shape with the pattern of expression of primary granules — in control iPSC–derived promyelocytes/myelocytes (39). However, NE and MPO in ELANE-mutant iPSC–derived promyelocytes/myelocytes presented a completely different pattern, with abundant NE located outside the primary granules, in subcortical regions without colocalization with MPO. Mislocalization of NE was not modified by the concentration of G-CSF, as no difference was found between promyelocytes/myelocytes from cultures containing 50 ng/ml or 1,000 ng/ml G-CSF (Figure 6, C–E). Interestingly, the mislocalization of NE is reversed by the addition of sivelestat at levels similar to those reached by the genetic correction of ELANE expression to cultures containing 50 ng/ml of G-CSF (Figure 6, C–E, and Supplemental Figure 13, A–C). The addition of sivelestat induced an approximately 3-fold increase in the correlation coefficients of colocalization between NE and MPO in the primary granules of ELANE-mutant–derived promyelocytes at levels similar to those reached by the genetic correction of ELANE expression (Figure 6, C–E, and Supplemental Figure 13, A–C). Furthermore, the addition of sivelestat markedly reduced the mRNA expression level of ER stress–induced UPR pathway genes BIP and ATF6, and of BIP protein expression in promyelocytes/myelocytes derived from ELANE-mutant iPSCs (Supplemental Figure 13, D–F) at levels similar to those reached by the genetic correction of ELANE expression (Supplemental Figure 13, G–H). At the same time, sivelestat also increased the survival and, modestly, the cell expansion of SCN iPSC–derived postmitotic granulocytic precursors (Supplemental Figure 14, A–C) and the expression level of the prosurvival gene BCL2 in the ELANE-mutant iPSC–derived promyelocytes (Supplemental Figure 14D) at levels similar to those reached by the genetic correction of ELANE expression (Supplemental Figure 14E), indicating that sivelestat ameliorates the ER stress and enhances cellular survival. These results indicate that sivelestat corrects the mislocalization of NE, ameliorates ER stress, reduces the UPR, and upregulates the expression level of CEPBA and BCL2 and the survival of ELANE mutant granulocyte precursors.
Discussion
Animal models and in vitro cultures consisting of cells derived from patients are often used to investigate disease pathophysiology and to develop therapies. Unfortunately, mutant-ELANE knockin mice fail to reproduce the abnormal granulopoiesis, as observed in SCN patients (18, 19). Moreover, BM cells are not an ideal experimental tool, since it is difficult to obtain sufficient nucleated cells due to the invasiveness of the aspiration procedure in pediatric patients with rare BM-failure syndromes. Moreover, the analysis of pathobiological mechanisms occurring during different stages of granulopoietic differentiation is difficult to address in primary patient samples where most of myelopoiesis is already arrested in the promyelocyte stage.
iPSCs have been shown to be an excellent tool to analyze pathogenetic mechanisms and for drug discovery (40–42). The use of iPSCs to address mechanisms of disease in SCN has been documented by our group and other groups (21, 22, 43). In this report, we first demonstrate that SCN-associated single point mutations in one allele of ELANE are necessary to induce granulopoietic differentiation arrest and apoptosis of granulocytic progenitors and precursors. This model confirms some of the current concepts of pathogenesis induced by heterozygote mutations of ELANE. First, the correction of ELANE mutations corrects granulopoietic defects and progenitor differentiation biases in iPSC-derived myelopoiesis, indicating that the mutations in one allele of ELANE are crucial contributors to the development of the key pathobiological features of this disease. Of course, other modifier genes may ameliorate or worsen the pathobiological features of mutant-ELANE SCN. Second, the deficient survival of granulocytic precursors derived from mutant-ELANE iPSCs depends on functional G-CSF signaling and can be rescued by restoration of deficient downstream Akt/Bcl2 signaling, a common feature in apoptosis induced by ER stress (44, 45). This finding provides crucial information on the mechanisms controlling survival of diseased granulocytes. Notably, overexpression of activated Akt and downstream Bcl2 cannot increase the lifespan of normal granulocytes (46), but it does in other pathological situations, like sepsis (47). Third, high doses of G-CSF result in the amelioration of dysgranulopoiesis, granulocyte precursor survival defects, as previously shown in iPSC-derived granulopoiesis (21), and of CEBPB expression levels, as shown in primary myelopoiesis from patients (11, 48). Notably, our data cannot rule out the existence of genes that modify the clinical and biological features of ELANE mutations that would affect the severity of the disease; for instance, such mutations could induce SCN in some patients and cyclic neutropenia in others (6).
NE is a glycoprotein with a single peptide chain of 218 amino acid residues and 4 disulfide bridges. Mutant NE–expressing granulocytic precursors show aberrant expression of NE outside primary granulation, with accumulation in the ER and subcortical intracellular spaces. NE mistrafficking correlates with the accumulation of NE in the ER, which induces stress that can be corrected by the addition of the NE inhibitor sivelestat to cultures of iPSC-derived myelopoiesis. Sivelestat activity is facilitated by a catalytic triad conserved in all serine proteases that consists of His 57, Asp102, and Ser195 residues forming a charge relay system. In NE, the S1 pocket containing the primary sivelestat binding domain is specifically modified, is rather hydrophobic, and has a hemispheric shape, causing the sivelestat to dock to NE with a higher affinity than most other serine proteases (49). Neither of the 2 mutations analyzed in this study affect the catalytic site, and therefore, the NE activity is unaffected and similar to controls (as shown in Supplemental Figure 10). Changes in the S1 pocket structure, as induced by sivelestat (49), may result in increased proton relaxation of residues that are crucial for tertiary structure folding of NE and alternatively translated proteins (22). Our data demonstrate that both mutations of ELANE result in intracellular mistrafficking of NE and/or alternatively translated proteins, which conserve the C-terminus region recognized by the anti-NE antibody used in our microscopy and immunoblot experiments. In our studies, sivelestat correction of impaired intracellular traffic of NE in ELANE-mutant granulocytic precursors restored CEBPA expression, granulopoietic differentiation, and granulocytic survival at levels similar to those reached by the isogenic correction of the ELANE mutation and, unlike high-dose G-CSF, failed to use the CEBPB expression–associated pathway of emergent granulopoiesis. These data suggest that, in addition to its inhibitory activity, the binding of sivelestat to NE induces conformational changes that prevent its mislocalization and ameliorate ER stress, resulting in restoration of granulocytic precursor survival, granulocyte differentiation, and CEBPA expression. While the precise mechanism of sivelestat remains unknown, it can be postulated that the high-binding affinity of sivelestat for NE (reviewed in ref. 50) may result in acting as a pharmacological chaperone with activity on protein folding and proteostasis (51, 52). In any event, the cellular mechanism used by sivelestat is different from the one employed by high-dose G-CSF. Namely, while high-dose G-CSF induces emergency granulopoiesis through upregulation of C/EBPβ, sivelestat acts directly through the normal granulocytic differentiation pathway, as evidenced by normalization of C/EBPα expression levels. Thus, we propose that SCN disease pathogenesis pivots upon NE mislocalization, which triggers UPR/ER stress and dysfunctional differentiation and survival signaling. Our work provides a paradigm that can be clinically exploited to achieve therapeutic responses by using lower doses of G-CSF in combination with small-molecule targeting to correct NE mislocalization, easing the management of these patients and possibly impacting the probability of these patients to develop clonal evolution and malignant disease.
Methods
Origin of cellular material used for analysis.
No clonal cell lines were generated. All the lines used were expected to be polyclonal or, at the very minimum, oligoclonal. One line was generated from each of 2 healthy donors (n = 2) and from each of 2 patients with heterozygote point mutations in exon 3 of ELANE (n = 2) for the experiments described in the study. One noncloned, isogenic CRISPR/Cas9-corrected line was produced per patient-derived parental iPSC line by bulk FACS sorting of EGFP+ cells where EGFP expression was used as a reporter of corrected ELANE exon 3 (Supplemental Figure 9C). A total of 6 lines were studied in this report. No specific selection of clones was used during the process.
Generation of iPSCs from healthy donor and ELANE-mutant SCN patient PB MNCs.
PB from healthy donors and SCN patients was obtained at Cincinnati Children’s Hospital Medical Center through informed consent under an approved institutional review board research protocol. iPSCs were generated from PB MNCs as described (53). Briefly, PB MNCs were purified from PB by Ficoll separation (GE Healthcare) and plated at 5 × 106 cells/ml in X-VIVO 10 (Lonza) supplemented with 10% FCS, SCF (100 ng/ml), TPO (100 ng/ml), IL-3 (100 ng/ml), IL-6 (20 ng/ml), FLT3L (100 ng/ml), and GM-CSF (10 ng/ml) (all cytokines from PeproTech) in a low-attachment plate (Corning) and cultured at 37°C in 5% CO2. After 2 days of culture, cells were transduced with a lentivirus containing OCT4, KLF4, MYC, and SOX2 (54) using polybrene at a MOI of 20–40. Two days after lentiviral transduction, cells were plated onto irradiated mouse embryonic fibroblasts (MEF, GlobalStem) in X-VIVO 10 supplemented with 10% FCS, SCF (100 ng/ml), TPO (100 ng/ml), IL-3 (10 ng/ml), IL-6 (20 ng/ml), and FLT3L (100 ng/ml). After 2 days of culture on MEF, the media was changed to hESC media (DMEM F/12, 20% KnockOut Serum Replacement (KSR, Invitrogen), 1% l-glutamine, 1% NEAA, 0.1 mM β-mercaptoethanol) supplemented with 10 ng/ml bFGF (PeproTech), and a small-molecule iPSC enhancement cocktail containing SB431542 (2 μM, Miltenyi Biotec), PD0325901 (0.5 μM, Cayman Chemical), and Thiazovivin (2 μM, Stemgent). After 10 days, the small molecules were removed and the medium was changed every other day. Once iPS-like colonies were observed, they were plucked and passaged onto fresh MEF and cultured with hESC media or onto Matrigel (BD Biosciences) and cultured in mTeSR1 medium (StemCell Technologies Inc.).
Hematopoietic differentiation of healthy donor– and SCN patient–derived iPSCs and FACS characterization.
Hematopoietic differentiation was carried out as described (23) with minor modifications. Briefly, 5 × 105 cells were plated on growth factor–reduced Matrigel (BD Biosciences) and cultured for 24 hours in hES media supplemented with 10 mM ROCK inhibitor Y-27632 (Millipore). After 24 hours, media was changed to RPMI supplemented with l-glutamine, ascorbic acid (50 μg/ml), monothioglycerol (100 μM), BMP4 (5 ng/ml), CHIR99021 (930 ng/ml), and VEGF (50 ng/ml). Twenty-four hours after media change, CHIR99021 was removed from the culture medium and bFGF was added at 20 ng/ml. On day 3, medium was changed to StemPro 34 (Invitrogen) containing BMP4 (5 ng/ml), bFGF (20 ng/ml), and VEGF (50 ng/ml). On days 4–5, cells were cultured in StemPro34 containing VEGF (15 ng/ml) and bFGF (5 ng/ml). On day 6, media was changed to serum-free SFD media with VEGF (50 ng/ml), Flt3L (5 ng/ml), SCF (50 ng/ml), and bFGF (50 ng/ml). For days 7–10, cells were cultured in SFD with VEGF (50 ng/ml), Flt3L (5 ng/ml), SCF (50 ng/ml), TPO (50 ng/ml), and IL-6 (10 ng/ml). Nonadherent cells were collected from the media on days 8–10; stained with the antibodies (all from BD Biosciences) for CD45-PacificBlue (catalog 560367, clone HI30), CD34-PECy7 (catalog 560710, clone 581), CD33-APC (catalog 551378, clone WM-53), and CD43-FITC (catalog 555475, clone 1G10); and FACS analyzed.
CFU assays.
Progenitor content and activity were evaluated in CFU assays using standard conditions. In brief, 2,500 nonadherent cells were mixed in 1 ml of methyl cellulose semisolid medium containing cytokine mixture (MethoCult H4034 Optimum, StemCell Technologies), and seeded into a 35-mm gridded disc. The disc containing cells in methyl cellulose semisolid medium was incubated at 37°C in a humidified 5% CO2 incubator. The hematopoietic colonies (CFU-G, CFU-M, CFU-GM, and CFU-Mix) generated were scored at day 14 under an inverted microscope. In order to quantify the number of hematopoietic progenitors at the time of plating, an aliquot of the nonadherent cells from control and SCN iPSCs on day 10 of hematopoietic differentiation was analyzed for CD45+ and CD34+ expression by flow cytometry. The number of colonies was normalized per 10,000 CD45+CD34+ by multiplying the percentage of CD45+/CD34+ cells in the initiating cell population derived from each sample by the number of each colony type.
Myeloid cell expansion and granulopoiesis of iPSC-derived hematopoietic progenitors.
iPSCs were differentiated into hematopoietic progenitors by 10 days of culture in myeloid expansion medium (IMDM + Ham’s F12 at 3:1 ratio) containing 0.5% N2 supplement, 1% B27 supplement without vitamin A, 0.5% human serum albumin, 100 μM monothioglycerol, 50 μg/ml ascorbic acid, 100 ng/ml recombinant SCF, 10 ng/ml IL-3, and 10 ng/ml GM-CSF. The cultures were further differentiated using granulopoietic culture conditions (IMDM + Ham’s F12 at 3:1 ratio) containing 0.5% N2 supplement, 1% B27 supplement without vitamin A, 0.5% human serum albumin, 100 μM monothioglycerol, 50 μg/ml ascorbic acid, and 50 ng/ml G-CSF (Neupogen filgrastim) for 5 days. At the granulopoietic differentiation stage, cells were cultured at low (50 ng/ml) or high (1,000 ng/ml) G-CSF doses. During myeloid expansion and granulopoietic differentiation, cells were cultured in presence or absence of Sivelestat (Sigma-Aldrich) at a concentration of 230 nM (~5 times the IC50 for NE) (55). At the end of granulopoietic differentiation, cells were cytospun onto a Superfrost Plus Microscope slide (Fisher Scientific). The cells were Wright-Giemsa stained and then scored for myeloid cell types (promyelocytes, myelocytes, metamyelocytes, bands, neutrophils, and monocytes) using an upright microscope (Motic BA310). For sorting of the promyelocytes, cells at the end of myeloid expansion were stained for CD45-Pacific Blue, CD34-PECy7, CD33-APC, CD11b-APCCy7 (catalog 557754, clone ICRF44, BD Biosciences), and CD15-FITC (catalog 562370, clone W6D3, BD Biosciences). The promyelocytes/myelocyte population (defined as CD45+/CD34–/CD33+/CD11b–/CD15dim) was selected by FACS.
Isolation and transcriptome analysis of myeloid populations isolated from BM- or iPSC-derived cultures.
Hematopoietic progenitors derived from control and SCN iPSCs were cultured in myeloid expansion medium containing 50 ng/ml SCF, 10 ng/ml IL-3, and 10 ng/ml GM-CSF for 5 days. The cells at this point were stained for CD45-Pacific blue, CD34-PECy7, CD33-APC, CD11b-APC-Cy7, and CD15-FITC. 7-AAD was used to eliminate the dead cells. The promyelocytic population (CD45+CD34–CD33+CD11b–CD15+/lo) was sorted, and the RNA from control and SCN iPSC promyelocytes was isolated using QIAGEN RNeasy Mini Kit. Transcriptional comparisons with healthy subject–derived iPSC lines (n = 7), mesoderm (first 7 days of the differentiation protocol, as published elsewhere, from 4 healthy subject–derived iPSC lines, n = 4; ref. 24), sorted BM and iPSC-derived myeloid progenitors (CD45+CD34+CD45RA+ or CD45+CD34+CD33+, respectively, n = 2–3 per group), BM promyelocytes (n = 2), and control iPSC–derived promyelocytes (n = 2) were performed. Total RNA was extracted from sorted populations derived from BM or iPSCs using RNeasy Mini Kit (QIAGEN) or TRIzol (Invitrogen) as required for the number of cells available. RNA quality and concentration were measured by Bioanalyzer 2100 using the RNA 6000 Nano Assay (Agilent Technologies). For RNA specimens obtained from fewer than 50,000 cells (sorted BM populations and iPSC-derived promyelocytes and progenitors), a linear amplification sequencing protocol was performed (Illumina). RNA-seq libraries were prepared using the Illumina TruSeq RNA preparation kit and sequenced on the Illumina HiSeq 2000 using single-end 20 million–bp reads. Reads were aligned with TopHat software, using hg19 as the reference genome and mapping reads per kilobase per million mapped reads (RPKM) as output. RPKM were log2-transformed and base-lined to the median expression of the average of each class of samples (iPSC-derived myeloid progenitors, BM progenitor/promyelocytes, iPSCs, and iPSCs differentiated to mesoderm). Differentially expressed genes were identified from a starting pool of all transcripts with more than 3 fragments per kilobase of exon per million fragments mapped (FPKM) in at least 2 of the 9 myeloid samples (12,859 gene-level summarized transcripts). For all these populations, the RNA samples were processed for RNA-seq analyses using RNA-seq protocol from NuGEN and Illumina. The amplified products were sequenced to analyze the gene expression profile. All original microarray data were deposited in the NCBI’s Gene Expression Omnibus (GEO GSE69622).
Retroviral transduction of hematopoietic progenitor cells.
Hematopoietic progenitors (CD45+ CD34+) derived from control or SCN iPSCs at day 10 of hematopoietic differentiation were collected and cultured in myeloid expansion medium containing 100 ng/ml recombinant SCF, 10 ng/ml IL-3, and 10 ng/ml GM-CSF for 12 hours, on Retronectin-coated (CH-296, Takara Bio Inc.) nontissue culture dishes. Recombinant retroviral vector supernatant carrying ca-Akt, murine Bcl2, or empty vector control were added for 8-hour transduction. At the end of transduction, cells were washed and cultured in myeloid expansion medium containing 100 ng/ml recombinant SCF, 10 ng/ml IL-3, and 10 ng/ml GM-CSF for 4 days. Myeloid precursor cells were analyzed for apoptosis using an annexin V staining kit according to the manufacturer’s instructions (BD Pharmingen). Early apoptosis was determined as annexin V–positive events on CD45+ hematopoietic cells after gating out large residual dead (7-AAD+) iPSCs. Cells expressing CD90.1 were gated and analyzed, as the retroviruses express CD90.1 through internal ribosomal entry site (IRES). Ninety-six hours after transduction, cells expressing CD90.1 were sorted and cultured in granulocyte differentiation medium containing 50 ng/ml G-CSF (Neupogen filgrastim) for 5 days for the cytospin analyses of myeloid differentiated cells.
Quantitative PCR.
Total RNA was extracted from the promyelocytes derived from control SCN iPSCs, or SCN mutation–corrected iPSCs using RNeasy minikit (QIAGEN) following the manufacturer’s instructions, and cDNA was prepared using TaqMan reverse transcription reagent (Roche Applied Science). The mRNA expression levels of Bcl2, BIP, ATF6, ca-Akt, CEBPA, and CEBPB were analyzed by quantitative PCR (qPCR) assay using TaqMan Universal PCR master mix and TaqMan primers for specific genes (Roche Applied Science). The expression level was normalized with the expression of internal control gene GAPDH.
Western blot analyses.
Hematopoietic progenitors derived from control or SCN iPSCs were cultured in myeloid expansion medium containing 100 ng/ml recombinant SCF, 10 ng/ml IL-3, and 10 ng/ml GM-CSF for 5 days. Promyelocytes from these cultures were sorted, and protein samples were prepared by lysing them in 1× RIPA buffer containing protease inhibitor cocktail and phosphatase inhibitors. The protein samples were dissolved in Laemmli buffer and electrophoresed through 4%–15% SDS-PAGE gradient gel followed by transfer to PVDF membrane. The membrane was blocked and treated with primary antibodies against NE Chicken IgY (22), and the rabbit polyclonal antibodies (Cell Signaling Technology) against BAX (catalog 2772), BAD (catalog 9292), BIM (catalog 2819), and anti-BIP (catalog ab21685; Abcam) or β-ACTIN (catalog A5441, clone AC-15, Sigma-Aldrich) followed by washing and subsequent treatment with secondary antibodies tagged with HRP (anti–mouse Ig [catalog 70765, Cell Signaling Technology], anti–rabbit Ig [catalog 70745, Cell Signaling Technology], and goat anti–chicken Ig [catalog ab97135, Abcam]). The blots were developed using a chemiluminescence coupled reaction.
Confocal immunofluorescence microscopy.
The sorted promyelocytes were cytospun onto Superfrost Plus microscope slides (Fisher Scientific), airdried and fixed in 4% paraformaldehyde for 30 minutes at 4°C, permeabilized with 0.1% Triton X-100 (catalog T9284, Sigma-Aldrich) for 10 minutes, and blocked with 5% normal goat serum for 30 minutes. The slides were stained with primary antibodies anti-NE and the antibodies (from Abcam) anti-MPO (catalog ab134132, clone EPR4793) or anti-BIP (catalog ab21685) at 4°C overnight. They were washed and then treated with secondary antibodies (from Invitrogen) goat anti–rabbit Alexa Fluor 488 (catalog A11034) or donkey anti–chicken IgY Alexa Fluor 568 (catalog A11041) at 1:500 v/v concentration for 1 hour at room temperature. Cells were washed and mounted using Gold Antifade mounting media (catalog P36935, Invitrogen) containing DAPI. The stained cells were analyzed by a LSM 710 confocal microscope system (Zeiss) equipped with an inverted microscope (Observer Z1, Zeiss) using a Plan Apochromat ×63 1.4 NA oil immersion lens, and images were processed using Adobe Photoshop v7.
Correction of ELANE exon 3 mutation by CRISPR/Cas9.
The human codon-optimized Cas9 plasmids were obtained from Addgene (42230). Two guide RNAs to target 5′ and 3′ of ELANE exon 3 were designed using the Zhang lab CRISPR design tool (http://crispr.mit.edu/). The guide RNAs (5′-GAGCCCATAACCTCTCGCGG-3′ and 5′-GAAACGGGAAAATACCCGCCA-3′) were then cloned into the Cas9 vector using a published protocol (56). The donor plasmid with 5′ 0.8 kb and 3′ 1.2 kb homology arms was synthesized by GenScript. ELANE CRISPR and donor plasmids were purified using the EndoFree Maxi Prep Kit (QIAGEN). For genome editing, iPSCs were cultured on 6-well Matrigel-coated dishes in mTeSR1 until confluence (StemCell Technologies Inc.). Cells were dissociated into single cells using accutase (Sigma-Aldrich) and nucleofected using the Amaxa Nucleofector (Lonza) and kit P3 according to manufacturer’s instructions with 1 μg CRISPR and 1 μg donor construct. After nucleofection, cells were plated on Matrigel-coated dishes in mTeSR1 media supplemented with 10 μM ROCK inhibitor Y-27632 (Millipore). Cells were allowed to expand for 2–3 weeks after cell sorting to recover. Cells were then dissociated into single cells using accutase, and the GFP+ (donor-expressing) cells were sorted by FACS. GFP+ iPSCs were then plated onto Matrigel-coated culture dishes in mTeSR media supplemented with 10 μM ROCK inhibitor Y-27632 for 24 hours. Once iPSC sub-lines were established, iPSCs were transitioned to MEF culture for hematopoietic differentiation.
Statistics.
Data are presented as individual data and mean ± SD.
Study approval.
This study has been performed following Institutional Review Board approval by the Cincinnati Children’s Hospital Medical Center, Cincinnati, Ohio, USA; a written informed consent was received from participants prior to inclusion in the study.
Supplementary Material
Acknowledgments
This study was supported by the NIH (1R01GM110628), as well as funds from Hoxworth Blood Center (to J.A. Cancelas and C. Lutzko) and Cincinnati Children’s Hospital Medical Center (to H.L. Grimes and C. Lutzko). We thank Robin Schroll, Stacy Bush, and Nathaniel Barasa for technical assistance. We thank Cincinnati Children’s Research Foundation Mouse, Flow Cytometry, Immunobiology and Cell Processing Cores for their services, supported by the Center of Excellence in Molecular Hematology (P30 DK090971) and the support by an NHLBI Progenitor Cell Consortium Administrative grant that supported the Cincinnati Cell Characterization Core, C4 (5U01 HL099997). We want to thank Margaret O’Leary for editorial assistance.
Footnotes
Conflict of interest: The authors have declared that no conflict of interest exists.
Reference information:J Clin Invest. 2015;125(8):3103–3116. doi:10.1172/JCI80924.
References
- 1.Berliner N, Horwitz M, Loughran TP., Jr Congenital and acquired neutropenia. Hematology Am Soc Hematol Educ Program. 2004;2004:63–79. doi: 10.1182/asheducation-2004.1.63. [DOI] [PubMed] [Google Scholar]
- 2.Horwitz MS, Corey SJ, Grimes HL, Tidwell T. ELANE mutations in cyclic and severe congenital neutropenia: genetics and pathophysiology. Hematol Oncol Clin North Am. 2013;27(1):19–41. doi: 10.1016/j.hoc.2012.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Donini M, et al. G-CSF treatment of severe congenital neutropenia reverses neutropenia but does not correct the underlying functional deficiency of the neutrophil in defending against microorganisms. Blood. 2007;109(11):4716–4723. doi: 10.1182/blood-2006-09-045427. [DOI] [PubMed] [Google Scholar]
- 4.Rosenberg PS, et al. The incidence of leukemia and mortality from sepsis in patients with severe congenital neutropenia receiving long-term G-CSF therapy. Blood. 2006;107(12):4628–4635. doi: 10.1182/blood-2005-11-4370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Carlsson G, et al. Hematopoietic stem cell transplantation in severe congenital neutropenia. Pediatr Blood Cancer. 2011;56(3):444–451. doi: 10.1002/pbc.22836. [DOI] [PubMed] [Google Scholar]
- 6.Dale DC, et al. Mutations in the gene encoding neutrophil elastase in congenital and cyclic neutropenia. Blood. 2000;96(7):2317–2322. [PubMed] [Google Scholar]
- 7.Horwitz M, Benson KF, Person RE, Aprikyan AG, Dale DC. Mutations in ELA2, encoding neutrophil elastase, define a 21-day biological clock in cyclic haematopoiesis. Nat Genet. 1999;23(4):433–436. doi: 10.1038/70544. [DOI] [PubMed] [Google Scholar]
- 8.Huntington JA, Read RJ, Carrell RW. Structure of a serpin-protease complex shows inhibition by deformation. Nature. 2000;407(6806):923–926. doi: 10.1038/35038119. [DOI] [PubMed] [Google Scholar]
- 9.Skokowa J, et al. Interactions among HCLS1, HAX1 and LEF-1 proteins are essential for G-CSF-triggered granulopoiesis. Nat Med. 2012;18(10):1550–1559. doi: 10.1038/nm.2958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Skokowa J, et al. NAMPT is essential for the G-CSF-induced myeloid differentiation via a NAD(+)-sirtuin-1-dependent pathway. Nat Med. 2009;15(2):151–158. doi: 10.1038/nm.1913. [DOI] [PubMed] [Google Scholar]
- 11.Skokowa J, et al. LEF-1 is crucial for neutrophil granulocytopoiesis and its expression is severely reduced in congenital neutropenia. Nat Med. 2006;12(10):1191–1197. doi: 10.1038/nm1474. [DOI] [PubMed] [Google Scholar]
- 12.Salipante SJ, et al. Contributions to neutropenia from PFAAP5 (N4BP2L2), a novel protein mediating transcriptional repressor cooperation between Gfi1 and neutrophil elastase. Mol Cell Biol. 2009;29(16):4394–4405. doi: 10.1128/MCB.00596-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Newburger PE, et al. Cyclic neutropenia and severe congenital neutropenia in patients with a shared ELANE mutation and paternal haplotype: evidence for phenotype determination by modifying genes. Pediatr Blood Cancer. 2010;55(2):314–317. doi: 10.1002/pbc.22537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Benson KF, et al. Mutations associated with neutropenia in dogs and humans disrupt intracellular transport of neutrophil elastase. Nat Genet. 2003;35(1):90–96. doi: 10.1038/ng1224. [DOI] [PubMed] [Google Scholar]
- 15.Ron D, Walter P. Signal integration in the endoplasmic reticulum unfolded protein response. Nat Rev Mol Cell Biol. 2007;8(7):519–529. doi: 10.1038/nrm2199. [DOI] [PubMed] [Google Scholar]
- 16.Grenda DS, et al. Mutations of the ELA2 gene found in patients with severe congenital neutropenia induce the unfolded protein response and cellular apoptosis. Blood. 2007;110(13):4179–4187. doi: 10.1182/blood-2006-11-057299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kollner I, et al. Mutations in neutrophil elastase causing congenital neutropenia lead to cytoplasmic protein accumulation and induction of the unfolded protein response. Blood. 2006;108(2):493–500. doi: 10.1182/blood-2005-11-4689. [DOI] [PubMed] [Google Scholar]
- 18.Grenda DS, et al. Mice expressing a neutrophil elastase mutation derived from patients with severe congenital neutropenia have normal granulopoiesis. Blood. 2002;100(9):3221–3228. doi: 10.1182/blood-2002-05-1372. [DOI] [PubMed] [Google Scholar]
- 19.Nanua S, et al. Activation of the unfolded protein response is associated with impaired granulopoiesis in transgenic mice expressing mutant Elane. Blood. 2011;117(13):3539–3547. doi: 10.1182/blood-2010-10-311704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pomp O, Colman A. Disease modelling using induced pluripotent stem cells: status and prospects. BioEssays. 2013;35(3):271–280. doi: 10.1002/bies.201200088. [DOI] [PubMed] [Google Scholar]
- 21.Hiramoto T, et al. Wnt3a stimulates maturation of impaired neutrophils developed from severe congenital neutropenia patient-derived pluripotent stem cells. Proc Natl Acad Sci U S A. 2013;110(8):3023–3028. doi: 10.1073/pnas.1217039110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tidwell T, et al. Neutropenia-associated ELANE mutations disrupting translation initiation produce novel neutrophil elastase isoforms. Blood. 2014;123(4):562–569. doi: 10.1182/blood-2013-07-513242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mills JA, Paluru P, Weiss MJ, Gadue P, French DL. Hematopoietic differentiation of pluripotent stem cells in culture. Methods Mol Biol. 2014;1185:181–194. doi: 10.1007/978-1-4939-1133-2_12. [DOI] [PubMed] [Google Scholar]
- 24.Kattman SJ, et al. Stage-specific optimization of activin/nodal and BMP signaling promotes cardiac differentiation of mouse and human pluripotent stem cell lines. Cell Stem Cell. 2011;8(2):228–240. doi: 10.1016/j.stem.2010.12.008. [DOI] [PubMed] [Google Scholar]
- 25.Pincus SH, Boxer LA, Stossel TP. Chronic neutropenia in childhood. Analysis of 16 cases and a review of the literature. Am J Med. 1976;61(6):849–861. doi: 10.1016/0002-9343(76)90409-5. [DOI] [PubMed] [Google Scholar]
- 26.L’Esperance P, Brunning R, Good RA. Congenital neutropenia: in vitro growth of colonies mimicking the disease. Proc Natl Acad Sci U S A. 1973;70(3):669–672. doi: 10.1073/pnas.70.3.669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Takayama S, et al. Evolutionary conservation of function among mammalian, avian, and viral homologs of the Bcl-2 oncoprotein. DNA Cell Biol. 1994;13(7):679–692. doi: 10.1089/dna.1994.13.679. [DOI] [PubMed] [Google Scholar]
- 28.Hsu PD, et al. DNA targeting specificity of RNA-guided Cas9 nucleases. Nat Biotechnol. 2013;31(9):827–832. doi: 10.1038/nbt.2647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Horwitz MS, Duan Z, Korkmaz B, Lee HH, Mealiffe ME, Salipante SJ. Neutrophil elastase in cyclic and severe congenital neutropenia. Blood. 2007;109(5):1817–1824. doi: 10.1182/blood-2006-08-019166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Freedman MH, et al. Myelodysplasia syndrome and acute myeloid leukemia in patients with congenital neutropenia receiving G-CSF therapy. Blood. 2000;96(2):429–436. [PubMed] [Google Scholar]
- 31.Dahl R, et al. Regulation of macrophage and neutrophil cell fates by the PU.1:C/EBPα ratio and granulocyte colony-stimulating factor. Nat Immunol. 2003;4(10):1029–1036. doi: 10.1038/ni973. [DOI] [PubMed] [Google Scholar]
- 32.Hirai H, et al. C/EBPbeta is required for ‘emergency’ granulopoiesis. Nat Immunol. 2006;7(7):732–739. doi: 10.1038/ni1354. [DOI] [PubMed] [Google Scholar]
- 33.Skokowa J, Welte K. Defective G-CSFR signaling pathways in congenital neutropenia. Hematol Oncol Clin North Am. 2013;27(1):75–88. doi: 10.1016/j.hoc.2012.11.001. [DOI] [PubMed] [Google Scholar]
- 34.Borregaard N, Cowland JB. Granules of the human neutrophilic polymorphonuclear leukocyte. Blood. 1997;89(10):3503–3521. [PubMed] [Google Scholar]
- 35.Filali M, Cheng N, Abbott D, Leontiev V, Engelhardt JF. Wnt-3A/β-catenin signaling induces transcription from the LEF-1 promoter. J Biol Chem. 2002;277(36):33398–33410. doi: 10.1074/jbc.M107977200. [DOI] [PubMed] [Google Scholar]
- 36.Gullberg U, Bengtsson N, Bulow E, Garwicz D, Lindmark A, Olsson I. Processing and targeting of granule proteins in human neutrophils. J Immunol Methods. 1999;232(1–2):201–210. doi: 10.1016/s0022-1759(99)00177-5. [DOI] [PubMed] [Google Scholar]
- 37.Gullberg U, Lindmark A, Lindgren G, Persson AM, Nilsson E, Olsson I. Carboxyl-terminal prodomain-deleted human leukocyte elastase and cathepsin G are efficiently targeted to granules and enzymatically activated in the rat basophilic/mast cell line RBL. J Biol Chem. 1995;270(21):12912–12918. doi: 10.1074/jbc.270.21.12912. [DOI] [PubMed] [Google Scholar]
- 38.Massullo P, et al. Aberrant subcellular targeting of the G185R neutrophil elastase mutant associated with severe congenital neutropenia induces premature apoptosis of differentiating promyelocytes. Blood. 2005;105(9):3397–3404. doi: 10.1182/blood-2004-07-2618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Egesten A, Breton-Gorius J, Guichard J, Gullberg U, Olsson I. The heterogeneity of azurophil granules in neutrophil promyelocytes: immunogold localization of myeloperoxidase, cathepsin G, elastase, proteinase 3, and bactericidal/permeability increasing protein. Blood. 1994;83(10):2985–2994. [PubMed] [Google Scholar]
- 40.Grskovic M, Javaherian A, Strulovici B, Daley GQ. Induced pluripotent stem cells — opportunities for disease modelling and drug discovery. Nat Rev Drug Discov. 2011;10(12):915–929. doi: 10.1038/nrd3577. [DOI] [PubMed] [Google Scholar]
- 41.Gunaseeli I, Doss MX, Antzelevitch C, Hescheler J, Sachinidis A. Induced pluripotent stem cells as a model for accelerated patient- and disease-specific drug discovery. Curr Med Chem. 2010;17(8):759–766. doi: 10.2174/092986710790514480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Deng W. Induced pluripotent stem cells: paths to new medicines. A catalyst for disease modelling, drug discovery and regenerative therapy. EMBO Rep. 2010;11(3):161–165. doi: 10.1038/embor.2010.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Morishima T, et al. Genetic correction of HAX1 in induced pluripotent stem cells from a patient with severe congenital neutropenia improves defective granulopoiesis. Haematologica. 2014;99(1):19–27. doi: 10.3324/haematol.2013.083873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hu P, Han Z, Couvillon AD, Exton JH. Critical role of endogenous Akt/IAPs and MEK1/ERK pathways in counteracting endoplasmic reticulum stress-induced cell death. J Biol Chem. 2004;279(47):49420–49429. doi: 10.1074/jbc.M407700200. [DOI] [PubMed] [Google Scholar]
- 45.Murakami Y, Aizu-Yokota E, Sonoda Y, Ohta S, Kasahara T. Suppression of endoplasmic reticulum stress-induced caspase activation and cell death by the overexpression of Bcl-xL or Bcl-2. J Biochem. 2007;141(3):401–410. doi: 10.1093/jb/mvm044. [DOI] [PubMed] [Google Scholar]
- 46.Souza LR, Silva E, Calloway E, Cabrera C, McLemore ML. G-CSF activation of AKT is not sufficient to prolong neutrophil survival. J Leukoc Biol. 2013;93(6):883–893. doi: 10.1189/jlb.1211591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Iwata A, et al. Extracellular administration of BCL2 protein reduces apoptosis and improves survival in a murine model of sepsis. PLoS One. 2011;6(2):e14729. doi: 10.1371/journal.pone.0014729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Skokowa J, Welte K. Dysregulation of myeloid-specific transcription factors in congenital neutropenia. Ann N Y Acad Sci. 2009;1176:94–100. doi: 10.1111/j.1749-6632.2009.04963.x. [DOI] [PubMed] [Google Scholar]
- 49.Feng L, Zhu W, Huang C, Li Y. Direct interaction of ONO-5046 with human neutrophil elastase through (1)H NMR and molecular docking. Int J Biol Macromol. 2012;51(3):196–200. doi: 10.1016/j.ijbiomac.2012.04.023. [DOI] [PubMed] [Google Scholar]
- 50.Kunisato A, Wakatsuki M, Shinba H, Ota T, Ishida I, Nagao K. Direct generation of induced pluripotent stem cells from human nonmobilized blood. Stem Cells Dev. 2011;20(1):159–168. doi: 10.1089/scd.2010.0063. [DOI] [PubMed] [Google Scholar]
- 51.Kim YE, Hipp MS, Bracher A, Hayer-Hartl M, Hartl FU. Molecular chaperone functions in protein folding and proteostasis. Annu Rev Biochem. 2013;82:323–355. doi: 10.1146/annurev-biochem-060208-092442. [DOI] [PubMed] [Google Scholar]
- 52.Rajan RS, Tsumoto K, Tokunaga M, Tokunaga H, Kita Y, Arakawa T. Chemical and pharmacological chaperones: application for recombinant protein production and protein folding diseases. Curr Med Chem. 2011;18(1):1–15. doi: 10.2174/092986711793979698. [DOI] [PubMed] [Google Scholar]
- 53.Warlich E, et al. Lentiviral vector design and imaging approaches to visualize the early stages of cellular reprogramming. Mol Ther. 2011;19(4):782–789. doi: 10.1038/mt.2010.314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Lucas SD, Costa E, Guedes RC, Moreira R. Targeting COPD: advances on low-molecular-weight inhibitors of human neutrophil elastase. Med Res Rev. 2013;33(suppl 1):E73–E101. doi: 10.1002/med.20247. [DOI] [PubMed] [Google Scholar]
- 55.Kawabata K, Suzuki M, Sugitani M, Imaki K, Toda M, Miyamoto T. ONO-5046, a novel inhibitor of human neutrophil elastase. Biochem Biophys Res Comm. 1991;177(2):814–820. doi: 10.1016/0006-291X(91)91862-7. [DOI] [PubMed] [Google Scholar]
- 56.Ran FA, Hsu PD, Wright J, Agarwala V, Scott DA, Zhang F. Genome engineering using the CRISPR-Cas9 system. Nat Protoc. 2013;8(11):2281–2308. doi: 10.1038/nprot.2013.143. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.