Abstract
Mutations of the PARK2 and PINK1 genes, encoding the cytosolic E3 ubiquitin-protein ligase Parkin and the mitochondrial serine/threonine kinase PINK1, respectively, cause autosomal recessive early-onset Parkinson's disease (PD). Parkin and PINK1 cooperate in a biochemical mitochondrial quality control pathway regulating mitochondrial morphology, dynamics and clearance. This study identifies the multifunctional PD-related mitochondrial matrix enzyme 17-β hydroxysteroid dehydrogenase type 10 (HSD17B10) as a new Parkin substrate. Parkin overproduction in cells increased mitochondrial HSD17B10 abundance by a mechanism involving ubiquitin chain extension, whereas PARK2 downregulation or deficiency caused mitochondrial HSD17B10 depletion in cells and mice. HSD17B10 levels were also found to be low in the brains of PD patients with PARK2 mutations. Confocal and Förster resonance energy transfer (FRET) microscopy revealed that HSD17B10 recruited Parkin to the translocase of the outer membrane (TOM), close to PINK1, both in functional mitochondria and after the collapse of mitochondrial membrane potential (ΔΨm). PD-causing PARK2 mutations impaired interaction with HSD17B10 and the HSD17B10-dependent mitochondrial translocation of Parkin. HSD17B10 overproduction promoted mitochondrial elongation and mitigated CCCP-induced mitochondrial degradation independently of enzymatic activity. These effects were abolished by overproduction of the fission-promiting dynamin-related protein 1 (Drp1). By contrast, siRNA-mediated HSD17B10 silencing enhanced mitochondrial fission and mitophagy. These findings suggest that the maintenance of appropriate mitochondrial HSD17B10 levels is one of the mechanisms by which Parkin preserves mitochondrial quality. The loss of this protective mechanism may contribute to mitochondrial dysfunction and neuronal degeneration in autosomal recessive PD.
Parkinson's disease (PD) is the most common neurodegenerative movement disorder. It is caused by progressive loss of the dopaminergic neurons of the substantia nigra pars compacta. It is mostly sporadic, but about 10% of cases are familial forms with Mendelian inheritance.1 Autosomal recessive forms of PD are caused by mutations of PARK2/parkin, PINK1, DJ-1 and ATP13A2. PARK2 mutations are responsible for almost 40% of cases beginning before the age of 45. Parkin is a cytosolic RING-in-between-RING ubiquitin ligase with HECT-like enzyme activity;2 it has versatile ubiquitylation activities and various putative protein substrates with diverse functions, involved in different cellular processes.1 Genetic studies in Drosophila have shown Parkin and the mitochondrial serine/threonine kinase PINK1 to be components of a molecular pathway maintaining mitochondrial physiology.3, 4, 5, 6, 7 In mammalian cells, PINK1 and Parkin cooperate in the clearance of dysfunctional mitochondria. The disruption of mitochondrial membrane (MM) potential (ΔΨm) by uncouplers or oxidative stress leads to PINK1 accumulation on the outer MM (OMM), the PINK1-dependent recruitment and activation of Parkin, the ubiquitylation and proteasome-dependent degradation of nearby substrates, and the initiation of mitophagy.1, 8, 9, 10, 11, 12 A series of mitochondrial targets interact with or are ubiquitylated by Parkin early in mitochondrial clearance.9, 13, 14, 15, 16, 17, 18, 19 There is considerable interest in the possible direct role of some of these proteins in Parkin-dependent mitochondrial degradation, particularly for Mitofusin 1/2,13, 20 VDACs,9, 17 hexokinases 1/216, 19 and subunits of the translocase of the OMM (TOM).21, 22 Moreover, the constitutive association of some Parkin with the OMM raises the question of a potential role of this protein in functions unrelated to mitophagy, such as regulating the degradation or targeting of specific mitochondrial proteins.23
We report here the identification of a new mitochondrial Parkin substrate, the nucleus-encoded mitochondrial matrix 17-β hydroxysteroid dehydrogenase type 10 (HSD17B10), also known as endoplasmic reticulum-associated amyloid-beta-binding protein (ERAB), Aβ-binding alcohol dehydrogenase (ABAD) and 3-hydroxyacyl-CoA dehydrogenase type II (HADH2).24, 25 HSD17B10 is a ubiquitous NADH-dependent multifunctional enzyme essential for mitochondrial maintenance and cell viability.25, 26 It is involved primarily in isoleucine catabolism, in which it dehydrogenates 2-methyl-3-hydroxybutyryl-CoA (MHB),27 but it has other substrates, including fatty acids and steroids, in several pathways.25 It is one of the three protein components of human mitochondrial RNAse P, which is required for the processing of mtDNA transcripts.28 HSD17B10 mutations cause a rare X chromosome-linked inborn error of metabolism, with classical neonatal and infantile forms associated with rapid, progressive neurodegeneration, and atypical juvenile and asymptomatic forms.24, 27 HSD17B10 is also thought to have a role in adult-onset neurodegenerative disorders, such as Alzheimer's disease, in which it may mediate amyloid β-driven mitochondrial toxicity,25, 29, 30 and PD, in which it is downregulated in the ventral midbrain.31
We and others have generated and characterized PARK2 knockout mice. These models did not show overt signs of neurodegeneration, but they presented neurochemical, biochemical and behavioral alterations reminiscent of preclinical dysfunction.32, 33, 34, 35, 36, 37, 38 We identified HSD17B10 as one of the proteins differentially expressed between PARK2 knockout and wild-type mice.37 Here, we provide evidence that Parkin increases the mitochondrial availability of HSD17B10 in a ubiquitin-dependent manner. HSD17B10 promoted mitochondrial elongation, sparing these organelles from degradation triggered by the protonophore carbonyl cyanide m-chlorophenylhydrazone (CCCP). Our results suggest that the maintenance of appropriate HSD17B10 levels may be a new mechanism by which Parkin preserves mitochondrial homeostasis.
Results
HSD17B10, a mitochondrial enzyme displaying abundance changes in Parkin-deficient mice, is a Parkin substrate
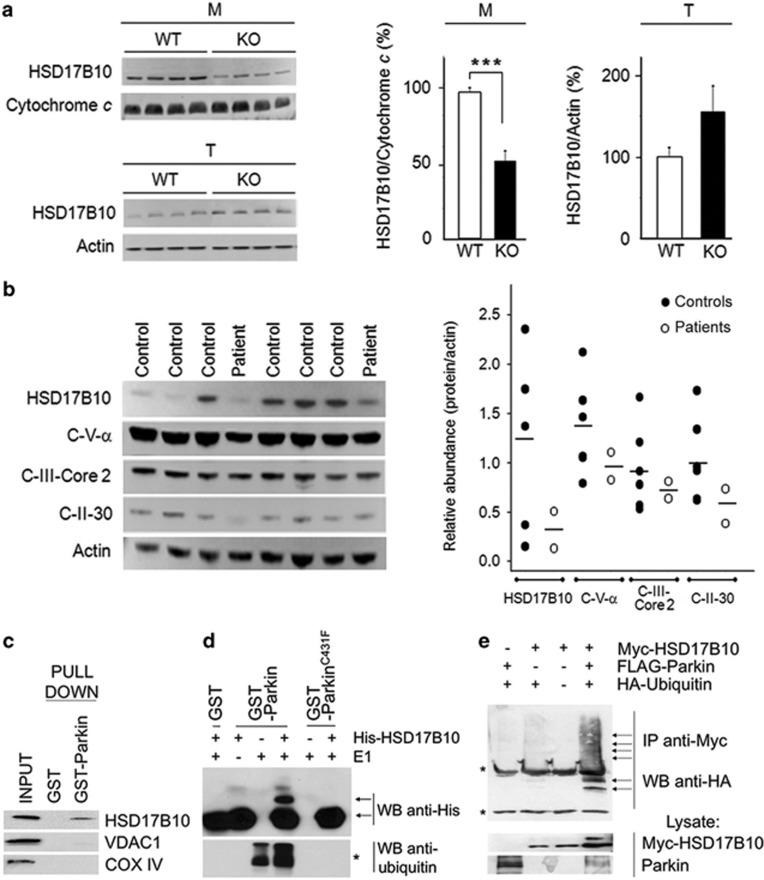
Using fluorescence two-dimensional difference gel electrophoresis and mass spectrometry, we previously identified HSD17B10 as a protein moderately upregulated (30%) in total fractions from the cortex and striatum of young and aged non-symptomatic PARK2 knockout mice (Periquet et al.37 and data not shown). For confirmation of these findings, we carried out western blots to compare the abundance of HSD17B10 in pure mitochondrial fractions from the whole brains of PARK2-deficient and wild-type mice. HSD17B10 levels in knockout mice were half those in wild-type mice, but were slightly higher in total brain fractions (Figure 1a), as in our previous proteomic study, possibly reflecting an adaptive mechanism aimed at restoring normal protein levels.37 Consistent with previous reports in sporadic PD,31 HSD17B10 levels also tended to be lower in total striatal extracts from two PD patients with PARK2 mutations than in those of unaffected subjects (Figure 1b), indicating that at this stage of overt neurodegeneration, compensation may become inefficient.
Figure 1.
Parkin modulates the levels of its substrate HSD17B10 in the brain. (a, b) Abundance of HSD17B10 or of respiratory chain complex subunits (Complex V subunit α, C-V-α; Complex III subunit Core 2, C-III-Core 2; Complex II subunit 30 KDa, C-II-30) in (a) pure brain mitochondria (M) or total brain extracts (T) from PARK2 knockout (KO) and wild-type (WT) mice (n=11–15) or (b) extracts from the striatum of PD patients with PARK2 mutations (n=2) and controls (n=6). Loading controls were cytochrome c and/or actin. The results in (a) are expressed as means±S.E.M. ***P<0.001. (c) GST pulldown assay illustrating the specific interaction between Parkin and HSD17B10 in lysates from COS7 cells. No interaction was observed between Parkin and VDAC1 or the subunit VIb of mitochondrial complex IV. (d) Purely in vitro ubiquitylation assay, showing the appearance of a HSD17B10 species compatible with monoubiquitylation in the presence of Parkin, but not in the presence of the enzymatically inactive C431F variant. This species is not observed in the absence of the ubiquitin-activating enzyme E1. *Anti-ubiquitin western blot showing autoubiquitylated Parkin. (e) Ubiquitylation of HSD17B10 by Parkin in COS7 cells overproducing the protein alone or with Parkin and/or HA-tagged ubiquitin. Ubiquitin-positive bands appear in affinity-purified HSD17B10 fractions only in the presence of Parkin (arrows). Asterisks indicate light and heavy IgG chains. IP, immunoprecipitation; WB, western blot
We then investigated whether Parkin interacted directly with HSD17B10, promoting its ubiquitylation. A recombinant GST-tagged Parkin protein precipitated endogenous HSD17B10 from whole-cell lysates (Figure 1c). In a purely in vitro assay, GST-Parkin promoted the generation of a HSD17B10-immunoreactive species with an apparent molecular mass compatible with monoubiquitylated HSD17B10 (Figure 1d). No such effect was observed with the enzymatically inactive C431F variant. COS7 cells coproducing HSD17B10, ubiquitin and Parkin, also generated ubiquitylated species of sizes consistent with mono- and polyubiquitylated HSD17B10 (Figure 1e).
Parkin increases the mitochondrial abundance of HSD17B10 in a ubiquitin-dependent manner
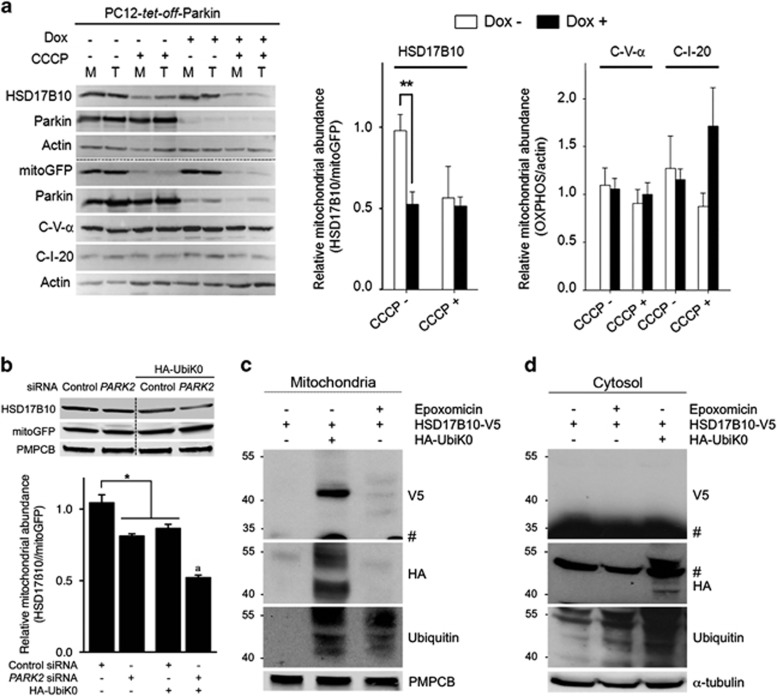
We investigated whether changes in HSD17B10 abundance reflected direct regulation by Parkin rather than compensatory mechanisms in response to chronic Parkin deficiency, by studying the effect of dynamic modulations of Parkin abundance on the mitochondrial targeting of exogenous HSD17B10 in mammalian cells. Human HSD17B10 and a mitochondrion-targeted green fluorescent protein (mitoGFP) were overproduced in rat PC12 cells producing human Parkin under the control of the tet-off gene regulatory system (PC12-tet-off-Parkin).23 Quantitative western blot analysis revealed a doubling of the ratio of HSD17B10 to mitoGFP levels in mitochondrion-enriched fractions from cells overproducing Parkin (Figure 2a, Dox−). This difference was abolished by treating the cells with CCCP, which dissipates the ΔΨm required for protein import.
Figure 2.
Mitochondrial HSD17B10 levels are regulated by Parkin and ubiquitin. (a) Effect of Parkin on the mitochondrial abundance (M/T; means±S.E.M.) of newly synthesized HSD17B10 relative to mitoGFP (n=4 independent experiments), or of endogenous subunits of respiratory chain complex I or V relative to actin in PC12-tet-off-Parkin cells (n=7 independent experiments). M, mitochondrion-enriched fractions; T, total cell fractions; Dox, doxycycline. (b) Effect of the siRNA-mediated downregulation of PARK2 or the overexpression of UbiK0 on the abundance of HSD17B10 relative to mitoGFP in mitochondrion-enriched fractions from HEK293T cells subjected to trypsin digestion. The mitochondrial matrix protease PMPCB was used as a loading control. n=4 independent experiments. Results are expressed as means±S.E.M.. *P<0.05, **P<0.01; aP<0.01 versus PARK2 siRNA and HA-UbiK0. (c, d) Western blot analysis showing the appearance of a HSD17B10 species compatible with monoubiquitylation specifically in mitochondrion-enriched fractions (1 min exposure) and not in cytosol (2 h exposure) from HEK293T cells overproducing UbiK0 but not in cells treated with epoxomicin. #Signal corresponding to unmodified HSD17B10 on the V5 blot (c, d), or residual signal corresponding to α-tubulin on the HA blot in (d)
Parkin silencing decreased intramitochondrial HSD17B10 levels by 15% in HEK293T cells (Figure 2b; Supplementary Figure S1a). PINK1 silencing resulted in a similar decrease (Supplementary Figures S1b and S2d). We investigated the role of ubiquitylation, by evaluating the impact of overproducing a lysine-less ubiquitin variant (UbiK0) inhibiting ubiquitin chain extension, or of the proteasome inhibitor epoxomicin. UbiK0 decreased mitochondrial HSD17B10 levels to a similar extent to PARK2 downregulation (Figure 2b). It also exacerbated the effect of Parkin depletion, resulting in a decrease in HSD17B10 levels of almost 50%. Epoxomicin treatment increased the abundance of HSD17B10 in the cytosol but had no effect on mitochondria (Supplementary Figure S1c).
UbiK0 overproduction led to the generation of an HSD17B10 protein with a molecular mass compatible with monoubiquitylation, consistent with impaired ubiquitin chain extension (Figure 2c). This protein was found in the mitochondrial fraction but not in the cytosol, and was absent from cells treated with epoxomicin, even after long exposure times (Figures 2c and d). It also disappeared following the limited trypsin digestion of mitochondrial fractions to eliminate proteins associated with the OMM (Supplementary Figure 1d).
HSD17B10 recruits Parkin to the mitochondrion in a PINK1-dependent manner, without promoting mitophagy
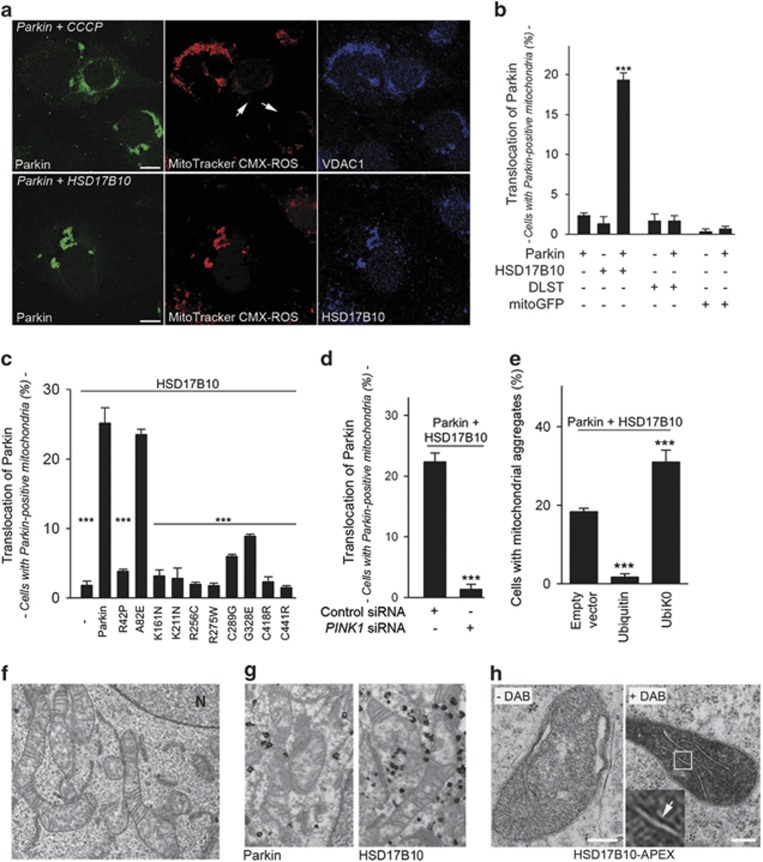
We then analyzed the subcellular distribution of HSD17B10 and Parkin in COS7 cells. When overproduced alone, HSD17B10 was associated with the mitochondrial reporter MitoTracker DeepRed, whereas exogenous Parkin was predominantly cytosolic (Supplementary Figure S2a). When coproduced with HSD17B10, but not with the Krebs cycle enzyme dihydrolipoamide S-succinyl transferase (DLST) or with mitoGFP, Parkin was recruited to the mitochondria in a large proportion of cells, resulting in the perinuclear aggregation and ubiquitylation of the organelles (Figures 3a and b; Supplementary Figures S2b and c). PD-causing PARK2 mutations prevented these events, possibly due to weakened interaction between HSD17B10 and Parkin, or lack of ubiquitylation (Figure 3c). Recruitment of Parkin by HSD17B10 was also abolished in cells depleted for PINK1 (Figure 3d; Supplementary Figure S2d). Moreover, ubiquitin overproduction mitigated mitochondrial aggregation, whereas UbiK0 enhanced this process, suggesting that this aggregation was due to the impairment of downstream mechanisms (Figure 3e).
Figure 3.
Parkin is recruited to the mitochondria by HSD17B10, in a PINK1-dependent manner. (a) Representative immunocytochemical staining showing the recruitment of exogenous Parkin to depolarized mitochondria in COS7 cells treated with CCCP for 6 h (upper panels; arrows), or to mitochondria with an intact membrane potential in cells overproducing HSD17B10 (lower panels). The treatments and transfection conditions are indicated in italics. Mitochondria were visualized with the potentiometric mitochondrial dye MitoTracker CMX-ROS and anti-VDAC1 or anti-HSD17B10 staining. Scale bar: 10 μm. (b) Mitochondrial translocation of Parkin, quantified as the proportion of cells with Parkin-positive mitochondria. COS7 cells were transfected to overproduce Parkin alone or together with HSD17B10, DLST or mitoGFP. (c, d) Effect of PD-causing PARK2 mutations (c) or PINK1 silencing (d) on the mitochondrial recruitment of Parkin by exogenous HSD17B10 in COS7 cells, quantified as in (b). (e) Effect of exogenous ubiquitin or UbiK0 on mitochondrial aggregation in COS7 cells overproducing Parkin and HSD17B10. Results are expressed as means±S.E.M. for one experiment representative of six (b, c) or three (d, e) carried out, with n=60–100 cells per condition. ***P<0.001 versus ‘+ Parkin' in (b); ‘Parkin' in (c); ‘Empty vector in (e). (f, g) Micrograph showing the ultrastructure of mitochondria (f) the localization of Parkin and HSD17B10 on the outer surface of aggregated mitochondria, as revealed by immunogold staining (g), and the localization of HSD17B10-APEX in isolated mitochondria, as revealed by incubation with the peroxidase substrate diaminobenzidine (DAB)58 (h) in COS7 cells. The arrow in the higher magnification of the framed region in (h) indicates the association of the staining with the inner MM. N, nucleus. Scale bar: 200 nm
The mitochondrial recruitment of Parkin and organelle clustering resemble the events observed after ΔΨm collapse.8 However, in cells overproducing HSD17B10, Parkin was translocated to mitochondria in which ΔΨm was preserved, as shown by MitoTracker CMX-ROS or TMRM staining (Figure 3a; Supplementary Figure S2e). Parkin is recruited to mitochondria with preserved ΔΨm by PINK1, particularly when mitochondrial import is blocked.22, 39, 40 PINK1 accumulated on mitochondria in cells overproducing HSD17B10, indicating that mitochondrial import mechanisms were overwhelmed (Supplementary Figure S2f). However, LC3-positive autophagic vesicles and immunoreactivity to the autophagic adaptor SQSTM1/p62 were rarely associated with mitochondrial clusters in cells coproducing HSD17B10 and Parkin, indicating an absence of mitophagy in these cells (Supplementary Figure S3). CCCP-induced mitochondrial aggregation is dependent on active transport along the microtubule network toward the perinuclear region, facilitating mitochondrial degradation by autophagy.12 By contrast, in cells producing Parkin and HSD17B10, mitochondrial aggregation was not affected by cytoskeleton disruption, suggesting a passive process driven by protein–protein interactions at the OMM, as previously described for the mitochondrial shaping protein Mitofusin 2 (Supplementary Figure S4).41 Moreover, ultrastructural analysis revealed morphologically normal mitochondria in these cells (Figure 3f). Parkin is not imported into mitochondria and HSD17B10 is synthesized in the cytosol.25 These proteins may therefore interact at the OMM, before HSD17B10 import. Both these proteins were detected at the OMM in mitochondrial aggregates, at the interface between adjacent organelles (Figure 3g). By contrast, outside mitochondrial aggregates, HSD17B10 was found in the mitochondrial matrix, associated with the inner MM in some cases, consistent with the expected final destination of this protein (Figure 3h).25, 31
Our observations suggest that HSD17B10 overproduction overwhelms mitochondrial protein import mechanisms, leading to PINK1 accumulation at the OMM. Parkin recruitment and ubiquitylation may, in turn, facilitate HSD17B10 import.
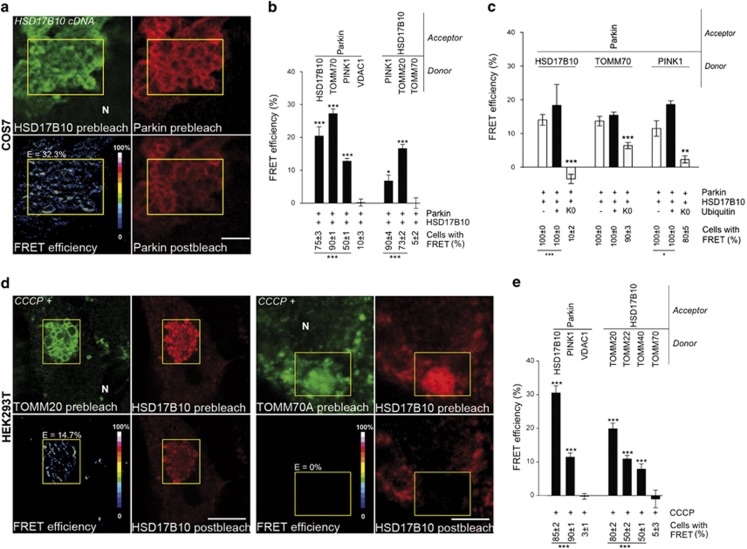
HSD17B10 interacts with Parkin and PINK1 at the TOM machinery and mitigates the Parkin-dependent mitophagy triggered by CCCP
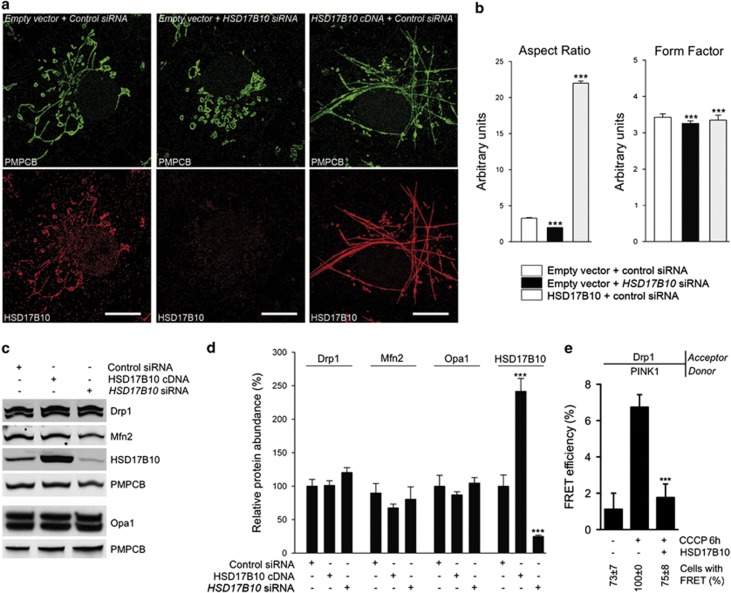
HSD17B10 is imported into mitochondria via the TOM complex, which is dedicated to the import of most nucleus-encoded mitochondrial proteins,42 and Parkin and PINK1 associate with this complex.21, 22 We investigated whether these proteins interacted close to the TOM machinery. We used Förster resonance energy transfer (FRET) microscopy, involving the detection of changes in donor fluorescence intensities after photobleaching of the acceptor, with donor/acceptor protein pairs labeled with appropriate secondary antibodies,22, 43 to check for physical proximity (≤10 nm). In tests of representative donor/acceptor pairs in cells coproducing Parkin and HSD17B10, we detected FRET compatible with the formation of a complex between these proteins and PINK1 at the TOM machinery (Figures 4a and b). UbiK0 overproduction compromised FRET between these molecular partners, demonstrating the need for ubiquitin chain extension (Figure 4c).
Figure 4.
Parkin interacts with HSD17B10 and PINK1 at the TOM machinery. (a) Representative FRET images for the indicated donor (green)–acceptor (red) pairs in COS7 cells overproducing Parkin and HSD17B10, Pseudocolor scale: pixel-by-pixel FRET efficiencies reflecting the increase in donor fluorescence after photobleaching of the acceptor. (b, c) Quantitative FRET analysis and percentage of cells displaying FRET for all donor–acceptor pairs analyzed in the indicated conditions. n=10–20 cells from one experiment representative of three (b) or two (c) carried out. (d, e) Representative FRET images (d) and quantitative FRET analysis for the indicated donor/acceptor pairs (e) in HEK293T cells treated with CCCP for 6 h. n=10–20 cells from one experiment representative of three carried out. E: Mean FRET efficiency within the region of interest (yellow, ROI). N: nucleus. Scale bar: 10 μm. Results are expressed as means±S.E.M. *P<0.05, **P<0.01, ***P<0.001 versus VDAC1/Parkin for donor/Parkin pairs and TOMM70/HSD17B10 for donor/HSD17B10 pairs in (b, e), or versus ‘−Ubiquitin' and ‘+Ubiquitin' in (c)
The association of Parkin and PINK1 at the TOM machinery after mitochondrial import blockade is a key step initiating the clearance of dysfunctional mitochondria after CCCP treatment.22 We therefore investigated whether HSD17B10 was also recruited to this complex in these conditions. Representative interactions between Parkin, HSD17B10, PINK1 and the TOM machinery were detected after CCCP treatment. These interactions concerned exclusively endogenous proteins in HEK293T cells, or exogenous Parkin and endogenous proteins in PC12-tet-off-Parkin cells differentiated into neurons (Figures 4d and e; Supplementary Figure S5a).23 Several PD-causing PARK2 mutations abolished or reduced FRET between endogenous HSD17B10 and Parkin, consistent with weakened protein–protein interactions or altered protein configurations within the complex (Supplementary Figure S5b).
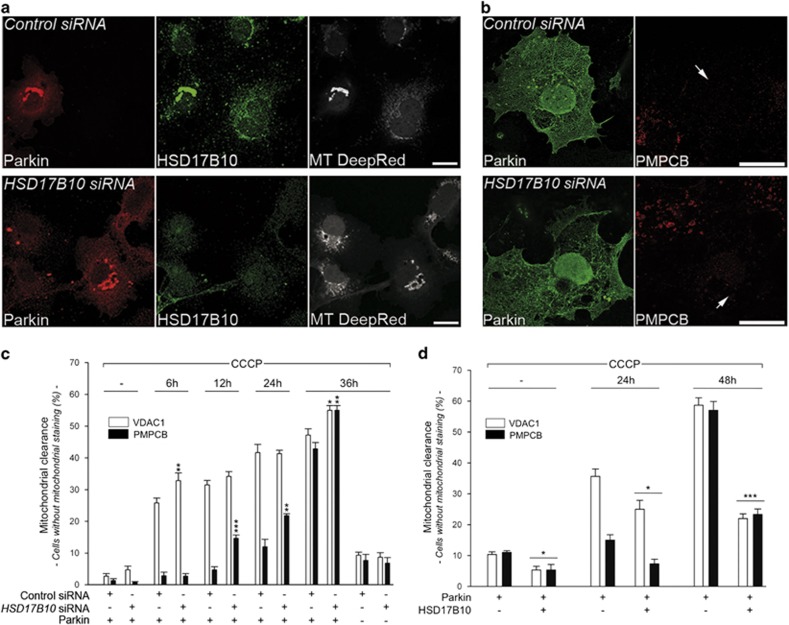
We then investigated the role of HSD17B10 in the degradation of dysfunctional mitochondria by Parkin. Parkin was recruited to mitochondria in cells lacking HSD17B10, so HSD17B10 was not required for this step (Figure 5a). Moreover, Parkin-dependent mitophagy occurred and was even enhanced following HSD17B10 depletion, resulting in a larger proportion of cells lacking the OMM marker VDAC1 soon after CCCP treatment, or lacking the matrix marker PMPCB at later time points than in control cells (Figures 5b and c). Conversely, HSD17B10 overproduction decreased mitochondrial clearance, as illustrated by the significantly smaller proportion of cells lacking mitochondrial markers after CCCP treatment (Figure 5d). Consistent with the lack of recruitment of autophagy markers (Supplementary Figure S3), no mitochondrial clearance was observed in cells overproducing Parkin and HSD17B10 in the absence of CCCP.
Figure 5.
HSD17B10 mitigates Parkin-dependent mitophagy. (a) Representative immunocytochemical staining illustrating the mitochondrial recruitment of Parkin triggered by CCCP (6 h) in COS7 cells with and without HSD17B10 silencing. Mitochondria were visualized by staining with MitoTracker DeepRed and for HSD17B10. (b) Representative immunofluorescence showing that mitochondrial loss triggered by CCCP (36 h) occurs in COS7 cells overproducing Parkin with and without the silencing of HSD17B10. Mitochondria were stained with anti-PMPCB antibodies. Scale bar: 10 μm. (c, d) Percentage of cells devoid of the OMM marker VDAC1 or the matrix marker PMPCB at various time points after the addition of CCCP to the culture medium, in the indicated transfection conditions. Results are expressed as means±S.E.M. n=3 independent experiments, with 180 cells scored per condition in each experiment (c) or n=3 independent wells from one experiment representative of five carried out, with 90 cells scored from each well (d). *P<0.05, **P<0.01, ***P<0.001 versus the ‘Control siRNA' (c) or versus the ‘Parkin only' condition (d) at each time point
HSD17B10 promotes mitochondrial elongation, protecting against mitochondrial damage following ΔΨm collapse
HSD17B10 overproduction was associated with a major change in mitochondrial morphology, with mitochondria appearing as long filaments extending across the cytoplasm (Figure 6a). We analyzed the morphology of the mitochondrial network by measuring two parameters widely used for this purpose: aspect ratio, providing an indication of mitochondrial length, and form factor, a combined evaluation of the length and degree of branching of the network.44 HSD17B10 overproduction led to a large increase in aspect ratio and a moderate decrease in form factor (Figure 6b). Conversely, HSD17B10 silencing resulted in mitochondrial fragmentation, as indicated by a significant decrease in both parameters (Figures 6a and b).
Figure 6.
HSD17B10 promotes mitochondrial elongation. (a) Representative immunocytochemical staining illustrating mitochondrial networks in COS7 cells transfected with siRNAs targeting the HSD17B10 gene or overproducing the protein. Mitochondria were visualized with anti-PMPCB and anti- HSD17B10 antibodies. Scale bar: 10 μm. (b) Quantitative confocal analysis of aspect ratio (length) and form factor (length and branching) of mitochondria in COS7 cells transfected as indicated. n=30 cells from three independent experiments. (c, d) Western blot analysis (c) and corresponding quantification (d) illustrating the effect of HSD17B10 downregulation or overproduction on the abundance of mitochondrial shaping proteins in COS7 cells. n=3 independent experiments. (e) Quantitative FRET analysis illustrating the effect of HSD17B10 overproduction on the molecular proximity between PINK1 and Drp1. n=15–20 cells from one experiment representative of three carried out. Results are expressed as means±S.E.M. ***P<0.001 versus ‘Empty vector+control siRNA' in (b); ‘control siRNA' in (d); and ‘- HSD17B10+CCCP' in (e)
Mitochondrial morphology is regulated by fusion and fission processes mediated by conserved dynamin-related GTPases:45 optic atrophy 1 (Opa1) and mitofusins (Mfn1/2), involved in fusion of the inner and outer MMs; and dynamin-related protein 1 (Drp1), which promotes mitochondrial fission. Defective mitochondria are isolated from the healthy network by the recruitment of Drp1 from the cytosol, triggering mitochondrial fragmentation and efficient clearance.13, 46 Using FRET microscopy, we showed that Drp1 was recruited to the proximity of PINK1 following mitochondrial depolarization.47 The changes in mitochondrial morphology observed when HSD17B10 levels were modulated were not associated with changes to steady-state levels of Opa1, Mfn2 and Drp1 (Figures 6c and d). However, lower levels of FRET between PINK1 and Drp1 were observed in cells overproducing HSD17B10, indicating interference with Drp1 recruitment to or assembly on depolarized mitochondria (Figure 6e; Supplementary Figure S6).
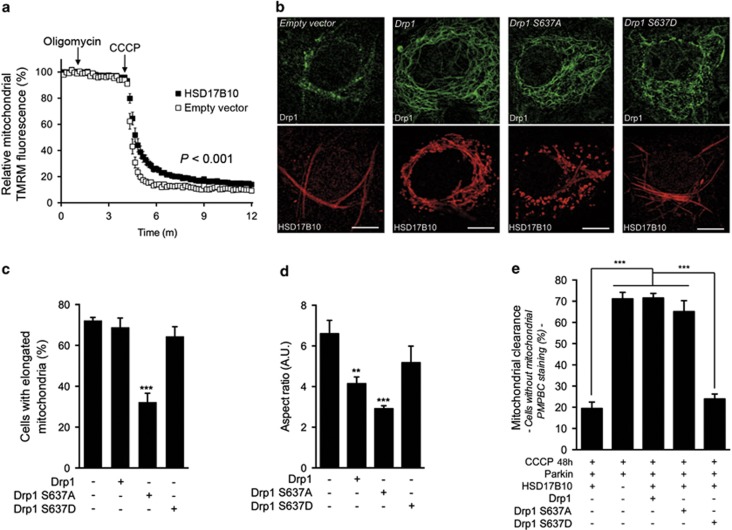
Mitochondrial elongation is an adaptive prosurvival mechanism associated with a better bioenergetic response to stress.48, 49, 50 In starved cells, it spares mitochondria from degradation by autophagy.49, 50 Analysis of the mean TMRM fluorescence intensity of mitochondria revealed consistently higher signals in cells overproducing HSD17B10 and a later, less severe decrease following CCCP treatment, consistent with better ΔΨm preservation (Figure 7a; Supplementary Figure S2e). HSD17B10 may, therefore, protect mitochondria by preserving a hyperfused network, resulting in lower levels of mitophagy, an indirect indicator of mitochondrial health. We explored this possibility by studying the effect of Drp1 on HSD17B10-mediated hyperfusion and sparing from mitophagy. We overproduced Drp1 or two Drp1 variants carrying amino-acid substitutions affecting serine 637, which is involved in regulating the subcellular distribution of the protein:51 Drp1 S637A, which mimics the calcineurin-dependent dephosphorylation of this residue, leading to association of the protein with mitochondria; and Drp1 S637D, which mimics phosphorylation of the residue by protein kinase A, sequestering the protein in the cytosol.51 Drp1 S637A and, to a lesser extent, Drp1, significantly reduced mitochondrial hyperfusion and decreased the ability of HSD17B10 to mitigate Parkin-dependent mitophagy in cells treated with CCCP, whereas Drp1 S637D had no effect (Figures 7b–e).
Figure 7.
HSD17B10-mediated mitochondrial hyperfusion protects mitochondria against CCCP, sparing them from Parkin-dependent clearance. (a) Analysis of the relative TMRM fluorescence of mitochondria in COS7 cells transfected with a control plasmid or a plasmid encoding HSD17B10, illustrating the HSD17B10-mediated attenuation of the decrease in ΔΨm triggered by CCCP. n=20–30 cells from one experiment representative of three carried out. (b–d) Representative immunofluorescence staining of COS7 cells, illustrating the effect of Drp1 variants on the ability of HSD17B10 to promote mitochondrial elongation (b), and quantification of these effects by evaluations of the proportion of cells with elongated mitochondria (n=200–300 cells from three independent experiments) (c) and the aspect ratio of mitochondria (n=20 cells from one experiment representative of three carried out) (d). Scale bar: 10 μm. (e) Quantification of Parkin-dependent mitophagy by analysis of the loss of the matrix marker PMPCB triggered by CCCP, in the indicated transfection conditions. n=100–200 cells from two independent experiments carried out. Results are expressed as means±S.E.M. **P<0.01, ***P<0.001 versus ‘−Drp1' in (c, d)
The mitochondrion-protecting function of HSD17B10 requires mitochondrial elongation and not the enzymatic activity of the protein
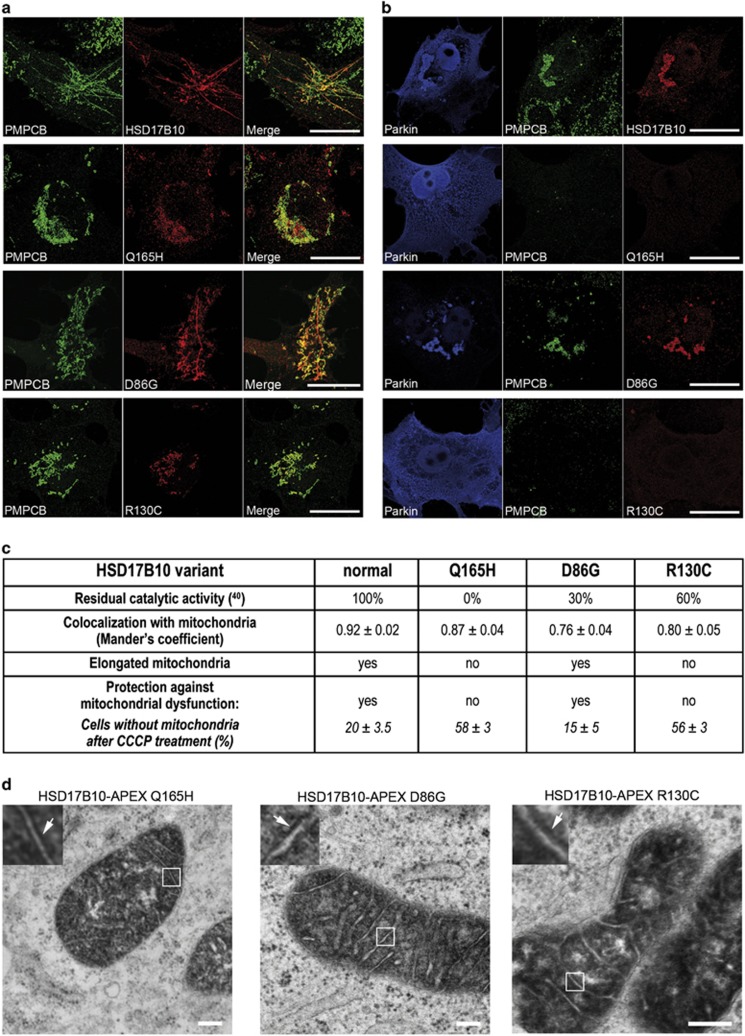
The clinical symptoms of patients with HSD17B10 mutations are not correlated with the activity of the encoded enzyme.24, 26 Moreover, the HSD17B10-dependent preservation of mitochondrial structural and functional integrity is independent of this activity, suggesting that another, non-enzymatic function of HSD17B10 is required.26 We explored the mechanisms by which HSD17B10 preserves mitochondrial function, by investigating the effects of three amino-acid substitutions in HSD17B10 with different impacts on catalytic activity (D86G, Q165H, R130C).26 The three protein variants were, as expected, located in the mitochondria, as indicated by the corresponding Mander's coefficients (Figures 8a and c); like the normal protein, they were found in the mitochondrial matrix, sometimes associated with the inner MM (Figure 8d). However, the Q165H and R130C variants, which had no enzymatic activity and a residual activity of 60%, respectively, were unable to promote mitochondrial elongation and to mitigate Parkin-dependent mitochondrial clearance. By contrast, the D86G variant (residual activity of 30%) behaved like the normal protein (Figures 8a and c). These results suggest that the mitochondrion-protecting function of HSD17B10 is directly correlated with the ability of this protein to promote mitochondrial elongation, and independent of its catalytic activity.
Figure 8.
Mitochondrial elongation rather than catalytic activity determines the ability of HSD17B10 to spare mitochondria from the degradation triggered by CCCP. (a–c) Representative immunofluorescence staining of COS7 cells, illustrating the degree to which HSD17B10 variants with various levels of residual activity can promote mitochondrial elongation (a), and protect against CCCP-induced mitochondrial dysfunction (b), as illustrated by the qualitative and quantitative estimates provided in the table (c). Mitochondria were visualized with HSD17B10 and PMPCB. Mander's coefficients indicate the colocalization of HSD17B10 and PMPCB staining (n=30 cells from three independent experiments). The percentages of COS7 cells coproducing Parkin and the indicated HSD17B10 variants displaying a loss of PMPCB 36 h after the addition of CCCP to the culture medium are represented as means±S.E.M. (n=3 independent experiments, with 60 cells scored per condition in each experiment). (d) Representative electron micrographs showing the submitochondrial localization of the HSD17B10 variants. Contrast was generated by treatment with diaminobenzidine. The arrow in the higher magnification of the framed region shows the association of HSD17B10 with the inner MM. Scale bar: 10 μm in (a, b) and 200 nm in (d)
Discussion
We identified the multifunctional mitochondrial matrix enzyme HSD17B10 as a new Parkin substrate. Using affinity purification-mass spectrometry to identify the repertoire of Parkin substrates in response to mitochondrial depolarization, Sarraf et al.18 also identified HSD17B10 as a protein directly interacting with Parkin. Contrary to reports for other mitochondrial proteins, this interaction was not amplified by CCCP treatment, suggesting that its occurrence is not related to the initiation of mitophagy. Instead, Parkin seems to maintain appropriate levels of HSD17B10 in the mitochondria by promoting the mitochondrial import of this protein: (i) the abundance of endogenous HSD17B10 was halved in mitochondrion-enriched fractions from Parkin-deficient mice; (ii) the mitochondrial targeting of an exogenous HSD17B10 protein was significantly reduced by PARK2 or PINK1 downregulation, whereas it was enhanced by Parkin overproduction in various cell models; (iii) Parkin and HSD17B10 associated with the TOM machinery, close to PINK1, on functional organelles that did not undergo mitophagy; (iv) interaction with the TOM complex was detected under conditions associated with PINK1 accumulation on the OMM (HSD17B10 overproduction or CCCP treatment), consistent with unmasking due to the mitochondrial import mechanism being overwhelmed. Whether other conditions of relevance for neurodegeneration in PD would also unmask this quality control function of Parkin – e.g. mitochondrial complex I inhibition, which has also been shown to trigger Parkin recruitment to mitochondria,52 – remains to be investigated.
The precise mechanisms by which Parkin facilitates the intramitochondrial translocation of HSD17B10 remain unclear, but our results suggest that the protein is ubiquitylated at the OMM before import. The cytosolic degradation, by the ubiquitin-proteasome system, of proteins targeted to the mitochondrial intermembrane space regulates the biogenesis of these proteins.53 Our data are not consistent with a role for the ubiquitin-proteasome system in regulating HSD17B10 levels in mitochondria, instead suggesting that modification by ubiquitin chains that are not recognized by the proteasome on the OMM upregulates HSD17B10 biogenesis. It remains unclear whether HS17B10 is imported in a ubiquitylated state by the TOM complex.
Ubiquitin-dependent regulation of the TOM-dependent import of HSD17B10 by Parkin, possibly in cooperation with PINK1, may ensure the correct maintenance of HSD17B10 homeostasis in conditions of low mitochondrial import efficiency. The impairment of this regulatory loop may have detrimental consequences, as HSD17B10 is essential for mitochondrial physiology: the in vivo depletion of HSD17B10 in Xenopus and mice leads to ultrastructural and functional abnormalities of the mitochondria, and constitutive knockout of the gene results in early embryonic lethality in mice.26 Conversely, mice transgenic for human HSD17B10 are protected against MPTP-mediated dopaminergic neurodegeneration.31 HSD17B10 levels are low in post-mortem ventral midbrain samples from patients with sporadic PD,31 and we show here that they also tend to be low in caudate and putamen samples from PD patients with PARK2 mutations.
HSD17B10 transcripts are abundant in various nuclei of the basal ganglia and in regions of the human brain enriched in dopaminergic neurons.54 These transcripts are more abundant in the substantia nigra than in the less vulnerable ventral tegmental area (Supplementary Figure S7). HSD17B10 expression levels are particularly high in the locus coeruleus and in the dorsal motor nucleus of the vagus nerve, which are also affected by neurodegeneration in PD. HSD17B10 depletion beyond a certain threshold may therefore contribute to neuronal vulnerability in PD. Consistent with this possibility, Tieu et al.31 showed that the period after MPTP intoxication in mice during which the apoptotic response was strongest was accompanied by a significant, but reversible decrease in HSD17B10 abundance.
Of note, HSD17B10 levels are increased in the brains of individuals with Alzheimer's disease, indicating that maintenance of HSD17B10 homeostasis is crucial to cell physiology.25, 29 The mechanisms by which HSD17B10 contributes to amyloid β-mediated neurotoxicity are poorly understood. However, it is not surprising that modest alterations of the levels or activity of this multifunctional enzyme can contribute to neurodegeneration in different pathological contexts.25
What molecular mechanisms underlie the mitochondrion-protecting function of HSD17B10? We provide strong evidence for a key role of mitochondrial elongation. HSD17B10 may promote this process, possibly by interfering with Drp1 recruitment to mitochondria. This morphological change has been shown to optimize ATP production under critical circumstances, but the underlying mechanisms remain unclear.48, 49, 50 Tieu et al.31 reported higher levels of mitochondrial respiration and ATP production in brain mitochondria from HSD17B10-transgenic mice than in those from wild-type mice, following the partial inhibition of mitochondrial complex I. This effect was not associated with changes to the enzymatic activities of the respiratory chain complexes, but was accompanied by a redistribution of HSD17B10 from the mitochondrial matrix to the inner MM. The authors suggested that HSD17B10 might associate with mitochondrial complex I under such conditions. The submitochondrial localization of the protein reported here is consistent with HSD17B10 having functions both in the mitochondrial matrix and in association with the inner MM. HSD17B10 may also participate in the activation of ATP synthase, which is also present in the inner MM and has been reported to be activated in elongated mitochondria resistant to autophagic degradation.49 Finally, HSD17B10 may protect mitochondria through its role as a component of the mitochondrial RNAse P complex, which promotes the processing of mitochondrial tRNAs through mechanisms independent of the catalytic activity of HSD17B10 and NADH binding.28, 55 The link between these functions and the mitochondrial elongation promoted by HSD17B10 remains unclear. Interestingly, a recent study reported identification of a new mitophagy-independent pathway linking Parkin function to linear ubiquitylation of the NF-kB modulator NEMO, upregulation of Opa1 and protection against stress.56 Although modulation of HSD17B10 expression did not modify Opa1 levels in our study, it will be important to evaluate whether HSD17B10 has a role in this pathway.
In summary, this work suggests that the maintenance of appropriate HSD17B10 levels in the mitochondria may be a novel mitochondrial quality control mechanism regulated by Parkin. The loss of this protective function may, together with the impaired degradation of dysfunctional mitochondria, contribute to neuronal dysfunction in autosomal recessive PD. Further studies are required to dissect the precise mechanisms by which Parkin preserves mitochondrial HSD17B10 levels, and to determine the extent to which they involve functional cooperation with PINK1 and the direct regulation of mitochondrial import through the TOM machinery.
Materials and Methods
Mammalian expression vectors, cell culture and transfection
The human HSD17B10 cDNA (UniProt Q99714) was inserted into pcDNA3; the HSD17B10 variants D86G, R130C and Q165H, kindly provided by J Zschocke, were inserted into pSMART. The other vectors used were pcDNA3 (Invitrogen, Life Technologies, Saint-Aubin, France) encoding human HA- or FLAG-tagged Parkin, Parkin variants, p38/JTV-1/AIMP,57 HA-, Myc- or V5-tagged HSD17B10, or FLAG-tagged DLST (provided by Z Ronai); pTrc–His (Invitrogen) encoding HSD17B10; pET (Clontech, Takara Bio Europe, Saint-Germain en-Laye, France) encoding HA-tagged ubiquitin; pDsRed-LC3 (provided by G Kroemer); pCB6-HA-mitoGFP (COX VIII MTS) and pCB6-Myc-Drp1 (provided by M Rojo); pCB6-Myc-Drp1 S637D/S637A, produced by site-directed mutagenesis from pCB6-Myc-Drp1; pCI (Promega, Charbonnières, France) encoding C-terminally APEX (pea L-ascorbate peroxidase K14D, W41F, E112K mutant)58-tagged HSD17B10 and HSD17B10 variants D86G, Q165H and R130C; pEF-HA-UbiK0/Ubi (provided by Y Yarden). The resulting cDNAs were then inserted into the pCI expression vector (Promega) via the NheI and NotI restriction sites. The sequences of three plasmid constructs were checked with an ABI3730 XL. The siRNAs used were validated HSD17B10-specific siRNAs and AllStars negative control siRNA (Qiagen, Courtaboeuf, France; 20 nM), PINK1- and PARK2- specific stealth siRNAs (Invitrogen, 20 nM). PC12-tet-off-Parkin, HEK293T and COS7 cells were grown as previously described.22, 23 Cells were transfected in the presence of DMRIE-C (Invitrogen; COS7, PC12) or in the presence of Lipofectamine 2000 (Invitrogen; COS7, HEK293T) for siRNA. Cells were incubated with 10 μM CCCP (Sigma-Aldrich, Saint Quentin Fallavier, France) for the indicated time points; 15 μg/ml nocodazole (Calbiochem, Millipore, Guyancourt, France) or 400 nM cytochalasin D (Sigma-Aldrich) for 16 h; 1 μM epoxomicin (Sigma-Aldrich) for 16 h; 50 nM doxycycline (Dox; Sigma-Aldrich); 5 μg/ml oligomycin (Sigma-Aldrich).
Quantitative real-time RT-PCR
We checked the efficacy of the siRNA-mediated silencing of endogenous PARK2 in HEK293T cells, by isolating total RNA from cells transfected with control or PARK2 siRNA, using the RNeasy plus Mini Kit (Qiagen). RNA from each sample (1 μg) was reverse-transcribed to generate cDNA. Real-time PCR was performed with the LightCycler 480 System (Roche Applied Science, Penzberg, Germany) and the LightCycler 480 DNA SYBR Green I Master kit (Roche Life Science, Meylan, France). Results were analyzed with LightCycler 480 sw 1.5 quantification software. We used the actin gene for normalization. The primers used for PARK2 were as follows: forward 1, 5′-acaccaagcatcttccagctcA-3′ reverse 1, 5′-tttctccacggtctctgcac-3′ forward 2, 5′-cgcaacaaatagtcggaac-3′ reverse 2, 5′-gagggtcgtgaacaaactg-3′.
GST pulldown, subcellular fractionation and western blot analyses
GST pulldown assays were carried out as previously described.57 Cells were lysed in 20 mM Hepes, 225 mM mannitol, 75 mM sucrose, 1 mM EGTA, 0.5 mM DTT, with phosphatase (0.2 mM Na3VO4, 1 mg/ml NaF, 5.4 mg/ml ß-glycerophosphate) and protease inhibitors (Complete Cocktail, Roche), pH 7.0 and the lysate was passed through a 22G needle to obtain total protein extract (COS7, HeLa cells). For ubiquitylation assays, COS7 cells were lysed 48 h after transfection, in a co-IP buffer (50 mM Tris–HCl pH 8, 150 mM NaCl, 5% glycerol, 0.5% NP40, 0.2 mM Na3VO4, 4 mg/ml NaF, Complete protease inhibitor cocktail; Roche). Lysates were centrifuged at 4 °C for 30 min at 13 000 × g. Supernatants were precleared by incubation with protein G Sepharose (Amersham, Dominique Dutscher, Brumath, France) for 1 h at 4 °C, and incubated with 2 μg of monoclonal anti-myc antibodies conjugated to protein G-Sepharose beads for 2 h at 4 °C, with gentle shaking. Immunoprecipitated proteins were washed four times in the co-IP buffer and once in PBS and analyzed by western blotting with monoclonal anti-HA antibodies. Lysates were subjected to western blotting with an anti-myc or a monoclonal anti-Parkin antibody. In vitro ubiquitylation assays were performed as previously described.59 Mitochondrion-enriched fractions were obtained by differential centrifugation, from cells homogenized with a Dounce homogenizer (80 manual strokes) as previously described,22 or with a mitochondrion isolation kit used according to the manufacturer's instructions (MACS, Miltenyi Biotec, Mercoeur, France; Pierce Mitochondria Isolation kit, Pierce, Thermo Fisher Scientific, Villebon-sur-Yvette, France). Cytosolic fractions were obtained by ultracentrifugation of the supernatants obtained after isolation of the mitochondrion-enriched fractions at 186 000 × g for 1.5 h at 4 °C. Pure mitochondrial fractions were obtained from mouse brains that had been ground, washed and homogenized with a Potter homogenizer, in 6 ml of isolation buffer (225 mM mannitol, 75 mM sucrose, 1 mM EGTA, 5 mM Hepes, 1 mg/ml BSA; pH 7.2), and centrifuged at 1700 × g for 10 min at 4 °C. The supernatant was layered on top of a Ficoll gradient (5 ml of 10% Ficoll and 5 ml of 7.5% Ficoll) in sucrose buffer (0.3 M sucrose, 25 mM EGTA, 5 mM Hepes) and centrifuged at 80 000 × g for 30 min at 4 °C. Mitochondrial pellets were resuspended in isolation buffer. Human brain protein extracts from PD patients and controls were obtained from lyophilized tissue37 resuspended in 50 mM HEPES pH 7.5, 150 mM NaCl, 1% Triton X-100, 10% glycerol, 100 mM MgCl2, 0.1 M Na4VO3, 10 mM Na4P2O7 × 10 H2O, 100 mM NaF, 1 mM EGTA, with protease inhibitors, homogenized in a Piston Pellet Eppendorf, and centrifuged at 4 °C for 5 min at 9000 × g and for 45 min at 100 000 × g. The mitochondrial abundance of exogenous HSD17B10 was calculated as the ratio of HSD17B10 to mitoGFP in mitochondrion-enriched fractions, divided by that in total cell fractions ([HSD17B10/mitoGFP]M/[HSD17B10/mitoGFP]T), and the mitochondrial abundance of subunits (S) of respiratory chain complexes was calculated as [S/actin]M/[S/actin]T. The mean values for the Dox – /CCCP – conditions were arbitrarily set at approximately 1. Mitochondrion-enriched fractions, obtained by differential centrifugation, were digested with 1 μg of trypsin (Sigma-Aldrich) per mg of protein for 20 min at 37 °C. The antibodies used for western blotting are indicated in Supplementary Table 1. Proteins were visualized with fluorescent antibodies or by enhanced chemiluminescence (Pierce). Chemiluminescence/fluorescence signals were detected on a film (ECL, Amersham Hyperfilm) or captured with a KODAK Image Station 4000 MM (KODAK, Rochester, NY, USA) or Odyssey Imaging (Li-COR Biosciences, Lincoln, NE, USA) system, and quantified with KODAK MI Software 4.0, ImageJ software (NIH, Bethesda, MD, USA) or Li-COR Image Studio.
Immunocytochemistry, epifluorescence, confocal and electron microscopy
Cells were fixed and stained by standard procedures. The antibodies used are indicated in Supplementary Table 2. MitoTracker (DeepRed/CMX-ROS; final concentration 500 nM) and TMRM (final concentration 10 nM) were purchased from Invitrogen and used according to the manufacturer's instructions. Mitochondrial aggregates, the translocation of p62, LC3-DsRed or Parkin, and mitophagy were quantified with a Zeiss Axioplan 2 imaging epifluorescence microscope equipped with ExploraNova FluoUp Imaging software (4.2.2, Explora Nova, La Rochelle, France) and a × 100 oil immersion objective (NA 1.40). Mitophagy and mitochondrial aggregation were assessed as the percentages of Parkin-positive or -negative cells devoid of VDAC1 and PMPCB staining (clearance), and cells with aggregates immunoreactive for Parkin and mitoGFP, DLST, p62, LC3-DsRed or HSD17B10 (aggregation). MitoTracker intensities were quantified on images of 10 cells per condition, in each of three independent experiments, with ImageJ analysis software (NIH). TMRM intensities were acquired with an inverted Zeiss Axiovert 200 videomicroscope (Zeiss, Jena, Germany) and a × 20 oil immersion objective (Supplementary Figure S2e), or an inverted Nikon Elipse Ti microscope with a × 20 objective (Figure 7a). We evaluated the effect of HSD17B10 overproduction on the response of ΔΨm to CCCP (2 μM) after addition of the complex V inhibitor oligomycin (5 μg/ml) by acquiring images at 10 s intervals over a 12-min period. The mean mitochondrial TMRM fluorescence intensity per cell was estimated with the ImageJ software, by selecting the entire mitochondrial network of each cell. Colocalization images were generated with a Leica SP2 or an Olympus FV-1000 confocal microscope (×60 oil immersion objective, NA 1.35). For FRET, images of donor and acceptor fluorescence were acquired with a Leica SP2 or an Olympus FV-1000 confocal microscope (×60 oil immersion objective, NA 1.35). A region of interest was drawn with Leica Confocal Software (Leica) or with Olympus FV-1000 software (Olympus, Tokyo, Japan), on randomly chosen areas of acceptor fluorescence. This region of interest was then photobleached with a 543 nm helium neon laser. Donor (D) fluorescence (F) intensities (I) were quantified with ImageJ, and FRET efficiencies were determined as follows: % FRET=(ID postbleach−ID prebleach)/ID postbleach=FD postbleach>FD prebleach. The morphology of the mitochondrial network was analyzed quantitatively with the ImageJ software, as previously described,44, 47 after the acquisition of images with the Zeiss Axioplan 2 imaging epifluorescence microscope (×63 oil immersion objective; Figure 6) or the Olympus FV-1000 confocal microscope (×60 oil immersion objective; Figure 7). Quantitative analysis of the morphology of the mitochondrial network, visualized by PMPCB or HSD17B10 staining, was performed on 20–30 pictures of cells overproducing the proteins of interest, with the ImageJ software (NIH), as previously described.44, 47 Standard and immunoelectron microscopy were carried out as previously described, with a JEOL 1200 EX2 electron microscope (Peabody, Peabody, MA, USA) at 80 kV (Figures 3f and g and Figure 8d).23, 60 For the localization of HSD17B10-APEX proteins, the cells were kept at room temperature for all steps from fixation to resin infiltration, and processed as previously described.58 They were then analyzed with a Hitachi 120 kV HT 7700 transmission electron microscope (Hitachi, Tokyo, Japan) at 70 kV.
Statistical analyses
Two-way ANOVA and the Holm-Sidak method were used to analyze the effects of Parkin and CCCP treatment or UbiK0 overproduction on HSD17B10 abundance (Figures 2a and b); the modulation of HSD17B10 levels on the clearance of mitochondrial markers (Figure 5) or mitochondrial TMRM fluorescence over time after CCCP treatment (Figure 7a). One-way ANOVA and the Holm-Sidak method were used to analyze the effects of HSD17B10, DLST or mitoGFP on the mitochondrial translocation of Parkin (Figure 3b); PARK2 variants on the mitochondrial translocation of Parkin triggered by HSD17B10 (Figure 3c); ubiquitin and UbiK0 on mitochondrial aggregation (Figure 3e); donors on FRET efficiencies for a given acceptor (Figures 4 and 6e; Supplementary Figure S5a); PARK2 mutations on FRET efficiencies (Supplementary Figure S5b); CCCP or the modulation of HSD17B10 levels on TMRM intensities (Supplementary Figure S2e); modulation of HSD17B10 levels on PINK1 levels (Supplementary Figure S2f) or relative abundance of Opa1, Mfn2 or Drp1 (Figure 6d); Drp1 or Drp1 variants on mitochondrial morphology (Figures 7c and d); HSD17B10 on Parkin-dependent mitophagy, or Drp1 or Drp1 variants on the mitophagy-mitigating effect of HSD17B10 (Figure 7e). ANOVA on ranks and Dunn's method were used to compare the effects of transfection condition or CCCP treatment on the mitochondrial recruitment of p62 or LC3-DsRed (Supplementary Figure S3); PARK2 silencing or UbiK0 on HSD17B10 abundance (Figure 2d); modulation of HSD17B10 levels on mitochondrial network complexity (Figure 6b). Student's t test was used to analyze the effects of drugs on mitochondrial aggregation (Supplementary Figure S4); PINK1 silencing on the mitochondrial translocation of Parkin (Figure 3d); Parkin deficiency on HSD17B10 abundance (Figure 1a); PARK2 siRNA on PARK2 mRNA levels (Supplementary Figure S1a); PARK2 or PINK1 silencing on HSD17B10 abundance (Supplementary Figure S1b); epoxomicin on HSD17B10 abundance in the cytosol or mitochondria (Supplementary Figure S1c).
Acknowledgments
We thank M Rojo and A Lombès for helpful discussions, S Jacquier for technical assistance, the Plate-Forme d'Imagerie Cellulaire de la Pitié-Salpêtrière (PICPS), G Kroemer Y, Z Ronai, Y Yarden, M Rojo, J Zschocke for providing vectors, and the Queen Square Brain Bank (QSBB, Institute of Neurology, London) for providing human brain tissue. This work was supported by Institut national de la santé et de la recherche médicale, Association France Parkinson, Fondation de France (Engt 2007 006456, Engt 2012 00034508), Agence Nationale de la Recherche (ANR-08-MNPS-02-01) APOPIS (funded by the EU under the Sixth Framework Programme, Grant Agreement LSHM-CT-2003-503330), MEFOPA (funded by the EU under the 7th Framework Programme, Grant Agreement HEALTH-2009-241791), Fondation ICM, ‘Investissements d'avenir' ANR-10-IAIHU-06. GB and MJ were supported by a fellowship from the French Ministry of Higher Education and Research. GB was also supported by Fondation de France.
Author Contributions
GB and MJ designed, performed and analyzed the experiments and wrote the manuscript; ST designed, performed and analyzed the experiments; RF-M, HA-O, TSG and M-PM performed and analyzed the experiments; HT and AJL provided neuropathologically characterized human brain tissue; FC, CG and DG designed and provided APEX constructs and advice; KG and EAF performed the experiments, shared protocols and provided advice; AB jointly supervised the work and provided funding; OC supervised and coordinated the work, provided funding and wrote the manuscript.
Glossary
- APEX
pea L-ascorbate peroxidase (K14D, W41F, E112K) variant
- CCCP
carbonyl cyanide m-chlorophenylhydrazone
- ΔΨm
mitochondrial membrane potential
- DAB
diaminobenzidine
- DLST
dihydrolipoamide S-succinyl transferase
- Dox
doxycycline
- Drp1
Dynamin-related protein 1
- FRET
Förster's resonance energy transfer
- HSD17B10
17-β hydroxysteroid dehydrogenase type 10
- OMM
outer mitochondrial membrane
- MM
mitochondrial membrane
- PINK1
PTEN-induced putative kinase 1
- PMPCB
mitochondrial processing peptidase subunit B
- TOM
translocase of the outer membrane
- VDAC1
voltage-dependent anion channel 1
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies this paper on Cell Death and Differentiation website (http://www.nature.com/cdd)
Edited by L Scorrano
Supplementary Material
References
- Corti O, Lesage S, Brice A. What genetics tells us about the causes and mechanisms of Parkinson's disease. Physiol Rev. 2011;91:1161–1218. doi: 10.1152/physrev.00022.2010. [DOI] [PubMed] [Google Scholar]
- Byrd RA, Weissman AM. Compact Parkin only: insights into the structure of an autoinhibited ubiquitin ligase. EMBO J. 2013;32:2087–2089. doi: 10.1038/emboj.2013.158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark IE, Dodson MW, Jiang C, Cao JH, Huh JR, Seol JH, et al. Drosophila pink1 is required for mitochondrial function and interacts genetically with parkin. Nature. 2006;441:1162–1166. doi: 10.1038/nature04779. [DOI] [PubMed] [Google Scholar]
- Park J, Lee SB, Lee S, Kim Y, Song S, Kim S, et al. Mitochondrial dysfunction in Drosophila PINK1 mutants is complemented by parkin. Nature. 2006;441:1157–1161. doi: 10.1038/nature04788. [DOI] [PubMed] [Google Scholar]
- Deng H, Dodson MW, Huang H, Guo M. The Parkinson's disease genes pink1 and parkin promote mitochondrial fission and/or inhibit fusion in Drosophila. Proc Natl Acad Sci USA. 2008;105:14503–14508. doi: 10.1073/pnas.0803998105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poole AC, Thomas RE, Andrews LA, McBride HM, Whitworth AJ, Pallanck LJ. The PINK1/Parkin pathway regulates mitochondrial morphology. Proc Natl Acad Sci USA. 2008;105:1638–1643. doi: 10.1073/pnas.0709336105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y, Ouyang Y, Yang L, Beal MF, McQuibban A, Vogel H, et al. Pink1 regulates mitochondrial dynamics through interaction with the fission/fusion machinery. Proc Natl Acad Sci USA. 2008;105:7070–7075. doi: 10.1073/pnas.0711845105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narendra D, Tanaka A, Suen DF, Youle RJ. Parkin is recruited selectively to impaired mitochondria and promotes their autophagy. J Cell Biol. 2008;183:795–803. doi: 10.1083/jcb.200809125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geisler S, Holmstrom KM, Skujat D, Fiesel FC, Rothfuss OC, Kahle PJ, et al. PINK1/Parkin-mediated mitophagy is dependent on VDAC1 and p62/SQSTM1. Nat Cell Biol. 2010;12:119–131. doi: 10.1038/ncb2012. [DOI] [PubMed] [Google Scholar]
- Matsuda N, Sato S, Shiba K, Okatsu K, Saisho K, Gautier CA, et al. PINK1 stabilized by mitochondrial depolarization recruits Parkin to damaged mitochondria and activates latent Parkin for mitophagy. J Cell Biol. 2010;189:211–221. doi: 10.1083/jcb.200910140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narendra DP, Jin SM, Tanaka A, Suen DF, Gautier CA, Shen J, et al. PINK1 is selectively stabilized on impaired mitochondria to activate Parkin. PLoS Biol. 2010;8:e1000298. doi: 10.1371/journal.pbio.1000298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vives-Bauza C, Zhou C, Huang Y, Cui M, de Vries RL, Kim J, et al. PINK1-dependent recruitment of Parkin to mitochondria in mitophagy. Proc Natl Acad Sci USA. 2010;107:378–383. doi: 10.1073/pnas.0911187107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka A, Cleland MM, Xu S, Narendra DP, Suen DF, Karbowski M, et al. Proteasome and p97 mediate mitophagy and degradation of mitofusins induced by Parkin. J Cell Biol. 2010;191:1367–1380. doi: 10.1083/jcb.201007013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan NC, Salazar AM, Pham AH, Sweredoski MJ, Kolawa NJ, Graham RL, et al. Broad activation of the ubiquitin-proteasome system by Parkin is critical for mitophagy. Hum Mol Genet. 2011;20:1726–1737. doi: 10.1093/hmg/ddr048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshii SR, Kishi C, Ishihara N, Mizushima N. Parkin mediates proteasome-dependent protein degradation and rupture of the outer mitochondrial membrane. J Biol Chem. 2011;286:19630–19640. doi: 10.1074/jbc.M110.209338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okatsu K, Iemura S, Koyano F, Go E, Kimura M, Natsume T, et al. Mitochondrial hexokinase HKI is a novel substrate of the Parkin ubiquitin ligase. Biochem Biophys Res Commun. 2012;428:197–202. doi: 10.1016/j.bbrc.2012.10.041. [DOI] [PubMed] [Google Scholar]
- Sun Y, Vashisht AA, Tchieu J, Wohlschlegel JA, Dreier L. Voltage-dependent anion channels (VDACs) recruit Parkin to defective mitochondria to promote mitochondrial autophagy. J Biol Chem. 2012;287:40652–40660. doi: 10.1074/jbc.M112.419721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarraf SA, Raman M, Guarani-Pereira V, Sowa ME, Huttlin EL, Gygi SP, et al. Landscape of the PARKIN-dependent ubiquitylome in response to mitochondrial depolarization. Nature. 2013;496:372–376. doi: 10.1038/nature12043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCoy MK, Kaganovich A, Rudenko IN, Ding J, Cookson MR. Hexokinase activity is required for recruitment of parkin to depolarized mitochondria. Hum Mol Genet. 2014;23:145–156. doi: 10.1093/hmg/ddt407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y, Dorn GW., 2nd PINK1-phosphorylated mitofusin 2 is a Parkin receptor for culling damaged mitochondria. Science. 2013;340:471–475. doi: 10.1126/science.1231031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lazarou M, Jin SM, Kane LA, Youle RJ. Role of PINK1 binding to the TOM complex and alternate intracellular membranes in recruitment and activation of the E3 ligase Parkin. Dev Cell. 2012;22:320–333. doi: 10.1016/j.devcel.2011.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertolin G, Ferrando-Miguel R, Jacoupy M, Traver S, Grenier K, Greene AW, et al. The TOMM machinery is a molecular switch in PINK1 and PARK2/PARKIN-dependent mitochondrial clearance. Autophagy. 2013;9:1801–1817. doi: 10.4161/auto.25884. [DOI] [PubMed] [Google Scholar]
- Darios F, Corti O, Lucking CB, Hampe C, Muriel MP, Abbas N, et al. Parkin prevents mitochondrial swelling and cytochrome c release in mitochondria-dependent cell death. Hum Mol Genet. 2003;12:517–526. doi: 10.1093/hmg/ddg044. [DOI] [PubMed] [Google Scholar]
- Zschocke J. HSD10 disease: clinical consequences of mutations in the HSD17B10 gene. J Inherit Metab Dis. 2012;35:81–89. doi: 10.1007/s10545-011-9415-4. [DOI] [PubMed] [Google Scholar]
- Yang SY, He XY, Isaacs C, Dobkin C, Miller D, Philipp M. Roles of 17beta-hydroxysteroid dehydrogenase type 10 in neurodegenerative disorders. J Steroid Biochem Mol Biol. 2014;143:460–472. doi: 10.1016/j.jsbmb.2014.07.001. [DOI] [PubMed] [Google Scholar]
- Rauschenberger K, Scholer K, Sass JO, Sauer S, Djuric Z, Rumig C, et al. A non-enzymatic function of 17beta-hydroxysteroid dehydrogenase type 10 is required for mitochondrial integrity and cell survival. EMBO Mol Med. 2010;2:51–62. doi: 10.1002/emmm.200900055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zschocke J, Ruiter JP, Brand J, Lindner M, Hoffmann GF, Wanders RJ, et al. Progressive infantile neurodegeneration caused by 2-methyl-3-hydroxybutyryl-CoA dehydrogenase deficiency: a novel inborn error of branched-chain fatty acid and isoleucine metabolism. Pediatr Res. 2000;48:852–855. doi: 10.1203/00006450-200012000-00025. [DOI] [PubMed] [Google Scholar]
- Holzmann J, Frank P, Loffler E, Bennett KL, Gerner C, Rossmanith W. RNase P without RNA: identification and functional reconstitution of the human mitochondrial tRNA processing enzyme. Cell. 2008;135:462–474. doi: 10.1016/j.cell.2008.09.013. [DOI] [PubMed] [Google Scholar]
- Yan SD, Fu J, Soto C, Chen X, Zhu H, Al-Mohanna F, et al. An intracellular protein that binds amyloid-beta peptide and mediates neurotoxicity in Alzheimer's disease. Nature. 1997;389:689–695. doi: 10.1038/39522. [DOI] [PubMed] [Google Scholar]
- Lustbader JW, Cirilli M, Lin C, Xu HW, Takuma K, Wang N, et al. ABAD directly links Abeta to mitochondrial toxicity in Alzheimer's disease. Science. 2004;304:448–452. doi: 10.1126/science.1091230. [DOI] [PubMed] [Google Scholar]
- Tieu K, Perier C, Vila M, Caspersen C, Zhang HP, Teismann P, et al. L-3-hydroxyacyl-CoA dehydrogenase II protects in a model of Parkinson's disease. Ann Neurol. 2004;56:51–60. doi: 10.1002/ana.20133. [DOI] [PubMed] [Google Scholar]
- Goldberg MS, Fleming SM, Palacino JJ, Cepeda C, Lam HA, Bhatnagar A, et al. Parkin-deficient mice exhibit nigrostriatal deficits but not loss of dopaminergic neurons. J Biol Chem. 2003;278:43628–43635. doi: 10.1074/jbc.M308947200. [DOI] [PubMed] [Google Scholar]
- Itier JM, Ibanez P, Mena MA, Abbas N, Cohen-Salmon C, Bohme GA, et al. Parkin gene inactivation alters behaviour and dopamine neurotransmission in the mouse. Hum Mol Genet. 2003;12:2277–2291. doi: 10.1093/hmg/ddg239. [DOI] [PubMed] [Google Scholar]
- Palacino JJ, Sagi D, Goldberg MS, Krauss S, Motz C, Wacker M, et al. Mitochondrial dysfunction and oxidative damage in parkin-deficient mice. J Biol Chem. 2004;279:18614–18622. doi: 10.1074/jbc.M401135200. [DOI] [PubMed] [Google Scholar]
- Von Coelln R, Thomas B, Savitt JM, Lim KL, Sasaki M, Hess EJ, et al. Loss of locus coeruleus neurons and reduced startle in parkin null mice. Proc Natl Acad Sci USA. 2004;101:10744–10749. doi: 10.1073/pnas.0401297101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perez FA, Palmiter RD. Parkin-deficient mice are not a robust model of parkinsonism. Proc Natl Acad Sci USA. 2005;102:2174–2179. doi: 10.1073/pnas.0409598102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Periquet M, Corti O, Jacquier S, Brice A. Proteomic analysis of parkin knockout mice: alterations in energy metabolism, protein handling and synaptic function. J Neurochem. 2005;95:1259–1276. doi: 10.1111/j.1471-4159.2005.03442.x. [DOI] [PubMed] [Google Scholar]
- Damiano M, Gautier CA, Bulteau AL, Ferrando-Miguel R, Gouarne C, Paoli MG, et al. Tissue- and cell-specific mitochondrial defect in Parkin-deficient mice. PLoS One. 2014;9:e99898. doi: 10.1371/journal.pone.0099898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greene AW, Grenier K, Aguileta MA, Muise S, Farazifard R, Haque ME, et al. Mitochondrial processing peptidase regulates PINK1 processing, import and Parkin recruitment. EMBO Rep. 2012;13:378–385. doi: 10.1038/embor.2012.14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin SM, Youle RJ. The accumulation of misfolded proteins in the mitochondrial matrix is sensed by PINK1 to induce PARK2/Parkin-mediated mitophagy of polarized mitochondria. Autophagy. 2013;9:1750–1757. doi: 10.4161/auto.26122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rojo M, Legros F, Chateau D, Lombes A. Membrane topology and mitochondrial targeting of mitofusins, ubiquitous mammalian homologs of the transmembrane GTPase Fzo. J Cell Sci. 2002;115:1663–1674. doi: 10.1242/jcs.115.8.1663. [DOI] [PubMed] [Google Scholar]
- Chacinska A, Koehler CM, Milenkovic D, Lithgow T, Pfanner N. Importing mitochondrial proteins: machineries and mechanisms. Cell. 2009;138:628–644. doi: 10.1016/j.cell.2009.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snapp EL, Hegde RS. Rational design and evaluation of FRET experiments to measure protein proximities in cells. Curr Protoc Cell Biol. 2006;Chapter 17:Unit 17 19. doi: 10.1002/0471143030.cb1709s32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koopman WJ, Visch HJ, Verkaart S, van den Heuvel LW, Smeitink JA, Willems PH. Mitochondrial network complexity and pathological decrease in complex I activity are tightly correlated in isolated human complex I deficiency. Am J Physiol Cell Physiol. 2005;289:C881–C890. doi: 10.1152/ajpcell.00104.2005. [DOI] [PubMed] [Google Scholar]
- Detmer SA, Chan DC. Functions and dysfunctions of mitochondrial dynamics. Nat Rev Mol Cell Biol. 2007;8:870–879. doi: 10.1038/nrm2275. [DOI] [PubMed] [Google Scholar]
- Twig G, Elorza A, Molina AJ, Mohamed H, Wikstrom JD, Walzer G, et al. Fission and selective fusion govern mitochondrial segregation and elimination by autophagy. EMBO J. 2008;27:433–446. doi: 10.1038/sj.emboj.7601963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buhlman L, Damiano M, Bertolin G, Ferrando-Miguel R, Lombes A, Brice A, et al. Functional interplay between Parkin and Drp1 in mitochondrial fission and clearance. Biochim Biophys Acta. 2014;1843:2012–2026. doi: 10.1016/j.bbamcr.2014.05.012. [DOI] [PubMed] [Google Scholar]
- Tondera D, Grandemange S, Jourdain A, Karbowski M, Mattenberger Y, Herzig S, et al. SLP-2 is required for stress-induced mitochondrial hyperfusion. EMBO J. 2009;28:1589–1600. doi: 10.1038/emboj.2009.89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomes LC, Di Benedetto G, Scorrano L. During autophagy mitochondria elongate, are spared from degradation and sustain cell viability. Nat Cell Biol. 2011;13:589–598. doi: 10.1038/ncb2220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rambold AS, Kostelecky B, Elia N, Lippincott-Schwartz J. Tubular network formation protects mitochondria from autophagosomal degradation during nutrient starvation. Proc Natl Acad Sci USA. 2011;108:10190–10195. doi: 10.1073/pnas.1107402108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cereghetti GM, Stangherlin A, Martins de Brito O, Chang CR, Blackstone C, Bernardi P, et al. Dephosphorylation by calcineurin regulates translocation of Drp1 to mitochondria. Proc Natl Acad Sci USA. 2008;105:15803–15808. doi: 10.1073/pnas.0808249105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joselin AP, Hewitt SJ, Callaghan SM, Kim RH, Chung YH, Mak TW, et al. ROS-dependent regulation of Parkin and DJ-1 localization during oxidative stress in neurons. Hum Mol Genet. 2012;21:4888–4903. doi: 10.1093/hmg/dds325. [DOI] [PubMed] [Google Scholar]
- Bragoszewski P, Gornicka A, Sztolsztener ME, Chacinska A. The ubiquitin-proteasome system regulates mitochondrial intermembrane space proteins. Mol Cell Biol. 2013;33:2136–2148. doi: 10.1128/MCB.01579-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hawrylycz MJ, Lein ES, Guillozet-Bongaarts AL, Shen EH, Ng L, Miller JA, et al. An anatomically comprehensive atlas of the adult human brain transcriptome. Nature. 2012;489:391–399. doi: 10.1038/nature11405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vilardo E, Nachbagauer C, Buzet A, Taschner A, Holzmann J, Rossmanith W. A subcomplex of human mitochondrial RNase P is a bifunctional methyltransferase—extensive moonlighting in mitochondrial tRNA biogenesis. Nucleic Acids Res. 2012;40:11583–11593. doi: 10.1093/nar/gks910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muller-Rischart AK, Pilsl A, Beaudette P, Patra M, Hadian K, Funke M, et al. The E3 ligase parkin maintains mitochondrial integrity by increasing linear ubiquitination of NEMO. Mol Cell. 2013;49:908–921. doi: 10.1016/j.molcel.2013.01.036. [DOI] [PubMed] [Google Scholar]
- Hampe C, Ardila-Osorio H, Fournier M, Brice A, Corti O. Biochemical analysis of Parkinson's disease-causing variants of Parkin, an E3 ubiquitin-protein ligase with monoubiquitylation capacity. Hum Mol Genet. 2006;15:2059–2075. doi: 10.1093/hmg/ddl131. [DOI] [PubMed] [Google Scholar]
- Martell JD, Deerinck TJ, Sancak Y, Poulos TL, Mootha VK, Sosinsky GE, et al. Engineered ascorbate peroxidase as a genetically encoded reporter for electron microscopy. Nat Biotechnol. 2012;30:1143–1148. doi: 10.1038/nbt.2375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fallon L, Belanger CM, Corera AT, Kontogiannea M, Regan-Klapisz E, Moreau F, et al. A regulated interaction with the UIM protein Eps15 implicates parkin in EGF receptor trafficking and PI(3)K-Akt signalling. Nat Cell Biol. 2006;8:834–842. doi: 10.1038/ncb1441. [DOI] [PubMed] [Google Scholar]
- Escobar-Khondiker M, Hollerhage M, Muriel MP, Champy P, Bach A, Depienne C, et al. Annonacin, a natural mitochondrial complex I inhibitor, causes tau pathology in cultured neurons. J Neurosci. 2007;27:7827–7837. doi: 10.1523/JNEUROSCI.1644-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.