Abstract
Uterine leiomyomas are benign tumors that can cause pain, bleeding, and infertility in some women. Mediator complex subunit 12 (MED12) exon 2 variants are associated with uterine leiomyomas; however, the causality of MED12 variants, their genetic mode of action, and their role in genomic instability have not been established. Here, we generated a mouse model that conditionally expresses a Med12 missense variant (c.131G>A) in the uterus and demonstrated that this alteration alone promotes uterine leiomyoma formation and hyperplasia in both WT mice and animals harboring a uterine mesenchymal cell–specific Med12 deletion. Compared with WT animals, expression of Med12 c.131G>A in conditional Med12–KO mice resulted in earlier onset of leiomyoma lesions that were also greater in size. Moreover, leiomyomatous, Med12 c.131G>A variant–expressing uteri developed chromosomal rearrangements. Together, our results show that the common human leiomyoma–associated MED12 variant can cause leiomyomas in mice via a gain of function that drives genomic instability, which is frequently observed in human leiomyomas.
Introduction
Uterine leiomyomas, or fibroids, are benign tumors arising from smooth muscle cells of the uterus. They are clinically diagnosed in 25% of women of reproductive age and are often associated with dysmenorrhea, dyspareunia, menorrhagia, infertility, and miscarriages (1, 2) and are the single largest cause of hysterectomy. Leiomyomas are monoclonal in origin, and 40% of the tumors have karyotypic abnormalities, including deletions in chromosome 7, trisomy of chromosome 12, and rearrangements involving the HMGA1 (6p21) and HMGA2 (12q14) genes (3–5). Whole-exome approaches have identified heterozygous somatic mutations in the mediator complex subunit 12 (MED12, Online Mendelian Inheritance in Man [OMIM] 300188) in approximately 70% of leiomyomas in patients from various ethnic and racial groups (6, 7). The majority of identified mutations occur in exon 2 of MED12.
MED12 is located on the X chromosome and encodes a 250-kDa protein that is a subunit of the large mediator complex and is involved in transcriptional regulation of the RNA polymerase II complex. The MED12 protein is highly conserved among eukaryotes (8) and plays an important role during embryogenesis, as Med12-null mouse embryos arrest at E7.5 due to impaired mesoderm formation (9). Despite the high prevalence of MED12 mutations within human uterine leiomyomas, their causality and mode of action are not well understood. Here, we show that the common Med12 variant associated with human leiomyomas, Med12 c.131G>A, can drive tumor formation alone in a gain-of-function manner and causes genomic instability.
Results and Discussion
Conditional loss of function of Med12 does not lead to uterine hyperplasia or leiomyomas.
We first determined whether the conditional inactivation of Med12 causes leiomyomas. Since Med12 is expressed from the X chromosome, random X chromosome inactivation will lead to random expression of either the paternal or maternal Med12 locus in uterine myometrial cells. We crossed anti-Mullerian hormone receptor type II–driven Cre (Amhr2-Cre) (10) with Med12fl/fl animals (9) to generate Med12fl/+ Amhr2-Cre animals and studied the effects of Med12 deficiency in a subpopulation of uterine mesenchymal cells. The use of Med12fl/+ animals, in which 1 allele is floxed and the other is WT, allowed us, in the presence of Amhr2-Cre recombinase, to generate a mosaic population of cells that either express or lack Med12.
Since Amhr2-Cre acts well after X chromosome inactivation is established (E6.5) (11), loss of Med12 function will not lead to skewed X inactivation in mouse uteri. To assess the Cre recombination in our hands, we crossed Amhr2-Cre mice with double-fluorescent Cre-reporter mT/mG mice (12), which express red fluorescence in all tissues and green fluorescence upon Cre recombination. Given our results (Supplemental Figure 1, A and B; supplemental material available online with this article; doi:10.1172/JCI81534DS1), we determined that approximately 60% of uterine mesenchymal cells underwent Cre-mediated excision. Recombination of the Med12fl allele and reduction of Med12 mRNA transcripts were confirmed in Med12fl/+ Amhr2-Cre uteri (Supplemental Figure 1, C–E). Neither leiomyoma formation nor hyperplasia were observed in adult uteri of Med12fl/+ Amhr2-Cre mice (Supplemental Figure 1, G and I). These results indicate that Med12 loss of function is not a mechanism of leiomyoma formation.
Expression of the Med12 c.131G>A variant on a background of conditional Med12 KO causes leiomyomas.
The most common MED12 mutation in leiomyomas among American women is a nonsynonymous variant, c.131G>A, predicted to substitute a highly conserved glycine with aspartic amino acid (p.Gly44Asp) (7). We investigated whether this Med12 mutation causes leiomyoma formation by generating a floxed Med12-mutant knockin mouse model (Supplemental Figure 2, A and B). We engineered the c.131G>A variant into the mouse Med12 cDNA (Med12mt) fused with a FLAG tag, subcloned it into the pROSA26-DV1 vector, and integrated it into the autosomal ROSA26 genomic locus. The presence of the FLAG reporter allowed us to distinguish the expression of mutant Med12 from WT Med12 (Supplemental Figure 2, D and F). The mice generated were heterozygous for mutant Med12 cDNA at the ROSA26 locus (Med12Rmt/+). We mated Med12Rmt/+ mice with Amhr2-Cre mice to conditionally express the mutated Med12 (c.131G>A) as early as E13.5 in the mouse uterine mesenchyme (10).
We investigated whether uterine leiomyomas will form in mice that express the Med12 c.131G>A variant on a conditional KO background (Figure 1A). In this model, Med12fl/y Amhr2-Cre males were bred with Med12Rmt/+ females to generate Med12fl/+ Med12Rmt/+ Amhr2-Cre females. A subset of uterine cells will express Med12 c.131G>A on an X chromosome Med12-null background (Figure 1B). We analyzed the Med12fl/+ Med12Rmt/+ Amhr2-Cre female reproductive tracts at 8 weeks and beyond 12 weeks of age. Nulliparous Med12fl/+ Med12Rmt/+ Amhr2-Cre female mice presented with pathological changes associated with leiomyoma formation at as early as 8 weeks of age (Supplemental Figure 3, B and D). Histological evaluation revealed that, beyond 12 weeks of age, 80% of the uteri contained lesions consistent with leiomyomas. These lesions consisted of extracellular matrix (ECM) deposits, accompanied by infiltration of fibroblasts and macrophages, hyperplasia, and disorganized muscle fiber arrangement, leading to complete destruction of myometrial architecture. Tumors formed in Med12fl/+ Med12Rmt/+ Amhr2-Cre–mutant uteri expressed mutant Med12, as shown by the expression of FLAG, which was fused to mutant Med12 in our ROSA construct (Supplemental Figure 2F).
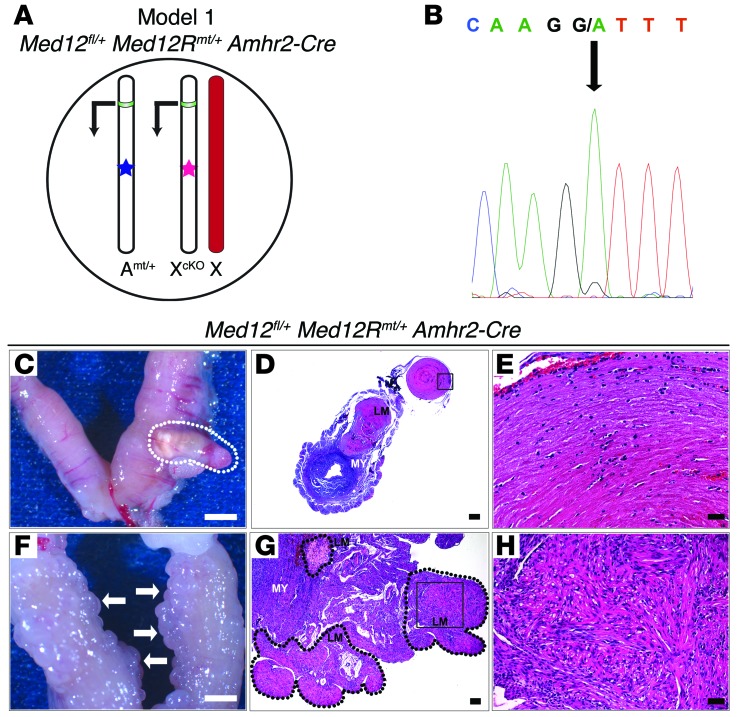
Figure 1. Med12fl/+Med12Rmt/+Amhr2-Cre mice develop uterine leiomyomas.
(A) Mouse model 1 (Med12fl/+ Med12Rmt/+ Amhr2-Cre). A subset of cells express the Med12 c.131G>A variant from the autosomal ROSA locus, while X chromosome–derived Med12 will either be conditionally excised at 1 locus, or silenced by X chromosome inactivation at the other locus. Transcription from the mutant autosome (Amt/+) is indicated with an arrow, and the promoter region is depicted in green. The blue star indicates the Med12 c.131G>A variant. The pink star indicates the floxed Med12 allele on the X chromosome (Xcko), which, in the presence of Amhr2-Cre, will lose exons 1–7. In the cells in which the Med12 floxed allele is inactivated, WT Med12 will be expressed. In cells with an active Med12 floxed allele, only mutant Med12 will be expressed. The red chromosome indicates the inactivated X chromosome. (B) cDNA sequencing from tumor tissue showing the presence of the mutant c.131G>A variant (green chromatogram peak, black arrow) in the absence of the WT Med12 allele, consistent with the monoclonal composition of tumor cells. (C) Eighteen-week-old multiparous Med12fl/+ Med12Rmt/+ Amhr2-Cre uterus revealed a 4-mm tumorous lesion (outlined by white dotted lines). (D) Histological examination confirmed the presence of a large leiomyoma nodule growing from the smooth muscle layer of the uterus. A higher-magnification image of the black-boxed neoplastic area appears in E and shows the presence of fascicles with plump spindle cells, eosinophilic cytoplasm, and ECM deposits. (F) Twenty-four-week-old Med12fl/+ Med12Rmt/+ Amhr2-Cre multiparous female uterus showing multiple nodules (white arrows). (G) Multiple leiomyoma nodules are outlined by black dotted lines, and the black box, shown at higher magnification in H, highlights fibrosis and ECM deposition. Approximately 80% (16 of 20 females) of the uteri exhibited leiomyoma-like lesions and hyperplasia. LM, leiomyoma; ES, endometrial stroma; MY, myometrium. Scale bars: 2,000 μm (C and F), 1,000 μm (D), 500 μm (G), 100 μm (H), 50 μm (E).
It has been noticed that estrogen and progesterone promote leiomyomatous growth, and 30% of leiomyomas in human pregnancies increase in volume (13). To corroborate these observations, we studied the effects of mouse parturition on leiomyomatous growth. Multiparous Med12fl/+ Med12Rmt/+ Amhr2-Cre females often had either grossly visible large leiomyomas (Figure 1C) or multiple small leiomyoma-like nodules (Figure 1F). Histology confirmed that these tumors arose from the smooth muscle layer and consisted of whorled fascicles of fusiform smooth muscle cells with an abundance of eosinophilic cytoplasm and ECM deposits (Figure 1, D, E, G, and H), consistent with the pathology seen in human uterine leiomyomas. Large tumors were often necrotic, hemorrhagic, and fibrotic. In addition, characteristic of leiomyomas, all tumors stained positive for smooth muscle actin and showed an abundance of collagen deposits when stained with Masson’s trichrome stain (Supplemental Figure 4, A–C). Eighty percent of multiparous Med12fl/+ Med12Rmt/+ Amhr2-Cre females had leiomyoma-like lesions. These results indicate that the Med12 c.131G>A variant causes leiomyoma-like lesions in mice.
The Med12 c.131G>A variant can cause uterine leiomyomas in mice on a WT background.
We investigated whether leiomyoma-like lesions were also present when the Med12 c.131G>A variant was expressed in mice on a WT background (Figure 2A). We generated animals coexpressing mutant Med12 from the autosome and a WT Med12 from the X chromosome (Med12Rmt/+ Amhr2-Cre) by crossing Med12Rmt/mt and Amhr2-Cre mice. Uteri from nulliparous Med12Rmt/+ Amhr2-Cre and control mice (Med12Rmt/+) were examined at 8 weeks of age and after 12 weeks of age.
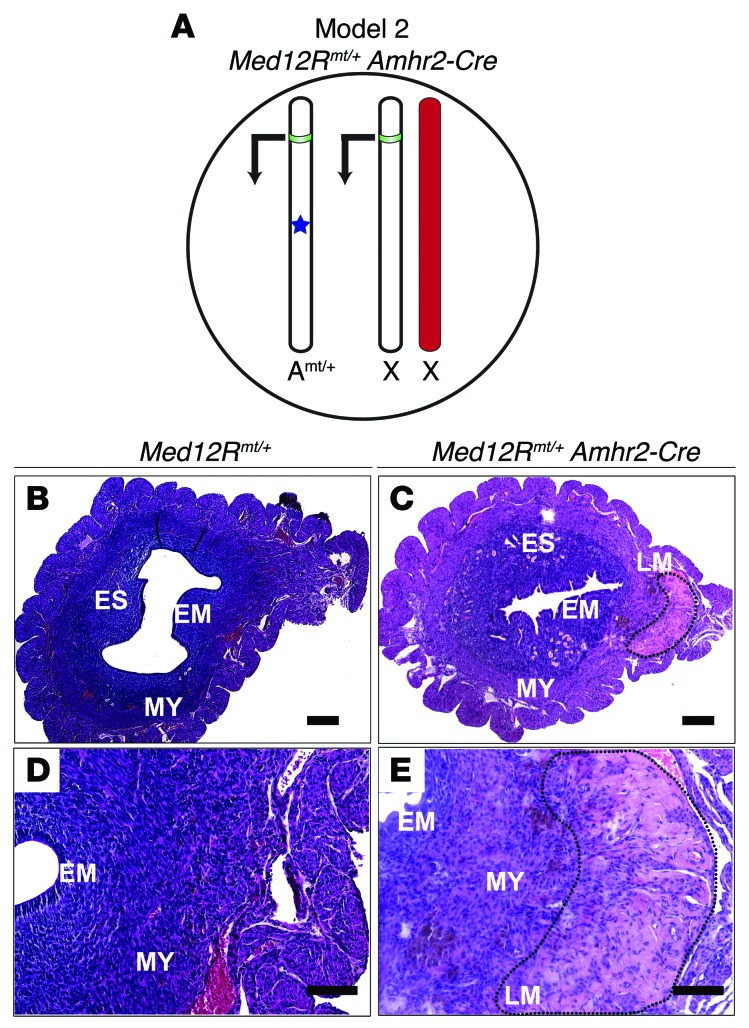
Figure 2. Med12Rmt/+Amhr2-Cre uteri develop prominent leiomyomas.
(A) Mouse model 2 (Med12Rmt/+ Amhr2-Cre). A subset of cells that express Amhr2-Cre will express the Med12 c.131G>A variant from the autosomal ROSA locus in the presence of X chromosome WT Med12. Transcription from a mutant autosome (Amt/+) is shown with an arrow, and the promoter region is depicted in green. The Med12 c.131G>A variant is depicted with a blue star. The red chromosome indicates the inactivated X chromosome. (B and D) Uteri from Med12Rmt/+ control mice that, in the absence of Amhr2-Cre, did not express the Med12 c.131G>A variant and showed normal cross-sectional histology. (C and E) Uteri from Med12Rmt/+ Amhr2-Cre mice that expressed the Med12 c.131G>A variant and revealed leiomyoma-like lesions in approximately 47% (8 of 17) of the females, with a typically sparse nuclear arrangement, a nodular pattern of cellular growth, and ECM deposition (black dotted lines). EM, endometrium. Scale bars: 500 μm (B and C), 100 μm (D and E).
In 8-week-old Med12Rmt/+ Amhr2-Cre mice, no leiomyoma-like lesions were observed (Supplemental Figure 5B). Fifty percent of the uteri from Med12Rmt/+ Amhr2-Cre–mutant mice that were over 12 weeks of age showed hyperplasia and leiomyomas, characterized by ECM deposition and a disorganized pattern of smooth muscle fiber arrangement (Supplemental Figure 5D). Uteri that expressed mutant Med12 weighed 20% to 30% more than did control uteri (P < 0.05) (Supplemental Figure 5E).
Examination of uteri from mice that were beyond 12 weeks of age revealed nodules that histologically resembled human leiomyomas due to deposition of ECM, whorl formation, and fewer nuclei (Figure 2, C and E). Our results show that the Med12 missense variant c.131G>A causes uterine hyperplasia and leiomyomas in both Med12 WT (Med12Rmt/+ Amhr2-Cre) and conditional KO (Med12fl/+ Med12Rmt/+ Amhr2-Cre) mice. Med12 c.131G>A variant penetrance was 47% in mice on a WT background, while it reached 80% in mice on the conditional KO background. In mice on the conditional Med12 deletion background, leiomyoma-like lesions tended to have earlier onset and achieve greater size. The Med12 missense c.131G>A variant, therefore, acts as a gain-of-function mutation.
Med12 mouse mutations and genomic instability.
Chromosomal rearrangements occur in 40% of human leiomyomas, and our data indicate that over 60% of uterine leiomyomas with an abnormal karyotype harbor MED12 mutations (7). To assess the genomic profiles of the Med12-mutated mouse tumors, we conducted array comparative genomic hybridization (aCGH) on 4 uteri with leiomyoma-like lesions (Figure 1) and compared the profiles with those of uteri from littermate controls without Cre (Med12fl/+ Med12Rmt/+). All 4 tumors showed genomic copy number gains and losses (40 per tumor), with mouse chromosomes 2, 7, 14, and 17 being most frequently affected. The affected regions often consisted of genes targeting cell cycle checkpoints or tumor pathways such as Ras, Wnt/β-catenin, Tp53/Rb, NF-κB, and TGF-β signaling. The complete list of aberrations in the uteri of Med12fl/+ Med12Rmt/+ Amhr2-Cre females is shown in Supplemental Table 1. Microarray analysis of Med12fl/+ Med12Rmt/+ Amhr2-Cre uteri also showed a few genomic regions with a pattern consistent with focal chromothripsis-like alterations (ref. 14 and Supplemental Figure 6A).
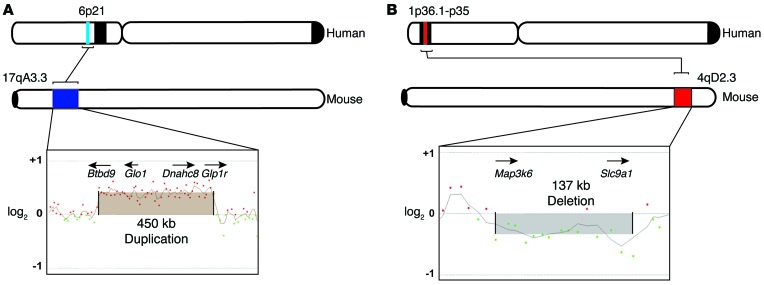
Approximately 50% of the mouse aberrations had syntenic counterparts on human chromosomes (Supplemental Table 2), and a number of these regions are known to be rearranged in human leiomyomas (Table 1). For example, mouse chromosome 17qA3.3, duplicated in Med12fl/+ Med12Rmt/+ Amhr2-Cre uteri (Figure 3A), maps to the human 6p21 locus. Similarly, in another Med12fl/+ Med12Rmt/+ Amhr2-Cre uterus, a deletion of the 4qD2.3 locus is syntenic to the human 1p36.1–p35 region (Figure 3B). Genomic rearrangements in 6p21 and 1p36.1–p35 are common in human leiomyomas. These results indicate that Med12 mutations can cause the genomic instability frequently seen in human leiomyomas.
Table 1. Med12fl/+ Med12Rmt/+ Amhr2-Cre uteri chromosomal aberrations and corresponding human syntenic regions implicated in human leiomyomas.
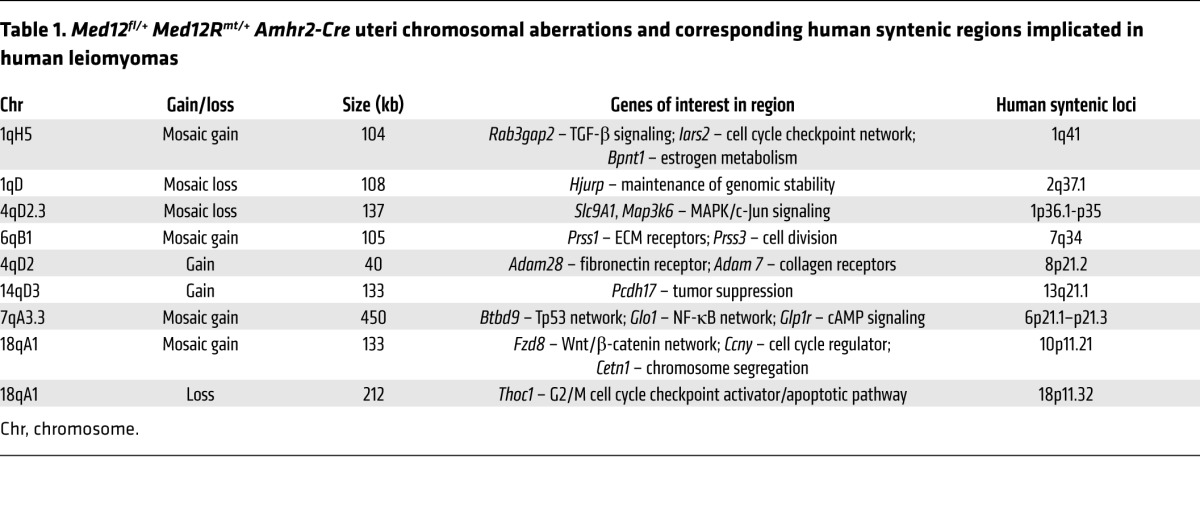
Figure 3. Representation of syntenic mapping of uterine rearrangements in Med12fl/+ Med12Rmt/+ Amhr2-Cre female mice on human chromosomal loci.
(A) Genomic duplication observed on mouse chromosomal locus 17qA3.3 is syntenic to the human 6p21 locus (shown in blue). A representative array profile of the 17qA3.3 region, highlighting the 450-kb duplication (chr17: 30586287–31049473), is also shown. (B) Genomic deletion observed on the mouse 4qD2.3 locus is syntenic to the human chromosomal locus 1p36.1–p35. The mouse deletion encompasses 137 kb and is shown in the respective array profile (chr4: 132799884–132936192). Positions are displayed approximately to scale according to the hg19 and mm9 physical maps, respectively.
Recurrent human MED12 exon 2 mutations have been associated with benign tumors such as uterine leiomyomas (7) and breast fibroadenomas (15); however, their etiology, genetic mechanism of action, and role in genomic instability are unknown. We generated mouse models with Med12 loss of function and the most common human MED12 c.131G>A variant and studied their effects in mouse uteri. Our mouse models show that the Med12 c.131G>A variant alone drives tumor formation via a gain-of-function mechanism. These tumors histologically mimic human uterine leiomyomas, and their growth is affected by pregnancy, in accordance with human data.
There are 3 published Tg mouse models of uterine leiomyomas; these include Tg overexpression of hGPR10 driven with the calbindin-D9K promoter (16), conditional deletion of Tsc2 (17), and conditional expression of a gain-of-function mutant form of β‑catenin (18). The phenotype in these mice is confined to increased myometrial thickness and formation of small nodules, but none of these mice show the dramatic tumors we report here (Figure 1). Interestingly, Amhr2-Cre drives mutant Med12 expression in both uterine smooth muscle (myometrium) and stroma, yet we only observed tumors derived from the mouse myometrium. In contrast, Amhr2-Cre–driven expression of the gain-of-function mutant form of β-catenin causes tumors in both the mouse myometrium and the stroma (18). These results indicate that Med12 exon 2 mutations have specific tumorigenic effects in smooth muscle cells.
aCGH of Med12 c.131G>A mouse uteri not only revealed genome-wide aberrations, but also showed complex chromosomal alterations such as chromothripsis. Recently, chromothripsis was reported in human leiomyomas and proposed as a possible mechanism of tumor progression (19). Chromosomal aberrations in mice also occur in regions that are syntenic to human 1p, 1q, 2q, 6p21, and 18p regions, also rearranged in human leiomyomas. It was previously shown that 60% of human leiomyomas with 6p21 rearrangements harbored MED12 exon 2 mutations (20). These data suggest that Med12 exon 2 mutations are precursors to genomic rearrangements and, hence, can cause genomic instability and drive tumor progression.
The limitations of our model include regulatory differences that may exist in the expression of Med12 on the X chromosome versus on an autosome. Autosomal Med12 is under the control of the ROSA promoter, which probably differs from the native Med12 promoter. Nonetheless, our model mimics the human condition and shows that Med12 variants can act through a gain-of-function mechanism. In the future, such models will provide a valuable tool for studying the role of MED12 in the genesis of uterine leiomyomas and the specificity of its effects on smooth muscle cells.
Methods
Further information can be found in the Supplemental Methods.
Materials.
Med12fl/fl mice were a gift of Heinrich Schrewe (Max-Planck Institute, Berlin, Germany), and the Amhr2-Cre mice were a gift of Richard Behringer (The University of Texas, Houston, Texas, USA).
Statistics.
A 2-tailed Student’s t test was applied to determine the difference of means among groups using GraphPad Prism 4.0 (GraphPad Software). Statistical significance was defined at a P value of less than 0.05.
Study approval.
All procedures were approved by the IACUC of the University of Pittsburgh and conducted in accordance with NIH guidelines for the care and use of laboratory animals.
Supplementary Material
Acknowledgments
We thank Y. Ren, A. Kishore, and K.J. Golnoski for their technical assistance and B. Campbell for his editorial comments. We thank Heinrich Schrewe and Richard Behringer for donating the mice. We thank the Magee-Womens Research Institute (MWRI) Transgenic Core for generation of the Med12Rmt mice and the Cytogenetics Laboratory for aCGH studies. This work was supported by a grant from the MWRI.
Footnotes
Conflict of interest: The authors have declared that no conflict of interest exists.
Reference information:J Clin Invest. 2015;125(8):3280–3284. doi:10.1172/JCI81534.
References
- 1.Stewart EA. Uterine fibroids. Lancet. 2001;357(9252):293–298. doi: 10.1016/S0140-6736(00)03622-9. [DOI] [PubMed] [Google Scholar]
- 2.Cramer SF, Patel A. The frequency of uterine leiomyomas. Am J Clin Pathol. 1990;94(4):435–438. doi: 10.1093/ajcp/94.4.435. [DOI] [PubMed] [Google Scholar]
- 3.Sandberg AA. Updates on the cytogenetics and molecular genetics of bone and soft tissue tumors: leiomyoma. Cancer Genet Cytogenet. 2005;158(1):1–26. doi: 10.1016/j.cancergencyto.2004.08.025. [DOI] [PubMed] [Google Scholar]
- 4.Nilbert M, Heim S, Mandahl N, Floderus UM, Willen H, Mitelman F. Characteristic chromosome abnormalities, including rearrangements of 6p, del(7q), +12, and t(12;14), in 44 uterine leiomyomas. Hum Genet. 1990;85(6):605–611. doi: 10.1007/BF00193583. [DOI] [PubMed] [Google Scholar]
- 5.Canevari RA, Pontes A, Rosa FE, Rainho CA, Rogatto SR. Independent clonal origin of multiple uterine leiomyomas that was determined by X chromosome inactivation and microsatellite analysis. Am J Obstet Gynecol. 2005;193(4):1395–1403. doi: 10.1016/j.ajog.2005.02.097. [DOI] [PubMed] [Google Scholar]
- 6.Makinen N, et al. MED12, the mediator complex subunit 12 gene, is mutated at high frequency in uterine leiomyomas. Science. 2011;334(6053):252–255. doi: 10.1126/science.1208930. [DOI] [PubMed] [Google Scholar]
- 7.McGuire MM, Yatsenko A, Hoffner L, Jones M, Surti U, Rajkovic A. Whole exome sequencing in a random sample of North American women with leiomyomas identifies MED12 mutations in majority of uterine leiomyomas. PLoS One. 2012;7(3):e33251. doi: 10.1371/journal.pone.0033251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bourbon HM. Comparative genomics supports a deep evolutionary origin for the large, four-module transcriptional mediator complex. Nucleic Acids Res. 2008;36(12):3993–4008. doi: 10.1093/nar/gkn349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rocha PP, Scholze M, Bleiss W, Schrewe H. Med12 is essential for early mouse development and for canonical Wnt and Wnt/PCP signaling. Development. 2010;137(16):2723–2731. doi: 10.1242/dev.053660. [DOI] [PubMed] [Google Scholar]
- 10.Jamin SP, Arango NA, Mishina Y, Hanks MC, Behringer RR. Requirement of Bmpr1a for Mullerian duct regression during male sexual development. Nat Genet. 2002;32(3):408–410. doi: 10.1038/ng1003. [DOI] [PubMed] [Google Scholar]
- 11.Wutz A. Gene silencing in X-chromosome inactivation: advances in understanding facultative heterochromatin formation. Nat Rev Genet. 2011;12(8):542–553. doi: 10.1038/nrg3035. [DOI] [PubMed] [Google Scholar]
- 12.Muzumdar MD, Tasic B, Miyamichi K, Li L, Luo L. A global double-fluorescent Cre reporter mouse. Genesis. 2007;45(9):593–605. doi: 10.1002/dvg.20335. [DOI] [PubMed] [Google Scholar]
- 13.Bulun SE. Uterine fibroids. N Engl J Med. 2013;369(14):1344–1355. doi: 10.1056/NEJMra1209993. [DOI] [PubMed] [Google Scholar]
- 14.Forment JV, Kaidi A, Jackson SP. Chromothripsis and cancer: causes and consequences of chromosome shattering. Nat Rev Cancer. 2012;12(10):663–670. doi: 10.1038/nrc3352. [DOI] [PubMed] [Google Scholar]
- 15.Lim WK, et al. Exome sequencing identifies highly recurrent MED12 somatic mutations in breast fibroadenoma. Nat Genet. 2014;46(8):877–880. doi: 10.1038/ng.3037. [DOI] [PubMed] [Google Scholar]
- 16.Varghese BV, et al. Loss of the repressor REST in uterine fibroids promotes aberrant G protein-coupled receptor 10 expression and activates mammalian target of rapamycin pathway. Proc Natl Acad Sci U S A. 2013;110(6):2187–2192. doi: 10.1073/pnas.1215759110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Prizant H, et al. Uterine-specific loss of Tsc2 leads to myometrial tumors in both the uterus and lungs. Mol Endocrinol. 2013;27(9):1403–1414. doi: 10.1210/me.2013-1059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tanwar PS, et al. Constitutive activation of Beta-catenin in uterine stroma and smooth muscle leads to the development of mesenchymal tumors in mice. Biol Reprod. 2009;81(3):545–552. doi: 10.1095/biolreprod.108.075648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mehine M, et al. Characterization of Uterine Leiomyomas by Whole-Genome Sequencing. N Engl J Med. 2013;369(1):43–53. doi: 10.1056/NEJMoa1302736. [DOI] [PubMed] [Google Scholar]
- 20.Markowski DN, Bartnitzke S, Loning T, Drieschner N, Helmke BM, Bullerdiek J. MED12 mutations in uterine fibroids-their relationship to cytogenetic subgroups. Int J Cancer. 2012;131(7):1528–1536. doi: 10.1002/ijc.27424. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.