Abstract
Technologies and experimental strategies have improved dramatically in the field of genomics and proteomics facilitating analysis of cellular and biochemical processes, as well as of proteins networks. Based on numerous such analyses, there has been a significant increase of publications in life sciences and biomedicine. In this respect, knowledge bases are struggling to cope with the literature volume and they may not be able to capture in detail certain aspects of proteins and genes. One important aspect of proteins is their phosphorylated states and their implication in protein function and protein interacting networks. For this reason, we developed eFIP, a web-based tool, which aids scientists to find quickly abstracts mentioning phosphorylation of a given protein (including site and kinase), coupled with mentions of interactions and functional aspects of the protein. eFIP combines information provided by applications such as eGRAB, RLIMS-P, eGIFT and AIIAGMT, to rank abstracts mentioning phosphorylation, and to display the results in a highlighted and tabular format for a quick inspection. In this chapter, we present a case study of results returned by eFIP for the protein BAD, which is a key regulator of apoptosis that is posttranslationally modified by phosphorylation.
Keywords: Text mining, BioNLP, Information extraction, Phosphorylation, Protein–protein interaction, PPI, Knowledge discovery
1. Introduction
There has been a general shift in paradigm from dedicating a lifetime’s work to analyzing of a single protein to the analysis of cellular and biochemical processes and networks. This has been made possible by a dramatic improvement in technologies and experimental strategies in the fields of genomics and proteomics (1). Although bioinformatics tools have greatly assisted in data analysis, both protein identification and functional interpretation are still major bottlenecks (2). In this regard, public knowledge bases constitute a valuable source of such information, but the manual curation of experimentally determined biological events is slow compared to the rapid increase in the body of knowledge represented in the literature. Hence, literature still continues to be a primary source of biological data. Nevertheless, manually finding the relevant articles is not a trivial task, with issues ranging from the ambiguity of some names to the identification of those articles that contain the specific information of interest.
Fortunately, the text mining community has recognized in recent years the opportunities and challenges of natural language processing (NLP) in the biomedical field (3), and has developed a number of resources for providing access to information contained in life sciences and biomedical literature. Table 1 lists a sampling of freely-available tools that address the various BioNLP applications. In addition, there are a large number of papers discussing research and techniques for these applications. For an indepth overview of these topics, please refer to review articles by Krallinger et al. (4) and Jensen et al. (5).
Table 1.
Biological applications and a sampling of available resources
| Biological applications | Resources |
|---|---|
| Protein–protein interaction | iHOP, Chilibot, KinasePathway, PPI Finder, Protein Corral |
| Gene name recognition/mention/tagger | ABNER, AIIAGMT, ABGene, BANNER, BIGNER, GAPSCORE, KEX, LingPipe, SciMiner |
| Acronym expansion and disambiguation | Acromine, AcroTagger, ADAM, ALICE, ARGH, Biomedical Abbreviation |
| Protein sequence | Mutation Finder, MeInfoText, mSTRAP, MutationFinder, PepBank, RLIMS-P |
| Text-mining search aids | Anne O’Tate, e-LiSe, FABLE, GoPubMed, MedEvi, NextBio |
However, BioNLP tools are only useful if they are designed to meet real-life tasks (4). In fact, this has been one of the obstacles for the general adoption of BioNLP tools by biologists, because many of these applications perform individual tasks (like gene/protein mention, phosphorylation, or protein–protein interaction), thus providing only one piece of information, which in itself might not be enough to describe the biology. To address this issue, we have designed eFIP (extraction of Functional Impact of Phosphorylation), a system that combines several publicly available tools to allow identification of abstracts that contain protein phosphorylation mentions (including the site and the kinase), coupled with mentions of functional implications (such as protein–protein interaction, function, process, localization, and disease). In addition, eFIP ranks these abstracts and presents the information in a user-friendly format for a quick inspection.
The rationale for performing this particular task relies on at least three aspects:
Phosphorylation is one of the most common protein post-translational modifications (PTMs). Phosphorylation of specific intracellular proteins/enzymes by protein kinases and dephosphorylation by phosphatases provides information of both activation and deactivation of critical cellular pathways, including regulatory mechanisms of metabolism, cell division, cell growth and differentiation (6).
Often protein phosphorylation has some functional impact. Proteins can be phosphorylated on different residues, leading to activation or down-regulation of their activity, alternative subcellular location, and binding partners. One such example is protein Smad2, whose phosphorylation state determines its interaction partners, its subcellular location, and its cofactor activity (7).
Currently, protein–protein interaction (PPI) data involving phosphorylated proteins is not yet well represented in the public databases. Thus, extracting this information is critical to the interpretation of PPI and prediction of the functional outcomes.
1.1. Goal of This Chapter
As mentioned before, interesting and important real-life tasks would require the combination of multiple individual tasks. A major focus of this chapter is to highlight how the combination of existing BioNLP tools can reveal some interesting biology about a protein. The specific goal is to describe eFIP, a tool that can assist a researcher in finding information in the literature about protein phosphorylation mentions that have some biological implication, such as PPI, localization, function, and disease.
1.2. The Approach
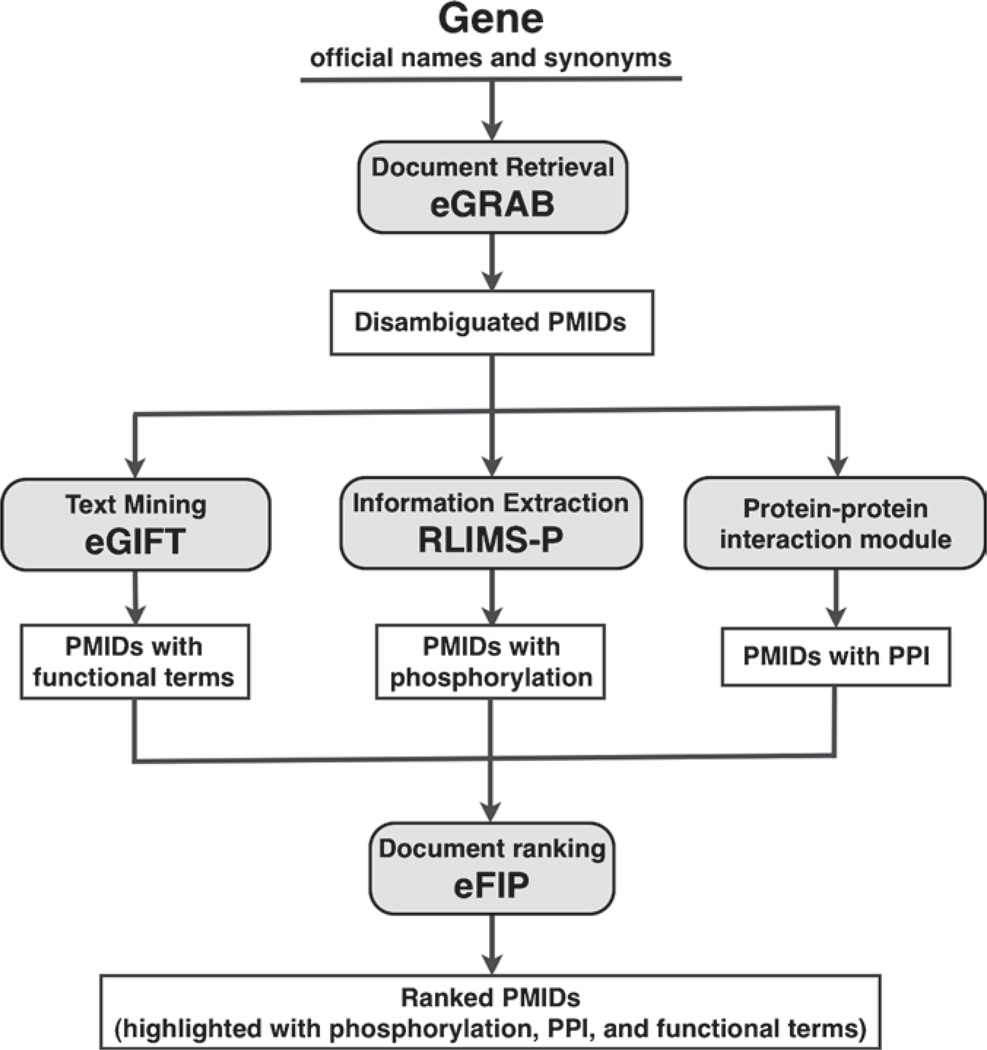
The BioNLP tasks behind eFIP include (1) document retrieval –selection of relevant scientific publications, and gene name disambiguation (eGRAB); (2) text mining – detection of functional terms (eGIFT); (3) information extraction – identification of substrate, phosphorylation sites, and kinase (RLIMSP); (4) protein– protein interaction identification (PPI module) and gene name recognition (AIIAGMT); and (5) document and sentence ranking – integration of text mining results with ranking and summarization (eFIP’s ranking module) (Fig. 1).
Fig. 1.
General pipeline of BioNLP tasks, including specific tools used in our approach. The protein–protein interaction module includes the gene name recognition tool (AIIAGMT).
For details regarding each individual tool mentioned here, please refer to Subheading 2. In Subheading 3, we will provide the user with a protocol to find relevant articles using the protein BAD as an example.
2. Materials
In this section, we briefly describe the tools depicted in Fig. 1.
2.1. Extractor of Gene-Related Abstracts
Extractor of Gene-Related ABstracts (eGRAB) is used to gather the literature for a given gene/protein. To retrieve all Medline abstracts relevant to a given gene/protein requires expanding the PubMed search query with all the synonyms of the gene/protein, as this is often mentioned in text by short names (acronyms and abbreviations) and gene symbols, with or without the accompanying long names. Searching short names and abbreviations is challenging as these names tend to be highly ambiguous, resulting in the retrieval of many irrelevant documents. Although augmenting the query using NOT operators, to disallow irrelevant expansions of the short names, may help in some cases with document retrieval, it does not circumvent the problem altogether. Short forms can be mentioned in text without the accompanying long form, thus making it impossible to automatically detect the relevance of the text based solely on the query.
For example, consider protein Carbamoyl-phosphate synthetase 1, whose short names are CPS1 and CPSI. The latter could also be an abbreviation for “cancer prevention study I,” “chronic prostatis symptom index,” and “chronic pain sleep inventory”. Equally ambiguous are non abbreviated short names. The task of disambiguating words with multiple senses dates back to Bruce and Wiebe (8) and Yarowsky (9), who proposed a word sense disambiguation (WSD) technique for English words with multiple definitions (e.g., “bank” in the context of “river,” and “bank” in the context of “financial institution”).
eGRAB starts by gathering all possible names and synonyms of a gene/protein from knowledge bases of genes and proteins (such as Entrez Gene, Uniprot, or BioThesaurus), searches PubMed using these names, and returns a set of disambiguated Medline abstracts to serve as the gene’s literature. This technique filters potentially irrelevant documents that mention the gene names in some other context, by creating language models for all the senses and assigning the closest sense to an ambiguous name. Similar methods have been described for disambiguating biomedical abbreviations by taking into consideration the context in which the abbreviations occur (10–13).
2.2. Extracting Genic Information from Text
Extracting Genic Information from Text (eGIFT) (14, 15) is a new, freely available online tool (http://biotm.cis.udel.edu/eGIFT/), which aims to link genes/proteins to key descriptors. The user can search for the gene/protein of interest and see its concepts grouped in categories: processes and functions, diseases, cellular components, motifs/domains, taxons, drugs, and genes. In eGIFT these concepts are extracted from the gene’s literature when they are statistically more frequent in this set of abstracts, as compared to abstracts about genes in general. For example, given the protein BAD and its literature identified by eGRAB, eGIFT focuses on the abstracts that are mainly about BAD, and identify concepts, such as “apoptosis,” “cell death,” and “dephosphorylation” as highly relevant to this gene. Although different in the overall approach, scoring formula, redundancy detection, multiword concept retrieval, and evaluation technique, eGIFT can be compared with methods described by Andrade and Valencia (16), XplorMed (17, 18), Liu et al. (19), and Shatkay and Wilbur (20).
2.3. Rule-Based Literature Mining System for Protein Phosphorylation
Rule-based LIterature Mining System for Protein Phosphorylation (RLIMS-P) (21, 22) is a system designed for extracting protein phosphorylation information from MEDLINE abstracts. Its unique features, which distinguishes it from other BioNLP systems, include the extraction of information about protein phosphorylation, along with the three objects involved in this process – the protein kinase, the phosphorylated protein (substrate), and the phosphorylation site (residue/position being phosphorylated). RLIMS-P employs techniques to combine information found in different sentences, because rarely are the three objects (kinase, substrate, and site) found in the same sentence. For this, RLIMS-P utilizes extraction rules that cover a wide range of patterns, including some specialized terms used only with phosphorylation. RLIMS-P was benchmarked using PIR annotated literature data from iProLINK (21). The online tool is available at http://www.proteininformationresource.org/pirwww/iprolink/rlimsp.shtml.
2.4. PPI Module
The PPI module is an internal implementation designed to detect mentions of PPI in text. This tool extracts text fragments, or text evidence, that explicitly describe a type of PPI (such as binding and dissociation), as well as the interacting partners. The primary engine of this tool is an extensive set of rules specialized to detect patterns of PPI mentions (manuscript in preparation).
The interacting partners identified are further sent to AIIAGMT, a gene/protein mention tool (described in more detail in the next sub-section), to confirm whether they are genuine protein mentions. Consider the sample phrase “several proapoptotic proteins commonly become associated with 14-3-3.” “14-3-3” is a protein, whereas “several proapoptotic proteins” prompts the need to further identify the actual proteins (Bad and FOXO3a) that interact with 14-3-3. Our PPI module can be compared to other systems that also extract text evidence of PPI from literature, such as PIE (23), BIOSMILE (24, 25), Chilibot (26) and iHOP (27).
2.5. AIIAGMT
As mentioned previously in this chapter, genes and proteins often have many synonyms that come in short and long forms. To aid the PPI module to confirm whether an interacting partner in a PPI mention is indeed a protein, we employ AIIAGMT (28). AIIAGMT is a gene/protein mention tagger that detects all the proteins mentioned in some given text. The tool ranked second in the BioCreative II competition (29) for the gene mention task (F-score of 87.21) (30). Other systems that also extract gene and protein mentions from text are ABGene (31), BIGNER (32), GAPSCORE (33), T2K Gene Tagger (34), and LingPipe (35).
2.6. eFIP’s Ranking Module
eFIP ranks abstracts mentioning a given protein based on three features: phosphorylation, functional terms, and proteins with which the given protein interacts. Because our main goal is to find information about a particular protein when it is in its phosphorylated state, we disregard abstracts that do not contain phosphorylation information. The next step is to distinguish the set of abstracts that mention a phosphorylation site for the given protein from the set of abstracts that mention only that the protein is phosphorylated. We rank the former set higher than the latter. Within these sets, a second ranking is performed, based on the following criteria (1) highly ranked are abstracts that include all three features, mentioned in one or two consecutive sentences; (2) following these are abstracts mentioning phosphorylation together with one other feature, in one or two consecutive sentences. When the features are found in the same sentence these abstracts are ranked higher than when they are found in two consecutive ones. Intuitively, the closer the two pieces of information, the higher the likelihood that they are related. We also consider the confidence level of rules or patterns matched for the PPI. For instance, “protein A binds to protein B” strongly indicates a PPI, whereas “the colocalization of proteins C and D” may suggest, but does not imply, a physical interaction. Some examples of the types of sentences mentioned above are depicted in Fig. 2. Based on our ranking, PMID:15161349 (A) would rank higher than PMID:12049737 (B).
Fig. 2.
Examples of sentences with different co-occurrence of ranked features. (a) Co-occurrence of the three features in one sentence (sentence 13); (b) Co-occurrence of phosphorylation and functional terms (sentences 4 and 5, respectively).
3. Methods
We present a use case on abstracts for the protein BAD (Bcl2-associated agonist of cell death). This protein is a key regulator of apoptosis that is posttranslationally modified by phosphorylation, which, in turn, defines BAD’s binding partners and localization, as well as its function as an antiapoptotic or proapoptotic molecule. Ideally, we want to find papers about BAD that describe, together, phosphorylation and its functional consequence. Typically, we would start by searching PubMed using the protein/gene names (including/excluding its synonyms), coupled with phosphory* fuzzy search to retrieve abstracts that mention the given protein and its phosphorylation. For example, we might search using the following query (BAD AND phosphory*), which retrieves 1,050 papers. However, based on this search, some irrelevant abstracts may be retrieved (e.g., PMID: 8755886, where BAD is mentioned as an adjective). This example reflects the ambiguity problem mentioned before. From the list of abstracts obtained, we then need to check manually those for which phosphorylation has some implication on BAD biology. As an alternative to this approach, we present eFIP, a system that allows, in one step, document retrieval, disambiguation of names, and extraction of information.
eFIP combines information that is output by tools described in Subheading 2. Initially, eGRAB gathers abstracts specific to the gene/protein. These abstracts are input to (1) eGIFT, which mines, from this set of abstracts, terms that are highly related to the given gene/protein (e.g., “apoptosis” and “cell survival” for protein BAD); (2) RLIMS-P, which detects protein phosphorylation information from these abstracts; and (3) PPI module, which identifies interacting proteins. eFIP uses this information to rank abstracts mentioning a given protein of interest. However, these detailed steps are hidden from the user. eFIP combines these tools and requires only the following steps from its users:
3.1. Accessing eFIP’s Website at http://biotm.cis.udel.edu/eFIP/
The search for a gene/protein is initiated from the Search eFIP link. Here, the gene/protein name or part of the name can be entered in the search box, and results are displayed for the search. For example, word BAD can be entered in the search box, and only one result is obtained for gene BAD. However, if a partial name is entered, such as bcl2 (initial part of one of BAD’s name), many results are retrieved. In this case selecting the gene corresponding to BAD is required (Fig. 3).
Fig. 3.
eFIP search page. The screenshot shows the list of possible gene/protein names when using bcl2 as a query. The user needs to select BAD to inspect its specific literature.
3.2. Inspecting the Result Page
- The primary result page contains the following information (Fig. 4):
- Names, synonyms and statistics: The result page shows the names and synonyms used for retrieving the articles. It also shows the number of articles that contain phosphorylation mentions as evaluated by the RLIMS-P tool (791 in BAD’s case). Note that the number of total articles disambiguated by eGRAB is 1,331.
- Ranked PMIDs, along with the information content of the abstract, are listed. Because all the abstracts have phosphorylation mentions by default, only the PPI and/or functional feature labels are displayed. Note that based on our ranking criteria, the first set of abstracts displayed are those that mention phosphorylation site information (206 abstracts).
-
Selecting a PMID leads to the abstract page (Fig. 5).
This page contains the summary table, with information extracted for phosphorylation and the predicted impact on function. We emphasize predicted here, because BioNLP tools are intended to assist the user by pointing to articles or sentences that are more likely to have the information needed. However, there is always a need to check the correctness of the information. The summary table, displayed on this page, consists of three main columns. The first column shows the number of the sentence that contains the evidence, thus facilitating its quick location within the abstract. The second column contains the phosphorylation information, as provided by RLIMS-P tool. Three different types of information are listed here: the substrate, the site, and the kinase. The third column provides information about the impact on phosphorylation. Here, we list the functional terms and/or interaction information provided by eGIFT and the PPI module, respectively. In this column, we also include action words (e.g., regulates, promotes, blocks), present in the text, to point to the modification or to the influence on the meaning of the functional term. These action words, provided by the PPI module, provide a more accurate result. Listed below the table is the corresponding abstract, with highlighted information. Note that each type of information has a distinct color, and for each color there is a dark and a light version, to give different confidence levels to the prediction (the dark color hints to a higher likelihood of the prediction). At the bottom of the abstract, you can select which information to include in the highlighting.
Fig. 4.
Result page for the protein BAD.
Fig. 5.
Summary table and highlighted information for PMID 10837486. The different features are color coded.
4. Discussion
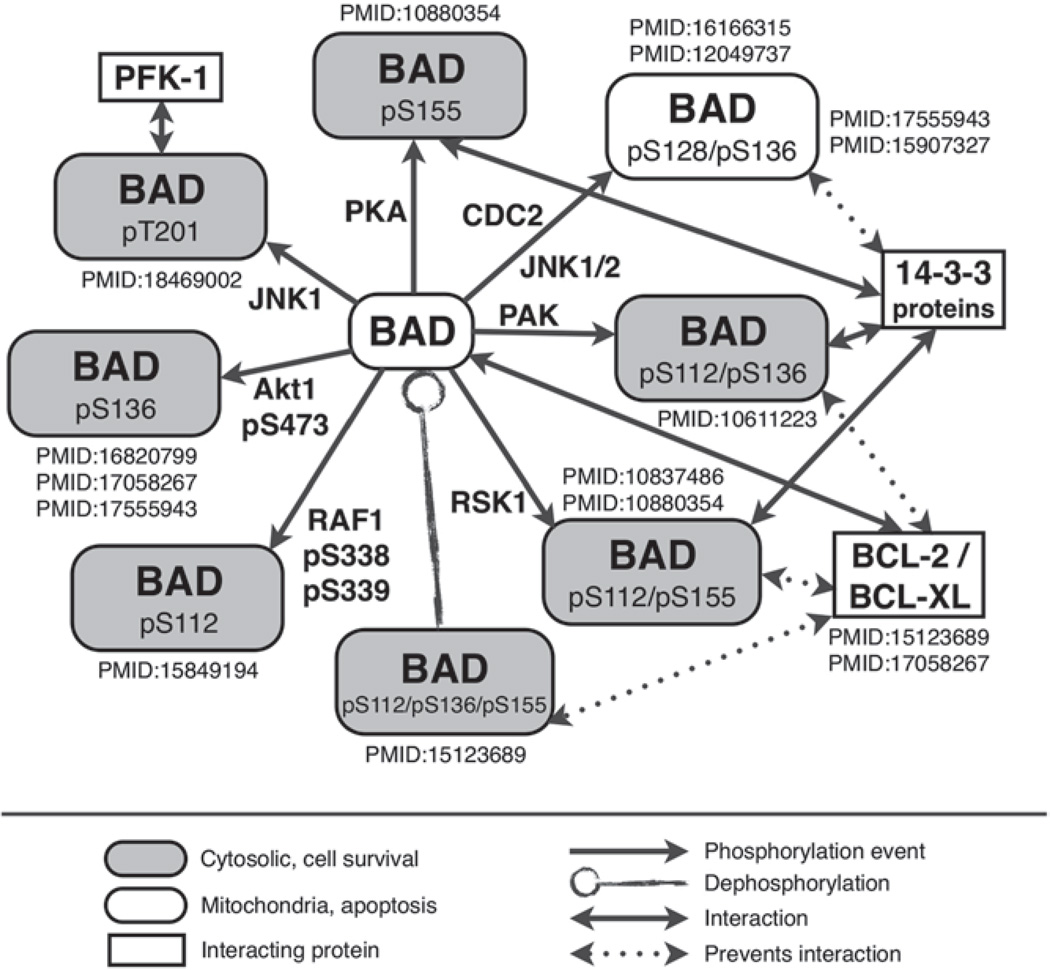
Using protein BAD and the information displayed in eFIP for this protein, we show in Fig. 6 the different phosphorylated forms of BAD, their functions, and their implication in PPI. The information depicted here is extracted from a subset of the highest ranked abstracts, as provided by eFIP. The rich information from the eFIP text mining tool uncovers interesting facts about BAD (1) BAD is a common hub for several pathways to regulate apoptosis, as evidenced by the various kinases that are able to phosphorylate this protein; (2) BAD has specific partners for its distinct phosphorylated forms; and (3) phosphorylation on BAD may have two opposing effects: apoptosis (through phosphorylation at Ser128) and cell survival (phosphorylation on other residues), which is mainly dictated by the association/disassociation to 14-3-3 proteins and BCL-2/BCL-XL proteins. This example highlights the importance of detecting more than just the phosphorylation mention. The phosphorylation site, as well as the kinase that links to the pathway, are important aspects in understanding the regulation of BAD. The majority of abstracts describing BAD focus on BAD’s interaction with apoptotic and antiapoptotic proteins. However, in this figure, we also point to an example in which phosphorylated BAD (Thr-201) leads to binding to phosphofructokinase (PFK-1), and the subsequent activation of glycolysis (a pathway that is key to cell survival).
Fig. 6.
Representation of different forms of phosphorylated BAD based on eFIP’s results (only a subset is shown). Note that from information listed by eFIP, we are able to represent the impact (cytosolic vs. mitochondria; apoptosis vs. cell survival) for the different forms of BAD. Moreover, the kinases, which accompany the phosphorylation arrows, help to link BAD to pathways. Whenever available, the phosphorylation state of the kinase is extracted and displayed here, as in the case of RAF1 p338/pS339.
Thus, we show that eFIP provides the means to find the most relevant papers about BAD phosphorylation, interaction partners, and its functions. Based on the literature data collected from eFIP for BAD protein, it is possible to predict, for example, how the regulation or inhibition of a certain pathway may affect the cell fate.
References
- 1.Preisinger C, von Kriegsheim A, Matallanas D, Kolch W. Proteomics and phosphoproteomics for the mapping of cellular signalling networks. Proteomics. 2008;8:4402–4415. doi: 10.1002/pmic.200800136. [DOI] [PubMed] [Google Scholar]
- 2.Huang H, Hu ZZ, Arighi C, Wu CH. Integration of bioinformatics resources for functional analysis of gene expression and proteomic data. Front Biosci. 2007;12:5071–5088. doi: 10.2741/2449. [DOI] [PubMed] [Google Scholar]
- 3.Hirschman L, Park JC, Tsujii J, Wong L, Wu CH. Accomplishments and challenges in literature data mining for biology. Bioinformatics. 2002;18:1553–1561. doi: 10.1093/bioinformatics/18.12.1553. [DOI] [PubMed] [Google Scholar]
- 4.Krallinger M, Morgan A, Smith L, Leitner F, Tanabe L, Wilbur J, Hirschman L, Valencia A. Evaluation of text-mining systems for biology: overview of the second BioCreative community challenge. Genome Biol. 2008;9:S1. doi: 10.1186/gb-2008-9-s2-s1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jensen LJ, Saric J, Bork P. Literature mining for the biologist: from information retrieval to biological discovery. Nat Rev Genet. 2006;7:119–129. doi: 10.1038/nrg1768. [DOI] [PubMed] [Google Scholar]
- 6.Salih E. Phosphoproteomics by mass spectrometry and classical protein chemistry approaches. Mass Spectrom Rev. 2005;24:828–846. doi: 10.1002/mas.20042. [DOI] [PubMed] [Google Scholar]
- 7.Wicks SJ, Lui S, Abdel-Wahab N, Mason RM, Chantry A. Inactivation of smad-transforming growth factor beta signaling by Ca(2+)-calmodulin-dependent protein kinase II. Mol Cell Biol. 2000;20:8103–8111. doi: 10.1128/mcb.20.21.8103-8111.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bruce R, Wiebe J. Word-sense disambiguation using decomposable models; Proceedings of the 32nd Annual Meeting on ACL; 1994. pp. 139–146. [Google Scholar]
- 9.Yarowsky D. Unsupervised word sense disambiguation rivaling supervised methods; Proceedings of the 33rd Annual Meeting on ACL; 1995. pp. 189–196. [Google Scholar]
- 10.Pakhomov S. Semi-supervised maximum entropy based approach to acronym and abbreviation normalization in texts; Proceedings of 40th Annual Meeting on ACL; 2001. p. 2001. [Google Scholar]
- 11.Yu Z, Tsuruoka Y, Tsujii J. Automatic resolution of ambiguous abbreviations in biomedical texts using support vector machines and one sense per discourse hypothesis; SIGIR’03 Workshop on Text Analysis and Search for Bioinformatics; 2003. [Google Scholar]
- 12.Gaudan S, Kirsch H, Rebholz-Schuhmann D. Resolving abbreviations to their senses in Medline. Bioinformatics. 2005;21:3658–3664. doi: 10.1093/bioinformatics/bti586. [DOI] [PubMed] [Google Scholar]
- 13.Stevenson M, Guo Y, Amri AA, Gaizauskas R. Disambiguation of biomedical abbreviations; Proceedings of the BioNLP 2009 Workshop, ACL; 2009. pp. 71–79. [Google Scholar]
- 14.Tudor CO, Vijay-Shanker K, Schmidt CJ. Mining the biomedical literature for genic information; Proceedings of Workshop on Current Trends in BioNLP, ACL; 2008. pp. 28–29. [Google Scholar]
- 15.Tudor CO, Schmidt CJ, Vijay-Shanker K. Mining for gene-related key terms: where do we find them?; Proceedings of the Third International Symposium on Semantic Mining in Biomedicine (SMBM); 2008. pp. 157–160. [Google Scholar]
- 16.Andrade MA, Valencia A. Automatic extraction of keywords from scientific text: application to the knowledge domain of protein families. Bioinformatics. 1998;14:600–607. doi: 10.1093/bioinformatics/14.7.600. [DOI] [PubMed] [Google Scholar]
- 17.Perez-Iratxeta C, Keer HS, Bork P, Andrade MA. Computing fuzzy associations for the analysis of biomedical literature. BioTechniques. 2002;32:1380–1385. doi: 10.2144/02326bc03. [DOI] [PubMed] [Google Scholar]
- 18.Perez-Iratxeta C, Perez AJ, Bork P, Andrade MA. Update on XplorMed: a web server for exploring scientific literature. Nucleic Acid Res. 2003;31:3866–3868. doi: 10.1093/nar/gkg538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Liu Y, Brandon M, Navathe S, Dingledine R, Ciliax BJ. Text mining functional keywords associated with genes. MedInfo. 2004:292–296. [PubMed] [Google Scholar]
- 20.Shatkay H, Wilbur WJ. Finding themes in medline documents: probabilistic similarity search; Proceedings of the Seventh IEEE Advances in Digital Libraries (ADL’00); 2000. pp. 183–192. [Google Scholar]
- 21.Hu ZZ, Narayanaswamy M, Ravikumar KE, Vijay-Shanker K, Wu CH. Literature mining and database annotation of protein phosphorylation using a rule-based system. Bioinformatics. 2005;21:2759–2765. doi: 10.1093/bioinformatics/bti390. [DOI] [PubMed] [Google Scholar]
- 22.Narayanaswamy M, Ravikumar KE, Vijay-Shanker K. Beyond the clause: extraction of phosphorylation information from Medline abstracts. Bioinformatics. 2005;21(Suppl 1):i319–i327. doi: 10.1093/bioinformatics/bti1011. [DOI] [PubMed] [Google Scholar]
- 23.Kim S, Shin SY, Lee IH, Kim SJ, Sriram R, Zhang BT. PIE: an online prediction system for protein–protein interactions from text. Nucleic Acids Res. 2008;36:W411–W415. doi: 10.1093/nar/gkn281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dai HJ, Huang CH, Lin RT, Tsai RT, Hsu WL. BIOSMILE web search: a web application for annotating biomedical entities and relations. Nucleic Acids Res. 2008;36:W390–W398. doi: 10.1093/nar/gkn319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tsai RTH, Chou WC, Su YS, Lin YC, Sung CL, Dai HJ, Yeh ITH, Ku W, Sung TY, Hsu WL. BIOSMILE: a semantic role labeling system for biomedical verbs using a maximum-entropy model with automatically generated template features. BMC Bioinformatics. 2007;8:325. doi: 10.1186/1471-2105-8-325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chen H, Sharp BM. Content-rich biological network constructed by mining PubMed abstracts. BMC Bioinformatics. 2004;5:147. doi: 10.1186/1471-2105-5-147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hoffmann R, Valencia A. Implementing the iHOP concept for navigation of biomedical literature. Bioinformatics. 2005;21:ii252–ii258. doi: 10.1093/bioinformatics/bti1142. [DOI] [PubMed] [Google Scholar]
- 28.Hsu CN, Chang YM, Kuo CJ, Lin YS, Huang HS, Chung IF. Integrating high dimensional bi-directional parsing models for gene mention tagging. Bioinformatics. 2008;24:i286–i294. doi: 10.1093/bioinformatics/btn183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Morgan AA, Lu Z, Wang X, Cohen AM, Fluck J, Ruch P, Divoli A, Fundel K, Leaman R, Hakenberg J, Sun C, Liu HH, Torres R, Krauthammer M, Lau WW, Liu H, Hsu CN, Schuemie M, Cohen KB, Hirschman L. Overview of BioCreative II gene normalization. Genome Biol. 2008;9(Suppl 2):S3. doi: 10.1186/gb-2008-9-s2-s3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. URL: http://www.bcsp1.iis.sinica.edu.tw:8080/aiiagmt/. [Google Scholar]
- 31.Tanabe L, Wilbur WJ. Tagging gene and protein names in biomedical text. Bioinformatics. 2004;20:216–225. doi: 10.1093/bioinformatics/18.8.1124. [DOI] [PubMed] [Google Scholar]
- 32.Li Y, Lin H, Yang Z. Incorporating rich background knowledge for gene named entity classification and recognition. BMC Bioinformatics. 2009;10:223. doi: 10.1186/1471-2105-10-223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chang JT, Schütze H, Altman RB. GAPSCORE: finding gene and protein names one word at a time. Bioinformatics. 2004;20:216–225. doi: 10.1093/bioinformatics/btg393. [DOI] [PubMed] [Google Scholar]
- 34. URL: http://www.bioinformatics.org/~hyy/textknowledge/genetag.php. [Google Scholar]
- 35. URL: http://www.alias-i.com/lingpipe/. [Google Scholar]