Abstract
Nuclear factor one (NFI) transcription factors are a group of site-specific DNA-binding proteins that are emerging as critical regulators of stem cell biology. During development NFIs promote the production of differentiated progeny at the expense of stem cell fate, with Nfi null mice exhibiting defects such as severely delayed brain and lung maturation, skeletomuscular defects and renal abnormalities, phenotypes that are often consistent with patients with congenital Nfi mutations. Intriguingly, recent research suggests that in adult tissues NFI factors play a qualitatively different role than during development, with NFIs serving to promote the survival and maintenance of slow-cycling adult stem cell populations rather than their differentiation. Here we review the role of NFI factors in development, largely focusing on their role as promoters of stem cell differentiation, and attempt to reconcile this with the emerging role of NFIs in adult stem cell niches.
Keywords: organogenesis, nuclear factor one, transcription factor, stem cell, niche, embryonic, development, adult
Introduction
A stem cell is an undifferentiated cell that has the capacity to retain stem cell identity through self-renewal, and to differentiate to generate multiple cell types (Weissman, 2000). Broadly speaking, there are two classes of stem cells, those present in the developing embryo and those resident in adult tissues. In the developing embryo, each organ system has a set of lineage restricted stem cells that proliferate in a continuous manner and which, through their differentiation, produce the post-mitotic cellular components of the particular organ system in a relatively short period of time. In contrast, resident adult tissue stem cells, which function to replace and repair tissue throughout life, comprise a relatively scarce and long-lived cellular population. Because of this, a proportion of these cells exist in a non-proliferative state (quiescence) for prolonged periods, thereby preserving the adult stem cell pool by preventing proliferative stress and precocious commitment to differentiation (Valcourt et al., 2012; Cheung and Rando, 2013).
A key determinant of stem cell proliferation and differentiation are the gene regulatory networks governed by transcription factors. One group of transcription factors that is highly expressed by stem cells during development, as well as by adult stem cells across a range of tissue types, is the NFI family. The first NFI factor isolated was described as a host-encoded protein required for the initiation of adenovirus replication (Nagata et al., 1982). Subsequently, four genes encoding Nfi family members in mammals were isolated, namely Nfia, Nfib, Nfic and Nfix (Rupp et al., 1990; Kruse et al., 1991). NFIs interact with double-stranded DNA as either hetero- or homodimers by binding to the palindromic sequence TTGGC(N5)GCCAA with high affinity, thereby activating or repressing gene transcription depending on the cellular context and gene promoter (for an in depth review of these topics see (Gronostajski, 2000)).
The expression pattern of NFIs during development was characterized over 15 years ago (Chaudhry et al., 1997); from this study and subsequent analyses it has been demonstrated that NFIs are highly expressed by embryonic stem and progenitor cells within the central nervous system (CNS), lung and skeletomuscular tissue, amongst others. This initial expression analysis, combined with the generation of Nfia null mice, provided the first indicators that Nfi genes are important regulators of stem cell biology during development (das Neves et al., 1999). Subsequent cellular and molecular characterization of Nfia, Nfib, Nfic and Nfix null mice (see Table 1) demonstrated that NFI factors play multiple roles during development that ultimately promote cellular differentiation, including activating cell-type specific programs of gene expression and repressing the transcription of genes encoding factors mediating stem cell self-renewal (Messina et al., 2010; Piper et al., 2010; Lajoie et al., 2014). Consistent with their role in promoting stem and progenitor cell differentiation during development, NFIs have been implicated in a number of developmental disorders (Lu et al., 2007; Malan et al., 2010; Priolo et al., 2012; Yoneda et al., 2012), and have been reported to act as tumor suppressors in some cancers, including medulloblastoma (Genovesi et al., 2013).
Table 1.
Summary of major phenotypes identified in Nfi null mice.
| Phenotype | Nfia KO | Nfib KO | Nfic KO | Nfvc KO |
|---|---|---|---|---|
| Survival | Majority die at birth (das Neves et al., 1999). | All die at birth (Grunder et al., 2002). | Survive to adulthood (Steele-Perkins et al., 2003). | Postnatal lethal (3–4 week) |
| CNS | Delayed glial and neuronal differentiation (Piper et al., 2010). Corpus callosum agenesis. | Delayed glial and neuronal differentiation (Barry et al., 2008; Betancourt et al., 2014; Piper et al., 2014). | Phenotype not examined. Weak expression detected in developing CNS (Chaudhry et al., 1997). | Delayed glial and neuronal differentiation (Heng et al., 2014). |
| Communicating hydrocephalus (das Neves et al., 1999). | Corpus callosum dysgenesis (Steele-Perkins et al., 2005). | Corpus callosum dysgenesis (Plachez et al., 2008). | ||
| Lung | No obvious phenotype. Expressed in developing lung (Steele-Perkins et al., 2005). | Severe lung hyperplasia. Die from respiratory defects (Steele-Perkins et al., 2005). | Phenotype not examined. Expressed in developing lung (Steele-Perkins et al., 2005). | Phenotype not examined. Expressed in developing lung (Steele-Perkins et al., 2005). |
| Muscle and skeletal tissue | No gross skeletal defects (das Neves et al., 1999). | Skeletal and muscular phenotype not examined. Expressed by muscle progenitors (Biressi et al., 2007b). | Skeletal and muscular phenotype not examined. Expressed by muscle progenitors (Biressi et al., 2007b). | Reduced and disorganized musculature (Messina et al., 2010). |
| Musculature not examined, expressed by muscle progenitors (Biressi et al., 2007b). | Kyphosis and reduced bone density (Driller et al., 2007). | |||
| Other | Abnormal ureteropelvic and ureterovesical junctions, bifid and megaureter (Lu et al., 2007). | - | Multiple tooth pathologies (Steele-Perkins et al., 2003). | - |
| Wound healing defects (Plasari et al., 2009). |
Recently, the development of novel in vitro models and the use of conditional knockout technologies have shown that NFIs are also important regulators of stem cell biology in adult tissues, including melanocyte stem cells within the hair follicle niche (Chang et al., 2013) and hematopoietic stem cells in adult bone marrow (Holmfeldt et al., 2013). Intriguingly, in these studies, loss-of-function analyses did not lead to a delay in the differentiation of the adult stem cell populations as would be expected based on the role of NFIs during development. Instead, the removal of NFI function led to the loss of stem cell quiescence, precocious differentiation and the loss, or cellular death, of the stem population. In this review we discuss the role of NFI factors in development, largely as promoters of differentiated states, and reconcile this with the emerging evidence for NFIs as mediators of quiescence and survival within adult stem cell niches.
NFIs drive stem and progenitor cell differentiation during development
Strong evidence points to a major role for NFIs during development in promoting differentiation at the expense of stem cell self-renewal. NFIs carry out this role by exerting multiple effects on stem cell populations that act cumulatively to promote differentiation. Evidence for this role is found across a range of tissue types, including the CNS (Barry et al., 2008; Piper et al., 2010; Heng et al., 2014), musculoskeletal system (Messina et al., 2010; Pistocchi et al., 2013) and lung (Hsu et al., 2011) as well as in a range of other contexts such as in the development of teeth (Park et al., 2007) and the mammary gland (Murtagh et al., 2003; Nilsson et al., 2006).
NFIs regulate both neuronal and glial lineages during CNS development
Within the developing CNS, neural stem cells give rise to post-mitotic cells in a temporally distinct manner, first generating neurons and subsequently glia. These post-mitotic cells then migrate away from the germinal zones of the developing brain and integrate into the emerging cellular layers, where they terminally differentiate (Kriegstein and Alvarez-Buylla, 2009). Results to date have shown that NFIA, NFIB and NFIX all have multifaceted roles in regulating neural stem and progenitor cell differentiation during development, including driving the differentiation of stem cells within the developing cerebral cortex and neuronal progenitors within the nascent cerebellum.
NFIs promote cortical neural stem cell differentiation
Radial glial cells are the principal class of stem cell in the developing cerebral cortex (dorsal telencephalon), generating the majority of the post-mitotic neurons and glia present in the mature cortex (Casper and McCarthy, 2006). During early development, radial glial cells, which are located in the cortical ventricular zone, predominantly divide symmetrically to expand the population of stem cells. As development progresses, radial glial cells switch to dividing asymmetrically, generating a secondary pool of progenitor cells called transit amplifying cells (or basal progenitors) that migrate to the subventricular zone region, followed by glia such as astrocytes and oligodendrocytes (Kriegstein and Alvarez-Buylla, 2009).
NFIs were first implicated in regulating the differentiation of radial glial cells by expression analyses. These studies revealed that NFIA, NFIB (Plachez et al., 2008), and NFIX (Campbell et al., 2008) are all expressed within the ventricular zone of the telencephalon from embryonic day 12 (E12) in mice until the end of corticogenesis. Subsequent examination of the neocortical and hippocampal phenotype of Nfia and Nfix null mice revealed that there was an expansion of the pool of radial glia from approximately E16 onwards, as identified through immunohistochemical staining for stem cell markers such as PAX6 and SOX2 (Piper et al., 2010; Heng et al., 2014). Similarly, in a recent study examining the neocortical and hippocampal phenotype of Nfib null mice, more PAX6-positive stem cells were identified within the ventricular zone, indicating that in mice lacking Nfib, cortical stem cell populations are also expanded (Betancourt et al., 2014). Despite the greater number of cortical radial glia within Nfia, Nfib and Nfix null mice at E16, these mouse lines do not display a concomitant increase in the expression of the intermediate progenitor cell marker TBR2 at this age (Piper et al., 2010; Betancourt et al., 2014; Heng et al., 2014); nor do they display increased expression of the astrocytic protein GFAP, which is instead dramatically reduced (Shu et al., 2003; Piper et al., 2010; Heng et al., 2014). Therefore, these data argue that in the absence of NFIs radial glial cells fail to differentiate down neuronal and glial lineages according to normal developmental timelines. This results in severe morphological defects such as the dramatically reduced size of the hippocampal dentate gyrus in postnatal Nfia and Nfix null animals (das Neves et al., 1999; Heng et al., 2014).
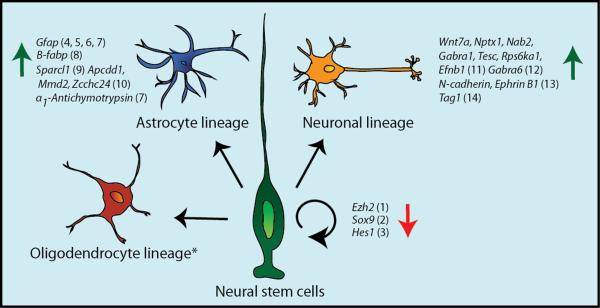
Molecular studies and expression analyses suggest that NFIs may promote the differentiation of radial glial cells into neurons and glia through a dual mechanism of directly inducing glial (and potentially neuronal) specific gene expression, while repressing the transcription of genes associated with the maintenance of stem cell populations (Fig. 1). While earlier studies showed a decrease in glial-specific markers in Nfia (das Neves et al., 1999) and Nfib null mice (Steele-Perkins et al., 2005), the first evidence that NFIs directly induce glial-specific gene expression came from rat cortical cell cultures. Here, a pan-NFI antibody was used to demonstrate that NFIs occupy the Gfap promoter prior to the induction of astrocyte differentiation, and that mutation of the NFI binding site correlates with reduced GFAP expression (Cebolla and Vallejo, 2006). Subsequent to this, using an in vitro model of cortical neural stem cells derived from mouse, Namihira et al. (2009) implicated NFIA as an intermediary factor in the canonical Notch/JAK/STAT pathway, which functions instructively during development to drive glial differentiation within the cortex. Specifically, they demonstrated that activation of Notch signaling induced NFIA expression in cortical neural stem cells, and that subsequent NFIA expression was correlated with disassociation of the inhibitory factor DNA methyltransferase 1 from the Gfap regulatory region, and with STAT3 binding site to the Gfap promoter. These findings illustrate that the regulation of promoter methylation is another means by which NFIs may induce expression of the Gfap locus. Intriguingly, recent studies in human neural progenitor cell cultures have further shown that the NFIX-3 splice variant is also a potent activator of the GFAP promoter, in this case through regulating nucleosome architecture and the recruitment of RNA polymerase to the transcription complex rather than via the modulation of DNA methylation (Singh et al., 2011). Together these findings illustrate the different ways in which NFIs can regulate the molecular pathway driving Gfap expression during glial differentiation. The precise mechanisms governing how NFI factors regulate the expression of other astrocyte specific genes currently thought to be downstream of NFIs, such as B-fabp (Bisgrove et al., 2000), Sparcl1 (Wilczynska et al., 2009), Apcdd1, Mmd2 and Zcchc24 (Kang et al., 2012) are yet to be determined.
Figure 1.
NFIs promote neural stem cell differentiation
NFIs repress the expression of genes associated with self-renewal of neural stem cells, and activate the expression of genes associated with neuronal and astrocyte differentiation. In this instance, NFI target genes were defined as misregulated genes identified via loss- or gain-of-function experiments which were further validated as downstream NFI targets via techniques such as ChIP-PCR, luciferase assays and gel-shift assays. *No direct NFI targets associated with the oligodendrocyte lineage have yet been identified. References: 1(Piper et al., 2014), 2(Heng et al., 2014), 3(Piper et al., 2010), 4(Cebolla and Vallejo, 2006), 5(Namihira et al., 2009), 6(Singh et al., 2011), 7(Gopalan et al., 2006), 8(Bisgrove et al., 2000), 9(Wilczynska et al., 2009), 10(Kang et al., 2012), 11(Ding et al., 2013), 12(Wang et al., 2004), 13(Wang et al., 2007), 14(Wang et al., 2010).
Repression of stem cell self-renewal pathways represents a second, complementary mechanism that may contribute to the delays in neuronal and glial development seen within Nfi null mice. Work from our group has shown that two genes implicated in cortical stem cell self-renewal, the epigenetic factor Ezh2 (Pereira et al., 2010) and the transcription factor Sox9 (Scott et al., 2010), are repressed by NFIB (Piper et al., 2014) and NFIX (Heng et al., 2014) respectively in vitro, and that the expression of these factors is upregulated in null mice. Similarly, we have also demonstrated that expression of the Hairy-Enhancer-of-Split (Hes) genes Hes1 and Hes5, which are key regulators of Notch signaling-induced stem cell self-renewal, are upregulated in Nfia and Nfib null mice (Piper et al., 2010), suggesting that NFIs may repress elements of the Notch pathway associated with self-renewal, while simultaneously co-operating with the JAK/STAT element of the Notch pathway that promotes glial differentiation (Figure 1). Collectively, these findings strongly support the idea that NFIs promote cortical stem cell differentiation through direct activation of cell-type-specific genes, while repressing the expression of loci that maintain stem cell fate. Further studies are required to delineate the specific functions of each individual NFI factor regulating the differentiation of cortical stem cell populations. Such studies will provide insight into whether it is the overall level of NFI factors that is crucial, or whether each individual factor regulates a unique set of genes. Next-generation genomic techniques such as chromatin immunoprecipitation followed by sequencing (ChIP-seq) using antibodies specific to individual NFI factors, combined with RNA sequencing (RNA-seq) of the different Nfi null mouse lines, have the capacity to provide these answers.
NFIs promote cerebellar granule neuron development
NFI proteins also play key roles as transcriptional regulators during the development of the most abundant neuron of the brain, the cerebellar granule neuron (reviewed in (Kilpatrick et al., 2012)). Towards the end of embryonic mouse development (~E18), granule neuron progenitors (GNPs) surround the primordial cerebellum, comprising a cell layer known as the external granular layer (EGL). This represents the main secondary germinal site for neurogenesis in the postnatal cerebellum. Granule neuron development from the EGL provides an elegant system in which to explore the molecules that mediate the switch from a proliferating neuronal progenitor cell to a terminally differentiated granule neuron (reviewed in (Martinez et al., 2013)). Specifically, GNPs proliferate in the outer EGL before exiting the cell cycle and commencing their migration inwards towards the internal granule layer (IGL). Within the EGL, GNPs begin to differentiate, extending bipolar axons that form fascicles of parallel fibres, and subsequently migrate tangentially before reaching the pre-migratory zone of the EGL. Here, GNPs extend long radial processes and migrate down through the molecular layer, to form the IGL. Within the IGL, GNPs complete the final stages of maturation and terminally differentiate, extending dendrites that form synapses with mossy fibres and additional neurons. Although a number of studies have contributed to our current understanding of the signaling pathways responsible for driving the proliferation of GNPs in the EGL (Dahmane and Ruiz i Altaba, 1999; Wallace, 1999; Lewis et al., 2004), what drives these cells to exit the cell cycle and ultimately differentiate to granule neurons remains poorly understood.
The NFI transcription factors have now been shown to play key roles in the progressive stages of post-mitotic GNP migration and differentiation. The identification of NFI proteins as directly regulating the transcription of Gabra6, a gene expressed in differentiated granule neurons residing in the IGL of the cerebellum (Kato, 1990), provided the first evidence of a role for the NFI family as transcriptional regulators in GNPs (Wang et al., 2004). Following this, expression analyses of cerebellar tissue isolated from post-natal day 7 (P7) mice suggested a role for these factors in the migration of GNPs to the IGL, with NFIA, NFIB and NFIX being expressed by GNPs residing in the deeper pre-migratory zone of the EGL, as well as by cells migrating through the molecular layer to the IGL (Wang et al., 2007). Using a variety of tools to manipulate NFI function in GNP culture in vitro, Wang et al. (2007) demonstrated that NFI proteins are critical for axon outgrowth and migration of post-mitotic GNPs. These findings were confirmed in P6 cerebellar slice cultures, in which the inhibition of NFI function induced the defasciculation of previously formed parallel fibres within the pre-migratory zone of the EGL, and subsequently impeded the radial migration of GNPs from the pre-migratory zone of the EGL to the IGL. In addition to these GNP differentiation events within the pre-migratory zone of the EGL, studies in ex vivo cerebellar slice cultures also revealed that the length and the number of dendrites formed from GNPs once they had migrated to the IGL was disrupted by blocking NFI function. The cerebellar phenotype of Nfia null mice is consistent with in vitro and ex vivo findings, with P17 Nfia null mice exhibiting shortened and disorientated parallel fibres and retarded migration of GNP cells within the cerebellum (Wang et al., 2007). Similarly, delayed GNP maturation was also observed in Nfix-deficient mice; however this was not thought to be a consequence of migratory defect of GNPs per se, but rather an overall delay in the development and differentiation of GNPs (Piper et al., 2011). Together these data strongly implicate NFI transcription factors in regulating various aspects of GNP maturation, including the switch from an immature, proliferating GNP to a post-mitotic GNP that has commenced its migration.
More recent studies have begun to elucidate the transcriptional targets of the NFIs that mediate both the early and late stages of GNP maturation. The cell adhesion molecules N-cadherin and ephrin-B1 have been identified as NFI gene targets responsible for mediating a range of differentiation defects, consistent with previous studies demonstrating their role in the migration of GNPs (Karam et al., 2000; Taniguchi et al., 2006). Functional inhibition of ephrin B1 or N-cadherin in GNPs in vitro and in ex vivo cerebellar slice cultures disrupts axon extension, migration and dendrite formation, with both proteins also reduced in migrating GNPs in the molecular layer and IGL of Nfia null mice (Wang et al., 2007). One subsequent study has identified an additional cell adhesion molecule, Tag1, as an NFI downstream target implicated in post-mitotic GNP differentiation (Wang et al., 2010). More recently, the NFI family has been demonstrated to play a central role in regulating a developmental switch program in GNPs during late stages of maturation, whereby genes required for mature granule neuron function, including dendrite and synapse formation, are up-regulated at the expense of genes expressed in more immature GNPs (Ding et al., 2013). This NFI-mediated late developmental switch was shown to be linked to the membrane potential of GNPs, whereby depolarization maintains immature GNPs via the activation of the calcineurin/ nuclear factor of activated T-cells, cytoplasmic (NFATc) pathway. NFATc was found to occupy and block the promoters of the late-expressed NFI genes. However, as GNPs begin to mature and undergo a hyperpolarizing shift in membrane potential, NFATc binding subsides, allowing the NFI family of proteins to bind to promoters of late-expressing genes in GNPs and induce dendrite formation.
Together, these data indicate the importance of NFI family members as critical regulators of a gene network that orchestrates several stages of GNP maturation, including axonogenesis, radial migration, dendritogenesis and synaptogenesis. Further advances are required to identify additional target genes of NFI family members in GNP development and to delineate the overlapping and non-overlapping roles of each NFI family member.
NFIs are implicated in medulloblastoma tumorigenesis
Given the central role of NFI family members in GNP differentiation, it is perhaps not surprising that NFI family members have recently been implicated in medulloblastoma, a disorder of GNP proliferation and differentiation. Medulloblastoma is a common malignant brain tumour of childhood, and certain subtypes of this disease have long been known to arise from GNPs of the EGL (Marino et al., 2000; Oliver et al., 2005; Schuller et al., 2008). Recently, a number of studies have identified NFI factors as being involved in medulloblastoma tumorigenesis, with transposon-mediated inactivation of Nfia, Nfib and/or Nfix shown to accelerate medulloblastoma initiation and/or progression in mouse models of medulloblastoma derived from GNPs (Wu et al., 2012; Genovesi et al., 2013; Lastowska et al., 2013). Indeed, a role for Nfia in medulloblastoma formation in vivo was recently supported by data showing that combined haploinsufficiency for both Nfia and Ptch1 exacerbated tumour development compared to the Ptch1 haploinsufficiency alone (Genovesi et al., 2013). Given the role of NFIA in the regulation of GNP differentiation, these findings further emphasize the link between the differentiation status of GNPs and the development of medulloblastoma. A similar role was previously observed for proneural transcription factor Math1, whereby the overexpression of Math1 blocked the trajectory of GNP differentiation, thereby significantly increasing tumour incidence and reducing tumour latency (Ayrault et al., 2010). Collectively, these studies demonstrate that medulloblastoma development is strongly linked to the differentiation status of GNPs. As such, further studies are required to assess the relevance of other NFI family members in medulloblastoma and to define the repertoire of target genes regulated by this family of transcription factors relevant to tumour development.
NFIX promotes the embryonic to fetal myoblast transition during musculoskeletal development
Another developmental context in which NFIs have been shown to drive the differentiation of stem and progenitor cells by activating cell-type-specific programs, while repressing the undifferentiated states of parental cells, is in the formation of the musculoskeletal system. With respect to muscle development, NFIX has been shown to activate an important transcriptional transition within muscle progenitors (myoblasts). Muscle development occurs in two stages, with each stage requiring distinct myoblast populations (Tajbakhsh, 2005; Biressi et al., 2007a). The first stage of muscle development or `primary myogenesis' occurs from E10–E12.5 in mice. During this initial phase a small fraction of the myoblast pool, referred to as `embryonic myoblasts', terminally differentiate, fuse and give rise to the multinucleated muscle fibers. The remaining myoblast population remains committed to the muscle lineage but exists in an undifferentiated state until secondary myogenesis takes place between E14.5 and E17.5 (Relaix et al., 2005). Myoblasts that participate in secondary myogenesis are called fetal myoblasts. Embryonic myoblasts and fetal myoblasts represent two distinct progenitor cell populations that produce muscle fibers that differ in morphology and in the myosin heavy chain (MyHC) isoforms and enzymes that they express (Barbieri et al., 1990; Zappelli et al., 1996; Ferrari et al., 1997; Biressi et al., 2007b).
To determine the molecular players that regulate this embryonic to fetal myoblast transcriptional switch, Biressi et al. (2007b) performed a genome wide-expression analysis on purified embryonic and fetal myoblasts and identified Nfi genes as potential candidates for this process. The expression of all four Nfi genes was robustly induced in fetal myoblasts, particularly Nfix, which was highly expressed in fetal myoblasts but virtually absent from the embryonic myoblast population. Functional analyses revealed that, similar to the role of NFIs in the development of the CNS, NFIX acted as a binary switch during the embryonic-fetal myoblast transition, inducing fetal-specific gene transcription while repressing the expression of genes associated with embryonic myoblasts. More specifically, conditional deletion of Nfix using a muscle-specific MyoD promoter prevented the initiation of fetal-specific transcription at E16; whereas in a gain-of-function mouse line overexpressing NFIX, fetal-specific genes were precociously expressed and embryonic myoblast gene expression was down-regulated (Messina et al., 2010). Molecular analysis demonstrated that these changes in gene expression were due to direct regulation by NFIX, with NFIX activating the expression of the fetal-specific gene Mck by forming a transcriptional complex with MEF2 and the kinase PKC-theta, and blocking the expression of the embryonic myoblast protein, slow MyHC, by suppressing its transcriptional activator NFATc4 (Calabria et al., 2009).
The musculature of Nfix loss- or gain-of-function embryos demonstrated the importance of correctly coordinating the embryonic-to-fetal transcriptional switch. Loss-of-function embryos were much smaller and the muscle fibers in hind-limb sections were disorganized, whereas in gain-of-function embryos the reciprocal phenotype was observed. Collectively, these molecular and anatomical experiments showed that NFIX plays a vital role during muscle development by activating the expression of fetal genes and repressing embryonic gene expression, a function that has recently been shown to be mostly conserved across evolutionary phyla (Pistocchi et al., 2013). Future experiments examining the role of NFIX in muscle development should examine whether NFIX regulates the differentiation of a third type of myogenic progenitor, the satellite cell. These cells act as adult stem cells that regenerate muscular tissue in response to injury or exercise (Collins et al., 2005; Biressi et al., 2007a). Due to the emerging role of Nfi genes in other adult stem cell niches (see Nfis in adult stem cell niches), this avenue of research is of particular interest.
NFIX also appears to have a role in the development of the skeleton. Mis-sense or dominant-negative mutations in Nfix are causative factors of Sotos syndrome and Marshall-Smith syndrome respectively (Malan et al., 2010; Yoneda et al., 2012). Both of these disorders are characterized by overgrowth and cognitive deficits, as well as skeletal defects such as kyphosis (curvature of the spine), advanced bone age and reduced bone density with increased propensity to fracturing. Although there is not currently a mechanistic understanding of how NFIX confers the skeletal phenotype of these disorders, similar skeletal defects including kyphosis and reduced bone density are observed in Nfix null mice; this mouse line may therefore provide a suitable model to probe this aspect of NFIX function (Driller et al., 2007; Harris et al., 2013).
Similarly, in addition to Nfix, Nfic has recently been shown to play an important role in postnatal bone formation. Nfic−/− mice were shown initially to have severe defects in postnatal tooth development (Steele-Perkins et al., 2003). These postnatal tooth defects appear to be mediated at least in part by antagonistic interactions between NFIC and the TGF-β signaling pathway during tooth development (Lee et al., 2011). In addition, NFIC interacts with TGF-β signaling during postnatal wound healing (Plasari et al., 2010) suggesting that NFIC-TGF-β interactions may represent a recurrent theme in postnatal NFIC function. More recently, NFIC has been shown to play a major role in postnatal osteoblast maturation through the regulation of the key osteoblast transcription factor Osterix (Lee et al., 2014). Of interest, no such effect on osteoblast formation was seen during fetal bone formation. Thus NFIC appears to have important roles in development primarily during the postnatal period.
Mesenchymal NFIB regulates lung development
NFIs have also been implicated in the development of a range of epithelial appendages, including the formation of teeth, and the development of the mammary glands (Kannius-Janson et al., 2002; Murtagh et al., 2003; Johansson et al., 2005), hair follicles (Plasari et al., 2010) and digestive tract (Driller et al., 2007). However, the best understood function of NFIs during epithelial appendage formation is during lung development, where NFIB has recently been shown to drive lung epithelial cell differentiation at the expense of stem and progenitor cell proliferation.
NFIB was linked to lung development by the observation that Nfib null mice die within 15 minutes postpartum due to respiratory stress, a phenotype not seen in other Nfi null mice (Grunder et al., 2002). Prenatal mouse lung development has four distinct stages. In the embryonic state (~E9.5–E10.5) the lung bud extends from the primitive gut endoderm and bifurcates. In the pseudoglandular stage (~E11–E16.5), the paired lung buds invade the mesenchyme to form an undifferentiated primordial bronchiole tree, and in the canalicular stage (E16.5–E17.5), the terminal sacs lined with epithelial cells develop, a process that is accelerated during the saccular stage (~E17.5–P5), during which further differentiation and diversification of epithelial cell sub-types occurs (Costa et al., 2001). Initial analyses of Nfib null mice demonstrated that lung development was apparently normal until E15.5, at which point development was arrested, with mutant lungs failing to form lung saccules (Steele-Perkins et al., 2005). Underpinning this gross histological phenotype was an increased proportion of proliferating cells in mutant lungs (Hsu et al., 2011; Holmfeldt et al., 2013) and a dramatic failure of differentiation of lung epithelial cell sub-types, including alveolar epithelial cells and bronchiolar exocrine cells in the distal lung, and ciliated cells in the proximal lung (Hsu et al., 2011). Collectively, these lines of evidence pointed to NFIB having a role in promoting lung epithelial differentiation at the expense of stem and progenitor proliferation during development.
Although the precise mechanism by which NFIB exerts this function is not clear, two recent studies have provided significant insights into this. Firstly, evidence from Hsu et al. (2011) suggests that NFIB may regulate lung epithelial cell differentiation though a non-cell-autonomous means. In this study the authors demonstrated that NFIB was predominantly expressed in the lung mesenchyme during development, and that conditional deletion of Nfib from the mesenchyme almost entirely recapitulated the defects in lung epithelial cell differentiation observed in Nfib null mice. Secondly, Lajoie et al. (2014) hypothesized that mesenchymal NFIB might regulate lung development in conjunction with the glucocorticoid receptor (GR). Similar to the phenotype in Nfib mutants, disruption of the gene encoding GR, Nr3c1, results in an immature lung phenotype in mice, with excess cellular proliferation and reduced expression of markers of epithelial cell differentiation (Cole et al., 1995). Using microarray data from lung tissue derived from both Nfib and Nr3c1 null mice at E18.5 the authors demonstrated that of the mis-regulated genes, 52 were under-expressed in both null lines, an overlap that was approximately 13 times larger than that expected by chance, suggesting that a subset of these genes may be directly activated by the coordinated activity of NFIB and GR proteins (Lajoie et al., 2014). These results are particularly significant as prenatal administration of glucocorticoids can stimulate lung maturation in premature infants (Seckl, 2004), suggesting that further characterization of the regulatory relationship between NFIB and GR may be of clinical value in treating lung immaturity.
Genetic redundancy of Nfis during development
Genetic redundancy is where two or more genes perform similar functions so that inactivation of one of these genes has no or little phenotypic effect. So far to date there is limited evidence that NFIs have redundant functions. For example, in the developing CNS inactivation of Nfia, Nfib or Nfix alone, is sufficient to render severe forebrain defects (Shu et al., 2003; Steele-Perkins et al., 2005; Campbell et al., 2008). Likewise in the developing cerebellum, NFIA, NFIB and NFIX are alone functionally required for normal development (Steele-Perkins et al., 2005; Wang et al., 2010; Piper et al., 2011). Interestingly however, these knockout mice still exhibit broadly similar phenotypes, suggesting that the overall level of NFI expression is more important than the function of individual Nfi genes during CNS development. In support of this, haploinsufficiency for NFIA or NFIX has been implicated as causative factors in neurodevelopmental disorders, and mice lacking one functional copy of Nfix display cognitive and neuroanatomical defects (Lu et al., 2007; Malan et al., 2010; Harris et al., 2013).
One potential scenario where NFIs may function redundantly is during lung development. Nfib null mice die from severe respiratory distress (Grunder et al., 2002), and, although Nfia, Nfic and Nfix are all expressed during lung development, none of these knockout mouse lines exhibit obvious lung defects. This indicates that loss of these factors alone is insufficient to adversely affect lung development, which could be due to genetic redundancy. One way to test this hypothesis would be to closely examine the lung phenotype of Nfia and Nfic (or Nfix) null mice. If neither of these null lines display clear lung phenotypes, the generation of a double Nfia/Nfic knockout mouse line with a subsequent lung phenotype would indicate redundancy.
NFIs in adult stem cell niches: a putative function in cell-cycle regulation and survival
In addition to being important regulators of stem cell biology during development, NFIs are highly expressed by adult tissue stem cells (see Table 2.). Most mammalian adult tissues contain a resident stem cell population, which functions to repair and regenerate tissue in response to stress or injury throughout the life of the organism (Cheung and Rando, 2013). A cardinal feature of adult tissue stem cells, even in high-turnover tissue such as the skin and the intestines, is that a proportion of the stem cell population resides outside the cell-cycle (being quiescent) for long periods of time, preventing metabolic and proliferative stress and thereby preserving genomic integrity and stem cell function (Valcourt et al., 2012).
Table 2.
Summary of known expression and/or function of NFIs in adult stem cell populations.
| Nfia | Nfib | Nfic | Nfix | |
|---|---|---|---|---|
| Adult neural stem cells | Expressed in stem cell niches of adult CNS. (Plachez et al., 2008; Plachez et al., 2012) | Expressed in stem cell niches of adult CNS. (Plachez et al., 2008; Plachez et al., 2012) | Expression not examined. | Expressed in stem cell niches of adult CNS. Implicated in neural stem cell quiescence. (Campbell et al., 2008; Martynoga et al., 2013) |
| Adult haematopoietic stem cells | Moderate-high expression in adult whole bone marrow. Loss of repopulating potential after knockdown (Holmfeldt et al., 2013). | Low expression. (Holmfeldt et al., 2013). | Moderate-high expression in adult whole bone marrow. (Holmfeldt et al., 2013). | Highly expressed in adult bone marrow. Promotes survival of HSCs and thereby repopulating potential. (Holmfeldt et al., 2013). |
| Hair follicle | No increase in transcript levels within HFSCs relative to progeny (Tumbar et al., 2004). | Transcript levels are elevated in HFSCs relative to progeny. (Tumbar et al., 2004). (Tumbar et al). | No increase in transcript levels within HFSCs relative to progeny (Tumbar et al., 2004). | No increase in transcript levels within HFSCs relative to progeny (Tumbar et al., 2004). |
| Coordinates quiescence in melanocyte stem cells (Chang et al., 2013). | ||||
| Adult muscle progenitors (satellite cells) | Expression unknown. | Expression unknown. | Expression unknown. | Expression unknown. |
The perinatal lethality of Nfia and Nfib null mice, and the severe developmental defects of viable Nfix null pups had until recently precluded analysis of NFI function in adult stem cell compartments. However, studies employing conditional knockout strategies and/or in vitro models have begun to uncover NFI function in these cellular populations. Intriguingly, these studies suggest that, in adult tissue, NFI factors play a contrasting role to that during development, serving to promote quiescence and/or survival of stem cells, rather than their differentiation (Chang et al., 2013; Holmfeldt et al., 2013; Martynoga et al., 2013).
NFIB coordinates quiescence in melanocyte stem cells
The first of these studies (Chang et al., 2013) examined the role of NFIB in stem cells of the hair follicle niche. Previously, NFIB expression had been shown to be elevated in hair follicle stem cells (HFSCs) relative to their differentiated progeny (hair cell), which led the authors to hypothesize that NFIB might be a key regulator of this niche (Tumbar et al., 2004). Normal hair production occurs in three phases: anagen (growth), catagen (cessation) and telogen (quiescence). Upon initiation of a new hair cycle (anagen phase), HFSCs and melanocyte stem cells enter the cell cycle in synchrony, enabling hair growth and pigmentation. Conditional deletion of Nfib from HFSCs failed to lead to HFSC stem cell lineage defects; instead, surprisingly, it led to melanocyte lineage abnormalities at the niche base. Specifically, differentiated melanocytes were ectopically found in the stem niche during the quiescent telogen phase. The authors demonstrated that these ectopic cells were caused by the precocious proliferation and differentiation of melanocyte stem cells, demonstrating that NFIB expression in HFSCs inductively coordinates quiescence in the melanocyte stem cell population during hair cycling, significantly increasing our understanding of how synchronous stem cell proliferation and differentiation within this adult hair follicle niche is regulated. Interestingly the authors found that upregulation of a single NFIB target, endothelin 2 (Edn2), identified through RNA-seq and ChIP-seq analyses was sufficient to phenocopy the effect of conditional Nfib deletion. As some endothelins such as Edn3 are crucial molecules for melanocyte specification during embryogenesis (Baynash et al., 1994), it will be important to examine whether NFIs also function in melanocyte development. Moreover, defining the role of NFIs within epithelial cancers remains an open question, with an initial study having found NFIB to be amplified and/or present at oncogenic chromosomal breakpoints in the epithelial based small cell lung cancer (Dooley et al., 2011).
NFIX is a mediator of quiescence in neural stem cells
As with NFIB within the hair follicle niche, in vitro studies have recently implicated NFIX in mediating quiescence in neural stem cells (Martynoga et al., 2013). Due to the relatively low turnover of stem cells in the adult brain, the majority of stem cells in the adult brain are quiescent (Suh et al., 2007). In an attempt to identify the transcription factors that regulate quiescence in adult neural stem cells, Martynoga and colleagues (2013), employed an in vitro model of neural stem cell quiescence. Specifically, they artificially induced a state of cellular quiescence by exposing a highly proliferative neural stem cell line derived from pluripotent embryonic stem cells to BMP4, a known cell-extrinsic mediator of adult neural stem cell quiescence (Mira et al., 2010; Sun et al., 2011). Epigenomic profiling was then used to identify active enhancer regions in both proliferating and quiescent neural stem cells. Subsequent motif analysis demonstrated that the binding domain of NFI transcription factors was highly enriched in active enhancer regions of the quiescent neural stem cell population. Next, using ChIP-seq with a pan-NFI antibody, the authors demonstrated that NFI proteins bound to 73% of the quiescent specific-enhancers, and that one member of the NFI family, NFIX, was robustly induced in quiescent neural stem cells, whereas NFIA and NFIB were down-regulated (Martynoga et al., 2013). These findings provided tantalizing evidence that NFIX actively regulates cellular quiescence within this paradigm, a hypothesis that was further supported by both gain-of-function experiments and by knockdown of Nfix expression through the use of RNAi molecules.
While this study strongly implicates NFIX as a central regulator of quiescence in neural stem cells, there are caveats to these findings. Firstly, modeling neural stem cell quiescence in vitro removes the influence of the cellular environment, which can dramatically alter stem cell behavior. Indeed, signals within the stem cell niches of the adult brain have been shown to greatly influence proliferation and differentiation of neural progenitor cells (Ming and Song, 2011). Moreover, as neural stem cell quiescence was modeled using a neural stem cell line derived from pluripotent embryonic stem cells, it is difficult to know how closely these findings recapitulate the epigenomic landscape of adult neural stem cells in vivo. The authors did attempt to address this limitation by examining the phenotype of Nfix null mice, and found a significantly reduced proportion of quiescent stem cells within the hippocampal dentate gyrus at P20. However, due to the significant developmental defects of this mutant line (see Table 1), it is difficult to definitively determine if the phenotype observed was due to a reduction in stem cell quiescence in the mutant, or was a reflection of the delayed maturation of the mutant dentate gyrus (Heng et al., 2014). Conditional knockout models to specifically delete Nfix from quiescent adult neural stem cell populations in vivo will provide the data required to determine if the findings in this in vitro model accurately reflect processes in the adult brain.
NFIX promotes survival of hematopoietic stem and progenitor cells
NFIX has also been shown to have a critical role in the survival of hematopoietic stem cells (HSCs) in adult bone (Holmfeldt et al., 2013). HSCs maintain hematopoiesis throughout life, self-renewing and also differentiating to give rise to all major lineages of the peripheral blood. Furthermore, after chemotherapy or irradiation, infused HSCs have the remarkable capacity to target the bone marrow stem cell niche and to repopulate the entire hematopoetic cellular cohort. Holmfeldt and colleagues (2013) found that NFIX was highly expressed in HSCs and that silencing of NFIX expression in HSCs greatly reduced repopulation capacity in lethally irradiated mice. Molecular analyses demonstrated that this phenotype was not due to the inability of NFIX-depleted HSCs to target the niche, but rather was a result of increased levels of apoptotic cell death after establishment within the niche. Consistent with this, Nfix knockdown led to significant down-regulation of genes associated with HSPC survival demonstrating that NFIX maintains the adult HSPC population post-transplantation by preventing apoptotic death through a cell-autonomous means. While these data clearly indicate a role for NFIX in survival of HSCs post-transplantation it is unclear whether NFIX is functionally important in steady-state (homeostatic) blood production. Indeed, because Nfix null mice survive up to 3 weeks after birth (see Table 1) this suggests that NFIX may be dispensable for homeostatic blood production. Conditional deletion of Nfix from HSCs in vivo, which will circumvent the premature lethality and skeletal defects of the Nfix null line would provide a suitable model to address this question.
NFI function during development versus NFI function in adult stem cells
While further testing of the different NFI factors across a broader range of adult stem cell niches is required, initial loss- or gain-of-function experiments suggest that NFIs serve to mediate quiescence and/or survival of adult stem cell populations (Chang et al., 2013; Holmfeldt et al., 2013; Martynoga et al., 2013). These findings appear inconsistent with one of the major functions of NFI factors during development, which is to promote stem and progenitor cell differentiation. What could account for the unexpected findings from these studies? The simplest explanation is that NFI factors or a subset of NFI factors regulate different sets of genes in adult stem cells compared with stem and progenitor populations during development. A powerful way to test this hypothesis could use next-generation sequencing techniques such as ChIP-seq on embryonically derived stem and progenitor cell populations, and compare these datasets with those ChIP-seq experiments performed on stem and progenitor populations isolated from adult tissue. A second potential way to test the hypothesis that NFI factors function differently in developmental versus adult contexts would be to examine pathologies in cancers of differential origin. For example, if NFIs drive differentiation during development, then it is a strong possibility that inactivating mutations in Nfi genes will be identified in developmental based cancers. Indeed, as discussed in this review, there are a number of recent studies that have shown a potential role for inactivating mutations in NFIs in contributing to medulloblastoma, the most common childhood brain cancer (Wu et al., 2012; Genovesi et al., 2013; Lastowska et al., 2013). It might therefore be interesting to determine whether NFIs act as oncogenes in adult-derived cancers, particularly as these cancers are thought to be caused by deregulated or chronic repair mechanisms, whereby stem cells are recruited to repopulate tissues. Currently, however, there are only a limited number of examples in which NFIs have been identified as putative oncogenes such as small cell lung cancer (Dooley et al., 2011) and glioblastoma (Glasgow et al., 2013). Furthermore, this approach is limited in that the high degree of heterogeneity within many forms of cancer may make the contribution of misregulated NFI expression to cancer progression difficult to ascertain. Therefore, a more fruitful approach to understand the differences in NFI function across developmental and adult contexts might be to introduce targeted mutations in a temporally controlled manner using conditional and inducible technologies.
Are there any phenotypic similarities in the NFI loss-of-function experiments performed during development versus those in adult stem cell niches? A common phenotype observed in Nfi null mice during development and in loss-of-function experiments performed in adult tissue is an initial increase in the number of proliferating cells. Considering the stem cell differentiation defects of Nfi null mice, this increase in proliferating cells during development is at least in part due to increased self-renewal, and delayed differentiation. However, it is yet to be directly tested as to whether the cell cycle is also deregulated amongst these stem cell populations, which could also contribute to the overall developmental phenotype of the null mice. Future research aimed at establishing to what extent NFIs directly regulate cell-cycle dynamics during development, as is observed in the adult, should yield further insight into the differences and similarities in NFI function across early developmental and adult contexts. This will enhance our understanding of the role of NFIs in stem cell biology more broadly, and the involvement of these transcription factors in developmental disorders and cancer.
Acknowledgements
We thank Rowan Tweedale for critical analysis of the manuscript.
Funding: This work was supported by National Health and Medical Research Council project grants (grant numbers 1003462, 1057751 and 1022308) to MP and by National Institute of Health and NYSTEM grants (grant numbers HL080624, C026714 and C026429 to RMG). The following authors were supported by fellowships: MP (Australian Research Council Future Fellowship; FT120100170); LH (Australian Postgraduate Award).
References
- Ayrault O, Zhao H, Zindy F, Qu C, Sherr CJ, Roussel MF. Atoh1 inhibits neuronal differentiation and collaborates with Gli1 to generate medulloblastoma-initiating cells. Cancer Res. 2010;70:5618–5627. doi: 10.1158/0008-5472.CAN-09-3740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barbieri G, De Angelis L, Feo S, Cossu G, Giallongo A. Differential expression of muscle-specific enolase in embryonic and fetal myogenic cells during mouse development. Differentiation. 1990;45:179–184. doi: 10.1111/j.1432-0436.1990.tb00471.x. [DOI] [PubMed] [Google Scholar]
- Barry G, Piper M, Lindwall C, Moldrich R, Mason S, Little E, Sarkar A, Tole S, Gronostajski RM, Richards LJ. Specific glial populations regulate hippocampal morphogenesis. J Neurosci. 2008;28:12328–12340. doi: 10.1523/JNEUROSCI.4000-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baynash AG, Hosoda K, Giaid A, Richardson JA, Emoto N, Hammer RE, Yanagisawa M. Interaction of endothelin-3 with endothelin-B receptor is essential for development of epidermal melanocytes and enteric neurons. Cell. 1994;79:1277–1285. doi: 10.1016/0092-8674(94)90018-3. [DOI] [PubMed] [Google Scholar]
- Betancourt J, Katzman S, Chen B. Nuclear factor one B regulates neural stem cell differentiation and axonal projection of corticofugal neurons. J Comp Neurol. 2014;522:6–35. doi: 10.1002/cne.23373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biressi S, Molinaro M, Cossu G. Cellular heterogeneity during vertebrate skeletal muscle development. Dev Biol. 2007a;308:281–293. doi: 10.1016/j.ydbio.2007.06.006. [DOI] [PubMed] [Google Scholar]
- Biressi S, Tagliafico E, Lamorte G, Monteverde S, Tenedini E, Roncaglia E, Ferrari S, Ferrari S, Cusella-De Angelis MG, Tajbakhsh S, Cossu G. Intrinsic phenotypic diversity of embryonic and fetal myoblasts is revealed by genome-wide gene expression analysis on purified cells. Dev Biol. 2007b;304:633–651. doi: 10.1016/j.ydbio.2007.01.016. [DOI] [PubMed] [Google Scholar]
- Bisgrove DA, Monckton EA, Packer M, Godbout R. Regulation of brain fatty acid-binding protein expression by differential phosphorylation of nuclear factor I in malignant glioma cell lines. J Biol Chem. 2000;275:30668–30676. doi: 10.1074/jbc.M003828200. [DOI] [PubMed] [Google Scholar]
- Calabria E, Ciciliot S, Moretti I, Garcia M, Picard A, Dyar KA, Pallafacchina G, Tothova J, Schiaffino S, Murgia M. NFAT isoforms control activity-dependent muscle fiber type specification. Proc Natl Acad Sci U S A. 2009;106:13335–13340. doi: 10.1073/pnas.0812911106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell CE, Piper M, Plachez C, Yeh YT, Baizer JS, Osinski JM, Litwack ED, Richards LJ, Gronostajski RM. The transcription factor Nfix is essential for normal brain development. BMC Dev Biol. 2008;8:52. doi: 10.1186/1471-213X-8-52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casper KB, McCarthy KD. GFAP-positive progenitor cells produce neurons and oligodendrocytes throughout the CNS. Mol Cell Neurosci. 2006;31:676–684. doi: 10.1016/j.mcn.2005.12.006. [DOI] [PubMed] [Google Scholar]
- Cebolla B, Vallejo M. Nuclear factor-I regulates glial fibrillary acidic protein gene expression in astrocytes differentiated from cortical precursor cells. J Neurochem. 2006;97:1057–1070. doi: 10.1111/j.1471-4159.2006.03804.x. [DOI] [PubMed] [Google Scholar]
- Chang CY, Pasolli HA, Giannopoulou EG, Guasch G, Gronostajski RM, Elemento O, Fuchs E. NFIB is a governor of epithelial-melanocyte stem cell behaviour in a shared niche. Nature. 2013;495:98–102. doi: 10.1038/nature11847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaudhry AZ, Lyons GE, Gronostajski RM. Expression patterns of the four nuclear factor I genes during mouse embryogenesis indicate a potential role in development. Dev Dyn. 1997;208:313–325. doi: 10.1002/(SICI)1097-0177(199703)208:3<313::AID-AJA3>3.0.CO;2-L. [DOI] [PubMed] [Google Scholar]
- Cheung TH, Rando TA. Molecular regulation of stem cell quiescence. Nat Rev Mol Cell Biol. 2013;14:329–340. doi: 10.1038/nrm3591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cole TJ, Blendy JA, Monaghan AP, Krieglstein K, Schmid W, Aguzzi A, Fantuzzi G, Hummler E, Unsicker K, Schutz G. Targeted disruption of the glucocorticoid receptor gene blocks adrenergic chromaffin cell development and severely retards lung maturation. Genes Dev. 1995;9:1608–1621. doi: 10.1101/gad.9.13.1608. [DOI] [PubMed] [Google Scholar]
- Collins CA, Olsen I, Zammit PS, Heslop L, Petrie A, Partridge TA, Morgan JE. Stem cell function, self-renewal, and behavioral heterogeneity of cells from the adult muscle satellite cell niche. Cell. 2005;122:289–301. doi: 10.1016/j.cell.2005.05.010. [DOI] [PubMed] [Google Scholar]
- Costa RH, Kalinichenko VV, Lim L. Transcription factors in mouse lung development and function. Am J Physiol Lung Cell Mol Physiol. 2001;280:L823–838. doi: 10.1152/ajplung.2001.280.5.L823. [DOI] [PubMed] [Google Scholar]
- Dahmane N, Ruiz i Altaba A. Sonic hedgehog regulates the growth and patterning of the cerebellum. Development. 1999;126:3089–3100. doi: 10.1242/dev.126.14.3089. [DOI] [PubMed] [Google Scholar]
- das Neves L, Duchala CS, Tolentino-Silva F, Haxhiu MA, Colmenares C, Macklin WB, Campbell CE, Butz KG, Gronostajski RM. Disruption of the murine nuclear factor I-A gene (Nfia) results in perinatal lethality, hydrocephalus, and agenesis of the corpus callosum. Proc Natl Acad Sci USA. 1999;96:11946–11951. doi: 10.1073/pnas.96.21.11946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding B, Wang W, Selvakumar T, Xi HS, Zhu H, Chow CW, Horton JD, Gronostajski RM, Kilpatrick DL. Temporal regulation of nuclear factor one occupancy by calcineurin/NFAT governs a voltage-sensitive developmental switch in late maturing neurons. J Neurosci. 2013;33:2860–2872. doi: 10.1523/JNEUROSCI.3533-12.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dooley AL, Winslow MM, Chiang DY, Banerji S, Stransky N, Dayton TL, Snyder EL, Senna S, Whittaker CA, Bronson RT, Crowley D, Barretina J, Garraway L, Meyerson M, Jacks T. Nuclear factor I/B is an oncogene in small cell lung cancer. Genes Dev. 2011;25:1470–1475. doi: 10.1101/gad.2046711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Driller K, Pagenstecher A, Uhl M, Omran H, Berlis A, Grunder A, Sippel AE. Nuclear factor I X deficiency causes brain malformation and severe skeletal defects. Mol Cell Biol. 2007;27:3855–3867. doi: 10.1128/MCB.02293-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferrari S, Molinari S, Melchionna R, Cusella-De Angelis MG, Battini R, De Angelis L, Kelly R, Cossu G. Absence of MEF2 binding to the A/T-rich element in the muscle creatine kinase (MCK) enhancer correlates with lack of early expression of the MCK gene in embryonic mammalian muscle. Cell Growth Differ. 1997;8:23–34. [PubMed] [Google Scholar]
- Genovesi LA, Ng CG, Davis MJ, Remke M, Taylor MD, Adams DJ, Rust AG, Ward JM, Ban KH, Jenkins NA, Copeland NG, Wainwright BJ. Sleeping Beauty mutagenesis in a mouse medulloblastoma model defines networks that discriminate between human molecular subgroups. Proc Natl Acad Sci U S A. 2013;110:E4325–4334. doi: 10.1073/pnas.1318639110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glasgow SM, Laug D, Brawley VS, Zhang Z, Corder A, Yin Z, Wong ST, Li XN, Foster AE, Ahmed N, Deneen B. The miR-223/nuclear factor I-A axis regulates glial precursor proliferation and tumorigenesis in the CNS. J Neurosci. 2013;33:13560–13568. doi: 10.1523/JNEUROSCI.0321-13.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gopalan SM, Wilczynska KM, Konik BS, Bryan L, Kordula T. Nuclear factor-1-X regulates astrocyte-specific expression of the alpha1-antichymotrypsin and glial fibrillary acidic protein genes. J Biol Chem. 2006;281:13126–13133. doi: 10.1074/jbc.M601194200. [DOI] [PubMed] [Google Scholar]
- Gronostajski RM. Roles of the NFI/CTF gene family in transcription and development. Gene. 2000;249:31–45. doi: 10.1016/s0378-1119(00)00140-2. [DOI] [PubMed] [Google Scholar]
- Grunder A, Ebel TT, Mallo M, Schwarzkopf G, Shimizu T, Sippel AE, Schrewe H. Nuclear factor I-B (Nfib) deficient mice have severe lung hypoplasia. Mech Dev. 2002;112:69–77. doi: 10.1016/s0925-4773(01)00640-2. [DOI] [PubMed] [Google Scholar]
- Harris L, Dixon C, Cato K, Heng YH, Kurniawan ND, Ullmann JF, Janke AL, Gronostajski RM, Richards LJ, Burne TH, Piper M. Heterozygosity for nuclear factor one x affects hippocampal-dependent behaviour in mice. PLoS One. 2013;8:e65478. doi: 10.1371/journal.pone.0065478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heng YH, McLeay RC, Harvey TJ, Smith AG, Barry G, Cato K, Plachez C, Little E, Mason S, Dixon C, Gronostajski RM, Bailey TL, Richards LJ, Piper M. NFIX regulates neural progenitor cell differentiation during hippocampal morphogenesis. Cereb Cortex. 2014;24:261–279. doi: 10.1093/cercor/bhs307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holmfeldt P, Pardieck J, Saulsberry AC, Nandakumar SK, Finkelstein D, Gray JT, Persons DA, McKinney-Freeman S. Nfix is a novel regulator of murine hematopoietic stem and progenitor cell survival. Blood. 2013;122:2987–2996. doi: 10.1182/blood-2013-04-493973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu YC, Osinski J, Campbell CE, Litwack ED, Wang D, Liu S, Bachurski CJ, Gronostajski RM. Mesenchymal nuclear factor I B regulates cell proliferation and epithelial differentiation during lung maturation. Dev Biol. 2011;354:242–252. doi: 10.1016/j.ydbio.2011.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johansson EM, Kannius-Janson M, Gritli-Linde A, Bjursell G, Nilsson J. Nuclear factor 1-C2 is regulated by prolactin and shows a distinct expression pattern in the mouse mammary epithelial cells during development. Mol Endocrinol. 2005;19:992–1003. doi: 10.1210/me.2004-0359. [DOI] [PubMed] [Google Scholar]
- Kang P, Lee HK, Glasgow SM, Finley M, Donti T, Gaber ZB, Graham BH, Foster AE, Novitch BG, Gronostajski RM, Deneen B. Sox9 and NFIA coordinate a transcriptional regulatory cascade during the initiation of gliogenesis. Neuron. 2012;74:79–94. doi: 10.1016/j.neuron.2012.01.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kannius-Janson M, Johansson EM, Bjursell G, Nilsson J. Nuclear factor 1-C2 contributes to the tissue-specific activation of a milk protein gene in the differentiating mammary gland. J Biol Chem. 2002;277:17589–17596. doi: 10.1074/jbc.M105979200. [DOI] [PubMed] [Google Scholar]
- Karam SD, Burrows RC, Logan C, Koblar S, Pasquale EB, Bothwell M. Eph receptors and ephrins in the developing chick cerebellum: relationship to sagittal patterning and granule cell migration. J Neurosci. 2000;20:6488–6500. doi: 10.1523/JNEUROSCI.20-17-06488.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato K. Novel GABAA receptor alpha subunit is expressed only in cerebellar granule cells. J Mol Biol. 1990;214:619–624. doi: 10.1016/0022-2836(90)90276-r. [DOI] [PubMed] [Google Scholar]
- Kilpatrick DL, Wang W, Gronostajski R, Litwack ED. Nuclear factor I and cerebellar granule neuron development: an intrinsic-extrinsic interplay. Cerebellum. 2012;11:41–49. doi: 10.1007/s12311-010-0227-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kriegstein A, Alvarez-Buylla A. The glial nature of embryonic and adult neural stem cells. Annu Rev Neurosci. 2009;32:149–184. doi: 10.1146/annurev.neuro.051508.135600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kruse U, Qian F, Sippel AE. Identification of a fourth nuclear factor I gene in chicken by cDNA cloning: NFI-X. Nucleic Acids Res. 1991;19:6641. doi: 10.1093/nar/19.23.6641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lajoie M, Hsu YC, Gronostajski RM, Bailey TL. An overlapping set of genes is regulated by both NFIB and the glucocorticoid receptor during lung maturation. BMC Genomics. 2014;15:231. doi: 10.1186/1471-2164-15-231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lastowska M, Al-Afghani H, Al-Balool HH, Sheth H, Mercer E, Coxhead JM, Redfern CP, Peters H, Burt AD, Santibanez-Koref M, Bacon CM, Chesler L, Rust AG, Adams DJ, Williamson D, Clifford SC, Jackson MS. Identification of a neuronal transcription factor network involved in medulloblastoma development. Acta Neuropathol Commun. 2013;1:35. doi: 10.1186/2051-5960-1-35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee DS, Choung HW, Kim HJ, Gronostajski RM, Yang YI, Ryoo HM, Lee ZH, Kim HH, Cho ES, Park JC. NFI-C regulates osteoblast differentiation via control of Osterix expression. Stem Cells. 2014;32:2467–2479. doi: 10.1002/stem.1733. [DOI] [PubMed] [Google Scholar]
- Lee DS, Yoon WJ, Cho ES, Kim HJ, Gronostajski RM, Cho MI, Park JC. Crosstalk between nuclear factor I-C and transforming growth factor-beta1 signaling regulates odontoblast differentiation and homeostasis. PLoS One. 2011;6:e29160. doi: 10.1371/journal.pone.0029160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis PM, Gritli-Linde A, Smeyne R, Kottmann A, McMahon AP. Sonic hedgehog signaling is required for expansion of granule neuron precursors and patterning of the mouse cerebellum. Dev Biol. 2004;270:393–410. doi: 10.1016/j.ydbio.2004.03.007. [DOI] [PubMed] [Google Scholar]
- Lu W, Quintero-Rivera F, Fan Y, Alkuraya FS, Donovan DJ, Xi Q, Turbe-Doan A, Li QG, Campbell CG, Shanske AL, Sherr EH, Ahmad A, Peters R, Rilliet B, Parvex P, Bassuk AG, Harris DJ, Ferguson H, Kelly C, Walsh CA, Gronostajski RM, Devriendt K, Higgins A, Ligon AH, Quade BJ, Morton CC, Gusella JF, Maas RL. NFIA haploinsufficiency is associated with a CNS malformation syndrome and urinary tract defects. PLoS Genet. 2007;3:e80. doi: 10.1371/journal.pgen.0030080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malan V, Rajan D, Thomas S, Shaw AC, Louis Dit Picard H, Layet V, Till M, van Haeringen A, Mortier G, Nampoothiri S, Puseljic S, Legeai-Mallet L, Carter NP, Vekemans M, Munnich A, Hennekam RC, Colleaux L, Cormier-Daire V. Distinct effects of allelic NFIX mutations on nonsense-mediated mRNA decay engender either a Sotos-like or a Marshall-Smith syndrome. Am J Hum Genet. 2010;87:189–198. doi: 10.1016/j.ajhg.2010.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marino S, Vooijs M, van Der Gulden H, Jonkers J, Berns A. Induction of medulloblastomas in p53-null mutant mice by somatic inactivation of Rb in the external granular layer cells of the cerebellum. Genes Dev. 2000;14:994–1004. [PMC free article] [PubMed] [Google Scholar]
- Martinez S, Andreu A, Mecklenburg N, Echevarria D. Cellular and molecular basis of cerebellar development. Front Neuroanat. 2013;7:18. doi: 10.3389/fnana.2013.00018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martynoga B, Mateo JL, Zhou B, Andersen J, Achimastou A, Urban N, van den Berg D, Georgopoulou D, Hadjur S, Wittbrodt J, Ettwiller L, Piper M, Gronostajski RM, Guillemot F. Epigenomic enhancer annotation reveals a key role for NFIX in neural stem cell quiescence. Genes Dev. 2013;27:1769–1786. doi: 10.1101/gad.216804.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Messina G, Biressi S, Monteverde S, Magli A, Cassano M, Perani L, Roncaglia E, Tagliafico E, Starnes L, Campbell CE, Grossi M, Goldhamer DJ, Gronostajski RM, Cossu G. Nfix regulates fetal-specific transcription in developing skeletal muscle. Cell. 2010;140:554–566. doi: 10.1016/j.cell.2010.01.027. [DOI] [PubMed] [Google Scholar]
- Ming GL, Song H. Adult neurogenesis in the mammalian brain: significant answers and significant questions. Neuron. 2011;70:687–702. doi: 10.1016/j.neuron.2011.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mira H, Andreu Z, Suh H, Lie DC, Jessberger S, Consiglio A, San Emeterio J, Hortiguela R, Marques-Torrejon MA, Nakashima K, Colak D, Gotz M, Farinas I, Gage FH. Signaling through BMPR-IA regulates quiescence and long-term activity of neural stem cells in the adult hippocampus. Cell Stem Cell. 2010;7:78–89. doi: 10.1016/j.stem.2010.04.016. [DOI] [PubMed] [Google Scholar]
- Murtagh J, Martin F, Gronostajski RM. The Nuclear Factor I (NFI) gene family in mammary gland development and function. J Mammary Gland Biol Neoplasia. 2003;8:241–254. doi: 10.1023/a:1025909109843. [DOI] [PubMed] [Google Scholar]
- Nagata K, Guggenheimer RA, Enomoto T, Lichy JH, Hurwitz J. Adenovirus DNA replication in vitro: identification of a host factor that stimulates synthesis of the preterminal protein-dCMP complex. Proc Natl Acad Sci U S A. 1982;79:6438–6442. doi: 10.1073/pnas.79.21.6438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Namihira M, Kohyama J, Semi K, Sanosaka T, Deneen B, Taga T, Nakashima K. Committed neuronal precursors confer astrocytic potential on residual neural precursor cells. Dev Cell. 2009;16:245–255. doi: 10.1016/j.devcel.2008.12.014. [DOI] [PubMed] [Google Scholar]
- Nilsson J, Bjursell G, Kannius-Janson M. Nuclear Jak2 and transcription factor NF1-C2: a novel mechanism of prolactin signaling in mammary epithelial cells. Mol Cell Biol. 2006;26:5663–5674. doi: 10.1128/MCB.02095-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oliver TG, Read TA, Kessler JD, Mehmeti A, Wells JF, Huynh TT, Lin SM, Wechsler-Reya RJ. Loss of patched and disruption of granule cell development in a pre-neoplastic stage of medulloblastoma. Development. 2005;132:2425–2439. doi: 10.1242/dev.01793. [DOI] [PubMed] [Google Scholar]
- Park JC, Herr Y, Kim HJ, Gronostajski RM, Cho MI. Nfic gene disruption inhibits differentiation of odontoblasts responsible for root formation and results in formation of short and abnormal roots in mice. J Periodontol. 2007;78:1795–1802. doi: 10.1902/jop.2007.060363. [DOI] [PubMed] [Google Scholar]
- Pereira JD, Sansom SN, Smith J, Dobenecker MW, Tarakhovsky A, Livesey FJ. Ezh2, the histone methyltransferase of PRC2, regulates the balance between self-renewal and differentiation in the cerebral cortex. Proc Natl Acad Sci U S A. 2010;107:15957–15962. doi: 10.1073/pnas.1002530107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piper M, Barry G, Harvey TJ, McLeay R, Smith AG, Harris L, Mason S, Stringer BW, Day BW, Wray NR, Gronostajski RM, Bailey TL, Boyd AW, Richards LJ. NFIB-mediated repression of the epigenetic factor Ezh2 regulates cortical development. J Neurosci. 2014;34:2921–2930. doi: 10.1523/JNEUROSCI.2319-13.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piper M, Barry G, Hawkins J, Mason S, Lindwall C, Little E, Sarkar A, Smith AG, Moldrich RX, Boyle GM, Tole S, Gronostajski RM, Bailey TL, Richards LJ. NFIA controls telencephalic progenitor cell differentiation through repression of the Notch effector Hes1. J Neurosci. 2010;30:9127–9139. doi: 10.1523/JNEUROSCI.6167-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piper M, Harris L, Barry G, Heng YH, Plachez C, Gronostajski RM, Richards LJ. Nuclear factor one X regulates the development of multiple cellular populations in the postnatal cerebellum. J Comp Neurol. 2011;519:3532–3548. doi: 10.1002/cne.22721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pistocchi A, Gaudenzi G, Foglia E, Monteverde S, Moreno-Fortuny A, Pianca A, Cossu G, Cotelli F, Messina G. Conserved and divergent functions of Nfix in skeletal muscle development during vertebrate evolution. Development. 2013;140:1528–1536. doi: 10.1242/dev.076315. [DOI] [PubMed] [Google Scholar]
- Plachez C, Cato K, McLeay RC, Heng YH, Bailey TL, Gronostasjki RM, Richards LJ, Puche AC, Piper M. Expression of nuclear factor one A and -B in the olfactory bulb. J Comp Neurol. 2012;520:3135–3149. doi: 10.1002/cne.23081. [DOI] [PubMed] [Google Scholar]
- Plachez C, Lindwall C, Sunn N, Piper M, Moldrich RX, Campbell CE, Osinski JM, Gronostajski RM, Richards LJ. Nuclear factor I gene expression in the developing forebrain. J Comp Neurol. 2008;508:385–401. doi: 10.1002/cne.21645. [DOI] [PubMed] [Google Scholar]
- Plasari G, Calabrese A, Dusserre Y, Gronostajski RM, McNair A, Michalik L, Mermod N. Nuclear factor I-C links platelet-derived growth factor and transforming growth factor beta1 signaling to skin wound healing progression. Mol Cell Biol. 2009;29:6006–6017. doi: 10.1128/MCB.01921-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plasari G, Edelmann S, Hogger F, Dusserre Y, Mermod N, Calabrese A. Nuclear factor I-C regulates TGF-{beta}-dependent hair follicle cycling. J Biol Chem. 2010;285:34115–34125. doi: 10.1074/jbc.M110.120659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Priolo M, Grosso E, Mammi C, Labate C, Naretto VG, Vacalebre C, Caridi P, Lagana C. A peculiar mutation in the DNA-binding/dimerization domain of NFIX causes Sotos-like overgrowth syndrome: a new case. Gene. 2012;511:103–105. doi: 10.1016/j.gene.2012.08.040. [DOI] [PubMed] [Google Scholar]
- Relaix F, Rocancourt D, Mansouri A, Buckingham M. A Pax3/Pax7-dependent population of skeletal muscle progenitor cells. Nature. 2005;435:948–953. doi: 10.1038/nature03594. [DOI] [PubMed] [Google Scholar]
- Rupp RA, Kruse U, Multhaup G, Gobel U, Beyreuther K, Sippel AE. Chicken NFI/TGGCA proteins are encoded by at least three independent genes: NFI-A, NFI-B and NFI-C with homologues in mammalian genomes. Nucleic Acids Res. 1990;18:2607–2616. doi: 10.1093/nar/18.9.2607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuller U, Heine VM, Mao J, Kho AT, Dillon AK, Han YG, Huillard E, Sun T, Ligon AH, Qian Y, Ma Q, Alvarez-Buylla A, McMahon AP, Rowitch DH, Ligon KL. Acquisition of granule neuron precursor identity is a critical determinant of progenitor cell competence to form Shh-induced medulloblastoma. Cancer Cell. 2008;14:123–134. doi: 10.1016/j.ccr.2008.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott CE, Wynn SL, Sesay A, Cruz C, Cheung M, Gomez Gaviro MV, Booth S, Gao B, Cheah KS, Lovell-Badge R, Briscoe J. SOX9 induces and maintains neural stem cells. Nat Neurosci. 2010;13:1181–1189. doi: 10.1038/nn.2646. [DOI] [PubMed] [Google Scholar]
- Seckl JR. Prenatal glucocorticoids and long-term programming. Eur J Endocrinol. 2004;151(Suppl 3):U49–62. doi: 10.1530/eje.0.151u049. [DOI] [PubMed] [Google Scholar]
- Shu T, Butz KG, Plachez C, Gronostajski RM, Richards LJ. Abnormal development of forebrain midline glia and commissural projections in Nfia knock-out mice. J Neurosci. 2003;23:203–212. doi: 10.1523/JNEUROSCI.23-01-00203.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh SK, Wilczynska KM, Grzybowski A, Yester J, Osrah B, Bryan L, Wright S, Griswold-Prenner I, Kordula T. The unique transcriptional activation domain of nuclear factor-I-X3 is critical to specifically induce marker gene expression in astrocytes. J Biol Chem. 2011;286:7315–7326. doi: 10.1074/jbc.M110.152421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steele-Perkins G, Butz KG, Lyons GE, Zeichner-David M, Kim HJ, Cho MI, Gronostajski RM. Essential role for NFI-C/CTF transcription-replication factor in tooth root development. Mol Cell Biol. 2003;23:1075–1084. doi: 10.1128/MCB.23.3.1075-1084.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steele-Perkins G, Plachez C, Butz KG, Yang G, Bachurski CJ, Kinsman SL, Litwack ED, Richards LJ, Gronostajski RM. The transcription factor gene Nfib is essential for both lung maturation and brain development. Mol Cell Biol. 2005;25:685–698. doi: 10.1128/MCB.25.2.685-698.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suh H, Consiglio A, Ray J, Sawai T, D'Amour KA, Gage FH. In vivo fate analysis reveals the multipotent and self-renewal capacities of Sox2+ neural stem cells in the adult hippocampus. Cell Stem Cell. 2007;1:515–528. doi: 10.1016/j.stem.2007.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Y, Hu J, Zhou L, Pollard SM, Smith A. Interplay between FGF2 and BMP controls the self-renewal, dormancy and differentiation of rat neural stem cells. J Cell Sci. 2011;124:1867–1877. doi: 10.1242/jcs.085506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tajbakhsh S. Skeletal muscle stem and progenitor cells: reconciling genetics and lineage. Exp Cell Res. 2005;306:364–372. doi: 10.1016/j.yexcr.2005.03.033. [DOI] [PubMed] [Google Scholar]
- Taniguchi H, Kawauchi D, Nishida K, Murakami F. Classic cadherins regulate tangential migration of precerebellar neurons in the caudal hindbrain. Development. 2006;133:1923–1931. doi: 10.1242/dev.02354. [DOI] [PubMed] [Google Scholar]
- Tumbar T, Guasch G, Greco V, Blanpain C, Lowry WE, Rendl M, Fuchs E. Defining the epithelial stem cell niche in skin. Science. 2004;303:359–363. doi: 10.1126/science.1092436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valcourt JR, Lemons JM, Haley EM, Kojima M, Demuren OO, Coller HA. Staying alive: metabolic adaptations to quiescence. Cell Cycle. 2012;11:1680–1696. doi: 10.4161/cc.19879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wallace VA. Purkinje-cell-derived Sonic hedgehog regulates granule neuron precursor cell proliferation in the developing mouse cerebellum. Curr Biol. 1999;9:445–448. doi: 10.1016/s0960-9822(99)80195-x. [DOI] [PubMed] [Google Scholar]
- Wang W, Crandall JE, Litwack ED, Gronostajski RM, Kilpatrick DL. Targets of the nuclear factor I regulon involved in early and late development of postmitotic cerebellar granule neurons. J Neurosci Res. 2010;88:258–265. doi: 10.1002/jnr.22199. [DOI] [PubMed] [Google Scholar]
- Wang W, Mullikin-Kilpatrick D, Crandall JE, Gronostajski RM, Litwack ED, Kilpatrick DL. Nuclear factor I coordinates multiple phases of cerebellar granule cell development via regulation of cell adhesion molecules. J Neurosci. 2007;27:6115–6127. doi: 10.1523/JNEUROSCI.0180-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W, Stock RE, Gronostajski RM, Wong YW, Schachner M, Kilpatrick DL. A role for nuclear factor I in the intrinsic control of cerebellar granule neuron gene expression. J Biol Chem. 2004;279:53491–53497. doi: 10.1074/jbc.M410370200. [DOI] [PubMed] [Google Scholar]
- Weissman IL. Stem cells: units of development, units of regeneration, and units in evolution. Cell. 2000;100:157–168. doi: 10.1016/s0092-8674(00)81692-x. [DOI] [PubMed] [Google Scholar]
- Wilczynska KM, Singh SK, Adams B, Bryan L, Rao RR, Valerie K, Wright S, Griswold-Prenner I, Kordula T. Nuclear factor I isoforms regulate gene expression during the differentiation of human neural progenitors to astrocytes. Stem Cells. 2009;27:1173–1181. doi: 10.1002/stem.35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu X, Northcott PA, Dubuc A, Dupuy AJ, Shih DJ, Witt H, Croul S, Bouffet E, Fults DW, Eberhart CG, Garzia L, Van Meter T, Zagzag D, Jabado N, Schwartzentruber J, Majewski J, Scheetz TE, Pfister SM, Korshunov A, Li XN, Scherer SW, Cho YJ, Akagi K, MacDonald TJ, Koster J, McCabe MG, Sarver AL, Collins VP, Weiss WA, Largaespada DA, Collier LS, Taylor MD. Clonal selection drives genetic divergence of metastatic medulloblastoma. Nature. 2012;482:529–533. doi: 10.1038/nature10825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoneda Y, Saitsu H, Touyama M, Makita Y, Miyamoto A, Hamada K, Kurotaki N, Tomita H, Nishiyama K, Tsurusaki Y, Doi H, Miyake N, Ogata K, Naritomi K, Matsumoto N. Missense mutations in the DNA-binding/dimerization domain of NFIX cause Sotos-like features. J Hum Genet. 2012;57:207–211. doi: 10.1038/jhg.2012.7. [DOI] [PubMed] [Google Scholar]
- Zappelli F, Willems D, Osada S, Ohno S, Wetsel WC, Molinaro M, Cossu G, Bouche M. The inhibition of differentiation caused by TGFbeta in fetal myoblasts is dependent upon selective expression of PKCtheta: a possible molecular basis for myoblast diversification during limb histogenesis. Dev Biol. 1996;180:156–164. doi: 10.1006/dbio.1996.0292. [DOI] [PubMed] [Google Scholar]