Abstract
Biophysical properties including particle size distribution, integrity, and shape of whole virus vaccine particles at different stages in tick-borne encephalitis (TBE) vaccines formulation were analyzed by a new set of methods. Size-exclusion chromatography (SEC) was used as a conservative sample preparation for vaccine particle fractionation and gas-phase electrophoretic mobility macromolecular analyzer (GEMMA) for analyzing electrophoretic mobility diameters of isolated TBE virions. The derived particle diameter was then correlated with molecular weight. The diameter of the TBE virions determined after SEC by GEMMA instrumentation was 46.8 ± 1.1 nm. Atomic force microscopy (AFM) and transmission electron microscopy (TEM) were implemented for comparison purposes and to gain morphological information on the virion particle. Western blotting (Dot Blot) as an immunological method confirmed biological activity of the particles at various stages of the developed analytical strategy. AFM and TEM measurements revealed higher diameters with much higher SD for a limited number of virions, 60.4 ± 8.5 and 53.5 ± 5.3 nm, respectively. GEMMA instrumentation was also used for fractionation of virions with specifically selected diameters in the gas-phase, which were finally collected by means of an electrostatic sampler. At that point (i.e., after particle collection), AFM and TEM showed that the sampled virions were still intact, exhibiting a narrow size distribution (i.e., 59.8 ± 7.8 nm for AFM and 47.5 ± 5.2 nm for TEM images), and most importantly, dot blotting confirmed immunological activity of the collected samples. Furthermore dimers and virion artifacts were detected, too.
For whole virus particles which are in the size range of about 50 nm and with a molecular weight of several MDa, just a few analytical methods are available for characterization purposes. Transmission electron microscopy (TEM) is used to determine particle morphology and size analysis of virus particles; however, the application is limited by the TEM resolution and possible artifacts due to sample preparation methods (fixing and staining). Morphological studies are also conducted by cryo-electron microscopy in combination with specific software tools revealing high-resolution images from different virus samples.1,2 For cryo-electron microscopy, sample preparation and handling during analysis is even more sophisticated and critical than for TEM measurements.3,4 Another commonly used method for detecting and separating virus fragments, whole virus particles, and aggregates is analytical ultracentrifugation (UC) and preparative UC.5 In general, electron microscopy and UC methods are not suitable for routine characterization of virus particles, because these methods are time- and labor-intensive as well as requiring high investments into equipment. Dynamic light-scattering (DLS) and multiple-angle light-scattering (MALS) measurement are used for particle size and distribution determination, but these methods encounter serious bias with heterogeneous samples.6,7 Recently, DLS and MALS have been combined with a fractionation method, known as FFF (field flow fractionation), whereby the particles are separated due to their gyro-dynamic diameter before detection.7,8 FFF rapidly analyses fragments, monomers, dimers and aggregates in a certain diameter range, but the resolution and accuracy is not sufficient for some samples containing a mixture of proteins or aggregates.9,10 Lately, atomic force microscopy (AFM) has been used with increasing frequency for biological material, even viruses and substructure of virus particles.11 For AFM, the sample preparation is not as elaborated as for TEM recordings, as the sample can be visualized without any staining procedure, and the measurements can be performed at ambient conditions. AFM images also give three-dimensional information on the samples.12,13
In comparison to the methods mentioned above, the gas-phase electrophoretic mobility macromolecular analyzer (GEMMA) applied in this study makes use of a completely different physical principle for obtaining size-related information. Within the GEMMA instrument, proteins, complexes, or other nanosize particles are separated due to different EM in an electric field, subsequently detected, and information on particle size distributions (in the range from 3 nm to 1 μm) is gained from analytes provided in an originally liquid phase. Details on GEMMA are described elsewhere.14-17 For GEMMA, a volatile solution containing the nonvolatile particles is electrosprayed in the nanoelectrospray (nanoES) device. The formed charged droplets are then charge-reduced in a bipolar atmosphere produced by a 210Po source to gain mainly singly charged positive as well as negative ions and neutral nanoparticles. The aerosol particles are transferred into a differential mobility analyzer (DMA). In the DMA, the single-positively or negatively charged particles are then classified according to the electrophoretic mobility in an electric field. Particles which passed the DMA are then detected in a condensation particle counter. The electrophoretic mobility diameter is calculated by using the Cunningham–Knudsen–Weber–Millikan equation.18 The molecular weight of the analyzed nanoparticles can be calculated from a calibration function, which is valid for up to at least 2 MDa for proteins.17
Instead of a detector, an electrostatic nanoparticle sampler (ENPS) can be connected, which can furthermore contain a substrate usable for subsequent microscopic (e.g., AFM mica plate or EM grid) or immunological methods (e.g., blotting membrane) mounted on an electrode located in the center of the ENPS. The separated charged particles leaving the DMA are guided into the ENPS. After entering the inner chamber of the ENPS, the velocity of the aerosol flow is reduced, and the electrostatic field between the electrode and the walls direct the charged particles toward the collecting surface. During sampling, a voltage of 0–10 000 V is applied, and the positively charged particles are directed toward the center electrode where the particles are sampled. For a set flow rate and voltage, the deposition efficiency decreases with particle size. The deposition efficiency is increased for equally charged, larger particles by (1) lowering the flow rate, which increases the residence time of the particles in the chamber, and by (2) increasing the applied voltage of the center electrode leading to a stronger electric field.19 The ENPS is usually used for collecting inorganic or polystyrene particles for microscopic analysis (e.g., scanning EM, TEM, or total reflection X-ray fluorescence).20-22 Nevertheless, it was shown that the biological activity of the enzyme β-galactosidase was preserved throughout the passage of an electrospray process and a DMA by collection of the enzyme in solution with a similar device.23
Within this study, we were interested in developing the GEMMA method to huge and inhomogeneous biological structures as intact enveloped virus particles. The whole virus particles used for this study was the tick-borne encephalitis virus (TBEV), which belongs to the family Flaviridae, genus Flavivirus. This virus is transmitted via ticks to humans similarly to the well-known Lyme disease. The mature virions of all subtypes are about 50 nm in diameter, consisting of a viral capsid of 30 nm diameter surrounded by a lipid bilayer membrane with incorporated glycoprotein M (membrane) and glycoprotein E (envelope). The capsid encloses the TBEV genome, which consists of a single-stranded, positive-sense RNA of approximately 11 kb in length. TBEV is the cause of a neuroinfectious disease, tick-borne encephalitis (TBE), endemic in Europe and Asia. For TBE, no curative medication is available, but prevention by vaccination is highly effective and safe.24
In our work, we report a strategy for analyzing differently inactivated whole TBEV formulations containing protein additives and different stabilizing agents. The virus particle size distributions within all samples were of interest. We developed a SEC method for virus purification and buffer exchange to the volatile buffer system necessary for the nanoES process of the GEMMA system in one step and representing an off-line combination of SEC with GEMMA. Such a potential approach was briefly mentioned for protein aggregates by Zachariah et al.25 A similar approach (SEC preseparation followed by fraction collection and GEMMA analysis) was developed for the separation and sizing of single-walled carbon nanotubes, which are quite rigid structures.26 Even earlier, an attempt was made to online couple SEC with a GEMMA system for standard proteins but without sizing capability (i.e., without an DMA).32 The GEMMA instrument was then used for two purposes: (1) measurement of the EMD (i.e., nanoparticle size) of viral vaccine particles and (2) collection of size-selected viral vaccine particles for further analysis. The collected samples were investigated by the imaging techniques AFM and TEM. Both orthogonal methods allowed the size comparison with GEMMA data. Moreover, an immunological test (dot blot) was performed to prove the biological activity of the collected GEMMA separated virions.
EXPERIMENTAL SECTION
Chemicals and Reagents
Antimouse IgG (whole molecule) - alkaline phosphatase antibody (goat) and Tween 20 were obtained from Sigma-Aldrich (St. Louis, MO, U.S.A.). The mouse-derived antivirus antibody and the human serum albumin (HSA)-containing PBS buffer without TBE virions were kindly provided by Baxalta Innovations (Orth/Donau, Austria; previously Baxter Innovations). The sucrose concentration was determined with an analytical UV-test for sucrose/d-glucose purchased from R-Biopharm (Darmstadt, Germany).27 All buffers and solutions were prepared with water of ultrahigh quality with a specific resistivity of 18.2 MΩ × cm at 25 °C delivered by a Simplicity UV apparatus (Millipore, Billerica, MA, U.S.A.). A detailed description of all other chemicals is provided in the supplement.
Samples
Tick-Borne Encephalitis Virus (TBEV) samples were provided by Baxalta Innovations (Orth/Donau, Austria) as inactivated virus vaccine particles. Two different sample formulations were provided: inactivated TBEV virions in approximately 40% sucrose in phosphate-buffered saline (PBS) buffer and a PBS dilution of the previous formulation containing also 0.1% HSA provided by Baxalta Innovations (Orth/Donau, Austria).28,29
Instrumentation
The SEC-System (Pharmacia, Uppsala, Sweden) was equipped with a prepacked Superdex 200 10/300 GL size exclusion column (GE Healthcare Bio-Sciences, Uppsala, Sweden). The gas-phase electrophoretic mobility macromolecular analyzer (GEMMA TSI Inc., Shoreview, MN, U.S.A.) instrument consisted of an electrospray aerosol generator, an electrostatic classifier control unit equipped with a nano differential mass analyzer (nano DMA) and an ultrafine condensation particle counter (CPC) for detecting the analyte or alternatively an electrostatic nano particle sampler (ENPS) for collecting the analyte on a selected substrate. A fused-silica capillary with an ID of 25 μm (polyimide coated, OD: 150 μm) was used for the spray process. The AFM images were recorded on a NanoScope III Multimode SPM instrument (Veeco Instruments, Santa Barbara, CA, U.S.A.) using silicon cantilevers with integrated silicon tips (NanoWorld, Neuchâtel, Switzerland, Arrow type: NC) and mica platelets for AFM (Plano, Wetzlar, Germany). Transmission electron microscopy was performed on a Tecnai G2 20 instrument (FEI, Hillsboro, OR, U.S.A.) on 300 mesh copper grids with carbon and Formvar coating purchased from Plano (Wetzlar, Germany).
GEMMA Operating Conditions
For operation in detection and in collection mode, the filtered air flow was set to 8.3 × 10−6 m3/sec, the concentric sheath gas CO2 flow to 1.7 × 10−6 m3/sec, and a differential chamber pressure of 29 kPa was used for the electrospray aerosol generator. For every sample, the voltage was set to operate the electrospray process in cone jet mode:30 the voltage was 1.00–2.00 kV. A sheath gas flow for the DMA of 1.7 × 10−4 m3/sec was selected to gain a measuring range of the electrophoretic mobility diameter (EMD) starting at 3.1 to 80.6 nm in automatic scanning mode. The median spectrum of 10 scans of each sample was calculated for data evaluation. Peak areas in GEMMA spectra were calculated with the software OriginPro 8.0 (OriginLab, Northampton, MA, U.S.A.).
For collecting particles, the DMA was set to a voltage correlating to a selected EM diameter. In the ENPS TEM grids, mica platelets for AFM images or nitrocellulose membranes for immunological testing were mounted on the center electrode. The ENPS was operated with a voltage of −7.0 kV and a gas flow of 1.7 × 10−5 m3/sec.
TEM and AFM Operating Conditions
For TEM, samples were negatively stained with 1% uranyl acetate for 1 min. The AFM images were recorded in air in tapping, constant amplitude mode. More details can be found in the supplement.
Dot Blot Conditions
Nitrocellulose membranes mounted in the ENPS were removed from the instrument after collection and washed with TBS-buffer with Tween 20 for 30 min followed by incubation with a mouse-derived anti-TBE virus antibody.
RESULTS AND DISCUSSION
In this study, an alternative method, namely, GEMMA, for virus characterization is presented to gain better statistics on particle sizes than commonly used microscopic methods, as thousands of particles can be analyzed in one run. Whole and inhomogeneous virions assembled from nucleic acids, proteins, and lipids can be investigated thanks to this size-separation-based analysis. This enables the recording of size-distribution patterns of candidate vaccine solutions. Formulated vaccines and earlier production stages usually contain different substances besides the immunogenic active pharmaceutical ingredient, which make analysis difficult and adequate sample preparation steps are necessary.
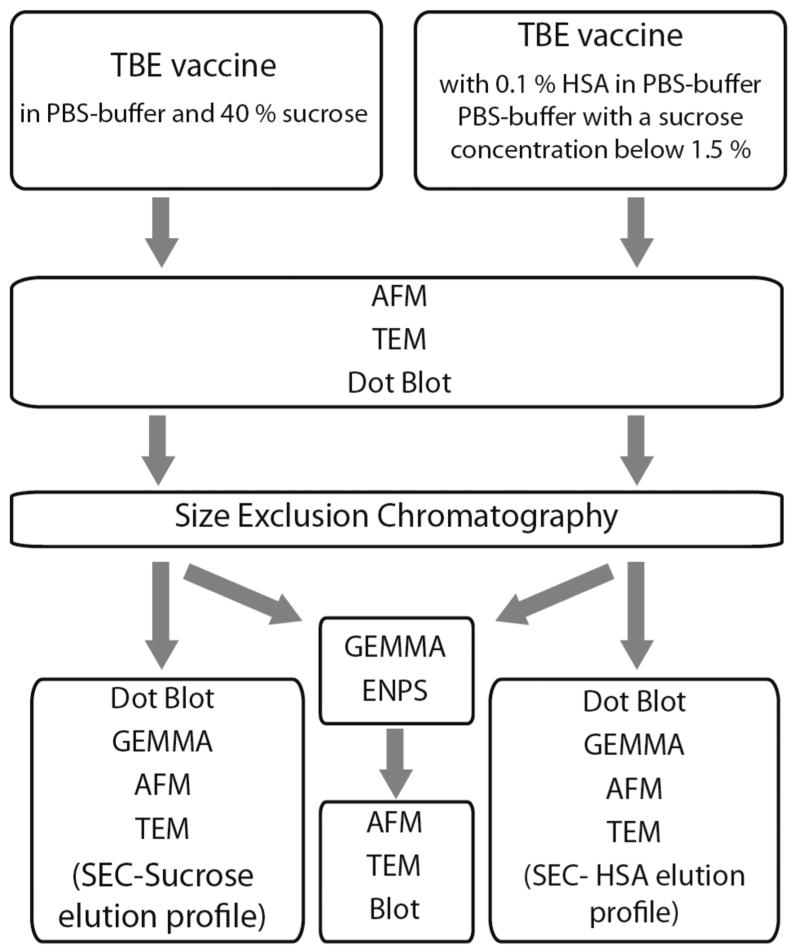
Scheme 1 shows a strategy on how to analyze different vaccine formulations containing buffer and protein components besides the actual virions of interest. In the case of TBEV vaccine production, the whole inactivated virus particles are prepared by sucrose gradient ultracentrifugation resulting in a solution containing about 40% sucrose and PBS buffer. In the subsequent downstream process, a dilution step with PBS-buffer and HSA-solution is implemented, resulting in 0.1% HSA and a sucrose concentration below 1.5% in the final vaccine formulation.28,29
Scheme 1. Strategy for the Analysis of Two Types of Tick-Borne Encephalitis Virus Vaccine Samples Containing Salts, Sucrose, and HSA Besides Vaccine Particles.
Microscopic methods (AFM and TEM) and the immunological dot blot method are capable of giving morphological and immunological information directly within the vaccine’s native buffer system, without buffer exchange or virion purification. In AFM and TEM images, a differentiation of virus particles and protein or salt structures is feasible, although the high protein concentration (HSA) resulted in structural artifacts for AFM and TEM recordings. The presence of salts, sugars, and proteins resulted in a lower contrast and diffuser boundaries of the viral particles in TEM and AFM images. This may complicate virion size measurements, and therefore, a load of single particles has to be evaluated for statistical significant results. The before-mentioned dot blot is insensitive to components such as salts, sugars or additional proteins, as the specificity of the antibody to the analyte is very high, enabling us to obtain clear results in all the investigated samples.
A SEC column was used for both types of TBEV samples as a purification step to eliminate interfering salts, sugars, and proteins, which in our case is PBS-buffer, sucrose, and HSA. The buffer was also exchanged to a volatile buffer system essential for the nanoES process within the GEMMA instrument during the same step. For the collected SEC-fractions, the different before-mentioned analytical methods were applied—dot blot, AFM and TEM—to elucidate the influence of the column-based separation step. Additionally, sucrose and HSA SEC elution profiles (in plain form) were obtained to confirm the efficiency of virion purification performed by the SEC.
After the SEC step, the use of the ENPS coupled with the GEMMA instrument provides the possibility to collect nanoparticles with a selected diameter for further investigations. The integrity of the viral particles after passage through the GEMMA instrument was investigated, either by performing dot blots on a membrane coated with the ENPS-collected viral particles to check epitope integrity, or by analyzing the shape of the sampled virions with AFM on mica platelets and with TEM on copper grids.
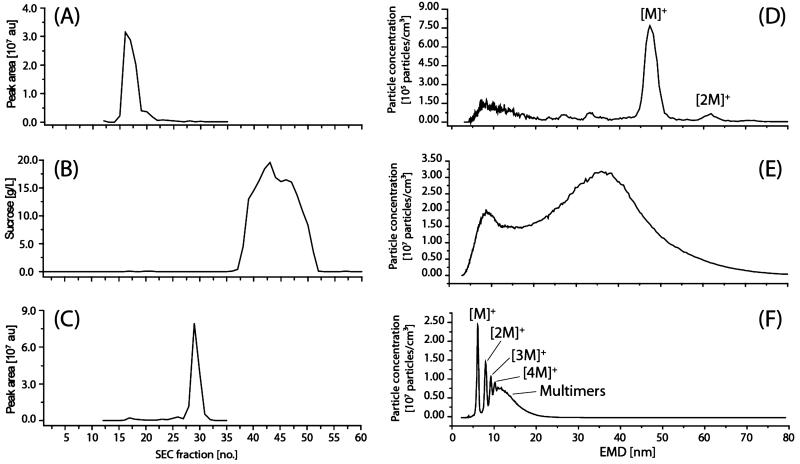
GEMMA spectra were recorded from selected SEC fractions, in which virus particles and HSA were expected to elute. The areas of the peaks in the spectra were integrated for each selected fraction. Figure 1 demonstrates that with the selected SEC column, a separation of the virions from the additives was achieved. Figure 1A shows a plot of peak areas of integrated TBEV relevant GEMMA signals versus elution time after measuring single SEC fractions on the GEMMA system (Figure 1D). The virus peak elutes in fraction 16–20 after separating a TBEV formulation containing 40% sucrose (total elution volume 8–10 mL). As an example in Figure 1D, a GEMMA analysis of SEC fraction 17 of the undiluted TBEV sample with 40% sucrose is presented. A most abundant singly charged monomer peak [M]+ and a singly charged dimer peak [2M]+ of the TBEV virion was observed. A theory for the formation of artificial dimers and oligomers supported by different experiments was described by a number of papers but only for well-defined proteins (60 to 160 kDa) and gold nanoparticles, which are quite different to vaccine particles.31 It was observed that high sample concentration gave rise to artifacts like nonspecific dimer or trimer formation.32 To prove whether the multimer peaks result from high analyte concentration or from native multimers, the analyte solution was diluted several times (dilution factor 5, 10, and 75 (v/v)) prior to GEMMA analysis. If the multimers already existed in solution—at least due to some specificity—and were not introduced during the spray process, the peak size positions of the monomer and all the multimers would stay at the same positions after dilution and reanalysis. If the multimer peaks disappear during subsequent dilution steps they are clearly derived from the spray process and a too high sample concentration. The dilution of this sample showed that the [2M]+ peak is not resulting from gas-phase dimers (data not shown), as the peak of the dimer does not disappear after dilution—this might be indicative of a specific dimeric aggregate.
Figure 1.
Size exclusion separations for buffer exchange and vaccine particles purification: (A) SEC elution profile of TBEV in PBS buffer without HSA with 40% sucrose created from calculated peak areas (GEMMA spectrum) of individual fractions versus eluted SEC fraction number; (B) SEC elution profile of PBS buffer with 40% sucrose; (C) SEC elution profile of HSA-containing PBS buffer; (D) GEMMA spectrum of TBEV in PBS buffer without HSA with 40% sucrose: SEC fraction 17; (E) GEMMA spectrum of sucrose SEC fraction 43; (F) GEMMA spectrum of HSA-containing buffer of SEC fraction 29 (see also C). au: arbitrary units.
The sucrose concentration of each fraction was determined (photometrically) and also plotted against SEC fractions (Figure 1B), showing that the highest sucrose concentration can be determined in fractions 36–51 (total elution volume 18–25.5 mL). The effect of high sucrose concentration on GEMMA analysis is shown Figure 1E. Only a broad signal of unspecific gas-phase multimerization of sucrose was observed. Fraction 43 shows that higher sucrose concentrations lead to larger sucrose particles with an increasing amount of so-called satellite droplets giving a peak at smaller EMD. Satellite droplets arise during the electrospray process as a consequence of a jet break up with very high sucrose concentration.33
It can be clearly seen (Figure 1A,B) that the virus particles elute in fractions 16–20 and the sucrose in 36–51. This shows very plainly that the separation of sucrose from the viral particles with SEC is necessary and simultaneously sufficient to get clear virions spectra and to avoid an increased EMD for the virus peak resulting from sucrose residues in the same droplet as the virus particles. It was already shown for proteins that higher sucrose concentrations lead to a shift to larger diameters, which can be explained as a dry crust enclosing the analytes. Studying the elution profile of HSA for the sample containing sucrose and 0.1% HSA, it was observed that HSA eluted as a rather sharp peak in the fractions 25–31 (total elution volume 12.5–15.5 mL). Again HSA peaks detected by GEMMA analysis (Figure 1F) were manually integrated, and peak areas were plotted against SEC fractions. As the elution of HSA starts in fraction 25 and the viral particles elute in fractions 16–20, the separation of these constituents is sufficient for following GEMMA analysis. Fraction 29 provides the highest amount of HSA; the GEMMA spectrum of this fraction shows several multimer peaks in addition to the singly charged monomer peak. This (Figure 1F) shows the effect of a too-high HSA concentration, leaving more than one analyte per solvent droplet after the nanoES process, resulting in singly charged multimers, which can be separated partly by GEMMA.16,32
The immunologic integrity of the viral particles was checked before and after SEC treatment of both samples (TBEV in 40% sucrose and TBEV with HSA addition) with a Dot Blot experiment. Starting material samples and SEC fractions 16–20 (which exhibit a virus vaccine peak in the GEMMA spectrum) showed a clear color development in the Dot Blot. This indicates that the antibody epitope on the viral particle stays intact and that the immunological activity is preserved during the SEC step (Figure 1-S).
Different parameters of the SEC process were tested for the influence on the measured virus particle diameter and the calculated peak area for the TBEV formulation containing 40% sucrose (Table 1). To increase the virion particle concentration in the collected SEC fractions, a vacuum centrifugation step was added to the analytical procedure to reduce the sample volume to one tenth. It was observed that using a 100 mM ammonium acetate buffer, pH = 7.4 for the SEC elution resulted in higher virus peak areas but lower diameters for the detected particles. The larger virus peak area is a consequence of the higher viral particle concentration. As an explanation for the decreasing diameter after the vacuum centrifugation step, it can be assumed that during the vacuum centrifugation step, the present volatiles (e.g., ammonium and acetic acid) are evaporating due to the vacuum, leaving fewer contaminants in the solution. Fewer contaminants in the solution during the nanoES process leads again to a thinner or no coating of the virion and to a shift of the measured viral particle to lower diameter values. With increasing the ammonium acetate concentration during SEC, the measured diameter increased (Table 1). This increase can be explained again by Dole’s evaporating droplet model, stating that residues present along with the analyte of interest in an evaporating droplet, remain with the particle in the droplet, hence leading to a shift in the observed diameter.33
Table 1. Influence of Different Parameters on TBEV Vaccine Particles Peak Diameter and Peak Areas Determined by GEMMAa.
| measured sample containing TBEV vaccine particle |
measured diameter [nm]b |
standard deviation [nm]b |
peak area [au]c |
standard deviation [au]c |
|---|---|---|---|---|
| with vacuum centrifugation step during sample preparation |
47.4 (N = 20) | ± 0.6 | 2.7 × 106 (N = 5) | 4.9 × 105 |
| without vacuum centrifugation step during sample preparation |
48.4 (N = 20) | ± 0.9 | 1.5 × 105 (N = 5) | 7.5 × 103 |
| with 20 mM ammonium acetate |
45.6 (N = 30) | ± 0.5 | 8.3 × 105 (N = 10) | 8.7 × 104 |
| with 50 mM ammonium acetate |
46.8 (N = 30) | ± 1.1 | 7.2 × 107 (N = 10) | 2.0 × 105 |
| with 100 mM ammonium acetate |
47.3 (N = 30) | ± 0.7 | 1.8 × 106 (N = 10) | 3.4 × 105 |
| no Tween 20 added in 50 mM ammonium acetate |
46.2 (N = 20) | ± 0.9 | 1.6 × 106 (N = 5) | 1.4 × 105 |
| with 0.001% Tween 20 in 50 mM ammonium acetate |
46.3 (N = 20) | ± 0.4 | 1.0 × 106 (N = 5) | 1.5 × 105 |
| with 0.01% Tween 20 in 50 mM ammonium acetate |
47.3 (N = 20) | ± 0.4 | 2.7 × 106 (N = 5) | 4.9 × 105 |
Numbers are calculated from N numbers of analysis representing therefore mean values.
Measured in fraction 16 to 18.
Measured in fraction 17.
It appears from the peak areas calculated from the data that an ammonium acetate concentration of 50 mM gives a higher number of virus particles in GEMMA measurements in comparison to 20 mM and 100 mM ammonium acetate. This indicates that the ionic strength of the buffer plays a crucial role during the high-yield purification of the virions in SEC. For further investigation, the stability of the whole virus particle had to be guaranteed, so Tween 20 was added to the solution after SEC fractionation up to a final concentration of 0.01%. Tween 20 is commonly used as nonionic surfactant in pharmaceutical formulations of protein and vaccine solutions to aid in solubility.34,35
The influence of Tween 20 was studied using a 50 mM ammonium acetate solution with various concentrations of the detergent. Again, higher concentrations led to larger diameters. However, 0.001% Tween 20 had no remarkable influence on the results (increase in particle diameter of 0.1 nm, which is below the accuracy of the technique). Again it can be assumed that higher Tween 20 concentrations leave more surfactant molecules in the same droplet with the virion, shifting the EMD to higher values (coating of virion by Tween 20 molecules). The peak area deduced from the data was not influenced by different Tween 20 concentrations, pointing out similar stability and/or solubility of the virus particles independently of the amount of Tween 20. Nevertheless, adding Tween 20 leads to better long-term stability of the samples.
From all these results, we concluded that 50 mM ammonium acetate, pH = 7.4 as elution buffer is most suitable for the SEC step. A vacuum centrifugation step can be part of sample preparation for intact virus analysis to reduce the volume to one tenth if Tween 20 at a concentration of 0.001% is added to the collection tube (final Tween 20 concentration after volume reduction is 0.01%). Under these conditions, a TBE particle diameter of 46.8 ± 1.1 nm was determined. To convert the detected EMD to molecular mass, it has to be stated that the calibration curve is validated only up to 22 nm (2 MDa) by measuring well-defined proteinous analytes. Nevertheless, the molecular weight can be calculated by extrapolation of the existing curve, giving a molecular weight of about 19.5 MDa.17 This finding fits nicely to molecular weight information available in literature, where the calculated molecular weight for flavivirus nanoparticles is stated to be around 22 MDa.36
SEC is a method where particles are forced through a chromatographic medium by applying pressure. Shear forces can therefore alter the morphology of larger particles leading furthermore to altered biological activity. To prove particle integrity during the developed SEC methodology, AFM and TEM images were recorded before and after SEC (Figure 1-S) of the TBEV sample containing 40% sucrose but no HSA. TEM analysis shows that the image background and the distribution of the negative stain are affected by the different buffer systems. Before separation, 40% sucrose is present in a PBS buffer, and the image shows a dark background with bright virions; however, after SEC, a 50 mM ammonium acetate buffer containing 0.01% Tween 20 is used, bringing about a bright background and dark areas around the virions. The viral particles themselves show more contrast between the membrane (bright) and the inner core (slightly darker) after SEC. Nevertheless, the morphology of the virions did not differ before and after SEC fractionation showing particles of spherical shape with homogeneous size distribution (Table 1-S). From these images, it can be concluded that there was no obvious damage done to the TBEV particles during SEC, resulting in whole virus particles eluting from the column. The immunological evaluation via dot blot testing also showed biological activity after SEC elution of the virions.
AFM images confirmed these findings, although a low tendency to aggregation of virions after SEC was observed, which may potentially result from the exchanged buffer system (different ionic strength, pH value, etc.). Furthermore, from AFM images, the average particle size of the virions was statistically evaluated using specialized software (see Experimental Section). Quantified numbers were compared with GEMMA results. The particle sizes gained by the three different methods are summarized in Table 1-S. The diameter for the TBEV particles before SEC was determined to be 58.2 ± 4.4 nm and 60.4 ± 8.5 nm after SEC by AFM. For the size determination before and after SEC, 84 and 125 particles, respectively, were evaluated with the software. The diameters determined by TEM for these two samples were 51.8 ± 5.4 nm and 53.5 ± 5.3 nm, respectively. For these values, 37 and 33 intact viral particles were evaluated manually with an image editing program, allowing us to put three diameter axes in three different angles (0°, 45°, and 90°) as the virions were not strictly round. GEMMA measurement is only possible after SEC fractionation as elaborated before and was determined to be 46.8 ± 1.1 nm.
Obviously, diameters revealed by AFM are larger than diameters determined by TEM and GEMMA. Two effects can explain this increase of the diameter: virus particles are adsorbed/immobilized on the freshly split mica (inorganic material) surfaces before analysis. This adsorption in combination with the drying process of the sample can adversely affect the globular structure of the virus (multiple sites of interactions between the virus particle and mica surface are given, flattening the nanoobject).37 Additionally, the mode of measurement (tapping mode) can influence the shape of the particle as the AFM tip constantly dilates the particle by putting sometimes too much pressure on top of the biological construct. This assumption is furthermore corroborated by the fact that the absolute particle enlargement is proportional to the size of the measured sphere.38
The evaluation of the TEM images revealed virion diameters slightly smaller than those deduced from AFM but still larger than those deduced from GEMMA results. Again sample preparation for TEM analysis can be considered critical. Artifacts in TEM images may be induced by dehydration and therefore flattening, shrinkage or distortion of specimens are possible consequences. The inhomogeneous precipitation of the negative stain may introduce further changes to the EM image, making interpretation difficult.39,40 Differences in TEM and AFM data can only result from different sample preparation methods, different principles of image creation, and different data as well as statistical processing.41 Nevertheless, both microscopic methods showed that the average TBEV particle diameter is smaller before SEC than after SEC. This may result from the different buffer used in SEC, because differences in ionic strength and osmolarity are able to introduce shrinkage or enlargement of biological systems.
In contrast to the number of particles, which were taken into account for the particle sizing with TEM and AFM, the number of virions that were evaluated for recording one GEMMA spectrum is huge (see Figure 1D). About 105 to 106 viral particles were classified and counted in one run. For every sample, the GEMMA run was repeated at least five times, and a mean spectrum was calculated to eliminate errors in the spectra. The values measured with the GEMMA are in good agreement with the diameter of approximately 50 nm mentioned in literature.42
For any type of analyte, the integrity of the shape and morphology of the particle is very often discussed along with biological activity. Can the biological activity be retained during the whole analytical procedure especially if gas-phase separation at atmospheric pressure is involved, where an analyte is presumed to be dehydrated? With respect to the presented study, the biological activity of TBEV is of interest after any separation step.
To show the biological activity of GEMMA separated-particles, a nitrocellulose membrane was mounted on the center electrode of the ENPS. In a second experiment, the nitrocellulose membrane was substituted by a freshly cleaved mica platelet. Particles of specific EMDs were collected and tested either for activity using an anti-TBEV antibody on the nitrocellulose membrane or studied according for integrity using the mica platelet and AFM. For a detailed investigation of the recorded GEMMA spectrum (Figure 2) of the TBEV-sample, separated peaks were selected with the DMA to collect these particles of the chosen EMD. The DMA was set to transfer particles with an EMD of 8.6, 27.9, 33.0, 47.6, and 61.0 nm (±5 nm approximation) to the ENPS.43 The particles of an EMD of 10 nm or less were assumed to be buffer residues and the detergent. Particles with an EMD of 8.6 nm did not show any color reaction on the membrane, due to the lack of antibody recognition in this fraction, and no particle was visualized by AFM. Nevertheless it has to be stated that the fact that the mica platelet showed an uneven and flat coating in comparison to a freshly cleaved platelet confirmed the assumption that this GEMMA signal results from buffer constituents and detergents (Tween 20). Particles collected at 27.0 nm were also inactive toward anti-TBEV antibodies, but AFM images definitely showed particles. It is assumed that these particles may represent remaining fragments of capsids which are detected after the virus has lost its envelope, for example. The antibody used is not specific for the capsids giving therefore no positive immunological assay (activity is directed toward a protein in the outer envelope). Collecting particles of 33.0 nm resulted in a faint color development on the nitrocellulose membrane, indicating low biological activity, and only a low number of particles in the AFM images were seen. It is assumed that these particles represent doubly charged TBEV species, as there is a small chance of lucky survivors (0.01% of original particle number) during the charge reduction process.44 Calculating the EMD of the doubly charged TBEV particle led to a theoretical diameter of 33.2 nm, which fits nicely to this assumption. The blot of the most abundant peak at 47.6 nm showed a clear color development, indicating the successful binding of the anti-TBEV antibody to the 47.6 nm particles. The AFM images of this GEMMA fraction showed particles in the size range of the virus particles and did not exhibit damage of their spherical shape. The size of viral particles in this GEMMA fraction was 59.8 ± 7.8 nm for AFM and 47.5 ± 5.2 nm for TEM analysis, albeit based on a relatively low number (see Table 1-S) of investigated nanoparticles. Particles at 61.0 nm also showed a slight color development on the nitrocellulose membrane, and AFM images clearly indicate the presence of particles. One explanation for these particles can be multimerization: The diameter of the singly charged dimer is calculated to be 21/3 of the diameter of the singly charged monomer (47.6 nm × 1.26 (21/3) = 60.0 nm), which fits again.15 Therefore, we concluded that the 61.0 nm peak is the singly charged dimer peak of the TBEV. A clear statement on whether this is a method-generated artifact (gas-phase dimer)31 or a native-specific dimer (aggregate) cannot be provided so far.
Figure 2.
GEMMA spectrum of SEC fraction 17 with relevant AFM images (squares) and Dot Blot (circles) images of collected size fractions with selected particle diameters (after GEMMA separation and ENPS collection obtained).
CONCLUSIONS
In conclusion, we have developed a workflow for analyzing different stages of vaccine production containing different additives like inorganic salts, sugars, and even proteins. By means of the GEMMA technology, a particle size distribution pattern of viral solutions could be gained at high statistical significance, which represents an advantage over microscopic methods. The use of a SEC column for preparative purposes was sufficient for purifying the virions, retaining the different ingredients and exchanging the buffer to a volatile buffer system. We have also demonstrated that the size determination of intact, whole TBEV particles, consisting of RNA, capsid proteins, and an envelope (lipids and proteins) with the GEMMA instrument is feasible. The AFM size determination showed larger diameters due to the lateral tip dilation and TBEV particle flattening on the mica surface. The collection of particles with a specific diameter/EMD could be performed to obtain detailed immunological and morphological information about these specific particles. The so captured virions were of correct size and shape in the AFM recordings. The immunological test after collection showed that the epitopes of the vaccine particles remain immunological intact during passage through the GEMMA instrument and “landing” on the substrate.
Supplementary Material
ACKNOWLEDGMENTS
We thank E. Eitenberger (Vienna University of Technology) and C. Dworak (Baxalta Innovations) for technical support in some experiments.
ABBREVIATIONS
- CPC
condensation particle counter
- DMA
differential mass analyzer
- EMD
electrophoretic mobility diameter
- ENPS
electrostatic nano particle sampler
- GEMMA
gas-phase electrophoretic mobility macromolecular analyzer
- HSA
human serum albumin
- TBEV
tick borne encephalitis virus
Footnotes
ASSOCIATED CONTENT
The Supporting Information is available free of charge on the ACS Publications website at DOI: 10.1021/acs.anal-chem.5b01198.
Further description of experimental methods; supplemental data including microscopic AFM and TEM images of TBEV samples, influence of SEC separation on mean TBE vaccine particle diameter (PDF)
The authors declare no competing financial interest.
REFERENCES
- (1).Tang L, Johnson JE. Biochemistry. 2002;41:11517–11524. doi: 10.1021/bi020170j. [DOI] [PubMed] [Google Scholar]
- (2).Zhang W, Heil M, Kuhn RJ, Baker TS. Virology. 2005;332:511–518. doi: 10.1016/j.virol.2004.11.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (3).Subramaniam S, Bartesaghi A, Liu J, Bennett AE, Sougrat R. Curr. Opin. Struct. Biol. 2007;17:596–602. doi: 10.1016/j.sbi.2007.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (4).Gastaminza P, Dryden KA, Boyd B, Wood MR, Law M, Yeager M, Chisari FV. J. Virol. 2010;84:10999–11009. doi: 10.1128/JVI.00526-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (5).Liu J, Andya J, Shire S. AAPS J. 2006;8:E580–E589. doi: 10.1208/aapsj080367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (6).Boyd RD, Pichaimuthu SK, Cuenat A. Colloids Surf., A. 2011;387:35–42. [Google Scholar]
- (7).Chuan YP, Fan YY, Lua L, Middelberg AP. J. Biotechnol. Bioeng. 2008;99:1425–1433. doi: 10.1002/bit.21710. [DOI] [PubMed] [Google Scholar]
- (8).Reschiglian P, Zattoni A, Roda B, Michelini E, Roda A. Trends Biotechnol. 2005;23:475–483. doi: 10.1016/j.tibtech.2005.07.008. [DOI] [PubMed] [Google Scholar]
- (9).Wei Z, McEvoy M, Razinkov V, Polozova A, Li E, Casas-Finet J, Tous GI, Balu P, Pan AA, Mehta H, Schenerman MA. J. Virol. Methods. 2007;144:122–132. doi: 10.1016/j.jviromet.2007.04.008. [DOI] [PubMed] [Google Scholar]
- (10).McEvoy M, Razinkov V, Wei Z, Casas-Finet JR, Tous GI, Schenerman MA. Biotechnol. Prog. 2011;27:547–554. doi: 10.1002/btpr.499. [DOI] [PubMed] [Google Scholar]
- (11).Giocondi M-C, Ronzon F, Nicolai MC, Dosset P, Milhiet P-E, Chevalier M, Le Grimellec C. J. Gen. Virol. 2010;91:329–338. doi: 10.1099/vir.0.015156-0. [DOI] [PubMed] [Google Scholar]
- (12).Kuznetsov YG, Malkin AJ, Lucas RW, Plomp M, McPherson AJ. Gen. Virol. 2001;82:2025–2034. doi: 10.1099/0022-1317-82-9-2025. [DOI] [PubMed] [Google Scholar]
- (13).Ferreira GP, Trindade GS, Vilela JM, Da Silva MI, Andrade MS, Kroon EG. J. Microsc. 2008;231:180–185. doi: 10.1111/j.1365-2818.2008.02028.x. [DOI] [PubMed] [Google Scholar]
- (14).de la Mora JF, de Juan L, Eichler T, Rosell J. TrAC, Trends Anal. Chem. 1998;17:328–339. [Google Scholar]
- (15).Kaufman SL, Skogen JW, Dorman FD, Zarrin F, Lewis KC. Anal. Chem. 1996;68:1895–1904. doi: 10.1021/ac951128f. [DOI] [PubMed] [Google Scholar]
- (16).Kaufman SL. J. Aerosol Sci. 1998;29:537–552. [Google Scholar]
- (17).Bacher G, Szymanski WW, Kaufman SL, Zöllner P, Blaas D, Allmaier G. J. Mass Spectrom. 2001;36:1038–1052. doi: 10.1002/jms.208. [DOI] [PubMed] [Google Scholar]
- (18).Tammet HJ. Aerosol Sci. 1995;26:459–475. [Google Scholar]
- (19).Dixkens J, Fissan H. Aerosol Sci. Technol. 1999;30:438–453. [Google Scholar]
- (20).Krinke TJ, Fissan H, Deppert K. Phase Transitions. 2003;76:333–345. [Google Scholar]
- (21).Bogan MJ, Henry Benner W, Hau-Riege SP, Chapman HN, Frank M. J. Aerosol Sci. 2007;38:1119–1128. [Google Scholar]
- (22).Yook S-J, Fissan H, Engelke T, Asbach C, van der Zwaag T, Kim JH, Wang J, Pui DYH. J. Aerosol Sci. 2008;39:537–548. [Google Scholar]
- (23).Allmaier G, Maißer A, Laschober C, Messner P, Szymanski WW. TrAC, Trends Anal. Chem. 2011;30:123–132. doi: 10.1016/j.trac.2010.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (24).Kiermayr S, Kofler RM, Mandl CW, Messner P, Heinz FX. J. Virol. 2004;78:8078–8084. doi: 10.1128/JVI.78.15.8078-8084.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (25).Guha S, Li M, Tarlov MJ, Zachariah MR. Trends Biotechnol. 2012;30:291–300. doi: 10.1016/j.tibtech.2012.02.003. [DOI] [PubMed] [Google Scholar]
- (26).Pease LF, Tsai D-H, Fagan JA, Bauer BJ, Zangmeister RA, Tarlov MJ, Zachariah MR. Small. 2009;5:2894–2901. doi: 10.1002/smll.200900928. [DOI] [PubMed] [Google Scholar]
- (27).Schöenrock KWR, Costesso D. J. Am. Soc. Sugar Beet Techn. 1975;18:349–359. [Google Scholar]
- (28).Barrett PN, Schober-Bendixen S, Ehrlich H. J. Vaccine. 2003;21:S41–S49. doi: 10.1016/s0264-410x(02)00814-9. [DOI] [PubMed] [Google Scholar]
- (29).Pöllabauer EM, Fritsch S, Pavlova BG, Löw-Baselli A, Firth C, Koska M, Maritsch F, Barrett PN, Ehrlich H. J. Vaccine. 2010;28:4558–4565. doi: 10.1016/j.vaccine.2010.04.075. [DOI] [PubMed] [Google Scholar]
- (30).Chen D-R, Pui DYH, Kaufman SL. J. Aerosol Sci. 1995;26:963–977. [Google Scholar]
- (31).Li M, Tan J, Tarlov MJ, Zachariah MR. Anal. Chem. 2014;86:12130–12137. doi: 10.1021/ac5030123. [DOI] [PubMed] [Google Scholar]; Li M, Guha S, Zangmeister R, Tarlov MJ, Zachariah MR. Aerosol Sci. Technol. 2011;45:849–860. [Google Scholar]; Li M, Guha S, Zangmeister R, Tarlov MJ, Zachariah MR. Langmuir. 2011;27:14732–14739. doi: 10.1021/la202177s. [DOI] [PubMed] [Google Scholar]
- (32).Lewis KC, Dohmeier DM, Jorgenson JW, Kaufman SL, Zarrin F, Dorman FD. Anal. Chem. 1994;66:2285–2292. doi: 10.1021/ac00086a014. [DOI] [PubMed] [Google Scholar]
- (33).Kaufman SL. Anal. Chim. Acta. 2000;406:3–10. [Google Scholar]
- (34).Chi EY, Krishnan S, Randolph TW, Carpenter JF. Pharm. Res. 2003;20:1325–1336. doi: 10.1023/a:1025771421906. [DOI] [PubMed] [Google Scholar]
- (35).Kamerzell TJ, Esfandiary R, Joshi SB, Middaugh CR, Volkin DB. Adv. Drug Delivery Rev. 2011;63:1118–1159. doi: 10.1016/j.addr.2011.07.006. [DOI] [PubMed] [Google Scholar]
- (36).Kuhn RJ, Zhang W, Rossmann MG, Pletnev SV, Corver J, Lenches E, Jones CT, Mukhopadhyay S, Chipman PR, Strauss EG, Baker TS, Strauss JH. Cell. 2002;108:717–725. doi: 10.1016/s0092-8674(02)00660-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (37).Moloney M, McDonnell L, O’Shea H. Ultramicroscopy. 2002;91:275–279. doi: 10.1016/s0304-3991(02)00109-2. [DOI] [PubMed] [Google Scholar]
- (38).Zeng Z-G, Zhu G-D, Guo Z, Zhang L, Yan X.-j., Du Q-G, Liu R. Ultramicroscopy. 2008;108:975–980. doi: 10.1016/j.ultramic.2008.04.001. [DOI] [PubMed] [Google Scholar]
- (39).Mast J, Demeestere L. Diagn. Pathol. 2009;4:5. doi: 10.1186/1746-1596-4-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (40).Weston A, Armer H, Collinson LJ. Chem. Biol. 2010;3:101–112. doi: 10.1007/s12154-009-0033-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (41).Chen H. Microsc. Microanal. 2007;13:384–389. doi: 10.1017/S1431927607070808. [DOI] [PubMed] [Google Scholar]
- (42).Schalich J, Allison S, Stiasny K, Mandl C, Kunz C, Heinz FJ. Virol. 1996;70:4549–4557. doi: 10.1128/jvi.70.7.4549-4557.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (43).Szymanski WW, Allmaier G. In: Nano - Chancen und Risiken aktueller Technologien. Gazsó A, Greßler S, Schiemer F, editors. Springer; Vienna, New York: 2007. pp. 21–38. [Google Scholar]
- (44).Wiedensohler AJ. Aerosol Sci. 1988;19:387–389. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.